Abstract
Reactive oxygen species (ROS) contribute to cellular damage in several different contexts, but their role during chilling damage is poorly defined. Chilling sensitivity both limits the distribution of plant species and causes devastating crop losses worldwide. Our screen of chilling-tolerant Arabidopsis (Arabidopsis thaliana) for mutants that suffer chilling damage identified a gene (At4g03410) encoding a chloroplast Mpv17_PMP22 protein, MPD1, with no previous connection to chilling. The chilling-sensitive mpd1-1 mutant is an overexpression allele that we successfully phenocopied by creating transgenic lines with a similar level of MPD1 overexpression. In mammals and yeast, MPD1 homologs are associated with ROS management. In chilling conditions, Arabidopsis overexpressing MPD1 accumulated H2O2 to higher levels than wild-type controls and exhibited stronger induction of ROS response genes. Paraquat application exacerbated chilling damage, confirming that the phenotype occurs due to ROS dysregulation. We conclude that at low temperature increased MPD1 expression results in increased ROS production, causing chilling damage. Our discovery of the effect of MPD1 overexpression on ROS production under chilling stress implies that investigation of the nine other members of the Mpv17_PMP22 family in Arabidopsis may lead to new discoveries regarding ROS signaling and management in plants.
Keywords: Arabidopsis, chilling damage, Mpv17_PMP22 proteins, oxidative stress, photosynthesis, temperature response
The discovery that overexpression of the chloroplast channel protein MPD1 increases reactive oxygen species accumulation and chilling damage reveals the importance of plant Mpv17_PMP22 proteins in oxidative stress.
Introduction
With the world population predicted to exceed 9.7 billion by 2050, global food security presents a significant challenge for humanity. The United Nations estimates that this will require global agricultural output to increase by 50% over the next 20 years (FAO, 2017). One way to achieve this goal is by developing crops with enhanced tolerance to environmental stresses, which account for 50% of crop yield losses worldwide (Rodziewicz et al., 2014). Despite the onset of global warming, cold stress is currently the most significant environmental factor limiting crop yields (Tripathi et al., 2016). Plant species originating in temperate regions accommodate to low temperature and typically survive even when exposed to prolonged freezing conditions (Thomashow, 1999), but more than half of plant species on earth have origins in tropical regions and suffer serious damage when exposed to low, non-freezing temperatures between 0 °C and 10 °C (Lyons, 1973).
The diverse external symptoms of chilling injury in plants include cessation of growth, wilting, chlorosis, necrosis, and eventual death. Essential food crops such as maize (Zea mays), soybean (Glycine max), and rice (Oryza sativa) are sensitive to low temperature. Chilling impairment reduces germination, restricts root development, lowers vegetative growth, and reduces fruit and seed production (Lukatkin et al., 2012). While many tissues suffer chilling damage, in leaves damage arises from reduced photosynthesis, including disrupted photosystems and chloroplast structures (Allen and Ort, 2001), producing chlorosis, development of necrotic lesions, and death of leaf tissue (Hussain et al., 2018).
Chilling sensitivity is distinct from freezing damage, which creates different types of cellular injury. However, gene expression responses that prepare plants for freezing (termed cold acclimation) do occur at chilling temperatures; >1000 genes are induced or repressed after as little as 6 h of chilling in Arabidopsis (Arabidopsis thaliana) (Usadel et al., 2008; Meissner et al., 2013; Zandalinas et al., 2020). These changes are regulated by layers of transcription factors acting in a cascade to induce or repress downstream gene expression, and a substantial proportion of them are necessary for acclimation to subsequent freezing temperatures (Zhu et al., 2007; Usadel et al., 2008; Thomashow, 2010; Zuther et al., 2018) but not for chilling tolerance itself, so cold acclimation confounds the use of transcriptional profiling to identify genes specific to preventing chilling damage. Nevertheless, consideration of particular processes affected by chilling, such as the production and turnover of reactive oxygen species (ROS), has identified altered gene expression that contributes to protection from chilling damage.
Production of ROS occurs in several subcellular organelles, including chloroplasts, mitochondria, and peroxisomes, and ROS are also produced by plasma membrane NAPH oxidases for pathogen defense and signaling functions (Fang et al., 2019; Smirnoff and Arnaud, 2019). In chloroplasts, generation of ROS occurs naturally as a byproduct of electron transfer through an oxygen-rich environment. While these ROS were once considered mainly disruptive to cell metabolism, recent studies show that they are essential in many developmental processes (Mhamdi and Van Breusegem, 2018). Further, these molecules act as signals regulating expression of genes encoding enzymes involved in ROS management (Dietz et al., 2016). Therefore, it is more accurate to say that unregulated production and persistence of ROS are disruptive. In chloroplasts, superoxide dismutase (SOD) enzymes convert superoxide anion radicals to the more stable hydrogen peroxide (H2O2) (Dreyer and Dietz, 2018), which acts as an intermediate in ROS metabolism, communicating chloroplast ROS status through interaction with peroxiredoxins and other proteins (Dreyer and Dietz, 2018). As environmental stresses increase ROS levels, H2O2 acts as a retrograde signal, triggering expression changes in nuclear genes mediating ROS breakdown and response (Zandalinas et al., 2020).
In both chilling-tolerant and chilling-sensitive plants, the biochemical reactions of photosynthetic carbon metabolism are more greatly reduced at low temperature than are biophysical light capture processes. This creates an excess of absorbed light energy that needs to be dissipated through non-photochemical quenching, but low temperatures also compromise non-photochemical quenching mechanisms (Latowski et al., 2011), leading to photoinhibition. As a result, over-reduction of the photosystems leads to substantial ROS production, including singlet oxygen, superoxide, and hydroxyl radicals that can damage protein, lipid, and nucleotide components of the cell unless the changes are successfully managed. Photoinhibition occurs because the D1 protein in the PSII reaction center suffers ongoing damage during light capture, requiring continuous replacement (Li et al., 2018). Replacement is temperature sensitive and, at low temperatures, especially below 5 °C, both chilling-tolerant and chilling-sensitive plants suffer photoinhibition, manifesting as a decline in the potential quantum yield of PSII, Fv/Fm, and contributing to excess energy accumulation in photosynthetic antennae and production of ROS (Wise, 1995; Li et al., 2018).
In chilling-tolerant plants, low temperature induces a critical set of genes and processes to modulate ROS levels and prevent cellular damage. In Arabidopsis, low temperature and other conditions induce robust expression of the gene ZINC FINGER OF ARABIDOPSIS THALIANA 12 (ZAT12, At5g59820), encoding a key regulator of transcriptional responses to increased ROS (Davletova et al., 2005). Downstream genes in the ROS response include SOD genes encoding superoxide dismutases, and CAT genes encoding catalases, which convert H2O2 to H2O and O2. Additionally, ascorbate peroxidase (APX) scavenges ROS by oxidizing ascorbate to dehydroascorbate, which is reduced back to ascorbate through the Foyer–Halliwell–Asada cycle including the glutathione-dependent dehydroascorbate reductase (DHAR) (Foyer and Shigeoka, 2011). Chilling-sensitive plants can be damaged through poor induction of such protective systems, rendering them unable to adequately process increased ROS produced at low temperatures (Dreyer and Dietz, 2018). Thus, ROS production and metabolism are competing processes that are key to understanding the different fates of chilling-sensitive and chilling-tolerant plants at low temperature.
All aerobic organisms must tightly regulate the production and breakdown of ROS because of their potential toxicity. In mammals, proteins containing an Mpv17_PMP22 domain have well-established links to ROS stress (Löllgen and Weiher, 2015). Mpv17_PMP22 domain proteins localize to mitochondria and peroxisomes, major sites of ROS generation and regulation, in humans (Spinazzola et al., 2006) and mice (Zwacka et al., 1994). Altered expression of Mpv17_PMP22 family members results in dysregulation of ROS balance, transcript changes of genes involved in ROS generation and breakdown, and the incidence of several rare genetic diseases (Löllgen and Weiher, 2015).
As a chilling-tolerant plant with powerful genetic resources, Arabidopsis is an excellent model for studying low-temperature stress. Here, we report our identification of a gene encoding a chloroplast Mpv17_PMP22 protein, MPD1 (At4g03410), with no previous association with chilling sensitivity. Analysis revealed that the chilling-sensitive mpd1-1 mutant was an overexpression allele, which we phenocopied by producing transgenic lines with similar elevated transcript levels. Each overexpression line was indistinguishable from the wild type when grown at 22 °C, but when transferred to 2 °C a significant loss of photosynthetic capacity resulted, coupled to visible leaf chlorosis and necrosis. In the cold, plants overexpressing MPD1 accumulated higher H2O2 levels than Col-0 and more strongly expressed genes associated with ROS stress. We conclude that high expression of MPD1 increased ROS production at low temperatures. This discovery suggests that investigation of the other members of the Mpv17_PMP22 family in Arabidopsis will contribute to understanding ROS production, signaling, and management in plants.
Materials and methods
High-throughput screen of T-DNA insertion lines
We acquired a pool of known homozygous insertional mutants labeled CS27942 from the Arabidopsis Biological Resource Center (https://www.arabidopsis.org). We sowed three plants into peat pods from each of 1244 randomly chosen lines. After 48 h of stratification at 4 °C, the plants were cultivated for 2 weeks at 22 °C under 24 h light of 150 μmol m–2 s–1 intensity. After removing lines with any visible defect, we subjected the remaining plants to 4 weeks of low-temperature growth at 4 °C under the same light intensity. After this chilling treatment, lines that showed a consistent phenotype across all three replicate plants were noted and selected for further cultivation at 2 °C to confirm their chilling sensitivity.
Plant growth conditions
Each experiment used 10 individual plants, with each line interplanted with three independent replicates and kept at 4 °C for 48 h to ensure even germination. Plant were cultivated in Percival growth chambers under constant light of 150 µE m–2 s–1 at 22 °C for 2 weeks and then transferred to 2 °C for 8 weeks before returning to the previous conditions for recovery. Plants undergoing paraquat treatment were grown at 22 °C for 2 weeks and then transferred to 2 °C and sprayed with 1 ml of either 5 mM paraquat or distilled water using a perfume atomizer. The same treatments were repeated every 48 h throughout the 2 week chilling period.
Locations of T-DNA insertion in mpd1-1 and mpd1-2
We extracted genomic DNA using the DNeasy Plant Mini Kit (Qiagen) and performed PCR using primers for the coding sequence of MPD1 and either the Salk pROK2 (mpd1-1) (Forward, 5ʹ TTGCAAAGATCACCCGATTT 3ʹ; Reverse, 5ʹ CTTTAGGGTTCCGATTTAGTGCTTTA 3ʹ) or Sail pDAP101 (mpd1-2) (Forward, 5ʹ CCCTGTGTCTTCGTGATCATA 3ʹ; Reverse, 5ʹ TAGCATCTGAATTTCATAACCAATCTCGATACAC 3ʹ). After sequencing the amplicon, we confirmed the location by alignment to the genome.
Plant growth measurements
We quantified plant growth by recording rosette diameters after cultivating plants for 2 weeks at 22 °C, and after 4 and 8 weeks at 2 °C. After 2 weeks of recovery at 22 °C, we determined each plant’s above-soil dry weight. All measurements occurred at midday, with care taken not to disrupt plant development, as previously described (Lunn et al., 2015). Rosette diameter was calculated by averaging the distance between the tips of four pairs of true leaves measured on each plant. After recovery, we cut the rosettes from the root at soil level and placed the biomass in aluminum for drying overnight at 120 °C before weighing.
In silico protein analysis and phylogeny
All Arabidopsis protein sequences and predicted sizes were obtained from the Arabidopsis database at TAIR (https://www.arabidopsis.org/). The consensus sequence of the Mpv17_PMP22 domain was obtained from the Pfam database (https://pfam.xfam.org/). Homologous proteins of other plant species were identified through NCBI protein BLAST analysis (https://blast.ncbi.nlm.nih.gov/). Members of the Arabidopsis MPD family were identified by searching the TAIR database using the site’s BLAST tool (https://www.arabidopsis.org/Blast/index.jsp). Protein and domain relationships were analyzed by Geneious v. R6, Biomatters Inc. New Zealand, using default alignment settings. Predicted protein localizations for the Arabidopsis MPD proteins are those listed in the SUBA4 database (https://suba.live/) (Hooper et al., 2017). The putative plastid targeting sequence for MPD1 was obtained by analysis with Ipsort_(http://ipsort.hgc.jp).
Subcellular localization of MPD1
We amplified the MPD1 coding sequence without a stop codon and cloned it into pENTR-d-TOPO (Invitrogen). We recombined the expression cassette into vector pB7FGW2 with the Gateway cloning system (Invitrogen) (Karimi et al., 2002), fusing the ORF to the enhanced green fluorescent protein (eGFP) fluorescent marker. Using Agrobacterium strain GV3101, we infiltrated Nicotiana benthamiana leaves employing a solution containing 10 mM magnesium chloride and 100 µM acetosyringone. After 48 h, we examined leaves with a Leica SP8 laser-scanning confocal microscope at emission wavelengths of 509 nm for GFP and 650 nm for chlorophyll autofluorescence.
Generation of the MPD1-OX3 line
The coding sequence of MPD1 was cloned into pENTR and sequenced for fidelity. We transferred the gene into the binary plasmid PB2GW7 under control of the Cauliflower mosaic virus (CaMV) 35S promoter (Karimi et al., 2002) and transformed it into Col-0 using floral dip via Agrobacterium tumefaciens GV3102 (Clough and Bent, 1998). Using BASTA resistance and pedigree analysis, we identified homozygous T2 progeny with MPD1 expression 3-fold higher than Col-0 for detailed investigation.
Photosynthetic fluorescence analysis
Photosynthetic parameters of plants were determined using a FluroCam FC 800-C (Photon Systems Instruments). For Fv/Fm, all plants were dark-adapted for 1 h before analysis while for ΦPSII we directly transferred plants to the FluroCam. Chlorophyll concentration was measured by homogenizing true leaves in 80% (v/v) acetone and then measuring absorption at 663 nm and 645 nm, before calculating by established methods (Porra, 2002).
Determination of H2O2 concentrations
For measurements of tissue H2O2, we used an Amplex Red™ Hydrogen Peroxide Assay Kit (Invitrogen) according to the manufacturer’s instructions. Leaf samples (~250 mg) were harvested from three plants in each of three independent experiments. Samples were immediately weighed and frozen in liquid nitrogen. After grinding in liquid nitrogen, 1 ml of ice-cold phosphate buffer (50 mM KH2PO4, pH 7.4) was added and the samples were incubated at 4 °C with shaking for 30 min. This extraction protocol is similar to others using pH 7.4 phosphate buffer (Chakraborty et al., 2016) but maintains samples at 0–4 °C to minimize post-harvest changes in H2O2. After extraction, samples were centrifuged at 12 000 g at 4 °C for 10 min. Three replicate, 100 µl aliquots of each supernatant were immediately transferred to wells of a 96-well plate containing 100 µl of freshly prepared Amplex Red™ mixture (100 µM 10-acetyl-3,7-dihydroxyphenoxazine, 0.2 U ml–1 horseradish peroxidase) and incubated in the dark at room temperature for 30 min. Each plate also contained H2O2 standards (0, 1, 2, 3, 4, 5, and 6 µM H2O2 in phosphate buffer). Quantitation of the standards and samples was made by fluorescence (excitation at 530 nm; detection at 580 nm) in a microplate reader. The concentration of H2O2 in each replicate was calculated from the standard curve and averaged to give the value for each biological replicate.
Quantitative PCR for gene transcription levels
To measure transcript levels of MPD1 and ROS marker genes, we harvested leaves from three independent replicates of each of the experimental lines into liquid nitrogen and stored them at –80 °C. We extracted RNA with the RNeasy Plant Mini Kit (Qiagen), digested with DNase I, then assessed RNA quality and standardized the concentration to 100 ng μl–1. We synthesized cDNA with the Superscript III First-Strand Synthesis System (Life Technologies) and verified equivalence by amplification with the reference tubulin beta chain 2 gene (At5g62690; TUB2). Quantitative PCR was performed by transferring aliquots of a master mix of each cDNA plus Platinum SYBR Green (Invitrogen) into tubes containing appropriate specific primers followed by amplification on a RealPlex Mastercycler (Eppendorf) with melting curve analysis, then by size verification by gel electrophoresis. We calculated transcript abundance for each gene analyzed relative to TUB2 using the primers listed in Supplementary Table S1.
Accession numbers
Sequence data can be found in the Arabidopsis Genome Initiative database (arabidopsis.org): MPD1 (At4g03410), Arabidopsis MPD1 homologs (At4g14305, At1g52870, At5g19750, At2g42770, At2g14860, At4g33905, At3g24570, At5g43140, and At4g04470), ZAT12 (At5g18340), tAPX (At1g77490), DHAR3 (At5g16710), SOD2 (At2g28190), DHAR1 (At1g19570), SOD1 (At1g08830), and CAT1 (At1g20630).
Results
Identification of a chilling-sensitive allele in Arabidopsis
To identify new genes involved in chilling, we screened 1244 independent single homozygous T-DNA SALK insertion lines generated in the Col-0 background from the Arabidopsis Biological Resource Center (ABRC) (Alonso et al., 2003) for reduced chilling tolerance. We grew three plants from each line for 2 weeks at 22 °C, removed lines with visible defects, then transferred the remaining lines to 4 °C, monitoring their growth. After 4 weeks chilling, all three SALK_031932C plants were smaller than the wild type and their leaves displayed yellow patches. We transferred these to 22 °C and grew them to maturity, harvesting seed to propagate for further analysis.
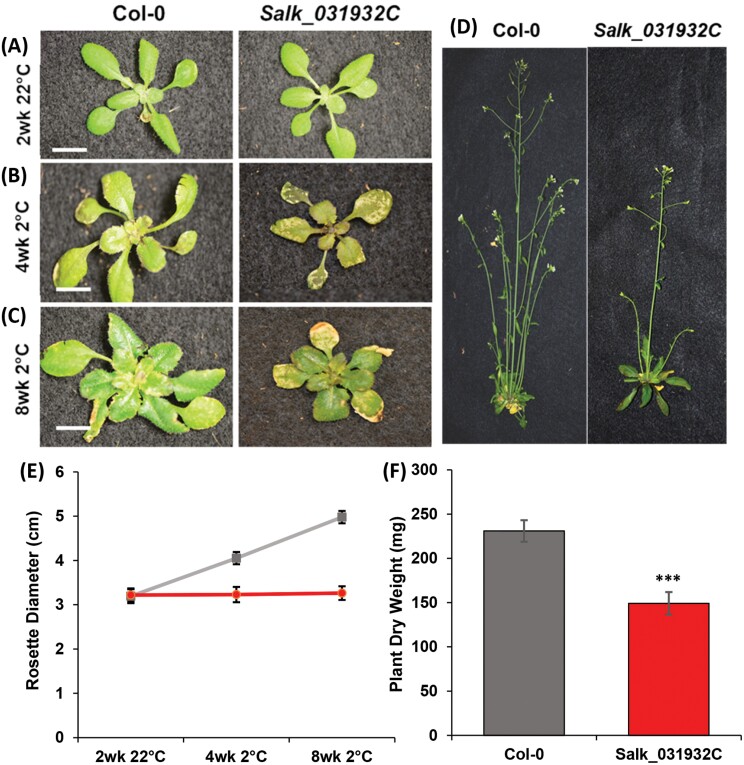
We investigated the SALK_031932C phenotype by cultivating three independent batches of 10 plants alongside Col-0 controls with a regimen of 2 weeks at 22 °C, followed by 8 weeks at 2 °C, and then recovery for 2 weeks at 22 °C. After the initial 2 weeks at 22 °C, no difference between mutant and control plants was visible, all having healthy green leaves of similar size (Fig. 1A). During the 8 weeks at 2 °C, Col-0 plants produced healthy leaves and expanded rosettes, but the rosettes of SALK_031932C did not expand and the leaves showed extensive chlorosis with necrotic areas evident after 4 weeks of chilling (Fig. 1B). Mutant plants remained smaller than Col-0 through 8 weeks of growth at 2 °C and, in contrast to healthy expanding new Col-0 leaves, emerging leaves of SALK_031932C developed chlorotic patches and remained small (Fig. 1C). After returning the plants to 22 °C for 2 weeks, SALK_031932C plants showed delayed flowering and seed production relative to Col-0 (Fig. 1D).
Fig. 1.
Chilling-sensitive phenotype of SALK_031932C. (A) SALK_031932C plants are similar to Col-0 after 2 weeks growth at 22 °C. (B) Mutant plants exhibit extensive damage 4 weeks after transfer to 2 °C. (C) Mutant plants remain small after 8 weeks at 2 °C, with new leaves having chlorotic and necrotic areas. (D) After 2 weeks recovery at 22 °C, mutant plants produce seeds but remain smaller than Col-0. (E) Rosette diameters of Col-0 (gray) and SALK_031932C (red) plants. (F) Dry weights after 2 weeks recovery at 22 °C. (A–C) Scale bar is 1 cm. (E, F) Means ±SD for n=30 plants (10 in each of three independent replicates); Student’s t-test confirmed equivalence at 22 °C and difference at 2 °C (P<0.001) in (E), with ∗∗∗ denoting significance in (F).
When we measured rosette diameters and dry weights of SALK_031932C and Col-0 during 8 weeks normal growth at 22 °C, mutant plants were similar to the wild type (Supplementary Table S2). In our chilling experiments, all plants likewise had equal rosette diameters after 2 weeks at 22 °C, both SALK_031932C and Col-0 plants averaging 3.22 ± 0.14 cm (Fig. 1E). After transfer to 2 °C, however, rosette diameters diverged steadily. After 8 weeks at 2 °C, Col-0 rosette diameter had increased by 1.5-fold to 4.98 ± 0.14 cm, whereas SALK_031932C rosettes showed no significant expansion, with a mean of 3.26 ± 0.10 cm (Fig. 1E). After their return to 22 °C, the mutant plants remained smaller than controls. After 2 weeks recovery at 22 °C, the dry weight of SALK_031932C plants averaged only 149 ± 12 mg, 64% of the 231 ± 12 mg of Col-0 (Fig. 1F), indicating that the retarded growth of SALK_031932C plants under chilling fails to recover on return to 22 °C.
Arabidopsis At4g03410 encodes a chloroplast-localized Mpv17_PMP22 domain protein
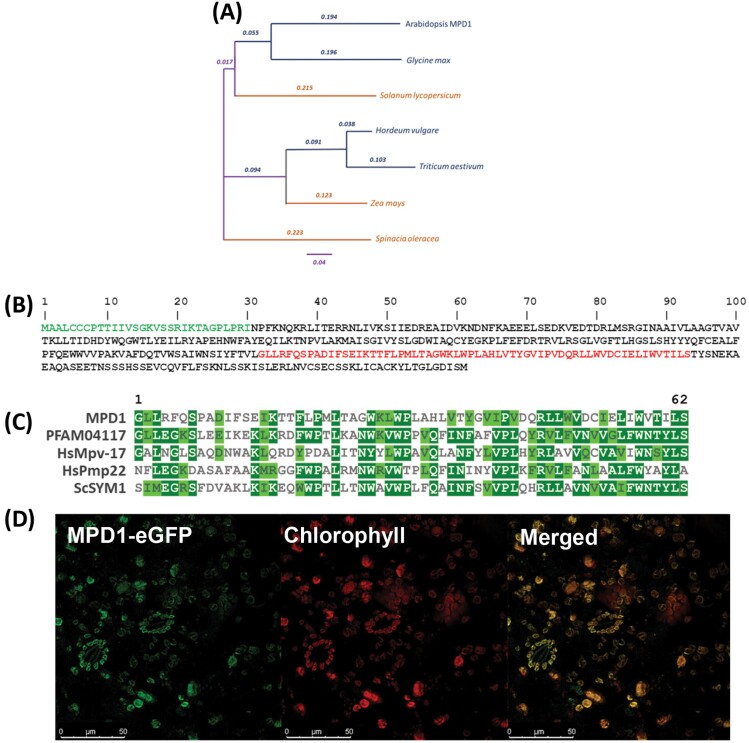
The T-DNA insertion site in SALK_031932C is in At4g03410, 123 bp upstream of the start codon and interrupting the transcript coding sequence (https://seqviewer.arabidopsis.org). Arabidopsis At4g03410 consists of six exons and five introns encoding a predicted protein of 361 amino acids (~41 kDa). A BLAST search of the NCBI protein database with this sequence revealed homologous sequences in a wide variety of both chilling-tolerant and chilling-sensitive plant species (Fig. 2A). A search of the NCBI Conserved Domains Database discovered that a Mpv17_PMP22 domain of 62 residues, Pfam 04117, was included in the predicted protein of At4g03410 (Fig. 2B). This domain, widely conserved in eukaryotes, is named after two mammalian proteins, Mpv17, a mouse mitochondrial inner membrane protein, and PMP22 (also Pxmp2; Brosius et al. 2002) a human peroxisomal membrane protein; a yeast mitochondrial protein (Sym1p) has also been characterized (Trott and Morano, 2004) (Fig. 2C). All characterized Mpv17_PMP22 domain proteins are integral membrane proteins, and analysis of the At4g03410 protein product with HMMTOP revealed five likely transmembrane regions (Supplementary Fig. S1). Accordingly, we named At4g03410 Mpv17 PMP22 Domain 1 (MPD1) and the SALK_031932C mutant line mpd1-1.
Fig. 2.
At4g03410 encodes a chloroplast-localized Mpv17_PMP22 domain protein. (A) Protein homologs of MPD1 in chilling-tolerant (blue) and chilling-sensitive (orange) plants. (B) MPD1 protein sequence with predicted chloroplast targeting sequence in green and the Mpv17_PMP22 domain in red. (C) Arabidopsis MPD1 Mpv17_PMP22 domain sequence homology to human HsMpv-17, HsPMP22, yeast ScSYM1, and Pfam 04117 consensus. (D) MPD1 C-terminal fused eGFP localization in transfected N. benthamiana leaves. Representative false-color image of MPD1–eGFP (509 nm emission, green), chloroplast autofluorescence (650 nm emission, red), and merged channels. Scale bars are given at the bottom left of each panel.
The BLAST tool at TAIR (http://arabidopsis.org) revealed nine other Arabidopsis proteins with significant homology to MPD1 (At4g14305, At1g52870, At5g19750, At2g42770, At2g14860, At4g33905, At3g24570, At5g43140, and At4g04470). While each of these proteins contain the Mpv17_PMP22 domain, their size, predicted subcellular targeting, and sequence similarity vary significantly (Supplementary Table S3). Consistent with homology to characterized Mpv17 and PMP22 proteins, the 10 Arabidopsis proteins include several putative mitochondrial and peroxisomal members, two of which (At4g04470 and At3g24570) have been described in biochemical studies (Tugal et al., 1999; Wi et al., 2020). Three family members are predicted to be chloroplast localized; MPD1, At1g52870, and At5g19750 (Supplementary Table S3).
In the first 30 residues of the MPD1 gene product, a putative chloroplast targeting signal was predicted by iPSORT (http://ipsort.hgc.jp) (Fig. 2B), and the Subcellular Location of Proteins in Arabidopsis Database (SUBA; https://suba.live/aboutSUBA4.html) (Hooper et al., 2017) predicted plastid localization for MPD1. To determine protein localization, we amplified the MPD1 ORF without a stop codon and fused this, at its C-terminus, to a DNA sequence encoding the fluorescent eGFP marker protein. After expression of the fusion under viral CaMV 35S promoter control in infiltrated N. benthamiana leaves, we examined both GFP fluorescence and chlorophyll autofluorescence with a confocal laser-scanning microscope. When the two channels were merged, the fluorescence signals overlapped, indicating that MPD1:eGFP localized to chloroplasts (Fig. 2D), confirming the in silico predictions and demonstrating that MPD1 is a chloroplast-localized protein.
Transcript analysis reveals that mpd1-1 is an overexpression mutant
We identified a second T-DNA line, Sail_583_B07, with an insertion in the fifth exon of the MPD1 gene, designating it mpd1-2, and after confirmation of the T-DNA insertion site we identified a homozygous subline by progeny analysis. However, when tested under our chilling treatment, this second allele did not exhibit a chilling phenotype. After 4 weeks at 2 °C, mpd1-2 plants were similar to Col-0 in appearance, not showing chlorosis and necrosis evident in mpd1-1 (Supplementary Fig. S2A). At the end of the experiment, following 2 weeks recovery at 22 °C, the appearance and dry weights of mpd1-2 and Col-0 plants were indistinguishable, while mpd1-1 were smaller with significantly lower dry weight (Supplementary Fig. S2B, C).
These results indicated that mpd1-1 chilling sensitivity is not caused by loss of At4g03410 expression. The T-DNA insertion in mpd1-1, upstream of the ATG start codon, probably uncouples the MPD1 ORF from endogenous promoter control, leaving the possibility that MPD1 is produced under control of a promoter within the T-DNA. Transcript expression is sometimes driven by a promoter in an upstream T-DNA (Okuley et al., 1994); one study concluded that it occurs in 5% of upstream insertions (Wang, 2008). To test this, we extracted RNA from leaves cultivated for 2 weeks at 22 °C, or after 4 weeks of chilling, and measured transcript abundance by quantitative reverse transcription–PCR (qRT–PCR) of the MPD1 coding region. At 22 °C, transcript levels of MPD1 in Col-0 averaged 0.98 ± 0.01 [normalized to tubulin beta chain 2 (TUB2, At5g62690)]. In contrast, expression levels from mpd1-1 leaves averaged 3.77 ± 0.34, a 3.8-fold increase above Col-0 (Supplementary Fig. S2D). Expression in the mpd1-2 mutant was only 0.06 ± 0.01. Neither Col-0 nor mpd1-1 expression changed significantly when chilled, averaging 0.94 ± 0.16 and 3.63 ± 0.45 (3.9-fold above Col-0). Thus, mpd1-1 is a constitutive overexpression allele of At4g03410 and we inferred that this overexpression was the likely cause of the observed phenotypes.
Overexpressing MPD1 causes chilling sensitivity in Arabidopsis
To test whether overexpression caused chilling sensitivity, we created additional constitutive overexpression alleles of At4g03410 by expressing the MPD1 coding sequence under CaMV 35S promoter control. After Agrobacterium-mediated transformation of Col-0 plants, followed by Basta selection, we identified lines with overexpression equivalent to mpd1-1 for analysis. We screened homozygous T2 lines by qRT–PCR and identified two lines, MPD1-OX3 and MPD1-OX7, with 3-fold overexpression of MPD1 (Supplementary Fig. S3A), and from the MPD1-OX3 parent we retained an untransformed isogenic sibling segregant (iso-OX3) to serve as an additional control. Plants of all three overexpression lines looked like Col-0 controls after 2 weeks growth at 22 °C (Supplementary Fig. 3A), but the MPD1-OX3, MPD1-OX7, and mpd1-1 lines all exhibited reduced growth and tissue damage 4 weeks after transfer to 2 °C (Supplementary Fig. S3B). We proceeded to characterize the MPD1-OX3 line in detail.
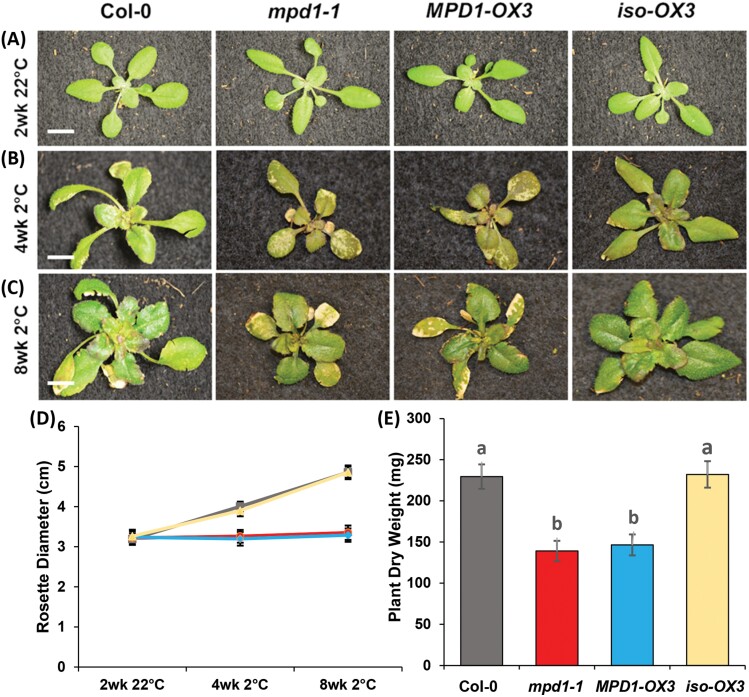
To compare phenotypes among Col-0, mpd1-1, MPD1-OX3, and iso-OX3, we subjected 10 plants of each to our established chilling regimens in three independent experiments. After 2 weeks at 22 °C, all plant lines were similar in growth, with healthy green leaves (Fig. 3A). The Col-0 plants continued healthy growth for 4 weeks after transfer to 2 °C, while mpd1-1 plants were smaller with yellow patches on their leaves as seen before. The MPD1-OX3 overexpression plants were similar to mpd1-1 plants, with unexpanded rosettes and yellow spots across their leaves, but untransformed iso-OX3 plants were like Col-0 (Fig. 3B). These differences continued for the 8 weeks chilling regime, as Col-0 and iso-OX3 expanded, but both mpd1-1 and MPD1-OX3 overexpression alleles remained small, with leaf chlorosis and necrosis (Fig. 3C). After transfer to 22 °C, mpd1-1 and MPD1-OX3 plants remained smaller than the Col-0 and iso-OX3 controls (Supplementary Fig. S4).
Fig. 3.
Overexpression of MPD1 causes chilling damage. (A) After 2 weeks growth at 22 °C, plants of mpd1-1 and MPD1-OX3 are similar to Col-0 and the isogenic segregant iso-OX3. (B, C) After 4 (B) and 8 (C) weeks at 2 °C, mpd1-1 and MPD1-OX3 exhibit chilling damage. (D) Rosette diameters of Col-0 (gray), mpd1-1 (red), MPD1-OX3 (blue), and iso-OX3 (yellow). (E) Dry weights after 2 weeks recovery at 22 °C. (A–C) Scale bar is 1 cm. (D, E) Means ±SD for n=30 plants (10 each in three independent replicates); analysis by one-way ANOVA with post-hoc Tukey test confirmed equivalences at 22 °C and differences at 2 °C (P<0.001) in (D), in (E) letters denote significance.
When we quantified phenotypes, plants from all the lines grown only at 22 °C remained similar throughout development (Supplementary Table S4). In the chilling experiment, after 2 weeks at 22 °C, rosettes of Col-0 averaged 3.19 ± 0.14 cm, and all the others were similar (Fig. 3D). After 8 weeks at 2 °C, Col-0 and iso-OX3 plants expanded to 4.86 ± 0.17 cm and 4.86 ± 0.14 cm, respectively. However, rosettes of mpd1-1 and MPD1-OX3 grew very little at low temperature, averaging 3.28 ± 0.15 cm and 3.23 ± 0.17 cm, respectively (Fig. 3D). After recovery at 22 °C, all lines reached maturity, but mpd1-1 and MPD1-OX3 plants weighed considerably less than the controls, averaging 139 ± 12 mg and 147 ± 13 mg, respectively, compared with 230 ± 15 mg for Col-0 and 232 ± 16 mg for iso-OX3 (Fig. 3E). Collectively, analyses of the T-DNA insertion and overexpression lines we generated establish that overexpression of MPD1 renders Arabidopsis chilling sensitive.
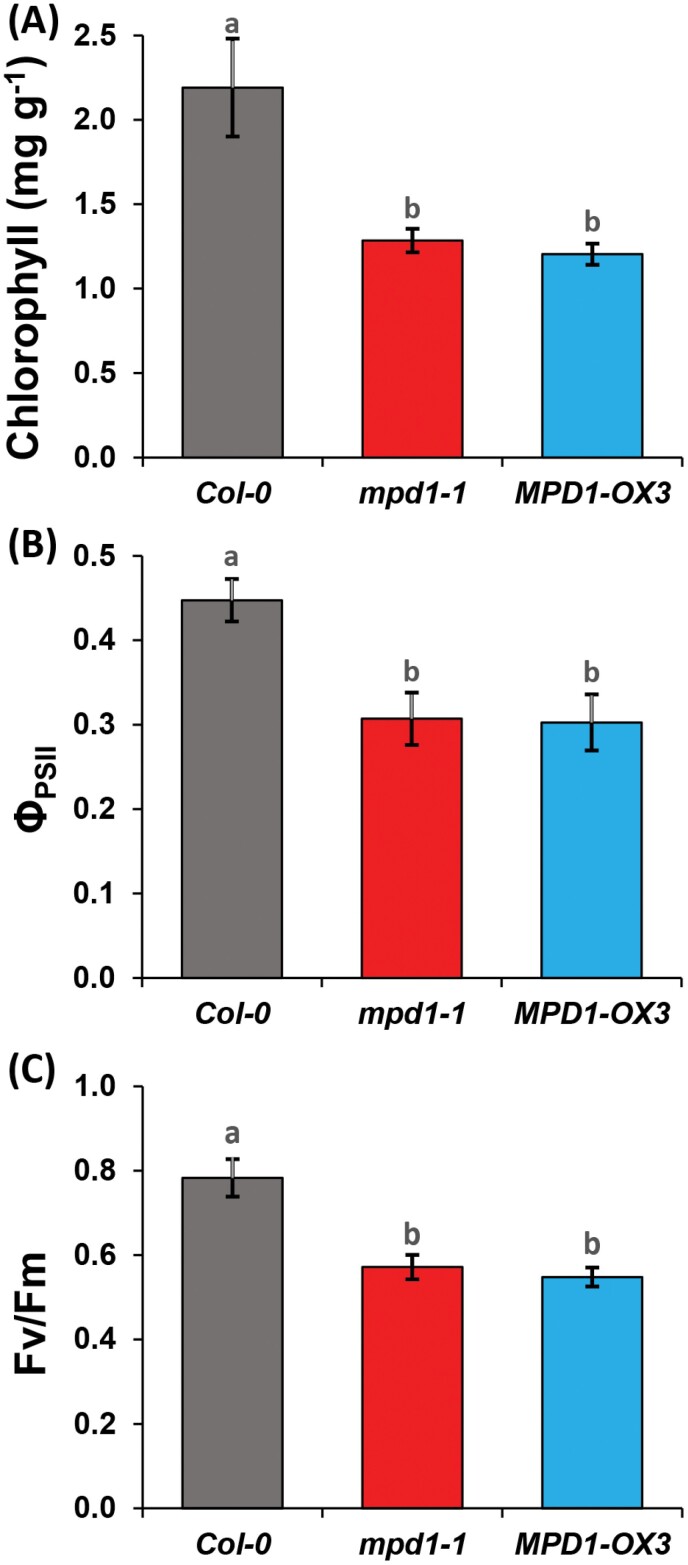
Overexpressing MPD1 reduces photosynthetic capacity during chilling
The chlorosis of mpd1-1 leaves under chilling conditions suggested impaired photosynthetic capacity. We evaluated photosynthesis after 4 weeks at 2 °C by first measuring chlorophyll content, which was 2.19 ± 0.29 mg g–1 of leaf in the Col-0 parent, but mpd1-1 and MPD1-OX3 were ~40% lower, averaging 1.28 ± 0.07 mg g–1 and 1.20 ± 0.06 mg g–1, respectively (Fig. 4A). The steady-state efficiency of PSII (ΦPSII) for Col-0 was 0.44 ± 0.02, but ΦPSII for mpd1-1 and MPD1-OX3 were 27% lower than for Col-0 and equivalent to each other, averaging 0.32 ± 0.03 and 0.35 ± 0.02 (Fig. 4B). When we measured the maximum quantum efficiency of dark-adapted plants (Fv/Fm), that for Col-0 was 0.78 ± 0.04, but mpd1-1 and MPD1-OX3 were >25% lower, at 0.57 ± 0.02 and 0.54 ± 0.02, respectively (Fig. 4C). Therefore, under chilling conditions, both MPD1 overexpression alleles suffer greatly reduced photosynthetic capacity.
Fig. 4.
Overexpression of MPD1 reduces photosynthesis at low temperature. (A) Chlorophyll contents of mpd1-1 and MPD1-OX3 are reduced relative to Col-0 and mpd1-2 after 4 weeks at 2 °C. (B, C) Fluorescence parameters ΦPSII (B) and Fv/Fm (C) are reduced in mpd1-1 and MPD1-OX3 relative to Col-0. (A–C) Means ±SD for n=30 plants (10 each in three independent replicates) analysis by one-way ANOVA and post-hoc Tukey test; letters denote significance (P<0.001).
Overexpression of MPD1 increases H2O2 accumulation during chilling
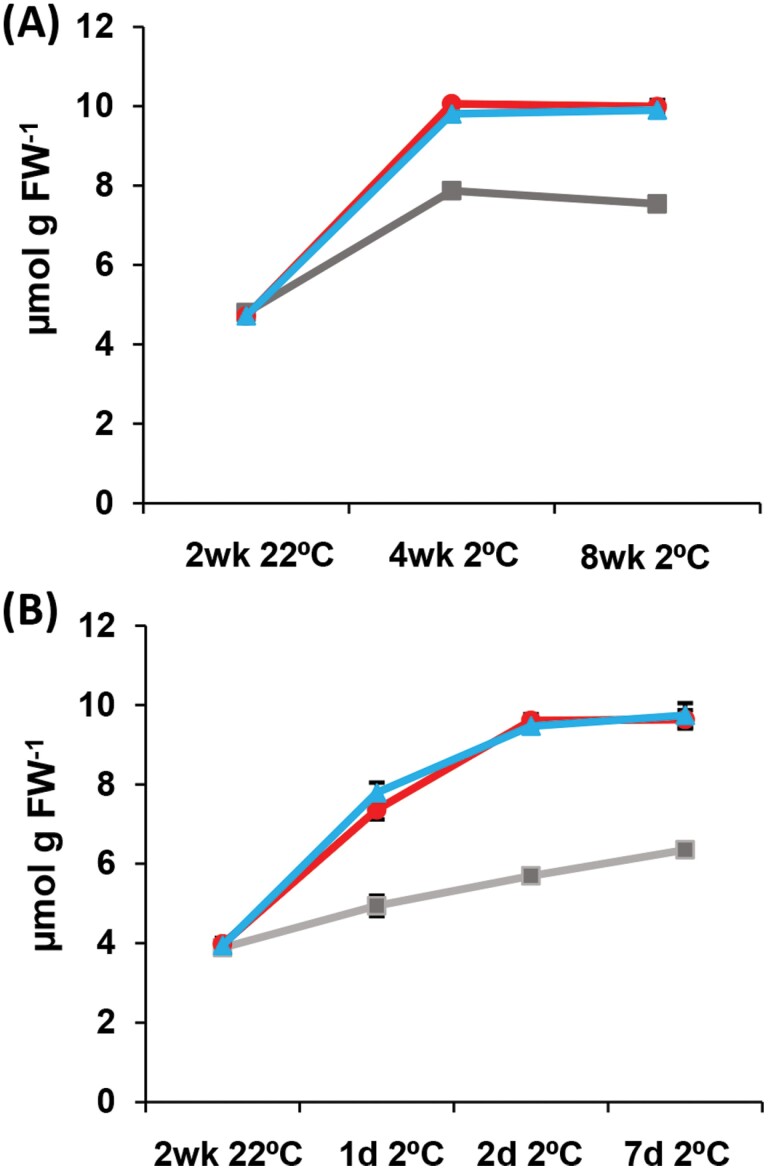
High levels of ROS can produce leaf damage, lower chlorophyll content, and reduce fluorescence parameters like those measured at low temperature in mpd1-1 and MPD1-OX3 plants. High levels of H2O2 are diagnostic of ROS excess, as this intermediate accumulates under high ROS stress (Smirnoff and Arnaud, 2019), so we measured H2O2 levels of leaves in Col-0, mpd1-1, and MPD1-OX3 lines. After 2 weeks at 22 °C, H2O2 levels in leaves of all lines were similar to those of Col-0, which averaged 4.80 ± 0.12 μmol g–1 FW (Fig. 5A). In Col-0, H2O2 concentration predictably rose under chilling conditions to 7.53 ± 0.14 μmol g–1 FW after 8 weeks. The H2O2 concentration in mpd1-1 leaves was greater, reaching 9.99 ± 0.16 μmol g–1 FW, 1.3-fold higher than that of Col-0 at 8 weeks, and the increase was matched in the second overexpression allele, MPD1-OX3, at 9.90 ± 0.18 μmol g-1 FW (Fig. 5A).
Fig. 5.
H2O2 concentration increases during chilling of plants overexpressing MPD1. (A) H2O2 concentrations in leaf tissue of Col-0 (gray), mpd1-1 (red), and MPD1-OX3 (blue) over 8 weeks at 2 °C. (B) H2O2 concentration over 7 d at 2 °C. (A, B) Means ±SD for n=30 plants (10 each in three independent replicates); analysis by one-way ANOVA with post-hoc Tukey test confirmed equivalences at 22 °C and differences at 2 °C (P<0.001).
If increased ROS stress causes leaf damage in the overexpression lines, we reasoned that the H2O2 levels would rise before onset of visible symptoms. We measured H2O2 again, sampling the first true leaf before the plants were transferred to the chilling treatment, and sampling again after 1, 2, and 7 d at 2 °C. All lines grown at 22 °C were similar to the Col-0 average of 3.88 ± 0.13 μmol g–1 FW H2O2 (Fig. 5B). H2O2 in Col-0 leaves increased by 65% during chilling, averaging 6.35 ± 0.13 μmol g–1 FW by day seven. However, chilling produced a 2.5-fold increase in H2O2 in mpd1-1 and MPD1-OX3, as leaf concentrations averaged 9.64 ± 0.23 μmol g–1 FW and 9.74 ± 0.30 μmol g–1 FW (Fig. 5B), a 1.5-fold increase above Col-0 levels. These results suggest that overexpression of MPD1 may either cause increased ROS production or adversely affect the signaling processes and gene induction that is required for protection against ROS stress.
Strong induction of ROS management genes during chilling in plants overexpressing MPD1
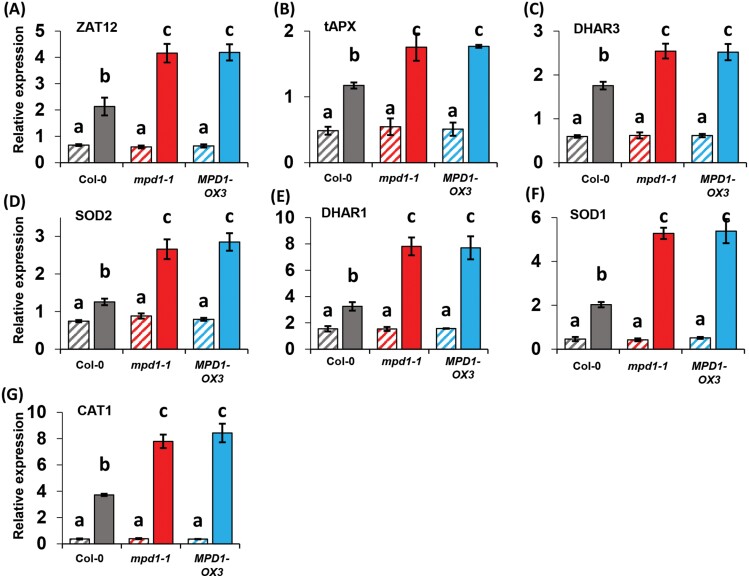
Increased H2O2 concentrations during ROS stress act as a signal, inducing expression of transcription factors and downstream genes that ameliorate ROS stress and avoid cellular damage (Smirnoff and Arnaud, 2019). A transcription factor gene central to this response is ZAT12 that is strongly induced by H2O2 signaling (Davletova et al., 2005). The transcriptional cascade induces expression of genes encoding enzymes important to ROS breakdown including SOD, APX, DHAR, and CAT (Dietz et al., 2016; Dreyer and Dietz, 2018; Smirnoff and Arnaud, 2019). To investigate gene expression responses to chilling-induced H2O2 accumulation in our lines, we first measured the transcript levels of ZAT12 and six structural genes of the ROS response in plants grown at 22 °C for 2 weeks. Col-0 plants expressed ZAT12 at a level of 0.67 ± 0.03 relative to the TUB2 gene used as internal control, and ZAT12 expression in the overexpression alleles was indistinguishable (Fig. 6A). Transcript levels of the six ROS metabolism genes that we measured exhibited a similar pattern of uniform, modest expression across the three plant lines (Fig. 6B–G).
Fig. 6.
Increased expression of ROS management genes in plants overexpressing MPD1. (A) For Col-0, mpd1-1, and MPD1-OX3 grown at 22 °C, ZAT12 expression is comparable (hatched bars). After transfer to 2 °C for 2 weeks, transcript levels are higher in mpd1-1 and MPD1-OX3 than in Col-0 (solid bars). (B–G) Genes encoding enzymes of ROS metabolism similarly show greater induction by chilling in mpd1-1 and MPD1-OX3 than in Col-0. The genes encode CAT1, catalase; DHAR1 and 3, dehydroascorbate reductase isozymes; SOD1 and 2, superoxide dismutase isozymes; and tAPX, thylakoid ascorbate peroxidase. Data are means ±SD for n=3 samples (10 plants in each of three independent replicates). Statistical analysis: one-way ANOVA and post-hoc Tukey test, letters denote significance (P<0.001).
The transcript level of ZAT12 increased 3.2-fold in Col-0 under chilling treatment, consistent with its role in coordinating responses to ROS stress and increased H2O2 concentration. In both lines overexpressing MPD1, ZAT12 was induced more than twice as strongly as in Col-0, averaging a 6.9-fold increase in mpd1-1 and 6.7-fold in MPD1-OX3 (Fig. 6A). The six other ROS response genes were also induced in Col-0 plants at 2 °C, ranging from ~1.3-fold (for SOD2) to >10-fold (for CAT1) above levels measured at 22 °C (Fig. 6B–G). However, for every gene analyzed, the level of induction was considerably higher in MPD1 overexpression lines. Compared with Col-0, the mpd1-1 and MPD1-OX3 plants ranged between 30% and 140% stronger induction of these six genes (Fig. 6B–G). Our data indicate that accumulation of higher H2O2 levels in chilled plants overexpressing MPD1 compared with Col-0 controls is associated with a more robust transcriptional response, both for the gene encoding the ZAT12 transcription factor and for genes encoding enzymes required for removal of excess ROS. We infer that these lines have an appropriate response to increased tissue concentrations of H2O2.
Overexpression of MPD1 increases paraquat susceptibility
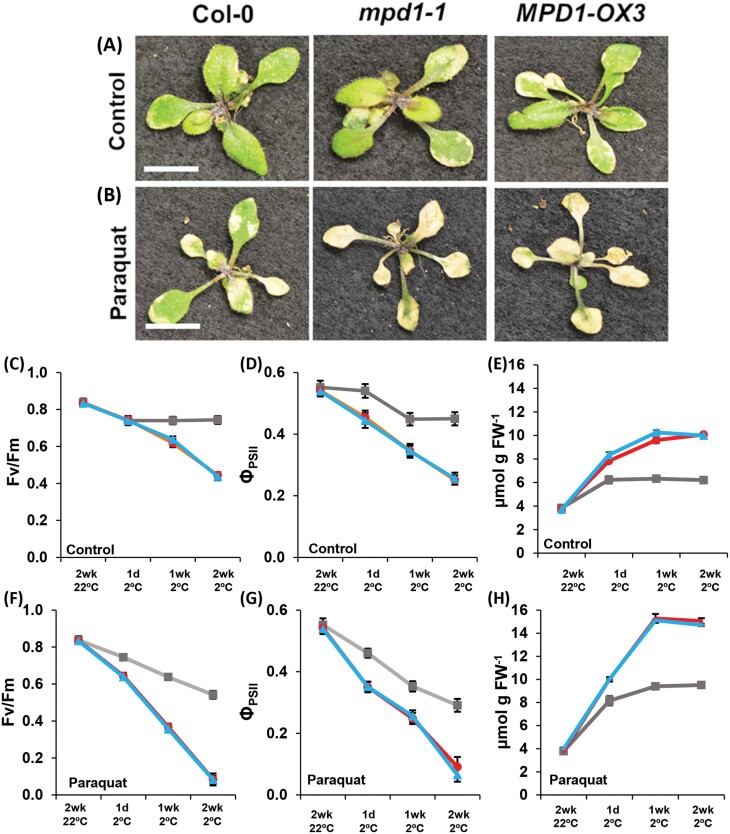
Our results show that plants overexpressing MPD1 produce high levels of ROS when chilled, overwhelming the management systems induced in mpd1-1 and MPD1-OX3 leaves. To explore ROS effects in Col-0 and the overexpression lines, we treated plants with a low concentration of paraquat (N,Nʹ-dimethyl-4,4ʹ-bipyridinium dichloride), a redox compound that interacts with electron transfer chains in chloroplasts and mitochondria to produce superoxide. High concentrations of paraquat disrupt cellular function and result in tissue death (Hawkes, 2014); however, lower concentrations cause more gradual damage, allowing for comparisons of ROS susceptibility between plants (Moustaka and Moustakas, 2014). We applied either distilled water or 5 mM paraquat every 2 d during 2 weeks of chilling. Col-0 plants remained healthy after distilled water application, while mpd1-1 and MPD1-OX3 displayed the same symptoms of leaf chlorosis and necrosis as observed before (Fig. 7A). With paraquat application, Col-0 developed chlorotic patches similar to the two MPD1-overexpressing lines in the absence of paraquat. Both mpd1-1 and MPD1-OX3 treated with paraquat suffered even more severe damage than Col-0, exhibiting completely yellow leaves after 2 weeks of treatment at 2 °C (Fig. 7B).
Fig. 7.
Paraquat treatment phenocopies chilling damage in Col-0 plants. (A) Representative images from the three experimental lines sprayed with water during 2 weeks at 2 °C (control treatment). (B) Increased tissue damage, relative to controls, in plants sprayed with 5 mM paraquat. (C–E) Fv/Fm (C), ΦPSII (D), and H2O2 concentration (E) from control plants after 2 weeks at 2 °C. (F–H) Fv/Fm (F), ΦPSII (G), and H2O2 concentration (H) from paraquat-treated plants after 2 weeks 2 °C. (A, B) Scale bars are 1 cm. (C–H) Col-0 (gray), mpd1-1 (red), and MPD1-OX3 (blue). (C–H) Means ±SD for n=30 plants (10 in each of three independent replicates); analysis by one-way ANOVA with post-hoc Tukey test confirmed equivalences at 22 °C and differences at 2 °C (P<0.001).
To quantify the treatment effects, we measured Fv/Fm, ΦPSII, and H2O2. After 2 weeks of growth at 22 °C, all values were similar to those of Col-0. The average values across all three lines were Fv/Fm=0.84 ± 0.02, ΦPSI =0.55 ± 0.02, and H2O2=3.76 ± 0.05 μmol g–1 FW (Fig. 7C–E). Treated with water only, Col-0 at 2 °C exhibited a modest initial decline in Fv/Fm and ΦPSII before stabilizing, averaging 0.74 ± 0.02 (Fv/Fm) and 0.45 ± 0.02 (ΦPSII) after 2 weeks chilling, while leaf H2O2 levels increased to 6.21 ± 0.20 μmol g–1 FW. Consistent with earlier results (Figs 5, 6), the fluorescent parameters of mpd1-1 declined to 0.45 ± 0.04 (Fv/Fm) and 0.25 ± 0.02 (ΦPSII), and leaf H2O2 concentration increased from 3.76 ± 0.08 μmol g–1 FW before transfer from 22 °C to 10.08 ± 0.11 μmol g–1 FW at 2°C. As before, the MPD1-OX3 line was similar to mpd1-1 for all three parameters (Fig. 7C–E).
With paraquat treatment, fluorescent parameters in Col-0 declined continuously (Fig. 7F, G); Fv/Fm decreased to 0.54 ± 0.02 and ΦPSII to 0.29 ± 0.02, while the concentration of H2O2 in leaves of Col-0 plants treated with paraquat increased to 9.51 ± 0.24 μmol g–1 FW (Fig. 7H). Significantly, these values approach those of the lines overexpressing MPD1 in the absence of paraquat, indicating a similar level of ROS stress in these differently treated plants. In mpd1-1 and MPD1-OX3 plants, paraquat treatment resulted in almost complete loss of photosynthetic function, with both Fv/Fm and ΦPSII ratios falling below 0.1 after 2 weeks (Fig. 7F, G). Paraquat application increased H2O2 levels in both overexpression lines, to 15.07 ± 0.23 μmol g–1 FW (mpd1-1) and 14.72 ± 0.11 μmol g–1 FW (MPD1-OX3) (Fig. 7E). These data demonstrate that paraquat treatment of Col-0 produces biochemical and physiological effects that mimic MPD1 overexpression in the absence of paraquat, while greatly exacerbating ROS stress in lines with increased MPD1 expression.
Discussion
When crops native to tropical or subtropical habitats are introduced into temperate regions, they typically suffer reduced growth and tissue damage at non-freezing temperatures between 0 °C and 10 °C (Lyons, 1973; Lukatkin et al., 2012; Barrero-Gil et al., 2016). Determining the biochemical and molecular processes required for chilling tolerance is key to improving our biological understanding of chilling-sensitive species and to enabling amelioration of chilling damage, through either marker-assisted breeding or genetic engineering. Previous screens for chilling-sensitive Arabidopsis mutants have successfully identified genes specifically required for chilling tolerance (Tokuhisa et al., 1997, 1998; Gao et al., 2017). To extend this approach, we screened a collection of homozygous, sequence-verified T-DNA insertion Arabidopsis mutants for loss of chilling tolerance. One mutant line identified from our screen, SALK_031932C, has a T-DNA insertion at locus At4g03410 encoding a protein containing an Mpv17_PMP22 domain (Pfam 04117). This domain was first identified in mice, where misexpression or mutation of the Mpv17 locus led to kidney disease (Weiher et al., 1990). Studies in mammals have identified proteins with this domain localized to peroxisomes and mitochondria (Zwacka et al., 1994; Rokka et al., 2009; Spinazzola et al., 2006; Antonenkov et al., 2015), primary locations for ROS generation and regulation. In yeast, the Mpv17_PMP22 domain protein, Sym1p, localizes to the mitochondria (Trott and Morano, 2004). For different members of the Mpv17_PMP22 family in mammals, both loss-of-function mutations (Trott and Morano, 2004; Casalena et al., 2014) and overexpression (Zwacka et al., 1994; Iida et al., 2006) are associated with dysregulation of ROS balance, changed expression of ROS-related genes, and increased susceptibility to ROS-inducing treatments.
The SALK T-DNA line with an insertion in the 5ʹ region of At4g03410 exhibited dramatic sensitivity to chilling conditions (Fig. 1). When a separate line with an insertion in an exon of the gene failed to replicate the phenotype, transcript analysis revealed that the original insertion line, mpd1-1, was an overexpression allele of At4g03410, and we phenocopied the original isolate by transgenic overexpression of the MPD1 ORF (Fig. 3; Supplementary Fig. S3). These intentional overexpression lines duplicated the phenotype of mpd1-1, and the line we characterized in detail, MPD-OX3, faithfully duplicated every phenotypic and biochemical characteristic of the original mpd1-1 mutant. Gene overexpression screens have produced many important discoveries, particularly when redundancy within a gene family means that loss-of-function approaches are unrewarding (Nakazawa et al., 2001; Bolle, 2011; Xiao et al., 2017). Our identification of chilling sensitivity caused by overexpression of MPD1 testifies to the power of genetic screens to discover unanticipated gene activities and reveal new facets of plant biology.
In mammals, the Mpv17_PMP22 family is small, consisting of MPV17 (Weiher et al., 1990) with two MPV17-like proteins (Iida et al., 2006), plus PMP22 (Pxmp2; Brosius et al. 2002). Our discovery of the sizeable family of homologous proteins in Arabidopsis that all contain a Mpv17_PMP22 domain suggests that more diverse roles have evolved for these proteins in plants (Supplementary Table S3). Notably, all are predicted to reside in the mitochondria, peroxisomes, or chloroplasts, major sites of ROS production in plants. Protein targeting analyses and proteomic experiments indicate that three MPD family members in addition to MPD1 are chloroplast proteins (At1g52870, At5g19750, and At2g42770). Our finding that insertion in an exon of At4g03410 (mpd1-2) had no visible phenotype, either at 22 °C or at 2 °C (Supplementary Fig. S3), may result from partial redundancy of function with these other chloroplast MPD proteins. Future characterization of multiple mutant lines will explore this possibility.
Plants of both mpd1-1 and MPD1-OX3 lines grew like Col-0 and appeared wild type at 22 °C (Figs 1–3; Supplementary Tables S3, S4) and were likewise indistinguishable from Col-0 in quantum efficiency of photosynthesis (Fv/Fm and ΦPSII; Fig. 7C, D), in concentration of H2O2 in leaf tissue (Figs 5, 7E), and in transcript levels of genes encoding proteins of the ROS response (Fig. 6). However, after transfer to 2 °C, plants overexpressing MPD1 exhibited multiple symptoms typical of chilling-sensitive plants. Both chlorosis and necrotic lesions that developed on leaves of mpd1-1 and MPD1-OX3 over the first 4 weeks of chilling (Figs 1B, 3B) are common in crops damaged by low temperature, and are symptomatic of cellular damage and reduced photosynthetic capacity (Lukatkin et al., 2012). Growth inhibition at low temperatures is common in chilling-sensitive crops (Hussain et al., 2018) and, like mpd1-1 and MPD1-OX3, other chilling-sensitive Arabidopsis mutants are typically smaller in chilling conditions (Tokuhisa et al., 1997, 1998; Chen and Thelen, 2013; Gao et al., 2017, 2020). Also characteristic of chilling-sensitive crops is failure to fully recover after chilling, like the MPD1 overexpression lines (Figs 1E–3E). Therefore, while MPD1 overexpression has no effect under our normal growth conditions, it renders Arabidopsis sensitive to chilling.
After 4 weeks at 2 °C, lines overexpressing MPD1 had a 40% reduction in chlorophyll content relative to Col-0 (Fig. 4A), which correlated with leaf chlorosis and necrosis (Figs 1B, 3B). MPD1 localizes to the chloroplasts (Fig. 2D), and symptoms of chilling sensitivity in mpd1-1 and MPD1-OX3 suggested damage to photosynthetic machinery, which we confirmed by measuring PSII by fluorescence parameters. At 2 °C, both maximum photochemical efficiency (Fv/Fm) and actual photochemical efficiency (ΦPSII) of MPD1-overexpressing plants were reduced by 25–30% compared with Col-0 (Fig. 4B, C). This reduction in photosynthetic efficiency probably results from photosystem damage beyond the loss of chlorophyll, due to excess accumulation of ROS (Li et al., 2018). Indeed, measurements of the most readily assayed ROS, H2O2, found dramatically higher levels under chilling conditions in our experimental lines. While at 22 °C both control and experimental plants had ~4 μmol g–1 FW H2O2 in leaves (Figs 6, 7E–H), after a week at 2 °C Col-0 H2O2 increased to a steady 6–7 μmol g–1 FW. In mpd1-1 and MPD1-OX3 plants, the H2O2 levels indicated a substantially higher ROS stress during the first week of chilling, increasing to ~10 μmol g–1 FW (Figs 6, 7E–H).
In plants, photosynthesis is the largest source of ROS, and chloroplasts actively manage ROS levels through biochemical and biophysical mechanisms to prevent damage to photosystem and chloroplast components (Dietz et al., 2016). In both the absence and presence of ROS stress, H2O2 acts as a retrograde signal modulating expression of transcription factors such as ZAT12 (Davletova et al., 2005) to control genes encoding enzymes that metabolize ROS compounds (Zandalinas et al., 2020). Plants of all our lines grew uniformly at 22 °C, when photosynthesis parameters indicated adequate management of ROS production by modest expression of the ROS response genes that we measured (Fig. 6). At 22 °C, transcript levels for Col-0 and MPD1 overexpression lines were the same. After transfer to 2 °C, Col-0 plants exhibited an ~50% increase in tissue H2O2 concentrations, triggering enhanced transcription evident in the 1.3- to 10-fold induction of all our marker genes (Fig. 6). For the mpd1-1 and MPD1-OX3 overexpression plants, the greater increases in H2O2 during chilling were associated with stronger induction of all seven of the marker genes. For example, ZAT12 transcript increases 6.9-fold in mpd1-1 and 6.7-fold in MPD1-OX3 compared with only 3.7-fold in Col-0. The transcript of SOD2 increased 1.3-fold in Col-0 after 4 weeks at 2 °C, but 3.0-fold in mpd1-1 and 3.6-fold in MPD1-OX3 (Fig. 6). Evidently, ROS signaling and response are working appropriately in MPD1 overexpression lines.
The growth phenotype, photosynthesis analysis, increased H2O2 concentrations, and marker gene analysis all indicate that increased MPD1 expression causes large increases in ROS production at chilling temperatures, overwhelming the antioxidant systems. We found additional confirmation for this hypothesis by treating plants with a low concentration of paraquat to induce ROS production. Paraquat treatment of wild-type Col-0 plants during chilling produced concentrations of H2O2 approaching those of untreated MPD1 overexpression plants after 2 weeks at 2 °C and resulted in declines in Fv/Fm and ΦPSII in the treated Col-0 plants that were similar to the corresponding data for untreated mpd1-1 and MPD1-OX3 plants (Fig. 7). These results support our conclusion that overexpression of MPD1 in mpd1-1 and MPD1-OX3 results in intense ROS stress at 2 °C that forms the basis of chilling damage in these lines.
Animal and yeast Mpv17-PMP22 proteins contain multiple membrane-spanning domains like MPD1 (Supplementary Fig. S1), form channels by homotrimer association in recombinant expression systems (Rokka et al., 2009; Antonenkov et al., 2015), and have been shown to transfer small anionic metabolites including pyruvate, glycolate, and α-ketoglutarate (Rokka et al., 2009; Dallabona et al., 2010). A detailed characterization of the human Mpv17 channel found that transport activity is under strict control modulated by membrane potential, pH, and redox conditions, suggesting that its trafficking function might be a direct response to redox changes acting on the Mpv17_PMP22 domain (Antonenkov et al., 2015).
These features of the animal Mpv17_PMP22 proteins suggest a possible mechanism by which overexpression of MPD1 results in chilling sensitivity in Arabidopsis. In this model MPD1 overexpression at normal growth temperatures causes no changes in phenotypic, biochemical, or gene expression parameters because chloroplast redox conditions allow tight biochemical regulation of channel activity. However, at low temperature, an initial increase in ROS results in increased MPD1 channel activity that, in the overexpression lines, leads to a feed-forward loop of ROS production and channel activity that overwhelms ROS control despite the robust genetic response that we observed in mpd1-1 and MPD1-OX3 plants (Figs 5, 6).
Our characterization of MPD1 overexpression lines and wild-type Arabidopsis clearly establishes that increased ROS stress is a key factor in the development of chilling damage. Future analysis of expression patterns and isoforms of MPD1 between chilling-sensitive and resistant species may further illuminate the roles of MPD1 proteins in plant chilling responses and lead to the development of strategies to assist molecular breeding of crop varieties tolerant to chilling stress. In addition, our discovery suggests that biochemical, physiological, and genetic studies of all 10 members of the Arabidopsis MPD family will further contribute to our understanding of abiotic stress and ROS production, signaling, and management in plants.
Supplementary data
The following supplementary data are available at JXB online.
Table S1. List of primers used for quantitative PCR analysis.
Table S2. Growth measurements of mpd1-1 and mpd1-2 at 22 °C.
Table S3. The Mpv17_PMP22 family in Arabidopsis.
Table S4. Growth measurements of MPD1-OX3 at 22 °C.
Fig. S1. Transmembrane domains of MPD1.
Fig. S2. The mpd1-1 mutant is an overexpression allele.
Fig. S3. Level of MPD1 transcript and phenotype for two MPD1 overexpression lines.
Fig. S4. Phenotype of MPD1-OX3 lines after recovery.
Glossary
Abbreviations
- eGFP
enhanced green fluorescent protein
- MPD1
Mpv17_PMP22 protein 1
- ROS
reactive oxygen species
Author contributions
DL, GS, JW, and JB: conceptualization and design; DL and GS: performing and interpreting experinents. All authors contributed to data analysis and manuscript preparation.
Conflict of interest
The authors declare no competing financial interests.
Funding
This work has been funded by the US National Science Foundation (Grant IOS-1555581) and by the Agricultural Research Center at Washington State University. GS was partially supported by the NIH, National Institute for General Medical Sciences through institutional training Grant T32-GM008336.
Data availability
Original data from experiments reported herein have been archived on the Research Exchange at Washington State University and are available at http://doi.org/10.7273/000001123
References
- Allen DJ, Ort DR.. 2001. Impacts of chilling temperatures on photosynthesis in warm-climate plants. Trends in Plant Science 6, 36–42. [DOI] [PubMed] [Google Scholar]
- Alonso JM, Stepanova AN, Leisse TJ, et al. 2003. Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301, 653–657. [DOI] [PubMed] [Google Scholar]
- Antonenkov VD, Isomursu A, Mennerich D, Vapola MH, Weiher H, Kietzmann T, Hiltunen JK.. 2015. The human mitochondrial DNA depletion syndrome gene MPV17 encodes a non-selective channel that modulates membrane potential. Journal of Biological Chemistry 290, 13840–13861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrero-Gil J, Huertas R, Rambla JL, Granell A, Salinas J.. 2016. Tomato plants increase their tolerance to low temperature in a chilling acclimation process entailing comprehensive transcriptional and metabolic adjustments. Plant, Cell & Environment 39, 2303–2318. [DOI] [PubMed] [Google Scholar]
- Bolle C, Schneider A, Leister D.. 2011. Perspectives on systematic analyses of gene function in Arabidopsis thaliana: new tools, topics and trends. Current Genomics 12, 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brosius U, Dehmel T, Gärtner J.. 2002. Two different targeting signals direct human peroxisomal membrane protein 22 to peroxisomes. Journal of Biological Chemistry 277, 774–784. [DOI] [PubMed] [Google Scholar]
- Casalena G, Krick S, Daehn I, et al. 2014. Mpv17 in mitochondria protects podocytes against mitochondrial dysfunction and apoptosis in vivo and in vitro. American Journal of Physiology. Renal Physiology 306, F1372–F1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakraborty S, Hill AL, Shirsekar G, Afzal AJ, Wang GL, Mackey D, Bonello P.. 2016. Quantification of hydrogen peroxide in plant tissues using Amplex Red. Methods 109, 105–113. [DOI] [PubMed] [Google Scholar]
- Chen M, Thelen JJ.. 2013. ACYL-LIPID DESATURASE2 is required for chilling and freezing tolerance in Arabidopsis. The Plant Cell 25, 1430–1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF.. 1998. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. The Plant Journal 16, 735–743. [DOI] [PubMed] [Google Scholar]
- Dallabona C, Marsano RM, Arzuffi P, Ghezzi D, Mancini P, Zeviani M, Ferrero I, Donnini C.. 2010. Sym1, the yeast ortholog of the MPV17 human disease protein, is a stress-induced bioenergetic and morphogenetic mitochondrial modulator. Human Molecular Genetics 19, 1098–1107. [DOI] [PubMed] [Google Scholar]
- Davletova S, Schlauch K, Coutu J, Mittler R.. 2005. The zinc-finger protein Zat12 plays a central role in reactive oxygen and abiotic stress signaling in Arabidopsis. Plant Physiology 139, 847–856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietz KJ, Turkan I, Krieger-Liszkay A.. 2016. Redox- and reactive oxygen species-dependent signaling into and out of the photosynthesizing chloroplast. Plant Physiology 171, 1541–1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreyer A, Dietz KJ.. 2018. Reactive oxygen species and the redox-regulatory network in cold stress acclimation. Antioxidants 7, 169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang P, Yan M, Chi C, Wang M, Zhou Y, Zhou J, Shi K, Xia X, Foyer CH, Yu J.. 2019. Brassinosteroids act as a positive regulator of photoprotection in response to chilling stress. Plant Physiology 180, 2061–2076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FAO. 2017. The future of food and agriculture–trends and challenges. Rome: Food and Agriculture Organisation. [Google Scholar]
- Foyer CH, Shigeoka S.. 2011. Understanding oxidative stress and antioxidant functions to enhance photosynthesis. Plant Physiology 155, 93–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao J, Lunn D, Wallis JG, Browse J.. 2020. Phosphatidylglycerol composition is central to chilling damage in the Arabidopsis fab1 mutant. Plant Physiology 184, 1717–1730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao J, Wallis JG, Jewell JB, Browse J.. 2017. Trimethylguanosine synthase1 (TGS1) is essential for chilling tolerance. Plant Physiology 174, 1713–1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawkes TR. 2014. Mechanisms of resistance to paraquat in plants. Pest Management Science 70, 1316–1323. [DOI] [PubMed] [Google Scholar]
- Hooper CM, Castleden IR, Tanz SK, Aryamanesh N, Millar AH.. 2017. SUBA4: the interactive data analysis centre for Arabidopsis subcellular protein locations. Nucleic Acids Research 45, D1064–D1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussain HA, Hussain S, Khaliq A, Ashraf U, Anjum SA, Men S, Wang L.. 2018. Chilling and drought stresses in crop plants: implications, cross talk, and potential management opportunities. Frontiers in Plant Science 9, 393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iida R, Yasuda T, Tsubota E, Takatsuka H, Matsuki T, Kishi K.. 2006. Human Mpv17-like protein is localized in peroxisomes and regulates expression of antioxidant enzymes. Biochemical and Biophysical Research Communications 344, 948–954. [DOI] [PubMed] [Google Scholar]
- Karimi M, Inzé D, Depicker A.. 2002. GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends in Plant Science 7, 193–195. [DOI] [PubMed] [Google Scholar]
- Latowski D, Kuczyńska P, Strzałka K.. 2011. Xanthophyll cycle—a mechanism protecting plants against oxidative stress. Redox Report 16, 78–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Aro EM, Millar AH.. 2018. Mechanisms of photodamage and protein turnover in photoinhibition. Trends in Plant Science 23, 667–676. [DOI] [PubMed] [Google Scholar]
- Löllgen S, Weiher H.. 2015. The role of the Mpv17 protein mutations of which cause mitochondrial DNA depletion syndrome (MDDS): lessons from homologs in different species. Biological Chemistry 396, 13–25. [DOI] [PubMed] [Google Scholar]
- Lukatkin AS, Brazaityte A, Bobinas C, Duchovskis P.. 2012. Chilling injury in chilling-sensitive plants: a review. Zemdirbyste 99, 111–124 [Google Scholar]
- Lunn D, Ibbett R, Tucker GA, Lycett GW.. 2015. Impact of altered cell wall composition on saccharification efficiency in stem tissue of Arabidopsis RABA GTPase-deficient knockout mutants. Bioenergy Research 8, 1362–1370 [Google Scholar]
- Lyons JM. 1973. Chilling injury in plants. Annual Review of Plant Physiology 24, 445–466 [Google Scholar]
- Meissner M, Orsini E, Ruschhaupt M, Melchinger AE, Hincha DK, Heyer AG.. 2013. Mapping quantitative trait loci for freezing tolerance in a recombinant inbred line population of Arabidopsis thaliana accessions Tenela and C24 reveals REVEILLE1 as negative regulator of cold acclimation. Plant, Cell & Environment 36, 1256–1267. [DOI] [PubMed] [Google Scholar]
- Mhamdi A, Van Breusegem F.. 2018. Reactive oxygen species in plant development. Development 145, 164376. [DOI] [PubMed] [Google Scholar]
- Moustaka J, Moustakas M.. 2014. Photoprotective mechanism of the non-target organism Arabidopsis thaliana to paraquat exposure. Pesticide Biochemistry and Physiology 111, 1–6. [DOI] [PubMed] [Google Scholar]
- Nakazawa M, Yabe N, Ichikawa T, Yamamoto YY, Yoshizumi T, Hasunuma K, Matsui M.. 2001. DFL1, an auxin-responsive GH3 gene homologue, negatively regulates shoot cell elongation and lateral root formation, and positively regulates the light response of hypocotyl length. The Plant Journal 25, 213–221. [DOI] [PubMed] [Google Scholar]
- Okuley J, Lightner J, Feldmann K, Yadav N, Lark E, Browse J.. 1994. Arabidopsis FAD2 gene encodes the enzyme that is essential for polyunsaturated lipid synthesis. The Plant Cell 6, 147–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porra RJ. 2002. The chequered history of the development and use of simultaneous equations for the accurate determination of chlorophylls a and b. Photosynthesis Research 73, 149–156. [DOI] [PubMed] [Google Scholar]
- Rodziewicz P, Swarcewicz B, Chmielewska K, Wojakowska A, Stobiecki M.. 2014. Influence of abiotic stresses on plant proteome and metabolome changes. Acta Physiologiae Plantarum 36, 1–19 [Google Scholar]
- Rokka A, Antonenkov VD, Soininen R, Immonen HL, Pirilä PL, Bergmann U, Sormunen RT, Weckström M, Benz R, Hiltunen JK.. 2009. Pxmp2 is a channel-forming protein in mammalian peroxisomal membrane. PLos One 4, e5090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smirnoff N, Arnaud D.. 2019. Hydrogen peroxide metabolism and functions in plants. New Phytologist 221, 1197–1214. [DOI] [PubMed] [Google Scholar]
- Spinazzola A, Viscomi C, Fernandez-Vizarra E, et al. 2006. MPV17 encodes an inner mitochondrial membrane protein and is mutated in infantile hepatic mitochondrial DNA depletion. Nature Genetics 38, 570–575. [DOI] [PubMed] [Google Scholar]
- Thomashow MF. 1999. Plant cold acclimation: freezing tolerance genes and regulatory mechanisms. Annual Review of Plant Physiology and Plant Molecular Biology 50, 571–599. [DOI] [PubMed] [Google Scholar]
- Thomashow MF. 2010. Molecular basis of plant cold acclimation: insights gained from studying the CBF cold response pathway. Plant Physiology 154, 571–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokuhisa JG, Feldmann KA, LaBrie ST, Browse J.. 1997. Mutational analysis of chilling tolerance in plants. Plant, Cell & Environment 20, 1391–1400 [Google Scholar]
- Tokuhisa JG, Vijayan P, Feldmann KA, Browse JA.. 1998. Chloroplast development at low temperatures requires a homolog of DIM1, a yeast gene encoding the 18S rRNA dimethylase. The Plant Cell 10, 699–711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripathi A, Tripathi DK, Chauhan DK, Kumar N, Singh GS.. 2016. Paradigms of climate change impacts on some major food sources of the world: a review on current knowledge and future prospects. Agricultural Ecosystems and Environment 216, 356–373 [Google Scholar]
- Trott A, Morano KA.. 2004. SYM1 is the stress-induced Saccharomyces cerevisiae ortholog of the mammalian kidney disease gene Mpv17 and is required for ethanol metabolism and tolerance during heat shock. Eukaryotic Cell 3, 620–631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tugal HB, Pool M, Baker A.. 1999. Arabidopsis 22-kilodalton peroxisomal membrane protein. Nucleotide sequence analysis and biochemical characterization. Plant Physiology 120, 309–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usadel B, Bläsing OE, Gibon Y, Poree F, Höhne M, Günter M, Trethewey R, Kamlage B, Poorter H, Stitt M.. 2008. Multilevel genomic analysis of the response of transcripts, enzyme activities and metabolites in Arabidopsis rosettes to a progressive decrease of temperature in the non-freezing range. Plant, Cell & Environment 31, 518–547. [DOI] [PubMed] [Google Scholar]
- Wang YH. 2008. How effective is T-DNA insertional mutagenesis in Arabidopsis? Journal of Biochemical Technology 1, 11–20 [Google Scholar]
- Weiher H, Noda T, Gray DA, Sharpe AH, Jaenisch R.. 1990. Transgenic mouse model of kidney disease: insertional inactivation of ubiquitously expressed gene leads to nephrotic syndrome. Cell 62, 425–434. [DOI] [PubMed] [Google Scholar]
- Wi J, Na Y, Yang E, Lee JH, Jeong WJ, Choi DW.. 2020. Arabidopsis AtMPV17, a homolog of mice MPV17, enhances osmotic stress tolerance. Physiology and Molecular Biology of Plants 26, 1341–1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wise RR. 1995. Chilling-enhanced photooxidation: the production, action and study of reactive oxygen species produced during chilling in the light. Photosynthesis Research 45, 79–97. [DOI] [PubMed] [Google Scholar]
- Xiao C, Barnes WJ, Zamil MS, Yi H, Puri VM, Anderson CT.. 2017. Activation tagging of Arabidopsis POLYGALACTURONASE INVOLVED IN EXPANSION2 promotes hypocotyl elongation, leaf expansion, stem lignification, mechanical stiffening, and lodging. The Plant Journal 89, 1159–1173. [DOI] [PubMed] [Google Scholar]
- Zandalinas SI, Fritschi FB, Mittler R.. 2020. Signal transduction networks during stress combination. Journal of Experimental Botany 71, 1734–1741. [DOI] [PubMed] [Google Scholar]
- Zhu J, Dong CH, Zhu JK.. 2007. Interplay between cold-responsive gene regulation, metabolism and RNA processing during plant cold acclimation. Current Opinion in Plant Biology 10, 290–295. [DOI] [PubMed] [Google Scholar]
- Zuther E, Lee YP, Erban A, Kopka J, Hincha DK.. 2018. Natural variation in freezing tolerance and cold acclimation response in Arabidopsis thaliana and related species. Advances in Experimental Medicine and Biology 1081, 81–98. [DOI] [PubMed] [Google Scholar]
- Zwacka RM, Reuter A, Pfaff E, Moll J, Gorgas K, Karasawa M, Weiher H.. 1994. The glomerulosclerosis gene Mpv17 encodes a peroxisomal protein producing reactive oxygen species. The EMBO Journal 13, 5129–5134. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Original data from experiments reported herein have been archived on the Research Exchange at Washington State University and are available at http://doi.org/10.7273/000001123