Abstract
Background/Aim: Our aim was to investigate possible influences of genetic variants in genes involved in the G1/S transition [cyclin-dependent kinase-2 (CDK2), cyclin E1 (CCNE1) and cyclin-dependent kinase inhibitor 1B (p27KIP1)] on the expression/activity of their corresponding proteins and to assess the functional impact of these variants on the risk of prostate cancer.
Materials and Methods: We genotyped 530 cases and 562 healthy controls for two relevant single nucleotide polymorphisms (CDK2 rs2069408 and CCNE1 rs997669) by TaqMan genotyping assay. p27KIP1 rs2066827 polymorphisms were studied by polymerase chain reaction-restriction fragment length polymorphism assay. In addition, the expression of CDK2, CCNE1 and p27KIP1 was evaluated by quantitative real-time-polymerase chain reaction and western blotting in 44 prostate cancer tissues and 31 benign prostatic hyperplasia tissues.
Results: No association was found between CDK2 rs2069408, CCNE1 rs997669 or p27KIP1 rs2066827 polymorphisms and an increased risk of prostate cancer development. Higher CDK2 expression was more prevalent in those with rs2069408 GG genotype than in AA carriers (p>0.05). We also noted reduced p27KIP1 protein expression in those with the p27KIP1 G109 allele. No difference was observed for CCNE1 expression in relation to the risky genotype (CC). A significant association was detected between CCNE1 mRNA overexpression and development of higher-grade carcinomas (Gleason score >7, p<0.05).
Conclusion: Polymorphisms CDK2 rs2069408, CCNE1 rs997669 and p27KIP1 rs2066827 have no significant impact on prostate cancer risk nor on the gene and protein expression of CDK2, CCNE1 and p27KIP1, although high CCNE1 expression was significantly associated with a higher tumour grade in patients with prostate cancer.
Keywords: CDK2, CCNE1, p27KIP1, G1/S transition, cell cycle, polymorphism, expression, prostate cancer, Slovak population
Prostate cancer is one of the most frequently diagnosed types of cancer, a leading cause of cancer-related deaths in men and displays racial disparity (1). It is a highly heterogeneous disease characterized by a complex etiopathogenesis in which familial, hormonal, and dietary factors play an important role (2,3). Genetic and epigenetic alterations also participate in the initiation and progression of prostate cancer by modifying its morphological model and influencing its biological behaviour (4).
For the physiological development of the prostate, a balance has to be maintained between the factors regulating normal cell growth, proliferation and apoptosis (5). One of the characteristics of tumorigenesis is the occurrence of genetic variations in the genes which regulate the cell cycle and, in particular, the checkpoint genes (6). The G1/S transition phase defines the fate of a cell based on proliferative or apoptotic signals. The main key regulatory complexes of the G1/S transition phase are two cyclin/cyclin-dependent kinase (CDK) complexes, namely cyclin D–CDK4/6 and cyclin E–CDK2, which have been reported to trigger the G1/S transition by promoting phosphorylation of retinoblastoma protein family proteins (7-10). Under physiological conditions, cyclin E–CDK2 complex activity is associated with DNA replication and centrosome duplication during the G1/S transition in the cell cycle (11). This complex is primarily inhibited by CDK inhibitor 1B, p27KIP1 (12,13). In contrast, the cyclin E–CDK2 complex can phosphorylate p27KIP1, ultimately leading to its degradation (14). Overexpression of cyclin E, high activity of cyclin E–CDK2, and a deficiency of p27KIP1 have been reported in several human cancer types (15-17).
We have hypothesized that common genetic variations, such as single nucleotide polymorphisms (SNPs), in the genes that control the cell cycle influence the expression or activity of the corresponding proteins and have a functional impact on the risk of prostate cancer. Previous studies have concentrated on the study of one of the p27KIP1 polymorphisms, namely p27KIP1V109G (rs2066827), which occurs at codon 109 (T>G) and causes the substitution of glycine for valine. This substitution affects the normal function of the p27KIP1 gene through altered transcription and leads to the dysregulation of the cell cycle and to the induction of tumorigenesis (18). The intron SNPs rs2069408 in CDK2 and rs997669 in CCNE1 have not been characterized functionally. However, there are no studies reporting association between CDK2 rs2069408, CCNE1 rs997669 and p27KIP1 rs2066827 polymorphisms and expression changes of the respective genes in prostate tumours. Thus, our aim was to analyse for the first time the way in which the selected SNPs influence the dynamic changes in gene and protein expression of CDK2, CCNE1 and p27KIP1 in tumour tissue of patients with prostate cancer and thus to determine any important roles that they might play in prostate carcinogenesis.
Materials and Methods
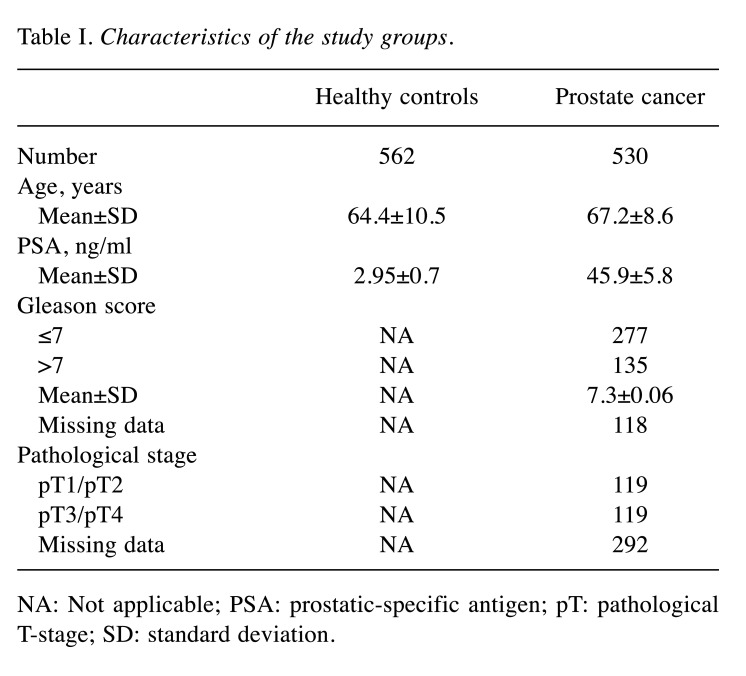
Study population. The Ethical Boards of the Jessenius Faculty of Medicine in Martin, Comenius University, approved the proposal of the present study. Briefly, 1,092 men (530 patients, 562 healthy individuals) who were recruited at the Department of Urology of University Hospital in Martin and the Jessenius Faculty of Medicine in Martin, Comenius University between May 2005 to May 2020 participated in the study. The patients with prostate cancer, healthy controls, and patients with benign prostatic hyperplasia (BPH) studied were all Caucasian. All of the cases were confirmed by histological diagnosis. The healthy controls had no previous history of any cancer and their prostate-specific antigen (PSA) levels were within the normal limit (<4 ng/ml). The clinical characteristics of the participants are summarized in Table I.
Table I. Characteristics of the study groups.
NA: Not applicable; PSA: prostatic-specific antigen; pT: pathological T-stage; SD: standard deviation.
Informed written consent was obtained from all individuals prior to their inclusion in the study.
Whole blood collected from each individual was used for DNA isolation. Tumour tissues were obtained from 44 patients with histologically confirmed prostate cancer and 31 patients with BPH (controls) and were stored in mRNA-stabilizing solution (RNAlater®; Applied Biosystems/Ambion, Waltham, MA, USA) at –80˚C until processed.
Genomic DNA was isolated from peripheral blood samples using a Wizard® Genomic DNA Purification Kit (Promega, Madison, WI, USA) and ethanol precipitation according to the manufacturer’s protocol.
CDK2 (rs2069408) and CCNE1 (rs997669) genotyping with TaqMan assays. The CDK2 rs2069408 and CCNE1 rs997669 polymorphisms were genotyped using the TaqMan SNP Genotyping Assay (Thermo Fisher Scientific, Waltham, MA, USA) in which a fluorogenic probe, consisting of an oligonucleotide labelled with both a fluorescent reporter dye [fluorescein amidite or 2’-chloro-7’phenyl-1,4-dichloro-6-carboxy-fluorescein] and a quencher dye, was included in a typical polymerase chain reaction (PCR). Amplification of the probe-specific product cleaved the probe generating an increase in reporter fluorescence (19). The PCR was performed according to the manufacturer’s instructions (provided by Thermo Fisher Scientific) on a Viia7 Real-Time PCR System (Thermo Fisher Scientific). The PCR program consisted of the following steps: Pre-read stage for 30s at 60˚C, polymerase activation and DNA denaturation for 3 min at 95˚C, followed by repetition of 40 amplification cycles for 15 s at 95˚C, 1 min at 60˚C and a final post-read step for 30 s at 60˚C. The 10 μl PCR mixture contained the CDK2 or CCNE1 probe, SsoAdvanced Universal Probes Supermix (BioRad Laboratories, Hercules, CA, USA) and 1×TE buffer (AppliChem, Darmstadt, Germany). The genotypes were determined visually based on the fluorescent emission data of the dye component as depicted in the X-Y scatter-plot of the Viia7 software (Thermo Fischer Scientific).
p27KIP1 (rs2066827) genotyping. The polymorphic site of the p27KIP1 V109G gene was determined by PCR–restriction fragment length polymorphism assays (20). The PCR primers used for amplification of a 454-bp region were: 5’-TGC AGA CCC GGG AGA AAG-3’ (forward) and 5’-CTA ACC CCG TCT GGC-3’ (reverse). The PCR program consisted of the following steps: initial denaturation at 95˚C for 5 min, followed by 35 cycles at 94˚C for 15 s, 60˚C for 30 s, 72˚C for 1 min and a final 5 min extension step at 72˚C. The 454-bp PCR fragment of p27KIP1 was digested with BglII restriction enzyme (10 U, 16 h, 37˚C). The resulting fragments were separated on a 3% agarose gel with ethidium bromide. The T allele lacks a BglII site that is present in the G allele. Therefore, digestion results in fragments of 76 and 377 bp (T allele) or 76, 116 and 262 bp (G allele). Individuals were classified as TT, TG or GG.
RNA extraction, cDNA synthesis and quantitative real-time PCR. Total RNA and cDNA were obtained and processed as described previously (21). A custom RT2 Profiler PCR array (Qiagen GmbH, Hilden, Germany) was used for quantitative PCR analysis on a Viia7 Real-Time PCR System (Thermo Fisher Scientific) according to the manufacturer’s protocol. Data were analysed according to the classic 2–ΔΔCt method and normalized to the average of glyceraldehyde-3-phosphate dehydrogenase and β-actin expression in each sample (21).
Western blot analysis. In brief, 14 prostate cancer tumour tissues were used to analyse the correlation between the CDK2 rs2069408, CCNE1 rs997669 and p27KIP1 rs2066827 polymorphisms and protein expression levels. Isolation of proteins from the tumour tissues and determination of protein concentrations were carried out as previously described (21). Protein samples (30 μg/μl) were separated by 1D electrophoresis in a 12% sodium dodecyl sulfate–polyacrylamide gel electrophoresis. The separated proteins were then transferred onto nitrocellulose membranes by semi-dry transfer. Any binding of non-specific proteins was removed by washing the membranes with 2% bovine serum albumin (Tris solution, 0.05% Tween 20, 10% sodium azide) for 1 h. Subsequently, the membranes were incubated with primary antibodies overnight at 4˚C, namely mouse monoclonal anti-β-actin (1:1,000, CST-3700S) and anti-CCNE1 (1:750, CST-4129), rabbit monoclonal anti-CDK2 (1:500, CST-2546) and rabbit polyclonal anti-p27 (1:1,000, CST-2552) all from Cell Signaling Technology (Danvers, MA, USA). As a next step, the membranes were washed with 50 mM Tris-Cl, pH 7.5, 150 mM NaCl and 0.05% Tween 20 (TBS-T solution) and incubated with secondary antibodies (1:5,000; Santa Cruz Biotechnology, Dallas, Texas, USA) for 1 h. The membranes were then washed again with TBS-T solution and incubated in Clarity Western ECL Substrate (BioRad Laboratories) for 2 min in the dark. After analysis of the membranes with Chemidoc XRS (BioRad Laboratories), the intensities of the individual bands were quantified using Quantity One software (BioRad Laboratories). The intensities of the spotted bands were normalized to the congruent intensities of β-actin bands.
Statistical analysis. Data were analysed with R [1], ver. 4.0.5. The fold change (FC) in expression was computed using the standard formula. Normality of FC values was assessed by the quantile-quantile plot (qqPlot) with the 95% confidence band constructed by bootstrapping. Symbox was used to suggest a transformation to bring FC close to normality. Logarithmic transformation (with base 2) of FC values was used and normality of the log-transformed FC values was assessed by qqPlot. Log-transformed FC data were explored using boxplots, overlaid with swarmplot of the data points. The null hypothesis of the equality of the population means of log-FC values in the two subpopulations was tested by two-sample, two-sided t-test. In the genomic association study, the Haldane exact test was used to test the Hardy–Weinberg equilibrium, the chi-squared test with simulated p-values was used in the general, dominant, recessive and multiplicative models, and the Cochran–Armitage test of trend was used in the additive genetic model. The median-unbiased estimate and mid-point exact confidence interval were used to estimate the odds ratio and its 95% confidence interval. Findings with p-values below 0.05 were considered statistically significant.
Results
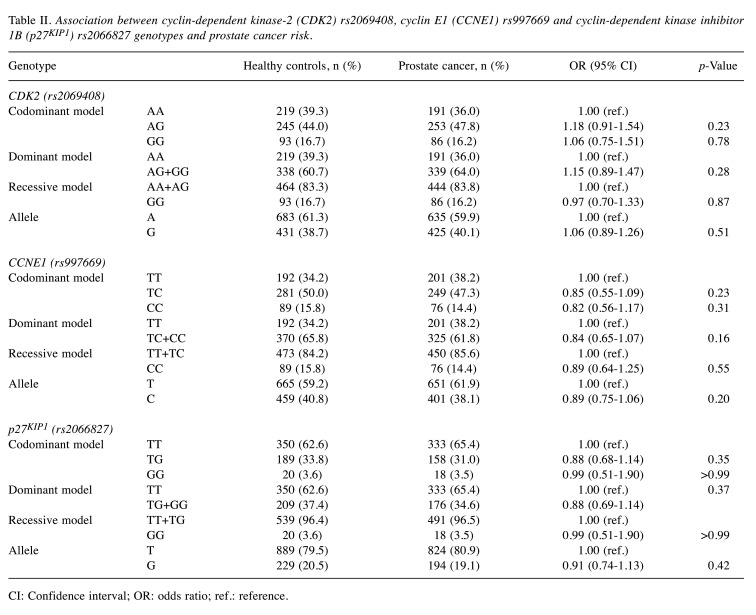
Firstly, in our case–control study, we investigated the association of the three selected SNPs, namely CDK2 rs2069408, CCNE1 rs997669 and p27KIP1 rs2066827, with prostate cancer risk in 530 patients with prostate cancer and 562 healthy controls. The distribution of genotypes and allelic frequencies of CDK2 rs2069408, CCNE1 rs997669 and p27KIP1 rs2066827 polymorphisms among patients and healthy controls were all in accordance with the Hardy–Weinberg equilibrium (p>0.05). The CDK2 rs2069408 genotype in 0.8% of healthy controls, the CCNE1 rs997669 genotype in 0.7% of cases and the genotype of p27KIP1 rs2066827 in 0.5% of healthy controls and 4% of cases were not determined because of the low concentration of DNA. The genotype-specific odds ratios for individual SNPs in selected regulatory genes of the cell cycle are shown in Table II. Overall, we found no significant association with susceptibility to prostate cancer for CDK2 rs2069408, CCNE1 rs997669 and p27KIP1 rs2066827 polymorphisms in codominant, dominant, and recessive genetic models.
Table II. Association between cyclin-dependent kinase-2 (CDK2) rs2069408, cyclin E1 (CCNE1) rs997669 and cyclin-dependent kinase inhibitor 1B (p27KIP1) rs2066827 genotypes and prostate cancer risk.
CI: Confidence interval; OR: odds ratio; ref.: reference.
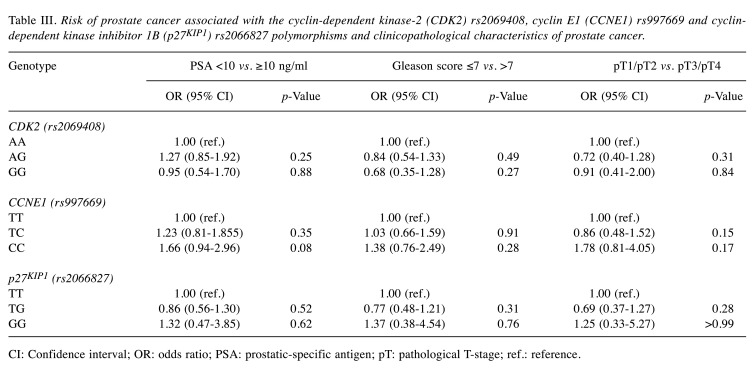
Furthermore, we analysed possible associations between CDK2 rs2069408, CCNE1 rs997669 and p27KIP1 rs2066827 polymorphisms and clinicopathological characteristics. We categorized the cases into groups according to the PSA level (<10 and ≥10 ng/ml), Gleason score (≤7 and >7) and pathological T-stage (pT1/pT2 and pT3/pT4). As shown in Table III, among those individuals with the CDK2 rs2069408 GG genotype and a Gleason score >7, the risk of prostate cancer was non-significantly reduced in comparison with individuals with the AA genotype and a Gleason score ≤7 (p>0.05). No significantly increased risk of prostate cancer was seen in association with CCNE1 rs997669 and p27KIP1 rs2066827 mutant genotypes (CC and GG, respectively) and clinicopathological parameters (p>0.05).
Table III. Risk of prostate cancer associated with the cyclin-dependent kinase-2 (CDK2) rs2069408, cyclin E1 (CCNE1) rs997669 and cyclindependent kinase inhibitor 1B (p27KIP1) rs2066827 polymorphisms and clinicopathological characteristics of prostate cancer.
CI: Confidence interval; OR: odds ratio; PSA: prostatic-specific antigen; pT: pathological T-stage; ref.: reference.
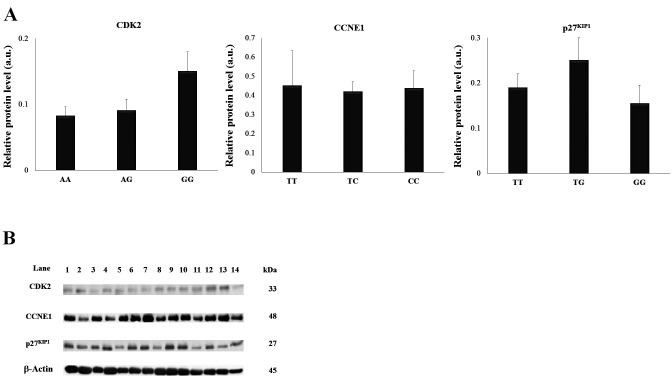
We selected 14 tumour tissues to confirm alterations in the expression of CDK2, CCNE1 and p27KIP1 associated with CDK2 rs2069408, CCNE1 rs997669 and p27KIP1 rs2066827 polymorphisms in prostate tissues by western blot analysis (Figure 1). The frequency distribution of CDK2 rs2069408, CCNE1 rs997669 and p27KIP1 rs2066827 homozygous, heterozygous, and mutant genotypes was 5, 5, and 4, respectively. We found that CDK2 protein levels were higher in individuals with the AG and GG genotypes than in those with the AA genotype. In the TC and CC genotypes for CCNE1 rs997669, the protein levels were not significantly different from those of the TT genotype. The protein levels of the p27KIP1 rs2066827 TG genotype were higher, whereas those for the GG genotype were reduced, when compared with those for TT genotype.
Figure 1. Association between alleles of cyclin-dependent kinase-2 (CDK2) rs2069408, cyclin E1 (CCNE1) rs997669 and cyclin-dependent kinase inhibitor 1B (p27KIP1) rs2066827 genes and their protein levels. A: Relative protein expression levels of CDK2, CCNE1 and p27KIP1 were calculated densitometrically in reference to the expression level of β-actin. Values are given as the means±SEM. B: Western blot analysis of CDK2, CCNE1 and p27KIP1 protein levels in prostate cancer tissues from individuals with different CDK2 rs2069408 CCNE1 rs997669 and p27KIP1 rs2066827 genotypes. Individual genotype designations: Lanes 1 to 5, homozygous wild-type genotype (n=5); lanes 5 to 10, heterozygous genotype (n=5); lanes 11 to 14, homozygous mutant genotype (n=4). SEM: standard error of the mean.
The expression of CDK2, CCNE1 and p27KIP1 mRNA was investigated by real-time PCR in prostate tumour tissues relative to that in BPH tissues. The relative CDK2 mRNA expression values were not significantly different in prostate cancer tissues [median log2(FC) value: 0.18; p=0.27] from those in the BPH tissues. A similar non-significant different was observed for CCNE1 expression [median log2(FC) value: 0.59; p=0.09]. The relative p27KIP1 mRNA expression was non-significantly reduced in prostate cancer tissues compared with BPH tissues [median log2(FC) value: –0.96; p=0.16].
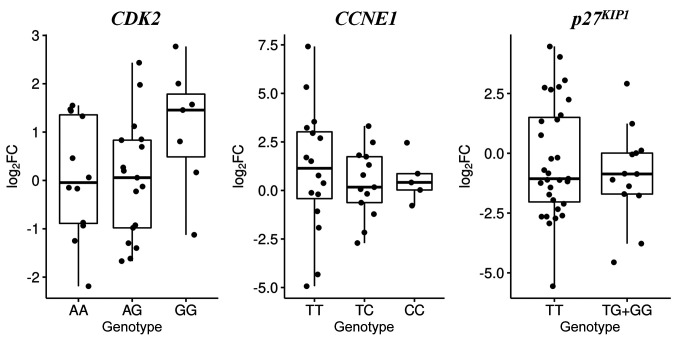
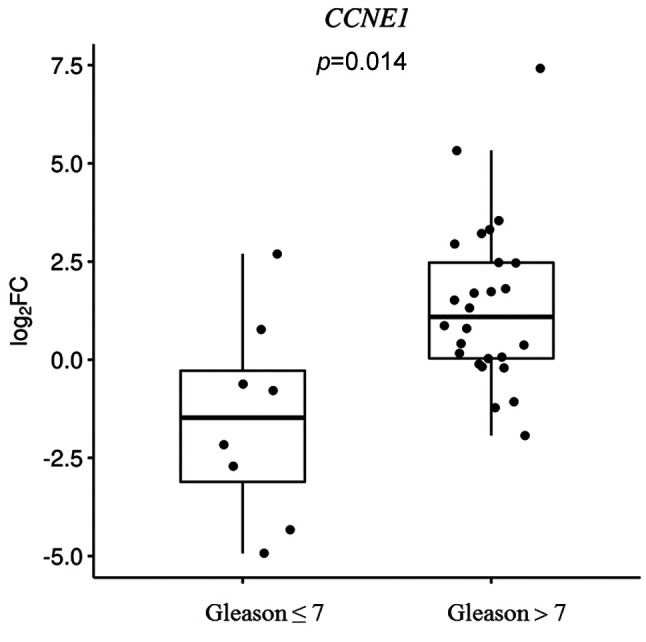
Furthermore, we detect an effect of the CDK2 rs2069408, CCNE1 rs997669 and p27KIP1 rs2066827 polymorphisms on the relative CDK2, CCNE1 and p27KIP1 mRNA expression levels (Figure 2). Individuals with the CDK2 rs2069408 GG genotype presented a higher relative CDK2 mRNA expression than those with AA genotype [median log2(FC) values: 1.46 and –0.05, respectively, p=0.27]. A non-significant lower relative expression of CCNE1 mRNA was observed in the CC genotype than in the TT genotype [median log2(FC) values: 0.41 and 1.14, respectively, p=0.09]. Additionally, no significant difference was seen between the median log2(FC) p27KIP1 values in the individuals who carried the TG/GG genotypes and those with the TT genotype [–0.87 and –1.06, respectively, p=0.38]. Relative CDK2, CCNE1 and p27KIP1 mRNA expression was also assessed according to clinicopathological features, such as PSA value, Gleason score and pathological T stage. The relative CCNE1 mRNA expression was statistically significantly elevated in individuals with a Gleason score >7 in comparison with Gleason score ≤7 [median log2(FC) values: 1.09 and –1.47, respectively, p=0.014, Figure 3]. For all other clinicopathological factors, no statistically significant correlation was detected with any of the polymorphisms.
Figure 2. Log2 fold change (FC) in the relative mRNA expression levels of cyclin-dependent kinase-2 (CDK2) rs2069408, cyclin E1 (CCNE1) rs997669 and cyclin-dependent kinase inhibitor 1B (p27KIP1) rs2066827 with respect to their genotypes among patients with prostate cancer. Boxplots depict the sample quartiles as the lower and upper edges of the box, and the sample median as a horizontal line inside the box. Whiskers denote the minimal and maximal values, unless there are outliers in the data.
Figure 3. Log2 fold change (FC) in the relative mRNA expression of cyclin E1 (CCNE1) with respect to the Gleason score among patients with prostate cancer. Boxplots depict the sample quartiles as the lower and upper edges of the box, and the sample median as a horizontal line inside the box. Whiskers denote the minimal and maximal values, unless there are outliers in the data. p-Value was derived from two-sample, two-sided t-test.
Discussion
Dysregulation of the cell cycle and alterations in regulatory proteins of the cell cycle, such as cyclins, CDKs and CDK inhibitors, lead to uncontrolled cell proliferation in many solid tumours. An understanding of the impact of the genetic alterations in these genes should therefore improve our ability to identify the interindividual variation in cancer development and progression. To our knowledge, no investigation has been reported the association of the CDK2 rs2069408, CCNE1 rs997669 and p27KIP1 rs2066827 polymorphisms with prostate cancer risk and their influence on corresponding mRNA and protein levels and this led to our decision to assess their functional relevance. Our results show that CDK2 rs2069408, CCNE1 rs997669 and p27KIP1 rs2066827 polymorphisms are not associated with an increased risk of prostate cancer development. Moreover, we found higher gene and protein expression of CDK2 in the presence of the risky genotype (GG) of rs2069408, no altered CCNE1 gene and protein expression in carriers of the CCNE1 rs997669 CC genotype, and reduced p27KIP1 protein expression in those with p27KIP1 G109 allele. In addition, we identified a significant association between the relative expression of CCNE1 mRNA and the development of higher-grade carcinomas (Gleason score >7).
The CDK2 rs2069408 and CCNE1 rs997669 polymorphisms have not yet been studied in prostate cancer. We detected no association between the CDK2 rs2069408 polymorphism and prostate cancer risk. Several studies have also found no association between this polymorphism and breast and ovarian cancer risk in various populations (22-26). With regard to CCNE1 rs997669, no association with prostate cancer was apparent in our study population. On the contrary, in a case–control study, Driver et al. reported an association of CCNE1 rs997669 with breast cancer in a British population (23). Moreover, genome-wide association studies have identified CCNE1 rs997669 as being associated with a risk for breast cancer and bladder cancer in Caucasian and Asian populations (27,28).
The exact mechanism by which the p27KIP1 rs2066827 polymorphism affects cancer susceptibility is not well understood. The V109 allele has been hypothesized to alter the interaction between p27KIP1 and its negative regulator p38jab1 because it is located on the interaction surface and possibly leads to p27KIP1 degradation and its reduced expression (29). Zhu et al., using Polyphen2 bioinformatics tools, to show that the p27KIP1 rs2066827 polymorphism is predicted to be benign and will not affect the protein function of p27KIP1 (30). During the past few years, several studies have focused on the effects of p27KIP1 rs2066827 polymorphism on prostate cancer risk within various ethnic populations (13,17,31-33). In the literature, both the T and G alleles of p27KIP1 rs2066827 have been shown to be associated with risk of but also protection against malignancy. One case–control study demonstrated that p27KIP1 rs2066827 polymorphism is associated with an increased risk of prostate cancer (31), whereas another study and a meta-analysis suggested that this polymorphism is associated with a reduced risk of prostate cancer (13,17). In addition, other studies have reported no association between the p27KIP1 rs2066827 polymorphism and prostate cancer risk (30,32,33), which our results are in agreement with. Several possible causes can be proposed related to the inconsistency between our study and the other investigations, such as sample sizes and heterogeneous populations/ethnic variance. Moreover, the p27KIP1 rs2066827 G allele might be functionally relevant or in linkage disequilibrium with alleles at other susceptibility loci.
The overexpression of CDK2 should cause abnormal regulation of the cell cycle and should influence human carcinogenesis (16,34). However, CDK2 activity has been shown to be altered not by up-regulation but by its binding partners (cyclins and CDK inhibitors) or by alterations to post-translational modifications (35). Furthermore, the expression of the major regulator of CDK2 activity, namely cyclin E1, is frequently observed at high levels in solid tumours (36,37). We have shown no altered relative CDK2 and CCNE1 mRNA expression in prostate tumour tissues compared with BPH. However, an increased expression of CDK2 and CCNE1 mRNA has been seen in several types of cancer, including primary hepatocellular carcinoma (38), advanced cervical cancer (39), breast (40) and ovarian (41) cancer.
Reduced expression of the CDK inhibitor, p27KIP1, is correlated with poor patient prognosis (42, 43). The translocation of p27KIP1 to the cytoplasm is hypothesized to result in the loss of its suppressive function in cell-cycle progression (44,45). However, other studies have been unable to identify a prognostic effect of reduced p27KIP1 expression (46,47). Zhu et al. used in-silico tools to show the expression of p27KIP1 to be reduced in prostate cancer tissue compared with paracancerous counterparts, especially in less advanced prostate cancer (Gleason score = 6 or 7), but not in more advanced prostate cancer (30). Our results revealed a similar trend for p27KIP1 mRNA levels in prostate tumour tissues when compared with BPH tissues. The differential expression of p27KIP1 mRNA has been also reported in various types of cancer (45,48,49).
Common SNPs might affect the normal function of proteins that play key roles in the G1/S transition by altering their transcriptional efficiency and protein levels, leading to cell-cycle dysregulation and cancer cell behaviour (29). In the present study, we evaluated whether CDK2, CCNE1 and p27KIP1 mRNA and protein expression in prostate tumours is influenced by the CDK2 rs2069408, CCNE1 rs997669 and p27KIP1 rs2066827 polymorphisms. To the best of our knowledge, we report the first analysis of this association in prostate cancer. We found a higher expression of CDK2 in the presence of the risky genotype (GG) of rs2069408 at both the mRNA and protein levels. We detected no significant difference in CCNE1 expression in carriers of the CCNE1 rs997669 CC genotype. Furthermore, our data suggest that the p27KIP1 rs2066827 G allele is associated with reduced p27KIP1 protein expression and no alterations in mRNA level. Han et al. observed that LNcap cells transfected with the p27KIP1 G allele exhibited a lower proliferative activity and expression level of p27KIP1 protein than cells transfected with the V allele (17).
We observed no significantly increased risk of prostate cancer for CDK2 rs2069408 and CCNE1 rs997669 mutant genotypes by all clinicopathological parameters. Furthermore, our data do not suggest that the p27KIP1 rs2066827 polymorphism is associated with PSA level, Gleason score or pathological T stage, thus confirming the overall null association with the clinicopathological characteristics of prostate cancer, as reported previously (32,33). Interestingly, we did find that CCNE1 mRNA expression was significantly increased in carcinomas with Gleason scores higher than 7.
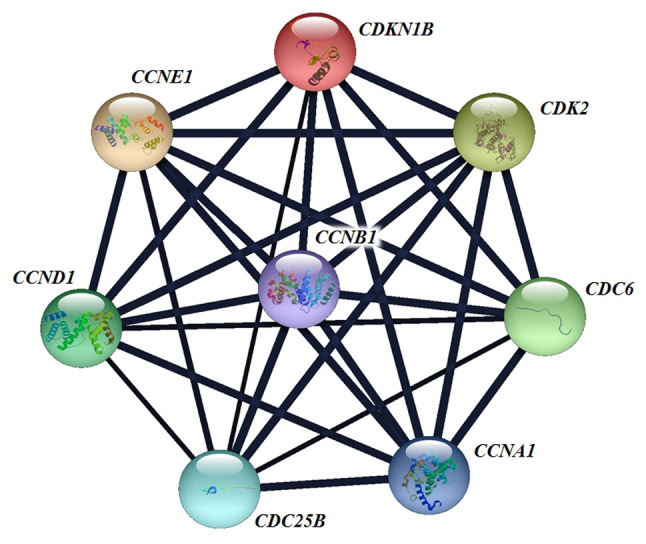
Moreover, we hypothesized that molecular mechanisms other than those of SNPs can affect alterations of regulatory proteins of the cell cycle in a cell-type and condition-specific manner such as by: a) Regulation of gene expression by transcription factor [e.g. FORKHEAD box transcription factor O1 (50)] or by microRNA (48); b) regulation of protein content by the control of proteolysis or mRNA translation (12); c) change in the cellular localization of cell-cycle regulators (51); and d) regulation of their expression by hormones [androgens and oestrogens (52)]. Because tumorigenesis and tumour progression are processes involving multiple genetic defects, other important genes that are involved in the CDK2–CCNE1–p27KIP1 gene–gene interaction might not have been efficiently tagged and might play important roles in prostate carcinogenesis (Figure 4).
Figure 4. Interactions of human cyclin-dependent kinase 2 (CDK2), cyclin E1 (CCNE1) and cyclin-dependent kinase inhibitor 1B (p27KIP1) with other genes generated by String software analysis. CCNA1: Cyclin A1; CCND1: cyclin D1; CCNB1: cyclin B1; CDC6: cell-division cycle 6; CDC25B: cell-division cycle 25B.
In summary, we observed no statistically significant overall associations of CDK2 rs2069408, CCNE1 rs997669 and p27KIP1 rs2066827 polymorphisms with prostate cancer risk and no significant effects of these polymorphisms on the gene and protein expression of CDK2, CCNE1 and p27KIP1. Significant interaction was found between the relative CCNE1 mRNA level and higher-grade carcinoma. Although the present study provides important insights into the role of genetic alterations in the regulators of the cell cycle in the development of prostate cancer, the strengths and limitations of our study should be mentioned. The major strength of our study is its large ethnically homogeneous sample size, which reduces allelic and genotypic heterogeneity found in case–control studies. We were however limited by the tissue samples available. Because this was a preliminary study, a more detailed understanding of the importance of polymorphisms in the regulation of the cell cycle and of gene–gene interactions can only be achieved by further studies.
Conflicts of Interest
The Authors declare no conflicts of interest.
Authors’ Contributions
MKS, MH, JJ and JK conceived the idea for this study and wrote the article. MH, JJ, MV, and KHH contributed to the design of the research. RD and DE were involved in the data collection. PK and MG analysed the data. All Authors edited and approved the final version of the article.
Acknowledgements
This work was supported by the Agency of Ministry of Education, Science, Research and Sport of the Slovak Republic under grants no. 1/0014/22 and APVV-15-0181.
References
- 1.Panigrahi GK, Praharaj PP, Kittaka H, Mridha AR, Black OM, Singh R, Mercer R, van Bokhoven A, Torkko KC, Agarwal C, Agarwal R, Abd Elmageed ZY, Yadav H, Mishra SK, Deep G. Exosome proteomic analyses identify inflammatory phenotype and novel biomarkers in African American prostate cancer patients. Cancer Med. 2019;8(3):1110–1123. doi: 10.1002/cam4.1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin. 2020;70(1):7–30. doi: 10.3322/caac.21590. [DOI] [PubMed] [Google Scholar]
- 3.Rawla P. Epidemiology of prostate cancer. World J Oncol. 2019;10(2):63–89. doi: 10.14740/wjon1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Attard G, Parker C, Eeles RA, Schröder F, Tomlins SA, Tannock I, Drake CG, de Bono JS. Prostate cancer. Lancet. 2016;387(10013):70–82. doi: 10.1016/S0140-6736(14)61947-4. [DOI] [PubMed] [Google Scholar]
- 5.Salinas CA, Tsodikov A, Ishak-Howard M, Cooney KA. Prostate cancer in young men: an important clinical entity. Nat Rev Urol. 2014;11(6):317–323. doi: 10.1038/nrurol.2014.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Möröy T, Geisen C. Cyclin E. Int J Biochem Cell Biol. 2004;36(8):1424–1439. doi: 10.1016/j.biocel.2003.12.005. [DOI] [PubMed] [Google Scholar]
- 7.Dong W, Zhu H, Gao H, Shi W, Zhang Y, Wang H, Li C, Song G, Zhang Y. Expression of Cyclin E/Cdk2/p27Kip1 in growth hormone adenomas. World Neurosurg. 2019;121:e45–e53. doi: 10.1016/j.wneu.2018.08.209. [DOI] [PubMed] [Google Scholar]
- 8.Asghar U, Witkiewicz AK, Turner NC, Knudsen ES. The history and future of targeting cyclin-dependent kinases in cancer therapy. Nat Rev Drug Discov. 2015;14(2):130–146. doi: 10.1038/nrd4504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Teixeira LK, Wang X, Li Y, Ekholm-Reed S, Wu X, Wang P, Reed SI. Cyclin E deregulation promotes loss of specific genomic regions. Curr Biol. 2015;25(10):1327–1333. doi: 10.1016/j.cub.2015.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Quereda V, Malumbres M. Cell cycle control of pituitary development and disease. J Mol Endocrinol. 2009;42(2):75–86. doi: 10.1677/JME-08-0146. [DOI] [PubMed] [Google Scholar]
- 11.Teixeira LK, Reed SI. Cyclin E Deregulation and Genomic Instability. Adv Exp Med Biol. 2017;1042:527–547. doi: 10.1007/978-981-10-6955-0_22. [DOI] [PubMed] [Google Scholar]
- 12.Bencivenga D, Caldarelli I, Stampone E, Mancini FP, Balestrieri ML, Della Ragione F, Borriello A. p27Kip1 and human cancers: A reappraisal of a still enigmatic protein. Cancer Lett. 2017;403:354–365. doi: 10.1016/j.canlet.2017.06.031. [DOI] [PubMed] [Google Scholar]
- 13.Wei F, Xu J, Tang L, Shao J, Wang Y, Chen L, Guan X. p27(Kip1) V109G polymorphism and cancer risk: a systematic review and meta-analysis. Cancer Biother Radiopharm. 2012;27(10):665–671. doi: 10.1089/cbr.2012.1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vlach J, Hennecke S, Amati B. Phosphorylation-dependent degradation of the cyclin-dependent kinase inhibitor p27. EMBO J. 1997;16(17):5334–5344. doi: 10.1093/emboj/16.17.5334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chu C, Geng Y, Zhou Y, Sicinski P. Cyclin E in normal physiology and disease states. Trends Cell Biol. 2021;31(9):732–746. doi: 10.1016/j.tcb.2021.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu Q, Gao J, Zhao C, Guo Y, Wang S, Shen F, Xing X, Luo Y. To control or to be controlled? Dual roles of CDK2 in DNA damage and DNA damage response. DNA Repair (Amst) 2020;85:102702. doi: 10.1016/j.dnarep.2019.102702. [DOI] [PubMed] [Google Scholar]
- 17.Han QH, Shan ZJ, Hu JT, Zhang N, Zhang XP. Relationship between gene polymorphisms and prostate cancer risk. Asian Pac J Trop Med. 2015;8(7):569–573. doi: 10.1016/j.apjtm.2015.06.005. [DOI] [PubMed] [Google Scholar]
- 18.Cavé H, Martin E, Devaux I, Grandchamp B. Identification of a polymorphism in the coding region of the p27Kip1 gene. Ann Genet. 1995;38(2):108. [PubMed] [Google Scholar]
- 19.Hui L, DelMonte T, Ranade K. Genotyping using the TaqMan assay. Curr Protoc Hum Genet. 2008;Chapter 2:Unit 2.10. doi: 10.1002/0471142905.hg0210s56. [DOI] [PubMed] [Google Scholar]
- 20.Li G, Sturgis EM, Wang LE, Chamberlain RM, Spitz MR, El-Naggar AK, Hong WK, Wei Q. Association between the V109G polymorphism of the p27 gene and the risk and progression of oral squamous cell carcinoma. Clin Cancer Res. 2004;10(12 Pt 1):3996–4002. doi: 10.1158/1078-0432.CCR-04-0089. [DOI] [PubMed] [Google Scholar]
- 21.Kmeťová Sivoňová M, Tatarková Z, Jurečeková J, Kliment J, Híveš M, Lichardusová L, Kaplán P. Differential profiling of prostate tumors versus benign prostatic tissues by using a 2DE-MALDI-TOF-based proteomic approach. Neoplasma. 2021;68(1):154–164. doi: 10.4149/neo_2020_200611N625. [DOI] [PubMed] [Google Scholar]
- 22.Han JY, Wang H, Xie YT, Li Y, Zheng LY, Ruan Y, Song AP, Tian XX, Fang WG. Association of germline variation in CCNE1 and CDK2 with breast cancer risk, progression and survival among Chinese Han women. PLoS One. 2012;7(11):e49296. doi: 10.1371/journal.pone.0049296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Driver KE, Song H, Lesueur F, Ahmed S, Barbosa-Morais NL, Tyrer JP, Ponder BA, Easton DF, Pharoah PD, Dunning AM, Studies in Epidemiology and Risks of Cancer Heredity (SEARCH) Team Association of single-nucleotide polymorphisms in the cell cycle genes with breast cancer in the British population. Carcinogenesis. 2008;29(2):333–341. doi: 10.1093/carcin/bgm284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gayther SA, Song H, Ramus SJ, Kjaer SK, Whittemore AS, Quaye L, Tyrer J, Shadforth D, Hogdall E, Hogdall C, Blaeker J, DiCioccio R, McGuire V, Webb PM, Beesley J, Green AC, Whiteman DC, Australian Ovarian Cancer Study Group , Goodman MT, Lurie G, Carney ME, Modugno F, Ness RB, Edwards RP, Moysich KB, Goode EL, Couch FJ, Cunningham JM, Sellers TA, Wu AH, Pike MC, Iversen ES, Marks JR, Garcia-Closas M, Brinton L, Lissowska J, Peplonska B, Easton DF, Jacobs I, Ponder BA, Schildkraut J, Pearce CL, Chenevix-Trench G, Berchuck A, Pharoah PD, Ovarian Cancer Association Consortium Tagging single nucleotide polymorphisms in cell cycle control genes and susceptibility to invasive epithelial ovarian cancer. Cancer Res. 2007;67(7):3027–3035. doi: 10.1158/0008-5472.CAN-06-3261. [DOI] [PubMed] [Google Scholar]
- 25.Song H, Hogdall E, Ramus SJ, Dicioccio RA, Hogdall C, Quaye L, McGuire V, Whittemore AS, Shah M, Greenberg D, Easton DF, Kjaer SK, Pharoah PD, Gayther SA. Effects of common germ-line genetic variation in cell cycle genes on ovarian cancer survival. Clin Cancer Res. 2008;14(4):1090–1095. doi: 10.1158/1078-0432.CCR-07-1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Goode EL, Fridley BL, Vierkant RA, Cunningham JM, Phelan CM, Anderson S, Rider DN, White KL, Pankratz VS, Song H, Hogdall E, Kjaer SK, Whittemore AS, DiCioccio R, Ramus SJ, Gayther SA, Schildkraut JM, Pharaoh PP, Sellers TA. Candidate gene analysis using imputed genotypes: cell cycle single-nucleotide polymorphisms and ovarian cancer risk. Cancer Epidemiol Biomarkers Prev. 2009;18(3):935–944. doi: 10.1158/1055-9965.EPI-08-0860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hicks C, Asfour R, Pannuti A, Miele L. An integrative genomics approach to biomarker discovery in breast cancer. Cancer Inform. 2011;10:185–204. doi: 10.4137/CIN.S6837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen M, Rothman N, Ye Y, Gu J, Scheet PA, Huang M, Chang DW, Dinney CP, Silverman DT, Figueroa JD, Chanock SJ, Wu X. Pathway analysis of bladder cancer genome-wide association study identifies novel pathways involved in bladder cancer development. Genes Cancer. 2016;7(7-8):229–239. doi: 10.18632/genesandcancer.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tomoda K, Kubota Y, Kato J. Degradation of the cyclin-dependent-kinase inhibitor p27Kip1 is instigated by Jab1. Nature. 1999;398(6723):160–165. doi: 10.1038/18230. [DOI] [PubMed] [Google Scholar]
- 30.Zhu L, Wang J, Yue C, Yuan W, Zhang W, Shi L, Mi Y, Wu X, Zhang LF, Zuo L. CDKN1B Val 109 Gly variant is not related to risk of prostate cancer. J Cell Biochem. 2019;120(10):18346–18356. doi: 10.1002/jcb.29144. [DOI] [PubMed] [Google Scholar]
- 31.Kibel AS, Suarez BK, Belani J, Oh J, Webster R, Brophy-Ebbers M, Guo C, Catalona WJ, Picus J, Goodfellow PJ. CDKN1A and CDKN1B polymorphisms and risk of advanced prostate carcinoma. Cancer Res. 2003;63(9):2033–2036. [PubMed] [Google Scholar]
- 32.Huang SP, Yu CC, Liu CC, Wu TT, Huang CH, Wu MT. CDKN1B V109G polymorphism frequency and prostate cancer risk in Taiwan. Urol Int. 2008;81(1):36–40. doi: 10.1159/000137638. [DOI] [PubMed] [Google Scholar]
- 33.Zhang M, Liang Q, Zhang L, Hao Z, Zhou J, Zhang L, Fan S, Liang C. p27-V109G polymorphism is not associated with the risk of prostate cancer: a case-control study of Han Chinese men in Central China. Dis Markers. 2018;2018:1418609. doi: 10.1155/2018/1418609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chohan TA, Qian H, Pan Y, Chen JZ. Cyclin-dependent kinase-2 as a target for cancer therapy: progress in the development of CDK2 inhibitors as anti-cancer agents. Curr Med Chem. 2015;22(2):237–263. doi: 10.2174/0929867321666141106113633. [DOI] [PubMed] [Google Scholar]
- 35.Tadesse S, Anshabo AT, Portman N, Lim E, Tilley W, Caldon CE, Wang S. Targeting CDK2 in cancer: challenges and opportunities for therapy. Drug Discov Today. 2020;25(2):406–413. doi: 10.1016/j.drudis.2019.12.001. [DOI] [PubMed] [Google Scholar]
- 36.Maddison LA, Huss WJ, Barrios RM, Greenberg NM. Differential expression of cell cycle regulatory molecules and evidence for a “cyclin switch” during progression of prostate cancer. Prostate. 2004;58(4):335–344. doi: 10.1002/pros.10341. [DOI] [PubMed] [Google Scholar]
- 37.Keyomarsi K, O’Leary N, Molnar G, Lees E, Fingert HJ, Pardee AB. Cyclin E, a potential prognostic marker for breast cancer. Cancer Res. 1994;54(2):380–385. [PubMed] [Google Scholar]
- 38.Lian J, Zhang X, Lu Y, Hao S, Zhang Z, Yang Y. Expression and significance of LncRNA-MINCR and CDK2 mRNA in primary hepatocellular carcinoma. Comb Chem High Throughput Screen. 2019;22(3):201–206. doi: 10.2174/1386207322666190404151020. [DOI] [PubMed] [Google Scholar]
- 39.Gao Y, Wang H, Zhong A, Yu T. Expression and prognosis of CyclinA and CDK2 in patients with advanced cervical cancer after chemotherapy. Cell Mol Biol (Noisy-le-grand) 2020;66(3):85–91. [PubMed] [Google Scholar]
- 40.Alexander A, Karakas C, Chen X, Carey JP, Yi M, Bondy M, Thompson P, Cheung KL, Ellis IO, Gong Y, Krishnamurthy S, Alvarez RH, Ueno NT, Hunt KK, Keyomarsi K. Cyclin E overexpression as a biomarker for combination treatment strategies in inflammatory breast cancer. Oncotarget. 2017;8(9):14897–14911. doi: 10.18632/oncotarget.14689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nakayama N, Nakayama K, Shamima Y, Ishikawa M, Katagiri A, Iida K, Miyazaki K. Gene amplification CCNE1 is related to poor survival and potential therapeutic target in ovarian cancer. Cancer. 2010;116(11):2621–2634. doi: 10.1002/cncr.24987. [DOI] [PubMed] [Google Scholar]
- 42.Yang RM, Naitoh J, Murphy M, Wang HJ, Phillipson J, deKernion JB, Loda M, Reiter RE. Low p27 expression predicts poor disease-free survival in patients with prostate cancer. J Urol. 1998;159(3):941–945. [PubMed] [Google Scholar]
- 43.Thomas GV, Schrage MI, Rosenfelt L, Kim JH, Salur G, deKernion JB, Dorey F, Said J, Reiter RE. Preoperative prostate needle biopsy p27 correlates with subsequent radical prostatectomy p27, Gleason grade and pathological stage. J Urol. 2000;164(6):1987–1991. [PubMed] [Google Scholar]
- 44.Kumari S, Kumar P, Kumar M, Singh S, Narayan G. Expression of p27 and p16 and their clinical significance in gastric cancer. Clin Transl Oncol. 2021;23(4):856–865. doi: 10.1007/s12094-020-02479-4. [DOI] [PubMed] [Google Scholar]
- 45.Chu IM, Hengst L, Slingerland JM. The Cdk inhibitor p27 in human cancer: prognostic potential and relevance to anticancer therapy. Nat Rev Cancer. 2008;8(4):253–267. doi: 10.1038/nrc2347. [DOI] [PubMed] [Google Scholar]
- 46.Sirma H, Broemel M, Stumm L, Tsourlakis T, Steurer S, Tennstedt P, Salomon G, Michl U, Haese A, Simon R, Sauter G, Schlomm T, Minner S. Loss of CDKN1B/p27Kip1 expression is associated with ERG fusion-negative prostate cancer, but is unrelated to patient prognosis. Oncol Lett. 2013;6(5):1245–1252. doi: 10.3892/ol.2013.1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nassif AE, Tâmbara Filho R. Immunohistochemistry expression of tumor markers CD34 and P27 as a prognostic factor of clinically localized prostate adenocarcinoma after radical prostatectomy. Rev Col Bras Cir. 2010;37(5):338–344. doi: 10.1590/s0100-69912010000500006. [DOI] [PubMed] [Google Scholar]
- 48.Zhan B, Huang L, Chen Y, Ye W, Li J, Chen J, Yang S, Jiang W. miR-196a-mediated downregulation of p27kip1 protein promotes prostate cancer proliferation and relates to biochemical recurrence after radical prostatectomy. Prostate. 2020;80(12):1024–1037. doi: 10.1002/pros.24036. [DOI] [PubMed] [Google Scholar]
- 49.Prasad SB, Yadav SS, Das M, Modi A, Kumari S, Pandey LK, Singh S, Pradhan S, Narayan G. PI3K/AKT pathway-mediated regulation of p27(Kip1) is associated with cell cycle arrest and apoptosis in cervical cancer. Cell Oncol (Dordr) 2015;38(3):215–225. doi: 10.1007/s13402-015-0224-x. [DOI] [PubMed] [Google Scholar]
- 50.Xing YQ, Li A, Yang Y, Li XX, Zhang LN, Guo HC. The regulation of FOXO1 and its role in disease progression. Life Sci. 2018;193:124–131. doi: 10.1016/j.lfs.2017.11.030. [DOI] [PubMed] [Google Scholar]
- 51.Hiromura K, Pippin JW, Blonski MJ, Roberts JM, Shankland SJ. The subcellular localization of cyclin dependent kinase 2 determines the fate of mesangial cells: role in apoptosis and proliferation. Oncogene. 2002;21(11):1750–1758. doi: 10.1038/sj.onc.1205238. [DOI] [PubMed] [Google Scholar]
- 52.Davis JN, Day ML. The convergence of hormone regulation and cell cycle in prostate physiology and prostate tumorigenesis. Mol Biotechnol. 2002;22(2):129–138. doi: 10.1385/MB:22:2:129. [DOI] [PubMed] [Google Scholar]