Abstract
Translational control of mRNAs is a point of convergence for many oncogenic signals through which cancer cells tune protein expression in tumorigenesis. Cancer cells rely on translational control to appropriately adapt to limited resources while maintaining cell growth and survival, which creates a selective therapeutic window compared to non‐transformed cells. In this review, we first discuss how cancer cells modulate the translational machinery to rapidly and selectively synthesize proteins in response to internal oncogenic demands and external factors in the tumor microenvironment. We highlight the clinical potential of compounds that target different translation factors as anti‐cancer therapies. Next, we detail how RNA sequence and structural elements interface with the translational machinery and RNA‐binding proteins to coordinate the translation of specific pro‐survival and pro‐growth programs. Finally, we provide an overview of the current and emerging technologies that can be used to illuminate the mechanisms of selective translational control in cancer cells as well as within the microenvironment.
Keywords: cancer, protein synthesis, translation and protein quality, translation inhibitors, translational control
Subject Categories: Cancer, Translation & Protein Quality
This review provides an overview of the cellular and molecular mechanisms cancer cells use to adjust mRNA translation and protein synthesis in response to internal and external demands of a dynamic tumor microenvironment.

Introduction
Cancer cells continually alter gene expression programs to adapt, grow, and survive in non‐physiological environments. It is now evident in the field that cancer cells can adapt to different stress conditions triggered by internal or external stimuli through the regulation of gene expression at the translational level (Truitt & Ruggero, 2016). Importantly, they can selectively synthesize proteins urgently needed on a rapid timescale (Shamir et al, 2016). While transcriptional regulation remains a major focus of cancer biologists, genome‐wide analyses have uncovered discrepancies between RNA abundance and corresponding protein levels, highlighting how quantification of RNA expression alone is insufficient to capture the actual protein levels in the cell (Liu et al, 2016; Buccitelli & Selbach, 2020).
Translation is a complex, multi‐step process which requires a multitude of factors—ribosomes, tRNAs, amino acids, and translation factors—working in concert to mediate protein synthesis. The process of translation is divided into different steps: initiation, elongation, termination, and ribosome recycling (Sonenberg & Hinnebusch, 2009; Jackson et al, 2010; Dever & Green, 2012; Robichaud et al, 2019). In this review, we will mainly focus on the initiation step, which is the rate‐limiting step controlling translation. Cancer cells tightly control this step, which impinges on selective translational control of specific mRNA networks. The untranslated regions (UTRs), which are the non‐coding regions of the mRNA flanking the coding sequence, are essential for the regulation of translation (Hinnebusch et al, 2016; Leppek et al, 2018). In particular, 5′UTRs contain several RNA sequence elements and secondary structures that provide a platform for trans element binding in order to modulate protein synthesis (Hinnebusch et al, 2016; Schuster & Hsieh, 2019). We will explore the different elements found in the 5′UTRs and 3′UTRs that cancer cells use to regulate gene expression and how these RNA regulons coordinate the expression of functionally related genes to steer many hallmarks of cancer development.
In this review, as a part of the Cancer Review Series 2021, we will highlight the important emerging concept that translation is selectively regulated to tailor a proteome in support of cancer initiation, progression, and metastasis. We will first discuss how cancer cells hijack different translation factors to drive translation of specific transcripts to maintain cancer cell fitness. Next, we will focus on how oncogenic pathways use trans and cis elements on specific transcripts to alter protein expression. Acting in concert, these factors promote expression of the mediators of nearly all hallmarks of cancer from “classical” hallmarks such as sustaining proliferation and control of cell survival to the “emerging” hallmark of avoiding immune destruction (Hanahan & Weinberg, 2011). Finally, we discuss current and developing technologies to study translational control in cancer. Through the application of these new techniques, we will continue to uncover the manifold ways cancer cells rely on translational control and how to exploit that unique vulnerability therapeutically.
Translation machinery and translational specificity
For many decades, translation factors were considered housekeeping proteins without any selectivity in promoting protein synthesis. However, more recent studies from several groups revealed that translation factors are hijacked by many oncogenes to drive transcript‐specific translation in order to maintain cancer cell fitness. In this section, we will focus on how different components of the translation machinery are involved in selective, pro‐oncogenic gene regulation and how they can be targeted therapeutically.
The eukaryotic initiation factor 4F (eIF4F) complex is the major node that oncogenic signaling pathways target to regulate gene expression at the translation level. The complex consists of the major cap‐binding protein, eIF4E, the scaffold protein, eIF4G, and the RNA helicase, eIF4A. Each component of the complex has been shown to be deregulated in different cancer types. Among them, eIF4E has emerged as a crucial nexus of translational control that is hyperactivated downstream of several oncogenic pathways (Fig 1A). While Myc promotes the transcription of eIF4E (Rosenwald et al, 1993; Jones et al, 1996), oncogenic Ras activates the phosphorylation of eIF4E by regulating the MAPK‐interacting serine/threonine kinase 1 (MNK1) (Waskiewicz et al, 1997; Furic et al, 2010), resulting in eIF4E hyperactivation. Moreover, eIF4E is also regulated via the mTOR pathway through eIF4E‐binding protein (4EBPs) suppressors, which inhibit eIF4E activity (Haghighat et al, 1995; Hsieh et al, 2012; Pourdehnad et al, 2013). All these oncogenic pathways converge to modulate eIF4E activity and highlight the importance of eIF4E in regulation of the cancer translatome. eIF4E‐dependent translation was shown to be essential in regulating selective translation involved in many diverse aspects of cancer from metabolism (Cunningham et al, 2014) to invasion (Robichaud et al, 2015). For example, tumor cells selectively exploit eIF4E to translate mRNAs needed to overcome an anti‐tumor immune response (Xu et al, 2019) or to make the tumor microenvironment more favorable for tumor growth (Bartish et al, 2020). One of the most surprising discoveries over the last several years is that, contrary to previous beliefs, eIF4E expression is not a limiting factor for overall protein synthesis. Reducing eIF4E levels by 50% does not perturb normal development and global protein synthesis; however, reduced eIF4E remarkably suppresses oncogenic transformation (Truitt et al, 2015). These findings uncovered that an excess amount of eIF4E is pro‐oncogenic, and importantly, specific eIF4E‐dependent translational control in cancer cells represents a new therapeutic vulnerability. Therefore, there is a growing interest in generating compounds that inhibit the activity of eIF4E (Fig 1B). Inhibitors that block the ability of eIF4E to recruit the pre‐initiation complex, such as the compounds 4EGI‐1 (Moerke et al, 2007), 4E1RCat (Cencic et al, 2011b), and 4E2RCat (Cencic et al, 2011a), have displayed anti‐tumor effects in pre‐clinical trials (Chen et al, 2012). Moreover, MNK1 inhibitors, such as cercosporamide and tomivosertib (also named eFT508), which block eIF4E phosphorylation, and hence the activity of eIF4E, suppress tumor progression and metastasis in both xenograft and genetically engineered mouse models (Konicek et al, 2011; Xu et al, 2019). Notably, eFT508 is currently in Phase II clinical trials (NCT03616834, NCT04622007).
Figure 1. Oncogenic regulation and therapeutic targeting of the eIF4F complex.
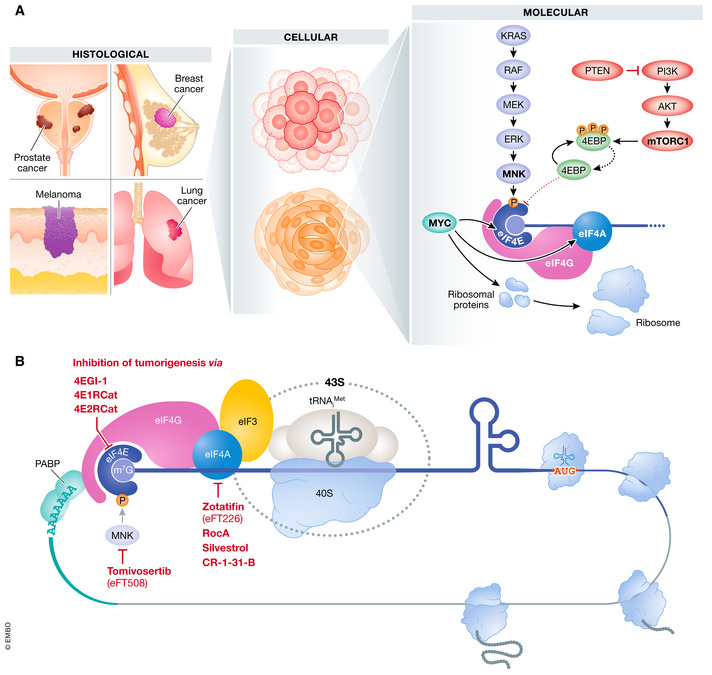
(A) In various cancer types, oncogenic pathways regulate the expression and activity of the translation machinery, converging on the eIF4F complex. The Myc oncogene promotes transcription of eIF4E, eIF4A, and several ribosomal proteins. MAPK‐interacting serine/threonine kinase MNK1, activated downstream of the RAS/ERK pathway, phosphorylates eIF4E which is crucial for the activity of the protein. In addition, mTORC1, which acts downstream of PI3K/AKT, phosphorylates 4E‐binding proteins, 4EBPs, which in turn release eIF4E to promote translation. Unphosphorylated 4EBPs compete with eIF4G to bind to eIF4E and inhibit translation. (B) Compounds targeting eIF4F complex inhibit cancer cell proliferation and tumorigenesis in vitro and in vivo. Tomivosertib (eFT508) inhibits MNK1, which regulates the activity of eIF4E through phosphorylation, and is showing promising results in clinical trials. 4EGI‐1, 4E1RCat, and 4E2RCat target the eIF4E‐eIF4G interaction to block cap‐dependent translation. Rocaglate derivatives, Zotatifin (eFT226), Rocaglamide (RocA), Silvestrol, and CR‐1‐31‐B, inhibit the activity of eIF4A by clamping the protein to polypurine stretches of the RNA. Among them, Zotatifin is already in Phase II clinical trials.
Although eIF4E has emerged as a high priority therapeutic target, the genetic interacting partners that act in concert with eIF4E‐dependent translational control to maintain cancer cell fitness are not well characterized. Our lab recently performed a genome‐wide CRISPRi screening to identify synthetic lethal partners of eIF4E, which uncovered more than 600 genetic interactions that sustain eIF4E oncogenic activity (Kuzuoglu‐Ozturk et al, 2021). Each interaction represents a potential target for combination therapy that can selectively target cancer cells at the post‐transcriptional level. Moreover, the screen unveiled novel functional connections between eIF4E and unexpected cellular processes, such as mitochondrial protein homeostasis. Specifically, cancer cells rely on selective eIF4E‐dependent translation to manage mitochondrial proteotoxic stress through increased translation of a master autophagy regulator and transcription factor, Tfeb, promoting cancer cell survival. These findings illustrate how the activity of a translation factor can orchestrate a wide variety of specific cellular processes genome wide to overcome oncogenic stress.
In addition to eIF4E, the DEAD‐box helicase eIF4A, a member of the eIF4F complex, is a key nexus in the regulation of pro‐cancerous signaling (Fig 1A). eIF4A unwinds secondary structures located in the 5′ untranslated regions (UTRs) to facilitate the scanning of the 43S ribosome complex for start codon recognition, and therefore, it is thought to be critical for the translation of mRNAs with long and complex 5′UTRs (Svitkin et al, 2001). There are two paralogs of eIF4A, eIF4A1 and eIF4A2, which are 90% homologous at the amino acid level (Nielsen & Trachsel, 1988). Interestingly, eIF4A1, a direct transcriptional target of Myc (Lin et al, 2008), is often overexpressed in a range of malignancies and has been shown to mediate selective translation of several oncogenes (Ji et al, 2003; Liang et al, 2014b; Modelska et al, 2015). In addition, a recent study showed that a decrease in eIF4A1 dosage suppresses lymphomagenesis in the Eμ‐Myc mouse model (Sénéchal et al, 2021). On the other hand, the overall role of eIF4A2 in translation and cancer is poorly understood. Ongoing research in cancer biology is mainly focused on eIF4A1‐dependent selective translation. Recent advancements employing ribosome profiling for transcriptome‐wide measurements of translational efficiency uncovered that mRNAs containing polypurine and GC‐rich sequence motifs in their 5′UTRs are specifically more sensitive to eIF4A1 activity (Rubio et al, 2014; Wolfe et al, 2014). Importantly, many oncogenes have complex 5′UTRs and have been shown to be dependent on eIF4A1 for efficient translation (Rubio et al, 2014; Steinhardt et al, 2014; Wolfe et al, 2014; Kong et al, 2019). Following these observations, numerous eIF4A inhibitors have been developed and have demonstrated potent anti‐tumorigenic effects in different pre‐clinical cancer models (Fig 1B). These data suggest that eIF4A1 is a very valuable target for cancer therapy and indeed one eIF4A inhibitor, eFT226 (known as zotatifin), is already in Phase I/II clinical trials (NCT04092673). The availability of eIF4A inhibitors has made it possible to investigate its targets. Specific mRNA targets of eIF4A1 have been identified through extensive studies in a variety of blood and solid tumor cancer models. These mRNAs vary from known oncogenes such as Myc and Mdm2 to key regulators of proliferation such as the cell cycle kinases Cdk6/Cdk10 (Table 1). Detailed information about the spectrum of eIF4A inhibitors can be found in reviews focused on this topic (Voss et al, 2017; Pal et al, 2019).
Table 1.
Transcripts dependent on eIF4A for efficient translation.
| Cancer model | eIF4A inhibitor | Specific mRNAs | Experimental method | Notes | Reference |
|---|---|---|---|---|---|
| T‐cell acute lymphoblastic leukemia | Silvestrol | Genome‐wide analysis identified 281 downregulated mRNAs including MYC, NOTCH1, MYB, CDK6, MDM2, CCND3, BCL2 and ETS1 | Ribosome Profiling | eIF4A1 helicase activity is essential to resolve G‐quadruplex structures at the 5′UTR of the target mRNAs | Wolfe et al (2014) |
| Triple‐negative breast cancer cell line MDA‐MB‐231 | Silvestrol | Genome‐wide analysis identified 284 downregulated mRNAs including CCND1, ARF6, BCL2, ROCK1, and CDK6 | Ribosome Profiling | Silvestrol sensitivity is dependent on 5′UTR complexity | Rubio et al (2014) |
| Diffuse large B‐cell lymphomas | Silvestrol | CARD11, BCL10, MALT1 | Targeted approach | Steinhardt et al (2014) | |
| BRAFV600E melanoma | Silvestrol | Genome‐wide analysis: CREBBP, HPRT, MLL3, NCOA6, ARID5B, and RICTOR | Polysome Profiling | Combination with BRAF and MEK inhibitors inhibits the emergence of melanoma persister cells | Shen et al (2019) |
| PDAC | Silvestrol | ARF6 | Targeted approach | KRAS promotes translation of ARF6 by suppressing Pdcd4, an inhibitor of eIF4A | Hashimoto et al (2019) |
| Breast cancer stem cells (BCSC) | Rocaglamide A (RocA) | NANOG, OCT4, and drug transporters | Targeted approach | RocA treatment reduces self‐renewal ability of BCSC and induces apoptosis | Sridharan et al (2019) |
| Prostatic ductal adenocarcinoma (PDAC) | CR‐1‐31‐B | mRNAs involved in redox and central carbon mechanism: CDK4/6 | Polysome Profiling | CR‐1‐31‐B suppresses PDA growth in vitro and in vivo | Chan et al (2019) |
| ER+ breast cancer, KRAS‐mutant NSCLC | CR‐1‐31‐B | mRNAs involved in the inhibition of apoptosis (MCL1 and BCL2) and in the regulation of cell cycle progression (CCND1, CCND3, CCNE1, and CDK6) | Targeted approach | Combination of CDK4/5 inhibitor palbociclib with CR‐1‐31‐B suppresses growth of the cells in vitro and in vivo | Kong et al (2019) |
| Breast Cancer Cells | Zotatifin (eFT226) | Receptor Tyrosine Kinases: ERBB2, FGFR1, and FGFR2 | Targeted Approach | Combination with PI3K/AKT inhibitors with Zotatifin suppresses cancer cell growth in vitro and in vivo | Gerson‐Gurwitz et al (2021) |
The structural component of the eIF4F complex, the scaffold protein eIF4GI, is upregulated in many different cancer types and is associated with increased metastases and higher tumor stage in prostate cancer and ovarian cancer, respectively (Braunstein et al, 2007; Comtesse et al, 2007; Attar‐Schneider et al, 2014; Li et al, 2016; Jaiswal et al, 2018; Valle et al, 2021). There are three members of the eIF4G protein family: eIF4GI (highest expression), eIF4GII (lowest expression), and DAP5 (known as eIF4G2, p97, and NAT1) (Parra et al, 2018). The majority of studies have focused on the role of eIF4GI and DAP5 in mediating selective translational control in cancer, while the function of eIF4GII remains largely unstudied. eIF4GI acts as a specific translation factor by modulating the stoichiometry of the eIF4F complex, and it also promotes selective translation through its ability to engage with internal ribosome entry sites (IRES) located in the 5′UTRs of key proangiogenic, hypoxia, and survival mRNAs (Braunstein et al, 2007; Silvera et al, 2009). eIF4GI can directly bind to the IRES elements in the 5′UTR of these mRNAs independently of eIF4E and drive cap‐independent translation by recruiting additional initiation factors and the ribosome to initiate translation. Similarly, DAP5 can also promote cap‐independent translation of mRNAs important for invasion, metastasis, and apoptosis, such as BCL2, APAF1, cIAP1, CDK1, and a (Hundsdoerfer et al, 2005; Marash et al, 2008; Weingarten‐Gabbay et al, 2014). Moreover, eIF4GI can also regulate translation of a subset of mRNAs important for survival and DNA damage response pathway in breast cancer (Badura et al, 2012). Additionally, a recent paper also showed that eIF4GI regulates expression of specific immunoregulatory proteins in non‐small cell lung cancer and may represent a therapeutic vulnerability for this cancer type (Valle et al, 2021).
The majority of prior research on translation initiation has focused on the eIF4F complex, consisting of only three proteins. However, exciting new research is drawing focus to other key components of the translation initiation machinery, in addition to the eIF4F complex. A multi‐protein complex eIF3, the largest initiation factor with 13 subunits, has been implicated in controlling the translation of mRNAs important for cellular proliferation (Fig 2A). eIF3 binds to 40S ribosomal subunit and promotes the binding of methionyl‐tRNAi and mRNA (Hershey, 2015). To date, evidence shows that overexpression of six individual subunits (3a, 3b, 3c, 3h, 3i, and 3m), while the repression of two others (3e and 3f), can cause malignant transformation (Hershey, 2015). Recent studies have uncovered the role of specific eIF3 subunits in selective translation and their function in different diseases, including cancer (Wolf et al, 2020; Fujii et al, 2021). Transcriptome‐wide assessment of eIF3 RNA binding with PAR‐CLIP showed that eIF3 binds the 5′UTRs of specific mRNAs associated with cancer‐related pathways, such as cell cycle control, differentiation, and apoptosis (Lee et al, 2015). Importantly, eIF3 binding to the oncogene JUN versus the tumor suppressor BTG1 mRNAs was found to have an opposite effect on translation (Lee et al, 2015). Additionally, eIF3d can bind the mRNA cap, in particular the cap of JUN mRNA, where it is essential for the assembly of the translation initiation complex independently of eIF4F (Lee et al, 2016). Expanding the role of eIF3d in selective translation, a recent paper demonstrated that Myc promotes the specific translation of the SF3A3 mRNA through an eIF3d‐mediated mechanism, which, in turn, regulates splicing and metabolic reprogramming that underlie Myc‐driven tumorigenesis (Cieśla et al, 2021). Additionally, a study showed that eIF3e promotes synthesis of the mitochondrial electron transport chain proteins through their 5′UTRs in the MCF7 breast cancer cell line (Shah et al, 2016). Although the current understanding of the role of individual subunits of the eIF3 complex is limited, these initiation factors are emerging as key players in directing the expression of the cancer‐specific proteome.
Figure 2. Roles of eIF3 and eIF5A in selective translational control.
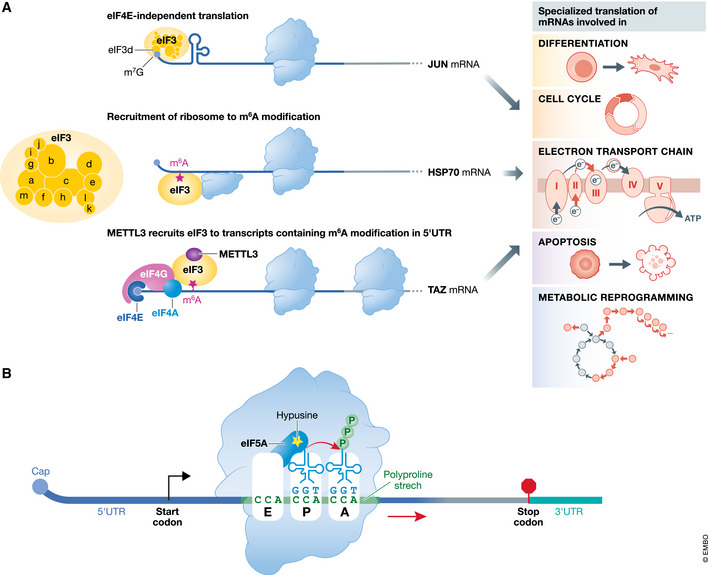
(A) The eIF3 complex with 13 subunits regulates specialized translation of mRNAs encoding proteins involved in differentiation, electron transport chain (ETC), cell cycle, apoptosis, and metabolic reprogramming. Different mechanisms of eIF3‐dependent translation are shown. eIF3 can directly bind to the cap via its 3D subunit and drive cap‐independent translation (JUN mRNA). eIF3 can recruit the ribosome to m6A modification containing mRNA for translation of specific mRNAs (HSP70 mRNA). In addition, METTL3 can recruit eIF3 to transcripts containing m6A modification in their 5′UTR and promote translation (TAZ mRNA). (B) eIF5A plays a role as ribosomal pause relief factor. eIF5A promotes peptide bond formation when the ribosome is stalled on a polyproline stretch. The unique post‐translational modification in eIF5A, hypusination, is required for the activity of the protein.
A growing body of evidence highlights the importance of the interaction between the eIF3 complex and the N6‐methyladenosine (m6A) machinery to specify selective translation (Fig 2A). Several excellent recent reviews provided an in‐depth perspective on the role of m6A and RNA modifications more broadly in the translational control of cancer (Barbieri & Kouzarides, 2020; He & He, 2021). An important study demonstrated that eIF3 can directly bind to m6A‐modified bases in the 5′UTR of select mRNA to recruit the 43S complex and initiate cap‐independent translation (Meyer et al, 2015). This mechanism is critical for maintaining translation of specific m6A‐containing mRNAs, such as HSP70, under stress conditions by bypassing cap‐binding proteins (Meyer et al, 2015). eIF3 was also shown to interact with the m6A “reader” protein YTHDF1 to promote the delivery of m6A‐containing mRNAs to the translational machinery, providing a mechanism to selectively increase translation efficiency (Wang et al, 2015). In addition, METTL3, an m6A “writer”, was shown to promote the translation of a subset of target mRNAs, including several oncogenes such as EGFR and TAZ, by recruiting eIF3 to the translation initiation complex, independently of its methyltransferase activity or m6A “reader” proteins, YTHDF1 or 2 (Lin et al, 2016). In fact, METTL3 interacts specifically with the eIF3h subunit to promote mRNA looping, enhancing ribosome recycling and the formation of densely packed polyribosomes, which together boost the translation of specific oncogenic mRNAs important for lung cancer (Choe et al, 2018). METTL3 can also interact with eIF3b to mediate translation of the YAP mRNA in lung cancer cells (Jin et al, 2019). Overall, the eIF3 complex is a multifaceted component of the translational machinery that integrates upstream oncogenic signals along with RNA structure and epigenetic modifications to exquisitely regulate selective post‐transcriptional gene expression.
In addition to initiation factors, exciting studies are highlighting the possible roles of elongation factors in translation specificity and cancer etiology (Knight et al, 2020). However, precisely how elongation factors impact cancer‐specific translation is poorly understood. One of the emerging elongation factors mediating selective translation is eIF5A. This protein was originally defined as an initiation factor; however, recent studies show its main role in translation elongation as a ribosomal pause relief factor (Fig 2B). In humans, there are two eIF5A isoforms, eIF5A1 and eIF5A2, both of which contain the amino acid hypusine formed by a post‐translational modification unique to a specific lysine residue in eIF5A. This modified amino acid is essential for the activity of eIF5A; therefore, it has attracted attention as a therapeutic target (Mathews & Hershey, 2015). While eIF5A1 is ubiquitously expressed in most cells and tissues, eIF5A2 is specifically expressed in the testes and brain (Jenkins et al, 2001; Clement et al, 2003). Interestingly, eIF5A2 is more broadly expressed in cancers of different tissues of origins (Caraglia et al, 2013; Wang et al, 2013; Mathews & Hershey, 2015; Wu et al, 2020a). While the role of eIF5A1 in cancer is still puzzling and requires further investigation, the majority of mechanistic studies on specificity of translation elongation have focused on eIF5A1. Therefore, we will refer to eIF5A1/2 jointly as eIF5A in this section. eIF5A is required to promote peptide bond formation when the ribosome is stalled (Gregio et al, 2009; Saini et al, 2009; Gutierrez et al, 2013; Pelechano & Alepuz, 2017; Schuller et al, 2017). Moreover, eIF5A depletion was shown to promote translation initiation at upstream near‐cognate start codons in yeast (Ivanov et al, 2018). An interesting recent study showed that a similar mechanism is conserved in human cells and, surprisingly, eIF5A regulates start codon selection of the MYC mRNA in cancer cells (Manjunath et al, 2019). In particular, loss of eIF5A promotes expression of an N‐terminally extended c‐Myc protein, demonstrating a novel translational regulation mechanism for MYC (Manjunath et al, 2019). Moreover, eIF5A may more generally regulate selective translation of oncogenes containing proline stretches or tripeptides (Met‐Phe‐Phe), which require eIF5A activity to prevent ribosome stalling (Saini et al, 2009; Gutierrez et al, 2013). In this way, eIF5A may be a critical factor for maintenance of cancer cell fitness by releasing stalled ribosomes to support the increased metabolic burden of oncogenic transformation. A recent study demonstrated that eIF5A promotes translation of specific mitochondrial transcripts involved in the tricarboxylic acid (TCA) cycle and oxidative phosphorylation, opening a new avenue of research into eIF5A function (Puleston et al, 2019). Further studies are required to elucidate the role of eIF5A in transcript‐specific translation that promotes cancer survival, in particular the connection between eIF5A and mitochondrial function. These initial studies into the functions of eIF5A in cancer demonstrate that eIF5A is an intriguing novel nexus of translational control and its role in MYC translation may represent a potential cancer cell selective therapeutic target.
Non‐coding RNAs (ncRNAs) also play a vital role as part of the translation machinery. The process of translation relies upon many different types of ncRNAs, such as ribosomal RNAs (rRNAs), transfer RNAs (tRNAs), microRNAs (miRNAs), and long ncRNAs (lncRNAs), whose functions and contributions to cancer are the subject of excellent reviews (Anastasiadou et al, 2018; Goodall & Wickramasinghe, 2021). Here, we will focus on how the availability of the decoding components of the translation machinery, tRNAs, regulates selective translation (Fig 3). As the decoders of the genetic code, tRNAs recognize specific triplet codons on the mRNA, mediating proper ribosomal incorporation of specific amino acids into the growing polypeptide. The human genome contains a total of 61 distinct sense codons, many of which encode the same amino acid. These 61 codons are recognized by a total of 49 tRNAs with unique anti‐codon sequences. tRNAs are found to be upregulated in different cancer types, which has historically been thought to promote global protein synthesis. Major oncogenic signals like the MAPK‐ERK and PI3K/mTOR pathways as well as Myc co‐opt the tRNA synthesis machinery, in particular RNA polymerase III activity, to modulate tRNA expression levels in support of cancer cell proliferation (Felton‐Edkins et al, 2003; Gomez‐Roman et al, 2003; Woiwode et al, 2008; Kantidakis et al, 2010). However, emerging evidence suggests that cancer cells selectively increase expression of specific tRNAs to enhance translation elongation efficiency for distinct subsets of mRNAs based on their codon composition (Dittmar et al, 2006; Pavon‐Eternod et al, 2009; Goodarzi et al, 2015). In breast cancer, upregulation of tRNAGlu UUC and tRNAArg CCG promote metastasis by enhancing expression of direct target genes, such as EXOSC2 and GRIPAP1, in a codon‐specific manner (Goodarzi et al, 2016) (Fig 3). Altered expression of specific tRNAs within the tumor microenvironment can also be a crucial driver of cancer progression. Increased levels of initiator methionine tRNA, particularly in stromal fibroblasts, are sufficient to promote tumor invasion, migration, and metastasis, specifically through altered translation of extracellular matrix components and dysregulated integrin signaling (Birch et al, 2016; Clarke et al, 2016). Moreover, cancer‐specific tRNA signatures can be crucial for the fate of the cells as they can coordinate the selective expression of different cellular programs, such as proliferation or differentiation (Gingold et al, 2014; Aharon‐Hefetz et al, 2020). In addition, tumor cells also mistranslate with higher frequency compared to non‐transformed cells and this translational error can promote tumor growth (Santos et al, 2018). Together these studies demonstrate that tRNAs are key regulatory components that shape the oncogenic translatome.
Figure 3. Codon usage and tRNAs in cancer.
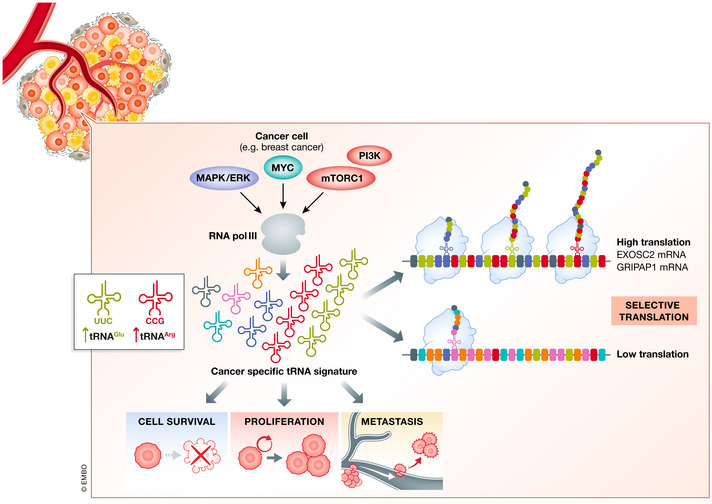
Cancer‐specific repertoire of tRNAs, the decoding components of the translation machinery, promote selective translation to maintain cancer cell fitness. RNA polymerase III, which is regulated by three major oncogenic pathways, MAPK/ERK, MYC, and PI3K/mTOR, can specifically alter the abundance and availability of specific tRNAs in cancer cells to promote translation elongation efficiency for specific subsets of mRNAs based on their codon composition. As an example, breast cancer cells express high levels of tRNAGlu UUC and tRNAArg CCG which enhance translation of EXOSC2 and GRIPAP1 mRNAs in a codon‐specific manner to promote tumorigenesis.
The relationship between tRNA availability and codon usage can also precisely tune the expression of specific cancer‐promoting genes. The effect of codon bias in cancer has mainly focused on the translation efficiency of different Ras oncogene isoforms. Two members of the family, KRAS and HRAS, have predominantly rare or common codons, respectively. This difference modulates their protein expression levels, although they share 85% identity at amino acid level (Lampson et al, 2013). Changing rare codons to common codons in KRAS not only increased the expression of K‐Ras but also enhanced its tumorigenic capacity (Lampson et al, 2013; Pershing et al, 2015). KRAS codon usage was also shown to affect transcription rate and protein conformation (Fu et al, 2018). Moreover, a recent study revealed that cancer mutational bias in codons within a family of oncogenes, such as Ras, may be related to how well the coding sequence is selectively and efficiently translated in proliferating cells compared to primary, non‐transformed cells (Benisty et al, 2020). The paradigm of Ras mRNA translation illustrates that neither mRNA transcript levels nor amino acid composition are sufficient predictors of oncogenic protein expression. The tunable and differential translation of Ras isoforms shows the centrality of translation in the process of oncogenic transformation and sustained tumorigenesis. In addition, the MAPK pathway can enhance expression of transcripts with rare codons, such as KRAS (Peterson et al, 2020), showing the complex, intertwined nature of translational regulation. Moreover, translational efficiency modulated by codon usage is not limited to RAS family members as similar codon usage patterns have also been reported for different oncogene protein families such as AKT, RAF, and FGFR (Benisty et al, 2020). From the classical initiation factors of the eIF4F complex to elongation factors like eIF5A to tRNA contributions, we are only beginning to appreciate how every facet of translational initiation works in concert to mediate the precise tuning of the cancer proteome to support cancer growth.
Specific regulation of mRNA transcripts to mediate expression of the cancer proteome
The translational machinery works in concert with the mRNA sequence and structure to tailor the composition of the pro‐tumorigenic proteome. Many oncogenic signaling programs converge on translational control, usurping functional RNA regulatory elements in mRNA to specify the translation of pro‐proliferative, pro‐survival, and anti‐apoptotic genetic programs to overcome the cellular stresses of oncogenic transformation, aberrant proliferation, and adaptation to the tumor microenvironment (Xu & Ruggero, 2019). In this way, cancer cells rely on altered translational activity, creating an addiction that distinguishes cancer cells from non‐transformed cells. Selective translation of cancer‐promoting mRNAs depends upon both cis‐ and trans‐regulatory factors. In trans, the translational machinery in conjunction with RNA‐binding proteins (RBPs) precisely control the translation of specific mRNAs, often regulating the expression of functionally linked groups of transcripts (for in‐depth reviews see (Pereira et al, 2017; Harvey et al, 2018; Qin et al, 2020)). Acting in cis, mRNA sequence features, such as RNA motif elements and alternative translation initiation sites (ATIS), as well as RNA structure integrate the upstream oncogenic signals to mediate the specificity and efficiency of translation.
Sequence features/motifs
Key features of mRNAs contribute to their selective translation downstream of oncogenic signaling programs. Sequence‐specific RNA elements in the untranslated regions of genes have been functionally associated with altered translational efficiency in response to pro‐tumorigenic signaling (Fig 4A). One of the first identified examples is the 5′‐terminal oligopyrimidine tract (TOP) motif, which regulates the translation efficiency of mRNAs encoding core components of the translational machinery, including ribosomal proteins (Levy et al, 1991) and translation factors (Iadevaia et al, 2008). The 5′ TOP motif is characterized by an invariable C residue proximal to the cap followed by an uninterrupted stretch of 4–15 pyrimidines (Perry, 2005). Critically, mTOR signaling regulates the translation of TOP motif‐containing mRNAs, enabling the coordinated expression of protein synthesis to control cell growth (Jefferies et al, 1994; Tang et al, 2001; Hsieh et al, 2012; Thoreen et al, 2012). In this context, mTOR activity integrates a wide variety of stress conditions or altered nutrient states common in cancer cells, such as hypoxia or amino acid starvation, to orchestrate a selective cellular response that promotes cancer cell survival (Tang et al, 2001; Miloslavski et al, 2014). Recent work has implicated additional oncogenic pathways in regulating 5′TOP‐specific translational control. In particular, loss of the tumor suppressor Arf promotes the translation of 5′TOP‐containing mRNAs, implicating other pathways that act alongside mTOR signaling to promote expression of pro‐growth protein repertoire (Cottrell et al, 2020).
Figure 4. RNA sequence elements regulate selective translation.
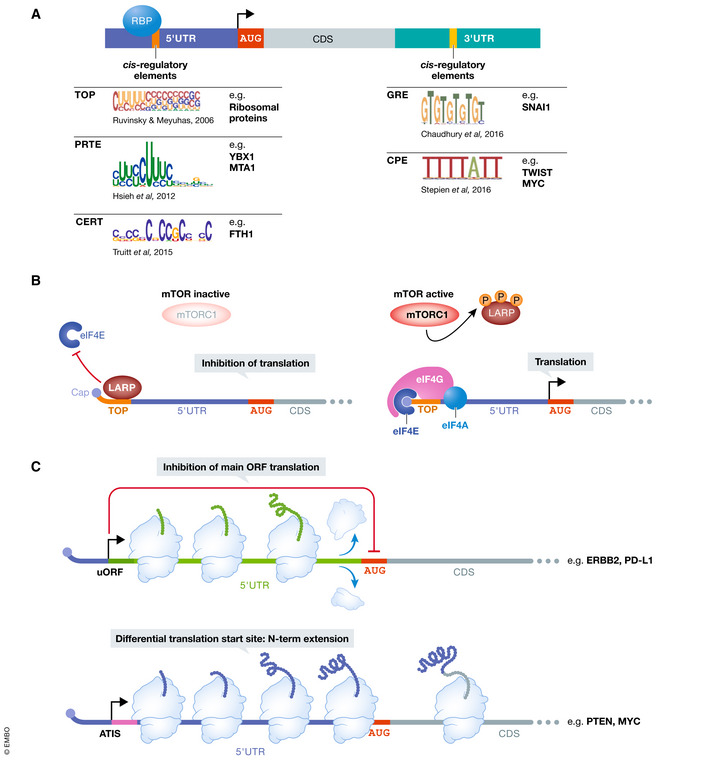
(A) RNA sequence cis‐regulatory elements in both the 5′UTR and 3′UTR of mRNAs play a role in specifying transcripts for translation downstream of certain oncogenic signals. For example, both the TOP and PRTE motifs mediate the translation of transcripts sensitive to mTOR activity. These RNA sequence elements function as part of a coordinated mechanism to regulate the translation of specific pro‐oncogenic programs, such as EMT (GRE), metabolic dysregulation (PRTE), and response to oxidative stress (CERT). Consensus sequences of each motif are shown. (B) RNA sequence elements interface with RNA‐binding proteins (RBPs) to regulate the translation of specific transcripts. A well‐studied example is the RBP LARP1’s modulation of the translation of 5′TOP motif containing transcripts. When mTORC1 is inactive, LARP1 binds the TOP motif to block eIF4F binding to the mRNA cap. However, active mTORC1 phosphorylates and physically binds LARP1 to allow eIF4F to access the cap and promote translation initiation. (C) RNA sequence features in the 5′UTR can alter translation of the main ORF (mORF). Under homeostatic conditions, the translation machinery engages upstream ORFs (uORFs) to diminish mORF translation. However, under oncogenic stress, uORF translation is suppressed to promote the translation of the mORF, which often encodes oncogenes or pro‐survival factors. In a similar way, cancer cell signaling can promote translation initiation at alternative start codons that are in‐frame with the main ORF to generate N‐terminally extended proteins. These alternative proteoforms possess different functions from the canonical protein as has been well described for the tumor suppressor PTEN.
Genome‐wide analysis of the oncogenic mTOR translation program has uncovered a broader landscape of “TOP‐like” sequence motifs. Ribosome profiling of hyperactive mTOR‐dependent translation in prostate cancer cells identified an enriched “TOP‐like” sequence, namely the pyrimidine‐rich translational element (PRTE), which is characterized by an invariant uridine at position 6 flanked by pyrimidines and, unlike the TOP motif, is located at variable positions within the 5′UTR (Hsieh et al, 2012). PRTE‐containing genes are enriched for key regulators of metastasis, such as YBX1 and MTA1, that coordinate cancer cell invasion downstream of oncogenic mTOR signaling. Additionally, the PRTE motif is necessary for the efficient translation of phosphoribosyl pyrophosphate synthetase 2 (PRPS2), a rate‐limiting nucleotide biosynthesis enzyme that maintains a sufficient nucleotide pool to promote MYC‐induced tumorigenesis (Cunningham et al, 2014). The TOP and PRTE motifs demonstrate the vital role of RNA sequence elements in integrating upstream oncogenic signals from mTOR to Myc to precisely respond to the unique metabolic demands of cancer cells.
Another recently identified RNA sequence element confers an even greater specificity to the translational regulation. Transcripts possessing the cytosine‐enriched regulator of translation (CERT) motif are sensitive to the expression level of the cap‐binding protein, eIF4E, during early tumorigenesis. The CERT motif mediates the selective translation initiation of mRNA networks that are essential for cellular transformation (Truitt et al, 2015). In particular, the CERT motif was shown to modulate the expression of key target genes of the antioxidant response, particularly ferritin and glutathione, to counteract reactive oxygen species and aid cancer cell survival (Truitt et al, 2015). Subsequent research has implicated CERT‐dependent translation as a pivotal response to cellular stress states, such as neuronal injury and regrowth as well as hyperactive mTOR signaling in epilepsy (Rozenbaum et al, 2018; Kim et al, 2019). Moreover, the CERT motif was found to be enriched in 4EBP1 target genes in prostate cancer cells, which is concordant with its initial identification in transcripts sensitive to eIF4E dosage (Jin et al, 2020). Intriguingly, recent work uncovered enrichment of the CERT sequence in the 5′UTR of genes dependent upon the DEAD‐box helicase DDX3 for efficient translation initiation (Calviello et al, 2021). These data suggest an intriguing relationship between RNA structures and sequences in selective translation initiation. Notably, DDX3 can mediate the selective translation of a mRNA network that contributes to tumor development (Oh et al, 2016). Overall, the CERT RNA element is emerging as a nexus of translational control, orchestrating a timely response to cellular stressors, particularly in cancer formation and progression.
Another type of cis‐regulatory element is the translation initiator of short 5′ UTR (TISU) regulatory motif that promotes accurate, efficient, and eIF4A‐independent translation of genes with short 5′UTRs without scanning (Elfakess & Dikstein, 2008; Elfakess et al, 2011). TISU element‐dependent translation mediates the synthesis of proteins involved in core biological processes such as protein biogenesis and degradation, RNA metabolism, and mitochondrial health (Sinvani et al, 2015; Haimov et al, 2017). Translation of TISU‐containing transcripts requires the cap‐binding complex eIF4F; however, the 48S ribosomal subunit recognizes and directly contacts the motif to promote the assembly of the complete 80S ribosomal complex and initiate translation without scanning to sustain translation of genes required for a cellular response to energy stress (Haimov et al, 2017). In line with its role in regulating core biosynthetic pathways, TISU‐dependent translation is sensitive to mTOR inhibition, implicating the TISU motif as another mediator of selective translation that promotes cancer cell growth and survival (Gandin et al, 2016). The TISU regulon demonstrates the capacity for 5′UTR sequence to selectively impinge on the translation of transcripts necessary to respond rapidly to maintain a pro‐growth cellular state.
Interactions between RNA regulatory elements and RNA‐binding proteins (RBPs) in the 3′UTR also contribute to the selective translational control of pro‐oncogenic programs (Fig 4A). One notable motif is the guanine/uridine‐rich element (GRE) found in the 3′UTR of genes important for epithelial‐to‐mesenchymal transition (EMT), including the EMT master regulator SNAI1 (Chaudhury et al, 2016). The GRE motif resembles the binding motif for the RBP CELF1 and indeed CELF1 directly binds the motif in the 3′UTR of these genes to regulate their translation downstream of TGFβ activation through an unknown mechanism. Another example of a translational control feature in the 3′UTR is the cytoplasmic polyadenylation element (CPE). The CPE was originally described as a regulator of protein synthesis in embryonic development, but evidence also suggests a role in modulating the translation of several pro‐tumoral mRNAs such as PLAT (encoding tPA), TWIST1, and MYC (Burns & Richter, 2008; Nairismägi et al, 2012; Ortiz‐Zapater et al, 2012). The CPE is bound by a family of RBPs known as CPE‐binding proteins (CPEBs) that differ in their affinities for the CPE consensus motifs and can act as either repressors or activators (Fernández‐Miranda & Méndez, 2012). Of note, CPEB4 is overexpressed in pancreatic cancer and glioblastoma and is a key regulator of oncogenic driver expression in early melanoma formation (Ortiz‐Zapater et al, 2012; Pérez‐Guijarro et al, 2016).
The tumor‐specific overexpression of certain CPEBs can be exploited to program cancer‐specific translation. For example, researchers have employed an oncolytic adenovirus with CPE regulatory sequences in the 3′UTR of the E1A gene to selectively kill tumor cells in an in vivo pancreatic cancer model with high CPEB4 protein levels (Villanueva et al, 2017). Although the precise mechanisms of 3′UTR‐driven translational control and potential collaborations with 5′UTR elements are only beginning to be revealed, they present great therapeutic potential for cancer‐specific treatments.
The mechanistic details of how RNA sequence elements interface with trans‐factors to control gene expression in cancer remains poorly understood. A working hypothesis is that upstream oncogenic pathways coordinate the recruitment of distinct RBPs and translation factors to these motifs (Fig 4B). A prime example is the proposed mechanism of how mTOR signaling modulates the interaction between the RBP LARP1 and the 5′ TOP motif to regulate translation (Tcherkezian et al, 2014; Fonseca et al, 2015). Detailed mechanistic experimentation has revealed that when mTOR is inactive, LARP1 competes with the eIF4F cap‐binding complex to block translation initiation. However, when activated, mTORC1 phosphorylates LARP1 to promote mRNA release and mTORC1 directly sequesters LARP1 via interaction with mTORC1 complex member raptor, thus promoting efficient translation initiation (Lahr et al, 2017; Philippe et al, 2017; Jia et al, 2021). Importantly, the expression of LARP1 itself is also altered in cancer and its dysregulation predicts poor survival in a broad array of cancer types, including lung, colorectal, ovarian, and hepatocellular carcinoma (Xie et al, 2013; Mura et al, 2015; Hopkins et al, 2016; Ye et al, 2016). The mechanism of LARP1 and 5′TOP‐dependent translation demonstrates the ability of oncogenic signaling to coordinate translation of core cell growth genes through the dynamic binding of a distinct RBP. More broadly, RBPs are emerging as key interpreters of upstream oncogenic signals and microenvironmental stressors conveying the message into rapid and specific translation of functionally related transcripts, as has been recently demonstrated downstream of Myc signaling and under hypoxic conditions (Ho et al, 2020). Current research has yet to uncover how the repertoire of RNA motifs in the 5′ and 3′UTR interface with these RBPs to coordinate translation of mRNA networks underlying diverse cellular processes associated with tumor development.
Alternative translation initiation sites
Another important 5′UTR feature that regulates selective translation initiation is the alternative translation initiation site (ATIS), which is located upstream of the start AUG initiation codon of the main open reading frame (Fig 4C). Many ATIS are part of small ORFs, called upstream ORFs (uORFs), which are found in nearly 50% of all human genes and typically act to repress translation of the downstream main ORF (Calvo et al, 2009). Genome‐wide ribosome profiling in mammalian cells has shown that many uORFs are associated with translating ribosomes (Ingolia et al, 2011). Mechanistically, uORFs modulate protein abundance through either ribosomal dissociation from the mRNA after termination of the uORF or through stalled ribosomes on the coding sequence of the uORF (Zhang et al, 2019). Therefore, efficient translation of the main ORF requires bypass of the uORF start codon, referred to as “leaky” scanning, where ribosomes can reinitiate downstream of the uORF through a mechanism that is poorly understood. Researchers have developed new selective mRNA footprinting techniques that allow comparison of the location of 40S versus 80S ribosomes and their association with initiation factors (eIF3B, eIFS21, eIF4G, and eIF4E) on mRNA transcripts in human cells (Bohlen et al, 2020; Wagner et al, 2020). The data show that 80S ribosomes located within uORFs can maintain contact with eIF4E and the translation initiation machinery to promote efficient downstream reinitiation at the AUG of the main ORF (Bohlen et al, 2020). Future studies using this technique to compare the footprints of different ribosomal complexes in non‐transformed and oncogenically transformed cells will help illuminate the mechanisms by which cancer cells rewire translational control through uORF regulation.
How do cancer cells exploit uORF regulation to selectively synthesize a pro‐tumorigenic proteome? The 5′UTRs of oncogenes are enriched for uORFs, enabling the fine tuning of their expression levels under homeostatic conditions (Kozak, 1987). Specifically, in non‐transformed cells, uORFs can repress the translation of mRNAs encoding oncogenes. However, cell stress conditions can favor uORF bypass and translation of the main ORF, enabling an appropriate, but temporally limited response until the cell regains homeostatic balance. A prominent set of genes under tight uORF translational control are the majority of human tyrosine kinase genes. Experiments have demonstrated that removal of their respective uORFs results in enhanced translation of the main ORF (Wethmar et al, 2016). One specific example is the translation of ERBB2, which encodes the HER2 receptor and is a common breast cancer driver. The 5′UTR of ERBB2 contains a uORF close to the main start codon that impairs main ORF translation under physiological conditions (Child et al, 1999). However, oncogenic stress can promote trans‐factor binding to an element in the 3′UTR to repress this inhibition and promote efficient translation of the main ORF (Mehta et al, 2006; Spevak et al, 2006). Additionally, cancer cells can selectively drive oncogene or tumor suppressor translation through the use of an alternative promoter/transcription start site such that the 5′UTR of the resulting mRNA isoform either includes uORFs upstream of tumor suppressors, such as in BRCA1, or excludes uORFs upstream of oncogenes such as in the negative p53 regulator MDM2 (Brown et al, 1999; Sobczak & Krzyzosiak, 2002 ). Additionally, changes in the cancer cell microenvironment can also suppress expression of uORF‐containing transcripts. In particular, a recent study showed that in response to hypoxia and anti‐cancer therapies, breast cancer cells preferentially express NANOG, SNAIL, and NODAL transcripts that lack repressive uORFs in their 5′UTRs, which facilitates a stem cell‐like state and cancer persistence in unfavorable conditions (Jewer et al, 2020). Cancer cells have found varied means to exploit and circumvent uORF‐mediated translational regulation to promote oncogenic growth. Cancer cells' unique reliance on uORF bypass to tailor their proteomes also provides a selective vulnerability, which can be targeted therapeutically.
Previous research has provided some insights into the mechanisms underlying uORF‐dependent selective translation. Intensive dissection of the translation of ATF4, a transcription factor that is the main downstream regulator of the integrated stress response (ISR), has shed light on factors that suppress uORF translation to promote main ORF translation. Under a variety of cellular stress conditions, global translation is repressed while the ATF4 protein is newly synthesized to orchestrate the cell’s transcriptional response to stress (Wortel et al, 2017). Work has shown that cancer cells depend on the translation of ATF4 and the integrated stress response to mitigate the persistent stress of both uncontrolled oncogenic signaling leading to an increased metabolic load as well as external stressors, such as hypoxia, to promote continued tumor growth and therapy resistance (Bi et al, 2005; Falletta et al, 2017; Nguyen et al, 2018). The ATF4 5′UTR contains two uORFs that under physiological conditions repress downstream translation. However, stress conditions cause a reduction in the eIF2‐GTP‐Met‐tRNA ternary complex resulting in “leaky” scanning such that ribosomes reinitiating after uORF1 bypass the uORF2 start codon to translate the main ORF encoding ATF4 (Vattem & Wek, 2004). Additionally, recent work demonstrated that the DENR‐MCTS1 ribosome recycling complex is necessary for this reinitiation downstream of uORF1 to translate ATF4 and may be a more general mechanism for selectively translating key oncogenes such as ARAF, RAF1, and CDK4 (Bohlen et al, 2020).
An important recent example of selective uORF‐dependent translation initiation during in vivo tumorigenesis is suppression of uORF translation to increase expression of the immune checkpoint protein PD‐L1 in aggressive cancers (Xu et al, 2019). This study suggests that similar mechanisms promoting ATF4 translation also act on the CD274 mRNA (encoding PD‐L1), thereby diminishing the anti‐tumor T‐cell response and fostering tumor immune evasion. Another study implicates eEF2K, an atypical protein kinase that negatively modulates the translation elongation stage, in fostering uORF bypass for increased PD‐L1 protein synthesis (Wu et al, 2020b). Other interesting research demonstrated that during the ISR, eIF5B plays a role in promoting efficient CD274 translation (Suresh et al, 2020). Intriguingly, a genome‐wide study of translation in a mouse model of skin squamous cell carcinoma formation demonstrated a counterintuitive increase in the occupancy of uORFs during early tumorigenesis, particularly in cancer‐related genes NRAS, CD44, Ki67, and RAC1 (Sendoel et al, 2017). The authors show that under stress conditions, alternative translation initiation factor, eIF2A, is required for the increase in the translation of the main downstream ORF in these uORF‐containing transcripts, although the mechanism of action is unclear. In support of the human disease relevance of this finding, eIF2A is upregulated in many squamous cancer types and correlates with poor patient outcomes (Sendoel et al, 2017). Further research into the functional impact of uORF number, length, and location will be key to understanding how oncogenic signaling regulates the translational control of cancer‐related genes to allow precise adaptation to rapidly changing internal and external cellular environments.
Many studies have documented genetic events that result in uORF gain or loss in specific pro‐tumorigenic genes, promoting oncogenic transformation and progression. A classic example is a hereditary mutation in the 5′UTR of CDKN2A that generates a de novo uORF, which decreases the expression of the encoded tumor suppressor p16(INK4A) and predisposes carriers to melanoma (Liu et al, 1999). A similar mechanism of action is observed with a deletion in the 5′UTR of CDKN1B, decreasing expression of the tumor suppressor p27(Kip1) to cause inherited multiple endocrine neoplasia syndrome type 4 (Occhi et al, 2013). Given the strong impact of inherited, germline mutations altering uORFs, researchers undertook a systematic search for cancer‐associated changes in uORFs that identified ~400 mutations that could impact uORF loss or gain of function (Schulz et al, 2018). They highlight loss‐of‐function uORF mutations in EPHB1 in breast and colon cancer, and in MAP2K6 in a colon adenocarcinoma sample, which functionally enhanced translation. With the increasing availability of whole‐genome sequences, new germline mutations that predispose individuals to cancer are being described, such as a study of 15,708 individuals that identified mutations in the 5′UTR of NF2 that cause loss of a uORF, causing neurofibromatosis (Whiffin et al, 2020). Interestingly, germline polymorphisms can generate de novo uORFs that influence response to therapy, such as the novel uORF in the 5′UTR of the DNA damage repair gene, ERCC5, that promotes its selective translation after treatment with platinum‐based chemotherapy to foster therapy resistance (Somers et al, 2015). Excitingly, the ever‐expanding whole‐genome sequencing of both normal and cancerous tissues will likely pinpoint more mutations that alter uORFs to modulate downstream translation, which will provide new means to identify oncogenic drivers as well as guide the selection of appropriate targeted therapeutics.
Finally, cancer can modulate the efficiency of translation from an upstream ATIS that is in frame with the main ORF to generate longer, alternative proteoforms that exhibit unique functions (Fig 4C). A paradigm of a gene which gives rise to N‐terminally extended isoforms is PTEN, a key negative regulator of PI3K/Akt signaling. At least four extended translational variants of PTEN have been described, although the nomenclature differs among publications (Malaney et al, 2017). The best studied is PTEN long (equivalent to PTENɑ), which is produced from an upstream CUG to generate a 173 amino acid N‐terminal extension (Hopkins et al, 2013; Liang et al, 2014a). The N‐terminal extension permits PTEN long secretion from the cell to non‐cell autonomously inhibit PI3K signaling and repress tumorigenesis (Hopkins et al, 2013). Additional work demonstrated a role for PTEN long/PTENɑ in promoting mitochondrial respiratory chain function and promoting ATP production; while PTENβ specifically localizes to the nucleolus where it suppresses ribosomal RNA processing and decreases cell proliferation (Liang et al, 2014a, 2017). Finally, recent research functionally described another ATIS‐encoded N‐terminal extended form, PTENε, which includes an additional 72 amino acids and acts to suppress filopodia formation and diminishes the invasion and migration of cancer cells (Zhang et al, 2021). The example of the intricate regulated expression of diverse PTEN proteoforms highlights the need to look beyond the mRNA transcript level towards the role of selective translation in shaping a diverse proteome suited to cancer cell survival and adaptation to stressors. Another critical cancer‐related gene that encodes an ATIS‐produced N‐terminal extension is the oncogene MYC. The long form of c‐Myc is produced from an alternative CUG upstream of the main AUG and its expression depends in part on the activity of the ribosomal pause relief factor eIF5A (Hann et al, 1988; Manjunath et al, 2019). This longer form of c‐Myc appears to have a tumor suppressive function via its occupancy of unique DNA‐binding sites and pro‐apoptotic properties (Hann et al, 1994; Benassayag et al, 2005). However, the role of the long form of c‐Myc in cancer initiation and progression in vivo remains unknown.
Structural regulatory elements
RNA structures play a powerful role in regulating the translation of key cancer‐related genes. Early on, it was noted that the majority of transcripts encoding oncogenic drivers had longer than average 5′UTRs, which promotes the formation of secondary structure and hinders efficient 40S ribosomal scanning (Kozak, 1987). Under homeostatic conditions, these structures precisely regulate translation initiation to selectively limit protein levels and maintain appropriate composition of the expressed proteome. Cancer cells can hyperactivate the eIF4F complex, in particular the eIF4A helicase, to promote unwinding of highly structured 5′UTRs and selectively increase the translation of pro‐oncogenic genes, such as cell cycle regulators and growth factor receptors (Wolfe et al, 2014). For example, the mRNA encoding the master transcription factor and proto‐oncogene c‐Myc has a highly structured 5′UTR that enables exquisite regulation of its translation (Stoneley et al, 2000; Cobbold et al, 2008). Although many RNA structures that drive the translation of cancer essential genes have been identified, research is only beginning to understand the mechanisms behind this very diverse class of RNA elements. However, it is clear that RNA structures in both the 5′ and 3′UTRs enable critical switching between modes and efficiency of translation to permit the sustained expression of vital cancer genes even under the many inhospitable conditions of the tumor microenvironment.
Many genes encoding the key effectors of the “hallmarks of cancer” contain structured elements in their 5′UTRs known as internal ribosome entry sites (IRES), which permit cap‐independent translation (Fig 5A). IRES‐dependent translation enables efficient translation of certain genes when cap‐dependent translation is downregulated, particularly under the stress conditions of hypoxia, low nutrient states (e.g., amino acid starvation), ER stress, and DNA damage, which are all common during cancer progression and/or after chemotherapeutic treatment (Kawai et al, 2004; Qin & Sarnow, 2004; Blais et al, 2006; Bushell et al, 2006; Thomas & Johannes, 2007). Important and diverse mediators of cancer progression contain cellular IRESs in their 5′UTRs, such as genes that promote cellular survival (e.g., BCL2, XIAP, and APAF‐1), metabolic rewiring (e.g., ferritin), angiogenesis (e.g., VEGF and FGF2), the response to hypoxia (e.g., HIF1A), and EMT (e.g., SNAIL and ZEB2) (Stein et al, 1998; Holcik et al, 1999; Coldwell et al, 2000; Lang et al, 2002; Sherrill et al, 2004; Braunstein et al, 2007; Beltran et al, 2008; Evdokimova et al, 2009; Daba et al, 2012; Morfoisse et al, 2014; Philippe et al, 2016). Genome‐wide polysome profiling analyses of different stress states as well as functional, high‐throughput discovery of cap‐independent translation elements in the human genome indicate that ~10–15% of all human 5′UTRs harbor the potential for cap‐independent translation through a “cellular IRES” (Spriggs et al, 2008; Weingarten‐Gabbay et al, 2016). Interestingly, some of the genes preferentially translated in a cap‐independent manner under different stress conditions are non‐overlapping, suggesting coordinated translation of subsets of IRES‐containing genes in response to unique pathophysiological states.
Figure 5. RNA structures mediate translational control.
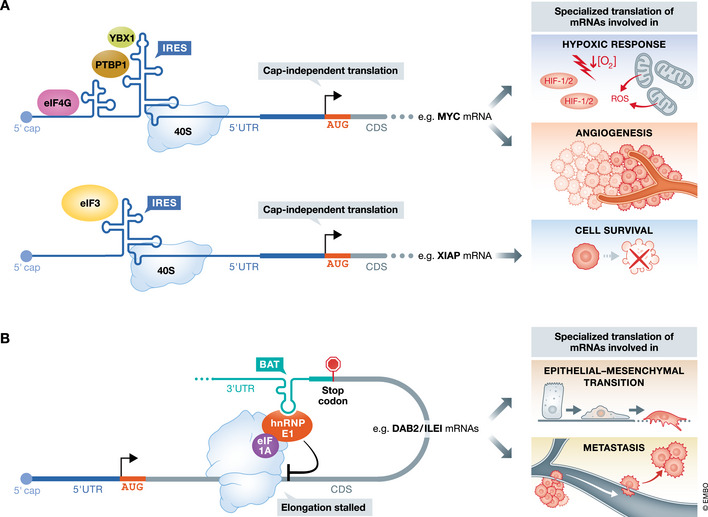
(A) The 5′UTRs of key pro‐tumorigenic transcripts contain RNA structures that promote selective translation initiation. The internal ribosome entry site (IRES) allows for cap‐independent translation of mRNAs critical to cancer cell growth and survival in the setting of decreased cap‐dependent translation, such as under low nutrient conditions or hypoxia. A key example of IRES‐dependent translational control is the oncogene MYC. The coordinated binding of the RBPs YBX1 and PTBP1 and the translation initiation machinery to the IRES‐like structure in the MYC 5′UTR can initiate cap‐independent translation in a cancer setting. Another example is the anti‐apoptotic factor XIAP. Binding of eIF3 to the cellular IRES located in the XIAP 5'UTR drives cap‐independent translation. (B) Structures in the 3′UTR of mRNAs can regulate translational elongation to coordinate the selective expression of key hallmarks of cancer. One important example is the function of the TGFβ‐activated translational (BAT) element in mediating the synthesis of proteins involved in EMT processes.
The precise mechanisms that mediate selective IRES‐dependent translation in cancer are only beginning to be elucidated. IRES structures are bound by RNA‐binding proteins referred to as IRES trans‐acting factors (ITAFs) as well as select translation factors, which, together, enable the recruitment of the 40S ribosome to promote cap‐independent translation initiation. For example, selective translation through the cellular IRES in the MYC 5′UTR is dependent upon a subset of canonical cap‐dependent factors, such as eIF4G and eIF3, as well as the RBPs PTBP1 and YBX1 (Fig 5A) (Spriggs et al, 2009; Cobbold et al, 2010). In multiple myloma, mutations within the MYC IRES strengthen the binding of PTBP1 and YBX1, upregulating c‐Myc protein expression, and promoting a feed‐forward loop wherein c‐Myc increases the transcription of YBX1 to drive tumorigenesis (Cobbold et al, 2010; Bommert et al, 2013). YBX1 has emerged as a key ITAF involved in the cap‐independent translation of genes with diverse, pro‐oncogenic functions. For example, YBX1 binds the 5′UTR of the gene encoding p16/INK4A to mediate response to hypoxia, to the IRES of SNAIL to coordinate an EMT translational program as well as mediating the cap‐independent translation of TGFB1 (Hu et al, 1999; Chappell et al, 2000, 2004). Additionally, the initiation complex eIF3 can bind directly to the mRNA of the anti‐apoptotic factor, XIAP, where it appears to function as a scaffold to recruit additional RBPs and the 40S ribosome to promote translation initiation (Thakor et al, 2016). Excitingly, inhibition of ITAF interaction with its corresponding IRES may be a potent anti‐cancer treatment. Research has demonstrated that inhibiting the binding of hnRNP A1 to both the MYC and CCND1 (Cyclin D1) transcripts may be a potential therapy for glioblastoma (Holmes et al, 2016). Finally, ITAFs can also act to repress the translation of their target transcripts, such as the tumor suppressor PDCD4. PDCD4, whose protein level is regulated by activated ribosomal protein S6 kinase 2 (S6K2), can bind XIAP and BCLXL mRNAs and directly repress their translation to block tumorigenesis (Liwak et al, 2012). Only a small subset of ITAFs and other factors that regulate IRES‐dependent translation have been identified. Hopefully, new technologies that enable high‐throughput, functional identification of proteins that bind to and regulate translation through these structures will shed light on the complex mechanisms that modulate the cancer translatome.
While many studies have focused on structural elements upstream of the start codon, RNA structures in the 3′UTR can also mediate the translational efficiency of critical oncogenic programs (Fig 5B). One notable structural RNA regulon in the 3′UTR is the TGFβ‐activated translational (BAT) element that consists of a stem loop with an asymmetrical bulge, which coordinates a translational program that promotes EMT. Active TGFβ signaling promotes phosphorylation and subsequent release of hnRNP E1 from the BAT element in the 3′UTRs of both DAB2 and ILEI, which are both necessary for the induction of EMT (Chaudhury et al, 2010). Under physiological conditions, hnRNP E1 blocks translational elongation by inhibiting the release of eEF1A from the ribosomal A site (Hussey et al, 2011). Consequently, depletion of hnRNP E1 is sufficient to promote EMT and metastasis of breast epithelial cells (Hussey et al, 2011). TGFβ is a fascinating nexus of post‐transcriptional regulation in that its own translation is also modulated in a cap‐independent fashion through an IRES (Kim et al, 1992; Jenkins et al, 2010). In the future, it would be interesting to assess whether and how the structure of 5′ and 3′UTR regulatory elements change in real time during different steps in cancer development. The results of these studies would be particularly important for the development of new small molecules that recognize specific RNA structures that can be exploited as cancer therapeutic interventions.
Current and emerging technologies to study translational control in cancer
Methods to study how RNA and proteins interact to modulate translation in non‐transformed cells, cancer cells, and within the tumor microenvironment have been pivotal in deciphering the oncogenic translational program. To directly assay the translation of specific mRNAs, the gold standard methodology remains polysome profiling. Polysome profiling entails the isolation of the cell type of interest and centrifugation of the cytoplasmic fraction on a sucrose gradient to enable separation of the free ribonuclear proteins (RNPs), mRNA occupied with few polysomes, and the high polysome fraction. Specific mRNA abundance within each fraction can be analyzed via qRT–PCR or high‐throughput sequencing. Alternatively, genome‐wide ribosome profiling, in which mature ribosomes are bulk isolated, treated with nuclease, and the resulting “footprints” are analyzed with high‐throughput sequencing can be employed (Ingolia et al, 2009; McGlincy & Ingolia, 2017). Ribosome profiling has contributed to both our fundamental understanding of selective translation initiation, for example, the use of alternative initiation codons and the role of uORFs in suppressing translation, as well as how cancer cells, anti‐cancer therapies, and the tumor microenvironment reshape the translation landscape (Ingolia et al, 2011, 2018; Hsieh et al, 2012; Xu et al, 2019). While ribosome profiling is an important tool for the study of translation control, the technique has certain limitations. Ribosome profiling data present a single snapshot in time and, for example, cannot differentiate a stalled from a translating ribosome or a bone fide start codon from an internal ATG. However, experimental modifications, such as harringtonine treatment, which immobilizes the ribosome immediately after initiation and results in footprint enrichment at initiation sites can provide more detailed insights (Ingolia et al, 2011; McGlincy & Ingolia, 2017). Additional considerations for ribosome profiling experiments versus other techniques have been reviewed elsewhere (Brar & Weissman, 2015; Ingolia et al, 2018). Overall, ribosome profiling remains a valuable technique to assess translation control in a relatively straightforward and unbiased manner.
While ribosome profiling remains an incredibly powerful tool for transcriptome‐wide analysis of translation efficiency, many groups have evolved the technique to provide more detailed insights into translational control (Table 2). One key new set of techniques enable the profiling specifically of the small ribosomal subunit during the course of translation initiation, extending beyond analysis of the complete 80S ribosomes assayed by the traditional method. Referred to as translation complex profile sequencing (TCP‐seq), this family of techniques enable the dissection of the steps of translation initiation at nucleotide resolution across the whole genome, which will be useful in understanding how oncogenic signaling programs direct the translation of specific pro‐tumorigenic RNA transcripts (Archer et al, 2016; Bohlen et al, 2020). Other groups have further modified ribosome profiling to assess both tissue‐specific translation from a mixed cell population as well as to measure translational efficiency in subcellular compartments (Sanz et al, 2009; Jan et al, 2014). In the future, both techniques can provide critical insights into the regulation of translation both within a cancer cell as well as in other cell types that constitute the tumor microenvironment. All of these existing technologies require material from many cells. However, ribosome profiling at the single cell level will shed light on how a cancer cell responds to the complex mixture of internal oncogenic stressors as well as the many external insults. Toward this goal, a technique called Ribo‐STAMP was recently published that harnesses an RNA editing enzyme coupled to a ribosomal protein to tag bound mRNAs, which can be analyzed with single‐cell RNA sequencing (scRNA‐seq) (Brannan et al, 2021). Excitingly, a new single‐cell ribosome profiling technique employs enzymatic reactions at the single‐cell scale, followed by pooled sequencing and machine learning‐based data analyses, which was successfully applied to specific, rare populations of primary colon cells (VanInsberghe et al, 2021). These constantly evolving ribosome profiling technologies will be powerful tools to precisely, yet in an unbiased manner, measure the translation of cancer cell‐specific genetic programs.
Table 2.
Methods to study translational control.
| Methods | Description | Reference | |
|---|---|---|---|
| Methods to study translational efficiency | Ribosome Profiling | Genome‐wide analysis of ribosome footprints. | Ingolia et al (2009) |
| Translation complex profile sequencing (TCP‐seq) | Specific profiling of the footprints of small ribosomal subunit during translation initiation. Adaptations of the technique isolate‐specific translation initiation complexes interacting with 40S ribosomes. | Sanz et al (2009), Jan et al (2014), Archer et al (2016), Bohlen et al (2020), Wagner et al (2020) | |
| Ribo‐STAMP | Exogenous expression of RNA editing enzyme coupled to a ribosomal protein to tag ribosome‐bound mRNAs, which are analyzed with single‐cell RNA sequencing (scRNA‐seq). | Brannan et al (2021) | |
| Single‐cell ribosome sequencing (scRibo‐seq) | Single‐cell‐level ribosomal footprinting and sequencing. Requires machine learning algorithms to provide codon resolution. Can be applied to rare populations from in vivo tissue samples. | VanInsberghe et al (2021) | |
| Protein Centric Methods to Study RNA–Protein Interactions | CLIP‐seq | UV Cross‐linking and immunoprecipitation followed by sequencing. Commonly used protocols include HITS‐CLIP, iCLIP, eCLIP, and irCLIP. Another variation, PAR‐CLIP, uses 4‐thiouridine and/or 5‐thioguanine as a nucleotide analog when UV penetration is insufficient. | Ramanathan et al (2019), Hafner et al (2021), Porter et al (2021), Licatalosi et al (2008), Konig et al (2011), Nostrand et al (2016), Hafner et al (2010), Zarnegar et al (2016) |
| Chemical cross‐linking Seq | Formaldehyde is used to cross‐link protein and RNA. Main protocols are fCLIP‐seq and xRIPiT‐Seq which are useful for RBPs that do not UV cross‐link to RNA, such as double stranded binding RBPs. Formaldehyde cross‐links protein–protein interactions producing indirect associations. | Singh et al (2014), Kim and Kim (2019) | |
| RNA Centric Methods to Study RNA–protein Interactions | Affinity capture of target mRNA | RNA is in vitro transcribed (IVT) and labeled with a molecular handle, such as biotin, for pull down after incubation with protein lysate. Alternatively, an aptamer (e.g., MS2, PP7, and S1m) can be appended to the RNA of interest, the RNA is expressed in the cell or IVT, incubated with protein lysate, and pulled down with cognate ligand. | Leppek and Stoecklin (2014), Zheng et al (2016), Gemmill et al (2020), Hogg and Collins (2007), Slobodin and Gerst (2010) |
| Endogenous RNA Pull down | ChIRP‐MS, CHART, TRIP, and RAP‐MS: these techniques use antisense oligos to capture the RNA of interest, followed by mass spectrometry for unbiased identification of proteins associated with the RNA. | Chu et al (2015), West et al (2014), Matia‐González et al (2017), McHugh and Guttman (2018) | |
| RNA‐based Proximity Labeling | Recruitment of promiscuous biotin ligase to RNA sequence of interest, which then tags proteins proximal to the RNA. The biotin tag enables purification of the proximal proteome with streptavidin beads. Most commonly BioID‐ (RaPID and CARPID) and APEX2‐based approaches. | Ramanathan et al (2018), Han et al (2020), Yi et al (2020) | |
| Methods to Study RNA Structure | DMS‐Seq/DMS‐MaP‐seq | DMS (dimethyl sulfate) modifies unpaired adenine and cytosine, which stop the reverse transcriptase (RT‐stops) during cDNA generation. High‐throughput sequencing can be used to identify the RT‐stops across the transcriptome. | Rouskin et al (2014), Zubradt et al (2017) |
| Selective 2‐hydroxyl acylation analyzed by primer extension (SHAPE) | SHAPE reagents react with and modify all four nucleotides, which are read out as RT‐stops; most common is the icSHAPE method. | Spitale et al (2015), Sun et al (2019) | |
| Mutate and Map | M2 and icM2 (in cell M2) entail systematic mutagenesis of nucleotides in an RNA sequence followed by chemical mapping to provide “two dimensional” base pairing information. | Kladwang et al (2011), Byeon et al (2021) | |
| Proteomic Methods to Measure Protein Synthesis | Stable isotope labeling with amino acids in cell culture (SILAC)‐based techniques | Pulsed treatment of cells with isotopically labeled amino acids followed by mass spectrometry analyses identifies and quantifies newly synthesized peptides during the labeling period (e.g., dynamic SILAC, p‐SILAC, & mePROD). Certain variations can be used in living organisms and for subcellular resolution. | Selbach et al (2008), Doherty et al (2009), Rhoads et al (2015), Mardakheh et al (2015), Baughman et al (2016), Klann et al (2020) |
| Non‐canonical amino acid labeling | Treatment with non‐canonical amino acids that can be incorporated into the nascent peptide chain, but are suitable for click chemistry addition of a fluorescent moiety for visualization (e.g., FUNCAT) or biotin “handle” for stringent purification and mass spectrometry analysis (e.g., BONCAT & HILAQ). | Dieterich et al (2006), Elliott et al (2014), Dieterich et al (2010), Calve et al (2016), Ma et al (2017), Evans et al (2019) | |
| Puromycin and puromycin‐ derivative labeling of nascent chains | Cells are treated with puromycin, biotinylated puromycin, or the cell‐permeable and azide‐reactive derivative OPP (O‐propargyl‐puromycin), which are incorporated into the nascent peptide chain to cause chain termination and can be visualized or purified and subjected to mass spectrometry analyses. Common variants include Puromycin‐associated nascent chain proteomics (PUNCH‐P) and OPP‐mediated protein identification (OPP‐ID). | Liu et al (2012), Aviner et al (2013), Forester et al (2018) | |
| Methods to Study Protein Abundance at Single‐Cell Level | Cytometry by Time of Flight (CyTOF) | Single‐cell‐level protein abundance detected through antibodies conjugated to rare heavy metal isotopes, which can be distinguished by mass cytometry. Can be used to detect 30–100 unique surface and intracellular proteins. | Spitzer and Nolan (2016) |
| Single‐cell mass spectrometry (scMS) | Single cells are isolated, each proteome is barcoded, and the samples are pooled for mass spectrometry analysis. Currently, not widely employed due to technical limitations of low input protein. | Budnik et al (2018), Schoof et al (2021) | |
| Methods to Measure Temporal and Spatial Dynamics of Translation | Single‐molecule FRET (smFRET) | Specific components of the translation machinery are labeled with fluorophores—ribosome, tRNA, other translation factors, or the mRNA. FRET is used to track the proximity of the two fluorophores and thus the relative positions of the labeled components. | Uemura et al (2010), Choi et al (2018), Lawson et al (2021) |
| Spatiotemporal translation of a single mRNA in a living cell | These approaches combine simultaneous labeling of nascent peptide formation and the RNA of interest in live cells to track subcellular location and rate of translation (e.g., SunTag system). | Wu et al (2016), Yan et al (2016), Wang et al (2016), Morisaki et al (2016) |
Many new techniques have been developed to capture RNA–protein interactions which will be very important in the study of different steps of tumor development and therapeutic response (Table 2). This family of approaches generally referred to as cross‐linking and immunoprecipitation followed by sequencing (CLIP‐seq) typically employ UV to cross‐link bound RNA to an RBP of interest in a specific cell or tissue type and have proven invaluable in assessing the compendium of RNAs bound by an RBP (Ramanathan et al, 2019; Hafner et al, 2021). This technique has also been extended to translation factors, such as eIF4E, and could be implemented to identify RNAs bound by the translation initiation machinery in situ within a cancer (Jensen et al, 2021). Enormous effort has been put into generating a baseline catalog of the RNAs bound by more than 100 different RBPs, which provides a starting point in understanding how RBPs orchestrate the translation of specific functional groupings of transcripts (Nostrand et al, 2020). Ultimately, the power of these techniques will be to illuminate how certain oncogenic signals direct RBPs to cooperate with each other and the translational machinery to promote cancer cell survival and growth.
Computational prediction of RNA structure based on phylogenetic comparison or free energy minimization has been the most readily available method to predict RNA secondary structures; however, the accuracy of these prediction algorithms is limited (Mailler et al, 2019). Biophysical approaches, such as X‐ray crystallography, nuclear magnetic resonance (NMR), and cryogenic electron microscopy (Cryo‐EM), can produce very powerful structural data but face many limitations with respect to RNA length, RNA flexibility, and lack of native cellular environment (Mailler et al, 2019). In the last decade, new RNA probing approaches to map nucleotide accessibility have dramatically improved the analysis of RNA structure in living cells (Table 2). Most techniques to probe RNA structure are based on chemical compounds that modify specific RNA bases in a manner that causes the reverse transcriptase (RT) enzyme to stop, which can be read out with high‐throughput sequencing as a shortened cDNA (RT stop), or cause a mutation at the modified base. For studies of single transcript or genome‐wide RNA structure in living cells, which requires cell permeable reagents, the most commonly used methods are dimethyl sulfate (DMS)‐Seq and selective 2‐hydroxyl acylation analyzed by primer extension (SHAPE) (Rouskin et al, 2014; Spitale et al, 2015). The nucleotide accessibility data can be incorporated into in silico predictions to generate high‐confidence RNA structure models. These structural predictions can then be verified through mutate and map strategies, in which mutations are introduced one at a time along the RNA sequence and the structural changes are analyzed to see if they match the prediction of the model (Kladwang et al, 2011). This technique has traditionally been applied in vitro but has recently been developed into an in vivo technique called in‐cell mutate and map (icM2) (Kladwang et al, 2011; Byeon et al, 2021). In the future, the combination of in cell RNA structural measurements with the rates of translational efficiency of those same transcripts will shed light on how RNA structures guide translational control under different oncogenic signaling programs.
Another important set of methodologies to assess the cancer‐specific translational landscape focus on proteomic techniques. It has been appreciated that mRNA transcript abundance does not strictly correlate with protein level across the entire genome (Buccitelli & Selbach, 2020). Therefore, quantitative proteomics is an important tool to assay the expression level of each protein in the proteome of different cell types and cell states (Table 2). On a single‐cell scale, proteomic technologies have lagged behind scRNA sequencing approaches, which has resulted in many scRNA‐seq experiments being used to infer protein expression on the single‐cell scale. Therefore, a great deal of effort is being invested in assaying protein composition at the single‐cell scale (Marx, 2019). Although not capable of measuring the whole proteome, cytometry by time of flight (CyTOF) has proved to be a very valuable tool to multiplex the quantification of proteins (30 to 100) at a single‐cell level (Spitzer & Nolan, 2016). Excitingly, very recently several groups have begun to push the boundaries to create relatively accessible techniques for single‐cell mass spectrometry (scMS). The most promising methods rely on scaled‐down sample prep in conjunction with a strategy of barcoding and multiplexing single cells for greater throughput and sensitivity (Budnik et al, 2018; Schoof et al, 2021). As scMS becomes a more widely available technique, it will be fascinating to examine how RNA and protein expression correlate at the single‐cell level within various cells of complex biological systems, such as the tumor and its surrounding microenvironment.
To further focus on the evaluation of rapid translational changes, proteomic technologies that capture nascent protein production can provide important insights into how cells respond to different environmental cues, such as response to cancer therapies (Aviner et al, 2013; Forester et al, 2018). Unbiased mass spectrometry analysis of the cellular proteome is a fundamental technique to understand how oncogenic signaling acts upon the transcriptome through translation to tailor protein expression. To assay nascent protein synthesis and protein turnover quantitatively, there are an array of proteomic techniques, employing alternatives to native amino acids to enable differentiation of newly synthesized peptides from the rest of the proteome. Briefly, cells or in some cases organisms can be pulsed with isotopically labeled amino acids (e.g., stable isotope labeling of amino acids in culture (SILAC)), non‐canonical amino acids, or peptide chain terminating chemicals like puromycin and O‐propargyl‐puromycin (OPP), which label newly synthesized peptides (Dieterich et al, 2006; Selbach et al, 2008; Doherty et al, 2009; Aviner et al, 2013; Ma et al, 2017; Forester et al, 2018). Subsequently, mass spectrometry can be used to distinguish and quantify the labeled, new synthesized peptides compared to the total proteome. Continued development and refinement of these techniques to a single‐cell scale will open a window into the heterogeneity of the proteome within the tumor, its microenvironment, and metastases that aids in tumorigenesis and progression.
Unbiased proteomics can also be applied to protein–RNA interactions to identify proteins bound to a specific transcript or RNA sequence (Table 2). A first set of methods use in vitro transcribed (IVT) RNA that is connected to a “molecular handle,” such as biotin or an RNA aptamer structure, which can be coupled to a matrix, mixed with protein lysate, and the bound proteins identified through mass spectrometry (Leppek & Stoecklin, 2014; Zheng et al, 2016). Additionally, to assay proteins bound to a specific RNA within cells, techniques have been developed that utilize chemical cross‐linking of the proteins to the RNA followed by antisense oligo pull down to isolate the RNA‐bound proteins, as exemplified by the ChIRP‐MS and CHART‐MS methods (West et al, 2014; Chu et al, 2015). As these methods require either abundant RNAs or high cell numbers, additional methods have been developed, such as RaPID, which exploits proximity biotinylation technology to covalently tag with biotin proteins that are in proximity to the RNA sequence of interest (Ramanathan et al, 2018). The biotin‐tagged proteins can then be easily extracted from the lysate with a streptavidin‐coated matrix and identified via mass spectrometry. All of these approaches can illuminate the identity of the proteins bound to and potentially mediating the selective translation of an mRNA of interest. Moreover, these proteomic methods can be applied to unique RNA sequence or structural elements determined through RNA‐sequencing–based technologies (e.g., CLIP or ribosome profiling) to reveal what proteins may be acting mechanistically to specify and coordinate translational programs.
Finally, there are a broad array of methods to assay the spatial and temporal dynamics of translation. These techniques are critical to understanding where within a cell and on what time scale translation is occurring (Table 2). A number of groups have developed techniques to assay the translation dynamics of single mRNAs. Single‐molecule Förster resonance energy transfer (smFRET) can be used to precisely monitor all stages of translation at the single codon level, which allows insights into the contributions various components of the translational machinery to the translation efficiency, for example, mRNA epigenetic modifications (Uemura et al, 2010; Choi et al, 2018; Lawson et al, 2021). Additionally, several techniques have been published that examine the spatiotemporal translation of a single mRNA in a living cell (Morisaki et al, 2016; Wang et al, 2016; Wu et al, 2016; Yan et al, 2016). All of these methods share a common approach where the target mRNA transcript is fused to RNA structures that can recruit a fluorescent protein while the translated nascent peptide is bound by a different fluorescent molecule such that the mRNA and the nascent peptide can be independently tracked. Importantly, these techniques can provide dual insight into the location of translation and the rate of translation and can even incorporate forced localization to the membrane or potentially another subcellular compartment (Yan et al, 2016). Further development of these technologies into methods that can be applied to more complex systems that contain multiple cell types will uncover how cancer cells and the microenvironmental cells precisely respond to signaling cues to synthesize just the right set of proteins at the right time and the right subcellular location.
Conclusion
Translational control has emerged as a critical nexus integrating upstream oncogenic signals into protein synthesis—the ultimate expression of the cellular proteome. For this reason, the selective control of protein synthesis is increasingly recognized as an important focus of cancer research. The rapidity of translational control provides a first line of action for cancer cells to respond to a myriad of cues including intra‐ and extracellular stressors, whereas transcriptional changes require a longer time scale. Importantly, through the coordination of the translational machinery, RNA‐binding proteins, and 5′ and 3′UTR RNA regulatory elements, translation can precisely, yet pervasively, mediate expression of an array of mRNA networks that are required for sustained tumorigenesis.
Building evidence suggests there is a “translation control code” composed of RNA sequence and structure regulatory elements contained in every mRNA in the cell. Each transcript’s unique set of RNA regulons—the “regulatory syntax” or “grammar”—allow the cell to selectively translate the appropriate genetic program in response to a variety of external and internal signals. Cancer cells can exploit this code to selectively modulate the expression of distinct transcripts that contribute to the hallmarks of cancer. It is already apparent that different RNA motifs often co‐occur within the same 5′UTR, such as the 5′TOP and PRTE motifs (Hsieh et al, 2012). We are only beginning to understand how cancer cells tailor networks of expressed RBPs and levels of translation factors to coordinate the translation of specific and functionally linked mRNAs to engage the genetic programs to respond to stress states, such as nutrient level changes or hypoxia (Ho et al, 2020). The integration of experimental methods to dissect the mechanisms of translational control will shed light on the rules of RNA regulatory element “grammar” that dictate the translational efficiency of a specific transcript.
Translational control also contributes to risk factors and predispositions for cancer formation. The translation machinery has emerged as a critical mediator of the cellular response in a variety of pathological states, for example, the metabolic dysregulation associated with obesity, and inflammation. Obesity is a significant risk factor for cancer development, particularly of the liver, as a result of non‐alcoholic fatty liver disease (NAFLD) (Font‐Burgada et al, 2016). Intriguingly, research has demonstrated that inhibition of eIF4E activity and other translation factors such as eIF6 through genetic or pharmacological means can diminish weight gain on a high nutrient diet, which may result in a reduced risk of cancer development (Moore et al, 2016; Conn et al, 2021; Scagliola et al, 2021). Translational control is also important in the inflammatory response, which is a key consequence of obesity as well as a major risk factor for developing other cancers, such as pancreatic adenocarcinoma (Mazumder et al, 2010; Font‐Burgada et al, 2016; Gameiro & Struhl, 2018; Klein, 2021). Translational control is at the heart of many disease states that can contribute to cancer formation and, therefore, is a potent therapeutic target to not only diminish tumorigenesis but potentially blunt cellular transformation.
Cancer cells hijack specific components of the translation machinery to regulate the synthesis of select proteins that are central to cancer‐specific processes, distinguishing cancer cells from non‐transformed cells. Therefore, compounds that target these translational components or the function of cis‐regulatory elements of key transcripts are attractive therapeutic opportunities. Already, several molecules that target translation factors are in clinical trials. Most prominently are several clinical trials for cancer patients that are testing compounds which inhibit the activity of eIF4E and eIF4A (NCT01675128; NCT03616834; NCT04622007; NCT04092673). Studies have identified small molecules that bind RNA structures in the 5′UTR of oncogenes, such as the Ras proteins, to block translation and promote cancer cell death; however, there has been limited success in translating this approach to the clinic (Katsuda et al, 2016; Miglietta et al, 2017). Additionally, the elongation and termination steps of translation are highly regulated. However, there is scant evidence regarding the mechanisms of selectivity of these processes, although the BAT structural regulon in the 3′UTR provides a window into the possibilities. Compounds targeting the translation elongation factor eEF1A, such as the synthetic ternatin variant dA3, have shown promise in pre‐clinical experiments, and another eEF1A inhibitor, Plitidepsin, has been clinically approved for the treatment of multiple myeloma (Carelli et al, 2015; Keysar et al, 2020). Research supports the robust efficacy of combinational therapies of translation inhibitors with known therapeutic compounds, such as inhibitors of anti‐apoptotic proteins and targeted therapeutics (Anderson et al, 2016; Kuzuoglu‐Ozturk et al, 2021; Thompson et al, 2021). Targeting translation from initiation through termination is an exciting new therapeutic avenue that may help overcome resistance to other anti‐cancer agents, providing more effective treatments.
Disclosure and competing interests statement
The authors declare that they have no conflict of interest.
Acknowledgements
J.R.K. was funded by the American Cancer Society postdoctoral fellowship (133966‐PF‐20‐009‐01‐RMC). D.K.‐O. was funded by a EMBO long‐term fellowship (ALTF 1005‐2015) and a Human Frontier Science Program long‐term fellowship (LT000711/2016‐L). This work was supported by NIH grant no. R35CA242986 (to D.R.) and by the American Cancer Society (RP‐19‐181‐01‐RMC; American Cancer Society Research Professor Award) (to D.R.).
The EMBO Journal (2022) 41: e109823.
This article is part of the Cancer Reviews series.
Contributor Information
Joanna R Kovalski, Email: Joanna.Kovalski@ucsf.edu.
Duygu Kuzuoglu‐Ozturk, Email: duygu.kuzuogluozturk@ucsf.edu.
Davide Ruggero, Email: davide.ruggero@ucsf.edu.
References
- Aharon‐Hefetz N, Frumkin I, Mayshar Y, Dahan O, Pilpel Y, Rak R (2020) Manipulation of the human tRNA pool reveals distinct tRNA sets that act in cellular proliferation or cell cycle arrest. Elife 9: e58461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anastasiadou E, Jacob LS, Slack FJ (2018) Non‐coding RNA networks in cancer. Nat Rev Cancer 18: 5–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson GR, Wardell SE, Cakir M, Crawford L, Leeds JC, Nussbaum DP, Shankar PS, Soderquist RS, Stein EM, Tingley JP et al (2016) PIK3CA mutations enable targeting of a breast tumor dependency through mTOR‐mediated MCL‐1 translation. Sci Transl Med 8: 369ra175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Archer SK, Shirokikh NE, Beilharz TH, Preiss T (2016) Dynamics of ribosome scanning and recycling revealed by translation complex profiling. Nature 535: 570–574 [DOI] [PubMed] [Google Scholar]
- Attar‐Schneider O, Drucker L, Zismanov V, Tartakover‐Matalon S, Lishner M (2014) Targeting eIF4GI translation initiation factor affords an attractive therapeutic strategy in multiple myeloma. Cell Signal 26: 1878–1887 [DOI] [PubMed] [Google Scholar]
- Aviner R, Geiger T, Elroy‐Stein O (2013) Novel proteomic approach (PUNCH‐P) reveals cell cycle‐specific fluctuations in mRNA translation. Gene Dev 27: 1834–1844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badura M, Braunstein S, Zavadil J, Schneider RJ (2012) DNA damage and eIF4G1 in breast cancer cells reprogram translation for survival and DNA repair mRNAs. Proc Natl Acad Sci USA 109: 18767–18772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbieri I, Kouzarides T (2020) Role of RNA modifications in cancer. Nat Rev Cancer 20: 303–322 [DOI] [PubMed] [Google Scholar]
- Bartish M, Tong D, Pan Y, Wallerius M, Liu H, Ristau J, de Souza Ferreira S, Wallmann T, van Hoef V, Masvidal L et al (2020) MNK2 governs the macrophage antiinflammatory phenotype. Proc Natl Acad Sci USA 117: 27556–27565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baughman JM, Rose CM, Kolumam G, Webster JD, Wilkerson EM, Merrill AE, Rhoads TW, Noubade R, Katavolos P, Lesch J et al (2016) NeuCode proteomics reveals Bap1 regulation of metabolism. Cell Rep 16: 583–595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beltran M, Puig I, Peña C, García JM, Álvarez AB, Peña R, Bonilla F, de Herreros AG (2008) A natural antisense transcript regulates Zeb2/Sip1 gene expression during Snail1‐induced epithelial–mesenchymal transition. Gene Dev 22: 756–769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benassayag C, Montero L, Colombié N, Gallant P, Cribbs D, Morello D (2005) Human c‐Myc isoforms differentially regulate cell growth and apoptosis in Drosophila melanogaster . Mol Cell Biol 25: 9897–9909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benisty H, Weber M, Hernandez‐Alias X, Schaefer MH, Serrano L (2020) Mutation bias within oncogene families is related to proliferation‐specific codon usage. Proc Natl Acad Sci USA 117: 30848–30856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi M, Naczki C, Koritzinsky M, Fels D, Blais J, Hu N, Harding H, Novoa I, Varia M, Raleigh J et al (2005) ER stress‐regulated translation increases tolerance to extreme hypoxia and promotes tumor growth. EMBO J 24: 3470–3481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birch J, Clarke CJ, Campbell AD, Campbell K, Mitchell L, Liko D, Kalna G, Strathdee D, Sansom OJ, Neilson M et al (2016) The initiator methionine tRNA drives cell migration and invasion leading to increased metastatic potential in melanoma. Biol Open 5: 1371–1379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blais JD, Addison CL, Edge R, Falls T, Zhao H, Wary K, Koumenis C, Harding HP, Ron D, Holcik M et al (2006) Perk‐dependent translational regulation promotes tumor cell adaptation and angiogenesis in response to hypoxic stress. Mol Cell Biol 26: 9517–9532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohlen J, Fenzl K, Kramer G, Bukau B, Teleman AA (2020) Selective 40S footprinting reveals cap‐tethered ribosome scanning in human cells. Mol Cell 79: 561–574.e5 [DOI] [PubMed] [Google Scholar]
- Bommert KS, Effenberger M, Leich E, Küspert M, Murphy D, Langer C, Moll R, Janz S, Mottok A, Weissbach S et al (2013) The feed‐forward loop between YB‐1 and MYC is essential for multiple myeloma cell survival. Leukemia 27: 441–450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brannan KW, Chaim IA, Marina RJ, Yee BA, Kofman ER, Lorenz DA, Jagannatha P, Dong KD, Madrigal AA, Underwood JG et al (2021) Robust single‐cell discovery of RNA targets of RNA‐binding proteins and ribosomes. Nat Methods 18: 507–519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brar GA, Weissman JS (2015) Ribosome profiling reveals the what, when, where and how of protein synthesis. Nat Rev Mol Cell Biol 16: 651–664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braunstein S, Karpisheva K, Pola C, Goldberg J, Hochman T, Yee H, Cangiarella J, Arju R, Formenti SC, Schneider RJ (2007) A hypoxia‐controlled cap‐dependent to cap‐independent translation switch in breast cancer. Mol Cell 28: 501–512 [DOI] [PubMed] [Google Scholar]
- Brown CY, Mize GJ, Pineda M, George DL, Morris DR (1999) Role of two upstream open reading frames in the translational control of oncogene mdm2. Oncogene 18: 5631–5637 [DOI] [PubMed] [Google Scholar]
- Buccitelli C, Selbach M (2020) mRNAs, proteins and the emerging principles of gene expression control. Nat Rev Genet 21: 630–644 [DOI] [PubMed] [Google Scholar]
- Budnik B, Levy E, Harmange G, Slavov N (2018) SCoPE‐MS: mass spectrometry of single mammalian cells quantifies proteome heterogeneity during cell differentiation. Genome Biol 19: 161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns DM, Richter JD (2008) CPEB regulation of human cellular senescence, energy metabolism, and p53 mRNA translation. Gene Dev 22: 3449–3460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bushell M, Stoneley M, Kong YW, Hamilton TL, Spriggs KA, Dobbyn HC, Qin X, Sarnow P, Willis AE (2006) Polypyrimidine tract binding protein regulates IRES‐mediated gene expression during apoptosis. Mol Cell 23: 401–412 [DOI] [PubMed] [Google Scholar]
- Byeon GW, Cenik ES, Jiang L, Tang H, Das R, Barna M (2021) Functional and structural basis of extreme conservation in vertebrate 5′ untranslated regions. Nat Genet 53: 729–741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calve S, Witten AJ, Ocken AR, Kinzer‐Ursem TL (2016) Incorporation of non‐canonical amino acids into the developing murine proteome. Sci Rep 6: 32377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calviello L, Venkataramanan S, Rogowski KJ, Wyler E, Wilkins K, Tejura M, Thai B, Krol J, Filipowicz W, Landthaler M et al (2021) DDX3 depletion represses translation of mRNAs with complex 5′ UTRs. Nucleic Acids Res 49: 5336–5350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvo SE, Pagliarini DJ, Mootha VK (2009) Upstream open reading frames cause widespread reduction of protein expression and are polymorphic among humans. Proc Natl Acad Sci USA 106: 7507–7512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caraglia M, Park MH, Wolff EC, Marra M, Abbruzzese A (2013) eIF5A isoforms and cancer: two brothers for two functions? Amino Acids 44: 103–109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carelli JD, Sethofer SG, Smith GA, Miller HR, Simard JL, Merrick WC, Jain RK, Ross NT, Taunton J (2015) Ternatin and improved synthetic variants kill cancer cells by targeting the elongation factor‐1A ternary complex. Elife 4: e10222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cencic R, Desforges M, Hall DR, Kozakov D, Du Y, Min J, Dingledine R, Fu H, Vajda S, Talbot PJ et al (2011a) Blocking eIF4E‐eIF4G interaction as a strategy to impair coronavirus replication. J Virol 85: 6381–6389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cencic R, Hall DR, Robert F, Du Y, Min J, Li L, Qui M, Lewis I, Kurtkaya S, Dingledine R et al (2011b) Reversing chemoresistance by small molecule inhibition of the translation initiation complex eIF4F. Proc Natl Acad Sci USA 108: 1046–1051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan K, Robert F, Oertlin C, Kapeller‐Libermann D, Avizonis D, Gutierrez J, Handly‐Santana A, Doubrovin M, Park J, Schoepfer C et al (2019) eIF4A supports an oncogenic translation program in pancreatic ductal adenocarcinoma. Nat Commun 10: 5151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chappell SA, Edelman GM, Mauro VP (2000) A 9‐nt segment of a cellular mRNA can function as an internal ribosome entry site (IRES) and when present in linked multiple copies greatly enhances IRES activity. Proc Natl Acad Sci USA 97: 1536–1541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chappell SA, Edelman GM, Mauro VP (2004) Biochemical and functional analysis of a 9‐nt RNA sequence that affects translation efficiency in eukaryotic cells. Proc Natl Acad Sci USA 101: 9590–9594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhury A, Cheema S, Fachini JM, Kongchan N, Lu G, Simon LM, Wang T, Mao S, Rosen DG, Ittmann MM et al (2016) CELF1 is a central node in post‐transcriptional regulatory programmes underlying EMT. Nat Commun 7: 13362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhury A, Hussey GS, Ray PS, Jin G, Fox PL, Howe PH (2010) TGF‐β‐mediated phosphorylation of hnRNP E1 induces EMT via transcript‐selective translational induction of Dab2 and ILEI. Nat Cell Biol 12: 286–293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Aktas BH, Wang Y, He X, Sahoo R, Zhang N, Denoyelle S, Kabha E, Yang H, Yefidoff Freedman R et al (2012) Tumor suppression by small molecule inhibitors of translation initiation. Oncotarget 3: 869–881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Child SJ, Miller MK, Geballe AP (1999) Translational control by an upstream open reading frame in the HER‐2/neu transcript*. J Biol Chem 274: 24335–24341 [DOI] [PubMed] [Google Scholar]
- Choe J, Lin S, Zhang W, Liu QI, Wang L, Ramirez‐Moya J, Du P, Kim W, Tang S, Sliz P et al (2018) mRNA circularization by METTL3–eIF3h enhances translation and promotes oncogenesis. Nature 561: 556–560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi J, Indrisiunaite G, DeMirci H, Ieong K‐W, Wang J, Petrov A, Prabhakar A, Rechavi G, Dominissini D, He C et al (2018) 2′‐O‐methylation in mRNA disrupts tRNA decoding during translation elongation. Nat Struct Mol Biol 25: 208–216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu C, Zhang QC, da Rocha ST, Flynn RA, Bharadwaj M, Calabrese JM, Magnuson T, Heard E, Chang HY (2015) Systematic discovery of Xist RNA binding proteins. Cell 161: 404–416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cieśla M, Ngoc PCT, Cordero E, Martinez ÁS, Morsing M, Muthukumar S, Beneventi G, Madej M, Munita R, Jönsson T et al (2021) Oncogenic translation directs spliceosome dynamics revealing an integral role for SF3A3 in breast cancer. Mol Cell 81: 1453–1468.e12 [DOI] [PubMed] [Google Scholar]
- Clarke C, Berg T, Birch J, Ennis D, Mitchell L, Cloix C, Campbell A, Sumpton D, Nixon C, Campbell K et al (2016) The initiator methionine tRNA drives secretion of type II collagen from stromal fibroblasts to promote tumor growth and angiogenesis. Curr Biol 26: 755–765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clement PMJ, Henderson CA, Jenkins ZA, Smit‐McBride Z, Wolff EC, Hershey JWB, Park MH, Johansson HE (2003) Identification and characterization of eukaryotic initiation factor 5A–2. Eur J Biochem 270: 4254–4263 [DOI] [PubMed] [Google Scholar]
- Cobbold LC, Spriggs KA, Haines SJ, Dobbyn HC, Hayes C, de Moor CH, Lilley KS, Bushell M, Willis AE (2008) Identification of internal ribosome entry segment (IRES)‐trans‐acting factors for the Myc family of iress. Mol Cell Biol 28: 40–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cobbold LC, Wilson LA, Sawicka K, King HA, Kondrashov AV, Spriggs KA, Bushell M, Willis AE (2010) Upregulated c‐myc expression in multiple myeloma by internal ribosome entry results from increased interactions with and expression of PTB‐1 and YB‐1. Oncogene 29: 2884–2891 [DOI] [PubMed] [Google Scholar]
- Coldwell MJ, Mitchell SA, Stoneley M, MacFarlane M, Willis AE (2000) Initiation of Apaf‐1 translation by internal ribosome entry. Oncogene 19: 899–905 [DOI] [PubMed] [Google Scholar]
- Comtesse N, Keller A, Diesinger I, Bauer C, Kayser K, Huwer H, Lenhof H, Meese E (2007) Frequent overexpression of the genes FXR1, CLAPM1 and EIF4G located on amplicon 3q26‐27 in squamous cell carcinoma of the lung. Int J Cancer 120: 2538–2544 [DOI] [PubMed] [Google Scholar]
- Conn CS, Yang H, Tom HJ, Ikeda K, Oses‐Prieto JA, Vu H, Oguri Y, Nair S, Gill RM, Kajimura S et al (2021) The major cap‐binding protein eIF4E regulates lipid homeostasis and diet‐induced obesity. Nat Metab 3: 244–257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cottrell KA, Chiou RC, Weber JD (2020) Upregulation of 5′‐terminal oligopyrimidine mRNA translation upon loss of the ARF tumor suppressor. Sci Rep 10: 22276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham JT, Moreno MV, Lodi A, Ronen SM, Ruggero D (2014) Protein and nucleotide biosynthesis are coupled by a single rate‐limiting enzyme, PRPS2, to drive cancer. Cell 157: 1088–1103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daba A, Koromilas AE, Pantopoulos K (2012) Alternative ferritin mRNA translation via internal initiation. RNA 18: 547–556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dever TE, Green R (2012) The elongation, termination, and recycling phases of translation in eukaryotes. Cold Spring Harb Perspect Biol 4: a013706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dieterich DC, Hodas JJL, Gouzer G, Shadrin IY, Ngo JT, Triller A, Tirrell DA, Schuman EM (2010) In situ visualization and dynamics of newly synthesized proteins in rat hippocampal neurons. Nat Neurosci 13: 897–905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dieterich DC, Link AJ, Graumann J, Tirrell DA, Schuman EM (2006) Selective identification of newly synthesized proteins in mammalian cells using bioorthogonal noncanonical amino acid tagging (BONCAT). Proc Natl Acad Sci USA 103: 9482–9487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dittmar KA, Goodenbour JM, Pan T (2006) Tissue‐specific differences in human transfer RNA expression. Plos Genet 2: e221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doherty MK, Hammond DE, Clague MJ, Gaskell SJ, Beynon RJ (2009) Turnover of the human proteome: determination of protein intracellular stability by dynamic SILAC. J Proteome Res 8: 104–112 [DOI] [PubMed] [Google Scholar]
- Elfakess R, Dikstein R (2008) A translation initiation element specific to mRNAs with very short 5′UTR that also regulates transcription. PLoS One 3: e3094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elfakess R, Sinvani H, Haimov O, Svitkin Y, Sonenberg N, Dikstein R (2011) Unique translation initiation of mRNAs‐containing TISU element. Nucleic Acids Res 39: 7598–7609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliott TS, Townsley FM, Bianco A, Ernst RJ, Sachdeva A, Elsässer SJ, Davis L, Lang K, Pisa R, Greiss S et al (2014) Proteome labeling and protein identification in specific tissues and at specific developmental stages in an animal. Nat Biotechnol 32: 465–472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans HT, Benetatos J, van Roijen M, Bodea L, Götz J (2019) Decreased synthesis of ribosomal proteins in tauopathy revealed by non‐canonical amino acid labelling. EMBO J 38: e101174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evdokimova V, Tognon C, Ng T, Ruzanov P, Melnyk N, Fink D, Sorokin A, Ovchinnikov LP, Davicioni E, Triche TJ et al (2009) Translational activation of Snail1 and other developmentally regulated transcription factors by YB‐1 promotes an epithelial‐mesenchymal transition. Cancer Cell 15: 402–415 [DOI] [PubMed] [Google Scholar]
- Falletta P, Sanchez‐del‐Campo L, Chauhan J, Effern M, Kenyon A, Kershaw CJ, Siddaway R, Lisle R, Freter R, Daniels MJ et al (2017) Translation reprogramming is an evolutionarily conserved driver of phenotypic plasticity and therapeutic resistance in melanoma. Gene Dev 31: 18–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felton‐Edkins ZA, Fairley JA, Graham EL, Johnston IM, White RJ, Scott PH (2003) The mitogen‐activated protein (MAP) kinase ERK induces tRNA synthesis by phosphorylating TFIIIB. EMBO J 22: 2422–2432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández‐Miranda G, Méndez R (2012) The CPEB‐family of proteins, translational control in senescence and cancer. Ageing Res Rev 11: 460–472 [DOI] [PubMed] [Google Scholar]
- Fonseca BD, Zakaria C, Jia J‐J, Graber TE, Svitkin Y, Tahmasebi S, Healy D, Hoang H‐D, Jensen JM, Diao IT et al (2015) La‐related protein 1 (LARP1) represses terminal oligopyrimidine (TOP) mRNA translation downstream of mTOR complex 1 (mTORC1)*. J Biol Chem 290: 15996–16020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Font‐Burgada J, Sun B, Karin M (2016) Obesity and cancer: the oil that feeds the flame. Cell Metab 23: 48–62 [DOI] [PubMed] [Google Scholar]
- Forester CM, Zhao Q, Phillips NJ, Urisman A, Chalkley RJ, Oses‐Prieto JA, Zhang L, Ruggero D, Burlingame AL (2018) Revealing nascent proteomics in signaling pathways and cell differentiation. Proc Natl Acad Sci USA 115: 201707514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu J, Dang Y, Counter C, Liu Y (2018) Codon usage regulates human KRAS expression at both transcriptional and translational levels. J Biol Chem 293: 17929–17940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii K, Zhulyn O, Byeon GW, Genuth NR, Kerr CH, Walsh EM, Barna M (2021) Controlling tissue patterning by translational regulation of signaling transcripts through the core translation factor eIF3c. Dev Cell 56: 2928–2937.e9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furic L, Rong L, Larsson O, Koumakpayi IH, Yoshida K, Brueschke A, Petroulakis E, Robichaud N, Pollak M, Gaboury LA et al (2010) eIF4E phosphorylation promotes tumorigenesis and is associated with prostate cancer progression. Proc Natl Acad Sci USA 107: 14134–14139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gameiro PA, Struhl K (2018) Nutrient deprivation elicits a transcriptional and translational inflammatory response coupled to decreased protein synthesis. Cell Rep 24: 1415–1424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gandin V, Masvidal L, Hulea L, Gravel S‐P, Cargnello M, McLaughlan S, Cai Y, Balanathan P, Morita M, Rajakumar A et al (2016) nanoCAGE reveals 5′ UTR features that define specific modes of translation of functionally related MTOR‐sensitive mRNAs. Genome Res 26: 636–648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gemmill D, D’souza S, Meier‐Stephenson V, Patel TR (2020) Current approaches for RNA‐labelling to identify RNA‐binding proteins1. Biochem Cell Biol 98: 31–41 [DOI] [PubMed] [Google Scholar]
- Gerson‐Gurwitz A, Young NP, Goel VK, Eam B, Stumpf CR, Chen J, Fish S, Barrera M, Sung E, Staunton J et al (2021) Zotatifin, an eIF4A‐selective inhibitor, blocks tumor growth in receptor tyrosine kinase driven tumors. Front Oncol 11: 766298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gingold H, Tehler D, Christoffersen N, Nielsen M, Asmar F, Kooistra S, Christophersen N, Christensen LL, Borre M, Sørensen K et al (2014) A dual program for translation regulation in cellular proliferation and differentiation. Cell 158: 1281–1292 [DOI] [PubMed] [Google Scholar]
- Gomez‐Roman N, Grandori C, Eisenman RN, White RJ (2003) Direct activation of RNA polymerase III transcription by c‐Myc. Nature 421: 290–294 [DOI] [PubMed] [Google Scholar]
- Goodall GJ, Wickramasinghe VO (2021) RNA in cancer. Nat Rev Cancer 21: 22–36 [DOI] [PubMed] [Google Scholar]
- Goodarzi H, Liu X, Nguyen HCB, Zhang S, Fish L, Tavazoie SF (2015) Endogenous tRNA‐derived fragments suppress breast cancer progression via YBX1 displacement. Cell 161: 790–802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodarzi H, Nguyen HCB, Zhang S, Dill BD, Molina H, Tavazoie SF (2016) Modulated expression of specific tRNAs drives gene expression and cancer progression. Cell 165: 1416–1427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregio APB, Cano VPS, Avaca JS, Valentini SR, Zanelli CF (2009) eIF5A has a function in the elongation step of translation in yeast. Biochem Biophys Res Commun 380: 785–790 [DOI] [PubMed] [Google Scholar]
- Gutierrez E, Shin B‐S, Woolstenhulme CJ, Kim J‐R, Saini P, Buskirk AR, Dever TE (2013) eIF5A promotes translation of polyproline motifs. Mol Cell 51: 35–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hafner M, Katsantoni M, Köster T, Marks J, Mukherjee J, Staiger D, Ule J, Zavolan M (2021) CLIP and complementary methods. Nat Rev Methods Primers 1: 20 [Google Scholar]
- Hafner M, Landthaler M, Burger L, Khorshid M, Hausser J, Berninger P, Rothballer A, Ascano M, Jungkamp A‐C, Munschauer M et al (2010) Transcriptome‐wide identification of RNA‐binding protein and microRNA target sites by PAR‐CLIP. Cell 141: 129–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haghighat A, Mader S, Pause A, Sonenberg N (1995) Repression of cap‐dependent translation by 4E‐binding protein 1: competition with p220 for binding to eukaryotic initiation factor‐4E. EMBO J 14: 5701–5709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haimov O, Sinvani H, Martin F, Ulitsky I, Emmanuel R, Tamarkin‐Ben‐Harush A, Vardy A, Dikstein R (2017) Efficient and accurate translation initiation directed by TISU involves RPS3 and RPS10e binding and differential eukaryotic initiation factor 1A regulation. Mol Cell Biol 37: e00150–e217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han S, Zhao BS, Myers SA, Carr SA, He C, Ting AY (2020) RNA–protein interaction mapping via MS2‐ or Cas13‐based APEX targeting. Proc Natl Acad Sci USA 117: 22068–22079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA (2011) Hallmarks of cancer: the next generation. Cell 144: 646–674 [DOI] [PubMed] [Google Scholar]
- Hann SR, Dixit M, Sears RC, Sealy L (1994) The alternatively initiated c‐Myc proteins differentially regulate transcription through a noncanonical DNA‐binding site. Gene Dev 8: 2441–2452 [DOI] [PubMed] [Google Scholar]
- Hann SR, King MW, Bentley DL, Anderson CW, Eisenman RN (1988) A non‐AUG translational initiation in c‐myc exon 1 generates an N‐terminally distinct protein whose synthesis is disrupted in Burkitt’s lymphomas. Cell 52: 185–195 [DOI] [PubMed] [Google Scholar]
- Harvey RF, Smith TS, Mulroney T, Queiroz RML, Pizzinga M, Dezi V, Villenueva E, Ramakrishna M, Lilley KS, Willis AE (2018) Trans‐acting translational regulatory RNA binding proteins. Wiley Interdiscip Rev RNA 9: e1465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto S, Furukawa S, Hashimoto A, Tsutaho A, Fukao A, Sakamura Y, Parajuli G, Onodera Y, Otsuka Y, Handa H et al (2019) ARF6 and AMAP1 are major targets of KRAS and TP53 mutations to promote invasion, PD‐L1 dynamics, and immune evasion of pancreatic cancer. Proc Natl Acad Sci USA 116: 17450–17459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He PC, He C (2021) m6A RNA methylation: from mechanisms to therapeutic potential. EMBO J 40: e105977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hershey JWB (2015) The role of eIF3 and its individual subunits in cancer. Biochim Biophys Acta BBA ‐ Gene Regul Mech 1849: 792–800 [DOI] [PubMed] [Google Scholar]
- Hinnebusch AG, Ivanov IP, Sonenberg N (2016) Translational control by 5′‐untranslated regions of eukaryotic mRNAs. Science 352: 1413–1416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho JJD, Balukoff NC, Theodoridis PR, Wang M, Krieger JR, Schatz JH, Lee S (2020) A network of RNA‐binding proteins controls translation efficiency to activate anaerobic metabolism. Nat Commun 11: 2677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogg JR, Collins K (2007) RNA‐based affinity purification reveals 7SK RNPs with distinct composition and regulation. RNA 13: 868–880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holcik M, Lefebvre C, Yeh C, Chow T, Korneluk RG (1999) A new internal‐ribosome‐entry‐site motif potentiates XIAP‐ mediated cytoprotection. Nat Cell Biol 1: 190–192 [DOI] [PubMed] [Google Scholar]
- Holmes B, Lee J, Landon KA, Benavides‐Serrato A, Bashir T, Jung ME, Lichtenstein A, Gera J (2016) Mechanistic target of rapamycin (mTOR) inhibition synergizes with reduced internal ribosome entry site (IRES)‐mediated translation of cyclin D1 and c‐MYC mRNAs to treat glioblastoma*. J Biol Chem 291: 14146–14159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopkins BD, Fine B, Steinbach N, Dendy M, Rapp Z, Shaw J, Pappas K, Yu JS, Hodakoski C, Mense S et al (2013) A secreted PTEN phosphatase that enters cells to alter signaling and survival. Science 341: 399–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopkins TG, Mura M, Al‐Ashtal HA, Lahr RM, Abd‐Latip N, Sweeney K, Lu H, Weir J, El‐Bahrawy M, Steel JH et al (2016) The RNA‐binding protein LARP1 is a post‐transcriptional regulator of survival and tumorigenesis in ovarian cancer. Nucleic Acids Res 44: 1227–1246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh AC, Liu YI, Edlind MP, Ingolia NT, Janes MR, Sher A, Shi EY, Stumpf CR, Christensen C, Bonham MJ et al (2012) The translational landscape of mTOR signalling steers cancer initiation and metastasis. Nature 485: 55–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu MC‐Y, Tranque P, Edelman GM, Mauro VP (1999) rRNA‐complementarity in the 5′ untranslated region of mRNA specifying the Gtx homeodomain protein: evidence that base‐ pairing to 18S rRNA affects translational efficiency. Proc Natl Acad Sci USA 96: 1339–1344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hundsdoerfer P, Thoma C, Hentze MW (2005) Eukaryotic translation initiation factor 4GI and p97 promote cellular internal ribosome entry sequence‐driven translation. Proc Natl Acad Sci USA 102: 13421–13426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussey GS, Chaudhury A, Dawson AE, Lindner DJ, Knudsen CR, Wilce MCJ, Merrick WC, Howe PH (2011) Identification of an mRNP complex regulating tumorigenesis at the translational elongation step. Mol Cell 41: 419–431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iadevaia V, Caldarola S, Tino E, Amaldi F, Loreni F (2008) All translation elongation factors and the e, f, and h subunits of translation initiation factor 3 are encoded by 5′‐terminal oligopyrimidine (TOP) mRNAs. RNA 14: 1730–1736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingolia NT, Ghaemmaghami S, Newman JRS, Weissman JS (2009) Genome‐wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science 324: 218–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingolia NT, Hussmann JA, Weissman JS (2018) Ribosome profiling: global views of translation. Csh Perspect Biol 11: a032698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingolia NT, Lareau LF, Weissman JS (2011) Ribosome profiling of mouse embryonic stem cells reveals the complexity and dynamics of mammalian proteomes. Cell 147: 789–802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov IP, Shin B‐S, Loughran G, Tzani I, Young‐Baird SK, Cao C, Atkins JF, Dever TE (2018) Polyamine control of translation elongation regulates start site selection on antizyme inhibitor mRNA via ribosome queuing. Mol Cell 70: 254–264.e6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson RJ, Hellen CUT, Pestova TV (2010) The mechanism of eukaryotic translation initiation and principles of its regulation. Nat Rev Mol Cell Biol 11: 113–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaiswal PK, Koul S, Shanmugam PST, Koul HK (2018) Eukaryotic translation initiation factor 4 gamma 1 (eIF4G1) is upregulated during prostate cancer progression and modulates cell growth and metastasis. Sci Rep 8: 7459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jan CH, Williams CC, Weissman JS (2014) Principles of ER cotranslational translocation revealed by proximity‐specific ribosome profiling. Science 346: 1257521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferies HB, Reinhard C, Kozma SC, Thomas G (1994) Rapamycin selectively represses translation of the “polypyrimidine tract” mRNA family. Proc Natl Acad Sci USA 91: 4441–4445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins RH, Bennagi R, Martin J, Phillips AO, Redman JE, Fraser DJ (2010) A conserved stem loop motif in the 5′untranslated region regulates transforming growth factor‐β1 translation. PLoS One 5: e12283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins ZA, Hååg PG, Johansson HE (2001) Human EIF5A2 on chromosome 3q25–q27 is a phylogenetically conserved vertebrate variant of eukaryotic translation initiation factor 5A with tissue‐specific expression. Genomics 71: 101–109 [DOI] [PubMed] [Google Scholar]
- Jensen KB, Dredge BK, Toubia J, Jin X, Iadevaia V, Goodall GJ, Proud CG (2021) capCLIP: a new tool to probe translational control in human cells through capture and identification of the eIF4E–mRNA interactome. Nucleic Acids Res 49: e105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jewer M, Lee L, Leibovitch M, Zhang G, Liu J, Findlay SD, Vincent KM, Tandoc K, Dieters‐Castator D, Quail DF et al (2020) Translational control of breast cancer plasticity. Nat Commun 11: 2498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji P, Diederichs S, Wang W, Böing S, Metzger R, Schneider PM, Tidow N, Brandt B, Buerger H, Bulk E et al (2003) MALAT‐1, a novel noncoding RNA, and thymosin β4 predict metastasis and survival in early‐stage non‐small cell lung cancer. Oncogene 22: 8031–8041 [DOI] [PubMed] [Google Scholar]
- Jia J‐J, Lahr RM, Solgaard MT, Moraes BJ, Pointet R, Yang A‐D, Celucci G, Graber TE, Hoang H‐D, Niklaus M et al (2021) mTORC1 promotes TOP mRNA translation through site‐specific phosphorylation of LARP1. Nucleic Acids Res 49: 3461–3489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin D, Guo J, Wu Y, Du J, Yang L, Wang X, Di W, Hu B, An J, Kong L et al (2019) m6A mRNA methylation initiated by METTL3 directly promotes YAP translation and increases YAP activity by regulating the MALAT1‐miR‐1914‐3p‐YAP axis to induce NSCLC drug resistance and metastasis. J Hematol Oncol 12: 135 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Jin H, Xu W, Rahman R, Na D, Fieldsend A, Song W, Liu S, Li C, Rosbash M (2020) TRIBE editing reveals specific mRNA targets of eIF4E‐BP in Drosophila and in mammals. Sci Adv 6: eabb8771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones RM, Branda J, Johnston KA, Polymenis M, Gadd M, Rustgi A, Callanan L, Schmidt EV (1996) An essential E box in the promoter of the gene encoding the mRNA cap‐binding protein (eukaryotic initiation factor 4E) is a target for activation by c‐myc. Mol Cell Biol 16: 4754–4764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kantidakis T, Ramsbottom BA, Birch JL, Dowding SN, White RJ (2010) mTOR associates with TFIIIC, is found at tRNA and 5S rRNA genes, and targets their repressor Maf1. Proc Natl Acad Sci USA 107: 11823–11828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsuda Y, Sato S, Asano L, Morimura Y, Furuta T, Sugiyama H, Hagihara M, Uesugi M (2016) A Small molecule that represses translation of G‐quadruplex‐containing mRNA. J Am Chem Soc 138: 9037–9040 [DOI] [PubMed] [Google Scholar]
- Kawai T, Fan J, Mazan‐Mamczarz K, Gorospe M (2004) Global mRNA stabilization preferentially linked to translational repression during the endoplasmic reticulum stress response. Mol Cell Biol 24: 6773–6787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keysar SB, Gomes N, Miller B, Jackson BC, Le PN, Morton JJ, Reisinger J, Chimed T‐S, Gomez KE, Nieto C et al (2020) Inhibiting translation elongation with SVC112 suppresses cancer stem cells and inhibits growth in head and neck squamous carcinoma. Cancer Res 80: 1183–1198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim B, Kim VN (2019) fCLIP‐seq for transcriptomic footprinting of dsRNA‐binding proteins: lessons from DROSHA. Methods 152: 3–11 [DOI] [PubMed] [Google Scholar]
- Kim JK, Cho J, Kim SH, Kang H‐C, Kim D‐S, Kim VN, Lee JH (2019) Brain somatic mutations in MTOR reveal translational dysregulations underlying intractable focal epilepsy. J Clin Invest 129: 4207–4223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SJ, Park K, Koeller D, Kim KY, Wakefield LM, Sporn MB, Roberts AB (1992) Post‐transcriptional regulation of the human transforming growth factor‐beta 1 gene. J Biol Chem 267: 13702–13707 [PubMed] [Google Scholar]
- Kladwang W, VanLang CC, Cordero P, Das R (2011) A two‐dimensional mutate‐and‐map strategy for non‐coding RNA structure. Nat Chem 3: 954–962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klann K, Tascher G, Münch C (2020) Functional translatome proteomics reveal converging and dose‐dependent regulation by mTORC1 and eIF2α. Mol Cell 77: 913–925.e4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein AP (2021) Pancreatic cancer epidemiology: understanding the role of lifestyle and inherited risk factors. Nat Rev Gastroenterol 18: 493–502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight JRP, Garland G, Pöyry T, Mead E, Vlahov N, Sfakianos A, Grosso S, De‐Lima‐Hedayioglu F, Mallucci GR, von der Haar T et al (2020) Control of translation elongation in health and disease. Dis Model Mech 13: dmm043208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong T, Xue Y, Cencic R, Zhu X, Monast A, Fu Z, Pilon V, Sangwan V, Guiot M‐C, Foulkes WD et al (2019) eIF4A inhibitors suppress cell‐cycle feedback response and acquired resistance to CDK4/6 inhibition in cancer. Mol Cancer Ther 18: 2158–2170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konicek BW, Stephens JR, McNulty AM, Robichaud N, Peery RB, Dumstorf CA, Dowless MS, Iversen PW, Parsons S, Ellis KE et al (2011) Therapeutic inhibition of MAP kinase interacting kinase blocks eukaryotic initiation factor 4E phosphorylation and suppresses outgrowth of experimental lung metastases. Can Res 71: 1849–1857 [DOI] [PubMed] [Google Scholar]
- Konig J, Zarnack K, Rot G, Curk T, Kayikci M, Zupan B, Turner DJ, Luscombe NM, Ule J (2011) iCLIP ‐ transcriptome‐wide mapping of protein‐RNA interactions with individual nucleotide resolution. J vis Exp 50: 2638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M (1987) An analysis of 5’‐noncoding sequences from 699 vertebrate messenger RNAs. Nucleic Acids Res 15: 8125–8148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuzuoglu‐Ozturk D, Hu Z, Rama M, Devericks E, Weiss J, Chiang GG, Worland ST, Brenner SE, Goodarzi H, Gilbert LA et al (2021) Revealing molecular pathways for cancer cell fitness through a genetic screen of the cancer translatome. Cell Rep 35: 109321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahr RM, Fonseca BD, Ciotti GE, Al‐Ashtal HA, Jia J‐J, Niklaus MR, Blagden SP, Alain T, Berman AJ (2017) La‐related protein 1 (LARP1) binds the mRNA cap, blocking eIF4F assembly on TOP mRNAs. Elife 6: e24146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lampson BL, Pershing NLK, Prinz JA, Lacsina JR, Marzluff WF, Nicchitta CV, MacAlpine DM, Counter CM (2013) Rare codons regulate KRas oncogenesis. Curr Biol 23: 70–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang KJD, Kappel A, Goodall GJ (2002) Hypoxia‐inducible factor‐1α mRNA contains an internal ribosome entry site that allows efficient translation during normoxia and hypoxia. Mol Biol Cell 13: 1792–1801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawson MR, Lessen LN, Wang J, Prabhakar A, Corsepius NC, Green R, Puglisi JD (2021) Mechanisms that ensure speed and fidelity in eukaryotic translation termination. Science 373: 876–882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee ASY, Kranzusch PJ, Cate JHD (2015) eIF3 targets cell‐proliferation messenger RNAs for translational activation or repression. Nature 522: 111–114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee ASY, Kranzusch PJ, Doudna JA, Cate JHD (2016) eIF3d is an mRNA cap‐binding protein that is required for specialized translation initiation. Nature 536: 96–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leppek K, Das R, Barna M (2018) Functional 5′ UTR mRNA structures in eukaryotic translation regulation and how to find them. Nat Rev Mol Cell Biol 19: 158–174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leppek K, Stoecklin G (2014) An optimized streptavidin‐binding RNA aptamer for purification of ribonucleoprotein complexes identifies novel ARE‐binding proteins. Nucleic Acids Res 42: e13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy S, Avni D, Hariharan N, Perry RP, Meyuhas O (1991) Oligopyrimidine tract at the 5′ end of mammalian ribosomal protein mRNAs is required for their translational control. Proc Natl Acad Sci USA 88: 3319–3323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Luo Q, Xie Z, Li G, Mao C, Liu YI, Wen X, Yin NA, Cao J, Wang J et al (2016) Characterization of the expression of the RNA binding protein eIF4G1 and its clinicopathological correlation with serous ovarian cancer. PLoS One 11: e0163447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang H, Chen X, Yin Q, Ruan D, Zhao X, Zhang C, McNutt MA, Yin Y (2017) PTENβ is an alternatively translated isoform of PTEN that regulates rDNA transcription. Nat Commun 8: 14771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang H, He S, Yang J, Jia X, Wang P, Chen XI, Zhang Z, Zou X, McNutt M, Shen W et al (2014a) PTENα, a PTEN isoform translated through alternative initiation, regulates mitochondrial function and energy metabolism. Cell Metab 19: 836–848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang S, Zhou Y, Chen Y, Ke G, Wen H, Wu X (2014b) Decreased expression of EIF4A1 after preoperative brachytherapy predicts better tumor‐specific survival in cervical cancer. Int J Gynecol Cancer 24: 908–915 [DOI] [PubMed] [Google Scholar]
- Licatalosi DD, Mele A, Fak JJ, Ule J, Kayikci M, Chi SW, Clark TA, Schweitzer AC, Blume JE, Wang X et al (2008) HITS‐CLIP yields genome‐wide insights into brain alternative RNA processing. Nature 456: 464–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C‐J, Cencic R, Mills JR, Robert F, Pelletier J (2008) c‐Myc and eIF4F are components of a feedforward loop that links transcription and translation. Cancer Res 68: 5326–5334 [DOI] [PubMed] [Google Scholar]
- Lin S, Choe J, Du P, Triboulet R, Gregory RI (2016) The m6A methyltransferase METTL3 promotes translation in human cancer cells. Mol Cell 62: 335–345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Xu Y, Stoleru D, Salic A (2012) Imaging protein synthesis in cells and tissues with an alkyne analog of puromycin. Proc Natl Acad Sci USA 109: 413–418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Dilworth D, Gao L, Monzon J, Summers A, Lassam N, Hogg D (1999) Mutation of the CDKN2A 5′ UTR creates an aberrant initiation codon and predisposes to melanoma. Nat Genet 21: 128–132 [DOI] [PubMed] [Google Scholar]
- Liu Y, Beyer A, Aebersold R (2016) On the dependency of cellular protein levels on mRNA abundance. Cell 165: 535–550 [DOI] [PubMed] [Google Scholar]
- Liwak U, Thakor N, Jordan LE, Roy R, Lewis SM, Pardo OE, Seckl M, Holcik M (2012) Tumor suppressor PDCD4 represses internal ribosome entry site‐mediated translation of antiapoptotic proteins and is regulated by S6 kinase 2. Mol Cell Biol 32: 1818–1829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y, McClatchy DB, Barkallah S, Wood WW, Yates JR (2017) HILAQ: a novel strategy for newly synthesized protein quantification. J Proteome Res 16: 2213–2220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mailler E, Paillart J, Marquet R, Smyth RP, Vivet‐Boudou V (2019) The evolution of RNA structural probing methods: from gels to next‐generation sequencing. Wiley Interdiscip Rev RNA 10: e1518 [DOI] [PubMed] [Google Scholar]
- Malaney P, Uversky VN, Davé V (2017) PTEN proteoforms in biology and disease. Cell Mol Life Sci 74: 2783–2794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manjunath H, Zhang H, Rehfeld F, Han J, Chang T‐C, Mendell JT (2019) Suppression of ribosomal pausing by eIF5A is necessary to maintain the fidelity of start codon selection. Cell Rep 29: 3134–3146.e6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marash L, Liberman N, Henis‐Korenblit S, Sivan G, Reem E, Elroy‐Stein O, Kimchi A (2008) DAP5 promotes cap‐independent translation of Bcl‐2 and CDK1 to facilitate cell survival during mitosis. Mol Cell 30: 447–459 [DOI] [PubMed] [Google Scholar]
- Mardakheh FK, Paul A, Kümper S, Sadok A, Paterson H, Mccarthy A, Yuan Y, Marshall CJ (2015) Global analysis of mRNA, translation, and protein localization: local translation is a key regulator of cell protrusions. Dev Cell 35: 344–357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marx V (2019) A dream of single‐cell proteomics. Nat Methods 16: 809–812 [DOI] [PubMed] [Google Scholar]
- Mathews MB, Hershey JWB (2015) The translation factor eIF5A and human cancer. Biochem Biophys Acta 1849: 836–844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matia‐González AM, Iadevaia V, Gerber AP (2017) A versatile tandem RNA isolation procedure to capture in vivo formed mRNA‐protein complexes. Methods 118: 93–100 [DOI] [PubMed] [Google Scholar]
- Mazumder B, Li X, Barik S (2010) Translation control: a multifaceted regulator of inflammatory response. J Immunol 184: 3311–3319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGlincy NJ, Ingolia NT (2017) Transcriptome‐wide measurement of translation by ribosome profiling. Methods 126: 112–129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McHugh CA, Guttman M (2018) RNA detection, methods and protocols. Methods Mol Biol 1649: 473–488 [DOI] [PubMed] [Google Scholar]
- Mehta A, Trotta CR, Peltz SW (2006) Derepression of the Her‐2 uORF is mediated by a novel post‐transcriptional control mechanism in cancer cells. Gene Dev 20: 939–953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer KD, Patil DP, Zhou J, Zinoviev A, Skabkin MA, Elemento O, Pestova TV, Qian S‐B, Jaffrey SR (2015) 5′ UTR m6A promotes cap‐independent translation. Cell 163: 999–1010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miglietta G, Cogoi S, Marinello J, Capranico G, Tikhomirov AS, Shchekotikhin A, Xodo LE (2017) RNA G‐quadruplexes in Kirsten Ras (KRAS) oncogene as targets for small molecules inhibiting translation. J Med Chem 60: 9448–9461 [DOI] [PubMed] [Google Scholar]
- Miloslavski R, Cohen E, Avraham A, Iluz Y, Hayouka Z, Kasir J, Mudhasani R, Jones SN, Cybulski N, Rüegg MA et al (2014) Oxygen sufficiency controls TOP mRNA translation via the TSC‐Rheb‐mTOR pathway in a 4E‐BP‐independent manner. J Mol Cell Biol 6: 255–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modelska A, Turro E, Russell R, Beaton J, Sbarrato T, Spriggs K, Miller J, Gräf S, Provenzano E, Blows F et al (2015) The malignant phenotype in breast cancer is driven by eIF4A1‐mediated changes in the translational landscape. Cell Death Dis 6: e1603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moerke NJ, Aktas H, Chen H, Cantel S, Reibarkh MY, Fahmy A, Gross J, Degterev A, Yuan J, Chorev M et al (2007) Small‐molecule inhibition of the interaction between the translation initiation factors eIF4E and eIF4G. Cell 128: 257–267 [DOI] [PubMed] [Google Scholar]
- Moore CEJ, Pickford J, Cagampang FR, Stead RL, Tian S, Zhao X, Tang X, Byrne CD, Proud CG (2016) MNK1 and MNK2 mediate adverse effects of high‐fat feeding in distinct ways. Sci Rep 6: 23476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morfoisse F, Kuchnio A, Frainay C, Gomez‐Brouchet A, Delisle M‐B, Marzi S, Helfer A‐C, Hantelys F, Pujol F, Guillermet‐Guibert J et al (2014) Hypoxia induces VEGF‐C expression in metastatic tumor cells via a HIF‐1α‐independent translation‐mediated mechanism. Cell Rep 6: 155–167 [DOI] [PubMed] [Google Scholar]
- Morisaki T, Lyon K, DeLuca KF, DeLuca JG, English BP, Zhang Z, Lavis LD, Grimm JB, Viswanathan S, Looger LL et al (2016) Real‐time quantification of single RNA translation dynamics in living cells. Science 352: 1425–1429 [DOI] [PubMed] [Google Scholar]
- Mura M, Hopkins TG, Michael T, Abd‐Latip N, Weir J, Aboagye E, Mauri F, Jameson C, Sturge J, Gabra H et al (2015) LARP1 post‐transcriptionally regulates mTOR and contributes to cancer progression. Oncogene 34: 5025–5036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nairismägi M‐L, Vislovukh A, Meng Q, Kratassiouk G, Beldiman C, Petretich M, Groisman R, Füchtbauer E‐M, Harel‐Bellan A, Groisman I (2012) Translational control of TWIST1 expression in MCF‐10A cell lines recapitulating breast cancer progression. Oncogene 31: 4960–4966 [DOI] [PubMed] [Google Scholar]
- Nguyen HG, Conn CS, Kye Y, Xue L, Forester CM, Cowan JE, Hsieh AC, Cunningham JT, Truillet C, Tameire F et al (2018) Development of a stress response therapy targeting aggressive prostate cancer. Sci Transl Med 10: eaar2036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen PJ, Trachsel H (1988) The mouse protein synthesis initiation factor 4A gene family includes two related functional genes which are differentially expressed. EMBO J 7: 2097–2105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Occhi G, Regazzo D, Trivellin G, Boaretto F, Ciato D, Bobisse S, Ferasin S, Cetani F, Pardi E, Korbonits M et al (2013) A novel mutation in the upstream open reading frame of the CDKN1B gene causes a MEN4 phenotype. Plos Genet 9: e1003350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh S, Flynn RA, Floor SN, Purzner J, Martin L, Do BT, Schubert S, Vaka D, Morrissy S, Li Y et al (2016) Medulloblastoma‐associated DDX3 variant selectively alters the translational response to stress. Oncotarget 7: 28169–28182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortiz‐Zapater E, Pineda D, Martínez‐Bosch N, Fernández‐Miranda G, Iglesias M, Alameda F, Moreno M, Eliscovich C, Eyras E, Real FX et al (2012) Key contribution of CPEB4‐mediated translational control to cancer progression. Nat Med 18: 83–90 [DOI] [PubMed] [Google Scholar]
- Pal I, Safari M, Jovanovic M, Bates SE, Deng C (2019) Targeting translation of mRNA as a therapeutic strategy in cancer. Curr Hematol Malig Rep 14: 219–227 [DOI] [PubMed] [Google Scholar]
- de la Parra C, Ernlund A, Alard A, Ruggles K, Ueberheide B, Schneider RJ (2018) A widespread alternate form of cap‐dependent mRNA translation initiation. Nat Commun 9: 3068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavon‐Eternod M, Gomes S, Geslain R, Dai Q, Rosner MR, Pan T (2009) tRNA over‐expression in breast cancer and functional consequences. Nucleic Acids Res 37: 7268–7280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelechano V, Alepuz P (2017) eIF5A facilitates translation termination globally and promotes the elongation of many non polyproline‐specific tripeptide sequences. Nucleic Acids Res 45: 7326–7338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira B, Billaud M, Almeida R (2017) RNA‐binding proteins in cancer: old players and new actors. Trends Cancer 3: 506–528 [DOI] [PubMed] [Google Scholar]
- Pérez‐Guijarro E, Karras P, Cifdaloz M, Martínez‐Herranz R, Cañón E, Graña O, Horcajada‐Reales C, Alonso‐Curbelo D, Calvo TG, Gómez‐López G et al (2016) Lineage‐specific roles of the cytoplasmic polyadenylation factor CPEB4 in the regulation of melanoma drivers. Nat Commun 7: 13418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry RP (2005) The architecture of mammalian ribosomal protein promoters. Bmc Evol Biol 5: 15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pershing NLK, Lampson BL, Belsky JA, Kaltenbrun E, MacAlpine DM, Counter CM (2015) Rare codons capacitate Kras‐driven de novo tumorigenesis. J Clin Invest 125: 222–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson J, Li S, Kaltenbrun E, Erdogan O, Counter CM (2020) Expression of transgenes enriched in rare codons is enhanced by the MAPK pathway. Sci Rep 10: 22166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philippe C, Dubrac A, Quelen C, Desquesnes A, Van Den Berghe L, Ségura C, Filleron T, Pyronnet S, Prats H, Brousset P et al (2016) PERK mediates the IRES‐dependent translational activation of mRNAs encoding angiogenic growth factors after ischemic stress. Sci Signal 9: ra44 [DOI] [PubMed] [Google Scholar]
- Philippe L, Vasseur J‐J, Debart F, Thoreen CC (2017) La‐related protein 1 (LARP1) repression of TOP mRNA translation is mediated through its cap‐binding domain and controlled by an adjacent regulatory region. Nucleic Acids Res 46: gkx1237‐ [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter DF, Miao W, Yang X, Goda GA, Ji AL, Donohue LKH, Aleman MM, Dominguez D, Khavari PA (2021) easyCLIP analysis of RNA‐protein interactions incorporating absolute quantification. Nat Commun 12: 1569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pourdehnad M, Truitt ML, Siddiqi IN, Ducker GS, Shokat KM, Ruggero D (2013) Myc and mTOR converge on a common node in protein synthesis control that confers synthetic lethality in Myc‐driven cancers. Proc Natl Acad Sci USA 110: 11988–11993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puleston DJ, Buck MD, Klein Geltink RI, Kyle RL, Caputa G, O’Sullivan D, Cameron AM, Castoldi A, Musa Y, Kabat AM et al (2019) Polyamines and eIF5A hypusination modulate mitochondrial respiration and macrophage activation. Cell Metab 30: 352–363.e8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin H, Ni H, Liu Y, Yuan Y, Xi T, Li X, Zheng L (2020) RNA‐binding proteins in tumor progression. J Hematol Oncol 13: 90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin X, Sarnow P (2004) Preferential translation of internal ribosome entry site‐containing mRNAs during the mitotic cycle in mammalian cells*. J Biol Chem 279: 13721–13728 [DOI] [PubMed] [Google Scholar]
- Ramanathan M, Majzoub K, Rao DS, Neela PH, Zarnegar BJ, Mondal S, Roth JG, Gai H, Kovalski JR, Siprashvili Z et al (2018) RNA–protein interaction detection in living cells. Nat Methods 15: 207–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramanathan M, Porter DF, Khavari PA (2019) Methods to study RNA–protein interactions. Nat Methods 16: 225–234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhoads TW, Prasad A, Kwiecien NW, Merrill AE, Zawack K, Westphall MS, Schroeder FC, Kimble J, Coon JJ (2015) NeuCode labeling in nematodes: proteomic and phosphoproteomic impact of ascaroside treatment in Caenorhabditis elegans*. Mol Cell Proteomics 14: 2922–2935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robichaud N, del Rincon SV, Huor B, Alain T, Petruccelli LA, Hearnden J, Goncalves C, Grotegut S, Spruck CH, Furic L et al (2015) Phosphorylation of eIF4E promotes EMT and metastasis via translational control of SNAIL and MMP‐3. Oncogene 34: 2032–2042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robichaud N, Sonenberg N, Ruggero D, Schneider RJ (2019) Translational control in cancer. Csh Perspect Biol 11: a032896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenwald IB, Rhoads DB, Callanan LD, Isselbacher KJ, Schmidt EV (1993) Increased expression of eukaryotic translation initiation factors eIF‐4E and eIF‐2 alpha in response to growth induction by c‐myc. Proc Natl Acad Sci USA 90: 6175–6178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouskin S, Zubradt M, Washietl S, Kellis M, Weissman JS (2014) Genome‐wide probing of RNA structure reveals active unfolding of mRNA structures in vivo . Nature 505: 701–705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozenbaum M, Rajman M, Rishal I, Koppel I, Koley S, Medzihradszky KF, Oses‐Prieto JA, Kawaguchi R, Amieux PS, Burlingame AL et al (2018) Translatome regulation in neuronal injury and axon regrowth. Eneuro 5: ENEURO.0276‐17.2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubio CA, Weisburd B, Holderfield M, Arias C, Fang E, DeRisi JL, Fanidi A (2014) Transcriptome‐wide characterization of the eIF4A signature highlights plasticity in translation regulation. Genome Biol 15: 476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saini P, Eyler DE, Green R, Dever TE (2009) Hypusine‐containing protein eIF5A promotes translation elongation. Nature 459: 118–121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos M, Pereira PM, Varanda AS, Carvalho J, Azevedo M, Mateus DD, Mendes N, Oliveira P, Trindade F, Pinto MT et al (2018) Codon misreading tRNAs promote tumor growth in mice. RNA Biol 15: 773–786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanz E, Yang L, Su T, Morris DR, McKnight GS, Amieux PS (2009) Cell‐type‐specific isolation of ribosome‐associated mRNA from complex tissues. Proc Natl Acad Sci USA 106: 13939–13944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scagliola A, Miluzio A, Ventura G, Oliveto S, Cordiglieri C, Manfrini N, Cirino D, Ricciardi S, Valenti L, Baselli G et al (2021) Targeting of eIF6‐driven translation induces a metabolic rewiring that reduces NAFLD and the consequent evolution to hepatocellular carcinoma. Nat Commun 12: 4878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoof EM, Furtwängler B, Üresin N, Rapin N, Savickas S, Gentil C, Lechman E, Keller UAD, Dick JE, Porse BT (2021) Quantitative single‐cell proteomics as a tool to characterize cellular hierarchies. Nat Commun 12: 3341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuller AP, Wu CC‐C, Dever TE, Buskirk AR, Green R (2017) eIF5A functions globally in translation elongation and termination. Mol Cell 66: 194–205.e5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz J, Mah N, Neuenschwander M, Kischka T, Ratei R, Schlag PM, Castaños‐Vélez E, Fichtner I, Tunn P‐U, Denkert C et al (2018) Loss‐of‐function uORF mutations in human malignancies. Sci Rep 8: 2395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuster SL, Hsieh AC (2019) The Untranslated regions of mRNAs in cancer. Trends Cancer 5: 245–262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selbach M, Schwanhäusser B, Thierfelder N, Fang Z, Khanin R, Rajewsky N (2008) Widespread changes in protein synthesis induced by microRNAs. Nature 455: 58–63 [DOI] [PubMed] [Google Scholar]
- Sendoel A, Dunn JG, Rodriguez EH, Naik S, Gomez NC, Hurwitz B, Levorse J, Dill BD, Schramek D, Molina H et al (2017) Translation from unconventional 5′ start sites drives tumour initiation. Nature 541: 494–499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sénéchal P, Robert F, Cencic R, Yanagiya A, Chu J, Sonenberg N, Paquet M, Pelletier J (2021) Assessing eukaryotic initiation factor 4F subunit essentiality by CRISPR‐induced gene ablation in the mouse. Cell Mol Life Sci 78: 6709–6719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah M, Su D, Scheliga J, Pluskal T, Boronat S, Motamedchaboki K, Campos A, Qi F, Hidalgo E, Yanagida M et al (2016) A transcript‐specific eIF3 complex mediates global translational control of energy metabolism. Cell Rep 16: 1891–1902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shamir M, Bar‐On Y, Phillips R, Milo R (2016) SnapShot: timescales in cell biology. Cell 164: 1302–1302.e1 [DOI] [PubMed] [Google Scholar]
- Shen S, Faouzi S, Bastide A, Martineau S, Malka‐Mahieu H, Fu YU, Sun X, Mateus C, Routier E, Roy S et al (2019) An epitranscriptomic mechanism underlies selective mRNA translation remodelling in melanoma persister cells. Nat Commun 10: 5713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherrill KW, Byrd MP, Eden MEV, Lloyd RE (2004) BCL‐2 translation is mediated via internal ribosome entry during cell stress*. J Biol Chem 279: 29066–29074 [DOI] [PubMed] [Google Scholar]
- Silvera D, Arju R, Darvishian F, Levine PH, Zolfaghari L, Goldberg J, Hochman T, Formenti SC, Schneider RJ (2009) Essential role for eIF4GI overexpression in the pathogenesis of inflammatory breast cancer. Nat Cell Biol 11: 903–908 [DOI] [PubMed] [Google Scholar]
- Singh G, Ricci EP, Moore MJ (2014) RIPiT‐Seq: a high‐throughput approach for footprinting RNA:protein complexes. Methods 65: 320–332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinvani H, Haimov O, Svitkin Y, Sonenberg N, Tamarkin‐Ben‐Harush A, Viollet B, Dikstein R (2015) Translational tolerance of mitochondrial genes to metabolic energy stress involves TISU and eIF1‐eIF4GI cooperation in start codon selection. Cell Metab 21: 479–492 [DOI] [PubMed] [Google Scholar]
- Slobodin B, Gerst JE (2010) A novel mRNA affinity purification technique for the identification of interacting proteins and transcripts in ribonucleoprotein complexes. RNA 16: 2277–2290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobczak K, Krzyzosiak WJ (2002) Structural determinants of BRCA1 translational regulation*. J Biol Chem 277: 17349–17358 [DOI] [PubMed] [Google Scholar]
- Somers J, Wilson LA, Kilday J‐P, Horvilleur E, Cannell IG, Pöyry TAA, Cobbold LC, Kondrashov A, Knight JRP, Puget S et al (2015) A common polymorphism in the 5′ UTR of ERCC5 creates an upstream ORF that confers resistance to platinum‐based chemotherapy. Gene Dev 29: 1891–1896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonenberg N, Hinnebusch AG (2009) Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell 136: 731–745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spevak CC, Park E‐H, Geballe AP, Pelletier J, Sachs MS (2006) her‐2 upstream open reading frame effects on the use of downstream initiation codons. Biochem Biophys Res Commun 350: 834–841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spitale RC, Flynn RA, Zhang QC, Crisalli P, Lee B, Jung J‐W, Kuchelmeister HY, Batista PJ, Torre EA, Kool ET et al (2015) Structural imprints in vivo decode RNA regulatory mechanisms. Nature 519: 486–490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spitzer MH, Nolan GP (2016) Mass cytometry: single cells, many features. Cell 165: 780–791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spriggs KA, Cobbold LC, Jopling CL, Cooper RE, Wilson LA, Stoneley M, Coldwell MJ, Poncet D, Shen Y‐C, Morley SJ et al (2009) Canonical initiation factor requirements of the Myc family of internal ribosome entry segments. Mol Cell Biol 29: 1565–1574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spriggs KA, Stoneley M, Bushell M, Willis AE (2008) Re‐programming of translation following cell stress allows IRES‐mediated translation to predominate. Biol Cell 100: 27–38 [DOI] [PubMed] [Google Scholar]
- Sridharan S, Robeson M, Bastihalli‐Tukaramrao D, Howard CM, Subramaniyan B, Tilley AMC, Tiwari AK, Raman D (2019) Targeting of the eukaryotic translation initiation factor 4A against breast cancer stemness. Front Oncol 9: 1311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein I, Itin A, Einat P, Skaliter R, Grossman Z, Keshet E (1998) Translation of vascular endothelial growth factor mRNA by internal ribosome entry: implications for translation under hypoxia. Mol Cell Biol 18: 3112–3119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinhardt JJ, Peroutka RJ, Mazan‐Mamczarz K, Chen Q, Houng S, Robles C, Barth RN, DuBose J, Bruns B, Tesoriero R et al (2014) Inhibiting CARD11 translation during BCR activation by targeting the eIF4A RNA helicase. Blood 124: 3758–3767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoneley M, Subkhankulova T, Quesne JPCL, Coldwell MJ, Jopling CL, Belsham GJ, Willis AE (2000) Analysis of the c‐myc IRES; a potential role for cell‐type specific trans‐acting factors and the nuclear compartment. Nucleic Acids Res 28: 687–694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L, Fazal FM, Li P, Broughton JP, Lee B, Tang L, Huang W, Kool ET, Chang HY, Zhang QC (2019) RNA structure maps across mammalian cellular compartments. Nat Struct Mol Biol 26: 322–330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suresh S, Chen BeiBei, Zhu J, Golden RJ, Lu C, Evers BM, Novaresi N, Smith B, Zhan X, Schmid V et al (2020) eIF5B drives integrated stress response‐dependent translation of PD‐L1 in lung cancer. Nat Cancer 1: 533–545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svitkin YV, Pause A, Haghighat A, Pyronnet S, Witherell G, Belsham GJ, Sonenberg N (2001) The requirement for eukaryotic initiation factor 4A (eIF4A) in translation is in direct proportion to the degree of mRNA 5′ secondary structure. RNA 7: 382–394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang H, Hornstein E, Stolovich M, Levy G, Livingstone M, Templeton D, Avruch J, Meyuhas O (2001) Amino acid‐induced translation of TOP mRNAs is fully dependent on phosphatidylinositol 3‐kinase‐mediated signaling, is partially inhibited by rapamycin, and is independent of S6K1 and rpS6 phosphorylation. Mol Cell Biol 21: 8671–8683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tcherkezian J, Cargnello M, Romeo Y, Huttlin EL, Lavoie G, Gygi SP, Roux PP (2014) Proteomic analysis of cap‐dependent translation identifies LARP1 as a key regulator of 5′TOP mRNA translation. Gene Dev 28: 357–371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thakor N, Smith MD, Roberts L, Faye MD, Patel H, Wieden H‐J, Cate JHD, Holcik M (2016) Cellular mRNA recruits the ribosome via eIF3‐PABP bridge to initiate internal translation. RNA Biol 14: 1–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas JD, Johannes GJ (2007) Identification of mRNAs that continue to associate with polysomes during hypoxia. RNA 13: 1116–1131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson PA, Eam B, Young NP, Fish S, Chen J, Barrera M, Howard H, Sung E, Parra A, Staunton J et al (2021) Targeting oncogene mRNA translation in B‐cell malignancies with eFT226, a potent and selective inhibitor of eIF4A. Mol Cancer Ther 20: 26–36 [DOI] [PubMed] [Google Scholar]
- Thoreen CC, Chantranupong L, Keys HR, Wang T, Gray NS, Sabatini DM (2012) A unifying model for mTORC1‐mediated regulation of mRNA translation. Nature 485: 109–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truitt ML, Conn CS, Shi Z, Pang X, Tokuyasu T, Coady AM, Seo Y, Barna M, Ruggero D (2015) Differential requirements for eIF4E dose in normal development and cancer. Cell 162: 59–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truitt ML, Ruggero D (2016) New frontiers in translational control of the cancer genome. Nat Rev Cancer 16: 288–304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uemura S, Aitken CE, Korlach J, Flusberg BA, Turner SW, Puglisi JD (2010) Real‐time tRNA transit on single translating ribosomes at codon resolution. Nature 464: 1012–1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valle LD, Dai L, Lin H, Lin Z, Chen J, Post SR, Qin Z (2021) Role of EIF4G1 network in non‐small cell lung cancers (NSCLC) cell survival and disease progression. J Cell Mol Med 25: 2795–2805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Nostrand EL, Freese P, Pratt GA, Wang X, Wei X, Xiao R, Blue SM, Chen J‐Y, Cody NAL, Dominguez D et al (2020) A large‐scale binding and functional map of human RNA‐binding proteins. Nature 583: 711–719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Nostrand EL, Pratt GA, Shishkin AA, Gelboin‐Burkhart C, Fang MY, Sundararaman B, Blue SM, Nguyen TB, Surka C, Elkins K et al (2016) Robust transcriptome‐wide discovery of RNA‐binding protein binding sites with enhanced CLIP (eCLIP). Nat Methods 13: 508–514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- VanInsberghe M, van den Berg J, Andersson‐Rolf A, Clevers H, van Oudenaarden A (2021) Single‐cell Ribo‐seq reveals cell cycle‐dependent translational pausing. Nature 597: 561–565 [DOI] [PubMed] [Google Scholar]
- Vattem KM, Wek RC (2004) Reinitiation involving upstream ORFs regulates ATF4 mRNA translation in mammalian cells. Proc Natl Acad Sci USA 101: 11269–11274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villanueva E, Navarro P, Rovira‐Rigau M, Sibilio A, Méndez R, Fillat C (2017) Translational reprogramming in tumour cells can generate oncoselectivity in viral therapies. Nat Commun 8: 14833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Voss MRH, van Diest PJ, Raman V (2017) Targeting RNA helicases in cancer: the translation trap. Biochim Biophys Acta BBA ‐ Rev Cancer 1868: 510–520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner S, Herrmannová A, Hronová V, Gunišová S, Sen ND, Hannan RD, Hinnebusch AG, Shirokikh NE, Preiss T, Valášek LS (2020) Selective translation complex profiling reveals staged initiation and co‐translational assembly of initiation factor complexes. Mol Cell 79: 546–560.e7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Han B, Zhou R, Zhuang X (2016) Real‐time imaging of translation on single mRNA transcripts in live cells. Cell 165: 990–1001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F, Guan X, Xie D (2013) Roles of eukaryotic initiation factor 5A2 in human cancer. Int J Biol Sci 9: 1013–1020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Zhao BS, Roundtree IA, Lu Z, Han D, Ma H, Weng X, Chen K, Shi H, He C (2015) N6‐methyladenosine modulates messenger RNA translation efficiency. Cell 161: 1388–1399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waskiewicz AJ, Flynn A, Proud CG, Cooper JA (1997) Mitogen‐activated protein kinases activate the serine/threonine kinases Mnk1 and Mnk2. EMBO J 16: 1909–1920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weingarten‐Gabbay S, Elias‐Kirma S, Nir R, Gritsenko AA, Stern‐Ginossar N, Yakhini Z, Weinberger A, Segal E (2016) Systematic discovery of cap‐independent translation sequences in human and viral genomes. Science 351: aad4939 [DOI] [PubMed] [Google Scholar]
- Weingarten‐Gabbay S, Khan D, Liberman N, Yoffe Y, Bialik S, Das S, Oren M, Kimchi A (2014) The translation initiation factor DAP5 promotes IRES‐driven translation of p53 mRNA. Oncogene 33: 611–618 [DOI] [PubMed] [Google Scholar]
- West JA, Davis CP, Sunwoo H, Simon MD, Sadreyev RI, Wang PI, Tolstorukov MY, Kingston RE (2014) The long noncoding RNAs NEAT1 and MALAT1 bind active chromatin sites. Mol Cell 55: 791–802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wethmar K, Schulz J, Muro EM, Talyan S, Andrade‐Navarro MA, Leutz A (2016) Comprehensive translational control of tyrosine kinase expression by upstream open reading frames. Oncogene 35: 1736–1742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whiffin N, Karczewski KJ, Zhang X, Chothani S, Smith MJ, Evans DG, Roberts AM, Quaife NM, Schafer S, Rackham O et al (2020) Characterising the loss‐of‐function impact of 5′ untranslated region variants in 15,708 individuals. Nat Commun 11: 2523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woiwode A, Johnson SAS, Zhong S, Zhang C, Roeder RG, Teichmann M, Johnson DL (2008) PTEN represses RNA polymerase iii‐dependent transcription by targeting the TFIIIB complex. Mol Cell Biol 28: 4204–4214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf DA, Lin Y, Duan H, Cheng Y (2020) eIF‐Three to Tango: emerging functions of translation initiation factor eIF3 in protein synthesis and disease. J Mol Cell Biol 12: 403–409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe AL, Singh K, Zhong YI, Drewe P, Rajasekhar VK, Sanghvi VR, Mavrakis KJ, Jiang M, Roderick JE, Van der Meulen J et al (2014) RNA G‐quadruplexes cause eIF4A‐dependent oncogene translation in cancer. Nature 513: 65–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wortel IMN, van der Meer LT, Kilberg MS, van Leeuwen FN (2017) Surviving stress: modulation of ATF4‐mediated stress responses in normal and malignant cells. Trends Endocrinol Metab 28: 794–806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu B, Eliscovich C, Yoon YJ, Singer RH (2016) Translation dynamics of single mRNAs in live cells and neurons. Science 352: 1430–1435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G‐Q, Xu Y‐M, Lau ATY (2020a) Recent insights into eukaryotic translation initiation factors 5A1 and 5A2 and their roles in human health and disease. Cancer Cell Int 20: 142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Xie J, Jin X, Lenchine RV, Wang X, Fang DM, Nassar ZD, Butler LM, Li J, Proud CG (2020b) eEF2K enhances expression of PD‐L1 by promoting the translation of its mRNA. Biochem J 477: 4367–4381 [DOI] [PubMed] [Google Scholar]
- Xie C, Huang L, Xie S, Xie D, Zhang G, Wang P, Peng L, Gao Z (2013) LARP1 predict the prognosis for early‐stage and AFP‐normal hepatocellular carcinoma. J Transl Med 11: 272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Poggio M, Jin HY, Shi Z, Forester CM, Wang Y, Stumpf CR, Xue L, Devericks E, So L et al (2019) Translation control of the immune checkpoint in cancer and its therapeutic targeting. Nat Med 25: 301–311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Ruggero D (2019) The role of translation control in tumorigenesis and its therapeutic implications. Annu Rev Cancer Biol 4: 1–21 [Google Scholar]
- Yan X, Hoek TA, Vale RD, Tanenbaum ME (2016) Dynamics of translation of single mRNA molecules In vivo . Cell 165: 976–989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye L, Lin S, Mi Y, Liu Y, Ma Y, Sun H, Peng Z, Fan J (2016) Overexpression of LARP1 predicts poor prognosis of colorectal cancer and is expected to be a potential therapeutic target. Tumor Biol 37: 14585–14594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi W, Li J, Zhu X, Wang XI, Fan L, Sun W, Liao L, Zhang J, Li X, Ye J et al (2020) CRISPR‐assisted detection of RNA–protein interactions in living cells. Nat Methods 17: 685–688 [DOI] [PubMed] [Google Scholar]
- Zarnegar BJ, Flynn RA, Shen Y, Do BT, Chang HY, Khavari PA (2016) irCLIP platform for efficient characterization of protein–RNA interactions. Nat Methods 13: 489–492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Wang Y, Lu J (2019) Function and evolution of upstream ORFs in eukaryotes. Trends Biochem Sci 44: 782–794 [DOI] [PubMed] [Google Scholar]
- Zhang Q, Liang H, Zhao X, Zheng L, Li Y, Gong J, Zhu Y, Jin Y, Yin Y (2021) PTENε suppresses tumor metastasis through regulation of filopodia formation. EMBO J 40: e105806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng X, Cho S, Moon H, Loh TJ, Jang HN, Shen H (2016) RNA‐protein complexes and interactions, methods and protocols. Methods Mol Biol 1421: 35–44 [DOI] [PubMed] [Google Scholar]
- Zubradt M, Gupta P, Persad S, Lambowitz AM, Weissman JS, Rouskin S (2017) DMS‐MaPseq for genome‐wide or targeted RNA structure probing in vivo . Nat Methods 14: 75–82 [DOI] [PMC free article] [PubMed] [Google Scholar]


