Abstract
In the study, ultraperformance liquid chromatography–quadrupole time-of-flight–mass spectrometry analysis of Leucosidea sericea leaf and stem extracts led to the identification of various classes of compounds. Further chromatographic purifications resulted in the isolation of 22 compounds that consisted of a new triterpenoid named leucosidic acid A (1), an acetophenone derivative 2, a phloroglucinol derivative 3, three chromones 4–6, seven pentacyclic triterpenoids 7–13, a phytosterol glucoside 14, a flavonoid 15, and seven flavonoid glycosides 16–22. Nineteen of these compounds including the previously undescribed triterpenoid 1 are isolated for the first time from L. sericea. The structures of the isolated compounds were assigned based on their high-resolution mass spectrometry and nuclear magnetic resonance data. Some of the isolated triterpenoids were evaluated for inhibitory activity against α-amylase, α-glucosidase, and pancreatic lipase. Of the tested compounds, 1-hydroxy-2-oxopomolic acid (7) and pomolic acid (13) showed higher potency on α-glucosidase than acarbose, which is used as a positive control in this study. The two compounds inhibited α-glucosidase with IC50 values of 192.1 ± 13.81 and 85.5 ± 6.87 μM, respectively.
1. Introduction
The Rosaceae plant family is a rose, medium-sized family of flowering plants, which encompasses greater than a hundred genera and more than 3000 recognized species.1,2 The plant family of Rosaceae is traditionally divided into four subfamilies depending on the type of fruits, which include Rosoideae, Prunoidae, Piraeoideae, and Maloideae.1,3 Various species belonging to this plant family are of monetary importance as food crops that encompass apples, almonds, cherries, pears, raspberries, and strawberries.1,4 Bioactive compounds that offer crucial health benefits have been identified from the Rosaceae plants.4,5 Owing to the health benefits associated with secondary metabolites from the Rosaceae family, most of the compounds and their enriched extracts find applications in the food industry, where they are used in the development of novel functional foods and nutraceuticals.5 As such, investigation of phytochemical composition of the Rosaceae species, especially those that are underutilized, can help to unveil the bioactive compounds that can confer favorable health benefits and unlock the potential applications and economic importance of these plants.5
Leucosidea sericea Eckl. & Zeyh is the only specie in the genus Leucosidea belonging to Rosaceae family and Rosoideae subfamily.6L. sericea is normally referred to as antique wood (English), cheche (Sesotho), munyonga (Tshivenda), umtshitshi (IsiZulu), ouhout (Afrikaans), and umtyityi (IsiXhosa).7 It usually grows in the highlands of South Africa as well as in Lesotho, and in Swaziland and Zimbabwe to a lesser degree.7 Different parts of the L. sericea plant are used for different ethnobotanical purposes, which include expulsion of parasitic worms of the intestines and treatment of asthma and aspiration mucus (phlegm).8 The ground paste of the leaves is used to ease severe inflammation of the eyeball.7,9 The leaves and stems have been used for the treatment of high blood pressure, cough, herpes, and HIV.9 The flowers and shoots are grazed by animals.10
As a result of the important pharmacological properties and uses of L. sericea, several scientific studies investigated the phytochemical composition and biological activities of the extracts of this plant. Earlier studies focused on the evaluation of the antimicrobial and antiparasitic activities of the plant.10,11 This led to the identification and isolation of the phloroglucinol derivatives as the potential active constituents responsible for the anthelmintic activity.10,11 Other phytochemicals such as phytosterols, fatty acids, chromones, triterpenoids, flavonoids, and essential oils have been reported from the plant.7,12 These constituents possess a wide range of biological activities including anti-inflammatory, antibacterial, antioxidant, and cytotoxic activities.7,12
The studies conducted on L. sericea evidence the great interest among the scientific communities on this plant. These studies are motivated by the untapped economic and commercial potential of L. sericea.7,13,14 Therefore, our aim was to study in detail the phytochemical composition of the leaf and stem extracts of L. Sericea. It is hoped that the knowledge of bioactive constituents of L. sericea will help to provide direction on further applications of the plant and aid to unlock its economic potential.
2. Results and Discussion
2.1. UPLC–MS Chemical Profiling
The ethyl acetate (EtOAc)-soluble extract was analyzed by ultraperformance liquid chromatography–quadrupole time-of-flight–mass spectrometry (UPLC–QTof–MS) for the identification of the compounds present. The analysis was performed in positive and negative electrospray ionization (ESI) resolution modes using a Waters MSE technology. The MSE method simultaneously records the MS data in low collision energy (function 1) and high collision energy (function 2) for the determination of molecular ions and fragment ions, respectively.15,16 Several peaks were observed from the EtOAc chromatogram (Supporting Information, Figure S1), indicative of the presence of various compounds with different polarities. The compounds were tentatively identified (Table S1) and classified as flavonoids, chromones, phloroglucinol derivatives, and triterpenoids.
2.1.1. Flavonoids and Chromones
Two isomeric compounds at rt 2.90 and 3.75 min (entries 2 and 5) showed the precursor ion at m/z 289.0705 [M – H]− in the negative ionization mode and 291.0871 [M + H]+ in the positive ionization mode, resulting in a chemical formula of C15H14O6. The fragment ions were observed at m/z 245, 203, 151, 137, and 125.17,18 Based on the parent and fragment ions observed, the compounds were tentatively identified as catechin and epicatechin.17,18 The compounds eluting at rt 2.59, 3.02, and 3.36 min (entries 1, 3 and 4) were identified as possible isomers of B-type procyanidin. They showed a precursor ion at m/z 577.1347 [M – H]− in the negative ionization mode and 579.1520 [M + H]+ in the positive ionization mode, giving a chemical formula of C30H26O12 for the compounds. The MSE spectra exhibited fragments at m/z 425 [M – H-152]−, 407 [M – H-152-18]−, and 289 [M – H-288]− indicative of retro Diels–Alder fragmentation, subsequent loss of an H2O molecule, and cleavage of epicatechin/catechin C–C linkage.19
A peak that eluted at rt 5.01 min (entry 6) in the positive ionization mode was identified as 5,7-dihydroxychromone with a chemical formula of C9H6O4 at m/z 179.0353 [M + H]+. The MSE indicated the presence of fragmentation peaks at m/z 163, 139, and 123. This compound was previously isolated and characterized by Pendota and colleagues from the leaves of L. sericea.12,20
A peak at rt 5.90 min (entry 7) showed a molecular ion of m/z 447.0927 [M – H]− in the negative ionization mode and 449.1100 [M + H]+ in the positive ionization mode. The chemical formula was determined to be C21H20O11. The main fragment appeared at m/z 284 [M – H-163]−, indicating the loss of a pyranose unit, and other fragments were observed at m/z 255, 145, 117, and 133.19,21 This was tentatively identified as kaempferol-3-O-glucoside.19,21 In L. sericea, the compounds with a similar precursor ion were previously identified as kaempferol galactopyranoside14 and kaempferol-7-O-β-D-glucopyranoside.20 A compound at rt 5.99 min (entry 8) with a precursor ion at m/z 463.0885 [M – H]− in the negative ionization mode and 465.1043 [M + H]+ in the positive ionization mode was identified as quercetin-3-O-glucoside.22 The chemical formula was established as C21H20O12. The MSE spectrum showed the presence of the main fragment at m/z 300 [M – H-163]− for quercetin aglycone, indicating the loss of a pyranose moiety. Other fragments were observed at m/z 271, 255, and 243.22 The compounds eluting at rt 6.04 and 6.08 min (entries 9 and 10) were identified as isomers of quercetin-3-O-rutinoside. The molecular ion peak was observed at m/z 609.1464 [M – H]− in the negative ionization mode and m/z 611.1646 [M + H]+ in the positive ionization mode, resulting in the chemical formula of C27H30O16. The MSE spectra showed the presence of the main fragments 463 [M – H-146]− and 301 [M – H-146-162]−, indicating the loss of two sugar units consisting of a rhamnose and a pyranose. Other fragments appeared at m/z 271, 255, 243, and 151.17 The compounds detected at rt 6.34, 6.43, and 6.50 min (entries 11–13) with a molecular ion peak at m/z 433.0782 [M – H]− in the negative ionization mode and 435.0939 [M – H]+ in the positive ionization mode and the chemical formula of C20H18O11 were identified as quercetin-3-O-pentoside isomers.17,23 The presence of the main fragment at m/z 300 [M – H-133]− indicated the loss of pentose unit. Other fragments appeared at m/z 271, 255, 243, and 151.17,23 The peaks eluting at rt 6.65 and 6.69 min (entries 14–15) were identified as kaempferol-3-O-rutinoside isomers with the chemical formula C27H30O15 at m/z 593.1504 [M – H]− in the negative ionization mode and m/z 595.1670 [M + H]+ in the positive ionization mode.16,20 The fragments at m/z 447 and 285 indicate the loss of rhamnose and glucose sugar units, and other fragments at m/z 255, 227, and 183 were observed from the spectra.16
A compound at rt 7.51 min (entry 16) in the positive ionization mode was identified as kaempferol-3-O-acetylglucoside with the chemical formula C23H22O12 at m/z 491.1203 [M + H]+ in the positive ionization mode and 489.1033 [M – H]− in the negative ionization mode. The high-energy MS spectrum showed the presence of m/z 287 and 147, indicating the loss of acetyl glucoside fragment.15,24 The compounds eluting at rt 8.12 and 8.25 min (entries 17 and 18) were identified as tiliroside isomers with the chemical formula C30H26O13 at m/z 593.1288 [M – H]− in the negative ionization mode and 595.1437 [M + H]+ in the positive ionization mode. The fragments m/z 285 [M – H-308]− indicate the loss of coumaric acid and the glucopyranoside moiety, and other fragments at m/z 255, 227, 161, and 125 were observed.12,20 Pendota and colleagues isolated this compound from the leaves of L. sericea, and the nuclear magnetic resonance (NMR) data indicated that this compound is a trans isomer.12 Therefore, the second isomer could be the cis isomer. A compound that eluted at rt 8.66 min (entry 19) was identified as kaempferol-3-O-[6″-O-acetyl-2″-O-p-coumaroyl]-glucoside with a chemical formula of C32H28O14.25 It gave an m/z 635.1372 [M – H]− in the negative ionization mode and 637.1570 [M + H]+ in the positive ionization mode. The fragmentation pattern indicated the presence of peaks at m/z 285 [M – H-350]−, indicating the loss of coumaroyl quinic acid, glucopyranoside sugar, and an acetyl group and other peaks at m/z 255, 227, and 145.25
2.1.2. Phloroglucinol Derivatives
The compounds at rt 9.54 and 9.56 min (entries 20 and 21) were tentatively identified as pilosanol B and epipilosanol B, while those at rt 9.75 and 9.77 min (entries 22 and 23) were tentatively identified as pilosanol C and epipilosanol C.26 The four isomeric compounds exhibited the molecular ions at m/z 525.1771 [M – H]− in the negative ionization mode and 527.1937 [M + H]+ in the positive ionization mode, which rendered the chemical formula of C28H30O10.26 The MSE spectra in the negative ionization mode indicated the presence of the main fragment at m/z 289 [M – H-236]−, indicating the cleavage of epicatechin/catechin C–C linkage in both epipilosanol B and C. Other fragments were observed at m/z 223 and 137.26 A compound that eluted at rt for 10.35 min (entry 24) was identified as 3-dimethylallyl-4-hydroxymandelic acid.27 It gave the chemical formula of C13H16O4, deduced from the sodium adduct at m/z 259.0965 [M + Na]+ and protonated molecular ion at m/z 237.1135 [M + H]+ in the positive ionization mode. The presence of a fragment at m/z 219.1031 indicated the loss of an H2O molecule. Other fragments were observed at m/z 191, 167, 137, 123, and 121 from the negative ionization mode spectrum.27 The compound that eluted at rt 11.54 (entry 26) was putatively identified as robustaol A with a chemical formula of C25H30O9 at m/z 475.1988 [M + H]+ and 473.1818 [M – H]− in the positive and negative ionization modes, respectively.28 The high-energy MS resulted in the fragmentation peaks at m/z 405, 345, 237, and 209 in the positive ionization mode. The fragments at m/z 235, 223, 195, 193, and 177 were observed in the negative ionization mode.28 A peak at a rt of 12.66 min (entry 28) was identified as α-kosin with a chemical formula of C25H22O8 at m/z 461.2184 [M + H]+. The MSE spectrum indicated the presence of fragment at m/z 237.20,29 The compounds at rt 11.97 and 13.31 min (entries 27 and 30) were identified as possible isomers of isomallotolerin.30,31 The chemical formula was determined to be C26H32O9 from the sodium adduct at m/z 511.1989 [M + Na]+ and protonated molecular ion peak at m/z 489.2154 [M + H]+.30,31 The fragmentation ions appeared at m/z 470, 417, 259, 237, and 209 in the MSE spectra.
In addition to the above-mentioned compounds, three unknown compounds that could possibly be phloroglucinol derivatives were identified at rt 10.75, 13.00, and 13.96 min (entries 25, 29, and 31). All these compounds showed characteristic fragments at m/z 237 (C13H17O4) and 209 (C12H17O3) observed in most phloroglucinol derivatives identified in this study. The phloroglucinol derivative at rt 10.75 min rendered a chemical formula of C20H24O6. The mass spectrum of the compound displayed a sodium adduct at m/z 383.1628 [M + Na]+ and a protonated molecular peak at m/z 361.1659 [M + H]+ in the positive ionization mode. The fragmentation peaks were present at m/z 405, 337, 237, 209, and 203. The second compound eluted at rt 13.00 min rendered a molecular formula of C29H30O6, deduced from the protonated molecular ion peak at m/z 475.2099 [M + H]+. The third unknown peak that eluted at rt 13.96 min gave a chemical formula of C24H30O8 at m/z 469.1852 [M + Na]+ and 447.2016 [M + H]+. The MSE showed the fragments at m/z 282, 237, 223, and 209.
2.1.3. Terpenoids
The compounds that eluted at rt 10.77 and 11.14 min (entries 32 and 35) were identified as possible isomers of 1-hydroxyeuscaphic acid with a molecular formula of C30H48O6 at m/z 503.3367 [M – H]− in the negative ionization mode and 505.3460 in the positive ionization mode.32 The fragments at m/z 485 and 439 indicated the loss of an H2O molecule and subsequent decarboxylation. Other fragments appeared at m/z 421, 287, 235, 179, 149, and 125.32 The compounds at rt 10.96 and 11.06 min (entries 33 and 34) were identified as 1-hydroxy-2-oxopomolic acid isomers with the molecular formula of C30H46O6 at m/z 501.3231 [M – H]− in the negative ionization mode.20 The fragments at m/z 483, 465, and 421 indicated loss of two H2O molecules and a CO2 molecule.12 An unknown triterpenoid eluted at rt 15.64 min (entry 37) in the positive ionization mode with a chemical formula of C29H48O3 at m/z 445.3685 [M + H]+. The MSE spectrum showed a fragment at m/z 427, indicating the loss of an H2O molecule, and other fragments at m/z 307 and 289. Based on the chemical formula, this compound could possibly be a phytosterol.
UPLC–QTof–MS analysis revealed that the L. sericea EtOAc fraction contains several compounds, mainly flavonoids, flavonoid glycosides, and phloroglucinol derivatives. In addition, a few triterpenoids were identified. Of the identified compounds, four (5,7-dihydroxychromone, tiliroside, α-kosin, and 1-hydroxy-2-oxopomolic) were previously isolated from the L. sericea extracts.12 Other flavonoid and flavonoid glycosides that include B-type procyanidin, isoquercitrin, kaempferol-3-O-glucoside, quercetin-3-O-rutinoside, and kaempferol-3-O-rutinoside were previously identified through LC–MS procedures.13,14,20 Additionally, the flavonoid compounds catechin, epicatechin, quercetin-3-O-pentoside, kaempferol-3-O-acetoglycoside, and kaempferol-3-O-[6″-O-acetyl-2″-O-p-coumaroyl]-glucoside were identified for the first time in this plant. All phloroglucinol derivatives except α-kosin were identified for the first time from the L sericea extracts. Pilosanol B, epipilosanol B, pilosanol C, and epipilosanol C were isolated in the Rosaceae family from the genus Agrimonia (Agrimonia pilosa specie).26,33 The pentacyclic triterpenoid 1-hydroxyeuscaphic acid is also reported for the first time from L. sericea.
2.2. Isolation of Compounds from L. sericea Extracts
Following tentative identification, the EtOAc-soluble fraction was further subjected to chromatographic purifications that included column chromatography, PTLC, LC-SPE, and prep-HPLC (Figure S2) to afford 22 compounds. The structures of the isolated compounds were mainly assigned based on the NMR and high-resolution mass spectrometry (HRMS) data and by comparison of their spectroscopic/spectrometric data with that reported in the literature. The isolated compounds were classified as acetophenones, phloroglucinols, chromones, triterpenoids, flavonoids, and flavonoid glycosides. They included one new triterpenoid 1 (Figure 1) and previously described compounds 2–22 (Figure S3).
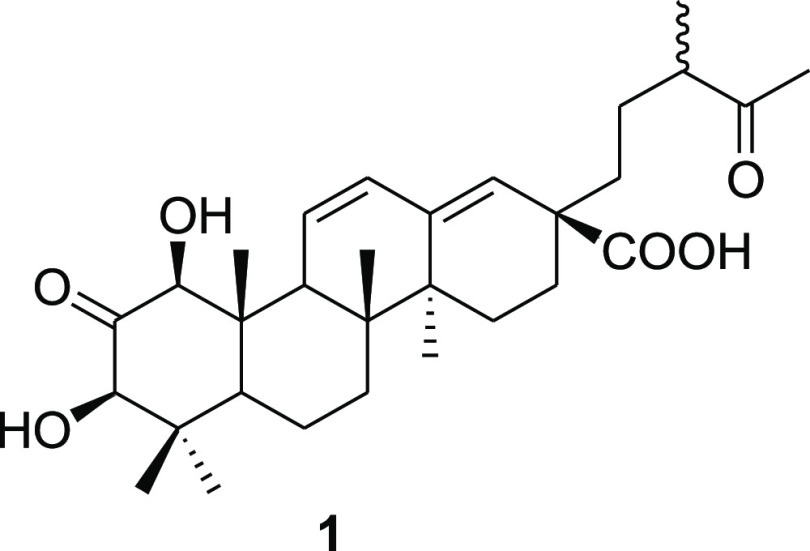
Figure 1.
Structure of the new compound 1 isolated from the L. sericea extract.
Compound 1 was isolated as a white amorphous solid. Its molecular formula C30H44O6 was deduced from the HRESIMS data, which showed a molecular ion peak at m/z 501.3211 [M + H]+ (calculated for C30H45O6, 501.3216 [M + H]+), as well as ammonium and sodium adducts at m/z 518.3456 [M + NH4]+ and 523.3043 [M + Na]+, respectively (Figure 2A). Furthermore, a fragment at m/z 455 [M + H–HCO2H]+ appeared as a base ion in the low collision energy mass spectrum (Figure 2A), showing that the molecule was highly susceptible to decarboxylation. This indicated the presence of a carboxyl functional group in the molecule. The high collision energy mass spectrum (Figure 2B) showed additional fragments at m/z 437 [M + H–HCO2H–H2O]+ and 383 [M + H–HCO2H–C4H8O]+ indicative of the presence of a hydroxy substituent and a side chain with a terminal ketocarbonyl group, respectively.
Figure 2.
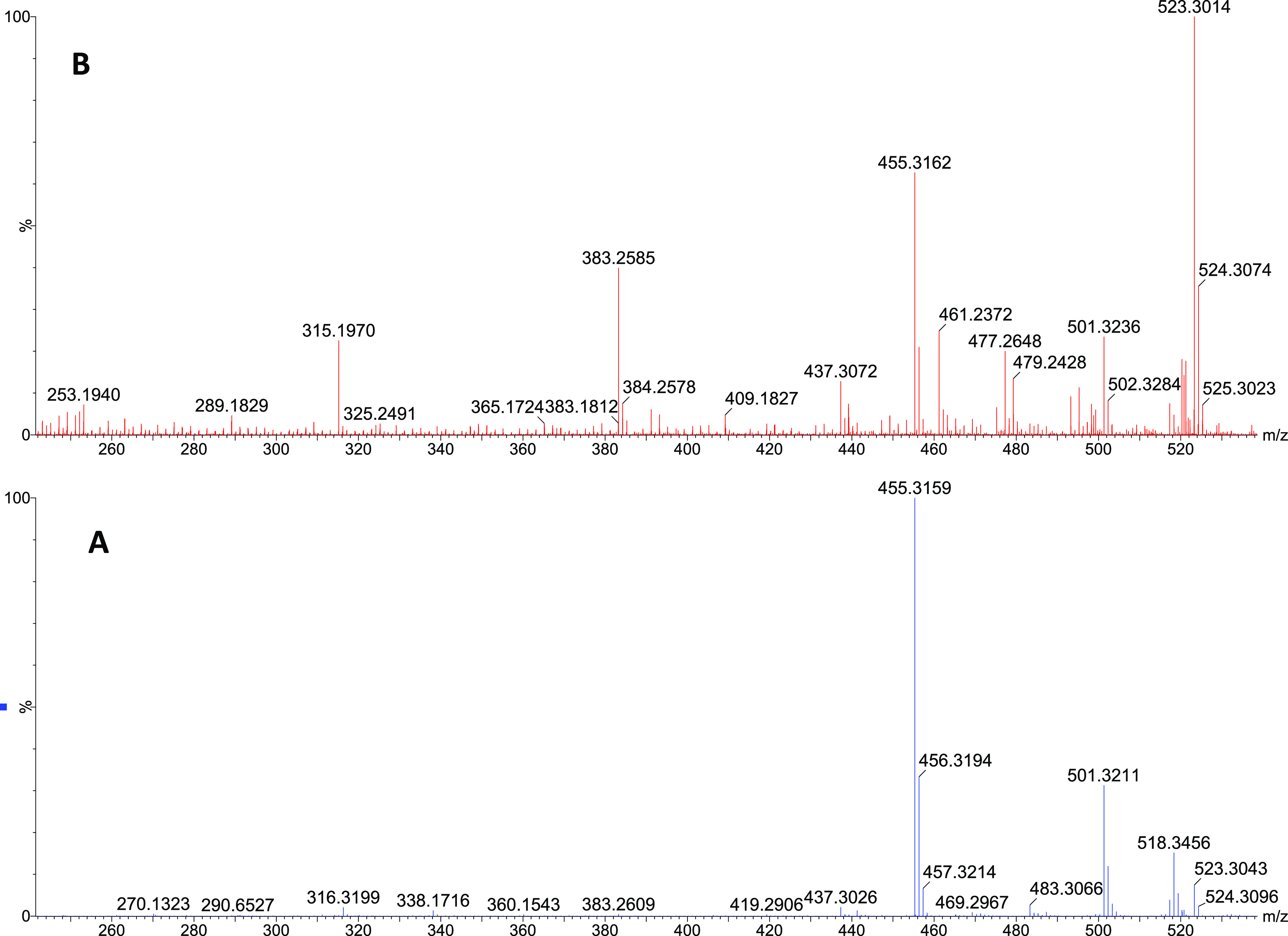
MS spectra of compound 1. (A) Low collision energy spectrum and (B) high collision energy spectrum.
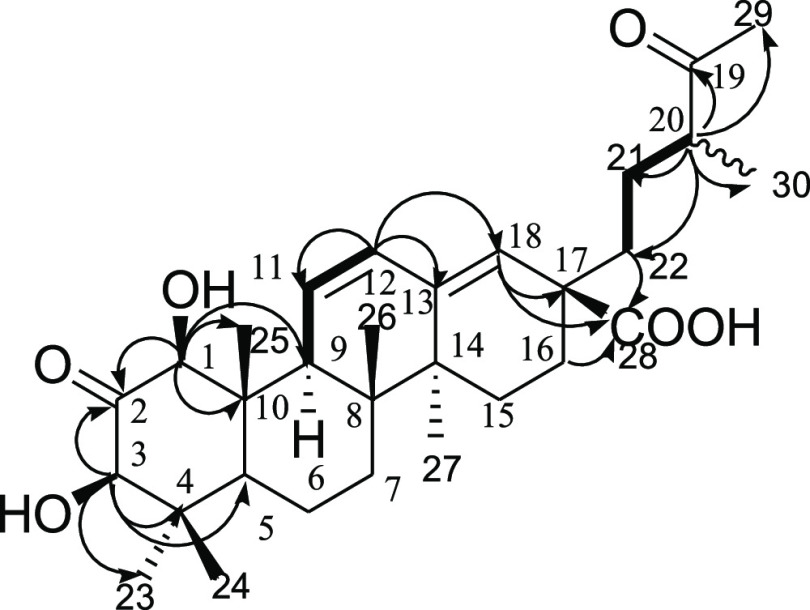
The NMR data of compound 1 is given in Table 1. The 1H NMR spectrum of 1 revealed the presence of two oxymethine protons at δH 4.22 (d, J = 1.5 Hz, H-1) and 4.06 (d, J = 1.5 Hz, H-3); three olefinic methines at δH 6.67 (1H, dd, J = 10.3 and 1.5 Hz, H-11), 5.91 (1H, d, J = 10.3 and 3.2 Hz, H-12), and 5.39 (1H, br S, H-18); and three methines at δH 1.56 (1H, br S, H-5), 2.59 (1H, m, H-9), and 2.52–2.57 (1H, m, H-20). Additionally, the 1H NMR spectrum of 1 displayed six methylene groups and seven methyl groups at δH 1.17, 0.69, 0.80, 0.74, 1.04, 1.08, and 2.14. These were assigned to five tertiary methyl groups (H3-23, H3-24, H3-25, H3-26, and H3-27), a secondary methyl group (H3-30), and finally a methyl ketone (H3-29). The 13C NMR and DEPT data revealed the presence of 30 carbons in the structure of 1 ascribed to two carbinol carbons at δC 85.6 (C-1) and 82.3 (C-3); two ketocarbonyls at δC 211.8 (C-2) and 215.3 (C-19); a carboxyl carbon at δC 179.4 (C-28); two sets of olefinic carbons at δC 131.2 (C-11), 130.4 (C-12), 143.6 (C-13), and 128.2 (C-18); three methine carbons at δC 51.8 (C-5), 56.1 (C-9), and 48.3 (C-20); five quaternary carbons at δC 46.2 (C-4), 42.4 (C-8), 49.4 (C-10), 42.3 (C-14), and 48.1 (C-17); six methylene group carbons at δC 18.8 (C-6), 32.8 (C-7), 27.3 (C-15), 27.9 (C-16), 28.7 (C-21), and 39.2 (C-22); and seven methyl group carbons at δC 29.1 (C-23), 16.4 (C-24), 15.2 (C-25), 16.9 (C-26), 20.2 (C-27), 28.3 (C-29), and 16.6 (C-30). The 1H–1H COSY cross-peak correlations of H-9 (δH 2.59)/H-11 (δH 6.67)/H-12 (δH 5.91), together with the HMBC correlations from H-11/C-13, C-9, C-10, and C-12 and that from H-12/C-9, C-11, and C-18, established that the two sets of olefins are conjugated to each other (Figure 3). The COSY correlations of H-20 to H2-21/H3-30, in addition to the HMBC correlations from H-20 to C-19, C-29, C-30, C-21, and C-22, suggest that the ketocarbonyl carbon is located at C-19 and that this part of the molecule may exist in a straight line chain. The HMBC spectrum of 1 showed key 2,3J correlations of H-1 (δH 4.22)/C-2, C-3, C-9, C-10, and C-25 and of H-3(δH 4.06)/C-1, C-2, C-5, and C-23, revealing that the A-ring of the triterpenoid is tetrasubstituted. The connectivities in the A-ring were further corroborated by the HMBC correlations from H3-24 to C-3, C-5, C-4, and C-23. The position of the carboxylic group was confirmed by the HMBC correlations from H-16 (δH 2.18)/C-18 and C-22 and to the carboxyl C-28. The relative configuration of 1 was assigned based on the 1H–1H NOESY data. The NOESY correlations from H-1 to H-3, H-9, and H-5; H-3 to H-1, H3-23, and H-5; and from H3-24 to H3-25 indicate that OH-1 and OH-3 are β-orientated. Thus, the structure of 1 was assigned as 1β,3β-dihydroxy-2,19-dioxo-18,19-seco-urs-11,13(18)-diene-28-oic acid and trivially named leucosidic A. The leucosidic A (1) compound is structurally similar to the known compound 1β,2α,3α-trihydroxy-19-oxo-18,19-seco-urs-11,13(18)-dien-28-oic acid.34 The unique features of this newly isolated 19-oxo-18,19-seco-ursane-type triterpene are the oxidation state at C-2 and the relative configurations at the 1 and 3 positions.
Table 1. 1H NMR [500 MHz, δH, Mult. (J in Hz)] and 13C NMR (125 MHz, δc) Data of the Triterpenoid 1 in CD3OD.
| position | 13C | 1H | position | 13C | 1H |
|---|---|---|---|---|---|
| 1 | 85.6, CH | 4.22, d (1.5) | 16 | 27.9, CH2 | 2.18, m; 1.49–1.56, m |
| 2 | 211.8, C | 17 | 48.1, C | ||
| 3 | 82.3, CH | 4.06, d (1.5) | 18 | 128.2, CH | 5.39, brs |
| 4 | 46.2, C | 19 | 215.3, C | ||
| 5 | 51.8, CH | 1.56, brs | 20 | 48.3, CH | 2.52–2.57, m |
| 6 | 18.8, CH2 | 1.65–1.83, m | 21 | 28.7, CH2 | 1.33, m; 1.65–1.72, m |
| 7 | 32.8, CH2 | 1.41–1.54, m | 22 | 39.5, CH2 | 1.49–1.56, m; 1.65–1.72, m |
| 8 | 42.4, Ca | 23 | 29.1, CH3 | 1.17, s | |
| 9 | 56.1, CH | 2.59, m | 24 | 16.4, CH3 | 0.69, s |
| 10 | 49.4, C | 25 | 15.2, CH3 | 0.80, s | |
| 11 | 131.2, CH | 6.67, dd (10.3, 1.5) | 26 | 16.9, CH3 | 0.74, s |
| 12 | 130.4, CH | 5.91, dd (10.3, 3.2) | 27 | 20.2, CH3 | 1.04, s |
| 13 | 143.6, C | 28 | 179.4, C | ||
| 14 | 42.3, Ca | 29 | 28.3, CH3 | 2.14, s | |
| 15 | 27.3, CH2 | 1.83–1.77, m | 30 | 16.6, CH3 | 1.08, d (7.0) |
Signal that can be interchanged.
Figure 3.
Key COSY (bold face) and HMBC (arrows) correlations of compound 1.
The structures of isolated acetophenone derivative, phloroglucinol derivative, and chromones are shown in Figure S3A. These are piceol (2), phlorisovalerophenone (3),35 and three chromones that include 5-hyroxy-7-methoxy-2-methylchromone also known as eugenin (4),36 5,7-dihydroxychromone (5),12,36 and 5-hydroxychromone-7-O-β-glucoside (6).37 Eight known triterpenoids were isolated, which included seven known ursene-type triterpenoids, namely, 1-hydroxy-2-oxopomolic acid (7),12 mixture of 2-oxopomolic acid (8)38 and its isomer 2α,19α-dihydroxy-3-oxo12-ursen-28-oic acid (2α-hydroxy-3-oxopomolic acid) (9),39 ursolic acid (10),38,40 mixture of corosolic acid (11)38,40 and tormentic acid (12),38 and pomolic acid (13),38 and a phytosterol glucoside, β-sitosterol glucoside (14) (Figure S3B).40,41 In addition, eight known flavonoids (Figure S3C), which are apigenin (15),42 kaempferol-3-O-α-arabinopyranoside (16),43 mixture of quercetin-3-O-α-arabinopyranoside (17)23 and astragalin (18),42 mixture of isoquercitrin (19)23 and its isomer hyperoside (20),23 tiliroside (21),12 and kaempferol-3-O-rutinoside (22),42 were isolated.
The isolation of three of the compounds, 5,7-dihydroxychromone (5), 1-hydroxy-2-oxopomolic acid (7), and tiliroside (21), from L. sericea extracts was already conducted in the previous studies.12 Therefore, this is the first study to report on the isolation of additional 19 compounds that include a new triterpenoid 1 from the L. sericea plant. Most of the flavonoid glycosides were tentatively identified in Section 2.1 and in other studies.13,14,20 Thus, the isolation of these compounds and the assignment of their structures based on both MS and NMR data confirm their presence in L. sericea extracts.
2.3. Inhibitory Activity of the Isolated Compounds on α-Amylase, α-Glucosidase, and Pancreatic Lipase
Flavonoids and triterpenoids are well known for their health-promoting benefits and are reported to have promising antidiabetic properties.44,45 The potential antidiabetic effects of dietary flavonoids such as apigenin (15), isoquercitrin (19), and tiliroside (21) have been well studied.46−48 In this study, some of the isolated triterpenoid compounds were evaluated for inhibitory effects against some diabetes-related enzymes that include α-amylase, α-glucosidase, and pancreatic lipase (Table 2). The compounds 1, 7, 8/9, 13, and 14 inhibited α-amylase activity with 50% inhibitory concentration (IC50) values ranging from 113.2 to 241 μM. The most active compound was β-sitosterol glucoside (14) with an IC50 value of 113.2 ± 3.51 μM. However, its potency was twice lower than that of the positive control acarbose, which is a known inhibitor of α-amylase. The compounds 1-hydroxy-2-oxopomolic acid (7) and pomolic acid (13) inhibited α-glucosidase with IC50 values of 192.1 ± 13.81 and 85.5 ± 6.87 μM, respectively. Both compounds showed higher potency than the positive control acarbose in this study. Evaluation of compounds 1, 7, 8/9, and 13 for the inhibition of pancreatic lipase revealed the mixture of 2-oxopomolic acid 8 and its isomer 9 to be more active (IC50 = 409.9 ± 26.33 μM) than all of the tested compounds, followed by pomolic acid (13) (IC50 = 457.7 ± 72.78 μM).
Table 2. Inhibitory Activity (IC50) of the Isolated Compounds (μM) on α-Amylase, α-Glucosidase, and Pancreatic Lipasea.
| compounds (μM) | α-amylase | α-glucosidase | pancreatic lipase |
|---|---|---|---|
| compound 1 | 241.8 ± 57.76 | 1151 ± 175.25 | |
| compound 7 | 130.2 ± 12.68 | 192.1 ± 13.81 | 981.5 ± 36.57 |
| compounds 8 and 9 | 123.0 ± 4.46 | 409.9 ± 26.33 | |
| compound 13 | 141.9 ± 6.15 | 85.5 ± 6.87 | 457.7 ± 72.78 |
| compound 14 | 113.2 ± 3.51 | ||
| acarbose | 54.2 ± 6.49 | 347.5 ± 46.23 | |
| orlistat | 3.123 ± 0.035 |
Data expressed as mean ± SD.
The results presented herein demonstrate that pentacyclic triterpenoids and phytosterol glucoside from L. sericea could modulate the key enzymes that are therapeutic targets in the management of diabetes and obesity. Inhibition of these enzymes indicates that the compounds can potentially retard dietary glucose and lipid liberation and delay their absorption, resulting in reduced postprandial hyperglycemia and hyperlipidemia. The results obtained in the present study are consistent with several reports that have shown the potential antidiabetic and antiobesity effects of triterpenoids.44,45,49
In conclusion, phytochemical profiling of the L. sericea extract led to the identification of several compounds, mainly flavonoids, flavonoid glycosides, and phloroglucinol derivatives. Subsequent chromatographic purifications of the EtOAc fraction resulted in the isolation of 22 compounds that included an acetophenone derivative, a phloroglucinol derivative, three chromones, nine triterpenoids, a flavonoid, and seven flavonoid glycosides. This is the first study to report a new triterpenoid leucosidic A (1) from the L. sericea plant. Evaluation of the isolated triterpenoids for inhibition of α-amylase, α-glucosidase, and pancreatic lipase activities revealed that the isolated triterpenoids had the ability to modulate these enzymes, which serve as therapeutic targets for diabetes and obesity. The identification and isolation of the wide variety of compounds signifies that L. sericea is a rich source of bioactive compounds. Most of the isolated compounds, particularly flavonoids and triterpenoids, are known for their putative health benefits relating to non-communicable diseases and find applications in the medical and food industries, where they are used as structural templates for the development of pharmaceutical agents and in the development of functional foods, nutraceuticals, and food supplements.
3. Materials and Methods
3.1. General Experimental Procedures
Optical rotations were recorded on a PerkinElmer 341 polarimeter. A Bruker Alpha II FT-IR spectrometer was used to acquire IR spectra. The NMR spectra were recorded at ambient temperature on a Bruker Avance III 400 MHz spectrometer with the BBI probe and the Avance III HD 500 MHz spectrometer with a Prodigy probe. 1H and 13C NMR chemical shifts were referenced to residual protonated solvent peaks at δH 7.26 and δC 77.0 for CDCl3; δH 2.05 and δC 29.84 for (CD3)2CO; δH 3.31 and δC 49.0 for CD3OD; and δH 2.50 and δC 39.52 for (CD3)2SO. The HRESIMS data were collected using a UPLC–QTof Synapt G2 HDMS (Waters Corp., MA, USA) in both positive and negative ESI modes. A hyphenated system consisting of an Agilent 1260 Infinity HPLC with a PDA detector, a Bruker AmaZon SL ion trap MS, a Bruker Prospect II SPE Interface, and a Bruker Sample Pro sample handler were used to conduct HPLC–MS–SPE purification of the fractions. Prep-HPLC purification of other fractions was done by a Waters chromatographic system with a PDA detector and a fraction collector (Waters, Milford, MA, USA). Column chromatography was performed using silica gel 0.063–0.200 mm (Sigma-Aldrich) and various solvent systems, and the collected fractions were spotted on aluminum-precoated TLC silica gel 60 F254 plates (Merck, Darmstadt, Germany) and visualized under 254 and 365 nm ultraviolet illumination. Preparative thin-layer chromatography was performed with preparative TLC silica gel 60 PF254 plates (Sigma-Aldrich Pty. Ltd., Johannesburg, South Africa).
3.2. Plant Material
The leaves and stems of L. sericea plants were collected from the rocky areas of QwaQwa, Free State Province, South Africa, in March 2019. A voucher specimen was deposited in the Schweickerdt Herbarium at the University of Pretoria and was assigned a voucher number PRU 127909.
3.3. Extraction
The leaves and stems of L. sericea were air-dried and ground to afford a fine powdered material. The plant material was split into three portions of 1.7 kg, and each was extracted with CH3OH (5 × 3 L). The solvent was combined and evaporated under reduced pressure using a rotary evaporator to afford the CH3OH extract. The CH3OH extract (503 g) was diluted with H2O and subjected to solvent–solvent extraction with EtOAc and BuOH (3 × 1 L) to afford three factions, the EtOAc-soluble faction (198 g), the BuOH fraction (62 g), and the H2O-soluble faction (50 g).
3.4. UPLC–QTof–MS Analysis
The UPLC was equipped with a kinetex 1.7 μm EVO C18, 2.1 mm × 100 mm column. The column temperature was set at 40 °C. The flow rate was kept constant at 0.35 mL/min. The mobile phase consisted of A: H2O + 0.1% HCO2H and B: CH3OH + 0.1% HCO2H. A total run time of 25 min was used following a gradient elution method as follows: 0.0 min—5% B, 14 min—95% B, 22 min—95% B, 23 min—5% B, and 25 min—5% B. An injection volume of 5 μL was used. The mass spectrometer was operated in positive and negative ESI resolution modes. N2 gas was used as the desolvation gas. The following parameters were then set: capillary voltage 2 400 V; sampling cone voltage 25 V; extraction cone 4 V; source temperature 120 °C; desolvation temperature 300 °C; desolvation gas 600 L/h; and cone gas flow 10.0 L/h. Throughout all acquisitions, a solution of leucine enkephalin was used as the lock spray solution that was constantly infused through a separate orthogonal ESI probe to compensate for experimental drift in mass accuracy. MS data was acquired between 50 and 1200 m/z in the resolution mode using the MSE centroid method. The MSE method simultaneously acquires MS data in two functions, namely function 1 (low collision energy) and function 2 (high collision energy). In function 1, the auto trap and transfer collision energies of 6 and 4 V were employed, respectively. The trap and the transfer collision energies were ramped from 20 to 40 V in function 2. The complete system was driven by Masslynx software.
3.5. Chemical Profiling
Molecular formulae (MF) of the pseudomolecular ions representing any possible compound were computed and selected based on the criterion that the mass difference between the measured and calculated mass was at or below 5 ppm and the lowest possible isotopic fit number for the identification of compounds. This MF was used to search for possible compound identity using freely available online databases such as Chemspider, Massbank, and METLIN. Fragmentation patterns were correlated with the proposed structures, and the data was also compared with that already published in the literature.
3.6. Isolation
EtOAc fraction (100 g) was subjected to silica gel column chromatography (CC) with gradient elution using 100% hexanes, hexanes/EtOAc (8:2 → 6:4 → 5:5 → 4:6 → 2:8), CH3OH/EtOAc (2:8 → 5:5), and 100% CH3OH to afford 8 main fractions (Fr. 1–Fr. 8). A portion of main Fr. 4 (20 mg) was also subjected to LC-SPE to afford compounds 2 (0.5 mg), 3 (0.6 mg), and 4 (0.6 mg). To accumulate more material, the same fraction Fr. 4 (200 mg) was subjected to preparative HPLC, resulting in an additional pure compound 10 (12 mg) and other fractions that contained mixtures of triterpenoids. To reduce the complexity of Fr. 4, 1 g of this fraction was subjected to CC eluted with hexanes/acetone (8:2 → 7:3 → 6:4 → 5:5) and 100% CH3OH to afford 16 subfractions. Subfraction 13 (463 mg) was subjected to LC-SPE and re-purified by prep-HPLC to afford compounds 5 (23 mg), 1 (5 mg), and 7 (100 mg) and a mixture of two isomers 8 and 9 (68 mg). The combined main fractions Fr. 5 + 6 (7 g) were subjected to CC eluted with hexanes/acetone (9:1 → 8:2 → 7:3), resulting in 16 fractions. Combined subfractions 9 and 10 (108 mg) were subjected to CC eluted with hexanes/acetone (8:2 → 7:3:5:5) to afford compound 13 (3.5 mg). Furthermore, subfraction 15 (144 mg) was then subjected to preparative TLC eluted with hexanes/acetone/formic acid (7:3:0.1) to afford four fractions 15 (f1–f4). Fraction 15f3 was subjected to LC-SPE affording compound 15 (0.5 mg). Main fraction 8 (22 g) was subjected to CC eluted with CH2Cl2/CH3OH (9:1 → 8.5:1.5 → 8:2 → 7:3 → 6:4 → 3:7) and 100% CH3OH to afford 11 subfractions (Fr. 1–Fr. 11). Subfraction 4 (805 mg) and subfraction 5 (466 mg) were subjected to solvent wash with 100% CH3OH to afford white powdered compounds 11 and 12 (400 mg) as well as compound 14 (300 mg), respectively. A portion of subfraction 8 (200 mg) was subjected to 2 × preparative TLC, resulting in fractions [Fr. 8 (f1–f8) × 2]. Similar fractions were combined, and Fr. 8–f5 was subjected to LC-SPE, resulting in compounds 6 (0.7 mg), 16 (0.3 mg), and 21 (1.2 mg). Fr. 8 (f6, f7) were individually subjected to LC-SPE, resulting in a mixture of compounds 17 and 18 (3.2 mg), a mixture of compounds 19 and 20 (1.3 mg), and compound 22 (0.3 mg).
3.7. LC-SPE and Preparative HPLC Purification
The LC-SPE purifications were performed as previously described50 with modifications of the HPLC method to facilitate the separation of the current fractions. To accumulate more material, some of the fractions were re-purified using prep-HPLC coupled to a PDA detector and a fraction collector (Waters, Milford, MA, USA). Each of the fraction (70 mg) was diluted with methanol (1 mL). An X-Bridge preparative C18 column (5 μm, 19 × 250 mm) was used for the separation. The mobile phase consisted of 0.1% formic acid in water (solvent A) and methanol (solvent B) and was pumped at a flow rate of 15 mL/min. Gradient elution was applied as follows: 20% B for 1.7 min, 20–80% B (1.7–15 min), 80% B (15–30 min), 80–95% B (30–33 min), 95% B (33–45 min), 95–100% B (45–52 min), 100% B (52–56 min), 100%–20% B (56–58 min), and 20% B (58–62 min). The injection volume was 300 μL. The samples were injected multiple times to accumulate the material. Data were collected using MassLynx4.1 (Waters, USA) software. The purified compounds were collected using a fraction collector and subsequently combined and concentrated using the speed vacuum.
3.8. In Vitro Enzyme Inhibition
3.8.1. α-Amylase Inhibition
The α-amylase inhibitory activity of the compounds was evaluated using the 3,5-dinitrosalicylic acid (DNSA) assay described by Elya et al. (2015) with some slight modifications.51 Briefly, 50 μL of porcine pancreatic amylase (2 U/mL) (Sigma-Aldrich) was preincubated with 50 μL of the test compound (10–400 μM) or acarbose (Glentham Life Sciences) at 25 °C for 10 min. This was followed by addition of 50 μL of 1% starch solution (Sigma-Aldrich) to initiate the reaction. The reaction mixture was allowed to stand at room temperature for a further 10 min before the reaction was stopped by the addition of 80 μL of DNSA (96 mM). The reaction mixture was then heated at 85 °C for 10 min and allowed to cool. Following dilution of the solution with 1.1 mL of double-distilled H2O, 200 μL was pipetted into 96-well plates (Greiner, clear F-bottom) to read absorbance at 540 nm using a Spectramax paradigm microplate reader (Molecular Devices Inc, San Jose, California).
3.8.2. α-Glucosidase Inhibition
The in vitro α-glucosidase inhibitory activity of the compounds was investigated using p-nitrophenyl-α-D-glucopyranoside (pNPG) (Sigma-Aldrich) as an artificial substrate.51 The working concentrations of the test compounds and enzyme were prepared in 100 mM phosphate buffer (pH 6.8). In a 96-well plate (Greiner, clear F-bottom), 25 μL of α-glucosidase from Saccharomyces cerevisiae (0.2 U/mL) (Sigma-Aldrich) was pre-incubated with 100 μL of the compound or acarbose (62.5–1000 μM) at 37 °C for 10 min. Thereafter, 25 μL of pNPG (5 mM) was added, and the reaction mixtures were incubated at 37 °C for 30 min. The reaction was stopped by adding 50 μL of NaOH (0.1 M). The enzyme activity was quantified by measuring absorbance at 405 nm using the Spectramax paradigm microplate reader.
3.8.3. Pancreatic Lipase Inhibition
The pancreatic lipase inhibitory activity of the isolated compounds was tested using p-nitrophenyl palmitate (p-NPP) (Sigma-Aldrich) as an artificial substrate.52 The reaction mixture consisted of 0.061 M of tris–HCl buffer, pH 7.4, 200 U/mL of lipase from porcine pancreas (Sigma-Aldrich), and test compound (62.5–1000 μM). The reaction was initiated by adding 10 mM p-NPP, and the reaction mixture was incubated at 37 °C for 20 min. Orlistat (Glentham Life Sciences) was used as a standard drug. The enzyme activity was monitored by colorimetrically measuring the release of p-nitrophenol at 405 nm using the Spectramax paradigm microplate reader.
Acknowledgments
This work was based on the research supported by the National Research Foundation of South Africa (grant numbers UID 113964, UID 117853, and UID 121928), the University of Pretoria, and the South African Nuclear Energy Corporation Ltd. We thank Dr. Madelien Wooding for assisting with the acquisition of the UPLC–QT of–MS data.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsomega.2c00096.
UPLC–QTof–MS chromatogram, tentatively identified compounds, flow diagram for isolation procedure, structures of the isolated compounds, analytical data of the isolated compounds, and NMR (1D and 2D) and MS spectra of the new compound 1 (PDF)
The authors declare no competing financial interest.
Supplementary Material
References
- Shulaev V.; Korban S. S.; Sosinski B.; Abbott A. G.; Aldwinckle H. S.; Folta K. M.; Iezzoni A.; Main D.; Arús P.; Dandekar A. M.; Lewers K.; Brown S. K.; Davis T. M.; Gardiner S. E.; Potter D.; Veilleux R. E. Multiple Models for Rosaceae Genomics. Plant Physiol. 2008, 147, 985–1003. 10.1104/pp.107.115618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hummer K. E.; Janick J.. Rosaceae: Taxonomy, Economic Importance, Genomics. Genetics and Genomics of Rosaceae; Folta K. M., Gardiner S. E., Eds.; Springer New York: New York, NY, 2009; pp 1–17. [Google Scholar]
- Potter D.; Eriksson T.; Evans R. C.; Oh S.; Smedmark J. E. E.; Morgan D. R.; Kerr M.; Robertson K. R.; Arsenault M.; Dickinson T. A.; Campbell C. S. Phylogeny and Classification of Rosaceae. Plant Syst. Evol. 2007, 266, 5–43. 10.1007/s00606-007-0539-9. [DOI] [Google Scholar]
- Ogah O.; Watkins C. S.; Ubi B. E.; Oraguzie N. C. Phenolic Compounds in Rosaceae Fruit and Nut Crops. J. Agric. Food Chem. 2014, 62, 9369–9386. 10.1021/jf501574q. [DOI] [PubMed] [Google Scholar]
- Garcia-Oliveira P.; Fraga-Corral M.; Pereira A. G.; Lourenço-Lopes C.; Jimenez-Lopez C.; Prieto M. A.; Simal-Gandara J. Scientific Basis for the Industrialization of Traditionally Used Plants of the Rosaceae Family. Food Chem. 2020, 330, 127197. 10.1016/j.foodchem.2020.127197. [DOI] [PubMed] [Google Scholar]
- Eriksson T.; Hibbs M. S.; Yoder A. D.; Delwiche C. F.; Donoghue M. J. The Phylogeny of Rosoideae (Rosaceae) Based on Sequences of the Internal Transcribed Spacers (ITS) of Nuclear Ribosomal DNA and the trnl/F Region of Chloroplast DNA. Int. J. Plant Sci. 2003, 164, 197–211. 10.1086/346163. [DOI] [Google Scholar]
- Mafole T. C.; Aremu A. O.; Mthethwa T.; Moyo M. An Overview on Leucosidea sericea Eckl. & Zeyh.: A Multi-Purpose Tree with Potential as a Phytomedicine. J. Ethnopharmacol. 2017, 203, 288–303. 10.1016/j.jep.2017.03.044. [DOI] [PubMed] [Google Scholar]
- Hutchings A. A Survey and Analysis of Traditional Medicinal Plants as Used by the Zulu; Xhosa and Sotho. Bothalia 1989, 19, 112–123. 10.4102/abc.v19i1.947. [DOI] [Google Scholar]
- Seleteng Kose L.; Moteetee A.; Van Vuuren S. Ethnobotanical Survey of Medicinal Plants Used in the Maseru District of Lesotho. J. Ethnopharmacol. 2015, 170, 184–200. 10.1016/j.jep.2015.04.047. [DOI] [PubMed] [Google Scholar]
- Bosman A. A.; Combrinck S.; Roux-van der Merwe R.; Botha B. M.; McCrindle R. I.; Houghton P. J. Isolation of an Anthelmintic Compound from Leucosidae sericea. S. Afr. J. Bot. 2004, 70, 509–511. 10.1016/s0254-6299(15)30189-7. [DOI] [Google Scholar]
- Adamu M.The Efficacy of Traditionally Used Leucosidea sericea (Rosaceae) against Haemonchus contortus and Microbial Pathogens; University of Pretoria, 2012. [Google Scholar]
- Pendota S. C.; Aremu A. O.; Slavětínská L. P.; Rárová L.; Grúz J.; Doležal K.; Van Staden J. Identification and Characterization of Potential Bioactive Compounds from the Leaves of Leucosidea sericea. J. Ethnopharmacol. 2018, 220, 169–176. 10.1016/j.jep.2018.03.035. [DOI] [PubMed] [Google Scholar]
- Badeggi U. M.; Badmus J. A.; Botha S. S.; Ismail E.; Marnewick J. L.; Africa C. W. J.; Hussein A. A. Biosynthesis, Characterization, and Biological Activities of Procyanidin Capped Silver Nanoparticles. J. Funct. Biomater. 2020, 11, 66. 10.3390/jfb11030066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badeggi U. M.; Ismail E.; Adeloye A. O.; Botha S.; Badmus J. A.; Marnewick J. L.; Cupido C. N.; Hussein A. A. Green Synthesis of Gold Nanoparticles Capped with Procyanidins from Leucosidea Sericea as Potential Antidiabetic and Antioxidant Agents. Biomolecules 2020, 10, 452. 10.3390/biom10030452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garzón G. A.; Manns D. C.; Riedl K.; Schwartz S. J.; Padilla-Zakour O. Identification of Phenolic Compounds in Petals of Nasturtium Flowers (Tropaeolum Majus) by High-Performance Liquid Chromatography Coupled to Mass Spectrometry and Determination of Oxygen Radical Absorbance Capacity (ORAC). J. Agric. Food Chem. 2015, 63, 1803–1811. 10.1021/jf503366c. [DOI] [PubMed] [Google Scholar]
- Morcol T. B.; Matthews P. D.; Kennelly E. J. Differences in Leaf Chemistry and Glandular Trichome Density between Wild Southwestern American Hop (Humulus neomexicanus) and Commercial Hop Cultivars. J. Agric. Food Chem. 2021, 69, 7798–7814. 10.1021/acs.jafc.1c02710. [DOI] [PubMed] [Google Scholar]
- Masike K.; de Villiers A.; Hoffman E. W.; Brand D. J.; Causon T.; Stander M. A. Detailed Phenolic Characterization of Protea Pure and Hybrid Cultivars by Liquid Chromatography-Ion Mobility-High Resolution Mass Spectrometry (LC-IM-HR-MS). J. Agric. Food Chem. 2020, 68, 485–502. 10.1021/acs.jafc.9b06361. [DOI] [PubMed] [Google Scholar]
- Qi D.; Li J.; Qiao X.; Lu M.; Chen W.; Miao A.; Guo W.; Ma C. Non-Targeted Metabolomic Analysis Based on Ultra-High-Performance Liquid Chromatography Quadrupole Time-of-Flight Tandem Mass Spectrometry Reveals the Effects of Grafting on Non-Volatile Metabolites in Fresh Tea Leaves (Camellia sinensis L.). J. Agric. Food Chem. 2019, 67, 6672–6682. 10.1021/acs.jafc.9b01001. [DOI] [PubMed] [Google Scholar]
- Fromm M.; Loos H. M.; Bayha S.; Carle R.; Kammerer D. R. Recovery and Characterisation of Coloured Phenolic Preparations from Apple Seeds. Food Chem. 2013, 136, 1277–1287. 10.1016/j.foodchem.2012.09.042. [DOI] [PubMed] [Google Scholar]
- Sehlakgwe P. F.; Lall N.; Prinsloo G. 1H-NMR Metabolomics and LC-MS Analysis to Determine Seasonal Variation in a Cosmeceutical Plant Leucosidea sericea. Front. Pharmacol. 2020, 11, 219. 10.3389/fphar.2020.00219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J.; Yu Q.; Cheng H.; Ge Y.; Liu H.; Ye X.; Chen Y. Metabolomic Approach for the Authentication of Berry Fruit Juice by Liquid Chromatography Quadrupole Time-of-Flight Mass Spectrometry Coupled to Chemometrics. J. Agric. Food Chem. 2018, 66, 8199–8208. 10.1021/acs.jafc.8b01682. [DOI] [PubMed] [Google Scholar]
- Monzón Daza G.; Meneses Macías C.; Forero A. M.; Rodríguez J.; Aragón M.; Jiménez C.; Ramos F. A.; Castellanos L. Identification of α-Amylase and α-Glucosidase Inhibitors and Ligularoside A, a New Triterpenoid Saponin from Passiflora ligularis Juss (Sweet Granadilla) Leaves, by a Nuclear Magnetic Resonance-Based Metabolomic Study. J. Agric. Food Chem. 2021, 69, 2919–2931. 10.1021/acs.jafc.0c07850. [DOI] [PubMed] [Google Scholar]
- Vvedenskaya I. O.; Rosen R. T.; Guido J. E.; Russell D. J.; Mills K. A.; Vorsa N. Characterization of Flavonols in Cranberry (Vaccinium macrocarpon) Powder. J. Agric. Food Chem. 2004, 52, 188–195. 10.1021/jf034970s. [DOI] [PubMed] [Google Scholar]
- Kajdzanoska M.; Gjamovski V.; Stefova M. HPLC-DAD-ESI-MSn Identification of Phenolic Compounds in Cultivated Strawberries from Macedonia. Maced. J. Chem. Chem. Eng. 2010, 29, 181–194. 10.20450/mjcce.2010.165. [DOI] [Google Scholar]
- Ouyang H.; Li T.; He M.; Li Z.; Tan T.; Zhang W.; Li Y.; Feng Y.; Yang S. Identification and Quantification Analysis on the Chemical Constituents from Traditional Mongolian Medicine Flos Scabiosae Using UHPLC-DAD-Q-TOF-MS Combined with UHPLC-QqQ-MS. J. Chromatogr. Sci. 2016, 54, 1028–1036. 10.1093/chromsci/bmw041. [DOI] [PubMed] [Google Scholar]
- Kim H. W.; Choi S. Y.; Jang H. S.; Ryu B.; Sung S. H.; Yang H. Exploring Novel Secondary Metabolites from Natural Products Using Pre-Processed Mass Spectral Data. Sci. Rep. 2019, 9, 1–11. 10.1038/s41598-019-54078-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pojer F.; Kahlich R.; Kammerer B.; Li S.-M.; Heide L. Clor, A Bifunctional Non-Heme Iron Oxygenase Involved in Clorobiocin Biosynthesis*. J. Biol. Chem. 2003, 278, 30661–30668. 10.1074/jbc.m303190200. [DOI] [PubMed] [Google Scholar]
- Qin G.-W.; Chen Z.-X.; Wang H.-C.; Qian M.-K. Structure and Synthesis of Robustaol A. Acta Chim. Sin. 1981, 39, 83–89. [Google Scholar]
- Sharma R.; Kishore N.; Hussein A.; Lall N. The Potential of Leucosidea sericea against Propionibacterium acnes. Phytochem. Lett. 2014, 7, 124–129. 10.1016/j.phytol.2013.11.005. [DOI] [Google Scholar]
- Saito Y.; Iga S.; Nakashima K.; Okamoto Y.; Gong X.; Kuroda C.; Tori M. Terpenoids from Ligularia virgaurea Collected in China: The First Example of Two Bakkane Derivatives with an Anhydride-Type Ring C and Nineteen New Chemical Constituents. Tetrahedron 2015, 71, 8428–8435. 10.1016/j.tet.2015.09.011. [DOI] [Google Scholar]
- Arisawa M.; Fujita A.; Hayashi T.; Morita N.; Kikuchi T.; Tezuka Y. Studies on Cytotoxic Constituents in Pericarps of Mallotus japonicus. IV. Chem. Pharm. Bull. 1990, 38, 698–700. 10.1248/cpb.38.698. [DOI] [PubMed] [Google Scholar]
- Wu C.; Cui B.; Dong H.; Ren Y.; Yang C.; Yao M.; Li W.; Gan C. Simultaneous Determination and Pharmacokinetics Study of Six Triterpenes in Rat Plasma by UHPLC-MS/MS after Oral Administration of Sanguisorba officinalis L. Extract. Molecules 2018, 23, 2980. 10.3390/molecules23112980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H. W.; Park J.; Kang K. B.; Kim T. B.; Oh W. K.; Kim J.; Sung S. H. Acylphloroglucinolated Catechin and Phenylethyl Isocoumarin Derivatives from Agrimonia pilosa. J. Nat. Prod. 2016, 79, 2376–2383. 10.1021/acs.jnatprod.6b00566. [DOI] [PubMed] [Google Scholar]
- Chen Z.; Tong L.; Feng Y.; Wu J.; Zhao X.; Ruan H.; Pi H.; Zhang P. Ursane-Type Nortriterpenes with a Five-Membered a-Ring from Rubus innominatus. Phytochemistry 2015, 116, 329–336. 10.1016/j.phytochem.2015.04.006. [DOI] [PubMed] [Google Scholar]
- Zhou W.; Zhuang Y.; Bai Y.; Bi H.; Liu T.; Ma Y. Biosynthesis of Phlorisovalerophenone and 4-Hydroxy-6-Isobutyl-2-Pyrone in Escherichia coli from Glucose. Microb. Cell Fact. 2016, 15, 149. 10.1186/s12934-016-0549-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pendse R.; Rao A. V. R.; Venkataraman K. 5,7-Dihydroxychromone from Arachis hypogoea Shells. Phytochemistry 1973, 12, 2033–2034. 10.1016/s0031-9422(00)91529-2. [DOI] [Google Scholar]
- Simon A.; Chulia A. J.; Kaouadji M.; Delage C. Quercetin 3-[Triacetylarabinosyl(1→6)Galactoside] and Chromones from Calluna vulgaris. Phytochemistry 1994, 36, 1043–1045. 10.1016/s0031-9422(00)90488-6. [DOI] [PubMed] [Google Scholar]
- Liu W.-j.; Hou X.-q.; Chen H.; Liang J.-y.; Sun J.-b. Chemical Constituents from Agrimonia pilosa Ledeb. And Their Chemotaxonomic Significance. Nat. Prod. Res. 2016, 30, 2495–2499. 10.1080/14786419.2016.1198351. [DOI] [PubMed] [Google Scholar]
- Xu H.-X.; Zeng F.-Q.; Wan M.; Sim K.-Y. Anti-HIV Triterpene Acids from Geum japonicum. J. Nat. Prod. 1996, 59, 643–645. 10.1021/np960165e. [DOI] [PubMed] [Google Scholar]
- Sun S.; Huang S.; Shi Y.; Shao Y.; Qiu J.; Sedjoah R.-C. A. -A.; Yan Z.; Ding L.; Zou D.; Xin Z. Extraction, Isolation, Characterization and Antimicrobial Activities of Non-Extractable Polyphenols from Pomegranate Peel. Food Chem. 2021, 351, 129232. 10.1016/j.foodchem.2021.129232. [DOI] [PubMed] [Google Scholar]
- Faizi S.; Ali M.; Saleem R.; Irfanullah; Bibi S. Complete 1H and 13C NMR Assignments of Stigma-5-en-3-O-Β-glucoside and its Acetyl Derivative. Magn. Reson. Chem. 2001, 39, 399–405. 10.1002/mrc.855. [DOI] [Google Scholar]
- Markham K. R.; Ternai B.; Stanley R.; Geiger H.; Mabry T. J. Carbon-13 NMR Studies of Flavonoids. III. Naturally Occurring Flavonoid Glycosides and their Acylated Derivatives. Tetrahedron 1978, 34, 1389–1397. 10.1016/0040-4020(78)88336-7. [DOI] [Google Scholar]
- De Cássia Lemos Lima R.; Kongstad K. T.; Kato L.; José das Silva M.; Franzyk H.; Staerk D.; Staerk D. High-Resolution PTP1B Inhibition Profiling Combined with HPLC-HRMS-SPE-NMR for Identification of PTP1B Inhibitors from Miconia albicans. Molecules 2018, 23, 1755. 10.3390/molecules23071755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nazaruk J.; Borzym-Kluczyk M. The Role of Triterpenes in the Management of Diabetes Mellitus and its Complications. Phytochem. Rev. 2015, 14, 675–690. 10.1007/s11101-014-9369-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva F. S. G.; Oliveira P. J.; Duarte M. F. Oleanolic, Ursolic, and Betulinic Acids as Food Supplements or Pharmaceutical Agents for Type 2 Diabetes: Promise or Illusion?. J. Agric. Food Chem. 2016, 64, 2991–3008. 10.1021/acs.jafc.5b06021. [DOI] [PubMed] [Google Scholar]
- Zhang L.; Zhang S.-T.; Yin Y.-C.; Xing S.; Li W.-N.; Fu X.-Q. Hypoglycemic Effect and Mechanism of Isoquercitrin as an Inhibitor of Dipeptidyl Peptidase-4 in Type 2 Diabetic Mice. RSC Adv. 2018, 8, 14967–14974. 10.1039/c8ra00675j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suh K. S.; Oh S.; Woo J.-T.; Kim S.-W.; Kim J.-W.; Kim Y. S.; Chon S. Apigenin Attenuates 2-Deoxy-D-Ribose-Induced Oxidative Cell Damage in Hit-T15 Pancreatic Β-Cells. Biol. Pharm. Bull. 2012, 35, 121–126. 10.1248/bpb.35.121. [DOI] [PubMed] [Google Scholar]
- Grochowski D. M.; Locatelli M.; Granica S.; Cacciagrano F.; Tomczyk M. A Review on the Dietary Flavonoid Tiliroside. Compr. Rev. Food Sci. Food Saf. 2018, 17, 1395–1421. 10.1111/1541-4337.12389. [DOI] [PubMed] [Google Scholar]
- Mosa R.; Naidoo J.; Nkomo F.; Mazibuko S.; Muller C.; Opoku A. In Vitro Antihyperlipidemic Potential of Triterpenes from Stem Bark of Protorhus longifolia. Planta Med. 2014, 80, 1685–1691. 10.1055/s-0034-1383262. [DOI] [PubMed] [Google Scholar]
- Ramabulana T.; Scheepers L.-M.; Moodley T.; Maharaj V. J.; Stander A.; Gama N.; Ferreira D.; Sonopo M. S.; Selepe M. A. Bioactive Lignans from Hypoestes aristata. J. Nat. Prod. 2020, 83, 2483–2489. 10.1021/acs.jnatprod.0c00443. [DOI] [PubMed] [Google Scholar]
- Elya B.; Handayani R.; Sauriasari R.; Azizahwati; Hasyyati U. S.; Permana I. T.; Permatasar Y. I. Antidiabetic Activity and Phytochemical Screening of Extracts from Indonesian Plants by Inhibition of Alpha Amylase, Alpha Glucosidase and Dipeptidyl Peptidase IV. Pak. J. Biol. Sci. 2015, 18, 279. 10.3923/pjbs.2015.279.284. [DOI] [Google Scholar]
- Mendis Abeysekera W. P. K.; Arachchige S. P. G.; Ratnasooriya W. D. Bark Extracts of Ceylon Cinnamon Possess Antilipidemic Activities and Bind Bile Acids in Vitro. J. Evidence-Based Complementary Altern. Med. 2017, 2017, 1–10. 10.1155/2017/7347219. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.