Abstract
Background
Emerged coronavirus disease 2019 (COVID-19) is a pandemic caused by the severe acute respiratory syndrome coronavirus 2 (SARS-COV-2). Disease severity is associated with elevated levels of proinflammatory cytokines, such as interleukin-6 (IL-6). Genetic polymorphisms in the regulatory regions of cytokine genes may be associated with differential cytokine production in COVID-19 patients. This study aimed to investigate the association between three potentially functional single-nucleotide polymorphisms (SNPs) in the promoter region of IL-6 and the severity of susceptibility to COVID-19 in an Iranian population.
Methods
In total, 346 individuals (175 patients with severe COVID-19 and 171 patients with mild COVID-19) were recruited for this cohort study. Genomic DNA was extracted from peripheral blood leukocytes of patients to determine the genotypes of three selected SNPs (rs1800795 (−174 G > C), rs1800796 (−572 G > C), and rs1800797 (−597 G > A)) in the promoter region of the IL-6 gene using polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP) method.
Results
There were no significant differences in the genotype or allele distribution of selected SNPs (rs1800795 (−174 G > C), rs1800796 (−572 G > C), and rs1800797 (−597 G > A)) in the promoter region of the IL-6 gene in patients with severe COVID-19 and patients with mild COVID-19.
Discussion
Our study indicated that these SNPs are not associated with COVID-19 severity in the Kurdish population from Kermanshah, Iran.
Keywords: Coronavirus disease 2019 (COVID-19), IL-6, Single nucleotide polymorphism, PCR-RFLP
1. Introduction
At the end of December 2019, in Wuhan, China, an outbreak of a novel coronavirus belonging to the beta-coronavirus subfamily was reported as a major threat to global public health [1], [2], [3]. On February 11, 2020, the World Health Organization (WHO) named the new coronavirus severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the causative agent of coronavirus disease 2019″ (COVID-19) [4], [5]. Although many patients with COVID-19 remain asymptomatic or experience mild-to-moderate disease, more than 20% of SARS-CoV-2 infections lead to severe acute respiratory distress syndrome (ARDS) with severe pneumonia and alveolar damage and, in worse cases, even death [6].
COVID-19 infection is accompanied by excessive inflammatory responses associated with the release of large amounts of pro-inflammatory cytokines such as interleukin-6 (IL-6), IL-1β, IL-10, IL-18, IL-4, IL-33, interferon (IFN)-γ, and tumor necrosis factor alpha (TNF-α), which are also known as cytokine storms [7]. The cytokine storm is the critical immunopathological mechanism underlying a more severe clinical course in cases of COVID-19 and is the ultimate cause of death [8]. According to known evidence, IL-6 is an important inflammatory cytokine that is superior to C-reactive protein (CRP) and other prognostic parameters such as leukopenia, fibrinogen, ferritin, prothrombin time, and D-dimer in predicting and progression of Covid-19 [9], [10], [11], [12]. IL-6 is produced by a subset of immune and non-immune cells in lung tissue, including T lymphocytes, resident alveolar macrophages, alveolar type II epithelial cells (ECs), and lung fibroblasts [13]. This cytokine plays an important role in the development of lymphopenia in COVID-19 patients by inducing lymphocytic apoptosis [14], [15]. In addition, high levels of IL6 have been shown to significantly affect lymphocyte function by significantly reducing human leukocyte D antigen expression (HLA-DR) combined with the depletion of natural killer (NK) cells, CD4 + lymphocytes, and CD19 + lymphocytes [16]. Moreover, IL-6 is thought to be involved in COVID-19-associated coagulopathy through the generation of tissue factors and thrombin, stimulation of platelet activation, and induction of endothelial dysfunction [17], [18], [19], [20]. Previous studies have demonstrated that serum levels of IL-6 are increased in patients with severe COVID-19, which is significantly correlated with adverse clinical consequences, including admission to the intensive care unit (ICU), ARDS, and death [21], [22], [23], [24].
The gene encoding human IL-6 is located on chromosome 7p21–14, and several single-nucleotide polymorphisms (SNPs) in the coding and non-coding regions of this gene have been reported [25]. The differences in cytokine production among different individuals may be due to the presence of SNPs that occur in critical regulatory regions, such as promoters, introns, and the 5′- UTR and 3′- UTR regulatory regions, which may affect the expression level of cytokines, whereas genetic polymorphisms in the gene-coding regions can lead to loss or change of function in the expressed proteins [26]. Many studies have demonstrated that the genetic polymorphisms at rs1800795 (−174 G > C), rs1800796 (−572 G > C), and rs1800797 (−597 G > A) of the IL-6 gene promoter are associated with serum levels of IL-6, prevalence, incidence, and/or progression of various diseases, such as sepsis, chronic obstructive pulmonary disease (COPD), hepatocellular carcinoma (HCC), and cancers [27], [28], [29], [30]. The role of polymorphisms in genes encoding IL-6 in the severity of COVID-19 is unclear. This study aimed to investigate the possible association between genetic polymorphisms at positions rs1800795 (−174 G > C), rs1800796 (−572 G > C), and rs1800797 (−597 G > A) of the IL-6 gene promoter and the severity of susceptibility to COVID-19 in the Kurdish population from Kermanshah, Iran.
2. Method and material
2.1. Patients
A total of 346 individuals (175 patients with severe COVID-19 and 171 patients with mild COVID-19) were enrolled in this cohort study. All patients were diagnosed with SARA-COV-2 infection using a positive nasopharyngeal RT-PCR test for COVID-19. The mild COVID-19 group consisted of 72 women and 99 men with a mean age of 40.46 ± 12.82 years, who were been referred to Samen AL-Aeme Medical Clinic in Kermanshah with mild symptoms such as malaise, sore throat, arthralgia, and anosmia. The severe COVID-19 group included 80 women and 95 men with a mean age of 60.78 ± 16.24 years, who were admitted to the intensive care unit of Imam Reza Hospital in Kermanshah with any of the following conditions: respiratory distress or respiratory failure, mechanical ventilation, oxygen partial pressure, and low oxygen concentration in arterial blood. All patients were from the Kurdish population of Kermanshah, Iran and from the same region in western Iran. Informed consent was obtained from all participants, and the study was approved by the Ethics Committee of Kermanshah University of Medical Science (IR.KUMS.REC.1399.967).
2.2. DNA extraction and genotyping
Genomic DNA was extracted from 2 mL EDTA anticoagulated peripheral blood samples using the salting-out method and stored at −20 °C for further use [31]. Genotyping of three SNPs, rs1800795 (−174 G > C), rs1800796 (−572 G > C), and rs1800797 (−597 G > A), in the promoter region of the IL-6 gene was determined using the polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP) method. The primers used for genotyping the IL-6 polymorphisms in the PCR-RFLP are shown in Table 1 .
Table 1.
PCR and RFLP conditions for IL-6 polymorphisms identifications.
| SNP locus | Primer sequences | Product size | Tm | Restriction enzyme | Fragment size |
|---|---|---|---|---|---|
|
rs1800795 −174 G > C |
F: TGCACTTTTCCCCCTAGTTGTGTCTTTC | 202 bp | 58 °C | Taq1 | C allele: 202 bp Gallele:173 bp + 30 bp |
| R: GAGCCTCAGACATCTCCAGTCCTAT | |||||
| rs1800796 − 634 G > C |
F: GACTCAGTGGCAATGGGGAGAGC | 606 bp | 61 °C | BsrbI | Gallele:262 bp + 344 bp C allele: 606 bp |
| R: CGCTAAGAAGCAGAACCACTCTTCC | |||||
| rs1800797 −597 A > G |
F: GACTCAGTGGCAATGGGGAGAGC | 606 bp | 61 °C | BtscI | Aallele:236 bp + 370 bp G allele: 606 bp |
| R: CGCTAAGAAGCAGAACCACTCTTCC |
DNA fragments comprising IL-6 polymorphisms were amplified in a final volume of 15 µL reaction mix containing 5.5 µL distilled water, 7 µL master mix (Sinaclon, Tehran, Iran), 1 µL of each primer, and 0.5 µL extracted DNA. The PCR conditions for the three SNPs were as follows: an initial denaturation step at 95 °C for 3 min, followed by 30 cycles of denaturation at 94 °C for 45 s, annealing temperature of 30 s at 58 °C for rs1800795 and 20 s at 61 °C for rs1800796 and rs1800797, and a final extension step at 72 °C for 10 min. PCR was performed using an iCycler C1000 (Bio-Rad Life Sciences, Hercules, CA, USA). The specificity of the PCR fragments for rs1800795, rs1800796, and rs1800797 polymorphisms, which were 202 bp, 606 bp, and 606 bp in length, respectively, was analyzed by electrophoresis on 2% agarose gel stained with 2 µL Green Viewer (Parstous, Mashhad, Iran).
RFLP digestion was performed with specific enzyme restriction for each genetic variant according to the manufacturer’s instructions, and the digested products were visualized by electrophoresis on a 2% agarose gel.
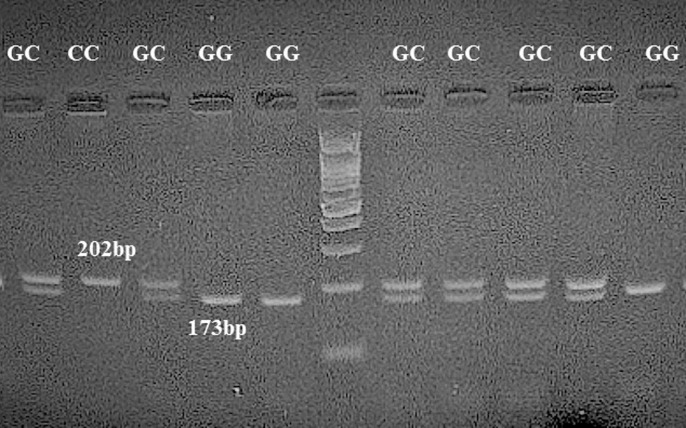
For rs1800795 (−174 G > C), the 202 bp PCR product was digested with Taq1 restriction enzyme (Fermentase, Thermo Fisher Scientific, USA) for 16 h at 37 °C. The G allele is cuttable and leads to fragments of 173 bp and 30 bp, while the C allele remains resistant to restriction enzymes, and the fragment is still 202 bp. Samples displaying 374 bp and 127 bp bands were typed as homozygote GG; samples exhibiting 173 bp and 30 bp and 202 bp bands were typed as GC heterozygotes; and samples showing one fragment of 202 bp were reported as homozygous CC (Fig. 1 ).
Fig. 1.
Restriction digestion (Taq1) products of the IL-6 rs1800795 G > C polymorphism in the promoter region on a 1% agarose gel. Homozygous wild-type GG genotype (173 bp + 30 bp); heterozygous GC genotype (202 bp + 173 bp + 30 bp); and homozygous mutant CC genotype (202 bp).
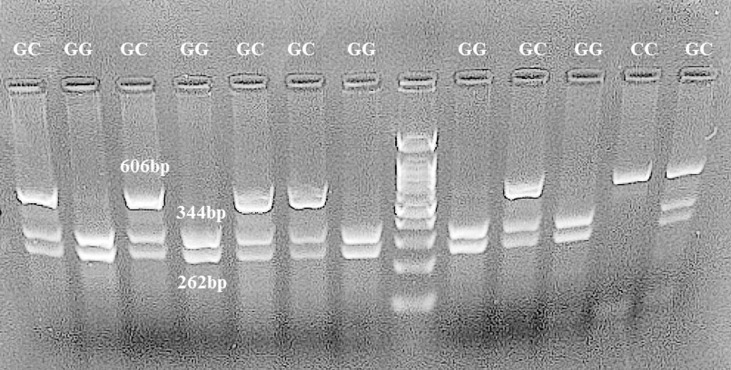
For rs1800796 (−572 G > C), the 606 bp PCR product was digested with BsrbI restriction enzyme (Fermentase, Thermo Fisher Scientific, USA) for 16 h at 37 °C. The G allele is cuttable and leads to fragments of 344 bp and 262 bp, while the C allele remains resistant to restriction enzymes, and the fragment is still 606 bp. Samples displaying 344 bp and 262 bp bands were typed as homozygote GG; samples exhibiting 344 bp and 262 bp and 606 bp bands were typed as GC heterozygotes; and samples showing one fragment of 606 bp were reported as homozygous CC (Fig. 2 ).
Fig. 2.
Restriction digestion (BsrBI) products of the IL-6 rs1800796 G > C polymorphism in the promoter region on 1% agarose gel. Homozygous wild-type GG genotype (262 bp + 344 bp); heterozygous GC genotype (606 bp + 262 bp + 344 bp); homozygous mutant CC genotype (606 bp).
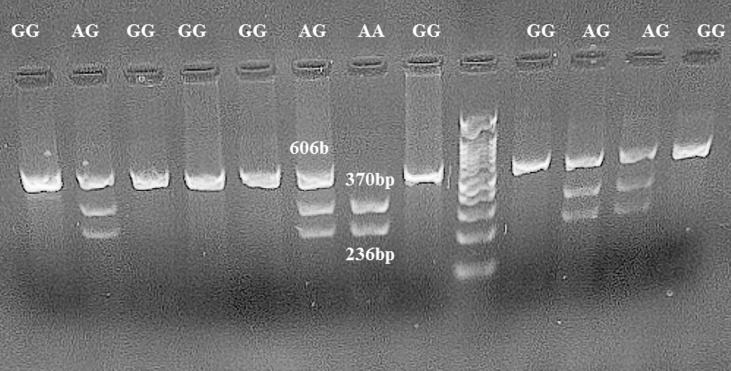
For rs1800797(−597 G > A), the 606 bp PCR product was digested with BtscI restriction enzyme (Fermentase, Thermo Fisher Scientific, USA) for 16 h at 55 °C. Allele A is cuttable and leads to fragments of 236 bp and 370 bp, while allele G remains resistant to restriction enzymes, and the fragment is still 606 bp. Samples displaying 236 bp and 370 bp bands were typed as homozygote GG, samples exhibiting 236 bp and 370 bp and 606 bp bands were typed as GC heterozygotes. Samples showing one fragment of 606 bp were reported as homozygous CC (Fig. 3 ).
Fig. 3.
Restriction digestion (BseGI) products of the IL-6 rs1800797 A > G polymorphism in the promoter region on a 1% agarose gel. Homozygous wild-type AA genotype (370 bp + 236 bp); heterozygous AG genotype (606 bp + 370 bp + 236 bp); homozygous mutant GG genotype (606 bp).
2.3. Statistical analysis
All statistical analyses were performed using the IBM SPSS software package ver. 22. Chi-square test was used to compare the genotype and allele frequencies of the rs1800795 (−174 G > C), rs1800796 (−572 G > C), and rs180097 (−597 G > A) SNPs between the severe COVID-19 and mild COVID-19 groups. In addition, the Hardy–Weinberg equilibrium for the three SNPs in the severe COVID-19 and mild COVID-19 groups was evaluated using the chi-square test. Odds ratios (ORs) and 95% confidence intervals (CIs) were calculated using logistic regression to evaluate the effects of these differences. P values of 0.05 or less were considered statistically significant.
3. Results
3.1. Demographic characteristic of patients with COVID-19
175 severe COVID-19(80 women, 95 men) with a mean age of 60.78 ± 16.24 years and 171 mild COVID-19(72 women, 99 men) with a mean age of 40.46 ± 12.82 years were studied. No statistical difference was observed between the two groups of severe COVID-19 patients and mild COVID-19 patients according to sex (p = 0.499). However, there was a significant difference between the severe COVID-19 and mild COVID-19 groups based on age (p = 0.00), mild COVID-19 patients were younger than severe COVID-19 patients.
3.2. Association of IL-6 gene polymorphisms with severity of COVID-19
To determine the association between IL-6 promoter polymorphisms and susceptibility to COVID-19 severity, three distinct polymorphic regions, rs1800795 (−174 G > C), rs1800796 (−572 G > C), and rs180097 (−597 G > A), were investigated. The distributions of genotypes, allele frequencies, and different genetic models (dominant, additive, and recessive) in patients with severe and mild COVID-19 are presented in Table 2 . No significant deviation from the Hardy–Weinberg equilibrium (HWE) was observed in any case (P > 0.05).
Table 2.
Distribution of allele and genotype frequencies of chemR23 gene polymorphisms in patients with AR and controls.
| SNP | Sever (N = 175) |
Mild (N = 171) |
P- value | OR(95% CI) |
|---|---|---|---|---|
| n (%) | n (%) | |||
| rs1800795 G > C | ||||
| Allele frequency | ||||
| G | 269 (76.9%) | 260 (76%) | 0.796 | Reference |
| C | 81 (23.1%) | 82 (24%) | 0.955 (0.672–1.356) | |
| Genotype frequency | ||||
| GG | 106 (60.6 %) | 103 (60.2%) | Reference | |
| GC | 57 (32.6 %) | 54 (31.6%) | 0.914 | 1.026 (0.647–1.626) |
| CC | 12 (6.9 %) | 14 (8.2%) | 0.661 | 0.833 (0.363–1.886) |
| Dominant model | ||||
| GG + GC | 163 (93.14 %) | 157 (39.76%) | 0.639 | 0.211 (0.543–2.700) |
| CC | 12 (6.85 %) | 14 (60.23%) | Reference | |
| Additive model | ||||
| GC | 57 (30.85%) | 54 (31.57%) | 0.843 | 1.047 (0.666–1.644) |
| GG + GC | 118 (67.42%) | 117 (68.42%) | Reference | |
| Recessive model | ||||
| CC | 12 (6.85%) | 14 (8.18 %) | 0.639 | 0.826 (0.370–1.840) |
| GG + GC | 163 (93.14%) | 157 (91.81%) | Reference | |
| HWE | 0.8 | 0.5 | ||
| rs1800796 G > C | ||||
| Allele frequency | ||||
| G | 291 (83.14%) | 295 (86.25%) | 0.256 | Reference |
| C | 59 (16.85 %) | 47 (13.74%) | 1.273 (0.839–1.929) | |
| Genotype frequency | ||||
| GG | 123 (70.3%) | 127 (74.3%) | Reference | |
| GC | 45 (25.7%) | 41 (24%) | 0.617 | 1.133 (0.694–1.851) |
| CC | 7 (4%) | 3 (1.8%) | 0.197 | 2.409(0.609–9.529) |
| Dominant model | ||||
| GG + GC | 168 (96%) | 168 (98.24%) | 0.213 | 0.429 (0.129–1.685) |
| CC | 7 (4.11%) | 3 (1.75%) | Reference | |
| Additive model | ||||
| GC | 45 (25.71%) | 41(23.97%) | 0.709 | 1.098 (0.674–1.788) |
| GG + CC | 130 (74.28 %) | 130 (17.54%) | Reference | |
| Recessive model | ||||
| CC | 7(4%) | 3(1.75 %) | 0.235 | 2.333 (0.593–9.176) |
| GG + GC | 168 (96%) | 168 (98.24%) | Reference | |
| HWE | 0.7 | 1 | ||
| rs1800797A > G | ||||
| Allele frequency | ||||
| A | 73 (20.85%) | 77 (22.51%) | 0.597 | Reference |
| G | 277 (79.14%) | 265 (77.48%) | 1.103 (0.768–1.583) | |
| Genotype frequency | ||||
| AA | 10 (5.7%) | 11(6.4.%) | Reference | |
| AG | 53 (30.3%) | 55 (32.2 %) | 0.903 | 1.060 (0.416–2.702) |
| GG | 112 (64%) | 105 (61.4%) | 0.727 | 1.173 (0.479–2.877) |
| Dominant model | ||||
| AA + AG | 63 (36%) | 66 (35.59%) | 0.618 | 0.895 (0.579–1.384) |
| GG | 112 (64 %) | 105 (61.40%) | Reference | |
| Additive model | ||||
| AG | 53 (30.3 %) | 55 (32.2%) | 0.706 | 0.916 (0.581–1.444) |
| AA + GG | 122 (69.7%) | 116 (67.8%) | Reference | |
| Recessive model | ||||
| GG | 112 (64%) | 105 (61.4%) | 0.618 | 1.117 (0.723–1.728) |
| AA + AG | 63 (36 %) | 66 (38.6 %) | Reference | |
| HWE | 0.7 | 0.8 | ||
Our results showed no statistically significant differences in allele and genotype distributions or different genetic models (dominant, additive, and recessive) between patients with severe COVID-19 and patients with mild COVID-19.
4. Discussion
In this study, we investigated the possible association between three potentially functional SNPs in the promoter region of the IL-6 gene (rs1800795 (−174 G > C), rs1800796 (−572 G > C), and rs1800797 (−597 G > A)) and the severity of susceptibility to COVID-19 between two patient groups consisting of 178 patients with severe COVID-19 and 175 patients with mild COVID-19 in an Iranian population. Our results showed no significant difference in the genotype and allele frequencies of these SNPs between the two groups of COVID-19 patients.
IL-6 is a pleiotropic cytokine that can act as a double-edged sword that affects both inflammatory and anti-inflammatory responses, depending on the activated signalling pathway [32], [33]. This cytokine is involved in critical cellular events such as survival, proliferation, differentiation, and trafficking of leukocytes [32]. IL-6 is produced and secreted by a wide range of cells such as macrophages, fibroblasts, keratinocytes, mast cells, DCs, monocytes, mesangial cells, T and B lymphocytes, and vascular endothelial cells (ECs) following tissue damage or infections [33], [34]. IL-6, along with other inflammatory cytokines, such as IL-10, IL-8, IL-4, and TNF-α, can cause cytokine storms that disrupt immune response regulation, leading to tissue damage [35]. Another adverse function of IL-6 is to disrupt the effective immune response to viral infections and cancers by inducing increased expression of inhibitory molecules, such as programmed cell death protein 1 (PD-1) and programmed death ligand 1 (PDL-1) [36]. In the case of viral infections such as SARS-CoV, MERS-CoV, and SARS-CoV-2, it has been demonstrated that cytokine storm and lymphopenia are two significant immunopathologic features in these patients [4], [37], [38]. Moreover, elevated levels of IL-6 have been reported in patients with severe COVID-19, which are positively correlated with damage to the lung tissue and infection progression [39], [40], [41]. Additionally, it is suggested that levels of IL-6 may be used as an inflammatory factor to predict the transition from mild to severe infection [42]. Genetic polymorphisms in the regulatory regions of cytokine genes may be associated with differential cytokine production [26]. Many studies have reported an association between some SNPs in the promoter region of the IL-6 gene, serum IL-6 levels, and the risk of developing different inflammatory diseases.
One recent study showed that there was a significant association between the IL-6 polymorphism at position −174 G/C (rs1800795) and the risk of developing COVID-19 in the Turkish population, which is in contrast with our study. Their findings showed that the frequency of the GG genotype and G allele was significantly higher in the macrophage activation syndrome (MAV) group than in the non-MAS group, and the G allele was reported as a risk factor for increased serum levels of IL-6 and progression to MAV [43]. In our study, the frequency of G allele and GG genotype SNPs −174 G / C (rs1800795) were higher in both patient groups than C allele and CC genotype. However, no difference was observed between severe COVID-19 patients and mild COVID-19 patients (p > 0.05). In contrast, Fishchuk et al. showed a significant increase in the frequency of the CC genotype and C allele of the −174 G/C (rs1800795) SNP in the development of the risk and course of COVID-19 in 31 patients with COVID-19 pneumonia compared to the population frequency [44]. In addition, one study reported that the −174 G/C (rs1800795) SNP was significantly associated with susceptibility to chronic obstructive pulmonary disease (COPD) in different Caucasian populations whereas this SNP was not found to be significantly associated with COPD in the North Indian population [28], [45]. Furthermore, a study of European Caucasian patients who underwent major cardiac or abdominal surgery revealed that the IL-6 rs1800795 CC genotype was associated with a higher risk of septic shock-related death [27]. Additionally, a Chinese study indicated that subjects with the IL-6–174 CC genotype had a higher risk of pneumonia-induced sepsis and higher mRNA levels [46]. Moreover, a meta-analysis suggested that the C allele of the −174 G/C (rs1800795) SNP is related to higher IL-6 production and pneumonia severity [47]. In contrast, a study showed that the GG genotype of the rs1800795 −174 G/C polymorphism is associated with high serum levels of IL-6 and the likelihood of sustained virologic response (SVR) in patients co-infected with HCV and HIV [48]. Similarly, other studies have demonstrated that the CC genotype of the rs1800795 −174 G/C polymorphism is associated with low production of IL-6 and an attenuated immune response against chronic HCV [49], [50]. In contrast, it has also been reported that genetic polymorphisms of IL-6 in rs1800796 (-572 G > C) were not associated with HCV infection and development of hepatocellular carcinoma (HCC) in an Egyptian population [51]. Also, several studies have indicated a significant role of rs1800796 polymorphism in the development of HBV [29], [52]. In addition, a meta-analysis confirmed the role of rs1800796 as a determining factor in hepatocellular carcinoma (HCC) [53]. Moreover, another study assessing two genetic polymorphisms of IL-6 (rs1800796 and rs1800795) showed a significant association between the CC genotype of IL-6 rs1800795 SNPs, and AA genotypes of IL-6 rs1800797 SNPs and susceptibility to the development of cervical cancer in the Lithuanian population [30].
This inconsistency in the results reported by different studies may be related to differences in the sample size, inclusion and exclusion criteria of patients, pathogenies of various inflammatory diseases, geographic area, ethnicity, and racial heterogeneity.
Our study showed that these three SNPs are not associated with COVID-19 in the Kurdish population from Kermanshah, Iran. However, there are some potential limitations to our study that should be noted. First, we did not measure RNA expression or IL-6 protein levels to assess the effects of different genotypes of these three SNPs on IL-6 expression levels. Second, the sample size was relatively small, and the patients were selected from only one Iranian population. Third, we did not investigate further SNPs in other IL-6 regions. Finally, to confirm these results, we propose that more extensive studies should be performed in different ethnic populations to investigate the association between genetic polymorphisms in IL-6 and the pathogenesis of COVID-19.
5. Conclusion
Considering the limitations of ethnicity, sample size, and genetic variant selection in this study, we could not demonstrate any significant association between three potential SNPs in the promoter region of the IL-6 gene (rs1800795 (−174 G > C), rs1800796 (−572 G > C), and rs1800797 (−597 G > A)) and susceptibility to COVID-19 severity in the Kurdish population from Kermanshah, Iran. Further genetic studies involving more SNPs and a larger sample size are required to clarify and confirm the association between genetic polymorphisms in IL-6 and SARS-COV-2 infection.
Credit authors statement
A. G.K. was involved in the concept and design of the study. Sara F. drafted the manuscript. All authors were involved in data collection, analysis, and interpretation and approved the final manuscript.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- 1.Millán-Oñate J., Rodriguez-Morales A.J., Camacho-Moreno G., Mendoza-Ramírez H., Rodríguez-Sabogal I.A., Álvarez-Moreno C. A new emerging zoonotic virus of concern: the 2019 novel Coronavirus (SARS CoV-2) Infectio. 2020;24(3):187–192. [Google Scholar]
- 2.Wu F., Zhao S.u., Yu B., Chen Y.-M., Wang W., Song Z.-G., Hu Y.i., Tao Z.-W., Tian J.-H., Pei Y.-Y., Yuan M.-L., Zhang Y.-L., Dai F.-H., Liu Y.i., Wang Q.-M., Zheng J.-J., Xu L., Holmes E.C., Zhang Y.-Z. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579(7798):265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhou P., Yang X.L., Wang X.G., Hu B., Zhang L., Zhang W., Si H.-R., Zhu Y., Li B., Huang C.-L., Chen H.D., Chen J., Luo Y., Guo H., Jiang R.-D., Liu M.Q., Chen Y., Shen X.R., Wang X., Zheng X.-S., Zhao K., Chen Q.J., Deng F., Liu L.L., Yan B., Zhan F.X., Wang Y.Y., Xiao G.F., Shi Z.L. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579(7798):270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yang X., Yu Y., Xu J., Shu H., Liu H., Wu Y., Zhang L., Yu Z., Fang M., Yu T. Clinical course and outcomes of critically ill patients with SARS-CoV-2 pneumonia in Wuhan, China: a single-centered, retrospective, observational study. Lancet Respir. Med. 2020;8(5):475–481. doi: 10.1016/S2213-2600(20)30079-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wu Z., McGoogan J.M. Characteristics of and important lessons from the coronavirus disease 2019 (COVID-19) outbreak in China: summary of a report of 72 314 cases from the Chinese Center for Disease Control and Prevention. JAMA. 2020;323(13):1239–1242. doi: 10.1001/jama.2020.2648. [DOI] [PubMed] [Google Scholar]
- 6.J. Brábek, D. Rosel, M. Fernandes, Repurposing of bazedoxifene to prevent cytokine storm in COVID-19 patients.
- 7.Costela-Ruiz V.J., Illescas-Montes R., Puerta-Puerta J.M., Ruiz C., Melguizo-Rodríguez L. SARS-CoV-2 infection: The role of cytokines in COVID-19 disease. Cytokine Growth Factor Rev. 2020;54:62–75. doi: 10.1016/j.cytogfr.2020.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bhaskar S., Sinha A., Banach M., Mittoo S., Weissert R., Kass J.S., Rajagopal S., Pai A.R., Kutty S. Cytokine storm in COVID-19—immunopathological mechanisms, clinical considerations, and therapeutic approaches: the REPROGRAM consortium position paper. Front. Immunol. 2020;11:1648. doi: 10.3389/fimmu.2020.01648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ponti G., Maccaferri M., Ruini C., Tomasi A., Ozben T. Biomarkers associated with COVID-19 disease progression. Crit. Rev. Clin. Lab. Sci. 2020;57(6):389–399. doi: 10.1080/10408363.2020.1770685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Velavan T.P., Meyer C.G. Mild versus severe COVID-19: laboratory markers. Int. J. Infect. Dis. 2020;95:304–307. doi: 10.1016/j.ijid.2020.04.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Herold T., Jurinovic V., Arnreich C., Hellmuth J.C., von Bergwelt-Baildon M., Klein M., Weinberger T. Level of IL-6 predicts respiratory failure in hospitalized symptomatic COVID-19 patients. MedRxiv. 2020 [Google Scholar]
- 12.Zeng F., Huang Y., Guo Y., Yin M., Chen X., Xiao L., Deng G. Association of inflammatory markers with the severity of COVID-19: a meta-analysis. Int. J. Infect. Dis. 2020;96:467–474. doi: 10.1016/j.ijid.2020.05.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Z. Chi, W. Zhao, L. Jia-Wen, Z. Hong, W. Gui-Qiang, The Cytokine Release Syndrome (CRS) of Severe COVID-19 and Interleukin-6 Receptor (IL-6R) Antagonist Tocilizumab man be the Key to Reduce the Mortality, https://www. ncbi. nlm. nih. gov/pmc/articles/PMC7118634/pdf/main. pdf (2020).
- 14.Tan L., Wang Q., Zhang D., Ding J., Huang Q., Tang Y.-Q., Wang Q., Miao H. Lymphopenia predicts disease severity of COVID-19: a descriptive and predictive study. Signal Transduction Targeted Ther. 2020;5(1):1–3. doi: 10.1038/s41392-020-0148-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Abbasifard M., Khorramdelazad H. The bio-mission of interleukin-6 in the pathogenesis of COVID-19: A brief look at potential therapeutic tactics. Life Sci. 2020;257:118097. doi: 10.1016/j.lfs.2020.118097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Giamarellos-Bourboulis E.J., Netea M.G., Rovina N., Akinosoglou K., Antoniadou A., Antonakos N., Damoraki G., Gkavogianni T., Adami M.-E., Katsaounou P., Ntaganou M., Kyriakopoulou M., Dimopoulos G., Koutsodimitropoulos I., Velissaris D., Koufargyris P., Karageorgos A., Katrini K., Lekakis V., Lupse M., Kotsaki A., Renieris G., Theodoulou D., Panou V., Koukaki E., Koulouris N., Gogos C., Koutsoukou A. Complex Immune Dysregulation in COVID-19 Patients with Severe Respiratory Failure. Cell Host Microbe. 2020;27(6):992–1000.e3. doi: 10.1016/j.chom.2020.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang D., Zhou X., Yan S., Tian R., Su L., Ding X., Xiao M., Chen Y.u., Zhao H., Chen H., Zhang H., Li Z., Li Q., Xu Y., Yan X., Li Y., Zhang S. Correlation between cytokines and coagulation-related parameters in patients with coronavirus disease 2019 admitted to ICU. Clin. Chim. Acta. 2020;510:47–53. doi: 10.1016/j.cca.2020.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Levi M., Van der Poll T., Ten Cate H., Van Deventer S. The cytokine-mediated imbalance between coagulant and anticoagulant mechanisms in sepsis and endotoxaemia. Eur. J. Clin. Invest. 1997;27(1):3–9. doi: 10.1046/j.1365-2362.1997.570614.x. [DOI] [PubMed] [Google Scholar]
- 19.Levi M., van der Poll T. Two-way interactions between inflammation and coagulation. Trends Cardiovasc. Med. 2005;15(7):254–259. doi: 10.1016/j.tcm.2005.07.004. [DOI] [PubMed] [Google Scholar]
- 20.Du F., Liu B., Zhang S. COVID-19: the role of excessive cytokine release and potential ACE2 down-regulation in promoting hypercoagulable state associated with severe illness. J. Thromb. Thrombolysis. 2021;51(2):313–329. doi: 10.1007/s11239-020-02224-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen X., Zhao B., Qu Y., Chen Y., Xiong J., Feng Y., Men D., Huang Q., Liu Y., Yang B. Detectable serum SARS-CoV-2 viral load (RNAaemia) is closely correlated with drastically elevated interleukin 6 (IL-6) level in critically ill COVID-19 patients. Clin. Infect. Dis. 2020 doi: 10.1093/cid/ciaa449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhou F., Yu T., Du R., Fan G., Liu Y., Liu Z., Xiang J., Wang Y., Song B., Gu X. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet. 2020;395(10229):1054–1062. doi: 10.1016/S0140-6736(20)30566-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gong J., Dong H., Xia S.Q., Huang Y.Z., Wang D., Zhao Y., Liu W., Tu S., Zhang M., Wang Q. Correlation analysis between disease severity and inflammation-related parameters in patients with COVID-19 pneumonia. MedRxiv. 2020 doi: 10.1186/s12879-020-05681-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu X., Wang H., Shi S., Xiao J. Association between IL-6 and severe disease and mortality in COVID-19 disease: a systematic review and meta-analysis. Postgrad. Med. J. 2021 doi: 10.1136/postgradmedj-2021-139939. [DOI] [PubMed] [Google Scholar]
- 25.Jia W., Fei G.-H., Hu J.-G., Hu X.-W. A study on the effect of IL-6 gene polymorphism on the prognosis of non-small-cell lung cancer. OncoTargets Therapy. 2015;8:2699. doi: 10.2147/OTT.S84636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Haukim N., Bidwell J., Smith A., Keen L., Gallagher G., Kimberly R., Huizinga T., McDermott M., Oksenberg J., McNicholl J. Cytokine gene polymorphism in human disease: on-line databases, supplement 2. Genes Immun. 2002;3(6):313–330. doi: 10.1038/sj.gene.6363881. [DOI] [PubMed] [Google Scholar]
- 27.Jiménez-Sousa M.A., Medrano L.M., Liu P., Fernandez-Rodriguez A., Almansa R., Gomez-Sanchez E., Ortega A., Heredia-Rodríguez M., Gómez-Pesquera E., Tamayo E. IL-6 rs1800795 polymorphism is associated with septic shock-related death in patients who underwent major surgery: a preliminary retrospective study. Ann. Intensive Care. 2017;7(1):1–9. doi: 10.1186/s13613-017-0247-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.He J.-Q., Foreman M.G., Shumansky K., Zhang X., Akhabir L., Sin D.D., Man S.F.P., DeMeo D.L., Litonjua A.A., Silverman E.K., Connett J.E., Anthonisen N.R., Wise R.A., Pare P.D., Sandford A.J. Associations of IL6 polymorphisms with lung function decline and COPD. Thorax. 2009;64(8):698–704. doi: 10.1136/thx.2008.111278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chang L., Lan T., Wu L., Li C., Yuan Y., Liu Z. The association between three IL-6 polymorphisms and HBV-related liver diseases: a meta-analysis. Int. J. Clin. Exp. Med. 2015;8(10):17036. [PMC free article] [PubMed] [Google Scholar]
- 30.Vitkauskaite A., Celiesiute J., Juseviciute V., Jariene K., Skrodeniene E., Samuolyte G., Nadisauskiene R.J., Vaitkiene D. IL-6 597A/G (rs1800797) and 174G/C (rs1800795) gene polymorphisms in the development of cervical cancer in Lithuanian women. Medicina. 2021;57(10):1025. doi: 10.3390/medicina57101025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zaveri P., Patel R., Patel M., Sarodia D., Munshi N.S. Modification of extraction method for community DNA isolation from salt affected compact wasteland soil samples. MethodsX. 2017;4:63–67. doi: 10.1016/j.mex.2017.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Scheller J., Chalaris A., Schmidt-Arras D., Rose-John S. The pro-and anti-inflammatory properties of the cytokine interleukin-6, Biochimica et Biophysica Acta (BBA)-Molecular. Cell Res. 2011;1813(5):878–888. doi: 10.1016/j.bbamcr.2011.01.034. [DOI] [PubMed] [Google Scholar]
- 33.T. Tanaka, M. Narazaki, T. Kishimoto, IL-6 in inflammation, immunity, and disease, Cold Spring Harbor perspectives in biology 6(10) (2014) a016295. [DOI] [PMC free article] [PubMed]
- 34.Mauer J., Denson J.L., Brüning J.C. Versatile functions for IL-6 in metabolism and cancer. Trends Immunol. 2015;36(2):92–101. doi: 10.1016/j.it.2014.12.008. [DOI] [PubMed] [Google Scholar]
- 35.Barrett D. In: Cytokine Storm Syndrome. Cron R.Q., Behrens E.M., editors. Springer International Publishing; Cham: 2019. IL-6 blockade in cytokine storm syndromes; pp. 561–568. [Google Scholar]
- 36.Bardhan K., Anagnostou T., Boussiotis V.A. The PD1: PD-L1/2 pathway from discovery to clinical implementation. Front. Immunol. 2016;7:550. doi: 10.3389/fimmu.2016.00550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gupta K.K., Khan M.A., Singh S.K. Constitutive inflammatory cytokine storm: a major threat to human health. J. Interferon Cytokine Res. 2020;40(1):19–23. doi: 10.1089/jir.2019.0085. [DOI] [PubMed] [Google Scholar]
- 38.Channappanavar R., Perlman S. Pathogenic human coronavirus infections: causes and consequences of cytokine storm and immunopathology. Seminars Immunopathol., Springer. 2017;39(5):529–539. doi: 10.1007/s00281-017-0629-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu F., Li L., Xu M., Wu J., Luo D., Zhu Y., Li B., Song X., Zhou X. Prognostic value of interleukin-6, C-reactive protein, and procalcitonin in patients with COVID-19. J. Clin. Virol. 2020;127 doi: 10.1016/j.jcv.2020.104370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Han H., Ma Q., Li C., Liu R., Zhao L.i., Wang W., Zhang P., Liu X., Gao G., Liu F., Jiang Y., Cheng X., Zhu C., Xia Y. Profiling serum cytokines in COVID-19 patients reveals IL-6 and IL-10 are disease severity predictors. Emerging Microbes Infect. 2020;9(1):1123–1130. doi: 10.1080/22221751.2020.1770129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lu R., Zhao X., Li J., Niu P., Yang B., Wu H., Wang W., Song H., Huang B., Zhu N. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395(10224):565–574. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang F., Nie J., Wang H., Zhao Q., Xiong Y., Deng L., Song S., Ma Z., Mo P., Zhang Y. Characteristics of peripheral lymphocyte subset alteration in COVID-19 pneumonia. J. Infect. Dis. 2020;221(11):1762–1769. doi: 10.1093/infdis/jiaa150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.F. Kerget, B. Kerget, Frequency of interleukin-6 rs1800795 (-174G/C) and rs1800797 (-597G/A) polymorphisms in COVID-19 patients in Turkey who develop macrophage activation syndrome, Japanese journal of infectious diseases (2021) JJID. 2021.046. [DOI] [PubMed]
- 44.Fishchuk L., Rossokha Z., Pokhylko V., Cherniavska Y., Tsvirenko S., Kovtun S., Medvedieva N., Vershyhora V., Gorovenko N. Modifying effects of TNF-α, IL-6 and VDR genes on the development risk and the course of COVID-19. Pilot study, Drug Metabolism and Personalized Therapy. 2021 doi: 10.1515/dmpt-2021-0127. [DOI] [PubMed] [Google Scholar]
- 45.Kirtipal N., Thakur H., Sobti R.C., Janmeja A.K. Association between IL6 gene polymorphism and the risk of chronic obstructive pulmonary disease in the north Indian population. Mol. Biol. Res. Commun. 2020;9(2):41. doi: 10.22099/mbrc.2019.34594.1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mao Z.-R., Zhang S.-L., Feng B. Association of IL-10 (-819T/C,-592A/C and-1082A/G) and IL-6-174G/C gene polymorphism and the risk of pneumonia-induced sepsis. Biomarkers. 2017;22(2):106–112. doi: 10.1080/1354750X.2016.1210677. [DOI] [PubMed] [Google Scholar]
- 47.Ulhaq Z.S., Soraya G.V. Anti-IL-6 receptor antibody treatment for severe COVID-19 and the potential implication of IL-6 gene polymorphisms in novel coronavirus pneumonia. Medicina clinica (English ed.) 2020;155(12):548–556. doi: 10.1016/j.medcle.2020.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nattermann J., Vogel M., Berg T., Danta M., Axel B., Mayr C., Bruno R., Tural C., Klausen G., Clotet B., Lutz T., Grünhage F., Rausch M., Nischalke H.D., Schewe K., Bienek B., Haerter G., Sauerbruch T., Rockstroh J.K., Spengler U. Effect of the interleukin-6 C174G gene polymorphism on treatment of acute and chronic hepatitis C in human immunodeficiency virus coinfected patients. Hepatology. 2007;46(4):1016–1025. doi: 10.1002/hep.21778. [DOI] [PubMed] [Google Scholar]
- 49.Bogdanović Z., Marinović-Terzić I., Kuret S., Jerončić A., Bradarić N., Forempoher G., Polašek O., Anđelinović Š., Terzić J. The impact of IL-6 and IL-28B gene polymorphisms on treatment outcome of chronic hepatitis C infection among intravenous drug users in Croatia. PeerJ. 2016;4 doi: 10.7717/peerj.2576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Barrett S., Goh J., Coughlan B., Ryan E., Stewart S., Cockram A.E.A., O’keane J., Crowe J. The natural course of hepatitis C virus infection after 22 years in a unique homogenous cohort: spontaneous viral clearance and chronic HCV infection. Gut. 2001;49(3):423–430. doi: 10.1136/gut.49.3.423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Madkour B., Gad A., Hamdy M.S., Zahran N., Aboul-Ezz M. Interleukin-6-572 promoter gene polymorphism and its association with chronic hepatitis C-induced hepatocellular carcinoma: an Egyptian study. Comp. Clin. Pathol. 2018;27(1):161–165. [Google Scholar]
- 52.Saxena R., Chawla Y.K., Verma I., Kaur J. IL-6 (− 572/− 597) polymorphism and expression in HBV disease chronicity in an Indian population. Am. J. Hum. Biol. 2014;26(4):549–555. doi: 10.1002/ajhb.22562. [DOI] [PubMed] [Google Scholar]
- 53.Aleagha O.E., Oltulu P., Sadeghi M. Association between interleukin 6 polymorphisms (rs1800796, rs1800795, rs2069837, rs17147230, and rs1800797) and hepatocellular carcinoma susceptibility: a meta-analysis. Clin. Experimental Hepatol. 2020;6(4):359. doi: 10.5114/ceh.2020.102171. [DOI] [PMC free article] [PubMed] [Google Scholar]