Abstract
Background
We investigated the pathogenesis of pneumococcal pneumonia using clinical specimens collected for pneumonia surveillance in The Gambia.
Methods
Lung aspirates and nasopharyngeal swabs from 31 patients were examined by culture, quantitative polymerase chain reaction (qPCR), whole genome sequencing, serotyping, and reverse-transcription qPCR.
Results
Five lung aspirates cultured pneumococci, with a matching strain identified in the nasopharynx. Three virulence genes including ply (pneumolysin) were upregulated >20-fold in the lung compared with the nasopharynx. Nasopharyngeal pneumococcal density was higher in pediatric pneumonia patients compared with controls (P < .0001).
Conclusions
Findings suggest that changes in pneumococcal gene expression occurring in the lung environment may be important in pathogenesis.
Keywords: lung, nasopharynx, pneumococcus, pneumonia, Streptococcus pneumoniae
We investigated the pathogenesis of pneumococcal pneumonia using clinical samples. Results demonstrated that pneumococci in the lung originated from the nasopharynx and suggested that changes in pneumococcal gene expression in the lung are a feature of pneumococcal pneumonia.
Streptococcus pneumoniae (the pneumococcus) is a common cause of community-acquired pneumonia, a major public health problem that primarily affects young children and the elderly. Pneumococcal pneumonia is believed to occur when pneumococci colonizing the upper respiratory tract are aspirated into the lung, where a failure to clear the bacteria leads to replication and triggers a damaging immune response. Supporting clinical data are limited because lung aspirates are not routinely collected in most settings; however, a previous study from The Gambia found that 85% of pediatric pneumonia patients with pneumococci identified in lung aspirates had the same serotype detected in a nasopharyngeal swab [1]. Studies have found higher pneumococcal loads in the nasopharynx in children with pneumonia compared with healthy controls, suggesting that increased pneumococcal density in the nasopharynx may facilitate bacterial spread to the lung, or alternatively, may be due to pneumonia [2, 3]. The processes by which pneumococci transition from a colonizing to a pathogenic state are not well understood.
Pneumococci possess a variety of virulence factors, including polysaccharide capsule, the toxin pneumolysin, adhesins, and surface proteins that allow them to cause disease [4]. After examining pneumococcal gene expression in a range of in vitro conditions designed to mimic host microenvironments, Aprianto et al [5] identified 498 conditionally expressed genes, highlighting the dynamic transcriptional ability of the pneumococcus. Experiments using animal models demonstrated that the pneumococcus modifies its gene expression in response to different niches within the host [6, 7]. To date, only a single published study has examined pneumococcal gene expression in clinical samples. Using nasopharyngeal swabs collected from healthy children, Sakai et al [8] found different levels of expression for 11 pneumococcal genes, demonstrating the feasibility of evaluating pneumococcal gene expression directly from clinical samples.
Using a unique set of samples collected as part of pneumonia surveillance in The Gambia, we sought to investigate the pathogenesis of pneumococcal pneumonia by comparing pneumococcal loads in the nasopharynx of pneumonia patients and healthy carriers, conducting whole genome sequencing of paired pneumococcal isolates obtained from the nasopharynx and lung of pneumonia patients, and analyzing pneumococcal gene expression directly from nasopharyngeal swabs and lung aspirates.
METHODS
Detailed methods are provided in the Supplementary Information. This study was nested within population-based surveillance for suspected pneumonia, septicemia, and meningitis among patients aged 2 months or greater in the Basse Health and Demographic surveillance system in The Gambia, as described previously [9]. All patients with suspected pneumonia who had a lung aspiration from April 2015 to May 2016 were included in this study. Nasopharyngeal swabs were collected from pneumonia patients and community controls. For all swabs and lung aspirates, an aliquot for ribonucleic acid (RNA) analysis was prepared by adding 2-fold volume of RNAprotect Bacteria Reagent (QIAGEN) to the clinical sample.
The surveillance project and this substudy were approved by The Gambia Government/Medical Research Council Joint Ethics Committee (approval numbers 1087 and L2016.09v2). Patients or their parent or guardian provided written informed consent for enrolment in the surveillance project and lung aspiration.
Laboratory Procedures
In brief, nasopharyngeal swab samples underwent deoxyribonucleic acid (DNA) extraction and lytA real-time quantitative polymerase chain reaction (qPCR) to detect and quantify pneumococci. Pneumococcal density data are reported as genome equivalents/mL (GE/mL). Molecular serotyping by microarray was conducted on nasopharyngeal swabs. Pneumococci isolated from the lung and nasopharynx were serotyped by traditional antibody-based methods and whole genome sequencing conducted using the MiSeq platform. Expression of 9 pneumococcal genes selected from the literature was examined by reverse-transcription qPCR after RNA extraction from clinical specimens. Genes and primer sequences are provided in Supplementary Table 1.
Data analysis was performed using STATA version 15.1 and GraphPad Prism version 7.03. Categorical data were compared using the χ 2 test. Pneumococcal density data were log10 transformed before analysis and examined using non-parametric methods. Densities were compared using the Mann-Whitney test, and multivariable quantile regression models included age, sex, and Pneumococcal conjugate vaccine (PCV) status as variables selected a priori.
RESULTS
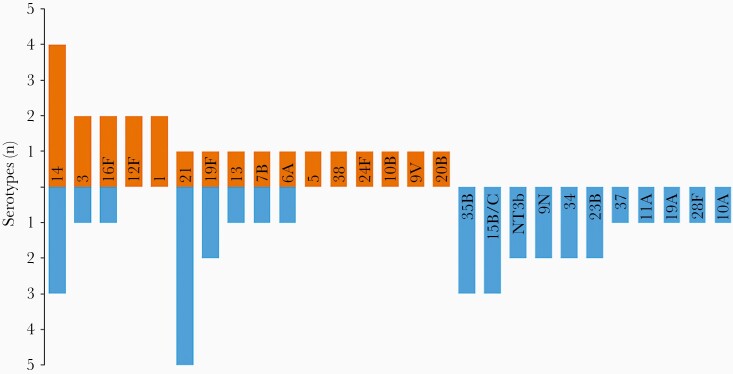
Samples from 31 pneumonia patients enrolled in surveillance were included in this study. Patient ages ranged from 2 months to 73 years, with a median of 3.8 years (interquartile range [IQR], 1.7–7.0). The majority were children, with 20 (65%) aged <5 years, 6 (19%) from 5 to 18 years, and 5 (16%) >18 years, and 20 of 31 (65%) were male. Examination of nasopharyngeal swabs from patients of all ages found that 27 (87%) contained pneumococci. We compared the prevalence, density, and serotypes of pneumococci in the nasopharynx of patients <5 years of age with community controls. Participant characteristics are shown in Supplementary Table 2. Pneumococcal prevalence was similar in pneumonia patients (18 of 20; 90%) and community controls (21 of 22, 96%, P = .493, χ 2 test). Among children with pneumococci detected in the nasopharynx, median density was higher in pneumonia patients (7.77 log10 GE/mL; IQR, 7.33–7.98) compared with controls (6.05; IQR, 5.67–6.47; P < .0001). When adjusted for age, sex, and PCV status, the difference remained significant, with a coefficient (difference between medians) of 1.61 log10 GE/mL (95% confidence interval, 0.96–2.26; P < .001). Serotypes identified in nasopharyngeal swabs from pneumonia patients and controls are shown in Figure 1. Eight serotypes were found in both participant groups, with 8 additional serotypes found only in pneumonia patients and 11 only found in community controls.
Figure 1.
Mirror plot of pneumococcal serotypes identified in nasopharyngeal swabs of children <5 years of age hospitalized with pneumonia (orange bars) compared with pneumococcal serotypes found in nasopharyngeal swabs collected from community controls (blue bars). NT3b refers to a lineage of nonencapsulated Streptococcus pneumoniae.
Culture of lung aspirates from 31 pneumonia patients of all ages identified pneumococci as the causative organism in 5 patients (serotypes 3, 14, 1, 12F, and 32A; n = 1 each) and Staphylococcus aureus in 1. All 5 patients who had pneumococci cultured from the lung aspirate had the same serotype identified in their nasopharyngeal swab. Pneumococcal lytA qPCR was conducted for the lung aspirates with sufficient volume remaining for analysis (n = 25). Ten (40%) lung aspirates tested positive by qPCR, including the 5 samples that were also positive by culture. Median pneumococcal density was higher in the culture-positive lung aspirates (5.74 log10 GE/mL; IQR, 4.93–5.93) compared with the culture-negative lung aspirates (3.32 log10 GE/mL; IQR, 2.86–3.88; P = .008).
To identify within-host genomic changes, in particular DNA mutations that may contribute to pneumococcal pathogenesis, whole genome sequencing was conducted on the 5 paired pneumococcal isolates from the lung and nasopharynx of the same patient. For all 5 patients, the isolate from the nasopharynx was the same as that from the lung, with >99% sequence similarity and matching multilocus sequence types (Supplementary Table 3). Three mutations were identified in isolates from lung aspirates (Supplementary Table 4).
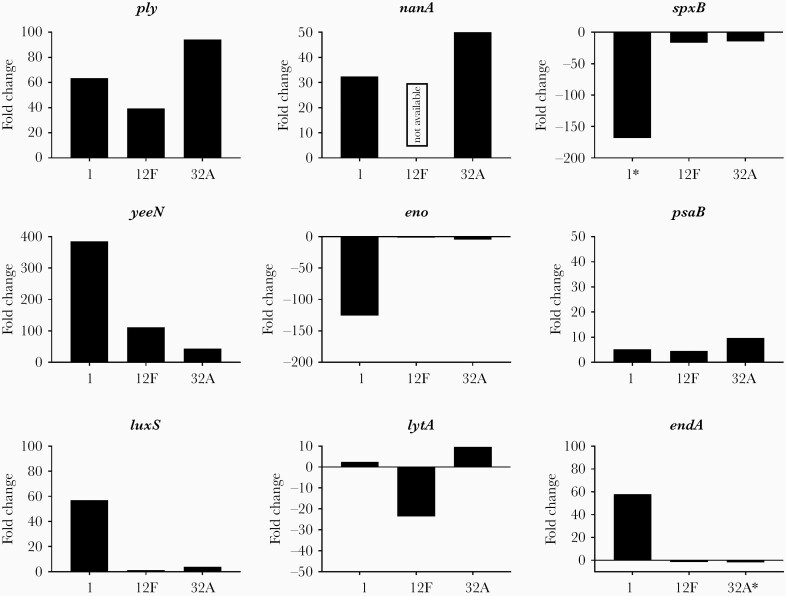
We investigated differences in pneumococcal gene expression by examining RNA extracted directly from nasopharyngeal swabs and lung aspirates from the same 5 patients. After evaluation of RNA concentration and quality, samples containing serotype 3 were excluded from analysis due to DNA contamination, and samples containing serotype 14 were excluded due to insufficient RNA yield from the lung aspirate. Differences in expression of 9 genes in the lung aspirate compared with the nasopharynx are shown in Figure 2. Virulence genes ply (pneumolysin), nanA (neuraminidase A), and the putative virulence regulator yeeN were consistently and strongly upregulated (fold change >20) in the lung compared with the nasopharynx. In contrast, psaB (pneumococcal surface antigen B), endA (endonuclease), and the quorum sensing gene luxS were upregulated to a lesser extent, or varied by strain. Genes encoding pyruvate oxidase and enolase (spxB, and eno, respectively) were downregulated in the lung, most prominently in serotype 1. Expression of lytA (autolysin) was downregulated in 12F but upregulated in serotypes 1 and 32A.
Figure 2.
Differential expression of pneumococcal genes in lung aspirates compared with nasopharyngeal swabs collected from the same patient by reverse-transcription quantitative polymerase chain reaction. Fold change in gene expression was derived using the 2−ΔΔCt method with gyrA as the reference gene. Each bar represents data from one gene in 1 set of patient samples. * indicates an imputed Cycle threshold (Ct) value of 40 used for the lung aspirate sample. For serotype 12F, data for nanA are not available because no transcripts were detected in either sample.
DISCUSSION
We investigated the microbiological features of pneumococcal pneumonia using a unique set of clinical specimens obtained as part of pneumonia surveillance in The Gambia. This is a high pneumococcal carriage setting, reflected in the high colonization rates observed in both pneumonia patients and community controls. In children under 5 years of age, pneumococcal densities in the nasopharynx were over 1 log higher in pneumonia patients compared with controls. These results are consistent with data from Vietnam and the multicenter Pneumonia Etiology Research for Child Health (PERCH) study [2, 3]. Using whole genome sequencing, we demonstrated that pneumococci present in the lung of pneumonia patients originated from the nasopharynx. Although our study numbers were small, a previous study in The Gambia found the same serotype in the nasopharynx and lung for 23 of 27 (85%) pneumonia cases [1]. A multicenter study of children <5 years hospitalized for pneumonia reported that for 34 cases with pneumococci detected in blood, 27 (79%) had the same serotype detected in the nasopharynx [10]. These findings, taken together, provide further evidence that pneumococcal pneumonia originates from bacteria present in the upper respiratory tract, and they suggest that sampling the nasopharynx may provide some insight into pneumonia etiology.
A major finding from our study is that few DNA mutations were acquired by the pneumococcus during disease, whereas prominent differences in pneumococcal gene expression were observed between the lung and the nasopharynx. Although it is possible that mutations, for example in the psaB promoter region, may have impacted virulence, our results suggest that transcriptional differences rather than genomic changes underpin pneumococcal pathogenesis within the host. The ply gene, which encodes the pore-forming toxin pneumolysin, was strongly upregulated in the lung for all 3 isolates examined. Expression of endA, which encodes an endonuclease that degrades neutrophil extracellular traps (NETs), was upregulated in the lung, whereas eno, which encodes an enolase that can induce NET formulation, was downregulated [11, 12]. These results suggest that avoidance of NETs may facilitate pneumococcal survival in the lung. SpxB has been linked to multiple pneumococcal activities, including virulence, colonization, capsule synthesis, and transmission. Downregulation of spxB in the lung may relate to one of these functions or its metabolic role in oxygen sensing and carbon source utilization [13]. It is interesting to note that the serotype 1 isolate displayed the most prominent changes in gene expression in the lung. This isolate belongs to sequence type 3081, a clone that is the leading cause of invasive pneumococcal disease in The Gambia and is highly virulent in animal models [14].
Viral testing was not conducted as part of this study. Not all lung aspirates underwent lytA qPCR, and serotyping was not conducted on the 5 culture-negative, lytA-positive lung aspirates. One key limitation is that gene expression studies were restricted to 3 paired specimens. We did not evaluate the expression of capsule, an important virulence factor. Certain capsular serotypes have been linked to pneumonia: serotypes 1, 5, 22F, 7F, and 14 were more likely to be found in the nasopharynx of children with pneumonia than in healthy controls in a study conducted before PCV use [15]. A similar study conducted in a post-PCV population would provide useful data for consideration of serotype coverage of next-generation PCVs. Although our sample size was small, data suggested a differing serotype distribution between pneumonia patients and community carriers, with serotypes 1 and 5 identified solely in patients.
CONCLUSIONS
There is a paucity of data on pneumococcal gene expression from clinical samples, with 1 study to date examining pneumococcal RNA in the nasopharynx of healthy carriers [8]. Due to technical challenges, only 3 paired samples from our study were suitable for analysis of pneumococcal gene expression. Nevertheless, this study represents a proof-of-concept for conducting pneumococcal gene expression analysis directly from patient clinical specimens, identifying considerable changes in expression that occur within the lung. Although we hypothesize that the lung environment triggered these changes, other factors may have facilitated pneumococcal spread to the lung, and evaluating additional isolates is warranted. More important, specimens were stored in an RNA-stabilizing reagent promptly after collection. Future studies may expand upon the current findings by examining other sample types, such as sputum or pleural fluid, or applying transcriptomic approaches such as RNA sequencing (RNA-seq) to examine global changes in pneumococcal gene expression. In an experimental model, dual RNA-seq has been used to identify transcriptional changes in both pneumococci and the host associated with pneumococcal invasion into the pleural space [6]. The identification of molecular signatures associated with pneumococcal pneumonia could inform development of novel diagnostics and protein-based vaccines. We envision that future studies building upon this approach will extend our understanding of the pathogenesis of pneumococcal pneumonia.
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Notes
Disclaimer. Funders had no role in study design, analysis, and interpretation of data, or manuscript preparation.
Financial support. Pneumonia surveillance in The Gambia was funded by Gavi, the Vaccine Alliance, the Bill & Melinda Gates Foundation (OPP1020327), and the UK Medical Research Council. This study was funded by the Murdoch Children’s Research Institute, which was supported by the Victorian Government’s Operational Infrastructure Support Program. C. S. was supported by an Australian National Health Medical Research Council Career Development Fellowship (1087957) and a Veski Inspiring Women Fellowship. Y. H. was supported by a student research award from the Australian Society for Microbiology Victorian Branch.
Potential conflicts of interest. C. S., E. M. D., and E. K. M. are investigators on an unrelated study funded by Pfizer. J. H. is an investigator on studies undertaken on behalf of St George’s, University of London or BUGS Bioscience that are sponsored or funded by vaccine manufacturers, including Pfizer, GlaxoSmithKline and Sanofi Pasteur. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
Presented in part: 2019 Australian Society for Microbiology Conference, June 30–July 3, 2019, Adelaide, Australia.
References
- 1. Tokarz R, Briese T, Morris G, et al. Serotype analysis of Streptococcus pneumoniae in lung and nasopharyngeal aspirates from children in the Gambia by MassTag PCR. J Clin Microbiol 2013; 51:995–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Vu HT, Yoshida LM, Suzuki M, et al. Association between nasopharyngeal load of Streptococcus pneumoniae, viral coinfection, and radiologically confirmed pneumonia in Vietnamese children. Pediatr Infect Dis J 2011; 30:11–8. [DOI] [PubMed] [Google Scholar]
- 3. Baggett HC, Watson NL, Deloria Knoll M, et al. ; PERCH Study Group . Density of upper respiratory colonization with Streptococcus pneumoniae and its role in the diagnosis of pneumococcal pneumonia among children aged <5 years in the PERCH Study. Clin Infect Dis 2017; 64:317–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Weiser JN, Ferreira DM, Paton JC. Streptococcus pneumoniae: transmission, colonization and invasion. Nat Rev Microbiol 2018; 16:355–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Aprianto R, Slager J, Holsappel S, Veening JW. High-resolution analysis of the pneumococcal transcriptome under a wide range of infection-relevant conditions. Nucleic Acids Res 2018; 46:9990–10006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ritchie ND, Evans TJ. Dual RNA-seq in Streptococcus pneumoniae infection reveals compartmentalized neutrophil responses in lung and pleural space. mSystems 2019; 4:e00216–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ogunniyi AD, Mahdi LK, Trappetti C, et al. Identification of genes that contribute to the pathogenesis of invasive pneumococcal disease by in vivo transcriptomic analysis. Infect Immun 2012; 80:3268–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Sakai F, Talekar SJ, Klugman KP, Vidal JE. Expression of Streptococcus pneumoniae virulence-related genes in the nasopharynx of healthy children. PLoS One 2013; 8:e67147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Mackenzie GA, Plumb ID, Sambou S, et al. Monitoring the introduction of pneumococcal conjugate vaccines into West Africa: design and implementation of a population-based surveillance system. PLoS Med 2012; 9:e1001161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Dananché C, Paranhos-Baccalà G, Messaoudi M, et al. et al. Serotypes of Streptococcus pneumoniae in children aged <5 years hospitalized with or without pneumonia in developing and emerging countries: a descriptive, multicenter study. Clin Infect Dis 2020; 70:875–83. [DOI] [PubMed] [Google Scholar]
- 11. Zhu L, Kuang Z, Wilson BA, Lau GW. Competence-Independent Activity of Pneumococcal Enda Mediates Degradation of Extracellular DNA and Nets and Is Important for Virulence. PLoS One 2013; 8:e70363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Mori Y, Yamaguchi M, Terao Y, Hamada S, Ooshima T, Kawabata S. α-Enolase of Streptococcus pneumoniae induces formation of neutrophil extracellular traps. J Biol Chem 2012; 287:10472–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Carvalho SM, Farshchi Andisi V, Gradstedt H, et al. Pyruvate oxidase influences the sugar utilization pattern and capsule production in Streptococcus pneumoniae. PLoS One 2013; 8:e68277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Bricio-Moreno L, Ebruke C, Chaguza C, et al. Comparative genomic analysis and in vivo modeling of Streptococcus pneumoniae ST3081 and ST618 isolates reveal key genetic and phenotypic differences contributing to clonal replacement of serotype 1 in The Gambia. J Infect Dis 2017; 216:1318–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Greenberg D, Givon-Lavi N, Newman N, Bar-Ziv J, Dagan R. Nasopharyngeal carriage of individual Streptococcus pneumoniae serotypes during pediatric pneumonia as a means to estimate serotype disease potential. Pediatr Infect Dis J 2011; 30:227–33. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.