Dear Editor,
Based on the recommendation of the WHO's Technical Advisory Group on Virus Evolution, WHO designated the variant B.1.1.529 as a variant of concern (VOC) and named Omicron on 26 November, 2021. Omicron was first observed in Africa in mid‐November 2021. 1 The infection by this variant has been rapidly spreading and 85 countries have already reported the cases of human infection with this variant as of 15 December 2021. 2 The rapid spread of Omicron has again fueled the fears of COVID‐19 all around the world like other four VOCs (Alpha, Beta, Gamma, and Delta).
Recently, the Omicron variant has classified into two different lineages BA.1 and BA.2 based on the mutations, some of which are common and some are unique to both lineages. 3 , 4 Still, there is no clear evidence or published article on the mutational diversity and phylogenetic analysis of these two lineages. Therefore, in the present study, we have performed the whole‐genome mutational mapping and phylogenetic analysis of BA.1 and BA.2 lineages. We have downloaded 6 genome sequences each of BA.1 and BA.2, and also the genome sequence of the prototype strain (hCoV‐19/Wuhan/WIV04/2019) from the Global Initiative on Sharing All Influenza Data (GISAID) and performed whole‐genome mutational analysis according to the protocol described in Sarkar et al. 5 , 6 Each of the six genomes of both BA.1 and BA.2 lineages was found to have 51 mutations dispersed throughout the genome, 32 of which are common to both lineages, whereas each lineage has 19 signature mutations. Among 32 common mutations, 21 are present in the S glycoprotein and the rest 11 are present in the other four coding regions (ORF1ab, E, M, and N). Nineteen unique mutations of BA.1 include 13 in the S glycoprotein and that of BA.2 includes 7 in the S glycoprotein (Table 1). Phylogenetic analysis of 12 genome sequences of Omicron variant, encompassing 6 genomes each of BA.1 and BA.2, along with the 2000 genomes of 25 different clades by Ultrafast Sample placement of Existing tRees, 6 , 7 revealed that genomes of Omicron variant formed a new cluster that emerged from the 20B clade (also known as GR) and also subdivided into two different subclusters (BA.1 and BA.2) based on the unique mutations (Figure 1).
Table 1.
List of common and unique mutations present within the genome of BA.1 and BA.2 lineages
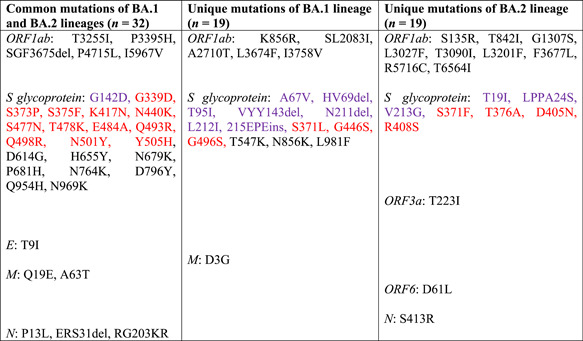
|
Note: Red color indicates the mutations of the receptor‐binding domain (RBD) of S glycoprotein. Violet color indicates the mutation of the N‐terminal domain (NTD) of S glycoprotein.
Figure 1.
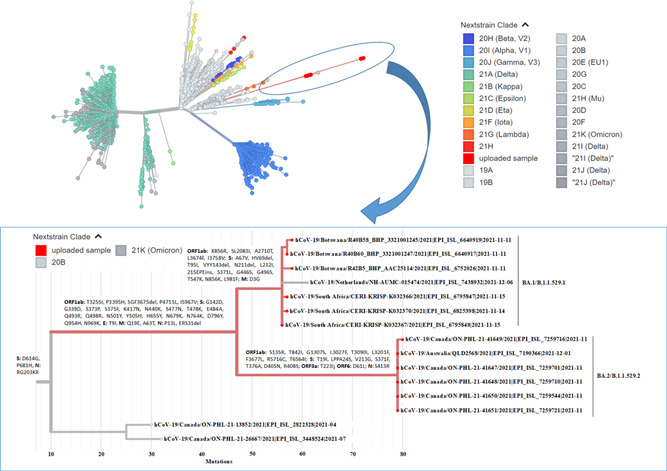
Phylogenetic analysis of the Omicron variant by Ultrafast Sample placement of Existing tRee (UShER). The phylogenetic tree (unrooted tree) was constructed with a total of 2012 SARS‐CoV‐2 strains involving 12 strains of the Omicron variant (red color) and 2000 strains from 25 different clades. Each color in the tree is representing a different clade/lineage
The S glycoprotein mediates virus attachment to ACE2 receptor, membrane fusion, and entry into the host cell, and also acts as a primary target for neutralizing antibodies elicited by the host immune response. 8 Presence of 34 and 28 mutations in the S glycoprotein of BA.1 and BA.2, respectively, raising concern whether these lineages have increased transmissibility, immune escape potential, and virulence compared to other circulating SARS‐CoV‐2 strains especially Delta which is currently dominating worldwide. Seven mutations of both BA.1 and BA.2 (G142D, K417N, T478K, N501Y, D614G, H655Y, and P681H) and three mutations of BA.1 (∆HV69del, T95I, and ∆YY144del) overlap four other VOCs (Alpha, Beta, Gamma, and Delta) and have previously been linked with high transmissibility, increased viral binding affinity, and immune evasion. 9 , 10 , 11 , 12 Functional implication of the remaining mutations of the S glycoprotein and other coding regions of BA.1 and BA.2 still unknown, leaving a question of how the whole set of mutations of the two lineages will affect viral fitness.
Preliminary evidence indicated that Omicron has increased infectivity and a high transmission rate compared to Delta. 13 , 14 , 15 However, whether the rapid spread of Omicron in countries with increased population immunity is due to increased transmissibility and/or immune evasion remains unclear. Though, some recent studies have claimed the immune evasion properties of the Omicron. 14 , 15 , 16 , 17 Based on this existing evidence, Omicron is anticipated to overtake Delta in areas where community transmission occurs. The severity of Omicron infection still remains elusive. Preliminary studies from South Africa suggested that Omicron may be less severe than Delta, 18 and all COVID‐19 patients, infected with Omicron, from countries of EU and EEA either showed mild symptoms or were asymptomatic. 19 Detection accuracy of routinely used PCR and antigen‐based rapid diagnostic test (Ag‐RDT) assays was not found to be influenced by most of the Omicron strains. However, the BA.1 lineage showed S gene target failure (SGTF) in RT‐PCR assay due to multiple deletions in the NTD of S glycoprotein, whereas BA.2 lineage may skip SGTF due to lack of deletions in the NTD. Overall, the global threat related to Omicron remains very high for its potential to escape humoral immune response and high transmissibility, which may lead to another wave of COVID‐19 with severe consequences. 19
CONFLICT OF INTERESTS
The authors declare that there are no conflict of interests.
AUTHOR CONTRIBUTIONS
Swagata Majumdar and Rakesh Sarkar conceived the study. Swagata Majumdar performed sequence retrieval of SARS‐CoV‐2 and whole‐genome mutational analysis. Rakesh Sarkar performed the phylogenetic analysis and drafted the manuscript. All authors revised the manuscript and approved the final manuscript for submission.
ACKNOWLEDGMENTS
The authors acknowledge the University Grants Commission (UGC) of India for providing fellowship to Swagata Majumdar. The authors would also like to acknowledge GISAID (https://www.gisaid.org/) for facilitating open data sharing.
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from the corresponding author upon reasonable request.
REFERENCES
- 1. World Health Organization . Classification of Omicron (B. 1.1. 529): SARS‐CoV‐2 variant of concern.
- 2. World Health Organization . Weekly operational update on COVID‐19. 12 December 2021.
- 3. Wang L, Cheng G. Sequence analysis of the emerging SARS‐CoV‐2 variant Omicron in South Africa. J Med Virol. 2021. [DOI] [PubMed] [Google Scholar]
- 4.Proposal to split B.1.1.529 to incorporate a newly characterised sibling lineage. Accessed December 15, 2021. https://github.com/cov-lineages/pango-designation/issues/361
- 5. Sarkar R, Mitra S, Chandra P, et al. Comprehensive analysis of genomic diversity of SARS‐CoV‐2 in different geographic regions of India: an endeavour to classify Indian SARS‐CoV‐2 strains on the basis of co‐existing mutations. Arch Virol. 2021;166(3):801‐812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Sarkar R, Saha R, Mallick P, et al. Emergence of a novel SARS‐CoV‐2 Pango Lineage B.1.1.526 in West Bengal, India. J Infect Public Health. 2021;15:42‐50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Turakhia Y, Thornlow B, Hinrichs AS, et al. Ultrafast Sample placement on Existing tRees (UShER) enables real‐time phylogenetics for the SARS‐CoV‐2 pandemic. Nat Genet. 2021;53(6):809‐816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Walls AC, Park YJ, Tortorici MA, Wall A, McGuire AT, Veesler D. Structure, function, and antigenicity of the SARS‐CoV‐2 spike glycoprotein. Cell. 2020;181(2):281‐292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Harvey WT, Carabelli AM, Jackson B, et al. SARS‐CoV‐2 variants, spike mutations and immune escape. Nat Rev Microbiol. 2021;19(7):409‐424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Greaney AJ, Starr TN, Gilchuk P, et al. Complete mapping of mutations to the SARS‐CoV‐2 spike receptor‐binding domain that escape antibody recognition. Cell Host Microbe. 2021;29(1):44‐57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Arora P, Pöhlmann S, Hoffmann M. Mutation D614G increases SARS‐CoV‐2 transmission. Signal Transduct Target Ther. 2021;6:101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Plante JA, Liu Y, Liu J, et al. Spike mutation D614G alters SARS‐CoV‐2 fitness. Nature. 2021;592(7852):116‐121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Yang W, Shaman J. SARS‐CoV‐2 transmission dynamics in South Africa and epidemiological characteristics of the Omicron variant. medRxiv. 2021. [Google Scholar]
- 14. Syed AM, Ciling A, Khalid MM, et al. Omicron mutations enhance infectivity and reduce antibody neutralization of SARS‐CoV‐2 virus‐like particles. medRxiv. 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zhang X, Wu S, Wu B, et al. SARS‐CoV‐2 Omicron strain exhibits potent capabilities for immune evasion and viral entrance. Signal Transduct Target Ther. 2021;6:430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Pulliam JR, van Schalkwyk C, Govender N, et al. Increased risk of SARS‐CoV‐2 reinfection associated with emergence of the Omicron variant in South Africa. medRxiv. 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Dejnirattisai W, Shaw RH, Supasa P, et al. Reduced neutralisation of SARS‐CoV‐2 omicron B.1.1. 529 variant by post‐immunisation serum. Lancet. 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wolter N, Jassat W, Walaza S, et al. Early assessment of the clinical severity of the SARS‐CoV‐2 Omicron variant in South Africa. medRxiv. 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. World Health Organization . Weekly operational update on COVID‐19. 13 December 2021.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.


