Abstract
The cell entry of SARS-CoV-2 has emerged as an attractive drug development target. We previously reported that the entry of SARS-CoV-2 depends on the cell surface heparan sulfate proteoglycan (HSPG) and the cortex actin, which can be targeted by therapeutic agents identified by conventional drug repurposing screens. However, this drug identification strategy requires laborious library screening, which is time-consuming and often limited number of compounds can be screened. As an alternative approach, we developed and trained a graph convolutional network (GCN)-based classification model using information extracted from experimentally identified HSPG and actin inhibitors. This method allowed us to virtually screen 170,000 compounds, resulting in ~2000 potential hits. A hit confirmation assay with the uptake of a fluorescently labeled HSPG cargo further shortlisted 256 active compounds. Among them, 16 compounds had modest to strong inhibitory activities against the entry of SARS-CoV-2 pseudotyped particles into Vero E6 cells. These results establish a GCN-based virtual screen workflow for rapid identification of new small molecule inhibitors against validated drug targets.
Graphical Abstract
Introduction
Since the outbreak of the COVID19 pandemic, global communities have suffered a significant loss of lives and economic growth. Although the development of COVID vaccines can significantly contain the spreading of SARS-CoV-2, the virus is constantly evolving into more infectious and transmissible variants (e.g., the delta strain), resulting in frequent breakthrough infections among vaccinated people.1-5 The constant increase of hospitalized patients in the USA and around the world despite the rollout of the vaccination programs has summoned the need to develop potent small molecule therapeutics for COVID patients.
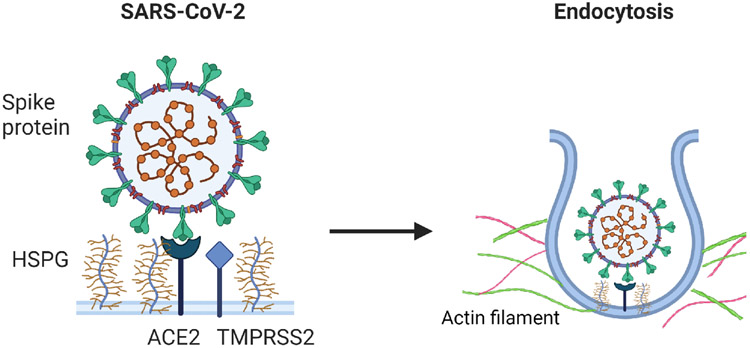
The cellular entry of SARS-CoV-2 is one of the key steps in the viral life cycle that represents a hot target for small molecule inhibitors.6,7 To target SARS-Cov-2 viral entry, the most popular target is the viral Spike protein given that drugs targeting Spike are less likely to disrupt cellular processes in the host cells. On the host side, a variety of proteins are being evaluated as potential anti-SARS-CoV-2 targets, which include angiotensin-converting enzyme 2 (ACE2), the major receptor for Spike on the cell surface,8-12 and transmembrane serine protease 2 (TMSSPR2), the protease involved in activating Spike and other factors regulating the endocytosis of virion particles.13 In addition, a recent report also discussed that an alternative entry points have been identified using neuropilin-1 (NRP1), which was found to significantly enhance the infectivity of SARS-CoV-2 by increasing viral entry into host cells rather than strengthening viral binding.14 Previously, we and others identified the cell surface heparan sulfate proteoglycans (HSPGs) as a critical cofactor that facilitate the entry of SARS-CoV-2 virions.8,9 We further showed that HSPGs, as negatively charged biopolymers, also facilitate the uptake of other positive charge-bearing endocytic cargos such as supercharged green fluorescence protein (GFP) and preformed -Synuclein pathogenic fibrils.15 HSPGs are a family of glycoproteins bearing one or more negatively charged polysaccharide chains consisting of repeated heparan sulfate disaccharide units. Most HSPG family members are anchored to the cell surface either as a single spanning membrane protein (e.g., Syndecans) or Glycosylphosphatidylinositol (GPI)-anchored protein (e.g., Glypicans). Due to the enrichment of negatively charged sulfate groups, HSPGs can effectively serve as an attachment anchor to increase the surface dwell time for endocytic cargos bearing positive charges, facilitating their engagement with a downstream receptor.6,15,16 The internalization of HSPG cargos also requires the cortex actin, a specialized layer of proteins associated with the inner surface of the plasma membrane. A major component of the cortex is actin filament, which associates with myosin motor proteins and other actin-binding proteins. These proteins together maintain plasma membrane dynamics to promote the maturation of clathrin-coated pits.15 The specific mechanism of the cellular entry of SARS-CoV-2 was presented in Figure 1.
Figure 1:
The schematic description of cellular entry of SARS-CoV-2.
We recently conducted a drug repurposing screen based on our previous study,6 and identified 8 drugs that inhibited HSPG-dependent entry of SARS-CoV-2 virions.6 Intriguingly, despite structural dissimilarity, several of the identified drugs display high potential of binding with heparin, a heparan sulfate analogue, suggesting that they may target the polysaccharide chain on the cell surface of HSPG to inhibit viral entry; however, alternative host protein targets may also exist.14 In addition to heparin-binding drugs, two structurally unrelated drugs, Sunitinib and BNTX, can both effectively disrupt the actin filaments underlying the plasma membrane (cortex actin) to inhibit HSPG-mediated endocytosis.9-12 Compared to drugs that target viral proteins, drugs targeting host factors essential for viral entry and replication are less likely to generate drug resistance as a result of viral mutations. While drug repurposing screen is an effective strategy to rapidly adopt existing drugs for new therapeutic uses, the original target(s) of the approved drugs often reduces their therapeutic specificity, which may cause undesired side effects for treating viral infection. For example, as a heparan sulfate binding compound, mitoxantrone delivers the most potent antiviral activity in vitro. However, because mitoxantrone was originally approved as anti-cancer chemotherapy via targeting DNA topoisomerases,17 cytotoxicity associated with DNA replication inhibition is an obvious concern.
To identify additional inhibitors targeting HSPG-mediated viral entry, we developed a graph convolutional network (GCN)-based classification approach. GCN can efficiently translate 3D structures into molecular graphs composed of nodes and edges, and then utilize these graphs to extract spatial information to achieve accurate molecular classification and properties predictions.18-21 Compared to other traditional computational methods based on molecular dynamics (MD) simulations or density functional theory (DFT), the computational cost of GCN is substantially lower. These features allowed us to rapidly screen 17,000 compounds in several NCATS libraries. From these libraries, we identified and confirmed a set of compounds (256) as inhibitors of HSPG-dependent endocytosis with the most potent IC50 value at 0.95 μM. Further testing with a SARS-CoV-2 pseudotyped particle entry assay confirmed 16 compounds as entry inhibitors.
Methods
Computational details
GCN model
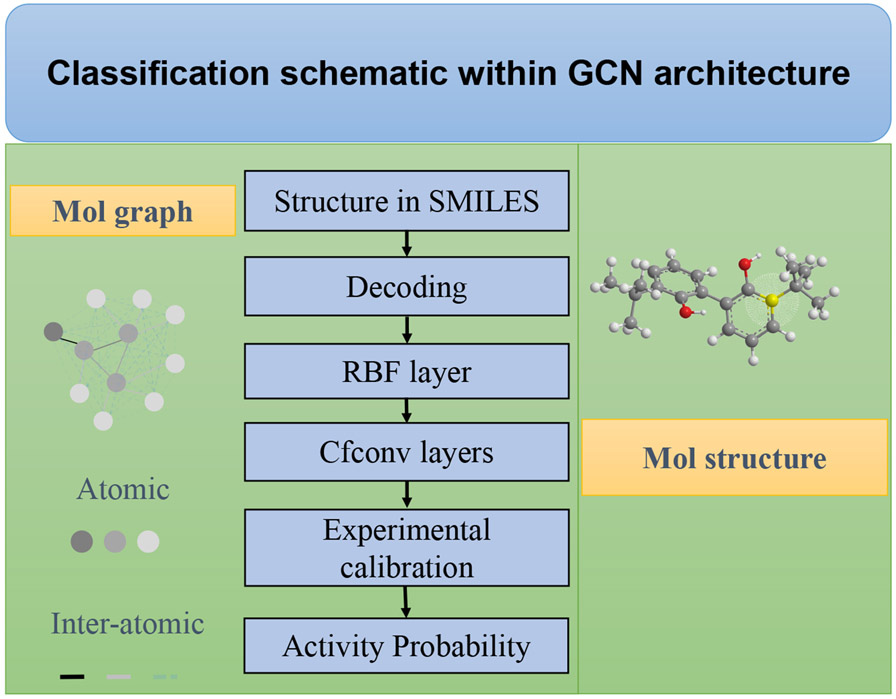
GCN-based approaches display considerable robustness for structural elucidations,18-21 because it could fully utilize the molecular graphs for information extraction with substantially reduced computational cost, compared to MD or DFT based methods.22-29 In addition, such an architecture can directly work on graph based inputs, instead of depending on collected descriptors, thus it is more suitable for spatial information processing.30 At the same time, it is still flexible enough to include different chemical knowledge as extra descriptors for specific assignments.22,31-40 In this study, we employed the self-developed GCN package for activity classifications, and the SchNet architecture was applied.41 The workflow of the applied GCN was described in Figure 2. For any given drug molecule, its structural information was contained in the simplified molecular-input line-entry system (SMILES) string, and GCN can transform the molecular graph into a set of numerical descriptors for computational processing.
Figure 2:
The architecture of GCN classification model for virtual screenings.
All the collected SMILES strings of drug molecules were first translated into molecular graphs through the TencentAlchemyDataset within Deep Graph Library (DGL) library.42,43 Each drug molecule is composed of edges and nodes within 3D space. Within the framework of GCN, the generated nodes represent atomic points within molecule, while the edges are corresponding to inter-atomic connections. And with these numerically encoded features, structure similarity can be well summarized, and related molecular properties can be mapped correspondingly. In fact, within any molecular or fragmentary graphs, all the connections between every two atoms are fully utilized for information extraction; the specific values were recorded in distance tensors at the radial basis function (RBF) layer, guaranteeing there is no omission of important structure information. In addition, within GCN model, to decently solve molecular graphs at atomic level, multiple continuous-filter convolutions (cfconv) layers were employed to optimize and record the inter-atomic evolution. For instance, at layer, the atom’s evolution can be expressed with the following equation:
| (1) |
in which, represents element-wise multiplication, and is the filter-generation that can map the atoms’ descriptions to the filter bank. To efficiently control the evolution accuracy via the applied the filter values, a Gaussian-type function, , was employed, which can be expressed with the following equation:
| (2) |
where, is the pre-set value of cutoff, and represents the bonding distance among the atom and atom. The is attributed to hyper parameters, and it was set to 0.1 in this study.41
For any predictive property or classification task, the computed value, , by GCN model is calibrated with respect to experimental measurement, , and the accuracy can be well indicated by the squared loss function, as shown below:
| (3) |
In this study, we applied the developed GCN package for drugs activity classification; however, it is worth noting that this promising architecture is also able to include various kinds of chemical & physical knowledge for more challenging structural assignments.
Data set
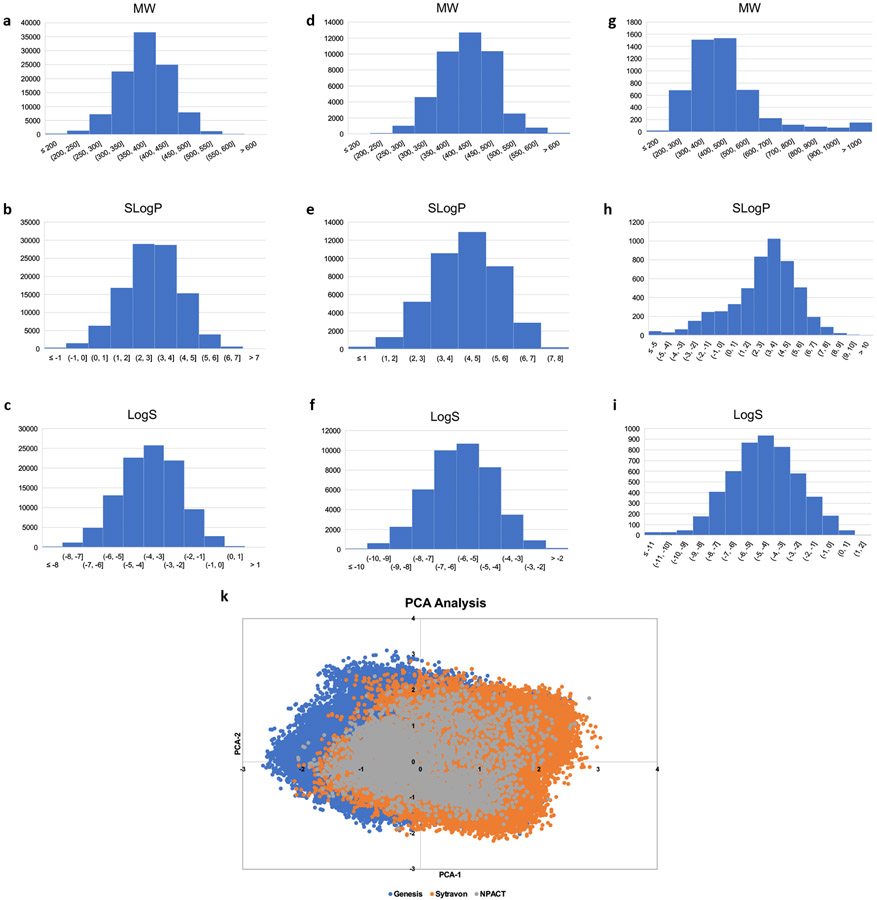
We applied the above-described GCN model to a previously reported COVID-19 related drug screening, which identified drugs that block HSPG-dependent entry of -Synuclein fibrils. Classification algorithm was based on NCATS’ collected activity values. The model was first trained by the collected data, which consisted of 3,832 compounds.6 Among them, 367 compounds show activities and 3,465 are inactive. These compounds were randomly divided with a ratio of 9:1; and 90% was used as the training set, and the remaining 10% as the test set. The trained GCN model was validated by the compounds in the test set, which scored an accuracy of 99.5%. The trained model was then used to screen more than 170,000 compounds contained in three independent libraries, Genesis, Sytravon, and NCATS Pharmacologically Active Chemical Toolbox (NPACT), none of which had been experimentally screened by endocytosis or SARS-CoV-2 PP entry assays. NCATS has assembled the Genesis collection with 100,000 compounds to provide a novel modern chemical library that emphasizes high-quality chemical starting points, sp3-enriched chemotypes, and core scaffolds that enable rapid purchase and derivatization via medicinal chemistry. The Sytravon library is a retired Pharma screening collection that contains 44,000 diverse and novel small molecules, with an emphasis on medicinal chemistry-tractable scaffolds. The NPACT is a library of 5,000 annotated compounds that inform on novel phenotypes, biological pathways and cellular processes. There are more than 7,000 mechanisms and phenotypes identified in the literature and worldwide patents that cover biological interactions within mammalian, microbial, plant and other model systems. The physicochemical properties and the chemical diversity coverage of these three libraries were shown in Figure 3.
Figure 3:
(a) to (c). Distribution of the chemical properties, molecular weight (MW), SlogP, and logS for Genesis library; (d) to (f). Distribution of the chemical properties, molecular weight (MW), SlogP, and logS for Sytravon library; (g) to (i). Distribution of the chemical properties, molecular weight (MW), SlogP, and logS for NPACT library; (k). The PCA analysis for the three independent libraries, Genesis, Sytravon, and NPACT.
-Synuclein fibrils uptake assay and drug verification
Fluorescence labeled -synuclein fibrils were generated as previously described.15 HEK293T cells were dispensed into black, clear-bottom 1536-well microplates (Greiner BioOne, # 789092-F)) at 5000cells/well in 5L media with 200nM pHrodo red-labeled -Syn fibrils and incubated at 37°C, 5% CO2, 85% humidity overnight (~16 h). Compounds picked from the virtual screen were titrated 1:3 with 11 points in DMSO and transferred to assay plates at a volume of 23 nl/well by an automated pintool workstation (Wako Automation, San Diego, CA). After 24 h of incubation, the fluorescence intensity of pHrodo red was measured by a CLARIOstar Plus plate reader (BMG Labtech). Data was normalized using the wells with cells containing 200nMpHrodo red-labeled Syn fibrils as 100% and the wells without cells as 0%.
Image processing and statistical analyses
Confocal images were processed using the Zeiss Zen software. To measure fluorescence intensity, we used the Fiji software. Images were converted to individual channels and regions of interest were drawn for measurement. Statistical analyses were performed using either Excel or GraphPad Prism 9. Data are presented as means ± SEM, which was calculated by GraphPad Prism 9. P values were calculated by Student’s t-test using Excel. Nonlinear curve fitting and IC50 calculation was done with GraphPad Prism 9 using the inhibitor response three variable model or the exponential decay model. Images were prepared with Adobe Photoshop and assembled in Adobe Illustrator. All experiments presented were repeated at least twice independently. Data processing and reporting are adherent to the community standards.
SARS-CoV-2 PP assay
HEK293T-ACE2-GFP cells seeded in white, solid bottom 384-well microplates (Greiner BioOne) at 6,000 cells/well in 15 μL medium were incubated at 37°C with 5% CO2 overnight (~16 h). Compounds were titrated 1:3 with 11 points in DMSO and dispensed into the assay plate at 23 nl/well via pintool. Cells were incubated with compounds for 1h at 37°C with 5% CO2 before 15 μl/well of PPs were added. The plates were then spinoculated by centrifugation at 1,500 rpm (453 x g) for 45 min and incubated for 48h at 37°C 5% CO2 to allow cell entry of PPs and the expression of luciferase. After the incubation, the supernatant was removed with gentle centrifugation using a Blue Washer (BlueCat Bio). Then 20 μL/well of Bright-Glo luciferase detection reagent (Promega) was added to assay plates and incubated for 5 min at room temperature. The luminescence signal was measured using a PHERAStar plate reader (BMG Labtech). Data were normalized with wells containing PPs as 100% and wells containing control DEnv PP as 0%.
ATP content cytotoxicity assay
HEK293T-ACE2-GFP cells were seeded in white, solid bottom 384-well microplates (Greiner BioOne) at 6,000 cells/well in 15 μl medium and incubated at 37°C with 5% CO2 overnight (~16 h). Compounds were titrated 1:3 in DMSO and dispensed via pintool at 23 nl/well to assay plates. Cells were incubated for 1 h at 37°C 5% CO2 before 15 μl/well of media was added. The plates were then incubated at 37°C for 48h at 37°C 5% CO2. After incubation, 30 μl/well of ATPLite (PerkinElmer) was added to assay plates and incubated for 15 min at room temperature. The luminescence signal was measured using a Viewlux plate reader (PerkinElmer). Data were normalized with wells containing cells as 100%, and wells containing media only as 0%.
Results and discussion
The overall performance of the GCN model
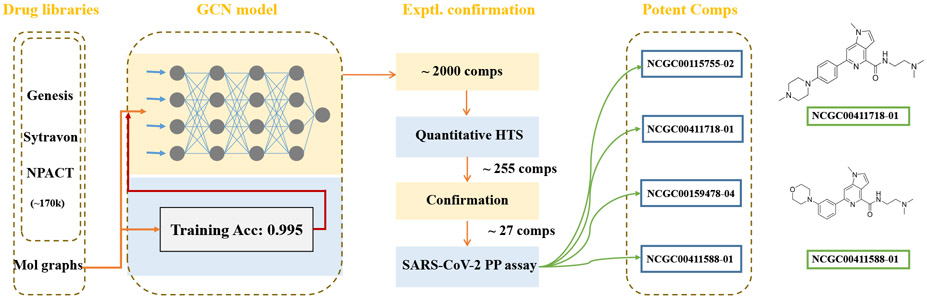
Unlike traditional computational drug discovery methods such as structural homology-based drug search, the GCN classification model utilizes molecular graphs to extract spatial information. The modeling process computes in bonding environment at atomic or inter-atomic level within a fully connected framework as opposed to utilizing simple descriptors. As a result, the structural features of drug molecules can be well captured and built from low-level logic,38,41 making no omission of important possibilities. This method results in a robust performance with the classification accuracy as high as 99.5% for training set (the workflow was described in Figure 4).
Figure 4:
The workflow of GCN classification model upon endocytosis screenings.
We also did a benchmarking analysis by comparing the classification accuracy of GCN with several popular machine learning algorithms. From the calculated AUC values, GCN performs comparably better than other methods (Table 1), especially for the case, in which the ratio of active/inactive compounds is small; in another aspect, from the confusion matrix of validation set, it is notable that GCN is advantageous to exclude inactive compounds with a high accuracy (more details can be found in supplementary Table S2). At the same time, based on Y-randomization test using a random counter training set (the activity was randomly shuffled in the training data set), we noticed that AUC value of GCN is largely reduced, indicating its dependence on highly solved structure similarity for active compounds detection. Additionally, the identified new compounds from our in-house libraries also to some degree show structural novelty to the active compounds in our training set, further highlighting its high applicability in real practice of decent drug screenings, compared to other structural assignment-based approaches.
Table 1:
The calculated AUC scores of the machine learning based classification methods upon the applied data set.
| Methoda) | AUC score | |
|---|---|---|
| 1 | Random forest | 0.585 |
| 2 | AdaBoost | 0.589 |
| 3 | GaussianNB | 0.537 |
| 4 | LogisticRegression | 0.604 |
| 5 | GradientBoosting | 0.575 |
| 6 | SVM | 0.594 |
| 7 | GCN (SchNet) | 0.683 |
The technical details can be found in our GitHub page: github.com/tcsnfrank0177/Graph-convolutional-network-DrugScreening.git.
Identification of inhibitors for HSPG-mediated endocytosis
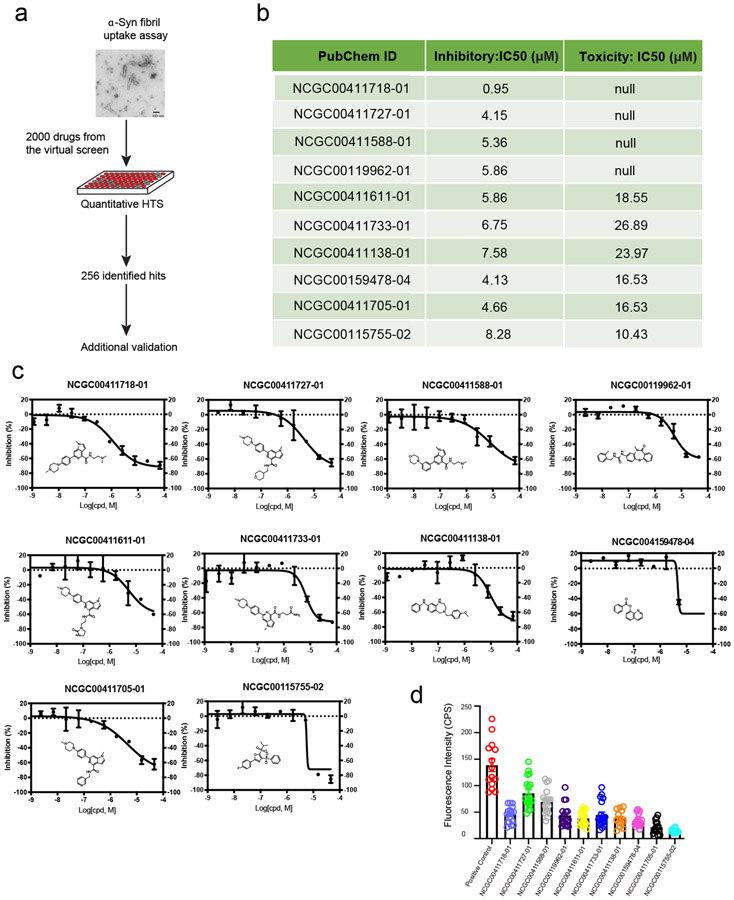
We used the GCN-based model to screen 170,000 compounds. ~2000 compounds were shortlisted by the virtual screen, which generated a small library that could be rapidly processed by a conventional quantitative high-throughput screen (qHTS) (Figure 5a). We then employed pHrodo red labeled -Synuclein fibrils as an HSPG cargo in a combination screen because -Synuclein fibrils share a similar entry mechanism as SARS-CoV-2.15 Importantly, the fluorescence intensity of cells treated with pHrodo-labeled -Synuclein fibrils is only dependent on the amount of internalized cargo and the endolysosomal pH. By comparison, the luciferase-based pseudoviral entry assay can be influenced not only by the level of viral entry, but also by other factors that impact mRNA expression, translation, and luciferase stability. The screen identified 256 active compounds with most potent IC50 value of 0.95 uM. We cherry-picked 10 top compounds based on their potency and structural novelty (Figure 5b), and measured their cytotoxicity by an ATP content assay. The results showed that for 4 out of the 10 compounds, the IC50 for cytotoxicity was at least 10-fold larger than that for the inhibition of -Synuclein fibril uptake (Figure 5b and c), suggesting a safety window for the usage of these drugs as endocytosis inhibitors. These newly identified compounds displayed significant structural novelty when compared to drugs in the training set. 6 This can be verified by the Tanimoto similarity analysis (please see Support Information for more details); Interestingly, among the 10 confirmed drugs, six of them share some common structural characteristics (NCGC00411611-01, NCGC00411727-01, NCGC00411705-01, NCGC00411733-01, NCGC00411718-01, NCGC00411588-01), suggesting a possible common mechanism for inhibiting HSPG-mediated endocytosis.
Figure 5:
Identification of inhibitors for HSPG-mediated endocytosis: (a). The workflow of -Synuclein fibrils uptake assay for confirmation of hits from virtual compound screen. (b). A summary of the activities of the top 10 compounds, IC50 was determined by titration experiments. (c). Dose-response curves of compound’s inhibitory effect on -Synuclein fibrils uptake. (d). Measured fluorescence intensity of internalized -Synuclein fibril-Alexa596 by U2OS cells treated with compound at 2-fold of its IC50. The experimental repeat number is 3.
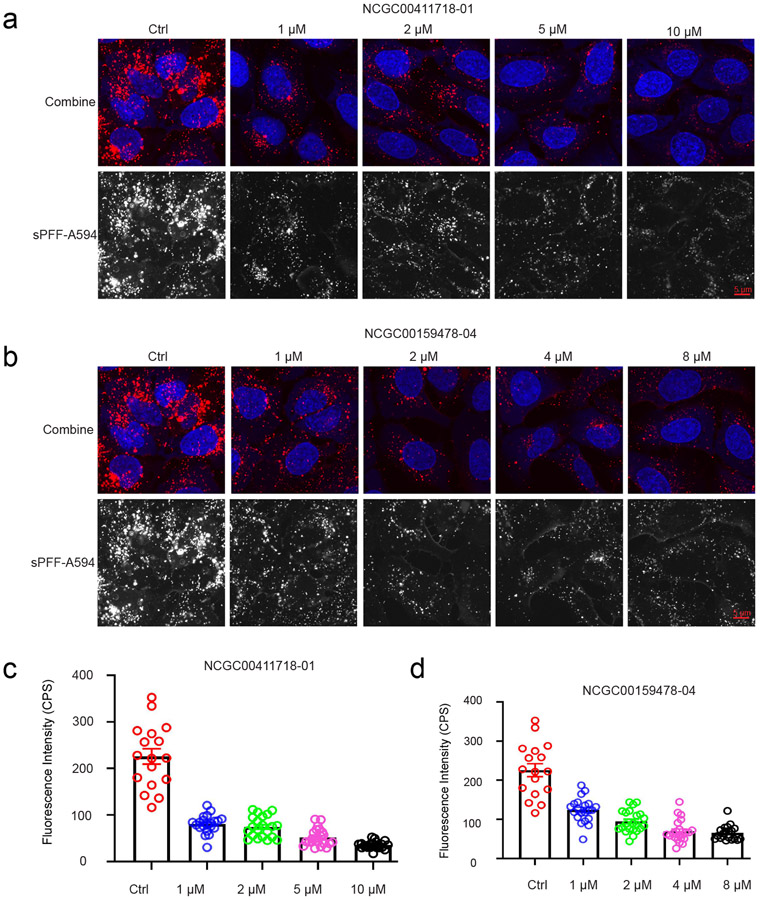
To rule out false-positive hits due to compound-induced changes in lysosomal pH, which could reduce the fluorescence of internalized -Synuclein fibrils, we measured the uptakes of -Synuclein fibrils labeled with a pH-insensitive dye (Alexa596) in U2OS cells. When cells were treated with the top 10 inhibitors at concentrations 2-fold higher than their respective IC50 values, we found that all compounds tested could significantly inhibit the uptake of -Synuclein fibrils compared to control treated cells (Figure 5d). These results suggest that these chemicals are indeed endocytosis inhibitors that block HSPG-mediated entry of -Synuclein fibrils. We then treated cells with increased concentrations of NCGC00411718 and NCGC00159478, which showed the highest inhibition on the entry of pHrodo-labeled -Synuclein fibrils. Drug-treated cells were incubated with Alexa596-labeled -Synuclein fibrils in the presence of the inhibitor for 2 hours and imaged by a confocal microscope. The results suggest that both compounds inhibit -Synuclein fibril uptake in a dose dependent manner with IC50 comparable to that measured by pHrodo-labeled -Synuclein fibrils (Figure 6a-d).
Figure 6:
Identification of new endocytosis inhibitors targeting HSPG-mediated endocytosis. (a). NCGC00411718-01 inhibits of -Synuclein fibril-Alexa594 uptake by U2OS cells in a dose dependent manner. (b). NCGC00159478-04 inhibits of -Synuclein hbril-Alexa594 uptake by U2OS cells in a dose dependent manner. (c and d). Quantification of internalized -Synuclein fibril-Alexa594 fluorescence intensity with compound treatment. Error bars indicate SEM. The experimental repeat number is 2.
Identification of SARS-CoV-2 entry inhibitors
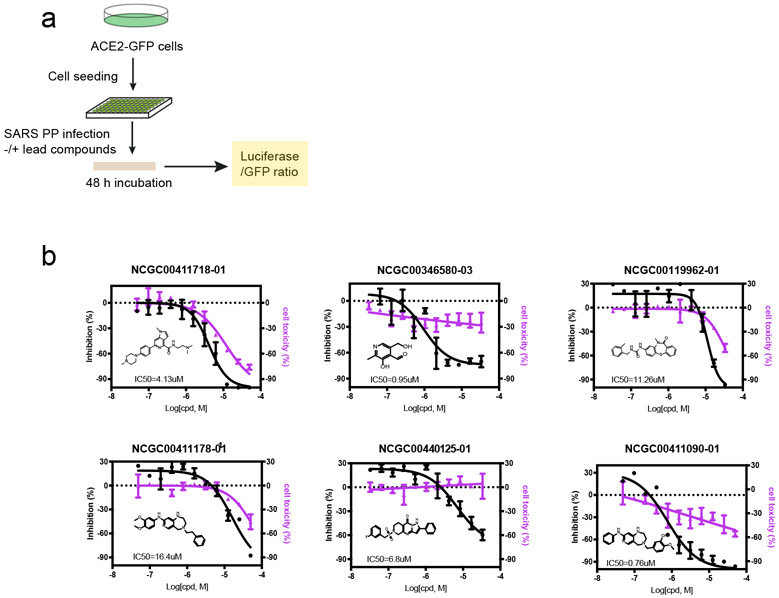
To test whether the newly identified endocytosis inhibitors could inhibit the entry of SARS-CoV-2, we used a previously established pseudotyped particle entry assay (Figure 7a). As shown previously,6 the entry of the pseudoviral particles into cells results in the expression of the luciferase reporter. To control the impact of ACE2-GFP expression levels on viral entry under drug-treated conditions, we normalized the luciferase signals by the ACE2-GFP level. We also measured the cytotoxicity of these chemicals in ACE2-GFP expressing cells using an ATP-based cell viability assay. We analyzed the top 27 compounds from the 256 inhibitors identified from the -Synuclein fibril uptake screen. Among them, 16 in total showed an inhibitory activity against the viral entry with the most potent IC50 value of 0.76 μM. It is notable that some toxicity was observed for these compounds in HEK293T-ACE2-GFP cells after 48 hr treatment. The viral inhibition and cytotoxicity curves of the top 6 compounds are shown in Figure 7b.
Figure 7:
Identification of SARS-CoV-2 entry inhibitors: (a). The experimental scheme for inhibitor testing in HEK293T-ACE2-GFP cells. (b). Dose-responsive titration of compound’s inhibitory effect on SARS-CoV-2 entry and cytotoxicity. The experimental repeat number is 3.
NCGC00115755 inhibited SARS-CoV-2 pseudotyped particle entry by disrupting actin filaments
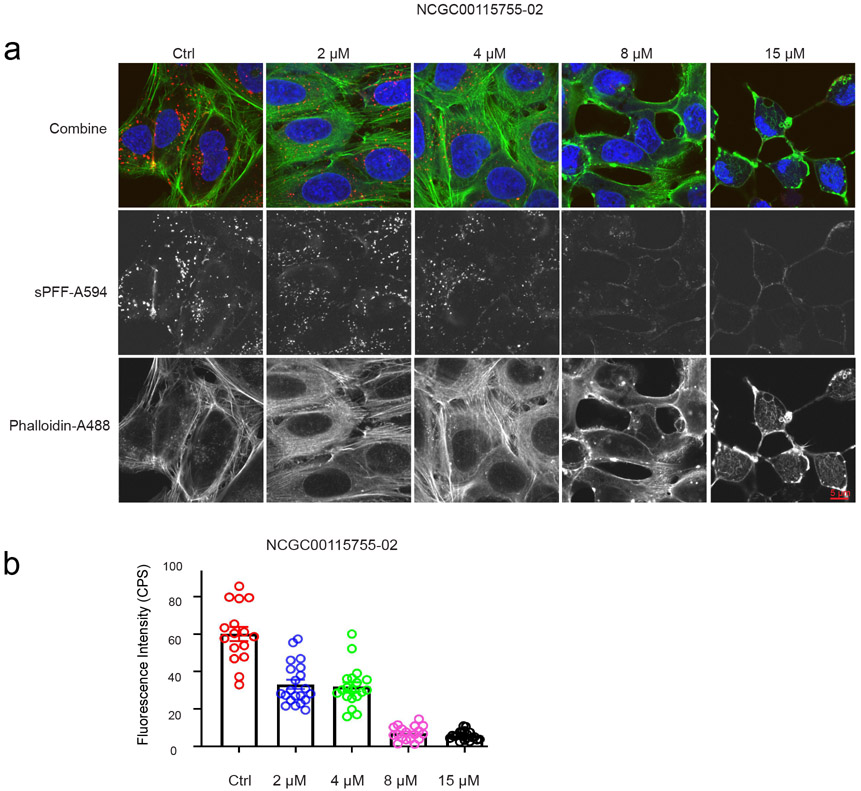
We previously showed that the actin network under the plasma membrane is critical for the entry of HSPG-dependent endocytosis cargos including SARS-CoV-2.6,15 We therefore asked whether any of the newly identified endocytosis could inhibit the actin cytoskeleton. To this end, we stained U2OS cells with Alexa488-labeled phalloidin, an actin binding dye. In control-treated cells, actin filaments were readily detected, which often run in parallel (Figure 8a). When cells treated with the top 10 endocytosis inhibitors were stained by Alexa488-labeled phalloidin, we observed dose-dependent disruption of cortex actin filaments only in NCGC00115755-02-treated cells by confocal fluorescence microscopy (Figure 8a) and it has anti-pseudotyped particle activity at IC50 of 5 M. Live cell imaging of cells expressing GFP-tagged Tractin, an actin binding reporter showed that untreated cells contain, in addition to stress fibers, many actin nucleation sites near the plasma membrane, which assemble comet tails (Supplementary videos). By contrast, in drug treated cells, the number of actin stress fibers were significantly reduced and actin comet tails were barely detectable (Supplementary videos). Altogether, these findings suggest that NCGC00115755-02 disrupts actin filament assembly, resulting in an endocytosis defect.
Figure 8:
NCGC00115755-02 targets cellular actin cytoskeleton: (a). Cells treated with NCGC00115755-02 at the indicated concentrations were incubated with Alexa594-labeled -Synuclein fibrils for 2 hours. Cells were stained with Phalloidin-Alexa488 in green to detect actin filaments and DAPI in blue to reveal the nuclei and then imaged. Note that cells treated with the drug has reduced level of internalized -Synuclein fibrils. NCGC00115755-02 treatment also causes the disassembly of actin stress fiber and generates large actin aggregates. (b). Quantification of Alexa594-labeled -Synuclein fluorescence intensity in a. Error bars indicate SEM. The experimental repeat number is 2.
Conclusion
Machine learning-based virtual screening technologies have the potential to efficiently select drug candidates for specific targets with high accuracy at an affordable cost, and therefore, is an important complementary to conventional high-throughput small molecule screening (HTS). SARS-CoV-2 viruses co-opt a cellular endocytosis pathway to enter human airway epithelial cells. This key viral entry step has been subjected to conventional drug repurposing screens and computer docking-based screens, yielding several viral entry inhibitors.6,14 In this study, we developed and trained a GCN model using the structural information from previously identified SARS-CoV-2 entry inhibitors. When this model was applied to untested chemical libraries, it can efficiently select compounds with high probability of showing an anti-SARS-CoV-2 activity. This model, when combined with conventional drug screening assays, generates a powerful strategy that allows rapid identification of new SARS-CoV-2 entry inhibitors. In principle, this strategy can be applied to any drug targets, which can quickly expand the existing inhibitor repertoire of any class. The findings shown in this study have revealed a promising venue for accelerated drug development.
Supplementary Material
Acknowledgement
The work was supported by the intramural research program of the National Institute of Diabetes, Digestive & Kidney Diseases (Y.Y.) and by the National Center for Advancing Translational Sciences (W.Z.) in the National Institutes of Health.
Footnotes
Supporting Information Available
Detailed similarity analysis can be found in Support Information for reference.
Data and software availability
Technical details of the developed package can be found on our GitHub page: github.com/tcsnfrank0177/Graph-convolutional-network-DrugScreening.git. Programming environment: Python 3.6 or higher is recommended. Supplementary videos are provided as attachment.
References
- (1).Kim D; Lee J; Yang J; Kim JW; Kim VN; Chang H The Architecture of SARS-CoV-2 Transcriptome. Cell 2020, 181, 914–921.e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Amanat F; Krammer F SARS-CoV-2 Vaccines: Status Report. Immunity 2020, 52, 583–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Krammer F SARS-CoV-2 vaccines in development. Nature 2020, 586, 516–527. [DOI] [PubMed] [Google Scholar]
- (4).Wu D; Wu T; Liu Q; Yang Z The SARS-CoV-2 outbreak: What we know. International Journal of Infectious Diseases 2020, 94, 44–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Clausen TM; Sandoval DR; Spliid CB; Pihl J; Perrett HR; Painter CD; Narayanan A; Majowicz SA; Kwong EM; McVicar RN; Thacker BE; Glass CA; Yang Z; Torres JL; Golden GJ; Bartels PL; Porell RN; Garretson AF; Laubach L; Feldman J; Yin X; Pu Y; Hauser BM; Caradonna TM; Kellman BP; Martino C; Gordts PLSM; and Chanda SK; Schmidt AG; Godula K; Leibel SL; Jose J; Corbett KD; Ward AB; Carlin AF; Esko JD SARS-CoV-2 Infection Depends on Cellular Heparan Sulfate and ACE2. Cell 2020, 183, 1043–1057.e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Zhang Q; Chen CZ; Swaroop M; Xu M; Wang L; Lee J; Wang AQ; Pradhan M; Hagen N; Chen L; Shen M; Luo Z; Xu X; Xu Y; Huang W; Zheng W; Ye Y Heparan sulfate assists SARS-CoV-2 in cell entry and can be targeted by approved drugs in vitro. Cell Discovery 2020, 6, 80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Haniff HS; Tong Y; Liu X; Chen JL; Suresh BM; Andrews RJ; Peterson JM; O’Leary CA; Benhamou RI; Moss WN; Disney MD Targeting the SARS-CoV-2 RNA Genome with Small Molecule Binders and Ribonuclease Targeting Chimera (RIBOTAC) Degraders. ACS Central Science 2020, 6, 1713–1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Huang Y; Yang C; Xu X; Xu W; Liu S Structural and functional properties of SARS-CoV-2 spike protein: potential antivirus drug development for COVID-19. Acta Pharmacologica Sinica 2020, 41, 1141–1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Tortorici MA; Veesler D In Complementary Strategies to Understand Virus Structure and Function; Rey FA, Ed.; Advances in Virus Research; Academic Press, 2019; Vol. 105; pp 93–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Burkard C; Verheije MH; Wicht O; van Kasteren SI; van Kuppeveld FJ; Haagmans BL; Pelkmans L; Rottier PJM; Bosch BJ; de Haan CAM Coronavirus Cell Entry Occurs through the Endo-/Lysosomal Pathway in a Proteolysis-Dependent Manner. PLOS Pathogens 2014, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Belouzard S; Millet JK; Licitra BN; Whittaker GR Mechanisms of Coronavirus Cell Entry Mediated by the Viral Spike Protein. Viruses 2012, 4, 1011–1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Inoue Y; Tanaka N; Tanaka Y; Inoue S; Morita K; Zhuang M; Hattori T; Sugamura K Clathrin-Dependent Entry of Severe Acute Respiratory Syndrome Coronavirus into Target Cells Expressing ACE2 with the Cytoplasmic Tail Deleted. Journal of Virology 2007, 81, 8722–8729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Hoffmann M; Kleine-Weber H; Schroeder Simon andKrüger N; Herrler T; Erichsen S; Schiergens TS; Herrler G; Wu N-H; Nitsche A; Müller MA; Drosten C; Pöhlmann S SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell 2020, 181, 271–280.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Kolarič A; Jukič M; Bren U Novel Small-Molecule Inhibitors of the SARS-CoV-2 Spike Protein Binding to Neuropilin 1. Pharmaceuticals 2022, 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Zhang Q; Xu Y; Lee J; Jarnik M; Wu X; Bonifacino JS; Shen J; Ye Y A myosin-7B–dependent endocytosis pathway mediates cellular entry of -synuclein fibrils and polycation-bearing cargos. Proceedings of the National Academy of Sciences 2020, 117, 10865–10875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Sarrazin S; Lamanna WC; Esko JD Heparan Sulfate Proteoglycans. Cold Spring Harbor Perspectives in Biology 2011, 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Wu C; MacLeod I; Su AI BioGPS and MyGene.info: organizing online, genecentric information. Nucleic Acids Research 2012, 41, D561–D565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).St. John PC; Guan Y; Kim Y; Kim S; Paton RS Prediction of organic homolytic bond dissociation enthalpies at near chemical accuracy with sub-second computational cost. Nat Commun 2020, 11, 2328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Kwon Y; Lee D; Choi Y; Kang M; Kang S Neural Message Passing for NMR Chemical Shift Prediction. Journal of Chemical Information and Modeling 2020, 60, 2024–2030. [DOI] [PubMed] [Google Scholar]
- (20).Gerrard W; Bratholm LA; Packer MJ; Mulholland AJ; Glowacki DR; Butts CP IMPRESSION – prediction of NMR parameters for 3-dimensional chemical structures using machine learning with near quantum chemical accuracy. Chem. Sci 2020, 11, 508–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Scarselli F; Gori M; Tsoi AC; Hagenbuchner M; Monfardini G The Graph Neural Network Model. IEEE Transactions on Neural Networks 2009, 20, 61–80. [DOI] [PubMed] [Google Scholar]
- (22).Sørensen KH; Jørgensen MS; Bruix A; Hammer B Accelerating atomic structure search with cluster regularization. The Journal of Chemical Physics 2018, 148, 241734. [DOI] [PubMed] [Google Scholar]
- (23).Wexler RB; Martirez JMP; Rappe AM Chemical Pressure-Driven Enhancement of the Hydrogen Evolving Activity of Ni2P from Nonmetal Surface Doping Interpreted via Machine Learning. Journal of the American Chemical Society 2018, 140, 4678–4683. [DOI] [PubMed] [Google Scholar]
- (24).Mansouri Tehrani A; Oliynyk AO; Parry M; Rizvi Z; Couper S; Lin F; Miyagi L; Sparks TD; Brgoch J Machine Learning Directed Search for Ultrain-compressible, Superhard Materials. Journal of the American Chemical Society 2018, 140, 9844–9853. [DOI] [PubMed] [Google Scholar]
- (25).Panapitiya G; Avendaño-Franco G; Ren P; Wen X; Li Y; Lewis JP Machine-Learning Prediction of CO Adsorption in Thiolated, Ag-Alloyed Au Nanoclusters. Journal of the American Chemical Society 2018, 140, 17508–17514. [DOI] [PubMed] [Google Scholar]
- (26).Rupp M; Ramakrishnan R; von Lilienfeld OA Machine Learning for Quantum Mechanical Properties of Atoms in Molecules. The Journal of Physical Chemistry Letters 2015, 6, 3309–3313. [Google Scholar]
- (27).Bai Y; Wilbraham L; Slater BJ; Zwijnenburg MA; Sprick RS; Cooper AI Accelerated Discovery of Organic Polymer Photocatalysts for Hydrogen Evolution from Water through the Integration of Experiment and Theory. Journal of the American Chemical Society 2019, 141, 9063–9071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Mater AC; Coote ML Deep Learning in Chemistry. Journal of Chemical Information and Modeling 2019, 59, 2545–2559. [DOI] [PubMed] [Google Scholar]
- (29).Faber FA; Hutchison L; Huang B; Gilmer J; Schoenholz SS; Dahl GE; Vinyals O; Kearnes S; Riley PF; von Lilienfeld OA Prediction Errors of Molecular Machine Learning Models Lower than Hybrid DFT Error. Journal of Chemical Theory and Computation 2017, 13, 5255–5264. [DOI] [PubMed] [Google Scholar]
- (30).Gao P; Zhang J; Sun Y; Yu J Accurate predictions of aqueous solubility of drug molecules via the multilevel graph convolutional network (MGCN) and SchNet architectures. Phys. Chem. Chem. Phys 2020, 22, 23766–23772. [DOI] [PubMed] [Google Scholar]
- (31).Behler J Perspective: Machine learning potentials for atomistic simulations. The Journal of Chemical Physics 2016, 145, 170901. [DOI] [PubMed] [Google Scholar]
- (32).Behler J First Principles Neural Network Potentials for Reactive Simulations of Large Molecular and Condensed Systems. Angewandte Chemie International Edition 2017, 56, 12828–12840. [DOI] [PubMed] [Google Scholar]
- (33).Gao P; Zhang J; Sun Y; Yu J Toward Accurate Predictions of Atomic Properties via Quantum Mechanics Descriptors Augmented Graph Convolutional Neural Network: Application of This Novel Approach in NMR Chemical Shifts Predictions. The Journal of Physical Chemistry Letters 2020, 11, 9812–9818. [DOI] [PubMed] [Google Scholar]
- (34).Wang J; Olsson S; Wehmeyer C; Pérez A; Charron NE; de Fabritiis G; Noé F; Clementi C Machine Learning of Coarse-Grained Molecular Dynamics Force Fields. ACS Central Science 2019, 5, 755–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Botu V; Batra R; Chapman J; Ramprasad R Machine Learning Force Fields: Construction, Validation, and Outlook. The Journal of Physical Chemistry C 2017, 121, 511–522. [Google Scholar]
- (36).Meldgaard SA; Kolsbjerg EL; Hammer B Machine learning enhanced global optimization by clustering local environments to enable bundled atomic energies. The Journal of Chemical Physics 2018, 149, 134104. [DOI] [PubMed] [Google Scholar]
- (37).Ouyang R; Xie Y; Jiang D.-e. Global minimization of gold clusters by combining neural network potentials and the basin-hopping method. Nanoscale 2015, 7, 14817–14821. [DOI] [PubMed] [Google Scholar]
- (38).Lu C; Liu Q; Wang C; Huang Z; Lin P; He L Molecular Property Prediction: A Multilevel Quantum Interactions Modeling Perspective. arXiv 2019, 1906.11081. [Google Scholar]
- (39).Gao P; Zhang J; Peng Q; Zhang J; Glezakou V-A General Protocol for the Accurate Prediction of Molecular 13C/1H NMR Chemical Shifts via Machine Learning Augmented DFT. Journal of Chemical Information and Modeling 2020, 60, 3746–3754. [DOI] [PubMed] [Google Scholar]
- (40).Gao P; Zhang J; Qiu H; Zhao S A general QSPR protocol for the prediction of atomic/inter-atomic properties: a fragment based graph convolutional neural network (F-GCN). Phys. Chem. Chem. Phys 2021, 23, 13242–13249. [DOI] [PubMed] [Google Scholar]
- (41).Schütt KT; Sauceda HE; Kindermans P-J; Tkatchenko A; Müller K-R SchNet - A deep learning architecture for molecules and materials. The Journal of Chemical Physics 2018, 148, 241722. [DOI] [PubMed] [Google Scholar]
- (42).Wang M; Zheng D; Ye Z; Gan Q; Li M; Song X; Zhou J; Ma C; Yu L; Gai Y; Xiao T; He T; Karypis G; Li J; Zhang Z Deep Graph Library: A Graph-Centric, Highly-Performant Package for Graph Neural Networks. 2019. [Google Scholar]
- (43).Chen G; Chen P; Hsieh C-Y; Lee C-K; Liao B; Liao R; Liu W; Qiu J; Sun Q; Tang J; Zemel R; Zhang S Alchemy: A Quantum Chemistry Dataset for Benchmarking AI Models. arXiv preprint arXiv:1906.09427 2019, [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.