Abstract
Encephalitis mediated by autoantibodies against neuronal antigens and herpes simplex encephalitis (HSE) are seemingly separate causes of encephalopathy in adults. Autoimmune encephalitis (AE) is autoimmune in origin, and herpes simplex encephalitis is infectious. The purpose of this study was to examine the role of cerebrospinal fluid (CSF) exosomes from patients with antibody-positive AE and HSE. Towards this, exosomes were isolated from CSF from 13 patients with anti-N-methyl-d-aspartate receptor (NMDAR) encephalitis, 11 patients with anti-gamma-aminobutyric acid-B (GABAB) receptor encephalitis, 9 patients with anti-leucine-rich glioma-inactivated 1 (LGI1) encephalitis, and 8 patients with anti-contactin-associated protein-like 2 (CASPR2) encephalitis, and 12 control individuals negative of antibodies against neuronal autoantigens. There were ten miRNAs highly expressed in patients with anti-NMDAR encephalitis compared to those in control subjects. Eight miRNAs were found to be lower expressed in anti-NMDAR encephalitis CSF-derived exosomes. Furthermore, Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways enriched by AE differential expressed exosomic miRNAs demonstrated that AE-related exosomic miRNAs may participate as a feedback regulation in cancer development. In addition, the exosome concentration in CSF of 9 HSE patients was significantly higher compared to those from 9 HSV( −) patients. This observation was consistent with the results that exosome concentration was found to be higher in the animal model which was inoculated intranasally with HSV-1 compared to controls. Furthermore, western blot demonstrated that the subunits of NMDAR, GABABR, and AMPAR were detected highly expressed in exosomes derived from sera of HSV-1-treated animal model compared to controls. More importantly, exosomes isolated from CSF of HSE patients contained higher expression levels of two miRNAs encoded by HSV, miR-H2-3p, and miR-H4-3p compared to those from HSV( −) patients. In summary, HSV may trigger brain autoimmunity in HSE by presentation of surface autoantigens via exosomes.
Supplementary Information
The online version contains supplementary material available at 10.1007/s12035-021-02705-2.
Keywords: Exosomes, microRNAs, Autoimmune encephalitis (AE), Herpes simplex encephalitis (HSE)
Introduction
Encephalitis is one type of brain inflammation caused by virus infection or autoimmune-mediated reactions. Several types of encephalitis, including the classic paraneoplastic encephalitis syndromes, are immune mediated and often associated with autoantibodies against autoantigens, including neuronal surface proteins [1]. Dalmau et al. reported that half of women with anti-N-methyl-d-aspartate receptor (NMDAR) encephalitis had underlying mature or immature ovarian teratomas [2]. Dalmau et al. also confirmed that ovarian teratomas containing nervous tissue exhibited NMDAR subunits [2]. Several other autoantibodies against multiple neuronal autoantigens have been discovered in autoimmune encephalitis (AE) disease [3–5], including anti-gamma-aminobutyric acid-B receptor (GABABR) encephalitis, anti-leucine-rich glioma-inactivated 1 (LGI1) encephalitis, and anti-contactin-associated protein-like 2 (CASPR2) encephalitis. The most frequent neoplasms associated with paraneoplastic limbic encephalitis are small cell lung cancer (SCLC), testicular tumors, and Hodgkin lymphoma. The type of associated autoantibody varies with tumor type. Most SCLC patients have anti-Hu (also known as anti-neuronal nuclear antibody type 1 [ANNA-1]) or CV2/collapsin-responsive mediator protein-5 (CRMP5) antibodies in their serum and cerebrospinal fluid (CSF) [6, 7], and these patients are more likely to develop other manifestations of paraneoplastic encephalomyelitis. Anti-NMDAR encephalitis can be associated with an underlying tumor that stimulates the production of anti-NMDAR antibodies. Ovarian teratoma is the most commonly associated tumor and is reported to be present in over half of adult females [8]. Half of adult cases with anti-GABAB receptor encephalitis have associated small cell lung carcinoma. Most adult cases with anti-AMPA receptor encephalitis are associated with neoplasms (including SCLC, thymoma, ovarian teratoma, and breast cancer) [9, 10].
Herpes simplex virus (HSV)-1 is neurotropic and highly destructive when it infects the brain, and it was reported to be associated with the development of encephalitis [11]. HSV encephalitis (HSE) has a critical mortality rate of nearly 30% if left untreated [12, 13]. The consequence of HSE despite treatment leads to devastating sequelae, including memory deficits and seizures [12, 13]. Although some patients were negative for the detection of HSV after treatment, retrospective analyses [14, 15] demonstrated that anti-NMDAR antibodies were detected in the sera and/or CSF, suggesting that early viral infection with HSV sowed the seeds of autoimmune encephalitis. Nearly 30% of patients with HSE develop autoimmune encephalitis, usually associated with the development of anti-NMDAR antibodies in a prospective study [16]. A study from Linnoila et al. reported an endogenous rodent model of post-HSE NMDAR encephalitis to clinically phenocopy the pathogenesis of autoimmune encephalitis [17]. The authors further found that anti-NMDAR antibodies were detected in the serum isolated from mice inoculated intranasally with HSV-1 [17]. However, the causality between HSV infection and the development of anti-NMDAR antibody encephalitis remains elusive.
Exosomes are membrane-bound nanostructure vesicles shed by endocytic pathways [18, 19]. Most cells in the CNS, including neurons, astrocytes, oligodendrocytes, and microglia, shed exosomes under both normal and pathological conditions [20] [21]. Exosomes are confirmed to mediate several biological/cellular processes required for normal brain function [22–24], as well as the pathogenesis of some neuroinflammatory disorders [25] and neurodegenerative diseases [26–28]. Exosomes are also found to serve as carriers of misfolded and pathogenic proteins [25, 29]. Aberrantly expressed cellular miRNAs were further validated to be selectively packaged and transported in exosomes to neighboring cells [21, 30]. Recent studies from our laboratory demonstrated that exosomes were present in the CSF of AE patients. In addition, specific neuronal autoantigens, including NMDAR, GABABR, LGI1, CASPR2, and AMPAR, were found to be expressed in CSF- or serum-derived exosomes from AE patients. Importantly, we found that AE-derived exosome immunization in mice led to an increased frequency of neuronal autoantigen–specific IL-17 and IFN-γ [31]. Therefore, these data indicated that exosomal presentation of neuronal autoantigens would cause autoimmunity, which is usually recognized as a novel and critical pathway in AE pathogenesis.
Thus far, noncoding RNAs in CSF exosomes from AE have not been explored. Toward this, we attempted to determine the miRNA profiles in exosomes secreted from local sites in AE brain, and we correlated the dysregulation of miRNAs with clinical features such as paraneoplastic occurrence. In addition, the mechanisms that allow the establishment of HSV-mediated encephalitis remain poorly understood. We hypothesized that HSV infection increased the presentation of surface autoantigens via exosomes, which would further cause the autoimmune response during HSE development. To this end, we investigated the exosome cargo including neuronal antigens and miRNAs encoded by HSV in HSE patients as well as the animal model of HSE.
Materials and Methods
Ethics and Subjects
All subjects consented to participate in the study, and the work received approval from the institutional ethics committee of Sir Run Run Shaw Hospital affiliated with Zhejiang University School of Medicine and was conducted in accordance with the tenets of the Declaration of Helsinki. AE patients were diagnosed according to the autoimmune encephalitis diagnostic criteria published in The Lancet Neurology [1]. The laboratory used indirect immunofluorescence assay for CSF and/or sera antibody detection as previously reported [1]. The control individuals were diagnosed without antibodies against neuronal autoantigens. All the AE patients and control individuals show herpes simplex virus type 1 (HSV-1) PCR negative. CSF samples from these subjects were further subject to identify CSF-specific oligoclonal IgG bands (OCBs). Positive OCBs (IgG) were identified in 53.8% of patients with anti-NMDAR (7/13) and 54.5% anti-GABAbR (6/11); 4 patient with anti-LGI1 antibodies (44.4%) and 3 patients with anti-CASPR2 antibodies (37.5%) revealed positive OCBs. Clinical information for the discovery cohort, including demographic features and symptoms, is included in Table 1. HSE patients were diagnosed by two senior neurologists, and HSV DNA PCR from CSF samples was positive in these patients. HSV-negative controls came from the emergency department. They presented with headache and fever, and encephalitis was excluded after CSF examination and magnetic resonance imaging scanning. Clinical information of HSV-positive/negative patients is included in Table 2 and Supplemental Table 2 (HSV-negative patients).
Table 1.
Demographic features and symptoms of cohorts with antibody-positive paraneoplastic autoimmune encephalitis
| Anti-NMDAR | Anti-GABAbR | Anti-LGI1 | Anti-CASPR2 | Control Subjects | |
|---|---|---|---|---|---|
| No. of patients | 13 | 11 | 9 | 8 | 12 |
| Sex (F/M) | 9/4 | 6/5 | 3/6 | 4/4 | 6/6 |
| Age (year) (mean (Q1–Q3)) | 28 (18–51) | 57 (42–72) | 62 (60–72) | 61 (55–69) | 43 (22–63) |
| Neurological syndrome | |||||
| Limbic encephalitis | 12 | 7 | 7 | 1 | 0 |
| Brainstem encephalitis | 1 | 0 | 1 | 1 | 0 |
| Cerebellar degeneration | 0 | 3 | 5 | 3 | 0 |
| Peripheral nerve disorders | 0 | 1 | 0 | 4 | 0 |
| CSF analyses | |||||
| Any abnormality | 10 | 9 | 7 | 4 | 0 |
| Elevated protein | 5 | 4 | 3 | 1 | 0 |
| Lymphocytic pleocytosis | 5 | 2 | 1 | 1 | 0 |
| Oligoclonal IgG bands | 7 | 6 | 4 | 3 | 0 |
| Cancer | |||||
| Small cell lung cancer | 0 | 2 | 2 | 0 | 0 |
| Non-small cell lung cancer | 0 | 0 | 2 | 1 | 0 |
| Testicular | 0 | 0 | 0 | 0 | 0 |
| Breast | 1 | 1 | 0 | 0 | 0 |
| Gynecological | 8 | 3 | 0 | 0 | 0 |
| Hodgkin | 0 | 0 | 0 | 0 | 0 |
| No tumor | 4 | 5 | 5 | 7 | 12 |
Q1, first quartile; Q3, third quartile
Table 2.
Demographic features and symptoms of herpes simplex encephalitis patients
| Patient no | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 |
|---|---|---|---|---|---|---|---|---|---|
| Age (year) | 23 | 18 | 35 | 29 | 42 | 27 | 39 | 26 | 20 |
| Sex | Male | Male | Female | Male | Male | Female | Female | Male | Male |
| Neurological syndrome | |||||||||
| Decreased level of consciousness | + | + | - | + | - | + | + | + | - |
| Mental symptoms | + | - | - | + | + | + | + | - | - |
| Fever | + | + | + | + | + | + | + | + | + |
| Headache | + | + | - | + | + | + | - | + | - |
| Seizure | - | + | - | + | - | + | + | - | - |
| Focal sign | + | + | + | - | + | + | + | + | + |
| CSF analyses | |||||||||
| Increased opening pressure | - | + | - | + | + | + | + | + | - |
| Elevated white cells | + | + | + | + | + | + | + | + | + |
| Elevated red cells | - | - | + | + | - | + | - | - | - |
| Elevated protein | - | + | + | + | - | + | + | + | - |
| MRI T2 lesions | |||||||||
| Temporal lobe | - | + | - | - | + | + | + | - | - |
| Medial/inferior frontal lobe | - | - | + | + | - | - | - | + | + |
| Cingulate/insula cortex | + | - | + | - | - | - | - | - | + |
| Hippocampal | - | + | - | + | - | - | - | - | - |
| Other | - | - | - | - | + | - | + | - | - |
| HSV PCR | + | + | + | + | + | + | + | + | + |
CSF, cerebrospinal fluid; MRI, magnetic resonance imaging; HSV, herpes simplex virus; PCR, polymerase chain reaction
Isolation and Purification of Exosomes
Cerebrospinal fluid was obtained by lumbar puncture following a standard protocol. CSF exosomes were isolated from CSF samples as previously reported from our group [32]. In brief, CSF was subjected to successive centrifugations of 3,000 × g (15 min) and 10,000 × g (30 min). Exosomes were then pelleted at 100,000 × g for 1 h, using an SW28 rotor (Beckman, Brea, CA). Exosome pellets were further purified with sucrose cushion (30%) ultracentrifugation to eliminate protein aggregates as previously reported [33]. Exosome pellets were resuspended in 0.32 M sucrose and centrifuged at 100,000 × g for 1 h (SW60Ti rotor; Beckman), and then resuspended in TRIzol for mRNA extraction or denatured in the protein loading buffer.
Transmission Electron Microscopy (TEM)
Exosome samples were first loaded onto Grids-Formvar/Carbon Coated, fixed in 2% paraformaldehyde (PFA), and washed with Gibco phosphate-buffered saline (PBS) of high purity. Samples were further processed under 2.5% glutaraldehyde fixative, washed with PBS, contrasted in 2% uranyl acetate, and embedded in a mixture of uranyl acetate (0.4%) and methyl cellulose (0.13%). The samples were finally subjected to observation and imaging by electron microscopy (Carl Zeiss NTS).
Nanoparticle Tracking Analysis (NTA)
Briefly, approximately 100 µL of exosome sample was loaded into the chamber of an LM10 unit (Nanosight), and three videos of every 30 s were recorded for each sample. Data analysis was performed with NTA software (Nanosight).
Western Blotting
Briefly, 20 μg of quantified exosomal protein was denatured by using 2 × SDS Page Buffer (Santa Cruz Biotechnology), treated at 100 °C for 5 min, separated by polyacrylamide gel electrophoresis, and transferred to membranes that were pretreated with decoloration with methanol. Blotting was performed with anti-TSG101, anti-CD9 antibody, and anti-C63 antibodies (Abcam, Cambridge, MA) and anti-cytochrome c antibody (BD Pharmingen). Antibodies against NR2B subunits of NMDAR (Upstate Biotechnology, Lake Placid, NY) and GABAB1R (Molecular Probes, Eugene, OR), and the GluR1 subunits of AMPAR (Chemicon, Temecula, CA) were used. Goat anti-rabbit/mouse horseradish peroxidase was used as a secondary antibody. The blots were developed with enhanced chemiluminescence (ECL) and exposed with iBright CL1000 imaging system (Invitrogen). Protein quantification was performed by bandscan and densitometry analysis with optical density for NR2B, GABAb1R, GluR1, TSG101, and CD9.
nCounter Human miRNA Expression Assay
The nCounter human v3 miRNA expression assay designed for miRNA profiling (NanoString Technologies) was applied. The raw data (the counts for each miRNA in a sample) produced by the nCounter Digital Analyzer were subjected to technical and biological normalization using nSolver software version 2.5. DIANA-mirPath [34] was employed to perform the enrichment analysis of predicted target genes by one or more miRNAs in biological pathways.
TaqMan miRNA Assay for Individual miRNAs
Exosomal small RNAs were isolated using the Qiagen miRNeasy Serum/Plasma Kit (Qiagen, Valencia, CA). The TaqMan miRNA Assay (Applied Biosystems, Foster City, CA) was chosen for the individual miRNA real-time PCR validation performed as the company’s protocol recommended.
Animal Model of Post-infectious Anti-NMDAR Encephalitis
All animal procedures were approved by our Institutional Animal Care and Use Committee. Balb/c female mice, ~ 12 weeks of age, were purchased from Shanghai Laboratory Animal Center. Six mice were inoculated intranasally with HSV-1 for 2 weeks. 1 × 106 plaque-forming units of HSV-1 (strain 17 syn +)6 were applied once daily. Blood/serum was collected at 3, 6, and 8 weeks post-inoculation and tested for anti-NMDAR antibodies through a cell-based assay as previously reported [8]. HEK293 cells transfected with subunits of NMDA receptor were fixed in 4% paraformaldehyde, permeabilized with 0.2% Triton X-100, and co-incubated with mouse sera (diluted 1:100) along with a rabbit monoclonal antibody against rabbit polyclonal antibodies against NR2B (1:200, Upstate Biotechnology) followed by the appropriate fluorescent secondary antibodies. Exosomes were isolated from pooled sera, the structures of exosomes were characterized by EM, and the concentration of exosomes was counted by NTA.
Statistical Analysis
Data were analyzed using GraphPad Prism. One-way ANOVA followed by the Newman–Keuls post hoc test for intergroup comparisons or the nonparametric unpaired Mann–Whitney test for group comparisons was used. p < 0.05 was deemed statistically significant.
Results
Characterization of CSF Exosomes in Antibody-Positive AE Patients
We isolated the exosomes from CSF fluid of 13 patients with anti-NMDA receptor encephalitis, 11 patients with anti-GABAB receptor encephalitis, 9 patients with anti-LGI1 encephalitis, 8 patients with anti-CASPR2 encephalitis, and 12 control individuals. Exosomes showed a spherical morphology with an average size of 94 ± 42 nm under electron microscopy (Fig. 1A). Western blot was performed using antibodies specific to CD63 and CD9. An enrichment of CD63 and CD9 was found in all exosome samples (Fig. 1B), while an absence of mitochondrial cytochrome C. These results clearly indicate that this vesicle population isolated from CSF of antibody-positive AE patients is enriched in exosomes.
Fig. 1.
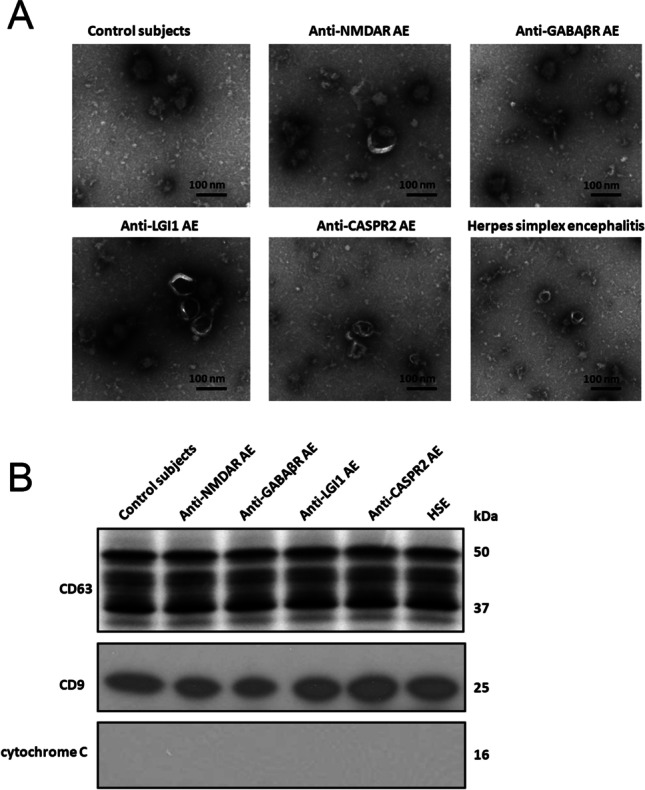
Exosome characterization. Characterization of exosome-like vesicles released from cerebrospinal fluid by A transmission electron microscopy of cerebrospinal fluid–derived exosomes from patients with anti-NMDA receptor encephalitis, patients with anti-GABAB receptor encephalitis, patients with anti-LGI1 encephalitis, patients with anti-CASPR2 encephalitis, herpes simplex encephalitis patients, and control subjects. The scale bar indicates 100 nm. B Western blotting. Presence of exosomal markers, CD63 and CD9, in protein lysates from cerebrospinal fluid–derived exosomes from antibody-positive autoimmune encephalitis patients, herpes simplex encephalitis patients, and control subjects
CSF-Derived Exosome miRNAs were Differentially Expressed in Antibody-Positive AE Patients
To identify the differentially expressed exosomal miRNAs derived from CSF, we performed miRNA profiling of purified exosomes isolated from CSF fluid of 13 patients with anti-NMDA receptor encephalitis, 11 patients with anti- GABAB receptor encephalitis, 9 patients with anti-LGI1 encephalitis, 8 patients with anti-CASPR2 encephalitis, 9 patients with HSE, and control subjects (n = 12), using the NanoString Technology platform. Comparison shows that 18 miRNAs with the most significant differences in expression in AE patients with anti-NMDAR Abs compared to those of control subjects (p < 0.05, the unadjusted p value) were selected using a t test. Specifically, 10 miRNAs were upregulated (hsa-miR-301a-5p, hsa-miR-21-5p, hsa-miR-128-5p, hsa-miR-155-5p, hsa-miR-34a-5p, hsa-miR-326-5p, hsa-miR-132-5p, hsa-miR-29b-5p, hsa-miR-340-5p, and hsa-miR-27b-5p) and 8 downregulated (hsa-let-7d-5p, hsa-miR-15b-5p, hsa-23b-5p, hsa-miR-10a-5p, hsa-miR-146a-5p, hsa-miR-20a-5p, hsa-miR-17-5p, hsa-miR-20b-5p) in CSF-derived exosomes of AE patients (NMDAR Abs positive) versus control subjects (Table 3). These 18 exosomal microRNAs were further compared in AE patients with GABABR Abs, LGI1 Ab, and CASPR2 Ab, with HSE and control subjects. These results defined a distinct signature of differential exosomal miRNAs related to the pathogenesis of antibody-positive AE. We further identified which biologic pathways were affected during the development of antibody-positive AE, we applied DIANA-mirPath on the dysregulated miRNA signature, and 48 Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways were significantly enriched (p < 0.05; Supplementary Table 1) after false discovery rate was corrected. The results demonstrated that fatty acid biosynthesis, mucin type O-glycan biosynthesis, signaling pathways regulating pluripotency of stem cells, TGF-beta signaling pathway, Hippo signaling pathway, Wnt signaling pathway, Ras signaling pathway, and ErbB signaling pathway were significantly enriched in differentially expressed exosomic miRNAs from anti-NMDA receptor encephalitis (all p < 0.001). For example, the KEGG pathway “Axon guidance” (hsa04360, p = 0.00027; Supplementary Fig. 1) was significantly altered in differentially expressed miRNAs between anti-NMDAR encephalitis patients and control subjects at baseline, with 13 miRNAs (hsa-miR-340-5p, hsa-miR-34a-5p, hsa-miR-15b-5p, hsa-miR-17-5p, hsa-miR-20b-5p, hsa-miR-146a-5p, hsa-miR-20a-5p, hsa-miR-10a-5p, hsa-miR-155-5p, hsa-let-7d-5p, hsa-miR-21-5p, hsa-miR-301a-5p, hsa-miR-27b-5p) targeting 56 genes, including EFNB2, GSK3B, MET, ROCK1, NRAS, PAK2, NGEF, and ARHGEF12. These results demonstrated that anti-NMDAR encephalitis associated miRNAs could dysregulate key cellular signal pathways, further may predispose AE.
Table 3.
miRNAs upregulated and downregulated in antibody-positive autoimmune encephalitis subtypes
| miRNA ID | Counts in NMDAR group | Counts in control group | Fold change (95%CI) | p value | Counts in GABAβR Group | Fold change (95%CI) | p value | Counts in LGI1 group | Fold change (95%CI) | p value | Counts in CASPR2 group | Fold change (95%CI) | p value | Counts in HSE Group | Fold change (95%CI) | p value |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| hsa-miR-301a-5p | 221.58 ± 27.89 | 21.58 ± 3.58 | 10.997 (6.391–13.225) | 0.014 | 45.33 ± 10.04 | 2.114(1.002–3.279) | 0.021 | 77.68 ± 15.55 | 3.658(2.687–5.074) | 0.173 | 210.73 ± 25.86 | 9.928(7.312–12.354) | 0.002 | 25.36 ± 4.55 | 1.175(0.859–1.521) | 0.228 |
| hsa-miR-21-5p | 231 ± 20.87 | 27.23 ± 7.94 | 8.387(5.063–11.118) | 0.034 | 94.35 ± 18.73 | 3.417(2.016–3.214) | 0.034 | 32.06 ± 6.56 | 1.186(0.759–1.689) | 0.611 | 105.44 ± 29.52 | 4.646(3.013–6.041) | 0.001 | 30.36 ± 8.56 | 1.115(0.841–1.355) | 0.556 |
| hsa-miR-128-5p | 93.05 ± 11.70 | 14.25 ± 2.06 | 7.876(5.138–10.242) | 0.020 | 51.63 ± 14.48 | 3.567(1.087–5.045) | 0.036 | 135.10 ± 19.33 | 9.47(7.146–11.287) | 0.021 | 4.09 ± 1.28 | 0.295(0.137–0.412) | 0.021 | 18.56 ± 3.33 | 1.302(1.025–1.639) | 0.057 |
| hsa-miR-155-5p | 325 ± 39.19 | 47.08 ± 8.17 | 6.616(4.089–9.411) | 0.040 | 248.62 ± 33.22 | 5.287(3.416–7.102) | 0.053 | 19.11 ± 5.27 | 0.417(0.189–0.682) | 0.505 | 465.33 ± 121.23 | 9.529(7.513–12.347) | 0.023 | 56.87 ± 10.23 | 1.208(0.987–1.553) | 0.126 |
| hsa-miR-34a-5p | 146.77 ± 42.10 | 27.77 ± 5.99 | 5.577(3.093–8.084) | 0.009 | 116.55 ± 25.14 | 4.208(3.091–6.156) | 0.042 | 235.47 ± 89.24 | 4.623(2.986–6.354) | 0.195 | 281.37 ± 46.57 | 10.575(8.541–13.245) | 0.003 | 36.75 ± 7.44 | 1.323(0.997–1.753) | 0.432 |
| hsa-miR-326-5p | 105.36 ± 11.58 | 20.18 ± 4.58 | 4.986(2.346–6.117) | 0.011 | 193.22 ± 45.17 | 9.721(7.057–11.751) | 0.097 | 85.21 ± 11.13 | 4.359(2.986–5.894) | 0.253 | 14.23 ± 5.89 | 0.629(0.421–0.911) | 0.265 | 30.54 ± 5.97 | 1.513(1.089–1.917) | 0.032 |
| hsa-miR-132-5p | 44.58 ± 8.30 | 10.25 ± 5.26 | 3.526(2.200–5.007) | 0.020 | 97.15 ± 10.48 | 9.537(7.024–11.762) | 0.095 | 90.41 ± 24.83 | 8.565(7.011–10.547) | 0.000 | 76.01 ± 14.68 | 6.811(5.017–8.412) | 0.004 | 8.55 ± 2.12 | 0.834(0.611–1.013) | 0.551 |
| hsa-miR-29b-5p | 61.58 ± 9.45 | 28.05 ± 5.62 | 2.196(1.287–3.042) | 0.020 | 94.42 ± 25.83 | 3.488(1.341–5.172) | 0.035 | 208.36 ± 70.77 | 7.52(6.342–8.024) | 0.550 | 177.30 ± 36.45 | 6.516(5.023–8.151) | 0.013 | 25.56 ± 4.11 | 0.911(0.751–1.059) | 0.178 |
| hsa-miR-340-5p | 110.47 ± 14.95 | 60.22 ± 9.49 | 1.827(0.927–3.041) | 0.012 | 356.86 ± 51.95 | 5.947(4.159–6.853) | 0.060 | 248.78 ± 51.82 | 4.032(2.698–5.898) | 0.514 | 19.09 ± 7.93 | 0.302(0.112–0.511) | 0.007 | 70.54 ± 11.54 | 1.171(0.895–1.425) | 0.522 |
| hsa-miR-27b-5p | 62.16 ± 7.80 | 43.25 ± 9.18 | 1.436(0.732–2.039) | 0.020 | 35.11 ± 7.51 | 0.6(0.343–0.956) | 0.206 | 298.50 ± 36.06 | 7.661(5.995–8.942) | 0.002 | 25.37 ± 7.39 | 0.594(0.405–0.714) | 0.017 | 59.45 ± 10.27 | 1.374(1.024–1.785) | 0.054 |
| hsa-let-7d-5p | 81.33 ± 12.72 | 132.76 ± 20.68 | 0.62(0.311–1.184) | 0.012 | 40.81 ± 9.60 | 0.295(0.002–0.461) | 0.073 | 31.42 ± 8.24 | 0.188(0.042–0.287) | 0.075 | 130.77 ± 27.43 | 0.999(0.741–1.245) | 0.854 | 68.23 ± 10.74 | 0.514(0.214–0.714) | 0.031 |
| hsa-miR-15b-5p | 51.29 ± 10.53 | 108.56 ± 31.33 | 0.474(0.155–0.844) | 0.013 | 354.31 ± 41.63 | 3.4(1.642–5.317) | 0.034 | 65.14 ± 9.42 | 0.588(0.237–0.724) | 0.017 | 162.32 ± 25.12 | 1.553(1.142–1.872) | 0.201 | 97.52 ± 21.54 | 0.898(0.714–1.157) | 0.558 |
| hsa-miR-23b-5p | 39.20 ± 7.28 | 133.35 ± 18.21 | 0.294(0.06–0.596) | 0.002 | 94.64 ± 17.57 | 0.719(0.314–1.023) | 0.087 | 94.51 ± 27.48 | 0.682(0.428–0.855) | 0.022 | 51.18 ± 9.63 | 0.423(0.215–0.711) | 0.024 | 100.27 ± 15.21 | 0.751(0.539–0.974) | 0.609 |
| hsa-miR-10a-5p | 25.10 ± 3.24 | 109.65 ± 26.63 | 0.229(0.049–0.479) | 0.002 | 683.99 ± 95.09 | 5.92(4.145–7.245) | 0.059 | 35.45 ± 9.49 | 0.226(0.024–0.405) | 0.004 | 36.09 ± 6.24 | 0.329(0.157–0.503) | 0.063 | 98.54 ± 15.24 | 0.898(0.642–1.224) | 0.430 |
| hsa-miR-146a-5p | 66.35 ± 8.63 | 403 ± 57.62 | 0.164(0.004–0.279) | 0.020 | 201.42 ± 74.92 | 0.486(0.123–0.841) | 0.305 | 361.06 ± 58.67 | 0.858(0.655–1.056) | 0.230 | 361.63 ± 52.02 | 0.872(0.701–1.051) | 0.789 | 355.21 ± 44.17 | 0.880(0.678–1.057) | 0.237 |
| hsa-miR-20a-5p | 104.72 ± 15.90 | 677.62 ± 78.66 | 0.155(0.004–0.294) | 0.019 | 601.80 ± 98.28 | 0.915(0.566–1.345) | 0.009 | 4.82 ± 1.46 | 0.006(0–0.019) | 0.004 | 541.61 ± 163.12 | 0.86(0.674–1.019) | 0.587 | 547.25 ± 65.89 | 0.807(0.614–1.051) | 0.358 |
| hsa-miR-17-5p | 93.66 ± 20.57 | 633.91 ± 70.95 | 0.147(0.003–0.381) | 0.014 | 59.75 ± 7.26 | 0.085(0.001–1.187) | 0.059 | 565.65 ± 140.96 | 0.842(0.697–1.120) | 0.045 | 497.49 ± 82.34 | 0.745(0.598–0.914) | 0.644 | 532.37 ± 52.38 | 0.839(0.689–1.041) | 0.657 |
| hsa-miR-20b-5p | 84.01 ± 26.62 | 603.07 ± 75.71 | 0.136(0.001–0.269) | 0.019 | 595.74 ± 95.12 | 0.989(0.489–1.489) | 0.113 | 358.41 ± 39.89 | 0.602(0.432–0.896) | 0.000 | 132.40 ± 37.32 | 0.166(0.032–0.305) | 0.155 | 421.57 ± 41.87 | 0.699(0.435–0.987) | 0.276 |
The p value describes significance of difference between AE subtype in comparison to healthy controls using nSolver Software. miRNA ID, official microRNA name according to miRBASE. p value = the unadjusted p value from the t test. Bold values represent higher expression; italicized values represent lower expression
Cancer-Associated Pathways Enriched in AE Dysregulated miRNAs
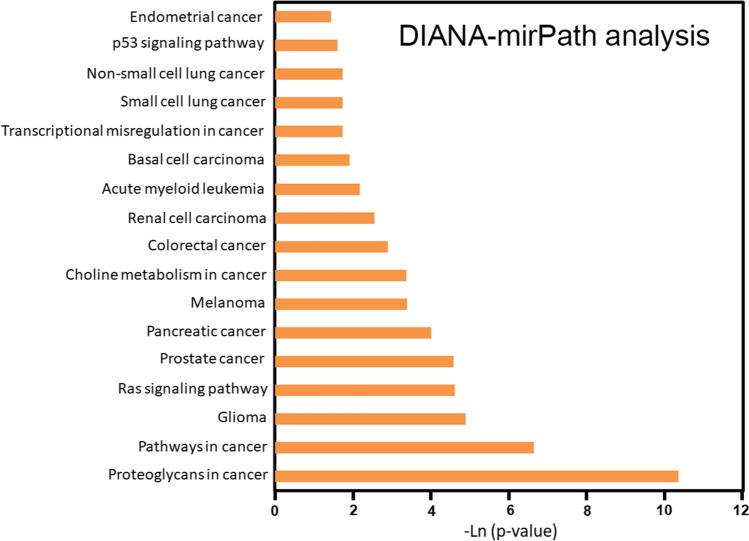
The presence of an occult tumor was found in the AE patients that serves as a stimulus for autoantibody production [35, 36]. Interestingly, there were 17 cancer-associated pathways enriched into the dysregulated 18 miRNAs, including endometrial cancer, p53 signaling pathway, non-small cell lung cancer, small cell lung cancer, transcriptional misregulation in cancer, basal cell carcinoma, acute myeloid leukemia, renal cell carcinoma, colorectal cancer, choline metabolism in cancer, melanoma, pancreatic cancer, prostate cancer, Ras signaling pathway, glioma, pathways in cancer, and proteoglycans in cancer (p < 0.02; Fig. 2). For example, the KEGG pathway “Pathways in cancer” (hsa05200, p = 2.312e − 07, Supplementary Fig. 2) was significantly altered in differentially expressed miRNAs between anti-NMDAR encephalitis patients and control subjects at baseline, with 15 miRNAs (hsa-miR-20a-5p, hsa-miR-17-5p, hsa-miR-20b-5p, hsa-miR-340-5p, hsa-miR-21-5p, hsa-let-7d-5p, hsa-miR-15b-5p, hsa-miR-155-5p, hsa-miR-27b-5p, hsa-miR-34a-5p, hsa-miR-146a-5p, hsa-miR-10a-5p, hsa-miR-301a-5p, hsa-miR-132-5p, hsa-miR-23b-5p) targeting 160 genes, including FZD7, BRAF, FGF12, FOS, GSK3B, PRKCA, STAT3, PDGFRA, FZD5, and E2F1. Data from these experiments demonstrated that exosomes expressing specific miRNAs may participate in cancer development, which indicates a feedback regulation from antibody-positive AE.
Fig. 2.
Cancer-associated Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways enriched in anti-NMDAR autoimmune encephalitis-associated dysregulated miRNAs. Graphical representation of anti-NMDAR‐mediated targeted pathways generated using DIANA‐miRPath (y‐axis: p values; x‐axis: miRNA‐associated pathways). p values for pathway analysis were obtained by Fisher’s exact test as enrichment analysis method and the false discovery rate (FDR) was estimated using the Benjamini and Hochberg method
Validation of miRNA Expression Using Independent Cohorts of Antibody-Positive AE Patients
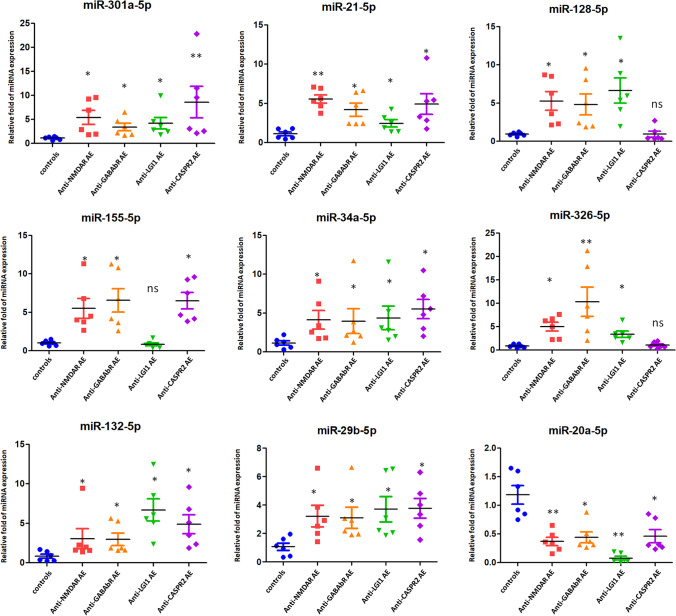
The miRNAs were selected for validation since they were found to relate with neuroinflammation and encephalitis [37–42]. Nine miRNAs (miR-301a-5p, miR-21-5p, miR-128-5p, miR-155-5p, miR-34a-5p, miR-326-5p, miR-132-5p, miR-29b-5p, and miR-20a-5p) were selected for further validation using an independent cohort of 6 patients with anti-NMDA receptor encephalitis, 6 patients with anti-GABAB receptor encephalitis, 6 patients with anti-LGI1 encephalitis, 6 patients with anti-CASPR2 encephalitis, and 6 control individuals. We found that miR-301a-5p, miR-21-5p, miR-34a-5p, miR-132-5p, and miR-29b-5p were significantly highly expressed in CSF exosomes in anti-NMDAR, GABABR, LGI1, and CASPR2 AE patients when compared with healthy controls (p < 0.05), while miR-20a-5p was significantly lowly expressed in CSF exosomes (Fig. 3). In addition, we investigated another three miRNAs (miR-128-5p, miR-155-5p, and miR-326-5p) using individual CSF exosomes. miR-128-5p and miR-326-5p were significantly increased in CSF exosomes isolated from anti-NMDAR, GABABR, and LGI1 AE patients when compared with healthy controls (p < 0.05); however, there was no significant difference of these two miRNAs between anti-CASPR2 AE patients and healthy controls (Fig. 3). Exosomal miR-155-5p was significantly highly expressed in anti-NMDAR, GABABR, and CASPR2 AE patients when compared with healthy controls (p < 0.05); however, there was no significant difference of exosomal miR-155-5p between anti-LGI1 AE patients and healthy controls (Fig. 3). Taken together, these data confirmed the validity of differentially expressed exosomal miRNAs in CSF related to the pathogenesis of antibody-positive AE.
Fig. 3.
Validation of miRNA expression. TaqMan real-time PCR to validate the expression levels of nine miRNAs (miR-301a-5p, miR-21-5p, miR-128-5p, miR-155-5p, miR-34a-5p, miR-326-5p, miR-132-5p, miR-29b-5p, and miR-20a-5p) selected for further validation using an independent cohort of 6 patients with anti-NMDA receptor encephalitis, 6 patients with anti-GABA receptor encephalitis, 6 patients with anti-LGI1 encephalitis, 6 patients with anti-CASPR2 encephalitis, and 6 control individuals. Data shown are as mean ± SEM. *p < 0.05; **p < 0.01; ***p < 0.001
Exosome Concentration Was Higher in HSE Patients Compared to HSV-Negative Patients
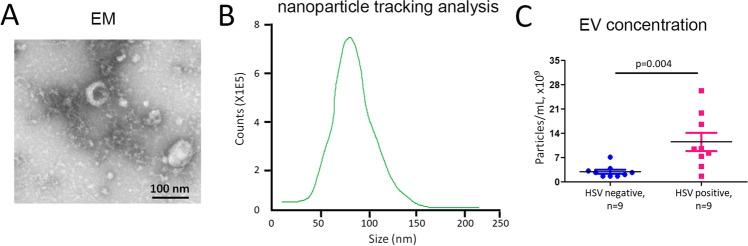
In order to determine whether the abundance of exosomes is changed in HSE and to define their compositions and roles, exosomes were isolated from CSF from 9 HSE patients and 9 HSV( −) patients. Exosomic structure was examined by TEM, and exosomic concentration was measured by NTA. The electron micrographs of the microvesicles revealed rounded structures with a size of 85–120 nm (Fig. 4A), similar to previously described exosomes. A similar size distribution was also confirmed by NTA with a peak around 93 nm (Fig. 4B). We sought to determine whether there were differences in the abundance of exosome production isolated from CSF in HSE patients compared to HSV( −) patients. An analysis of a cross-sectional cohort revealed a significantly higher EV concentration in 9 HSE patients compared to 9 HSV( −) patients (HSV( +): 11.65 × 109 ± 2.33 × 109 vs. HSV( −): 3.81 × 109 ± 0.54 × 109 particles/mL; p = 0.004; Fig. 4C). These data indicated that HSV infection would increase the production of exosomes secreted into CSF in HSE, suggesting a potential role of exosomes in HSE.
Fig. 4.
Exosome was induced for secretion into CSF in HSE compared to HSV( −) patients. A The electron micrographs of the microvesicles. B Microvesicle concentration was measured by nanoparticle tracking analysis (NTA). C Comparison of exosome concentration between 9 HSE patients compared to 9 HSV( −) patients
Exosome Was Induced in Mouse Model of HSE
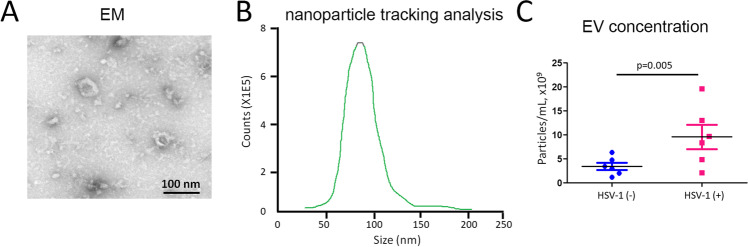
In order to validate the results obtained from clinical subjects, we developed an endogenous rodent model of post-infectious encephalitis. Six mice were inoculated intranasally with HSV-1. Another six age- and sex-matched control Balb/c mice were inoculated with vehicle solution. Serum was collected at 8 weeks post-inoculation and tested for exosome content. The electron micrographs of the exosomes revealed rounded structures with a size of 65–94 nm (Fig. 5A). NTA demonstrated a similar size distribution with a peak around 90 nm (Fig. 5B). We compared the exosome production between HSV-treated mice and control group. Results revealed a significantly higher exosome concentration in HSV-treated mice (n = 6) compared to the control group (n = 6) (HSV-1( +): 10.22 × 109 ± 2.19 × 109 vs. HSV-1( −): 4.55 × 109 ± 0.33 × 109 particles/mL; p = 0.005; Fig. 5C). These data confirmed that exosomes were induced for secretion after HSV infection, providing more evidence of the pathogenetic role of exosomes in HSE.
Fig. 5.
Exosome was induced in mouse model of HSE. A The electron micrographs of the exosomes derived from sera of mouse model of HSE. B Nanoparticle tracking analysis (NTA) to show size distribution of exosomes derived from sera of mouse model of herpes simplex encephalitis. C Comparison of the difference of exosome concentration in HSV-treated mice (n = 6) compared to control group (n = 6)
Exosomes Isolated from HSE Patients Expressed Higher NMDAR, GABAR, and AMPAR Subunits
We next examine the exosomal protein composition, especially neuronal autoantigens. Exosomes isolated from pooled sera collected from 6 HSV-treated mice were subjected for western blot, and results demonstrated that NR2B subunits of NMDAR and GABAB1R, and the GluR1 subunits of AMPAR were present and highly expressed in HSV-1-treated mice compared to those in control group (Fig. 6). TSG101 and CD9 were detected as the exosome markers and loading control.
Fig. 6.
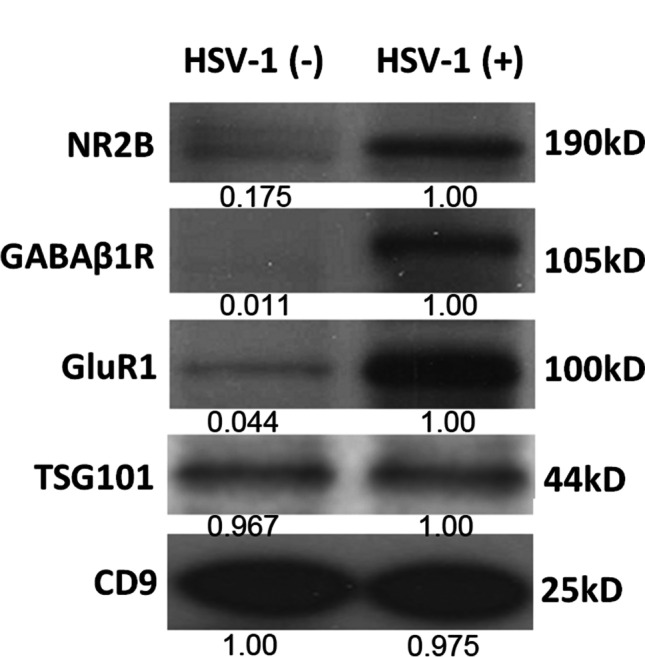
Exosome expressed higher NMDAR, GABAR, and AMPAR subunits in HSV-induced animal model. Western blot to show the protein level of NR2B subunits of NMDAR and GABAB1R, and the GluR1 subunits of AMPAR on the pooled sera exosomes from 6 HSV-treated mice compared to those in control group (n = 6). Protein quantification was performed by bandscan and densitometry analysis with optical density for NR2B, GABAb1R, GluR1, TSG101, and CD9. TSG101 and CD9 were detected as the exosome markers and loading control
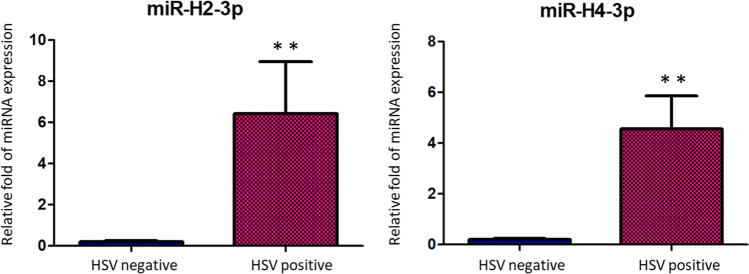
miR-H2-3p and miR-H4-3p Encoded by HSV Were Dysregulated in Exosomes Isolated from CSF of HSE Patients
We further examined the presence of miRNAs encoded and expressed by HSV in exosomes isolated from CSF of patients with HSE. miR-H2-3p, miR-H4-3p, miR-H3, miR-H4-5p, miR-H5, and miR-H6 were chosen for TaqMan microRNA detection assay. Only miR-H2-3p and miR-H4-3p were detectable in CSF exosomes from HSE. However, miR-H3, miR-H4-5p, miR-H5, and miR-H6 were not detectable in both groups. After comparison, we found that miR-H2-3p and miR-H4-3p were significantly highly expressed in CSF-derived exosomes isolated from HSE patients (n = 5) when compared to those in HSV( −) subjects (n = 5) (miR-H2-3p, p < 0.01; miR-H4-3p, p < 0.01; Fig. 7). These data suggested that HSV-encoded miRNAs may potentially target host genes to regulate HSE by the way of exosomal genetic transfer.
Fig. 7.
Exosomal miR-H2-3p and miR-H4-3p miRNAs expressed by HSV. miR-H2-3p and miR-H4-3p were compared for expression from cerebrospinal fluid exosomes isolated from 5 HSE patients compared to 5 HSV( −) patients. Relative expression of miR-H2-3p and miR-H4-3p was calculated and observed statistically differentially expressed compared to the control group. *p < 0.05, **p < 0.01
Discussion
Concurrent inflammatory findings in the CSF, including the presence of OCBs, lymphocytic pleocytosis, and elevated protein levels, were found in antibody-mediated AE [43]. In this study, we presented the evidence to support the role of exosomes and exosomal expression of miRNAs and neuronal antigens to the pathophysiology of HSE and AE. Results from the current study demonstrated dysregulated miRNAs in CSF-derived exosomes of AE patients (NMDAR Abs positive) versus control subjects. Similar expression patterns were demonstrated in AE patients with anti-GABABR Abs, anti-LGI1 Abs, and anti-CASPR2 Abs (Table 3). Further study demonstrated important enriched cancer-associated pathways related to the dysregulated miRNAs, including endometrial cancer, the p53 signaling pathway, non-small cell lung cancer, small cell lung cancer, transcriptional dysregulation in cancer, basal cell carcinoma, acute myeloid leukemia, renal cell carcinoma, colorectal cancer, choline metabolism in cancer, melanoma, pancreatic cancer, prostate cancer, the Ras signaling pathway, glioma, pathways in cancer, and proteoglycans in cancer (Fig. 2). These results indicated that exosomes expressing specific miRNAs may participate in cancer development, which indicates a feedback regulation from antibody-positive AE. Ectopic expression of neuronal antigens in various types of malignant tumors could trigger the anti-tumor immune response cross-reacting with the cognate CNS protein, which often leads to the paraneoplastic neurological syndromes [44].
MicroRNAs are a group of short, noncoding RNA molecules that post-transcriptionally regulate the expression of target mRNAs. Inflammatory processes may be promoted by miRNAs [45], such as miR-155, or suppressed by miRNAs, including miR-146a, miR-124, and miR-21. miR-155 is a central proinflammatory mediator of the CNS in response to TLR signaling [46–48]. miR-155 targets include SOCS1 [48], SHIP1 [49], C/EBP-β [50], and IL13Rα1 [51]. In addition, miR-155 expression is induced by p53 in immune cells, and miR-155 subsequently targets c-Maf, which induces differentiation and anti-inflammatory responses [52]. In our study, we found that miR-155 was significantly more highly expressed in anti-NMDAR, GABABR, and CASPR2 AE patients than in control subjects (p < 0.05; Fig. 3). These data demonstrated that miR-155 may participate in autoimmune encephalitis by regulating the SOCS1, SHIP1, C/EBP-β, and IL13Rα1 signaling cascades in microglia. Anti-voltage-gated potassium channel (VGKC) antibodies include antibodies against associated proteins LGI1 and CASPR2 in limbic encephalitis [53]. Results in our study demonstrated that miRNAs 128-5p was found to be over-expressed in CASPR2 and LGI1 AE cases, and miR-155-5p was found highly expressed in CASPR2 encephalitis, while not in LGI1 encephalitis (Fig. 3). These data indicated that both miRNAs 128-5p and 155-5p participated in the encephalitis caused by anti-VGKC antibodies. Pathways associated with SOCS1, SHIP1, C/EBP-β, and IL13Rα1 signaling cascades might regulate the development CASPR2 encephalitis but not LGI1 encephalitis. miR-128 was reported to participate in the inflammation pathogenesis by regulating the p38α/M-CSF inflammatory signaling pathway [54]. Thus, p38α/M-CSF inflammatory signaling pathway might fine-tune anti-LGI1- and anti-CASPR2-associated encephalitis. Furthermore, p53 also suppresses c-Maf through the induction of two additional microRNAs, miR-34a and miR-145, which target Twist2, an activator of c-Maf expression [52]. The results of our study demonstrated that miR-34a-5p was significantly more highly expressed in CSF exosomes in anti-NMDAR, GABABR, LGI1, and CASPR2 AE patients than in control subjects (p < 0.05; Fig. 3), which indicated that miR-34a could inhibit the anti-inflammatory effect in AE by regulating Twist2 mRNA. Additionally, we found that miR-301a-5p was significantly more highly expressed in CSF exosomes in anti-NMDAR, GABABR, LGI1, and CASPR2 AE patients than in control subjects. There are several potential mechanisms underlying the role of miR-301a in the development of AE. Bibhabasu Hazra et al. [55] reported that the expression of miR-301a is increased in Japanese encephalitis virus (JEV)–infected microglial cells and the human brain. Overexpression of miR-301a augments the JEV-induced inflammatory response, whereas inhibition of miR-301a completely reverses the effects. They further found that miR-301a regulates the inflammatory response to JEV infection via suppression of NKRF activity [55]. We have not assessed the effect on the targeted exosomal miRNA pathways in the study. Future functional study on animal model of AE should be employed by targeting the specific miRNAs or its regulated pathways.
Infection with HSV leads to blisters or sores in the epithelia, and HSV further survives in sensory neurons via a life-long latent reservoir [56]. Although asymptomatic pathology usually occurs after infection with HSV, periodic reactivation of HSV within latently infected neurons may lead to recurrent infections [57]. HSV is an important cause of encephalitis, which is the most common type of sporadic encephalitis in humans, with a high mortality rate of up to 70% if left untreated [13]. HSE has common and even severe symptoms, including fever, headaches, vomiting, neurological deficits, and seizures [58]. Recent studies have identified the specific types of immune cells that induce brain inflammation in HSE [59]. Using a mouse model, the authors showed that neutrophils increased the permeability of the blood–brain barrier and further led to brain damage, which is commonly found in HSE [59]. At the same time, monocytes were found to play a protective role by clearing HSV and limiting viral replication, previously evidenced as an important way to prevent brain damage [59]. Therefore, immune cells infiltrate the CNS, and disruption of the blood–brain barrier causes a neuroinflammatory response during HSE [60]. However, the mechanisms by which HSV causes encephalitis remain poorly understood. The results of this study demonstrated that exosomes were induced for secretion into local brain sites and even into sera in HSE (Fig. 4). Viral infections are considered one more trigger of antibody-mediated encephalitis [61]. NMDAR autoantibodies in patients were firstly discovered in patients with HSE, which was leading to the hypothesis that autoimmune-mediated clinical symptoms happened after HSV infection [15]. Further studies validated that there was a frequency of almost 1/3 of patients with HSE developing NMDAR encephalitis thereafter [14, 16]. In addition, HSV-induced exosome production was validated in an animal model of post-HSV encephalitis (Fig. 5). More importantly, exosomes isolated from pooled sera collected from 6 HSV-treated mice demonstrated that NR2B subunits of NMDAR and GABAB1R, and the GluR1 subunits of AMPAR were present and highly expressed in HSV-1-treated mice (Fig. 6). These data indicated that exosomes play critical roles in the presentation of surface autoantigens, inducing autoimmunity and inflammation, which could be recognized as a novel mechanism for HSE pathogenesis. Due to the challenge in the collection of sufficient CSF samples from HSE patients, exosomes isolated from the CSF of HSV-positive patients are required to compare the protein expression levels of NMDAR, GABAR, and AMPAR subunits from those of HSV( −) patients. It was reported that experimental mice inoculated intranasally with HSV-1 developed serum NMDAR antibodies [17]. Autoimmunity also leads to development of autoantibodies to GABAa, AMPA, and dopamine D2 receptors [62]. The type of autoimmunity effect extended to other viruses, such as Epstein-Barr virus, varicella-zoster virus, herpesvirus, hepatitis viruses, and Japanese encephalitis B virus, which indicated that virus-induced autoantibody generation could be a broad mechanism of pathology in autoimmune neurological disease [63]. However, it requires more investigation to prove the causal relationship.
The only abundant viral gene product from HSV-1 after it established latency in neurons is the latency-associated transcript (LAT), which has been proven to be a noncoding RNA [12, 64]. Umbach et al. reported that LAT encoded four distinct miRNAs. Among them, miR-H2-3p was transcribed complementarily to mRNA of infected cell protein 0 (ICP0), a viral immediate-early transcriptional activator in HSV-1 replication and latency [65]. A fifth HSV-1 miRNA, miR-H6, was identified in latently infected trigeminal ganglia [66]. miR-H6 inhibits ICP4 mRNA translation by seeding region complementarity to the target mRNA; thus, miR-H6 is required for HSV-1 productive infection [66]. Latency HSV-encoded miRNAs, miR-H2-3p and miR-H4-3p, coordinate with human miR-138, which is highly abundant in neurons, to promote latency during lytic infection [67, 68]. The results from our current study demonstrated that miR-H2-3p and miR-H4-3p were significantly more highly expressed in CSF-derived exosomes isolated from HSE patients than those isolated from HSV( −) subjects (miR-H2-3p, p < 0.01; miR-H4-3p, p < 0.01; Fig. 7). Therefore, exosomal expression of miR-H2-3p may further inhibit ICP0 protein expression, thus decreasing HSV entry into the productive replication cycle and latency [65, 69, 70], which may be further considered a potential mechanism for HSE in patients. Increased exosomal miR-H4-3p expression may inhibit ICP34.5 expression via an “antisense” mechanism [71, 72]. In addition, we found that miR-H3, miR-H4-5p, miR-H5, and miR-H6 were not detectable in exosomes isolated from the CSF of all patients, indicating that these viral miRNAs could not be taken up by exosomes shed by brain cells.
There are several limitations in this study. First, caution should be taken to understand the results because of the relatively small sample size in the current study. A larger cohort study for validation will be required in the future. Second, abolishment of exosome secretion induced by HSV is needed to confirm its in vivo role in HSE. The detailed molecular mechanisms underlying HSV-induced exosomes during encephalitis require further investigation.
In conclusion, our results demonstrated that there was a specific miRNA signature that was differentially expressed in antibody-positive AE patients. Functionally, the dysregulated miRNAs were enriched in cancer-associated pathways, indicating there was a feedback regulation of AE in cancer development. Exosomes could be induced during HSV infection. These secreted exosomes contained higher expression of NMDAR, GABAR, and AMPAR subunits as well as miR-H2-3p and miR-H4-3p encoded by HSV in encephalitis patients as well as HSV-induced animal model. Therefore, we concluded that HSV-induced exosomes are critical players in HSE development via the presentation of surface/cellular neuronal autoantigens that trigger brain autoimmunity. These findings provide potential novel strategies for therapeutic intervention.
Supplementary Information
Below is the link to the electronic supplementary material.
Abbreviations
- HSE
Herpes simplex encephalitis
- AE
Autoimmune encephalitis
- CSF
Cerebrospinal fluid
- NMDAR
N-Methyl-d-aspartate receptor
- GABABR
Gamma-aminobutyric acid-B receptor
- LGI
Leucine-rich glioma-inactivated 1
- CASPR2
Contactin-associated protein-like 2 receptor
- ANNA-1
Anti-neuronal nuclear antibody type 1
- CRMP5
Collapsin-responsive mediator protein-5
Author Contribution
JCG and YXG designed the research. YAL, JCG, YBM, XJW, ZSL, and XMX performed research. YAL, JCG, and HMC collected subjects and analyzed data. YXG supervised the study. YAL, JCG, and YXG contributed to the manuscript writing.
Funding
This research was supported by Wenling Social Development Science and Technology Project (2021S00202), Wenling Science and Technology Plan Social Project (2018C31BA0084), Zhejiang Provincial Natural Science Foundation of China under Grant No. LY19H090018, Project 81401038 supported by National Natural Science Foundation of China, and Zhejiang Province Medicine Health General Research Program (2020KY602). The funders had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Data Availability
The datasets generated and/or analyzed during the current study are available from the corresponding author on reasonable request.
The study was approved by the institutional ethics committee of Sir Run Run Shaw Hospital affiliated with Zhejiang University School of Medicine, and the need for informed consent was received from all subjects. The study was performed in accordance with the 1964 Declaration of Helsinki and its later amendments. All animal procedures were approved by our Institutional Animal Care and Use Committee at Sir Run Run Shaw Hospital affiliated with Zhejiang University.
Declarations
Consent to Participate
The informed consent was received from all subjects.
Consent for Publication
The manuscript has been seen and approved by all authors as given on the title page of the manuscript.
Conflict of Interest
The authors declare no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Graus F, Titulaer MJ, Balu R, Benseler S, Bien CG, Cellucci T, Cortese I, Dale RC, Gelfand JM, Geschwind M, Glaser CA, Honnorat J, Hoftberger R, Iizuka T, Irani SR, Lancaster E, Leypoldt F, Pruss H, Rae-Grant A, Reindl M, Rosenfeld MR, Rostasy K, Saiz A, Venkatesan A, Vincent A, Wandinger KP, Waters P, Dalmau J. A clinical approach to diagnosis of autoimmune encephalitis. The Lancet Neurology. 2016;15(4):391–404. doi: 10.1016/S1474-4422(15)00401-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sansing LH, Tuzun E, Ko MW, Baccon J, Lynch DR, Dalmau J. A patient with encephalitis associated with NMDA receptor antibodies. Nat Clin Pract Neurol. 2007;3(5):291–296. doi: 10.1038/ncpneuro0493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brenton JN, Goodkin HP. Antibody-mediated autoimmune encephalitis in childhood. Pediatr Neurol. 2016;60:13–23. doi: 10.1016/j.pediatrneurol.2016.04.004. [DOI] [PubMed] [Google Scholar]
- 4.Gu Y, Zhong M, He L, Li W, Huang Y, Liu J, Chen Y, Xiao Z. Epidemiology of antibody-positive autoimmune encephalitis in Southwest China: a multicenter study. Front Immunol. 2019;10:2611. doi: 10.3389/fimmu.2019.02611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lancaster E, Dalmau J. Neuronal autoantigens–pathogenesis, associated disorders and antibody testing. Nat Rev Neurol. 2012;8(7):380–390. doi: 10.1038/nrneurol.2012.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Alamowitch S, Graus F, Uchuya M, Rene R, Bescansa E, Delattre JY. Limbic encephalitis and small cell lung cancer. Clinical and immunological features. Brain : a journal of neurology. 1997;120(Pt 6):923–928. doi: 10.1093/brain/120.6.923. [DOI] [PubMed] [Google Scholar]
- 7.Lawn ND, Westmoreland BF, Kiely MJ, Lennon VA, Vernino S. Clinical, magnetic resonance imaging, and electroencephalographic findings in paraneoplastic limbic encephalitis. Mayo Clin Proc. 2003;78(11):1363–1368. doi: 10.4065/78.11.1363. [DOI] [PubMed] [Google Scholar]
- 8.Dalmau J, Gleichman AJ, Hughes EG, Rossi JE, Peng X, Lai M, Dessain SK, Rosenfeld MR, Balice-Gordon R, Lynch DR. Anti-NMDA-receptor encephalitis: case series and analysis of the effects of antibodies. The Lancet Neurology. 2008;7(12):1091–1098. doi: 10.1016/S1474-4422(08)70224-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hoftberger R, van Sonderen A, Leypoldt F, Houghton D, Geschwind M, Gelfand J, Paredes M, Sabater L, Saiz A, Titulaer MJ, Graus F, Dalmau J. Encephalitis and AMPA receptor antibodies: novel findings in a case series of 22 patients. Neurology. 2015;84(24):2403–2412. doi: 10.1212/WNL.0000000000001682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Joubert B, Kerschen P, Zekeridou A, Desestret V, Rogemond V, Chaffois MO, Ducray F, Larrue V, Daubail B, Idbaih A, Psimaras D, Antoine JC, Delattre JY, Honnorat J. Clinical spectrum of encephalitis associated with antibodies against the alpha-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid receptor: case series and review of the literature. JAMA Neurol. 2015;72(10):1163–1169. doi: 10.1001/jamaneurol.2015.1715. [DOI] [PubMed] [Google Scholar]
- 11.Braun E, Zimmerman T, Hur TB, Reinhartz E, Fellig Y, Panet A, Steiner I. Neurotropism of herpes simplex virus type 1 in brain organ cultures. J Gen Virol. 2006;87(Pt 10):2827–2837. doi: 10.1099/vir.0.81850-0. [DOI] [PubMed] [Google Scholar]
- 12.Bloom DC. HSV LAT and neuronal survival. Int Rev Immunol. 2004;23(1–2):187–198. doi: 10.1080/08830180490265592. [DOI] [PubMed] [Google Scholar]
- 13.Bradshaw MJ, Venkatesan A. Herpes simplex virus-1 encephalitis in adults: pathophysiology, diagnosis, and management. Neurotherapeutics : the journal of the American Society for Experimental NeuroTherapeutics. 2016;13(3):493–508. doi: 10.1007/s13311-016-0433-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Armangue T, Leypoldt F, Malaga I, Raspall-Chaure M, Marti I, Nichter C, Pugh J, Vicente-Rasoamalala M, Lafuente-Hidalgo M, Macaya A, Ke M, Titulaer MJ, Hoftberger R, Sheriff H, Glaser C, Dalmau J. Herpes simplex virus encephalitis is a trigger of brain autoimmunity. Ann Neurol. 2014;75(2):317–323. doi: 10.1002/ana.24083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pruss H, Finke C, Holtje M, Hofmann J, Klingbeil C, Probst C, Borowski K, Ahnert-Hilger G, Harms L, Schwab JM, Ploner CJ, Komorowski L, Stoecker W, Dalmau J, Wandinger KP. N-methyl-D-aspartate receptor antibodies in herpes simplex encephalitis. Ann Neurol. 2012;72(6):902–911. doi: 10.1002/ana.23689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Armangue T, Spatola M, Vlagea A, Mattozzi S, Carceles-Cordon M, Martinez-Heras E, Llufriu S, Muchart J, Erro ME, Abraira L, Moris G, Monros-Gimenez L, Corral-Corral I, Montejo C, Toledo M, Bataller L, Secondi G, Arino H, Martinez-Hernandez E, Juan M, Marcos MA, Alsina L, Saiz A, Rosenfeld MR, Graus F, Dalmau J, Spanish Herpes Simplex Encephalitis Study G Frequency, symptoms, risk factors, and outcomes of autoimmune encephalitis after herpes simplex encephalitis: a prospective observational study and retrospective analysis. The Lancet Neurology. 2018;17(9):760–772. doi: 10.1016/S1474-4422(18)30244-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Linnoila J, Pulli B, Armangue T, Planaguma J, Narsimhan R, Schob S, Zeller MWG, Dalmau J, Chen J. Mouse model of anti-NMDA receptor post-herpes simplex encephalitis. Neurology(R) neuroimmunology & neuroinflammation. 2019;6(2):e529. doi: 10.1212/NXI.0000000000000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cheng L, Sun X, Scicluna BJ, Coleman BM, Hill AF. Characterization and deep sequencing analysis of exosomal and non-exosomal miRNA in human urine. Kidney Int. 2014;86(2):433–444. doi: 10.1038/ki.2013.502. [DOI] [PubMed] [Google Scholar]
- 19.Manterola L, Guruceaga E, Gallego Perez-Larraya J, Gonzalez-Huarriz M, Jauregui P, Tejada S, Diez-Valle R, Segura V, Sampron N, Barrena C, Ruiz I, Agirre A, Ayuso A, Rodriguez J, Gonzalez A, Xipell E, Matheu A, Lopez de Munain A, Tunon T, Zazpe I, Garcia-Foncillas J, Paris S, Delattre JY, Alonso MM. A small noncoding RNA signature found in exosomes of GBM patient serum as a diagnostic tool. Neuro Oncol. 2014;16(4):520–527. doi: 10.1093/neuonc/not218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Street JM, Barran PE, Mackay CL, Weidt S, Balmforth C, Walsh TS, Chalmers RT, Webb DJ, Dear JW. Identification and proteomic profiling of exosomes in human cerebrospinal fluid. J Transl Med. 2012;10:5. doi: 10.1186/1479-5876-10-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Banigan MG, Kao PF, Kozubek JA, Winslow AR, Medina J, Costa J, Schmitt A, Schneider A, Cabral H, Cagsal-Getkin O, Vanderburg CR, Delalle I. Differential expression of exosomal microRNAs in prefrontal cortices of schizophrenia and bipolar disorder patients. PLoS ONE. 2013;8(1):e48814. doi: 10.1371/journal.pone.0048814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Antonucci F, Turola E, Riganti L, Caleo M, Gabrielli M, Perrotta C, Novellino L, Clementi E, Giussani P, Viani P, Matteoli M, Verderio C. Microvesicles released from microglia stimulate synaptic activity via enhanced sphingolipid metabolism. EMBO J. 2012;31(5):1231–1240. doi: 10.1038/emboj.2011.489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fruhbeis C, Frohlich D, Kuo WP, Amphornrat J, Thilemann S, Saab AS, Kirchhoff F, Mobius W, Goebbels S, Nave KA, Schneider A, Simons M, Klugmann M, Trotter J, Kramer-Albers EM. Neurotransmitter-triggered transfer of exosomes mediates oligodendrocyte-neuron communication. PLoS Biol. 2013;11(7):e1001604. doi: 10.1371/journal.pbio.1001604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lopez-Verrilli MA, Picou F, Court FA. Schwann cell-derived exosomes enhance axonal regeneration in the peripheral nervous system. Glia. 2013;61(11):1795–1806. doi: 10.1002/glia.22558. [DOI] [PubMed] [Google Scholar]
- 25.Fevrier B, Vilette D, Archer F, Loew D, Faigle W, Vidal M, Laude H, Raposo G. Cells release prions in association with exosomes. Proc Natl Acad Sci USA. 2004;101(26):9683–9688. doi: 10.1073/pnas.0308413101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Emmanouilidou E, Melachroinou K, Roumeliotis T, Garbis SD, Ntzouni M, Margaritis LH, Stefanis L, Vekrellis K. Cell-produced alpha-synuclein is secreted in a calcium-dependent manner by exosomes and impacts neuronal survival. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2010;30(20):6838–6851. doi: 10.1523/JNEUROSCI.5699-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rajendran L, Honsho M, Zahn TR, Keller P, Geiger KD, Verkade P, Simons K. Alzheimer’s disease beta-amyloid peptides are released in association with exosomes. Proc Natl Acad Sci USA. 2006;103(30):11172–11177. doi: 10.1073/pnas.0603838103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang G, Dinkins M, He Q, Zhu G, Poirier C, Campbell A, Mayer-Proschel M, Bieberich E. Astrocytes secrete exosomes enriched with proapoptotic ceramide and prostate apoptosis response 4 (PAR-4): potential mechanism of apoptosis induction in Alzheimer disease (AD) J Biol Chem. 2012;287(25):21384–21395. doi: 10.1074/jbc.M112.340513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vella LJ, Sharples RA, Lawson VA, Masters CL, Cappai R, Hill AF. Packaging of prions into exosomes is associated with a novel pathway of PrP processing. J Pathol. 2007;211(5):582–590. doi: 10.1002/path.2145. [DOI] [PubMed] [Google Scholar]
- 30.Bellingham SA, Coleman BM, Hill AF. Small RNA deep sequencing reveals a distinct miRNA signature released in exosomes from prion-infected neuronal cells. Nucleic Acids Res. 2012;40(21):10937–10949. doi: 10.1093/nar/gks832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gu J, Jin T, Li Z, Chen H, Xia H, Xu X, Gui Y. Exosomes expressing neuronal autoantigens induced immune response in antibody-positive autoimmune encephalitis. Mol Immunol. 2021;131:164–170. doi: 10.1016/j.molimm.2020.12.034. [DOI] [PubMed] [Google Scholar]
- 32.Gui Y, Liu H, Zhang L, Lv W, Hu X. Altered microRNA profiles in cerebrospinal fluid exosome in Parkinson disease and Alzheimer disease. Oncotarget. 2015;6(35):37043–37053. doi: 10.18632/oncotarget.6158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gunasekaran M, Xu Z, Nayak DK, Sharma M, Hachem R, Walia R, Bremner RM, Smith MA, Mohanakumar T. Donor-derived exosomes with lung self-antigens in human lung allograft rejection. Am J Transplant Off J Am Soc Transplant Am Soc Transplant Surg. 2017;17(2):474–484. doi: 10.1111/ajt.13915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Papadopoulos GL, Alexiou P, Maragkakis M, Reczko M, Hatzigeorgiou AG. DIANA-mirPath: integrating human and mouse microRNAs in pathways. Bioinformatics. 2009;25(15):1991–1993. doi: 10.1093/bioinformatics/btp299. [DOI] [PubMed] [Google Scholar]
- 35.Tanaka K, Kawamura M, Sakimura K, Kato N (2020) Significance of autoantibodies in autoimmune encephalitis in relation to antigen localization: an outline of frequently reported autoantibodies with a non-systematic review. International journal of molecular sciences 21 (14). doi:10.3390/ijms21144941 [DOI] [PMC free article] [PubMed]
- 36.Tirthani E, Said MS, Smith RG, Jadhav N, Shanina E (2020) Paraneoplastic encephalomyelitis. In: StatPearls. Treasure Island (FL), [PubMed]
- 37.Ksiazek-Winiarek DJ, Kacperska MJ, Glabinski A. MicroRNAs as novel regulators of neuroinflammation. Mediators Inflamm. 2013;2013:172351. doi: 10.1155/2013/172351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang Z, Brandt S, Medeiros A, Wang S, Wu H, Dent A, Serezani CH. MicroRNA 21 is a homeostatic regulator of macrophage polarization and prevents prostaglandin E2-mediated M2 generation. PLoS ONE. 2015;10(2):e0115855. doi: 10.1371/journal.pone.0115855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.O’Connell RM, Taganov KD, Boldin MP, Cheng G, Baltimore D. MicroRNA-155 is induced during the macrophage inflammatory response. Proc Natl Acad Sci USA. 2007;104(5):1604–1609. doi: 10.1073/pnas.0610731104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Thounaojam MC, Kaushik DK, Kundu K, Basu A. MicroRNA-29b modulates Japanese encephalitis virus-induced microglia activation by targeting tumor necrosis factor alpha-induced protein 3. J Neurochem. 2014;129(1):143–154. doi: 10.1111/jnc.12609. [DOI] [PubMed] [Google Scholar]
- 41.Slota JA, Booth SA (2019) MicroRNAs in neuroinflammation: implications in disease pathogenesis, biomarker discovery and therapeutic applications. Non-coding RNA 5 (2). doi:10.3390/ncrna5020035 [DOI] [PMC free article] [PubMed]
- 42.Gaudet AD, Fonken LK, Watkins LR, Nelson RJ, Popovich PG. MicroRNAs: roles in regulating neuroinflammation. The Neuroscientist : a review journal bringing neurobiology, neurology and psychiatry. 2018;24(3):221–245. doi: 10.1177/1073858417721150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Blinder T, Lewerenz J. Cerebrospinal fluid findings in patients with autoimmune encephalitis-a systematic analysis. Front Neurol. 2019;10:804. doi: 10.3389/fneur.2019.00804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Darnell RB, Posner JB. Paraneoplastic syndromes involving the nervous system. N Engl J Med. 2003;349(16):1543–1554. doi: 10.1056/NEJMra023009. [DOI] [PubMed] [Google Scholar]
- 45.Macfarlane LA, Murphy PR. MicroRNA: biogenesis, function and role in cancer. Curr Genomics. 2010;11(7):537–561. doi: 10.2174/138920210793175895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bala S, Marcos M, Kodys K, Csak T, Catalano D, Mandrekar P, Szabo G. Up-regulation of microRNA-155 in macrophages contributes to increased tumor necrosis factor alpha (TNF{alpha}) production via increased mRNA half-life in alcoholic liver disease. J Biol Chem. 2011;286(2):1436–1444. doi: 10.1074/jbc.M110.145870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cardoso AL, Guedes JR, Pereira de Almeida L, Pedroso de Lima MC. miR-155 modulates microglia-mediated immune response by down-regulating SOCS-1 and promoting cytokine and nitric oxide production. Immunology. 2012;135(1):73–88. doi: 10.1111/j.1365-2567.2011.03514.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang P, Hou J, Lin L, Wang C, Liu X, Li D, Ma F, Wang Z, Cao X. Inducible microRNA-155 feedback promotes type I IFN signaling in antiviral innate immunity by targeting suppressor of cytokine signaling 1. J Immunol. 2010;185(10):6226–6233. doi: 10.4049/jimmunol.1000491. [DOI] [PubMed] [Google Scholar]
- 49.O’Connell RM, Chaudhuri AA, Rao DS, Baltimore D. Inositol phosphatase SHIP1 is a primary target of miR-155. Proc Natl Acad Sci USA. 2009;106(17):7113–7118. doi: 10.1073/pnas.0902636106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Worm J, Stenvang J, Petri A, Frederiksen KS, Obad S, Elmen J, Hedtjarn M, Straarup EM, Hansen JB, Kauppinen S. Silencing of microRNA-155 in mice during acute inflammatory response leads to derepression of c/ebp Beta and down-regulation of G-CSF. Nucleic Acids Res. 2009;37(17):5784–5792. doi: 10.1093/nar/gkp577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Martinez-Nunez RT, Louafi F, Sanchez-Elsner T. The interleukin 13 (IL-13) pathway in human macrophages is modulated by microRNA-155 via direct targeting of interleukin 13 receptor alpha1 (IL13Ralpha1) J Biol Chem. 2011;286(3):1786–1794. doi: 10.1074/jbc.M110.169367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Su W, Hopkins S, Nesser NK, Sopher B, Silvestroni A, Ammanuel S, Jayadev S, Moller T, Weinstein J, Garden GA. The p53 transcription factor modulates microglia behavior through microRNA-dependent regulation of c-Maf. J Immunol. 2014;192(1):358–366. doi: 10.4049/jimmunol.1301397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tanaka K. Anti-VGKC complex antibodies and limbic encephalitis. Brain and nerve = Shinkei kenkyu no shinpo. 2021;73(5):605–610. doi: 10.11477/mf.1416201804. [DOI] [PubMed] [Google Scholar]
- 54.Hong Y, Ren X, Liu W, Sun K, Chen B, Liu B, Yu X, Chen Q, Qian Q, Xie X, Jiang C. miR-128 participates in the pathogenesis of chronic constipation by regulating the p38alpha/M-CSF inflammatory signaling pathway. Am J Physiol Gastrointest Liver Physiol. 2021;321(4):G436–G447. doi: 10.1152/ajpgi.00114.2021. [DOI] [PubMed] [Google Scholar]
- 55.Hazra B, Chakraborty S, Bhaskar M, Mukherjee S, Mahadevan A, Basu A. miR-301a regulates inflammatory response to Japanese encephalitis virus infection via suppression of NKRF activity. J Immunol. 2019;203(8):2222–2238. doi: 10.4049/jimmunol.1900003. [DOI] [PubMed] [Google Scholar]
- 56.Rechenchoski DZ, Faccin-Galhardi LC, Linhares REC, Nozawa C. Herpesvirus: an underestimated virus. Folia Microbiol. 2017;62(2):151–156. doi: 10.1007/s12223-016-0482-7. [DOI] [PubMed] [Google Scholar]
- 57.Cliffe AR, Wilson AC (2017) Restarting lytic gene transcription at the onset of herpes simplex virus reactivation. Journal of virology 91 (2). doi:10.1128/JVI.01419-16 [DOI] [PMC free article] [PubMed]
- 58.Rabinstein AA. Herpes virus encephalitis in adults: current knowledge and old myths. Neurol Clin. 2017;35(4):695–705. doi: 10.1016/j.ncl.2017.06.006. [DOI] [PubMed] [Google Scholar]
- 59.Michael BD, Bricio-Moreno L, Sorensen EW, Miyabe Y, Lian J, Solomon T, Kurt-Jones EA, Luster AD. Astrocyte- and neuron-derived CXCL1 drives neutrophil transmigration and blood-brain barrier permeability in viral encephalitis. Cell Rep. 2020;32(11):108150. doi: 10.1016/j.celrep.2020.108150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mancini M, Vidal SM. Insights into the pathogenesis of herpes simplex encephalitis from mouse models. Mammalian genome : official journal of the International Mammalian Genome Society. 2018;29(7–8):425–445. doi: 10.1007/s00335-018-9772-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pruss H (2021) Autoantibodies in neurological disease. Nature reviews Immunology.10.1038/s41577-021-00543-w [DOI] [PMC free article] [PubMed]
- 62.Linnoila JJ, Binnicker MJ, Majed M, Klein CJ, McKeon A. CSF herpes virus and autoantibody profiles in the evaluation of encephalitis. Neurology(R) neuroimmunology & neuroinflammation. 2016;3(4):e245. doi: 10.1212/NXI.0000000000000245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pruss H. Postviral autoimmune encephalitis: manifestations in children and adults. Curr Opin Neurol. 2017;30(3):327–333. doi: 10.1097/WCO.0000000000000445. [DOI] [PubMed] [Google Scholar]
- 64.Stevens JG, Wagner EK, Devi-Rao GB, Cook ML, Feldman LT. RNA complementary to a herpesvirus alpha gene mRNA is prominent in latently infected neurons. Science. 1987;235(4792):1056–1059. doi: 10.1126/science.2434993. [DOI] [PubMed] [Google Scholar]
- 65.Everett RD. ICP0, a regulator of herpes simplex virus during lytic and latent infection. BioEssays : news and reviews in molecular, cellular and developmental biology. 2000;22(8):761–770. doi: 10.1002/1521-1878(200008)22:8<761::AID-BIES10>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- 66.Preston CM. Control of herpes simplex virus type 1 mRNA synthesis in cells infected with wild-type virus or the temperature-sensitive mutant tsK. J Virol. 1979;29(1):275–284. doi: 10.1128/JVI.29.1.275-284.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Pan D, Flores O, Umbach JL, Pesola JM, Bentley P, Rosato PC, Leib DA, Cullen BR, Coen DM. A neuron-specific host microRNA targets herpes simplex virus-1 ICP0 expression and promotes latency. Cell Host Microbe. 2014;15(4):446–456. doi: 10.1016/j.chom.2014.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sun L, Li Q. The miRNAs of herpes simplex virus (HSV) Virologica Sinica. 2012;27(6):333–338. doi: 10.1007/s12250-012-3266-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Cai W, Astor TL, Liptak LM, Cho C, Coen DM, Schaffer PA. The herpes simplex virus type 1 regulatory protein ICP0 enhances virus replication during acute infection and reactivation from latency. J Virol. 1993;67(12):7501–7512. doi: 10.1128/JVI.67.12.7501-7512.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Halford WP, Kemp CD, Isler JA, Davido DJ, Schaffer PA. ICP0, ICP4, or VP16 expressed from adenovirus vectors induces reactivation of latent herpes simplex virus type 1 in primary cultures of latently infected trigeminal ganglion cells. J Virol. 2001;75(13):6143–6153. doi: 10.1128/JVI.75.13.6143-6153.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lee LY, Schaffer PA. A virus with a mutation in the ICP4-binding site in the L/ST promoter of herpes simplex virus type 1, but not a virus with a mutation in open reading frame P, exhibits cell-type-specific expression of gamma(1)34.5 transcripts and latency-associated transcripts. Journal of virology. 1998;72(5):4250–4264. doi: 10.1128/JVI.72.5.4250-4264.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Randall G, Roizman B. Transcription of the derepressed open reading frame P of herpes simplex virus 1 precludes the expression of the antisense gamma(1)34.5 gene and may account for the attenuation of the mutant virus. Journal of virology. 1997;71(10):7750–7757. doi: 10.1128/JVI.71.10.7750-7757.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated and/or analyzed during the current study are available from the corresponding author on reasonable request.
The study was approved by the institutional ethics committee of Sir Run Run Shaw Hospital affiliated with Zhejiang University School of Medicine, and the need for informed consent was received from all subjects. The study was performed in accordance with the 1964 Declaration of Helsinki and its later amendments. All animal procedures were approved by our Institutional Animal Care and Use Committee at Sir Run Run Shaw Hospital affiliated with Zhejiang University.