Abstract
Background
Blastocystis is a common gut protistan parasite in humans and animals worldwide, but its interrelationship with the host gut microbiota and mucosal immune responses remains poorly understood. Different murine models of Blastocystis colonization were used to examine the effect of a common Blastocystis subtype (ST4) on host gut microbial community and adaptive immune system.
Results
Blastocystis ST4-colonized normal healthy mice and Rag1−/− mice asymptomatically and was able to alter the microbial community composition, mainly leading to increases in the proportion of Clostridia vadinBB60 group and Lachnospiraceae NK4A136 group, respectively. Blastocystis ST4 colonization promoted T helper 2 (Th2) response defined by interleukin (IL)-5 and IL-13 cytokine production, and T regulatory (Treg) induction from colonic lamina propria in normal healthy mice. Additionally, we observed that Blastocystis ST4 colonization can maintain the stability of bacterial community composition and induce Th2 and Treg immune responses to promote faster recovery from experimentally induced colitis. Furthermore, fecal microbiota transplantation of Blastocystis ST4-altered gut microbiome to colitis mice reduced the severity of colitis, which was associated with increased production of short-chain fat acids (SCFAs) and anti-inflammatory cytokine IL-10.
Conclusions
The data confirm our hypothesis that Blastocystis ST4 is a beneficial commensal, and the beneficial effects of Blastocystis ST4 colonization is mediated through modulating of the host gut bacterial composition, SCFAs production, and Th2 and Treg responses in different murine colonization models.
Supplementary Information
The online version contains supplementary material available at 10.1007/s00018-022-04271-9.
Keywords: Blastocystis, Gut microbiota, Th2, Treg, Colitis
Background
Blastocystis, classified under the stramenopile phylum, is the most common unicellular intestinal parasite found in humans and various animals, with an estimate of more than 1 billion people colonized worldwide [1]. The clinical significance of Blastocystis remains controversial, although it has been widely studied for more than 100 years [2]. Blastocystis has been associated with inflammatory bowel disease (IBD) and irritable bowel syndrome (IBS) [3, 4]. Some microbiome studies have reported that Blastocystis colonization decreases the abundance of beneficial bacterial Bifidobacterium in humans and mouse models [5, 6]. However, most Blastocystis–gut microbiota association studies have revealed that Blastocystis-colonized individuals have a higher gut bacterial diversity and lower levels of Bacteroides compared to Blastocystis-free individuals, suggesting that Blastocystis may be a beneficial commensal rather than a pathogen [7, 8]. These discrepancies may be influenced by the complex nature of Blastocystis wherein several genetically distinct subtypes exist [1]. Different subtypes exhibit extensive differences in genome size, effects on gut microbiota, and immune responses [8–10]. However, there is still no consensus on the existence of pathogenic and non-pathogenic subtypes to date, although some in vitro Blastocystis–host studies reveal subtype-associated pathobiological outcomes [5, 11].
Based on analyses of the small subunit (SSU) rRNA gene, 25 subtypes have been identified in humans and a wide range of animals [12]. Among them, ST1–4 are the most reported subtypes in humans, accounting for around 90% of human infected cases [13]. The prevalence of ST4 appears to be more geographically variable, as it is mainly reported in Europe and rarely found in South America, Africa and Asia [14, 15]. Interactions among host, gut microbiota, and Blastocystis can drive the development of the immune system in colonized individuals and play a role in maintaining intestinal homeostasis. Indeed, it has been reported in vitro and in mouse models that Blastocystis is involved in the host’s innate and adaptive immunity in regulating the function and differentiation of the immune cell repertoire of the gut [16, 17]. T helper 2 (Th2) cells and T regulatory (Treg) cells are well known for their crucial roles in fighting extracellular parasite infection and suppressing intestinal inflammation [18]. However, it is unclear whether these immune cells are involved in the process of Blastocystis colonization.
Although several studies have investigated the association between Blastocystis and the gut microbial composition, only limited studies have analyzed this association at the subtype level. Blastocystis ST4 was originally isolated from a Wistar rat [19] and is the most prevalent subtype observed in the Flemish Gut Flora Project (FGFP) [8], TwinsUK [20] and American Gut Project (AGP) [21]. In the current study, we explored the interactions between Blastocystis ST4, gut microbiota, and host immunity in different mice models. Furthermore, we evaluated the impact of Blastocystis ST4 colonization on experimentally induced colitis. Our findings showed that Blastocystis ST4 colonization not only alters the gut microbial composition, but also enhances the accumulation of Th2 and Treg cells in the colonic mucosa. Additionally, Blastocystis ST4 colonization prevents loss of microbiota diversity and contributes towards attenuation of disease in a murine model of experimental colitis. These results revealed a previously unrecognized mutualistic relationship between Blastocystis ST4, gut microbiota and host immunity.
Methods
Culture of Blastocystis
The axenized Blastocystis isolate WR1 belonging to ST4 was used in this study. ST4-WR1 was originally isolated from a healthy Wistar rat during an animal survey at National University of Singapore [19]. ST4 is a common zoonotic subtype frequently detected in humans and a wide range of animals, including nonhuman primates, bovines, goats, dogs, rodents, and birds [22]. Blastocystis was maintained in 10 ml of pre-reduced Iscove’s modified Dulbecco’s medium (IMDM) (Gibco) supplemented with heat-inactivated 10% horse serum (Gibco). Cultures were incubated under anaerobic conditions in an Anaerojar (Oxoid) with gas pack (Oxoid) at 37 °C and subcultured every 3–4 days. Blastocystis cell counts were assessed manually using hemocytometer (Kova International).
Mice and treatments
The animal experiments were performed according to the Singapore National Advisory Committee for Laboratory Animal Research guidelines. All animal procedures performed in this study were approved by the Institutional Animal Care and Use Committee of National University of Singapore. C57BL/6 and Rag1−/− (Rag1tm1Mom) mice of 8–12 weeks of age were bred and maintained in the animal facilities of the National University of Singapore (NUS). Littermates of the same sex and age were randomly assigned to the different experimental groups. Mice were colonized with Blastocystis ST4 via oral gavage with 5 × 107 live Blastocystis cells suspended in sterile phosphate-buffered saline (PBS) three times per week before euthanization at day 3 post last gavage. The control mice were orally gavaged with equal amounts of PBS at the same time. The mice used in all the experiments were age and sex matched.
For dextran sulfate sodium (DSS)-induced colitis with ST4 colonization model: C57BL/6 mice were administered 2% DSS (molecular mass = 36,000–50,000 Da; MP Biomedicals) w/v in drinking water for 7 days. Mice were then orally gavaged with 5 × 107 live Blastocystis cells three times per week for two consecutive weeks. In all experiments, mice were monitored daily for changes in body weight, stool consistency and presence of fecal blood. Disease activity index (DAI) was used to assess the severity of colitis as previously described [23]. In brief, daily calculation of DAI for each mouse was based on weight loss, occult blood, and stool consistency/diarrhea. Each parameter was scored on a scale of 1–4, with a maximum DAI score of 12. Score 0: no weight loss, normal stool, no blood; score 1: 1–3% weight loss, softer stool; score 2: 3–6% weight loss, loose stool, blood visible in stool; score 3: 6–9% weight loss, diarrhea, blood visible in stool; score 4: > 9% weight loss, diarrhea, gross bleeding.
Scanning electron microscopy
Mouse cecum and colon tissues were processed by opening the gut longitudinally without disturbing the intestinal contents with the help of binocular dissecting microscope. The opened tissues were pinned down to a silicone mat in four corners and fixed in 2.5% glutaraldehyde at 4 °C overnight as previously described [24]. The overnight fixed samples were washed two times (20 min each) with PBS and kept at 4 °C until further processing. Afterward, they were processed by post-fixing in 1% osmium tetroxide for 1 h, followed by dehydration with increasing concentrations of ethanol and critical-point dried. The dried samples were coated with 25 nm of gold and imaged on a field emission JSM-6701F Scanning Electron Microscopy (SEM) at a voltage of 10 kV.
DNA extraction and real-time quantitative PCR
DNA of the mouse fecal microbiota was isolated by using QIAamp Fast DNA Stool Mini Kit (Qiagen, Germany) according to the manufacturers. Real-time quantitative PCR (qPCR) to estimate the number of Blastocystis cells in feces was performed in accordance with a previously published protocol [25]. Briefly, 2 µl of the extracted DNA was added to a mixture of 5 µl SsoAdvanced Universal SYBR Green Supermix and 0.5 μM forward primer BL18SPPF1 (5‵- AGTAGTCATACGCTCGTCTCAAA -3‵) and 0.5 μM reverse primer BL18SR2PP (5‵- TCTTCGTTACCCGTTACTGC -3‵) for the SSU rRNA gene of Blastocystis. The qPCR was performed on an ABI 7500 real-time PCR system instrument (Life Technologies) at 95 °C for 5 min followed by 45 cycles of 95 °C for 15 s, 68 °C for 10 s, and 72 °C for 15 s, and completed with melting curve analysis. Each sample was quantified in triplicate. A standard curve was produced with a dilution series (107, 106, 105, 104, 103, 102, 101 cells/ml) of Blastocystis ST4. The Ct values of each sample were compared with that of the standard curve and the number of Blastocystis cells was calculated.
All mice were examined for the presence of Blastocystis by SEM and qPCR amplification of the Blastocystis SSU rRNA gene. Mice were considered to be successfully colonized after Blastocystis was observed in the gut lumen by SEM, and SSU rRNA gene was successfully amplified from the stool samples.
Fecal microbiota transplantation
Fecal microbiota transplantation (FMT) was performed through oral gavage of feces preparations from donor mice as previously described [26]. In brief, microbiota for FMT was obtained from control (PBS-gavaged) Rag1−/− mice or ST4-colonized Rag1−/− mice administered with Blastocystis ST4. Feces were collected, diluted with PBS (50 mg/ml), and then administered to recipients by oral gavage (10 mg/mice) three times a week.
Histology
For histological studies, the small intestine (SI), cecum and colon tissues were fixed in 4% neutral buffered formalin before processing and embedding in paraffin based on standard protocols. 4.5 μm sections were prepared and stained with hematoxylin and eosin (H&E). Histology scoring was performed in a blinded fashion, whereby changes in intestinal crypt architecture, level of tissue damage, goblet cell loss, and inflammatory cell infiltrates were scored as previously described [27].
16S rRNA gene sequencing
The V3–V4 region of the 16S rRNA gene was amplified using the 341-F (CCTAYGGGRBGCASCAG) and 806-R (GGACTACNNGGGTATCTAAT) primers. Gene amplification was carried out using Phusion High-Fidelity PCR Master Mix (New England Biolabs). Single amplifications were performed in 50 μl reactions with 50 ng of template DNA. Cycling protocol consisted of 94 °C for 4 min, followed by 30 cycles of 94 °C for 30 s, 54 °C for 30 s, and 72 °C for 30 s, with a final extension of 72 °C for 5 min. The size of the amplicons was determined using 2% agarose gel electrophoresis. Samples with size between 400 and 450 bp were extracted and purified from agarose gel using Qiagen Gel Extraction kit (Qiagen, Germany). Sequencing libraries were generated from the amplicons using NEBNext Ultra DNA Library Pre® Kit for Illumina, following manufacturer's recommendations and index codes were added. The library quality was assessed on the Qubit@ 2.0 Fluorometer (Thermo Scientific) and Agilent Bioanalyzer 2100 system. Qualified library was sequenced on an Illumina Novaseq platform (Illumina, San Diego CA, USA) and 250 bp paired-end reads were generated.
Bioinformatic and statistical analysis
Paired-end reads were assigned to samples based on their unique barcodes and truncated by cutting off the barcode and primer sequences. Paired-end clean reads were merged using FLASH version 1.2.7, which was designed to merge paired-end reads when the reads overlap the read generated from the opposite end of the same DNA fragment [28], and the splicing sequences were called tags. The tags were compared with the reference database using UCHIME algorithm to detect chimera sequences [29], and these chimera sequences were subsequently removed, resulting in effective tags. Next, effective tags were trimmed to 200 bp to remove the low-quality portion of the sequences (mean quality score < 20) using the DADA2 plugin for Quantitative Insights into Microbial Ecology (QIIME2 version 2021.02) [30, 31]. Taxonomic assignment was performed using the BLAST fitted classifier trained on the SILVA 138 reference database with the feature-classifier plugin for QIIME2 [32] based on 100% similarity. Biodiversity index analysis was calculated using QIIME2 and displayed with R software (Version R-4.0.3). Alpha diversity analysis was done using the metrics Shannon, Simpson, and Richness index. Pairwise comparisons of microbial communities in different groups were carried out using permutational multivariate analysis of variance (PERMANOVA, Bray–Curtis distance) in the q2-diversity-plugin in QIIME. Principal coordinate analysis (PCoA) and heatmap analysis were performed using R package.
Isolation of lamina propria cells
To analyze intestinal lymphocytes, the intestines were longitudinally opened and washed with ice-cold PBS to remove luminal contents. The tissues were cut into 1 cm pieces and incubated in Roswell Park Memorial Institute (RPMI) 1640 medium (Sigma-Aldrich) containing 1 mM EDTA (Sigma-Aldrich) and 1 mM DTT (Sigma-Aldrich) at room temperature for 20 min under slow rotation and spun down to remove the supernatant. The remaining pieces were incubated in RPMI containing 25% HEPES, 10% fetal calf serum (FCS), 1 mM EDTA, and 1 mM DTT at 37 ℃ for 1 h under slow rotation and then washed by PBS to remove epithelial cells and intraepithelial lymphocytes. Tissue pieces were digested with 0.3 mg/ml collagenase D (Sigma-Aldrich), 0.4 mg/ml dispase (Gibco) and 40 µg/ml DNase I (Roche) at 37 ℃ for 30 min under slow rotation. The digested tissue pieces and supernatants were filtered by 70 μm cell strainer and glass wool separately. After centrifugation, pellets containing the lamina propria (LP) lymphocytes were harvested.
Flow cytometric analysis
Lymphocytes were stimulated for 6 h with a cell stimulation cocktail of PMA (50 ng/ml), ionomycin (750 ng/ml) and 0.7 μl/ml GolgiStop (monensin, BD Biosciences). Live/dead stain was used to evaluate the viability of the cells. For surface staining, stimulated cells were stained with anti-CD4 (APC/FITC; Biolegend). Fixation and permeabilization buffers from Biolegend were used for intracellular cytokine staining. Fixed and permeabilized cells were stained with fluorochrome-conjugated anti-mouse antibodies against interleukin (IL)-4 (BV421; Biolegend), IL-5 (PE; Biolegend), IL-13 (PE; Biolegend), IL-10 (BV421; Biolegend), IL-17 (PerCP-Cy5.5; eBioscience, CA, USA), interferon gamma (IFN-γ) (BV711; Biolegend), and tumor necrosis factor (TNF-α) (APC; eBioscience) at 4 ℃ for 10 h. Flow cytometric analysis was performed on Fortessa X-20 (BD biosciences) and the data were analyzed using FlowJo_V10 software. The gating strategies are shown in Supplementary Fig. 4.
LC/MS/MS assay
Liquid chromatography/tandem mass spectrometry (LC/MS/MS) was carried out for analysis of short-chain fatty acids (SCFAs) in derivatized stool extracts as previously described [33]. In brief, 500 μl of ice-cold extraction solvent containing 10 μM of d5-benzoic acid as internal standard (IS) was added to 250 mg of stool sample and subjected to vortex mixing for 5 min at ambient temperature. The suspension was then centrifuged at 18,000g for 10 min at 4 °C. The supernatant was carefully removed and centrifuged again at 18,000g for 10 min at 4 °C. An aliquot of 100 μl was subsequently derivatized using a final concentration of 10 mM aniline and 5 mM EDC for 2 h at 4 °C. The derivatization reaction was quenched using a final concentration of 18 mM succinic acid and 4.6 mM 2-mercaptoethanol for 2 h at 4 °C. All samples were stored at 4 °C until analysis on the same day. Analysis was performed using an Agilent 1290 Infinity LC system (Agilent Technologies, Santa Clara, CA, USA) interfaced with an AB Sciex QTRAP 5500 hybrid linear ion-trap quadrupole mass spectrometer equipped with a TurboIonSpray source (Applied Biosystems, Foster City, CA). Details of the LC/MS/MS and calibration methods were similar as previously described [33].
Statistical analysis
Statistical analysis was performed using R-4.0.3 software and GraphPad Prism 8 software (GraphPad Software, CA, USA). Two independent replicates were performed for each experiment. The unpaired two-tailed Student’s t test was used to evaluate differences between two groups. Two-way ANOVA and one-way ANOVA analysis with Tukey multiple comparisons test was used for comparison of more than two groups. Graphs show mean ± SEM. * p < 0.05, ** p < 0.01, *** p < 0.001.
Results
Blastocystis ST4 colonization exerts no harmful effects on normal healthy mice
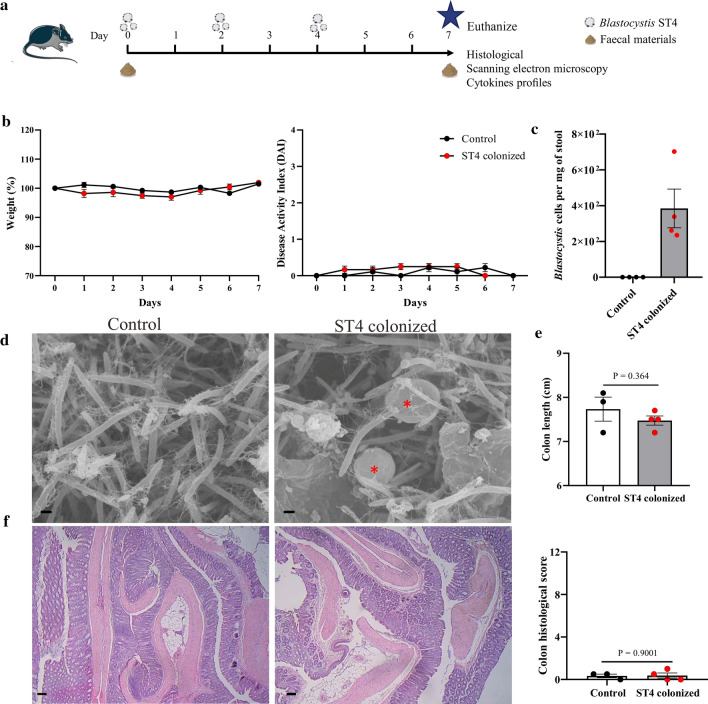
Blastocystis ST4 is commonly found in humans and a variety of animals worldwide, predominantly in rats [34]. Although some in vitro studies have shown that ST4 infection can increase epithelial permeability and release pro-inflammatory cytokines [16, 35], the effects of ST4 in vivo are less well understood. To determine the effects of Blastocystis ST4 colonization on host intestinal bacterial communities and immune responses, we established a mouse model of oral Blastocystis colonization. Specifically, C57BL/6 mice were orally inoculated with 5 ×107 cells of Blastocystis three times per week, and the mice were euthanized 3 days after the final gavage with Blastocystis (Fig. 1a). Interestingly, we observed that Blastocystis ST4 colonization did not cause any abnormalities, characterized by no significant difference in weight change and DAI between ST4-colonized and non-colonized control mice (Fig. 1b). The number of Blastocystis ST4 cells was quantitated by qPCR analysis (Fig. 1c), and scanning electron microscopic analysis of the contents of the cecum and colon from ST4-clonized mice revealed the presence of Blastocystis, which colonizes the intestinal lumen and closely adheres to intestinal microbes (Fig. 1d). Additionally, we examined the histopathology of SI, cecum and colon, and scored these based on the degree of inflammation and tissue damage. The mice colonized with ST4 showed intact mucosal epithelium without any ulcerated lesions or an abnormal level of inflammatory cell infiltration (Fig. 1e, f; Supplementary Fig. 1), which is consistent with previous findings in rats experimentally colonized with Blastocystis ST4 [17, 36]. Overall, these findings indicate that Blastocystis ST4 colonization did not cause abnormal phenotypic changes or any gut histopathology within the duration of the experiment.
Fig. 1.
Blastocystis ST4 colonization did not induce any abnormal effects on normal healthy mice. a Experimental design. b Weight changes and disease activity index (DAI) between control and ST4-colonized mice. c Blastocystis ST4 cells per milligram of stool in ST4-clonized mice. d Scanning electron microscopy (SEM) of colonic and cecum tissues from control and ST4-clonized mice, Blastocystis are indicated with red asterisk (*). Scale bar = 1 μm. e Colon length at endpoint. f Representative micrographs of H&E-stained colon sections from control and ST4-colonized mice, and colonic histological scores at day 7. Scale bar = 100 μm
Blastocystis ST4 colonization alters the bacterial community composition in normal healthy mice
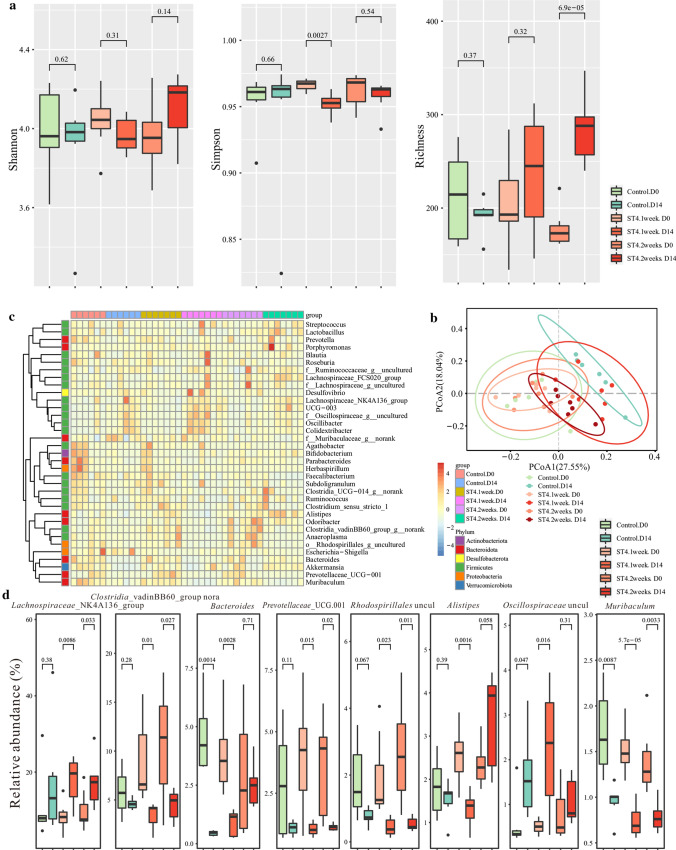
Blastocystis-colonized individuals showed distinct difference in bacterial community structure when compared to non-colonized individuals [37], suggesting Blastocystis has the ability to modulate intestinal microbiota. To investigate the effects of Blastocystis ST4 colonization on gut bacterial communities, fecal samples of control and ST4-colonized mice were collected at day 0 and day 7, and bacterial compositions were investigated using 16S rRNA gene sequencing. Rarefaction analysis was used to estimate the total number of observed features that could be identified from the samples; this showed that a credible number of reads (average 101, 487) had been measured in each group (Supplementary Fig. 2). No significant differences in bacterial diversity and richness were detected in ST4-colonized and control mice over time, as measured by Shannon, Simpson and Richness indices (Fig. 2a). However, we observed significant difference in bacterial community composition in the ST4-colonized group between day 0 and day 7, measured by beta diversity of Bray–Curtis dissimilarity (PERMANOVA p = 0.026; Fig. 2b; Supplementary Table 1). The heatmap showed that differences in the relative abundances of various taxa between control and ST4-colonized mice (Fig. 2c). Specifically, we observed higher levels of unclassified Clostridia vadinBB60 group, Tuzzerella, and Peptococcaceae uncultured, and lower levels of Odoribacter, and Lachnospiraceae ASF356 at day 7 post-Blastocystis ST4 colonization (Fig. 2d, Supplementary Fig. 3). In contrast, the significantly reduced bacterial taxa in ST4-free mice were Lachnospiraceae NK4A136 group, Odoribacter, Lachnospiraceae uncultured, Blautia and Oscillibacter, while Alloprevotella, Bacteroides, and Paraprevotella were the most significantly increased (Fig. 2d, Supplementary Fig. 3). Overall, these data indicated that Blastocystis ST4 colonization alters the bacterial community compositions in normal healthy mice.
Fig. 2.
Blastocystis ST4 colonization alters the murine fecal bacterial community compositions. a Alpha diversity was measured by Shannon, Simpson, and Richness indexes in the fecal samples of control and ST4-colonized mice (n = 4 mice per group). b PCoA of fecal gut microbiota in control and ST4-clonized mice at day 0 and day 7. c Heatmap of ST4 colonization-associated taxonomic markers at day 7. d Bacterial genera (relative abundance in the top 35) showing significant differences in their relative abundance between control and ST4-colonized mice
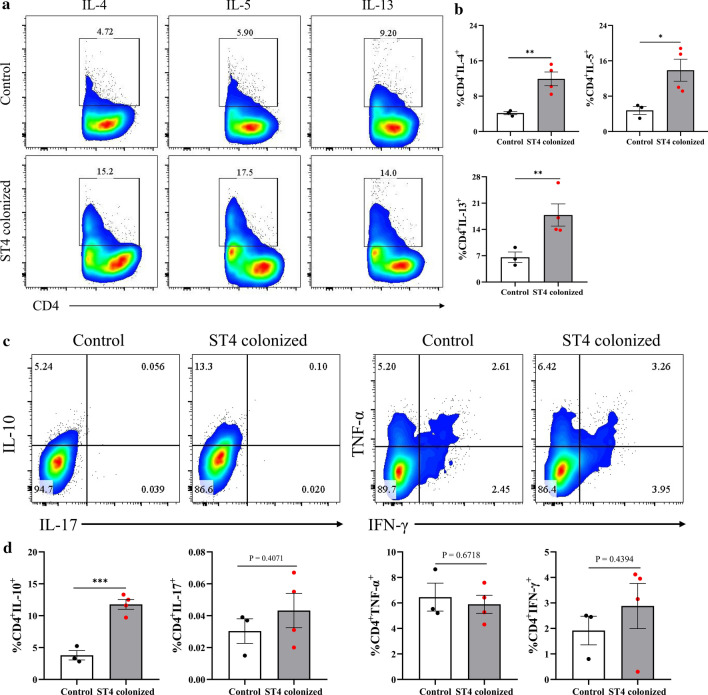
Blastocystis ST4 colonization induces the accumulation of Th2 and Treg cells
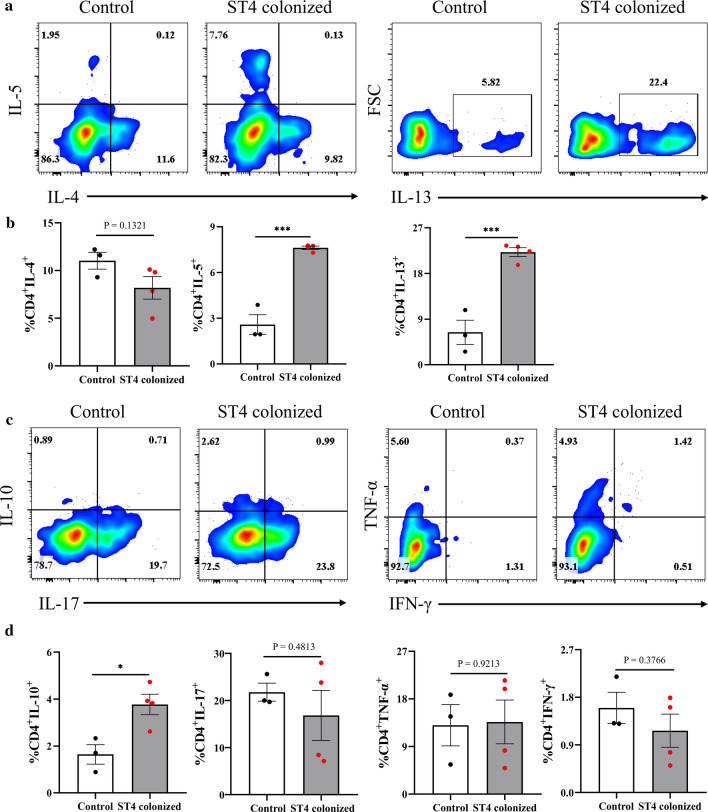
Blastocystis has been reported to involve the host adaptive immune responses. However, these effects are mainly based on in vitro studies [38]. To comprehensively investigate the effect of Blastocystis ST4 colonization on adaptive immune responses in a murine model, we examined the polarization status of T cell subsets in the colonic LP from control and ST4-colonized mice (Supplementary Fig. 4). The CD4 T cell population from the Blastocystis-associated group was enriched with IL-5-producing cells, reflecting a Th2 differentiated phenotype [39] (Fig. 3a, b). Furthermore, a substantial increase of the Th2 compartment (as defined by the IL-13 expression) was observed in the ST4-colonized group (average 6% and 22% of IL-13-positive cells within the CD4 subset in the control and ST4 group, respectively) (Fig. 3a, b). Notably, together with an increase of the colonic Th2 compartment, the ST4-colonized group displayed a substantial increase in IL-10-producing CD4+ Treg cells (Fig. 3c). In contrast, the Th1 compartment (defined by the IFN-γ and TNF-α cytokines) and the Th17 subset (expressing IL-17) were comparable in ST4-colonized mice and control mice (Fig. 3c, d). These data suggest that colonic Th2 and Treg responses are induced by Blastocystis ST4 colonization.
Fig. 3.
Colonization with Blastocystis ST4 induces accumulation of Th2 and Treg cells in the colonic lamina propria (LP). a Colored contour plots show staining for IL-4, IL-5, and IL-13 within CD4+ cells. b Bar charts show the percentage of IL-4, IL-5, and IL-13 expressing CD4+ T cells. c Colored contour plots show staining for IL-10, IL-17, TNF-α, and IFN-γ within CD4+ cells. d Bar charts show the percentage of IL-10, IL-17, TNF-α, and IFN-γ expressing CD4+ T cells. Statistical significance is indicated by *p < 0.05, and ***p < 0.001, unpaired Student’s t test
Blastocystis ST4 colonizes asymptomatically in Rag1−/− mice
Th2 and Treg cells play a crucial role in the host's resistance to parasite infection and in limiting intestinal inflammation [40]. Blastocystis ST4 could asymptomatically colonize normal healthy mice, possibly because it activates the host Th2 and Treg cells that promote mucus production, and increases gut motility to maintain intestinal homeostasis [41]. To investigate the role of adaptive immunity in Blastocystis ST4 colonization, we carried out experimental infections in immunodeficient Rag1−/− mice that lack all mature lymphocytes. Rag1−/− mice were orally inoculated with Blastocystis ST4, with extension of the inoculation duration, from 7 to 14 days, to further investigate the effect of Blastocystis ST4 colonization in mice (Fig. 4a). Microscopic analysis showed the presence of Blastocystis in both ST41week and ST42weeks colonized mice (Fig. 4b). Histological examination of the colon tissues revealed no difference in histological scores, and showed intact structure and no inflammatory cell filtrate among the three groups (Fig. 4c, d). Similarly, there was no significant difference in colon length between groups (Fig. 4d). The number of Blastocystis cells was determined by qPCR amplification of SSU rRNA gene, and the mice from ST42weeks colonized group showed higher proportion of Blastocystis than ST41week colonized group (Fig. 4c). These data showed that Blastocystis ST4 could asymptomatically colonize Rag1−/− mice.
Fig. 4.
Blastocystis ST4 colonizes asymptomatically in Rag1−/− mice. a Experimental design. b Scanning electron microscopy (SEM) of colonic and cecum tissues from control and ST4-clonized mice, Blastocystis are indicated with red asterisk (*). Scale bar = 1 μm. c Representative micrographs of H&E-stained colon sections from ST4-colonized and control mice at day 14. Scale bar = 100 μm. d Colon length and colonic histological scores at endpoint. Blastocystis ST4 cells per milligram of stool in ST4-clonized mice
Impact of Blastocystis ST4 colonization on the bacterial microbiome of Rag1−/− mice
Host adaptive immune responses can regulate the gut microbial compositions [42]. It is unclear whether the changes of intestinal microbiome are caused by the ST4 itself or the interaction of the ST4 and host adaptive immune system. Thus, we further explored the impact of Blastocystis ST4 colonization on gut microbial composition in Rag1−/− mice. Rarefaction curves showed that most of the reads were obtained, thus allowing us to undertake further analysis (Supplementary Fig. 5). There was no significant difference in Shannon index among control, ST41week, and ST42weeks groups during the experimental period, whereas ST41week mice revealed decreased Simpson index and ST42weeks mice increased Richness index at day 14 post-colonization (Fig. 5a). Bacterial community composition changed significantly over time for both ST4-colonized groups (PERMANOVA p < 0.01; Fig. 5b; Supplementary Table 1). The relative abundance of various taxa among different groups at day 0 and day 14 are presented as a heatmap (Fig. 5c). Of these bacterial taxa, six genera Prevotellaceae UCG.001, unclassified Clostridia vadinBB60 group, unclassified Rhodospirillales, Muribaculum, Anaeroplasma, and Escherichia–Shigella were significantly decreased in the fecal microbiota of both ST41week and ST42weeks groups following Blastocystis ST4 colonization (Fig. 5d; Supplementary Fig. 6). In contrast, two genera Lachnospiraceae NK4A136 group and Oscillospiraceae UCG-003 were significantly enriched in ST4-colonizd groups (Fig. 5d; Supplementary Fig. 6). These data collectively suggest that Blastocystis ST4 can modulate the gut microbiota in Rag1−/− mice, even without the participation of B and T immune cells.
Fig. 5.
Effect of Blastocystis ST4 colonization on fecal bacterial community composition in Rag1−/− mice. a Alpha diversity was measured by Shannon, Simpson, and Richness indexes (n = 6 in the control group and n = 7 in the ST4-colonized group). b PCoA of fecal gut microbiota in control and ST4-clonized mice at day 0 and day 14. c Heatmap of ST4 colonization-associated taxonomic markers at day 14. d Bacterial genera (relative abundance in the top 35) showing significant differences in their relative abundance between ST4-colonized and control mice
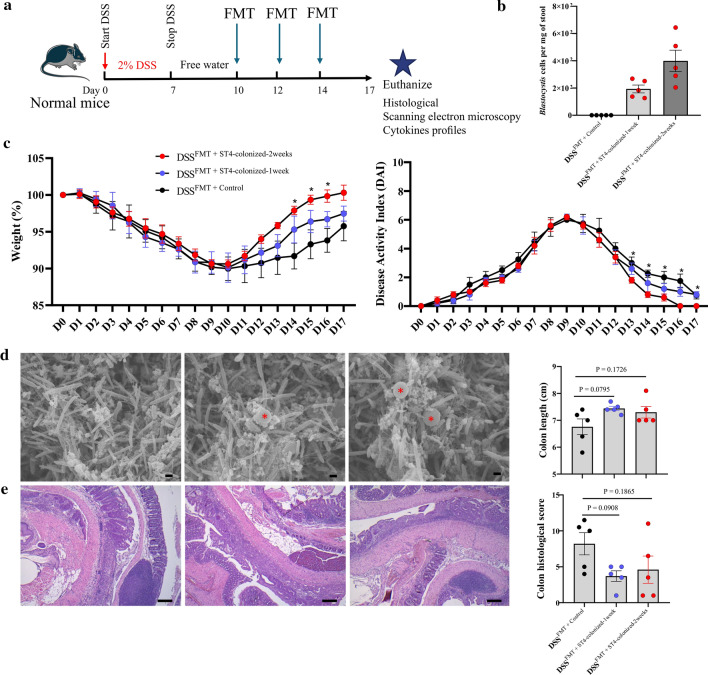
Blastocystis ST4 colonization promotes recovery from DSS-induced colitis
Blastocystis ST7 infection caused colonic tissue damage and ulceration in DSS-induced colitis mice [5], while long-term colonization with Blastocystis ST3 promotes a faster recovery from colitis in rats [43], suggesting different subtypes exert differential effects on the host. To further explore whether Blastocystis ST4 colonization can modulate the severity of disease in an experiment model of DSS-induced colitis, DSS-treated mice were orally inoculated with Blastocystis ST4 three times per week for two consecutive weeks (Fig. 6a, b). Mice with Blastocystis ST4 colonization showed faster recovery after DSS-induced colitis, as compared to that in Blastocystis-free mice, as reflected by weight changes and DAI (Fig. 6c). Histological examination of the colon revealed reduced mucosal ulceration and damage, and lower histological scores in ST4-clonized mice (Fig. 6d). Similarly, there was significant difference in colon lengths between ST4-colonized mice and control mice (Fig. 6d). These data indicate that Blastocystis ST4 colonization promotes recovery of mice from DSS-induced colitis.
Fig. 6.
Blastocystis ST4 colonization contributes to mice recovery from experimentally induced colitis. a Experimental design. b Blastocystis ST4 cells per milligram of stool in ST4-clonized mice. c Weight changes and DAI between control and ST4-colonized mice during treatment. d Representative micrographs of H&E-stained colon sections from control and ST4-colonized mice at day 21. Colon length and colonic histological at end point. Scale bar = 100 μm. *p < 0.05, unpaired Student’s t test
Gut microbiota changes after Blastocystis ST4 colonization in DSS-induced colitis mice
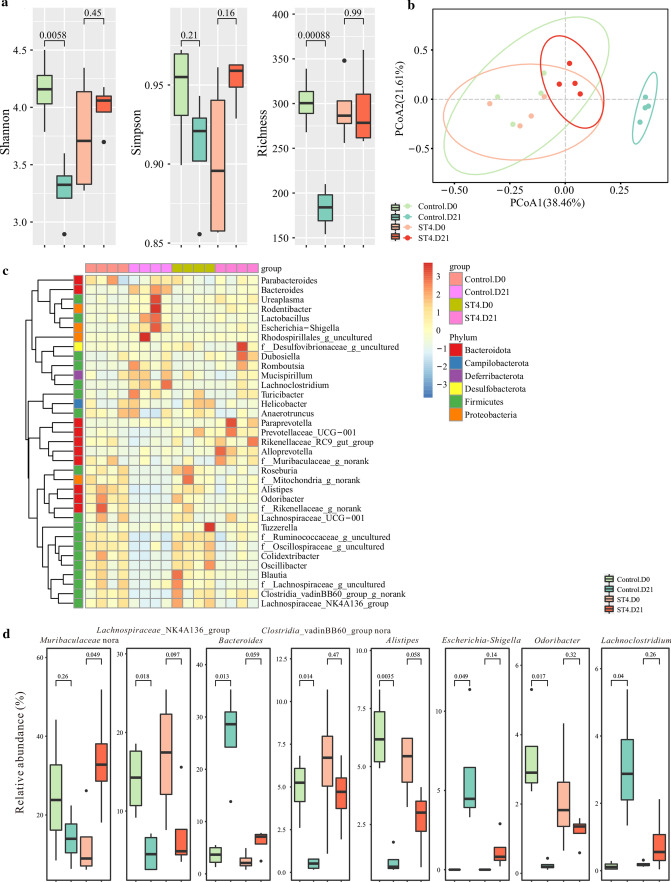
Accumulating evidence indicates that the gut microbiome plays a pivotal role in host health and immune homeostasis [44]. The changes in gut microbial composition induced by helminth infection have the ability to reduce intestinal inflammation in an experimental mouse model [45]. To investigate if the protective effect of Blastocystis ST4 on DSS-induced colitis mice is related to alterations in the gut bacterial community, we compared the variations in the microbial communities in ST4-colonized and control mice over time. Reduced alpha diversity (Shannon and Richness indexes) was observed in control mice, whereas Blastocytsis ST4 colonization maintained a stable alpha diversity over time (Supplementary Fig. 7; Fig. 7a). Both ST4-colonized and control group showed pronounced differences in bacterial composition during the experiment period (PERMANOVA p < 0.05; Fig. 7b; Supplementary Table 1). The relative abundance of various taxa are shown as a heatmap among groups (Fig. 7c). In particular, the genera Lachnospiraceae NK4A136 group, unclassified Clostridia vadinBB60 group, Alistipes, Odribacter, Oscillibacter, Colidextribacter, unclassified Oscillospiraceae, unclassified Ruminococcaceae, Roseburia, and uncultured Mitochondria were significantly reduced in control mice over experimental time, whereas the genera Bacteroides, Escherichia–Shigella, Lachnoclostridium, and Paraprevotella were progressively expanded in control mice over time (Fig. 7d; Supplementary Fig. 8). Conversely, colonization with Blastocystis ST4 appears to maintain a relatively stable microbiota communities compared to Blastocystis-free mice after DSS administration. Higher proportions of unclassified Muribaculaceae and lower abundance of Oscillibacter, Colidextribacter, unclassified Oscillospiraceae and unclassified Ruminococcaceae were observed in ST4-colonized mice over time (Fig. 7d, Supplementary Fig. 8). Overall, these results suggest that the protective effect of Blastocystis ST4 on DSS-induced colitis mice was associated with a stable bacterial diversity and microbiota communities.
Fig. 7.
Improvement of experimentally induced colitis is associated with gut microbiota. a Alpha diversity was measured by Shannon, Simpson, and Richness indexes (n = 4 per group). b PCoA of fecal gut microbiota in control and ST4-clonized mice at day 0 and day 21. c Heatmap of ST4 colonization-associated taxonomic markers. d Bacterial genera showing significant differences in their relative abundance between ST4-colonized and control mice
Protective effects of Blastocystis ST4 in DSS-induced colitis mice may be mediated by activation of Th2 and Treg cells responses
Helminth-mediated Th2 or Treg cell responses have been exploited to ameliorate experimental colitis in a mouse model [46, 47]. We asked if Blastocystis ST4 also regulates these immune responses to confer protection from DSS-induced colitis. Colonic tissues from DSS-induced colitis mice with Blastocystis ST4 colonization demonstrated increased expression of Th2 (IL-4, IL-5, and IL-13) cytokines relative to control mice (Fig. 8a, b). The immunophenotype of ST4-colonized DSS-treated mice was consistent with the previous observations on normal mice (Fig. 3a, b). Importantly, the size of the anti-inflammatory cytokine IL-10-producing Treg cell subset in colonic LP was also elevated in ST4-colonized mice (Fig. 8c, d). Similar to what we observed in normal healthy mice, the numbers of Th1-asscociated cytokine expressing cells (IFN-γ+ and TNF-α+), as wells as Th17 cells (IL-17) were similar in ST4-colonized and control mice (Fig. 8c, d). Collectively, the composition of the Blastocystis-associated CD4 compartments in colonic LP from the DSS-induced colitis model indicated this organism's direct interaction with the host’s adaptive immune system. Furthermore, Blastocystis-driven changes in the gut microbiome with the accompanying shift within immune compartments are plausible key factors that account for attenuation in the severity of DSS-mediated colitis.
Fig. 8.
Blastocystis ST4 colonization activates Th2 and Treg cells responses in colonic mucosa of DSS-induced colitis mice. a Colored contour plots show staining for IL-4, IL-5, and IL-13 within CD4+ cells. b Bar charts show the percentage of IL-4, IL-5, and IL-13 expressing CD4+ T cells. c Colored contour plots show staining for IL-10, IL-17, TNF-α, and IFN-γ within CD4+ cells. d Bar charts show the percentage of IL-10, IL-17, TNF-α, and IFN-γ expressing CD4+ T cells. Statistical significance is indicated by *p < 0.05, **p < 0.01, and ***p < 0.001, unpaired Student’s t test
Transfer of fecal microbiota from ST4-colonized Rag1−/− mice reduces inflammation in experiment-induced colitis
Next, to determine the effect of Blastocystis ST4-altered microbial communities that was independent of adaptive immunity-mediated microbiota changes on experiment-induced colitis, we performed FMT from ST4-colonized Rag1−/− mice into DSS-treated mice (Fig. 9a). DSSFMT + ST4−colonized mice showed faster recovery and better health status, as measured by weight changes and DAI (Fig. 9c), compared to DSSFMT + control. In addition, lower levels of intestinal inflammation and higher colon length were detected in DSSFMT + ST4−colonized mice, although did not reach significance difference (Fig. 9e). We further evaluated the bacterial communities in experiment-induced mice upon different FMT conditions. Rarefaction curves showed that the depth of sequencing was sufficient for analysis (Supplementary Fig. 9). DSSFMT+ST4−colonized mice showed a stable bacterial α-diversity when compared to DSSFMT+Control mice, which showed significant decreases, as indicated by the Shannon, Simpson and Richness indices (Fig. 10a). The gut microbial communities changed significantly over time for DSSFMT + ST4−colonized and DSSFMT+Control groups (PERMANOVA p < 0.05; Fig. 10b; Supplementary Table 1). The differences in the relative abundance of various taxa were observed in heatmap format (Fig. 10c). We observed higher levels of commensals and SCFA-producing taxa, Akkermansia, unclassified Clostridia vadinBB60 group, uncultured Rhodospirillales, and Clostridia UCG.014 in DSSFMT + ST4−colonized mice (Fig. 10d; Supplementary Fig. 10). DSSFMT + Control mice showed the enrichment of the genera Lachnospiraceae NK4A136 group, unclassified Clostridia vadinBB60 group, Prevotellaceae UCG.001, Bacteroides, Blautia, and Clostridia UCG.014 (Fig. 10d; Supplementary Fig. 10). Altogether, these findings suggest that transfer of ST4-altered gut microbiome are beneficial to the recovery of experimental colitis.
Fig. 9.
Transfer of ST4-altered microbiome to colitis mice reduces colonic inflammation. a Experimental design. b Blastocystis ST4 cells per milligram of stool in DSSFMT + ST4−clonized mice. c Weight changes and DAI between DSSFMT + Control mice and DSSFMT + ST4−clonized mice. d Scanning electron microscopy (SEM) of colonic and cecum tissues from DSSFMT + Control mice and DSSFMT + ST4−clonized mice, Blastocystis are indicated with red asterisk (*). Scale bar = 1 μm. e Representative micrographs of H&E-stained colon sections from DSSFMT + Control mice and DSSFMT + ST4−clonized mice at day 17. Scale bar = 100 μm. Colon length and colonic histological scores at end point
Fig. 10.
Gut microbiota analysis upon different FMT conditions treatment in DSS-induced colitis mice. a Alpha diversity was measured by Shannon, Simpson, and Richness indexes (n = 4 in control group and n = 5 in ST4-colonized group). b PCoA of fecal gut microbiota in control and ST4-clonized mice at day 0 and day 17. c Heatmap of taxonomic markers among different groups. d Bacterial genera showing significant differences in their relative abundance among groups
FMT influences SCFAs and Treg cells IL-10 production in DSS-induced colitis recipients
The host microbiome plays an important role in regulating physiology through microbiota-derived metabolites, especially SCFAs, during host–microbiome interactions [48]. To gain mechanistic insight into the faster recovery from intestinal inflammation in DSSFMT +ST4−colonized mice, we firstly quantitated eight SCFAs (acetic, propionic, butyric, isobutyric, valeric acid, isovaleric, 2-methylbutyric, and caproic acid) of feces by LC/MS/MS. The levels of five SCFAs in fecal samples of DSSFMT +ST4−colonized−2 weeks mice exhibited significantly higher concentration changes than DSSFMT +control mice (Fig. 11a). SCFAs are critical to the immune system and can serve as substrates for host energy metabolism [49]. We also detected increased SCFAs in Rag1−/− mice after Blastocystis ST4 colonization for one or two weeks (Supplementary Fig. 11). Besides, it has been determined that microbiota-derived SCFAs have the ability to modulate Treg cell function and can alleviate the colonic inflammation [50, 51]. We then assessed whether re-colonization of DSS treated mice with microbiota from ST4-colonized mice via FMT influences colonic immune cells. Interestingly, we observed an increased number of CD4+ cells expressing IL-10 and decreased number of CD4+ cells expressing TNF-α in DSSFMT + ST4−colonized mice (Fig. 11b, c). Taken together, these data indicate that transfer of ST4-altered gut microbiota in DSS-induced colitis recipients increases SCFAs production and induces accumulation of IL-10-producing Treg cells.
Fig. 11.
FMT influences SCFAs and Treg cells IL-10 production in DSS-induced colitis recipients. a Fold change of each SCFA relative to levels at day 0 from DSSFMT mice (recipients). b Colored contour plots show staining for IL-10 within CD4+ cells, and bar chart shows the percentage of IL-10 expressing CD4+ T cells. c Colored contour plots show staining for TNF-α within CD4+ cells, and bar chart shows the percentage of TNF-α expressing CD4+ T cells. *p < 0.05, **p < 0.01, ***p < 0.001, Two-way ANOVA (a) and one-way ANOVA (b, c) analysis with Tukey multiple comparison test
Discussion
Although the pathogenicity of Blastocystis is controversial, accumulating evidence shows that it is often present in asymptomatic individuals and is associated with healthy gut microbiota [52]. To understand the causal role of Blastocystis ST4 on the host gut microbiota and mucosal immune responses, we established murine models of oral colonization to investigate the effects of Blastocystis colonization on gut microbiota, and adaptive immune responses. Our results suggest that Blastocystis ST4 colonization in normal healthy and Rag1−/− mice did not cause any pathological lesions or inflammatory cell infiltration in colonic mucosa. Furthermore, Blastocystis ST4 colonization and transfer of ST4 colonization-altered gut microbiota to experimentally induced colitis mice promoted faster recovery from experimental caused colitis.
Microbial alpha diversity is considered an important marker for gut health, and high bacterial diversity implies stability and resilience of the gut ecosystem [53]. We monitored changes in the alpha diversity over the course of colonization with Blastocystis ST4 in several mouse models. Interestingly, alpha diversity (measured by Richness index) was significantly increased in Rag1−/− mice colonization with Blastocystis ST4 at day 14 compared with day 0 (Fig. 5a). Similarly, although it's a different model of infection, a previous study showed infection with ST4 cysts increase in bacterial richness in rats [36]. The higher bacterial richness was also observed in individuals with Blastocystis colonization in the majority of microbiome studies [54, 55]. On the other hand, colonization with ST4 appears to maintain a stable fecal bacterial alpha diversity in normal healthy and DSS-induced colitis mice, while a significant decrease in alpha diversity was detected in control mice after DSS administration (Figs. 2a, 7a, and 10a). Loss of alpha diversity has been implicated in IBD patients [56, 57], and it is also a sign of dysbiosis in many other human diseases [58]. Thus, the decreased alpha diversity in control mice but not ST4-colonized mice may explain the differences in recovery from colitis.
Blastocystis and gut microbiota co-inhabit the host intestinal tract and are capable of interacting with each other. We observed that ST4 colonization significantly changed the bacterial community compositions, but there were some differences in specific taxa, depending on the mouse model utilized. For example, ST4 colonization mainly increased the abundance of Clostridia vadinBB60 group, belonging to the Clostridia class, in normal healthy mice, which was positively correlated with the Treg cell counts [59]. Lachnospiraceae NK4A136 group, one of the known short-chain fatty acid (SCFA)-producing bacteria that degrade complex polysaccharides [60], was the most enriched bacteria in ST4-colonized Rag1−/− mice. Hu et al. reported that the elevated abundance of Lachnospiraceae NK4A136 group showed anti-inflammation effects in obese mice [61]. The initial abundances of Clostridia vadinBB60 group and Lachnospiraceae NK4A136 group across different mouse models were different, which may have contributed to the inconsistent results across different mouse models (Figs. 2d, 5d). Furthermore, host adaptive immune responses can also regulate gut microbial compositions [42]. In addition to the direct effect of Blastocystis ST4 on the gut microbiota in normal healthy mice and Rag1−/− mice (which lack mature lymphocytes), the host's adaptive immune response would have influenced the composition of gut microbiota, as evidenced in the day 0 data.
Notably, we observed Blastocystis ST4 colonization in DSS-induced colitis inhibits the expansion of Bacteroides and Escherichia–Shigella. It has been determined that inhibition of expansion of Bacteroides can alleviate intestinal inflammation in Nod2−/− mice [62]. Escherichia–Shigella, belonging to the family Enterobacteriaceae, are one of the most important enteric pathogens causing gastroenteritis worldwide [63]. A previous study reported that the expansion of Escherichia–Shigella was associated with lower bacterial diversity and pro-inflammatory effects [64]. Therefore, our study demonstrated that the improvement of intestinal inflammation by Blastocystis ST4 colonization may be related to the inhibition of these pathobiont bacteria.
FMT has been successfully used in the treatment of Clostridium difficile infection (CDI) and IBD [65, 66]. An interesting study included Blastocystis-positive (ST1 and ST3) donor samples for FMT treatment of recurrent CDI, and demonstrated that the presence of Blastocystis ST1 and ST3 from donors did not cause any adverse gastrointestinal symptoms [67]. In our study, we found that transfer ST4-altered microbiota from Rag1−/− mice reduces inflammation in experiment-induced colitis through an increase in “beneficial” microbes such as Akkermansia. Bacteria belonging to Akkermansia are associated with gut health, and the expansion of Akkermansia can increase mucus production to ameliorate intestinal inflammation [68]. Moreover, FMT from ST4-colonized mice increased SCFAs production and the proportion of anti-inflammatory cytokine IL-10 more profoundly than FMT from control mice. This is the first study to demonstrate that FMT from a donor colonized with Blastocystis ST4 improves the intestinal inflammation in a mouse model. Although we did not dissociate the effect of ST4 together with altered microbiota from the effect of the ST4 itself during the FMT, we determined that the beneficial effects of ST4 on the host are transferable.
The gut microbial community plays an important role in the development and modulation of the immune system [69]. We observed that Blastocystis ST4 colonization activates Th2 immune responses in normal healthy mice and DSS-induced colitis mice. It has been determined that Th2 cells are important sources of type 2 cytokines (IL-4, IL-5 and IL-13) and are also important effector cells during the inflammatory process [70]. Recent data showed an expansion of IL-13- and IL-4-producing CD4+ T cells in Nod2−/− mice contributes to ameliorating the intestinal injury response [62]. Another interesting study in primates demonstrated that the experimental administration of Trichuris trichiura can ameliorate colitis by both induction the colonic CD4+ T cells producing IL-4 and modulation of microbial populations [71].
In addition, we also found that Blastocystis ST4 colonization increases the number of IL-10-producing Treg in the colonic mucosa of DSS-induced mice. The cytokine IL-10 produced by Treg cells is required for containment of inflammatory responses in mucosal tissues [72]. Both humans and mice deficient in IL-10 or IL-10 receptor (IL-10R) are prone to develop severe intestinal inflammation [73–75], highlighting the importance of IL-10 in preventing this disease process. On the other hand, the gut microbe-derived SCFAs can enhance the expression of Foxp3 and IL-10-expressing colonic Tregs by inhibition of histone deacetylase (HDAC) or in a GPR43-dependent manner [50, 76, 77]. Although the three most abundant SCFAs, acetate, propionate, and butyrate, did not change significantly after FMT and ST4 colonization, there was elevation in the other five SCFAs, isobutyric acid, valeric acid, isovaleric acid, 2-methylbutyric acid, and caproic acid, which have been reported to induce the accumulation of anti-inflammatory cytokine IL-10 [78–80]. However, it is unclear whether the increased production of IL-10 is directly caused by Blastocystis ST4 or indirectly through regulating the gut microbiota-derived SCFAs. Future studies should focus on understanding the mechanistic connection between Blastocystis ST4 colonization, IL-10 signaling, and bacterial-derived SCFAs using relevant animal models.
Conclusions
We demonstrated that Blastocystis ST4 colonization is able to alter the gut bacterial community composition in an adaptive immunity-independent fashion, evidenced through the use of Rag1−/− mice. In models with intact immune systems, Blastocystis ST4 induces the expression of Th2 and Treg cytokines. Notably, alterations in gut microbiota composition mediated by Blastocystis ST4 colonization is associated with amelioration of colonic inflammation, likely through immunomodulatory effects of SCFAs, Th2 and Treg effectors. These findings represent an important contribution toward the elucidation of the complex interplay between Blastocystis ST4, gut microbiota, and host adaptive immune responses.
Supplementary Information
Below is the link to the electronic supplementary material.
Supplementary file1 (TIF 8999 KB) Figure S1. Blastocystis ST4 colonization did not induce any abnormal effects on C57BL/6 mice. a, Representative micrographs of H&E-stained caecum sections and histological scores from ST4-colonized and control mice at day 7. b, Representative micrographs of H&E-stained small intestine sections and histological scores from ST4-colonized and control mice at day 7. Scale bar = 100 μm.
Supplementary file2 (TIF 482 KB) Figure S2. Rarefaction curves (threshold is 90,000) showing microbial diversity based on the Shannon index (upper panel) and Observed features (bottom panel) from normal healthy mice.
Supplementary file3 (TIF 184 KB) Figure S3. Comparison of relative abundancies of different taxa between control and ST4-colonized mice.
Supplementary file4 (TIF 9690 KB) Figure S4. Gating strategy of the immune compartments isolated from colonic lamina propria.
Supplementary file5 (TIF 580 KB) Figure S5. Rarefaction curves (threshold is 24,000) showing microbial diversity based on the Shannon index (upper panel) and Observed features (bottom panel) from Rag1−/− mice.
Supplementary file6 (TIF 311 KB) Figure S6. Comparison of relative abundancies of different taxa between control and ST4-colonized mice.
Supplementary file7 (TIF 526 KB) Figure S7. Rarefaction curves (threshold is 58,000) showing microbial diversity based on the Shannon index (upper panel) and Observed features (bottom panel) from DSS-induced colitis mice.
Supplementary file8 (TIF 236 KB) Figure S8. Comparison of relative abundancies of different taxa between control and ST4-colonized mice.
Supplementary file9 (TIF 826 KB) Figure S9. Rarefaction curves (threshold is 26,000) showing microbial diversity based on the Shannon index (upper panel) and Observed features (bottom panel) from DSSFMT mice.
Supplementary file10 (TIF 290 KB) Figure S10. Comparison of relative abundancies of different taxa between control and ST4-colonized mice.
Supplementary file11 (TIF 1457 KB) Figure S11. Fold-change of each SCFA relative to levels at day 0 from Rag1-/- mice (donor mice).
Supplementary file12 (XLSX 10 KB) Table S1. PERMANOVA of beta-diversity analysis as measured by Bray-Curtis dissimilarity.
Supplementary file13 (XLSX 620 KB) Table S2. Sample metadata, feature table and taxonomic classifications.
Acknowledgements
We would like to thank Dr. Rohan Williams for assistance in microbiome data analysis and Dr. Tong Jiexin for critical editing and comments on our manuscript.
Abbreviations
- IBD
Inflammatory bowel disease
- IBS
Irritable bowel syndrome
- SSU
Small subunit
- Th2
T helper 2
- Treg
T regulatory
- FGFP
Flemish Gut Flora Project
- AGP
American Gut Project
- IMDM
Iscove’s modified Dulbecco’s medium
- PBS
Phosphate-buffered saline
- DSS
Dextran sulfate sodium
- DAI
Disease activity index
- H&E
Hematoxylin and eosin
- SI
Small intestine
- SEM
Scanning electron microscopy
- QIIME
Quantitative Insights into Microbial Ecology
- RPMI
Roswell Park Memorial Institute
- FCS
Fetal calf serum
- FBS
Fetal bovine serum
- IL
Interleukin
- IFN-γ
Interferon gamma
- TNF-α
Tumor necrosis factor alpha
- LP
Lamina propria
- SCFAs
Short-chain fatty acids
- CDI
Clostridium difficile Infection
- CD
Crohn’s disease
- HDAC
Histone deacetylase
Author contributions
KSWT conceived and designed the study. CWP and LD performed animal experiments. CWP and YZ performed histological analyses. EYK and LD performed 16S rRNA gene sequencing and analyzed the data. LW and LD performed immunological experiments and analyses. TTA and BM performed scanning electron microscopy experiments. DYQK and ECYC performed LC/MS/MS assays. YZ, GP, and NRJG provided scientific insights and critically reviewed the manuscript. The authors read and approved the final manuscript.
Funding
LD was the recipient of a scholarship from the Chinese Scholarship Council (CSC). LW and TTA are recipients of the NUSMed Postdoctoral Fellowship. This work was supported by research grants R-571–000-061–114 to NRJG, R-148–000-277–114 to ECYC, R-571-000-044-133 to BM, and R-571–000-037–114 to KSWT, and supported by Chengdu Giant Panda Breeding Research Foundation (CPF2017–12).
Availability of data and materials
The datasets generated and analyzed in the current study are available in the Sequence Read Archive (SRA) database at NCBI under BioProject ID PRJNA669121 (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA669121).
Declarations
Ethics approval and consent to participate
All animal handling and procedures were performed in accordance with the Institutional Animal Care and Use Committee of National University of Singapore (protocol no. R19-1259).
Consent for publication
Not applicable.
Conflicts of interest
The authors declare no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Guangneng Peng, Email: pgn.sicau@163.com.
Kevin Shyong Wei Tan, Email: mictank@nus.edu.sg.
References
- 1.Andersen LO, Stensvold CR. Blastocystis in health and disease: are we moving from a clinical to a public health perspective? J Clin Microbiol. 2016;54(3):524–528. doi: 10.1128/JCM.02520-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Clark CG, van der Giezen M, Alfellani MA, Stensvold CR. Recent developments in Blastocystis research. Adv Parasitol. 2013;82:1–32. doi: 10.1016/B978-0-12-407706-5.00001-0. [DOI] [PubMed] [Google Scholar]
- 3.Shirvani G, Fasihi-Harandi M, Raiesi O, Bazargan N, Zahedi MJ, Sharifi I, Kalantari-Khandani B, Nooshadokht M, Shabandoust H, Mohammadi MA, et al. Prevalence and molecular subtyping of Blastocystis from patients with irritable bowel syndrome, inflammatory bowel disease and chronic urticaria in Iran. Acta Parasitol. 2020;65(1):90–96. doi: 10.2478/s11686-019-00131-y. [DOI] [PubMed] [Google Scholar]
- 4.Peña S, Carrasco G, Rojas P, Castillo D, Ozaki LS, Mercado R. Determination of subtypes of Blastocystis sp. in chilean patients with and without inflammatory bowel syndrome, a preliminary report. Parasite Epidemiol Control. 2020;8:e00125. doi: 10.1016/j.parepi.2019.e00125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yason JA, Liang YR, Png CW, Zhang Y, Tan KSW. Interactions between a pathogenic Blastocystis subtype and gut microbiota: in vitro and in vivo studies. Microbiome. 2019;7(1):30. doi: 10.1186/s40168-019-0644-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Céline N, Julien S, Bruno P, Christina NM, Ivan W, Amandine C, Eric V, Valérie L, Frédéric D, Michel D. Blastocystis is associated with decrease of fecal microbiota protective bacteria: comparative analysis between patients with irritable bowel syndrome and control subjects. PLoS ONE. 2014;9(11):e111868. doi: 10.1371/journal.pone.0111868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Audebert C, Even G, Cian A, Group BI, Loywick A, Merlin S, Viscogliosi E, Chabé M Colonization with the enteric protozoa Blastocystis is associated with increased diversity of human gut bacterial microbiota. Sci Rep. 2016;6:25255. doi: 10.1038/srep25255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tito RY, Chaffron S, Caenepeel C, Lima-Mendez G, Wang J, Vieira-Silva S, Falony G, Hildebrand F, Darzi Y, Rymenans L, et al. Population-level analysis of Blastocystis subtype prevalence and variation in the human gut microbiota. Gut. 2019;68(7):1180–1189. doi: 10.1136/gutjnl-2018-316106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gentekaki E, Curtis BA, Stairs CW, Klimeš V, Eliáš M, Salas-Leiva DE, Herman EK, Eme L, Arias MC, Henrissat B. Extreme genome diversity in the hyper-prevalent parasitic eukaryote Blastocystis. Plos Biol. 2017;15(9):e2003769. doi: 10.1371/journal.pbio.2003769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ajjampur SS, Tan KS. Pathogenic mechanisms in Blastocystis spp. - interpreting results from in vitro and in vivo studies. Parasitol Int. 2016;65:772–779. doi: 10.1016/j.parint.2016.05.007. [DOI] [PubMed] [Google Scholar]
- 11.Wu Z, Mirza H, Tan KS. Intra-subtype variation in enteroadhesion accounts for differences in epithelial barrier disruption and is associated with metronidazole resistance in Blastocystis subtype-7. PLoS Negl Trop Dis. 2014;8(5):e28 85. doi: 10.1371/journal.pntd.0002885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Maloney JG, da Cunha MJR, Molokin A, Cury MC, Santin M. Next-generation sequencing reveals wide genetic diversity of Blastocystis subtypes in chickens including potentially zoonotic subtypes. Parasitol Res. 2021;120(6):2219–2231. doi: 10.1007/s00436-021-07170-3. [DOI] [PubMed] [Google Scholar]
- 13.Stensvold CR, Tan KSW, Clark CG. Blastocystis. Trends Parasitol. 2020;36(3):315–316. doi: 10.1016/j.pt.2019.12.008. [DOI] [PubMed] [Google Scholar]
- 14.Beghini F, Pasolli E, Truong TD, Putignani L, Caccio SM, Segata N. Large-scale comparative metagenomics of Blastocystis, a common member of the human gut microbiome. ISME J. 2017;11(12):2848–2863. doi: 10.1038/ismej.2017.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Alfellani MA, Stensvold CR, Vidal-Lapiedra A, Onuoha ESU, Fagbenro-Beyioku AF, Clark CG. Variable geographic distribution of Blastocystis subtypes and its potential implications. Acta Trop. 2013;126(1):11–18. doi: 10.1016/j.actatropica.2012.12.011. [DOI] [PubMed] [Google Scholar]
- 16.Puthia MK, Lu J, Tan KS. Blastocystis ratti contains cysteine proteases that mediate interleukin-8 response from human intestinal epithelial cells in an NF-kappaB-dependent manner. Eukaryot Cell. 2008;7(3):435–443. doi: 10.1128/EC.00371-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Iguchi A, Yoshikawa H, Yamada M, Kimata I, Arizono N. Expression of interferon gamma and proinflammatory cytokines in the cecal mucosa of rats experimentally infected with Blastocystis sp strain RN94-9. Parasitol Res. 2009;105(1):135–140. doi: 10.1007/s00436-009-1373-5. [DOI] [PubMed] [Google Scholar]
- 18.Filyk HA, Osborne LC. The Multibiome. The intestinal ecosystem's influence on immune homeostasis, health, and disease. EBioMedicine. 2016;13:46–54. doi: 10.1016/j.ebiom.2016.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen XQ, Singh M, Ho LC, Moe KT, Tan SW, Yap EH. A survey of Blastocystis sp. in rodents. Lab Anim Sci. 1997;47(1):91–94. [PubMed] [Google Scholar]
- 20.Goodrich JK, Waters JL, Poole AC, Sutter JL, Koren O, Blekhman R, Beaumont M, Van Treuren W, Knight R, Bell JT, et al. Human genetics shape the gut microbiome. Cell. 2014;159(4):789–799. doi: 10.1016/j.cell.2014.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McDonald D, Hyde E, Debelius JW, Morton JT, Gonzalez A, Ackermann G, Aksenov AA, Behsaz B, Brennan C, Chen Y, et al. American gut: an open platform for citizen science microbiome research. Systems. 2018;3(3):17004. doi: 10.1128/mSystems.00031-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hublin JSY, Maloney JG, Santin M. Blastocystis in domesticated and wild mammals and birds. Res Vet Sci. 2020;135:260–282. doi: 10.1016/j.rvsc.2020.09.031. [DOI] [PubMed] [Google Scholar]
- 23.Cooper HS, Murthy SN, Shah RS, Sedergran DJ. Clinicopathologic study of dextran sulfate sodium experimental murine colitis. Lab Invest. 1993;69(2):238–249. [PubMed] [Google Scholar]
- 24.Mishra A, Lai GC, Yao LJ, Aung TT, Shental N, Rotter-Maskowitz A, Shepherdson E, Singh GSN, Pai R, Shanti A, et al. Microbial exposure during early human development primes fetal immune cells. Cell. 2021;S0092–8674(21):00574–582. doi: 10.1016/j.cell.2021.04.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Philippe P, Ivan W, Aurélie A, Hicham EA, Frédéric D, Valérie L. Development and evaluation of a real-time PCR assay for detection and quantification of Blastocystis parasites in human stool samples: prospective study of patients with hematological malignancies. J Clin Microbiol. 2011;49(3):975–983. doi: 10.1128/JCM.01392-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Burrello C, Garavaglia F, Cribiù FM, Ercoli G, Lopez G, Troisi J, Colucci A, Guglietta S, Carloni S, Guglielmetti S, et al. Therapeutic faecal microbiota transplantation controls intestinal inflammation through IL10 secretion by immune cells. Nat Commun. 2018;9(1):5184. doi: 10.1038/s41467-018-07359-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Melgar S, Karlsson L, Rehnström E, Karlsson A, Utkovic H, Jansson L, Michaëlsson E. Validation of murine dextran sulfate sodium-induced colitis using four therapeutic agents for human inflammatory bowel disease. Intern Immunopharmacol. 2008;8(6):836–844. doi: 10.1016/j.intimp.2008.01.036. [DOI] [PubMed] [Google Scholar]
- 28.Magoč T, Salzberg SL. FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics. 2011;27(21):2957–2963. doi: 10.1093/bioinformatics/btr507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics. 2011;27(16):2194–2200. doi: 10.1093/bioinformatics/btr381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bolyen E, Rideout JR, Dillon MR, Bokulich NA, Abnet CC, Al-Ghalith GA, et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat Biotech. 2019;37(8):852–857. doi: 10.1038/s41587-019-0209-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJ, Holmes SP. DADA2: High-resolution sample inference from Illumina amplicon data. Nat Methods. 2016;13(7):581–583. doi: 10.1038/nmeth.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, et al. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 2013;41:590–596. doi: 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chan JC, Kioh DY, Yap GC, Lee BW, Chan EC. A novel LCMSMS method for quantitative measurement of short-chain fatty acids in human stool derivatized with (12)C- and (13)C-labelled aniline. J Pharm Biomed Anal. 2017;138:43–53. doi: 10.1016/j.jpba.2017.01.044. [DOI] [PubMed] [Google Scholar]
- 34.Chai Y, Deng L, Liu H, Yao J, Zhong Z, Fu H, Shen L, Zhou Z, Deng J, Hu Y, et al. First subtyping of Blastocystis sp. from pet rodents in southwestern china. Intern J Parasitol Parasites and Wildl. 2020 doi: 10.1016/j.ijppaw.2020.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Puthia MK, Sio SW, Lu J, Tan KS. Blastocystis ratti induces contact-independent apoptosis, F-actin rearrangement, and barrier function disruption in IEC-6 cells. Infect Immun. 2006;74(7):4114–4123. doi: 10.1128/IAI.00328-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Defaye M, Nourrisson C, Baudu E, Lashermes A, Meynier M, Meleine M, Wawrzyniak I, Bonnin V, Barbier J, Chassaing B, et al. Fecal dysbiosis associated with colonic hypersensitivity and behavioral alterations in chronically Blastocystis-infected rats. Sci Rep. 2020;10(1):9146. doi: 10.1038/s41598-020-66156-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kodio A, Coulibaly D, Koné AK, Konaté S, Doumbo S, Guindo A, Bittar F, Gouriet F, Raoult D, Thera MA, et al. Blastocystis colonization is associated with increased diversity and altered gut bacterial communities in healthy malian children. Microorganisms. 2019 doi: 10.3390/microorganisms7120649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Deng L, Wojciech L, Gascoigne NRJ, Peng G, Tan KSW. New insights into the interactions between Blastocystis, the gut microbiota, and host immunity. PLoS Pathog. 2021;17(2):e1009–e1253. doi: 10.1371/journal.ppat.1009253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Upadhyaya B, Yin Y, Hill BJ, Douek DC, Prussin C. Hierarchical IL-5 expression defines a subpopulation of highly differentiated human Th2 cells. J Immunol. 2011;187(6):3111–3120. doi: 10.4049/jimmunol.1101283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Smith P, Mangan NE, Walsh CM, Fallon RE, McKenzie AN, van Rooijen N, Fallon PG. Infection with a helminth parasite prevents experimental colitis via a macrophage-mediated mechanism. J Immunol. 2007;178(7):4557–4566. doi: 10.4049/jimmunol.178.7.4557. [DOI] [PubMed] [Google Scholar]
- 41.Allen JE, Maizels RM. Diversity and dialogue in immunity to helminths. Nature Rev Immunol. 2011;11(6):375–388. doi: 10.1038/nri2992. [DOI] [PubMed] [Google Scholar]
- 42.Zhang H, Sparks JB, Karyala SV, Settlage R, Luo XM. Host adaptive immunity alters gut microbiota. ISME J. 2015;9(3):770–781. doi: 10.1038/ismej.2014.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Billy V, Lhotská Z, Jirků M, Kadlecová O, Frgelecová L, Parfrey LW, Pomajbíková KJ. Blastocystis colonization alters the gut microbiome and in some cases, promotes faster recovery from induced colitis. Front Microbiol. 2021;12:641–483. doi: 10.3389/fmicb.2021.641483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zheng D, Liwinski T, Elinav E. Interaction between microbiota and immunity in health and disease. Cell Res. 2020;30(6):492–506. doi: 10.1038/s41422-020-0332-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.White EC, Houlden A, Bancroft AJ, Hayes KS, Goldrick M, Grencis RK, Roberts IS. Manipulation of host and parasite microbiotas: survival strategies during chronic nematode infection. Sci Adv. 2018;4(3):eaap7399. doi: 10.1126/sciadv.aap7399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Leung J, Hang L, Blum A, Setiawan T, Stoyanoff K, Weinstock J. Heligmosomoides polygyrus abrogates antigen-specific gut injury in a murine model of inflammatory bowel disease. Inflam Bowel Dis. 2012;18(8):1447–1455. doi: 10.1002/ibd.22858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Adalid-Peralta L, Fragoso G, Fleury A, Sciutto E. Mechanisms underlying the induction of regulatory T cells and its relevance in the adaptive immune response in parasitic infections. Intern J Biol Sci. 2011;7(9):1412–1426. doi: 10.7150/ijbs.7.1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wojciech L, Tan KSW, Gascoigne NRJ. Taming the sentinels: microbiome-derived metabolites and polarization of T cells. Intern J Mol Sci. 2020 doi: 10.3390/ijms21207740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schönfeld P, Wojtczak L. Short- and medium-chain fatty acids in energy metabolism: the cellular perspective. J Lipid Res. 2016;57(6):943–954. doi: 10.1194/jlr.R067629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Arpaia N, Campbell C, Fan X, Dikiy S, van der Veeken J, deRoos P, Liu H, Cross JR, Pfeffer K, Coffer PJ, et al. Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature. 2013;504(7480):451–455. doi: 10.1038/nature12726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Carretta MD, Quiroga J, López R, Hidalgo MA, Burgos RA. Participation of short-chain fatty acids and their receptors in gut inflammation and colon cancer. Front Physiol. 2021;12:662–739. doi: 10.3389/fphys.2021.662739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gabrielli S, Furzi F, Fontanelli Sulekova L, Taliani G, Mattiucci S. Occurrence of Blastocystis-subtypes in patients from Italy revealed association of ST3 with a healthy gut microbiota. Parasite Epidemiol Control. 2020;9:e00134. doi: 10.1016/j.parepi.2020.e00134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lloyd-Price J, Abu-Ali G, Huttenhower C. The healthy human microbiome. Genome Med. 2016;8(1):51. doi: 10.1186/s13073-016-0307-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nieves-Ramírez ME, Partida-Rodríguez O, Laforest-Lapointe I, Reynolds LA, Brown EM, Valdez-Salazar A, Morán-Silva P, Rojas-Velázquez L, Morien E, Parfrey LW, et al. Asymptomatic intestinal colonization with protist Blastocystis is strongly associated with distinct microbiome ecological patterns. mSystems. 2018 doi: 10.1128/mSystems.00007-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Andersen LO, Bonde I, Nielsen HB, Stensvold CR. A retrospective metagenomics approach to studying Blastocystis. FEMS Microbiol Ecol. 2015 doi: 10.1093/femsec/fiv072. [DOI] [PubMed] [Google Scholar]
- 56.Franzosa EA, Sirota-Madi A, Avila-Pacheco J, Fornelos N, Haiser HJ, Reinker S, et al. Gut microbiome structure and metabolic activity in inflammatory bowel disease. Nat Microbiol. 2019;4(2):293–305. doi: 10.1038/s41564-018-0306-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Huttenhower C, Kostic AD, Xavier RJ. Inflammatory bowel disease as a model for translating the microbiome. Immunity. 2014;40(6):843–854. doi: 10.1016/j.immuni.2014.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lozupone CA, Stombaugh JI, Gordon JI, Jansson JK, Knight R. Diversity, stability and resilience of the human gut microbiota. Nature. 2012;489(7415):220–230. doi: 10.1038/nature11550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Atarashi K, Tanoue T, Oshima K, Suda W, Nagano Y, Nishikawa H, et al. Treg induction by a rationally selected mixture of Clostridia strains from the human microbiota. Nature. 2013;500(7461):232–236. doi: 10.1038/nature12331. [DOI] [PubMed] [Google Scholar]
- 60.Org E, Blum Y, Kasela S, Mehrabian M, Kuusisto J, Kangas AJ, et al. Relationships between gut microbiota, plasma metabolites, and metabolic syndrome traits in the METSIM cohort. Genome Biol. 2017;18(1):70. doi: 10.1186/s13059-017-1194-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hu S, Wang J, Xu Y, Yang H, Wang J, Xue C, et al. Anti-inflammation effects of fucosylated chondroitin sulphate from Acaudina molpadioides by altering gut microbiota in obese mice. Food Funct. 2019;10(3):1736–1746. doi: 10.1039/C8FO02364F. [DOI] [PubMed] [Google Scholar]
- 62.Ramanan D, Bowcutt R, Lee SC, Tang MS, Kurtz ZD, Ding Y, Honda K, Gause WC, Blaser MJ, Bonneau RA, et al. Helminth infection promotes colonization resistance via type 2 immunity. Science. 2016;352(6285):608–612. doi: 10.1126/science.aaf3229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Devanga Ragupathi NK, Muthuirulandi Sethuvel DP, Inbanathan FY, Veeraraghavan B. Accurate differentiation of Escherichia coli and Shigella serogroups: challenges and strategies. New Microb New Infect. 2017;21:58–62. doi: 10.1016/j.nmni.2017.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Castaño-Rodríguez N, Underwood AP, Merif J, Riordan SM, Rawlinson WD, Mitchell HM, Kaakoush NO. Gut microbiome analysis identifies potential etiological factors in acute gastroenteritis. Infect Immun. 2018 doi: 10.1128/IAI.00060-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cheng S, Ma X, Geng S, Jiang X, Li Y, Hu L, et al. Fecal microbiota transplantation beneficially regulates intestinal mucosal autophagy and alleviates gut barrier injury. mSystems. 2018 doi: 10.1128/mSystems.00137-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Khanna S. My treatment approach to Clostridioides difficile infection. Mayo Clin Proc. 2021;96(8):2192–2204. doi: 10.1016/j.mayocp.2021.03.033. [DOI] [PubMed] [Google Scholar]
- 67.Terveer EM, van Gool T, Ooijevaar RE, Sanders I, Boeije-Koppenol E, Keller JJ, et al. Human transmission of Blastocystis by fecal microbiota transplantation without development of gastrointestinal symptoms in recipients. Clin Infect Dis. 2019;71(10):2630–2636. doi: 10.1093/cid/ciz1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Everard A, Belzer C, Geurts L, Ouwerkerk JP, Druart C, Bindels LB, et al. Cross-talk between Akkermansia muciniphila and intestinal epithelium controls diet-induced obesity. Proc Natl Acad Sci U S A. 2013;110(22):9066–9071. doi: 10.1073/pnas.1219451110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hmp C. Structure, function and diversity of the healthy human microbiome. Nature. 2012;486(7402):207–214. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gause WC, Rothlin C, Loke P. Heterogeneity in the initiation, development and function of type 2 immunity. Nat Rev Immunol. 2020;20(10):603–614. doi: 10.1038/s41577-020-0301-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Broadhurst MJ, Ardeshir A, Kanwar B, Mirpuri J, Gundra UM, Leung JM, Wiens KE, Vujkovic-Cvijin I, Kim CC, Yarovinsky F, et al. Therapeutic helminth infection of macaques with idiopathic chronic diarrhea alters the inflammatory signature and mucosal microbiota of the colon. PLoS Pathog. 2012;8(11):e1003000. doi: 10.1371/journal.ppat.1003000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Rubtsov YP, Rasmussen JP, Chi EY, Fontenot J, Castelli L, Ye X, Treuting P, Siewe L, Roers A, Henderson WRJr, et al. Regulatory T cell-derived interleukin-10 limits inflammation at environmental interfaces. Immunity. 2008;28(4):546–558. doi: 10.1016/j.immuni.2008.02.017. [DOI] [PubMed] [Google Scholar]
- 73.Kühn R, Löhler J, Rennick D, Rajewsky K, Müller W. Interleukin-10-deficient mice develop chronic enterocolitis. Cell. 1993;75(2):263–274. doi: 10.1016/0092-8674(93)80068-P. [DOI] [PubMed] [Google Scholar]
- 74.Begue B, Verdier J, Rieux-Laucat F, Goulet O, Morali A, Canioni D, Hugot JP, Daussy C, Verkarre V, Pigneur B, et al. Defective IL10 signaling defining a subgroup of patients with inflammatory bowel disease. Am J Gastroenterol. 2011;106(8):1544–1555. doi: 10.1038/ajg.2011.112. [DOI] [PubMed] [Google Scholar]
- 75.Glocker EO, Kotlarz D, Boztug K, Gertz EM, Schäffer AA, Noyan F, Perro M, Diestelhorst J, Allroth A, Murugan D, et al. Inflammatory bowel disease and mutations affecting the interleukin-10 receptor. N Engl J Med. 2009;361(21):2033–2045. doi: 10.1056/NEJMoa0907206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Furusawa Y, Obata Y, Fukuda S, Endo TA, Nakato G, Takahashi D, Nakanishi Y, Uetake C, Kato K, Kato T, et al. Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature. 2013;504(7480):446–450. doi: 10.1038/nature12721. [DOI] [PubMed] [Google Scholar]
- 77.Smith PM, Howitt MR, Panikov N, Michaud M, Gallini CA, Bohlooly YM, Glickman JN, Garrett WS. The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis. Science. 2013;341(6145):569–573. doi: 10.1126/science.1241165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Nakkarach A, Foo HL, Song AA, Mutalib NEA, Nitisinprasert S, Withayagiat U. Anti-cancer and anti-inflammatory effects elicited by short chain fatty acids produced by Escherichia coli isolated from healthy human gut microbiota. MicrobCell Fact. 2021;20(1):36. doi: 10.1186/s12934-020-01477-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Luu M, Pautz S, Kohl V, Singh R, Romero R, Lucas S, et al. The short-chain fatty acid pentanoate suppresses autoimmunity by modulating the metabolic-epigenetic crosstalk in lymphocytes. Nature Commun. 2019;10(1):760. doi: 10.1038/s41467-019-08711-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wang Z, Zhang X, Zhu L, Yang X, He F, Wang T, et al. Inulin alleviates inflammation of alcoholic liver disease via SCFAs-inducing suppression of M1 and facilitation of M2 macrophages in mice. Int Immunopharmacol. 2020;78:106–62. doi: 10.1016/j.intimp.2019.106062. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary file1 (TIF 8999 KB) Figure S1. Blastocystis ST4 colonization did not induce any abnormal effects on C57BL/6 mice. a, Representative micrographs of H&E-stained caecum sections and histological scores from ST4-colonized and control mice at day 7. b, Representative micrographs of H&E-stained small intestine sections and histological scores from ST4-colonized and control mice at day 7. Scale bar = 100 μm.
Supplementary file2 (TIF 482 KB) Figure S2. Rarefaction curves (threshold is 90,000) showing microbial diversity based on the Shannon index (upper panel) and Observed features (bottom panel) from normal healthy mice.
Supplementary file3 (TIF 184 KB) Figure S3. Comparison of relative abundancies of different taxa between control and ST4-colonized mice.
Supplementary file4 (TIF 9690 KB) Figure S4. Gating strategy of the immune compartments isolated from colonic lamina propria.
Supplementary file5 (TIF 580 KB) Figure S5. Rarefaction curves (threshold is 24,000) showing microbial diversity based on the Shannon index (upper panel) and Observed features (bottom panel) from Rag1−/− mice.
Supplementary file6 (TIF 311 KB) Figure S6. Comparison of relative abundancies of different taxa between control and ST4-colonized mice.
Supplementary file7 (TIF 526 KB) Figure S7. Rarefaction curves (threshold is 58,000) showing microbial diversity based on the Shannon index (upper panel) and Observed features (bottom panel) from DSS-induced colitis mice.
Supplementary file8 (TIF 236 KB) Figure S8. Comparison of relative abundancies of different taxa between control and ST4-colonized mice.
Supplementary file9 (TIF 826 KB) Figure S9. Rarefaction curves (threshold is 26,000) showing microbial diversity based on the Shannon index (upper panel) and Observed features (bottom panel) from DSSFMT mice.
Supplementary file10 (TIF 290 KB) Figure S10. Comparison of relative abundancies of different taxa between control and ST4-colonized mice.
Supplementary file11 (TIF 1457 KB) Figure S11. Fold-change of each SCFA relative to levels at day 0 from Rag1-/- mice (donor mice).
Supplementary file12 (XLSX 10 KB) Table S1. PERMANOVA of beta-diversity analysis as measured by Bray-Curtis dissimilarity.
Supplementary file13 (XLSX 620 KB) Table S2. Sample metadata, feature table and taxonomic classifications.
Data Availability Statement
The datasets generated and analyzed in the current study are available in the Sequence Read Archive (SRA) database at NCBI under BioProject ID PRJNA669121 (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA669121).