Abstract
Context
Anemia during early pregnancy (EP) is common in developing countries and is associated with adverse health consequences for both mothers and children. Offspring of women with EP anemia often have low birth weight, which increases risk for cardiometabolic diseases, including type 2 diabetes (T2D), later in life.
Objective
We aimed to elucidate mechanisms underlying developmental programming of adult cardiometabolic disease, including epigenetic and transcriptional alterations potentially detectable in umbilical cord blood (UCB) at time of birth.
Methods
We leveraged global transcriptome- and accompanying epigenome-wide changes in 48 UCB from newborns of EP anemic Tanzanian mothers and 50 controls to identify differentially expressed genes (DEGs) in UCB exposed to maternal EP anemia. DEGs were assessed for association with neonatal anthropometry and cord insulin levels. These genes were further studied in expression data from human fetal pancreas and adult islets to understand their role in beta-cell development and/or function.
Results
The expression of 137 genes was altered in UCB of newborns exposed to maternal EP anemia. These putative signatures of fetal programming, which included the birth weight locus LCORL, were potentially mediated by epigenetic changes in 27 genes and associated with neonatal anthropometry. Among the DEGs were P2RX7, PIK3C2B, and NUMBL, which potentially influence beta-cell development. Insulin levels were lower in EP anemia–exposed UCB, supporting the notion of developmental programming of pancreatic beta-cell dysfunction and subsequently increased risk of T2D in offspring of mothers with EP anemia.
Conclusions
Our data provide proof-of-concept on distinct transcriptional and epigenetic changes detectable in UCB from newborns exposed to maternal EP anemia.
Keywords: developmental programming, maternal early pregnancy anemia, beta-cell function, beta-cell development, type 2 diabetes, epigenetic programming
Anemia is a major health problem globally, especially among pregnant women. About 42% of pregnant women worldwide suffer from anemia defined as hemoglobin (Hb) < 11 g/dL in the first and third trimesters and < 10.5 g/dL in the second trimester (1). The prevalence of anemia during early pregnancy (EP) is about 18% in developed countries, but 35% to 75% in developing countries (2). Sub-Saharan Africa has a high incidence of anemia and in Tanzania, the prevalence was reported to be 57% in 2015-2016 (3). Anemia can be caused by several factors including malnutrition, infections such as malaria, and hereditary disorders (4-6). In Tanzania, malaria together with iron deficiency are common causes of anemia during pregnancy (7-9).
Anemia during pregnancy can have severe effects on the health of the offspring. Moderate to severe anemia (Hb < 8-9 g/dL) has been associated with decreased birth weight (BW) (10, 11), and first and second trimester anemia may be especially detrimental (12-14). Barker and Hales hypothesized that insults during fetal life causing decreased BW could predispose an individual to conditions like diabetes and cardiovascular disease (CVD) later in life (15-18). How and when anemia affects fetal growth is not well established, but poor growth prior to the third trimester of pregnancy could represent a particularly vulnerable time-window of pregnancy for fetal programming predisposing to diabetes and CVD (19). In Tanzania, anemia and infectious diseases remain high in children, while diabetes and CVD increase rapidly in older generations (20, 21), facilitating a need to explore the role of maternal anemia on the epidemic of diabetes and CVD.
The impact of anemia during EP on the risk of chronic diseases in the offspring may potentially be mediated by epigenetic mechanisms, for example, DNA methylation, which can influence gene expression in the offspring, here represented by umbilical cord blood (UCB). Therefore, examination of gene expression alterations in the UCB from offspring of mothers who had anemia early in pregnancy could provide insights on how this sensitive fetal period may predispose the offspring to chronic disease later in life. To address these questions, we performed RNA sequencing of UCB from offspring of Tanzanian mothers with EP anemia from the FOETALforNCD study (22). We furthermore studied the accompanying changes in epigenetic patterns among selected genes whose expression levels were altered by early maternal anemia. Subsequently, we assessed if the expression levels of these genes were associated with short and/or long-term outcomes in the offspring. We studied the expression levels of the same genes in fetal and adult liver as well as pancreatic samples from another study (Swedish, GTEx) to determine if they had any potential roles in development and/or function of these organs. We compared insulin levels in UCB from EP anemic mothers to controls to identify early signs of impaired insulin secretion or action among EP anemic offspring. We then tested if any of the genes whose expression was altered in UCB of early-anemic mothers could have an impact on beta-cell development/function, which could then explain the lower cord insulin levels and higher type 2 diabetes (T2D) risk in later life.
Methods
Study Participants
The FOETALforNCD cohort profile has been described in detail previously (22). In brief, the field study was conducted in a rural region of Northeast Tanzania, in Korogwe District and adjacent villages in Handeni District, Tanga Region, from July 2014 to December 2016. The central field site was the maternity ward and the reproductive and child health clinic (RCH clinic) at Korogwe District Hospital and mobile outreach clinics were set up in 48 villages. Participants were recruited prior to conception or in early pregnancy and followed throughout pregnancy until birth and 1 month postnatally. In total, 1415 nonpregnant women participated in the preconception study, 538 in the pregnancy study, and 427 in the birth cohort study. Data collection included: maternal blood, screening for noncommunicable diseases and malaria, ultrasound in each trimester, neonatal anthropometry at birth and at 1 month of age, and UCB (22). Briefly, UCB samples from 366 participants were collected in EDTA, serum, and PAXgene tubes as described previously (22). The study was approved by regional ethical committees and all participants gave written informed consent.
Study Design and Maternal Diagnosis
Maternal hemoglobin (Hb) was measured at the time of inclusion, all antenatal visits, and at delivery. Inclusion criteria for the main cohort in the presented analyses were: availability of both UCB and UCB collected in PAXgene tubes (for RNAseq), malaria-negative throughout pregnancy, BW measured within 24 hours of delivery, gestational age of > 22 weeks at delivery (≥ 155 days), HIV negative (unknown HIV status excluded), a live birth, not having any hypertensive disorders, not having high Hb (>14 g/dL) at any point during pregnancy, and only singleton births (ie, no twins or triplets).
Anemia cases
Anemic cases comprised 50 women with early anemia defined as Hb ≤ 10.9 g/dL at the earliest time point during the first and second trimesters. We selected 48 cases based on those who had the lowest Hb levels at the lowest gestational age and who were consistently anemic during early pregnancy, and reached 50 cases when the cutoff at a gestational age of 21 weeks + 5 days was set (Supplementary Table S1 (23)).
Controls
The 50 controls were selected on the condition that they (1) did not have anemia up to 21 weeks + 4 days of gestational age (GA) during all the Hb measurements, (2) Hb was within normal range (ie, Hb not > 15, since Hb levels up to 15 are within normal range for women; all mothers had Hb < 14, with the exception of 1 mother who had a Hb = 14.4 only at delivery and was included in the study) (3) did not show any drop of an empirically selected > 2.3 g/dL from the lowest Hb in the GA interval (<21 weeks + 4 days) and until delivery, (4) did not, at any time point in pregnancy, have an Hb < 10.0 g/dL. Based on these criteria, 50 women with the highest Hb levels were selected as controls (Supplementary Table S1 (23)).
RNA Isolation and DNA Isolation and Quantification
DNA was extracted using the QIAamp DNA Blood Midi Kit (Qiagen, catalog # 51185, Sollentuna). DNA was quantified using picogreen assay (Promega). RNA was extracted from cord blood stored in PAXgene tubes using the PAXgene Blood RNA Kit (Qiagen, catalog# 762174, Sollentuna). RNA quantity was assessed using the Agilent 4200 Tape station system and quantified using the Qubit assays (Thermofisher Scientific).
RNA Sequencing
Library preparation, sequencing, and alignment
A quantity of 1 μg of RNA with RNA integrity number (RIN) ≥ 7 was used for library preparation using the TruSeq Stranded Total RNA Library Prep Globin (Illumina). RNA sequencing was performed using the 75-bp paired-end protocol on a Nextseq 500 platform (Illumina). Each individual transcriptome yielded, on average, 24.3 ± 10 (mean ± SD) million paired-end reads mapped to the human genome (NCBI build 38). Paired-end 101-bp length output reads were aligned to the human reference genome (hg19) using STAR (2.4.1a) (24) with the following parameters:
STAR –genomeDir ../Human/STAR_Genomes/hg38_Gencode22 –sjdbGTFfile genes.gtf –readFilesIn file.fastq.gz –out FilterType BySJout –runThreadN 12 –outFilterMultimapNmax 20 –alignSJDBoverhangMin 1 –sjdbOverhang 99 –outFilterMismatchNoverLmax 0.04 –alignIntronMin 20 –alignIntronMax 1000000 –outSAMstrandFieldintronMotif –outReadsUnmappedFastx –readFilesCommandzcat
The annotated RefSeq GTF transcript and fasta genome files were downloaded from Ensembl (https://www.ensembl.org/index.html). Gene expression was measured as the normalized sum of expression of all exons. Exons were defined as nonoverlapping unique exonic units, as described previously (25). The dexseq_count python script (https://www.huber.embl.de/pub/DEXSeq/) was used by counting uniquely mapped reads in each exon. The htseq_count python script (http://www-huber.embl.de/users/anders/HTSeq/doc/counting.html) was used for counting uniquely mapped reads for each gene (26).
Preprocessing and statistical analyses
Gene and exon expression matrices were compiled after concatenating the gene and exon level counts, respectively, from all individuals and converting them to a list-based data object called a DGEList using edgeR (27). Expression was normalized for library sizes and expressed in terms of counts per million (cpm). Very low counts across all libraries provide very little information on differences in gene expression between different phenotypes; therefore, such genes were filtered out: the criteria of at least 5 cpm in at least 10% of the cases was retained. The quantile-adjusted conditional maximum likelihood (qCML) method was used to calculate the likelihood by conditioning on the total counts for each tag and using pseudo counts after adjusting for library sizes. The exact test based on the qCML methods was used to compute differences in gene expression in UCB between cases and controls adjusted for maternal age, gestational age at delivery, sex, batch, maternal body mass index (BMI) at inclusion, and UCB composition. UCB composition comprised cord Hb levels, T cells (CD8, CD4), B cells, large granular lymphocytes (NK cells), monocytes, and granulocytes derived from methylation profiles in UCB (28).
For assessment of correlation with phenotypes, 137 genes showing altered expression in UCB from newborns of early-anemic mothers were selected. Correlation of gene expression of the selected genes with maternal Hb levels in EP and maternal Hb levels at delivery was performed using the partial correlation test by applying the “ppcor” package in R (29). The correlations were adjusted for maternal age, gestational age at delivery, sex, batch, maternal BMI at inclusion, and UCB composition.
Association with placental weight, placental size, birth weight, waist-to-length ratio, and cord insulin and c-peptide levels was performed using Pearson correlation test after log normal transformation of the data in R or python.
Global DNA Methylation
Genome-wide DNA methylation analyses were performed on DNA extracted from buffy coats on the Illumina Infinium EPIC Bead Chip (WG-317-1001) with Infinium assay using the standard Infinium HD Assay Methylation Protocol Guide (part number 15019519, Illumina). Samples were randomly distributed on the arrays. For these assays, 1 µg of DNA was bisulfite-treated according to protocol (EZ DNA Methylation Kit, Zymo Research), hybridized to the Illumina Infinium EPIC Bead Chips and scanned on Illumina iScan according to protocol. The Illumina Infinium EPIC Bead Chip contains > 850 000 probes with 99% coverage of RefSeq genes with the capacity for 12 samples per chip.
The Genome Studio methylation module software of the GenomeStudio Genome Browser (NCBI build 38) was used to calculate the raw methylation score for each DNA methylation site, which is represented as the methylation β value. The β values were calculated as:
β = intensity of the methylated allele [M] ÷ (Intensity of the unmethylated allele [U] + intensity of the methylated allele [M] + 100)
All samples passed GenomeStudio quality control steps based on built-in control probes for staining, hybridization, extension, and specificity and displayed high quality bisulfite conversion efficiency with an intensity signal above 4000 (30). Probes with detection P > 0.01, < 3 beads in at least 5% of samples per probe, Non-CpG probes, single-nucleotide polymorphism (SNP)-related probes, multi-hit probes, and allosomal CpG probes were filtered out. In total, DNA methylation data were obtained for 763 541 probes. Background correction and beta mixture quantile normalization (BMIQ) to normalize the type I and type II probes was implemented using ChAMP (31). The singular value decomposition (SVD) method was used to assess batch effects and ComBat (32) was implemented for correction of multiple batch effects. Since genome-wide methylation analysis was performed on DNA obtained from buffy coats, their methylation status could potentially reflect the combination of blood cell types. RefbaseEWAS was implemented to correct for changes in distribution of white blood cells between different subpopulations using DNA methylation signatures in combination with a previously obtained external validation set consisting of signatures from purified leukocyte samples (28, 33). To reduce heteroscedasticity for highly methylated or unmethylated sites, β values were converted to M values in the lumi package (34) for further analysis calculated as (M = log2(β/(1−β))) (35). For association with maternal Hb levels, partial correlation with the aforementioned covariates was implemented.
Correlation of CpG methylation with gene expression
For correlation of CpG methylation with gene expression, data-frames were created for each gene tested with their respective CpG sites based on the annotation provided for the Illumina EPIC chip. The correlation of CpG methylation with gene expression was performed using Pearson correlation test in R after log transformation of expression CPM and methylation beta values. Gene-wise, false discovery rate (FDR) correction was applied by correcting the P values for the number of CpG sites tested for each gene.
Correlation of CpG methylation with phenotypes
Pearson correlation test was applied to assess the correlation of CpG methylation with phenotypes like placental weight, placental size, birth weight, waist-to-length ratio, cord insulin, and c-peptide levels after data normalization using log transformation. Only CpG sites whose methylation levels associated at least nominally with gene expression of their respective genes were selected for the analysis. Additionally, only genes showing at least nominal correlation with their respective trait were considered and FDR correction was applied by taking into account only the CpG sites tested.
Genome-Wide Association Study
DNA samples were genotyped on the Illumina Infinium OmniExpressExome-8 v1.4 using Illumina iScan at the Department of Clinical Sciences, Clinical Research Centre, Lund University, Malmö, Sweden.
Variants with call rate < 99%, extreme heterozygosity (> mean ± [3 × SD]), monomorphic variants or with low minor allele frequency (MAF < 1%) were removed before imputation. A cutoff of Hardy-Weinberg equilibrium (exact P < 5.7 × 10-8) and (exact was P < 10-4) was used for common and rare variants, respectively. Quality control was performed using the PLINK 1.9 software package (28). After quality control, 281 589 variants were left for imputation. The genome-wide association studies (GWAS) scaffold was mapped to NCBI build 37 of the human genome, and imputation to the 1000G reference panel (Phase 3- http://www.well.ox.ac.uk/~wrayner/tools/) for European ancestry SNPs were available after imputation. All SNPs with MAF < 0.01 and imputation quality < 0.4 were removed from analysis. Expression quantitative trait locus (eQTL) analysis was implemented in MATRIX-QTL after the stipulated transformation of data.
RNAseq and GWAS of Fetal Tissues
Human fetal pancreas (n = 16) and liver (n = 11) tissue was obtained from material available following elective termination of pregnancy during the first trimester at the University Hospital in Malmö, Sweden. The study was approved by the national ethics committee (permit number 2018-579), written and oral consent was obtained prior to collection. Tissue samples were dissected and homogenized in Trizol and further extracted using RNeasy columns for bulk extraction. The RNA integrity number (RIN) was > 7 and approximately 1 microgram of RNA was obtained upon Trizol extraction. RNA was purified and TruSeq Stranded Total RNA Ribo-Zero H/M/R Gold (Illumina) was used for library preparation. RNA sequencing was performed on the Hiseq 2000 (Illumina). Data were processed as per the GTEX pipeline (36). Briefly, the reads obtained were aligned to the human reference genome hg19/GRCh37 using STAR v2.4.2a based on GENCODE v19 annotation and gene level counts were obtained using RNA-SeQC v1.1.8 (37). The TPM values obtained were normalized and differential expression analysis was performed between embryonic and adult RNAseq data using edgeR, with focus on the specified genes whose expression was altered in UCB from early-anemic mothers.
Simultaneously GWAS was run on DNA extracted from the same embryos using the Illumina Omni Express Exome chip (Illumina). GWAS data was processed as described earlier and Matrix eQTL was run to identify genetic variation altering gene expression in each of the tissues (38).
Immunohistochemistry Assays
For immunochemistry, 6- to 8-month-old pancreata from human terminated fetuses were processed for paraffin (6-μm sections) and cryo-embedding (10-µm sections), respectively, and immunostained as described previously (39) with the following antibodies: guinea pig α-insulin (1:2000, Millipore/1:800, AB_433703), mouse α-glucagon (1:2000, AB_259852), mouse NUMBL (1:2000, AB_2851987), rabbit PIK3C2B (1:500, AB_2893430), rabbit P2RX7 (AB_2545496) antibodies. Nuclear counterstaining was performed using 4′,6-diamidino-2-phenylindole (DAPI; 1:6000, Invitrogen).
Gene Set Enrichment Analysis
Gene set enrichment analysis was performed using the Kolmogorov-Smirnoff tests to assess if the expression of a particular set of genes was enriched in EP anemia–exposed UCB compared with those from nonanemic mothers. For the gene sets, we tested for the enrichment of eGenes, or target genes from eQTLs in the UCB for SNPs previously associated with T2D, BW, and obesity. We also used the KEGG pathway gene lists to test for enrichment of hematopoietic pathways genes (40, 41).
Results
Expression Landscapes of Cord Blood From Offspring of Mothers With Anemia During Early Pregnancy Are Distinct From Controls
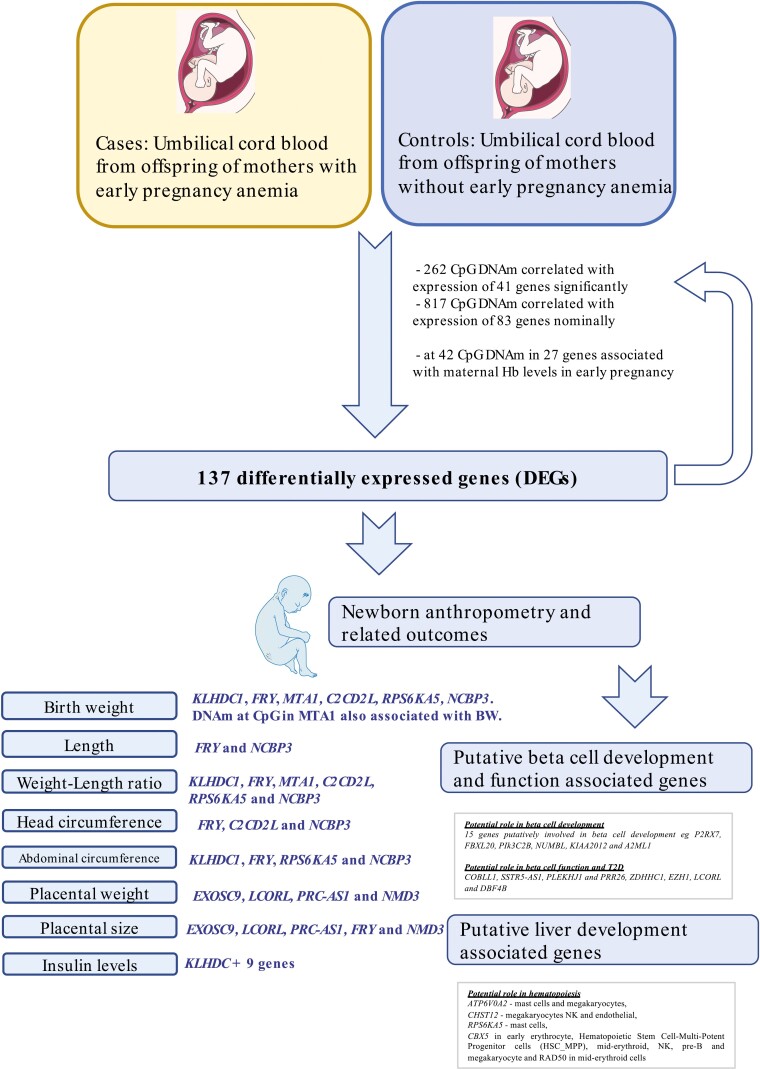
Mothers consistently having low Hb levels (Hb ≤ 10.9 g/dL) in early pregnancy (EP) defined as the first and second trimesters (21 weeks + 5 days) comprised EP anemic cases. These women were older, and had lower BMI (kg/m2) than control mothers without EP anemia (Fig. 1, Supplementary Table S1 (23)).
Figure 1.
Overview of study design and findings.
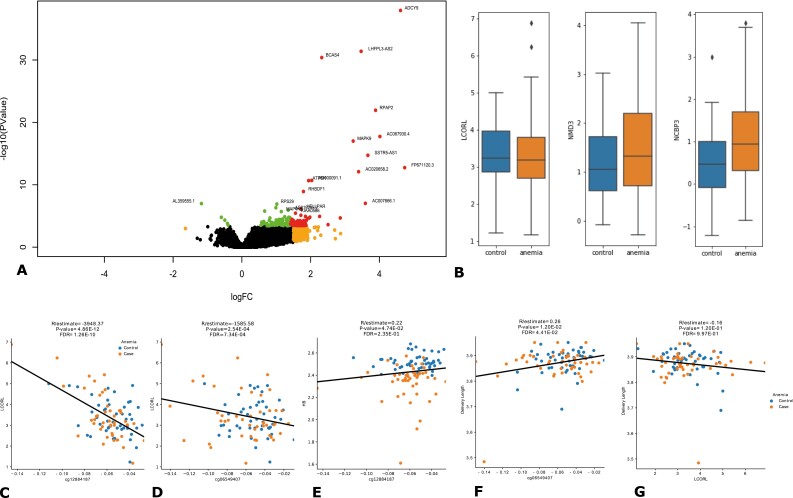
To examine differences in gene expression in UCB from newborns who had been exposed to these conditions, we performed RNA sequencing (RNAseq) from 48 UCB samples from newborns of mothers with who had EP anemia and 50 controls without EP anemia. We identified 137 differentially expressed genes, including 111 protein-coding genes and 26 non-coding and antisense RNAs including the LCORL, NMD3, and NMBP3 genes (Figs. 1, 2A, 2B, Supplementary Table S2 (23)). The majority of the genes were upregulated in UCB from newborns exposed to early maternal anemia.
Figure 2.
(A) Volcano plot showing differentially expressed genes (DEGs) between cord blood from mothers with early pregnancy anemia compared to controls. All genes showing FDR < 0.05 are presented in green. DEGs showing logFC > 1.5 are presented in orange. DEGs with FDR < 0.05 and log FC > 1 are presented in red while those with FDR < 0.005 and logFC > 1 are labeled. (B) The expression of LCORL, NMD3 and NCBP1 genes was upregulated in umbilical cord blood from offspring of mothers with early pregnancy anemia compared to controls. (C) cg12884187 and (D) cp06549407 DNAm negatively correlated with LCORL expression (E) cg12884187 DNAm correlated with maternal HB levels in early pregnancy (F) cg06549407 DNAm correlated positively with delivery length (G) LCORL expression correlated with delivery length.
We next examined if expression of these genes was also associated with maternal Hb levels at delivery. One gene, ZNF417, was significantly (FDR < 0.05) associated, whereas 29 other genes were nominally (P < 0.05) associated with maternal Hb at delivery, and the remaining 107 genes did not show any association (Supplementary Table S3 (23)).
In the search for pathways mediating the effect of anemia on gene expression in UCB, we performed pathway analysis using PANTHER(v14) (42). This analysis yielded an overrepresentation of genes involved in developmental, immune, and metabolic processes (Supplementary Figure S1a), including genes involved in integrin, cytokine and chemokine, endothelial, and P53 signaling, and apoptosis pathways (Supplementary Figure S1b (23)).
Epigenetic patterns accompany changes in gene expression in cord blood from mothers with EP anemia
EP anemia can modify fetal programming through epigenetic changes in the fetus, which in turn leads to changes in gene expression. We first aimed to identify CpG sites with DNA methylation (DNAm) associated with the expression of the 137 DEGs. The DNAm at 262 CpG sites in 41 genes were significantly associated with expression of their respective genes (FDR < 0.05). Some of these included cg11265882, cg12884187, and cg06549407 (Fig. 2C, 2D) which negatively correlated with LCORL expression, and CpG sites cg13580487 and cg08510057 in NMD3, which negatively correlated with NMD3 expression (Supplementary Table S4 (23)).
We then sought to investigate if the CpG DNAm which had correlated with gene expression here also associated with maternal Hb levels at the first contact (at GA 60.46 days [± 19.09]) in the first trimester. DNAm at these sites correlating with maternal Hb levels in the first trimester would indicate that epigenetic signatures that were altered due to early maternal anemia exposure and could be potential epigenetic signatures of fetal programming. When all CpG sites with nominal methylation-expression correlation were considered, DNAm at 42 CpG sites in 27 genes was also associated with maternal Hb levels in EP (Supplementary Table S5 (23)). Of particular interest was the CpG site cg12884187 whose methylation was negatively correlated with LCORL expression. Methylation at the same CpG site here correlated positively with maternal Hb levels in EP (Fig. 2E, Supplementary Table S5 (23)). DNAm at CpG sites in COBLL1, PIK3C2B, PRC1-AS1, P2RX7, FRY, and CIT was associated with gene expression levels of their respective genes and correlated with maternal Hb levels in EP (Table S5 (23)).
Genes Showing Altered Expression in EP Anemia–Exposed Cord Blood Correlates With Offspring Birth Weight
Maternal anemia in EP is associated with low BW (43, 44). Consistent with previous findings, maternal Hb levels at first contact (GA 60.46 [± 19.09] days) in our study were positively correlated with BW of the offspring (r2 = 0.26, P = 0.009) (Supplementary Figure S2A (23)) whereas maternal Hb at delivery was not (r2 = 0.17, P = 0.86) (Supplementary Figure S2B (23)).
We first investigated the correlation of LCORL methylation and expression with BW and placental weight and size, since: (a) LCORL showed differential expression (DE); (b) DNAm at CpG sites in LCORL associated with its expression; (c) DNAm at the same CpG site was also associated with maternal Hb levels in EP; and (d) LCORL is a known BW locus.
DNAm at 11 CpG sites correlated with LCORL expression (Supplementary Table S4 (23)). Of these, DNAm at 2 CpG sites, including cg10374588 and cg06549407 (Fig. 2F) (23), also correlated with length of the neonate. DNAm at CpG sites including cg11265882 and cg01535003 correlated significantly whereas DNAm at cg10374588, cg12884187, cg27176246, and cg01089331 correlated nominally with placental size (Supplementary Table S6) (23). DNAm at cg01535003 correlated significantly, whereas DNAm at cg06549407 and cg20358902 correlated nominally with placental weight (Supplementary Table S7 (23)). Expression of the LCORL gene was significantly associated with length of the neonate at birth (Fig. 2G), placental weight and size but not with BW (Supplementary Table S6 (23)).
We then tested the 137 DEGs for their correlation with BW; the expression of KLHDC1 significantly correlated with BW (Supplementary Figure S3A (23)) whereas FRY, MTA1, C2CD2L, RPS6KA5, and NCBP3 associated nominally (Supplementary Table S6 (23)). Methylation at one CpG site which previously correlated with MTA1 expression here nominally correlated with BW (Table S7 (23)).
Next, we then explored if any of these genes correlated with placental weight, placental size, and neonatal anthropometry. The expression of all 6 genes including KLHDC1 (Supplementary Figure S3B (23)) was also correlated with neonatal weight-length ratio, while that of KLHDC1, FRY, RPS6KA5, and NCBP3 correlated with abdominal circumference. FRY, C2CD2L, and NCBP3 correlated with head circumference while FRY and NCBP3 expression correlated with length of the neonate. In support of this, DNAm at cg07808960 and cg15212210 nominally correlated with weight to length ratio and head circumference, respectively.
Expression of 4 genes including EXOSC9, LCORL, PRC-AS1, and NMD3, which earlier showed differential expression in both studies, here correlated nominally with placental size as well as placental weight. FRY expression was also nominally correlated with placental size (Supplementary Table S6 (23)). Among the 9 CpG sites whose DNAm correlated with NMD3 expression, 3 CpG sites correlated with placental weight. Finally, CpG sites in C2CD2L and FBXL20 also correlated with placental size (Table S7 (23)).
Genes Showing Altered Expression in EP Anemia–Exposed Cord Blood Correlates With Insulin and C-Peptide Levels in Cord Blood
Insulin is key in regulating glucose homeostasis, and fetal insulin levels are also known to stimulate fetal growth (45). In our study, insulin levels were lower in UCB from EP anemic mothers compared with the controls, indicative of impaired insulin secretion among EP anemic offspring (Supplementary Table S1 (23)). We therefore sought to determine which of the genes previously consistently correlated with neonatal outcomes (BW, weight to length ratio, and others: KLHDC1, FRY, MTA1, C2CD2L, RPS6KA5, NCBP3, PRC1-AS1, and NMD3), also correlated with cord insulin levels.
Of the above listed 8 genes, expression of KLHDC1 significantly correlated with insulin levels (Supplementary Figure S3C) whereas NMD3 correlated nominally (Supplementary Table S8 (23)).
Assessing KLHDC1 expression in RNAseq data from 191 donor human pancreatic islets from another study, we found that KLHDC1 expression was not associated with that of insulin expression (Figure S3D) and was downregulated in T2D donor islets (Figure S3E).
The expression of NMD3 was nominally downregulated in T2D islets and negatively correlated with INS, the latter in both islets well as in cord blood (Table S8, S9) (23).
Genes Showing Altered Expression in EP Anemia–Exposed Cord Blood Associates With Insulin Secretion and Beta-Cell Function
Next, we examined if any of the 137 genes showed potential roles in insulin secretion and beta-cell function.
Look up of expression in RNAseq from 191 pancreatic donor islets
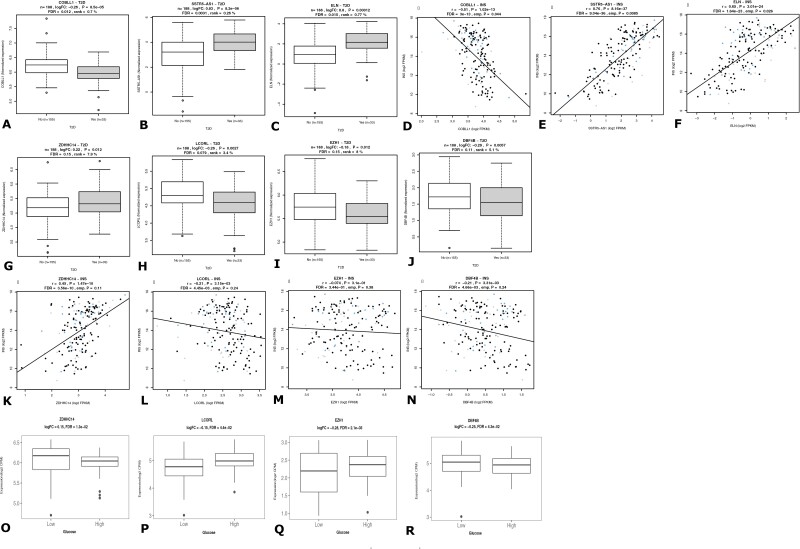
We first looked up the expression of all 137 DEGs in RNAseq data from islets from 191 donors (Islet GeneView). The expression of the COBLL1 gene was significantly downregulated in islets from T2D donors (n = 191) and correlated negatively with INS expression. SSTR5-AS1 and ELN expression was upregulated in T2D islets and correlated positively with INS (Fig. 3A-3F, Supplementary Table S9 (23)).
Figure 3.
RNAseq from human pancreatic islets (n = 188). COBLL1 (A), SSTR5-AS1 (B), and ELN (C) were differentially expressed between diabetic donor islets compared to controls. The expression of COBLL1 (D) was negatively whereas those of SSTR5-AS1 (E) and ELN (F) were positively correlated with INS expression. ZDHHC14 (G) expression was upregulated whereas LCORL (H), EZH1 (I) and DBF4B (J) expression was downregulated in T2D compared to non-T2D donor islets, ZDDHC14 (K) expression correlated positively with insulin expression whereas EZHI (M) and DBF4B (N) correlated negatively. LCORL (L) showed no correlation. Normoglycemic islets (n = 31) were exposed to normal (5.5 mmol/L) and high (18.9 mmol/L) for 24 hours. ZDHHC14(O) expression was significantly upregulated whereas that of LCORL (P), EZH1 (Q) and DBF4B (R) was downregulated upon high glucose stimulation.
The expression levels of 14 genes including COBLL1, BRCA1, FBXL20, PIK3C2B, SDHC, NMD3, EZH1, and DBF4B were negatively correlated whereas 5 genes including BRD9, NUMBL, and ZDHHC14 were positively correlated with INS expression (Fig. 3K-3N), Supplementary Table S9 (23)).
Look up of expression in RNAseq from glucose-treated pancreatic donor islets from hyperglycemic and normoglycemic donors
In order to test if any of these genes were also responsive to glucose, we then looked up these genes in our RNAseq data from normal (5.5 mmol/L) and high glucose (18.9 mmol/L) treated islets from normoglycemic donors and hyperglycemic donors from our previous study (46).
COBLL1 and SSTR5-AS1, whose expression was altered in T2D donor islets, here showed no response to acute glucose exposure in either hyperglycemic or normoglycemic donor islets. These could therefore be considered to have causal involvement in T2D pathogenesis.
Among the 14 genes that negatively correlated with INS expression, the expression of CATSPER2 and DBF4B was downregulated in 18.5 mM glucose-treated islets from normoglycemic donors. Among the genes that were positively correlated with INS, PLEKHJ1, and PRR26 were upregulated. Since these genes were expressed in response to acute glucose in the short term but not the long term, and they correlated with INS expression in islets, they could therefore be directly involved in insulin secretion (Supplementary Table S9 (23)).
Among the 12 genes showing nominal differential expression in T2D donor islets, 10 were also associated with INS expression. Among these, ZDHHC1 expression was upregulated whereas EZH1, LCORL, and DBF4B was downregulated upon glucose treatment (Fig. 4O-4R, Supplementary Table S9 (23)). These are potential key players in beta-cell function and likely to be important in the pathogenesis of T2D.
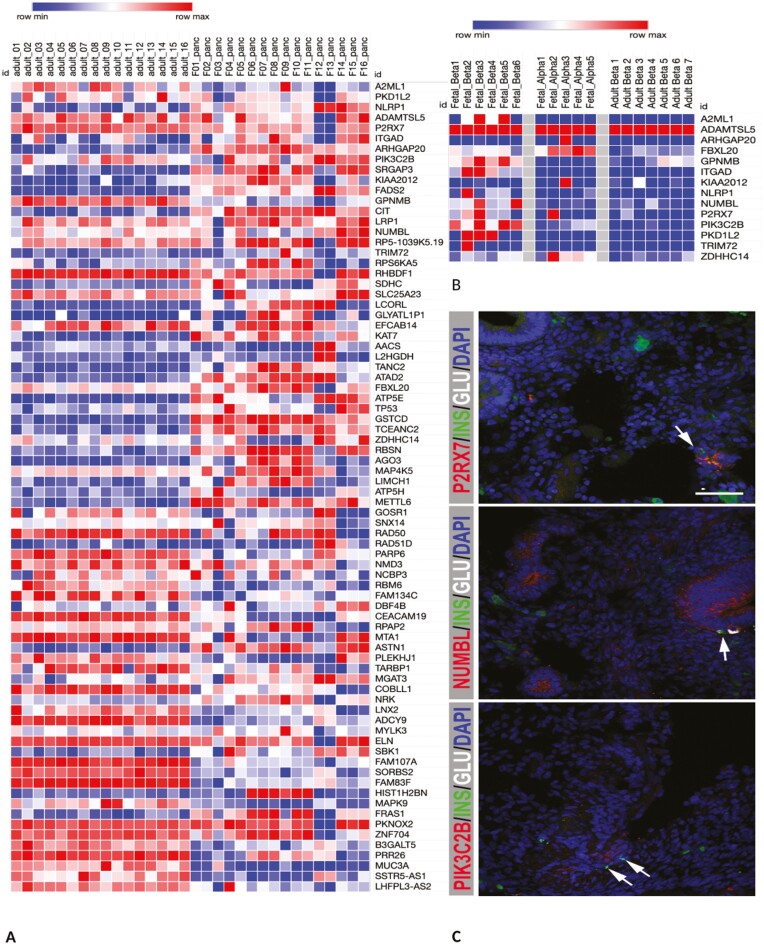
Figure 4.
(A) Expression of genes whose expression in altered in cord blood from mothers with early anemia in fetal vs adult pancreas. (B) Expression of selected genes in sorted fetal beta, alpha and adult beta-cells (C) Immunohistochemical staining of 8-week fetal pancreas was performed for P2XR7 (red), NUMBL (red), PIK3C2B (red), INS (green), and GCG (white). Scale bar indicates 50 µm, pictures were taken with a 20× objective. Arrows denote insulin positive cells. Note: False positive green staining outside pancreatic epithelium is due to auto-fluorescence from red blood cells.
Gene Expression in Embryonic Pancreas and Liver
Based on the hypothesis that genes whose expression was enriched in embryonic/ fetal tissue are more likely to be involved in development, we then examined the expression of these 137 DEGs in another study from Sweden where we had access to RNAseq data for fetal pancreas and liver samples. Here, we assessed their expression in embryonic pancreas and liver compared to expression in adult pancreas and liver respectively, using the publicly available GTEX database (36).
Pancreas
We next examined the expression of the 137 genes in 16 fetal (Swedish) and 328 adult pancreas (GTEx) to understand their role in development and/or function. Beta-cells in the pancreatic islets produce insulin. We reasoned that genes whose expression was high in fetal pancreas, and higher in fetal beta-cells compared to adult pancreas, and adult beta and fetal alpha cells respectively potentially reflect those that influence differentiation and ultimately beta-cell mass. Lower beta-cell mass is unable to compensate for the higher insulin demands due to insulin resistance, potentially due to obesity in later life, resulting in the development of diabetes (47, 48). To achieve this, we first performed differential analysis between fetal and adult pancreas to identify genes showing higher expression in the respective tissues. Next, we assessed their expression in human pancreatic islets, as well as sorted alpha and beta-cells from adult and fetal islets.
Discernable levels of expression in either the fetal or adult pancreas was determined for 121 out of the 137 genes. Among these, the expression of 43 genes was upregulated in the fetal relative to the adult pancreas, suggestive of their role in development, which included the LCORL, ATP5H, RPS6KA5, and other genes associated with neonatal outcomes, including FRY (Fig. 4A), Supplementary Table S10 (23)). Of these, the HELLPAR, ARHGAP20, and ZBTB8B genes showed fetal pancreas specific expression, showing ≥ 1 cpm in 75% of the samples in fetal pancreas but ≤ 1 cpm in at least 75% of the adult pancreas samples.
Expression in pancreatic donor islets
A total of 12 genes eg, ITGAD, KIAA2012, A2ML1, and P2RX7 showed very low to no expression in adult islets (43) (Supplementary Table S11 (23)). LCORL expression was downregulated in T2D islets (Fig. 3F).
Expression in sorted fetal and adult alpha and beta-cells
Next, we performed a selection of genes based on expression in RNAseq data fetal and adult sorted alpha and beta-cells reported by Blodgett et al (49) to focus on genes potentially involved in alpha and beta-cell development. We first compared our list of 43 genes upregulated in fetal pancreas with clusters that defined enrichment of fetal beta, alpha, or fetal in general. The NUMBL, PIK3C2B and TP53 genes were among a cluster of genes defined to be beta-cell specific whereas the LCORL was alpha-cell enriched. FBXL20 and MAP4K5 were fetal enriched (Fig. 4B).
Next, we assessed if our set of 43 genes showed higher expression fetal beta-cells compared to adult beta as well as adult alpha cells. The expression of 18 genes eg, A2ML1, FBXL20, NUMBL, P2RX7, PIK3C2B genes was higher in fetal beta compared to adult beta-cells (Blodgett et al (49), Table S12, Fig. 4B). Moreover, the expression of A2ML1, GPNMB, NUMBL, P2RX7, PIK3C2B, ADAMTSL5, ITGAD, NLRP1 and PKD1L2 was higher in fetal beta compared to alpha cells (Fig. 4B, Supplementary Table S12 (23), Blodgett et al (49)).
Identification of genes potentially impacting beta-cell mass
In total, we found 15 genes based on: (1) higher expression in fetal pancreas compared to adult; (2) very low levels of expression in adult pancreatic islets; (3) being part of a cluster of genes defining fetal beta-cells; or (4) upregulated in fetal beta compared to adult beta-cells (Fig. 3B, Supplementary Table S13 (23)). Immunohistochemical staining of 8 weeks fetal pancreas showed that P2XR7 is mainly expressed in endocrine progenitors, NUMBL, in the whole pancreatic epithelium, including some insulin and glucagon producing cells, whereas PIK3C2B expression was detected in the epithelium (Fig. 3C).
Potential long-term impact on insulin secretion and T2D
We next examined if these genes had eQTLs in human pancreatic islets (n = 191) and found 3 genes to have genome-wide significant eQTLs (Supplementary Table S14a (23)). Next, we examined the loci spanning the genes for genetic associations with indices of insulin secretion and T2D in the AMP knowledge portal. Of these, the rs34815312 SNP, which is an eQTL for the P2RX7 gene in islets, was associated with insulin secretion indices in a previous intravenous glucose tolerance test GWAS study (P = 0.03), with T2D in FinnGen GWAS (P = 0.01), HbA1c in TOPMed meta-analysis (P = 0.01) and a significant association with red blood cell count in the PheWAS scan of the UK biobank (P = 0.00002). The eQTLs for A2ML1 associated with type 1 diabetes and T2D from previous GWAS (Supplementary Table S13 (23)). Multiple nominal eQTLS for these genes also associated with insulin secretion indices in the MAGIC dataset (Supplementary Table S14b (23)).
We suggest that these genes can potentially lower beta-cell mass, which may underlie lower insulin secretion in response to higher demands for insulin in later life, thus leading to development of T2D.
Liver
Fetal liver is the site of hematopoiesis in mammals. Therefore, we compared RNA expression data from 11 human embryonic/fetal (7-14 weeks postconception) and 226 adult liver (GTEx database), and observed that for the 137 above genes, 39 genes, for example, TP53, RPS6KA5, and PRC-AS1, were expressed in fetal liver, but showed low to no expression in adult liver. In contrast, 23 genes showed higher expression in adult liver compared to fetal liver (Supplementary Table S15, Figure S4A (23)). We then compared the expression of the 39 genes showing higher expression in fetal liver to gene profiles that modulated specification of erythroid, megakaryocyte and mast cell lineages (50). TARBP1, TP53, RPS29, ATP5E, and EXOSC9 showed almost ubiquitous expression, whereas ATP6V0A2 expression was seen in mast cells and megakaryocytes, CHST12 in megakaryocytes natural killer (NK) and endothelial cells, RPS6KA5 in mast cells, CBX5 in early erythrocyte, Hematopoietic Stem Cell-Multi-Potent Progenitor cells (HSC_MPP), mid-erythroid, NK, pre-B, and megakaryocytes, and RAD50 in mid-erythroid cells (Supplementary Figure S4B, S4C (23)).
T2D-Associated Genes Overrepresented Among Genes Expressed in Cord Blood From Mothers With Early Anemia
Poor in utero conditions have been associated with long-term outcomes in the offspring for eg, T2D, obesity, and retinopathy. In order to evaluate if maternal EP anemia alters expression of genes associated with long-term outcomes, we first identified if the known T2D, obesity, and hematopoietic genes had eQTLs in the UCB and if their expression was impacted due to maternal early anemia.
Previous GWAS have identified 403 loci associated with T2D risk (Supplementary Table S16 (23)). Among them, 252 SNPs showed nominal cis-eQTL effects on 359 genes in UCB (Supplementary Table S17 (23)). Of them, 72 genes showed high expression in fetal compared to adult pancreas (Supplementary Table S18 (23)). Gene set enrichment analysis showed that these 72 genes were significantly overrepresented in UCB from mothers with EP anemia (P < 0.021, Supplementary Figure S5 (23)).
Gene set enrichment analysis using eGenes in UCB from previous GWAS associated BW and obesity loci as well as genes involved in hematopoiesis from KEGG pathways did not show any significant enrichment (Supplementary Figures S6-S9 (23)).
Discussion
By leveraging global gene expression, DNA methylation, and genotyping on UCB obtained from offspring of mothers who had anemia during EP, we provide novel insights into fetal programming signatures and molecular pathways related to metabolism and T2D. Maternal anemia during EP is associated with low BW, small for gestational age, and preterm delivery (12, 51, 52). Through differential expression analysis, we found the expression of 137 genes altered and 8 of these changes were also observed in our second study. The expression of some of these genes also associated with maternal Hb levels at delivery, and it is possible that these expression changes initiated in EP are retained throughout pregnancy and are also modulated by maternal Hb levels at a later stage.
The developmental plasticity in response to nutritional depletion in EP due to maternal anemia is speculated to alter fetal programming and cause adverse consequences in later life (53). This was suggested to be mediated by epigenetic changes in developmental genes (53). We here also show accompanying epigenetic signatures that correlated with gene expression that are potentially altered in UCB of offspring due to early maternal anemia in both the cohorts.
In accordance with previous reports showing an association between anemia and small for gestational age and low BW (43, 51, 54, 55), we observed an association between maternal Hb levels in early anemia and BW of the offspring. We here provide data showing how epigenetic mechanisms could be involved in this scenario. The Ligand Dependent Nuclear Receptor Corepressor Like (LCORL) gene is a component of the polycomb repression complex-2 (PRC2), which mediates methylation of histone H3lys27 (H3K27), and is important for determining methylation status and regulation of cellular identity during fetal development (56, 57). DNA methylation at CpG sites in the LCORL gene was robustly associated with LCORL expression, and also with maternal Hb levels in EP. The epigenetic signature is a potential consequence of maternal anemia in EP and could therefore represent an example of a signature of fetal programming. Moreover, LCORL expression was associated with placental size and weight, which in turn could affect fetal growth and BW (58-60). Furthermore, CpG methylation correlated with LCORL expression, and also correlated with length of the newborn, placental size, and placental weight. This provides further evidence for the mechanism by which altered fetal programming could affect the outcomes for the newborn with potential for long-term consequences.
Interestingly, insulin levels were lower in UCB from neonates of EP anemic mothers compared to those without. An explanation for this could be that undernutrition associated with EP anemia evokes an impaired fetal pancreatic development and maturation resulting in reduced in vivo insulin secretion and subsequently increased lifetime risk of T2D. Insulin is furthermore an important growth hormone, which has a significant effect on fetal growth, mediated through altered gene expression. KLHDC1 represents such an example, whose expression was increased in UCB from early-anemic mothers, and was negatively correlated with BW, weight to length ratio of the neonate, and with UCB insulin levels. In adult pancreatic islets, KLHDC1 expression correlated negatively with BMI and did not associate with insulin expression. We hypothesized that higher levels of KLHDC1 expression could be a result of lower insulin and therefore result in poor fetal growth leading to lower BW and weight to length ratio. While the KLHDC1 gene is expressed in embryonic and adult liver and pancreas, its expression is markedly higher in the adult tissues. The KLHDC1 gene encodes the Kelch domain containing 1 protein, which is highly expressed in pancreas, liver, and skeletal muscle where it is suggested to have roles in skeletal muscle development (61). Low BW has been clearly associated with higher risk of T2D and CVD (62). We hypothesized that KLHDC1 is involved in fetal programming induced by early anemia during pregnancy that alters future risk of metabolic disease through BW and fetal development, and this is potentially mediated by insulin.
Indeed, we found many of the genes whose expressions were altered in UCB from newborns of EP anemic mothers to also correlate with insulin levels in human pancreatic islets, and some whose expression was also altered in islets from T2D donors. Given the importance of insulin as a growth factor, we can therefore speculate that these genes affect fetal growth through altered insulin secretion.
Many of the DEGs have established roles in development. In support of this, multiple genes whose expression was altered in UCB with anemia exposure during EP also showed elevated expression in human embryonic pancreas and liver and low expression in their adult tissues. Of particular interest were genes potentially involved in regulating beta-cell mass. We show how some genes showing altered expression in UCB of offspring with early maternal anemia are also expressed in fetal pancreas, but show low to no expression in adults, and are enriched in fetal beta-cells, and associating with fetal insulin levels. These are the most likely candidates to influence beta-cell mass. People with T2D have deficits of beta-cell mass ranging from 20% to 65% (63, 64). While beta-cell death might explain this deficit, it has been hypothesized that insufficient islet development during the prenatal and postnatal period could play a key role. While this might not manifest during early adulthood, insufficient insulin secretion in later life in response to developing obesity and increased insulin demands lead to the development of T2D (65). Indeed, some of these dysregulated loci including the P2RX7, KIAA2012, A2ML1, and other genes potentially involved in beta-cell development also associated with indices of insulin secretion and T2D risk in GWAS studies (66, 67). Moreover, some genes have previously been shown to have developmental roles. NUMB Like Endocytic Adaptor Protein coding NUMBL is an important gene for cell differentiation and embryonic neurogenesis by inhibiting NOTCH signaling (68). ZDHHC14, a protein-cysteine S-palmitoyltransferase coding gene, is a known tumor suppressor. PIK3C2B plays significant roles in signaling pathways involved in cell proliferation, oncogenic transformation, cell survival, cell migration, and intracellular protein trafficking (69). This lower beta-cell mass could potentially explain the lower cord insulin levels in offspring of EP anemic mothers.
The liver is the site of hematopoiesis in the mammalian embryo up until the middle of the second trimester (50) and some of our putative fetal programming genes were also expressed in precursors of hematopoiesis, although none of them were key players reported in Popescu et al (50).
Hb is the key molecular transporter of oxygen from lungs to all tissues. Under normal conditions, the amount of oxygen delivered far exceeds the demand. During pregnancy, oxygen demands of the fetus are substantially high and hypoxia-inducible factor-1α (HIF-1α) protein has been shown to accumulate in placentae and hearts of anemic fetuses of animal models (70, 71). The ATP5H gene coding the subunit D of mitochondrial H(+)-ATP synthase, a multi-subunit complex, and ADCY9 gene encoding the enzyme adenylate cyclase type 9 are genes important in ATP formation, and both are upregulated in early maternal anemia–exposed UCB. These genes are vital for mitochondrial energy production (72). Therefore, we speculated that ATP5H upregulation is a consequence of hypoxia subsequent to anemia during EP potentially as a compensatory mechanism to combat hypoxia. In addition, cyclic AMP (cAMP) is an important player in the regulatory mechanism of hypoxia response by downregulating the activity of the master regulator of hypoxic activation of genes for angiogenesis, hormone synthesis, glycolysis, and cell survival, the hypoxia-inducible factor 1 (HIF1) gene (73). Interestingly, ADCY9 belongs to the protein kinase A signaling pathway and was deregulated leading to transgenerational impairment of ovarian development in fish (74).
In the absence of data on development of metabolic disease in the offspring, we examined expression levels in the UCB samples of validated T2D susceptibility genes (derived from eQTL SNPs of T2D loci) (66). Indeed, we observed significant enrichment of T2D-associated genes among the genes that in UCB samples were differentially expressed in cord blood from neonates exposed to EP maternal anemia. This provides compelling evidence of a potential overlap between mechanisms involved in developmental programming of T2D by anemia in pregnancy, on one side, and genetic mechanisms underlying T2D susceptibility on the other. In further support of this idea, the expression of some of these T2D susceptibility genes was altered in pancreatic islets from T2D donors.
Our studies were focused on –omics signatures in UCB, since UCB is representative of the intrauterine environment and relatively easily accessible. Moreover, UCB DNAm and changes in gene expression have been associated with several outcomes linked to early childhood weight and adiposity (75) and autism spectrum disorder (76). Finally, only few whole transcriptomic studies on UCB have been performed to date, and none of these have taken maternal EP anemia into account (77).
This study has potential limitations. One such limitation is the limited power and lack of precise replication cohorts. Another limitation is the lack of long-term outcome data in the offspring. This limitation, however, we share with most other studies, particularly those based in developing countries in the field. As a surrogate, we use BW as a predictor of long-term outcome as previously robustly established. Altered expression of important developmental genes, like imprinted genes, could have potential effects on long-term outcomes. Interestingly, the expression of T2D-associated genes was enriched in UCB from anemic mothers. A potential limitation is that most of our data are from UCB and it is not entirely clear how well UCB reflects processes in other organs.
We cannot in the current study design rule out any association between anemia diagnosis in the pregnant women, and whether the women also were subjected to undernutrition. Indeed, the association between undernutrition and fetal programming mechanisms is one of the most studied within the Developmental Origins of Health and Disease hypothesis. Anemia is highly linked to low iron intake, and that can possibly be associated with malnutrition. Several studies have investigated the relationship between maternal undernutrition and low BW, differential expression, and methylation in offspring. However, in our thorough literature search we did not detect any publications that described association between maternal undernutrition and the identified gene candidates (LCORL, P2RX7, PIK3C2B, and NUMBL).
The women participating in the study were all given offered daily iron and folic acid supplementation from pregnancy recognition till delivery. If anemia was diagnosed, it was treated with increased supplementation with either 2 to 3 combination tablets of iron-folic acid per day or with Hemovit multivitamin syrup (200 mg ferrous sulfate, 0.5 mg vitamin B6, 50 µg B12, 1.5 mg folic acid, and 2.33 mg zinc sulfate per 5 mL [Shelys Pharmaceuticals, Dar es Salaam, Tanzania]) in a dose of 10 mL 2 to 3 times daily depending on severity (22). Therefore, this could be a potential confounder and its effects in gene expression and DNA methylation cannot be ruled out.
In conclusion, we provide a catalog of effects on how maternal early anemia in pregnancy causes changes in the expression of key genes and affects BW with potential impact on future metabolic disease. The study was carried out in Korogwe, Tanzania, where a high incidence of anemia and infectious diseases in the parent generation is followed by higher rates of noncommunicable diseases in subsequent generations (78). The intrauterine period represents a key trigger of later events and therefore provides a window of opportunity for interventions, aiming at reducing untoward effects in the offspring.
Acknowledgments
We thank all the Tanzanian women who participated in the study. We are grateful to the local community leaders in Korogwe and Handeni Districts, and the Health Management team of Korogwe District. We are also grateful to the research team in Tanzania, including clinicians, nurses, laboratory technicians, data entry clerks and data manager, project drivers, and field workers for their hard work. Furthermore, we are grateful to the entire administration at NIMR Tanga Center—Korogwe Research Station for their continued support.
We thank Mattias Borell, Johan Hultman, Maria Sterner, Malin Neptin, Esa Laurila, and Gabriella Gremsperger (Lund University) for technical and laboratory support.
Human pancreatic islets, muscle, fat, and liver samples were provided by The Nordic Network for Clinical Islet Transplantation. We also want to express our deepest gratitude to the deceased organ donors as well as to their relatives.
Glossary
Abbreviations
- BW
birth weight
- cpm
counts per million
- CVD
cardiovascular disease
- DEGs
differentially expressed genes
- DNAm
DNA methylation
- EP
early pregnancy
- eQTL
expression quantitative trait locus
- FDR
false discovery rate
- GA
gestational age
- GWAS
genome-wide association studies
- Hb
hemoglobin
- SNP
single-nucleotide polymorphism
- T2D
type 2 diabetes
- UCB
umbilical cord blood
Financial Support
The work is this paper has been financially supported by grants from The Danish Council for Strategic Research (1309-00003B) to Ib Christian Bygbjerg, (Partners: University of Copenhagen, Institute of International Health, Immunology and Microbiology; Rigshospitalet, Department for Endocrinology; Lund University, Department of Clinical Research; Aarhus University Hospital, Department of Obstetrics and Gynecology), Swedish Research Council strategic research environment grant (EXODIAB, 2009-1039) to Leif Groop, Diabetes Wellness Sverige (720-858-16JDWG), Hjelt Foundation, Heart Lung Foundation (20180522) and Crafoord foundation (20200891) to Rashmi B Prasad. Line Hjort was funded by the Danish Diabetes Academy supported by the Novo Nordisk Foundation, and The Danish Diabetes association. Christentze Schemiegelow was funded by the Lundbeck Foundation (R209-2015-3580) and Laege Sofus Carl Emil Friis og Hustru Olga Doris Friis’ scholarship.
Author Contributions
R.P.B., L.H., A.V., L.G., I.B., and C.S. made substantial contributions to the conception and design of the study. R.P.B., L.H., and I.A. performed experiments. Data analysis was performed by R.P.B., G.H., and O.A. In addition, R.P.B., L.H., D.M., O.M., S.M., L.G.G., C.S., and I.B. contributed significantly to data acquisition. All authors contributed to interpretation of data. The manuscript was drafted by R.B.P., then revised and approved by all authors.
Conflict of Interests
L.G.G. has stocks in NovoNordisk. The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest. All authors declare no conflict of interest.
Ethical Approval
The study was approved by the regional and institutional review boards at NIMR, Tanzania and University of Copenhagen.
Consent to Participate
Written informed consent was obtained from all study participants.
Clinical Trial Information
ClinicalTrials.gov registration no. NCT02191683; registered July 16, 2014.
Data Availability
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
References
- 1. McLean E, Cogswell M, Egli I, Wojdyla D, de Benoist B. Worldwide prevalence of anaemia, WHO Vitamin and Mineral Nutrition Information System, 1993-2005. Public Health Nutr. 2009;12(4):444-454. [DOI] [PubMed] [Google Scholar]
- 2. Ozturk M, Ozturk O, Ulubay M, et al. Anemia prevalence at the time of pregnancy detection. Turk J Obstet Gynecol. 2017;14(3):176-180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Statistics NNB. Tanzania demographic and health survey and malaria indicator survey 2015-2016.2016. Accessed June 21, 2020. https://www.nbs.go.tz/index.php/en/census-surveys/health-statistics/demographic-and-health-survey-dhs
- 4. Tolentino K, Friedman JF. An update on anemia in less developed countries. Am J Trop Med Hyg. 2007;77(1):44-51. [PubMed] [Google Scholar]
- 5. Sliwa K. The multifactorial burden of anaemia in Africa. S Afr Med J. 2009;99(12):864-865. [PubMed] [Google Scholar]
- 6. Tine RC, Ndiaye M, Hansson HH, et al. The association between malaria parasitaemia, erythrocyte polymorphisms, malnutrition and anaemia in children less than 10 years in Senegal: a case control study. BMC Res Notes. 2012;5:565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. McClure EM, Goldenberg RL, Dent AE, Meshnick SR. A systematic review of the impact of malaria prevention in pregnancy on low birth weight and maternal anemia. Int J Gynaecol Obstet. 2013;121(2):103-109. [DOI] [PubMed] [Google Scholar]
- 8. Stephen G, Mgongo M, Hussein Hashim T, Katanga J, Stray-Pedersen B, Msuya SE. Anaemia in pregnancy: prevalence, risk factors, and adverse perinatal outcomes in Northern Tanzania. Anemia. 2018;2018:1846280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wirth JP, Woodruff BA, Engle-Stone R, et al. Predictors of anemia in women of reproductive age: biomarkers reflecting inflammation and nutritional determinants of anemia (BRINDA) project. Am J Clin Nutr. 2017;106(Suppl 1):416S-427S. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kozuki N, Lee AC, Katz J, Child Health Epidemiology Reference Group. . Moderate to severe, but not mild, maternal anemia is associated with increased risk of small-for-gestational-age outcomes. J Nutr. 2012;142(2):358-362. [DOI] [PubMed] [Google Scholar]
- 11. Mahajan S, Aalinkeel R, Shah P, Singh S, Gupta N, Kochupillai N. Nutritional anaemia dysregulates endocrine control of fetal growth. Br J Nutr. 2008;100(2):408-417. [DOI] [PubMed] [Google Scholar]
- 12. Dewey KG, Oaks BM. U-shaped curve for risk associated with maternal hemoglobin, iron status, or iron supplementation. Am J Clin Nutr. 2017;106(Suppl 6):1694S-1702S. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hamalainen H, Hakkarainen K, Heinonen S. Anaemia in the first but not in the second or third trimester is a risk factor for low birth weight. Clin Nutr. 2003;22(3):271-275. [DOI] [PubMed] [Google Scholar]
- 14. Scanlon KS, Yip R, Schieve LA, Cogswell ME. High and low hemoglobin levels during pregnancy: differential risks for preterm birth and small for gestational age. Obstet Gynecol. 2000;96(5 Pt 1):741-748. [DOI] [PubMed] [Google Scholar]
- 15. Hales CN. Fetal and infant growth and impaired glucose tolerance in adulthood: the “thrifty phenotype” hypothesis revisited. Acta Paediatr Suppl. 1997;422(S422):73-77. [DOI] [PubMed] [Google Scholar]
- 16. Vaag AA, Grunnet LG, Arora GP, Brons C. The thrifty phenotype hypothesis revisited. Diabetologia. 2012;55(8):2085-2088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wang C, Lin L, Su R, et al. Hemoglobin levels during the first trimester of pregnancy are associated with the risk of gestational diabetes mellitus, pre-eclampsia and preterm birth in Chinese women: a retrospective study. BMC Pregnancy Childbirth. 2018;18(1):263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Azulay CE, Pariente G, Shoham-Vardi I, Kessous R, Sergienko R, Sheiner E. Maternal anemia during pregnancy and subsequent risk for cardiovascular disease. J Matern Fetal Neonatal Med. 2015;28(15):1762-1765. [DOI] [PubMed] [Google Scholar]
- 19. Vielwerth SE, Jensen RB, Larsen T, et al. The effect of birthweight upon insulin resistance and associated cardiovascular risk factors in adolescence is not explained by fetal growth velocity in the third trimester as measured by repeated ultrasound fetometry. Diabetologia. 2008;51(8):1483-1492. [DOI] [PubMed] [Google Scholar]
- 20. Kalage R, Blomstedt Y, Preet R, Hoffman K, Bangha M, Kinsman J. INDEPTH training and research centres of excellence (INTREC) -Tanzania country report.2012. http://www.intrec.info/index.html
- 21. Preet R. INDEPTH training and research centres of excellence (INTREC): building research capacity in social determinants of health in low- and middle-income countries.2015. http://www.intrec.info/index.html
- 22. Hjort L, Lykke Moller S, Minja D, et al. FOETAL for NCD-FOetal exposure and epidemiological transitions: the role of Anaemia in early Life for non-communicable diseases in later life: a prospective preconception study in rural Tanzania. BMJ Open. 2019;9(5):e024861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Prasad RB, Hatem G, Hjort L, et al. Data for: Mapping the cord blood transcriptome of pregnancies affected by early maternal anemia to identify signatures of fetal programming. LUDC repository. Deposited December 3, 2021. https://www.ludc.lu.se/resources/ludc-repository; Direct data link https://lu.box.com/s/i2om6qxxo1jzsvnve9xrs4qmrxdpeu0x. doi: 10.6084/m9.figshare.17129543 [DOI]
- 24. Dobin A, Davis CA, Schlesinger F, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29(1):15-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Anders S, Reyes A, Huber W. Detecting differential usage of exons from RNA-seq data. Genome Res. 2012;22(10):2008-2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Anders S, Pyl PT, Huber W. HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31(2):166-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26(1):139-140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Houseman EA, Accomando WP, Koestler DC, et al. DNA methylation arrays as surrogate measures of cell mixture distribution. BMC Bioinf. 2012;13:86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kim S. ppcor: an R package for a fast calculation to semi-partial correlation coefficients. Commun Stat Appl Methods. 2015;22(6):665-674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Teschendorff AE, Menon U, Gentry-Maharaj A, et al. An epigenetic signature in peripheral blood predicts active ovarian cancer. PLoS One. 2009;4(12):e8274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Teschendorff AE, Marabita F, Lechner M, et al. A beta-mixture quantile normalization method for correcting probe design bias in Illumina Infinium 450 k DNA methylation data. Bioinformatics. 2013;29(2):189-196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Johnson WE, Li C, Rabinovic A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics. 2007;8(1):118-127. [DOI] [PubMed] [Google Scholar]
- 33. Houseman EA, Kim S, Kelsey KT, Wiencke JK. DNA methylation in whole blood: uses and challenges. Curr Environ Health Rep. 2015;2(2):145-154. [DOI] [PubMed] [Google Scholar]
- 34. Du P, Kibbe WA, Lin SM. lumi: a pipeline for processing Illumina microarray. Bioinformatics. 2008;24(13):1547-1548. [DOI] [PubMed] [Google Scholar]
- 35. Du P, Zhang X, Huang CC, et al. Comparison of Beta-value and M-value methods for quantifying methylation levels by microarray analysis. BMC Bioinf. 2010;11:587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Carithers LJ, Moore HM. The Genotype-Tissue Expression (GTEx) Project. Biopreserv Biobank 2015;13(5):307-308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. DeLuca DS, Levin JZ, Sivachenko A, et al. RNA-SeQC: RNA-seq metrics for quality control and process optimization. Bioinformatics. 2012;28(11):1530-1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Shabalin AA. Matrix eQTL: ultra fast eQTL analysis via large matrix operations. Bioinformatics. 2012;28(10):1353-1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Matsuoka TA, Zhao L, Artner I, et al. Members of the large Maf transcription family regulate insulin gene transcription in islet beta cells. Mol Cell Biol. 2003;23(17):6049-6062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102(43):15545-15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Mootha VK, Lindgren CM, Eriksson KF, et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet. 2003;34(3):267-273. [DOI] [PubMed] [Google Scholar]
- 42. Mi H, Muruganujan A, Ebert D, Huang X, Thomas PD. PANTHER version 14: more genomes, a new PANTHER GO-slim and improvements in enrichment analysis tools. Nucleic Acids Res. 2019;47(D1):D419-D426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Rahmati S, Delpishe A, Azami M, Hafezi Ahmadi MR, Sayehmiri K. Maternal Anemia during pregnancy and infant low birth weight: A systematic review and Meta-analysis. Int J Reprod Biomed (Yazd). 2017;15(3):125-134. [PMC free article] [PubMed] [Google Scholar]
- 44. Sukrat B, Wilasrusmee C, Siribumrungwong B, et al. Hemoglobin concentration and pregnancy outcomes: a systematic review and meta-analysis. Biomed Res Int. 2013;2013:769057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Fowden AL. The role of insulin in fetal growth. Early Hum Dev. 1992;29(1-3):177-181. [DOI] [PubMed] [Google Scholar]
- 46. Ottosson-Laakso E, Krus U, Storm P, et al. Glucose-induced changes in gene expression in human pancreatic islets: causes or consequences of chronic hyperglycemia. Diabetes. 2017;66(12):3013-3028. [DOI] [PubMed] [Google Scholar]
- 47. Prasad RB, Groop L. Genetics of type 2 diabetes-pitfalls and possibilities. Genes (Basel). 2015;6(1):87-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Lyssenko V, Groop L, Prasad RB. Genetics of type 2 diabetes: it matters from which parent we inherit the risk. Rev Diabet Stud. 2015;12(3-4):233-242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Blodgett DM, Nowosielska A, Afik S, et al. Novel observations from next-generation RNA sequencing of highly purified human adult and fetal islet cell subsets. Diabetes. 2015;64(9):3172-3181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Popescu DM, Botting RA, Stephenson E, et al. Decoding human fetal liver haematopoiesis. Nature. 2019;574(7778):365-371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Xiong X, Buekens P, Alexander S, Demianczuk N, Wollast E. Anemia during pregnancy and birth outcome: a meta-analysis. Am J Perinatol. 2000;17(3):137-146. [DOI] [PubMed] [Google Scholar]
- 52. Ren A, Wang J, Ye RW, Li S, Liu JM, Li Z. Low first-trimester hemoglobin and low birth weight, preterm birth and small for gestational age newborns. Int J Gynaecol Obstet. 2007;98(2):124-128. [DOI] [PubMed] [Google Scholar]
- 53. Gicquel C, El-Osta A, Le Bouc Y. Epigenetic regulation and fetal programming. Best Pract Res Clin Endocrinol Metab. 2008;22(1):1-16. [DOI] [PubMed] [Google Scholar]
- 54. Haider BA, Olofin I, Wang M, Spiegelman D, Ezzati M, Fawzi WW; on behalf of Nutrition Impact Model Study Group (anaemia). Anaemia, prenatal iron use, and risk of adverse pregnancy outcomes: systematic review and meta-analysis. BMJ. 2013;346:f3443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Figueiredo A, Gomes-Filho IS, Silva RB, et al. Maternal anemia and low birth weight: a systematic review and meta-analysis. Nutrients. 2018;10(5):601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Conway E, Jerman E, Healy E, et al. A family of vertebrate-specific polycombs encoded by the LCOR/LCORL genes balance PRC2 subtype activities. Mol Cell. 2018;70(3):408-421.e8. [DOI] [PubMed] [Google Scholar]
- 57. Gudbjartsson DF, Walters GB, Thorleifsson G, et al. Many sequence variants affecting diversity of adult human height. Nat Genet. 2008;40(5):609-615. [DOI] [PubMed] [Google Scholar]
- 58. Salavati N, Smies M, Ganzevoort W, et al. The possible role of placental morphometry in the detection of fetal growth restriction. Front Physiol. 2018;9:1884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Roland MC, Friis CM, Voldner N, et al. Fetal growth versus birthweight: the role of placenta versus other determinants. PLoS One. 2012;7(6):e39324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Salavati N, Sovio U, Mayo RP, Charnock-Jones DS, Smith GC. The relationship between human placental morphometry and ultrasonic measurements of utero-placental blood flow and fetal growth. Placenta. 2016;38:41-48. [DOI] [PubMed] [Google Scholar]
- 61. Chin KT, Xu HT, Ching YP, Jin DY. Differential subcellular localization and activity of kelch repeat proteins KLHDC1 and KLHDC2. Mol Cell Biochem. 2007;296(1-2):109-119. [DOI] [PubMed] [Google Scholar]
- 62. Knop MR, Geng TT, Gorny AW, et al. Birth weight and risk of type 2 diabetes mellitus, cardiovascular disease, and hypertension in adults: a meta-analysis of 7 646 267 participants from 135 studies. J Am Heart Assoc. 2018;7(23):e008870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Butler AE, Janson J, Bonner-Weir S, Ritzel R, Rizza RA, Butler PC. Beta-cell deficit and increased beta-cell apoptosis in humans with type 2 diabetes. Diabetes. 2003;52(1):102-110. [DOI] [PubMed] [Google Scholar]
- 64. Meier JJ, Bonadonna RC. Role of reduced beta-cell mass versus impaired beta-cell function in the pathogenesis of type 2 diabetes. Diabetes Care. 2013;36(Suppl 2):S113-S119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Meier JJ. Linking the genetics of type 2 diabetes with low birth weight: a role for prenatal islet maldevelopment? Diabetes. 2009;58(6):1255-1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Mahajan A, Taliun D, Thurner M, et al. Fine-mapping type 2 diabetes loci to single-variant resolution using high-density imputation and islet-specific epigenome maps. Nat Genet. 2018;50(11):1505-1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Prokopenko I, Poon W, Magi R, et al. A central role for GRB10 in regulation of islet function in man. PLoS Genet. 2014;10(4):e1004235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Petersen PH, Zou K, Hwang JK, Jan YN, Zhong W. Progenitor cell maintenance requires numb and numblike during mouse neurogenesis. Nature. 2002;419(6910):929-934. [DOI] [PubMed] [Google Scholar]
- 69. Maffucci T, Cooke FT, Foster FM, Traer CJ, Fry MJ, Falasca M. Class II phosphoinositide 3-kinase defines a novel signaling pathway in cell migration. J Cell Biol. 2005;169(5):789-799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Toblli JE, Cao G, Oliveri L, Angerosa M. Effects of iron deficiency anemia and its treatment with iron polymaltose complex in pregnant rats, their fetuses and placentas: oxidative stress markers and pregnancy outcome. Placenta. 2012;33(2):81-87. [DOI] [PubMed] [Google Scholar]
- 71. Mascio CE, Olison AK, Ralphe JC, Tomanek RJ, Scholz TD, Segar JL. Myocardial vascular and metabolic adaptations in chronically anemic fetal sheep. Am J Physiol Regul Integr Comp Physiol. 2005;289(6):R1736-R1745. [DOI] [PubMed] [Google Scholar]
- 72. Song KH, Kim JH, Lee YH, et al. Mitochondrial reprogramming via ATP5H loss promotes multimodal cancer therapy resistance. J Clin Invest. 2018;128(9):4098-4114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Torii S, Okamura N, Suzuki Y, Ishizawa T, Yasumoto K, Sogawa K. Cyclic AMP represses the hypoxic induction of hypoxia-inducible factors in PC12 cells. J Biochem. 2009;146(6):839-844. [DOI] [PubMed] [Google Scholar]
- 74. Lai KP, Wang SY, Li JW, et al. Hypoxia causes transgenerational impairment of ovarian development and hatching success in fish. Environ Sci Technol. 2019;53(7):3917-3928. [DOI] [PubMed] [Google Scholar]
- 75. Kresovich JK, Zheng Y, Cardenas A, et al. Cord blood DNA methylation and adiposity measures in early and mid-childhood. Clin Epigenetics. 2017;9:86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Andrews SV, Ellis SE, Bakulski KM, et al. Cross-tissue integration of genetic and epigenetic data offers insight into autism spectrum disorder. Nat Commun. 2017;8(1):1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Breen MS, Wingo AP, Koen N, et al. Gene expression in cord blood links genetic risk for neurodevelopmental disorders with maternal psychological distress and adverse childhood outcomes. Brain Behav Immun. 2018;73:320-330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Siddharthan T, Ramaiya K, Yonga G, et al. Noncommunicable diseases in East Africa: assessing the gaps in care and identifying opportunities for improvement. Health Aff (Millwood). 2015;34(9):1506-1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.