Dear Editor,
In November 2021, Omicron, discovered in Botswana 1 and classified as the fifth variant of concern 2 , 3 by the World Health Organization on November 26, 2021, the most mutated variant of SARS‐CoV‐2, has now circulated in 150 countries/territories until January 8, 2022, with 552 191 confirmed cases and causing 115 deaths. 4 Omicron was recently divided into three lineages (BA.1, BA.2, and BA.3). 5 The differences between the BA.1 and BA.2 lineages are explored. 6 Here we describe how these three lineages differ in their spike protein. Our study found that there were no specific mutations for the BA.3 lineage in spike protein. Instead, it is a combination of mutations in BA.1 and BA.2 spike proteins.
All three lineages were first detected at approximately the same time and from the same place: BA.1 [hCoV‐19/Botswana/R40B59_BHP_3321001248/2021 (EPI_ISL_6640916) (2021‐11‐11) (Botswana/South East/Greater Gaborone/Gaborone)], BA.2 [hCoV‐19/South Africa/CERI‐KRISP‐K032307/2021 (2021‐11‐17) (South Africa/Gauteng/Tshwane)], and BA.3 [hCoV‐19/South Africa/NICD‐N22163/2021 (EPI_ISL_7605713) (2021‐11‐18) (South Africa/North West)]. Therefore, viruses that develop simultaneously and from the same place have equal chances of spreading worldwide. Though all the three lineages have spread worldwide, the rate of spread of these three lineages is different. A total of 258 129 complete genome sequences up to January 11, 2022, have been submitted to the GISAID database. It contains BA.1 99.13% (255 898/258 129), BA.2 0.85% (2198/258 129), and BA.3 0.013% (33/258 129). Of these three lineages, it is questionable why only BA.1 dominates much more than the other lineages. This is likely due to differences in mutations in the spike protein required for virus transmission and host cell entry. 7 , 8
We analyzed whether there were any specific mutations in the spike protein required for virus transmission and entry in these three lineages of the Omicron variant. We analyzed these three lineages' spike protein mutations in the online tool “SARS‐CoV‐2 (hCoV‐19) Mutation Reports” 9 (Figure 1A). We have analyzed almost all BA.1 (289,761 sequences), BA.2 (3,562 sequences), and BA.3 (39 sequences) sequences available in GISAID on January 12, 2022. This study revealed 37 mutations in the spike protein of BA.1, 31 mutations in BA.2, and 33 mutations in BA.3 (Figure 1B). Of these mutations, 21 mutations (G142D, G339D, S373P, S375F, K417N, N440K, S477N, T478K, E484A, Q493R, Q498R, N501Y, Y505H, D614G, H655Y, N679K, P681H, N764K, D796Y Q954H, and N969K) (Figure 1C) are the most common mutations in all three lineages. These 21 mutations include the N501Y and Q498R mutations, which are expected to enhance binding to the ACE2 receptor, and the H655Y, N679K, and P681H mutations, which are believed to increase spike cleavage and facilitate virus transmission. 10 , 11 , 12
Figure 1.
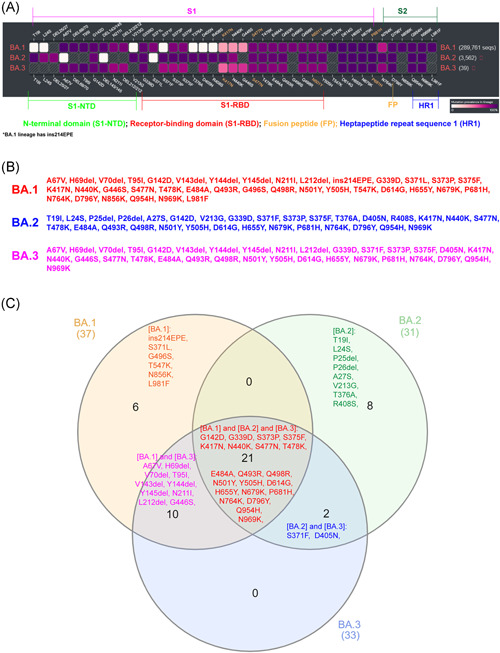
Comparison of spike protein mutations of the BA.1, BA.2, and BA.3 lineages in the Omicron variant. (A) Diagram depicts the comparison of spike protein mutations of the BA.1, BA.2, and BA.3 lineages in the Omicron variant. Furthermore, it represents the comparison of mutations in different functional domains of the spike protein. The color given to each mutation indicates the percentage of this particular mutation in the total available sequences of that lineage (as of January 12, 2022). The scale bar depicts this percentage. (B) Mutations in the BA.1, BA.2, and BA.3 lineages spike protein are shown. The mutation positions in white color in (A) are represented in less than 0.5% of the analyzed sequences, and therefore they were omitted in the analysis. The online tool “SARS‐CoV‐2 (hCoV‐19) Mutation Reports” does not analyze insertions. Also, the BA.1 lineage contains ins214EPE in significant sequences. (C) The Venn diagram shows mutations in the spike protein of BA.1, BA.2, and BA.3 lineages
Comparing BA.1 (289 761 sequences) and BA.2 (3562 sequences) lineages, spike protein revealed 16 (ins214EPE, S371L, G496S, T547K, N856K, L981F, A67V, H69del, V70del, T95I, V143del, Y144del, Y145del, N211I, L212Idel, and G446S) specific mutations in BA.1 and 10 (T19I, L24S, P25del, P26del, A27S, V213G, T376A, and R408S) specific mutations in BA.2 (Figure 1C). The results of this study are almost identical to the results of the previously predicted study with fewer sequences (available sequences at that time). 6 From these, it can be inferred that these 16 specific mutations may play a key role in BA.1 becoming the dominant Omicron lineage that propagates faster than BA.2. However, with 21 common mutations, BA.3 shares ten mutations (A67V, H69del, V70del, T95I, V143del, Y144del, Y145del, N211I, L212del, and G446S) from BA.1 and two (S371F and D405N) mutations from BA.2 to form its spike protein (Figure 1C). In other words, of the 33 mutations in the BA.3 lineage spike protein, 31 mutations are common to BA.1. It is noteworthy that this BA.3 lineage caused the lowest number of cases in these three lineages. Therefore, it can be speculated that the reason for the BA.3 lineage spreading at very low speeds and causing fewer cases may have been due to the loss of six mutations (ins214EPE, S371L, G496S, T547K, N856K, and L981F) from BA.1 or obtaining two mutations from BA.2 (S371F and D405N). Of these six mutations, no studies have been reported on the ins214EPE (S1‐N Terminal domain) mutation. However, it is noteworthy that ins214EPE has not been found in the significant number of BA.1 lineage virus sequences that have recently spread in India. Mutations such as S371L (S1‐receptor‐binding domain), T547K, N856K, and L981F (S2‐ Heptapeptide repeat sequence 1) may require viral oligomerization interfaces. 13 The oligomerization of spike protein is considered to be the most critical factor necessary for the fusion of the virus with the host cell and the formation of the fission pore. 14 , 15 Furthermore, S371L mutation is binding with NAG ligand and antibodies. 13 G496S mutation is present in the host surface receptor binding, antibody recognition sites. 13 Notably, the G496S mildly increases the surface expression of RBD in the yeast‐display study. 16
Experimental studies to discover antivirals against these six mutations in the BA.1 lineage virus may reduce the spread of Omicron in the future. So far, Omicron has been thought to cause mild disease, but it is also possible to develop some mutations that can lead to serious illness. Therefore, we hope that studies and steps to reduce the Omicron spreading rate will be more optimal in controlling more casualties in the future.
CONFLICT OF INTEREST
The authors declare no conflict of interest.
AUTHOR CONTRIBUTIONS
All the authors contributed significantly to this manuscript. Perumal A. Desingu analyzed and wrote the first draft, K. Nagarajan and Kuldeep Dhama reviewed the manuscript. All the authors reviewed and approved the final submission.
ACKNOWLEDGMENTS
Perumal A. Desingu is a DST‐INSPIRE faculty is supported by research funding from the Department of Science and Technology (DST/INSPIRE/04/2016/001067), Government of India, and Core grant from the Science and Engineering Research Board (SERB) (CRG/2018/002192), Department of Science and Technology (DST), Government of India. All the authors acknowledge and thank their respective institutes and universities of affiliation.
Contributor Information
Perumal A. Desingu, Email: perumald@iisc.ac.in, Email: padesingu@gmail.com.
Kuldeep Dhama, Email: kdhama@rediffmail.com.
DATA AVAILABILITY STATEMENT
Not applicable.
REFERENCES
- 1. Lawler DF, Bebiak DM. Nutrition and management of reproduction in the cat. Vet Clin North Am Small Anim Pract. 1986;16(3):495‐519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Mohapatra RK, Tiwari R, Sarangi AK, et al. Twin combination of Omicron and Delta variant triggering a Tsunami wave of ever high surges in COVID‐19 cases: a challenging global threat with a special focus on Indian sub‐continent. J Med Virol. 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kandeel M, Mohamed MEM, Abd El‐Lateef HM, Venugopala KN, El‐Beltagi HS. Omicron variant genome evolution and phylogenetics. J Med Virol. 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. https://newsnodes.com/nu_tracker. Accessed January 11, 2022.
- 5.Enhancing Readiness for Omicron (B.1.1.529): Technical Brief and Priority Actions for Member States. Accessed January 11, 2022. https://www.who.int/docs/default-source/coronaviruse/20211217-global-technical-brief-and-priority-action-on-omicron_latest-2.pdf?sfvrsn=bdd8297c_12
- 6. Majumdar S, Sarkar R. Mutational and phylogenetic analyses of the two lineages of the Omicron variant. J Med Virol. 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Harvey WT, Carabelli AM, Jackson B, et al. SARS‐CoV‐2 variants, spike mutations and immune escape. Nat Rev Microbiol. 2021;19(7):409‐424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ou X, Liu Y, Lei X, et al. Characterization of spike glycoprotein of SARS‐CoV‐2 on virus entry and its immune cross‐reactivity with SARS‐CoV. Nat Commun. 2020;11(1):1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Latif AA, Mullen JL, Alkuzweny M, et al. The Center for Viral Systems Biology. outbreak.info. Accessed January 14, 2022. https://outbreak.info/compare-lineages?pango=BA.1%26pango=BA.2%26pango=BA.3%26pango=B.1.1.529%26gene=S%26threshold=0.2%26nthresh=1%26sub=false%26dark=true
- 10.Science Brief: Omicron (B.1.1.529) Variant. Accessed January 11, 2022. https://www.cdc.gov/coronavirus/2019-ncov/science/science-briefs/scientific-brief-omicron-variant.html
- 11. Andreata‐Santos R, Janini LMR, Duraes‐Carvalho R. From Alpha to Omicron SARS‐CoV‐2 variants: what their evolutionary signatures can tell us? J Med Virol. 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wang L, Cheng G. Sequence analysis of the emerging SARS‐CoV‐2 variant Omicron in South Africa. J Med Virol. 2021;11:59. [DOI] [PubMed] [Google Scholar]
- 13. https://www.gisaid.org/epiflu-applications/covsurver-mutations-app/. Accessed January 11, 2022.
- 14. Tai L, Zhu G, Yang M, et al. Nanometer‐resolution in situ structure of the SARS‐CoV‐2 postfusion spike protein. Proc Natl Acad Sci U S A. 2021;118(48):e2112703118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Xiong X, Qu K, Ciazynska KA, et al. A thermostable, closed SARS‐CoV‐2 spike protein trimer. Nat Struct Mol Biol. 2020;27(10):934‐941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Starr TN, Greaney AJ, Hilton SK, et al. Deep mutational scanning of SARS‐CoV‐2 receptor binding domain reveals constraints on folding and ACE2 binding. Cell. 2020;182(5):1295‐1310.e20. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.


