Abstract
The novel Severe acute respiratory syndrome coronavirus‐2 (SARS‐CoV‐2) variant, Omicron (PANGO lineage B.1.1.529) is being reported from all around the world. The WHO has categorized Omicron as a Variant of Concern (VOC) considering its higher transmissibility and infectivity, vaccine breakthrough cases. As of January 6, 2022, Omicron has been reported in at least 149 countries. Therefore, this study was planned to investigate the transmission dynamics and mutational prevalence of the novel SARS‐CoV‐2 Omicron variant. The transmission dynamics and Omicron SARS‐CoV‐2 divergence was studied using GISAID and Nextstrain which provides information about the genetic sequences, epidemiological, geographical, and species‐specific data of human, avian, and animal viruses. Further, the mutation prevalence in spike glycoprotein of Omicron was studied, and the frequency of the crucial mutations was compared with the other prevalent VOCs. The transmission dynamics suggest that the Omicron was first identified in South Africa and then it was reported in the United Kingdom followed by the United States and Australia. Further, our phylogenetic analysis suggests that Omicron (BA.1) was clustered distinctly from the other VOCs. In the Spike glycoprotein, the Omicron (B.1.1.529) demonstrates critical 32 amino acid changes. This study may help us to understand mutational hotspots, transmission dynamics, phylogenetic divergence, effect on testing and immunity, which shall promote the progress of the clinical application and basic research.
Keywords: COVID‐19, Delta, Omicron, B.1.1.529, receptor‐binding domain, SARS‐CoV‐2, Variants of Concern
Highlights
First report showing the transmissibility of the novel sSevere acute respiratory syndrome coronavirus‐2 (SARS‐CoV‐2) Omicron (B.1.1.529) Variant of Concern.
Transmission dynamics analysis suggests that Omicron is more transmissible than other Variant of Concerns (VOCs).
Omicron (BA.1) is clustered distinctly from the other VOCs in a monophyletic clade.
The K417N, N440K, and G446S are the less prevalent mutations identified in the RBD of the Omicron SARS‐CoV‐2.
1. INTRODUCTION
The world is facing the recurrent emergence of Severe acute respiratory syndrome coronavirus‐2 (SARS‐CoV‐2) variants. 1 So far, at least 312 million cases and 5.5 million fatalities have been documented globally. Since January 2021, many SARS‐CoV‐2 variants have emerged and transmitted across several countries. According to the WHO, SARS‐CoV‐2 variants have been categorized based on the transmission ability, infectivity, and diagnostic and immune escape ability. SARS‐CoV‐2 variants are categorized as Variant of Concern (VOC), Variant of Interest (VOI), and Variants under monitoring (VUM) to focus global research and awareness, as well as to lead the on‐going worldwide responses to the COVID‐19 pandemic. 2 , 3 Additionally, the variants have been classified based on the genomic sequencing data available at The Phylogenetic Assignment of Named Global Outbreak Lineages (PANGOLIN), Global Initiative on Sharing All Influenza Data (GISAID), and Nextstrain. The Alpha (Pango lineage B.1.1.7), Beta (Pango lineage B.1.351), Gamma (Pango lineage P.1), and Delta (Pango lineage B.1.617.2) variants have been the VOCs till the recent emergence of novel SARS‐CoV‐2 variant Omicron (Pango lineage B.1.1.529). 4
On November 24, 2021, from South Africa, the Omicron was first reported to the WHO. The initial rapid transmission of Omicron in South Africa has put the worldwide health systems and WHO on red alert. The B.1.1.529 lineage has been subsequently sub‐classified into BA.1 (B.1.1.529.1), BA.2 (B.1.1.529.2), and BA.3 (B.1.1.529.3). Importantly, the most prominent sub‐lineage among the Omicron is BA.1 (B.1.1.529.1). 5 Subsequently, the Omicron has been transmitted to Belgium, Israel, Botswana, and Hong Kong. By November 29, 2021, the Omicron was spread in Australia, Austria, the Czech Republic, Belgium, Canada, Italy, Denmark, France, Germany, the Netherlands, and the United Kingdom. 6 , 7 According to the WHO, as of January 6, 2022, the Omicron variant is reported in at least 149 countries. 8 Omicron has a significant growth advantage over the Delta variant and it spreads rapidly than Delta, in countries with known community transmission with a doubling period of 1.5–3 days. 9 This causes worries about the novel variant's greater transmissibility and infectivity, as well as its ability to evade immunity established by natural infections or vaccination. 10 Therefore, the present study was planned to investigate the transmission dynamics, phylogenetic analysis, and mutation prevalence of Omicron SARS‐CoV‐2 that may help us establish effective preventive and therapeutic strategies in the forthcoming COVID‐19 waves. 11 Considering the higher transmissibility, the management of the forthcoming pandemic is crucial to achieve 12 along with implementation of restriction guidelines as it was implemented in the previous outbreaks. 13
2. MATERIALS AND METHOD
2.1. Transmission dynamics of Omicron
To study the transmission dynamics of Omicron, we have used GISAID (https://www.gisaid.org/hcov19-variants/). GISAID provides information about the genetic sequences, epidemiological, geographical, and species‐specific data of human, avian, and animal viruses. The transmission lines were analyzed to understand the transmission dynamics of Omicron in various countries. 14 , 15
2.2. Phylogenetic analysis of Omicron
To determine the Omicron SARS‐CoV‐2 divergence, we employed the Nextstrain, which offers the most recent worldwide genomic sequencing of the SARS‐CoV‐2 data as soon as it is released by GISAID (https://www.gisaid.org/). Phylogenetic tree analysis was performed to observe the mutational divergence of Omicron SARS‐CoV‐2 in the unrooted tree. 16
2.3. Mutation prevalence of Omicron versus other VOCs
To determine the mutation prevalence in spike glycoprotein of Omicron, we have analyzed the frequencies of amino acid substitutions reported on SARS‐CoV‐2 (hCoV‐19) Mutation Reports (https://outbreak.info/compare-lineages?pango=Alpha%26gene=S%26threshold=0.2). The frequency of the crucial mutations was compared with the other prevalent VOCs. 17
2.4. Statistical analysis of mutational prevalence
To identify the most significant mutations in the Omicron, we have taken 60 410 Omicron sequences and performed the χ 2 test analysis where the prevalence of specific mutation was considered as the observed value and the expected value was considered to be 100. If the null hypothesis is true, the observed and expected frequencies will be close in value and the χ 2 statistic will be close to zero. If the null hypothesis is false, then the χ 2 statistic will be large.
3. RESULTS AND DISCUSSION
3.1. Transmission dynamics of Omicron
Omicron‐specific cases increase exponentially and spread worldwide within days of initial identification. 18 Transmission dynamics showing the widespread transmission of novel SARS‐CoV‐2 Omicron (Figure 1A). The transmission dynamics suggest that the Omicron was first identified in South Africa (Figure 1B) and then it was reported in the United Kingdom (Figure 1C) followed by the United States (Figure 1D). Subsequently, the cases were reported from various countries (Figure 1E). As of December 28, 2021, globally the total number of Omicron cases is 53 695 and the highest number of Omicron cases have been identified in the United Kingdom (34 573), USA (8311), and in Denmark (2001), South Africa (1643), Australia (859), Belgium (609), Canada (586), and in Switzerland is (471), and so on. Considering the rapid transmission of Omicron in a short span of time, the Omicron is expected to be more transmissible than other VOCs. 19 , 20 The temporal change in the number of Omicron cases in a population was measured with or with pre‐existing immunity.
Figure 1.

Transmission dynamics of SARS‐CoV‐2 Omicron. (A) Transmission dynamics showing the widespread transmission of novel SARS‐CoV‐2 Omicron, where the color lines are showing the timeline of Omicron transmission. 16 (B) SARS‐CoV‐2 Omicron variant first reported in South Africa followed by the United Kingdom (C) and subsequently spread to the United States (D). 17 (E) Relative SARS‐CoV‐2 Omicron genome frequency per region (exponentially smoothed alpha = 0.3) showing the prevalence of Omicron in South America, Oceania, Europe‐UK, North America, Asia, Africa, and Europe‐noUK 16
3.2. Phylogenetic analysis of Omicron
To study the evolutionary links between the Omicron variant and the recently emerged SARS‐CoV‐2 variants, we have analyzed the phylogenetic data based on the genomic sequencing of SARS‐CoV‐2. When the phylogenetic tree is plotted in unrooted style, the divergence between SARS‐CoV‐2 variants is obvious. We have found that the Omicron (BA.1) was clustered distinctly from the other VOCs producing a monophyletic clade (Figure 2A), based on the recent submission of continent wise Omicron genomic data (Figure 2B). Omicron is distinct from other SARS‐CoV‐2 VOCs because of mutations in the spike suggesting that Omicron formed a new emergent group that was not originating with other variants. Mutations in the spike protein, and more specifically in receptor‐binding domain (RBD) cause the SARS‐CoV‐2 variant's critical infectivity and antibody resistance, which may affect COVID‐19 transmissibility, infectivity, neutralizing antibody escapes, and vaccination breakthrough cases. 22 Alpha, Beta, Gamma, and Delta are four VOCs, and Eta, Iota, Kappa, and Lambda are four VOIs that are now in use across the world. 23 In terms of immune evasion and transmission, Alpha, Beta, and Delta variants of SARS‐CoV2 had a large effect globally, with Delta quickly replacing other variants to prevail globally, including in South Africa. 24
Figure 2.

Mutations and phylogenetic analysis of SARS‐CoV‐2 Omicron. (A) Phylogenetic tree (unrooted) showing the distinct clusters acquired by the novel SARS‐CoV‐2 Omicron. (B) Clade representing continent wise recent submission of Omicron genomic data. 21 (C) Mutations in RBD domain of SARS‐CoV‐2 VOCs in compassion with the novel SARS‐CoV‐2 Omicron 17
3.3. Mutations in Omicron SARS‐CoV‐2
The SARS‐CoV‐2 spike glycoprotein interacts with human ACE2 receptors for attachment and internalization. 25 For emergency use, most medicines and vaccinations authorized have been developed to counteract the spikes and ACE2 interactions to prevent attachment. 26 , 27 Apart from the mutations in the spike glycoprotein, Omicron variant mutations are found in many different SARS‐CoV‐2 proteins, particularly NSP3 (K38R, SΔ1265, A1892T, L1266I), NSP4 (T492I), NSP5 (P132H), NSP6 (I189V, Δ105‐107), NSP12 (P323L), NSP14 (I42V), nucleocapsid protein, envelope protein, and membrane protein. In the S protein receptor‐binding domain (RBD) alterations are being studied for their possible influence on antibody resistance and infectivity.
3.4. Mutation in the RBD of Omicron and its implication
In the spike glycoprotein, the variant of Omicron includes 32 amino acid alterations, three minor deletions in the N terminus domain (NTD), three deletions in Omicron spike that is, Δ143–145, Δ69–70, and Δ211, and one minuscule insertion in spike at 214 positions, that is, ins214EPE, with 15 mutations occurring in the receptor‐binding domain (RBD). 21 Mutations are referred to as Δ69–70, Ala67Val, Thr95Ile, Δ143–145, Δ211, Gly142Asp, ins214EPE, Leu212Ile, Gly339Asp, Ser373Pro, Ser371Leu, Lys417Asn, Ser375Phe, Asn440Lys, Ser477Asn, Gly446Ser, Glu484Ala, Thr478Lys, Gln493Arg Gln498Arg, Gly496Ser, Tyr505His, Asn501Tyr, Asp614Gly, Thr547Lys, Asn679Lys, His655Tyr, Asn764Lys, Pro681His, Asn856Lys, Asp796Tyr, Asn969Lys, Gln954His, Leu981Phe, The RBD has 15 of these changes (residues 319–541). 17 , 25 , 28
Prominently, the 15 RBD mutations are as G339D, S371L, S373P, S375F, K417N, N440K, G440K, G446S, S477N, T478K, E484A, Q493R, G496S, N501Y, and Y505H (Figure 2C). Because of its RBD mutations N440K, T478K, and N501Y, Omicron may be nearly 10 times more transmissible than the original SARS‐CoV‐2. 29 Because of its RBD mutations K417N, E484A, and Y505H, the Omicron has a higher potential to impact the interaction of most 132 antibodies with the S protein, indicating that it has a higher vaccination escape capacity than the Delta variant. K417N, which is also part of the Beta variant that arose in South Africa, produces the most substantial breakdown of recognized antibodies amongst the 15 RBD mutations. E484A is another mutation that has a significant impact on several known antibodies. The variant has several changes and deletions in additional genomic locations. 30 Nonstructural protein modifications (NSP)3 as Lys38Arg, SΔ1265, Ala1892Thr; Leu1266Ile and NSP4 as Thr492Ile; NSP5 as Phe132His; NSP6 as Ile189Val; Δ105–107, NSP12 as Pro323Leu; NSP14‐Ile42Val; membrane (M) as Gln19Glu, Asp3Gly, Ala63Thr; envelope (E) as Thr9Ile nucleocapsid (N) as Gly204Arg, Δ31–33, Pro13Leu, Arg203Lys. In contrast to other common VOCs, such as Delta (B.1.617.2), Omicron has a high mutation rate. Moreover, such Omicron alterations may be associated with increased infectivity, higher viral binding affinity, and antibody evasion. 31 , 32
3.5. Mutational prevalence of Omicron
To observe the mutational prevalence of Omicron, we have compared the prevalence of crucial mutations observed in the Omicron in comparison with the other VOCs. Our analysis suggests that the prevalence of the mutation in RBD as K417N, N440K, and G446S is 22.53%, 25.08%, and 25.73% with χ 2 test values are 60.01, 56.11, and 55.14, respectively. Similarly, one another mutation in spike observed as N764K exhibits 55.77% prevalence with χ 2 test values of 19.55 among the Omicron genome sequencing data submitted (Table 1). These data suggest that these mutations are not prevalent in Omicron.
Table 1.
Mutational prevalence of Omicron
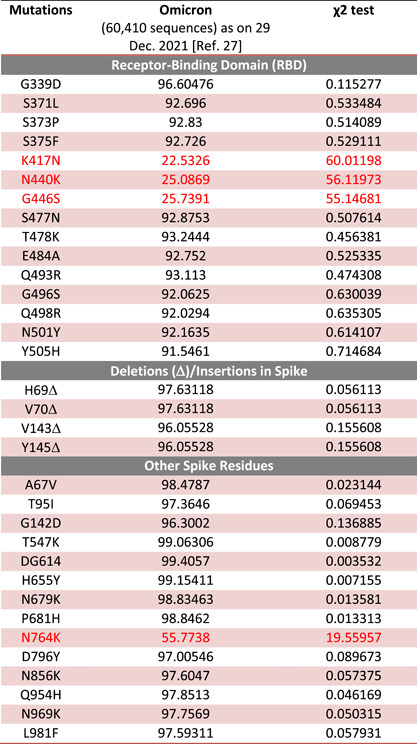
|
Note: Red color showing the less prevalent mutations in Omicron.
4. CONCLUSIONS AND FUTURE PERSPECTIVES
The WHO has categorized the COVID‐19 Omicron variant as a variant of concern, based on evidence that it has various alterations that might affect its transmissibility and pathogenicity, vaccine effectiveness, and monoclonal antibody‐mediated protection. The specificity and efficacy of the variant monitoring system, as well as infectious preventive measures in each nation, are critical for efficient prevention and therapeutic management of Omicron. The most efficient way for COVID‐19 protection and control has already been demonstrated via vaccination. Live attenuated vaccines, replicating and non‐replicating viral vector vaccines, DNA/RNA vaccines, and protein‐subunit vaccines are the major types of vaccines that are currently available for human use. The 32 amino acid alterations in the spike protein of Omicron, which include three modest deletions and one short insertion, some of which are concerning and may be related to humoral immune escape potential and higher transmissibility. These alterations may increase the potential to escape existing immunizations. Our findings show the transmission dynamics of the novel SARS‐CoV‐2 Omicron which shows that it's highly transmissible. Moreover, the mutational comparison of Omicron's spike glycoprotein with other VOCs of SARS‐CoV‐2 helps us to better understand the worldwide transmissibility, pathophysiology, and the development of efficient prevention and treatment strategies toward novel SARS‐COV‐2 variants.
CONFLICT OF INTERESTS
The authors declare that there are no conflict of interests.
AUTHOR CONTRIBUTIONS
Shailendra K. Saxena conceived the idea and planned the study. Swatantra Kumar, Saniya Ansari, and Shailendra K. Saxena collected the data, devised the initial draft, reviewed the final draft, and contributed equally to this study as the first author. Shailendra K. Saxena, Saniya Ansari, Swatantra Kumar, Janusz T. Paweska, Vimal K. Maurya, Anil K. Tripathi, and Ahmed S. Abdel‐Moneim finalized the draft for submission. All authors read and approved the final version of the manuscript.
ACKNOWLEDGMENTS
The authors are grateful to the Vice Chancellor, King George's Medical University (KGMU) Lucknow, for the encouragement for this study. Ahmed S.Abdel‐Moneim also acknowledges the support of Taif University Researchers Supporting Project No. TURSP‐2020/11. The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
Saxena SK, Kumar S, Ansari S, et al. Transmission dynamics and mutational prevalence of the novel Severe acute respiratory syndrome coronavirus‐2 Omicron Variant of Concern. J Med Virol. 2022;94:2160‐2166. 10.1002/jmv.27611
DATA AVAILABILITY STATEMENT
The authors confirm that the data supporting the findings of this study are available within the article.
REFERENCES
- 1. Vaughan A. Omicron emerges. New Sci. 2021;252(3363):7. 10.1016/S0262-4079(21)02140-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Saxena SK, Kumar S, Ansari S, et al. Characterization of the novel SARS‐CoV‐2 Omicron (B.1.1.529) Variant of Concern and its global perspective. J Med Virol. 2022. 1–7. 10.1002/jmv.27524 [DOI] [PubMed] [Google Scholar]
- 3. World Health Organization . Naming SARS‐CoV‐2 Variants. Accessed on 30 December 2021. https://www.who.int/en/activities/tracking-SARS-CoV-2-variants/
- 4. Centers of Disease Control and Prevention . Science Brief: Omicron (B.1.1.529) Variant. Accessed on 28 December 2021. https://www.cdc.gov/coronavirus/2019-ncov/science/science-briefs/scientific-brief-Omicron-variant.html [PubMed]
- 5. World Health Organization . Classification of Omicron (B.1.1.529): SARS‐CoV‐2 Variant of Concern. Accessed on 31 December 2021. https://www.who.int/news/item/26-11-2021-classification-of-Omicron-(B.1.1.529)-sars-cov-2-variant-of-concern
- 6. Wong SC, Au AK, Chen H, et al. Transmission of Omicron (B.1.1.529) ‐ SARS‐CoV‐2 Variant of Concern in a designated quarantine hotel for travelers: a challenge of elimination strategy of COVID‐19. Lancet Reg Health West Pac. 2021:100360. 10.1016/j.lanwpc.2021.100360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Harvey WT, Carabelli AM, Jackson B, et al. COVID‐19 Genomics UK (COG‐UK) Consortium, Peacock SJ, Robertson DL. SARS‐CoV‐2 variants, spike mutations and immune escape. Nat Rev Microbiol. 2021;19(7):409‐424. 10.1038/s41579-021-00573-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. World Health Organization . Enhancing Response to Omicron (COVID‐19 Variant B.1.1.529): Technical Brief and Priority Actions for Member States. Accessed on 13 January 2021. https://www.who.int/publications/m/item/enhancing-readiness-for-Omicron-(B.1.1.529)-technical-brief-and-priority-actions-for-member-states
- 9. Petersen E, Ntoumi F, Hui DS, et al. Emergence of new SARS‐CoV‐2 Variant of Concern Omicron (B.1.1.529) ‐ highlights Africa's research capabilities, but exposes major knowledge gaps, inequities of vaccine distribution, inadequacies in global COVID‐19 response and control efforts. Int J Infect Dis. 2021;(21):00888. 10.1016/j.ijid.2021.11.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kumar S, Saxena SK. Structural and molecular perspectives of SARS‐CoV‐2. Methods. 2021;195:23‐28. 10.1016/j.ymeth.2021.03.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gupta A, Pradhan A, Maurya VK, et al. Therapeutic approaches for SARS‐CoV‐2 infection. Methods. 2021;195:29‐43. 10.1016/j.ymeth.2021.04.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Sathian B, Banerjee I, Mekkodathil AA, et al. Epidemiologic characteristics, clinical management, and public health implications of Coronavirus disease 2019 (COVID‐19) in pregnancy: a systematic review and meta‐analysis. Nepal J Epidemiol. 2021;11:1103‐1125. 10.3126/nje.v11i4.41911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Shimul SN, Alradie‐Mohamed A, Kabir R, Al‐Mohaimeed A, Mahmud I. Effect of easing lockdown and restriction measures on COVID‐19 epidemic projection: a case study of Saudi Arabia. PLoS One. 2021;16(9):e0256958. 10.1371/journal.pone.0256958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Khare S, Gurry C, Freitas L, et al. GISAID's role in pandemic response. China CDC Wkly. 2021;3(49):1049‐1051. 10.46234/ccdcw2021.255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Elbe S, Buckland‐Merrett G. Data, disease and diplomacy: GISAID's innovative contribution to global health. Glob Chall. 2017;1(1):33‐46. 10.1002/gch2.1018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Shu Y, McCauley J. GISAID: Global Initiative on Sharing All Influenza Data—from vision to reality. Euro Surveill. 2017;22(13):30494. 10.2807/1560-7917.ES.2017.22.13.30494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Latif AA, Mullen JL & Alkuzweny M Omicron Variant Report Hughes and the Center for Viral Systems Biology 2022. https://outbreak.info/
- 18. Schmidt F, Weisblum Y, Rutkowska M, et al. High genetic barrier to SARS‐CoV‐2 polyclonal neutralizing antibody escape. Nature. 2021;600(7889):512‐516. 10.1038/s41586-021-04005-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Chen RE, Zhang X, Case JB, et al. Resistance of SARS‐CoV‐2 variants to neutralization by monoclonal and serum‐derived polyclonal antibodies. Nat Med. 2021;27(4):717‐726. 10.1038/s41591-021-01294-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Tao K, Tzou PL, Nouhin J, et al. The biological and clinical significance of emerging SARS‐CoV‐2 variants. Nat Rev Genet. 2021;12:757‐773. 10.1038/s41576-021-00408-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Genomic Epidemiology of Novel Coronavirus ‐ Global Subsampling 2022. Accessed on 4 January. https://nextstrain.org/ncov/gisaid/global?branchLabel=emerging_lineage%26l=scatter%26p=full%26scatterX=S1_mutations%26tl=pango_lineage%26transmissions=show [Google Scholar]
- 22. Ali F, Kasry A, Amin M. The new SARS‐CoV‐2 strain shows a stronger binding affinity to ACE2 due to N501Y mutant. Med Drug Discov. 2021;10:100086. 10.1016/j.medidd.2021.100086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Centers of Disease Control and Prevention . Science Brief: Omicron (B.1.1.529) Variant. Accessed on 02 January 2022. https://www.cdc.gov/coronavirus/2019-ncov/science/science-briefs/scientific-brief-Omicron-variant.html. [PubMed]
- 24. World Health Organization . Tracking SARS‐CoV‐2 variants. Accessed 26 December 2021. https://www.who.int/en/activities/tracking-SARS-CoV-2-variants/
- 25. Wang L, Cheng G. Sequence analysis of the emerging SARS‐CoV‐2 Variant Omicron in South Africa. J Med Virol. 2021. 1–6. 10.1002/jmv.27516 [DOI] [PubMed] [Google Scholar]
- 26. Zhang Q, Xiang R, Huo S, et al. Molecular mechanism of interaction between SARS‐CoV‐2 and host cells and interventional therapy. Signal Transduct Target Ther. 2021;6(1):233. 10.1038/s41392-021-00653-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kumar S, Maurya VK, Prasad AK, Bhatt MLB, Saxena SK. Structural, glycosylation and antigenic variation between 2019 novel coronavirus (2019‐nCoV) and SARS coronavirus (SARS‐CoV). VirusDisease. 2020;31(1):13‐21. 10.1007/s13337-020-00571-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. O'Toole Á, Hill V, Pybus OG, et al. Tracking the international spread of SARS‐CoV‐2 lineages B.1.1.7 and B.1.351/501Y‐V2 with Grinch. Wellcome Open Res. 2021;6:121. 10.12688/wellcomeopenres [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Chen J, Wang R, Gilby NB, Wei GW. Omicron (B.1.1.529): infectivity, vaccine breakthrough, and antibody resistance. ArXiv [Preprint]. 2021;arXiv:2112.01318v1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Somerville M, Curran JA, Dol J, et al. Public health implications of SARS‐CoV‐2 variants of concern: a rapid scoping review. BMJ Open. 2021;11(12):e055781. 10.1136/bmjopen-2021-055781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wang Y, Zhang L, Li Q, et al. The significant immune escape of pseudotyped SARS‐CoV‐2 variant Omicron. Emerg Microbes Infect. 2022;1:1‐5. 10.1080/22221751.2021.2017757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. European Centre for Disease Prevention and Control . Implications of the Emergence and Spread of the SARS‐CoV‐2 B.1.1. 529 Variant of Concern (Omicron), for the EU/EEA. 26 December 2021. ECDC: Stockholm; 2021. https://www.ecdc.europa.eu/en/publications-data/threat-assessment-brief-emergence-sars-cov-2-variant-b.1.1.529
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The authors confirm that the data supporting the findings of this study are available within the article.


