Abstract
Esophageal squamous cell carcinoma (ESCC) is a cancer type with limited treatment options. The present study aimed to screen for small molecules that may inhibit ESCC cell viability. The small-molecule-perturbed signatures were extrapolated from the library of integrated network-based cellular signatures (LINCS) database. Since LINCS does not include small-molecule-perturbed signatures of ESCC cells, it was hypothesized that non-ESCC cell lines that display transcriptome profiles similar to those of ESCC may have similar small-molecule-perturbated responses to ESCC cells and that identifying small molecules that inhibit the viability of these non-ESCC cells may also inhibit the viability of ESCC cells. The transcriptomes of >1,000 cancer cell lines from the Cancer Cell Line Encyclopedia database were analyzed and 70 non-ESCC cell lines exhibiting similar transcriptome profiles to those of ESCC cells were identified. Among them, six cell lines with transcriptome signatures upon drug perturbation were available in the LINCS, which were used as reference signatures. A total of 20 ESCC datasets were analyzed and 522 downregulated and 461 upregulated differentially expressed genes (DEGs) that were consistently altered across >50% of the datasets were identified. These DEGs together with the reference signatures were then used as inputs of the ZhangScore method to score small molecules that may reverse transcriptome alterations of ESCC. Among the top-ranked 50 molecules identified by the ZhangScore, four candidates that may inhibit ESCC cell viability were experimentally verified. Furthermore, 2-[(aminocarbonyl)amino]-5-(4-fluorophenyl)-3-thiophenecarboxamide (TPCA-1), an inhibitor of the NF-κB pathway, was able to preferentially inhibit the viability of ESCC cells compared with non-tumorigenic epithelial Het-1A cells. Mechanistically, TPCA-1 induced ESCC KYSE-450 cell apoptosis by inhibiting the phosphorylation of inhibitor of NF-κB kinase subunit β, leading to IκBα stabilization and NF-κB signaling pathway inhibition. Collectively, these results demonstrated that LINCS-based drug repositioning may facilitate drug discovery and that TPCA-1 may be a promising candidate molecule in the treatment of ESCC.
Keywords: esophageal squamous cell carcinoma, library of integrated network-based cellular signatures, gene expression profiling, ZhangScore, drug discovery
Introduction
Esophageal cancer is the 7th most common type of cancer with ~572,000 estimated new cases and ~509,000 deaths in 2018 (1) and a five-year survival rate of <20% (2). The predominant histological subtype is esophageal squamous cell carcinoma (ESCC), which accounts for >85% of esophageal cancers (3). There are various known risk factors for ESCC, including alcohol consumption, cigarette smoking, chewing tobacco, hot drinks, indoor air pollution, pickles, preserved meat, a low intake of fruits and vegetables (4). The mainstay treatment for early-stage ESCC is surgery (endoscopic resection or esophagectomy). For advanced ESCC, surgery is frequently combined with neoadjuvant treatments, such as chemotherapy or chemoradiotherapy (5). However, the poor prognosis of ESCC highlights the urgent requirement for the development of novel therapeutics.
To aid the discovery of novel drug candidates or the repurposing of approved drugs, the National Institutes of Health (NIH) have launched the Library of Integrated Network-based Cellular Signatures (LINCS) (6). Using L1000 technology, LINCS has detected transcriptional signatures for 1.4 million genetic (CRISPR knock-out, shRNA knockdown and open reading frame overexpression) and small-molecule (drug and tool compound) perturbations spanning 50 cell types of varied lineages (6). It has been proven by several studies that LINCS is useful for drug discovery. For head and neck squamous cell carcinoma with mutant p53, the phosphatidylinositol-3-kinase α-selective inhibitor alpelisib was found, using the LINCS approach, to disrupt the interaction of MYC, mutant p53 and Yes-associated protein with MYC target promoters (7). A recent study indicated that crizotinib, celastrol and GSK1059615 may be combined with erlotinib to inhibit erlotinib-resistant PC9 lung cancer cells (8). In addition, it was discovered that aurora inhibitors and JQ1 synergistically inhibit glioblastoma and that mitoxantrone or imatinib may synergize with gemcitabine (9). ZhangScore is an algorithm that is able to evaluate the similarity of transcriptomes based on drug-perturbated gene expression data. Lin et al (10) indicated that the ZhangScore method (11), which may provide the significance level of a small molecule by bootstrapping, is superior to other methods and has high accuracy in identifying potential drugs.
NF-κB signaling is an important pathway that regulates various biological processes, including inflammation, survival, proliferation and immune response (12). NF-κB signaling is frequently constitutively activated in numerous cancer types, which relies on the phosphorylation and degradation of its specific inhibitor IκB protein by the IκB kinase (IKK) complex (13-15). Degradation of IκB protein leads to the release of the p65/RELA-p50 (canonical) or p52-RELB complexes into the nucleus, inducing the transcription of multiple target genes involved in pro-inflammation, angiogenesis, cell proliferation, anti-apoptosis and cell-matrix remodeling (16). Thus, targeting NF-κB signaling is considered a promising strategy for cancer treatment. 2-[(Aminocarbonyl)amino]-5-(4-fluorop henyl)-3-thiophenecarboxamide (TPCA-1) is a potent selective inhibitor of IKKβ (17). It was reported that flubendazole, an inhibitor of NF-κB signaling, may induce apoptosis in ESCC cells (15). However, whether TPCA-1 is able to inhibit ESCC has remained elusive.
In the present study, the extensive data on the small moleculeperturbed transcription signatures in the LINCS project were used to identify potential small inhibitory molecules for ESCC. A list of 50 small molecules was compiled and prioritized, and the four top-scoring candidate molecules (nutlin-3a, vemurafenib, TPCA-1 and CID2858522) were experimentally verified to inhibit ESCC cell viability. In particular, it was determined that TPCA-1, which targets the IKKβ protein, was able to inhibit the viability of ESCC cells more effectively than that of non-tumorigenic esophageal epithelial cells.
Materials and methods
Dataset collection
The analytical workflow of the present study is illustrated in Fig. 1. A total of 20 gene expression datasets of ESCC were obtained from the Xena (https://xena.ucsc.edu/), Sequence Read Archive (SRA, https://www.ncbi.nlm.nih.gov/sra) and Gene Expression Omnibus (GEO, https://www.ncbi.nlm.nih.gov/geo) databases. A total of 911 samples were included, of which 515 were tumor samples or tumor cell lines and 396 were normal esophagus or nontumorigenic cell lines. Gene expression data and corresponding annotation files for >1,000 human cancer cell lines were downloaded from the Cancer Cell Line Encyclopedia (CCLE, https://portals.broadinstitute.org/ccle). The preprocessed level 5 GCTX format data of small-molecule-perturbed gene expression files, as well as the corresponding cell annotation and experimental annotation files, were downloaded from the GEO database (accession no. GSE92742).
Figure 1.
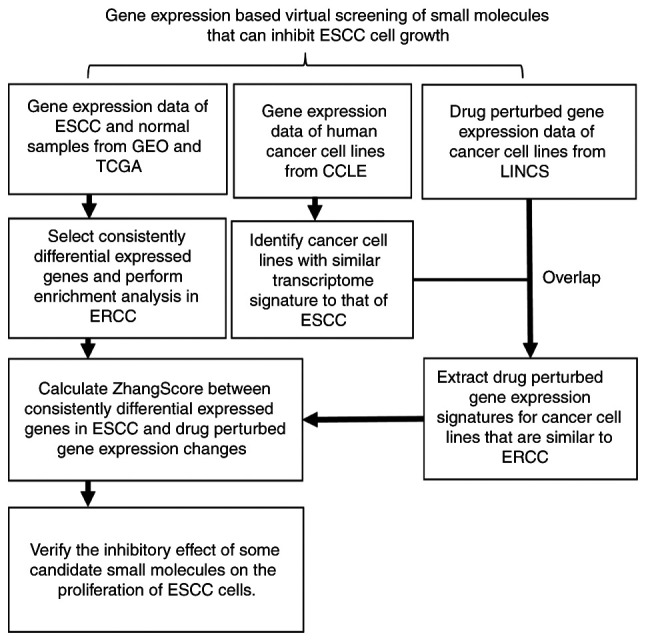
Workflow for screening small molecules that can inhibit ESCC cell growth. ESCC, esophageal squamous cell carcinoma; TCGA, The Cancer Genome Atlas; LINCS, Library of Integrated Network-based Cellular Signatures; CCLE, Cancer Cell Line Encyclopedia; GEO, gene expression omnibus.
Identification of the consensus differentially expressed genes (DEGs) in ESCC
For ESCC expression chips, the Limma package (18) was used to perform background correction, standardization and differential expression analysis. For RNAseq data from the SRA database, align-free aligner Salmon (19) was used to perform quantification. Gene-level expression data were then imported into the R environment by the tximport package (20) and DEG analysis was performed using the DESeq2 (21) package. For RNAseq data from The Cancer Genome Atlas (TCGA), the expression data were obtained from Xena and gene expression data were extracted for ESCC and normal tissues. DESeq2 was then used to perform DEG analysis. DEGs were defined as adjusted P<0.05 and |log2 FoldChange|>1 in each dataset. To obtain the consistent transcriptional changes in ESCC, DEGs that were shared by >50% of the 20 datasets were further identified as consensus DEGs. The ClusterProfiler (22) package was used to perform Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genome (KEGG) enrichment analysis and visualization for DEGs.
Screening for the cancer cells most similar to ESCC cells at the gene expression level
As the current version of LINCS has no small-molecule-perturbed experiment performed on ESCC cells, the cell lines most similar to ESCC cells at the transcriptome level were first identified using the CCLE dataset. The R-package GGally (23) was used to first identify the most representative ESCC cell lines with a high pair-wise Pearson correlation coefficient (PCC). The mean PCC between one ESCC cell line and all other representative ESCC cell lines was >0.7. The mean value for each gene was then calculated among the representative cell lines as the representative profile of ESCC. The PCCs between the representative profile of ESCC and no-ESCC cell lines were computed in LINCS. The cancer cell lines with a PCC >0.83 (determined by the inflection point presented in Fig. S1B) were defined as similar to ESCC cells at the transcriptome level. Finally, six cell lines (A375, CORL23, HT115, HT29, SNUC5 and SW620) were indicated to be similar to ESCC cells and to have small-molecule perturbation data in LINCS.
Candidate small molecule screening using LINCS data
The SignatureSearch (24) package was used to parse and extract gene expression data prior to and after small molecule treatment for the six cell lines similar to ESCC cells in the LINCS. Next, the RCSM (10) package was used to calculate the ZhangScore (11) between DEGs of ESCC (query gene signatures) and the small-molecule-perturbed gene expression (reference gene signatures). The molecules were ordered by ZhangScore in ascending order for each of the six cell lines. Finally, RobustRankAggreg (RRA) (25) was used to integrate the ranking results of molecules from the six cell lines. The significant level of each of the top-ranking small molecules among all six rank lists was obtained. The final list was ranked based on the sum of the ZhangScore across the six cell lines in ascending order.
Cell culture
KYSE-150, KYSE-180 and KYSE-450 cell lines were from German collection of microorganisms and cell cultures. TE-1 cell line was from the RIKEN BRC cell bank. Eca-109 cell line was from China center for type culture collection. The immortalized human esophageal epithelial cell line, Het-1A was from American type culture collection. All cell lines used in the present study were cultured at 37°C in a thermostatic cell incubator (Thermo Fisher Scientific, Inc.) with saturated humidity and 5% CO2. Cells were cultured in RPMI-1640 medium (Gibco; Thermo Fisher Scientific, Inc.) supplemented with 10% FBS (HyClone; Cytiva) and 1% double resistance (100 U/ml penicillin and 100 µg/ml streptomycin; HyClone; Cytiva).
Cell viability assay
ESCC cells were seeded in 96-well plates at a density of 2,000 cells/100 µl and cultured overnight at 37°C with 5% CO2. Following treatment with the indicated concentrations of small molecules for different durations, their survival was measured with an MTS kit (Thermo Fisher Scientific, Inc.) using an enzyme labeler (Varioskan Flash; Thermo Fisher Scientific, Inc.). The absorbance was measured at a wavelength of 490 nm and cell viability was calculated as follows: Viability=[Optical density (OD)Drug−ODBlank]/(ODControl−ODBlank).
Colony-forming assay
Cells (700/well) were exposed to various concentrations (0, 0.5, 1, 2 and 4 µM) of TPCA-1 (MedChemExpress, Inc.) in 6-well plates and then cultured for 2 weeks at 37°C in an incubator with 5% CO2. Following gentle washing with PBS once at room temperature, 1 ml methanol was used to fix each well for 10 min at room temperature. Each well was then dyed with 1 ml crystalline purple for 10 min at room temperature. Finally, each well was washed several times with PBS. A gel imager (ChemiDoc XRS Plus; Bio-Rad Laboratories, Inc.) with bright light optics was used to acquire images.
Cell apoptosis analysis
KYSE-450 cells were treated with different concentrations of TPCA-1 (0, 0.5, 1, 2 and 4 µM) for 48 h. Cells were washed twice with pre-cooled PBS, digested with trypsin (Gibco; Thermo Fisher Scientific, Inc.) and centrifuged at 1,000 × g for 5 min at room temperature. The supernatant was then discarded and 195 ml Annexin V-FITC binding buffer was added to gently resuspend the cells. Finally, 5 ml Annexin V-FITC and 10 ml propidium iodide staining solution (Beyotime Institute of Biotechnology) were added to the cell suspension with gentle mixing. Following incubation at room temperature (20-25°C) in the dark for 10 min, stained cells were ana lyzed using a flow cytometer (FACSCalibur, BD Biosciences).
Western blot analysis
KYSE-450 and Het-1A cells were treated with different concentrations of TPCA-1 (0, 0.5, 1, 2 and 4 µM) for 48 h. Cells were scraped down and centrifuged in pre-warmed PBS. After extracting the cell lysate with lysis buffer (50 mM Tris·HCl pH 8.0, 250 mM NaCl, 40 mM NaF, 0.5 mM NaVO4, 0.5% Nonidet P-40, 1 mM PMSF, 2 µg/ml leupeptin and 2 µg/ml aprotinin), the total protein concentration was measured with Bradford reagent (B6916; MilliporeSigma) using a spectrophotometer (Thermo Fisher Scientific, Inc.). Equal amounts of total protein (50 µg for each lane) were subjected to SDS-PAGE separation with 10% polyacrylamide gel, followed by electro-transfer to a polyvinylidene difluoride membrane (Bio-Rad Laboratories, Inc.) in a Mini-PROTEAN Tetra vertical electrophoresis cell (Bio-Rad Laboratories, Inc.). The membrane was then blocked with 5% skimmed milk (cat. no. P0126; Beyotime Institute of Biotechnology) for 1 h at room temperature and then incubated with specific antibodies at 4°C for 16 h. The primary antibodies for IκBα (cat. no. 4814; 1:1,000 dilution), phosphorylated (p)-IKKβ (cat. no. 2697; 1:1,000 dilution), IKKβ (cat. no. 2678; 1:1,000 dilution), p-P65 (cat. no. 3033; 1:1,000 dilution), P65 (cat. no. 8242; 1:1,000 dilution), caspase 3 (cat. no. 9662; 1:1,000 dilution) and cleaved caspase 3 (cat. no. 9662; 1:1,000 dilution) were purchased from Cell Signaling Technology, Inc. Antibodies for GAPDH (cat. no. 0037; 1:3,000 dilution) and Bim (cat. no. AY1380; 1:1,000 dilution) were purchased from Abways Technology. The HRP-conjugated goat anti-mouse (cat. no. sc-2005; 1:3,000 dilution) and goat anti-rabbit IgG antibodies (cat. no. sc-2004; 1:1,000 dilution) were purchased from Santa Cruz Biotechnology, Inc. The bands were visualized by enhanced chemiluminescence (cat. no. P10100; New Cell & Molecular Biotech Co., Ltd.) in a gel imager (ChemiDoc XRS Plus; Bio-Rad Laboratories, Inc.).
Statistical analysis
All bioinformatics analyses and calculations were performed in R. Values are expressed as the mean ± standard error of the mean. All statistical analysis was performed using GraphPad Prism 8 software (GraphPad Software, Inc.). An unpaired Student's t-test was used to determine the significance of differences between groups with and without TCPA-1 treatment in each cell line. Two-way ANOVA followed by Tukey's post-hoc test was used to determine the significance of differential responses of KYSE-450 and Het-1A cells to TPCA-1 treatment. P<0.05 was considered to indicate a statistically significant difference.
Results
Consensus DEGs in ESCC
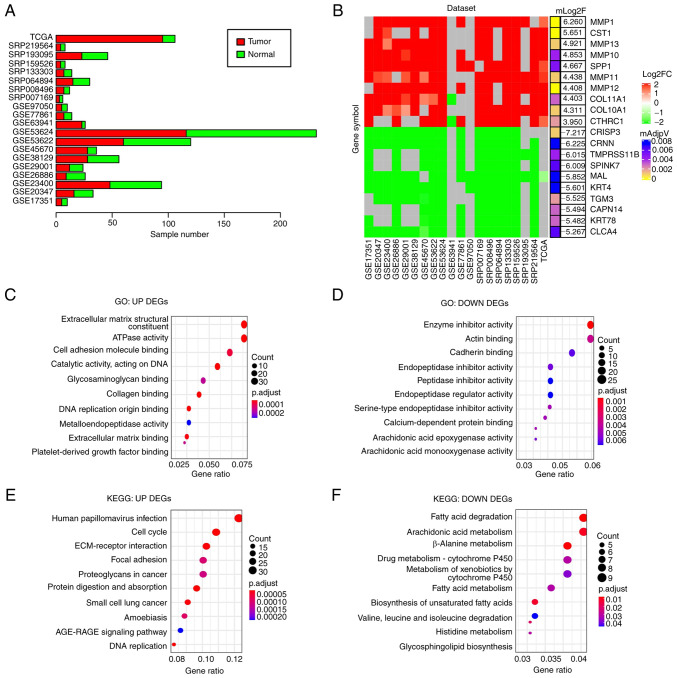
To obtain DEGs in ESCC compared with normal controls, 20 ESCC gene expression datasets from the GEO, SRA or Xena databases consisting of 911 samples (515 tumor and 396 normal samples) were analyzed (Fig. 2A) and DEGs were identified in each dataset (adjusted P<0.05 and |log2FoldChange|>1). DEGs identified in at least 10 datasets (≥50%) were considered as DEGs of ESCC in the present study. In total, 521 downregulated and 460 upregulated genes were identified (Table SI). Of note, several cell matrix-related proteins, including MMP-1, -13 and -10, were among the top 10 upregulated DEGs (Fig. 2B), suggesting that the alteration of cell matrix-related gene expression may have an important role in ESCC development.
Figure 2.
Screening for consistent DEGs in esophageal squamous cell carcinoma and enrichment analysis. (A) Bar chart indicating the number of tumors and normal samples in the 20 different ESCC datasets. (B) Heat map presenting the fold changes of the top 10 upregulated and top 10 downregulated genes that were common in the 20 datasets. Gray colour indicates the gene was not detected in the corresponding dataset. (C and D) GO enrichment analysis of (C) upregulated DEGs and (D) downregulated DEGs. (E and F) Enrichment analyses of KEGG pathways for (E) upregulated DEGs and (F) downregulated DEGs. KEGG, Kyoto Encyclopedia of Genes and Genomes; GO, Gene Ontology; DEG, differentially expressed gene; TCGA, The Cancer Genome Atlas.
GO enrichment analyses indicated that the upregulated DEGs were mainly related to extracellular matrix (Fig. 2C) and the downregulated DEGs were mainly associated with peptidase inhibitor and arachidonic acid epoxygenase (Fig. 2D). KEGG pathway analysis suggested that the upregulated DEGs were primarily enriched in extracellular matrix-receptor interaction and cell cycle regulators (Fig. 2E) and the downregulated DEGs in fatty acid degradation and cytochrome P450 (Fig. 2F). Both the arachidonic acid epoxygenase and fatty acid degradation pathways are associated with inflammation (26,27), suggesting that disrupted immune response may be involved in ESCC development.
Identification of cancer cell lines with similar gene expression patterns to ESCC cells
Although LINCS (28) has transcriptome profiles derived from a variety of human cancer cells treated with various small molecules, there is a lack of perturbation data for ESCC cells. Thus, it was hypothesized that non-ESCC cell lines that display transcriptome profiles similar to that of ESCC in LINCS may have similar cellular responses to treatment of ESCC cells with small molecules and that the small molecules that inhibit the viability of these non-ESCC cells may also inhibit the viability of ESCC cells. To do this, the gene expression data of >1,000 human cancer cell lines from CCLE were analyzed (29), including 22 ESCC cell lines. Gene expression datasets derived from both RNAseq and microarrays were used to calculate the PCC in each pair from a total of 22 ESCC cell lines. As presented in Fig. S1A, the PCCs of seven ESCC cell lines, including OE21, KYSE-510, TE-11, KYSE-70, KYSE-520, KYSE-180 and TE-14, exhibited high pairwise similarities (PCC>0.7). The mean expression values of each gene in the seven cell lines were calculated and then used as a reference profile for ESCC cells to calculate the PCCs between this reference profile and the transcriptome profiles of non-ESCC cancer cells in CCLE. A total of 70 non-ESCC cancer cell lines with a PCC>0.83 (determined by inflection point) exhibited a similar transcriptome profile to that of ESCC cells (Fig. S1B). Among them, transcriptome profiles of small-molecule perturbations were available for six cell lines, including A375 human melanoma, CORL23 human lung large cell carcinoma, HT115 human colon carcinoma, HT29 human colorectal cancer, SNUC5 human cecum adenocarcinoma and SW620 human colon adenocarcinoma cell lines (Fig. S1C) in LINCS. Of note, data for 31,398 perturbations were available for the six cell lines (Fig. S1D).
ZhangScore and RRA methods for ranking candidate small molecules that may reverse the aberrant gene expression of ESCC
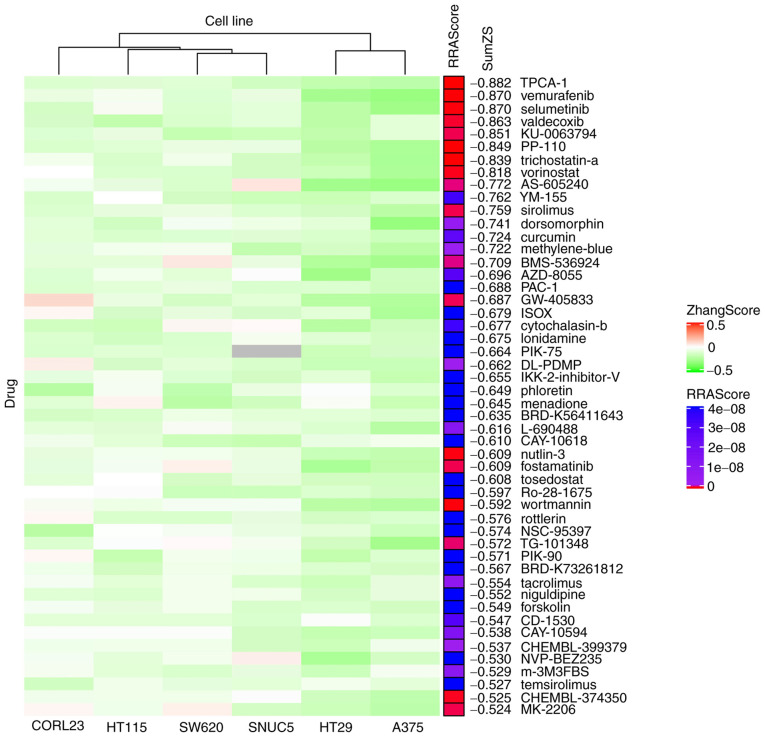
The ZhangScore algorithm is able to calculate the reversing extent of a small-molecule-perturbed signature, termed reference signature, compared with a disease signature, termed query signature. The more negative the ZhangScore, the higher the potential of the small molecule to reverse the altered expression of malignant cells back to their non-malignant state (11). Therefore, DEGs shared in ESCC were used as the query signature and small-molecule-perturbed signatures were used as the reference signatures (each reference signature was derived from the transcriptome of cells treated with a combination of small molecule, concentration and duration) from six cell lines identified above. The ZhangScore was first calculated for each small-molecule-perturbed signature from each cell line. The smallest negative ZhangScore of each small molecule, among various combinations of concentrations and durations, was used for candidates ranking in each cell line (Table SII). The RRA method (25) was then used to integrate the ranking results from the six cell lines. The final ranking was based on the sum of ZhangScores (SumZS) derived from the six cell lines and presented in ascending order (Table I presents the top 50 molecules). In Fig. 3, the heatmap illustrates that the majority of the top 50 small molecules had a negative ZhangScore (in green color) in all the six cell lines, suggesting that these small molecules were likely able to reverse the expression of DEGs upon small molecule perturbation. The RRA scores labeled in red indicate the top-ranking small molecules that had the most consistent effects among all six cell lines.
Table I.
Targets and current status in clinical trials of the top 50 small molecules.
| Rank | Molecule | Target protein (classes) | PMID | Indications/clinical trials/classification |
|---|---|---|---|---|
| 1 | TPCA-1 | IKKβ (inhibitor) | 15316093 | Unknown |
| 2 | Vemurafenib | BRAFV600E (inhibitor) | 35045748 | Melanoma |
| 3 | Selumetinib | MAP2K1 (inhibitor) | 32504375 | Melanoma/neurofibromatosis type 1/thyroid cancer |
| 4 | Valdecoxib | PTGS2 (inhibitor) | 23517091 | Rheumatoid arthritis [withdrawn] |
| 5 | KU-0063794 | mTOR (inhibitor) | 19402821 | Mild atopic asthma (Phase 2) |
| 6 | PP-110 | Unknown | Not available | Unknown |
| 7 | Trichostatin-a | HDAC (inhibitor) | 25923331 | Relapsed or refractory hematologic malignancies (Phase 1) |
| 8 | Vorinostat | HDAC (inhibitor) | 35158962 | Cutaneous T-cell lymphoma/ESCC (NCT00537121, Phase 1) |
| 9 | AS-605240 | PIK3CG (inhibitor) | 24935930 | Unknown |
| 10 | YM-155 | Survivin (inhibitor) | 21737502 | Melanoma (Phase 2)/metastatic breast cancer (Phase 2) |
| 11 | Sirolimus | mTOR (inhibitor) | 34415233 | Organ rejection/prostate cancer (Phase 2)/pancreatic cancer (Phase 2) |
| 12 | Dorsomorphin | AMPK (inhibitor) | 18026094 | Unknown |
| 13 | Curcumin | PPARG (inhibitor) | 23534763 | Periodontitis (Phase 4)/advanced pancreatic cancer (Phase 2)/prostate cancer (Phase 3) |
| 14 | Methylene-blue | GUCY1A2 (inhibitor) | 30666126 | Methaemoglobinaemia/colorectal cancer (Phase 3) |
| 15 | BMS-536924 | IGF1R (inhibitor) | 16134929 | Unknown |
| 16 | AZD-8055 | mTOR (inhibitor) | 21333749 | Recurrent Gliomas (Phase 1)/advanced tumors (Phase 1) |
| 17 | PAC-1 | Caspase-3 (activator) | 19281821 | Uveal melanoma (Phase 2)/advanced malignancies (Phase 1) |
| 18 | GW-405833 | Unknown | 20863899 | Unknown |
| 19 | ISOX | HDAC (inhibitor) | 29867749 | Unknown |
| 20 | Cytochalasin-b | Unknown | Not available | Unknown |
| 21 | lonidamine | HK (inactivator) | 16986056 | Prostatic hyperplasia (Phase 2/3, terminated) |
| 22 | PIK-75 | PI3K (inhibitor) | 17362206 | Unknown |
| 23 | DL-PDMP | Unknown | 21571032 | Unknown |
| 24 | IKK-2-inhibitor-V | IKKβ (inhibitor) | 23211970 | Unknown |
| 25 | Phloretin | Unknown | 14019989 | Flavors |
| 26 | Menadione | Cytochrome P450 (suppressor) | 27167070 | Vitamins |
| 27 | BRD-K56411643 | Unknown | Not available | Unknown |
| 28 | L-690488 | Unknown | Not available | Unknown |
| 29 | CAY-10618 | Unknown | Not available | Unknown |
| 30 | Nutlin-3a | MDM2 (antagonist) | 14704432 | Unknown |
| 31 | Fostamatinib | SYK (inhibitor) | 16946104 | Advanced colorectal, non-small cell lung, head and neck hepatocellular and renal cell carcinomas among other cancers (Phase 2)/thrombocytopenia (Phase 2)/COVID-19 (Phase 2/3) |
| 32 | Tosedostat | aminopeptidase (inhibitor) | 18701491 | Acute myeloid leukaemia (phase 1/2) |
| 33 | Ro-28-1675 | Unknown | 20161845 | Unknown |
| 34 | Wortmannin | PIK3CG (inhibitor) | 24654606 | Radiation-sensitizing/insulin antagonists/immunosuppressive |
| 35 | Rottlerin | Protein kinase (inhibitor) | 8123051 | Unknown |
| 36 | NSC-95397 | Cdc25 (inhibitor) | 11901209 | Unknown |
| 37 | TG-101348 | JAK2 (inhibitor) | 32346607 | Primary or secondary myelofibrosis |
| 38 | PIK-90 | PI3K (inhibitor) | 16864657 | Unknown |
| 39 | BRD-K73261812 | β-catenin (inhibitor) | 19382889 | Unknown |
| 40 | Tacrolimus | FKBP1A (inhibitor) | 23228564 | Transplant rejection/leukemia or lymphoma (Phase 3) |
| 41 | Niguldipine | Calcium Channel (Blockers) | 2548881 | Antineoplastic/antihypertensive/calcium channel blockers |
| 42 | Forskolin | AC (activator) | 24559688 | Cardiotonic/vasodilator/bronchodilator/immunologic |
| 43 | CD-1530 | RARG (agonist) | 27336223 | Unknown |
| 44 | CAY-10594 | Phospholipase D2 inhibitor | 31076618 | Unknown |
| 45 | CHEMBL-399379 | Unknown | Not available | Unknown |
| 46 | NVP-BEZ235 | mTOR (inhibitor) | 20804212 | Orally bioavailable antineoplastic agents |
| 47 | m-3M3FBS | PLCβ (activator) | 34625112 | Unknown |
| 48 | Temsirolimus | mTOR (inhibitor) | 18543327 | Advanced renal-cell carcinoma/mantle cell lymphoma |
| 49 | CID2858522 | PKC (inhibitor) | 23469385 | Unknown |
| 50 | MK-2206 | Akt (inhibitor) | 23917345 | Orally bioavailable antineoplastic activity |
PMID, PubMed ID for related papers.
Figure 3.
Heatmap displaying the top 50 small molecules that may potentially reverse the expression changes in esophageal squamous cell carcinoma. Rows represent small molecules and columns represent cell lines (clustered by column). Red indicates positive ZhangScores, green indicates negative ZhangScores and gray indicates a lack of experimental data. The RRAScore indicates the level of significance (P-value) calculated using the RobustRankAggreg package based on the ranking results of the six cell lines. SumZS is the sum of the ZhangScore calculated in the six cell lines for each listed molecule.
The top-ranking small molecules included several known and clinically used anti-cancer drugs, such as vemurafenib (ranked 2nd) for the treatment of melanoma (30) and vorinostat (ranked 8th) for the treatment of cutaneous T-cell lymphoma (31). Several drugs identified are involved in immune regulation, such as sirolimus (ranked 11th) and tacrolimus (ranked 40th), which are used to counteract organ rejection (32). Of note, certain natural products from edible plants or their analogs, such as curcumin (ranked 13th), phloretin (ranked 25th) and menadione (ranked 26th), were also identified.
Experimental validation of the effects of selective molecules on ESCC cell viability
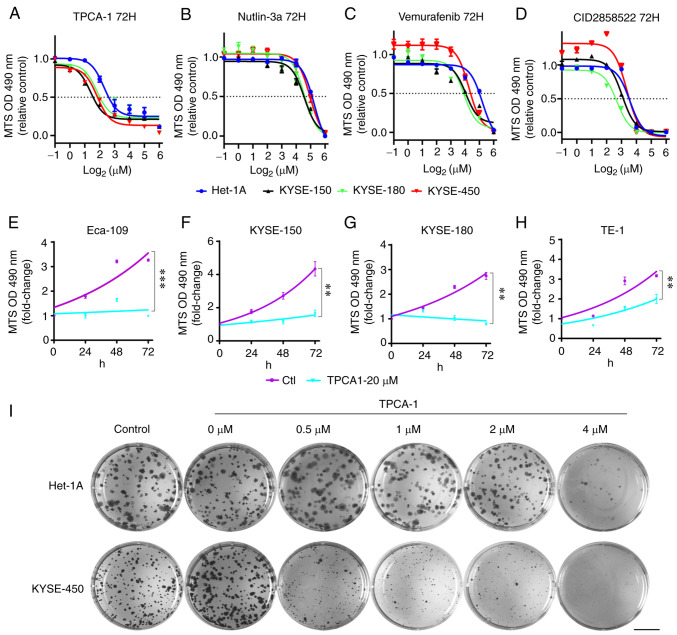
To verify the effectiveness of the screening methods used in the present study, the effects of TPCA-1 (IKKβ inhibitor; ranked 1st), vemurafenib (BRAFV600E inhibitor, Food and Drug Administration-approved for the treatment of melanoma; ranked 2nd), nutlin-3a (MDM2 antagonists; ranked 30th) and CID2858522 (PKC inhibitor; ranked 49th) on the viability of ESCC cells and a non-tumorigenic human epithelial Het-1A cells were examined. These results indicated that, although different ESCC cells exhibited variable sensitivity to TPCA-1, nutlin-3a, vemurafenib or CID2858522, these four small molecules markedly inhibited ESCC cell viability in a dose-dependent manner (Fig. 4A-D, Table II). Of note, while KYSE-450 cells appeared to be somewhat resistant, Het-1A cells exhibited marked resistance to the treatment with these molecules (Table II). Furthermore, TPCA-1 was more potent in inhibiting ESCC cell viability than nutlin-3a, vemurafenib or CID2858522, as evidenced by the lowest IC50 values (Table II). Of note, TPCA-1 significantly inhibited the growth of ESCC cells (Fig. 4E-H) and had higher selectivity for inducing death of KYSE-450 cells compared with non-tumorigenic Het-1A cells (Fig. S2). To substantiate the differential inhibitory effects of TPCA-1 on ESCC and non-tumorigenic epithelial cells, colony formation assays were performed. As indicated in Fig. 4I, 0.5 µM TPCA-1 significantly inhibited the colony formation ability of KYSE-450 cells. By contrast, the same dose of TPCA-1 only had a minor effect on the colony formation ability of Het-1A cells. These results indicated that TPCA-1 inhibits KYSE-450 cells more significantly than non-tumorigenic epithelial Het-1A cells.
Figure 4.
Effects of TPCA-1, nutlin-3a, vemurafenib or CID2858522 on the viability and growth of ESCC cells and Het-1A cells. (A-D) Reduction of the viability of ESCC or Het-1A cells by indicated small molecules. (A) TPCA-1, (B) nutlin-3a, (C) vemurafenib or (D) CID2858522. IC50 values are listed in Table II. (E-H) Inhibition of ESCC cell growth by TPCA-1, a potent inhibitor of IKKβ protein. (E) Eca-109, (F) KYSE-150, (G) KYSE-180 and (H) TE-1. P-values were calculated based on the fold changes of cell viability between 20 µM TPCA-1 treated cells and untreated cells at 72 h. (I) Effects of TPCA-1 on the growth of Het-1A or KYSE-450 cells by colony-formation assay. Images of cells in each well (35 mm in diameter) are provided (scale bar, 10 mm). Data are derived from three independent experiments and presented as the mean ± standard error of the mean. **P<0.01, ***P<0.001. TPCA-1, 2-[(aminocarbonyl) amino]-5-(4-fluorophenyl)-3-thiophenecarboxamide; ESCC, esophageal squamous cell carcinoma; OD490, optical density at 490 nm.
Table II.
IC50 values (µM) of esophageal squamous cell carcinoma cell lines treated with the indicated small molecules for 72 h.
| Molecule | Het-1A | KYSE-150 | KYSE-180 | KYSE-450 |
|---|---|---|---|---|
| TPCA-1 | 4.26 (3.43-5.41) | 2.63 (2.30-3.03) | 3.45 (2.62-4.61) | 3.56 (3.00-4.15) |
| Nutlin-3a | 38.30 (35.68-41.28) | 25.43 (23.73-27.19) | 26.84 (24.67-29.11) | 35.38 (31.59-39.64) |
| Vemurafenib | 37.17 (34.08-40.66) | 18.51 (13.30-27.42) | 15.37 (12.34-18.70) | 19.36 (17.50-21.51) |
| CID2858522 | 11.77 (10.85-13.05) | 8.26 (7.59-9.01) | 6.45 (5.80-7.12) | 10.28 (8.93-11.85) |
The 95% confidence interval is presented in parentheses.
TPCA-1 induced apoptosis in ESCC cells through inhibition of NF-κB signaling
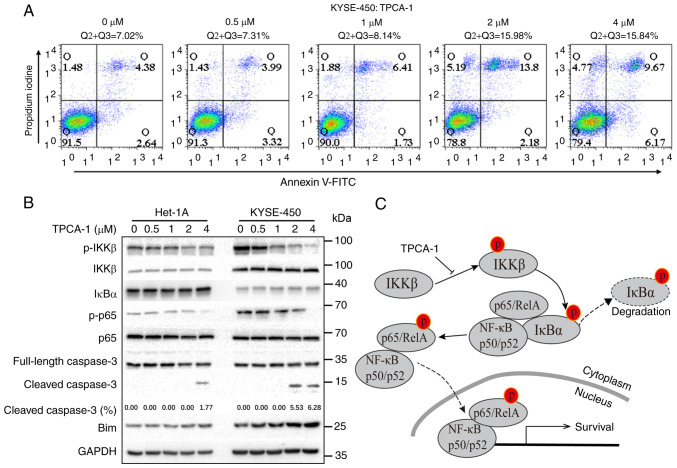
To explore the mechanisms through which TPCA-1 inhibits the growth of ESCC cells, the effects of TPCA-1 on KYSE-450 cell apoptosis were examined using flow cytometry. As presented in Fig. 5A, TPCA-1 significantly induced KYSE-450 cell apoptosis in a dose-dependent manner. Of note, as presented in Fig. 5B, the NF-κB pathway was significantly activated in KYSE-450 but not in Het-1A cells, as evidenced by the lower IκBα expression and higher levels of p-KKβ and p-p65 compared with Het-1A cells. Treatment of KYSE-450 cells with TPCA-1 led to the inhibition of both p-IKKβ and p-p65, ultimately resulting in increased Bim protein expression, cleaved caspase 3 and apoptosis. Despite the observation that TPCA-1 also inhibited IKKβ phosphorylation in Het-1A cells, the expression level of IκBα remained high and suppression of p-p65 was barely observed. Of note, treatment with 4 µM TPCA-1 was still able to trigger apoptosis in Het-1A cells, suggesting that TPCA-1 promotes apoptosis in Het-1A cells only when used at higher doses. Collectively, these results indicated that Het-1A cells are less dependent on the activation of NF-κB signaling for survival and are thus less sensitive to the TPCA-1-mediated inhibition of IKKβ.
Figure 5.
TPCA-1, a potent inhibitor of IKKβ protein, induces Het-1A and KYSE-450 cell apoptosis. (A) Effects of TPCA-1 on KYSE-450 cell apoptosis, as indicated by flow cytometric analyses. (B) Western blot analysis of the expression of proteins in the NF-κB signaling pathway and apoptosis markers in Het-1A and KYSE-450 cells. The effects of short- and long-term exposures on IKKβ and IκBα are presented. The percentage of cleaved caspase 3 was measured by ImageJ with images of full-length and cleaved caspase 3 at the same exposure duration. (C) Mechanistic model for TPCA-1-induced apoptosis through the inhibition of the NF-κB pathway. TPCA-1, 2-[(aminocarbonyl)amino]-5-(4-fluorophenyl)-3-thiophenecarboxamide; Q, quadrant; p-p65, phosphorylated p65.
Discussion
The LINCS project has generated millions of small-molecule-perturbed transcription signatures in a variety of different cell lines (28,33). Using a LINCS-based approach, it was previously determined that crizotinib, celastrol or GSK1059615 may be combined with erlotinib to inhibit the growth of erlotinib-tolerant PC9 lung cancer cells (8), while mitoxantrone or imatinib may synergize with gemcitabine to inhibit glioblastoma (9). However, LINCS does not currently contain the transcription signatures of small-molecule perturbations in the ESCC cell lines. Thus, the small-molecule-perturbed transcription signatures cannot be used to screen for putative drugs against ESCC directly using the LINCS datasets. It may therefore be hypothesized that non-ESCC cell lines that display transcriptome profiles similar to that of ESCC in the LINCS may have similar cellular responses to small-molecule treatment in ESCC cells and that identifying small molecules that inhibit the viability of these non-ESCC cells may also inhibit the viability of ESCC cells. In the present study, the LINCS datasets, Zhang Score and RRA methods were used to screen for the small molecules that may reverse the transcriptome aberration of ESCC cells. Among the 50 top-ranking molecules, four molecules were selected for experimental verification: TPCA-1 (IKKβ inhibitor; ranked 1st), vemurafenib (BRAFV600E inhibitor; ranked 2nd), nutlin-3a (MDM2 antagonists; ranked 30th) and CID2858522 (PKC inhibitor; ranked 49th). It was indicated that all four molecules were able to significantly inhibit the growth and viability of multiple ESCC cells, suggesting that the approach used in the present study may be applicable in small molecule screening for different cancer types without small-molecule-perturbated profiles in the LINCS.
The top-ranking small molecules may be divided into several categories, including inhibitors of growth signaling pathways, molecules targeting epigenetic modification and immune modulation drugs. These pathways are known to be important for cancer progression and treatment. Of note, several small molecules are clinically approved drugs or currently part of a clinical trial for cancer treatment. For instance, vemurafenib (ranked 2nd) is a BRAFV600E inhibitor used for the treatment of melanoma (34). Selumetinib (ranked 3rd) is a MAP2K1 inhibitor used for the treatment of melanoma, neurofibromatosis type 1 and thyroid cancer (35). Vorinostat (ranked 8th) is an HDAC inhibitor that has completed a phase I clinical trial on cutaneous T-cell lymphoma (31). Of note, several further drugs have been approved for the treatment of other diseases, such as valdecoxib (ranked 4th) which targets PTGS2 and is used for the treatment of rheumatoid arthritis (36). Furthermore, it has been reported that certain molecules may inhibit ESCC cells in vitro or in animal models, such as curcumin (37,38) and menadione (39).
In the present study, it was determined that TPCA-1, a molecule targeting IKKβ, exhibits a greater inhibitory effect on ESCC KYSE-450 cells than non-tumorigenic epithelial Het-1A cells. The results indicated that NF-κB signaling was activated in KYSE-450 cells, in keeping with the observation that NF-κB signaling is frequently constitutively activated in ESCC (40). The activation of NF-κB signaling relies on the phosphorylation of IKKβ, which in turn promotes the phosphorylation and degradation of IκBα, resulting in the activation and nuclear localization of the NF-κB complex, thus facilitating cell survival (14,15). The present study demonstrated that TPCA-1 potently inhibits NF-κB signaling in KYSE-450 cells, resulting in apoptosis. By contrast, treatment with low-dose TPCA-1 (0.5-2 µM) failed to influence the levels of IκBα protein and phosphorylated p65 in Het-1A cells, which may explain why TPCA-1 is more potent in killing KYSE-450 cells than Het-1A cells. Of note, a higher dose of TPCA-1 may still trigger apoptosis of Het-1A.
Among the tested ESCC cell lines, KYSE-450 cells exhibited a certain amount of resistance to the four tested molecules. By searching the cancer dependency map (41), a unique missense mutation of SMAD4, SMAD4D255N, was identified in KYSE-450, but not in KYSE-150 or KYSE-180 cells. SMAD4 is a critical tumor suppressor. The inactivation of SMAD4 by point mutations or deletions has been indicated to cause drug resistance in colon cancer (42) and pancreatic ductal adenocarcinoma (43). Low SMAD4 expression is also associated with cetuximab resistance in head and neck squamous cell carcinoma, a cancer type similar to ESCC (44). Taken together, the SMAD4D255N mutation may contribute to the drug resistance observed in KYSE-450 cells, which should be further investigated.
Several limitations of the approach used in the present study require to be noted. First, the ranking of small molecules in the present study was primarily based on the drugs' potential to reverse the transcriptome change of ESCC. This strategy does not consider the regulation of gene expression levels other than transcription, such as mRNA translation, post-translational modification and protein stability. Furthermore, the screening process is unable to take the targets of drugs into consideration, which may lead to the truly valuable drugs for clinical application being missed.
In conclusion, based on small-molecule-perturbed transcriptome profiling of cell lines that are similar to ESCC cells, candidate molecules that may perturb the transcriptome signatures in ESCC were identified. A list of the top-ranking 50 small molecules that may potentially reverse the aberrant transcriptome alterations of ESCC was compiled. Numerous candidates are known to target crucial pathways in ESCC, including the RAS/RAF/MEK/MAPK, PI3K/AKT/mTOR and NF-κB pathways. It was experimentally verified that four candidates, TPCA-1, nutlin-3a, vemurafenib and CID2858522, were able to inhibit the viability of multiple ESCC cell lines. TPCA-1 (ranked 1st), a potent inhibitor of IKKβ, exerted a higher inhibitory effect on ESCC cells compared with non-tumorigenic epithelial Het-1A cells. Mechanistically, TPCA-1 exerts its anti-tumor activity by suppressing the NF-κB signaling pathway. Of note, NF-κB signaling is activated in ESCC cells, which are sensitive to the inhibition of TPCA-1. The high expression of IκBα in Het-1A cells may confer resistance to drug treatment. Thus, the potent and selective killing of ESCC cells may make TPCA-1 a potential small molecule in ESCC treatment. The screening procedure used in the present study may serve as a novel virtual screening method for drug discovery.
Supplementary Data
Acknowledgments
The authors would like to thank Professor Haoyang Cai of Sichuan University (Chengdu, China) for providing the computational resources.
Funding Statement
This research was funded in part by the National Natural Science Foundation of China (grant nos. 32100441 and 81830108) and the Sichuan University Postdoctoral Interdisciplinary Innovation Fund (grant no. 0020404153020).
Availability of data and materials
The raw data used in the present study were derived from the following public repositories: Xena (https://xena.ucsc.edu), the Sequence Read Archive (https://www.ncbi.nlm.nih.gov/sra), the Library of Integrated Network-based Cellular Signatures (https://lincsproject.org/LINCS/data/overview), Gene Expression Omnibus (https://www.ncbi.nlm.nih.gov/geo) and the Cancer Cell Line Encyclopedia (https://portals.broadinstitute.org/ccle) databases. The datasets generated during the present study are available from the corresponding author on reasonable request.
Authors' contributions
LJZ and JY prepared the draft of the original manuscript. ZYL, JY and ZXX participated in writing, reviewing and editing the manuscript. LJZ and ZYL performed data curation, experiments and formal analyses. ZXX supervised the study. ZXX and JY were responsible for project administration. ZXX and JY acquired funding. ZXX, JY and ZYL confirm the authenticity of all the raw data. All authors have read and agreed to the final version of the manuscript to be published.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Montgomery EA. Oesophageal cancer. In: Stewart BW, Wild CP, editors. World Cancer Report 2014. World Health Organization, International Agency for Research on Cancer; Lyon: 2014. pp. 528–543. [Google Scholar]
- 3.Thrift AP. Global burden and epidemiology of Barrett oesophagus and oesophageal cancer. Nat Rev Gastroenterol Hepatol. 2021;18:432–443. doi: 10.1038/s41575-021-00419-3. [DOI] [PubMed] [Google Scholar]
- 4.Sheikh M, Poustchi H, Pourshams A, Etemadi A, Islami F, Khoshnia M, Gharavi A, Hashemian M, Roshandel G, Khademi H, et al. Individual and combined effects of environmental risk factors for esophageal cancer based on results from the golestan cohort study. Gastroenterology. 2019;156:1416–1427. doi: 10.1053/j.gastro.2018.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Watanabe M, Otake R, Kozuki R, Toihata T, Takahashi K, Okamura A, Imamura Y. Recent progress in multidisciplinary treatment for patients with esophageal cancer. Surg Today. 2020;50:12–20. doi: 10.1007/s00595-019-01878-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Subramanian A, Narayan R, Corsello SM, Peck DD, Natoli TE, Lu X, Gould J, Davis JF, Tubelli AA, Asiedu JK, et al. A next generation connectivity map: L1000 platform and the first 1000000 profiles. Cell. 2017;171:1437–1452. doi: 10.1016/j.cell.2017.10.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ganci F, Pulito C, Valsoni S, Sacconi A, Turco C, Vahabi M, Manciocco V, Mazza EMC, Meens J, Karamboulas C, et al. PI3K inhibitors curtail myc-dependent mutant p53 gain-of-function in head and neck squamous cell carcinoma. Clin Cancer Res. 2020;26:2956–2971. doi: 10.1158/1078-0432.CCR-19-2485. [DOI] [PubMed] [Google Scholar]
- 8.Aissa AF, Islam A, Ariss MM, Go CC, Rader AE, Conrardy RD, Gajda AM, Rubio-Perez C, Valyi-Nagy K, Pasquinelli M, et al. Single-cell transcriptional changes associated with drug tolerance and response to combination therapies in cancer. Nature Commun. 2021;12:1628. doi: 10.1038/s41467-021-21884-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stathias V, Jermakowicz AM, Maloof ME, Forlin M, Walters W, Suter RK, Durante MA, Williams SL, Harbour JW, Volmar CH, et al. Drug and disease signature integration identifies synergistic combinations in glioblastoma. Nat Commun. 2018;9:5315. doi: 10.1038/s41467-018-07659-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lin K, Li L, Dai Y, Wang H, Teng S, Bao X, Lu ZJ, Wang D. A comprehensive evaluation of connectivity methods for L1000 data. Brief Bioinform. 2020;21:2194–2205. doi: 10.1093/bib/bbz129. [DOI] [PubMed] [Google Scholar]
- 11.Zhang SD, Gant TW. A simple and robust method for connecting small-molecule drugs using gene-expression signatures. BMC Bioinformatics. 2008;9:258. doi: 10.1186/1471-2105-9-258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Oeckinghaus A, Ghosh S. The NF-kappaB family of transcription factors and its regulation. Cold Spring Harb Perspect Biol. 2009;1:a000034. doi: 10.1101/cshperspect.a000034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yu H, Lin L, Zhang Z, Zhang H, Hu H. Targeting NF-kappaB pathway for the therapy of diseases: Mechanism and clinical study. Signal Transduct Target Ther. 2020;5:209. doi: 10.1038/s41392-020-00312-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tian F, Fan T, Zhang Y, Jiang Y, Zhang X. Curcumin potentiates the antitumor effects of 5-FU in treatment of esophageal squamous carcinoma cells through downregulating the activation of NF-κB signaling pathway in vitro and in vivo. Acta Biochim Biophys Sin (Shanghai) 2012;44:847–855. doi: 10.1093/abbs/gms074. [DOI] [PubMed] [Google Scholar]
- 15.Tao J, Zhao H, Xie X, Luo M, Gao Z, Sun H, Huang Z. The anthelmintic drug flubendazole induces cell apoptosis and inhibits NF-kappaB signaling in esophageal squamous cell carcinoma. Onco Targets Ther. 2019;12:471–478. doi: 10.2147/OTT.S193206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Taniguchi K, Karin M. NF-κB, inflammation, immunity and cancer: Coming of age. Nat Rev Immunol. 2018;18:309–324. doi: 10.1038/nri.2017.142. [DOI] [PubMed] [Google Scholar]
- 17.Podolin PL, Callahan JF, Bolognese BJ, Li YH, Carlson K, Davis TG, Mellor GW, Evans C, Roshak AK. Attenuation of murine collagen-induced arthritis by a novel, potent, selective small molecule inhibitor of IkappaB kinase 2, TPCA-1 (2-[(aminocarbonyl)amino]-5-(4-fluorophenyl)-3-thiophenecarboxamide), occurs via reduction of proinflammatory cytokines and antigen-induced T cell proliferation. J Pharmacol Exp Ther. 2005;312:373–381. doi: 10.1124/jpet.104.074484. [DOI] [PubMed] [Google Scholar]
- 18.Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Patro R, Duggal G, Love MI, Irizarry RA, Kingsford C. Salmon provides fast and bias-aware quantification of transcript expression. Nat Methods. 2017;14:417–419. doi: 10.1038/nmeth.4197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Soneson C, Love MI, Robinson MD. Differential analyses for RNA-seq: Transcript-level estimates improve gene-level inferences. F1000Res. 2015;4:1521. doi: 10.12688/f1000research.7563.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yu G, Wang LG, Han Y, He QY. clusterProfiler: An R package for comparing biological themes among gene clusters. OMICS. 2012;16:284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Barret Schloerke DC, Joseph Larmarange, Briatte Francois, Marbach Moritz, Thoen Edwin, Amos Elberg, Crowley Jason. GGally: Extension to 'ggplot2'. R package version 2.0.0. https://CRAN.R-project.org/package=GGally.
- 24.Duan Y, Evans DS, Miller RA, Schork NJ, Cummings SR, Girke T. signatureSearch: Environment for gene expression signature searching and functional interpretation. Nucleic Acids Res. 2020;48:e124. doi: 10.1093/nar/gkaa878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kolde R, Laur S, Adler P, Vilo J. Robust rank aggregation for gene list integration and meta-analysis. Bioinformatics. 2012;28:573–580. doi: 10.1093/bioinformatics/btr709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Spector AA. Arachidonic acid cytochrome P450 epoxygenase pathway. J Lipid Res. 2009;50(Suppl):S52–S56. doi: 10.1194/jlr.R800038-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Khasawneh J, Schulz MD, Walch A, Rozman J, de Angelis M, Klingenspor M, Buck A, Schwaiger M, Saur D, Schmid RM, et al. Inflammation and mitochondrial fatty acid beta-oxidation link obesity to early tumor promotion. Proc Natl Acad Sci USA. 2009;106:3354–3359. doi: 10.1073/pnas.0802864106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Keenan AB, Jenkins SL, Jagodnik KM, Koplev S, He E, Torre D, Wang Z, Dohlman AB, Silverstein MC, Lachmann A, et al. The library of integrated network-based cellular signatures NIH program: System-level cataloging of human cells response to perturbations. Cell Syst. 2018;6:13–24. doi: 10.1016/j.cels.2017.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ghandi M, Huang FW, Jane-Valbuena J, Kryukov GV, Lo CC, McDonald ER, III, Barretina J, Gelfand ET, Bielski CM, Li H, et al. Next-generation characterization of the cancer cell line encyclopedia. Nature. 2019;569:503–508. doi: 10.1038/s41586-019-1186-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Seth R, Messersmith H, Kaur V, Kirkwood JM, Kudchadkar R, McQuade JL, Provenzano A, Swami U, Weber J, Alluri KC, et al. Systemic therapy for melanoma: ASCO guideline. J Clin Oncol. 2020;38:3947–3970. doi: 10.1200/JCO.20.00198. [DOI] [PubMed] [Google Scholar]
- 31.Argnani L, Broccoli A, Zinzani PL. Cutaneous T-cell lymphomas: Focusing on novel agents in relapsed and refractory disease. Cancer Treat Rev. 2017;61:61–69. doi: 10.1016/j.ctrv.2017.10.007. [DOI] [PubMed] [Google Scholar]
- 32.Ciancio G, Burke GW, Gaynor JJ, Mattiazzi A, Roth D, Kupin W, Nicolas M, Ruiz P, Rosen A, Miller J. A randomized long-term trial of tacrolimus and sirolimus versus tacrolimus and mycophenolate mofetil versus cyclosporine (NEORAL) and sirolimus in renal transplantation. I. Drug interactions and rejection at one year. Transplantation. 2004;77:244–251. doi: 10.1097/01.TP.0000101290.20629.DC. [DOI] [PubMed] [Google Scholar]
- 33.Cheng L, Li L. Systematic quality control analysis of LINCS data. CPT Pharmacometrics Syst Pharmacol. 2016;5:588–598. doi: 10.1002/psp4.12107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bollag G, Hirth P, Tsai J, Zhang J, Ibrahim PN, Cho H, Spevak W, Zhang C, Zhang Y, Habets G, et al. Clinical efficacy of a RAF inhibitor needs broad target blockade in BRAF-mutant melanoma. Nature. 2010;467:596–599. doi: 10.1038/nature09454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Markham A, Keam SJ. Selumetinib: First approval. Drugs. 2020;80:931–937. doi: 10.1007/s40265-020-01331-x. [DOI] [PubMed] [Google Scholar]
- 36.Bensen W, Weaver A, Espinoza L, Zhao WW, Riley W, Paperiello B, Recker DP. Efficacy and safety of valdecoxib in treating the signs and symptoms of rheumatoid arthritis: A randomized, controlled comparison with placebo and naproxen. Rheumatology (Oxford) 2002;41:1008–1016. doi: 10.1093/rheumatology/41.9.1008. [DOI] [PubMed] [Google Scholar]
- 37.Liu Y, Wang X, Zeng S, Zhang X, Zhao J, Zhang X, Chen X, Yang W, Yang Y, Dong Z, et al. The natural polyphenol curcumin induces apoptosis by suppressing STAT3 signaling in esophageal squamous cell carcinoma. J Exp Clin Cancer Res. 2018;37:303. doi: 10.1186/s13046-018-0959-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tung LN, Song S, Chan KT, Choi MY, Lam HY, Chan CM, Chen Z, Wang HK, Leung HT, Law S, et al. Preclinical study of novel curcumin analogue SSC-5 using orthotopic tumor xenograft model for esophageal squamous cell carcinoma. Cancer Res Treat. 2018;50:1362–1377. doi: 10.4143/crt.2017.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu W, Xie L, He YH, Wu ZY, Liu LX, Bai XF, Deng DX, Xu XE, Liao LD, Lin W, et al. Large-scale and high-resolution mass spectrometry-based proteomics profiling defines molecular subtypes of esophageal cancer for therapeutic targeting. Nat Commun. 2021;12:4961. doi: 10.1038/s41467-021-25202-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li B, Li YY, Tsao SW, Cheung AL. Targeting NF-kappaB signaling pathway suppresses tumor growth, angiogenesis, and metastasis of human esophageal cancer. Mol Cancer Ther. 2009;8:2635–2644. doi: 10.1158/1535-7163.MCT-09-0162. [DOI] [PubMed] [Google Scholar]
- 41.Boehm JS, Golub TR. An ecosystem of cancer cell line factories to support a cancer dependency map. Nat Rev Genet. 2015;16:373–374. doi: 10.1038/nrg3967. [DOI] [PubMed] [Google Scholar]
- 42.Papageorgis P, Cheng K, Ozturk S, Gong Y, Lambert AW, Abdolmaleky HM, Zhou JR, Thiagalingam S. Smad4 inactivation promotes malignancy and drug resistance of colon cancer. Cancer Res. 2011;71:998–1008. doi: 10.1158/0008-5472.CAN-09-3269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen YW, Hsiao PJ, Weng CC, Kuo KK, Kuo TL, Wu DC, Hung WC, Cheng KH. SMAD4 loss triggers the phenotypic changes of pancreatic ductal adenocarcinoma cells. BMC Cancer. 2014;14:181. doi: 10.1186/1471-2407-14-181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ozawa H, Ranaweera RS, Izumchenko E, Makarev E, Zhavoronkov A, Fertig EJ, Howard JD, Markovic A, Bedi A, Ravi R, et al. SMAD4 loss is associated with cetuximab resistance and induction of MAPK/JNK activation in head and neck cancer cells. Clin Cancer Res. 2017;23:5162–5175. doi: 10.1158/1078-0432.CCR-16-1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw data used in the present study were derived from the following public repositories: Xena (https://xena.ucsc.edu), the Sequence Read Archive (https://www.ncbi.nlm.nih.gov/sra), the Library of Integrated Network-based Cellular Signatures (https://lincsproject.org/LINCS/data/overview), Gene Expression Omnibus (https://www.ncbi.nlm.nih.gov/geo) and the Cancer Cell Line Encyclopedia (https://portals.broadinstitute.org/ccle) databases. The datasets generated during the present study are available from the corresponding author on reasonable request.