Abstract
Recently, the severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) Omicron variant (B.1.1.529) was first identified in Botswana in November 2021. It was first reported to the World Health Organization (WHO) on November 24. On November 26, 2021, according to the advice of scientists who are part of the WHO's Technical Advisory Group on SARS‐CoV‐2 Virus Evolution (TAG‐VE), the WHO defined the strain as a variant of concern (VOC) and named it Omicron. Compared to the other four VOCs (Alpha, Beta, Gamma, and Delta), the Omicron variant was the most highly mutated strain, with 50 mutations accumulated throughout the genome. The Omicron variant contains at least 32 mutations in the spike protein, which was twice as many as the Delta variant. Studies have shown that carrying many mutations can increase infectivity and immune escape of the Omicron variant compared with the early wild‐type strain and the other four VOCs. The Omicron variant is becoming the dominant strain in many countries worldwide and brings new challenges to preventing and controlling coronavirus disease 2019 (COVID‐19). The current review article aims to analyze and summarize information data about the biological characteristics of amino acid mutations, the epidemic characteristics, immune escape, and vaccine reactivity of the Omicron variant, hoping to provide a scientific reference for monitoring, prevention, and vaccine development strategies for the Omicron variant.
Keywords: B.1.1.529, COVID‐19, immune escape, Omicron, SARS‐CoV‐2
Highlights
The Omicron variant was the most highly mutated variant of concerns (VOCs).
The Omicron variant spread faster than the original virus strain and other VOCs.
The Omicron variant produces a higher risk of reinfection than the other variants.
1. INTRODUCTION
Coronavirus disease 2019 (COVID‐19) caused by severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) has occurred in many countries worldwide, causing severe damage to the medical system and human health. 1 , 2 , 3 , 4 , 5 As of December 20, 2021, there have been 273 900 334 confirmed cases of COVID‐19, including 5 351 812 deaths reported to the WHO. There are 40 countries with more than 1 million confirmed cases of COVID‐19, and 112 countries have more than 100 000 cases (https://covid19.who.int/).
RNA viruses have a higher mutation rate than DNA viruses. Like other RNA viruses, the large genome of SARS‐CoV‐2 (~30 kb) and low‐fidelity RNA‐dependent RNA polymerase (RdRp) are the reasons for the high adaptive mutation rate when adapting to new human hosts. 6 , 7 From January to September 2021, several SARS‐CoV‐2 variants appeared and became prominent epidemic strains in many countries, including four VOCs (Alpha, 8 Beta, 9 Gamma, 10 and Delta 11 ) defined by the WHO. Compared with the early wild‐type strain, the four VOCs accumulated many mutations in the spike protein, which led to these VOCs being more transmissible and producing immune escape. 12 , 13 , 14
B.1.1.529 was first detected in specimens collected on November 11, 2021, in Botswana and on November 14, 2021, in South Africa. It was first reported to the WHO on November 24. On November 26, the WHO defined it as the fifth variant of concern (VOC) and named it Omicron. 15 The Omicron variant is the most mutated strain among many SARS‐CoV‐2 variants (including VOCs and VOIs) during the COVID‐19 pandemic. The amino acid mutations of the Omicron variant are widely distributed on four structural proteins, including Spike (S), Envelope (E), Membrane (M), Nucleocapsid (N) proteins, and nonstructural proteins (NSPs) (NSP3, NSP4, NSP5, NSP6, NSP12, NSP1). 16
The SARS‐CoV‐2 spike protein consists of S1 and S2 subunits and furin protease cleavage sites. The S1 subunit consists of an N‐terminal domain (NTD) and receptor‐binding domain (RBD). The receptor‐binding motif (RBM) directly contacts the receptor (angiotensin‐converting enzyme‐2, ACE2) on the surface of human cells, which mediates the invasion of the virus into cells and determines the transmissibility of the virus. 1 , 7 , 17 In addition, the spike protein is the dominant neutralization target of convalescent plasma, vaccines, and monoclonal antibodies (mAbs). 18 , 19 Adaptive mutation of the SARS‐CoV‐2 genome can change the infectivity, immune evasion, and phenotypic characteristics of the virus. 13 , 14 , 20 The emergence of the Omicron variant has caused serious concern about the increased infectivity, immune escape ability, and reinfection risk. 21 Therefore, many countries have made travel restrictions to prevent the rapid spread of the Omicron variant.
1.1. Characteristics of amino acid mutations of highly mutated Omicron variants
Genome sequencing showed that the Omicron variant belonged to Pango line B.1.1.529, Nextstrain branch 21k, and GISAID branch GR/484A (https://www.gisaid.org/). The amino acid mutations of the Omicron(21K)spike protein contain 28 amino acid substitutions, three deletions, and one insertion (A67 V, Δ69–70, T95I, G142D, Δ143–145, Δ211, L212I, ins214EPE, G339D, S371 L, S373P, S375F, K417N, N440K, G446S, S477N, T478K, E484A, Q493R, G496S, Q498R, N501Y, Y505H, T547K, D614G, H655Y, N679K, P681H, N764K, D796Y, N856K, Q954H, N969K, L981F). 15 On December 7, the Omicron sublineage was detected in South Africa, and the Nextstrain defined it as 21 L, Omicron (21 L, BA.2). The Omicron (21 L, BA.2) spike protein contains 29 amino acid substitutions and one insertion (T19I, L24S, ins25PPA, D142D, V213G, G339D, S371 L, S373P, S375F, T376A, D405N, R408S, K417N, N440K, G446S, S477N, T478K, E484A, Q493R, Q498R, N501Y, Y505H, D614G, H655Y, N679K, P681H, N764K, D796Y, Q954H, N969K) (https://www.nicd.ac.za/diseases-a-z-index/disease-index-covid-19/sars-cov-2-genomic-surveillance-update/). Compared to the Omicron (21K, BA.1), the Omicron (21 L, BA.2) spike protein lacks Δ69–70, which is associated with S gene target failure (SGTF) and is not detected by SGTF.
The number of mutations of the Omicron variant spike protein is twice that of the Delta variant, which has spread in many countries worldwide. 14 Notably, approximately 15 of these mutations are located in RBD, a primary target of neutralizing antibodies (NAbs), 22 and are more common than other VOCs (Delta: L452R, T478K; Beta: K417N, E484K, N501Y; three Gama K417T, E484K, N501Y; Alpha: N501Y in RBD). 12 , 13 , 14
Omicron variant has critical mutations in the spike protein that were previously reported in other VOCs (Alpha, Beta, Gamma, and Delta) and VOIs (Kappa, Zeta, Lambda, and Mu), 23 including Δ69–70, P681H, N501Y, and D614G in Alpha, K417N in Beta, H655Y, K417N in Gamma and T478K mutation in Delta. Both structural modeling and pseudovirus experiments indicated that RBD with T478K, 24 N501Y, 25 D614G, 26 or Δ69–70 27 mutations could increase the tightness and affinity of binding with the hACE2 receptor or induce more rapid cell‐cell fusion and the formation of multi‐nucleated cells, thus increasing the infectivity of SARS‐CoV‐2 variants. Several clinical studies have shown that the transmissibility of Alpha is 50% higher than that of the early wild‐type strain 8 and that Delta seems to be approximately 60% more transmissible than Alpha. 28 In addition, the Omicron variant carrying “Q498R‐N501Y” double mutations may further improve the binding affinity of RBD to the hACE2 receptor. 29 It is worth noting that there are triple mutations “H655Y + N679K + P681H” near the furin cleavage site of the Omicron variant spike protein. Previous studies found that “H655Y + N679K + P681H” might accelerate S1/S2 cleavage through furin protease and enhance the fusion of the virus and host cell membrane, leading to enhanced replication ability and infectivity of the virus. 30 , 31
Molecular evidence provides a good explanation for how mutations in the spike protein (such as K417N, N440K, G446S, S4777N, T478K, and N501Y) 32 , 33 , 34 , 35 cause a reduction in the neutralizing activity of mAbs, convalescent plasma, and serum‐induced by vaccines. 36 Molecular dynamic simulations have pointed out that “K417N‐E484K‐N501Y” triple mutations induce spike protein conformational changes greater than those induced by N501Y or E484K alone. 36 Triple mutations allowed SARS‐CoV‐2 variants carrying “K417N‐E484K‐N501Y” mutations to more effectively escape neutralization activity, leading to the Beta variant that caused the second wave of outbreaks in South Africa to be more immune escape than other VOCs (Alpha, Gamma, and Delta). It is worth noting that the Omicron variant also has a similar triple mutation, “K417N + E484A + N501Y,” which may produce immune escape. 37 In addition, a team of virologist Paul Bienias of Rockefeller University, New York, found that the pseudovirus carrying 20 mutations PMS20 (all reported mutations) was resistant to the neutralization of the convalescent plasma and serum‐induced by mRNA vaccine. 38 One preprint from the Peking University research team reported that six single‐point mutations (K417N, N440K, G446S, E484A, Q493K, G496S) of the Omicron variant can have escaped neutralization activity. 39
The Omicron variant not only accumulated a large number of mutations in the spike protein but also in open reading frame 1ab (ORF1ab), an envelope protein (M), a nuclear envelope protein (E), and nucleocapsid protein (N) (including NSP3‐K38R, V1069I, Δ1265, L1266I, A1892T; NSP4‐T492I; NSP5‐P132H; NSP6‐Δ105–107, A189 V; NSP12‐P323 L; NSP14‐I42 V; E‐T9I; M‐D3G, Q19E, A63T; N‐P13 L, Δ31–33, R203K, G204R). 23 NSPs, particularly NSP12 and NSP14, of CoVs are indispensable for viral replication and transcription. It has been shown that the P323 L mutation in NSP12 can reduce the fidelity of viral gene replication, 40 , 41 the Δ105–107 mutation in NSP6 may affect the innate immune response and T cell immunity, 42 and the R203K/G204R mutation in the N protein can enhance the virulence of variants. 43
Altogether, compared to the early wild‐type strain and other VOCs (Alpha, Beta, Gamma, and Delta), the Omicron variant had the largest number of mutations, involving Spike (S), Envelope (E), Membrane (M), Nucleocapsid (N) proteins, and NSPs (NSP3, NSP4, NSP5, NSP6, NSP12, NSP1). These mutations can affect the biological characteristics of the Omicron variant, including increasing the transmissibility, causing immune escape, and enhancing the virulence of the Omicron variant, as shown in Figure 1.
Figure 1.
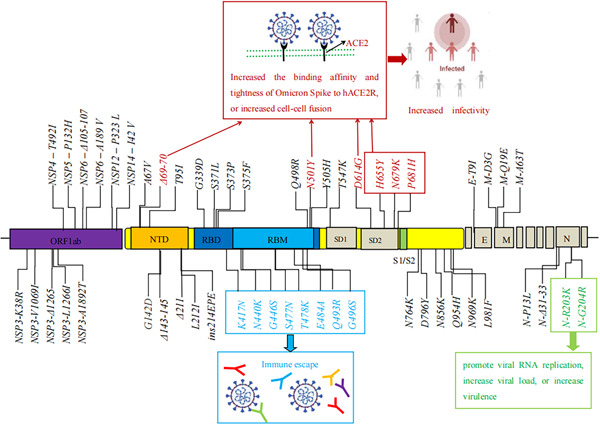
The biological characteristics of key amino acid mutations of the Omicron variant. E, envelope; M, membrane; N, nucleocapsid proteins; NTD, N‐terminal domain; ORF, open reading frame; RBD, receptor‐binding domain; RBM, receptor binding motif; S1/S2, furin cleavage site; SD1, subdomain 1; SD2, subdomain 2
1.2. Outbreak and infectivity
Compared to the Delta variant, Omicron has received much attention for just 2 weeks after its appearance. An in silico analysis showed that the infectivity of Omicron might be more than 10‐fold higher than that of the original virus and approximately twice as high as that of Delta. 38 According to reported data, researchers estimated that the Rt of Omicron is 1.4‐ to 3.1‐fold higher than that of Delta. As of November 26, 2021, travel‐related cases have also been detected in Belgium, Hong Kong, and Israel. On November 29, 2021, 3 days after the announcement by the WHO, cases of VOC Omicron were detected in Austria, Australia, Belgium, Canada, the Czech Republic, Denmark, France, Germany, Italy, the Netherlands, and the United Kingdom. 21
The Omicron variant spread rapidly in Gauteng Province, the largest city in South Africa. Genome sequencing analysis from the Tulio de Oliveira team, a bioinformatician at the University of KwaZulu‐natal province, found that all 77 virus samples detected in Gauteng Province from November 12 to 20 were from the B.1.1.529 variant. 44 Before the Omicron variant was discovered, the number of confirmed cases of COVID‐19 in South Africa was quite low every day. However, it rapidly increased from 273 cases on November 16 to more than 1200 cases on November 25, of which more than 80% occurred in Gauteng Province, 45 where the first Omicron variant cases were found. In the outbreak's epicenter—Gauteng—74% of samples sequenced from the last 3 weeks of November involved the new variant. This suggests that Omicron rapidly outcompetes the prevalent Delta variant, which had already replaced the local Beta variant in South Africa. In late November, South Africa's National Institute for Communicable Diseases (NICD) in Johannesburg determined that R was above 2 in Gauteng (R0: the average number of people infected with person infections). According to the data from NICD, the Delta variant is the main epidemic strain in August, September, and October in South Africa. Sequence analysis showed that 73% (228/312) were Omicron variants, while the Delta variant accounted for only 23% (72/312) in November. From November 30 to December 3, the number of confirmed cases of the Omicron variant in South Africa increased from 4373 to 16 055 (data available from https://www.nicd.ac.za/latest-confirmed-cases-of-covid-19-in-south-africa/). These data indicated that the Omicron variant has replaced Delta as the dominant epidemic strain in South Africa and is driving a new wave of outbreaks in South Africa. 46
The first Omicron‐infected case in Europe was diagnosed in Belgium on November 26, and the patient tested positive for COVID‐19 on November 22. On November 26, 2021, the European Centre for Disease Prevention and Control (ECDC) classified this variant as a VOC due to concerns regarding immune escape and potentially increased transmissibility compared to the Delta variant. As of December 1, 2021, a total of 70 confirmed cases of the Omicron variant have been reported in 13 countries in the European Union and European Economic Area (EU/EEA). As of December 19, 4691 confirmed cases had been reported in 28 countries in the EU/EEA, accounting for 67‐fold higher than at the beginning of the month (https://www.ecdc.europa.eu/en/news-events/epidemiological-update-omicron-variant-concern-voc-data-19-December-2021). Delta VOC remains currently the most prevalent variant in the EU/EEA. However, based on modeling predictions and depending on the growth advantage and level of immune escape, Omicron VOCs are likely to become the dominant variant in the EU/EEA within the first 2 months of 2022.
On November 30, 2021, the United States designated Omicron as a VOC. On December 1, 2021, the first case attributed to the Omicron variant was identified in the United States in a person who recently returned from travel to South Africa. On December 2, 2021, a second case was reported in a person with no international travel history who also attended a convention in the days preceding symptom onset. Nowcast, which is a model that estimates more recent proportions of circulating variants in the United States, showed that the sequence of Omicron accounts for 73.2% (95% PI: 34%–94.9%) of the total sequence and that Delta accounts for 26.6% (95% PI: 5.1%–65.8%) from 12 to December 18, 2021 (data available from https://www.cdc.gov/coronavirus/2019-ncov/variants/omicron-variant.html).
On November 26, 2021, the UK Health Security Agency (UKHSA) designated Omicron as the variety being monitored (VUI‐21‐NOV‐01). On November 27, two Omicron confirmed cases were first detected in the United Kingdom. 45 On December 3, UKHSA reported 75 new Omicron confirmed cases, and the total number of Omicron confirmed cases was 3.6‐fold (104 vs. 29) that of the previous day. According to the latest data released by the UKHSA, the number of Omicron confirmed cases in the United Kingdom rose from 4487 to 45 145 between 12 and 18 December 2021 (data available from https://www.gov.uk/government/news/covid-19-variants-identified-in-the-uk). Moreover, according to the Omicron monitoring data from COG‐UK, Trevor Bedford, an American evolutionary virologist, pointed out that the logistic growth rate of Alpha variants in the United Kingdom in January was 0.08 per day, while that of Omicron was 0.41 (https://pangolin.cog-uk.io/). These data indicated that community transmission of Omicron has occurred in the United Kingdom.
The above studies indicated that the Omicron variant spread faster and more widely than the original virus strain and other VOCs. As of December 20, 2021, there were 34 911 Omicron confirmed cases, including seven deaths, reported by 95 countries (https://www.ecdc.europa.eu/en/news-events/epidemiological-update-omicron-variant-concern-voc-data-19-December-2021).
1.3. Clinical severity
SARS‐CoV‐2 not only invades the respiratory system but also causes other organ injuries in severe cases, such as kidney injury, 47 liver injury, 48 myocardial injury, 49 coagulation dysfunction, 50 and gastrointestinal symptoms. 51 , 52 Compared with the early original strains, many clinical studies have suggested that Alpha, Beta, and Delta variants increase the risk of hospitalization, ICU admission, and death. 8 , 9 , 11 , 12
Previously reported cases from South Africa 46 and 43 cases of Omicron in the United States 53 , 54 found that the symptoms of confirmed cases were asymptomatic or had mild symptoms, and no deaths were reported (among the 43 confirmed cases in the United States, symptoms were 89% cough, 65% fatigue, 59% stuffy or runny nose, 14% fever, 8% nausea, 4% diarrhea, and 3% loss of taste or smell). Subsequently, UKHSA announced that ten individuals infected with Omicron were hospitalized, and one died. 55 As of December 20, 2021, seven deaths in those infected with Omicron have been reported. However, the impact of Omicron on the rate of hospitalization and mortality needs more cases and longer observation times to be determined.
1.4. Increase the risk of reinfections
South Africa has experienced three COVID‐19 outbreaks caused by the original virus, Beta variant, and Delta variant. 9 , 13 One preprint 56 from NCID showed that in the rapidly building Omicron surge, the proportion of new cases involving people who have already experienced COVID‐19 is more than three times higher than in the second (Beta) and third waves (Delta). Contrary to expectation, the estimated hazard ratio (HR) for reinfection versus primary infection was lower during waves driven by the Beta and Delta variants than for the first wave (HR for the second wave vs. the first wave: 0.75 [95% CI: 0.59–0.97]; for the third wave vs. the first wave: 0.71 [95% CI: 0.56–0.92]). In contrast, the recent spread of the Omicron variant has been associated with an increase in the reinfection hazard coefficient and a decrease in the hazard coefficient for primary infection. The estimated hazard ratio for reinfection versus primary infection from 1 to November 27, 2021, versus the first wave was 2.39 (95% CI: 1.88–3.11). 56 Similarly, in the United Kingdom, 5153 individuals with an Omicron infection between November 1 and December 11, 2021, were identified; 305 were linked to a previously confirmed infection and had an interval from the previous positive test of 90 days or more (latest updates December 12, 2021. on SARS‐CoV‐2 variants detected in the United Kingdom. https://www.gov.uk/government/news/covid-19-variants-identified-in-the-uk.2021). These data suggested that the Omicron variant produces immune escape, with a higher risk of reinfection than the Beta and Delta variants.
1.5. Immune escape
1.5.1. Resistance to neutralization of the Omicron variant by convalescent plasma and vaccine sera
Vaccines have played a huge role in the fight against COVID‐19, but the problem of vaccine escape has become more acute over time. 13 , 57 , 58 , 59 The first four cases of Omicron infection in Botswana were fully vaccinated, suggesting possible vaccine escape. 60 , 61 In addition, based on 132 three‐dimensional (3D) structures, models of antibody‐RBD complexes show that Omicron may be twice as likely to escape current vaccines than the Delta variant. 38
On December 7, 2021, the earliest serum neutralization assay showed that the neutralization activity of the Pfizer/BNT162b2 mRNA vaccine‐declined serum against the Omicron variant was significantly decreased by 41.4‐fold compared to the original strain (geometric mean titer [GMT] FRNT50: 1321 vs. 32, p = 0.0018). 62 The Pfizer official announced that the neutralization activity of two doses of the Pfizer vaccine against Omicron was significantly decreased. 63 Similarly, in vitro authentic Omicron virus experiments showed the neutralization activity of two BNT162b2 (nonboosted) doses and three BNT162b2 (0.5 or 3 months after boosting) against Omicron was decreased by 11.4‐, 37.0‐, and 24.5‐fold compared with the Delta variant, respectively. 64 Sera from double mRNA1273‐vaccinated (nonboosted) and BNT162b2‐boosted mice showed a 20‐ and 22.7‐fold reduction in neutralization capacity. 65
An in vitro study of pseudovirus Omicron showed that the neutralization activity of convalescent plasma (n = 28) against Omicron was reduced by 8.4‐fold, whereas the neutralization activity of convalescent plasma against other VOCs and VOIs pseudotyped was decreased by 1.2–4.5‐fold compared to the D614G strain. 64 In addition, one preprint from Peking University's research team found that Omicron can escape 85% of anti‐RBD neutralizing antibodies of diverse epitopes (n = 247) from convalescent plasma and serum‐induced by vaccine. 39
All of these data revealed that the Omicron variant more easily escaped the neutralization activity of convalescent plasma and two doses of vaccine‐induced serum than the original virus and other VOCs, including Beta and Delta.
1.5.2. Reduced neutralizing activity of the monoclonal antibody
The neutralization effect analysis of nine monoclonal antibody drugs on Omicron pseudovirus showed that seven monoclonal antibodies (LY‐CoV555, LY‐CoV016, REGN10933, REGN10987, AZD1061) were completely ineffective against Omicron (IC50 > 6.5 ng/L). Only two monoclonal antibodies can effectively neutralize against Omicron (IC50: 0.181 ng/L for Vir‐7831; 0.287 ng/µl for DXP‐604). 39 In addition, an in vitro study of authentic Omicron virus showed that imdevimab and casirivimab could effectively prevent Delta infection, while Omicron was resistant to casirivimab and imdevimab. 65 Similarly, Cameroni et al. 66 reported that 89.7% (26/29) of RBM‐directed monoclonal antibodies (mAbs) lost in vitro neutralizing activity against Omicron, with only 3 out of 29 mAbs retaining unaltered potency, including the ACE2‐mimicking S2K146 mAb. These results indicated that the magnitude of Omicron‐mediated immune evasion marks a major SARS‐CoV‐2 antigenic shift.
1.6. Vaccine effectiveness against Omicron variant
The serum neutralization assay found that the escape was incomplete, and 5/6 participants were vaccinated after the previous infection, showing relatively high neutralization titers against Omicron. 62 This suggests that previous infection followed by vaccination or booster is likely to increase the neutralization level and likely confer protection from severe disease in Omicron infection.
The Pfizer official announced that after the third dose of BNT162b2 injection, the titer of neutralizing antibody increased by 25‐fold, reaching the level of two doses of inoculation to protect the infection of the original virus strain. 63 One mRNA vaccine effectiveness evaluation 67 showed that two doses of the ChAdOx1 vaccine were ineffective against the Omicron variant. In the population who received two doses of BNT162b2, the vaccine effectiveness was 88.0% (95% CI: 65.9%–95.8%) 2–9 weeks after the second vaccination, and then it continued to decrease. The vaccine effectiveness dropped from 48.5% (95% CI: 24.3%–65.0%) to 34.2% (−5%–58.7%) from 10–14 weeks to 25 weeks after the second vaccination. However, after the third dose of BNT162b2 was vaccinated based on two doses of ChAdOx1 or BNT162b2, the vaccine effectiveness against Omicron was still high, exceeding 70% (71.4% and 75.5%, respectively) after 2 weeks. 67 Similarly, one study from Doria‐Rose NA's team showed that Omicron was 41–84‐fold and 5.3–7.4‐fold less sensitive to neutralization than D614G and Beta, respectively, when assayed with serum samples obtained 4 weeks after two standard inoculations with 100 µg mRNA‐1273 vaccine. However, a booster dose of mRNA‐1273 increases Omicron neutralization titers and may reduce the risk of symptomatic vaccine breakthrough infections. 68
Moreover, inactivated vaccine effectiveness evaluation demonstrated 69 that at 14 days post‐two‐dose BBIBP‐CorV vaccines, the GMTs were 67.40 (95% CI: 39.56–114.90) against prototype, 8.85 (4.26–18.40) against Beta, 35.07 (22.96–53.58) against Delta, and 6.04 (4.53–8.07) against Omicron, while neutralization activity against Omicron was below the lower limit of quantitation in 80% of the samples. The neutralization titers of Omicron exhibited a significant fold reduction compared to Beta and Delta compared to the prototype. (11.16‐fold vs. 7.62‐ and 1.92‐fold reduction). On Day 14, post booster vaccination with BBIBP‐CorV, GMTs were increased 13.78 (95% CI: 7.71–24.60)‐fold against the prototype, 10.63 (5.62–20.07)‐fold against Beta, and 8.56 (4.51–16.26)‐fold against Delta. The neutralization titer against Omicron at Day 14 post booster vaccination was 48.73 (28.61–82.99). At Day 28 post booster vaccination, GMTs remained stable or increased to 414.20 (284.20–603.60), 203.50 (123.90–334.10), 294.90 (164.10–529.80), and 47.69 (26.37–86.24) against the Beta, Delta, and Omicron prototypes, respectively. Post booster vaccination, 100% of samples showed positive neutralization activity against Omicron, albeit illustrating a significant reduction (5.86‐ to 14.98‐fold) against Omicron compared to the prototype at 14 days after the homologous or heterologous vaccine boosters.
This suggests that in individuals who have received two doses of mRNA vaccines (ChAdOx1, BNT162b2) or inactivated vaccines (BBIBP‐CorV), the third dose of vaccine booster injection could improve neutralization against Omicron.
2. CONCLUSION
To date, the Omicron variant is the highest mutated variant among VOCs. It has 50 mutations in the whole genome and 26–32 mutations in the spike protein. Many mutations have been found in VOCs (Alpha, Beta, Gamma, and Delta) and VOIs (Kappa, Zeta, Lambda, and Mu) and are linked to heightened infectivity and increased risk of increased risk reinfection. Moreover, these mutations give the Omicron variant the ability to evade convalescent plasma and vaccine sera and mAbs. The Omicron variant easily escaped immunity compared with other VOCs (Alpha, Beta, Gamma, and Delta). The Omicron variant may be the most transmissible VOC and is becoming the main epidemic variant in many countries worldwide.
CONFLICT OF INTERESTS
The authors declare no conflict of interest.
AUTHOR CONTRIBUTIONS
Dandan Tian conceived and wrote the manuscript and prepared the figures. Yanhong Sun and Huihong Xu contributed to the data and literature collection. Qing Ye conceived and contributed to the modification and revision of the manuscript. All authors contributed to this article and approved the submitted versions.
Tian D, Sun Y, Xu H, Ye Q. The emergence and epidemic characteristics of the highly mutated SARS‐CoV‐2 Omicron variant. J Med Virol. 2022;94:2376‐2383. 10.1002/jmv.27643
DATA AVAILABILITY STATEMENT
The data supporting this study's findings are available from the corresponding author upon reasonable request.
REFERENCES
- 1. Lu R, Zhao X, Li J, et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395(10224):565‐574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Zhou P, Yang XL, Wang XG, et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579(7798):270‐273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ye Q, Wang B, Mao J. The pathogenesis and treatment of the ‘Cytokine Storm’ in COVID‐19. J Infect. 2020;80(6):607‐613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ye Q, Wang B, Mao J, et al. Epidemiological analysis of COVID‐19 and practical experience from China. J Med Virol. 2020;92(7):755‐769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Chen Z, Wang B, Mao S, Ye Q. Assessment of global asymptomatic SARS‐CoV‐2 infection and management practices from China. Int J Biol Sci. 2021;17(4):1119‐1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Su S, Wong G, Shi W, et al. Epidemiology, genetic recombination, and pathogenesis of coronaviruses. Trends Microbiol. 2016;24(6):490‐502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Pachetti M, Marini B, Benedetti F, et al. Emerging SARS‐CoV‐2 mutation hot spots include a novel RNA‐dependent‐RNA polymerase variant. J Transl Med. 2020;18(1):179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Leung K, Shum MH, Leung GM, Lam TT, Wu JT. Early transmissibility assessment of the N501Y mutant strains of SARS‐CoV‐2 in the United Kingdom, October to November 2020. Euro Surveill. 2021;26(1):2002106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Makoni M. South Africa responds to new SARS‐CoV‐2 variant. Lancet. 2021;397(10271):267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Faria NR, Mellan TA, Whittaker C, et al. Genomics and epidemiology of the P.1 SARS‐CoV‐2 lineage in Manaus, Brazil. Science. 2021;372(6544):815‐821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Singh J, Rahman SA, Ehtesham NZ, Hira S, Hasnain SE. SARS‐CoV‐2 variants of concern are emerging in India. Nat Med. 2021;27(7):1131‐1133. [DOI] [PubMed] [Google Scholar]
- 12. Tian D, Sun Y, Zhou J, Ye Q. The global epidemic of SARS‐CoV‐2 variants and their mutational immune escape. J Med Virol. 2021:1‐11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Tian D, Sun Y, Zhou J, Ye Q. The global epidemic of the SARS‐CoV‐2 Delta variant, key spike mutations and immune escape. Front Immunol. 2021;12:751778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Harvey WT, Carabelli AM, Jackson B, et al. SARS‐CoV‐2 variants, spike mutations and immune escape. Nat Rev Microbiol. 2021;19(7):409‐424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Organization WH . Classification of Omicron (B.1.1.529): SARS‐CoV‐2 variant of concern. 2021. https://www.who.intnewsitem/26-11-2021-classification-of-omicron-(b.1.1.529)-sars-cov-2-variant-of-concern
- 16. Gu H, Krishnan P, Ng DYM, et al. Probable transmission of SARS‐CoV‐2 Omicron variant in quarantine hotel, Hong Kong, China, November 2021. Emerg Infect Dis. 2021;28(2):460‐462. 10.3201/eid2802.212422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ye Q, Lai EY, Luft FC, Persson PB, Mao J. SARS‐CoV‐2 effects on the renin‐angiotensin‐aldosterone system, therapeutic implications. Acta Physiol (Oxf). 2021;231(4):e13608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lan J, Ge J, Yu J, et al. Structure of the SARS‐CoV‐2 spike receptor‐binding domain bound to the ACE2 receptor. Nature. 2020;581(7807):215‐220. [DOI] [PubMed] [Google Scholar]
- 19. Hoffmann M, Kleine‐Weber H, Schroeder S, et al. SARS‐CoV‐2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181(2):271‐280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Volz E, Hill V, McCrone JT, et al. Evaluating the effects of SARS‐CoV‐2 spike mutation D614G on transmissibility and pathogenicity. Cell. 2021;184(1):64‐75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Islam R, Hossain J. Detection of SARS‐CoV‐2 Omicron (B.1.1.529) variant has created panic among the people across the world: what should we do right now?. J Med Virol. 2021;1‐2. 10.1002/jmv.27546 [DOI] [PubMed] [Google Scholar]
- 22. Chi X, Yan R, Zhang J, et al. A neutralizing human antibody binds to the N‐terminal domain of the Spike protein of SARS‐CoV‐2. Science. 2020;369(6504):650‐655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.University S coronavirus antiviral & resistance database. https://covdb.stanford.edu/page/susceptibility-data/
- 24. Starr TN, Greaney AJ, Hilton SK, et al. Deep mutational scanning of SARS‐CoV‐2 receptor binding domain reveals constraints on folding and ACE2 binding. Cell. 2020;182(5):1295‐1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Liu H, Zhang Q, Wei P, et al. The basis of a more contagious 501Y.V1 variant of SARS‐CoV‐2. Cell Res. 2021;31(6):720‐722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hou YJ, Chiba S, Halfmann P, et al. SARS‐CoV‐2 D614G variant exhibits efficient replication ex vivo and transmission in vivo. Science. 2020;370(6523):1464‐1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Meng B, Kemp SA, Papa G, et al. Recurrent emergence of SARS‐CoV‐2 spike deletion H69/V70 and its role in the Alpha variant B.1.1.7. Cell Rep. 2021;35(13):109292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zahradník J, Marciano S, Shemesh M, et al. SARS‐CoV‐2 variant prediction and antiviral drug design are enabled by RBD in vitro evolution. Nat Microbiol. 2021;6(9):1188‐1198. [DOI] [PubMed] [Google Scholar]
- 29. Callaway E. Delta coronavirus variant: scientists brace for impact. Nature. 2021;595(7865):17‐18. [DOI] [PubMed] [Google Scholar]
- 30. Sabir DK. Analysis of SARS‐COV2 spike protein variants among Iraqi isolates. Gene Rep. 2022;26:101420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Mertens J, Coppens J, Loens K, et al. Monitoring the SARS‐CoV‐2 pandemic: screening algorithm with single nucleotide polymorphism detection for the rapid identification of established and emerging variants. Clin Microbiol Infect. 2022;28(1):124‐129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Planas D, Bruel T, Grzelak L, et al. Sensitivity of infectious SARS‐CoV‐2 B.1.1.7 and B.1.351 variants to neutralizing antibodies. Nat Med. 2021;27(5):917‐924. [DOI] [PubMed] [Google Scholar]
- 33. Kemp SA, Collier DA, Datir RP, et al. SARS‐CoV‐2 evolution during treatment of chronic infection. Nature. 2021;592(7853):277‐282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Wang Z, Schmidt F, Weisblum Y, et al. mRNA vaccine‐elicited antibodies to SARS‐CoV‐2 and circulating variants. Nature. 2021;592(7855):616‐622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Li Q, Nie J, Wu J, et al. SARS‐CoV‐2 501Y.V2 variants lack higher infectivity but do have immune escape. Cell. 2021;184(9):2362‐2371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wang R, Zhang Q, Ge J, et al. Analysis of SARS‐CoV‐2 variant mutations reveals neutralization escape mechanisms and the ability to use ACE2 receptors from additional species. Immunity. 2021;54(7):1611‐1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Alenquer M, Ferreira F, Lousa D, et al. Signatures in SARS‐CoV‐2 spike protein conferring escape to neutralizing antibodies. PLoS Pathog. 2021;17(8):e1009772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Schmidt F, Weisblum Y, Rutkowska M, et al. High genetic barrier to SARS‐CoV‐2 polyclonal neutralizing antibody escape. Nature. 2021;600(7889):512‐516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Cao Y, Wang J, Jian F, et al. B.1.1.529 escape the majority of SARS‐CoV‐2 neutralizing antibodies of diverse epitopes. 2021. 10.21203/rs.3.rs-1148985/v1.preprint [DOI]
- 40. Chan WS, Au CH, Lam HY, et al. Evaluation on the use of Nanopore sequencing for direct characterization of coronaviruses from respiratory specimens, and a study on emerging missense mutations in partial RdRP gene of SARS‐CoV‐2. Virol J. 2020;17(1):183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Sahin E, Bozdayi G, Yigit S, et al. Genomic characterization of SARS‐CoV‐2 isolates from patients in Turkey reveals the presence of novel mutations in spike and nsp12 proteins. J Med Virol. 2021;93(10):6016‐6026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Benvenuto D, Angeletti S, Giovanetti M, et al. Evolutionary analysis of SARS‐CoV‐2: how mutation of Non‐Structural Protein 6 (NSP6) could affect viral autophagy. J Infect. 2020;81(1):e24‐e27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Maitra A, Sarkar MC, Raheja H, et al. Mutations in SARS‐CoV‐2 viral RNA identified in Eastern India: possible implications for the ongoing outbreak in India and impact on viral structure and host susceptibility. J Biosci. 2020;45(1):76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Guardian M. New COVID‐19 variant is a concern as cases show a sharp increase in Gauteng. 2021. https://mg.co.za/coronavirus-essential/2021-11-25-new-covid-19-variant-is-a-concern-as-cases-show-a-sharp-increase-in-gauteng/
- 45. Torjesen I. Covid‐19: Omicron may be more transmissible than other variants and partly resistant to existing vaccines, scientists fear. BMJ. 2021;375:n2943. [DOI] [PubMed] [Google Scholar]
- 46. Dyer O. Covid‐19: South Africa's surge in cases deepens alarm over Omicron variant. BMJ. 2021;375:n3013. [DOI] [PubMed] [Google Scholar]
- 47. Han X, Ye Q. Kidney involvement in COVID‐19 and its treatments. J Med Virol. 2021;93(3):1387‐1395. [DOI] [PubMed] [Google Scholar]
- 48. Tian D, Ye Q. Hepatic complications of COVID‐19 and its treatment. J Med Virol. 2020;92(10):1818‐1824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Qing Y, Shang S, Fu J, Gong F, Shu Q, Mao J. Crosstalk between coronavirus disease 2019 and cardiovascular disease and its treatment. ESC Heart Fail. 2020;7(6):3464‐3472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Luo HC, You CY, Lu SW, Fu YQ. Characteristics of coagulation alteration in patients with COVID‐19. Ann Hematol. 2021;100(1):45‐52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Ye Q, Wang B, Zhang T, Xu J, Shang S. The mechanism and treatment of gastrointestinal symptoms in patients with COVID‐19. Am J Physiol Gastrointest Liver Physiol. 2020;319(2):G245‐G252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Zhou X, Ye Q. Cellular immune response to COVID‐19 and potential immune modulators. Front Immunol. 2021;12:646333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Enhancing readiness for Omicron (B.1.1.529): technical brief and priority actions for member states. December 10, 2021. https://www.who.int/publications/m/item/enhancing-readiness-for-omicron-(b.1.1.529)-technical-brief-and-priority-actions-for-member-states
- 54.Omicron variant: what you need to know. Updated December 10, 2021. https://www.cdc.gov/coronavirus/2019-ncov/variants/omicron-variant.html
- 55. COVID‐19 variants identified in the UK. 2021. https://www.gov.uk/government/news/covid-19-variants-identified-in-the-uk.
- 56. Pulliam JRC, van Schalkwyk C, Govender N, et al. Increased risk of SARS‐CoV‐2 reinfection associated with emergence of the Omicron variant in South Africa. medRxiv. 2021. 10.1101/2021.11.11.21266068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Han X, Xu P, Ye Q. Analysis of COVID‐19 vaccines: types, thoughts, and application. J Clin Lab Anal. 2021;35(9):e23937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Han X, Ye Q. The variants of SARS‐CoV‐2 and the challenges of vaccines. J Med Virol. 2021. 10.1002/jmv.27513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Tian D, Sun Y, Zhou J, Ye Q. The global epidemic of SARS‐CoV‐2 variants and their mutational immune escape. J Med Virol. 2021:1‐11. 10.1002/jmv.27376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Chen J, Wang R, Gilby NB, et al. Omicron (B.1.1.529): infectivity, vaccine breakthrough, and antibody resistance. ArXiv. 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Gao SJ, Guo H, Luo G. Omicron variant (B.1.1.529) of SARS‐CoV‐2, a global urgent public health alert. J Med Virol. 2021. 10.1002/jmv.27491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Cele S, Jackson L, Khoury DS, et al. ARS‐CoV‐2 Omicron has extensive but incomplete escape of Pfizer BNT162b2 elicited neutralization and requires ACE2 for infection. medRxiv. 2021. 10.1101/2021.12.08.21267417 [DOI] [Google Scholar]
- 63. Pfizer B 2021. Pfizer and BioNTech provide update on Omicron variant. https://www.businesswire.com/news/home/1208005542/en/
- 64. Wang Y, Zhang L, Li Q, et al. The significant immune escape of pseudotyped SARS‐CoV‐2 variant Omicron. Emerg Microbes Infect. 2022;11(1):1‐5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Wilhelm A, Widera M, Grikscheit K, et al. Reduced neutralization of SARS‐CoV‐2 Omicron variant by vaccine sera and monoclonal antibodies. medRxiv. 2021. 10.1101/2021.12.07.21267432 [DOI] [Google Scholar]
- 66. Cameroni E, Saliba C, Bowen JE, et al. Broadly neutralizing antibodies overcome SARS‐CoV‐2 Omicron antigenic shift. Nature. 2021. 10.1038/s41586-021-04386-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Andrews N, Stowe J, Kirsebom F, et al. Effectiveness of COVID‐19 vaccines against the Omicron (B.1.1.529) variant of concern. Biomed Pharmacother. 2021. 10.1101/2021.12.14.21267615 [DOI] [Google Scholar]
- 68. Doria‐Rose NA, Shen X, Schmidt SD, et al. Booster of mRNA‐1273 strengthens SARS‐CoV‐2 Omicron neutralization. medRxiv. 10.1101/2021.12.15.21267805 [DOI] [Google Scholar]
- 69. Ai J, Zhang H, Zhang Y, et al. Omicron variant showed lower neutralizing sensitivity than other SARS‐CoV‐2 variants to immune sera elicited by vaccines after boost. Emerg Microbes Infect. 2021;11:1‐24. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data supporting this study's findings are available from the corresponding author upon reasonable request.


