Abstract
Background
The influenza A(H3N2) vaccine was updated from clade 3C.3a in 2015–2016 to 3C.2a for 2016–2017 and 2017–2018. Circulating 3C.2a viruses showed considerable hemagglutinin glycoprotein diversification and the egg-adapted vaccine also bore mutations.
Methods
Vaccine effectiveness (VE) in 2016–2017 and 2017–2018 was assessed by test-negative design, explored by A(H3N2) phylogenetic subcluster and prior season’s vaccination history.
Results
In 2016–2017, A(H3N2) VE was 36% (95% confidence interval [CI], 18%–50%), comparable with (43%; 95% CI, 24%–58%) or without (33%; 95% CI, −21% to 62%) prior season’s vaccination. In 2017–2018, VE was 14% (95% CI, −8% to 31%), lower with (9%; 95% CI, −18% to 30%) versus without (45%; 95% CI, −7% to 71%) prior season’s vaccination. In 2016–2017, VE against predominant clade 3C.2a1 viruses was 33% (95% CI, 11%–50%): 18% (95% CI, −40% to 52%) for 3C.2a1a defined by a pivotal T135K loss of glycosylation; 60% (95% CI, 19%–81%) for 3C.2a1b (without T135K); and 31% (95% CI, 2%–51%) for other 3C.2a1 variants (with/without T135K). VE against 3C.2a2 viruses was 45% (95% CI, 2%–70%) in 2016–2017 but 15% (95% CI, −7% to 33%) in 2017–2018 when 3C.2a2 predominated. VE against 3C.2a1b in 2017–2018 was 37% (95% CI, −57% to 75%), lower at 12% (95% CI, −129% to 67%) for a new 3C.2a1b subcluster (n = 28) also bearing T135K.
Conclusions
Exploring VE by phylogenetic subcluster and prior vaccination history reveals informative heterogeneity. Pivotal mutations affecting glycosylation sites, and repeat vaccination using unchanged antigen, may reduce VE.
Keywords: influenza virus, influenza vaccine, vaccine effectiveness, genomics, subtype, A(H3N2)
During 2016–2017 and 2017–2018 influenza A(H3N2) epidemics, updated clade 3C.2a vaccine bore egg-adaptation mutations and circulating 3C.2a viruses showed substantial genetic diversity. Vaccine effectiveness by phylogenetic subcluster and prior vaccination history reveal informative heterogeneity underpinning subtype-specific findings.
Both Canada and the United States experienced consecutive seasonal epidemics due to influenza A(H3N2) in 2016–2017 and 2017–2018 [1–4]. In the United States, the 2017–2018 epidemic was characterized as one of high severity, comparable to the 2014–2015 A(H3N2) epidemic, but more severe than any other of the prior decade [5].
The Canadian Sentinel Practitioner Surveillance Network (SPSN) is a community-based system for annual monitoring of influenza vaccine effectiveness (VE) against medically attended febrile respiratory illness due to laboratory-confirmed influenza. The SPSN uses a test-negative design that it first pioneered for this purpose during the 2004–2005 season [6]. Each year, the SPSN also genetically and antigenically characterizes contributing viruses as context for VE interpretation.
During the 2014–2015 A(H3N2) epidemic, the Canadian SPSN reported the lowest VE in more than a decade of annual monitoring (−17%; 95% confidence interval [CI], −50% to 9%) [6, 7]. A combination of agent-host factors was invoked to explain this finding. In 2014–2015, the A(H3N2) vaccine component was unchanged from the prior 2013–2014 season and belonged to clade 3C.1 whereas most circulating viruses belonged to the antigenically distinct clade 3C.2a (Table 1) [7]. The majority of participants vaccinated in 2014–2015 had also received the identical vaccination in 2013–2014 and the VE in 2014–2015 for these repeat recipients (−32%; 95% CI, −75% to 0%) was significantly lower than among participants reporting no prior vaccination in 2013–2014 (53%; 95% CI, 10%–75%). The antigenic-distance hypothesis was invoked to explain these observations; in particular, use of identical vaccine antigen the prior season, mismatched to circulating viruses, was hypothesized to have negatively interfered with vaccine performance in 2014–2015 [14–16].
Table 1.
Recommended Influenza Vaccine Strains and Distribution of Circulating Viruses by Clade, Canadian Sentinel Practitioner Surveillance Network (SPSN) Provinces, 2012–2013 to 2017–2018 Seasons
| Clade Distribution of Influenza A(H3N2) Viruses Sequenced by SPSN, No. (%)a | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Season [Ref] | Vaccine Clade(Representative Strain) | No. Sequences | 3C.1 | 3C.2 | 3C.3 | 3C.3a | 3C.3b | 3C.2a | 3C.2a1 | 3C.2a1a | 3C.2a1b | 3C.2a2 | 3C.2a3 | 3C.2a4 |
| 2012–2013 [8] | 3C (Victoria/361/2011) | 152b | 7(5) | 73(48) | 63(41) | − | − | − | − | − | − | − | − | − |
| 2013–2014 [9] | 3C.1(Texas/50/2012) | 11 | − | 2(18) | 5(45) | 1(9) | − | 3(27) | − | − | − | − | − | − |
| 2014–2015 [7] | 3C.1(Texas/50/2012) | 460 | − | − | 9(2) | 3(1) | 39(8) | 409(89) | − | − | − | − | − | − |
| 2015–2016 [10] | 3C.3a(Switzerland/9715293/2013) | 39c | − | − | − | 5(13) | − | 7(18) | 27(69) | − | − | − | − | − |
| 2016–2017 [see text] | 3C.2a(Hong Kong/4801/2014) | 574 | − | − | − | 21(4) | − | 7(1) | 257d(45) | 93(16) | 73(13) | 81(14) | 41(7) | 1(<1) |
| 2017–2018 [see text] | 3C.2a(Hong Kong/4801/2014) | 620 | − | − | − | 27(4) | − | 1(<1) | − | 5(1) | 34(5) | 540(87) | 10(2) | 3(<1) |
aThe sequences of SPSN viruses included in prior seasons’ vaccine effectiveness analyses were retrospectively reassessed for signature mutations and reclassified according to currently recognized clade nomenclature where applicable [11–13]. Reanalysis resulted in reclassification of viruses in 2012–2013 and 2015–2016 but not in 2013–2014 or 2014–2015, compared to the original publication. "−" indicates not detected.
bThere were 9 (6%) viruses in 2012–2013 that belonged to clade 6; no sequenced SPSN viruses in any season thereafter clustered with clade 6.
cFour additional A(H3N2) viruses were identified as clade 3C.2a but the sequences were not available for retrospective reanalysis.
dIncludes other 3C.2a viruses that do not cluster otherwise with the well-recognized 3C.2a1a and 3C.2a1b subgroups [11–13].
The World Health Organization (WHO) recommended a change from the clade 3C.1 vaccine strain used in 2013–2014 and 2014–2015 to a clade 3C.3a strain for 2015–2016, switching again in 2016–2017 to a clade 3C.2a strain (Table 1) [11–13, 17]. During the 2016–2017 A(H3N2) epidemic, circulating clade 3C.2a viruses showed considerable further diversification in the hemagglutinin (HA) glycoprotein (Table 1). The WHO, however, recommended no change to the A(H3N2) vaccine component for 2017–2018, again retaining the identical clade 3C.2a antigen as used in 2016–2017 [17]. With adaptation for egg-based manufacturing, the clade 3C.2a vaccine strain used both seasons acquired mutations affecting antigenicity and immunogenicity [18, 19].
Here, we estimate influenza VE for the 2016–2017 and 2017–2018 A(H3N2) epidemics in Canada. To assess the influence of genetic heterogeneity among circulating variants, we explore VE by phylogenetic subcluster, with particular reference to pivotal antigenic site mutations. To assess the effects of repeat vaccination with antigen that was changed (2016–2017) versus unchanged (2017–2018) from the prior season, we stratify VE estimates by prior season’s vaccination history. Finally, for comparison, we assess VE for influenza A(H1N1)pdm09 and influenza B during the same seasons.
METHODS
Canadian SPSN
The Canadian SPSN includes sentinel practitioner sites in the 4 most populous provinces including British Columbia and Alberta in western Canada and Ontario and Quebec in eastern Canada. Patients ≥1 year old presenting between November and April and within 7 days of onset of influenza-like illness (ILI) are eligible for inclusion in VE analyses. ILI is defined by self-reported fever and cough and at least 1 other symptom of sore throat, myalgia, arthralgia, or prostration; fever is not a requirement for elderly adults ≥65 years old. After obtaining verbal consent, sentinel practitioners document epidemiological information on a standard questionnaire completed at the time of nasal/nasopharyngeal specimen collection. Review boards in each province provided ethics approval.
Virus Detection and Characterization
Laboratory methods are detailed in Supplementary Material 1. Testing for influenza viruses was by real-time reverse-transcriptase polymerase chain reaction at provincial public health reference laboratories. A convenience sample of virus isolates was characterized antigenically by hemagglutination inhibition (HI) assay; by convention, ≥8-fold reduction in heterologous versus homologous HI titer was interpreted as antigenic distinction [20]. For A(H3N2) viruses, antigenic characterization was in relation to the cell-passaged clade 3C.2a vaccine reference strain. Given recognized issues with HI characterization of A(H3N2) viruses, notably the failure of clade 3C.2a strains to efficiently agglutinate erythrocytes [12, 13, 21], characterization was foremost undertaken genetically.
Sanger sequencing of the HA gene was conducted on primary patient specimens. Viruses were grouped based upon HA phylogenetic subclustering and signature amino acid substitutions informed by published sources [11–13, 17, 22, 23]. We specify these substitutions, indicating affected HA antigenic sites in parentheses (labeled A–E for H3N2). In parentheses we also annotate substitutions considered pivotal on the basis of their being associated with potential gain or loss of N-linked glycosylation (± CHO) and/or involving the receptor-binding site (RBS). We also indicate pivotal substitutions affecting HA positions historically implicated in major A(H3N2) antigenic-cluster transitions [24–29].
GenBank accession numbers for SPSN A(H3N2) sequences included in VE analyses in 2016–2017 are: KY583507–KY583727, MH216203–MH216548 (except MH216229, MH216330, MH216446). For 2017–2018, accession numbers are: MG889597–MG889825 (except MG889617, MG889637, MG889791, MG889809, MG889818, MG889819), MN538368–MN538764.
Vaccines
All vaccines used in Canada in 2016–2017 and 2017–2018 were egg based. Additional details related to influenza vaccine strains and products are provided in Supplementary Material 2.
The A(H3N2) clade 3C.2a vaccine strain used both seasons acquired egg-adaptation mutations recognized to have affected antigenicity and immunogenicity, namely N96S [D], L194P [B](RBS), and T160K [B](−CHO) [17–19]. Of note, the T160K substitution, associated with loss of glycosylation, exposes 2 major antigenic site B cluster transition positions (158 and 159) in proximity to the RBS [17–19, 29, 30].
Vaccine Effectiveness Estimation
VE was estimated by test-negative design as previously reported [6–10, 15]: cases tested influenza virus positive and controls tested negative for any influenza virus. VE was stratified by influenza type/subtype and further explored by phylogenetic subcluster for A(H3N2) viruses. Patients self-reporting at least 1 dose of influenza vaccine ≥2weeks before ILI onset were considered vaccinated.
The odds ratio (OR) for medically attended, laboratory-confirmed influenza illness between vaccinated versus unvaccinated participants was derived by logistic regression with VE = (1 – OR) x 100%. In primary analyses, ORs were adjusted for age group (1–8, 9–19, 20–49, 50–64, and ≥65 years), province, specimen collection interval (dichotomized as ≤4 days or 5–7 days), and calendar time (week of specimen collection modelled using a natural cubic spline function with 3 equally spaced knots). Participants missing information for vaccination, timing, or covariates were excluded. In sensitivity analyses, VE was further explored with sex and comorbidity as covariates and with restriction by age group and epidemic period (November/December; January/February; March/April). Repeat vaccination effects were assessed in the subset of participants ≥9 years old (the threshold age for 1- vs 2-dose priming recommendations) using a 4-level indicator variable defined by current season’s vaccination alone; current and prior seasons’ vaccination; or prior season’s vaccination alone; relative to participants unvaccinated both seasons (reference group). To address sparse data, all adjusted VE estimates were also checked using Firth’s method of penalized logistic regression [31–33].
RESULTS
A(H3N2) Virological Findings by Season
2016–2017
For 2016–2017, 2074 eligible specimens were included, with collection dates spanning November 1 to April 29. Of these, 818 (39%) tested influenza-positive including 693 (85%) influenza A, of which 684 (99%) with known subtype (n = 692) were H3N2 (Supplementary Table 3 and Supplementary Figure 4).
The majority (574/684; 84%) of A(H3N2) viruses detected by the SPSN in 2016–2017 were sequenced, most of which (423; 74%) belonged to clade 3C.2a1 (Table 1). Sequencing revealed substantial heterogeneity underlying the A(H3N2) epidemic, with multiple genetic variants contributing (Table 2, Figure 1, and Supplementary Figure 5A). At least 2 genetic subgroups of 3C.2a1 have been recognized by authoritative sources, namely 3C.2a1a and 3C.2a1b [11–13]. Clade 3C.2a1a is defined by a threonine to lysine substitution at position 135, resulting in loss of glycosylation at an historically recognized major antigenic site A cluster-transition position, also involving the RBS (ie, T135K [A](RBS)(−CHO)) [29]. Of 3C.2a1 viruses detected by the SPSN, 93/423 (22%) were 3C.2a1a and 73/423 (17%) were 3C.2a1b, the latter defined by other substitutions (not involving T135K). Most (257; 61%) 3C.2a1 viruses clustered outside of these 2 subgroups, with a few of these (16; 6%) also bearing the T135K substitution. The remaining 151 (26%) A(H3N2) sequences in 2016–2017 were distributed among other 3C.2a clades (eg, 3C.2a2 [81; 14%], 3C.2a3 [41; 7%]) or with clade 3C.3a (21; 4%).
Table 2.
Influenza A(H3N2) Phylogenetic Subclusters by Season, Canadian Sentinel Practitioner Surveillance Network 2016–2017 and 2017–2018
| Parent Groups With Defining Substitutions [Antigenic Site]a + Extra Substitutions [Antigenic Site]b | 2016–2017, n (%), N = 574 | 2017–2018, n (%), N = 620 |
|---|---|---|
| 3C.2a = 3C.2c+ L3I + N144S [A] + F159Y [B] + K160T [B] (+CHO) + N225D (RBS) + Q311H [C]d | 7 (1) | 1 (<1) |
| +T131K [A] + R142K [A]e | 5 | − |
| 3C.2a1 = 3C.2a + N171K [D]f | 38 (7) | − |
| + S47T [C] + G78S [E] | 14 | − |
| + R142G [A] | 10 | − |
| 3C.2a1 + N121K [D] | 5 (1) | − |
| + R142K [A] | 1 | |
| 3C.2a1 + N121K [D] + I140M [A] g | 7 (1) | − |
| 3C.2a1 + N121K [D] + R142G [A] | 94 (16) | − |
| + T135K [A] (RBS) (−CHO) | 16 | − |
| 3C.2a1 + N121K [D] + R142G [A] + I242V [D] g | 113 (20) | − |
| 3C.2a1a = 3C.2a1 + N121K [D] + T135K [A] (RBS) (−CHO)g | 93 (16) | 5 (1) |
| + T167S [D] (−CHO) | 19 | − |
| + N96S [D] | 17 | − |
| + S46T [C] (−CHO) | 8 | − |
| 3C.2a1b = 3C.2a1 + N121K [D] + K92R [E] + H311Q [C] | 73 (13) | 34 (5) |
| + Q197R [B] | 57 | 3 |
| + E62G [E] + T135K [A] (RBS) (−CHO) + R142E [A] h | − | 3 |
| + E62G [E] + T135K [A] (RBS) (−CHO) + R142G [A]g | − | 25 |
| + E62G [E] + R142G [A] | − | 1 |
| 3C.2a2 = 3C.2a + T131K [A] + R142K [A] + R261Q [E]i | 81 (14) | 540 (87) |
| + F193S [B] | 20 | 2 |
| + R201K [D] | 11 | 6 |
| + H156Q [B] | 6 | − |
| + A212T [D]j | − | 170 |
| + K92R [E]k | − | 67 |
| + S144R [A] | − | 10 |
| + T160K [B] (−CHO) | − | 11 |
| + Y94H [E] | − | 9 |
| 3C.2a3 = 3C.2a + N121K [D] + S144 K [A] | 41 (7) | 10 (2) |
| + S219Y [D] | 30 | − |
| + T135K [A] (RBS) (−CHO) + R150K [A] + R261Q [E] | − | 9 |
| 3C.2a4 = 3C.2a + N31S + D53N [C] + R142G [A] + S144R [A] + N171K [D] + I192T [B] + Q197H [B] | 1 (<1) | 3 (<1) |
| 3C.3a = 3C.3l+ A138S [A] (RBS) + F159S [B] + N225D (RBS) + K326Rm | 21 (4) | 27 (4) |
| + L3I + S91N [E] + N144K [A] (−CHO) + F193S [B]n, o | 20 | 27 |
Abbreviations: +/− CHO, gain/loss of potential N-linked glycosylation site; RBS, substitutions within the receptor binding site; "−" indicates not detected.
aParent genetic groups with their defining hemagglutinin (HA) substitutions and number of viruses are shown in bold font. The number of viruses with additional specified substitutions are shown in normal font. Substitutions are for HA1 unless specified as for HA2. Clades 3C.2a, 3C.2a1, 3C.2a1a, 3C.2a1b, 3C.2a2, 3C.2a3, 3C.2a4, and 3C.3a are well-recognized parent groupings [11–13, 17]. Additional 3C.2a1 parent groups specified here are based on phylogenetic analysis, additionally informed by published sources [22, 23]. Not all substitutions or potential subclusters are displayed; for additional details see phylogenetic trees in Supplementary Figure 5A and 5B. Note that the vaccine strain each season was clade 3C.2a.
bExtra substitutions in antigenic sites of at least 1% of all viruses either season are shown. The tally with T131K [A], T135K [A], or R142G/K/E [A] are also shown.
cClade 3C.2 defined by 3C + N145S [A] + HA2: D160N.
dAll clade 3C.2a viruses are also S96N [D], K160T [B] (+CHO) and P194L [B] (RBS) relative to the egg-adapted 3C.2a vaccine (ie, the latter is 96S, 160K, and 194P).
eOf these clade 3C.2a viruses bearing T131K [A] + R142K [A] substitutions, one also bears A212T [D] but none bear R261Q [E], the latter clade defining for 3C.2a2.
fClade 3C.2a1 defining substitutions additionally include HA2: G155E and HA2: I77V.
gAdditional defining substitution includes HA2: G150E.
hIn 2017–2018, one other 3C.2a1b virus bore T135N [A] (RBS) (−CHO). No 3C.2a1b virus bore T131K [A], subsequently prominent among 3C.2a1b viruses in 2018–2019 [11–13].
iAdditional substitutions enumerated for clade 3C.2a2 are not mutually exclusive. Two 3C.2a2 viruses in 2017–2018 are K142R. Further subclusters of clade 3C.2a2 have not been defined by published sources to date [11–13].
jIn 2017–2018, 8/170 of these viruses form a potential subcluster; the remainder are distributed and/or cluster phylogenetically with viruses that lack A212T [D].
kIn 2017–2018, 66/67 of these viruses form a potential subcluster.
lClade 3C.3 defined by 3C + T128A[B](−CHO) + R142G [A] + N145S[A].
mRelative to the clade 3C.2a vaccine strain, clade 3C.3a viruses are I3L, S144N [A], Y159S (rather than F159S owing to clade-defining F159Y substitution in 3C.2a viruses), H311Q [C], and HA2: N160D, but no longer N145S [A] and N225D (RBS) as a result of parallel substitutions in the 3C.2a vaccine strain (ie, the vaccine strain is also 145S and 225D). All clade 3C.3a viruses are also S96N [D] and P194L [B] (RBS) relative to the egg-adapted clade 3C.2a vaccine strain (ie, the egg-adapted vaccine is 96S and 194P). Unlike clade 3C.2a viruses, clade 3C.3a viruses are 160K (nonglycosylated), as is the egg-adapted clade 3C.2a vaccine strain.
nThese clade 3C.3a viruses are also HA2: D160N.
oRelative to the clade 3C.2a vaccine strain, these clade 3C.3a viruses are S144K (rather than N144K owing to N144S substitution in 3C.2a viruses) and no longer possess I3L and HA2: N160D (from footnote n) as a result of parallel substitutions in the 3C.2a vaccine strain (ie, the vaccine strain is 3I and HA2: 160N).
Figure 1.
Influenza A(H3N2) virus detection by genetic grouping and week of specimen collection, 2016–2017 and 2017–2018 seasons. Among eligible patients presenting with influenza-like illness, influenza A(H3N2) detections overall each season (A) and by phylogenetic subcluster (B) are displayed by week of specimen collection. The overall contribution by phylogenetic subcluster is shown in pie charts by season. Case counts by subcluster are shown in the legend by season as (n = 2016–2017, n 2017–2018).
Heterogeneity in circulating A(H3N2) variants was also observed by region, epidemic period, and age. For example, a single 3C.2a1 subcluster predominated during the earlier epidemic affecting the western province of Alberta but was scarce in other SPSN provinces (Supplementary Figure 6A). Clade 3C.3a contributed minimally overall, but later in the season and mostly in the eastern province of Quebec. Most of the clade 3C.3a cases were children 1–19 years old (62%; 13/21) whereas a smaller proportion of predominant 3C.2a1 cases (30%; 127/423; P = .002) or test-negative controls (25%; 309/1256; P < .001) were children (Supplementary Figure 7).
Antigenic characterization by HI assay was attempted on 49% (333/684) A(H3N2) viruses in 2016–2017, of which just 13% (43/333) were successful, including 9% (25/285) of the 3C.2a viruses but virtually all of the 3C.3a viruses (17/18) (Supplementary Table 8). Of A(H3N2) viruses successfully characterized, 42% (18/43) were antigenically distinct from the vaccine, including a minority of the 3C.2a variants overall (2/25) but virtually all of the 3C.3a (16/17) viruses.
2017–2018
For 2017–2018, 3483 eligible specimens were included, with collection dates spanning November 5 to April 28. Of these 1764 (51%) tested influenza positive including 793 (45%) influenza A, of which 639 (83%) with known subtype (n = 773) were H3N2 (Supplementary Table 3 and Supplementary Figure 4).
The majority (620/639; 97%) of A(H3N2) viruses detected by the SPSN in 2017–2018 were sequenced, most of which (540; 87%) belonged to clade 3C.2a2 (Table 1). Clade 3C.2a2 is defined by a T131K [A] substitution, which is located near 2 major antigenic site B cluster-transition positions (155 and 156) and the RBS [29]. Although additional substitutions and potential subclustering were observed among 3C.2a2 viruses (Table 2, and Supplementary Figure 5B), to date further 3C.2a2 subgroups have not been defined by authoritative sources [11–13]. In 2017, 3C.2a2 viruses were noted elsewhere to have reassorted and acquired the neuraminidase of 3C.2a1a viruses [11–13]; however, the SPSN does not routinely characterize the neuraminidase. Unlike 2016–2017, few (39; 6%) A(H3N2) viruses in 2017–2018 belonged to clade 3C.2a1, and these predominantly clustered with 3C.2a1b (34/39), most of which (28/34) comprised an emergent subcluster that newly acquired the pivotal T135K substitution. Other subclusters of 3C.2a1 prevalent in 2016–2017 were not detected at all in 2017–2018. The remaining 41 (7%) of A(H3N2) viruses clustered with 3C.2a (1; <1%), 3C.2a3 (10; 2%), 3C.2a4 (3; <1%), or with clade 3C.3a (27; 4%).
As in 2016–2017, the western province of Alberta showed an earlier epidemic peak but with subclade 3C.2a2 viruses predominating throughout (Supplementary Figure 6B). Clade 3C.3a viruses again contributed minimally and later in the season, foremost from the eastern SPSN provinces of Ontario (13/27) and Quebec (13/27) and with the same disproportionate involvement of children 1–19 years old (56%; 15/27) as in 2016–2017, whereas a smaller proportion of predominant 3C.2a2 cases (17%; 94/540; P < .001) or test-negative controls (20%; 336/1719; P < .001) were children (Supplementary Material 7).
Antigenic characterization by HI assay was attempted on 45% (287/639) A(H3N2) viruses, of which 14% (41/287) were successful, including 8% (21/260) of the 3C.2a viruses but virtually all (18/19) of the clade 3C.3a viruses (Supplementary Table 8). Of viruses characterized, 46% (19/41) were antigenically distinct from the vaccine, notably all of the 3C.3a viruses.
VE Against A(H3N2)
The profile of participants contributing to A(H3N2) VE analyses is provided in Supplementary Table 9. Adults 20–64 years old comprised 60%–65% of participants and about one-third of test-negative controls were considered vaccinated.
With Firth’s method, all VE point estimates remained within 3% (absolute) of estimates without Firth’s, including all subset and subcluster analyses. VE estimates are shown without Firth’s method unless specified.
2016–2017
In 2016–2017, VE against A(H3N2) was 36% (95% CI, 18%–50%) overall (Figure 2). VE was similar in sensitivity analyses additionally adjusting for comorbidity and sex but steadily declined across epidemic periods (Table 3). The western province of Alberta experienced higher VE associated with the earlier epidemic due to a unique 3C.2a1 subcluster (Supplementary Figure 6A).
Figure 2.
Influenza VE estimates against influenza A(H3N2), 2016–2017 and 2017–2018 seasons, are displayed overall and for select phylogenetic subclusters. Additional A(H3N2) VE findings overall are provided in Table 3 and by phylogenetic subcluster in Supplementary Table 11. All estimates were adjusted for age group (1–8, 9–19, 20–49, 50–64, ≥65 years), province (Alberta, British Columbia, Ontario, Quebec), specimen collection interval (≤4 days, 5–7 days), and calendar time (week of specimen collection modeled using natural cubic spline function with 3 equally spaced knots). All estimates displayed were additionally assessed using Firth’s penalized logistic regression [30–32], but remained within 1% (absolute) in 2016–2017 and within 3% (absolute) in 2017–2018. Abbreviations: +, means with the specified substitution; −, means without the specified substitution; CI, confidence interval; NE, not estimated; VE, vaccine effectiveness.
Table 3.
Influenza Vaccine Effectiveness Against Influenza A(H3N2), Canadian Sentinel Practitioner Surveillance Network 2016–2017 and 2017–2018
| 2016–2017 | 2017–2018 | |||||||
|---|---|---|---|---|---|---|---|---|
| Model and Covariates | A(H3N2) Cases n vac/ N Total (%) | Influenza Test- Negative Controls n vac/ N Total (%) | Unadjusted VE % (95% CI) | Adjusteda VE% (95% CI) | A(H3N2) Cases n vac/ N Total (%) | Influenza Test-Negative Controls n vac/ N Total (%) | Unadjusted VE % (95% CI) | Adjusteda VE % (95% CI) |
| Overall (age,b province,c interval,d calendar timee) | 163/684 (24) | 416/1256 (33) | 37 (22–49) | 36 (18–50) | 201/639 (31) | 596/1719 (35) | 14 (−5 to 29) | 14 (−8 to 31) |
| + Comorbidityb-f | 148/624 (24) | 392/1163 (34) | 39 (24–51) | 36 (17–50) | 197/611 (32) | 572/1642 (35) | 11 (−8 to 27) | 9 (–14 to 27) |
| + Comorbidity and sexb-f,g | 148/621 (24) | 392/1159 (34) | 39 (24−51) | 35 (16−50) | 195/605 (32) | 569/1627 (35) | 12 (–8 to 27) | 10 (−13 to 28) |
| Restricted by age subset | ||||||||
| 1–19 yearsc-e,h | 26/201 (13) | 57/309 (18) | 34 (−9 to 60) | 12 (−53 to 49) | 21/124 (17) | 65/336 (19) | 15 (−46 to 51) | −21 (−116 to 33) |
| 20–64 yearsc-e,i | 84/386 (22) | 240/770 (31) | 39 (18–54) | 41 (19–56) | 109/404 (27) | 353/1133 (31) | 18 (−5 to 37) | 15 (−12 to 35) |
| ≥ 65 yearsc-e | 53/97 (55) | 119/177 (67) | 41 (2–65) | 40 (−8 to 67) | 71/111 (64) | 178/250 (71) | 28 (−15 to 55) | 25 (−24 to 55) |
| Restricted by province | ||||||||
| Albertab,d,e | 27/136 (20) | 109/301 (36) | 56 (29–73) | 55 (22–74) | 48/159 (30) | 163/429 (38) | 29 (−4 to 52) | 43 (10–64) |
| British Columbiab,d,e | 62/212 (29) | 147/427 (34) | 21 (−13 to 45) | 23 (−19 to 50) | 24/70 (34) | 149/442 (34) | −3 (−75 to 40) | 5 (−70 to 47) |
| Ontariob,d,e | 49/195 (25) | 126/329 (38) | 46 (20–63) | 38 (1–62) | 100/236 (42) | 227/566 (40) | −10 (−49 to 19) | −11 (−56 to 21) |
| Quebecb,d,e | 25/141 (18) | 34/199 (17) | −5 (−85 to 41) | 22 (−50 to 59) | 29/174 (17) | 57/282 (20) | 21 (−29 to 52) | 18 (−46–54) |
| Restricted by epidemic period | ||||||||
| November/Decemberb-d | 35/183 (19) | 83/332 (25) | 29 (−11 to 55) | 42 (5–65) | 52/162 (32) | 129/396 (33) | 2 (−45 to 34) | 26 (−17 to 54) |
| January/Februaryb-d | 113/450 (25) | 211/610 (35) | 37 (17–52) | 30 (5–48) | 122/419 (29) | 311/900 (35) | 22 (0–40) | 15 (−12 to 36) |
| March/Aprilb-d | 15/51 (29) | 122/314 (39) | 34 (−25 to 66) | 18 (−73 to 61) | 27/58 (47) | 156/423 (37) | −49 (−159 to 14) | −20 (−129 to 37) |
Abbreviation: CI, confidence interval; n vac, number vaccinated; VE, vaccine effectiveness.
aAll estimates displayed were additionally assessed using Firth’s penalized logistic regression [30–32] but remained within 1% (absolute) in 2016–2017 and within 3% (absolute) in 2017–2018.
bAdjusted for age group specified as: 1–8, 9–19, 20–49, 50–64, ≥ 65 years.
cAdjusted for province specified as: Alberta, British Columbia, Ontario, Quebec.
dAdjusted for specimen collection interval specified as ≤4 days, 5–7 days.
eAdjusted for calendar time based on week of specimen collection modeled using natural cubic spline function with 3 equally spaced knots.
fAdditionally adjusted for comorbidity specified as yes or no (excluding those with missing information) for any chronic conditions that Canada’s National Advisory Committee on Immunization has indicated place individuals at higher risk of serious complications from influenza including: heart, pulmonary (including asthma), renal, metabolic (such as diabetes), blood, cancer, or immune comprising conditions; conditions that compromise management of respiratory secretions and increase risk of aspiration; or morbid obesity (body mass index ≥ 40).
gAdditionally adjusted for sex as male or female, excluding those with missing information.
hAdjusted for age group specified as: 1–8, 9–19 years.
iAdjusted for age group as 20–49, 50–64 years.
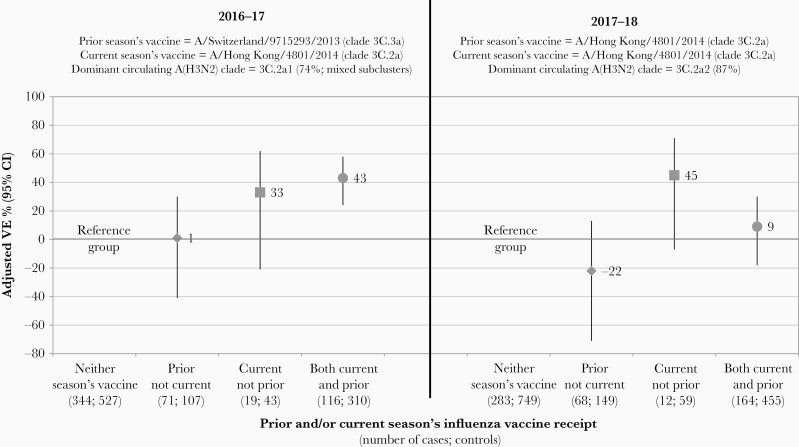
Most participants ≥9 years old who reported being vaccinated in 2016–2017 also reported vaccination in 2015–2016 (87%; 426/488) (Figure 3 and Supplementary Table 10). VE in 2016–2017 was comparable for participants with (43%; 95% CI, 24%–58%) or without (33%; 95% CI, −21% to 62%) prior vaccination in 2015–2016.
Figure 3.
Influenza vaccine effectiveness (VE) against influenza A(H3N2) by current and/or prior season’s influenza vaccine receipt, 2016–2017 and 2017–2018 seasons. Restricted to participants ≥9 years old and with complete data for current and prior season’s vaccine receipt. The prior and current seasons’ recommended vaccine strains are specified by season, alongside the dominant circulating influenza A(H3N2) clade. Note that the 2015–2016 3C.3a vaccine (cell or egg passaged) is antigenically distinct from the 2016–2017 3C.2a vaccine (cell or egg passaged); the 2016–2017/2017–2018 egg-adapted 3C.2a vaccine is antigenically distinct from 3C.2a2 viruses that predominated in 2017–2018. All estimates were adjusted for age group (9–19, 20–49, 50–64, ≥65 years), province (Alberta, British Columbia, Ontario, Quebec), specimen collection interval (≤4 days; 5–7 days), and calendar time (week of specimen collection modeled using natural cubic spline function with 3 equally spaced knots). All estimates displayed were additionally assessed using Firth’s penalized logistic regression [30–32], but were identical except for current season only vaccination (within 1% absolute in 2016–2017 and within 2% absolute in 2017–2018). Additional details are provided in Supplementary Table 10.
In 2016–2017, VE against predominant clade 3C.2a1 viruses was 33% (95% CI, 11%–50%) overall: 18% (95% CI, −40% to 52%) for 3C.2a1a viruses with T135K; 60% (95% CI, 19%–81%) for 3C.2a1b viruses (without T135K); and 31% (95% CI, 2%–51%) for other 3C.2a1 subclusters combined (with/without T135K) (Figure 2 and Supplementary Table 11). VE against minority 3C.2a2 viruses (bearing T131K) was 45% (95% CI, 2%–70%).
2017–2018
In 2017–2018, VE against A(H3N2) was 14% (95% CI, −8% to 31%) overall (Figure 2), VE was again similar in sensitivity analyses and declined across epidemic periods (Table 3). Alberta also experienced higher VE associated with an earlier epidemic but this was predominantly due to 3C.2a2 viruses as for other provinces.
Most participants ≥9 years old who reported being vaccinated in 2017–2018 also reported vaccination in 2016–2017 (90%; 619/690) (Figure 3 and Supplementary Table 10). VE in 2017–2018 was lower for participants with (9%; 95% CI, −18% to 30%) versus without (45%; 95% CI, −7% to 71%) prior vaccination in 2016–2017.
VE against predominant 3C.2a2 viruses in 2017–2018 was 15% (95% CI, −7% to 33%). VE against minority 3C.2a1b viruses was 37% (95% CI, −57% to 75%) overall but 12% (95% CI, −129% to 67%) for the 3C.2a1b subcluster newly bearing T135K (Figure 2 and Supplementary Table 11).
VE Against A(H1N1)pdm09 and Influenza B
VE and virological details for A(H1N1)pdm09 and influenza B are provided in Supplementary Material 12–14. With just 8 A(H1N1)pdm09 cases in 2016–2017, VE could not be estimated; with 134 cases in 2017–2018, VE against A(H1N1)pdm09 was 58% (95% CI, 29%–75%). Influenza B viruses, predominantly Yamagata lineage, contributed less in 2016–2017 (n = 123) than 2017–2018 (n = 977) when they showed earlier cocirculation with A(H3N2) (Supplementary Figure 4). VE against influenza B was 72% (95% CI, 52%–84%) in 2016–2017 and 46% (95% CI, 34%–56%) in 2017–2018.
DISCUSSION
According to estimates of the Canadian SPSN, during consecutive seasonal epidemics, VE against A(H3N2) overall was <40% in 2016–2017 and even lower in 2017–2018 at <20%. Globally, efforts are underway to determine why VE against A(H3N2) is recurrently lower than for other influenza types or subtypes, as also identified in the current analysis [6, 34]. We sought to learn from the heterogeneity underpinning subtype-specific results by exploring suboptimal VE for A(H3N2) more finely by phylogenetic subcluster and prior vaccination history.
In 2016–2017, the Canadian SPSN estimated VE of 36% against A(H3N2) overall, driven by the dominant 3C.2a1 clade for which VE was 33%. Although low, these point estimates are consistent with the historical average VE estimates for A(H3N2) reported in meta-analysis (33%; 95% CI, 26%–39%) [34]. The 2016–2017 end-of-season estimates from Canada are also similar to midseason report (42%; 95% CI, 18%–59%) [35] and to end-of-season estimates elsewhere that clade 3C.2a1 viruses predominated, including the United States (33%; 95% CI, 23%–41%) [36], United Kingdom (32%; 95% CI, 10%–48%) [37], and Europe (28%; 95% CI, 17%–38%) [38].
In 2017–2018, the Canadian SPSN estimated lower VE of 14% against A(H3N2), driven by the dominant 3C.2a2 clade for which VE was 15%. This is the second-lowest VE estimate against A(H3N2) reported by the SPSN in more than a decade of annual monitoring, worse only during the 2014–2015 epidemic (−17%; 95% CI, −50% to 9%) [6, 7]. The 2017–2018 end-of-season estimate is comparable to our midseason report (17%; 95% CI, −14% to 40%) [39], and to the VE reported by Australia for its severe 2017 A(H3N2) epidemic (10%; 95% CI, −16% to 31%) [23]. A greater mix of 3C.2a variants contributed in Australia with <10% 3C.2a2 and 30% 3C.2a1b, most of the latter (34/61; 56%) having picked up the extra T135K substitution also found among 3C.2a1b viruses (28/34; 82%) in Canada in 2017–2018 [23]. The Canadian VE estimate is also similar to Europe’s for 2017–2018 (13%; 95% CI, −15% to 34%) where a more equal mix of 3C.2a2 (167/312; 54%) and 3C.2a1b (132/312; 42%) viruses contributed, with 84% of the latter also bearing T135K as in Canada [38; personal communication; E Kissling, Epiconcept; March 2, 2020]. In the United Kingdom, VE against A(H3N2) was also low (−16%; 95% CI, −59% to 15%), with clade 3C.2a2 predominance (113/162; 70%) but greater contribution overall by 3C.2a1b viruses bearing T135K/N (25/162; 15%) compared to Canada (29/620; 5%) [40]. Finally, in the United States, VE against A(H3N2) was also low (22%; 95% CI, 12%–31%), particularly among adults aged 18–49 years (14%; 95% CI, −6% to 30%), but the mix of contributing 3C.2a variants was not specified [41].
Despite predominance and diversification of clade 3C.2a1 variants in 2016–2017 (Figure 1), clade 3C.2a2 viruses became predominant in 2017–2018. Thereafter, in 2018–2019, clade 3C.2a2 was largely displaced by further evolved clade 3C.2a1b and clade 3C.3a viruses [11–13]. Across these different evolutionary branches of A(H3N2), a large number of parallel (shared) substitutions were observed. Mutations that independently arise in separate subclusters may indicate common foci of immune selection pressure on the viral HA. In that regard, we highlight 2 parallel and pivotal substitutions that first arose in 2016–2017, affecting antigenic site A in proximity to the receptor-binding site, including: T135K, also constituting a loss of glycosylation; and T131K, also located near 2 major antigenic site B cluster-transition positions (155 and 156) [29]. Clade-defining for 3C.2a1a, T135K also subsequently appeared as extra substitution within a 3C.2a1b subcluster that emerged in 2017–2018 (ie, 3C.2a1b/T135K). Clade-defining for 3C.2a2, T131K also appeared in another 3C.2a1b subcluster in 2018–2019 (ie, 3C.2a1b/T131K). These 3C.2a1b/T135K and 3C.2a1b/T131K subclusters have since cocirculated as competing variants during the 2018–2019 and 2019–2020 seasons, comprising varying proportions of A(H3N2) viruses regionally [11–13]. Underscoring that, the WHO has recommended a clade 3C.2a1b/T131K virus as A(H3N2) component for the 2020 influenza vaccine for the southern hemisphere, but a 3C.2a1b/T135K virus for the 2020–2021 vaccine for the northern hemisphere [17].
In analysis by phylogenetic subcluster for the 2016–2017 season, we noted VE point estimates were lower for 3C.2a1a viruses defined by T135K (18%) than overall for A(H3N2) viruses (36%) or for 3C.2a1b without T135K (60%), other 3C.2a1 variants with/without T135K (31%) or 3C.2a2 viruses instead defined by T131K (45%). In 2017–2018, we noted that A(H3N2) VE was generally lower (14%), including for clade 3C.2a2 (15%) and 3C.2a1b (37%), but for the latter was particularly low for the newly emergent subcluster bearing T135K (12%). Lower VE for 3C.2a1b/T135K (7%; 95% CI, −52% to 43%) versus 3C.2a1b/T131K (57%; 95% CI, 16%–78%) has also since been reported from Europe for their 2018–2019 A(H3N2) epidemic [42]. Although associated with overlapping confidence intervals and constituting circumstantial evidence, these observations reinforce a potentially pivotal role for T135K loss of glycosylation affecting the receptor binding site and facilitating immunological escape.
During the 2016–2017 and 2017–2018 epidemics, we found only circulating clade 3C.3a viruses to be antigenically distinct from the cell-passaged clade 3C.2a vaccine strain. The limited number of clade 3C.3a viruses we identified overall were disproportionately found among children in both 2016–2017 and 2017–2018, foreshadowing a similar pattern for 3C.3a viruses during the 2018–2019 A(H3N2) epidemic in Canada [43]. Despite substantial genetic heterogeneity among predominant clade 3C.2a strains in 2016–2017 and 2017–2018, the Canadian SPSN could not attribute low VE to antigenic drift. Although based upon a small number of viruses antigenically characterized, our observations are similar to findings from the Crick Worldwide Influenza Centre, which summarized in its September reports that among viruses collected after January 31, >80% (47/58) of 3C.2a1 viruses in 2016–2017, and virtually all 3C.2a1b (32/33) and 3C.2a2 (24/25) viruses in 2017–2018, were antigenically similar to the cell-passaged 3C.2a vaccine strain [12]. Conversely, among the same isolates, nearly half of the 3C.2a1 (23/58), all 3C.2a1b, and all but one 3C.2a2 viruses were antigenically distinct from the egg-adapted 3C.2a vaccine strain [12]. Pivotal mutations in the egg-adapted vaccine in combination with mutations in circulating viruses may have increased antigenic distances and reduced VE both seasons.
The antigenic-distance hypothesis predicts minimal interference when the prior season’s vaccine antigen is distinct from the current season’s vaccine [15, 16]. Consistent with that hypothesis, we did not observe evidence that receipt of the 2015–2016 clade 3C.3a vaccine negatively interfered with effectiveness of the 2016–2017 clade 3C.2a vaccine. Conversely, the antigenic-distance hypothesis predicts pronounced negative interference when prior and current seasons’ vaccine strains are identical but are distinct from circulating viruses [15, 16]. Under those conditions, we observed lower VE point estimates among participants who received the identical 3C.2a vaccine in 2016–2017 and 2017–2018 (9%; 95% CI, −18% to 30%) compared to those vaccinated in 2017–2018 only (45%; 95% CI, −7% to 71%). Under similar conditions, investigators in Australia also reported lower VE point estimates among recipients of identical 3C.2a vaccine in 2016 and 2017 (3%; 95% CI, −29% to 27%) compared to those vaccinated in 2017 only (43%; 95% CI, −1% to 71%) [23]. Ferret experiments recapitulating these conditions have similarly shown reduced vaccine protection, and furthermore increased virus shedding, among animals vaccinated twice consecutively versus current season only with the identical 2016–2017/2017–2018 egg-adapted 3C.2a antigen [44].
In test-negative design analysis, test-negative controls should be representative of the source population. Among our test-negative controls ≥18 years old, 37% in 2016–2017 and 38% in 2017–2018 were considered vaccinated; this compares well with national coverage survey estimates of 36% and 38%, respectively, of Canadians ≥18 years old overall who reported being vaccinated. Similarly, among our controls ≥65 years old, 67% and 71%, respectively, were considered vaccinated in 2016–2017 and 2017–2018, which is also similar to national survey estimates of 69% and 71%, respectively [45, 46]. The proportion of our controls with comorbidity was also consistent in 2016–2017 (22%) and 2017–2018 (24%) with other national surveillance data indicating >20% of Canadians live with a major chronic disease [47]. These basic checks for potential selection bias are reassuring but we cannot rule out other biases, such as faulty recall of self-reported current or prior vaccination history, or residual confounding. We did not explore the effect of preexisting immunological landscapes shaped not only by prior season’s vaccination, but also by prior lifetime influenza infection history, recognizing that epitope-specific imprinting in childhood (ie, immunological cohort effects) may also differentially affect current vaccine protection by age [43]. Sample size constitutes the greatest limitation to our stratified analyses, resulting in broad and overlapping confidence intervals in subcluster and other subgroup analyses. Our comparisons based upon VE point estimates provide useful signals for further investigation but lack the statistical power and precision to be considered robust.
Better understanding of recurrently low VE against A(H3N2) will require subtype-specific analyses to become more granular. Empirical VE analyses stratified by phylogenetic subcluster, with reference to pivotal antigenic site mutations, could also help validate and refine theoretical models for epitope-based VE prediction [48–50]. Comprehensive understanding will likely require the integration not only of genomic tools, but also proteomic, glycomic, immunological, bioinformatic, and other advanced systems technology to synthesize the complex interactions underpinning influenza vaccine performance. Collaboration and investment sufficient to genetically characterize contributing viruses and to adequately power subset analyses should be a priority for influenza VE monitoring networks globally.
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Notes
Acknowledgments. The authors gratefully acknowledge the contribution of sentinel sites whose regular submission of specimens and data provide the basis of our analyses. We wish to acknowledge the administrative, coordination and data entry support in each participating province: Lisan Kwindt, Kaitlyn Shaw, and Shinhye Kim, British Columbia Centre for Disease Control; Elaine Douglas, Virginia Goetz, Dylan Kendrick, and Manish Ranpara, TARRANT, Alberta; Kathleen Parris, Public Health Ontario; and Sophie Auger, Institut National de Santé Publique du Québec. We thank those who provided laboratory and technical support in each province at the British Columbia Centre for Disease Control Public Health Laboratory, the Alberta Provincial Laboratory for Public Health, the Public Health Ontario Laboratory, the Laboratoire de Santé Publique du Québec, and the National Microbiology Laboratory. The authors acknowledge contributing laboratories and investigators of sequences uploaded to the Global Initiative on Sharing All Influenza Data (www.gisaid.org).
Disclaimer . The views expressed herein do not necessarily represent the view of the Public Health Agency of Canada.
Financial support. This work was supported by the British Columbia Centre for Disease Control; Alberta Health; Public Health Ontario; Ministère de la Santé et des Services Sociaux du Québec; l’Institut National de Santé Publique du Québec; and the Public Health Agency of Canada (grant to D. M. S.; arrangement number 1617-HQ-000083).
Potential conflicts of interest. G. D. S. has received grants for investigator-initiated studies unrelated to influenza vaccine from Pfizer and provided paid expert testimony for the Ontario Nurses Association, the Quebec Ministry of Justice and GSK. M. K. has received research grants from Roche and Hologic for unrelated studies. S. J. D. is a content expert consultant to Johnson and Johnson (Janssen) Pharmaceuticals on a literature search for point-of-care testing for respiratory viruses. Other authors report no potential conflicts.
All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
Presented in part: Canadian Immunization Conference, Ottawa, Ontario, Canada, 4–6 December 2018; and Options X for the Control of Influenza, Singapore, 29 August 2019.
References
- 1. Public Health Agency of Canada (PHAC). Weekly influenza reports. Ottawa: PHAC. https://www.canada.ca/en/public-health/services/diseases/flu-influenza/influenza-surveillance/weekly-influenza-reports.html. Accessed 2 October 2019. [Google Scholar]
- 2. Centers for Disease Control and Prevention (CDC). FluView: Weekly U.S. influenza surveillance report. Atlanta, GA: CDC. https://www.cdc.gov/flu/weekly/. Accessed 2 October 2019. [Google Scholar]
- 3. Blanton L, Alabi N, Mustaquim D, et al. Update: influenza activity in the United States during the 2016-17 season and composition of the 2017-18 influenza vaccine. MMWR Morb Mortal Wkly Rep 2017; 66:668–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Garten R, Blanton L, Elal AIA, et al. Update: influenza activity in the United States during the 2017-18 season and composition of the 2018-19 influenza vaccine. MMWR Morb Mortal Wkly Rep 2018; 67:634–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Centers for Disease Control and Prevention (CDC). How CDC classifies flu severity. Atlanta, GA: CDC. https://www.cdc.gov/flu/about/classifies-flu-severity.htm. Accessed 2 October 2019. [Google Scholar]
- 6. British Columbia Centre for Disease Control. Sentinel network (SPSN). http://www.bccdc.ca/health-info/diseases-conditions/influenza/sentinel-network-spsn. Accessed 2 October 2019.
- 7. Skowronski DM, Chambers C, Sabaiduc S, et al. A perfect storm: impact of genomic variation and serial vaccination on low influenza vaccine effectiveness during the 2014-2015 season. Clin Infect Dis 2016; 63:21–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Skowronski DM, Janjua NZ, De Serres G, et al. Low 2012-13 influenza vaccine effectiveness associated with mutation in the egg-adapted H3N2 vaccine strain not antigenic drift in circulating viruses. PLoS One 2014; 9:e92153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Skowronski DM, Chambers C, Sabaiduc S, et al. Integrated sentinel surveillance linking genetic, antigenic, and epidemiologica monitoring of influenza vaccine-virus relatedness and effectiveness during the 2013–2014 influenza season. J Infect Dis 2015; 12:726–39. [DOI] [PubMed] [Google Scholar]
- 10. Skowronski DM, Chambers C, Sabaiduc S, et al. Beyond antigenic match: possible agent-host and immuno-epidemiological influences on influenza vaccine effectiveness during the 2015-2016 season in Canada. J Infect Dis 2017; 216:1487–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Nextstrain. Real-time tracking of influenza A/H3N2 evolution using data from GISAID.https://nextstrain.org/flu/seasonal/h3n2/ha/6y. Accessed 2 October 2019.
- 12. Francis Crick Institute. Worldwide influenza centre. Annual and interim reports.https://www.crick.ac.uk/research/worldwide-influenza-centre/annual-and-interim-reports/. Accessed 2 October 2019.
- 13. European Centre for Disease Prevention and Control. Influenza virus characterization reports, summary Europe. https://ecdc.europa.eu/en/seasonal-influenza/surveillance-and-disease-data/influenza-virus-characterisation. Accessed 2 October 2019.
- 14. Belongia EA, Skowronski DM, McLean HQ, Chambers C, Sundaram ME, De Serres G. Repeated annual influenza vaccination and vaccine effectiveness: review of evidence. Expert Rev Vaccines 2017; 16:1–14. [DOI] [PubMed] [Google Scholar]
- 15. Skowronski DM, Chambers C, De Serres G, et al. Serial vaccination and the antigenic distance hypothesis: effects on influenza vaccine effectiveness during A(H3N2) epidemics in Canada, 2010-2011 to 2014-2015. J Infect Dis 2017; 215:1059–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Smith DJ, Forrest S, Ackley DH, Perelson AS. Variable efficacy of repeated annual influenza vaccination. Proc Natl Acad Sci U S A 1999; 96:14001–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. World Health Organization (WHO). WHO recommendations on the composition of influenza virus vaccines Geneva, Switzerland: WHO, 2019. https://www.who.int/influenza/vaccines/virus/recommendations/en/. Accessed 2 October 2019. [Google Scholar]
- 18. Zost SJ, Parkhouse K, Gumina ME, et al. Contemporary H3N2 influenza viruses have a glycosylation site that alters binding of antibodies elicited by egg-adapted vaccine strains. Proc Natl Acad Sci U S A 2017; 114:12578–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wu NC, Zost SJ, Thompson AJ, et al. A structural explanation for the low effectiveness of the seasonal influenza H3N2 vaccine. PLoS Pathog 2017; 13:e1006682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Katz JM, Hancock K, Xu X. Serologic assays for influenza surveillance, diagnosis and vaccine evaluation. Expert Rev Anti Infect Ther 2011; 9:669–83. [DOI] [PubMed] [Google Scholar]
- 21. Skowronski DM, Sabaiduc S, Chambers C, et al. Mutations acquired during cell culture isolation may affect antigenic characterisation of influenza A(H3N2) clade 3C.2a viruses. Euro Surveill 2016; 21:30112. [DOI] [PubMed] [Google Scholar]
- 22. Melidou A, Broberg E; European Region Influenza Surveillance Network . Predominance of influenza A(H3N2) virus genetic subclade 3C.2a1 during an early 2016/17 influenza season in Europe. Contribution of surveillance data from World Health Organization (WHO) European Region to the WHO vaccine composition consultation for northern hemisphere 2017/18. Vaccine 2017; 35:4828–35. [DOI] [PubMed] [Google Scholar]
- 23. Sullivan SG, Chilver MB, Carville KS, et al. Low interim influenza vaccine effectiveness, Australia, 1 May to 24 September 2017. Euro Surveill 2017; 22:17-00707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. An Y, McCullers JA, Alymova I, Parsons LM, Cipollo JF. Glycosylation analysis of engineered H3N2 influenza A virus hemagglutinins with sequentially added historically relevant glycosylation sites. J Proteome Res 2015; 14:3957–69. [DOI] [PubMed] [Google Scholar]
- 25. Tate MD, Job ER, Deng YM, Gunalan V, Maurer-Stroh S, Reading PC. Playing hide and seek: how glycosylation of the influenza virus hemagglutinin can modulate the immune response to infection. Viruses 2014; 6:1294–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Abe Y, Takashita E, Sugawara K, Matsuzaki Y, Muraki Y, Hongo S. Effect of the addition of oligosaccharides on the biological activities and antigenicity of influenza A/H3N2 virus hemagglutinin. J Virol 2004; 78:9605–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ndifon W, Wingreen NS, Levin SA. Differential neutralization efficiency of hemagglutinin epitopes, antibody interference, and the design of influenza vaccines. Proc Natl Acad Sci U S A 2009; 106:8701–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Popova L, Smith K, West AH, et al. Immunodominance of antigenic site B over site A of hemagglutinin of recent H3N2 influenza viruses. PLoS One 2012; 7:e41895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Koel BF, Burke DF, Bestebroer TM, et al. Substitutions near the receptor binding site determine major antigenic change during influenza virus evolution. Science 2013; 342:976–9. [DOI] [PubMed] [Google Scholar]
- 30. Altman MO, Angel M, Kosik I, et al. Human influenza A virus hemagglutinin glycan evolution follows a temporal pattern to a glycan limit. mBio 10:e00204-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Firth D. Bias reduction of maximum likelihood estimates. Biometrika 1993; 80:27–8. [Google Scholar]
- 32. Heinze G, Schemper M. A solution to the problem of separation in logistic regression. Stat Med 2002; 21:2409–19. [DOI] [PubMed] [Google Scholar]
- 33. Devika S, Jeyaseelan L, Sebastian G. Analysis of sparse data in logistic regression in medical research: a newer approach. J Postgrad Med 2016; 62:26–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Belongia EA, Simpson MD, King JP, et al. Variable influenza vaccine effectiveness by subtype: a systematic review and meta-analysis of test-negative design studies. Lancet Infect Dis 2016; 16:942–51. [DOI] [PubMed] [Google Scholar]
- 35. Skowronski DM, Chambers C, Sabaiduc S, et al. Interim estimates of 2016/17 vaccine effectiveness against influenza A(H3N2), Canada, January 2017. Euro Surveill 2017; 22:30460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Flannery B, Chung JR, Monto AS, et al. ; US Flu VE Investigators . Influenza vaccine effectiveness in the United States during the 2016-2017 Season. Clin Infect Dis 2019; 68:1798–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Pebody R, Warburton F, Ellis J, et al. End-of-season influenza vaccine effectiveness in adults and children, United Kingdom, 2016/17. Euro Surveill 2017; 22:17-00306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kissling E, Pozo F, Buda S, et al. ; I-MOVE/I-MOVE+ Study Team . Effectiveness of influenza vaccine against influenza A in Europe in seasons of different A(H1N1)pdm09 and the same A(H3N2) vaccine components (2016-17 and 2017-18). Vaccine X 2019; 3:100042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Skowronski DM, Chambers C, De Serres G, et al. Early season co-circulation of influenza A(H3N2) and B(Yamagata): interim estimates of 2017/18 vaccine effectiveness, Canada, January 2018. Euro Surveill 2018; 23:18-00035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Pebody R, Djennad A, Ellis J, et al. End of season influenza vaccine effectiveness in adults and children in the United Kingdom in 2017/18. Euro Surveill 2019; 24: 1800488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Rolfes MA, Flannery B, Chung JR, et al. ; US Influenza Vaccine Effectiveness (Flu VE) Network, the Influenza Hospitalization Surveillance Network, and the Assessment Branch, Immunization Services Division, Centers for Disease Control and Prevention . Effects of influenza vaccination in the United States during the 2017-2018 influenza season. Clin Infect Dis 2019; 69:1845–53.30715278 [Google Scholar]
- 42. Kissling E, Pozo F, Buda S, et al. Low 2018/19 vaccine effectiveness against influenza A(H3N2) among 15-64-year olds in Europe: exploration by birth cohort. Euro Surveill 2019; 24:1900604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Skowronski DM, Sabaiduc S, Leir S, et al. Paradoxical clade- and age-specific vaccine effectiveness during the 2018/19 influenza A(H3N2) epidemic in Canada: potential imprint-regulated effect of vaccine (I-REV). Euro Surveill 2019; 24:1900585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Music N, Tzeng WP, Liaini Gross F, et al. Repeated vaccination against matched H3N2 influenza virus gives less protection than single vaccination in ferrets. NPJ Vaccines 2019; 4:28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Public Health Agency of Canada. 2016/17 seasonal influenza vaccine coverage in Canada.Ottawa: Government of Canada, 2018. http://publications.gc.ca/collections/collection_2018/aspc-phac/HP40-198-2017-eng.pdf. Accessed 2 October 2019. [Google Scholar]
- 46. Public Health Agency of Canada. Seasonal influenza vaccine coverage in Canada, 2017–2018.Ottawa: Government of Canada, 2019. http://publications.gc.ca/collections/collection_2019/aspc-phac/HP40-198-2018-eng.pdf. Accessed 2 October 2019. [Google Scholar]
- 47. Public Health Agency of Canada. How healthy are Canadians?Ottawa: Government of Canada, 2016. https://www.canada.ca/en/public-health/services/publications/healthy-living/how-healthy-canadians.html#s3-3. Accessed 2 October 2019. [Google Scholar]
- 48. Gupta V, Earl DJ, Deem MW. Quantifying influenza vaccine efficacy and antigenic distance. Vaccine 2006; 24:3881–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Pan K, Subieta KC, Deem MW. A novel sequence-based antigenic distance measure for H1N1, with application to vaccine effectiveness and the selection of vaccine strains. Protein Eng Des Sel 2011; 24:291–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Deem MW, Pan K. The epitope regions of H1-subtype influenza A, with application to vaccine efficacy. Protein Eng Des Sel 2009; 22:543–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.