Abstract
COVID's Omicron variant has sparked a slew of concerns across the globe. This review aims to provide a brief overview of what we know about the Omicron variant right now. The new variant has been discovered in 149 countries across all six World Health Organization (WHO) regions since its discovery in South Africa on November 24, 2021 and became the dominant variant in the country in less than 3 weeks. The WHO has warned that the B.1.1.529 variant is spreading at an unprecedented rate, and has urged countries to prepare for the worst. Over the course of this time, researchers from Africa and around the world have uncovered a wealth of information about the virus's epidemiology and biological properties. Case numbers are increasing exponentially in hard‐hit areas such as South Africa, United Kingdom, and USA (overtaking the delta variant), implying that the variant is highly transmissible. Initial research has provided some insights into the efficacy of vaccines against the Omicron variant and whether it produces major illness, however, much remains unknown, and additional work is needed to investigate what the initial reports represent in real‐world situations.
Keywords: B.1.1.529 variant, COVID‐19, Omicron, SARS‐Cov‐2 coronavirus, transmission rate
Highlights
-
1.
The current Omicron (B.1.1.529) SARS‐CoV‐2 variant outbreak summary in the world is addressed.
-
2.
Up to date information on its emergence, epidemiology, genomics, replication, clinical manifestations, diagnosis, and management strategies are discussed here.
-
3.
Omicron variant spike protein has a higher affinity for the human ACE2 receptor.
-
4.
Omicron has a significant effect on vaccine‐induced neutralizing antibody titers.
-
5.
The mutations in the Omicron receptor‐binding domain (RBD) have possible implications for the ongoing COVID pandemic.
1. INTRODUCTION
The arrival of Omicron proves that we are not done with COVID yet, and that COVID is not done with us either. We need to know where Omicron came from to look ahead and see where the virus is progressing. One thing is certain: more variants will emerge from the same source as this one. Omicron is unlike any other variant in the market at the moment. More than 300 million people have been infected and over 5.5 million people have died as a result of the corona virus 2019 (COVID‐19) pandemic caused by SARS‐CoV‐2. Omicron has been found in 149 countries across all six World Health Organization (WHO) regions since it is identified in Botswana (Figure 1). In the ongoing SARS‐Cov‐2 coronavirus pandemic, the World Health Organization identified Omicron as a variant of concern (VOC). In less than one month of Omicron variant discovery, it is on rise in countries such as South Africa, United Kingdom, and Denmark. 1 , 2 The UK had recorded the largest daily number of new coronavirus cases since the outbreak began. In addition, the number of daily cases in South Africa reached a new high level. The dominance of the Omicron coronavirus variant in Denmark was confirmed by the Statens Serum Institute (10 000 cases of infection were reported in one day). 3
Figure 1.
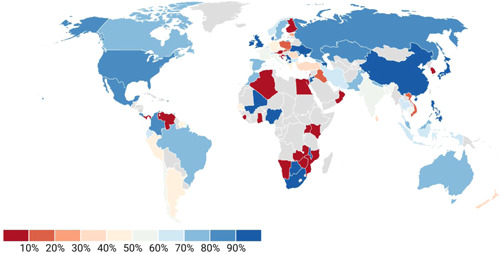
Worldwide percentage share of Omicron variant
Variants such as Omicron pose the greatest threat to the end of pandemic, and researchers alerted that this will continue to arise as the virus mutates. 4 As cases of coronavirus infection with the Omicron variant increase all around the world, researchers are racing to characterize the heavily mutated variant to know how it tends to spread and assess new risks throughout this wave of the global epidemic.
The Omicron variant contains over 30 mutations to the virus's spike proteins, which cover the virus's outside and are the primary targets of vaccines and treatments such as monoclonal antibodies. The Omicron variant multiplies about 70 times faster than the delta variant inside human respiratory tract tissue, according to researchers at the University of Hong Kong. 4 , 5 The variant also reaches higher levels in the tissue, compared with delta, 48 h after infection. Garcia‐Beltran and his colleagues also suggests Omicron is more infectious than delta. 6 , 7 The symptoms appeared to be similar to that of other coronavirus variants. The Omicron variant of the coronavirus continues to spread throughout the world with symptoms of runny nose, headache, fatigue (either mild or severe), sneezing, and sore throat‐like symptoms in infected people (Figure 2). 8
Figure 2.

Omicron outbreak: what do we know so far. WHO, World Health Organization
2. TIMELINE RECAPPED
The first genome which is now known as the Omicron variant was sequenced in early November 2021 from a viral isolate collected in Gauteng, South Africa, and made publicly available through a virus genome sharing database GISAID by the National Institute for Communicable Diseases, South Africa. Further investigation revealed that genomes from Botswana and Hong Kong had been deposited, each with a large number of mutations, particularly in the spike protein. The cluster of genomes was reported to the Pango Network, an open community of researchers working together to annotate SARS‐CoV‐2 lineages, and the lineage was given the name B.1.1.529. 6
The lineage was designated as a VOC due to the unique cluster, which was epidemiologically associated with an increase in cases in Gauteng, and the settings of mutations, many of which had previously been linked to immune escape and advice by the Technical Advisory Group on SARS‐CoV‐2 Virus Evolution (TAG‐VE) (Figure 3). 1
Figure 3.
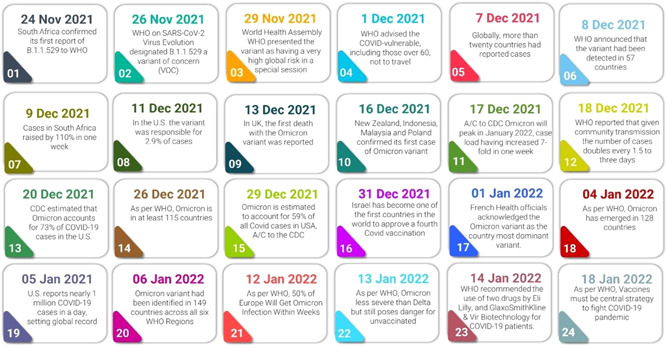
The SARS‐CoV‐2 Omicron variant's timeline
Over 2700 sequences of the Omicron variant are now available in GISAID, a database that shares genomic sequences deposited by researchers from around the world. As more sequences were submitted to GISAID, it became clear that some genomes lacked the complete set of mutations that define Omicron, despite having many of the signature mutations. The Pango Network split the initially designated B.1.1.529 lineage into two sister lineages, BA.1 and BA.2 (where BA is an alias for B.1.1.529). Whereas both of these lineages have most of the essential spike protein mutations identified for Omicron, the lineage BA.2 lacks a deletion in the spike protein found in the original lineage (BA.1). 1 , 6
The transmission rate, immune evasion, and the proportions of people who experience serious diseases and die are all useful parameters for determining how the variant will affect the population. 9 , 10 The findings for each of these parameters are discussed in detail in the following.
3. MECHANISM
In silico analysis revealed that the Omicron SARS‐CoV‐2 variant spike protein has a higher affinity for the human ACE2 receptor. 11 Omicron's spike protein had at least 30 amino acid substitutions, three small deletions, and one small insertion. 12 Omicron clings to cells more tightly, making them more resistant to antibodies. When the spike grabs onto a cell‐surface protein called angiotensin‐converting enzyme (ACE2), molecular interactions were discovered using computational methods of the spike protein on Omicron's surface (Figure 4). 13 , 14
Figure 4.
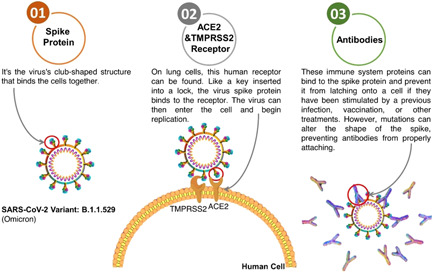
Omicron variant attaches to cells and antibodies sheds light on its behavior
4. IMMUNE EVASION
There are two parts to the immune system. Antibodies recognize and bind proteins on microorganisms' surfaces, and neutralize them. T cells, which can recognize and kill virus‐infected cells, are involved in the second type of infection. The antibody‐mediated response is thought to determine the initial barrier of infection, whereas the cellular response is crucial in the progression of disease severity in COVID‐19. 15 Antibodies and are the mainstays of Omicron's evidence. 16
The Omicron variant has approximately 32 mutations in the spike protein, most of which have been linked to antibody‐binding sites, implying that it would evade antibodies from earlier infections, vaccines, and several monoclonal antibodies used in diagnosis. 6 Whereas clinical assumptions of vaccine efficacy can only be made after a significant number of tract infections with clearly delineated vaccination status have occurred, initial assessments of immune evasion could be made in the lab using neutralization tests. Whole viruses or virus‐like particles (pseudoviruses) along with sera from persons who have been vaccinated or have been infected heretofore are assessed in these studies. This determines whether the virus can evade the antibodies found in these people's sera. While these are not exact indicators of vaccine effectiveness, they can only give us a heads‐up. 17
Many studies from around the world are available publicly. While the ranges of neutralization compared to the ancestral lineages and Delta are wide, it is clear that Omicron's neutralization is significantly less than that of the ancestral lineage (B.1, 20–40‐fold lower) or Delta (about fivefold lower). The only silver lining is that antibodies from booster shots or infections before vaccination (hybrid immunity) appear to neutralize the virus to some limited extent. 18
Evidence on vaccine efficacy was also reported from the United Kingdom Health Security Agency, who assessed vaccine efficacy against symptomatic infection and found that the findings of the lab matched up very well.
Two doses of AstraZeneca (ChAdOx1) have seemed to provide very few protections against symptomatic infection, whereas an added booster with mRNA vaccine appears to provide much greater protection. Because these are disrupted events, estimates for protection against severe disease and death would be much later. In terms of public health, this means that a large number of people with preexisting immunity from COVID‐19 infections or two doses of vaccines may still experience symptomatic re‐infections and vaccine breakthrough infections. 19 , 20
5. GENETIC MUTATIONS IN OMICRON
SARS‐CoV‐2 is still mutating, resulting in the emergence of several new variants. Gao et al. 21 found that it may have progressed in a large immunocompromised population with a lesser vaccination rate, where healthcare infrastructures are relatively weaker than in other countries. Koleya et al. explained the spread of Omicron by their investigation. Omicron spike glycoprotein has 30 mutations spread throughout all of the trimeric protein's domains. In comparison to wild‐type residues, mutant residues are involved in a greater number of intramolecular interactions. Seven mutations are found at the interacting interphase of receptor‐binding domain (RBD) with human ACE receptor, resulting in 17 interactions. The amount and quality of these interactions indicated that the Omicron RBD domain is more effective at binding to receptors. 22 In the Omicron variant, the RBD of Spike has 15 mutations. The ACE2 protein binds to mutated residues 2.5 times stronger. The binding domain's total charge shifts to a positive value. Omicron RBD and ACE2 have a significantly more relaxed dynamic in their complex. 23 The omicron variant's perplexing mutational sequence combines contradictory characteristics that may reduce (virological properties) or increase (immunological escape) the variant's transmission in the human population. Omicron is predicted to be less infective than delta due to an irregular distribution of its electrostatic potential and a defect in the S1–S2 cleavage. 24
In Omicron RBD, the S477N mutation has also been found in B.1.620. S477N has been shown to improve RBD's affinity for hACE2. 25 Han et al. discovered that N477, but not S477, produces an H‐bond with S19 from hACE2 to improve Omicron RBD binding to hACE2. E484Q mutation was found in Kappa variants, while E484K mutation was found in Beta, Gamma, Zeta, Eta, and Theta variants. 26 , 27 In the case of the Omicron variant, E484 became A484. In hACE2, E484 of the RBD prototype forms a weak contact with K31. The side chain is too short to make contact with hACE2, leading to decreased binding. Both Q493K and Q498H enhance SARS‐CoV‐2 RBD‐binding affinity to hACE2. 28 Both Q493 and Q498 in the Omicron RBD‐hACE2 complex were replaced with arginine (R), a positively charged amino acid, and R493 and R498 construct salt bridges with E35 and D38, respectively. Among the various naturally occurring mutations, the E484K mutation, which is carried by several variants and could potentially increase receptor‐binding affinity while reducing immune response, has raised serious concerns. Wang et al. found that the E484K mutation improves RBD‐binding affinity to the hACE2 receptor, implying greater transmissibility of E484K‐containing variants. However, RBD‐ binding affinity to the studied neutralizing antibodies/nanobodies is reduced, implying that these antibodies/nanobodies are less effective. 29 Numerous mutations, including the H69/V70 deletions and the T478K and E484A mutations shared by the Omicron variant and other VOCs, have previously been associated to immune escapes and increased neutralizing antibody resistance. 30
The developing variations have a number of critical changes in the RBD, allowing it to evade immune monitoring and so reduce vaccination protection. In comparison to other variants, Omicron has the most mutations in the RBD's receptor‐binding motif (RBM), where E484 and Q493 have been shown to play major roles in immune evasion. E484A, Q493K, and Q493R, which are responsible for immunological escape, have been consistently documented to develop in immune‐compromised patients or even during monoclonal antibody treatment. 31 , 32 Surprisingly, when compared to the prototype SARS‐CoV‐2, the neutralization capacity of sera inoculated with a double mRNA1273‐vaccine (non‐boosted) and a BNT162b2‐boosted vaccination was lowered by around 20 and 22.7 times for Omicron, respectively. 33
Nucleocapsid mutations in the SARS‐CoV‐2 virus B.1.1.7 origin is associated with R203K/G204R. SARS‐CoV‐2 variants with the 203K/204R mutation have higher transmission and virulence. Mutations in the nucleocapsid protein and spike protein are vital for viral spread during the pandemic. 34
In comparison to other mutating variants, the Omicron variant has an excessive number of mutations, particularly in the Spike gene. Spike gene mutations cause changes in 32 amino acid residues, which is more than in other SARS‐CoV‐2 variants. In the codons for the amino acid residues on the surface of the Spike protein, there are a lot of nonsynonymous mutations, which could affect SARS‐CoV‐2 replication, infectivity, and antigenicity. 35 Single point mutations within region increased the virus's infectivity significantly, up to 150‐fold in some cases. A strong interaction between mutations in the N and S proteins is the most likely explanation for both enhanced immune evasion and enhanced replication capacity of Omicron. 36 Due to multivalent charge‐charge interactions, increased occurrence of positively charged amino acids in some domains of the spike protein (Delta: +4; Omicron: +5 vs. wild type) enhanced binding to cellular polyanionic receptors. This finding sets the foundation for the development of targeted drugs. 37
The RBD of the Omicron variant has 15 mutations, with four of them (K417N, Q493R, N501Y, and Y505H) affecting nine key residues and potentially increasing the virus's ability to bind to hACE2 and infectivity. 38 The N501Y mutation, which has previously been identified in the Beta and Gamma variants of the RBD, is a common mutation. S protein with the N501Y mutation has a ninefold higher affinity for the hACE2 receptor than S protein without the mutation. 39 , 40 Multiple complex structures of SARS‐CoV‐2 RBD‐hACE2 possessing N501Y substitution show that Y501 forms π–π stacking interaction with Y41 and forms H‐bonds with K353 of hACE2. 41 , 42 Notably, Q498R and N501Y mutations of Omicron RBD were previously observed during in vitro evolution, which exhibited ~600‐fold higher affinity with hACE2. 43 K417N, G446S, E484A, G496S, and Y505H substitutions decrease the binding affinity of Omicron RBD with hACE2, whereas S477N and N501Y compensatively increases the binding affinity of Omicron RBD with hACE2.
Inside the Omicron lineage, Wang et al. investigated two subclades with either K417N or K440N and S446K mutations. K417N can boost RBD surface expression and resistance to neutralizing monoclonal antibodies in a mild way, similar to the Beta variant (20H, B.1.351). 44 A few substituted residues in the Omicron RBD‐hACE2 complex reduce the binding interactions between RBD and hACE2, whereas others increase it. Omicron RBDs with the K417N mutation were also found in Beta, Gamma, and even some Delta lineage RBDs. 26 Omicron has accumulated a number of mutations, including S477N, Q498R, and N501Y, which have previously been linked to increased ACE2 receptor binding, 45 potentially improving viral infectivity in host cells.
Omicron has a number of novel mutations in or near the NTD, RBD, RBM, furin cleavage site, and S2 domains, which are thought to affect ACE2 binding and/or antibody binding based on previous research on spike protein‐ACE2 or antibody interactions. 46 As a result, the Omicron variant could be more infectious or vaccine resistant than other VOCs. Omotuyi et al. 47 showed that Omicron is more transmissible and interacts less efficiently with neutralizing convalescent monoclonal antibody, which has implications for transmissibility if other mutations in the S protein induce cell fusion and viral entry. 47 However, the change to basic amino acids improved the transmissibility of the S‐RBD Omicron mutant. 48
6. TRANSMISSION RATE
One other important parameter of consideration is the rate of transmission, which can help determine how quickly a variant spread in a given area and is crucial in developing testing as well as management strategies, including hospitalization. The doubling time is a useful estimate for determining the rate of transmission. South Africa, the United Kingdom, and Denmark have all provided preliminary data in this regard. According to data from African countries, Omicron variant is more transmissible than Delta. The titers of vaccinated individuals' sera to neutralize Omicron were much lower than any of the other variants studied. 49 Omicron was found to have a significant ability to evade immunity from previous infections with other variants or vaccination. 50 In addition, Omicron infected people who were fully vaccinated as well as those who had additional booster shots. 51 The reproduction number of Omicron was 4.2 times greater than that of the Delta variant based on the frequencies of nucleotide sequences of Omicron in Gauteng province, South Africa. 52 Ito et al. 53 found that the effective reproduction number of Omicron is 3.19 times greater than that of Delta under the same epidemiological conditions in Denmark. 53 While in United Kingdom and India reproduction number of Omicron was found to be 4.0 and 2.5, respectively 54 , 55 (Figure 5). Initial estimates suggest that the Omicron variant's doubling time is 2.5–3 days, which is significantly less than the Delta variant. This significant advantage of Omicron over Delta indicates that in areas where the Delta variant is still being transmitted widely, such as the United Kingdom, the Omicron parentage would arise as the prominent lineage in 3–4 weeks. 38 , 56
Figure 5.
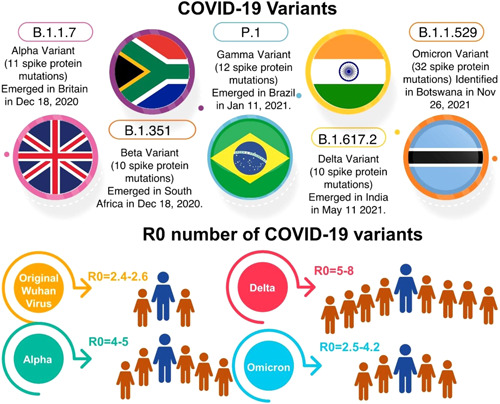
Comparison of COVID‐19 variants and its basic reproductive number
From a health standpoint, a higher rate of transmission would mean a large number of people could be exposed to the virus in a short time. This would have an impact on the system's ability to check as well as provide appropriate care to those in need. This is critical because a large wave of infections can rapidly overwhelm existing healthcare capacity. The majority of cases affected by this new coronavirus variant in the United States increased sevenfold in just a week, from 0.4% to 2.9%, according to the Centers for Disease Control and Prevention. 57 In England, the chance of exposure to the omicron variant to another family member is three times greater than the risk of spreading the delta variant. 58
7. DISEASE SEVERITY
Some other key consideration for public health is disease severity, which is presumably the most challenging to properly estimate because disease severity and deaths occur late in the phase of illness, making accurate assessment time‐consuming. 59 There could also be biases in the data, such as demographics, reinfections, or vaccination, making the findings nongeneralizable to other settings. 60 Among these constraints, beginning and preliminary figures from South Africa suggest that the proportion of Omicron variant patients requiring hospitalization in Gauteng province is much lower than in previous waves. 6 Oxygen demands and Intensive Care Unit admissions have both shown similar patterns. 61 While this is a positive aspect, a reasonably high rate of transmission could quickly exhaust existing healthcare ability, resulting in more deaths than predicted. Such healthcare stress may also lead to an increase in mortalities that are not linked to COVID‐19. 62 Omicron multiplies 70 times faster in tissues that line airway passages than the earlier Delta variant, potentially facilitating person‐to‐person spread, according to the researchers. However, Omicron replicates 10 times slower in lung tissues than the original coronavirus, which could contribute to less severe illness. 12 Amidst increased infectivity and transmissibility, Otaki et al. 63 found that the Omicron variant has developed to have greater antigenicity as well as less virulence in humans.
8. IMPACT IN DIAGNOSTICS
The broadly used reverse‐transcriptase polymerase chain reaction (RT‐PCR) test for diagnosing SARS‐CoV‐2 infectious diseases uses primers that bind to the virus's genome to amplify a specific genomic region. A typical RT‐PCR kit contains two or more sets of primers that target the genome at two or more genes to improve diagnostic sensitivity and specificity. 64 Mutations in the virus genome, which may be where the primers bind by chance, could render these primers ineffective. This is referred to as a dropout or target failure. 65 This would usually not affect the diagnosis because the other sites would continue to function normally. One of the mutations in the Omicron variant is right at the primer binding site, which is targeted by some of the most widely used kits around the world. As a result of the Omicron variant, the spike primers will not work properly. This is known as a Spike Gene Target Failure, or S‐dropout, and it is been used as a surrogate for Omicron variants in surveillance. 66 The important thing to remember is that the S‐dropout only applies to Omicron's BA.1 cluster; the BA.2 cluster, which makes up a tiny fraction of Omicron, does not cause S‐dropout and thus may be overlooked. This also emphasizes the significance of genomic surveillance to determine accurate surveillance assessments. 67
9. VACCINES
In humans, the severity of the disease is determined not only by virus replication but also by each individual's immune response to the infection, which can sometimes progress to life‐threatening inflammation. 68 However, if the virus itself is less harmful to humans, a highly infectious virus can infect a more serious disease and death by infecting a large number of people. 69 As a result, when combined with current studies indicating that the Omicron variant can partially evade vaccine and infection‐induced immunity, the overall threat posed by the Omicron variant is likely to be significant. So many studies have suggested that T cells in people who have been vaccinated can mount a strong defense against the variant, which could help prevent severity of disease, hospitalization, and death. 70 , 71
Scientists are concerned that the mutations in the variant could allow it to avoid some of the defensive antibodies produced by Covid vaccines. 72 Initial laboratory research suggests that it may be somewhat resistant to vaccines, though it is unlikely to be completely immune. Two shots of the Pfizer–BioNTech vaccine were found to be 70% effective in preventing hospitalization from infection with the omicron variant, compared to 90% protection against hospitalization from the delta variant, according to South Africa's leading healthcare administrator. 73 Based on laboratory studies, Pfizer released a statement that a third dose provides strong protection against the new variant. 74
Low levels of protective antibodies against omicron were found in China's Sinopharm vaccine, Russia's Sputnik vaccine, and Johnson & Johnson's vaccine. The study, led by Humabs Biomed SA scientists, provides initial findings on how numerous vaccines available around the world may fare against the heavily mutated variant. 75
Initial research supports vaccine‐producing companies' suspicions that the variant could circumvent some vaccine protection, but researchers have also pointed out that the immune system may have other tools that could help it understand and fight the virus although if antibody levels decline. Willett et al. 76 found that natural infection immunity is more protective than two vaccine doses, but inferior to three doses. Cele et al. showed that Omicron variant successfully evades the Pfizer vaccine regimen's neutralizing antibodies. They found that neutralization protection against the Omicron variant was 40 times lower than against the D614G Triad strain. This includes those who have had the best protection, having been infected and then vaccinated twice with the Pfizer vaccine. Pfizer claims in a recent press release that three doses provide significant protection, but there is no evidence to back up this claim. 77 The spread of SARS‐CoV‐2 Omicron variant has caused concern among cancer patients, according to Valanparambil et al. Non‐small cell lung cancer patients had lower binding and live‐virus‐neutralizing antibody titers to SARS‐CoV‐2 mRNA vaccines than healthy vaccines, with significantly decreased live‐virus neutralization of the Delta and Omicron variants than the wild‐type strain. 78 The neutralizing potential of vaccine‐induced and hybrid immunity‐induced antibodies is significantly reduced in the Omicron variant. This could justify immune escape and high transmission despite extensive vaccination coverage. 79 The BNT162b2 vaccine's third dosage (booster) may boost the level of cross‐neutralizing antibodies to the omicron variant. 80
10. SILVER LINING
Preliminary evidence suggests that people who have achieved hybrid immunity (infection plus vaccination) tend to be well protected against severe disease and death. 81 According to preliminary data from South Africa, the proportion of infected patients who need to be hospitalized is lower than in previous waves. While this does not necessarily imply that the variant is "milder" than other variants, it does suggest that populations similar to South Africa's young and with a high number of previous infections may be less influenced at this time. 6
11. STRATEGIES TO MANAGE THE UPCOMING WAVE OF INFECTIONS
The data show that vaccines do protect against severe death and disease, and therefore urgent need is to vaccinate as many people as possible, particularly those in high‐risk groups, with two doses of vaccines. 82 , 83 With such a high rate of transmission, even a small number of patients' hospitalization can put a huge strain on available resources, both for testing and for providing care to those in need. As a result, measures to slow transmission through coordinated public health strategies must be designed and executed ahead of time, minimizing the impact of such interventions on livelihoods. 84 , 85 , 86
The present crisis also necessitates a strong emphasis on tried‐and‐true prevention strategies. A growing body of evidence suggests that nonpharmacological interventions, such as masks and ventilation, are effective, despite their underappreciation. There is also compelling evidence that masks, particularly high‐quality masks (FFP2/N95), are highly effective in preventing infection. 87 Given the evidence of a high rate of transmission, it is critical to consider better safeguards to prevent vulnerable populations, such as those over 60, those with multiple comorbidities, and those on immunosuppression, with better masks. 88 , 89
Likewise, the significance of ventilation and social distance cannot be overstated, particularly in situations where a huge number of people are probably to congregate during the festive and wedding season. The primary focus on ventilation in places where numerous footfalls are expected, such as schools, government offices, and wedding halls, would go a long way toward limiting transmission during the season. 90 , 91
Molecular surveillance methods, such as whole‐genome sequencing and the use of spike‐dropouts as surrogates for assessing prevalence, are extremely useful in determining the prevalence in communities, assessing growth, and preparing healthcare systems to manage the onslaught of cases well ahead of time. 92
12. CONCLUSION AND FUTURE PERSPECTIVES
Omicron was discovered in South Africa on November 24, 2021, and it only took 3 weeks for it to become the dominant variant in the country. It is also on track to supplant the delta variant and take over as the dominant variant in the United Kingdom. The majority of early research indicates that the omicron variant is more contagious than previous strains of the coronavirus, but a definitive picture of its transmissibility is still unknown. Cases in hard‐hit areas, such as South Africa and the United Kingdom, are increasing at an exponential rate, implying that the variant is highly transmissible.
The new evidence on the omicron variant provides far too little solace. The rapid spread of the Omicron variant is likely to put public health systems to the test, particularly their ability to plan and implement strategies ahead of time, as well as their ability to respond efficiently and quickly. It is probably the best time for general public to get their vaccines, pull up their masks, let a lot of fresh air into their rooms, and avoid crowds. It is always preferable to be safe rather than sorry. Breakthrough infections have been seen in patients who have been vaccinated but have become infected afterward, according to global data. However, the severity of the infection is significantly reduced after vaccination. The mutations may aid in the spread of the variant and allow it to evade specific antibodies produced by vaccines or natural immunity resulting from past infections. Omicron is also a warm‐up for the next pandemic. The task at hand is to detect, track, and slow the spread of a health threat (epidemics which are beyond comprehension.). This is the type of work that will be required to avert the next pandemic outbreak.
CONFLICT OF INTERESTS
The authors declare that there are no conflict of interests.
ETHICS STATEMENT
Not applicable.
AUTHOR CONTRIBUTIONS
Conceptualization, investigation, writing‐original draft preparation, and software: Vineet Sharma; data curation and formal analysis: Himanshu Rai; editing: Dev Nath Singh Gautam and Pradeep Kumar Prajapati; writing‐review, editing, and supervision: Rohit Sharma.
Sharma V, Rai H, Gautam DNS, Prajapati PK, Sharma R. Emerging evidence on Omicron (B.1.1.529) SARS‐CoV‐2 variant. J Med Virol. 2022;94:1876‐1885. 10.1002/jmv.27626
DATA AVAILABILITY STATEMENT
Data sharing not applicable to this article as no datasets were generated or analyzed during the current study.
REFERENCES
- 1.Update on Omicron. WHO. Nov 28, 2021. Accessed January 18, 2022. https://www.who.int/news/item/28-11-2021-update-on-omicron
- 2. CDC COVID‐19 Response Team . SARS‐CoV‐2 B.1.1.529 (Omicron) variant—United States, December 1–8, 2021. MMWR Morb Mortal Wkly Rep. 2021;70(50):1731‐1734. 10.2807/1560-7917.ES.2015.20.34.30002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Espenhain L, Funk T, Overvad M, et al. Epidemiological characterisation of the first 785 SARS‐CoV‐2 Omicron variant cases in Denmark, December 2021. Euro Surveill. 2021;26(50):2101146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bai Y, Du Z, Xu M, et al. International risk of SARS‐CoV‐2 Omicron variant importations originating in South Africa. medRxiv. 2021. 10.1101/2021.12.07.21267410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Lu L, Mok BW, Chen LL, et al. Neutralization of SARS‐CoV‐2 Omicron variant by sera from BNT162b2 or Coronavac vaccine recipients. medRxiv. 2021. 10.1101/2021.12.13.21267668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tracking of variants. GISAID. 2021. Accessed November 30, 2021. https://www.gisaid.org/hcov19-variants/
- 7. Garcia‐Beltran WF, Lam EC, St. Denis K, et al. mRNA‐based COVID‐19 vaccine boosters induce neutralizing immunity against SARS‐CoV‐2 Omicron variant. Cell. 2021;184:2372‐2383.e9. 10.1016/j.cell.2021.12.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lacobucci G. Covid‐19: runny nose, headache, and fatigue are commonest symptoms of omicron, early data show. BMJ. 2021:375. 10.1136/bmj.n3103 [DOI] [PubMed] [Google Scholar]
- 9. Rao S, Singh M. The newly detected B.1.1.529 (Omicron) variant of SARS‐CoV‐2 with multiple mutations: implications for transmission, diagnostics, therapeutics, and immune evasion. DHR Proc. 2021;1(S5):7‐10. 10.47488/dhrp.v1iS5.35 [DOI] [Google Scholar]
- 10. He X, Hong W, Pan X, Lu G, Wei X. SARS‐CoV‐2 Omicron variant: characteristics and prevention. MedComm. 2021;2:838‐845. 10.1002/mco2.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ortega JT, Jastrzebska B, Rangel HR. Omicron SARS‐CoV‐2 variant spike protein shows an increased affinity to the human ACE2 receptor: an in silico analysis. Pathogens. 2022;11(1):45. 10.3390/pathogens11010045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Science brief: omicron (B.1.1.529) variant. Centers for Disease Control and Prevention (CDC). 2021. https://www.cdc.gov/coronavirus/2019-/science/science-briefs/scientific-brief-omicron-variant.html [PubMed]
- 13. Dyer O. Covid‐19: Omicron is causing more infections but fewer hospital admissions than delta, South African data show. BMJ. 2021;375:n3104. 10.1136/bmj.n3104 [DOI] [PubMed] [Google Scholar]
- 14. Lubin JH, Markosian C, Balamurugan D, et al. Structural models of SARS‐CoV‐2 Omicron variant in complex with ACE2 receptor or antibodies suggest altered binding interfaces. BioRxiv. 2021. 10.1101/2021.12.12.472313 [DOI] [Google Scholar]
- 15. Bartleson JM, Radenkovic D, Covarrubias AJ, Furman D, Winer DA, Verdin E. SARS‐CoV‐2, COVID‐19 and the aging immune system. Nat Aging. 2021;1(9):769‐782. 10.1038/s43587-021-00114-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Li CX, Noreen S, Zhang LX, et al. A critical analysis of the SARS‐CoV‐2 (COVID‐19) pandemic, emerging variants, therapeutic interventions, and vaccination strategies. Biomed Pharmacother. 2021;146:112550. 10.1016/j.biopha.2021.112550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wu Y, Huang X, Yuan L, et al. A recombinant spike protein subunit vaccine confers protective immunity against SARS‐CoV‐2 infection and transmission in hamsters. Sci Transl Med. 2021;13(606):eabg1143. 10.1126/scitranslmed.abg1143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Schmidt F, Muecksch F, Weisblum Y, et al. Plasma neutralization properties of the SARS‐CoV‐2 Omicron variant. MedRxiv. 2021. 10.1101/2021.12.12.21267646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hall V, Hopkins S. COV‐BOOST: evidence to support rapid booster deployment. Lancet. 2021;398(10318):2209‐2211. 10.1016/S0140-6736(21)02799-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. McIntyre PB, Aggarwal R, Jani I, et al. COVID‐19 vaccine strategies must focus on severe disease and global equity. Lancet. 2021. 10.1016/S0140-6736(21)02835-X [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Gao SJ, Guo H, Luo G. Omicron variant (B.1.1.529) of SARS‐CoV‐2, a global urgent public health alert! J Med Virol. 2021. 10.1002/jmv.27491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Koleya T, Kumara M, Goswami A, Ethayathulla AS, Hariprasada G. Structural modeling of Omicron spike protein and its complex with human ACE‐2 receptor: molecular basis for high transmissibility of the virus. Biochem Biophys Res Commun. 2022;592:51‐53. 10.1016/j.bbrc.2021.12.082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Rath SL, Padhi AK, Mandal N. Scanning the RBD‐ACE2 molecular interactions in Omicron variant. Biochem Biophys Res Commun. 2022;592:18‐23. 10.1016/j.bbrc.2022.01.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Fantini J, Yahi N, Colson P, Chahinian H, La Scola B, Raoult D. The puzzling mutational landscape of the SARS‐2‐variant Omicron. J Med Virol. 2022. 10.1002/jmv.27577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Singh A, Steinkellner G, Köchl K, Gruber K, Gruber CC. Serine 477 plays a crucial role in the interaction of the SARS‐CoV‐2 spike protein with the human receptor ACE2. Sci Rep. 2021;11(1):1. 10.1038/s41598-021-83761-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Han P, Li L, Liu S, et al. Receptor binding and complex structures of human ACE2 to spike RBD from Omicron and Delta SARS‐CoV‐2. Cell. 2022. 10.1016/j.cell.2022.01.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Thye AY, Law JW, Pusparajah P, Letchumanan V, Chan KG, Lee LH. Emerging SARS‐CoV‐2 variants of concern (VOCs): an impending global crisis. Biomedicines. 2021;9(10):1303. 10.3390/biomedicines9101303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Huang K, Zhang Y, Hui X, et al. Q493K and Q498H substitutions in Spike promote adaptation of SARS‐CoV‐2 in mice. EBioMedicine. 2021;67:103381. 10.1016/j.ebiom.2021.103381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wang P, Nair MS, Liu L, et al. Increased resistance of SARS‐CoV‐2 variants B.1.351 and B.1.1.7 to antibody neutralization. BioRxiv. 2021. 10.1101/2021.01.25.428137 [DOI] [Google Scholar]
- 30. Greaney AJ, Starr TN, Gilchuk P, et al. Complete mapping of mutations to the SARS‐CoV‐2 spike receptor‐binding domain that escape antibody recognition. Cell Host Microbe. 2021;29(1):44‐57. 10.1016/j.chom.2020.11.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Focosi D, Novazzi F, Genoni A, et al. Emergence of SARS‐CoV‐2 spike protein escape mutation Q493R after treatment for COVID‐19. Emerg Infect Dis. 2021;27(10):2728‐2731. 10.3201/eid2710.211538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Guigon A, Faure E, Lemaire C, et al. Emergence of Q493R mutation in SARS‐CoV‐2 spike protein during bamlanivimab/etesevimab treatment and resistance to viral clearance. J Infect. 2021. 10.1016/j.jinf.2021.08.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Wilhelm A, Widera M, Grikscheit K, et al. Reduced neutralization of SARS‐CoV‐2 omicron variant by vaccine sera and monoclonal antibodies. MedRxiv. 2021. 10.1101/2021.12.07.21267432 [DOI] [Google Scholar]
- 34. Wu H, Xing N, Meng K, et al. Nucleocapsid mutations R203K/G204R increase the infectivity, fitness, and virulence of SARS‐CoV‐2. Cell Host Microbe. 2021;29(12):1788‐1801. 10.1016/j.chom.2021.11.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ma W, Yang J, Fu H, et al. Genomic perspectives on the emerging SARS‐CoV‐2 Omicron variant. Genomics Proteomics Bioinformatics. 2022. 10.1016/j.gpb.2022.01.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Saito A, Irie T, Suzuki R, et al. Enhanced fusogenicity and pathogenicity of SARS‐CoV‐2 Delta P681R mutation. Nature. 2021:1‐10. 10.1038/s41586-021-04266-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Nie C, Sahoo AK, Herrmann A, Ballauff M, Netz RR, Haag R. Charge matters: mutations in Omicron variant favor binding to cells. ChemBioChem. 2022. 10.1002/cbic.202100681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kumar S, Thambiraja TS, Karuppanan K, Subramaniam G. Omicron and Delta variant of SARS‐CoV‐2: a comparative computational study of spike protein. J Med Virol. 2021. 10.1002/jmv.27526 [DOI] [PubMed] [Google Scholar]
- 39. Nasreen S, He S, Chung H, et al. Effectiveness of COVID‐19 vaccines against variants of concern. Medrxiv. 2021. 10.1101/2021.06.28.21259420 [DOI] [Google Scholar]
- 40. Ali F, Kasry A, Amin M. The new SARS‐CoV‐2 strain shows a stronger binding affinity to ACE2 due to N501Y mutant. Med Drug Discov. 2021;10:100086. 10.1016/j.medidd.2021.100086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Han P, Su C, Zhang Y, et al. Molecular insights into receptor binding of recent emerging SARS‐CoV‐2 variants. Nat Commun. 2021;12(1):1‐9. 10.1038/s41467-021-26401-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Zhu X, Mannar D, Srivastava SS, et al. Cryo‐electron microscopy structures of the N501Y SARS‐CoV‐2 spike protein in complex with ACE2 and 2 potent neutralizing antibodies. PLoS Biol. 2021;19(4):e3001237. 10.1371/journal.pbio.3001237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Zahradník J, Marciano S, Shemesh M, et al. SARS‐CoV‐2 variant prediction and antiviral drug design are enabled by RBD in vitro evolution. Nat Microbiol. 2021;6(9):1188‐1198. 10.1038/s41564-021-00954-4 [DOI] [PubMed] [Google Scholar]
- 44. Wang WB, Liang Y, Jin YQ, Zhang J, Su JG, Li QM. E484K mutation in SARS‐CoV‐2 RBD enhances binding affinity with hACE2 but reduces interactions with neutralizing antibodies and nanobodies: binding free energy calculation studies. J Mol Graph Model. 2021;109:108035. 10.1016/j.jmgm.2021.108035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Wang L, Cheng G. Sequence analysis of the emerging Sars‐CoV‐2 variant Omicron in South Africa. J Med Virol. 2021. 10.1002/jmv.27516 [DOI] [PubMed] [Google Scholar]
- 46. Harvey WT, Carabelli AM, Jackson B, et al. SARS‐CoV‐2 variants, spike mutations and immune escape. Nat Rev Microbiol. 2021;19(7):409‐424. 10.1038/s41579-021-00573-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Omotuyi IO, Afolabi EO, Oyinloye BE, Fatumo S, Femi‐Oyewo MN, Bogoro SE. SARS‐CoV‐2 Omicron spike glycoprotein receptor binding domain exhibits super‐binder ability with ACE2 but not convalescent monoclonal antibody. Comput Biol Med. 2022;142:105226. 10.1016/j.compbiomed.2022.105226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Hanai T. Quantitative in silico analysis of SARS‐CoV‐2 S‐RBD omicron mutant transmissibility. Talanta. 2022;240:123206. 10.1016/j.talanta.2022.123206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Rössler A, Riepler L, Bante D, von Laer D, Kimpel J. SARS‐CoV‐2 B.1.1.529 variant (Omicron) evades neutralization by sera from vaccinated and convalescent individuals. MedRxiv. 2021. 10.1101/2021.12.08.21267491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Pulliam JRC, van Schalkwyk C, Govender N, et al. Increased risk of SARS‐CoV‐2 reinfection associated with emergence of the Omicron variant in South Africa. MedRxiv. 2021. 10.1101/2021.11.11.21266068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Kuhlmann C, Mayer CK, Claassen M, et al. Breakthrough infections with SARS‐CoV‐2 Omicron variant despite booster dose of mRNA vaccine. SSRN Electron J. 2021. https://ssrn.com/abstract=3981711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Nishiura H, Ito K, Anzai A, Kobayashi T, Piantham C, Rodriguez‐Morales AJ. Relative reproduction number of SARS‐CoV‐2 Omicron (B.1.1.529) compared with Delta variant in South Africa. J Clin Med. 2022;11(1):30. 10.3390/jcm11010030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Ito K, Piantham C, Nishiura H. Relative instantaneous reproduction number of Omicron SARS‐CoV‐2 variant with respect to the Delta variant in Denmark. J Med Virol. 2021. 10.1002/jmv.27560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Barnard RC, Davies NG, Pearson CA, Jit M, Edmunds WJ. Projected epidemiological consequences of the Omicron SARS‐CoV‐2 variant in England, December 2021 to April 2022. MedRxiv. 2021. https://www.medrxiv.org/content/10.1101/2021.12.15.21267858v1 [Google Scholar]
- 55. Ranjan R. Omicron Impact in India: an early analysis of the ongoing COVID‐19 third wave. MedRxiv. 2022. 10.1101/2022.01.09.22268969 [DOI] [Google Scholar]
- 56. Karim SS, Karim QA. Omicron SARS‐CoV‐2 variant: a new chapter in the COVID‐19 pandemic. Lancet. 2021;398(10317):2126‐2128. 10.1016/S0140-6736(21)02758-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Centers for Disease Control (CDC). Accessed November 30, 2021. https://covid.cdc.gov/covid-datatracker/#datatracker-home
- 58.SARS‐CoV‐2 variants of concern and variants under investigation in England. UK Health Security Agency (UHSA). Accessed December 10, 2021. https://assets.publishing.service.gov.uk/government/uploads/system/uploads/attachment_data/file/1042367/technical_briefing-31-10-december-2021.pdf
- 59. Zhang H, Wang X, Fu Z, et al. Potential factors for prediction of disease severity of COVID‐19 patients. MedRxiv. 2020. 10.1101/2020.03.20.20039818 [DOI] [Google Scholar]
- 60. Randolph HE, Barreiro LB. Herd immunity: understanding COVID‐19. Immunity. 2020;52(5):737‐741. 10.1016/j.immuni.2020.04.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Bezuidenhout MC, Wiese OJ, Moodley D, et al. Correlating arterial blood gas, acid–base and blood pressure abnormalities with outcomes in COVID‐19 intensive care patients. Ann Clin Biochem. 2020;58(2):95‐101. 10.1177/F0004563220972539 [DOI] [PubMed] [Google Scholar]
- 62. Otaki JM, Nakasone W, Nakamura M. Nonself mutations in the spike protein suggest an increase in the antigenicity and a decrease in the virulence of the Omicron variant of SARS‐CoV‐2. BioRxiv. 2022. 10.1101/2021.12.30.474613 [DOI] [Google Scholar]
- 63. Standish K. A coming wave: suicide and gender after COVID‐19. J Gend Stud. 2021;30(1):114‐118. 10.1080/09589236.2020.1796608 [DOI] [Google Scholar]
- 64. Liu R, Han H, Liu F, et al. Positive rate of RT‐PCR detection of SARS‐CoV‐2 infection in 4880 cases from one hospital in Wuhan, China, from Jan to Feb 2020. Clin Chim Acta. 2020;505:172‐175. 10.1016/j.cca.2020.03.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Toyoshima Y, Nemoto K, Matsumoto S, Nakamura Y, Kiyotani K. SARS‐CoV‐2 genomic variations associated with mortality rate of COVID‐19. J Hum Genet. 2020;65(12):1075‐1082. 10.1038/s10038-020-0808-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Metzger C, Lienhard R, Seth‐Smith H, et al. PCR performance in the SARS‐CoV‐2 Omicron variant of concern? Swiss Med Wkly. 2021;151:w30120. 10.4414/smw.2021.w30120 [DOI] [PubMed] [Google Scholar]
- 67. Shi W, Wang Q, Zhang J, Zhang J, Tan F, Yu S. The first two imported cases of SARS‐CoV‐2 Omicron variant‐Tianjin Municipality, China, December 13, 2021. China CDC Weekly. 2021;3:1‐2. http://weekly.chinacdc.cn/en/article/doi/10.46234/ccdcw2021.266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Yongchen Z, Shen H, Wang X, et al. Different longitudinal patterns of nucleic acid and serology testing results based on disease severity of COVID‐19 patients. Emerg Microbes Infect. 2020;9(1):833‐836. 10.1080/22221751.2020.1756699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Hafeez A, Ahmad S, Siddqui SA, Ahmad M, Mishra S. A review of COVID‐19 (Coronavirus Disease‐2019) diagnosis, treatments and prevention. EJMO. 2020;4(2):116‐125. 10.14744/ejmo.2020.90853 [DOI] [Google Scholar]
- 70. Callaway E, Ledford H. How bad is Omicron? What scientists know so far. Nature. 2021;600(7888):197‐199. 10.1038/d41586-021-03614-z [DOI] [PubMed] [Google Scholar]
- 71. Redd AD, Nardin A, Kared H, et al. Minimal cross‐over between mutations associated with Omicron variant of SARS‐CoV‐2 and CD8+ T cell epitopes identified in COVID‐19 convalescent individuals. BioRxiv. 2021. 10.1101/2021.12.06.471446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Dolgin E. Omicron is supercharging the COVID vaccine booster debate. Nature. 2021. 10.1038/d41586-021-03592-2 [DOI] [PubMed] [Google Scholar]
- 73. Volkov O. Predicted symptomatic effectiveness of Pfizer‐BioNTech BNT162b2 vaccine against Omicron variant of SARS‐CoV‐2. MedRxiv. 2021. 10.1101/2021.12.09.21267556 [DOI] [Google Scholar]
- 74. Mahase E. 2021. Covid‐19: do vaccines work against Omicron—and other questions answered. 10.1136/bmj.n3062 [DOI] [PubMed]
- 75. Cameroni E, Saliba C, Bowen JE, et al. Broadly neutralizing antibodies overcome SARS‐CoV‐2 Omicron antigenic shift. BioRxiv. 2021. 10.1101/2021.12.12.472269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Willett BJ, Grove J, MacLean O, et al. The hyper‐transmissible SARS‐CoV‐2 Omicron variant exhibits significant antigenic change, vaccine escape and a switch in cell entry mechanism. MedRxiv. 2022. 10.1101/2022.01.03.21268111 [DOI] [Google Scholar]
- 77. Cele S, Jackson L, Khoury DS, et al. Omicron extensively but incompletely escapes Pfizer BNT162b2 neutralization. Nature. 2021:1‐5. 10.1038/s41586-021-04387-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Valanparambil RM, Carlisle J, Linderman SL, et al. Antibody response to SARS‐CoV‐2 mRNA vaccine in lung cancer patients: Reactivity to vaccine antigen and variants of concern. MedRxiv. 2022. 10.1101/2022.01.03.22268599 [DOI] [Google Scholar]
- 79. Medigeshi GR, Batra G, Murugesan DR, et al. Sub‐optimal neutralisation of Omicron (B.1.1.529) variant by antibodies induced by vaccine alone or SARS‐CoV‐2 infection plus vaccine (hybrid immunity) post 6‐months. MedRxiv. 2022. 10.1101/2022.01.04.22268747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Rössler A, Riepler L, Bante D, von Laer D, Kimpel J. SARS‐CoV‐2 Omicron variant neutralization in serum from vaccinated and convalescent persons. New Engl J. Med. 2022. 10.1056/NEJMc2119236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Gruell H, Vanshylla K, Tober‐Lau P, et al. mRNA booster immunization elicits potent neutralizing serum activity against the SARS‐CoV‐2 Omicron variant. MedRxiv. 2021. 10.1101/2021.12.14.21267769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Agrawal U, Katikireddi SV, McCowan C, et al. COVID‐19 hospital admissions and deaths after BNT162b2 and ChAdOx1 nCoV‐19 vaccinations in 2.57 million people in Scotland (EAVE II): a prospective cohort study. Lancet Respir Med. 2021;9(12):1439‐1449. 10.1016/S2213-2600(21)00380-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Ladhani SN. Crossing the Rubicon: a fine line between waiting and vaccinating adolescents against COVID‐19. J Infect. 2021;83(3):294‐297. 10.1016/j.jinf.2021.07.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Coccia M. The impact of first and second wave of the COVID‐19 pandemic in society: comparative analysis to support control measures to cope with negative effects of future infectious diseases. Environ Res. 2021;197:111099. 10.1016/j.envres.2021.111099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Cot C, Cacciapaglia G, Islind AS, Óskarsdóttir M, Sannino F. Impact of US vaccination strategy on COVID‐19 wave dynamics. Sci Rep. 2021;11(1):1‐11. 10.1038/s41598-021-90539-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Hale T, Angrist N, Hale AJ, et al. Government responses and COVID‐19 deaths: global evidence across multiple pandemic waves. PLoS One. 2021;16(7):e0253116. 10.1371/journal.pone.0253116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Peters A, Palomo R, Ney H, et al. The COVID‐19 pandemic and N95 masks: reusability and decontamination methods. Antimicrob Resist Infect Control. 2021;10(1):1‐6. 10.1186/s13756-021-00921-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Wang L, Sun Y, Yuan Y, Mei Q, Yuan X. Clinical challenges in cancer patients with COVID‐19: aging, immunosuppression, and comorbidities. Aging. 2020;12(23):24462‐24474. 10.18632/Faging.104205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Shah P, Owens J, Franklin J, et al. Demographics, comorbidities and outcomes in hospitalized Covid‐19 patients in rural southwest Georgia. Ann Med. 2020;52(7):354‐360. 10.1080/07853890.2020.1791356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Chen CY, Chen PH, Chen JK, Su TC. Recommendations for ventilation of indoor spaces to reduce COVID‐19 transmission. J Formos Med Assoc. 2021;120(12):2055‐2060. 10.1016/j.jfma.2021.08.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Pei G, Taylor M, Rim D. Human exposure to respiratory aerosols in a ventilated room: effects of ventilation condition, emission mode, and social distancing. Sustain Cities Soc. 2021;73:103090. 10.1016/j.scs.2021.103090 [DOI] [Google Scholar]
- 92. Oude Munnink BB, Worp N, Nieuwenhuijse DF, et al. The next phase of SARS‐CoV‐2 surveillance: real‐time molecular epidemiology. Nat Med. 2021;27(9):1518‐1524. 10.1038/s41591-021-01472-w [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data sharing not applicable to this article as no datasets were generated or analyzed during the current study.


