Abstract
Astrocytes have a prominent role in metabolic homeostasis of the brain and can signal to adjacent neurons by releasing glutamate via a process of regulated exocytosis. Astrocytes synthesize glutamate de novo owing to the pyruvate entry to the citric/tricarboxylic acid cycle via pyruvate carboxylase, an astrocyte specific enzyme. Pyruvate can be sourced from two metabolic fuels, glucose and lactate. Thus, we investigated the role of these energy/carbon sources in exocytotic glutamate release from astrocytes. Purified astrocyte cultures were acutely incubated (1 h) in glucose and/or lactate-containing media. Astrocytes were mechanically stimulated, a procedure known to increase intracellular Ca2+ levels and cause exocytotic glutamate release, the dynamics of which were monitored using single cell fluorescence microscopy. Our data indicate that glucose, either taken-up from the extracellular space or mobilized from the intracellular glycogen storage, sustained glutamate release, while the availability of lactate significantly reduced the release of glutamate from astrocytes. Based on further pharmacological manipulation during imaging along with tandem mass spectrometry (proteomics) analysis, lactate alone, but not in the hybrid fuel, caused metabolic changes consistent with an increased synthesis of fatty acids. Proteomics analysis further unveiled complex changes in protein profiles, which were condition-dependent and generally included changes in levels of cytoskeletal proteins, proteins of secretory organelle/vesicle traffic and recycling at the plasma membrane in aglycemic, lactate or hybrid-fueled astrocytes. These findings support the notion that the availability of energy sources and metabolic milieu play a significant role in gliotransmission.
Keywords: Astrocytes, Aglycemia, Ca2+ and glutamate imaging, Glucose, Lactate, Proteomics
Introduction
Astrocytic glutamate acts at the crossroads of signaling and metabolism in the mammalian brain (reviewed in [1]). Astrocytes have a prominent role in the brain operation as they are capable of gliotransmission, i.e. cell–cell, be that to nearby astrocytes or neurons, signaling by releasing a variety of transmitters, including glutamate, through a process of regulated exocytosis (reviewed in [2, 3]). As glutamate is the main excitatory transmitter in the brain, both its intra- and extracellular concentrations are tightly controlled (reviewed in [4, 5]). The intracellular glutamate concentration in astrocytes is dually regulated both by the action of cytosolic glutamine synthetase [6] and the mitochondrial oxidative metabolism, which mainly imparts the activity of glutamate dehydrogenase and the tricarboxylic acid (TCA) cycle enzymes (reviewed in [7-9]). A portion of cytosolic glutamate is concentrated into glutamatergic secretory vesicles. Indeed, astrocytes express all the three known types of vesicular glutamate transporters [10-12], of which VGLUT3, but not VGLUTs 1 and 2, along with the availability of cytosolic glutamate contribute to the regulation of exocytotic glutamate release from these glial cells [13].
As alluded to above, astrocytes serve as a metabolic hub whereby glutamate interlaces carbohydrate and amino acid metabolisms. Astrocytes are the lone cells in the brain that synthesize glutamate de novo, thereby controlling the overall glutamate availability within the brain. This astrocytic function critically depends on the activity of pyruvate carboxylase [14, 15], an astrocyte-specific enzyme carrying an important anaplerotic reaction that creates oxaloacetate from pyruvate. In turn, this feeds the production of α-ketoglutarate, another TCA cycle intermediate, which can exit the TCA cycle by transamination into glutamate due to the activity of mitochondrial aspartate and alanine aminotransferases [16, 17]. Consequently, two major cellular fuels, D-glucose and L-lactate, not only serve as sources of pyruvate but also as precursors of glutamate [18]. Yet, it is unknown whether and how the availability of these main energy substrates may affect exocytotic glutamate release from astrocytes, which is the very subject of the present investigation.
Using purified cultures of rat cortical astrocytes and single-cell fluorescence microscopy, we obtained data indicating that D-glucose, either taken-up from the extracellular space or mobilized from the intracellular glycogen storage, promoted exocytotic release of glutamate from astrocytes, while the availability of L-lactate, as either the sole or auxiliary (to glucose) fuel, hampered this process. Based on additional pharmacological manipulations, lone lactate action on astrocytic secretory function appears to be associated with metabolic changes consistent with an increased synthesis of fatty acids carried out by fatty acid synthase, an increased amount of which was detected using a tandem mass spectrometry (proteomics) analysis. Additional proteomics findings support the notion that the availability of specific energy source (D-glucose, L-lactate or their hybrid) results in the remodeling of the protein landscape in astrocytes, a condition-dependent process that generally included changes in levels of cytoskeletal proteins, proteins of secretory organelle/vesicle traffic and recycling at the plasma membrane in aglycemic, lactate or hybrid-fueled astrocytes when compared to normoglycemic cells. These metabolic and protein profile changes render astrocytes with a modifiable output via glutamatergic gliotransmission. As astrocytic metabolic, glucose- and lactate-based, networks play roles in sustainability of glutamatergic synaptic transmission [19] along with the requirement for long-term memory formation [20], our findings are of importance to brain operation in health and disease.
Materials and Methods
Ethical Approval
All procedures involving animals were in strict accordance with the National Institutes of Health Guide for Care and Use of Laboratory Animals and were approved by the University of Alabama at Birmingham Institutional Animal Care and Use Committee. The procedures also conform to the principles of UK regulations [21].
Cell Cultures
We prepared enriched astrocytic cultures using a modification [22] of the originally described shaking procedure [23]. Visual cortices isolated from 0- to 2-day-old Sprague Dawley rats were treated enzymatically in Hank’s Balanced Salt Solution (HBSS) (Life Technologies; Carlsbad, CA) containing 20 IU/ml papain and 0.2 mg/ml L-cysteine (1 h at 37 °C). After subsequent treatment with trypsin inhibitor (10 mg/ml; type II-O; 5 min at room temperature) to terminate the enzymatic reaction, tissue was dispersed mechanically by triturating through a glass pipette in cell culture medium containing α-minimum essential medium (Cat.No. 41061; without phenol red; Life Technologies, Carlsbad, CA) supplemented with fetal bovine serum (10% v/v, Hyclone, Cat. No. SV30014.03, Lot No. FTM33793; Logan, UT), sodium bicarbonate (14 mM), sodium pyruvate (1 mM), D-glucose (20 mM), L-glutamine (2 mM), penicillin (100 IU/ml) and streptomycin (100 μg/ml) (pH 7.4). Cells were initially plated into tissue culture flasks (25 cm2) and maintained at 37 °C in a humidified 5% CO2/95% air atmosphere in a complete culture medium. After 14–24 days in culture, the cells were shaken twice (260 rpm at 37 °C), first for 1.5–2 h and then, after exchange of complete medium, again for 18–20 h. At that time, the procedure diverged. For proteomics, the remaining attached cells, i.e. purified astrocytes were rinsed twice with HBSS, subjected to treatments (see below), then scraped off the flask, pelleted by centrifugation (100 × g, 10 min) and their pellets snap frozen and stored at −80 °C until used for mass spectrometry analysis (see below). For all other experiments, cells that remained adhered after shaking procedure were detached from flasks using trypsin [10000 Nα-benzoyl-L-arginine ethyl ester hydrochloride (BAEE) units/ml; Sigma-Aldrich] and replated onto round (12 mm in diameter) glass coverslips precoated with polyethyleneimine (1 mg/ml; Sigma-Aldrich). These purified astrocytes were kept in culture for 1–3 d (15–27 d after initial plating) until used in experiments. Our astrocytic cultures consistently reach the purity of well over 99%, as confirmed by glial fibrillary acidic protein immunoreactivity and visualization of accumulation of the dipeptide β-Ala-Lys conjugated to 7-amino-4-methylcoumarin-3-acetic acid, as we previously described [10].
Treatments
All (pre)treatments were done at room temperature (20–24 °C) for 60 min as follows: after three washes in HBSS, external solution containing (in mM) 140 NaCl, 5 KCl, 2 CaCl2, 2 MgCl2, and 10 HEPES, (pH 7.4) with (i) no (0 mM) D-glucose (Glc), (ii) 5 mM Glc, or (iii) 2.5 mM L-lactate (Lac) with or without 5 mM Glc was applied to astrocytes. Of note, the concentrations of D-glucose and L-lactate are selected within the upper level of the normal range values found in the blood/plasma with the rationale that astrocytes could experience such levels in the brain as they are intimately interfaced with the vasculature. In a subset of experiments, astrocytes in 0 mM Glc containing external solution were subjected to a glycogenolysis (GL) treatment by receiving 100 μM of norepinephrine/noradrenaline (NE) for 75 min, a period within which 1 mM 1,4-dideoxy-1,4-imino-d-arabinitol (DAB, Sigma-Aldrich) was added for the last 15 min [24]. Alternatively, astrocytes in external solution containing both 2.5 mM Lac and 5 mM Glc received 1 mM of phenylsuccinate (PS) or 5 μM quercetine (Q) for 1 h. All chemicals were obtained from Sigma-Aldrich (St. Louis, MO). For Ca2+ and glutamate measurement, done promptly after the treatment, astrocytes were washed with and then imaged in a fresh external solution as in the treatment (i-iii) but devoid of additive(s), i.e., NE, DAB, PS and Q, where applicable. For solution preparation, we used water as solvent, unless otherwise specified. Water was purified by the Milli-Q® Synthesis system (Millipore Corp). This ultra-pure water has 18.2 MΩ*cm resistivity, less than five parts per billion (ppb) of organics content and pyrogen content less than 0.001 EU/ml.
Ca2+ Measurements
We monitored cytosolic Ca2+ levels of solitary astrocytes, devoid of cell–cell contact to reduce intercellular signaling, using a Ca2+ indicator, fluo-3 [25]. Cells were loaded in external solution containing the acetoxymethyl (AM) ester derivative of fluo-3 (10 μg/ml; Molecular Probes) and pluronic acid (0.025% w/v; Molecular Probes), for 30 min at room temperature. After washing in external solution, de-esterification of the dye was permitted for 30 min at room temperature. Coverslips containing fluo-3-loaded cells were mounted into a recording chamber filled with external solution and imaged; an individual time-lapse experiment lasted 3 min. All data were background subtracted, using regions of the coverslip field containing no cells, and expressed as dF/Fo (percentage), where Fo represents the fluorescent level before cell stimulation, and dF represents the change in fluorescence. The dF/Fo of all groups were normalized to the control group median value. Data were expressed as a median ± interquartile range.
Glutamate Measurements
We optically monitored extracellular glutamate levels using an L-glutamate dehydrogenase (GDH)-linked assay [10, 26], in which GDH generates NADH from NAD+ (β-nicotinamide adenine dinucleotide) in the presence of glutamate. Astrocytes were bathed in an enzymatic assay solution containing external solution supplemented with NAD+ (1 mM; Sigma-Aldrich, Cat. No. N6522) and GDH (~ 53 IU/ ml; Sigma-Aldrich, Cat. No. G2626) (pH = 7.4). Glutamate released from solitary astrocytes in the extracellular space was detected as an increase in NADH (conversion from NAD+ as glutamate is converted to α-ketoglutarate) fluorescence in areas surrounding cells. Every experiment (3 min time-lapse) was preceded by a sham run (3 min time-lapse) on cells bathed in solution lacking GDH and NAD+, which was used to correct for photo-bleaching and background subtraction. Data were expressed as dF/Fo (percentage), in which dF represents the change of fluorescence, while Fo represents the fluorescence level of a region of the coverslip in the near vicinity of the astrocyte, immediately and laterally of its soma, before mechanical stimulation. The dF/Fo of all groups were normalized to the control group median value in order to allow comparisons between experimental batches and accommodate for variations in GDH concentration and culture conditions. Furthermore, we made ratios of ranked glutamate over Ca2+ peaks (15 pair of cells in each condition) which were then normalized to the median value of the control group (the latter with the inherently proportional ratio of 1) to gain an insight on whether the various treatments have a (dis)proportional effect on two contributing parameters. Data were expressed as a median ± interquartile range.
Mitochondrial NADH/NADPH Imaging
Changes in intracellular/mitochondrial NADH and NADPH content were determined by imaging astrocytes (solitary and/or in contact) prior to and after one of the three treatments (normoglycemia, aglycemia or GL) as described above. Analyzed regions of interest were defined as rectangles of minimum 6.4 μm × 6.4 μm in the perinuclear region containing mitochondria, as per image inspection [27, 28]. Each cell received three such regions of interest positioned to triangulate the nucleus, and the average fluorescence intensity of each cell reported. All imaging data were corrected for background subtraction, and expressed as F/Fo (percentage), in which F represents the fluorescence intensity after the treatment, while Fo represents the initial fluorescence intensity prior to the treatment. Data were expressed as a median ± interquartile range.
Imaging Acquisition and Processing
All experiments were done at room temperature. We used an inverted microscope (TE 300; Nikon, Melville, NY) equipped with differential interference contrast and widefield epifluorescence illumination (100-W halogen and 100-W xenon arc lamps, respectively). Images were captured through a 40X SFluor oil-immersion objective (1.3 numerical aperture; Nikon) using a CoolSNAP-HQ cooled CCD camera (Roper Scientific, Tucson, AZ) driven by V++ imaging software (Digital Optics, Auckland, New Zealand). For Ca2+ imaging, we used a standard fluorescein/FITC filter set (Chroma Technology, Rockingham, VT, USA), while for glutamate imaging experiments we used a DAPI filter set (Nikon). The later DAPI filter set was also used for detection of native cell autofluorescence representing intracellular, mainly mitochondrial, NADH [29-32]. For time-lapse image acquisition, a camera and an electronic shutter (Vincent Associates, Rochester, NY) inserted in the excitation pathway were controlled by software. All the raw fluorescence images had pixel intensities without saturation and within the camera’s dynamic range (0–4095).
Stimulation of Astrocytes
To evoke an increase in the cytosolic Ca2+ concentrations in astrocytes and consequential exocytotic/vesicular glutamate release from these cells, we mechanically stimulated astrocytes using patch pipettes [25, 33], a stimulus promoting vesicular fusions in these glial cells [34]. In addition, this approach allows spatial–temporal control of the stimulus application without affecting the plasma membrane integrity [34, 35]. Of note, Rose Bengal, an inhibitor of vesicular glutamate uptake abolishes mechanically-induced glutamate release from astrocytes [10]. To control for the contact between the pipette and the solitary astrocyte we monitored pipette resistance using a patch-clamp amplifier (PC-ONE; Dagan, Minneapolis, MN) to achieve the comparable strength of the stimulus (increase in resistance) under all conditions tested [36]
Gel-Based Liquid Chromatography-Tandem Mass Spectrometry (GeLC-MS/MS)
To study astrocyte proteomics, we used the GeLC-MS/MS approach [37], whereby SDS-PAGE is used to separate astrocytic protein lysate followed by in-gel digestion, and analysis by LC–MS/MS. Frozen astrocyte pellets were resuspended in ice-cold, 1.25 X buffer A (50 mM Tris–HCl pH 7.8 at 4 °C, 5 mM EDTA, 1 mM EGTA, 10% glycerol and 2 mM phenylmethanesulfonyl fluoride), solubilized by addition of 4% SDS, and subjected to in-gel tryptic digestion using a modified trypsin intended to minimize autolysis (Trypsin Gold, Promega, Cat. No. V5280), as previously described [38]. Mass spectrometry was performed at the UAB Targeted Metabolomics and Proteomics Laboratory core facility using a hybrid Triple TOF 5600 mass spectrometer (AB SCIEX, Toronto, ON) coupled online to a split-less NanoLC-1D Plus System (Eksigent Technologies, Dublin, CA). Peptides (200 ng) were loaded onto a Nano cHiPLC trap column (75 μm × 15 cm ChromXP C18-CL 3 μm 120 Å, Eksigent Technologies) at a constant flow rate of 2 μl/min using a NanoLC-AS1 autosampler (Eksigent Technologies). After washing the trap column for 4 min in solvent A (0.1% formic acid in water), peptides were loaded on a Nano cHiPLC analytical column (200 μm × 0.5 mm ChromXP C18-CL 3 μm 120 Å, Eksigent Technologies) at a flow rate of 300 nl/min and then eluted with a linear gradient from 95% solvent A and 5% solvent B (0.1% formic acid in acetonitrile) to 50% solvent A and 50% solvent B over 90 min. Peptides were then electrosprayed and analyzed on a hybrid Triple TOF 5600 mass spectrometer (AB SCIEX, Toronto, ON) operating in data-dependent mode using Analyst Software version 1.5 TF (AB SCIEX). Survey scans were acquired in 250 ms from 400 to 1250 m/z and 20 product ion scans were collected with 50 ms dwell times from 400 to 2000 m/z if exceeding a threshold of 100 counts per second. The data were centroided and de-isotoped using Analyst (AB SCIEX). Peptide matches were inferred using the in-house Mascot Server (version 2.2.07) to search the fragment ion (MS2) spectra against a concatenated reverse target-decoy database containing rat sequences extracted from the UniProtKB/Swiss-Prot protein database (release 2014_10). All searches were performed with full tryptic cleavage specificity. Carbamidomethyl (C) was set as a fixed modification; oxidation (M) and deamidation (N and Q) were set as variable modifications. The maximum allowed missed cleavages = 2, peptide mass tolerance = ± 0.05 Da, and MS/MS mass tolerance = ± 0.03 Da. MS/MS based peptide and protein identifications were validated with Scaffold (version Scaffold_4.2.1). Peptide identifications were accepted if their identities were established at greater than 90.0% probability by the Scaffold Local FDR algorithm. Protein identifications were accepted if their identities were established at greater than 99.0% probability and if containing at least two identified peptides. Protein probabilities were assigned by the Protein Prophet algorithm [39]. The estimated protein false discovery rate was 0.7% based on the number of decoy hits at the PeptideProphet cut-off score. Differences in protein abundance were assessed by spectral counting. Spectral counts per protein were normalized to total spectral counts per sample and then averaged across three (in some cases two) biological replicates.
Statistical Analysis
We used the GB-Stat software (version 6.5; Dynamic Microsystems Inc., Silver Spring, MD) and the SAS® software (version 9.2 for Windows; SAS Institute Inc., Cary, NC) for statistical analysis. All groups contain sample sizes (astrocytes or their frozen pellets) originating from at least three independent experimental runs/culture preparations, each preparation containing tissue pooled from two animals of either sexes. For an individual set of experiments, the number of subjects (astrocytes or their preparations) required for the study was estimated using power analysis (set at 80% and α = 0.05). As some of the imaging data did not conform to normality as per the Shapiro–Wilk or D’Agostino tests, for simplicity, we analyzed all data using non-parametric statistics. The increase in fluo-3 and exogenous NADH fluorescence due to mechanical stimulation was tested using Wilcoxon Signed Rank Test. The comparison of the effects caused by various treatments on mechanically-evoked changes in cytosolic Ca2+ and extracellular glutamate, as well as that of native intracellular/mitochondrial autofluorescence (NADH and NADPH) were done using Kruskal–Wallis One-Way ANOVA (KWA) followed by Newman-Keuls post-hoc test for multiple comparisons (NKA). Differences in average normalized spectral counts of proteins were identified by applying a likelihood ratio test for independence, i.e., G-test. The G-value was calculated as previously described [40] and then used to determine p-values according to the chi-square distribution table with one degree of freedom. Significance was established at **p ≤ 0.01, or *p ≤ 0.05.
Results
D-Glucose Promotes Exocytotic Glutamate Release from Astrocytes
D-glucose serves as a precursor of pyruvate, which is utilized in the TCA cycle not only to drive (additional to glycolysis) ATP synthesis but also for de novo glutamate synthesis [reviewed in [41] (Fig. 1)]. We assessed whether the availability of D-glucose, be that by the uptake from the extracellular source or by the recruitment from the intracellular glycogen store, may affect Ca2+-dependent exocytotic glutamate release from cultured astrocytes. For simplicity, from here on, when we refer to D-glucose we may omit its chirality, except when deemed necessary.
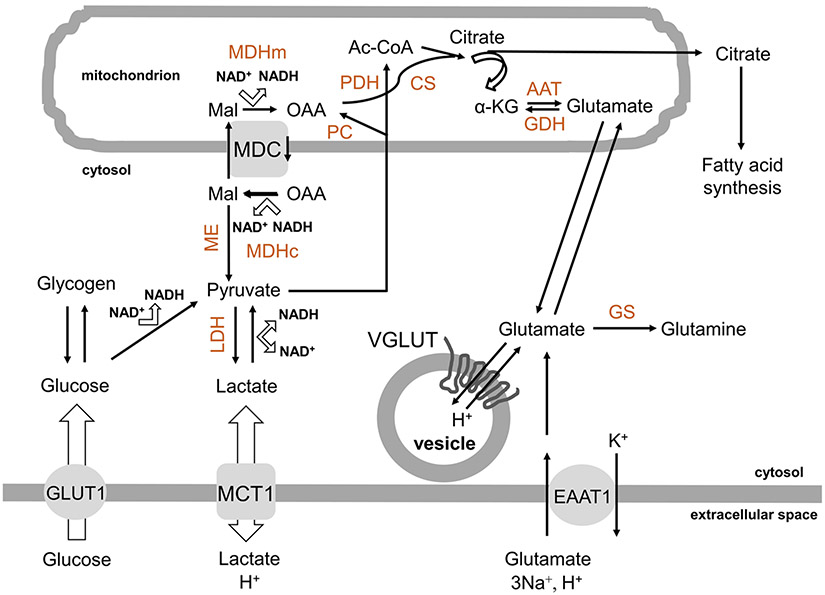
Fig. 1.
Metabolic sources for de novo synthesis of glutamate in astrocytes. Glucose can be used as a carbon source for de novo synthesis of glutamate in astrocytes. This sugar is taken-up from the extracellular space via the plasmalemmal glucose transporter GLUT1 into the cytosol and then either stored in the polymeric from as glycogen from which can be retrieved on demand, or consumed in glycolysis to pyruvate and further to lactate to recuperate NAD+; the latter reaction is bidirectional, so that lactate can be converted to pyruvate. Lactate can be bidirectionally transported down its concentration gradient out/in astrocytes by the plasmalemmal proton-linked mono-carboxylate transporter MCT1. Pyruvate can be transported into mitochondria where it dually enters the citric/tricarboxylic acid (TCA) cycle via: (1) astrocyte-specific pyruvate carboxylase (PC), which converts pyruvate to oxaloacetate (OAA), and (2) pyruvate dehydrogenase (PDH), the first component enzyme of pyruvate dehydrogenase complex, transforming pyruvate into acetyl-coenzyme A (Ac-CoA). In turn, OAA and Ac-CoA, are condensed to citrate by citrate synthase (CS). Citrate can exit mitochondria into the cytosol where it is utilized in fatty acid synthesis, or feeds into the TCA cycle, which generates ATP and glutamate, the latter by transamination of α-ketoglutarate (α-KG) mainly by aspartate amino transferase (AAT). Glutamate that leaves mitochondria into the cytosol can be converted to glutamine due to the activity of glutamine synthetase (GS) and can be transported to glutamatergic secretory organelles/vesicles via proton-dependent vesicular glutamate transporters, VGLUTs. Glutamate can be taken up from the extracellular space by the plasmalemmal excitatory amino acids transporter 1 (EAAT1), which for each glutamate molecule also transports 3Na+ and 1 proton in, while 1 K+ out of cell); the reversal operation of this transporter is not in the realm of physiology. As extracellular glutamate concentration increases (to ~ 0.5 mM) in the extracellular space, a higher proportion of cytosolic glutamate is converted to α–KG and oxidized in the TCA cycle for energy. This conversion is primarily, but not exclusively, done via mitochondrial glutamate dehydrogenase (GDH). Some of OAA is also used in the malate shuttle, which involves: A mitochondrial dicarboxylate carrier (MDC) which transports malate (Mal) from the cytosol to the mitochondrion, while OAA in the opposite direction, and B two forms of malate dehydrogenases, mitochondrial (MDHm) and cytosolic (MDHc), differentiated not only by their location, but also in structure and catalysis reactions in opposite directions in this process. Malic enzyme (ME) in the cytosol can convert malate to pyruvate. Additional details on NAD+/NADH ratio related to glycolysis and the malate shuttle are available in Results
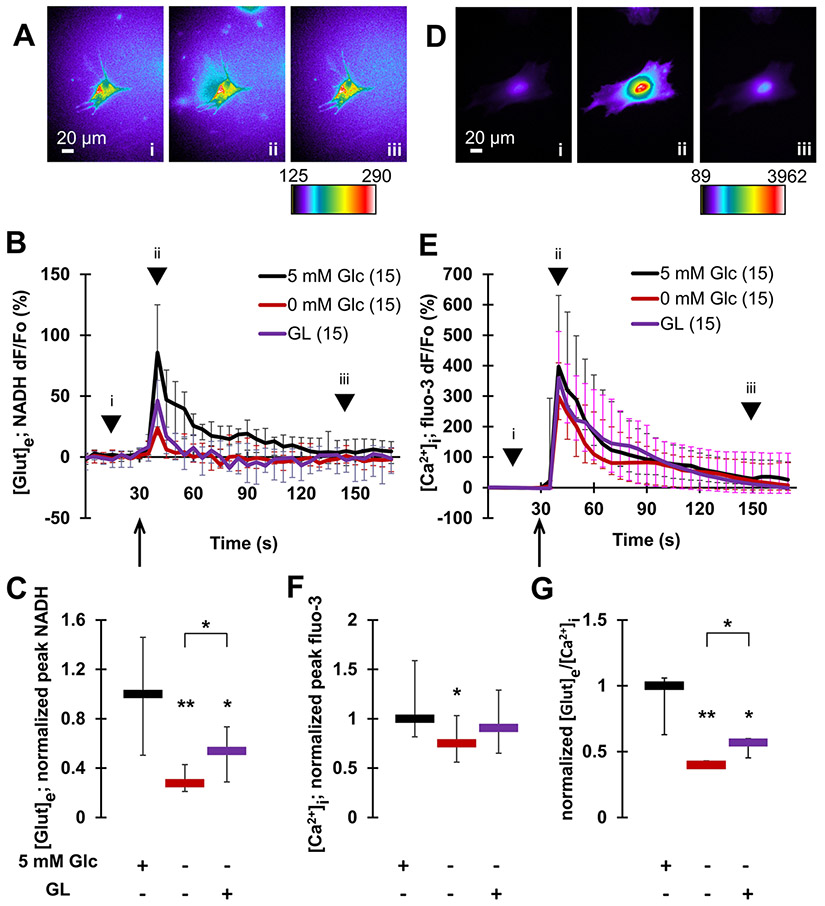
We imaged and measured the changes in extracellular glutamate concentration using the GDH-based assay in which the accumulation of NADH fluorescence in the extracellular space surrounding solitary astrocytes reports on glutamate released from these glial cells (Fig. 2A-C). Cytosolic Ca2+ dynamics in solitary astrocytes were monitored using fluo-3, a fluorescent Ca2+ indicator [25] (Fig. 2D-F). Mechanical stimulation of astrocytes in the control condition/normoglycemia, i.e. when cells were bathed in external solution containing 5 mM glucose, caused a significant increase in extracellular glutamate levels (NADH median dF/Fo = 85.7%; Wilcoxon Signed Rank Test, p ≤ 0.01) (Fig. 2A-B, and replotted in Fig. 4A) along with the underlying increase in cytosolic Ca2+ levels (fluo-3 dF/Fo = 397%; Wilcoxon Signed Rank Test, p ≤ 0.01) (Fig. 2D-E and replotted in Fig. 4C), consistent with our previous work (e.g., [10, 27]). For further comparison of the effects that various (pre)treatments may have on these measurements, we normalized the peak evoked responses obtained from astrocytes in all conditions to the median peak response obtained in the control condition (Fig. 2C, F; for details see materials and methods). In normoglycemic astrocytes inherent to the approach, this ratio equals 1 both for glutamate and Ca2+ measurements as well as for their ratio (Fig. 2G; see materials and methods for the normalization approach).
Fig. 2.
Glucose availability promotes exocytotic glutamate release from astrocytes. Mechanical stimulation induces glutamate release from (A–C), and underlying cytosolic Ca2+ elevations in (D–F), individual solitary astrocytes. A Images (raw data) of NADH signal before (i), at a peak response (ii) and after stimulation (iii). The pseudo color scale is a linear representation of the NADH fluorescence intensities ranging from 125 to 290 intensity units (i.u.). Scale bar, 20 μm. B Time lapse of extracellular NADH fluorescence dynamics in immediate vicinity of individual astrocytes, reporting on glutamate release from these glial cells. Changes in NADH fluorescence are shown as dF/Fo (percentage) after background subtraction and correction for bleaching. Mechanical stimulation of normoglycemic astrocytes (5 mM Glc) caused glutamate release from these cells. This release was greatly reduced in aglycemic astrocytes (0 mM Glc). There was a partial recovery from this reduction observed in astrocytes subjected to the glycogenolysis (GL) protocol, which recruited glucose from the intracellular glycogen store. Arrow indicates the time point when mechanical stimulation was applied, while arrowheads correspond to acquisition time points of images shown in (A). C Summary of normalized peak NADH responses shown in B. D In experiments parallel to those in A–C, astrocytes were mechanically stimulated while measuring cytosolic Ca2+ levels using fluo-3. This stimulation causes increases of cytosolic Ca2+ in astrocytes. Images (raw data) of cytosolic fluo-3 signal before (i), at a peak response (ii) and after stimulation (iii). The pseudo color scale is a linear representation of the fluo-3 fluorescence intensities in astrocytes ranging from 89 to 3962 i.u.; scale bar, 20 μm. E Time lapse of fluo-3 fluorescence dynamics reporting on cytosolic Ca2+ levels in astrocytes. Changes in fluo-3 fluorescence are shown as dF/Fo (percentage) after background subtraction. Mechanical stimulation caused an increase of cytosolic Ca2+ levels in normoglycemic astrocytes. This increase was marginally reduced in aglycemic astrocytes, but not in astrocytes lacking external glucose subjected to the GL protocol. Other annotations as in B. F Summary of normalized peak fluo-3 responses shown in B. G The ratio of glutamate release and cytosolic Ca2+ responses reveals a significant decrease in the ratio in aglycemic astrocytes when compared to normoglycemic controls. This decrease was partially rescued in astrocytes subjected to the GL protocol. All data points in graphs are shown as medians with interquartile ranges (IQRs). Number of astrocytes studied in each condition is given in parentheses in B and E. In C, F and G, charts below graphs indicate the presence (+) or absence (−) of a compound and/or a treatment. Asterisks indicate a statistical difference compared to the control/normoglycemic group. The brackets mark other differences. Significance was established using Kruskal–Wallis one-way ANOVA (KWA) followed by Newman-Keuls post-hoc test for multiple comparisons (NKT) **p ≤ 0.01, *p ≤ 0.05
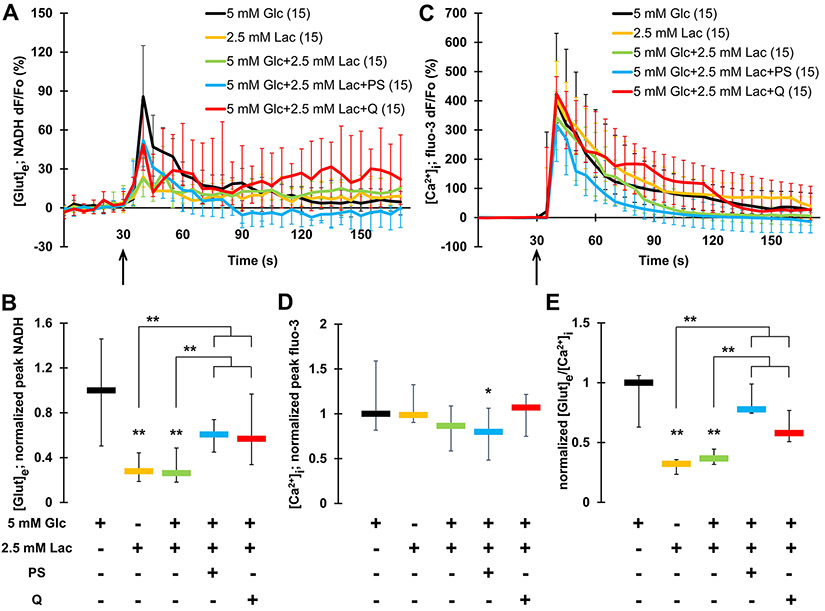
Fig. 4.
Lactate hampers exocytotic glutamate release from astrocytes. A Time lapse of NADH fluorescence, reporting on glutamate release from astrocytes. Mechanically-induced glutamate release from normoglycemic astrocytes (5 mM Glc; data replotted from Fig. 1B) is significantly higher than that from astrocytes bathed in alternative fuels, lactate (2.5 mM Lac), or the hybrid lactate-glucose fuel. Pharmacological interference with the malate shuttle using phenylsuccinate (PS), or manipulation of the plasmalemmal lactate transporter using its blocker quercetine (Q) resulted in partial rescue of the hampering effect of lactate on glutamate release from astrocytes bathed in the hybrid fuel. B Summary of normalized peak NADH responses shown in A and reporting on extracellular glutamate levels. C Time lapse of fluo-3 fluorescence, reporting on Ca2+ levels in astrocytes. Mechanically-induced Ca2+ increase in astrocytes (normoglycemic trace replotted from Fig. 1E) (black circles) was marginally reduced in PS-treated astrocytes bathed in the hybrid fuel. D Summary of normalized peak fluo-3 responses shown in C and reporting on cytosolic Ca2+ levels. E The ratio of extracellular glutamate release and cytosolic Ca2+ responses reveals a significant decrease in the ratio obtained from astrocytes using lactate or the hybrid fuel, the latter being partially rescued when astrocytes additionally received either PS or Q treatment. All data points in graphs are shown as medians with IQRs. Number of astrocytes studied in each condition is given in parentheses in A and C. Asterisks indicate a significant change of measurements assessed using a KWA and NKT, **p ≤ 0.01, *p ≤ 0.05. Other annotations as in Fig. 2
We deprived astrocytes of glucose (0 mM; 1 h) by pre-incubating them in external solution lacking this sugar, and then kept in that solution throughout the imaging paradigm. Such aglycemia leads to the reduction of cytosolic glucose level in astrocytes [42]. There was a dramatic decrease in mechanically-induced glutamate release [Fig. 2C; Kruskal–Wallis one-way ANOVA (KWA) followed by Newman–Keuls post-hoc test for multiple comparisons (NKT), p ≤ 0.01], accompanied with a significant decrease in cytosolic Ca2+ response when compared to control astrocytes in the normoglycemic (5 mM) condition (Fig. 2F; KWA and NKT, p ≤ 0.05). The reduced Ca2+ response in aglycemic astrocytes is expected as mechanical-stimulation recruits Ca2+ from the endoplasmic reticulum (ER) store, the (re) filling of which is glucose-dependent as it requires ATP for the activity of the store-specific Ca2+ ATPase [35]. However, exocytotic glutamate release is a process with a well-defined Ca2+-dependency [43]. Thus, the 2.5-fold disproportionate hampering of glutamate release vs. cytosolic Ca2+ in aglycemia, seen as a decrease in the ratio (0.40) of these two parameters (Fig. 2G), suggests a lack of secretory organellesvesicles available for fusion to the plasmalemma and/or a decrease in glutamate availability for packaging into secretory vesicles [13].
Next, we attempted to rescue the effect of external glucose deprivation by mobilizing cytosolic glucose from glycogen (Fig. 1). To that end, astrocytes were submitted to a glycogenolysis (GL) treatment whereby they were pre-incubated with norepinephrine (NE, 100 μM, 75 min) in bath solution lacking glucose to promote GL [24]. To prevent the cytosolic glucose re-entry to glycogen synthesis upon halting GL, we added 1,4-dideoxy-1,4-imino-d-arabinitol (DAB; 1 mM) during the last 15 min of GL to allow for DAB action. Of note, this inhibitor of glycogen synthesis also inhibits glycogen phosphorylase, an enzyme that catalyzes the rate-limiting step in GL; thus, by the end of our GL treatment we effectively blocked the so-called glycogen shunt activity [24], as demonstrated in single cultured astrocytes [44]. After this GL treatment, astrocytes were washed and then imaged in the absence of NE/DAB in external solution lacking glucose. Washing cells let them recover from the exposure to NE, known to cause an increase in their cytosolic Ca2+ levels [43]. The GL maneuver partially rescued the effect of extracellular glucose deprivation. This is evident by the intermediate mechanically-induced glutamate and Ca2+ responses, and their ratio, from GL-challenged astrocytes when compared to those obtained from glucose-deprived astrocytes and astrocytes under the normoglycemic condition (Fig. 2C, F for glutamate and Ca2+ response, respectively; Fig. 2G for the ratio of glutamate release and cytosolic Ca2+ responses).
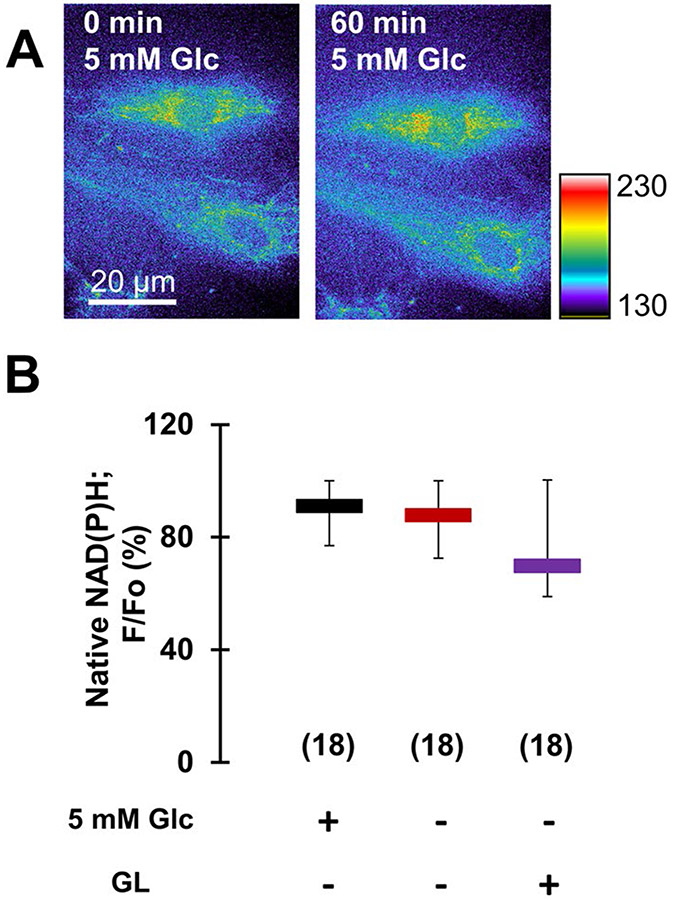
To evaluate whether the reduction in glutamate release due to aglycemia could result from consumption of glutamate in the TCA cycle, we implemented UV-induced autofluorescence imaging to assess the status of mitochondrial NAD(P)H. Namely, glutamate can be oxidized in mitochondria to α-ketoglutarate by GDH (Fig. 1), with concomitant reduction of non-fluorescent NAD(P)+ to fluorescent NAD(P)H; this is the very reaction that, in a simplified form, we have utilized for our extracellular glutamate release imaging. This method has been long used as a measure of cellular oxidative metabolism; increased glutamate oxidation in mitochondria results in an increase in the mitochondrial NAD(P)H autofluorescence signal [45, 46]. We imaged astrocytes in normoglycemic external solution, which was then replaced with and incubated in the same solution (sham treatment) or in external solution lacking glucose (aglycemic treatment), or astrocytes were subjected to the GL treatment. At the end of incubation/treatment, the same astrocytes were re-imaged and perinuclear regions containing mitochondria were analyzed for the change of NAD(P)H signal. In normoglycemic astrocytes, the NAD(P)H signal was well retained after 1 h of sham/normoglycemic treatment (Fig. 3A, B, median F/Fo = 91%). The signals originating from aglycemic or GL-treated astrocytes were not statistically different from that of normoglycemic cells (Fig. 3B). Thus, it appears that aglycemia or GL treatment do not statistically affect consumption of glutamate in oxidative metabolism in astrocyte.
Fig. 3.
Aglycemia of astrocytes does not significantly affect glutamate oxidation in their mitochondria. A UV-induced autofluorescence imaging of native NAD(P)H in astrocytes (raw data) before (left), and after incubation of cells for 60 min (right) in 5 mM glucose (control, normoglycemia). The pseudo color scale is a linear representation of the fluorescence intensities ranging from 130 to 230 i.u.; scale bar, 20 μm. B Analysis of intracellular/mitochondrial NAD(P)H signal, reporting on oxidative metabolism, expressed as percentage of fluorescence retained (F/Fo) after the incubation period, shows no significant difference between NAD(P)H fluorescence in normoglycemic and aglycemic astrocytes or astrocytes lacking external glucose subjected to the GL protocol. All data points in B are shown as medians with IQRs. Number of astrocytes studied in each condition is given in parentheses in B. Other annotations as in Fig. 2
Taken together these data indicate that glucose, sourced either from the extracellular space or from the intracellular glycogen store, is an important precursor of glutamate utilized in exocytotic release of this transmitter from astrocytes.
L-Lactate Hinders Exocytotic Glutamate Release from Astrocytes
As it is the case for D-glucose, L-lactate can be used as a metabolic fuel; it gets dehydrogenated to pyruvate, which enters the TCA cycle (reviewed in [41]) (Fig. 1). Thus, in parallel to experiments assessing the role of glucose in Ca2+-dependent exocytotic glutamate release from cultured astrocytes, we assessed the effect that L-lactate as the sole fuel or an additional fuel to glucose may have on this process. For simplicity, from here on, we mainly omit chirality referrals for L-lactate.
The replacement of extracellular glucose (5 mM) by lactate (2.5 mM; 1 h) caused a reduction in mechanically-induced glutamate release (median of normalized peak = 0.28) when compared to normoglycemic astrocytes (KWA and NKT, p ≤ 0.01) (Fig. 4A, B); this response from lactate-bathed astrocytes (Fig. 4B) was on par to that seen in aglycemic astrocytes (median of normalized peak = 0.28) (Figs. 2C). In stark contrast, the mechanically-induced Ca2+ response in astrocytes bathed in extracellular solution containing lactate as the sole energy substrate was unaffected when compared to that of the control astrocytes solely utilizing glucose (Fig. 4C, D), yielding a low ratio (0.32; 1.00 in normoglycemic astrocytes) between glutamate release and cytosolic Ca2+ response in lactate-bathed astrocytes (Fig. 4E). These data suggest that pyruvate generated from extracellular lactate does not as readily serve as a precursor for newly synthetized glutamate, while ATP synthesis remains sufficient to support Ca2+ dynamics during the time course of our experimental paradigms.
Next, we provided astrocytes with a hybrid fuel by bathing them (1 h) in the extracellular solution containing both glucose (5 mM) and lactate (2.5 mM). Surprisingly, normoglycemia in presence of lactate did not restore the level of mechanically-induced glutamate release from astrocytes (median normalized peak = 0.26) (Fig. 4A, B), while Ca2+ response was unaffected (Fig. 4C, D); the resulting ratio (0.37) between glutamate release and cytosolic Ca2+ response (Fig. 4E) was similar to that seen in glucose-deprived astrocytes with or without lactate (Fig. 2G). These data suggest that lactate as fuel alone does not readily serve as a precursor of glutamate, while in the hybrid glucose-lactate fuel, it may prohibit the glucose-powered production of glutamate; of course, treatments could also cause non-metabolic intracellular changes.
Normally, astrocytes convert glycolytically-derived pyruvate to lactate in the cytosol via lactate dehydrogenase (LDH) to retain high NAD+/NADH ratio in the cytosol and thus sustain glycolytic rate (Fig. 1). The concentration of lactate we provided extracellularly would lead to the “revert” operation of LDH, i.e. lactate oxidation, which competes with glycolysis in production of pyruvate and contributes (along with glycolysis) to reduction of the NAD+/NADH ratio. However, this ratio can be regenerated through the malate shuttle (Fig. 1), involving malate transport from the cytosol to the mitochondrion via the mitochondrial dicarboxylate carrier (MDC, solute carrier SLC25A10); malate in the cytosol gets generated from oxaloacetate with simultaneous conversion of NADH to NAD+, a reaction catalyzed by cytosolic malate dehydrogenase. In the cytosol, malate can also convert to pyruvate by activity of malic enzyme. Consequently, blocking the malate shuttle in the conditions of the hybrid glucose-lactate fuel provided in the extracellular space of astrocytes would allow for the formation of pyruvate from glucose; in turn, pyruvate would proceed to the TCA cycle and de novo glutamate synthesis. In order to assess this scenario, we pre-incubated astrocytes, in the hybrid glucose-lactate fuel containing external solution, with phenylsuccinate (PS; 1 mM, 1 h), a mitochondrial dicarboxylate carrier blocker and hence a malate shuttle blocker [47]. After washout of PS, astrocytes were imaged while bathed in external solution containing the hybrid fuel. This PS maneuver partially rescued the hindering effect of lactate on mechanically-induced glutamate release (normalized peak median = 0.61) (Fig. 4B), while Ca2+ responses remained unaffected when compared to the response of astrocytes bathed in the hybrid fuel without being exposed to PS (Fig. 4D). There was recovery of the ratio of glutamate release to cytosolic Ca2+ response (0.78; Fig. 4E). Of note, mechanically-induced Ca2+ response in PS-treated hybrid fueled astrocytes was marginally reduced when compared to those of normoglycemic astrocytes (Fig. 4D, KWA and NKT, p ≤ 0.05), perhaps indicating that the ER store-specific Ca 2+ ATPase is differentially powered by ATP generated from glycolysis then that from the TCA cycle. Nonetheless, these data are consistent with the fact that the malate shuttle is a main, but not the only mechanism that regenerates NAD+ from NADH; NAD+ could be also recuperated through the glycerol 3-phosphate shuttle [48].
Next, we pharmacologically manipulated the system by adding the plasmalemmal mono-carboxylate transporter (MCT) blocker quercetine to the bath solution containing both lactate and glucose to reduce the lactate entry into the cell. Similarly, this strategy also partially rescued mechanically-induced glutamate release (normalized peak median 0.57, KWA and NKT, p ≤ 0.01) (Fig. 4B), while Ca2+ responses were unaffected by this treatment (Fig. 4D). There was a partial recovery of the ratio of glutamate release to cytosolic Ca2+ response (0.58; Fig. 4E). This outcome supports the notion that the entry of lactate from the extracellular space leads to its hindering effect on glutamate release.
Taken together, this series of experiments imply that lactate alone does not serve well as a precursor for de novo glutamate synthesis in astrocytes and when utilized in presence of glucose, lactate interferes with glutamate production from glucose and/or perhaps with non-metabolic processes to consequently reduce vesicular glutamate release. Taken together, it appears as lactate causes a shift in metabolism perhaps by “shunting” TCA cycle prior to the production of α-ketoglutarate, presumably at the level of citrate, which would then drive cytosolic fatty acid synthesis (Fig. 1). As further pharmacological dissection of the TCA cycle to identify the site of possible action by lactate would require the usage of agents lacking specificity, we undertook a proteomics approach. Using matching conditions as used in the above glutamate release-cytosolic Ca2+ response study, we assessed whether astrocytes, utilizing lactate as fuel as opposed to glucose or the hybrid fuel, have an altered protein profile that would explain changes seen in glutamate release.
Lack of Glucose or Use of Lactate as Fuel Instead of, or in Addition to, Glucose Leads to an Altered Protein Profile in Astrocytes
To study astrocyte proteomics, we used gel-based liquid chromatography-tandem mass spectrometry (GeLC-MS/MS] [37], whereby SDS-PAGE was used to separate astrocytic protein lysate followed by in-gel digestion and analysis by LC–MS/MS. We made protein extractions from purified astrocytic cultures in four different conditions, i.e., bathed/treated for 1 h in external solution containing: (A) 5 mM glucose (normoglycemia), (B) no (0 mM) glucose (aglycemia), (C) 2.5 mM lactate as metabolic fuel instead of glucose and (D) the hybrid fuel comprised of combined (A + C) glucose and lactate. The relative abundance of each protein was assessed using label-free spectral counts recorded from protein extracts obtained from three independent purified astrocytic culture preparations. Out of a total of 1319 proteins identified (raw data available in Supplementary Table S1), we discuss below a subset of relevant proteins for this study, as listed in Table 1, which includes but is not limited to proteins that show statistically significant change in abundance due to a treatment. We provide concise function(s) for most of individual protein discussed; more detailed information on individual proteins is available at https://www.uniprot.org/uniprot/ web site by searching using UniProt ID and gene names, which we provide in Table 1. Although we only initially (to avoid congestion) refer to the Table 1 in the text, one should read the content of this entire subheading by following line items in Table 1 at all times. Also, we textually only refer to spectral counts of a subset of proteins, while all the values are readily available in Table 1 and Supplementary Table S1.
Table 1.
Analysis of protein profiles (label-free spectral counts) from (A) normoglycemic, (B) aglycemic, (C) lactate- and (D) hybrid-fueled astrocytes
| UniProt ID | Gene | Protein name | Conditions (A–D) |
p-value |
||||||
|---|---|---|---|---|---|---|---|---|---|---|
| (A) 5 mM Glc | (B) 0 mM Glc | (C) 2.5 mM Lac | (D) 5 mM Glc + 2.5 mM Lac |
A vs. B | A vs. C | A vs. D |
||||
| I | Trypsin digestion control | |||||||||
| TRYP_PIG | LOC100302368 | Trypsin (EC 3.4.21.4) $ | 48.4 ± 12.3 | 55.4 ± 3.9 | 55 ± 10.3 | 46 ± 6.2 | 0.49 | 0.52 | 0.81 | |
| II | Astrocyte characterization | |||||||||
| GFAP_RAT | Gfap | Glial fibrillary acidic protein, GFAP | 194.7 ± 40.3 | 186.3 ± 21.8 | 213.9 ± 31.7 | 204 ± 61.6 | 0.67 | 0.34 | 0.64 | |
| GLNA_RAT | Glul | Glutamine synthetase # | 12.1 ± 10.3 | 13.1 ± 4.8 | 9.1 ± 3.5 | 9.5 ± 9.1 | 0.85 | 0.52 | 0.57 | |
| AL1A1_RAT | Aldh1a1 | Retinal dehydrogenase 1 | 4.4 ± 0.7 | 4.0 ± 0.4 | 4.7 ± 2.3 | 4.4 ± 1.8 | 0.88 | 0.93 | 0.99 | |
| NDRG2_RAT | Ndrg2 | Protein NDRG2 | 6.2 ± 3.2 | 4.1 ± 0.4 | 5.1 ± 0.9 | 5.6 ± 0.0 | 0.529 | 0.758 | 0.877 | |
| RAB6A_RAT | Rab6a | Ras-related protein, Rab-6A # | 6.2 ± 1.7 | 5.3 ± 0.6 | 6.0 ± 2.6 | 7.6 ± 3.0 | 0.79 | 0.95 | 0.70 | |
| EAA1_RAT | Slc1a3 | Excitatory amino acid transporter 1, EAAT1 # | 21.9 ± 9.9 | 29.4 ± 7.1 | 26.5 ± 3.6 | 22.0 ± 9.0 | 0.29 | 0.51 | 0.99 | |
| CXA1_RAT | Gja1 | Gap junction alpha-1 protein, connexin 43 | 2.9 ± 0.8 | 6.2 ± 1.6 | 3.7 ± 0.3 | 4.7 ± 0.9 | 0.256 | 0.735 | 0.506 | |
| AQP4_RAT | Aqp4 | Aquaporin-4 | 4.1 ± 1.5 | 3.9 ± 1.2 | 5.1 ± 0.4 | 4.2 ± 1.4 | 0.939 | 0.748 | 0.966 | |
| NEST_RAT | Nes | Nestin | 17.3 ± 11.1 | 13.3 ± 7.3 | 19.1 ± 7.4 | 19.3 ± 11.7 | 0.475 | 0.762 | 0.740 | |
| TBB3_RAT | Tubb3 | Tubulin beta-3 chain | 90.2 ± 14.4 | 76.5 ± 14.5 | 93.5 ± 6.7 | 97.3 ± 20.1 | 0.287 | 0.810 | 0.608 | |
| VIME_RAT | Vim | Vimentin | 857.3 ± 180.5 | 871.4 ± 29.1 | 992.8 ± 31.9 | 898.1 ± 179.4 | 0.733 | 0.002 * | 0.330 | |
| III | Cytoskeleton (also see ad II for GFAP, nestin, beta-3 tubulin and vimentin) | |||||||||
| PLEC_RAT | Plec | Plectin | 121 ± 30.6 | 142.4 ± 65.6 | 107.0 ± 36.8 | 80.2 ± 53.6 | 0.187 | 0.355 | 0.004 * | |
| 1433E_RAT | Ywhae | 14–3-3 protein epsilon | 34.8 ± 8.9 | 31.0 ± 7.0 | 49.9 ± 5.3 | 44.4 ± 6.8 | 0.636 | 0.1 | 0.279 | |
| ACTG_RAT | Actg1 | Actin, cytoplasmic 2 (gamma) | 482.2 ± 90.7 | 416.3 ± 79.2 | 390.9 ± 33.2 | 466.1 ± 121.0 | 0.028 * | 0.002 * | 0.601 | |
| ACTC_RAT | Actc1 | Actin, alpha cardiac muscle 1 | 268.9 ± 21.8 | 266.1 ± 13.5 | 232.9 ± 14.5 | 289.4 ± 55.4 | 0.906 | 0.108 | 0.386 | |
| ACTN1_RAT | Actn1 | Alpha-actinin-1 | 175.7 ± 16.0 | 129.1 ± 19.8 | 145.2 ± 4.8 | 169.7 ± 26.4 | 0.008 * | 0.089 | 0.745 | |
| ACTN4_RAT | Actn4 | Alpha-actinin-4 | 150.7 ± 17.6 | 102.4 ± 18.6 | 115.7 ± 6.5 | 135.9 ± 24.2 | 0.002 * | 0.031 * | 0.379 | |
| DREB_RAT | Dbn1 | Drebrin | 5.4 ± 2.9 | 3.5 ± 1.7 | 4.1 ± 1.0 | 5.2 ± 2.1 | 0.514 | 0.667 | 0.932 | |
| EZRI_RAT | Ezr | Ezrin | 29.0 ± 6.4 | 43.2 ± 5.9 | 32.6 ± 11.9 | 30.4 ± 1.3 | 0.094 | 0.648 | 0.864 | |
| MOES_RAT | Msn | Moesin | 52.0 ± 13.7 | 73.0 ± 9.0 | 53.3 ± 9.9 | 64.7 ± 7.5 | 0.059 | 0.902 | 0.238 | |
| RAC1_RAT | Rac1 | Ras-related C3 botulinum toxin substrate 1 | 11.3 ± 0.6 | 9.8 ± 0.8 | 11.0 ± 0.8 | 9.0 ± 0.9 | 0.744 | 0.958 | 0.621 | |
| ROCK2_RAT | Rock2 | Rho-associated protein kinase 2 | 6.4 ± 1.4 | 4.8 ± 1.2 | 3.4 ± 1.0 | 12.3 ± 6.8 | 0.625 | 0.340 | 0.168 | |
| MYH9_RAT | Myh9 | Myosin-9 | 251.6 ± 62.2 | 213.5 ± 63.7 | 261.1 ± 36.6 | 245.8 ± 30.2 | 0.078 | 0.674 | 0.795 | |
| MYH10_RAT | Myh10 | Myosin-10 | 171.2 ± 26.6 | 154.0 ± 39.0 | 182.1 ± 9.2 | 167.9 ± 23.6 | 0.341 | 0.563 | 0.859 | |
| TPM1_RAT | Tpm1 | Tropomyosin alpha-1 chain | 46.8 ± 7.5 | 36.0 ± 5.6 | 43.8 ± 4.3 | 54.1 ± 5.7 | 0.238 | 0.755 | 0.463 | |
| TPM2_RAT | Tpm2 | Tropomyosin beta chain | 35.7 ± 3.3 | 26.5 ± 1.2 | 29.9 ± 0.5 | 30.5 ± 2.6 | 0.244 | 0.474 | 0.524 | |
| TPM3_RAT | Tpm3 | Tropomyosin alpha-3 chain | 30.3 ± 2.0 | 23.3 ± 5.8 | 28.0 ± 3.5 | 32.1 ± 3.0 | 0.341 | 0.763 | 0.816 | |
| TPM4_RAT | Tpm4 | Tropomyosin alpha-4 chain | 34.4 ± 6.5 | 26.3 ± 7.6 | 34.2 ± 3.5 | 38.9 ± 0.1 | 0.292 | 0.972 | 0.607 | |
| FINC_RAT | Fn1 | Fibronectin | 35.3 ± 27.6 | 15.8 ± 8.2 | 15.8 ± 3.1 | 28.5 ± 18.6 | 0.006 * | 0.006 * | 0.395 | |
| VINC_RAT | Vcl | Vinculin | 88.9 ± 9.8 | 60.3 ± 6.7 | 71.6 ± 2.4 | 66.5 ± 11.0 | 0.019 * | 0.173 | 0.073 | |
| CADH2_RAT | Cdh2 | Cadherin-2 | 14.9 ± 2.8 | 19.9 ± 5.8 | 13.6 ± 2.7 | 14.1 ± 1.3 | 0.391 | 0.815 | 0.891 | |
| ICAM1_RAT | Icam1 | Intercellular adhesion molecule 1 | 4.8 ± 1.7 | 5.3 ± 2.0 | 5.2 ± 2.5 | 5.9 ± 1.5 | 0.875 | 0.911 | 0.756 | |
| ARC1A_RAT | Arpc1a | Actin-related protein 2/3 complex subunit 1A | 2.5 ± 0.5 | 3.0 ± 0.5 | 2.2 ± 0.6 | 3.3 ± 0.5 | 0.826 | 0.925 | 0.729 | |
| ARPC2_RAT | Arpc2 | Actin-related protein 2/3 complex subunit 2 | 14.9 ± 1.9 | 15.9 ± 2.3 | 6.8 ± 2.6 | 14.2 ± 2.0 | 0.847 | 0.077 | 0.902 | |
| ARP2_RAT | Actr2 | Actin-related protein 2 | 15.1 ± 2.9 | 12.7 ± 2.6 | 13.1 ± 2.3 | 16.6 ± 2.0 | 0.649 | 0.698 | 0.795 | |
| ARP3_RAT | Actr3 | Actin-related protein 3 | 28.6 ± 5.0 | 27.5 ± 1.0 | 23.1 ± 2.1 | 29.1 ± 3.6 | 0.893 | 0.448 | 0.947 | |
| CAZA1_RAT | Capza1 | F-actin-capping protein subunit alpha-1 | 6.6 ± 1.3 | 4.7 ± 1.7 | 5.5 ± 0.8 | 6.6 ± 1.4 | 0.568 | 0.754 | 0.984 | |
| CAZA2_RAT | Capza2 | F-actin-capping protein subunit alpha-2 | 6.8 ± 1.5 | 5.3 ± 1.8 | 4.9 ± 1.0 | 7.0 ± 1.6 | 0.660 | 0.566 | 0.958 | |
| CAPZB_RAT | Capzb | F-actin-capping protein subunit beta | 11.0 ± 1.6 | 8.6 ± 2.0 | 9.2 ± 1.2 | 10.4 ± 3.7 | 0.586 | 0.689 | 0.897 | |
| GELS_RAT | Gsn | Gelsolin | 5.1 ± 0.9 | 3.5 ± 1.8 | 6.3 ± 1.9 | 6.0 ± 2.2 | 0.591 | 0.717 | 0.783 | |
| PROF1_RAT | Pfn1 | Profilin-1 | 29.6 ± 8.0 | 17.0 ± 8.9 | 27.0 ± 9.7 | 28.6 ± 3.6 | 0.063 | 0.725 | 0.897 | |
| SEPT2_RAT | Sept.2 | Septin-2 | 22.7 ± 0.3 | 24.6 ± 5.6 | 23.0 ± 0.9 | 20.5 ± 1.2 | 0.782 | 0.976 | 0.732 | |
| SEPT7_RAT | Sept.7 | Septin-7 | 25.9 ± 0.9 | 27.6 ± 6.1 | 22.2 ± 2.7 | 20.4 ± 4.0 | 0.818 | 0.588 | 0.416 | |
| SEPT8_RAT | Sept.8 | Septin-8 | 8.9 ± 1.1 | 8.3 ± 1.2 | 9.0 ± 0.3 | 7.5 ± 0.9 | 0.892 | 0.972 | 0.732 | |
| SEPT9_RAT | Sept.9 | Septin-9 | 11.5 ± 1.8 | 13.0 ± 2.3 | 8.7 ± 1.2 | 11.9 ± 4.5 | 0.754 | 0.537 | 0.940 | |
| SEP11_RAT | Sept.11 | Septin-11 | 24.9 ± 2.5 | 21.1 ± 3.7 | 20.1 ± 3.6 | 20.7 ± 2.5 | 0.572 | 0.472 | 0.535 | |
| TBA1A_RAT | Tuba1a | Tubulin alpha-1A chain | 125.2 ± 20.7 | 96 ± 11.7 | 120.3 ± 7.3 | 114.7 ± 17.8 | 0.050 * | 0.754 | 0.499 | |
| TBA1B_RAT | Tuba1b | Tubulin alpha-1B chain | 125.6 ± 14.8 | 82.4 ± 3.4 | 105.3 | 109.9 ± 15.1 | 0.003 * | NA | 0.305 | |
| TBA4A_RAT | Tuba4a | Tubulin alpha-4A chain | 100.3 ± 10.5 | 67.7 ± 8.5 | 90.7 ± 8.1 | 94.4 ± 18.0 | 0.012 * | 0.491 | 0.676 | |
| TBB2B_RAT | Tubb2b | Tubulin beta-2B chain | 148.6 ± 20.9 | 128.4 ± 26.9 | 149.2 ± 13.4 | 146.3 ± 32.3 | 0.224 | 0.971 | 0.894 | |
| TBB4B_RAT | Tubb4b | Tubulin beta-4B chain | 133.4 ± 15.2 | 119.6 ± 21.2 | 136.1 ± 11.1 | 139.2 ± 28.2 | 0.386 | 0.868 | 0.727 | |
| TBB5_RAT | Tubb5 | Tubulin beta-5 chain | 142.2 ± 16.6 | 124.6 ± 23.7 | 145.0 ± 10.6 | 146 ± 26.8 | 0.279 | 0.873 | 0.827 | |
| MACF1_RAT | Macf1 | Microtubule-actin cross-linking factor | 2.9 ± 0.8 | 5.7 ± 4.9 | 4.5 ± 2.3 | 7.0 | 0.332 | 0.543 | NA | |
| MAP1A_RAT | Map1a | Microtubule-associated protein 1A | 5.1 ± 0.2 | 5.1 ± 2.8 | 4.3 ± 0.3 | 5.6 ± 2.0 | 0.995 | 0.811 | 0.882 | |
| ACTZ_RAT | Actr1a | Alpha-centractin | 8.7 ± 1.4 | 9.5 ± 1.0 | 9.4 ± 1.1 | 8.3 ± 2.2 | 0.866 | 0.870 | 0.920 | |
| IV | Secretory organelle/vesicle trafficking | |||||||||
| MYO1C_RAT | Myo1c | Unconventional myosin-Ic | 33.3 ± 9.4 | 27.1 ± 12.8 | 18.5 ± 5.5 | 21.8 ± 8.9 | 0.421 | 0.037 * | 0.118 | |
| MYO1D_RAT | Myo1d | Unconventional myosin-Id | 6.4 ± 1.4 | 7.2 ± 1.6 | 5.4 | 3.9 ± 1.6 | 0.821 | NA | 0.443 | |
| MYO1E_RAT | Myo1e | Unconventional myosin-Ie | 7.8 ± 2.1 | 5.9 ± 3.3 | 5.0 ± 1.9 | 4.1 ± 1.2 | 0.619 | 0.431 | 0.282 | |
| DYHC1_RAT | Dync1h1 | Cytoplasmic dynein 1 heavy chain 1 | 59.8 ± 9.8 | 50.3 ± 31.1 | 41.3 ± 9.1 | 40.1 ± 15.5 | 0.366 | 0.065 | 0.048 * | |
| DC1I2_RAT | Dync1i2 | Cytoplasmic dynein 1 intermediate chain 2 | 3.0 ± 0.8 | 3.9 ± 1.1 | 6.2 | 3.4 ± 0.8 | 0.741 | NA | 0.866 | |
| DC1L2_RAT | Dync1li2 | Cytoplasmic dynein 1 light intermediate chain 2 | 4.3 ± 1.3 | 4.8 ± 1.9 | 4.8 ± 0.9 | 4.8 ± 2.1 | 0.881 | 0.883 | 0.881 | |
| KINH_RAT | Kif5b | Kinesin-1 heavy chain | 11.6 ± 4.7 | 10.3 ± 1.3 | 8.7 ± 3.2 | 10.2 ± 5.3 | 0.795 | 0.524 | 0.762 | |
| KLC1_RAT | Klc1 | Kinesin light chain 1 | 4.6 ± 0.4 | 4.4 ± 0.8 | 2.7 ± 0.3 | 4.8 ± 1.7 | 0.936 | 0.467 | 0.960 | |
| RAB7A_RAT | Rab7a | Ras-related protein Rab-7a | 11.6 ± 1.4 | 11.3 ± 2.5 | 11.7 ± 1.4 | 11.4 ± 1.7 | 0.948 | 0.986 | 0.970 | |
| RAB31_RAT | Rab31 | Ras-related protein Rab-31 | 4.1 ± 1.0 | 4.4 ± 1.8 | 5.8 ± 0.7 | 5.0 ± 0.9 | 0.904 | 0.592 | 0.759 | |
| V | Secretory organelle/vesicle recycling | |||||||||
| RAB6A_RAT | Rab6a | Ras-related protein, Rab-6A # | 6.2 ± 1.7 | 5.3 ± 0.6 | 6.0 ± 2.6 | 7.6 ± 3.0 | 0.79 | 0.95 | 0.70 | |
| SC22B_RAT | Sec22b | Vesicle-trafficking protein SEC22b | 6.1 ± 0.6 | 8.9 ± 2.7 | 8.1 ± 2.6 | 5.3 ± 1.9 | 0.463 | 0.580 | 0.825 | |
| USO1_RAT | Uso1 | General vesicular transport factor p115 | 5.7 ± 2.1 | 2.7 ± 0.7 | 4.7 ± 2.5 | 3.2 ± 1.3 | 0.291 | 0.765 | 0.410 | |
| RB11B_RAT | Rab11b | Ras-related protein, Rab-11B | 8.0 ± 3.1 | 6.5 ± 1.6 | 9.5 ± 1.0 | 8.1 ± 1.3 | 0.703 | 0.720 | 0.976 | |
| STX12_RAT | Stx12 | Syntaxin 12 | 3.3 ± 0.9 | 4.4 ± 0.1 | 3.2 ± 0.1 | 3.5 ± 0.6 | 0.69 | 0.96 | 0.95 | |
| VAMP2_RAT | Vamp2 | VAMP2/synaptobrevin 2 | 2.4 | 3.5 | ND | 2.3 | NA | NA | NA | |
| VAMP3_RAT | Vamp3 | VAMP3/cellulbrevin | 4 ± 1.0 | 5.9 ± 0.4 | 3.3 ± 1.5 | 4 ± 0.2 | 0.54 | 0.80 | 0.99 | |
| VAMP7_RAT | Vamp7 | Vesicle-associated membrane protein 7 | ND | 1.8 ± 0.0 | ND | ND | NA | NA | NA | |
| LAMP1_RAT | Lamp1 | Lysosome-associated membrane glycoprotein 1 | 8.2 ± 2.0 | 7.4 ± 2.4 | 7.0 ± 0.6 | 9.4 ± 2.4 | 0.852 | 0.754 | 0.765 | |
| VATB2_RAT | Atp6v1b2 | V-type proton ATPase subunit B, brain isoform | 4.7 ± 0.6 | 5.9 ± 1.1 | 5.5 ± 1.3 | 4.7 ± 1.4 | 0.72 | 0.83 | 0.99 | |
| SCRN1_RAT | Scrn1 | Secernin-1 | 7.4 ± 3.8 | 6.2 ± 0.6 | 11.5 ± 2.6 | 15.7 ± 0.4 | 0.751 | 0.337 | 0.080 | |
| SNAA_RAT | Napa | Alpha-soluble NSF attachment protein | 3.4 ± 1.4 | 2.6 ± 0.0 | 2.4 ± 0.8 | 2.3 ± 0.4 | 0.745 | 0.667 | 0.638 | |
| CAV1_RAT | Cav1 | Caveolin-1 | 8.2 ± 2.0 | 8.9 ± 2.6 | 8.2 ± 2.5 | 7.7 ± 1.5 | 0.855 | 0.999 | 0.912 | |
| CLH1_RAT | Cltc | Clathrin heavy chain 1 | 47.2 ± 22.2 | 69.0 ± 20.6 | 77.2 ± 5.6 | 77.1 ± 4.4 | 0.043 * | 0.007 * | 0.007 * | |
| AP2M1_RAT | Ap2m1 | AP-2 complex subunit mu | 9.6 ± 2.2 | 12.1 ± 2.7 | 10.4 ± 0.8 | 9.8 ± 2.2 | 0.587 | 0.848 | 0.951 | |
| DYN2_RAT | Dnm2 | Dynamin-2 | 5.9 ± 2.0 | 5.9 ± 3.7 | 6.8 ± 2.9 | 5.4 ± 1.1 | 0.997 | 0.799 | 0.880 | |
| ANXA1_RAT | Anxa1 | Annexin A1 | 50.1 ± 10.4 | 32.4 ± 8.0 | 57.2 ± 6.8 | 56.3 ± 11.1 | 0.050 * | 0.494 | 0.554 | |
| ANXA2_RAT | Anxa2 | Annexin A2 | 79.8 ± 8.6 | 67.4 ± 5.6 | 82.0 ± 7.7 | 89.2 ± 8.6 | 0.306 | 0.860 | 0.470 | |
| ANXA4_RAT | Anxa4 | Annexin A4 | 4.6 ± 0.8 | 2.1 ± 0.4 | 2.9 ± 0.4 | 3.6 ± 1.5 | 0.316 | 0.516 | 0.724 | |
| ANXA6_RAT | Anxa6 | Annexin A6 | 39.8 ± 4.3 | 28.0 ± 9.7 | 38.5 ± 9.7 | 53.1 ± 5.5 | 0.150 | 0.884 | 0.168 | |
| VI | Calcium dynamics | |||||||||
| AT2A2_RAT | Atp2a2 | Sarcoplasmic/endoplasmic reticulum calcium ATPase 2 | 24 ± 2.5 | 22 ± 5.3 | 16.7 ± 1.9 | 17 ± 4.7 | 0.77 | 0.25 | 0.27 | |
| ITPR2_RAT | Itpr2 | Inositol 1,4,5-trisphosphate receptor, type 2 | ND | 1.8 | 2.2 | ND | NA | NA | NA | |
| REVERSE_RYR2_RAT-DECOY | Ryr2 | Ryanodine receptor 2 | 1.9 | ND | ND | ND | NA | NA | NA | |
| NAC1_RAT | Slc8a1 | Sodium/calcium exchanger 1 | ND | 1.8 | ND | ND | NA | NA | NA | |
| STIM1_RAT | Stim1 | Stromal interaction molecule 1 | ND | 1.8 | ND | ND | NA | NA | NA | |
| ESYT1_RAT | Esyt1 | Extended synaptotagmin-1 | 13.6 ± 5.4 | 9.6 ± 5.0 | 6.8 ± 2.1 | 8.5 ± 4.6 | 0.40 | 0.13 | 0.28 | |
| VDAC1_RAT | Vdac1 | Voltage-dependent anion-selective channel protein 1 | 28.3 ± 1.9 | 34.8 ± 7.0 | 24.7 ± 4.1 | 29.7 ± 8.8 | 0.419 | 0.614 | 0.863 | |
| VDAC2_RAT | Vdac2 | Voltage-dependent anion-selective channel protein 2 | 14.1 ± 2.3 | 17.8 ± 2.2 | 10.3 ± 2.7 | 11.7 ± 2.3 | 0.51 | 0.43 | 0.63 | |
| VDAC3_RAT | Vdac3 | Voltage-dependent anion-selective channel protein 3 | 12.1 ± 2.5 | 16.9 ± 1.7 | 12.0 ± 0.9 | 12.0 ± 1.9 | 0.372 | 0.983 | 0.979 | |
| NAC1_RAT | Slc8a1 | Sodium/calcium exchanger 1, NCLX | ND | 1.8 | ND | ND | NA | NA | NA | |
| MPCP_RAT | Slc25a3 | Phosphate carrier protein, mitochondrial | 19.3 ± 4.0 | 23.7 ± 2.1 | 18.2 ± 0.2 | 20.6 ± 5.7 | 0.504 | 0.847 | 0.845 | |
| ADT2_RAT | Slc25a5 | ADP/ATP translocase 2 | 25.7 ± 2.6 | 28.5 ± 3.8 | 23.9 ± 2.8 | 25.9 ± 7.0 | 0.703 | 0.795 | 0.975 | |
| VII | Metabolism | |||||||||
| Glucose transport | ||||||||||
| GTR1_RAT | Slc2a1 | Solute carrier family 2, facilitated glucose transporter member 1 | 8.4 ± 1.8 | 2.6 | 6.9 ± 0.8 | 7.7 ± 0.8 | NA | 0.71 | 0.85 | |
| Lactate transport | ||||||||||
| MOT1_RAT | Slc16a1 | Monocarboxylate transporter 1 | 6.5 ± 1.9 | 8.0 ± 2.0 | 5.2 ± 1.8 | 5.8 ± 2.0 | 0.69 | 0.72 | 0.85 | |
| Glutamate metabolism | ||||||||||
| EAA1_RAT | Slc1a3 | Excitatory amino acid transporter 1, EAAT1 # | 21.9 ± 9.9 | 29.4 ± 7.1 | 26.5 ± 3.6 | 22 ± 9.0 | 0.29 | 0.51 | 0.99 | |
| GLNA_RAT | Glul | Glutamine synthetase # | 12.1 ± 10.3 | 13.1 ± 4.8 | 9.1 ± 3.5 | 9.5 ± 9.1 | 0.85 | 0.52 | 0.57 | |
| DHE3_RAT | Glud1 | Glutamate dehydrogenase 1, mitochondrial | 28 ± 3.6 | 31.8 ± 7.3 | 28.6 ± 3.1 | 32.6 ± 3.0 | 0.62 | 0.94 | 0.55 | |
| Glucose phosphorylation | ||||||||||
| HXK1_RAT | Hk1 | Hexokinase-1 | 50.5 ± 2.5 | 49.9 ± 9.6 | 50.1 ± 6.2 | 43.4 ± 2.4 | 0.95 | 0.97 | 0.46 | |
| Glycolysis | ||||||||||
| G6PI_RAT | Gpi | Glucose-6-phosphate isomerase | 25.2 ± 0.7 | 20.4 ± 4.0 | 34.7 ± 3.4 | 30.4 ± 4.0 | 0.48 | 0.22 | 0.48 | |
| K6PF_RAT | Pfkm | 6-phosphofructokinase, muscle type | 6.7 ± 1.0 | 9.5 ± 2.4 | 11.1 ± 3.3 | 13.9 ± 5.0 | 0.49 | 0.30 | 0.11 | |
| K6PL_RAT | Pfkl | 6-phosphofructokinase, liver type | 8.8 ± 1.7 | 8.0 ± 2.7 | 12.8 ± 5.6 | 11.2 ± 5.0 | 0.84 | 0.39 | 0.60 | |
| ALDOA_RAT | Aldoa | Fructose-bisphosphate aldolase A | 57.1 ± 10.7 | 58.5 ± 5.7 | 48.9 ± 4.3 | 55.0 ± 1.8 | 0.90 | 0.43 | 0.84 | |
| TPIS_RAT | Tpi1 | Triosephosphate isomerase | 19.3 ± 4.3 | 12.7 ± 1.3 | 20.8 ± 6.0 | 20.3 ± 0.5 | 0.25 | 0.80 | 0.87 | |
| G3P_RAT | Gapdh | Glyceraldehyde-3-phosphate dehydrogenase, GAPDH | 157.0 ± 10.0 | 135.5 ± 6.3 | 147.5 ± 27.5 | 149.6 ± 13.6 | 0.21 | 0.59 | 0.68 | |
| PGK1_RAT | Pgk1 | Phosphoglycerate kinase 1 | 31.1 ± 6.6 | 24.5 ± 4.1 | 32.0 ± 1.6 | 28.6 ± 8.0 | 0.38 | 0.90 | 0.75 | |
| PGM1_RAT | Pgm1 | Phosphoglucomutase-1 | 10.8 ± 7.0 | 13.5 ± 5.2 | 19.1 ± 7.1 | 14.8 ± 3.3 | 0.58 | 0.13 | 0.43 | |
| ENOA_RAT | Eno1 | Alpha-enolase | 83.8 ± 16.2 | 64.5 ± 19.7 | 98.2 ± 6.9 | 80.8 ± 17.3 | 0.11 | 0.28 | 0.82 | |
| KPYM_RAT | Pkm | Pyruvate kinase PKM | 74.2 ± 6.9 | 57.1 ± 16.0 | 85.5 ± 16.6 | 78.8 ± 16.7 | 0.14 | 0.37 | 0.71 | |
| LDHA_RAT | Ldha | L-lactate dehydrogenase A chain | 72.8 ± 9.3 | 58.7 ± 12.7 | 71.1 ± 14.4 | 75.5 ± 16.0 | 0.22 | 0.88 | 0.83 | |
| LDHB_RAT | Ldhb | L-lactate dehydrogenase B chain | 26.3 ± 1.4 | 23.9 ± 5.3 | 32.1 ± 2.9 | 34.6 ± 6.8 | 0.74 | 0.44 | 0.28 | |
| Pyruvate conversion | ||||||||||
| PYC_RAT | Pc | Pyruvate carboxylase, mitochondrial | 10.4 ± 4.3 | 10.7 ± 7.1 | 7.8 ± 2.0 | 4.0 ± 2.1 | 0.94 | 0.54 | 0.09 | |
| ODPB_RAT | Pdhb | Pyruvate dehydrogenase E1 component subunit beta, mitochondrial | 14.8 ± 0.2 | 10.1 ± 0.6 | 8.3 ± 0.7 | 12.7 ± 2.7 | 0.342 | 0.176 | 0.682 | |
| TCA cycle | ||||||||||
| CISY_RAT | Cs | Citrate synthase, mitochondrial | 18.5 ± 4.0 | 19.9 ± 4.0 | 14.1 ± 1.4 | 12.2 ± 1.4 | 0.82 | 0.44 | 0.25 | |
| ACON_RAT | Aco2 | Aconitate hydratase, mitochondrial | 27.8 ± 3.1 | 33.2 ± 3.5 | 28.3 ± 3.9 | 30.0 ± 3.9 | 0.49 | 0.95 | 0.78 | |
| IDHP_RAT | Idh2 | Isocitrate dehydrogenase [NADP], mitochondrial | 26.9 ± 5.4 | 33.0 ± 4.6 | 25.9 ± 4.3 | 22.2 ± 3.4 | 0.43 | 0.89 | 0.50 | |
| IDH3A_RAT | Idh3A | Isocitrate dehydrogenase [NAD] subunit alpha, mitochondrial | 11.9 ± 2.8 | 13.9 ± 2.5 | 14.4 ± 6.6 | 13.6 ± 1.7 | 0.69 | 0.62 | 0.73 | |
| IDH3B_RAT | Idh3B | Isocitrate dehydrogenase [NAD] subunit beta, mitochondrial | 4.3 ± 0.5 | 4.8 ± 1.2 | 3.7 ± 0.3 | 4.5 ± 1.7 | 0.88 | 0.96 | 0.96 | |
| ODO1_RAT | Ogdh | 2-oxoglutarate dehydrogenase, mitochondrial | 12.5 ± 6.1 | 18.7 ± 4.3 | 11.6 ± 2.8 | 9.5 ± 2.2 | 0.27 | 0.85 | 0.52 | |
| SUCA_RAT | Suclg1 | Succinyl-CoA ligase [ADP/GDP-forming] subunit alpha, mitochondrial | 4.9 ± 1.3 | 3.9 ± 1.9 | 3.8 ± 1.6 | 3.8 ± 0.3 | 0.74 | 0.71 | 0.71 | |
| DHSA_RAT | Sdha | Succinate dehydrogenase [ubiquinone] flavoprotein subunit, mitochondrial | 10.3 ± 2.4 | 10.7 ± 0.9 | 7.9 ± 0.7 | 11.0 ± 1.9 | 0.94 | 0.57 | 0.89 | |
| MDHM_RAT | Mdh2 | Malate dehydrogenase, mitochondrial # | 44.5 ± 6.4 | 40.0 ± 4.6 | 39.3 ± 8.7 | 47.8 ± 9.4 | 0.63 | 0.57 | 0.73 | |
| Malate shuttle | ||||||||||
| AATC_RAT | Got1 | Aspartate aminotransferase, cytoplasmic | ND | ND | 2.0 ± 0.3 | ND | NA | NA | NA | |
| MDHC_RAT | Mdh1 | Malate dehydrogenase, cytoplasmic | 21.8 ± 1.3 | 16.5 ± 2.4 | 22.2 ± 2.2 | 22.7 ± 2.9 | 0.40 | 0.95 | 0.89 | |
| MDHM_RAT | Mdh2 | Malate dehydrogenase, mitochondrial # | 44.0 ± 6.4 | 40.0 ± 4.6 | 39.0 ± 8.7 | 47.8 ± 9.4 | 0.63 | 0.57 | 0.73 | |
| AATM_RAT | Got2 | Aspartate aminotransferase, mitochondrial | 20.9 ± 0.8 | 26.7 ± 1.9 | 20.2 ± 1.2 | 23.7 ± 2.0 | 0.40 | 0.92 | 0.68 | |
| MAOX_RAT | Me1 | NADP-dependent malic enzyme | 5.2 ± 1.2 | 5.7 ± 3.9 | 8.4 ± 2.3 | 6.8 ± 2.6 | 0.888 | 0.383 | 0.641 | |
| Glycogenesis | ||||||||||
| PGM1_RAT | Pgm1 | Phosphoglucomutase-1 | 10.8 ± 7.0 | 13.5 ± 5.2 | 19.1 ± 7.1 | 14.8 ± 3.3 | 0.58 | 0.13 | 0.43 | |
| UGGG1_RAT Uggt1 | UDP-glucose:glycoprotein glucosyltransferase 1 | 7.8 ± 2.8 | 9.8 ± 3.7 | 7.3 ± 1.5 | 12.9 ± 6.1 | 0.65 | 0.89 | 0.26 | ||
| GYS1_RAT | Gys1 | Glycogen synthase | ND | 1.7 | ND | ND | NA | NA | NA | |
| Glycogenolysis | ||||||||||
| PYGB_RAT | Pygb | Glycogen phosphorylase, brain form (Fragment) | 39.7 ± 4.4 | 24.5 ± 8.4 | 33.3 ± 8.5 | 34.6 ± 5.1 | 0.06 | 0.45 | 0.55 | |
| Pentose phosphate pathway | ||||||||||
| G6PD_RAT | G6pdx | Glucose-6-phosphate 1-dehydrogenase | 13.9 ± 2.2 | 10.8 ± 7.0 | 18.6 ± 6.8 | 16.5 ± 3.7 | 0.53 | 0.41 | 0.65 | |
| 6PGD_RAT | Pgd | 6-phosphogluconate dehydrogenase, decarboxylating | 22.2 ± 2.1 | 20.4 ± 2.4 | 18.6 ± 4.0 | 17.1 ± 1.9 | 0.78 | 0.57 | 0.42 | |
| TALDO_RAT | Taldo1 | Transaldolase | 11.4 ± 2.4 | 8.5 ± 3.5 | 12.6 ± 2.5 | 9.7 ± 4.0 | 0.52 | 0.80 | 0.72 | |
| TKT_RAT | Tkt | Transketolase | 23.6 ± 4.8 | 26.9 ± 4.3 | 28.2 ± 3.0 | 25.7 ± 2.0 | 0.64 | 0.52 | 0.76 | |
| Fatty acid metabolism | ||||||||||
| ACLY_RAT | Acly | ATP-citrate synthase | 24.9 ± 7.8 | 21.4 ± 4.0 | 22.7 ± 1.5 | 18.0 ± 5.9 | 0.61 | 0.75 | 0.29 | |
| FAS_RAT | Fasn | Fatty acid synthase | 17.5 ± 9.1 | 28.7 ± 4.5 | 34.9 ± 13.8 | 16.4 ± 10.7 | 0.10 | 0.015 * | 0.85 | |
| VIII | Transcription | |||||||||
| SND1_RAT | Snd1 | Staphylococcal nuclease domain-containing protein 1 | 28.7 ± 7.3 | 24.6 ± 1.5 | 23.6 ± 5.2 | 15.2 ± 2.0 | 0.570 | 0.483 | 0.040 * | |
| U520_RAT | Snrnp200 | U5 small nuclear ribonucleoprotein 200 kDa helicase | 3.0 ± 1.0 | 12.5 ± 8.2 | 6.0 ± 1.9 | 5.2 ± 2.4 | 0.012 * | 0.309 | 0.436 | |
| HDAC1_RAT | Hdac1 | Histone deacetylase 1 | 3.0 ± 1.0 | 3.9 ± 1.1 | 2.9 ± 1.5 | 4.5 ± 1.1 | 0.734 | 0.985 | 0.574 | |
Classification categories of proteins are indicated in Roman numbers (I-VIII) and bold. Subclasses in the category VII are italicized
The relative abundance of each protein was assessed using label-free spectral counts, shown as mean of three (two for italicized entries) biological replicates ± SD. Single entries without SD indicate proteins that were detected in 1of 3 biological replicates from a given treatment
Bold p-values denote significant increase, while bold and underlined p-values denote significant decrease. G-test (likelihood ratio test for independence)
statistically significant at p ≤ 0.05 as compared to normoglycemic astrocytes (condition A)
Glc D-glucose; Lac L-lactate; ND not detected; NA not applicable
Currently only the temporary gene symbol assigned to the locus (LOC) is available for the trypsin pro-peptide (A0A4X1V2S2_PIG, Peptidase S1 domain-containing protein); obtained by BLAST search (https://blast.ncbi.nlm.nih.gov/Blast.cgi)
Indicates proteins listed in more than one pathway
Initially, we assessed whether our tryptic digestion is consistently carried out in preparations obtained from astrocytes subjected to different conditions/treatments. Albeit we used a modified trypsin for in-gel digestion to minimize autolytic activity to the arginine cutting sites, porcine trypsin fragments are inevitable and we used their spectral counts for this analysis. Astrocytic protein profiles in all conditions were similarly tainted with porcine trypsin fragments (Table 1), indicating that any difference in spectral counts for native (rat) proteins that we may find would be attributable to specific treatments(s) provided to astrocytes in comparison to protein levels found in normoglycemic/control astrocytes (5 mM Glc/condition A; Table 1).
We previously reported, and also routinely confirmed here (see “Materials and Methods” section), that our astrocytic cultures are of high purity, as per glial fibrillary acidic protein (GFAP) immunoreactivity in well over 99%, if not all, cells that also lack neuronal markers (e.g., [10, 34]). Indeed, using direct detection in our proteomics approach, we found our preparations devoid of neuronal contamination as per absence of neuron-specific enolase [49] and SNAP25 [10, 22, 34] (see Discussion on absence of protein detection). Similarly, purified astrocyte cultures were devoid of oligodendrocytes (olig1/2 and myelin basic protein) [50], microglia (ionized calcium binding adaptor molecule 1) [51] and NG2 (chondroitin sulfate proteoglycan 4) [52] cells as per lack of detection of their respective, parenthetically provided, cell markers. In stark contrast, we detected high and comparable levels (186.3–213.9 spectral counts, s.c., on average) of astrocytic marker GFAP [53], an intermediate cytoskeletal filament, in preparations from astrocytes of all four treatments. Astrocytic markers glutamine synthetase [54], Aldh1a1 [55], protein NDRG2 (N-myc downregulated gene 2)[56] and Rab6a [57] were similarly present in all conditions, albeit at a lower levels (9.1–13.1 s.c., 4.0–4.7 s.c., 4.1–6.2 s.c. and 5.3–7.6 s.c. on average, respectively). As our culture system yields astrocytes arrested in their developmental stage at the time of harvest from the brain [10, 58], we find expression of excitatory amino acid transporter 1 (EAAT1/GLAST), but not EAAT2/Glt-1, as previously described [59, 60]; levels of expression of EAAT1 (21.9–29.4 s.c. on average) were similar in all conditions. We also find connexin 43 as the only gap junctional protein, which is consistent with the notion that Cx43 is the sole connexin expressed in astrocytes within two postnatal weeks [61, 62]; again, with similar levels of expression across different groups. Astrocytes in all conditions similarly expressed aquaporin-4, which is particularly expressed on the astrocytic plasmalemma at the blood–brain and brain-liquor interfaces [63]. Our GFAP and EAAT1 positive astrocytes also showed expression of nestin, β-3-tubulin and vimentin, as seen in human fetal astrocytes [64]. While nestin (13.3–19.3 s.c. on average) and β-3 tubulin (76.5–97.3 s.c. on average) were expressed at similar levels in all conditions, expression of vimentin (857.3–992.8 s.c. on average), the most abundant protein overall, was increased in astrocytes utilizing lactate (2.5 mM Lac, condition C in Table 1), as compared to glucose, as the sole fuel.
Intermediary filaments of the cytoskeleton, GFAP and vimentin, represent cytoskeletal tracks for secretory vesicles and endosome trafficking in astrocytes [65], while nestin modulates secretory vesicle fusions, in particular fusion pore dynamics [66]. Interestingly, hybrid fueled astrocytes (5 mM Glc + 2.5 mM Lac, condition D in Table 1) had reduced levels of plectin, a cytolinker that interlinks intermediate filaments with microtubules and microfilaments, dysfunction of which results in neuronal tauopathy affecting organelle trafficking and reduced learning and memory in a mouse model [67]. We also observed several isoforms of the 14-3-3 protein (ε, α/β, δ/ζ, γ, θ and η; see Supplemental Table S1, UniProt ID: 1433E_RAT, 1433B_RAT, 1433Z_RAT and 1433G_RAT, 1433T_RAT, 1433T_RAT, and 1433F, respectively), a family of regulatory adaptor proteins implicated as mediators of diverse cellular functions including metabolism [68] and vesicular transport [69]. Quantities of individual isoforms were statistically similar across all conditions. Interestingly, the strongest signal we observed was for the epsilon isoform of 14-3-3 (Table 1) which has been reported to be increased in abundance in reactive astrocytes where it interacts with GFAP and vimentin [70].
We further analyzed the proteins of, or associated with, major cytoskeletal components, microtubules and actin filaments, known to play a role in vesicular trafficking in astrocytes [71, 72]. Indeed, the second most abundant protein in our astrocytes was γ-/cytoplasmic 2 actin (390.9–482.2 s.c. on average), the amount of which was significantly lower in aglycemic (0 mM Glc, condition B in Table 1) and lactate-fueled astrocytes when compared to normoglycemic/control astrocytes; this was unlike α-/cardiac 1 actin, the levels of which were unaffected by various treatments. Same treatment-dependent reduction pattern as with α-actin was seen in expression of α-actinin 4, a bundling protein thought to cross-link F-actin to a variety of intracellular structures; another isoform, α-actinin 1, was reduced only in aglycemic astrocytes.
Expression of drebrin, that links Cx43 to the actin cytoskeleton [73], was unaffected by treatments. Acting-binding proteins ezrin and radixin, but not moesin, of the ERM family proteins, which link actin cytoskeleton to the plasma membrane, have been described in GFAP-devoid peripheral astrocytic processes (PAPs), filopodia, lammelipoda and microvilli [74]. These PAPs surround synapses and can play a role in the tripartite synapse [75]. In our protein preparations, we found ezrin and moesin, but not radixin, and their levels were unaffected by the various conditions/treatments. Furthermore, small GTPase Rac 1 and its effector ROCK2, a key regulator of actin cytoskeleton, were present similarly in all conditions. Of note, the inhibition of ROCK2 causes restructuring of the astrocytic cytoskeleton [76] leading to astrocytic stellation, along with an increase in glutamate uptake via EAAT1/GLAST [60]. Expression of myosins 9 and 10 (that appear to play role in cell shape) and tropomyosin α −1, β, α-3 and α-4 chains (implicated in stabilizing actin filaments in non-muscle cells), were unaffected. However, there was significant reduction of both fibronectin and vinculin in aglycemic astrocytes, while lactate-fueled astrocytes had reduction of fibronectin, indicating that these treatments may result in decrease of cell adhesion. Expression of other cell adhesion proteins such as cadherin 2 and intercellular adhesion molecule 1, were unaffected.
Levels of acting-binding (actin-related protein 2/3 complex subunits 1A and 2) and ATP-binding (actin-related proteins 2 and 3) components of the Arp2/3 complex, which mediates actin polymerization, were unaltered by treatments of astrocytes, and so were the levels of Ca2+-independent (F-actin-capping protein, subunits α-1, α-2 and β) and Ca2+-dependent (gelsolin) actin-capping proteins, that bind to the plus/barbed end of F-actin. Similarly, levels of profillin-1, a concentration-dependent modulator of actin polymerization, were not affected by treatments. Detected septins 2, 7, 8, 9 and 11, filament-forming cytoskeletal GTPases necessary for normal organization of the actin cytoskeleton, were also unaffected by treatments. It should be noted that septin 8 can interact with vesicular SNARE synaptobrevin 2/VAMP2 [77].
Tubulins α-1A, α-1B and α-4A, which are major components of the microtubule α-chain containing a non-exchangeable GTP site, were significantly decreased in aglycemic astrocytes. However, levels of tubulins β-2B, β-4B and β-5, major components of the microtubule β-chain containing an exchangeable GTP site, were unaffected by any of the treatments. Level of microtubule-actin cross-linking factor, a protein with namesake function, was unaffected by treatments (in hybrid fueled astrocytes, however, it was only detected in one preparation prohibiting statistical testing), as were levels of microtubule-associated protein 1A, involved in cross-bridging microtubules and other skeletal elements, and of α-centractin, which is associated with the centrosome and plays a role in microtubule-based vesicle motility.
Movement, i.e. trafficking, of secretory organelle/vesicle along cytoskeletal tracks (made of actin, intermediary filaments and microtubules as discussed above) is powered by a variety of directional motors. Indeed, we have detected the presence of unconventional myosin-Ic, an actin-based motor, the level of which was reduced when astrocytes were solely fueled by lactate; the levels of unconventional myosin-Id and -Ie were unaltered, however (with the caveat that in lactate fueled astrocytes, myosin-Id was only detected in one preparation ruling out statistical testing). In hybrid-fueled astrocytes, we found significantly reduced levels of cytoplasmic dynein 1 heavy chain 1, which serves as a motor for the retrograde motility of vesicles and organelles along microtubules. Levels of cytoplasmic dynein 1 intermediate (in lactate fueled astrocytes, it was detected in one preparation, thus preventing statistical testing) and light intermediate chains type 2 were unaffected by various treatments. We also found no change in levels of kinesin-1 (heavy chain and light chain 1), microtubule-dependent motor required for normal distribution of mitochondria and lysosomes, and of Rab7a, a key regulator of endosomal-lysosomal trafficking. Similarly, levels of Rab31, which is necessary for mannose 6-phosphate receptors trafficking from the trans-Golgi network to endosomes in oligodendrocytes, were unaffected by the treatments [78].
Astrocyte release a variety of gliotransmitters, which fall into two main categories, small molecules, like amino acids and ATP, loaded locally by endomembrane transporters, and peptides which enter vesicles via the synthetic secretory pathway [2]. The latter pathway is also utilized to traffic plasma membrane proteins made in the ER and transiting Golgi compartments where they are sorted into organelles. At any juncture there is also retrograde transport/recycling of secretory organelles/vesicles. As already motioned, levels of pan-astrocytic marker Rab6a, a regulator of retrograde transport from the Golgi apparatus to the ER, was unaltered by the treatments, so were the levels of SEC22b involved in bidirectional ER-Golgi traffic and of general vesicular transport factor p115 required for intercisternal transport in the Golgi stack, for transcytotic fusion and/or subsequent binding of the vesicles to the target membrane. Similarly, levels of Rab11b that plays a role in endocytic recycling and of endosomal t-SNARE syntaxin 12 were unaffected by the treatments. The presence of the latter two endosomal SNARE proteins is well aligned with findings of relatively large, ~ 300 nm in diameter, synaptobrevin 2/VAMP2-laden secretory vesicles in astrocytes [34, 79]. Indeed, we detected by the proteomics assay secretory vesicle SNARE proteins synaptobrevin 2/VAMP2 and cellubrevin/VAMP3, which were some of the first v-SNAREs described in astrocytes [22], and appear to be functionally interchangeable in Ca2+-dependent exocytosis [80-82]. There was a lack of detection of synaptobrevin 2 in astrocyte fueled by lactate alone, a finding that should be cautiously interpreted as variable expression of this SNARE did not grant us statistical analysis. However, this variability piqued an assessment of the detection limit for our protein analysis method (see Discussion). Levels of cellubrevin were statistically unaffected by any of the treatments. We detected VAMP7, the vesicular SNARE which mediates secretory lysosome exocytosis of ATP and cathepsin B from astrocytes [83], only in aglycemic astrocytes; lysosome-associated membrane glycoprotein 1 (LAMP1), a late-endosomal/lysosomal marker, was similarly present in all the conditions. We found the levels of brain isoform of non-catalytic subunit B of the peripheral V1 complex of vacuolar-type proton ATPase (V-ATPase) unaffected by treatments, implying that acidification of secretory organelles/vesicles necessary for uptake of amino acid transmitters, including glutamate, into the vesicular lumen was also unaffected. We have not detected presence of vesicular glutamate transporters 1, 2 and 3 in any preparation, likely due to their low expression in these glial cells as we originally reported [10]. Expression of secernin-1, which increases extent of Ca2+-dependent secretion from peritoneal mast cells [84], was unaffected by treatments, so was the level of α-soluble NSF attachment protein, an adaptor protein that sustains membrane trafficking by disassembling SNARE complexes that form during membrane fusion [85]. In stark contrast to caveolin-1 expression, reporting on receptor-independent endocytosis, which was unaffected by the treatments, expression of clathrin heavy chain 1, reporting on receptor-dependent endocytosis, was statistically increased in aglycemic astrocytes as well as lactate- and hybrid-fueled astrocytes when compared to normoglycemic astrocytes. Linking clathrin scaffold to the vesicular membrane and mediating vesicle endocytosis from the plasmalemma requires adaptor protein complex 2 (AP2) [86, 87], the subunit μ of which was detected in our preparations and was unaffected by the treatments. Similarly unaffected were levels of dynamin-2, a large GTPase that plays a role in constitutive endocytosis [88]. However, there was reduction of annexin A1 expression in aglycemic astrocytes. This protein inhibits vesicular release of adrenocorticotropin from a neuroendocrine cell line through ROCK activation and downstream augmentation of actin polymerization, likely resulting in a filamentous actin mesh which serves a barrier for vesicular access to the plasma membrane [89]. Other annexins detected (A2, A4, and A6) in our preparations, that play role in exocytosis [90-92], were unaffected.
As exocytotic release of glutamate is a Ca2+-dependent process, we appraised the protein components known to support Ca2+ flux and transport across the plasmalemma and endomembranes of astrocytes (reviewed in e.g., [93, 94]). In all the conditions, we detected similar levels of the ER Ca2+ ATPase of the SERCA type 2, which is responsible for (re) filling the ER store with Ca2+. Expression of inositol 1,4,5 tris–phosphate (IP3R) and ryanodine (Ryr2) receptors, both sourcing Ca2+ for triggering astrocytic glutamate release [35], was variable and prohibited statistical testing. Ryr2 was only detected in the normoglycemic condition, while IP3R in aglycemic and lactate-fueled astrocytes.
Ca2+ necessary for the store replenishment for vesicular glutamate release is ultimately drawn from the extracellular Ca2+ via the store-operated Ca2+ entry [36, 58] and Na+/Ca2+ exchanger [95] at the plasma membrane of astrocytes. At the molecular level the store-operated plasmalemmal channels may involve formation of Orai/Stim1 protein complexes and/or some types of Transient Receptor Potential (TRP) proteins, in particular TRPC1 protein containing channels [93, 94]. We have not detected the presence of TRPC1 and Orai in any preparations/conditions, while Na+/Ca2+ exchanger 1 and STIM1 were only detected in a single preparation obtained from aglycemic astrocytes. Similarly, we have not detected the presence TRPA1 in any of the conditions/preparations, the channel activity of which contributed to setting the resting [Ca2+]i in hippocampal astrocytes. [96]. The release of Ca2+ from the ER in astrocytes occurs within structural sub-plasmalemmal microdomains correlated to vesicular fusion events [97]. The extended synaptotagmin 1 (Esyt-1) is an ER protein that functions as a Ca2+-dependent tether between the ER and the plasmalemma [98]. We found Esyt-1 expression at similar levels in all the conditions, implicating that the ER-plasmalemmal tethering is unaffected by glucose and/or lactate, or absence thereof.
Another organelle, the mitochondrion, participates in buffering cytosolic Ca2+ transients, which drive vesicular glutamate release [27]. At high cytosolic levels, mitochondrial Ca2+ uptake is mediated by the Ca2+ uniporter complex, containing voltage-dependent anion-selective channel protein (VDAC), that has considerable Ca2+ permeability, on the outer mitochondrial membrane and the highly selective mitochondrial Ca2+ uniporter complex (composed of mitochondrial Ca2+ uniporter MCU1/2, mitochondrial EF hand Ca2+ uniporter regulator MICU1/2, essential MCU regulator EMRE, and mitochondrial Ca2+ uniporter regulator MCUR1) on the inner membrane; see reviewed in [99]. While we detected VDAC 1, 2 and 3 at similar levels in all preparations, respectively, we have not detected any of the inner membrane components of this complex. At low cytosolic levels, free Ca2+ exits the mitochondrial matrix through the mitochondrial Na+/Ca2+ exchanger (NCLX), and via the formation of the mitochondrial permeability transition pore (MPTP) [27, 100]. We have detected the presence of NCLX only in a single preparation from aglycemic astrocytes. The molecular identity of MPTP remains elusive, however; mitochondrial phosphate carrier protein (SLC25a3) has been put forward as the main candidate for the transmembrane pore component, while adenine nucleotide translocase (SLC25a5) likely serves a regulatory role of MPTP [28, 101, 102]. These proteins were similarly present in astrocytes in all conditions.
There are structural–functional interactions between the ER and mitochondria [103]. For instance, the above reported IP3Rs of the ER and VDACs of the outer mitochondrial membrane interact to form a functional ER-mitochondria Ca2+ conduit [104]. Similar to variable expression of IP3Rs, we also detected variable expression (which did not allow statistical testing) of several other ER-mitochondria linkers, e.g., ATPase family AAA domain-containing protein 3, mitochondrial fission 1 protein and vesicle-associated membrane protein-associated protein B (see Supplementary Table S1).
We next analyzed metabolic pathways of glucose, lactate and glutamate. We detected plasmalemmal transporters for both metabolic fuels, glucose (solute carrier family 2, facilitated glucose transporter member 1, GLUT1) and lactate (monocarboxylate transporter 1), along with plasmalemmal glutamate transporter EAAT1/GLAST, the latter already disclosed above. The expression of these transporters, respectively, was statistically indiscriminate in all the treatments. The sole exception was GLUT1, the expression of which in aglycemic astrocytes was detected in one preparation and could not be statistically scrutinized (Table 1). We did not find expression of plasmalemmal lactate-responsive G-protein-coupled receptor (GPCR), i.e., hydroxycarboxylic acid receptor 1 (HCAR1) formerly known as GPR81, detected in the mouse cortex and expressed in astrocytes [105]. Similarly, we have not found expression of OR51E2, lactate-activated GPCR, and a proton-sensitive GPCR GPR4, to which lactate seems to be a negative allosteric modulator; based on RNA sequencing, GPR4 but not OR51E2 was found in astrocytes of the rat locus coeruleus [106]. As we reported above, glutamine synthetase was detected at similar levels in all conditions, so was mitochondrial glutamate dehydrogenase 1.
In agreement with previous reports [107, 108], we detected other enzymes crucial for metabolism of glucose, lactate and glutamate. These enzymes, classified by their contribution to processes of glucose phosphorylation, glycolysis, pyruvate conversion, the TCA cycle, the malate shuttle, glycogenesis, glycogenolysis and the pentose phosphate pathway, were generally expressed in astrocytes at levels statistically indiscriminate across all conditions. The exceptions were glycogen synthase and cytoplasmic aspartate aminotransferase, the (lack of) detection of which did not permit statistical testing. As pertains to fatty acid metabolism, we found expression of fatty acid synthase elevated in lactate-fueled astrocytes when compared to normoglycemic astrocytes, while aglycemia or exposure to the hybrid glucose-lactate fuel has not affected the expression of this enzyme in astrocytes.
Two proteins classified into the transcription/transcriptional modification pathway had significant changes. Staphylococcal nuclease domain-containing protein 1, an endonuclease that mediates microRNA decay and regulates mRNAs involved in G1-to-S phase transition, has been described elevated in several cancers and malignant astrocytes, i.e., glioma/astrocytoma cells [109]. The levels of this nuclease was reduced in astrocytes using the hybrid glucose-lactate fuel. U5 small nuclear ribonucleoprotein 200 kDa helicase as a key component of spliceosomes complexes plays a dual role in pre-mRNA splicing and cell cycle regulation, the later by promoting exit from the G2/M phase and advancing S phase [110]. The levels of this helicase was increased in aglycemic astrocytes. As lactate can be a transcriptional regulator linking cell metabolism and gene transcription through its action as an endogenous weak inhibitor of histone deacetylases (HDACs) [111], we assessed the presence of these enzymes, and found expression of HDAC1 at similar levels in all the conditions.
Taken together, the proteomics data add new dimension for interpretation of the Ca2+ and glutamate imaging data. Our working hypothesis prior to proteomics was that lactate does not readily serve as a precursor for de novo glutamate synthesis in astrocytes by “shunting” the TCA cycle prior to the production of α-ketoglutarate, presumably at level of citrate; this seems to be supported by proteomics data showing increased levels of fatty acid synthase in lactate-fueled astrocytes. However, this was not the case in the data set from astrocytes exposed to the hybrid glucose-lactate fuel. It appears that changes underlying altered exocytotic glutamate release from astrocytes utilizing different energy sources for their metabolism are much more complex than the sole change in metabolism. Indeed, proteomics brings to the limelight the disbalance between endocytosis and exocytosis as the common, but partial, explanation for the reduced amount of glutamate release in all treatments (aglycemia, lactate or glucose-lactate exposure of astrocytes) when compared to the control/normoglycemic condition; it is likely that is caused by increased clathrin-mediated endocytosis. Proteomics analysis paints a picture of condition-dependent changes in protein profiles encompassing proteins of the cytoskeleton, secretory organelle/vesicle traffic and recycling at the plasma membrane. These findings may have translational impact as, for example, glucose and lactate consumptions vary during the sleep–wake cycle related to lactate clearance via the glymphatic system [112].
Discussion
Glutamate has emerged as a versatile molecule used in the brain cellular metabolism and intercellular, both homotypic and heterotypic, communication. Its metabolic and signaling pathways are interlaced, which is the subject of our present work. Experimental set-ups to study metabolic regulation of signaling in the brain face many obstacles, as glutamate is abundant and omnipresent in all the cell types. A cell culture system can provide an opportunity to study this process in an experimentally well-controlled environment. Thus, we used purified cultures of astrocytes to observe astrocytes independently from other brain cells, and we focused on metabolic regulation of exocytotic glutamate release. To that end, cells were acutely (1 h) offered metabolic substrates, glucose and/or lactate, in the bath solution to drive de novo synthesis of glutamate in astrocytes. We recorded mechanically-induced exocytotic glutamate release from individual astrocytes, while, in parallel experiments, the same stimulus induced Ca2+ elevations in astrocytes. Our data imply a major role of glucose contribution in generating vesicular glutamate release, while the presence of lactate revealed a significant reduction in the release of glutamate. This unexpected discovery suggested that astrocytes utilizing extracellular lactate might undergo a metabolic shift and/ or changes in secretory organelle/vesicle trafficking and/or recycling. The complexity of proteins governing these pathways required us to use tandem mass spectrometry with an in-depth protein expression analysis. We showed specific changes in protein profiles in aglycemic astrocyte as well as those fueld by lactate alone or by the hybrid glucose-lactate fuel when compared to normoglycemic astrocytes.
As stated above, we used purified astrocytes in culture, which offered us spatiotemporal control of stimulation for cytosolic Ca2+ and extracellular glutamate imaging, and ensured cell-specify for biochemical, mass spectrometry analysis. These astrocytes appear as flat polygonal cells having less complex processes/morphological appearance than astrocytes in situ [10, 35, 58] (also see Figs. 2 and 3). Our culture system yields GFAP-positive astrocytes arrested in their developmental stage at the time of harvest from the brain [10, 58] as witnessed, in the present work (Table 1), by expression of connexin 43 (but not other connexins) and EAAT1 (but not EAAT2). Here, we further characterized these astrocytes showing that they express other astrocytic markers, glutamine synthetase, Aldh1a1, NDRG2 and Rab6a. As seen in human fetal astrocytes [64], our astrocytes also express nestin, β-III-tubulin and vimentin. The expression of β-III-tubulin has been generally used as a neuron-specific marker in developmental neurobiology and stem cell research. However, in our own previous work we used immunocytochemistry to report on abundant β-III-tubulin stains of cultured hippocampal neurons as well on a faint fluorescence emission from nearby astrocytes [113]. As we routinely check culture purity by the absence of neuronal marker SNAP25 by RT-PCR, and by antibodies used in immunocytochemistry and Western blotting [10, 34], we side with conclusion reached elsewhere [64], that β-III-tubulin cannot be considered neuron-specific label in all conditions.
Mechanical stimulation offers direct stimulation of astrocytes without receptor activation and mimics the action of endogenous ligands [114]. Visually, deformation of mechanically stimulated astrocytes appears to be far less than deformation seen in astrocytes at the interface with blood vessel that undergo diameter change [115, 116]. Thus, this stimulus may have physiological relevance, which we, ourselves, previously disregarded.
There are over 24000 different protein-coding genes in rat [117], although it is likely that individual cells express only a subset of such protein-coding genes at a given time. For example, the results of comprehensive, MS-based analyses suggest that cultured human cells express ~ 10000 proteins [118] or ~ 50% of the protein-coding genes in the human genome. In the present work, our proteomics profiling study identified 1319 proteins, and, thus, we estimate that we sampled ~ 10% of the proteome from our astrocyte cultures. This limitation was expected as our proteomics workflow was primarily intended to characterize enzymes involved in metabolism, which are generally among the most abundant—and, thus, easiest to detect—proteins present in a cell. Nonetheless, given that the sensitivity of our MS-based workflow precluded deeper sampling of the astrocyte proteome, we attempt here to provide an estimate of this limit. We deemed synaptobrevin 2 levels at the cusp of detection as we reported the dichotomous finding of its presence in some and absence in other samples. We estimate that: (a) an individual astrocyte likely contains around 350 secretory vesicles (probably an underestimate as 23 astrocytes contained 8042 synaptobrevin 2-laden vesicles within the sub-plasmalemmal space where cells adhered to coverslips, but not the entire cells [34]); and (b) there are ~ 25 synaptobrevin 2 molecules per astrocytic secretory vesicle [79]. Thus, protein detection limit in our proteomics approach is at ~ 8750 synaptobrevin 2 molecules per individual astrocyte. Therefore, the sensitivity of our proteomics approach cannot allow us to detect, for example, vesicular glutamate transporters (VGLUTs) that we and others readily detected in a variety of astrocytes and conditions (in vitro, freshly-isolated and in vivo) using immunocytochemistry and Western blots (reviewed in [4]), albeit at levels ~ 2–3 orders of magnitude lesser than in neurons [10]. This is likely due to much higher number of vesicles present in neurons, estimated at 70000–188000 (reviewed in [119]), and higher density of vesicular proteins in neuronal vesicles. For example, each neuronal secretory vesicle contains 70 molecules of synaptobrevin-2, about 3-times higher than that in astrocytic vesicle [120]. Consequently, it should come as no surprise that we did not detect the presence of VGLUTs in astrocytes using proteomics. The focus, however, should be on the differences we observed, and some caution is warranted when interpreting the absence of proteins.
Further, aside from the impact of protein abundance on the limit of detection, it is also recognized that certain proteins may be more difficult to detect by certain MS approaches based on other properties, such as the length and composition of their amino acid sequence. These properties may work alone or in concert to make detection by MS more difficult. First, the sequences of some proteins may not contain a suitable number or location of tryptic cleavage sites. This property can decrease the total number of peptides available for MS detection and/or result in the generation of short or long peptides that are more difficult to detect [121]. Second, the capability to detect peptides from membrane proteins using standard MS-based workflows has proved challenging because these proteins exhibit different inherent properties as compared to soluble proteins. For example, in addition to being relatively low in abundance, membrane proteins may be more difficult to detect using standard workflows because of their limited solubility and, relatedly, their constituent hydrophobic, transmembrane domains which can restrict protease accessibility and also result in a lack of suitable tryptic cleavage sites [122]. Thus, we suspect that our failure to detect certain proteins in the current proteomic study most likely results from a combination of factors, including their relatively low abundance and their inherent properties derived from their amino acid sequence.
We reported the presence of porcine trypsin fragments in all of our protein extracts (Table 1). As our culture procedure used for obtaining protein extracts did not utilized trypsin to detach cells, the source of tryptic fragments must come from autolysis of a modified trypsin intended to minimize autolysis. The amount of fragments was used for statistical analysis indicating that we similarly treated protein extracts obtained in various conditions. In terms of further assessing the activity of trypsin for quality control purposes, a typical approach is to look at missed cleavages, i.e., the number of peptides generated that have a trypsin cleavage site that was not cut by the enzyme. The samples used in this study all looked favorable in this respect, both in terms of absolute trypsin activity as well as consistency across the samples.
We ran additional searches for external/non-rat proteins in our preparations and found two contaminant protein groups, one sourced from fetal bovine serum used in our cell culture and the other sourced from human skin (Supplementary Table S2, also see raw data in Supplementary Table S1). These are expected contaminates for the proteomic approach used; the fragments/peptides of these contaminates are easily discernable from rat proteins and therefore do not obstruct our analysis.
Our data using aglycemia should be of interest for management of De Vivo disease, a rare autosomal dominant, genetic metabolic disorder associated with a deficiency of GLUT1. Astrocyte with their gliotransmission and metabolic-signaling networks may as well contribute to the phenotypic spectrum of this disease, in particular to seizures [123-125].
Astrocytes exhibit higher glycolytic rate when compared to neurons [126], but they also have a rather active TCA cycle. The critical finding that astrocytes, in contrast to neurons, express pyruvate carboxylase [14, 15] meant that astrocytes can have a dual entry of pyruvate into the TCA cycle granting them capability to synthesize de novo glutamate as the sole cell type in the brain. Both glucose and lactate can provide for generation of pyruvate and contribute to de novo glutamate synthesis [18]. However, our data implicates that glucose, external or internally recruited from glycogen (Fig. 2) provide much better for exocytotic glutamate release from astrocytes than lactate (Fig. 4). Additionally, lactate hampers glucose ability to provide for exocytotic glutamate release from astrocytes. Thus, it appears that there might be separate pools of pyruvate generated from various metabolic fuels and utilized in discriminate manner for glutamate release. This is an area of metabolomics that needs to be studied, especially that release of glutamate and/or lactate from astrocytes plays a role in modulation of synaptic transmission [75] and plasticity [20, 127] and in learning and memory [20, 75].
Brain lactate concentration varies during the sleep–wake cycle and is further increased during sleep deprivation [105]. Its parenchymal concentration is inversely correlated to its clearance via the glymphatic system, the operation of which critically depends on astrocytes and their aquaporin 4 [105]. Whether changes we observed in astrocytes utilizing lactate as fuel may undergo cyclical fluctuations, it represents another issue that should be studied in future.
Our finding that lactate hampers glutamate release could be at display: (a) in cases of lacticemia (and presumably increased lactate in the brain), which occurs during muscle damage/exercise [128] or can be associated with an acute overdose of metformin mainly in adults and as an adverse effect of this drug in diabetic patients with comorbidities [129]; or (b) in the perinatal brain development, when lactate is used as an energy source [130]. This finding may also be relevant in the context of metabolic interventions such as ketogenic diets and fasting which are reported to result in elevated concentrations of both brain lactate and ketone bodies (e.g., β-hydroxybutyrate)[131]. While it has been generally accepted that the liver, hepatocytes in particular, supplies/supply other organs with ketone bodies, it appears that astrocytes are also ketogenic cells [132]. Interestingly, the ketone body beta-hydroxybutyrate has also been demonstrated to inhibit glucose consumption in astrocytes [133]. The contribution of gliotransmission to these events, however, is presently unknown and warrants further investigation. Furthermore, a number of pathological conditions are associated with increased lactate in the brain. Lactate provides protection during ischemia [134-136]. Perhaps this might be by hampering de novo synthesis of glutamate, which could be a mechanism to prevent excitotoxicity. Astrocytes and their glutamate and energy metabolism are involved in epileptiform activity [137, 138]. Interestingly, blockade of LDH in neurons and astrocytes [139] has been proposed for treatment of epilepsy. Thus, the present work may provide a fertile ground for management of seizures. In addition, lactate is glycolytically produced (so called the Warburg effect) and released in large quantities by primary brain tumors [140, 141]; whether this lactate can affect nearby healthy astrocytes to affect gliotransmission is yet to be determined.
The finding that astrocytes fueled by lactate alone have increased level of fatty acid synthase has several implications. Expected augmented fatty acid synthesis would result in rapid formation of abundant lipids droplets (LDs), dynamic organelles that not only store/liberate lipids, but also contribute to membrane trafficking and cellular/lipid signaling [142]. These lipid- enriched organelles also contain a variety of proteins [143-145]. This protein repertoire includes: (i) small GTPases Rab18 and ADP-ribosylation factor 1, which warrant trafficking of LDs and are detected in our proteomics approach (former with a variable expression not allowing for statistical testing, while the latter at statistically similar levels in all conditions tested; see Supplementary Table S1); (ii) as well as caveolins, which are predominately associated with caveolae [146], but also bind to fatty acids in LDs [147, 148]. Thus, we suspect that lactate fueled astrocytes (which presumably accumulate fatty acids in LDs, while displaying similar levels of caveolin-1 as astrocytes in other conditions tested) would showed differential partitioning of caveolin-1 in LDs as compared to aglycemic, normoglycemic or hybrid-fueled astrocytes. This LD partitioning likely is not a simple redistribution of caveolin-1 into formed LDs, but also involves trafficking at the plasma membrane [147]. As caveolin-1 can bind to cholesterol and they can together get internalized from the plasma membrane, the associated cell-surface signaling via cholesterol-rich lipid-raft domains would likely be affected as well (reviewed in [142]). Through their role in the lipid synthesis, metabolism and signaling, LDs are closely associated with inflammatory responses, diabetes mellitus, obesity, atherosclerosis and cancer [149-151]. Moreover, microbial pathogens such as Mycobacterium tuberculosis, Mycobacterium laepre, the hepatitis C virus and the dengue virus all use LD components to advance their survival proliferation [152-154]. Whether glutamatergic gliotransmission plays a role in manifestations of these conditions/diseases in the brain remains to be elucidated. Finally, the increased cytosolic fatty acid synthesis in lactate-fueled astrocytes could be advantageous in stress when cells use fatty acids as an energetic substrate, a strategy successfully implemented by glioma cells for their survival [155]. Fatty acid/beta-oxidation in mitochondria requires the activity of carnitine palmitoylotransferases 1 and 2, both found at similar levels in all the conditions we tested (Supplementary Table S1).
L-lactate, long considered a waste product of glucose degradation, is emerging as an important metabolite in brain activity, not only as an energy substrate, but also as a signaling molecule (reviewed in [156]), referred to as “lactormone” [128]. Its signaling can occur through HCAR1 [105], expressed in cortical astrocytes, and the activation of which inhibits adenylyl-cyclase (AC) resulting in decreased intercellular 3′-5′-cyclic adenosine monophosphate (cAMP) levels. In our culture system, this second messenger causes maturation of astrocytes seen as an increase of GFAP levels and also as change in morphology, i.e. stellation [157]. In the present work, we have not detected the presence of this receptor (and other lactate receptors, OR51E2 and GPR4, which could be under our proteomics detection limit, however, as discussed above), but more importantly we have not detected changes in GFAP levels in lactate or hybrid-fueled astrocytes. The caveat here is that previous cAMP-dependent maturation/stellation changes occurred during the time course of 3 days, while a possible opposing effect (rounder cells with a decrease in GFAP associated with the dedifferentiation of astrocytes [158]) here would be of an acute nature (within 1 h). Thus, it is still possible, if our astrocyte express HCAR1, that some of the effects we have seen might be related to activity of this receptor; concentration of lactate used here (2.5 mM) would cause partial activation of HCAR1, given its half maximal effective concentration (EC50) for lactate at 4.2 mM in cortical neurons [159], Shall our astrocyte express GPR4, lactate effects should be similar to those in above scenario with HCAR (rounder cells), as lactate is a negative allosteric modulator of GPR4-mediated cAMP production [106]. If, however, our astrocytes would express OR51E2, which promotes AC resulting in increased intracellular cAMP [106], than the effect would be maturation/stellation of astrocytes. Interestingly, via the activation of a non-HCAR1 receptor, perhaps OR51E2, extracellular lactate caused an increase in cytosolic cAMP, which led to the enhancement of aerobic glycolysis in cultured astrocytes [160]. This signaling pathway was augmented in astrocytes lacking αGDI, a protein controlling the cycling of small GTPases [161]. Mutations of the human αGDI represent one of the many genetic alterations that cause X-linked intellectual disability (XLID) clinically characterized by cognitive deficits [162, 163]. Thus, our data is also relevant to this intellectual disability.
In our proteomic study, out of 1319 detected proteins in preparation from astrocytes, 17 proteins showed alteration, four up and 13 down regulated, in their levels in all the treatments combined when compared to normoglycemic astrocytes. The most diverse changes occurred in the cytoskeletal protein group as 10 of the above proteins were classified in this group. Remaining proteins (numbers in each group disclosed parenthetically) belonged to secretory organelle/vesicle trafficking (2), secretory organelle/vesicle recycling (2), metabolism (1) and transcription (2) groups. There was a total of 22 changes, as some proteins varied their expression upon more than one treatment. Only one protein, clathrin heavy chain 1, classified into the secretory organelle/vesicle recycling proteins category, showed ubiquitous change, i.e., increase in its level in all the three treatments when compared to the control/normoglycemic condition, indicating that likely enhanced receptor-mediated endocytosis plays a role in reduction of exocytosis glutamate release in aglycemic astrocytes and those fueled by lactate or the hybrid glucose-lactate fuel. The most diverse changes occurred in the group of cytoskeletal proteins. Aglycemia caused most changes in protein expression followed by lactate treatment, while the least changes were seen in hybrid-fueled astrocytes.
Aglycemic astrocytes showed changes in expression of 11 proteins as follows: decrease in 8 cytoskeletal proteins (cytoplasmic 2/γ-actin, α-actinin-1, α-actinin-4, fibronectin, vinculin, tubulin α-1A chain, tubulin α-1B chain, and tubulin α-4A chain), ambivalent change in secretory vesicle recycling proteins (beside increase in clathrin heavy chain 1, there was decrease in annexin A1 expression), and an increase in level of U5 small nuclear ribonucleoprotein 200 kDa helicase, a translational protein.
Lactate-fueled astrocytes show changes in expression of seven proteins. There was decrease in expression of three cytoskeletal proteins: cytoplasmic 2/γ-actin, γ-actinin-4 and fibronectin; however, there was the sole exception of an increase in level of vimentin. These lactate-fueled astrocytes also showed decrease in unconventional myosin-Ic (a secretory vesicle trafficking protein), and increases in already mentioned clathrin heavy chain 1 (a secretory vesicle recycling protein), as well as in fatty acid synthase (a metabolic protein).
Lactate- and glucose-, i.e., hybrid-fueled astrocytes showed changes in four proteins. Besides ubiquitous increase in clathrin heavy chain 1, there was decrease in expression of plectin (a cytoskeletal protein), cytoplasmic dynein 1 heavy chain 1 (a secretory vesicle trafficking) and staphylococcal nuclease domain-containing protein 1 (a transcription protein).
Overall, acute changes in metabolic fuel or lack thereof can lead to condition-dependent protein profile changes with the common denominator being altered levels of cytoskeletal proteins and those of secretory vesicles trafficking and recycling.
We found changes in levels of two proteins that can affect transcription; staphylococcal nuclease domain-containing protein was reduced in astrocytes using the hybrid glucose-lactate fuel, while U5 small nuclear ribonucleoprotein 200 kDa helicase was increased in aglycemic astrocytes. Other detected proteins in that group were rather unaffected (for example UniProt ID: SYDC_RAT, SYVC_RAT, SYSC_RAT, SYYC_RAT, SYTC_RAT, SYFA_RAT, SYG_RAT, SYAC_RAT, SYRC_RAT and RTCB_RAT; as disclosed in Supplementary Table S1). Also, we detected HDAC1 in all preparations, the activity of which lactate inhibits to promote expression of HDAC-associated genes; however, lactate EC50 of 40 mM in this action [111] is not within the realm of physiological event. Thus, it is more likely that significant changes in protein levels are rather a result of changes in the rate of protein degradation/turnover. Indeed, we previously described significant GFAP level dynamics in astrocyte residing in the supraoptic nucleus at even shorter timelines, with just 5 min [164]. However, we see no changes in expression levels of detected proteasome components in our present work (for example, UniProt ID: UFL1_RAT, NEDD4_RAT, UBE2N_RAT, PSME2_RAT, UB2V2_RAT, UCHL1_RAT, UBA1_RAT, RL40_RAT (+ 3), UCHL1_RAT, PRS6B_RAT, PSD13_RAT, PRS4_RAT, PRS6A_RAT, PSMD1_RAT, PSMD2_RAT, PRS7_RAT, PSD11_RAT and PRS8_RAT; as disclosed in Supplementary Table S1). It is still possible there could be some proteins that escaped our detection, albeit a more plausible action underlying changes in protein levels is an alteration in the activity of various degradation enzymes, Such possibility can be studied in future using a variety of functional assays available [165, 166].
In conclusion, we suggest that the availability of different energy sources and metabolic milieu play a significant role in exocytotic glutamate release from astrocytes and that changes in metabolic and protein profiles endow astrocytes with a modifiable signaling output via gliotransmission. Furthermore, our findings should be of importance in understanding the role of astrocytes in physiological (e.g., development, sleep–wake cycle, and learning and memory) and pathophysiological (e.g., metabolic disorders, ischemia, epilepsy and primary brain tumors) conditions.
Data Availability
At any request, we will freely provide the original data sets.
Supplementary Material
Acknowledgements
We would like to thank the UAB Targeted Metabolomics and Proteomics Laboratory Core facility for providing mass spectrometry services. VP is an Honorary Professor at University of Rijeka, Croatia.
Funding
This work is supported by a Grant from the National Institute of General Medical Sciences of the National Institutes of Health (R01GM123971 to VP).
Footnotes
Conflict of interest The authors declare no conflict and competing financial interest.
Supplementary Information The online version contains supplementary material available at https://doi.org/10.1007/s11064-021-03340-y.
Ethical Approval All procedures involving animals were approved by The University of Alabama at Birmingham Institutional Animal Care and Use Committee.
Consent for Publication All the authors agree to publish this work in Neurochemical Research.
References
- 1.Verkhratsky A, Schousboe A, Parpura V (2014) Glutamate and ATP: the crossroads of signaling and metabolism in the brain. Adv Neurobiol 11:1–12 [DOI] [PubMed] [Google Scholar]
- 2.Parpura V, Zorec R (2010) Gliotransmission: exocytotic release from astrocytes. Brain Res Rev 63:83–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zorec R, Araque A, Carmignoto G, Haydon PG, Verkhratsky A, Parpura V (2012) Astroglial excitability and gliotransmission: an appraisal of Ca2+ as a signalling route. ASN Neuro. 10.1042/AN20110061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Montana V, Malarkey EB, Verderio C, Matteoli M, Parpura V (2006) Vesicular transmitter release from astrocytes. Glia 54:700–715 [DOI] [PubMed] [Google Scholar]
- 5.Danbolt NC (2001) Glutamate uptake. Prog Neurobiol 65:1–105 [DOI] [PubMed] [Google Scholar]
- 6.Martinez-Hernandez A, Bell KP, Norenberg MD (1977) Glutamine synthetase: glial localization in brain. Science 195:1356–1358 [DOI] [PubMed] [Google Scholar]
- 7.McKenna MC (2013) Glutamate pays its own way in astrocytes. Front Endocrinol 4:191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kreft M, Bak LK, Waagepetersen HS, Schousboe A (2012) Aspects of astrocyte energy metabolism, amino acid neurotransmitter homoeostasis and metabolic compartmentation. ASN neuro 4:AN20120007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Obel LF, Andersen KM, Bak LK, Schousboe A, Waagepetersen HS (2012) Effects of adrenergic agents on intracellular Ca2+ homeostasis and metabolism of glucose in astrocytes with an emphasis on pyruvate carboxylation, oxidative decarboxylation and recycling: implications for glutamate neurotransmission and excitotoxicity. Neurotox Res 21:405–417 [DOI] [PubMed] [Google Scholar]
- 10.Montana V, Ni Y, Sunjara V, Hua X, Parpura V (2004) Vesicular glutamate transporter-dependent glutamate release from astrocytes. J Neurosci 24:2633–2642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang Q, Pangrsic T, Kreft M, Krzan M, Li N, Sul JY, Halassa M, Van Bockstaele E, Zorec R, Haydon PG (2004) Fusion-related release of glutamate from astrocytes. J Biol Chem 279:12724–12733 [DOI] [PubMed] [Google Scholar]
- 12.Bezzi P, Gundersen V, Galbete JL, Seifert G, Steinhauser C, Pilati E, Volterra A (2004) Astrocytes contain a vesicular compartment that is competent for regulated exocytosis of glutamate. Nat Neurosci 7:613–620 [DOI] [PubMed] [Google Scholar]
- 13.Ni Y, Parpura V (2009) Dual regulation of Ca2+-dependent glutamate release from astrocytes: vesicular glutamate transporters and cytosolic glutamate levels. Glia 57:1296–1305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yu AC, Drejer J, Hertz L, Schousboe A (1983) Pyruvate carboxylase activity in primary cultures of astrocytes and neurons. J Neurochem 41:1484–1487 [DOI] [PubMed] [Google Scholar]
- 15.Shank RP, Bennett GS, Freytag SO, Campbell GL (1985) Pyruvate carboxylase: an astrocyte-specific enzyme implicated in the replenishment of amino acid neurotransmitter pools. Brain Res 329:364–367 [DOI] [PubMed] [Google Scholar]
- 16.Westergaard N, Drejer J, Schousboe A, Sonnewald U (1996) Evaluation of the importance of transamination versus deamination in astrocytic metabolism of [U-13C]glutamate. Glia 17:160–168 [DOI] [PubMed] [Google Scholar]
- 17.Westergaard N, Varming T, Peng L, Sonnewald U, Hertz L, Schousboe A (1993) Uptake, release, and metabolism of alanine in neurons and astrocytes in primary cultures. J Neurosci Res 35:540–545 [DOI] [PubMed] [Google Scholar]
- 18.Waagepetersen HS, Bakken IJ, Larsson OM, Sonnewald U, Schousboe A (1998) Metabolism of lactate in cultured GABAergic neurons studied by 13C nuclear magnetic resonance spectroscopy. J Cereb Blood Flow Metab 18:109–117 [DOI] [PubMed] [Google Scholar]
- 19.Rouach N, Koulakoff A, Abudara V, Willecke K, Giaume C (2008) Astroglial metabolic networks sustain hippocampal synaptic transmission. Science 322:1551–1555 [DOI] [PubMed] [Google Scholar]
- 20.Suzuki A, Stern SA, Bozdagi O, Huntley GW, Walker RH, Magistretti PJ, Alberini CM (2011) Astrocyte-neuron lactate transport is required for long-term memory formation. Cell 144:810–823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Drummond GB (2009) Reporting ethical matters in the Journal of Physiology: standards and advice. J Physiol 587:713–719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Parpura V, Fang Y, Basarsky T, Jahn R, Haydon PG (1995) Expression of synaptobrevin II, cellubrevin and syntaxin but not SNAP-25 in cultured astrocytes. FEBS Lett 377:489–492 [DOI] [PubMed] [Google Scholar]
- 23.McCarthy KD, de Vellis J (1980) Preparation of separate astroglial and oligodendroglial cell cultures from rat cerebral tissue. J Cell Biol 85:890–902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Walls AB, Sickmann HM, Brown A, Bouman SD, Ransom B, Schousboe A, Waagepetersen HS (2008) Characterization of 1,4-dideoxy-1,4-imino-d-arabinitol (DAB) as an inhibitor of brain glycogen shunt activity. J Neurochem 105:1462–1470 [DOI] [PubMed] [Google Scholar]
- 25.Parpura V, Basarsky TA, Liu F, Jeftinija K, Jeftinija S, Haydon PG (1994) Glutamate-mediated astrocyte-neuron signalling. Nature 369:744–747 [DOI] [PubMed] [Google Scholar]
- 26.Innocenti B, Parpura V, Haydon PG (2000) Imaging extracellular waves of glutamate during calcium signaling in cultured astrocytes. J Neurosci 20:1800–1808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Reyes RC, Parpura V (2008) Mitochondria modulate Ca2+-dependent glutamate release from rat cortical astrocytes. J Neurosci 28:9682–9691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Reyes RC, Perry G, Lesort M, Parpura V (2011) Immunophilin deficiency augments Ca2+-dependent glutamate release from mouse cortical astrocytes. Cell Calcium 49:23–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chance B, Schoener B, Oshino R, Itshak F, Nakase Y (1979) Oxidation-reduction ratio studies of mitochondria in freeze-trapped samples. NADH and flavoprotein fluorescence signals. J Biol Chem 254:4764–4771 [PubMed] [Google Scholar]
- 30.Patterson GH, Knobel SM, Arkhammar P, Thastrup O, Piston DW (2000) Separation of the glucose-stimulated cytoplasmic and mitochondrial NAD(P)H responses in pancreatic islet beta cells. Proc Natl Acad Sci USA 97:5203–5207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Blacker TS, Mann ZF, Gale JE, Ziegler M, Bain AJ, Szabadkai G, Duchen MR (2014) Separating NADH and NADPH fluorescence in live cells and tissues using FLIM. Nat Commun 5:3936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.De Ruyck J, Famerée M, Wouters J, Perpète EA, Preat J, Jacquemin DJ (2007) Towards the understanding of the absorption spectra of NAD(P)H/NAD(P)+ as a common indicator of dehydrogenase enzymatic activity. Chem Phys Lett 450:119–122 [Google Scholar]
- 33.Araque A, Li N, Doyle RT, Haydon PG (2000) SNARE protein-dependent glutamate release from astrocytes. J Neurosci 20:666–673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Malarkey EB, Parpura V (2011) Temporal characteristics of vesicular fusion in astrocytes: examination of synaptobrevin 2-laden vesicles at single vesicle resolution. J Physiol 589:4271–4300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hua X, Malarkey EB, Sunjara V, Rosenwald SE, Li WH, Parpura V (2004) Ca2+-dependent glutamate release involves two classes of endoplasmic reticulum Ca2+ stores in astrocytes. J Neurosci Res 76:86–97 [DOI] [PubMed] [Google Scholar]
- 36.Reyes RC, Verkhratsky A, Parpura V (2013) TRPC1-mediated Ca2+ and Na+ signalling in astroglia: differential filtering of extracellular cations. Cell Calcium 54:120–125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Piersma SR, Warmoes MO, de Wit M, de Reus I, Knol JC, Jimenez CR (2013) Whole gel processing procedure for GeLC-MS/MS based proteomics. Proteome Sci 11:17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Heaven MR, Flint D, Randall SM, Sosunov AA, Wilson L, Barnes S, Goldman JE, Muddiman DC, Brenner M (2016) Composition of Rosenthal fibers, the protein aggregate hallmark of Alexander disease. J Proteome Res 15:2265–2282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nesvizhskii AI, Keller A, Kolker E, Aebersold R (2003) A statistical model for identifying proteins by tandem mass spectrometry. Anal Chem 75:4646–4658 [DOI] [PubMed] [Google Scholar]
- 40.Xia Q, Liao L, Cheng D, Duong DM, Gearing M, Lah JJ, Levey AI, Peng J (2008) Proteomic identification of novel proteins associated with Lewy bodies. Front Biosci 13:3850–3856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hertz L, Zielke HR (2004) Astrocytic control of glutamatergic activity: astrocytes as stars of the show. Trends Neurosci 27:735–743 [DOI] [PubMed] [Google Scholar]
- 42.Prebil M, Chowdhury HH, Zorec R, Kreft M (2011) Changes in cytosolic glucose level in ATP stimulated live astrocytes. Biochem Biophys Res Commun 405:308–313 [DOI] [PubMed] [Google Scholar]
- 43.Parpura V, Haydon PG (2000) Physiological astrocytic calcium levels stimulate glutamate release to modulate adjacent neurons. Proc Natl Acad Sci USA 97:8629–8634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fink K, Velebit J, Vardjan N, Zorec R, Kreft M (2021) Noradrenaline-induced l-lactate production requires D-glucose entry and transit through the glycogen shunt in single-cultured rat astrocytes. J Neurosci Res 99:1084–1098 [DOI] [PubMed] [Google Scholar]
- 45.Chance B, Cohen P, Jobsis F, Schoener B (1962) Intracellular oxidation-reduction states in vivo. Science 137:499–508 [DOI] [PubMed] [Google Scholar]
- 46.Connor JA, Kreulen DL, Prosser CL (1976) Relation between oxidative metabolism and slow rhythmic potentials in mammalian intestinal muscle. Proc Natl Acad Sci USA 73:4239–4243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Christensen T, Bruhn T, Diemer NH, Schousboe A (1991) Effect of phenylsuccinate on potassium- and ischemia-induced release of glutamate in rat hippocampus monitored by microdialysis. Neurosci Lett 134:71–74 [DOI] [PubMed] [Google Scholar]
- 48.Stryer L (1988) Biochemistry. W. H. Freeman, San Francisco [Google Scholar]
- 49.Isgrò MA, Bottoni P, Scatena R (2015) Neuron-specific enolase as a biomarker: biochemical and clinical aspects. Adv Exp Med Biol 867:125–143 [DOI] [PubMed] [Google Scholar]
- 50.Deloulme JC, Raponi E, Gentil BJ, Bertacchi N, Marks A, Labourdette G, Baudier J (2004) Nuclear expression of S100B in oligodendrocyte progenitor cells correlates with differentiation toward the oligodendroglial lineage and modulates oligodendrocytes maturation. Mol Cell Neurosci 27:453–465 [DOI] [PubMed] [Google Scholar]
- 51.Ito D, Imai Y, Ohsawa K, Nakajima K, Fukuuchi Y, Kohsaka S (1998) Microglia-specific localisation of a novel calcium binding protein, Iba1. Brain Res Mol Brain Res 57:1–9 [DOI] [PubMed] [Google Scholar]
- 52.Stallcup WB, Beasley L (1987) Bipotential glial precursor cells of the optic nerve express the NG2 proteoglycan. J Neurosci 7:2737–2744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Messing A, Brenner M (2020) GFAP at 50. ASN Neuro 12:1759091420949680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Norenberg MD (1979) Distribution of glutamine synthetase in the rat central nervous system. J Histochem Cytochem 27:756–762 [DOI] [PubMed] [Google Scholar]
- 55.Cahoy JD, Emery B, Kaushal A, Foo LC, Zamanian JL, Christopherson KS, Xing Y, Lubischer JL, Krieg PA, Krupenko SA, Thompson WJ, Barres BA (2008) A transcriptome database for astrocytes, neurons, and oligodendrocytes: a new resource for understanding brain development and function. J Neurosci 28:264–278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Flugge G, Araya-Callis C, Garea-Rodriguez E, Stadelmann-Nessler C, Fuchs E (2014) NDRG2 as a marker protein for brain astrocytes. Cell Tissue Res 357:31–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Melzer L, Freiman TM, Derouiche A (2021) Rab6A as a pan-astrocytic marker in mouse and human brain, and comparison with other glial markers (GFAP, GS, Aldh1L1, SOX9). Cells 10:72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Malarkey EB, Ni Y, Parpura V (2008) Ca2+ entry through TRPC1 channels contributes to intracellular Ca2+ dynamics and consequent glutamate release from rat astrocytes. Glia 56:821–835 [DOI] [PubMed] [Google Scholar]
- 59.Furuta A, Rothstein JD, Martin LJ (1997) Glutamate transporter protein subtypes are expressed differentially during rat CNS development. J Neurosci 17:8363–8375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gottipati MK, Bekyarova E, Haddon RC, Parpura V (2015) Chemically functionalized single-walled carbon nanotubes enhance the glutamate uptake characteristics of mouse cortical astrocytes. Amino Acids 47:1379–1388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kunzelmann P, Schroder W, Traub O, Steinhauser C, Dermietzel R, Willecke K (1999) Late onset and increasing expression of the gap junction protein connexin30 in adult murine brain and long-term cultured astrocytes. Glia 25:111–119 [DOI] [PubMed] [Google Scholar]
- 62.Nagy JI, Li X, Rempel J, Stelmack G, Patel D, Staines WA, Yasumura T, Rash JE (2001) Connexin26 in adult rodent central nervous system: demonstration at astrocytic gap junctions and colocalization with connexin30 and connexin43. J Comp Neurol 441:302–323 [DOI] [PubMed] [Google Scholar]
- 63.Nagelhus EA, Ottersen OP (2013) Physiological roles of aquaporin-4 in brain. Physiol Rev 93:1543–1562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Draberova E, Del Valle L, Gordon J, Markova V, Smejkalova B, Bertrand L, de Chadarevian JP, Agamanolis DP, Legido A, Khalili K, Draber P, Katsetos CD (2008) Class III beta-tubulin is constitutively coexpressed with glial fibrillary acidic protein and nestin in midgestational human fetal astrocytes: implications for phenotypic identity. J Neuropathol Exp Neurol 67:341–354 [DOI] [PubMed] [Google Scholar]
- 65.Potokar M, Stenovec M, Gabrijel M, Li L, Kreft M, Grilc S, Pekny M, Zorec R (2010) Intermediate filaments attenuate stimulation-dependent mobility of endosomes/lysosomes in astrocytes. Glia 58:1208–1219 [DOI] [PubMed] [Google Scholar]
- 66.Lasič E, Trkov Bobnar S, Wilhelmsson U, de Pablo Y, Pekny M, Zorec R, Stenovec M (2020) Nestin affects fusion pore dynamics in mouse astrocytes. Acta Physiol (Oxf, Engl) 228:e13399. [DOI] [PubMed] [Google Scholar]
- 67.Valencia RG, Mihailovska E, Winter L, Bauer K, Fischer I, Walko G, Jorgacevski J, Potokar M, Zorec R, Wiche G (2020) Plectin dysfunction in neurons leads to tau accumulation on microtubules affecting neuritogenesis, organelle trafficking, pain sensitivity and memory. Neuropathol Appl Neurobiol. 10.1111/nan.12635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kleppe R, Martinez A, Doskeland SO, Haavik J (2011) The 14-3-3 proteins in regulation of cellular metabolism. Semin Cell Dev Biol 22:713–719 [DOI] [PubMed] [Google Scholar]
- 69.Gelperin D, Weigle J, Nelson K, Roseboom P, Irie K, Matsumoto K, Lemmon S (1995) 14-3-3 proteins: potential roles in vesicular transport and Ras signaling in Saccharomyces cerevisiae. Proc Natl Acad Sci USA 92:11539–11543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Satoh J, Yamamura T, Arima K (2004) The 14-3-3 protein epsilon isoform expressed in reactive astrocytes in demyelinating lesions of multiple sclerosis binds to vimentin and glial fibrillary acidic protein in cultured human astrocytes. Am J Pathol 165:577–592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Potokar M, Kreft M, Li L, Daniel Andersson J, Pangrsic T, Chowdhury HH, Pekny M, Zorec R (2007) Cytoskeleton and vesicle mobility in astrocytes. Traffic (Cph, Denmark) 8:12–20 [DOI] [PubMed] [Google Scholar]
- 72.Osborne KD, Lee W, Malarkey EB, Irving AJ, Parpura V (2009) Dynamic imaging of cannabinoid receptor 1 vesicular trafficking in cultured astrocytes. ASN Neuro 1:AN20090040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Butkevich E, Hulsmann S, Wenzel D, Shirao T, Duden R, Majoul I (2004) Drebrin is a novel connexin-43 binding partner that links gap junctions to the submembrane cytoskeleton. Curr Biol 14:650–658 [DOI] [PubMed] [Google Scholar]
- 74.Derouiche A, Frotscher M (2001) Peripheral astrocyte processes: monitoring by selective immunostaining for the actin-binding ERM proteins. Glia 36:330–341 [DOI] [PubMed] [Google Scholar]
- 75.Araque A, Parpura V, Sanzgiri RP, Haydon PG (1999) Tripartite synapses: glia, the unacknowledged partner. Trends Neurosci 22:208–215 [DOI] [PubMed] [Google Scholar]
- 76.Racchetti G, D’Alessandro R, Meldolesi J (2012) Astrocyte stellation, a process dependent on Rac1 is sustained by the regulated exocytosis of enlargeosomes. Glia 60:465–475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ito H, Atsuzawa K, Morishita R, Usuda N, Sudo K, Iwamoto I, Mizutani K, Katoh-Semba R, Nozawa Y, Asano T, Nagata K (2009) Sept8 controls the binding of vesicle-associated membrane protein 2 to synaptophysin. J Neurochem 108:867–880 [DOI] [PubMed] [Google Scholar]
- 78.Rodriguez-Gabin AG, Ortiz E, Demoliner K, Si Q, Almazan G, Larocca JN (2010) Interaction of Rab31 and OCRL-1 in oligodendrocytes: its role in transport of mannose 6-phosphate receptors. J Neurosci Res 88:589–604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Singh P, Jorgacevski J, Kreft M, Grubisic V, Stout RF Jr, Potokar M, Parpura V, Zorec R (2014) Single-vesicle architecture of synaptobrevin2 in astrocytes. Nat Commun 5:3780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Bhattacharya S, Stewart BA, Niemeyer BA, Burgess RW, McCabe BD, Lin P, Boulianne G, O’Kane CJ, Schwarz TL (2002) Members of the synaptobrevin/vesicle-associated membrane protein (VAMP) family in Drosophila are functionally interchangeable in vivo for neurotransmitter release and cell viability. Proc Natl Acad Sci USA 99:13867–13872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Borisovska M, Zhao Y, Tsytsyura Y, Glyvuk N, Takamori S, Matti U, Rettig J, Sudhof T, Bruns D (2005) v-SNAREs control exocytosis of vesicles from priming to fusion. Embo J 24:2114–2126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Deak F, Shin OH, Kavalali ET, Sudhof TC (2006) Structural determinants of synaptobrevin 2 function in synaptic vesicle fusion. J Neurosci 26:6668–6676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Verderio C, Cagnoli C, Bergami M, Francolini M, Schenk U, Colombo A, Riganti L, Frassoni C, Zuccaro E, Danglot L, Wilhelm C, Galli T, Canossa M, Matteoli M (2012) TI-VAMP/VAMP7 is the SNARE of secretory lysosomes contributing to ATP secretion from astrocytes. Biol Cell 104:213–228 [DOI] [PubMed] [Google Scholar]
- 84.Way G, Morrice N, Smythe C, O’Sullivan AJ (2002) Purification and identification of secernin, a novel cytosolic protein that regulates exocytosis in mast cells. Mol Biol Cell 13:3344–3354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Marz KE, Lauer JM, Hanson PI (2003) Defining the SNARE complex binding surface of alpha-SNAP: implications for SNARE complex disassembly. J Biol Chem 278:27000–27008 [DOI] [PubMed] [Google Scholar]
- 86.Nakatsu F, Ohno H (2003) Adaptor protein complexes as the key regulators of protein sorting in the post-Golgi network. Cell Struct Funct 28:419–429 [DOI] [PubMed] [Google Scholar]
- 87.McGhie TK, Walton MC (2007) The bioavailability and absorption of anthocyanins: towards a better understanding. Mol Nutr Food Res 51:702–713 [DOI] [PubMed] [Google Scholar]
- 88.Cao H, Chen J, Krueger EW, McNiven MA (2010) SRC-mediated phosphorylation of dynamin and cortactin regulates the “constitutive” endocytosis of transferrin. Mol Cell Biol 30:781–792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.McArthur S, Yazid S, Christian H, Sirha R, Flower R, Buckingham J, Solito E (2009) Annexin A1 regulates hormone exocytosis through a mechanism involving actin reorganization. FASEB J 23:4000–4010 [DOI] [PubMed] [Google Scholar]
- 90.Podszywalow-Bartnicka P, Kosiorek M, Piwocka K, Sikora E, Zablocki K, Pikula S (2010) Role of annexin A6 isoforms in catecholamine secretion by PC12 cells: distinct influence on calcium response. J Cell Biochem 111:168–178 [DOI] [PubMed] [Google Scholar]
- 91.Ali SM, Burgoyne RD (1990) The stimulatory effect of calpactin (annexin II) on calcium-dependent exocytosis in chromaffin cells: requirement for both the N-terminal and core domains of p36 and ATP. Cell Signal 2:265–276 [DOI] [PubMed] [Google Scholar]
- 92.Willshaw A, Grant K, Yan J, Rockliffe N, Ambavarapu S, Burdyga G, Varro A, Fukuoka S, Gawler D (2004) Identification of a novel protein complex containing annexin A4, rabphilin and synaptotagmin. FEBS Lett 559:13–21 [DOI] [PubMed] [Google Scholar]
- 93.Verkhratsky A, Rodriguez JJ, Parpura V (2012) Calcium signalling in astroglia. Mol Cell Endocrinol 353:45–56 [DOI] [PubMed] [Google Scholar]
- 94.Verkhratsky A, Untiet V, Rose CR (2020) Ionic signalling in astroglia beyond calcium. J Physiol 598:1655–1670 [DOI] [PubMed] [Google Scholar]
- 95.Reyes RC, Verkhratsky A, Parpura V (2012) Plasmalemmal Na+/Ca2+ exchanger modulates Ca2+-dependent exocytotic release of glutamate from rat cortical astrocytes. ASN Neuro 4:AN20110059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Shigetomi E, Tong X, Kwan KY, Corey DP, Khakh BS (2012) TRPA1 channels regulate astrocyte resting calcium and inhibitory synapse efficacy through GAT-3. Nat Neurosci 15:70–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Marchaland J, Cali C, Voglmaier SM, Li H, Regazzi R, Edwards RH, Bezzi P (2008) Fast subplasma membrane Ca2+ transients control exo-endocytosis of synaptic-like microvesicles in astrocytes. J Neurosci 28:9122–9132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Idevall-Hagren O, Lu A, Xie B, De Camilli P (2015) Triggered Ca2+ influx is required for extended synaptotagmin 1-induced ER-plasma membrane tethering. EMBO J 34:2291–2305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Verkhratsky A, Parpura V (2015) Physiology of astroglia: channels, receptors, transporters, ion signaling and gliotransmission. Morgan & Claypool Publishers, San Rafael [Google Scholar]
- 100.Parnis J, Montana V, Delgado-Martinez I, Matyash V, Parpura V, Kettenmann H, Sekler I, Nolte C (2013) Mitochondrial exchanger NCLX plays a major role in the intracellular Ca2+ signaling, gliotransmission, and proliferation of astrocytes. J Neurosci 33:7206–7219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Leung AW, Halestrap AP (2008) Recent progress in elucidating the molecular mechanism of the mitochondrial permeability transition pore. Biochim Biophys Acta 1777:946–952 [DOI] [PubMed] [Google Scholar]
- 102.Basso E, Fante L, Fowlkes J, Petronilli V, Forte MA, Bernardi P (2005) Properties of the permeability transition pore in mitochondria devoid of Cyclophilin D. J Biol Chem 280:18558–18561 [DOI] [PubMed] [Google Scholar]
- 103.Csordas G, Weaver D, Hajnoczky G (2018) Endoplasmic reticulum-mitochondrial contactology: structure and signaling functions. Trends Cell Biol 28:523–540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Rapizzi E, Pinton P, Szabadkai G, Wieckowski MR, Vandecasteele G, Baird G, Tuft RA, Fogarty KE, Rizzuto R (2002) Recombinant expression of the voltage-dependent anion channel enhances the transfer of Ca2+ microdomains to mitochondria. J Cell Biol 159:613–624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Lauritzen KH, Morland C, Puchades M, Holm-Hansen S, Hagelin EM, Lauritzen F, Attramadal H, Storm-Mathisen J, Gjedde A, Bergersen LH (2014) Lactate receptor sites link neurotransmission, neurovascular coupling, and brain energy metabolism. Cereb Cortex 24:2784–2795 [DOI] [PubMed] [Google Scholar]
- 106.Mosienko V, Rasooli-Nejad S, Kishi K, De Both M, Jane D, Huentelman MJ, Kasparov S, Teschemacher AG (2018) Putative receptors underpinning l-lactate signalling in locus coeruleus. Neuroglia 1:365–380 [Google Scholar]
- 107.Yang JW, Suder P, Silberring J, Lubec G (2005) Proteome analysis of mouse primary astrocytes. Neurochem Int 47:159–172 [DOI] [PubMed] [Google Scholar]
- 108.Bentaib A, De Tullio P, Chneiweiss H, Hermans E, Junier MP, Leprince P (2015) Metabolic reprogramming in transformed mouse cortical astrocytes: a proteomic study. J Proteomics 113:292–314 [DOI] [PubMed] [Google Scholar]
- 109.Tong L, Wang C, Hu X, Pang B, Yang Z, He Z, He M, Wei L, Chu M (2016) Correlated overexpression of metadherin and SND1 in glioma cells. Biol Chem 397:57–65 [DOI] [PubMed] [Google Scholar]
- 110.Ehsani A, Alluin JV, Rossi JJ (2013) Cell cycle abnormalities associated with differential perturbations of the human U5 snRNP associated U5–200kD RNA helicase. PLoS ONE 8:e62125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Latham T, Mackay L, Sproul D, Karim M, Culley J, Harrison DJ, Hayward L, Langridge-Smith P, Gilbert N, Ramsahoye BH (2012) Lactate, a product of glycolytic metabolism, inhibits histone deacetylase activity and promotes changes in gene expression. Nucleic Acids Res 40:4794–4803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Lundgaard I, Lu ML, Yang E, Peng W, Mestre H, Hitomi E, Deane R, Nedergaard M (2017) Glymphatic clearance controls state-dependent changes in brain lactate concentration. J Cereb Blood Flow Metab 37:2112–2124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Malarkey EB, Reyes RC, Zhao B, Haddon RC, Parpura V (2008) Water soluble single-walled carbon nanotubes inhibit stimulated endocytosis in neurons. Nano Lett 8:3538–3542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Sanzgiri RP, Araque A, Haydon PG (1999) Prostaglandin E(2) stimulates glutamate receptor-dependent astrocyte neuromodulation in cultured hippocampal cells. J Neurobiol 41:221–229 [PubMed] [Google Scholar]
- 115.Gordon GR, Choi HB, Rungta RL, Ellis-Davies GC, MacVicar BA (2008) Brain metabolism dictates the polarity of astrocyte control over arterioles. Nature 456:745–749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Metea MR, Kofuji P, Newman EA (2007) Neurovascular coupling is not mediated by potassium siphoning from glial cells. J Neurosci 27:2468–2471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Shimoyama M, Smith JR, Bryda E, Kuramoto T, Saba L, Dwinell M (2017) Rat genome and model resources. ILAR J 58:42–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Beck M, Schmidt A, Malmstroem J, Claassen M, Ori A, Szymborska A, Herzog F, Rinner O, Ellenberg J, Aebersold R (2011) The quantitative proteome of a human cell line. Mol Syst Biol 7:549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Rizzoli SO, Betz WJ (2005) Synaptic vesicle pools. Nat Rev Neurosci 6:57–69 [DOI] [PubMed] [Google Scholar]
- 120.Takamori S, Holt M, Stenius K, Lemke EA, Gronborg M, Riedel D, Urlaub H, Schenck S, Brugger B, Ringler P, Muller SA, Rammner B, Grater F, Hub JS, De Groot BL, Mieskes G, Moriyama Y, Klingauf J, Grubmuller H, Heuser J, Wieland F, Jahn R (2006) Molecular anatomy of a trafficking organelle. Cell 127:831–846 [DOI] [PubMed] [Google Scholar]
- 121.Fricker LD (2015) Limitations of mass spectrometry-based peptidomic approaches. J Am Soc Mass Spectrom 26:1981–1991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Alfonso-Garrido J, Garcia-Calvo E, Luque-Garcia JL (2015) Sample preparation strategies for improving the identification of membrane proteins by mass spectrometry. Anal Bioanal Chem 407:4893–4905 [DOI] [PubMed] [Google Scholar]
- 123.Braakman HMH, Engelen M, Nicolai J, Willemsen M (2018) Stroke mimics add to the phenotypic spectrum of GLUT1 deficiency syndrome. J Neurol Neurosurg Psychiatry 89:668–670 [DOI] [PubMed] [Google Scholar]
- 124.De Vivo DC, Trifiletti RR, Jacobson RI, Ronen GM, Behmand RA, Harik SI (1991) Defective glucose transport across the blood-brain barrier as a cause of persistent hypoglycorrhachia, seizures, and developmental delay. N Engl J Med 325:703–709 [DOI] [PubMed] [Google Scholar]
- 125.Pearson TS, Akman C, Hinton VJ, Engelstad K, De Vivo DC (2013) Phenotypic spectrum of glucose transporter type 1 deficiency syndrome (Glut1 DS). Curr Neurol Neurosci Rep 13:342. [DOI] [PubMed] [Google Scholar]
- 126.Walz W, Mukerji S (1988) Lactate release from cultured astrocytes and neurons: a comparison. Glia 1:366–370 [DOI] [PubMed] [Google Scholar]
- 127.Perea G, Araque A (2007) Astrocytes potentiate transmitter release at single hippocampal synapses. Science 317:1083–1086 [DOI] [PubMed] [Google Scholar]
- 128.Ferguson BS, Rogatzki MJ, Goodwin ML, Kane DA, Rightmire Z, Gladden LB (2018) Lactate metabolism: historical context, prior misinterpretations, and current understanding. Eur J Appl Physiol 118:691–728 [DOI] [PubMed] [Google Scholar]
- 129.Bebarta VS, Pead J, Varney SM (2015) Lacticemia after acute overdose of metformin in an adolescent managed without intravenous sodium bicarbonate or extracorporeal therapy. Pediatr Emerg Care 31:589–590 [DOI] [PubMed] [Google Scholar]
- 130.Medina JM, Tabernero A (2005) Lactate utilization by brain cells and its role in CNS development. J Neurosci Res 79:2–10 [DOI] [PubMed] [Google Scholar]
- 131.Pan JW, Rothman TL, Behar KL, Stein DT, Hetherington HP (2000) Human brain beta-hydroxybutyrate and lactate increase in fasting-induced ketosis. J Cereb Blood Flow Metab 20:1502–1507 [DOI] [PubMed] [Google Scholar]
- 132.Guzman M, Blazquez C (2001) Is there an astrocyte-neuron ketone body shuttle? Trends Endocrinol Metab 12:169–173 [DOI] [PubMed] [Google Scholar]
- 133.Valdebenito R, Ruminot I, Garrido-Gerter P, Fernandez-Moncada I, Forero-Quintero L, Alegria K, Becker HM, Deitmer JW, Barros LF (2016) Targeting of astrocytic glucose metabolism by beta-hydroxybutyrate. J Cereb Blood Flow Metab 36:1813–1822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Schurr A, Payne RS, Miller JJ, Rigor BM (1997) Brain lactate is an obligatory aerobic energy substrate for functional recovery after hypoxia: further in vitro validation. J Neurochem 69:423–426 [DOI] [PubMed] [Google Scholar]
- 135.Schurr A, Payne RS, Miller JJ, Tseng MT, Rigor BM (2001) Blockade of lactate transport exacerbates delayed neuronal damage in a rat model of cerebral ischemia. Brain Res 895:268–272 [DOI] [PubMed] [Google Scholar]
- 136.Cureton EL, Kwan RO, Dozier KC, Sadjadi J, Pal JD, Victorino GP (2010) A different view of lactate in trauma patients: protecting the injured brain. J Surg Res 159:468–473 [DOI] [PubMed] [Google Scholar]
- 137.Reyes RC, Parpura V (2008) Models of astrocytic Ca dynamics and epilepsy. Drug Discov Today Dis Models 5:13–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Stout RF Jr, Spray DC, Parpura V (2009) Astrocytic “powergrid”: delivery upon neuronal demand. Cellscience 5:34–43 [PMC free article] [PubMed] [Google Scholar]
- 139.Sada N, Lee S, Katsu T, Otsuki T, Inoue T (2015) Epilepsy treatment. targeting LDH enzymes with a stiripentol analog to treat epilepsy. Science 347:1362–1367 [DOI] [PubMed] [Google Scholar]
- 140.Roslin M, Henriksson R, Bergstrom P, Ungerstedt U, Bergenheim AT (2003) Baseline levels of glucose metabolites, glutamate and glycerol in malignant glioma assessed by stereotactic microdialysis. J Neurooncol 61:151–160 [DOI] [PubMed] [Google Scholar]
- 141.Marcus HJ, Carpenter KL, Price SJ, Hutchinson PJ (2010) In vivo assessment of high-grade glioma biochemistry using microdialysis: a study of energy-related molecules, growth factors and cytokines. J Neurooncol 97:11–23 [DOI] [PubMed] [Google Scholar]
- 142.Martin S, Parton RG (2006) Lipid droplets: a unified view of a dynamic organelle. Nat Rev Mol Cell Biol 7:373–378 [DOI] [PubMed] [Google Scholar]
- 143.Liu P, Ying Y, Zhao Y, Mundy DI, Zhu M, Anderson RG (2004) Chinese hamster ovary K2 cell lipid droplets appear to be metabolic organelles involved in membrane traffic. J Biol Chem 279:3787–3792 [DOI] [PubMed] [Google Scholar]
- 144.Brasaemle DL, Dolios G, Shapiro L, Wang R (2004) Proteomic analysis of proteins associated with lipid droplets of basal and lipolytically stimulated 3T3-L1 adipocytes. J Biol Chem 279:46835–46842 [DOI] [PubMed] [Google Scholar]
- 145.Fujimoto Y, Itabe H, Sakai J, Makita M, Noda J, Mori M, Higashi Y, Kojima S, Takano T (2004) Identification of major proteins in the lipid droplet-enriched fraction isolated from the human hepatocyte cell line HuH7. Biochim Biophys Acta 1644:47–59 [DOI] [PubMed] [Google Scholar]
- 146.Parton RG (1996) Caveolae and caveolins. Curr Opin Cell Biol 8:542–548 [DOI] [PubMed] [Google Scholar]
- 147.Pol A, Martin S, Fernandez MA, Ferguson C, Carozzi A, Luetterforst R, Enrich C, Parton RG (2004) Dynamic and regulated association of caveolin with lipid bodies: modulation of lipid body motility and function by a dominant negative mutant. Mol Biol Cell 15:99–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Trigatti BL, Anderson RG, Gerber GE (1999) Identification of caveolin-1 as a fatty acid binding protein. Biochem Biophys Res Commun 255:34–39 [DOI] [PubMed] [Google Scholar]
- 149.Bozza PT, Viola JP (2010) Lipid droplets in inflammation and cancer. Prostaglandins Leukot Essent Fatty Acids 82:243–250 [DOI] [PubMed] [Google Scholar]
- 150.Greenberg AS, Coleman RA, Kraemer FB, McManaman JL, Obin MS, Puri V, Yan QW, Miyoshi H, Mashek DG (2011) The role of lipid droplets in metabolic disease in rodents and humans. J Clin Invest 121:2102–2110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Bosma M, Kersten S, Hesselink MK, Schrauwen P (2012) Re-evaluating lipotoxic triggers in skeletal muscle: relating intramyocellular lipid metabolism to insulin sensitivity. Prog Lipid Res 51:36–49 [DOI] [PubMed] [Google Scholar]
- 152.Heaton NS, Randall G (2010) Dengue virus-induced autophagy regulates lipid metabolism. Cell Host Microbe 8:422–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Suzuki M, Shinohara Y, Ohsaki Y, Fujimoto T (2011) Lipid droplets: size matters. J Electron Microsc (Tokyo) 60(Suppl 1):S101–116 [DOI] [PubMed] [Google Scholar]
- 154.Guo Y, Cordes KR, Farese RV Jr, Walther TC (2009) Lipid droplets at a glance. J Cell Sci 122:749–752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Juraszek B, Czarnecka-Herok J, Nalecz KA (2021) Glioma cells survival depends both on fatty acid oxidation and on functional carnitine transport by SLC22A5. J Neurochem 156:642–657 [DOI] [PubMed] [Google Scholar]
- 156.Magistretti PJ, Allaman I (2018) Lactate in the brain: from metabolic end-product to signalling molecule. Nat Rev Neurosci 19:235–249 [DOI] [PubMed] [Google Scholar]
- 157.Gottipati MK, Kalinina I, Bekyarova E, Haddon RC, Parpura V (2012) Chemically functionalized water-soluble single-walled carbon nanotubes modulate morpho-functional characteristics of astrocytes. Nano Lett 12:4742–4747 [DOI] [PubMed] [Google Scholar]
- 158.Gottipati MK, Samuelson JJ, Kalinina I, Bekyarova E, Haddon RC, Parpura V (2013) Chemically functionalized single-walled carbon nanotube films modulate the morpho-functional and proliferative characteristics of astrocytes. Nano Lett 13:4387–4392 [DOI] [PubMed] [Google Scholar]
- 159.Bozzo L, Puyal J, Chatton JY (2013) Lactate modulates the activity of primary cortical neurons through a receptor-mediated pathway. PLoS ONE 8:e71721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Vardjan N, Chowdhury HH, Horvat A, Velebit J, Malnar M, Muhic M, Kreft M, Krivec SG, Bobnar ST, Mis K, Pirkmajer S, Offermanns S, Henriksen G, Storm-Mathisen J, Bergersen LH, Zorec R (2018) Enhancement of astroglial aerobic glycolysis by extracellular lactate-mediated increase in cAMP. Front Mol Neurosci 11:148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.D’Adamo P, Horvat A, Gurgone A, Mignogna ML, Bianchi V, Masetti M, Ripamonti M, Taverna S, Velebit J, Malnar M, Muhic M, Fink K, Bachi A, Restuccia U, Belloli S, Moresco RM, Mercalli A, Piemonti L, Potokar M, Bobnar ST, Kreft M, Chowdhury HH, Stenovec M, Vardjan N, Zorec R (2021) Inhibiting glycolysis rescues memory impairment in an intellectual disability Gdi1-null mouse. Metabolism 116:154463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Tejada MI, Ibarluzea N (2020) Non-syndromic X linked intellectual disability: current knowledge in light of the recent advances in molecular and functional studies. Clin Genet 97:677–687 [DOI] [PubMed] [Google Scholar]
- 163.Curie A, Sacco S, Bussy G, de Saint MA, Boddaert N, Chanraud S, Meresse I, Chelly J, Zilbovicius M, des Portes V (2009) Impairment of cerebello-thalamo-frontal pathway in Rab-GDI mutated patients with pure mental deficiency. Eur J Med Genet 52:6–13 [DOI] [PubMed] [Google Scholar]
- 164.Wang YF, Sun MY, Hou Q, Parpura V (2013) Hyposmolality differentially and spatiotemporally modulates levels of glutamine synthetase and serine racemase in rat supraoptic nucleus. Glia 61:529–538 [DOI] [PubMed] [Google Scholar]
- 165.Vaden JH, Bhattacharyya BJ, Chen PC, Watson JA, Marshall AG, Phillips SE, Wilson JA, King GD, Miller RJ, Wilson SM (2015) Ubiquitin-specific protease 14 regulates c-Jun N-terminal kinase signaling at the neuromuscular junction. Mol Neurodegener 10:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Crimmins S, Jin Y, Wheeler C, Huffman AK, Chapman C, Dobrunz LE, Levey A, Roth KA, Wilson JA, Wilson SM (2006) Transgenic rescue of ataxia mice with neuronal-specific expression of ubiquitin-specific protease 14. J Neurosci 26:11423–11431 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
At any request, we will freely provide the original data sets.