Abstract
Mitogen-activated protein kinase (MAPK) cascades transmit environmental signals and induce stress and defence responses in plants. These signalling cascades are negatively controlled by specific Ser/Thr protein phosphatases of the type 2C (PP2C) and dual-specificity phosphatase (DSP) families that inactivate stress-induced MAPKs; however, the interplay between phosphatases of these different types has remained unknown. This work reveals that different Arabidopsis MAPK phosphatases, the PP2C-type AP2C1 and the DSP-type MKP1, exhibit both specific and overlapping functions in plant stress responses. Each single mutant, ap2c1 and mkp1, and the ap2c1 mkp1 double mutant displayed enhanced stress-induced activation of the MAPKs MPK3, MPK4, and MPK6, as well as induction of a set of transcription factors. Moreover, ap2c1 mkp1 double mutants showed an autoimmune-like response, associated with increased levels of the stress hormones salicylic acid and ethylene, and of the phytoalexin camalexin. This phenotype was reduced in the ap2c1 mkp1 mpk3 and ap2c1 mkp1 mpk6 triple mutants, suggesting that the autoimmune-like response is due to MAPK misregulation. We conclude that the evolutionarily distant MAPK phosphatases AP2C1 and MKP1 contribute crucially to the tight control of MAPK activities, ensuring appropriately balanced stress signalling and suppression of autoimmune-like responses during plant growth and development.
Keywords: ANAC, AP2C1, Arabidopsis, dual-specificity phosphatase, mitogen-activated protein kinase, MKP1, Ser/Thr protein phosphatase of the type 2C, WRKY
Mitogen-activated protein kinase (MAPK) phosphatase double mutant ap2c1 mkp1 Arabidopsis plants exhibit constitutive, autoimmune-like stress responses, dependent on their substrate MAPKs MPK3 and MPK6.
Introduction
Reversible protein phosphorylation is one of the most commonly used mechanisms for the molecular transmission of stress signals and developmental cues. This mechanism is based on the opposing actions of protein kinases and protein phosphatases. Mitogen-activated protein kinases (MAPKs) are highly conserved major components of developmental and stress signalling cascades in eukaryotes. MAPKs are activated by upstream MAPK kinases via phosphorylation of Thr and Tyr residues within their activation loop. This activation eventually leads to the reprogramming of cellular activities, including the modulation of gene expression, to generate appropriate responses. The activation of MAPKs does not represent a simple on/off switch, as both the magnitude and duration of activation are crucial for determining the signalling outcome (Marshall, 1995). Prolonged or constant activation of a MAPK cascade can have detrimental effects, as illustrated by the hypersensitive response-induced cell death in plants expressing a constitutively active version of MAPK kinase (Ren et al., 2002; Liu et al., 2007). Thus, negative regulation and inactivation mechanisms are important for the correct cellular response. Specific protein phosphatases can dephosphorylate and thereby inactivate MAPKs. As dual phosphorylation of the Thr-X-Tyr motif in the activation loop is required for MAPK activation (Caunt and Keyse, 2013), dephosphorylation of either phospho-amino acid residue inactivates the MAPK and inhibits downstream signalling. Interestingly, this inactivation can be accomplished by evolutionarily distant protein phosphatases, including Ser/Thr protein phosphatases of the type 2C (PP2C) MAPK phosphatases (Schweighofer et al., 2004; Schweighofer et al., 2007; Fuchs et al., 2013) and PTP-type dual specificity (Tyr and Ser/Thr) phosphatases (DSPs) (Bartels et al., 2010; Jiang et al., 2018). However, their interplay is presently unknown.
Arabidopsis thaliana DSP-type MAPK phosphatase 1 (MKP1) interacts with the stress-responsive MAPKs MPK3, MPK4, and MPK6, and controls their activities (Ulm et al., 2002; Bartels et al., 2009; Anderson et al., 2011). The mkp1 knockout mutant is hypersensitive to genotoxic stress, including UV-B radiation (Gonzalez Besteiro and Ulm, 2013a, b), but is more resistant than the wild type (WT) to the virulent bacterial pathogen Pseudomonas syringae pv. tomato (Pto) (Bartels et al., 2009; Anderson et al., 2011; Anderson et al., 2014; Escudero et al., 2019). Specifically in Arabidopsis Columbia accession, mkp1 shows an autoimmune-like growth phenotype dependent on the disease resistance gene homologue SUPPRESSOR OF npr1-1, CONSTITUTIVE 1 (SNC1), and is associated with enhanced MAPK activities (Bartels et al., 2009). The phospho-Tyr-specific PTP-type protein phosphatase PTP1 also interacts with MPK6 and MPK3 in transient assays. The mkp1 ptp1 double mutant exhibits up-regulation of MPK6-dependent plant defence responses and a further enhanced autoimmune-like phenotype (Bartels et al., 2009).
A group of PP2C-type phosphatases, including AP2C1, interacts with MAPKs and controls their activities (Schweighofer et al., 2007; Umbrasaite et al., 2010). AP2C1 is induced by wounding and biotic stress, and functions as a negative regulator of MPK3, MPK4, and MPK6, controlling levels of wound-induced jasmonate and ethylene (ET) as well as plant immunity (Schweighofer et al., 2007; Galletti et al., 2011; Sidonskaya et al., 2016; Shubchynskyy et al., 2017). ap2c1 plants do not display obvious developmental phenotypes under standard growth conditions (Schweighofer et al., 2007), implying a specific function of AP2C1 under stress conditions and the contribution of other, presently unknown, MAPK phosphatases for MAPK control in the absence of AP2C1.
Activation of transcription factors (TFs) and changes in gene expression are part of the cellular response to a perceived signal in order to reprogram cellular processes (Rauf et al., 2013). A number of TFs, including WRKY and AP2-domain/ethylene-responsive factor (AP2/ERF) family members, have been suggested or demonstrated to act downstream of MAPKs in plants (Asai et al., 2002; Kim and Zhang, 2004; Menke et al., 2005; Nakano et al., 2006; Popescu et al., 2009; Bethke et al., 2009; Mao et al., 2011; Li et al., 2012; Meng and Zhang, 2013; Guan et al., 2014). Subsequently, these proteins may constitute an important link between pathogen- or wound-induced MAPK signalling and downstream transcriptional reprogramming.
Considering the broad spectrum of signals transmitted by the same MAPKs (Rodriguez et al., 2010; Meng and Zhang, 2013), such as MPK6, it is puzzling how the specificity of the responses for perceived stimuli is generated. The phylogenetic diversity and distinct enzymatic mechanisms of protein phosphatases that are able to inactivate MAPKs support the idea of a contribution of MAPK phosphatases to the versatility and specificity of MAPK networks. Here, we investigate the roles of the phylogenetically distant Arabidopsis MAPK phosphatases AP2C1 and MKP1 and, in particular, their functional redundancies. We show that AP2C1 and MKP1 together repress plant autoimmune-like responses, including salicylic acid (SA) and ET accumulation, and early senescence. These observations in the ap2c1 mkp1 double mutant are underlined by the mis-expression of specific TF genes, including members of the WRKY, AP2/ERF, and Arabidopsis NAM, ATAF, and CUC (ANAC) families whose expression is at least partially mediated by MPK6 and MPK3.
Materials and methods
Plant lines, genetic crosses, and growth conditions
All plant lines used in this study were in the Arabidopsis thaliana accession Columbia (Col-0), with mkp1 being an introgression line from a Wassilewskija background (Bartels et al., 2009). The T-DNA insertion line ap2c1 (SALK_065126) (Schweighofer et al., 2007) was crossed with the T-DNA insertion lines ptp1 (SALK_118658) and mkp1 to generate the ap2c1 ptp1 and ap2c1 mkp1 double mutants, respectively. mpk6-2 (SALK_073907) and mpk3-1 (SALK_151594) were used for genetic crosses generating ap2c1 mkp1 mpk6 and ap2c1 mkp1 mpk3 triple mutants. The T-DNA insertion lines ap2c2 (GABI-Kat_316F11) and ap2c3 (SALK_109986) (Umbrasaite et al., 2010) were crossed with mkp1 to generate ap2c2 mkp1 and ap2c3 mkp1 double mutants. Combinatorial mutants were identified in the F2 generation and also confirmed in subsequent generations by PCR genotyping using T-DNA- and gene-specific primers (Schweighofer et al., 2007; Bartels et al., 2009; Umbrasaite et al., 2010; Shubchynskyy et al., 2017). For protein and RNA extraction, and for measurements of ET and SA, plants were grown on soil for 5–7 weeks in a phytotron chamber under short-day conditions (8 h light, 22 °C/16 h dark, 20 °C).
For experiments at the seedling stage, seeds were surface sterilized and spread on plates containing half-strength Murashige and Skoog (MS) medium (Duchefa), pH 5.7, 1% (w/v) sucrose, and 0.7% plant agar (w/v; Duchefa). Seedlings were grown under long-day conditions (16 h light/8 h dark) at 22 °C. For suppression of growth phenotypes, plants were kept under short-day conditions in a tray with closed lid at 24 °C with saturating relative humidity.
Ex vivo kinase activity assay and MAPK immunoblotting
Plant protein extraction and the ex vivo kinase assay were performed as described previously (Schweighofer et al., 2007; Schweighofer et al., 2009) using polyclonal antibodies for immunoprecipitation and myelin basic protein (MBP) as the in vitro substrate of immunoprecipitated MAPKs. A master mix containing MBP and γ-ATP was used for each series of kinase assays to ensure equal distribution of the substrate to all reactions. Treatment with the pathogen-associated molecular pattern (PAMP) flg22 was performed as described previously (Shubchynskyy et al., 2017). Signal intensities of phosphorylated MBP were quantified with ImageQuant software (version 5.1; Amersham). MAPK protein amounts were visualized with the antibodies Anti-AtMPK3 (M8318), Anti-MPK4 (A6979), and Anti-AtMPK6 (A7104) (all Sigma).
RNA extraction and quantitative reverse transcription–PCR
Total RNA from leaves was isolated with the RNeasy Plant Mini Kit (Qiagen) and treated with TURBO DNA-free DNase I (Ambion) according to the manufacturers’ instructions. RNA integrity was checked on 1% (w/v) agarose gels and the concentration was measured before and after DNAse I digestion. The absence of genomic DNA was verified by PCR using primers targeting an intron of the control gene At5g65080. cDNA synthesis was performed using the First Strand cDNA Synthesis Kit (Thermo Scientific). The efficiency of cDNA synthesis was estimated by quantitative reverse transcription–PCR (RT–qPCR) analysis using a primer pair amplifying the 3ʹ part of the control gene encoding GAPDH and a primer pair amplifying the 5ʹ part of the same gene. RT–qPCR reactions were performed as described previously (Balazadeh et al., 2008). ACTIN2 was selected as a reference gene for which four replicates were measured in each PCR run, and their average cycle threshold (CT) was used for relative expression analyses. TF expression data were normalized by subtracting the mean ACTIN2 gene CT value from the CT value (ΔCT) of each gene of interest. The expression value in the comparison between different genotypes was calculated using the expression 2–ΔΔCT, where ΔΔCT represents the ΔCT of the mutant of interest minus the ΔCT of the control (WT). For TF expression profiling, an advanced version of an expression profiling platform (Balazadeh et al., 2008) that was originally described by Czechowski et al. (2004) was used, covering 1880 Arabidopsis TF genes. Statistical analysis was performed with JASP software (https://jasp-stats.org; version 0.14.1).
ET measurements and quantification of total SA and camalexin
Measurements of ET were performed by gas chromatography (Hewlett Packard 5890 Series II) with an Al2O3 column (Agilent Technologies). Whole rosettes of 4-week-old plants grown under long-day conditions were collected, and leaves were wounded and transferred into 20 ml vials containing 4 ml half-strength MS medium with 0.8% (w/v) plant agar, to reduce the volume of the head space, and the vials were sealed so they were airtight. After 24 h, 100 µl of the gas phase was taken from the vials and analysed by gas chromatography–flame ionization detection. ET production was calculated per milligram of fresh tissue per hour.
Total SA was quantified as described previously (Rozhon et al., 2005) except that 20 µM EDTA was added to the HPLC eluent. Camalexin levels were determined as described previously (Shubchynskyy et al., 2017).
Results
ap2c1 mkp1 double mutant plants show growth and developmental defects that are at least partially mediated by MPK6 and MPK3
To investigate the specific and/or overlapping roles of the MAPK phosphatases AP2C1 and MKP1, we took advantage of the Arabidopsis T-DNA insertion knockout mutants ap2c1 and mkp1, respectively (Schweighofer et al., 2007; Bartels et al., 2009). Phenotypically, ap2c1 and mkp1 mutant plants did not show any difference from WT plants when grown for up to 5 weeks under short-day conditions (Fig. 1A). However, mkp1 plants grown under long-day conditions showed altered morphology, such as aberrant leaf development and early senescence, which appeared approximately 3 weeks after germination (Supplementary Fig. S1A), as described previously (Bartels et al., 2009).
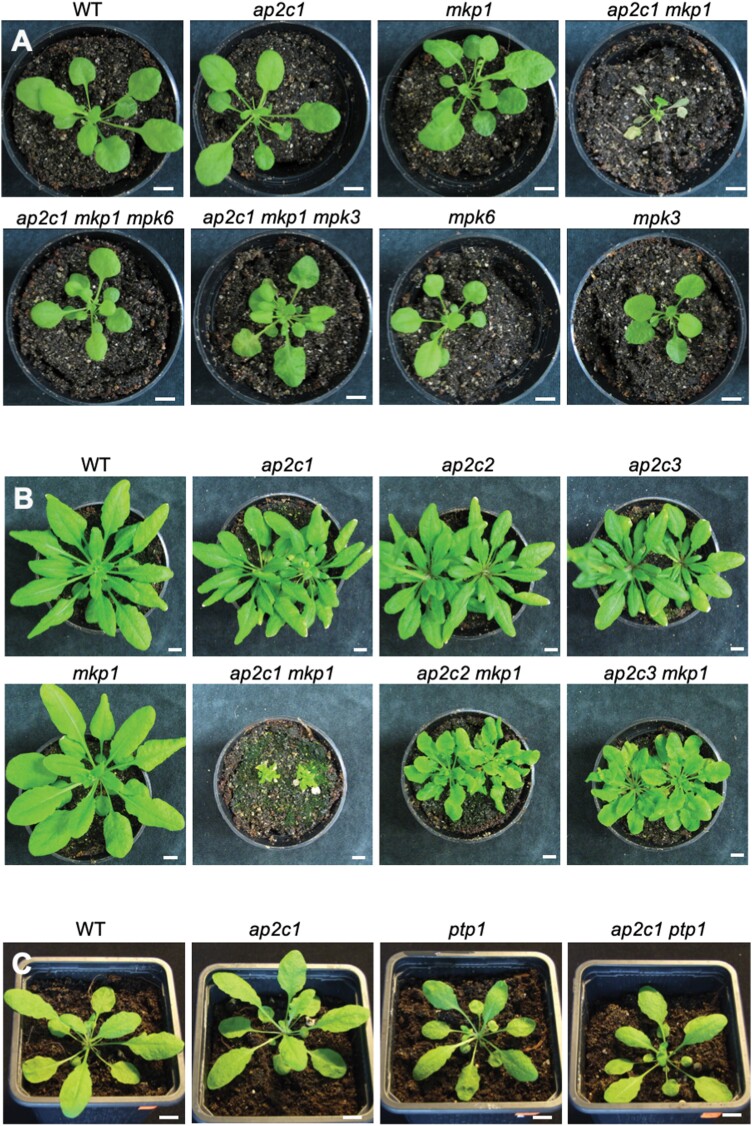
Fig. 1.
Loss of both AP2C1 and MKP1 causes developmental defects and precocious cell death, mediated by MPK6 and MPK3. (A) Phenotypes of 5-week-old plants of the indicated genotypes grown under standard short-day conditions. (B) Phenotypes of plants of the indicated genotypes grown for 4 weeks under short-day conditions followed by 3 weeks under long-day conditions. (C) Phenotypes of plants of the indicated genotypes grown for 6 weeks under short-day conditions. Scale bars=1 cm.
To further analyse the functions of AP2C1 and MKP1 in plants, we generated a double mutant by genetic crossing. ap2c1 mkp1 plants showed phenotypic differences from both the WT and the single mutants. These appeared about 2 weeks after germination under standard growth conditions in soil in all ap2c1 mkp1 double mutants; first their sizes started differing, and during further growth ap2c1 mkp1 plants developed more pronounced multiple defects, including severe dwarfism and aberrant leaf development (Fig. 1A, B; Supplementary Fig. S1B–G). Four weeks after germination, phenotypic abnormalities became even more evident, including early senescence, spontaneous macroscopic lesions, and abnormal leaf morphology. These developmental defects were suppressed when plants were grown under conditions of elevated humidity and increased temperature (Supplementary Fig. S2), indicating a dependency on environmental cues. However, during flowering, misshapen inflorescences and strongly reduced fertility were always observed (Supplementary Fig. S1E). These phenotypes were specific for ap2c1 mkp1 plants, as crossing ap2c1 with ptp1 did not lead to phenotypic alterations (Fig. 1C) compared with mkp1 ptp1 (Bartels et al., 2009). Crossing mkp1 with either of two other clade B AP2C mutants, ap2c2 or ap2c3 (Umbrasaite et al., 2010), led to only mild phenotypes compared with the strong defects of ap2c1 mkp1 plants (Fig. 1B).
Since MPK6 and MPK3 are targets of AP2C1 and MKP1 (Ulm et al., 2002; Schweighofer et al., 2007; Galletti et al., 2011), we addressed the impact of MPK6 and MPK3 on the phenotypic aberrations detected in ap2c1 mkp1 plants. To this end, ap2c1 mkp1 mpk6 and ap2c1 mkp1 mpk3 triple mutant plants were created and their phenotypes were compared with those of ap2c1 mkp1. The phenotype of ap2c1 mkp1 mpk6 plants was more similar to that of the WT than that of ap2c1 mkp1. The loss of MPK6 suppressed most phenotypic defects observed in ap2c1 mkp1, such as extreme dwarfism, aberrant leaf shapes, premature leaf senescence, and impaired fertility (Fig. 1A, Supplementary Fig. S3). However, at later developmental stages, ap2c1 mkp1 mpk6 plants still appeared overall smaller than WT and displayed senescence in the older leaves (Supplementary Fig. S3E, M). Similarly, ap2c1 mkp1 mpk3 plants suppressed the observed ap2c1 mkp1 phenotypes in early development (Fig. 1A) but to a lesser extent at later developmental stages compared with ap2c1 mkp1 mpk6 (Supplementary Fig. S3F, N). Both ap2c1 mkp1 mpk6 and ap2c1 mkp1 mpk3 developmental phenotypes were suppressed during growth in conditions of elevated temperature and increased humidity, as observed with ap2c1 mkp1 (Supplementary Fig. S2).
Overall, these results suggest that AP2C1 and MKP1 protein phosphatases act partially redundantly and that the presence of at least either gene is necessary for normal plant development. The phenotypes observed in ap2c1 mkp1 plants are predominantly MPK6- and MPK3-dependent.
Dual control of stress-induced MAPK activities by AP2C1 and MKP1
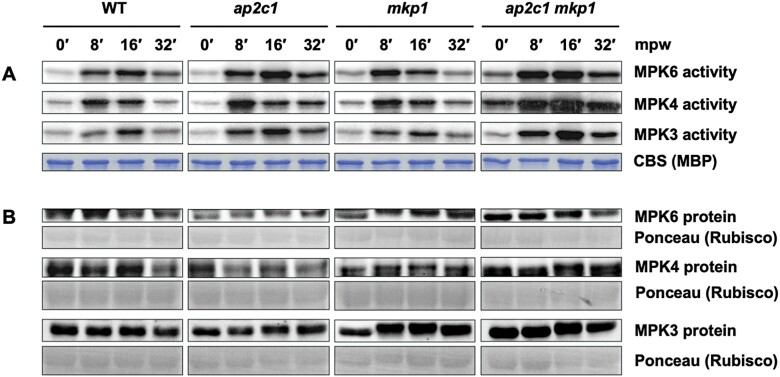
Our previous work revealed the involvement of AP2C1 in the regulation of MAPK activities induced by wounding, nematode feeding, and PAMPs (Schweighofer et al., 2007; Sidonskaya et al., 2016; Shubchynskyy et al., 2017). To check for a potential overlapping role of MKP1, we first analysed MPK3, MPK4, and MPK6 activities after wounding the leaves of WT plants and of the single mutant plants ap2c1 and mkp1. Kinase activities were assayed after immunoprecipitation from total protein extracts using specific antibodies. In agreement with our previous findings (Schweighofer et al., 2007), ap2c1 plants showed higher and more sustained wound-induced activities of MPK3, MPK4, and MPK6 compared with the WT (Fig. 2). Interestingly, MPK4 activity was more intense and sustained in ap2c1 compared with mkp1 plants, indicating a specific role of AP2C1 in the regulation of MPK4 during wounding. MPK3 activity in ap2c1 plants was slightly higher and more sustained in comparison to WT and mkp1. In mkp1 plants, the peak activity of MPK6 was shifted to an earlier time point compared with the WT. In ap2c1 mkp1 plants, we detected strongly and moderately enhanced basal activity of MPK4 and MPK6, respectively, whereas the basal activity of MPK3 was not affected in comparison to the WT or single mutant lines. The stronger and more sustained wound-induced activation of MAPKs observed in single-mutant plants was additionally enhanced in the double mutant ap2c1 mkp1 (Fig. 2, Supplementary Fig. S4A). The MPK3, MPK4, and MPK6 protein amounts were comparable in both the single and double mutants and the WT (Fig. 2). Treatment of WT, ap2c1 and mkp1 single and double mutants, and ap2c1 mkp1 mpk3 and ap2c1 mkp1 mpk6 triple mutant plants with the PAMP flg22 led to similar MAPK activation patterns as those observed after wounding, with increased and prolonged MAPK activities in the ap2c1 mkp1 plants compared with the single mutants and the WT (Supplementary Fig. S4B, C).
Fig. 2.
AP2C1 and MKP1 control wound-induced MAPK activities. Analysis of wound-induced MPK6, MPK4, and MPK3 kinase activities and protein amounts of leaves from 6-week-old WT, ap2c1, mkp1, and ap2c1 mkp1 plants grown under short-day conditions. (A) MAPK activities were determined after immunoprecipitation by phosphorylation of MBP detected by autoradiography. The entire kinase assay was based on one common master mix containing MBP and γ-ATP. Loading is demonstrated by Coomassie blue staining (CBS); representative lanes are shown. The experiment was repeated twice with similar results. (B) MAPK protein amounts before and after wounding demonstrated by immunoblotting of MPK3, MPK4, and MPK6 from total protein extract using specific antibodies. Loading is demonstrated by Ponceau S staining (Rubisco protein). mpw, Minutes post wounding.
MKP1 has been reported to be constitutively expressed (Ulm et al., 2002), whereas AP2C1 is transcriptionally responsive to stress (Schweighofer et al., 2007; Sidonskaya et al., 2016). We tested whether reciprocal compensational expression may occur under long-day conditions and thus analysed AP2C1 and MKP1 mRNA levels in the mkp1 and ap2c1 mutants, respectively. RT–qPCR analyses showed only very slightly increased expression of MKP1 in ap2c1 plants, whereas the expression of AP2C1 was approximately 160% of the WT level in the mkp1 plants (Supplementary Fig. S5), suggesting a compensatory transcriptional activation of AP2C1 in the absence of MKP1. Our results suggest cooperative action and partial redundancy in the regulation of MAPKs by these two evolutionarily distant and unrelated MAPK phosphatases.
AP2C1 and MKP1 play partially redundant roles in the control of wound-induced ET synthesis
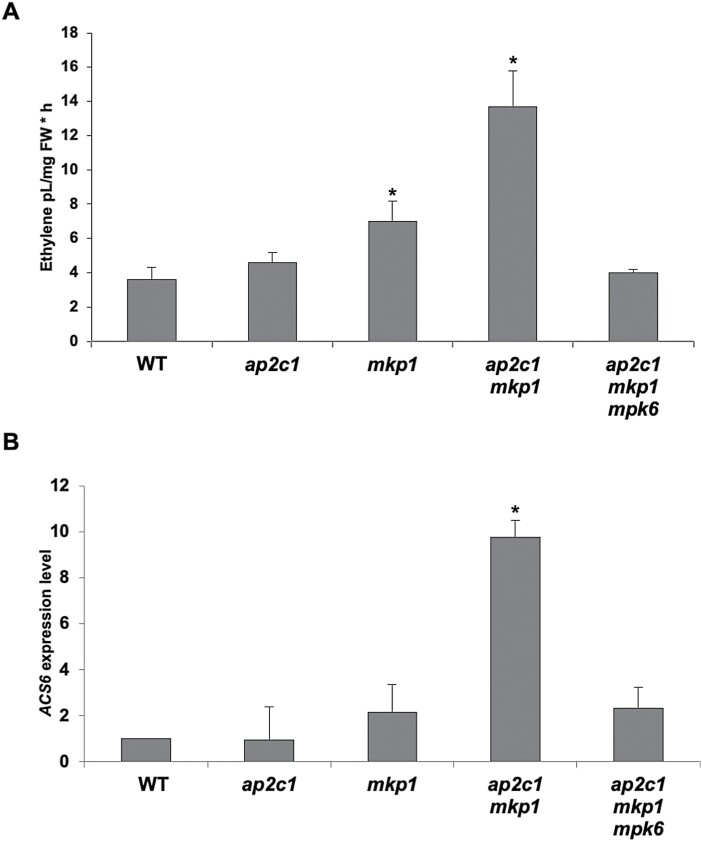
Enhanced ET production is an early response of plants subjected to biotic or abiotic stresses (Wang et al., 2002; Ju and Chang, 2012). We have previously shown that ectopic expression of AP2C1 suppresses MPK6 activation and wound-induced ET production in plant leaves (Schweighofer et al., 2007). Since both AP2C1 and MKP1 control MPK6 activity, a major determinant in the regulation of ET biosynthesis (Liu and Zhang, 2004; Li et al., 2012), we analysed the amounts of wound-induced ET in leaves of WT, ap2c1, mkp1, ap2c1 mkp1, and ap2c1 mkp1 mpk6 plants. As reported previously (Schweighofer et al., 2007), the wound-induced ET amounts were similar in ap2c1 and WT plants (Fig. 3A). However, significantly greater (P<0.05) amounts of ET accumulated in wounded mkp1 plants and even higher amounts in the ap2c1 mkp1 double mutant (Fig. 3A). Our data suggest a primary role of MKP1 in the control of wound-triggered ET production and that although disruption of AP2C1 alone is not sufficient to alter ET production upon wounding, it contributes significantly to the regulation of ET amounts in the absence of MKP1. Interestingly, and in agreement with the overall milder phenotype, wound-induced ET accumulation in ap2c1 mkp1 mpk6 plants was similar that detected in the WT (Fig. 3A).
Fig. 3.
ap2c1 mkp1 plants have higher ACS6 expression and produce more ET upon wounding than WT plants, mainly mediated by MPK6. (A) ET levels produced by 4-week-old plants of the indicated genotypes grown under standard long-day conditions. ET amounts are shown as pl mg–1 plant fresh weight (FW) per hour. Bars represent mean ±SD values of three biological replicates. ∗P<0.05 (Student’s t-test). (B) RT–qPCR analysis of ACS6 expression in leaves of 6-week-old plants of the indicated genotypes grown under short-day conditions; expression levels in the WT are set to 1. Bars represent mean ±SD values of three biological replicates. ∗P<0.05 (Student’s t-test).
The transcriptional regulation of 1-aminocyclopropane-1-carboxylic synthase (ACS) enzymes contributes to the control of ET production (Li et al., 2012). Therefore, we quantified the transcripts of ACS6, the expression of which is significantly induced after pathogen attack (Li et al., 2012) and wounding (Li et al., 2018). Compared with the WT, no changes in ACS6 transcript levels were detected in ap2c1, slightly higher levels in mkp1, and a 9-fold increase in ap2c1 mkp1, whereas in ap2c1 mkp1 mpk6 plants levels were not significantly different from those in WT plants (Fig. 3B). Thus, our data show that ACS6 is more strongly expressed in ap2c1 mkp1 plants, which likely contributes to the increased amounts of ET upon wounding, and that both effects are mediated by MPK6. Taken together, the wound-induced MAPK activities, expression patterns, and effects on ET production suggest that AP2C1 and MKP1 have both distinct and overlapping functions in wounded leaves.
Transcription factor gene expression is deregulated in ap2c1 mkp1 plants
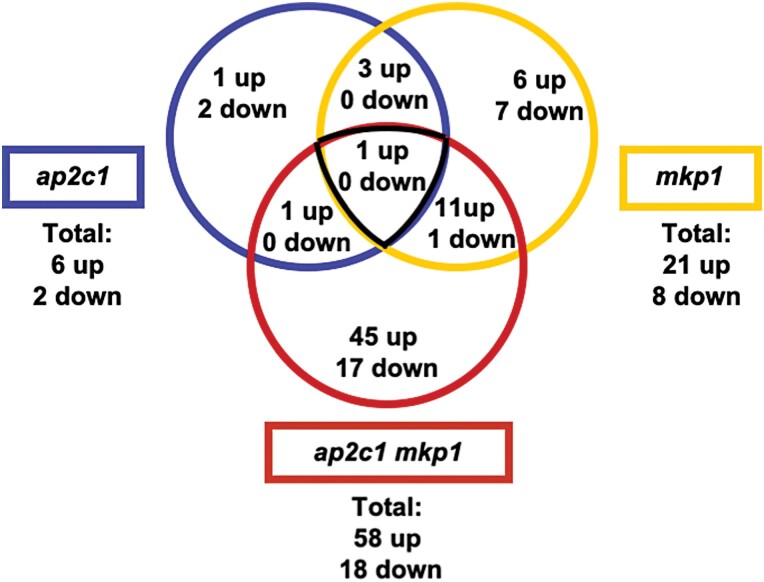
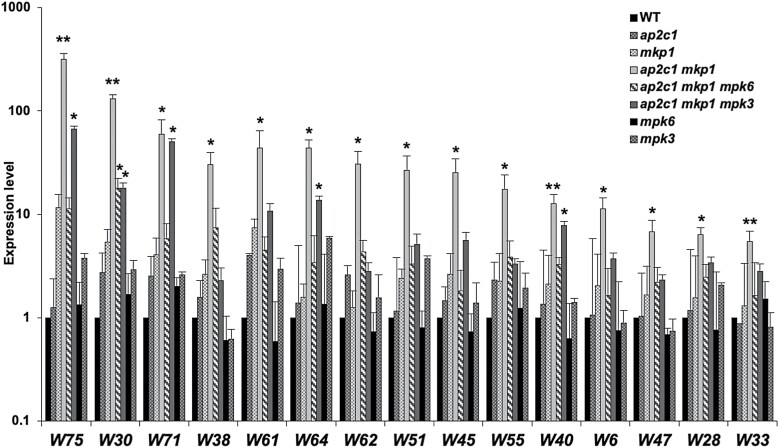
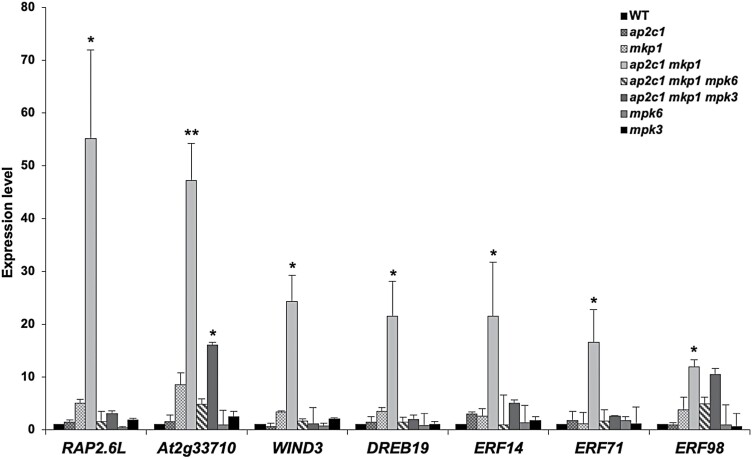
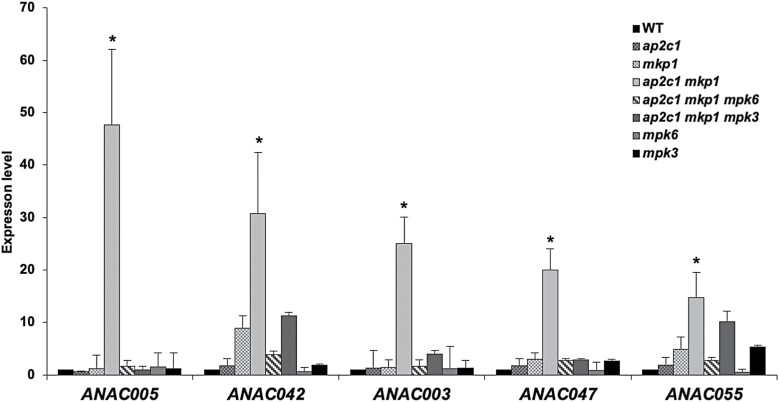
To investigate if and how AP2C1 and MKP1 influence the regulation of gene expression under standard growth conditions, we used an RT–qPCR platform for high-throughput expression profiling of 1880 Arabidopsis TF-encoding genes (Balazadeh et al., 2008). We selected genes showing an at least 3-fold mean difference of expression levels in ap2c1, mkp1, or ap2c1 mkp1 plants compared with the WT. We identified three genes encoding TFs that were deregulated in ap2c1 but not in mkp1 (Supplementary Table S1), 25 genes that were deregulated in mkp1 but not in ap2c1 (Supplementary Table S2), and four genes concomitantly regulated by AP2C1 and MKP1 (Supplementary Table S3). Figure 4 shows the number of genes whose expression levels were changed in ap2c1, mkp1, or ap2c1 mkp1 plants, compared with the WT. The TF genes dysregulated in the double mutant, and their expression values relative to the WT, are presented in Supplementary Table S4. The deregulation of 76 TF-encoding genes (58 up-regulated, 18 down-regulated) was found reproducibly in at least three different experiments in ap2c1 mkp1 double mutant plants. Among them, genes encoding members of the WRKY family were most abundant: 15 WRKY genes were up-regulated (Fig. 5, Supplementary Table S4) and one was down-regulated (Supplementary Table S4). A further prevalent group of TF-encoding genes affected in ap2c1 mkp1 plants includes AP2/ERF genes, described for their involvement in development, including RAP2.6L (Yang et al., 2018) and WIND3 (Smit et al., 2020) (Fig. 6). ANAC TF family members are implicated in senescence and stress-related processes (Bu et al., 2008; Jensen et al., 2010; Wu et al., 2012; Saga et al., 2012). Our results show that several ANAC TF-encoding genes were up-regulated in ap2c1 mkp1 plants (Fig. 7). Thus, our data suggest a cooperative function of AP2C1 and MKP1 in the transcriptional regulation of a set of WRKY, AP2/ERF, and ANAC genes in the WT.
Fig. 4.
Venn diagram of TF genes differentially expressed in the MAPK phosphatase mutant plants. The number of genes up- or down-regulated at least 3-fold in ap2c1, mkp1, and ap2c1 mkp1 plants compared with the WT in three biological replicates is indicated. The expression of 1880 TF-encoding genes was analysed.
Fig. 5.
Expression of WRKY-encoding genes. The transcript levels of WRKY-encoding (W) genes were quantified by RT–qPCR in plants of the indicated genotypes and compared with the WT (expression levels in the WT were set to 1). Bars represent mean ±SD values of three replicates. Data are plotted on a log10 scale after normalization over WT values. ∗P<0.05, ∗∗P<0.01 (Mann–Whitney U test).
Fig. 6.
Genes encoding members of the AP2/ERF TF family are highly up-regulated in ap2c1 mkp1 plants. Transcript levels of AP2/ERF-encoding genes were quantified by RT–qPCR in plants of the indicated genotypes and compared with the WT (expression levels in the WT were set to 1). Bars represent mean ±SD values of at least three replicates. ∗P<0.05, ∗∗P<0.01 (Mann–Whitney U test).
Fig. 7.
Genes encoding members of the ANAC TF family are highly up-regulated in ap2c1 mkp1 plants. Transcript levels of ANAC genes were quantified by RT–qPCR in plants of the indicated genotypes and compared with the WT (expression levels in the WT were set to 1). Bars represent mean ±SD values of at least three replicates. ∗P<0.05 (Mann-Whitney U test).
Our observation that ap2c1 mkp1 mpk6 and ap2c1 mkp1 mpk3 plants are phenotypically much less affected than the ap2c1 mkp1 double mutant suggested that severe phenotypic aberrations in the double mutant are mediated mainly by MPK6 and probably to a lesser extent by MPK3. This prompted us to investigate ap2c1 mkp1 mpk6 and ap2c1 mkp1 mpk3 plants for the expression of TFs misregulated in ap2c1 mkp1. Indeed, most of the TF genes strongly affected in ap2c1 mkp1 plants (Supplementary Table S4) were not significantly altered in their expression in ap2c1 mkp1 mpk6 and ap2c1 mkp1 mpk3, linking MAPK over-activation to the TF misexpression in the double mutant (Figs 5–7). Among these, the expression of WRKY75, WRKY71, WRKY64, and WRKY40 (Fig. 5) and of At2g33710 (Fig. 6) in the double mutant are more MPK3-independent, while the expression of other identified WRKYs (Fig. 5), the AP2/ERF family members RAP2.6L, WIND3, DREB19, ERF14, ERF17, ERF71, and ERF98 (Fig. 6), and several ANAC TF-encoding genes, such as ANAC005, ANAC042, ANAC003, ANAC047, and ANAC055 (Fig. 7), are mainly dependent on the presence of both MPK6 and MPK3.
Defence responses, camalexin, SA, and the senescence marker gene SAG12 are up-regulated in ap2c1 mkp1 plants
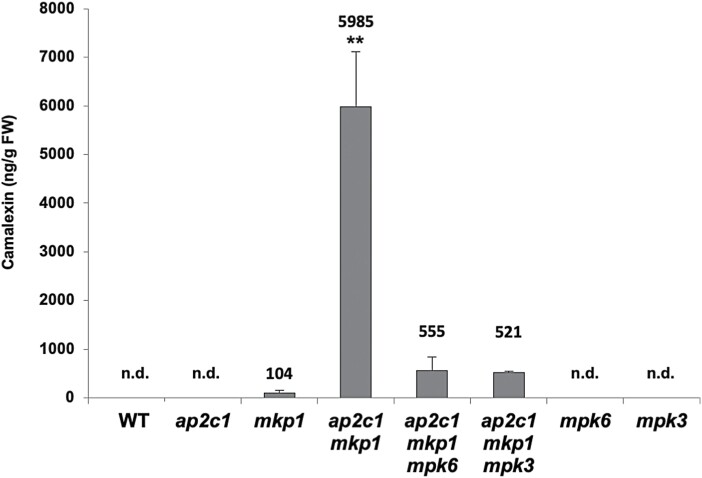
It has been shown previously that mkp1 plants accumulate higher levels of the phytoalexin camalexin (Bartels et al., 2009). To investigate whether the expression of genes encoding camalexin biosynthesis enzymes was affected in ap2c1 mkp1 plants, we studied the expression of a key gene in the pathway, CYP71B15/PAD3. A strong up-regulation (>300-fold) was detected in ap2c1 mkp1 plants compared with the WT (Supplementary Fig. S6). Moreover, ap2c1 mkp1 mpk6 plants still had remarkably high transcript levels (up-regulation >10-fold) of CYP71B15/PAD3. In addition, mkp1 single mutant plants showed a ~10-fold up-regulation of the gene (Supplementary Fig. S6).
To investigate whether the increased CYP71B15/PAD3 expression level correlates with camalexin accumulation, total camalexin was quantified in WT and mutant plants. Indeed, we found increased camalexin concentrations in mkp1 plants, in agreement with previous findings (Bartels et al., 2009), and very high camalexin accumulation in the ap2c1 mkp1 mutant, which was not solely dependent on MPK6 and MPK3 (Fig. 8).
Fig. 8.
Camalexin accumulation in ap2c1 mkp1 plants is mainly mediated by MPK6 and MPK3. Concentrations of total camalexin determined by HPLC in leaves of 4-week-old plants of the indicated genotypes. Results shown are means ±SE (n=4). n.d., Not detected. ∗∗P<0.01 (Student’s t-test).
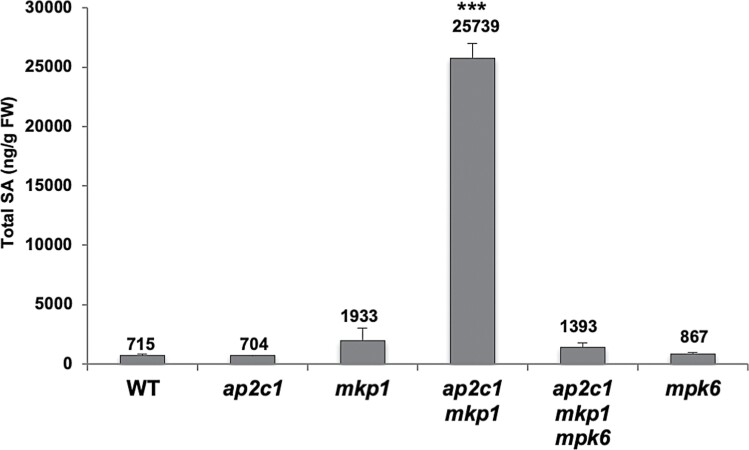
The up-regulation of MAPK activities and macroscopic lesion formation in leaves of ap2c1 mkp1 indicated the possible activation of a hypersensitive-like response in these plants. Since this response is associated with the accumulation of the stress hormone SA, we measured SA in leaves of ap2c1 mkp1 and ap2c1 mkp1 mpk6, as well as in WT and the single mutants. We found a 35-fold increase of SA in ap2c1 mkp1 plants compared with the WT (Fig. 9), whereas ap2c1, ap2c1 mkp1 mpk6, and mpk6 plants showed SA concentrations similar to the WT. In agreement with previous data (Bartels et al., 2009), we also detected enhanced total SA concentrations (>2-fold) in mkp1 plants compared with the WT (Fig. 9).
Fig. 9.
ap2c1 mkp1 plants accumulate high concentrations of SA in an MPK6-dependent manner. Total SA concentrations of 5-week-old plants of the indicated genotypes grown under standard short-day conditions were determined by HPLC and expressed as ng g–1 FW. Error bars represent the SD of four biological replicates. ∗∗∗P<0.001 (Student’s t-test).
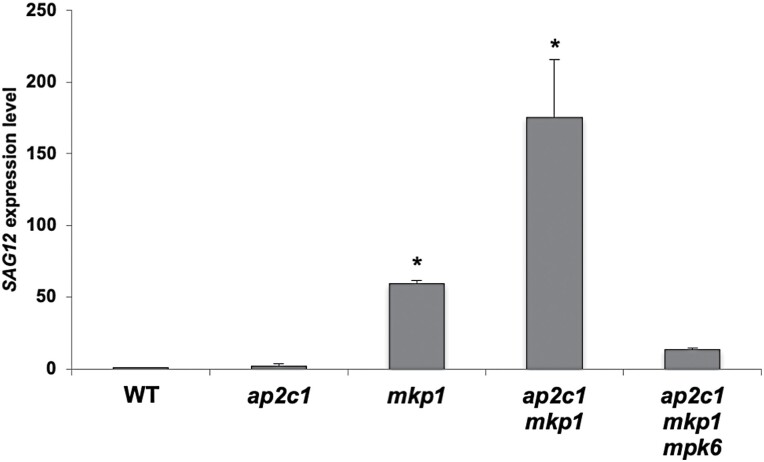
The leaf necrosis observed in ap2c1 mkp1 leaves (Fig. 1A; Supplementary Fig. S1G) and the up-regulation of WRKY6 (Fig. 5), which is a senescence-related marker gene (Rushton et al., 2010), suggested that early senescence was induced in these plants. Thus, we investigated the expression of the senescence-specific marker gene SENESCENCE-ASSOCIATED GENE12 (SAG12) (Noh and Amasino, 1999) and found that it was strongly up-regulated in ap2c1 mkp1 plants, dependent on MPK6 (Fig. 10). These data, along with the up-regulation of WRKY6 (Fig. 5), indicate aberrant early induction of senescence-related processes in the double phosphatase mutant.
Fig. 10.
Up-regulation of the senescence-marker gene SAG12 in ap2c1 mkp1 plants is mainly mediated by MPK6. RT–qPCR quantification of SAG12 transcript level in plants of the indicated genotypes compared with WT plants grown under standard short-day conditions. Error bars represent the SD of three biological replicates. ∗P<0.05 (Student’s t-test).
Discussion
Coordinated control of MAPK activities by AP2C1 and MKP1
Acclimation for survival is a fundamental principle that relies on intracellular signalling in every organism. Different signals converge at the level of MAPK cascades and from there diverge into a range of different downstream pathways and responses (Andreasson and Ellis, 2010; Rodriguez et al., 2010; Rasmussen et al., 2012). Considering the broad spectrum of signals transduced by overlapping players of MAPK pathways, it is puzzling how response specificity is attained (Lampard et al., 2009; Rodriguez et al., 2010; Meng and Zhang, 2013). Several signalling scenarios have been investigated that could help explain pathway specificity, including activity-dependent kinase distribution and localization, protein complex formation (e.g. interaction with scaffolding proteins), and dephosphorylation by protein phosphatases (Krysan and Colcombet, 2018). Over the past decades, mainly the functions of MPK3/MPK4/MPK6 in diverse pathways have been described, indicating them as both points of divergence and integration hubs in cellular signalling (Peng et al., 2018; Bigeard and Hirt, 2018).
Here, we provide evidence that two evolutionarily distinct MAPK phosphatases control stress-related signalling in Arabidopsis by inactivating an overlapping set of target MAPKs that mediate stress and defence responses. The Ser/Thr PP2C phosphatase AP2C1 and the DSP MKP1 contribute to ensure the appropriate inactivation of MAPKs during stress. Both AP2C1 and MKP1 target MPK3, MPK4, and MPK6 (Ulm et al., 2002; Schweighofer et al., 2007; Bartels et al., 2009; Anderson et al., 2011; Galletti et al., 2011; Sidonskaya et al., 2016; Shubchynskyy et al., 2017). The enhanced activation of MAPKs by wounding and by PAMP (flg22), and the constitutive stress signalling in the absence of stress in ap2c1 mkp1 plants, indicate that the lack of both MAPK phosphatases creates a shortfall downstream of MAPKs, exemplified by the deregulated expression of TF-encoding genes.
The increased kinase activity in mkp1 plants versus WT at the earlier time points after wounding and flg22 application compared with ap2c1 versus WT suggests that the contribution of MKP1 to inactivating MAPKs is already set before stress treatment, or during a very early stage of signalling. In agreement with this suggested function of MKP1 during early signalling, previous work has also shown that mkp1 and mkp1 ptp1 mutant plants demonstrate equally elevated MAPK activities without stress treatment, underlining the major role of MKP1 in MAPK regulation in ambient conditions (Bartels et al., 2009). On the contrary, AP2C1 adds to MAPK inactivation at later time points. It is possible that AP2C1 is primarily responsible for keeping the stress-induced activation below a certain threshold and controlling the duration of kinase activation during acute stress, acting as an ‘emergency brake’, while MKP1 is predominantly responsible for suppressing kinase activities under normal conditions, providing a ‘constitutive brake’. This hypothesis is supported by the induction of AP2C1 expression by a plethora of stresses, while MKP1 shows comparatively marginal changes in expression (https://www.genevestigator.com). These observations are also consistent with a recent comprehensive analysis of the Arabidopsis proteome, which covers more than 14 000 proteins and where in ambient conditions the overall MKP1 abundance exceeds by far that of AP2C1 (http://athena.proteomics.wzw.tum.de/) (Mergner et al., 2020), underlining the rather specific role of AP2C1 under stress conditions. The AP2C1 paralogues AP2C2 and AP2C3 (Umbrasaite et al., 2010; Umbrasaite et al., 2011; Schweighofer et al., 2014) as well as MKP1 and PTP1 interact with the same MAPKs and dephosphorylate them to various extents (Bartels et al., 2009). However, the rather mild phenotypes of ap2c2 mkp1 and ap2c3 mkp1 plants, and the WT-like appearance of ap2c1 ptp1 (Fig. 1C) compared with ap2c1 mkp1 plants, clearly indicate specific genetic interactions and redundant functions of the evolutionarily distant AP2C1 and MKP1 phosphatases in the regulation of signalling pathways.
Manifestation of cell death in ap2c1 mkp1 plants
The lesions in leaves of ap2c1 mkp1 plants suggest autoimmune-like responses, most likely caused by misregulation of MAPKs and/or failed control of guarding resistance (R) proteins (Rodriguez et al., 2016). AP2C1 and MKP1 share the target MAPKs MPK3, MPK4, and MPK6, where MPK4 and some of its upstream MAPK cascade members, for example, MEKK1 and MKK1/2, were originally described as negative regulators of plant immunity based on their mutant plant phenotypes (Petersen et al., 2000; Rasmussen et al., 2012). The improper activation of the R-gene SUMM2 is mainly responsible for the phenotypic defects of the mpk4 mutant and other mutant plants in the pathway, identifying the MEKK1-MKK1/2-MPK4 module as a positive regulator of stress responses (Zhang et al., 2012). Similar observations connecting phosphatase-targeted MAPKs with autoimmune-like phenotypes have been made by ectopically expressing constitutively active MPK3 (Genot et al., 2017) or by inducibly expressing MKK5, which activates MPK3 and MPK6 (Lassowskat et al., 2014). Both approaches led to a plethora of phenotypic and molecular changes, including dwarfism, lesion formation, de-repression of defence gene expression, and the accumulation of stress hormones, similar to the ap2c1 mkp1-related phenotypes described in this work (Figs 1–10; Supplementary Figs S1, S3, S4, S6; Supplementary Table S4).
The single mkp1 and double mkp1 ptp1 mutants show constitutive defence responses including increased levels of SA and camalexin, suggesting partially overlapping functions of MKP1 and PTP1 in repressing SA biosynthesis (Bartels et al., 2009). Similarly, the strong accumulation of SA and camalexin in ap2c1 mkp1 compared with mkp1 plants suggests a collaborative action of both AP2C1 and MKP1 as negative regulators of SA and camalexin production (Figs 8, 9; Supplementary Fig. S6). The camalexin accumulation in ap2c1 mkp1 is largely reduced in ap2c1 mkp1 mpk6 and ap2c1 mkp1 mpk3 triple mutants, indicating the dependency of camalexin biosynthesis on partially redundant actions of MPK6 and MPK3, which is in agreement with previous findings (Mao et al., 2011). The SA accumulation in ap2c1 mkp1 plants is probably mainly MPK6-dependent, as the introduction of the mpk6 mutation in ap2c1 mkp1 mpk6 plants restores SA concentrations similar to those of WT and mkp1 ptp1 mpk6 mutant plants (Bartels et al., 2009). Notably, rescue of the severe ap2c1 mkp1 growth phenotypes by elevated temperature is in accordance with the observed temperature dependency of SA-related phenotypes (Ichimura et al., 2006; Suarez-Rodriguez et al., 2007; Su et al., 2007), as well as with the suppression of SNC1 expression and reduction of SNC1 activity by high temperature (Yang and Hua, 2004; Zhu et al., 2010). The resistance protein SNC1 is a modifier of mkp1 in the Col-0 accession, where partial rescue of mkp1 and mkp1 ptp1 growth phenotypes by a loss-of-function snc1 mutation indicates a sensitized SNC1 signalling pathway in the absence of MKP1 (Bartels et al., 2009).
Previous findings that SA acts together with ET to regulate cell death (Rao et al., 2002), the requirement of ET biosynthesis for H2O2 accumulation and subsequent cell death (Overmyer et al., 2003), and the induction of cell death in Arabidopsis leaves by persistent activation of MAPKs with gain-of-function MKK4 and MKK5 (Ren et al., 2002) all correlate with the cell-death phenotype observed in the ap2c1 mkp1 mutant, where MAPKs—and other stress-related factors—may be (hyper)-activated. Therefore, we conclude that the majority of the phenotypes observed in ap2c1 mkp1 plants, both visible and molecular, are due to the misregulation of MAPK pathways, even in the absence of stress.
AP2C1 and MKP1 affect MAPK-regulated ET biosynthesis
Activated MPK6 controls ET levels by inducing the transcription of ACS family genes and by phosphorylating ACS proteins, the rate-limiting enzymes in ET biosynthesis. Phosphorylated ACSs become more stable and, thus, ET synthesis is increased by elevated MPK6 activity (Kim et al., 2003; Liu and Zhang, 2004; Xu et al., 2008; Li et al., 2012). In ap2c1 mkp1, the increased ET production is likely due to, at least in part, the highly increased expression of ACS6 compared with the WT. A considerable additive effect on ET overproduction in the double ap2c1 mkp1 mutant compared with the mkp1 single mutant suggests that even though MKP1 is a determining MAPK phosphatase affecting ET production, there are overlapping and non-redundant functions of AP2C1 and MKP1 in the regulation of stress-induced ET biosynthesis. The detection of increased and MPK6-dependent expression of WRKY33, which encodes a TF that binds to the promoter of ACS genes and is a substrate of MPK3 and MPK6, suggests an involvement of WKRY33 itself in ACS overexpression in ap2c1 mkp1 plants (Figs 3, 5; Li et al., 2012). The identification of genes encoding TFs of the AP2/ERF family (ET-responsive element-binding proteins) among the most strongly induced ones in ap2c1 mkp1 plants suggests a path to increased ET amounts in these plants.
AP2C1 and MKP1 control the expression of stress-responsive TF-encoding genes, predominantly via partially redundant actions of MPK6 and MPK3
Transcriptional reprogramming in response to activated MAPK signalling suggests an involvement of TFs. Our results indicate that the concomitant lack of the MAPK regulators AP2C1 and MKP1 results in elevated basal MAPK activities and leads to highly increased expression of WRKY TF genes, in some cases by more than 100-fold compared with the WT. The ap2c1 mkp1 mutant phenotypes and the described functions of some up-regulated WRKYs indicate that stress responses are constitutively active in these plants. This correlates with reports demonstrating an involvement of WRKYs in oxidative stress responses, in the induction of ET and camalexin biosynthesis (WRKY30, WRKY33), the response to pathogens (WRKY71, WRKY40), basal defence (WRKY38), and defence- and senescence-related processes (WRKY6) (Rushton et al., 2010).
Direct feedback mechanisms among WRKYs themselves have been demonstrated (Mao et al., 2011) and are generally proposed, where WRKYs positively auto-regulate their own gene expression and/or cross-regulate the expression of other WRKY genes (Pandey and Somssich, 2009; Mao et al., 2011; Birkenbihl et al., 2017). Thus, it could be that the increased activation of MAPKs in ap2c1 mkp1 plants leads to phosphorylation and thus activation of MAPK target WRKY proteins, which serve as activated TFs for a further series of WRKY genes. In any case, MPK6 and MPK3 seem to be major players responsible for mediating the up-regulation of several WRKYs, AP2/ERFs, ANACs, and other TF-encoding genes. Both MAPKs control the expression of several WRKYs to different extents, as shown in ap2c1 mkp1 plants compared with ap2c1 mkp1 mpk6 and ap2c1 mkp1 mpk3 plants (Fig. 5). These data also demonstrate that not only MAPKs but also other factor(s) affect WRKY gene expression. We confirmed MPK6- and MPK3-dependent WRKY33 expression (Mao et al., 2011); however, the higher MPK4 activities in ap2c1 mkp1 may also lead to higher amounts of active WRKY33 protein (Qiu et al., 2008; Birkenbihl et al., 2017). Our data suggest that AP2C1 and MKP1 may play a dual role in regulating camalexin biosynthesis, on the one hand by controlling MPK6 and MPK3 activities, which positively regulate WKRY33 expression, and on the other hand by controlling MPK4 activity, which in turn stimulates WRKY33, leading to the transactivation of CYP71B15/PAD3.
Remarkably, the concomitant absence of AP2C1 and MKP1 in non-stress conditions causes a distinct transcriptional activation of TFs compared with single mutant ap2c1 and mkp1 plants after stress treatments: challenging ap2c1 plants with Pto for 4 h led to 88 differentially regulated TFs (Shubchynskyy et al., 2017), whereas only four TFs from this set were up-regulated in untreated ap2c1 mkp1 plants (Supplementary Table S4).
Comparing transcriptional changes of PAMP-treated mkp1 plants (Jiang et al., 2017) with untreated ap2c1 mkp1 double mutant plants (Supplementary Table S4) revealed a more MKP1-specific TF induction: a next-generation sequencing (RNA-seq) transcriptome analysis of mkp1 plants 90 min after treatment with the PAMP elf26 revealed that from 67 identified TFs among the 1102 MKP1-dependent transcripts (Jiang et al., 2017) 21 TFs were also changed in untreated ap2c1 mkp1 plants (Supplementary Table S4). In accordance with the severe phenotype of mkp1 ptp1 mutant plants and its suppression by elevated growth temperature or crossing with mpk3 and mpk6 mutants (Bartels et al., 2009), the proposed predominant role of MKP1 under non-stress conditions and AP2C1 during stress conditions to regulate (MAPK) signalling are emphasized.
A PAMP flg22-activated MPK3/MPK6 pathway was previously reported to elevate WRKY22 and WRKY29 expression (Asai et al., 2002). Strongly enhanced MPK3/MPK6 activities, but unaffected expression of either WRKY22 or WRKY29 in untreated ap2c1 mkp1 plants, after PAMP elf26 and Pto treatment (Jiang et al., 2017; Shubchynskyy et al., 2017) show that for WRKY22/29 overexpression, MPK3 and MPK6 hyperactivation is necessary (Asai et al., 2002) but not sufficient (Fig. 5; Supplementary Table S4), and that other factors (possibly MAPKs) may be playing a role instead of MPK3 and MPK6.
Senescence is repressed by AP2C1 and MKP1 phosphatases in an MPK6-dependent way
Several lines of evidence indicate that the ap2c1 mkp1 mutant undergoes precocious senescence. Leaf senescence is a highly regulated process that finally leads to cell death and tissue disintegration, at the same time contributing to the fitness of the whole plant. Senescence is controlled by endogenous and environmental cues, and can be triggered prematurely by different abiotic/biotic stresses such as pathogen attack, wounding, UV light irradiation, or high ozone levels (Hanfrey et al., 1996; Miller et al., 1999; John et al., 2001; He et al., 2001; Lim et al., 2007). The MKK9-MPK6 cascade has been shown to positively regulate leaf senescence in Arabidopsis (Zhou et al., 2009). Hyperactivation of MPK6 and other MAPKs, in addition to autoimmune-like responses, also promotes senescence, which is very evident in older leaves of ap2c1 mkp1 plants and correlates with significant up-regulation of the senescence-specific marker gene SAG12 (Noh and Amasino, 1999; Guo and Gan, 2005). Partial suppression of SAG12 overexpression in ap2c1 mkp1 mpk6 suggests an MPK6-dependent regulation (possibly involving other MAPKs) in promoting plant senescence.
Genome-wide transcriptomics previously identified several senescence-related TFs from the ANAC family (Breeze et al., 2011). We identified strong MAPK-dependent induction of ANAC005, JUB1/ANAC042 (Wu et al., 2012; Saga et al., 2012; Shahnejat-Bushehri et al., 2016), ANAC003/XVP (Yang et al., 2020), ANAC047 (Mito et al., 2011), and ANAC055 (Tran et al., 2004; Bu et al., 2008; Hickman et al., 2013; Schweizer et al., 2013) in ap2c1 mkp1 plants. This induction of senescence-related TFs reveals a novel link between senescence-related processes and MAPK signalling.
We conclude that the induction of senescence processes as well as hypersensitive response-like cell death results in the premature death of leaves in ap2c1 mkp1 plants. The crosstalk between senescence and abiotic stress or pathogen responses is accentuated in ap2c1 mkp1 plants, where up-regulation of TFs involved in these processes occurs.
Taken together, our results show that two evolutionarily unrelated MAPK phosphatases, AP2C1 and MKP1, perform both distinct and overlapping functions in the regulation of stress-induced MPK3, MPK4, and MPK6 activities. Our genetic dissection indicates that the known roles of MPK6 and MPK3 in mediating cell death and ET-, SA- and senescence-related phenotypes are counter-balanced by both AP2C1 and MKP1. It also demonstrates that the expression of specific TF-encoding genes is affected by hyperactivation of MAPK(s) due to the lack of these two MAPK phosphatases in planta, revealing potential new signalling target genes downstream of MPK6 and MPK3. In the future, the study of individual and combinatorial mutants will allow us to genetically disentangle the contribution of specific protein kinases and phosphatases to complex signalling networks and downstream cell responses.
Supplementary data
The following supplementary data are available at JXB online.
Fig. S1. Loss of both AP2C1 and MKP1 leads to severe phenotypes in growth and development.
Fig. S2. Plant phenotypes grown in conditions with increased humidity and elevated temperature.
Fig. S3. Phenotypes of Arabidopsis single, double and triple mutant plants.
Fig. S4. AP2C1 and MKP1 control stress-induced MAPK activities.
Fig. S5. Detection of MKP1 and AP2C1 expression levels in ap2c1 and mkp1 mutants.
Fig. S6. Up-regulation of the camalexin biosynthetic gene CYP71B15/PAD3 is mainly mediated by MPK6.
Table S1. Expression of TF-encoding genes modulated by the absence of AP2C1 but not MKP1.
Table S2. Expression of TF-encoding genes modulated by the absence of MKP1 but not of AP2C1.
Table S3. Expression of TF-encoding genes modulated by the absence of both MKP1 and AP2C1.
Table S4. TF-encoding genes deregulated in ap2c1 mkp1 plants.
Table S5. The Arabidopsis Genome Initiative (AGI) codes of the genes analysed in this report.
Acknowledgements
We thank Verena Ibl (University of Vienna, Austria), Mary G. Wallis (University of Applied Sciences, FH Campus Vienna, Austria) and Francesca Cardinale (University of Turin, Italy) for critical reading of the manuscript, and the Nottingham Arabidopsis Stock Centre for providing SALK mutant lines.
We wish to dedicate this article to the co-authors who passed away too early: Felix Mauch (1955–2021), Irute Meskiene (1956–2017) (for obituary see Paškauskas et al., (2017)), and Manfred Schwanninger (1963–2013) (for obituary see Meder (2014)).
Author contributions
ZA, VK, VS, KK, FM, RU, SaB, BMR, IM, and AS designed the experiments; ZA, VK, VS, KK, MaS, WR, MiS, SeB, SaB, and AS performed the experiments; ZA, SaB, BMR, IM, and AS wrote the paper.
Conflict of interest
The authors declare no conflicts of interest.
Funding
This work was supported by the Austrian Science Fund (FWF) with the grants I-255, W1220-B09 to IM, and P27254 to IM/AS. SeB was supported by the German Research Foundation (DFG) through the Research Training Group GRK1305. Financial support was provided through the European Union’s Horizon 2020 research and innovation programme, project PlantaSYST (SGA-CSA no. 739582 under FPA no. 664620). BMR and SaB thank the MPI of Molecular Plant Physiology (Potsdam, Germany) for financial support.
Data availability
The data supporting the findings of this study are available from the corresponding author, Alois Schweighofer, upon request.
References
- Anderson JC, Bartels S, Gonzalez Besteiro MA, Shahollari B, Ulm R, Peck SC.. 2011. Arabidopsis MAP Kinase Phosphatase 1 (AtMKP1) negatively regulates MPK6-mediated PAMP responses and resistance against bacteria. The Plant Journal 67, 258–268. [DOI] [PubMed] [Google Scholar]
- Anderson JC, Wan Y, Kim YM, Pasa-Tolic L, Metz TO, Peck SC.. 2014. Decreased abundance of type III secretion system-inducing signals in Arabidopsis mkp1 enhances resistance against Pseudomonas syringae. Proceedings of the National Academy of Sciences, USA 111, 6846–6851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreasson E, Ellis B.. 2010. Convergence and specificity in the Arabidopsis MAPK nexus. Trends in Plant Science 15, 106–113. [DOI] [PubMed] [Google Scholar]
- Asai T, Tena G, Plotnikova J, Willmann MR, Chiu WL, Gomez-Gomez L, Boller T, Ausubel FM, Sheen J.. 2002. MAP kinase signalling cascade in Arabidopsis innate immunity. Nature 415, 977–983. [DOI] [PubMed] [Google Scholar]
- Balazadeh S, Riano-Pachon DM, Mueller-Roeber B.. 2008. Transcription factors regulating leaf senescence in Arabidopsis thaliana. Plant Biology 10 (Suppl 1), 63–75. [DOI] [PubMed] [Google Scholar]
- Bartels S, Anderson JC, Gonzalez Besteiro MA, Carreri A, Hirt H, Buchala A, Metraux JP, Peck SC, Ulm R.. 2009. MAP KINASE PHOSPHATASE 1 and PROTEIN TYROSINE PHOSPHATASE 1 are repressors of salicylic acid synthesis and SNC1-mediated responses in Arabidopsis. The Plant Cell 21, 2884–2897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartels S, Gonzalez Besteiro MA, Lang D, Ulm R.. 2010. Emerging functions for plant MAP kinase phosphatases. Trends in Plant Science 15, 322–329. [DOI] [PubMed] [Google Scholar]
- Bethke G, Unthan T, Uhrig JF, Poschl Y, Gust AA, Scheel D, Lee J.. 2009. Flg22 regulates the release of an ethylene response factor substrate from MAP kinase 6 in Arabidopsis thaliana via ethylene signaling. Proceedings of the National Academy of Sciences, USA 106, 8067–8072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bigeard J, Hirt H.. 2018. Nuclear signaling of plant MAPKs. Frontiers in Plant Science 9, 469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birkenbihl RP, Kracher B, Roccaro M, Somssich IE.. 2017. Induced genome-wide binding of three Arabidopsis WRKY transcription factors during early MAMP-triggered immunity. The Plant Cell 29, 20–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breeze E, Harrison E, McHattie S, et al. 2011. High-resolution temporal profiling of transcripts during Arabidopsis leaf senescence reveals a distinct chronology of processes and regulation. The Plant Cell 23, 873–894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bu Q, Jiang H, Li CB, Zhai Q, Zhang J, Wu X, Sun J, Xie Q, Li C.. 2008. Role of the Arabidopsis thaliana NAC transcription factors ANAC019 and ANAC055 in regulating jasmonic acid-signaled defense responses. Cell Research 18, 756–767. [DOI] [PubMed] [Google Scholar]
- Caunt CJ, Keyse SM.. 2013. Dual-specificity MAP kinase phosphatases (MKPs): shaping the outcome of MAP kinase signalling. FEBS Journal 280, 489–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czechowski T, Bari RP, Stitt M, Scheible WR, Udvardi MK.. 2004. Real-time RT-PCR profiling of over 1400 Arabidopsis transcription factors: unprecedented sensitivity reveals novel root- and shoot-specific genes. The Plant Journal 38, 366–379. [DOI] [PubMed] [Google Scholar]
- Escudero V, Torres MA, Delgado M, et al. 2019. Mitogen-activated protein kinase phosphatase 1 (MKP1) negatively regulates the production of reactive oxygen species during Arabidopsis immune responses. Molecular Plant-Microbe Interactions 32, 464–478. [DOI] [PubMed] [Google Scholar]
- Fuchs S, Grill E, Meskiene I, Schweighofer A.. 2013. Type 2C protein phosphatases in plants. FEBS Journal 280, 681–693. [DOI] [PubMed] [Google Scholar]
- Galletti R, Ferrari S, De Lorenzo G.. 2011. Arabidopsis MPK3 and MPK6 play different roles in basal and oligogalacturonide- or flagellin-induced resistance against Botrytis cinerea. Plant Physiology 157, 804–814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genot B, Lang J, Berriri S, Garmier M, Gilard F, Pateyron S, Haustraete K, Van Der Straeten D, Hirt H, Colcombet J.. 2017. Constitutively active Arabidopsis MAP kinase 3 triggers defense responses involving salicylic acid and SUMM2 resistance protein. Plant Physiology 174, 1238–1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez Besteiro MA, Bartels S, Albert A, Ulm R.. 2011. Arabidopsis MAP kinase phosphatase 1 and its target MAP kinases 3 and 6 antagonistically determine UV-B stress tolerance, independent of the UVR8 photoreceptor pathway. The Plant Journal 68, 727–737. [DOI] [PubMed] [Google Scholar]
- Gonzalez Besteiro MA, Ulm R.. 2013a. ATR and MKP1 play distinct roles in response to UV-B stress in Arabidopsis. The Plant Journal 73, 1034–1043. [DOI] [PubMed] [Google Scholar]
- Gonzalez Besteiro MA, Ulm R.. 2013b. Phosphorylation and stabilization of Arabidopsis MAP kinase phosphatase 1 in response to UV-B stress. Journal of Biological Chemistry 288, 480–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan Y, Meng X, Khanna R, LaMontagne E, Liu Y, Zhang S.. 2014. Phosphorylation of a WRKY transcription factor by MAPKs is required for pollen development and function in Arabidopsis. PLoS Genetics 10, e1004384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y, Gan S.. 2005. Leaf senescence: signals, execution, and regulation. Current Topics in Developmental Biology 71, 83–112. [DOI] [PubMed] [Google Scholar]
- Hanfrey C, Fife M, Buchanan-Wollaston V.. 1996. Leaf senescence in Brassica napus: expression of genes encoding pathogenesis-related proteins. Plant Molecular Biology 30, 597–609. [DOI] [PubMed] [Google Scholar]
- He Y, Tang W, Swain JD, Green AL, Jack TP, Gan S.. 2001. Networking senescence-regulating pathways by using Arabidopsis enhancer trap lines. Plant Physiology 126, 707–716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hickman R, Hill C, Penfold CA, et al. 2013. A local regulatory network around three NAC transcription factors in stress responses and senescence in Arabidopsis leaves. The Plant Journal 75, 26–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichimura K, Casais C, Peck SC, Shinozaki K, Shirasu K.. 2006. MEKK1 is required for MPK4 activation and regulates tissue-specific and temperature-dependent cell death in Arabidopsis. Journal of Biological Chemistry 281, 36969–36976. [DOI] [PubMed] [Google Scholar]
- Jensen MK, Kjaersgaard T, Nielsen MM, Galberg P, Petersen K, O’Shea C, Skriver K.. 2010. The Arabidopsis thaliana NAC transcription factor family: structure–function relationships and determinants of ANAC019 stress signalling. Biochemical Journal 426, 183–196. [DOI] [PubMed] [Google Scholar]
- Jiang L, Chen Y, Luo L, Peck SC.. 2018. Central roles and regulatory mechanisms of dual-specificity MAPK phosphatases in developmental and stress signaling. Frontiers in Plant Science 9, 1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang L, Wan Y, Anderson JC, Hou J, Islam SM, Cheng J, Peck SC.. 2017. Genetic dissection of Arabidopsis MAP kinase phosphatase 1-dependent PAMP-induced transcriptional responses. Journal of Experimental Botany 68, 5207–5220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- John CF, Morris K, Jordan BR, Thomas B, S AH-M.. 2001. Ultraviolet-B exposure leads to up-regulation of senescence-associated genes in Arabidopsis thaliana. Journal of Experimental Botany 52, 1367–1373. [PubMed] [Google Scholar]
- Ju C, Chang C.. 2012. Advances in ethylene signalling: protein complexes at the endoplasmic reticulum membrane. AoB Plants 2012, pls031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim CY, Liu Y, Thorne ET, Yang H, Fukushige H, Gassmann W, Hildebrand D, Sharp RE, Zhang S.. 2003. Activation of a stress-responsive mitogen-activated protein kinase cascade induces the biosynthesis of ethylene in plants. The Plant Cell 15, 2707–2718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim CY, Zhang S.. 2004. Activation of a mitogen-activated protein kinase cascade induces WRKY family of transcription factors and defense genes in tobacco. The Plant Journal 38, 142–151. [DOI] [PubMed] [Google Scholar]
- Krysan PJ, Colcombet J.. 2018. Cellular complexity in MAPK signaling in plants: questions and emerging tools to answer them. Frontiers in Plant Science 9, 1674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lampard GR, Lukowitz W, Ellis BE, Bergmann DC.. 2009. Novel and expanded roles for MAPK signaling in Arabidopsis stomatal cell fate revealed by cell type-specific manipulations. The Plant Cell 21, 3506–3517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lassowskat I, Bottcher C, Eschen-Lippold L, Scheel D, Lee J.. 2014. Sustained mitogen-activated protein kinase activation reprograms defense metabolism and phosphoprotein profile in Arabidopsis thaliana. Frontiers in Plant Science 5, 554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G, Meng X, Wang R, Mao G, Han L, Liu Y, Zhang S.. 2012. Dual-level regulation of ACC synthase activity by MPK3/MPK6 cascade and its downstream WRKY transcription factor during ethylene induction in Arabidopsis. PLoS Genetics 8, e1002767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Han X, Yang L, Deng X, Wu H, Zhang M, Liu Y, Zhang S, Xu J.. 2018. Mitogen-activated protein kinases and calcium-dependent protein kinases are involved in wounding-induced ethylene biosynthesis in Arabidopsis thaliana. Plant, Cell & Environment 41, 134–147. [DOI] [PubMed] [Google Scholar]
- Lim PO, Kim HJ, Nam HG.. 2007. Leaf senescence. Annual Review of Plant Biology 58, 115–136. [DOI] [PubMed] [Google Scholar]
- Liu Y, Ren D, Pike S, Pallardy S, Gassmann W, Zhang S.. 2007. Chloroplast-generated reactive oxygen species are involved in hypersensitive response-like cell death mediated by a mitogen-activated protein kinase cascade. The Plant Journal 51, 941–954. [DOI] [PubMed] [Google Scholar]
- Liu Y, Zhang S.. 2004. Phosphorylation of 1-aminocyclopropane-1-carboxylic acid synthase by MPK6, a stress-responsive mitogen-activated protein kinase, induces ethylene biosynthesis in Arabidopsis. The Plant Cell 16, 3386–3399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao G, Meng X, Liu Y, Zheng Z, Chen Z, Zhang S.. 2011. Phosphorylation of a WRKY transcription factor by two pathogen-responsive MAPKs drives phytoalexin biosynthesis in Arabidopsis. The Plant Cell 23, 1639–1653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall CJ. 1995. Specificity of receptor tyrosine kinase signaling: transient versus sustained extracellular signal-regulated kinase activation. Cell 80, 179–185. [DOI] [PubMed] [Google Scholar]
- Meder R. 2014. Manfred Schwanninger 4 September 1963–25 December 2013. NIR News 25, 18–19. [Google Scholar]
- Meng X, Zhang S.. 2013. MAPK cascades in plant disease resistance signaling. Annual Review of Phytopathology 51, 245–266. [DOI] [PubMed] [Google Scholar]
- Menke FL, Kang HG, Chen Z, Park JM, Kumar D, Klessig DF.. 2005. Tobacco transcription factor WRKY1 is phosphorylated by the MAP kinase SIPK and mediates HR-like cell death in tobacco. Molecular Plant-Microbe Interactions 18, 1027–1034. [DOI] [PubMed] [Google Scholar]
- Mergner J, Frejno M, List M, et al. 2020. Mass-spectrometry-based draft of the Arabidopsis proteome. Nature 579, 409–414. [DOI] [PubMed] [Google Scholar]
- Miller JD, Arteca RN, Pell EJ.. 1999. Senescence-associated gene expression during ozone-induced leaf senescence in Arabidopsis. Plant Physiology 120, 1015–1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mito T, Seki M, Shinozaki K, Ohme-Takagi M, Matsui K.. 2011. Generation of chimeric repressors that confer salt tolerance in Arabidopsis and rice. Plant Biotechnology Journal 9, 736–746. [DOI] [PubMed] [Google Scholar]
- Nakano T, Suzuki K, Fujimura T, Shinshi H.. 2006. Genome-wide analysis of the ERF gene family in Arabidopsis and rice. Plant Physiology 140, 411–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noh YS, Amasino RM.. 1999. Identification of a promoter region responsible for the senescence-specific expression of SAG12. Plant Molecular Biology 41, 181–194. [DOI] [PubMed] [Google Scholar]
- Overmyer K, Brosche M, Kangasjarvi J.. 2003. Reactive oxygen species and hormonal control of cell death. Trends in Plant Science 8, 335–342. [DOI] [PubMed] [Google Scholar]
- Pandey SP, Somssich IE.. 2009. The role of WRKY transcription factors in plant immunity. Plant Physiology 150, 1648–1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paškauskas R, Schweighofer A, Kvederavičiūtė K.. 2017. In memoriam Irutė Meškienė (1956–2017). Botanica 23, 178–182. [Google Scholar]
- Peng Y, van Wersch R, Zhang Y.. 2018. Convergent and divergent signaling in PAMP-triggered immunity and effector-triggered immunity. Molecular Plant-Microbe Interactions 31, 403–409. [DOI] [PubMed] [Google Scholar]
- Petersen M, Brodersen P, Naested H, et al. 2000. Arabidopsis MAP kinase 4 negatively regulates systemic acquired resistance. Cell 103, 1111–1120. [DOI] [PubMed] [Google Scholar]
- Popescu SC, Popescu GV, Bachan S, Zhang Z, Gerstein M, Snyder M, Dinesh-Kumar SP.. 2009. MAPK target networks in Arabidopsis thaliana revealed using functional protein microarrays. Genes & Development 23, 80–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu JL, Fiil BK, Petersen K, et al. 2008. Arabidopsis MAP kinase 4 regulates gene expression through transcription factor release in the nucleus. The EMBO Journal 27, 2214–2221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao MV, Lee HI, Davis KR.. 2002. Ozone-induced ethylene production is dependent on salicylic acid, and both salicylic acid and ethylene act in concert to regulate ozone-induced cell death. The Plant Journal 32, 447–456. [DOI] [PubMed] [Google Scholar]
- Rasmussen MW, Roux M, Petersen M, Mundy J.. 2012. MAP kinase cascades in Arabidopsis innate immunity. Frontiers in Plant Science 3, 169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rauf M, Arif M, Fisahn J, Xue GP, Balazadeh S, Mueller-Roeber B.. 2013. NAC transcription factor SPEEDY HYPONASTIC GROWTH regulates flooding-induced leaf movement in Arabidopsis. The Plant Cell 25, 4941–4955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren D, Yang H, Zhang S.. 2002. Cell death mediated by MAPK is associated with hydrogen peroxide production in Arabidopsis. Journal of Biological Chemistry 277, 559–565. [DOI] [PubMed] [Google Scholar]
- Rodriguez E, El Ghoul H, Mundy J, Petersen M.. 2016. Making sense of plant autoimmunity and ‘negative regulators’. FEBS Journal 283, 1385–1391. [DOI] [PubMed] [Google Scholar]
- Rodriguez MC, Petersen M, Mundy J.. 2010. Mitogen-activated protein kinase signaling in plants. Annual Review of Plant Biology 61, 621–649. [DOI] [PubMed] [Google Scholar]
- Rozhon W, Petutschnig E, Wrzaczek M, Jonak C.. 2005. Quantification of free and total salicylic acid in plants by solid-phase extraction and isocratic high-performance anion-exchange chromatography. Analytical and Bioanalytical Chemistry 382, 1620–1627. [DOI] [PubMed] [Google Scholar]
- Rushton PJ, Somssich IE, Ringler P, Shen QJ.. 2010. WRKY transcription factors. Trends in Plant Science 15, 247–258. [DOI] [PubMed] [Google Scholar]
- Saga H, Ogawa T, Kai K, Suzuki H, Ogata Y, Sakurai N, Shibata D, Ohta D.. 2012. Identification and characterization of ANAC042, a transcription factor family gene involved in the regulation of camalexin biosynthesis in Arabidopsis. Molecular Plant-Microbe Interactions 25, 684–696. [DOI] [PubMed] [Google Scholar]
- Schweighofer A, Ayatollahi Z, Meskiene I.. 2009. Phosphatase activities analyzed by in vivo expressions. Methods in Molecular Biology 479, 247–260. [DOI] [PubMed] [Google Scholar]
- Schweighofer A, Hirt H, Meskiene I.. 2004. Plant PP2C phosphatases: emerging functions in stress signaling. Trends in Plant Science 9, 236–243. [DOI] [PubMed] [Google Scholar]
- Schweighofer A, Kazanaviciute V, Scheikl E, et al. 2007. The PP2C-type phosphatase AP2C1, which negatively regulates MPK4 and MPK6, modulates innate immunity, jasmonic acid, and ethylene levels in Arabidopsis. The Plant Cell 19, 2213–2224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweighofer A, Shubchynskyy V, Kazanaviciute V, Djamei A, Meskiene I.. 2014. Bimolecular fluorescent complementation (BiFC) by MAP kinases and MAPK phosphatases. Methods in Molecular Biology 1171, 147–158. [DOI] [PubMed] [Google Scholar]
- Schweizer F, Bodenhausen N, Lassueur S, Masclaux FG, Reymond P.. 2013. Differential contribution of transcription factors to Arabidopsis thaliana defense against Spodoptera littoralis. Frontiers in Plant Science 4, 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahnejat-Bushehri S, Tarkowska D, Sakuraba Y, Balazadeh S.. 2016. Arabidopsis NAC transcription factor JUB1 regulates GA/BR metabolism and signalling. Nature Plants 2, 16013. [DOI] [PubMed] [Google Scholar]
- Shubchynskyy V, Boniecka J, Schweighofer A, et al. 2017. Protein phosphatase AP2C1 negatively regulates basal resistance and defense responses to Pseudomonas syringae. Journal of Experimental Botany 68, 1169–1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidonskaya E, Schweighofer A, Shubchynskyy V, Kammerhofer N, Hofmann J, Wieczorek K, Meskiene I.. 2016. Plant resistance against the parasitic nematode Heterodera schachtii is mediated by MPK3 and MPK6 kinases, which are controlled by the MAPK phosphatase AP2C1 in Arabidopsis. Journal of Experimental Botany 67, 107–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smit ME, McGregor SR, Sun H, et al. 2020. A PXY-mediated transcriptional network integrates signaling mechanisms to control vascular development in Arabidopsis. The Plant Cell 32, 319–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su SH, Suarez-Rodriguez MC, Krysan P.. 2007. Genetic interaction and phenotypic analysis of the Arabidopsis MAP kinase pathway mutations mekk1 and mpk4 suggests signaling pathway complexity. FEBS Letters 581, 3171–3177. [DOI] [PubMed] [Google Scholar]
- Suarez-Rodriguez MC, Adams-Phillips L, Liu Y, Wang H, Su SH, Jester PJ, Zhang S, Bent AF, Krysan PJ.. 2007. MEKK1 is required for flg22-induced MPK4 activation in Arabidopsis plants. Plant Physiology 143, 661–669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran LS, Nakashima K, Sakuma Y, Simpson SD, Fujita Y, Maruyama K, Fujita M, Seki M, Shinozaki K, Yamaguchi-Shinozaki K.. 2004. Isolation and functional analysis of Arabidopsis stress-inducible NAC transcription factors that bind to a drought-responsive cis-element in the early responsive to dehydration stress 1 promoter. The Plant Cell 16, 2481–2498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulm R, Ichimura K, Mizoguchi T, Peck SC, Zhu T, Wang X, Shinozaki K, Paszkowski J.. 2002. Distinct regulation of salinity and genotoxic stress responses by Arabidopsis MAP kinase phosphatase 1. The EMBO Journal 21, 6483–6493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulm R, Revenkova E, di Sansebastiano GP, Bechtold N, Paszkowski J.. 2001. Mitogen-activated protein kinase phosphatase is required for genotoxic stress relief in Arabidopsis. Genes & Development 15, 699–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umbrasaite J, Schweighofer A, Kazanaviciute V, et al. 2010. MAPK phosphatase AP2C3 induces ectopic proliferation of epidermal cells leading to stomata development in Arabidopsis. PLoS One 5, e15357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umbrasaite J, Schweighofer A, Meskiene I.. 2011. Substrate analysis of Arabidopsis PP2C-type protein phosphatases. Methods in Molecular Biology 779, 149–161. [DOI] [PubMed] [Google Scholar]
- Wang KLC, Li H, Ecker JR.. 2002. Ethylene biosynthesis and signaling networks. The Plant Cell 14 (Suppl), S131–S151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu A, Allu AD, Garapati P, et al. 2012. JUNGBRUNNEN1, a reactive oxygen species-responsive NAC transcription factor, regulates longevity in Arabidopsis. The Plant Cell 24, 482–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Li Y, Wang Y, Liu H, Lei L, Yang H, Liu G, Ren D.. 2008. Activation of MAPK kinase 9 induces ethylene and camalexin biosynthesis and enhances sensitivity to salt stress in Arabidopsis. Journal of Biological Chemistry 283, 26996–27006. [DOI] [PubMed] [Google Scholar]
- Yang JH, Lee KH, Du Q, Yang S, Yuan B, Qi L, Wang H.. 2020. A membrane-associated NAC domain transcription factor XVP interacts with TDIF co-receptor and regulates vascular meristem activity. New Phytologist 226, 59–74. [DOI] [PubMed] [Google Scholar]
- Yang S, Hua J.. 2004. A haplotype-specific Resistance gene regulated by BONZAI1 mediates temperature-dependent growth control in Arabidopsis. The Plant Cell 16, 1060–1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S, Poretska O, Sieberer T.. 2018. ALTERED MERISTEM PROGRAM1 restricts shoot meristem proliferation and regeneration by limiting HD-ZIP III-mediated expression of RAP2.6L. Plant Physiology 177, 1580–1594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Wu Y, Gao M, Zhang J, Kong Q, Liu Y, Ba H, Zhou J, Zhang Y.. 2012. Disruption of PAMP-induced MAP kinase cascade by a Pseudomonas syringae effector activates plant immunity mediated by the NB-LRR protein SUMM2. Cell Host and Microbe 11, 253–263. [DOI] [PubMed] [Google Scholar]
- Zhou C, Cai Z, Guo Y, Gan S.. 2009. An Arabidopsis mitogen-activated protein kinase cascade, MKK9-MPK6, plays a role in leaf senescence. Plant Physiology 150, 167–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y, Qian W, Hua J.. 2010. Temperature modulates plant defense responses through NB-LRR proteins. PLoS Pathogens 6, e1000844. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data supporting the findings of this study are available from the corresponding author, Alois Schweighofer, upon request.