Dear Editor,
Pepper (C. annuum L.) is an important vegetable crop worldwide [1], with remarkable diversity in morphology, nutrition, color, flavor, and yield. A great effort has been made to identify quantitative trait loci (QTLs) and genes that affect these traits [2, 3]. However, few effective combinatorial events in small population sizes have limited the mapping of high-confidence QTLs for marker-assisted breeding (MAB) and map-based gene cloning. The combined multi-omics approach has provided powerful tools for the rapid mining of candidate genes for MAB and for improving our understanding of candidate genes and their molecular regulation [4, 5].
To construct a pepper recombinant inbred line (RIL) population, we generated an F1 generation by crossing a late-maturing solitary pod pepper (16 L816) with a late-maturing sweet pepper (16 L15) and then selfed the F1 generation for an additional six rounds (Fig. S1). We cultivated the resultant 148 RIL plants and collected data on four important fruit-related traits: fruit color (FC), fruit width (FWD), fruit length (FL), and fruit weight (FWT). These four traits varied considerably among independent lines. Correlation analysis indicated that FWD had a significant positive correlation with FWT and a significant negative correlation with FL (Table S1).
By performing RNA sequencing, RNA-seq analysis, and SNP marker development on fruits from all 148 RIL progenies, we obtained high-quality bin markers consisting of 78,605 SNPs and divided them into 12 linkage groups (LGs) corresponding to 12 chromosomes (Fig. 1a). These SNPs were used to construct a high-density genetic linkage map for pepper. The total genetic distance of the map was 1737 cM, with an average distance of 0.73 cM between adjacent SNPs (Table S2). By comparing the genetic map with physical locations in the reference genome, we found that most segments had high collinearity, although a small number of abnormalities may have been caused by chromosome assembly errors (Fig. 1b). We used the R package qtl [6] to perform interval mapping with the expectation–maximization algorithm (EM) and to calculate the logarithm of odds (LOD). Genome-wide QTL analysis was performed using the high-density linkage map and the phenotypic data on four traits obtained from the RIL population. LOD scores above 3.3 were used to define effective QTLs, and we identified twelve significant QTLs for the four pepper traits (Fig. 1c, Table S3). One QTL for FC (qFC6.1) was located on LG 6 and contained 51 candidate genes, including capsanthin-capsorubin synthase (CCS) (Capana06g000615) [7]. There were two significant QTLs for FL (qFL3.1 on LG 3 and qFL7.1 on LG 7), four QTLs for FWD on LGs 2, 3, 6, and 11, and five QTLs for FWT (qFWT2.1 on LG 2, qFWT4.1 on LG 4, qFWT6.1 on LG 6, qFWT7.1 on LG 7, and qFWT7.2 on LG 7).
Figure 1.
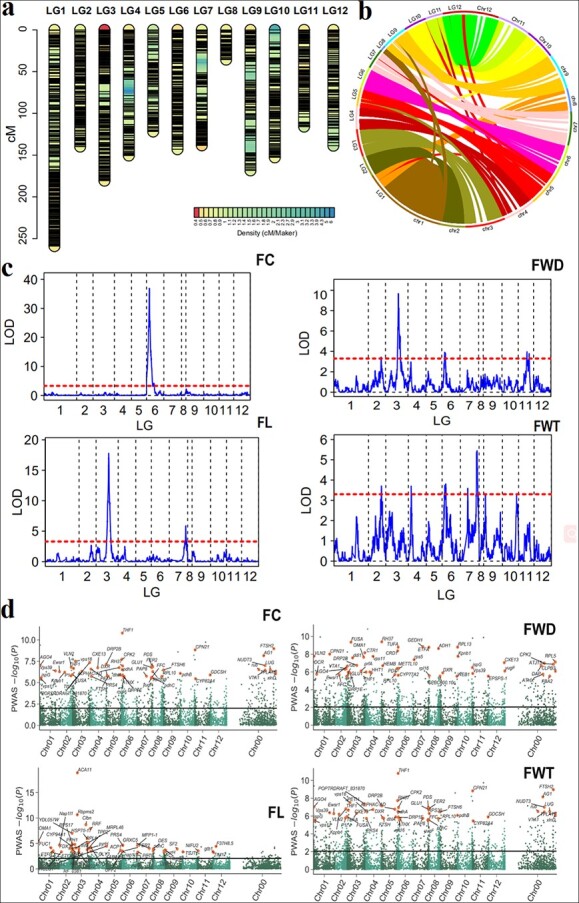
Transcriptome- and proteome-wide association of the recombinant inbred line population. (a) High-density genetic map of pepper. (b) Collinearity analysis of the genetic map with the physical positions of the reference genome. (c) Genome-wide scanning of QTLs for pepper fruit traits. The red lines indicate the significance threshold, with a LOD score of 3.3. Inner ticks on the x-axis depict the locations of observed markers. (c) Manhattan plot of TWAS for fruit traits. The black line represents the significance threshold calculated by 1000 permutations. FC, fruit color; FWD, fruit width; FL, fruit length; FWT, fruit weight.
To identify the genes that regulate these traits, we carried out transcriptome-wide association studies (TWAS) on all RILs to detect correlations between gene expression and phenotype using the limix module [8]. Among the 26 632 genes that passed quality control, 421,021, 1494, and 1594 significantly expressed genes (FDR < 0.05) were detected for FC, FWD, FL, and FWT, respectively (Fig.S2). By comparing the physical locations of all genes on the LG with the eQTL interval, we defined genes whose start interval intersected with the eQTL interval as cis-eQTLs and others as trans-eQTLs [9]. The expression levels of 2700 of the 6158 genes were regulated by cis-eQTLs, and those of 4082 genes were regulated by trans-eQTLs. A total of 574 genes had multiple trans-eQTLs, resulting in the detection of 4710 trans eQTLs, accounting for 63.6% of all eQTLs. Nine trans-eQTL hotspots were detected on the 12 LGs using the qtlhot package (https://CRAN.R-project.org/package=qtlhot) (LOD 3.43 and significance level of 0.01–0.1). They were located on LGs 1, 2, 3, 4, 6, 7, 9, 11, and 12, with LG6–1 containing a total of 645 genes.
We performed label-free proteome sequencing of all fruits in the RIL population and detected 7923 peptides, 7047 of which could be quantified. Based on proteome-wide association analysis (PWAS) of these peptides, 864 proteins were significantly associated with FC, and 855, 304, and 811 proteins were significantly associated with FWD, FL, and FWT, respectively (Fig. 1d). A similar pQTL analysis of the proteomic data revealed that 26.7% of the proteins were significantly associated with cis pQTL sites and 73.2% of those were significantly associated with trans-acting sites. Seventeen protein hotspots coincided with eQTLs. The hotspot locus LG6–1 contained 455 proteins, suggesting that the LG6–1 region has complex regulatory relationships at both transcriptional and translational levels during pepper fruit development.
To investigate the regulatory mechanisms of pepper fruit traits using multi-omics data, we performed a joint multiple histology analysis. We first identified the overlap of genes significantly associated with each trait in TWAS and PWAS and found that 37 genes were significantly associated with all four traits in the PWAS analysis (Table S4, Fig.S3), suggesting that some common regulatory pathways control these traits. We also compared the overlap of significant genes between TWAS and PWAS for each trait, and 2, 21, 15, and 42 overlapping genes were identified for FC, FWD, FL, and FWT, respectively (Table S5, Fig.S3). However, few genes overlapped between TWAS and PWAS for more than one trait.
In addition to PWAS and TWAS, we also used metabolic pathway analysis to successfully localize the pepper fruit color gene CCS. Combined with the results of PWAS and TWAS, this approach enabled us to identify the upstream carotenoid biosynthesis genes 1-Deoxy-D-xylulose 5-phosphate reductoisomerase (DXR) and phytoene desaturase (PDS) in the RIL population; these genes may also play an important role in the regulation of pepper fruit color.
In the present study, 12 significant QTL loci for pepper FC, FWD, FL, and FWT, and 80 significant trait-associated genes were identified through QTL mapping in an RIL population and association analysis of traits using multiple transcriptional and translational data. We used a relatively small population of RIL lines to effectively associate QTLs and important genes by TWAS and PWAS analysis. Integrated omics analyses can help us to understand the regulatory mechanism of pepper fruit development and to accelerate the mining and cloning of candidate genes. The QTLs detected in this study will be useful for selecting appropriate RILs for further marker-assisted selection when breeding hybrid peppers of diverse market types.
Supplementary Material
Acknowledgments
We thank the Jingjie PTM BioLab (HangZhou) Co., Ltd for sequencing service. This research was funded by the National Natural Science Foundation of China (U19A2028) and the Key R&D Projects of Hunan Province (2020NK2058).
Author contributions
Conceptualization, X.Z. and L.O.; methodology, B.Y. and H.S.; formal analysis, Z.L. and B.Y.; investigation, Z.Z. and W.C.; writing-original draft preparation, Z.L. and R.H.; writing-review and editing, Z.L. and X.D.; visualization, Z.L. and Y.H.; funding acquisition, X.Z. and L.O.
Data availability
The raw data required to reproduce these findings cannot be shared at this time as they also form part of an ongoing study.
Conflict of interest statement
The authors declare no competing interests.
Supplementary data
Supplementary data is available at Horticulture Research online.
Reference
- 1. Liu F, Yu H, Deng Yet al. PepperHub, an informatics hub for the chili pepper research community. Mol Plant. 2017;10:1129–32. [DOI] [PubMed] [Google Scholar]
- 2. Han K, Lee HY, Ro NYet al. QTL mapping and GWAS reveal candidate genes controlling capsaicinoid content in capsicum. Plant Biotechnol J. 2018;16:1546–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Li X, Zheng H, Wu Wet al. QTL mapping and candidate gene analysis for alkali tolerance in Japonica Rice at the bud stage based on linkage mapping and genome-wide association study. Rice. 2020;13:48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Zhang L, Yu Y, Shi Tet al. Genome-wide analysis of expression quantitative trait loci (eQTLs) reveals the regulatory architecture of gene expression variation in the storage roots of sweet potato. Hortic Res. 2020;7:90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ichihashi Y, Date Y, Shino Aet al. Multi-omics analysis on an agroecosystem reveals the significant role of organic nitrogen to increase agricultural crop yield. Proc Natl Acad Sci U S A. 2020;117:14552–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Broman KW, Wu H, Sen Set al. R/QTL: QTL mapping in experimental crosses. Bioinformatics. 2003;19:889–90. [DOI] [PubMed] [Google Scholar]
- 7. Ha S, Kim JB, Park JSet al. A comparison of the carotenoid accumulation in capsicum varieties that show different ripening colours: deletion of the capsanthin-capsorubin synthase gene is not a prerequisite for the formation of a yellow pepper. J Exp Bot. 2007;58:3135–44. [DOI] [PubMed] [Google Scholar]
- 8. Casale FP, Rakitsch B, Lippert Cet al. Efficient set tests for the genetic analysis of correlated traits. Nat Methods. 2015;12:755–8. [DOI] [PubMed] [Google Scholar]
- 9. Brynedal B, Choi JM, Raj Tet al. Large-scale trans-eQTLs affect hundreds of transcripts and mediate patterns of transcriptional co-regulation. Am J Hum Genet. 2017;100:581–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw data required to reproduce these findings cannot be shared at this time as they also form part of an ongoing study.


