Summary
We combine molecular dynamics, statistical mechanics, and hybrid quantum mechanics/molecular mechanics simulations to describe mechanistically the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) RNA-dependent RNA polymerase (RdRp). Our study analyzes the binding mode of both natural triphosphate substrates as well as remdesivir triphosphate (the active form of drug), which is bound preferentially over ATP by RdRp while being poorly recognized by human RNA polymerase II (RNA Pol II). A comparison of incorporation rates between natural and antiviral nucleotides shows that remdesivir is incorporated more slowly into the nascent RNA compared with ATP, leading to an RNA duplex that is structurally very similar to an unmodified one, arguing against the hypothesis that remdesivir is a competitive inhibitor of ATP. We characterize the entire mechanism of reaction, finding that viral RdRp is highly processive and displays a higher catalytic rate of incorporation than human RNA Pol II. Overall, our study provides the first detailed explanation of the replication mechanism of RdRp.
Keywords: SARS-CoV-2, viral replication, RNA-dependent RNA polymerase, biocatalysis, reaction mechanism, remdesivir, antivirals, free energy calculations, QM/MM
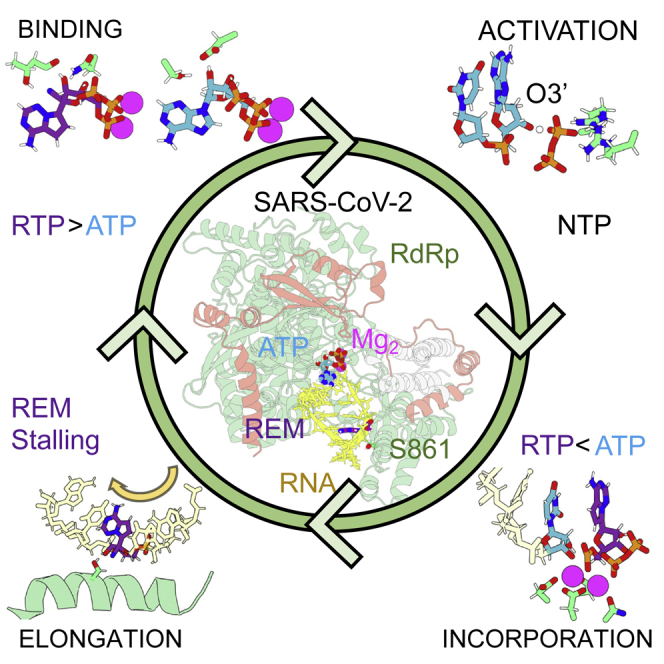
Graphical abstract
Highlights
-
•
SARS-CoV-2 RdRp and human RNA Pol II display opposite ATP and RTP binding preferences
-
•
Viral polymerase makes use of a self-activated mechanism
-
•
RTP is incorporated slower that its natural counterpart, ATP, inside RdRp
-
•
During RNA polymerization, remdesivir is stalled in a stabilizing trap
The bigger picture
RNA-dependent RNA polymerase (RdRp) from severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is an attractive target to attack the viral replication. Many efforts have been directed toward the design of effective inhibitors of RdRp. However, the mechanism of nucleoside-triphosphate binding, as well as nucleotide activation and incorporation at atomic resolution, has not been deciphered. Also, the molecular mechanism of the promising antiviral remdesivir, a nucleotide analog, is still under study. In this work, binding preferences between natural and remdesivir-TP molecules have been analyzed inside RdRp and human RNA polymerase II active sites. Also, the detailed reaction mechanism of nucleotide activation and incorporation inside the RdRp are characterized. Afterward, during RNA polymerization, remdesivir is stalled in a stabilizing trap. The characterization of the replication mechanism of SARS-CoV-2 RdRp provided by this study can help guide the design of next-generation antivirals.
Aranda et al. reveal the binding, activation, incorporation, and elongation of natural and remdesivir nucleotides inside SARS-CoV-2 RNA-dependent RNA polymerase. Reaction mechanisms are characterized at atomic resolutions and compared with human RNA polymerase II. During RNA polymerization, remdesivir is stalled in a stabilizing trap that prevents further translocation.
Introduction
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the virus that caused coronavirus disease 2019 (COVID-19), emerged in China in December 2019 and rapidly spread over the world, causing a worldwide health threat with more than 5 million fatalities.1 From a phylogenetic point of view, SARS-CoV-2 belongs to the β genus of the coronavirus family, which includes other highly infective pathogens such as the Middle East respiratory syndrome CoV (MERS-CoV) or the severe acute respiratory syndrome CoV (SARS-CoV).2 SARS-CoV-2 has a large (30 kb of positive RNA) genome, which forces it to strike a balance between high replication fidelity and genetic diversity.3, 4, 5, 6 A highly processive RNA-dependent RNA polymerase (RdRp) and a proof-reading exonuclease are the crucial elements for maintaining the stability of the viral genome, while at the same time enabling its mutation to adapt to new environments.
RdRp is the core of the replication machinery of the virus and one of the largest proteins in the viral genome (932 residues). It binds to nsp7 and nsp8 to form an active complex,7, 8, 9 one that first uses sense RNA as a template to generate a negative copy and then, in a second cycle, generates new copies of genomic and sub-genomic RNAs. At least two other proteins are involved in the replication process: a 601-residue helicase and a 527-residue proof-reading exonuclease.10,11 A simple BLAST12 query shows that SARS-CoV-2 RdRp is highly conserved within the coronavirus family, but homologs out of this family show a quite low identity (less than 25%), which suggests that we are faced with a quite new protein, but with a surprisingly high functional efficiency.
Due to its central role in the viral infection cycle, RNA polymerases are a major target for fighting RNA viruses.13, 14, 15 Although vaccines began to be available as of November 2021, 50% of world’s population still remains without any dose of it, and the use of effective antivirals is needed to prevent future fast-spreading coronavirus outbreaks due to ineffective immunizations or the emergence of new variants. As of today, the only FDA-approved drug for the treatment of SARS-CoV-2 infection16 is a RdRp-inhibitor, remdesivir (R), a C-nucleoside (see Figure S1) that was approved for the treatment of Ebola.17 Being a negative single-strand RNA virus, Ebola is very distant from SARS-CoV-2, but its replication is also dependent on the action of an RdRp.
Several SARS-CoV-2 RdRp cryoelectron microscopy (cryo-EM) structures have been reported since the beginning of the pandemic. The first structure comprised the RdRp complex.8 Afterward, structures including different RNA duplexes were resolved showing that RNA binding causes no drastic rearrangements in RdRp.9,18,19 W. Yin et al. were able to capture a structure where remdesivir has already been incorporated in a nascent RNA strand. Also, a pyrophosphate (PPi) molecule departing from the active site and two Mg2+ near it were found.18 Recently, structures of RdRp in complex with favipiravir triphosphate (TP) provided new insights of the pre-catalytic state. However, these structures were resolved with the drug in a non-productive conformation20 or without Mg2+ ions in the active site.21 So far, however, the limited amount of atomistic data on the mode of binding and reaction mechanism of incorporation of both natural substrates and remdesivir hampers our ability to develop new and more active compounds.
We present here a comprehensive study on the mechanism of action of SARS-CoV-2 RdRp. Atomistic simulations characterize binding interactions and substrate preferences in the active site of the viral polymerase. While remdesivir binds to the viral active site more strongly than its natural counterpart, the opposite is found for human RNA polymerase II. Molecular dynamics (MD) and quantum mechanics/molecular mechanics (QM/MM) simulations demonstrate that the enzyme follows a canonical 2-ion reaction mechanism with a catalytic efficiency higher than that of the highly evolved human RNA polymerase II (RNA Pol II). Calculations and biochemical experiments demonstrate that remdesivir triphosphate (RTP; the expected bioactive form of remdesivir) can be recognized and incorporated into nascent RNA with an efficiency only slightly lower than natural nucleotides, i.e., remdesivir is not an inhibitor of nucleotide incorporation. Extended MD simulations failed to detect any dramatic distortion in the RNA duplex due to the presence of remdesivir that would account for its inhibitory properties. Furthermore, no steric clashes were detected when the nascent RNA duplex was displaced along the exit channel (see below), which argues against the hypothesis that steric clashes are responsible for delayed inhibition, suggesting a more specific inhibitory mechanism related perhaps to a transient covalent bond with the enzyme. In summary, our results, obtained through thorough computational simulations, provide the first atomistic description of the mechanism of action for SARS-CoV-2 RNA polymerase and provide clues on the mysterious inhibition mechanism of remdesivir.
Results and discussion
SARS-CoV-2 RdRp active site architecture
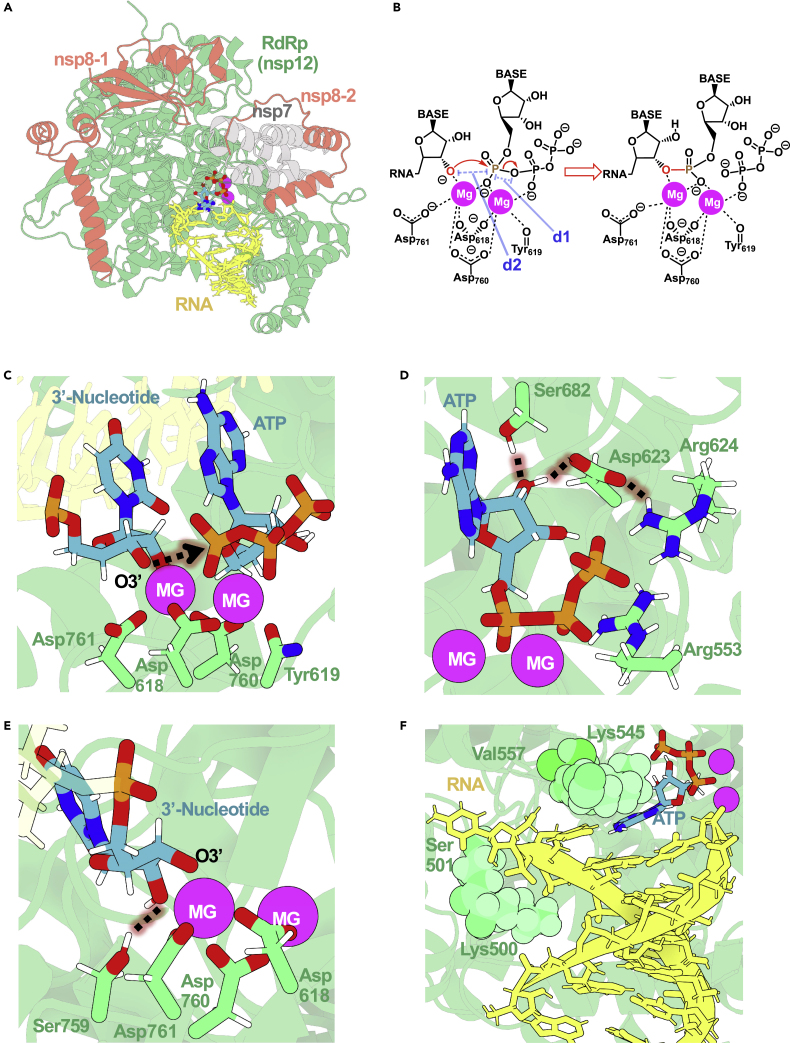
The viral protein (see Figure 1A) has a canonical active site, quite similar to those of other polymerases. The two essential Mg2+ are coordinated by the ⍺ and β phosphate groups of the incoming triphosphate nucleotide, as well as Asp618, Asp760, Asp761, Tyr619, and the O3′ terminal of the negative RNA strand (see Figures 1B and 1C), which—according to the circular reaction mechanism for polymerases by de Vivo and coworkers—is expected to be ionized.22 Classical molecular-interaction-potential calculations,23 using Mg2+ as the binding cation, further corroborate the two Mg2+ binding sites (see supplemental information and Figure S2). Compared with available structural data,8,18 our catalytically competent active site unveils the slight movement of the Asp618-Tyr619 loop (Figure S2) in order to create well-defined coordination spheres for the two Mg2+ and to be able to proceed with a phosphoryl transfer with high efficiency. Two arginine residues (Arg624 and Arg553) bind the phosphates of the incoming nucleotide, aligning the gamma phosphate for an effective transfer (see Figure 1D). The preferential affinity for ribonucleotide triphosphate substrates (as opposed to deoxyribonucleotide ones) can be explained by the need for north puckering that controls the alignment of the reactive groups, as well as the presence of specific H bonds between the 2′OH group of the nucleoside triphosphate (NTP) and the side chains of Asp623 and Ser683 in the catalytic site (see Figure 1D). Additional hydrogen bonds are found between i+1 2′OH and Ser759 (see Figure 1E). The base specificity is controlled by the complementarity of hydrogen bonding with the template nucleobase (see Figures 1F and S3) and by phosphate coordination, as well as by the residues surrounding the active site that mechanically introduce strong isosteric requirements, altogether making a non-Watson-Crick pairing scheme very unlikely (see Figure 1F). Overall, the structural picture of the active site emerging from EM structures8,9,18,19 and atomistic simulations strongly suggests that despite a short evolutionary history, SARS-CoV-2 RdRp has all the structural requirements to be an efficient RNA polymerase both in terms of catalysis rate and substrate specificity.
Figure 1.
Active site of SARS-CoV-2 RdRp makes it an efficient polymerase
(A) The replication complex of SARS-CoV-2 formed by the nsp12 RdRp enzyme (in green), the nsp8 and nsp7 cofactors (orange and gray, respectively), and an RNA template and nascent strands (yellow).
(B) Scheme depicting the two-metal-ion mechanism used by SARS-CoV-2 RdRp.
(C) The active site contains a well-defined coordination sphere of the two Mg2+ ions.
(D) NTP substrate recognition in the active site of RdRp is mediated by a pair of arginines, an aspartate, and a serine.
(E) The deprotonated 3′ terminal nucleotide is stabilized by one of the catalytic ions in the RdRp’s active site.
(F) RdRp active site pocket enables base specificity between the incoming nucleotide and the template.
Figures were prepared with 3D Protein Imager.24 A 3D structure representation can be accessed through https://mmb.irbbarcelona.org/3dRS/s/Y17cub.
We also analyzed the SARS-CoV RdRp active site through MD simulations (see experimental procedures and supplemental experimental procedures). SARS-CoV and SARS-CoV-2 RdRp (nsp12) share more than a 96% sequence similarity, and their structures’ root-mean-square deviation (RMSD) is 0.84 Å.8 In addition, the loops containing the catalytic residues Asp618, Asp760, Asp761, and Tyr619, involved in the coordination of the Mg2+s, are identical in sequence. During our simulations, SARS-CoV RdRp and SARS-CoV-2 RdRp Mg2+ coordination spheres remained almost identical (see Figure S4). The average distances between magnesium ions were found to be 3.9 ± 0.2 and 3.7 ± 0.1 Å for SARS-CoV and SARS-CoV-2, respectively. Also, the distance involved in nucleotide incorporation between Pα an O3′ atoms remained very similar at 3.6 ± 0.2 and 3.5 ± 0.1 Å, respectively. Thus, the observed difference in activity7 between them should be attributed to other factors.
SARS-CoV-2 RdRp and human RNA Pol II display opposite binding preferences with respect to RTP and ATP
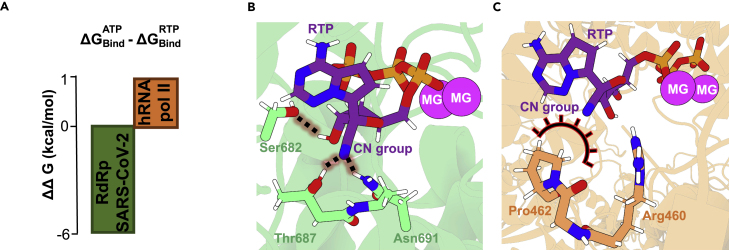
To directly compare the affinity of the binding sites of SARS-CoV-2 RdRp and human RNA Pol II, we performed alchemical free energy simulations, transforming ATP into RTP either in the binding pocket or in free solution. The obtained free-energy differences were directly translated into affinity differences (ΔΔG) using standard thermodynamic cycles as described above. As shown in Figure 2, the viral RdRp is characterized by a sizable preference for RTP, in agreement with available biochemical data,14 which is a serendipitous fact given that it was originally designed to block an evolutionarily distinct polymerase in the Ebola virus. On the other hand, the lower affinity for the active site of human RNA Pol II suggests that remdesivir will rarely be incorporated into nascent human mRNA, partially explaining the drug’s low toxicity in humans.25 The observed preferences can be rationalized by inspecting stabilizing interactions inside enzymes’ active sites. The nitrile group of RTP can accept two hydrogen bonds with Thr687 and Asn691 sidechains of RdRp of SARS-CoV-2 (Figure 2B). Similar interactions were observed in previous MD studies.26,27 On the contrary, inside RNA Pol II, no hydrogen-bond donors able to stabilize the nitrile group are found (Figure 2C).
Figure 2.
Binding preferences in viral and human RNA polymerases
(A) Binding free energies of ATP and RTP inside SARS-CoV-2 RdRp and human RNA Pol II (negative values corresponding to a preference for RTP over ATP).
(B) Most important interactions between RTP nitrile group and RdRp polymerase.
(C) RTP inside human RNA Pol II active site.
The SARS-CoV-2 RdRp activation mechanism
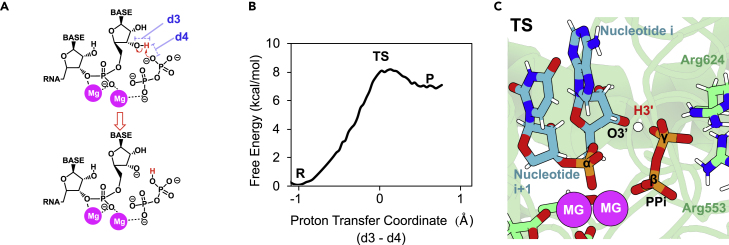
Before investigating the detailed mechanism of nucleotide incorporation, we explored the mechanism of O3′ deprotonation of the 3′-end ribose inside RdRp. This is a mandatory step of nucleophile formation that precedes nucleotide incorporation. We explored whether RdRp is able to activate the 3′-end nucleotide through a self-activated mechanism as has been proposed for other polymerases.22 As shown in Figure 3 (see also Video S1), once a nucleotide has been incorporated inside RdRp, the newly created PPi molecule is found in perfect arrangement to abstract the O3′ hydrogen atom. Our simulations confirm that one non-bridging oxygen atom of PPi’s γ phosphate group can abstract the proton of the 3′ hydroxyl with a free energy of activation of 8.1 kcal/mol. This barrier is ∼3 kcal/mol higher than the reported value for O3′ deprotonation inside DNA polymerase eta (Pol-η).28 Interestingly, the arrangement inside RdRp enables the direct transfer of the proton from the O3′ hydroxyl group to the γ phosphate group of the PPi, while in other polymerases, like the aforementioned DNA Pol-η, the β phosphate group firstly deprotonates the O3′ atom and then donates the proton to the γ group.22 O3′ atom’s deprotonation inside the RdRp step is found to be slightly endothermic as in DNA Pol’s two-metal mechanism,28 which, as described by us and others, is overcome by subsequent PPi–H release from the polymerase’s active site.29, 30, 31, 32, 33 Finally, following the replication cycle, nucleic-acid translocation and PPi–H departure from RdRp’s active site enable the binding of the subsequent NTP molecule.22
Figure 3.
Mechanism of activation through O3′ deprotonation inside RdRp
(A) Scheme depicting the activation mechanism used by SARS-CoV-2 RdRp.
(B) Free-energy profile as a function of the proton transfer coordinate (d3-d4), obtained for the proton transfer reaction between the O3′ of the just-incorporated nucleotide and the γ phosphate group of the newly formed PPi.
(C) Transition state (TS) where the H3′ proton belonging to O3′ atom of nucleotide “i” is halfway to being transferred to γ phosphate group of PPi. Reaction coordinate (RC) consisted of the distances between O3′ or Oγ atoms to the H atom to be transferred, displayed as d3 and d4, respectively.
The SARS-CoV-2 RdRp reaction mechanism of nucleotide incorporation
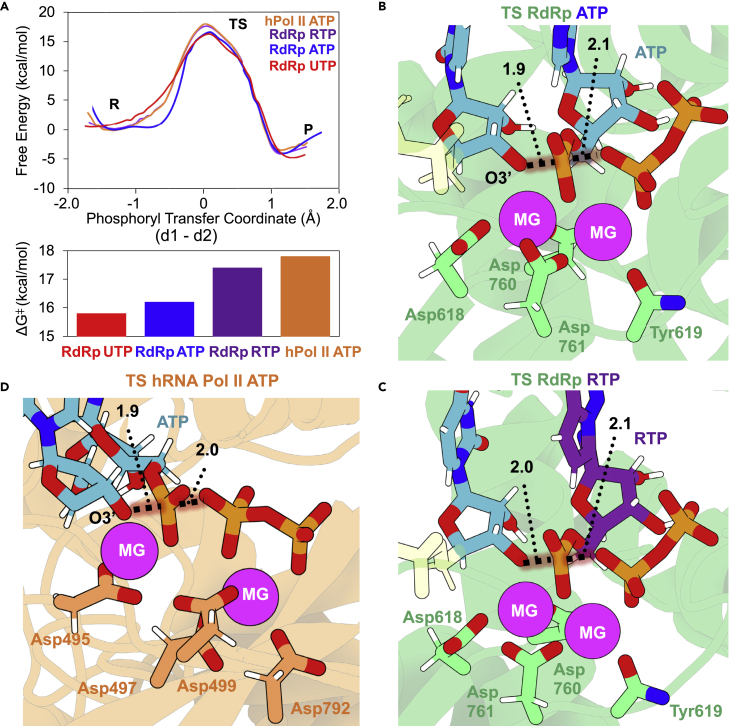
QM/MM simulations were used (see experimental procedures and supplemental experimental procedures) to study the ability of SARS-CoV-2 RdRp to incorporate either a natural triphosphate (NTP; exemplified here by uridine-5′-triphosphate [UTP] and ATP) or RTP (remdesivir-TP) into nascent RNA. In the QM/MM reactant state, the Pα atom of the ATP is 3.6 ± 0.2 Å far from O3′ atom of the terminal nucleotide (see Figure S5). In addition, the O3′ atom is in line to perform the nucleophilic attack on the Pα atom of the ATP, displaying an angle of 173° ± 5° between Oα-Pα-O3′ atoms. While one Mg2+ cation is coordinated by the O3′ atom of the 3′ terminal, the Pα atom of the ATP, and carboxylate atoms of Asp618, Asp760, and Asp761, the other Mg2+ cation is coordinated by the carbonyl group of Tyr619 and carboxylate atoms of Asp618 and Asp760. In addition, the Mg2+-Mg2+ distance is 3.6 ± 0.1 Å. Following the two-metal-ion mechanism scheme, one Mg2+ activates the O3′ atom toward the attack of Pα of NTP, and the other Mg2+ stabilizes the upcoming PPi leaving group, while both are in perfect disposition to stabilize the negatively charged transition state (TS). In the TS, the phosphoryl group is halfway to being transferred to the O3′ atom of the terminal nucleotide (see Figures 4B and S5). The O3′-Pα distance is 1.9 ± 0.1 Å, while the Pα-Oα distance is 2.1 ± 0.1 Å. Also, the Mg2+-Mg2+ distance is slightly reduced by 0.2 ± 0.1 Å, while Mg2+s maintained the same interactions with their respective coordination spheres. Metal-aided nucleotidyl transfer reactions can proceed through associative or dissociative TSs (SN2 or SN1, respectively) depending on the specific enzyme catalyzing the process.29 It has been observed that while CRISPR-Cas9 proceeds through a concerted associative mechanism,34 group II introns display a dissociative one.35 Thus, in light of our results, the reaction inside SARS-CoV-2 RdRp proceeds through a concerted associative TS where the breaking and forming bond lengths are found to be similarly extended. Finally, in the product state, the nucleotide has been fully transfer reflected in a Pα-Oα distance of 3.5 Å, and a PPi molecule is formed (see Figure S5).
Figure 4.
RNA elongation inside RdRp of SARS-CoV-2 and human RNA Pol II
(A) Top, free-energy profiles as a function of the phosphoryl transfer coordinate (d1-d2) for the incorporation of a U, an A, or an R to a nascent viral RNA strand. Profile for the incorporation of the inside human RNA Pol II is also shown. The phosphorylation reaction consists of a nucleophilic attack of the O3′ of the terminal nucleotide on the Pα of the triphosphate nucleotide. Bottom, bar plot displaying free energies of activation for the process for different NTPs and enzymes.
(B and C) Active site views of the TSs were found when ATP (B) and RTP (C) (purple C atoms) are the substrates for the elongation reaction inside RdRp. The phosphoryl group is half-way to being transferred. One Mg2+ activates the O3′ toward nucleophilic attack and stabilizes the negatively charged TS. The other Mg2+ stabilizes the charged TS, as well as the nascent negatively charged PPi molecule. Distances involved in the reaction are shown as dotted lines with their average values in Å.
(D) TS insight of the elongation reaction catalyzed by human RNA pol II.
A careful analysis of trajectories and the free-energy profiles shows that the formation of a phosphodiester bond proceeds through a single free-energy maximum (TS) corresponding to an activation barrier of 15.8 kcal mol−1 for UTP and 16.2 kcal mol−1 for ATP (see Figures 4, S5, and S6 and Videos S2 and S3) and is characterized by a negative free energy of ∼5 kcal/mol (prior to PPi release). To obtain an estimate of the relative efficiency of the viral enzyme, we studied the RNA polymerization reaction catalyzed by human RNA Pol II (see Figures 4D and S7 and Video S4). Recently published kinetic rate constants36 of nucleotide incorporation by SARS-CoV-2’s RdRp provide support for the quantitative accuracy of our estimates, giving confidence to the suggested reaction mechanism. To evaluate the degree of fitness of the enzyme, we used the same procedure to predict the RNA Pol II activation barrier (see Figure 4), getting a value of 17.8 kcal mol−1 for ATP, in good agreement with experimental estimates.37,38 This value is ∼1.5 kcal/mol higher than that of the viral RdRp (see Figure 4A), demonstrating that despite its short evolutionary trajectory, the efficiency of SARS-Cov-2 RdRp is at least similar to those of highly evolved eukaryotic polymerases.39, 40, 41
Our equilibrium trajectories demonstrate that RTP fits very well into the active site, showing strong canonical Watson-Crick interactions with the uridine in the template RNA. Being isosteric to adenosine, it achieves a perfect shape complementarity and arrangement of reactive groups in the binding site, predicting that it will act as a substrate rather than an inhibitor. This hypothesis is confirmed by QM/MM simulations showing that incorporation of RTP can happen with a free-energy barrier only slightly larger than that of a natural substrate (10% up to 17.4 kcal mol−1; see Figures 4C and S6 and Video S5). Such an increase is mostly due to a slight misalignment of the Oα-Pα-O3′ attack angle (161° ± 8° for RTP, 173° ± 5° for ATP, and 172° ± 5° for UTP) due to RTP nitrile’s group interactions (see Figures S8 and S9). Thus, we can rule out the possibility of remdesivir inhibiting RdRp by blocking the ATP-binding site of RNA polymerase. On the contrary, our simulation strongly suggests that RTP can be efficiently incorporated in front of uridine in a nascent RNA strand. Again, the order of incorporation predicted by our theoretical calculations, UTP > ATP > RTP, agrees perfectly with recent accurate pre-steady-state kinetic experiments,36 providing additional support to our calculations (see comparison in Figure S10). Dangerfield et al. also observed that although RTP is incorporated more slowly than ATP inside RdRp, it is incorporated more efficiently than its counterpart due to a higher specificity constant (kcat/Km),36 which also agrees with our estimates.
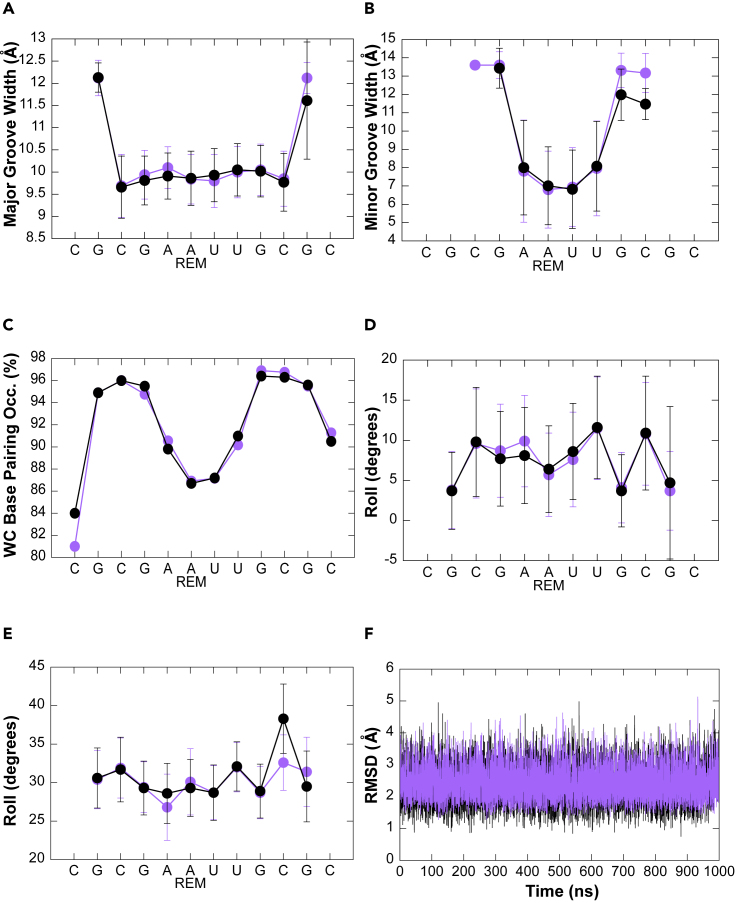
Remdesivir is well tolerated in an RNA duplex
Remdesivir is a C-nucleoside with an extra C1′ cyano group, and as such, it might distort the helix, causing delayed inhibition of the enzyme due to hampered displacement of the nascent duplex along the exit channel. To explore this possibility, we performed MD simulations of two RNA duplexes differing only in the substitution of a central r(A·U) pair by r(R·U) one (see experimental procedures). The results, summarized in Figure 5, strongly suggest that remdesivir is well tolerated in an RNA duplex and does not introduce any major structural distortion that would justify termination of RNA synthesis. In particular, there are no significant differences between the hydrogen-bonding stability of r(A·U) and r(R·U) pairs (see Figure 5C), and helical parameters of the duplexes are insensitive to the presence of remdesivir. In summary, our results strongly argue against the idea that a dramatic structural alteration of the RNA duplex is the key factor responsible for remdesivir-induced termination of RNA synthesis. Our results agree with recent cryo-EM structures where remdesivir was incorporated at different or multiple positions of the nascent RNA strand without altering RNA-duplex structure.42,43
Figure 5.
Remdesivir does not distort RNA structure
(A–F) Major (A) and minor (B) groove width, Watson-Crick base pairing (C), roll (D), twist (E), and RMSD (F) of the double helix are not affected when remdesivir is present (purple) with respect to control sequence (black) during MD simulations. The average values across the simulations are shown in black and purple dots for the control and remdesivir-containing sequences, respectively. Average standard deviations are shown as black and purple bars. The RMSD of one of the five replicas is shown in (F) (see also Figure S11).
Remdesivir does not block nascent-strand elongations through steric hindrance
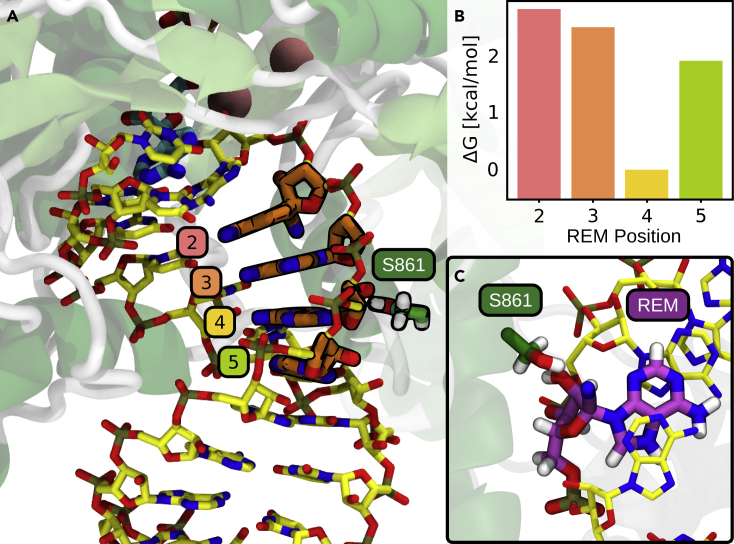
Trying to explore alternative reasons for the inhibitory properties of remdesivir, we slid the nascent RNA duplex along the exit tunnel of RdRp to make the r(R·U) pair by simulating the addition of extra nucleotides, which allowed us to scan interactions of the RNA at several positions along the exit tunnel. After R was incorporated, we were not able to detect any point of steric clash that could justify stopping the polymerase progression (see Figure S12). Interestingly, when three more nucleotides were incorporated, we found Ser861, whose sidechain is located at around 3.7 ± 0.3 Å from the nitrile group of remdesivir (see Figure S13A and S13B). However, considering the flexibility of the Ser sidechain, especially in a well-solvated microenvironment (see Figures S13A, S13C, and S13D), this strongly argues against the hypothesis that nitrile-Ser861 steric clash can explain the inhibitory properties of remdesivir.14,42,43 To further discard the steric-clash hypothesis, we performed an alchemical mutational (A→R) scan along the nascent strand, looking at relative preferences of adenosine and remdesivir at positions from (i+2) to (i+5) in a poly-A RNA duplex embedded in the RdRp channel (see Figure 6). Very interestingly, A/R ΔG differences were very similar for positions i+2, i+3, and i+5, suggesting that there are not specific interactions between the incorporated nucleotide and the tunnel residues (as expected for an RNA polymerase designed to have a continuum output flux). On the contrary, significant differences are found at position (i+4), where a local free-energy minimum would hinder the movement of the nascent RNA toward the exit of the tunnel. The stabilization of remdesivir at position i+4 seems related to the formation of direct or water-mediated hydrogen bonds between the cyano-group nitrogen and side-chain hydroxyl of Ser861 (see Figure S13).
Figure 6.
Remdesivir elongation along RdRp’s exit channel
(A and B) Alchemically slided remdesivir along the i+2 to i+5 positions of the nascent RNA strand (A) and its associated free energies (B). In the i+4 position, remdesivir is energetically preferred with respect to neighboring positions.
(C) Insight displaying a stabilizing interaction between Ser861 and remdesivir is shown. Alchemically mutated adenines to remdesvir molecules are depicted in cyan, and the two Mg2+ cations of the active site are depicted as pink balls. The Ser861 residue, which is hypothesized to be involved in a steric clash, is shown.
Current biochemical and structural data show that remdesivir is stalled after 3 or 4 nucleotides are incorporated, but the reasons are unclear.14,42, 43, 44, 45 Also, biochemical experiments have showed that inhibition can be overcome with higher NTP concentrations.14,42 In order to dissect the mechanism, structures of RdRp in post- and pre-translocated states with remdesivir in different positions of the nascent RNA strand have been resolved.42,43 A previous MD study also suggested that the delayed termination mechanism could be due to remdesivir’s destabilization of base pair interactions as well as of the nucleotide located in the active site.26 Other experiments have suggested that when remdesivir is present in the template RNA strand, it would also inhibit RdRp through a secondary mechanism.44 Some authors have noted as key for main inhibition a steric clash of the nitrile group with Ser861,14,43, 44, 45 but our calculations failed to detect any steric clashes, suggesting the opposite, i.e., the vicinities of Ser861 are quite stabilizing for remdesivir. This would suggest that rather than being sterically unable to reach the position, remdesivir-containing RNA might be trapped in this position, hampering further sliding. Analysis of the recognition site (see Figures 6, S13C, and S13D) suggests that Ser861 could even be involved in a water-catalyzed Pinner’s reaction,46 which would lead to a transient covalent bond of RNA to the enzyme. The lack of stability of the resulting complex47, 48, 49, 50 precludes its experimental detection, but a transient covalent bond could contribute to trap remdesivir at the i+4 position, requiring an extra addition of NTP to escape from this stalling situation.14
To further explore the role of Ser861 as a stalling element in the sliding of remdesivir-containing RNA, we explored the exit channels of human RNA polymerases and CoV RdRps (see Figures S14 and S15). In the case of human RNA polymerases, no serine (or similar residue) was found in the expected displacement path of the remdesivir nitrile group (see Figure S14), suggesting that, if incorporated by human polymerases, remdesivir will not stop sliding of the nascent RNA. On the contrary, the same exercise made with other CoV RdRps (see Figure S15) found an amphipathic α-helix with a conserved Ser, which can play the same role of Ser861 in other coronaviruses. We can speculate that the same could happen for Ebola virus, the original targeted virus for remdesivir, but lack of structural information on Ebola RdRp precludes a detailed analysis.
Conclusions
SARS-CoV-2 RdRp is a protein common to other coronaviruses but shows little homology outside the family. As CoVs are just a few thousand years old, the protein has had a limited evolutionary period, and we could expect low efficiency. However, and quite surprisingly, our calculations demonstrate that the enzyme is very efficient, even more than eukaryotic polymerases. The viral enzyme follows a mechanism that is similar to that of bacterial or eukaryotic polymerases with the transferred phosphate being stabilized by 2 Mg2+ ions exquisitely coordinated by acidic residues of the catalytic site, while the phosphates of the incoming nucleotide are stabilized by a network of basic residues. The SARS-Cov-2 RdRp makes use of a self-activated mechanism where the gamma-phosphate group of PPi molecule deprotonates the hydroxylic 3′ terminal, generating the nucleophile that participates in the subsequent incorporation of a nucleotide.
Quite surprisingly, RTP is an excellent substitute of ATP, although it is not well recognized by human RNA Pol II. Simulations also show that after being bound to the viral active site, RTP does not block RdRp, but it is incorporated into the nascent RNA in front of a uridine. The resulting duplex does not show dramatic structural changes, which would hinder displacement of the nascent duplex along the exit channel. In fact, analysis of the displacement of the RNA-containing remdesivir along the exit tunnel of RdRp fails to detect points of steric clashes. Moreover, free-energy calculations show that the i+4 position is quite a favorable site for remdesivir. This suggests that the protein environment around i+4, particularly a hydrated serine, can act as a trap for the nascent RNA duplex, stalling the displacement of the helix by stabilizing remdesivir by either non-covalent or transient-covalent contacts.
Experimental procedures
Resource availability
Lead contact
Further information and requests for resources should be directed to and will be fulfilled by the lead contact, Modesto Orozco (modesto.orozco@irbbarcelona.org).
Materials availability
This study did not generate new unique reagents.
RdRp complex setup
The cryo-EM structure of the RdRp (nsp12) of SARS-CoV-2 complexed to nsp8 and nsp78 cofactors and the cryo-EM structure also complexed with an RNA duplex18 were used as our starting models. The first model was obtained when no other structure was available46 by aligning the SARS-CoV-2 RdRp with RdRp from a hepatitis C virus X-ray structure,51 from which RNA, the two catalytic metal ions, and the diphosphate group of an NTP analog were extracted (see supplemental experimental procedures for details). The structure was refined by comparing the cryo-EM structure of RdRp in complex with a full RNA strand18 (see Figure S16). Finally, we built an ATP, UTP, or RTP molecule inside the active site of RdRp in the expected orientation required for incorporation into the nascent RNA strand. The systems were solvated and neutralized prior to optimization, thermalization (T = 298 K), and equilibration (see supplemental experimental procedures for details). The final equilibrated structures were the starting points for further MD simulations. The SARS-CoV RdRp starting model, including an RNA duplex and an ATP molecule, was built from an available cryo-EM structure52 and subjected to the same modeling and equilibration protocols as described above. The progression of the polymerization process was simulated by adding additional base pair steps to the RNA duplex, moving the r(R·U) pair from i+1 to i+5 position while keeping constant the reactive alignments at the i position. These structures allowed us to trace the sliding of the nascent RNA duplex along the exit path and check for potential reasons for the R-induced delayed termination of the polymerization reaction.
Human RNA Pol II complex setup
The X-ray structure of human RNA Pol II (PDB: 5FLM)53 consisting on a polymerase protein complex (composed on 12 subunits), a DNA template, and an RNA transcript were taken and submitted to further modeling. In order to obtain a fully processive protein enzyme, we aligned the aforementioned human RNA Pol II, whose trigger loop is in an opened conformation, to an RNA Pol II X-ray structure (PDB: 2E2H and 2E2J)54 in the “closed” state. Finally, we extracted one Mg2+ cation and the triphosphate moiety, absents in the X-ray structure (PDB: 5FLM), from the closed-state X-ray structures (PDB: 2E2H and 2E2J) and introduced them to the final model of human RNA Pol II. The final structure was subjected to the same equilibration protocol as described above (see supplemental experimental procedures).
MD simulations on RdRp and human RNA Pol II
Classical trajectories were used to refine and check the stability of complexes prior to running QM/MM simulations, as well as to determine the binding mode and perform free-energy calculations. Minimization, thermalization, and equilibration were performed using standard procedures, as described in supplemental experimental procedures. Production simulations were carried out using the AMBER 19 program55 and state-of-the-art conditions for a total time of at least 0.5 μs. Water molecules were described through the TIP3P56 model, parameters of magnesium ions were taken from Allner et al.,57 and Carlson et al.58 parameters were used for triphosphate groups, PARMBSC1 for DNA,59 PARMBSC0-chiOL3 for RNA,60, 61, 62, 63 and ff14SB64 for the proteins. Parameters and charges of ATP, UTP, RTP, remdesivir nucleotide, and 3′ terminal nucleotides and R were derived to be compatible with the force fields making use of the RED server.65 Additional details of the simulation setups can be found in supplemental experimental procedures and Figure S17.
QM/MM exploration of the minimum free-energy paths and potential of mean force
QM subsystems used in reactivity calculations for RdRp and RNA Pol II are shown in Figure S18. The link-atom method was employed to join QM and MM regions.55 The hybrid QM/MM models were built using randomly selected snapshots obtained in the last ns of unrestrained MD simulations, which were then minimized and re-equilibrated at a QM/MM-hybrid level of theory.
QM/MM-MD simulations were performed to obtain minimum free-energy paths (MFEPs) by means of the string method.66,67 This method allowed us to explore different reaction mechanisms and select the preferred one in terms of free energy. Sixty to one-hundred twenty string nodes were used. Afterward, a path collective variable (CV)67 was defined to obtain the potential of mean force (PMF) using 60 to 120 umbrella sampling68 windows. The chosen set of CVs that followed the progress of the reactions and the breaking and forming bonds are shown in Figures 1B and 3A and supplemental experimental procedures (see Figure S18). MFEPs were obtained at the DFTB369,70/MM level, and the PMFs were corrected at the B3LYP/6-311++G∗∗ level. Each of the sampling windows consisted on 20 ps of equilibration followed by 200 ps of production. We checked that the length of the production for the PMFs was sufficient to reduce the statistical error to the order of 1 kcal mol−1. The error of all free-energy barriers and profiles was calculated as 95% confidence intervals and reached error values within ±1 kcal mol−1. The AMBER program55 with electrostatic embedding was used for the QM/MM calculations. Corrections at the high level of theory were made with Gaussian1671 (see supplemental experimental procedures).
Binding free-energy calculations
The difference binding free energy of ATP and RTP to polymerases was determined by computing the differences in the free energy associated with the ATP/RTP change in protein-complexed and isolated states.72 The human and viral polymerases, complexed with an alchemical ATP/RTP residue in the binding site, were simulated in NPT conditions for 250 ns in both physical endpoints corresponding to ATP and RTP, and the initial 50 ns were considered equilibration. As a reference state, a single solvated ATP/RTP residue complexed with Mg2+ was simulated in the same manner. The resulting 200 ns trajectories were then used to extract seeding frames for the non-equilibrium free-energy protocol based on the Crooks theorem.73 To obtain the binding free energies, 200 short 1 ns simulations were launched in each direction, with λ changing steadily from 0 to 1 or vice versa. Values of non-equilibrium work were computed for each run and converted to free energies using the Bennett acceptance ratio (BAR) method,74 all using an in-house implementation of the protocol (gitlab.com/KomBioMol/crooks).
Free energy of remdesivir progression
To verify whether steric hindrance could be responsible for the stalling of RNA extension by RdRP, we performed alchemical free-energy simulations in which an adenine was mutated into remdesivir within the poly-A RNA duplex at four different positions (from i+2 to i+5). A non-equilibrium free-energy protocol virtually identical to the one outlined above was used to obtain relative free energies.
MD simulations on nascent RNA
We performed MD simulations on two duplexes, r(CGCGAAUUGCGC)·r(GCGCAAUUCGCG) and r(CGCGARUUGCGC)·r(GCGCAAUUCGCG), to determine the structural impact of the introduction of a remdesivir in a canonical RNA duplex. Starting structures were those expected for a canonical RNA duplex as implemented in AMBER. Systems were hydrated, minimized, thermalized, and equilibrated using standard protocols75,76 before MD were performed at constant temperature (T = 298 K) and pressure (p = 1 atm). A total of 5 μs cumulative simulation time was sampled for each system. Details of simulations are shown in supplemental experimental procedures.
Acknowledgments
This work has been supported by the Spanish Ministry of Science (BFU2014-61670-EXP), the Catalan SGR, the Instituto Nacional de Bioinformática, the European Research Council (ERC SimDNA), the European Union’s Horizon 2020 research and innovation program under grant agreement no. 676556, the Biomolecular and Bioinformatics Resources Platform (ISCIII PT 13/0001/0030) cofunded by the Fondo Europeo de Desarrollo Regional (FEDER), and the MINECO Severo Ochoa Award of Excellence (Government of Spain) (awarded to IRB Barcelona). M.O. is an ICREA academia researcher. J.A. acknowledges the Spanish Ministry of Science for a Juan de la Cierva post-doctoral grant. J.A. thanks Dr. K. Zinovjev for the string-method technical support. J.A. thanks the support team of the MareNostrum supercomputer and the support team at IRB Barcelona, specially R. Ramos. M.T. thanks the Instituto de Salud Carlos III for a Miguel Servet for a grant (CPII18/00032). Calculations were performed on the MareNostrum supercomputer at the Barcelona Supercomputer Center and in MMB cluster. M.O. and J.A. thank Prof. Núria López-Bigas and Dr. Francisco Martínez-Jiménez for their help in filtering data of mutations.
Author contributions
J.A. and M.O. conceived the project. J.A. conceptualized the goals and aims of the project. J.A. and M.W. performed simulations and analyzed the data. J.A. and M.O. supervised the project. M.O. acquired funding support for this project. J.A. prepared the initial manuscript. M.O., M.W., and J.A. reviewed and edited the initial manuscript and provided critical commentary and revisions. J.A. and M.O. prepared the final versions of the manuscript. M.T. and I.B.-H. commented on and discussed the experimental support of the project. All authors discussed, commented on, and revised the final version of the manuscript.
Declaration of interests
The authors declare no competing interests.
Published: April 19, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.checat.2022.03.019.
Contributor Information
Juan Aranda, Email: juan.aranda@irbbarcelona.org.
Modesto Orozco, Email: modesto.orozco@irbbarcelona.org.
Supplemental information
Data and code availability
The data that support the plots within this paper are available from the corresponding authors upon request. Trajectories will be deposited in the BIOEXCEL-COVID-19 database: https://bioexcel-cv19.bsc.es and will be accesible upon publication.
References
- 1.WHO . WHO; 2021. Novel Coronavirus (COVID-19) Situation. [Google Scholar]
- 2.De Wit E., Van Doremalen N., Falzarano D., Munster V.J. SARS and MERS: recent insights into emerging coronaviruses. Nat. Rev. Microbiol. 2016;14:523–534. doi: 10.1038/nrmicro.2016.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Robson F., Khan K.S., Le T.K., Paris C., Demirbag S., Barfuss P., Rocchi P., Ng W.L. Coronavirus RNA proofreading: molecular basis and therapeutic targeting. Mol. Cell. 2020;79:710–727. doi: 10.1016/j.molcel.2020.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hadfield J., Megill C., Bell S.M., Huddleston J., Potter B., Callender C., Sagulenko P., Bedford T., Neher R.A. Genomic epidemiology of novel coronavirus - global subsampling. Nextstrain real-time Track. Pathog. Evol. 2020;26:2854. [Google Scholar]
- 5.Lu R., Zhao X., Li J., Niu P., Yang B., Wu H., Wang W., Song H., Huang B., Zhu N., et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395:565–574. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rausch J.W., Capoferri A.A., Katusiime M.G., Patro S.C., Kearney M.F. Low genetic diversitymay be an Achilles heel of SARS-CoV-2. Proc. Natl. Acad. Sci. U S A. 2020;117:24614–24616. doi: 10.1073/pnas.2017726117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Peng Q., Peng R., Yuan B., Zhao J., Wang M., Wang X., Wang Q., Sun Y., Fan Z., Qi J., et al. Structural and biochemical characterization of the nsp12-nsp7-nsp8 core polymerase complex from SARS-CoV-2. Cell Rep. 2020;31:107774. doi: 10.1016/j.celrep.2020.107774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gao Y., Yan L., Huang Y., Liu F., Zhao Y., Cao L., Wang T., Sun Q., Ming Z., Zhang L., et al. Structure of the RNA-dependent RNA polymerase from COVID-19 virus. Science. 2020;368:779–782. doi: 10.1126/science.abb7498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hillen H.S., Kokic G., Farnung L., Dienemann C., Tegunov D., Cramer P. Structure of replicating SARS-CoV-2 polymerase. Nature. 2020;584:154–156. doi: 10.1038/s41586-020-2368-8. [DOI] [PubMed] [Google Scholar]
- 10.Romano M., Ruggiero A., Squeglia F., Maga G., Berisio R. A structural view of SARS-CoV-2 RNA replication machinery: RNA synthesis, proofreading and final capping. Cells. 2020;9:1267. doi: 10.3390/cells9051267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.V’kovski P., Kratzel A., Steiner S., Stalder H., Thiel V. Coronavirus biology and replication: implications for SARS-CoV-2. Nat. Rev. Microbiol. 2020;193:155–170. doi: 10.1038/s41579-020-00468-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McGinnis S., Madden T.L. BLAST: At the core of a powerful and diverse set of sequence analysis tools. Nucleic Acids Res. 2004;32:W20–W25. doi: 10.1093/nar/gkh435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gordon C.J., Tchesnokov E.P., Feng J.Y., Porter D.P., Götte M. The antiviral compound remdesivir potently inhibits RNAdependent RNA polymerase from Middle East respiratory syndrome coronavirus. J. Biol. Chem. 2020;295:4773–4779. doi: 10.1074/jbc.AC120.013056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gordon C.J., Tchesnokov E.P., Woolner E., Perry J.K., Feng J.Y., Porter D.P., Götte M. Remdesivir is a direct-acting antiviral that inhibits RNA-dependent RNA polymerase from severe acute respiratory syndrome coronavirus 2 with high potency. J. Biol. Chem. 2020;295:6785–6797. doi: 10.1074/jbc.RA120.013679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang H., Rao Z. Structural biology of SARS-CoV-2 and implications for therapeutic development. Nat. Rev. Microbiol. 2021;19:685–700. doi: 10.1038/s41579-021-00630-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.U.S. National Library of Medicine . U.S. National Library of Medicine; 2020. Expanded Access Treatment Protocol: Remdesivir (RDV; GS-5734) for the Treatment of SARS-CoV2 (CoV) Infection ( NCT04323761) [Google Scholar]
- 17. NCT03719586 Investigational Therapeutics for the Treatment of People with Ebola Virus Disease. 2018. https://clinicaltrials.gov/show/nct03719586
- 18.Yin W., Mao C., Luan X., Shen D.D., Shen Q., Su H., Wang X., Zhou F., Zhao W., Gao M., et al. Structural basis for inhibition of the RNA-dependent RNA polymerase from SARS-CoV-2 by remdesivir. Science. 2020;368:1499–1504. doi: 10.1126/science.abc1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang Q., Wu J., Wang H., Gao Y., Liu Q., Mu A., Ji W., Yan L., Zhu Y., Zhu C., et al. Structural basis for RNA replication by the SARS-CoV-2 polymerase. Cell. 2020;182:417–428.e13. doi: 10.1016/j.cell.2020.05.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Naydenova K., Muir K.W., Wu L.F., Zhang Z., Coscia F., Peet M.J., Castro-Hartmann P., Qian P., Sader K., Dent K., et al. Structure of the SARS-CoV-2 RNA-dependent RNA polymerase in the presence of favipiravir-RTP. Proc. Natl. Acad. Sci. U S A. 2021;118 doi: 10.1073/pnas.2021946118. e2021946118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Peng Q., Peng R., Yuan B., Wang M., Zhao J., Fu L., Qi J., Shi Y. Structural basis of SARS-CoV-2 polymerase inhibition by favipiravir. Innovation (N Y) 2021;2:100080. doi: 10.1016/j.xinn.2021.100080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Genna V., Vidossich P., Ippoliti E., Carloni P., Vivo M.D. A self-activated mechanism for nucleic acid polymerization catalyzed by DNA/RNA polymerases. J. Am. Chem. Soc. 2016;138:14592–14598. doi: 10.1021/jacs.6b05475. [DOI] [PubMed] [Google Scholar]
- 23.Gelpí J.L., Kalko S.G., Barril X., Cirera J., De La Cruz X., Luque F.J., Orozco M. Classical molecular interaction potentials: improved setup procedure in molecular dynamics simulations of proteins. Proteins Struct. Funct. Genet. 2001;45:428–437. doi: 10.1002/prot.1159. [DOI] [PubMed] [Google Scholar]
- 24.Tomasello G., Armenia I., Molla G. The Protein Imager: a full-featured online molecular viewer interface with server-side HQ-rendering capabilities. Bioinformatics. 2020;36:2909–2911. doi: 10.1093/bioinformatics/btaa009. [DOI] [PubMed] [Google Scholar]
- 25.Warren T.K., Jordan R., Lo M.K., Ray A.S., Mackman R.L., Soloveva V., Siegel D., Perron M., Bannister R., Hui H.C., et al. Therapeutic efficacy of the small molecule GS-5734 against Ebola virus in rhesus monkeys. Nature. 2016;531:381–385. doi: 10.1038/nature17180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Byléhn F., Menéndez C.A., Perez-Lemus G.R., Alvarado W., De Pablo J.J. Modeling the binding mechanism of remdesivir, favilavir, and ribavirin to SARS-CoV-2 RNA-dependent RNA polymerase. ACS Cent. Sci. 2021;7:164–174. doi: 10.1021/acscentsci.0c01242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wakchaure P.D., Ghosh S., Ganguly B. Revealing the inhibition mechanism of RNA-dependent RNA polymerase (RdRp) of SARS-CoV-2 by remdesivir and nucleotide analogues: a molecular dynamics simulation study. J. Phys. Chem. B. 2020;124:10641–10652. doi: 10.1021/acs.jpcb.0c06747. [DOI] [PubMed] [Google Scholar]
- 28.Stevens D.R., Hammes-Schiffer S. Exploring the role of the third active site metal ion in DNA polymerase η with QM/MM free energy simulations. J. Am. Chem. Soc. 2018;140:8965–8969. doi: 10.1021/jacs.8b05177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Genna V., Donati E., De Vivo M. The catalytic mechanism of DNA and RNA polymerases. ACS Catal. 2018;8:11103–11118. [Google Scholar]
- 30.Silva D.A., Weiss D.R., Avila F.P., Da L.T., Levitt M., Wang D., Huang X. Millisecond dynamics of RNA polymerase II translocation at atomic resolution. Proc. Natl. Acad. Sci. U S A. 2014;111:7665–7670. doi: 10.1073/pnas.1315751111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Genna V., Gaspari R., Dal Peraro M., De Vivo M. Cooperative motion of a key positively charged residue and metal ions for DNA replication catalyzed by human DNA polymerase-η. Nucleic Acids Res. 2016;44:2827–2836. doi: 10.1093/nar/gkw128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Da L.T., Wang D., Huang X. Dynamics of pyrophosphate ion release and its coupled trigger loop motion from closed to open state in RNA polymerase II. J. Am. Chem. Soc. 2012;134:2399–2406. doi: 10.1021/ja210656k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Aranda J., Terrazas M., Gómez H., Villegas N., Orozco M. An artificial DNAzyme RNA ligase shows a reaction mechanism resembling that of cellular polymerases. Nat. Catal. 2019;2:544–552. [Google Scholar]
- 34.Casalino L., Nierzwicki Ł., Jinek M., Palermo G. Catalytic mechanism of non-target DNA cleavage in CRISPR-cas9 revealed by ab initio molecular dynamics. ACS Catal. 2020;10:13596–13605. doi: 10.1021/acscatal.0c03566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Casalino L., Palermo G., Rothlisberger U., Magistrato A. Who activates the nucleophile in ribozyme catalysis? An answer from the splicing mechanism of group II introns. J. Am. Chem. Soc. 2016;138:10374–10377. doi: 10.1021/jacs.6b01363. [DOI] [PubMed] [Google Scholar]
- 36.Dangerfield T.L., Huang N.Z., Johnson K.A. Remdesivir is effective in combating COVID-19 because it is a better substrate than ATP for the viral RNA-dependent RNA polymerase. iScience. 2020;23:101849. doi: 10.1016/j.isci.2020.101849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kellinger M.W., Ulrich S., Chong J., Kool E.T., Wang D. Dissecting chemical interactions governing RNA polymerase II transcriptional fidelity. J. Am. Chem. Soc. 2012;134:8231–8240. doi: 10.1021/ja302077d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Geronimo I., Vidossich P., Donati E., De Vivo M. Computational investigations of polymerase enzymes: structure, function, inhibition, and biotechnology. Wiley Interdiscip. Rev. Comput. Mol. Sci. 2021;11:e1534. [Google Scholar]
- 39.Berdis A.J. Mechanisms of DNA polymerases. Chem. Rev. 2009;109:2862–2879. doi: 10.1021/cr800530b. [DOI] [PubMed] [Google Scholar]
- 40.Castro C., Smidansky E.D., Arnold J.J., Maksimchuk K.R., Moustafa I., Uchida A., Götte M., Konigsberg W., Cameron C.E. Nucleic acid polymerases use a general acid for nucleotidyl transfer. Nat. Struct. Mol. Biol. 2009;16:212–218. doi: 10.1038/nsmb.1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wu W.J., Yang W., Tsai M.D. How DNA polymerases catalyse replication and repair with contrasting fidelity. Nat. Rev. Chem. 2017;1:1–16. [Google Scholar]
- 42.Bravo J.P.K., Dangerfield T.L., Taylor D.W., Johnson K.A. Remdesivir is a delayed translocation inhibitor of SARS-CoV-2 replication. Mol. Cell. 2021;81:1548–1552.e4. doi: 10.1016/j.molcel.2021.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kokic G., Hillen H.S., Tegunov D., Dienemann C., Seitz F., Schmitzova J., Farnung L., Siewert A., Höbartner C., Cramer P. Mechanism of SARS-CoV-2 polymerase stalling by remdesivir. Nat. Commun. 2021;12:1–7. doi: 10.1038/s41467-020-20542-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tchesnokov E.P., Gordon C.J., Woolner E., Kocinkova D., Perry J.K., Feng J.Y., Porter D.P., Götte M. Template-dependent inhibition of coronavirus RNA-dependent RNA polymerase by remdesivir reveals a second mechanism of action. J. Biol. Chem. 2020;295:16156–16165. doi: 10.1074/jbc.AC120.015720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Seifert M., Bera S.C., van Nies P., Kirchdoerfer R.N., Shannon A., Le T.-T.-N., Meng X., Xia H., Wood J.M., Harris L.D., et al. Inhibition of SARS-CoV-2 polymerase by nucleotide analogs from a single-molecule perspective. Elife. 2021;10:e70968. doi: 10.7554/eLife.70968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Aranda J., Orozco M. RNA-dependent RNA polymerase from SARS-CoV-2. Mechanism of reaction and inhibition by remdesivir. bioRxiv. 2020 doi: 10.1101/2020.06.21.163592. Preprint at. [DOI] [Google Scholar]
- 47.Moon J.B., Coleman R.S., Hanzlik R.P. Reversible covalent inhibition of papain by a peptide nitrile. 13C NMR evidence for a thioimidate ester adduct. J. Am. Chem. Soc. 1986;108:1350–1351. [Google Scholar]
- 48.Kalgutkar A.S., Dalvie D.K. Drug discovery for a new generation of covalent drugs. Expert Opin. Drug Discov. 2012;7:561–581. doi: 10.1517/17460441.2012.688744. [DOI] [PubMed] [Google Scholar]
- 49.Devkota A.K., Edupuganti R., Yan C., Shi Y., Jose J., Wang Q., Kaoud T.S., Cho E.J., Ren P., Dalby K.N. Reversible covalent inhibition of eEF-2K by carbonitriles. ChemBioChem. 2014;15:2435–2442. doi: 10.1002/cbic.201402321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Berteotti A., Vacondio F., Lodola A., Bassi M., Silva C., Mor M., Cavalli A. Predicting the reactivity of nitrile-carrying compounds with cysteine: a combined computational and experimental study. ACS Med. Chem. Lett. 2014;5:501–505. doi: 10.1021/ml400489b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Appleby T.C., Perry J.K., Murakami E., Barauskas O., Feng J., Cho A., Fox D., Wetmore D.R., McGrath M.E., Ray A.S., et al. Structural basis for RNA replication by the hepatitis C virus polymerase. Science. 2015;347:771–775. doi: 10.1126/science.1259210. [DOI] [PubMed] [Google Scholar]
- 52.Kirchdoerfer R.N., Ward A.B. Structure of the SARS-CoV nsp12 polymerase bound to nsp7 and nsp8 co-factors. Nat. Commun. 2019;10:1–9. doi: 10.1038/s41467-019-10280-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bernecky C., Herzog F., Baumeister W., Plitzko J.M., Cramer P. Structure of transcribing mammalian RNA polymerase II. Nature. 2016;529:551–554. doi: 10.1038/nature16482. [DOI] [PubMed] [Google Scholar]
- 54.Wang D., Bushnell D.A., Westover K.D., Kaplan C.D., Kornberg R.D. Structural basis of transcription: role of the trigger loop in substrate specificity and catalysis. Cell. 2006;127:941–954. doi: 10.1016/j.cell.2006.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Case D.A., Babin V., Berryman J.T., Betz R.M., Cai Q., Cerutti D.S., Cheatham T.E., Darden T.A., Duke R.E., Gohlke H., et al. University of California; 2019. Amber 19. [Google Scholar]
- 56.Jorgensen W.L., Chandrasekhar J., Madura J.D., Impey R.W., Klein M.L. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983;79:926. [Google Scholar]
- 57.Allnér O., Nilsson L., Villa A. Magnesium ion-water coordination and exchange in biomolecular simulations. J. Chem. Theor. Comput. 2012;8:1493–1502. doi: 10.1021/ct3000734. [DOI] [PubMed] [Google Scholar]
- 58.Meagher K.L., Redman L.T., Carlson H.A. Development of polyphosphate parameters for use with the AMBER force field. J. Comput. Chem. 2003;24:1016–1025. doi: 10.1002/jcc.10262. [DOI] [PubMed] [Google Scholar]
- 59.Ivani I., Dans P.D., Noy A., Pérez A., Faustino I., Hospital A., Walther J., Andrio P., Goñi R., Balaceanu A., et al. Parmbsc1: a refined force field for DNA simulations. Nat. Methods. 2015;13:55–58. doi: 10.1038/nmeth.3658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cheatham T.E., Cieplak P., Kollman P.A. A modified version of the Cornell et al. Force field with improved sugar pucker phases and helical repeat. J. Biomol. Struct. Dyn. 1999;16:845–862. doi: 10.1080/07391102.1999.10508297. [DOI] [PubMed] [Google Scholar]
- 61.Wang J., Cieplak P., Kollman P.A. How well does a restrained electrostatic potential (RESP) model perform in calculating conformational energies of organic and biological molecules? J. Comput. Chem. 2000;21:1049–1074. [Google Scholar]
- 62.Pérez A., Marchán I., Svozil D., Sponer J., Cheatham T.E., Laughton C.A., Orozco M. Refinement of the AMBER force field for nucleic acids: improving the description of α/γ conformers. Biophys. J. 2007;92:3817–3829. doi: 10.1529/biophysj.106.097782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zgarbová M., Otyepka M., Šponer J., Mládek A., Banáš P., Cheatham T.E., Jurečka P. Refinement of the Cornell et al. Nucleic acids force field based on reference quantum chemical calculations of glycosidic torsion profiles. J. Chem. Theor. Comput. 2011;7:2886–2902. doi: 10.1021/ct200162x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Maier J.A., Martinez C., Kasavajhala K., Wickstrom L., Hauser K.E., Simmerling C. ff14SB: improving the accuracy of protein side chain and backbone parameters from ff99SB. J. Chem. Theor. Comput. 2015;11:3696–3713. doi: 10.1021/acs.jctc.5b00255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Vanquelef E., Simon S., Marquant G., Garcia E., Klimerak G., Delepine J.C., Cieplak P., Dupradeau F.Y. R.E.D. Server: a web service for deriving RESP and ESP charges and building force field libraries for new molecules and molecular fragments. Nucleic Acids Res. 2011;39:W511–W517. doi: 10.1093/nar/gkr288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Vanden-Eijnden E., Venturoli M. Revisiting the finite temperature string method for the calculation of reaction tubes and free energies. J. Chem. Phys. 2009;130:05B605. doi: 10.1063/1.3130083. [DOI] [PubMed] [Google Scholar]
- 67.Zinovjev K., Tuñón I. Adaptive finite temperature string method in collective variables. J. Phys. Chem. A. 2017;121:9764–9772. doi: 10.1021/acs.jpca.7b10842. [DOI] [PubMed] [Google Scholar]
- 68.Torrie G.M., Valleau J.P. Nonphysical sampling distributions in Monte Carlo free-energy estimation: umbrella sampling. J. Comput. Phys. 1977;23:187–199. [Google Scholar]
- 69.Gaus M., Cui Q., Elstner M. DFTB3: extension of the self-consistent-charge density-functional tight-binding method (SCC-DFTB) J. Chem. Theor. Comput. 2011;7:931–948. doi: 10.1021/ct100684s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gaus M., Lu X., Elstner M., Cui Q. Parameterization of DFTB3/3OB for sulfur and phosphorus for chemical and biological applications. J. Chem. Theor. Comput. 2014;10:1518–1537. doi: 10.1021/ct401002w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Frisch G.W., Schlegel H.B., Scuseria G.E., Robb M.A., Cheeseman J.R., Scalmani G., Barone V., Petersson G.A., Nakatsuji H., Li X., et al. Gaussian, Inc.; 2016. Gaussian 16, Rev. B.01. [Google Scholar]
- 72.Lybrand T.P., McCammon J.A., Wipff G. Theoretical calculation of relative binding affinity in host-guest systems. Proc. Natl. Acad. Sci. U S A. 1986;83:833–835. doi: 10.1073/pnas.83.4.833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Crooks G.E. Entropy production fluctuation theorem and the nonequilibrium work relation for free energy differences. Phys. Rev. E. 1999;60:2721. doi: 10.1103/physreve.60.2721. [DOI] [PubMed] [Google Scholar]
- 74.Bennett C.H. Efficient estimation of free energy differences from Monte Carlo data. J. Comput. Phys. 1976;22:245–268. [Google Scholar]
- 75.Dans P.D., Walther J., Gómez H., Orozco M. Multiscale simulation of DNA. Curr. Opin. Struct. Biol. 2016;37:29–45. doi: 10.1016/j.sbi.2015.11.011. [DOI] [PubMed] [Google Scholar]
- 76.Dans P.D., Ivani I., Hospital A., Portella G., González C., Orozco M. How accurate are accurate force-fields for B-DNA? Nucleic Acids Res. 2017;45:4217–4230. doi: 10.1093/nar/gkw1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the plots within this paper are available from the corresponding authors upon request. Trajectories will be deposited in the BIOEXCEL-COVID-19 database: https://bioexcel-cv19.bsc.es and will be accesible upon publication.