Abstract
Background/Aim: Methionine addiction is a fundamental and general hallmark of cancer cells, which require exogenous methionine, despite large amounts of methionine synthesized endogenously. 5-Methylthioadenosine phospho-rylase (MTAP) plays a principal role as an enzyme in the methionine-salvage pathway, which produces methionine and adenine from methylthioadenosine and is deleted in 27.5% to 37.5% of osteosarcoma patients.
Materials and Methods: Human osteosarcoma cell lines U2OS, SaOS2, MNNG/HOS (HOS) and 143B, were used. The MTAP gene was knocked out in U2OS with CRISPR/Cas9. 143B and HOS have an MTAP deletion and SaOS2 is positive for MTAP. MTAP was determined by western blotting. The four cell lines were compared for sensitivity to recombinant methioninase (rMETase).
Results: MTAP-deleted osteosarcoma cell lines MNNG/HOS and 143B were significantly more sensitive to rMETase than MTAP-positive osteosarcoma cell lines U2OS and SaOS2. In addition, MTAP knock-out U2OS cells were more sensitive to rMETase than the parental MTAP-positive U2OS cells.
Conclusion: The present results demonstrated that the absence of MTAP sensitizes osteosarcoma cells to methionine restriction by rMETase, a promising clinical strategy.
Keywords: Osteosarcoma, methionine addiction, Hoffman efffect, recombinant methioninase, methionine restriction, MTAP, deletion, knockout, CRISPR/Cas9
Methionine addiction, discovered by one of us (RMH) in 1976 (1), is a general and fundamental hallmark of cancer cells and is termed the Hoffman effect. Methionine addiction occurs because cancer cells have highly overactive transmethylation reactions (1-5).
Cancer cells under methionine restriction arrest in S/G2 phase in contrast to normal cells, which arrest in G0 phase (6).
5-Methylthioadenosine phosphorylase (MTAP) is a rate-limiting enzyme in the methionine salvage pathway. This pathway produces methionine and adenine from 5'-methylthioadenosine (MTA) (7,8). Between 27.5% to 37.5% of patients with osteosarcoma have a homozygous MTAP deletion (9,10).
Cancer cells with MTAP deletion are sensitive to methionine restriction (11-19) but have not been tested with recombinant methioninase (rMETase), the most efficient means of methionine restriction, which is the topic of the present report.
Materials and Methods
Cell culture. U2OS, SaOS2, MNNG/HOS (HOS), 143B human osteosarcoma cell lines were obtained from the American Type Culture Collection (Manassas, VA, USA). Cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) (10-013-CV; Corning, Corning, NY, USA) with 10% fetal bovine serum (FBS) and 1 IU/ml penicillin/streptomycin.
Recombinant methioninase. rMETase is a homotetrameric enzyme, with a 172-kDa molecular mass. Production of rMETase was as previously reported, using fermentation, a heat step, polyethylene-glycol precipitation and gel filtration (20).
MTAP gene-knockout. U2OS cells were transfected using CRISPR/Cas9 (Santa Cruz Biotechnology, Santa Cruz, CA, USA) according to the protocol provided by the company. Briefly, cells were cultured in six-well plates (2.5×105 cells/well) in DMEM (1 ml/well) and incubated overnight. After cells grew to approximately 60% confluence, DMEM was aspirated and DMEM containing 10% FBS, but no antibiotics, were added and cells were incubated for 1 h. MTAP was knocked out in U2OS cells using CRISPR/Cas9 with the following materials and methods: MTAP Double Nickase plasmid (2 μg/20 μl) (sc-406223-NIC; Santa Cruz Biotechnology) or Control Double Nickase Plasmid (2 μg/20 μl) (sc-437281; Santa Cruz Biotechnology) were added to 130 μl Plasmid Transfection Medium (sc-108062; Santa Cruz Biotechnology), bringing the final volume to 150 μl (solution A). UltraCruz Transfection Reagent (sc-395739, 10 μl; Santa Cruz Biotechnology) was added to 140 μl Plasmid Transfection Medium (sc-108062; Santa Cruz Biotechnology), bringing the final volume to 150 μl (solution B). Solution A was added dropwise to solution B, bringing the total volume to 300 μl, immediately vortexed, and incubated at room temperature for 20 min (solution C). DMEM, which contains 10% FBS but no antibiotics, was aspirated from the wells and a total of 300 μl of solution C was added to the wells and the plates were incubated for 72 h at 37˚C with 5% CO2. After incubation, solution C was aspirated, 2 ml of DMEM containing 10% FBS and 5 μg/ml puromycin dihydrochloride (sc-108071; Santa Cruz Biotechnology) was added and plates were then incubated for 7 days at 37˚C with 5% CO2, at which time MTAP gene knocked-out cells were selected by resistance to puromycin (see below). During the incubation, the medium was changed once.
Limiting-dilution cloning of MTAP gene-knockout cells. After cell selection with puromycin for 7 days, the puromycin-containing medium was aspirated, and cells were cultured with DMEM containing 10% FBS and 1 IU/ml penicillin/streptomycin. At the end of the 10-day culture period, cells were harvested with trypsin and suspended at a density of 10 cells/ml in DMEM. The cells were then added to 96-well plates (1 cell/well, 100 μl) and incubated at 37˚C with 5% CO2. Wells containing a single-cell colony were marked and were allowed to incubate for 2 more weeks to expand the colony. The individual colonies were then cultured in T25 cell-culture flasks for 7 days. The cells were harvested with trypsin and proteins were extracted. Immunoblotting for MTAP was performed to confirm complete knock-out of the MTAP gene.
Immunoblotting. Cells with and without the MTAP gene were cultured in T25 cell-culture flasks. They were then lysed, and protein was extracted using RIPA Lysis and Extraction buffer (Thermo Fisher Scientific, Waltham, MA, USA) with 1% Halt Protease Inhibitor Cocktail (Thermo Fisher Scientific). Protein extract samples were loaded onto 10% sodium dodecyl sulfate–polyacrylamide gel electrophoresis gels and transferred to 0.45 μm polyvinylidene difluoride membranes (GE10600023; GE Healthcare, Chicago, IL, USA) after electrophoresis. The membranes were blocked using Bullet Blocking One for Western Blotting (Nakalai Tesque Inc., Kyoto, Japan). Antibody to MTAP (ab126770, 1:10,000; Abcam, Cambridge, UK) and anti-β actin (20536-1-AP, 1:1,500; Proteintech, Rosemont, IL, USA) were used. β-Actin was used as a loading control. As a second antibody, horseradish-peroxidase-conjugated anti-rabbit IgG (SA00001-2, 1:20,000; Proteintech, Rosemont, IL, USA) was used. The signals were detected with a UVP ChemStudio (Analytik Jena, Upland, CA, USA) using a Clarity Western ECL Substrate (Bio-Rad Laboratories, Hercules, CA, USA) to qualitatively visualize immunoreactivity.
rMETase sensitivity assay. Sensitivity to rMETase of osteosarcoma cell lines was assessed using the WST-8 reagent (Dojindo Laboratory, Kumamoto, Japan). The cells were cultured in 96-well plates at the following densities for each cell line: U2OS: 1.5×103 cells/well; SaOS2: 2.5×103 cells/well; HOS: 1.0×103 cells/well; and 143B: 7.5×102 cells/well; in DMEM (100 μl/well) and incubated overnight at 37˚C with 5% CO2. Cells were treated with rMETase at different concentrations, between 0.25 U/ml and 4.0 U/ml, for 96 h at 37˚C with 5% CO2. After rMETase treatment, 10 μl WST-8 solution was added to each well and the cells were incubated for an additional 1 h under the same conditions. Absorbance was then measured with a microplate reader (Sunrise; Tecan, Männedorf, Switzerland), at a wavelength of 450 nm. Drug-sensitivity curves were constructed using Microsoft Excel for Mac 2016 ver. 15.52 (Microsoft, Redmond, WA, USA) and half-maximal inhibitory-concentration (IC50) values were calculated with ImageJ ver. 1.53k (National Institutes of Health, Bethesda, MD, USA). Experiments were repeated twice for each cell line, each in triplicate.
Statistical analysis. All statistical analyses were performed with JMP pro ver. 15.0.0 (SAS Institute, Cary, NC, USA). Tukey-Kramer analysis was performed to compare each group separately for a significant difference. The Dunnett test was performed to compare each of the means with the control. Bar graphs show the mean and error bars show standard deviation of the mean. A probability value of ≤0.05 was defined as statistically significant.
Results
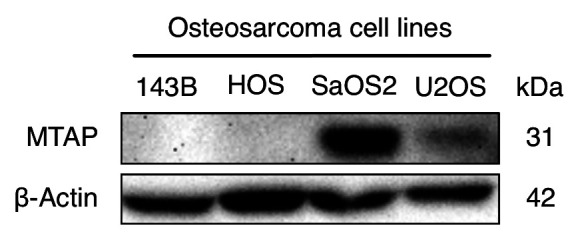
The MTAP gene is expressed in U2OS and SaOS2 but not in HOS and 143B osteosarcoma cell lines. MTAP gene expression was evaluated by immunoblotting in osteosarcoma cell lines. MTAP expression in U2OS and SaOS2, but not in HOS and 143B observed in the present report, is consistent with previous reports (9,10) (Figure 1).
Figure 1. Immunoblotting of 5-methylthioadenosine phosphorylase (MTAP) and β-actin in osteosarcoma cell lines U2OS, SaOS2, HOS and 143B.
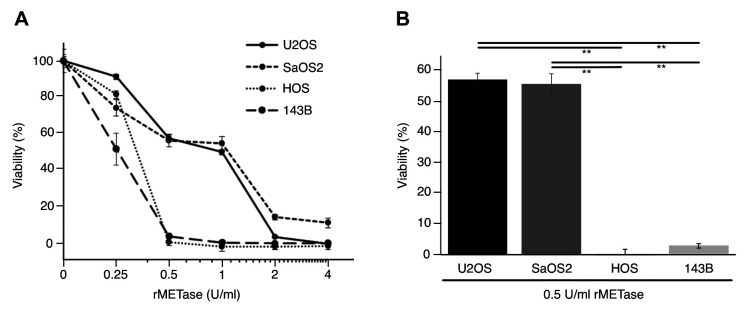
Osteosarcoma cell lines with an MTAP deletion are more sensitive to rMETase than MTAP-positive cells. rMETase inhibited the proliferation of MTAP-positive osteosarcoma cell lines with the following IC50 values: U2OS: 0.74 U/ml and SaOS2: 0.72 U/ml. In contrast, rMETase inhibited MTAP-negative cells with the following IC50 values: HOS: 0.22 U/ml and 143B: 0.24 U/ml, demonstrating that MTAP-negative cancer cells were much more sensitive to rMETase than MTAP-positive cells (p<0.001) (Figure 2).
Figure 2. A: Sensitivity of human osteosarcoma cell line U2OS, SaOS2, HOS and 143B to treatment with various concentrations of recombinant methioninase (rMETase) (mean±SD, n=3). B: Comparison of the efficacy of rMETase (0.5 U/ml) on the osteosarcoma cell lines. **Significantly different at p<0.001.
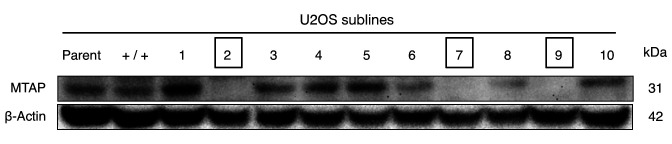
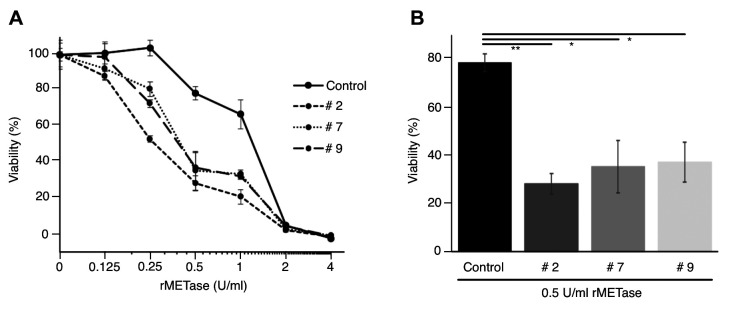
MTAP knock-out U2OS cells are more sensitive to rMETase than parental MTAP-positive cells. The MTAP gene was knocked out in U2OS cells (Figure 3). Three new independent MTAP-knockout sublines were designated as U2OS MTAP−/−#2; U2OS MTAP−/−#7; and U2OS MTAP−/−#9. U2OS MTAP+/+, which was treated with a control plasmid, was therefore used as the control (Figure 3). Compared to the U2OS MTAP+/+ control, all three MTAP knock-out sublines were more sensitive to rMETase, with the following IC50 values: U2OS MTAP+/+ control: 1.14 U/ml; U2OS MTAP−/−#2: 0.31 U/ml; U2OS MTAP−/−#7: 0.47 U/ml; and U2OS MTAP−/−#9: 0.45 U/ml. U2OS MTAP−/−#2 (p<0.001), U2OS MTAP−/−#7 (p=0.001), U2OS MTAP−/−#9 (p=0.0013) cells were much more sensitive to rMETase at 0.5 U/ml than the U2OS MTAP+/+ control (Figure 4).
Figure 3. Immunoblotting of 5-methylthioadenosine phosphorylase (MTAP) and β-actin in parent, control, and MTAP knock-out U2OS cell lines. U2OS sublines numerated in squares have MTAP knocked out.
Figure 4. A: Sensitivity to recombinant methioninase (rMETase) of 5-methylthioadenosine phosphorylase (MTAP)+/+ control and MTAP knock-out U2OS sublines #2, #7 and #9 (mean±SD, n=3). B: Comparison of the efficacy of 0.5 U/ml rMETase against MTAP+/+ control cells and MTAP−/− sublines #2, #7 and #9. Significantly different at *p<0.01 and **p<0.001.
Discussion
MTAP is the rate-limiting enzyme in the methionine-salvage pathway and has global effects on cellular methylation due to the utilization of S-adenosyl methionine in the methionine salvage pathway (11-19). In addition, the MTAP gene is adjacent to the tumor suppressor P16 (13,19) and often both are deleted together.
The present study showed osteosarcoma cell lines with MTAP deletion to be significantly more sensitive to rMETase than MTAP-positive ones. MTAP-knock-out U2OS cells were more sensitive to rMETase than parental MTAP-positive U2OS cells, which supports the concept that MTAP deletion sensitizes osteosarcoma cell lines to rMETase. The present study is the first to show that the absence of MTAP highly sensitizes cancer cells to rMETase. This was shown by direct comparison of sensitivity to rMETase of an MTAP-positive cell line with its sublines in which MTAP was knocked out; the MTAP knockout sublines of U2OS cells were significantly more sensitive to rMETase than the parental MTAP-positive cells. In addition, when the MTAP-positive and MTAP-negative cell lines were compared, it was shown that the MTAP-negative cell lines, HOS and 143B, were more sensitive to rMETase.
A previous study by Tang et al. suggested that MTAP is related to methionine dependence (addiction) in that MTAP-negative cells were unable to proliferate when homocysteine replaced methionine in the culture medium, while MTAP-positive cancer cells were still able to proliferate (12). However, their studies were defective since the FBS used in the medium was not dialyzed, and thereby actually contributed significant amounts of methionine to the ‘methionine-free’ culture medium. It was previously shown that even a very small amount of methionine (1 μM) highly stimulated cancer-cell proliferation in culture medium containing homocysteine instead of methionine (1). Previous studies also showed that MTAP replacement in MTAP-negative cancer cells did not revert methionine dependence to methionine independenc(12). Although the study of Tang et al. (12) is an old one, it demonstrates that care must be taken to ensure that medium termed ‘methionine-free’ is indeed free of methionine. The present study demonstrates that it is much more straight forward to determine methionine addiction with rMETase. Methionine dependence (addiction) is due to excessive transmethylation in cancer cells and is independent of MTAP (1-5).
Future studies will focus on potential therapy for patients with tumors with MTAP deletions with rMETase, using patient-derived orthotopic xenograft mouse models of osteosarcoma, which we have established to identify potential effective treatment strategies in our laboratory (21-23).
MTAP-deficient cancer cells are also sensitive to purine inhibitors, such as 6-mercaptopurine, methotrexate, pemetrexed, azaserine, azathioprine, L-alanosine and mizoribine (11,24-28). Future studies will evaluate the efficacy of rMETase in combination with inhibitors of purine synthesis, in both MTAP-positive cancer and MTAP-negative cancer, which may be more malignant (29) possibly due to co-deletion of P16 with the MTAP gene (19). Methionine addiction is a fundamental (1-5,30-37) and general (35-37) hallmark of cancer which is enhanced as a therapeutic target (38) by MTAP deletion.
Conflicts of Interest
YA, JY, KH, NM, YK and RMH are or were unsalaried associates of AntiCancer Inc. QH is an employee of AntiCancer Inc. The Authors have no conflicts of interest to declare in relation to this study.
Authors’ Contributions
YA, YT and RMH were involved in study conception and design. YA, JY, KH, NM and YK were involved in acquisition of data. YA, YT, JY, KH, NM, YK and RMH analyzed and interpreted data. YA, YT and RMH wrote the article. All Authors reviewed and approved the article.
Acknowledgements
This study was funded in part by the Robert M. Hoffman Foundation for Cancer Research. This article is dedicated to the memory of A. R. Moossa, MD, Sun Lee, MD, Professor Li Jiaxi, Masaki Kitajima, MD, Joseph R. Bertino, MD and Shigeo Yagi, PhD.
References
- 1.Hoffman RM, Erbe RW. High in vivo rates of methionine biosynthesis in transformed human and malignant rat cells auxotrophic for methionine. Proc Natl Acad Sci USA. 1976;73(5):1523–1527. doi: 10.1073/pnas.73.5.1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stern PH, Hoffman RM. Elevated overall rates of transmethylation in cell lines from diverse human tumors. In Vitro. 1984;20(8):663–670. doi: 10.1007/BF02619617. [DOI] [PubMed] [Google Scholar]
- 3.Hoffman RM, Jacobsen SJ. Reversible growth arrest in simian virus 40-transformed human fibroblasts. Proc Natl Acad Sci USA. 1980;77(12):7306–7310. doi: 10.1073/pnas.77.12.7306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Coalson DW, Mecham JO, Stern PH, Hoffman RM. Reduced availability of endogenously synthesized methionine for S-adenosylmethionine formation in methionine-dependent cancer cells. Proc Natl Acad Sci USA. 1982;79(14):4248–4251. doi: 10.1073/pnas.79.14.4248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hoffman RM, Jacobsen SJ, Erbe RW. Reversion to methionine independence in simian virus 40-transformed human and malignant rat fibroblasts is associated with altered ploidy and altered properties of transformation. Proc Natl Acad Sci USA. 1979;76(3):1313–1317. doi: 10.1073/pnas.76.3.1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stern PH, Hoffman RM. Enhanced in vitro selective toxicity of chemotherapeutic agents for human cancer cells based on a metabolic defect. J Natl Cancer Inst. 1986;76(4):629–639. doi: 10.1093/jnci/76.4.629. [DOI] [PubMed] [Google Scholar]
- 7.Tisdale MJ. Methionine synthesis from 5’-methylthioadenosine by tumour cells. Biochem Pharmacol. 1983;32(19):2915–2920. doi: 10.1016/0006-2952(83)90396-9. [DOI] [PubMed] [Google Scholar]
- 8.Toohey JI. Methylthioadenosine nucleoside phosphorylase deficiency in methylthio-dependent cancer cells. Biochem Biophys Res Commun. 1978;83(1):27–35. doi: 10.1016/0006-291x(78)90393-5. [DOI] [PubMed] [Google Scholar]
- 9.García-Castellano JM, Villanueva A, Healey JH, Sowers R, Cordon-Cardo C, Huvos A, Bertino JR, Meyers P, Gorlick R. Methylthioadenosine phosphorylase gene deletions are common in osteosarcoma. Clin Cancer Res. 2002;8(3):782–787. [PubMed] [Google Scholar]
- 10.Miyazaki S, Nishioka J, Shiraishi T, Matsumine A, Uchida A, Nobori T. Methylthioadenosine phosphorylase deficiency in Japanese osteosarcoma patients. Int J Oncol. 2007;31(5):1069–1076. [PubMed] [Google Scholar]
- 11.Hori H, Tran P, Carrera CJ, Hori Y, Rosenbach MD, Carson DA, Nobori T. Methylthioadenosine phosphorylase cDNA transfection alters sensitivity to depletion of purine and methionine in A549 lung cancer cells. Cancer Res. 1996;56(24):5653–5658. [PubMed] [Google Scholar]
- 12.Tang B, Li YN, Kruger WD. Defects in methylthioadenosine phosphorylase are associated with but not responsible for methionine-dependent tumor cell growth. Cancer Res. 2000;60(19):5543–5547. [PubMed] [Google Scholar]
- 13.Christopher SA, Diegelman P, Porter CW, Kruger WD. Methylthioadenosine phosphorylase, a gene frequently codeleted with p16(cdkN2a/ARF), acts as a tumor suppressor in a breast cancer cell line. Cancer Res. 2002;62(22):6639–6644. [PubMed] [Google Scholar]
- 14.Kirovski G, Stevens AP, Czech B, Dettmer K, Weiss TS, Wild P, Hartmann A, Bosserhoff AK, Oefner PJ, Hellerbrand C. Down-regulation of methylthioadenosine phosphorylase (MTAP) induces progression of hepatocellular carcinoma via accumulation of 5’-deoxy-5’-methylthioadenosine (MTA) Am J Pathol. 2011;178(3):1145–1152. doi: 10.1016/j.ajpath.2010.11.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kryukov GV, Wilson FH, Ruth JR, Paulk J, Tsherniak A, Marlow SE, Vazquez F, Weir BA, Fitzgerald ME, Tanaka M, Bielski CM, Scott JM, Dennis C, Cowley GS, Boehm JS, Root DE, Golub TR, Clish CB, Bradner JE, Hahn WC, Garraway LA. MTAP deletion confers enhanced dependency on the PRMT5 arginine methyltransferase in cancer cells. Science. 2016;351(6278):1214–1218. doi: 10.1126/science.aad5214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mavrakis KJ, McDonald ER 3rd, Schlabach MR, Billy E, Hoffman GR, deWeck A, Ruddy DA, Venkatesan K, Yu J, McAllister G, Stump M, deBeaumont R, Ho S, Yue Y, Liu Y, Yan-Neale Y, Yang G, Lin F, Yin H, Gao H, Kipp DR, Zhao S, McNamara JT, Sprague ER, Zheng B, Lin Y, Cho YS, Gu J, Crawford K, Ciccone D, Vitari AC, Lai A, Capka V, Hurov K, Porter JA, Tallarico J, Mickanin C, Lees E, Pagliarini R, Keen N, Schmelzle T, Hofmann F, Stegmeier F, Sellers WR. Disordered methionine metabolism in MTAP/CDKN2A-deleted cancers leads to dependence on PRMT5. Science. 2016;351(6278):1208–1213. doi: 10.1126/science.aad5944. [DOI] [PubMed] [Google Scholar]
- 17.Marjon K, Cameron MJ, Quang P, Clasquin MF, Mandley E, Kunii K, McVay M, Choe S, Kernytsky A, Gross S, Konteatis Z, Murtie J, Blake ML, Travins J, Dorsch M, Biller SA, Marks KM. MTAP deletions in cancer create vulnerability to targeting of the MAT2A/PRMT5/RIOK1 axis. Cell Rep. 2016;15(3):574–587. doi: 10.1016/j.celrep.2016.03.043. [DOI] [PubMed] [Google Scholar]
- 18.Kalev P, Hyer ML, Gross S, Konteatis Z, Chen CC, Fletcher M, Lein M, Aguado-Fraile E, Frank V, Barnett A, Mandley E, Goldford J, Chen Y, Sellers K, Hayes S, Lizotte K, Quang P, Tuncay Y, Clasquin M, Peters R, Weier J, Simone E, Murtie J, Liu W, Nagaraja R, Dang L, Sui Z, Biller SA, Travins J, Marks KM, Marjon K. MAT2A inhibition blocks the growth of MTAP-deleted cancer cells by reducing PRMT5-dependent mRNA Splicing and Inducing DNA Damage. Cancer Cell. 2021;39(2):209–224.e11. doi: 10.1016/j.ccell.2020.12.010. [DOI] [PubMed] [Google Scholar]
- 19.Sanderson SM, Mikhael PG, Ramesh V, Dai Z, Locasale JW. Nutrient availability shapes methionine metabolism in p16/MTAP-deleted cells. Sci Adv. 2019;5(6):eaav7769. doi: 10.1126/sciadv.aav7769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tan Y, Xu M, Tan X, Tan X, Wang X, Saikawa Y, Nagahama T, Sun X, Lenz M, Hoffman RM. Overexpression and large-scale production of recombinant L-methionine-alpha-deamino-gamma-mercaptomethane-lyase for novel anticancer therapy. Protein Expr Purif. 1997;9(2):233–245. doi: 10.1006/prep.1996.0700. [DOI] [PubMed] [Google Scholar]
- 21.Igarashi K, Kawaguchi K, Yamamoto N, Hayashi K, Kimura H, Miwa S, Higuchi T, Taniguchi Y, Yonezawa H, Araki Y, Morinaga S, Misra S, Nelson SD, Dry SM, Li Y, Odani A, Singh SR, Tsuchiya H, Hoffman RM. A Novel anionic-phosphate-platinum complex effectively targets a cisplatinum-resistant osteosarcoma in a patient-derived orthotopic xenograft mouse model. Cancer Genomics Proteomics. 2020;17(3):217–223. doi: 10.21873/cgp.20182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Aoki Y, Tome Y, Wu NF, Yamamoto J, Hamada K, Han Q, Bouvet M, Nishida K, Hoffman RM. Oral-recombinant methioninase converts an osteosarcoma from docetaxel-resistant to -sensitive in a clinically-relevant patient-derived orthotopic-xenograft (PDOX) mouse model. Anticancer Res. 2021;41(4):1745–1751. doi: 10.21873/anticanres.14939. [DOI] [PubMed] [Google Scholar]
- 23.Higuchi T, Igarashi K, Yamamoto N, Hayashi K, Kimura H, Miwa S, Bouvet M, Tsuchiya H, Hoffman RM. Osteosarcoma patient-derived orthotopic xenograft (PDOX) models used to identify novel and effective therapeutics: a review. Anticancer Res. 2021;41(12):5865–5871. doi: 10.21873/anticanres.15406. [DOI] [PubMed] [Google Scholar]
- 24.Dervieux T, Brenner TL, Hon YY, Zhou Y, Hancock ML, Sandlund JT, Rivera GK, Ribeiro RC, Boyett JM, Pui CH, Relling MV, Evans WE. De novo purine synthesis inhibition and antileukemic effects of mercaptopurine alone or in combination with methotrexate in vivo. Blood. 2002;100(4):1240–1247. doi: 10.1182/blood-2002-02-0495. [DOI] [PubMed] [Google Scholar]
- 25.Bertino JR, Waud WR, Parker WB, Lubin M. Targeting tumors that lack methylthioadenosine phosphorylase (MTAP) activity: current strategies. Cancer Biol Ther. 2011;11(7):627–632. doi: 10.4161/cbt.11.7.14948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jing W, Zhu H, Liu W, Zhai X, Tian H, Yu J. MTAP-deficiency could predict better treatment response in advanced lung adenocarcinoma patients initially treated with pemetrexed-platinum chemotherapy and bevacizumab. Sci Rep. 2020;10(1):843. doi: 10.1038/s41598-020-57812-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li W, Su D, Mizobuchi H, Martin DS, Gu B, Gorlick R, Cole P, Bertino JR. Status of methylthioadenosine phosphorylase and its impact on cellular response to L-alanosine and methylmercaptopurine riboside in human soft tissue sarcoma cells. Oncol Res. 2004;14(7-8):373–379. doi: 10.3727/0965040041292332. [DOI] [PubMed] [Google Scholar]
- 28.Batova A, Diccianni MB, Omura-Minamisawa M, Yu J, Carrera CJ, Bridgeman LJ, Kung FH, Pullen J, Amylon MD, Yu AL. Use of alanosine as a methylthioadenosine phosphorylase-selective therapy for T-cell acute lymphoblastic leukemia in vitro. Cancer Res. 1999;59(7):1492–1497. [PubMed] [Google Scholar]
- 29.Li CF, Fang FM, Kung HJ, Chen LT, Wang JW, Tsai JW, Yu SC, Wang YH, Li SH, Huang HY. Downregulated MTAP expression in myxofibrosarcoma: A characterization of inactivating mechanisms, tumor suppressive function, and therapeutic relevance. Oncotarget. 2014;5(22):11428–11441. doi: 10.18632/oncotarget.2552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stern PH, Mecham JO, Wallace CD, Hoffman RM. Reduced free-methionine in methionine-dependent SV40-transformed human fibroblasts synthesizing apparently normal amounts of methionine. J Cell Physiol. 1983;117(1):9–14. doi: 10.1002/jcp.1041170103. [DOI] [PubMed] [Google Scholar]
- 31.Yamamoto J, Han Q, Inubushi S, Sugisawa N, Hamada K, Nishino H, Miyake K, Kumamoto T, Matsuyama R, Bouvet M, Endo I, Hoffman RM. Histone methylation status of H3K4me3 and H3K9me3 under methionine restriction is unstable in methionine-addicted cancer cells, but stable in normal cells. Biochem Biophys Res Commun. 2020;533(4):1034–1038. doi: 10.1016/j.bbrc.2020.09.108. [DOI] [PubMed] [Google Scholar]
- 32.Yamamoto J, Aoki Y, Inubushi S, Han Q, Hamada K, Tashiro Y, Miyake K, Matsuyama R, Bouvet M, Clarke SG, Endo I, Hoffman RM. Extent and Instability of Trimethylation of Histone H3 Lysine Increases With Degree of Malignancy and Methionine Addiction. Cancer Genomics Proteomics. 2022;19(1):12–18. doi: 10.21873/cgp.20299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hoffman RM, Jacobsen SJ, Erbe RW. Reversion to methionine independence in simian virus 40-transformed human and malignant rat fibroblasts is associated with altered ploidy and altered properties of transformation. Proc Natl Acad Sci USA. 1979;76(3):1313–1317. doi: 10.1073/pnas.76.3.1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Aoki Y, Yamamoto J, Tome Y, Hamada K, Masaki N, Inubushi S, Tashiro Y, Bouvet M, Endo I, Nishida K, Hoffman RM. Over-methylation of histone H3 lysines is a common molecular change among the three major types of soft-tissue sarcoma in patient-derived xenograft (PDX) mouse models. Cancer Genomics Proteomics. 2021;18(6):715–721. doi: 10.21873/cgp.20292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mecham JO, Rowitch D, Wallace CD, Stern PH, Hoffman RM. The metabolic defect of methionine dependence occurs frequently in human tumor cell lines. Biochem Biophys Res Commun. 1983;117(2):429–434. doi: 10.1016/0006-291x(83)91218-4. [DOI] [PubMed] [Google Scholar]
- 36.Stern PH, Wallace CD, Hoffman RM. Altered methionine metabolism occurs in all members of a set of diverse human tumor cell lines. J Cell Physiol. 1984;119(1):29–34. doi: 10.1002/jcp.1041190106. [DOI] [PubMed] [Google Scholar]
- 37.Tan Y, Xu M, Hoffman RM. Broad selective efficacy of recombinant methioninase and polyethylene glycol-modified recombinant methioninase on cancer cells in vitro. Anticancer Res. 2010;30(4):1041–1046. [PubMed] [Google Scholar]
- 38.Yamamoto J, Han Q, Simon M, Thomas D, Hoffman RM. Methionine Restriction: Ready for Prime Time in the Cancer Clinic. Anticancer Res. 2022;42(2):641–644. doi: 10.21873/anticanres.15521. [DOI] [PubMed] [Google Scholar]