ABSTRACT
Endogenous retroviruses (ERVs) are fossils left in our genome from retrovirus infections of the past. Their sequences are part of every vertebrate genome and their random integrations are thought to have contributed to evolution. Although ERVs are mainly silenced by the host genome, they have been found to be activated in multiple disease states, such as auto-inflammatory disorders and neurological diseases. However, the numerous copies in mammalian genomes and the lack of tools to study them make defining their role in health and diseases challenging. In this study, we identified eight copies of the zebrafish endogenous retrovirus zferv. We created and characterised the first in vivo ERV reporter line in any species. Using a combination of live imaging, flow cytometry and single-cell RNA sequencing, we mapped zferv expression to early T cells and neurons. Thus, this new tool identified tissues expressing ERV in zebrafish, highlighting a potential role of ERV during brain development and strengthening the hypothesis that ERV play a role in immunity and neurological diseases. This transgenic line is therefore a suitable tool to study the function of ERV in health and diseases.
KEY WORDS: Retroelement, Zebrafish, Endogenous retrovirus, Reporter line, zferv, LTR
Summary: Characterisation of the zebrafish endogenous retrovirus zferv reveals expression in early T cells and neurons, highlighting a potential role of endogenous retroviruses in brain development and immune disorders.
INTRODUCTION
Over 40% of the human genome consists of endogenous transposable elements capable of recombination and disruption of genes and modification of their expression (Lander et al., 2001). Endogenous retroviruses (ERVs) are transposable elements originating from old integrations of retroviruses so successful that they have become part of most vertebrate genomes studied. ERVs replicate autonomously using a ‘copy-and-paste’ mechanism and, although they constitute a smaller percentage of all retroelements, they still represent 8% of the human genome (Bourque et al., 2018; Lander et al., 2001). The majority of known ERVs have lost their ability to replicate, but those most recently acquired still have intact genomes with the ability to produce viral RNA and particles. However, many competent ERVs are under strict transcriptional suppression by epigenetic mechanisms (Maksakova et al., 2008; Rowe et al., 2010; Turelli et al., 2014), protecting the host organisms against potential retroviral insertions and viral activities.
Although immobilised by mutations or transcriptionally repressed, ERVs have a complex relationship with the human genome, which they can regulate by providing cis-regulatory elements to surrounding genes and by lifting their transcriptionally repressed state. Through these mechanisms, it is believed that transposable elements have fuelled some of the necessary genetic changes for evolution (Feschotte, 2008; Kunarso et al., 2010). Syncytin-1, an ERV envelope gene essential for the vascularisation of the placenta, is at the origin of evolutionary diversification of the placenta (Chuong, 2018; Mi et al., 2000). During human embryogenesis, expression of specific ERV families have been associated with cell identity and cell potency in early stem cells (Göke et al., 2015). Additionally, ERV expression has been reported in healthy human tissues, such as ovary and testis for ERV-9 (Pi et al., 2004), pancreas (Shiroma et al., 2001), breast (Tavakolian et al., 2019), stomach and small intestine (Okahara et al., 2004). Mainly based on transcriptional studies, the expression of different families of ERV is likely to be extended to more tissues; however, their role in tissue development and function remains largely unknown.
ERVs have been linked directly and indirectly to the evolution and functioning of the immune system. Enhancer regions of interferon-stimulated genes key to the interferon pathway, such as IRF1 and STAT1, were found introduced and amplified by ERV elements, with the human inflammasome failing to form upon the deletion of a subset of ERVs (Chuong et al., 2016). Adaptive immunity also benefits from the presence of ERVs. Indeed, ERV peptide recognition is used in T-cell selection to optimise antigen recognition and T cells have a higher sensitivity to exogenous virus infection when presented with ERV peptides during their initial thymic selection (Mandl et al., 2013; Young et al., 2012). The human ERV (HERV) envelope gene contains immunosuppressive domains that reduce the Th1 response during pregnancy and therefore promote foetal development (Knerr et al., 2004; Lokossou et al., 2020). The role of ERVs in our immune system, particularly in the training of our adaptive immunity, can be a double-edged sword as ERVs are linked to a range of different disease states, including autoimmunity.
Aberrant expression of ERVs contributes to multiple pathologies. ERVs are found in abundance in multiple forms of cancers and are considered tumour-promoting factors (extensively reviewed by Bermejo et al., 2020). The pathology of autoinflammatory diseases has also been strongly associated with ERVs. Systemic lupus erythematosus (SLE) is an autoimmune disorder with increased levels of autoantigen for an ERV (Baudino et al., 2014). Recently, one murine SLE susceptibility locus was identified as a key suppressor of ERV expression, consolidating the role of ERVs in the pathogenesis of SLE (Treger et al., 2019). A similar disorder is Aicardi–Goutières Syndrome (AGS), a type 1 interferonopathy that resembles congenital cytomegalovirus infection and is caused by mutations in several genes encoding enzymes responsible for nucleic acid metabolism (Crow et al., 2015). Mutations in some of these genes, such as TREX1, MDA5 (IFIH1) and ADAR1 (ADAR), trigger an antiviral immune response due to self-reactivity of retroelements (Ahmad et al., 2018; Benitez-Guijarro et al., 2018; Chung et al., 2018; Li et al., 2017; Thomas et al., 2017). Interestingly, anti-reverse transcriptase therapy in AGS patients can decrease the interferon response, highlighting the role of aberrant presence of ERVs in triggering an immune response (Rice et al., 2018). Increased expression of ERVs has been found in brains of patient suffering from neurodegenerative diseases, such as motor neuron disease (Li et al., 2015) and multiple sclerosis (MS) (Johnston et al., 2001; Mameli et al., 2007). Overexpression of a human ERV in neurons was shown to be neurotoxic, suggesting a potential role of ERVs in triggering neuronal toxicity (Li et al., 2015). A direct link to the pathology of these disorders has yet to be made, but, nonetheless, ERVs appear to be strong causal factors for autoimmune and neurological disorders.
Although ERV enrichment has been detected in neurological pathologies, little is known about the function of ERV in healthy tissues. The exact role of ERV in our immune system and brain pathologies has yet to be understood and there is no model system specifically looking at ERV function in vivo. Zebrafish is already established as a model to study the immune system, with significant homology with mammals in innate and adaptive immunity (Renshaw and Trede, 2012; Trede et al., 2004). The genetic tractability and transparency of the zebrafish embryos have allowed the creation of transgenic reporter lines, some of which have elucidated the role of immune cell behaviour in vivo (Renshaw et al., 2006). The tractability of the zebrafish has already been exploited to visualise the expression of the human ERV-9 in oocytes, similarly to human expression (Pi et al., 2004).
In this study, we used the zebrafish as a tractable in vivo model to characterise the zebrafish endogenous retrovirus zferv. We identified multiple zferv family members, including two complete genomes, we named zferv1a and zferv1b. Using the transparency of the zebrafish larvae, we developed a reporter line for zferv1a and imaged for the first time ERV activation in healthy tissues in vivo. We showed that zferv1a is expressed in the thymus and in the brain. Colocalisation analysis and single-cell RNA sequencing revealed expression of zferv1a specifically in T cells, suggesting a potential role for ERV in lymphocyte function or development. Brain expression of zferv1a appears to include neuronal expression. This transgenic line can be used as a tool to investigate further the role of ERVs in immunity and in neurological disorders.
RESULTS
The zebrafish genome contains multiple endogenous retrovirus integrations
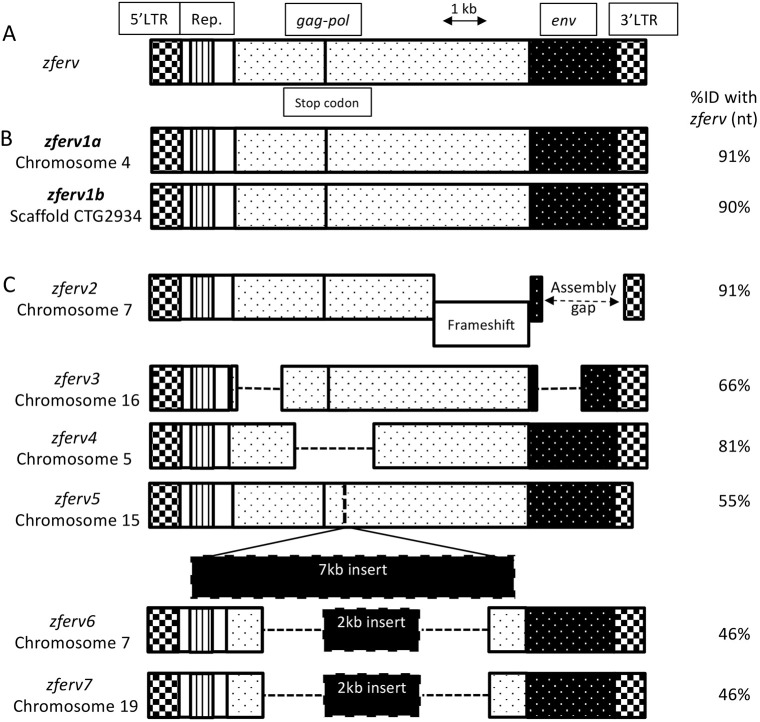
The presence of an ERV in zebrafish, named zferv, was reported by Shen and Steiner while screening a thymic cDNA library (Shen and Steiner, 2004) (Fig. 1A). We searched for related sequences in the most recent reference zebrafish genome (GRCz11, Tü strain) using BLASTN searches, with the original zferv sequence as a query. Limiting ourselves to sequences flanked by long terminal repeats (LTRs) on both sides, we retrieved eight hits scattered in the zebrafish genome, but not the exact zferv sequence, possibly because of strain difference (Fig. 1, Table S1). We identified two sequences encoding apparently fully functional ERVs, with 91% and 90% identity to zferv, which we named zferv1a and zferv1b, respectively (Fig. 1B). These two ERVs encode almost identical proteins (95-97% identity at the amino-acid level) and have highly homologous LTR promoter regions (95% identity at the nucleotide level). An additional six pseudo zferv genes (here called zferv2 to zferv7) were identified (Fig. 1C), zferv2 containing a frameshift, zferv3 and zferv4 with large deletions, and zferv5, zferv6 and zferv7 containing a large insertion, all resulting in a degenerated ERV genome.
Fig. 1.
Multiple copies of zferv are present in the zebrafish genome. (A) Diagram of the original zferv genes found by Shen and Steiner (2004) used as a reference for nucleotide identity (ID% nt). (B) Diagram representing the two closest related zferv genes found in most recent zebrafish genome GRCz11, named zferv1a and zferv1b. (C) Diagram of six pseudo zferv genes with degenerated genome (dashed line represents insertions). 3′LTR, 3′ long terminal repeat; 5′LTR, 5′ long terminal repeat; env, envelope gene; gag-pol, genes encoding polyprotein and reverse transcriptase; Rep., repetitive element.
zferv1a is actively expressed in the brain and in the thymus
zferv expression was initially found in the thymus at 5 days post-fertilisation (dpf) using an RNA probe against the envelope (env) gene of the original zferv (Shen and Steiner, 2004). To verify this observation, the entire zferv1a genome was subcloned from a fosmid provided by the Sanger Center (Cambridge, UK) and used to create an in situ hybridisation (ISH) RNA probe against the envelope gene of zferv1a, here called env1a. ISH performed on a time course starting from two-cell-stage embryos until 5 dpf confirmed a strong signal in the thymus at 5 dpf (Fig. 2A), similarly to what was previously reported (Shen and Steiner, 2004). Individually labelled cells were visible around the thymus following the branchial arches and around the ear (Fig. 2A, black box). Additionally, we identified a signal at an earlier time point during gastrulation and a clear signal in the brain, which was particularly strong at 3 dpf (Fig. 2A). Neuromasts were labelled after 24 h exposure to the probe (Fig. S1). There was some signal in the thymus using the sense probe signal, suggesting some bidirectional expression of zferv1a, as reported for some human ERVs (Chiappinelli et al., 2015).
Fig. 2.
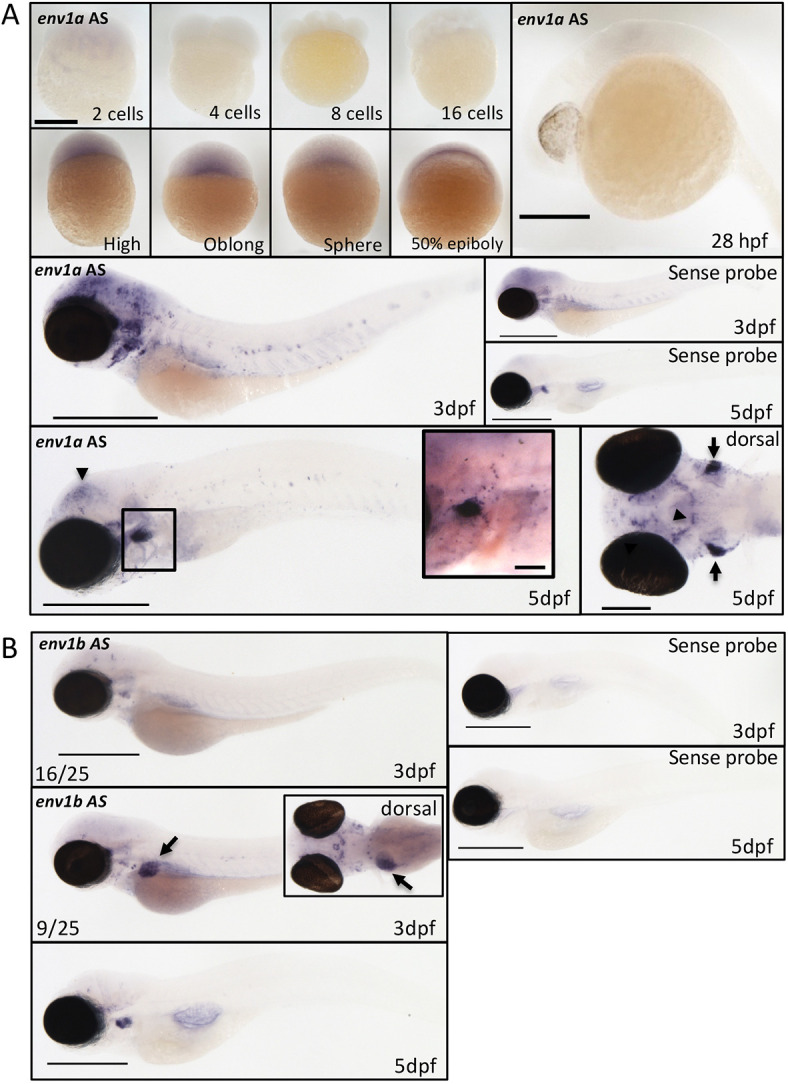
Reporter line for zferv1a recapitulates endogenous expression. (A) Expression of the envelope gene (env) of zferv1a (called env1a here) using antisense (AS) and sense in situ hybridisation RNA probes from the two-cell stage until 5 dpf (black arrowheads point to brain expression). Scale bars: 500 µm unless specified otherwise. Bottom left panel shows dorsal view of env1aAS expression in the brain (black arrowheads) and thymus (black arrows). Scale bar: 200 µm. Inset shows a magnified image of the thymus area (boxed), highlighting strong expression around the thymus with single positive cells visible in the vicinity of the thymus around the ear and alongside the branchial arches. Scale bar: 70 µm. (B) Expression of the envelope gene (env) of zferv1b (called env1b AS here) by in situ hybridisation at 3 dpf and 5 dpf. Liver expression is highlighted by black arrows. Inset shows dorsal view of the asymmetric liver expression. Scale bars: 500 µm.
To ensure the specificity of this probe to the zferv1a gene, we created a probe against the reverse transcriptase gene of zferv1a (pol1a) and showed similar labelling of the brain and thymus (Fig. S2). We additionally created a probe against another zferv isoform, using specific primers (Fig. S3) to the envelope gene of zferv1b (called here env1b). Although we still identified a signal in the thymus at 5 dpf, we observed a different expression pattern from env1a with no brain expression and a strong labelling of the liver in some embryos at 3 dpf (Fig. 2B).
zferv1a reporter line recapitulates endogenous expression
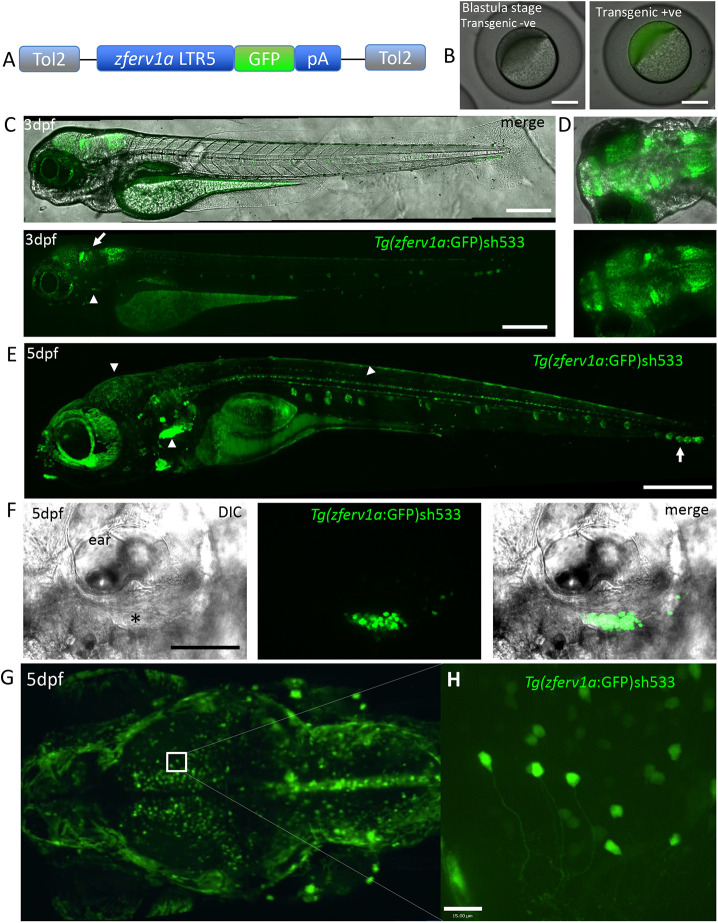
To follow the expression of the zebrafish endogenous retrovirus we identified as zferv1a, we took a transgenic approach taking advantage of the promoter activity of retroviral LTR. We used the 5′ viral promoter ltr5 from zferv1a to drive GFP expression by Gateway recombination (Fig. 3A). Injected embryos showing expression in their thymus were raised and screened in adulthood for germline transmission; one founder was selected to generate the transgenic line bearing the sh533 allele, which was analysed at the F2 generation. Comparable to observations by ISH, transgenic embryos (obtained from transgenic mothers) displayed GFP signal at the blastula stage and developed a strong GFP signal in the brain by 3 dpf (Fig. 3C,D). GFP signal observed in the eye was caused by the reflective nature of the retina and not by expression of the transgene, as shown using illumination from different laser also highlighting the eye and in situ hybridisation labelling with the env1a probe in a 5 dpf embryo with pigment partially removed in the eye using a tyrosinase crispant (Fig. S4). A weak signal in the thymus could be observed at 3 dpf (Fig. 3C, white arrowheads) as the thymus started to develop. At 5 dpf, a strong signal was observed in the thymus (Fig. 3E,F), comparable to that observed by ISH (Fig. 2A). Consistent with ISH, our reporter line also labelled the optic tectum region of the brain and the spinal cord at 5 dpf, with axons labelled in individual neurons (Fig. 3G,H). GFP signal was also found in neuromasts (Fig. 3E, white arrow), which we also observed in a small proportion of 5 dpf embryos by ISH with a longer development time (Fig. S1). The transmission rate of 50% to F3 embryos of the brain signal was variable depending on which F2 animal was used, including complete and partial brain labelling, whereas the thymic expression remained constant throughout the generations.
Fig. 3.
Reporter line for zferv1a recapitulates endogenous expression. (A) Diagram of the Gateway construct used to create the Tg(zferv1a:GFP) reporter line using the pDestCryCFP Tol2 backbone. (B) Representative images of non-transgenic and Tg(zferv1a:GFP:pA)sh533 transgenic embryos at the blastula stage. (C) Lateral view of DIC and GFP high-resolution images of a 3 dpf Tg(zferv1a:GFP:pA)sh533 embryo, displaying high expression of the transgene in the brain (white arrow) and the start of a signal in the thymus (white arrowheads). Scale bars: 400 µm. (D) Dorsal view of the brain at 3 dpf, showing GFP expression alone or merged with brightfield image. (E) High-resolution image of a 5 dpf Tg(zferv1a:GFP:pA)sh533 embryo, displaying high expression of the transgene in the thymus, brain and spinal cord (white arrowheads). Note the signal in neuromasts alongside the trunk (white arrow). Scale bar: 500 µm. (F) Single-slice DIC and confocal GFP fluorescence merged image of the thymus (black asterisk) situated underneath the ear in 5 dpf embryos and the signal from the Tg(zferv1a:GFP)sh533 reporter. Scale bar: 100 µm. (G) Dorsal view of a maximum projection of the brain from a 5 dpf Tg(zferv1a:GFP) embryo acquired with a lightsheet microscope. (H) High-resolution single-slice image from the optic tectum of the same Tg(zferv1a:GFP) embryo. Scale bar: 15 µm.
This transgenic line was not the first attempt to create a fluorescent reporter for zferv1a. Our initial reporter line was made using both 5′ (ltr5) and 3′ (ltr3) promoter regions in order to resemble the genome structure of an ERV. This line, called Tg(ltr5:GFP:ltr3)sh500, contained the CryCFP eye marker to help with selection of positive embryos. Although outcrossed F1 animals showed a normal transmission rate (50% of positive embryos in F2, with CFP in lens and GFP in thymus), the F2 transmission rate became unstable. Selected F2-positive embryos grown to adulthood failed to transmit the same signal pattern, with often reduced or complete loss of thymus labelling and random brain expression pattern despite transmission of the CFP lens phenotype at expected Mendelian ratios. We also observed a dampening of the GFP signal in F3 and complete loss in F4 animals. This suggested that the transgene was behaving as a transposable element and was ultimately silenced by the host or lost in the germline. This line was abandoned, and we continued our study with the sh533 line.
Zebrafish endogenous retrovirus zferv1a expression is restricted to lymphoid cells
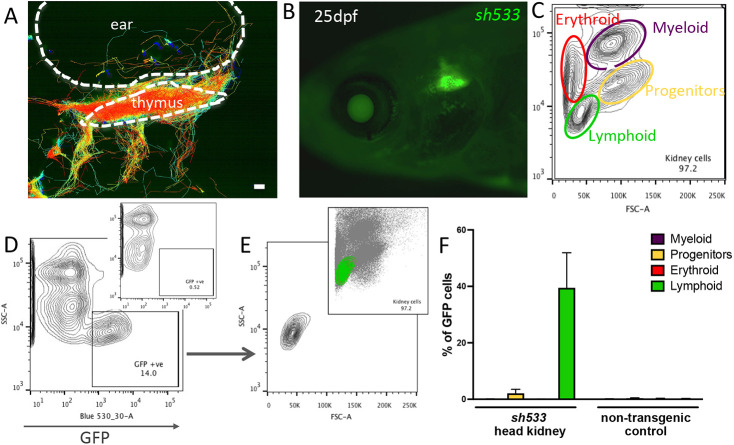
To study further the nature of the cells labelled in the thymus in the sh533 line, we took advantage of the transparency and ease of access of the thymic tissue to perform live imaging. Cell tracking of time-lapse imaging of the sh533 line revealed a dynamic behaviour of GFP-labelled cells, approaching and exiting the thymus following the ear and the branchial arches (Fig. 4A, Movie 1), as previously described for thymocytes (Dee et al., 2016; Kissa et al., 2008). This thymic signal persisted beyond embryonic stages and was still visible in juveniles at 25 dpf (Fig. 4B). To analyse the origin of the zferv1a cell population, we dissociated three head kidneys from Tg(zferv1a:GFP)sh533 and non-transgenic control adults and used size and granularity scatter by flow cytometry to distinguish between the various haematopoietic lineages (Fig. 4C). According to these criteria and using a back-gating method for quantification (Fig. 4D,E, Fig. S5), we found that GFP-positive cells have a typical lymphocyte profile (Fig. 4F).
Fig. 4.
zferv1a is expressed in cells from the lymphoid lineage in adult haematopoietic tissues. (A) Cell tracking reveals dynamic behaviour of entry and exit of the thymus by GFP-positive cells in Tg(zferv1a:GFP)sh533. Whole-stack analysis of cell movement using TrackMate in Fiji. Scale bar: 10 µm. (B) Expression of Tg(zferv1a:GFP)sh533 in a 25 dpf zebrafish highlighting the triangular shaped thymus. (C) Forward scatter (FSC)/side scatter (SSC) plot of a whole kidney marrow separating different haematopoietic lineages by size and granularity. (D) SSC and GFP plot from a single-cell suspension of a whole kidney marrow from an adult Tg(zferv1a:GFP) with gate selecting GFP-positive cells. Inset shows the distribution of cells from a non-transgenic adult whole kidney marrow. (E) FSC/SSC plot of selected GFP-positive cells from D. Inset shows the location of the GFP-positive cells in the haematopoietic lineage FSC/SSC plot. (F) Quantification of GFP-positive cells in myeloid, erythroid, lymphoid lineages and progenitors from dissected sh533 and non-transgenic control adult head kidneys (n=3).
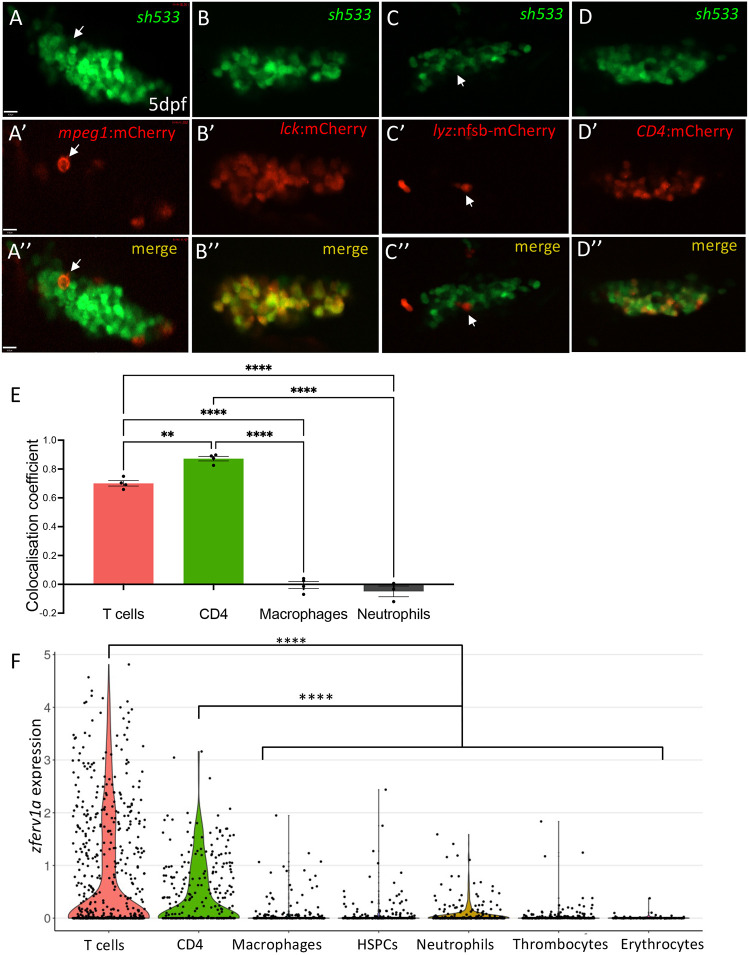
To confirm the lymphoid identity of zferv1a-expressing cells, we performed in vivo colocalisation analysis in 5 dpf embryos by crossing the Tg(zferv1a:GFP)sh533 line to different reporter lines labelling a wide spectrum of immune cell populations. We crossed F1 Tg(zferv1a:GFP)sh533 to reporter lines labelling lymphoid cells, such as Tg(lck:mCherry), labelling T cells (Langenau et al., 2004) as well as natural killer (NK) and innate lymphoid cells (Hernández et al., 2018), or Tg(CD4:mCherry), labelling CD4+ T cells and some macrophage/dendritic cells (Dee et al., 2016), and showed significant colocalisation between GFP- and mCherry-expressing cells by colocalisation analysis (Fig. 5B-B″,D-E). By contrast, crossing of Tg(zferv1a:GFP)sh533 to macrophage Tg(mpeg1:mCherryCAAX)sh378 (Fig. 5A-A″) or neutrophil Tg(lyz:nfsb-mCherry)sh260 (Fig. 5C-C″) (Buchan et al., 2019) reporter lines did not reveal significant colocalisation (Fig. 5E). To validate these results, we used single-cell RNA-sequencing datasets available on zebrafish blood cells (ArrayExpress accession numbers: MTAB-5530 and E-MTAB-4617) as a robust assay to analyse gene expression in specific cell types and tissues. Using the extensive gene expression dataset available, we mapped the expression of the entire genome of zferv1a (ENSDARG00000110878) in erythrocytes, haematopoietic stem and progenitor cells, macrophages, neutrophils, CD4+ cells, T cells and thrombocytes. Only sporadic expression was observed in all cell lineages except for the T-cell subsets ‘CD4+’ and ‘T cells’, where significant expression of env was detected (Fig. 5F). Altogether, these data are consistent with immune expression of zferv1a being restricted to T cells.
Fig. 5.
Immune expression of zferv1a is restricted to CD4+ cells and T cells. (A-D″) Single-slice confocal image of the thymus from double-transgenic Tg(zferv1a:GFP)sh533 and Tg(mpeg1:mCherryCAAX)sh378, which labels macrophage membranes (A-A″, white arrows point to a single macrophage), Tg(lck:mCherry), which labels T cells (B-B″), Tg(lyz:nfsb-mCherry)sh260, which labels neutrophils (C-C″, white arrows point to a single neutrophil), and Tg(CD4:mCherryCAAX), which labels CD4+ cells (D-D″) with merge image used for colocalisation analysis. (E) Histogram of ImageJ-generated colocalisation coefficients of Tg(zferv1a:GFP)sh533 crossed to Tg(mpeg1:mCherryCAAX)sh378 for macrophages, Tg(lyz:nfsb-mCherry)sh260 for neutrophils, Tg(CD4:mCherry) for CD4+ cells and Tg(lck:mCherry) for T cells (one-way ANOVA, n=4). (F) Violin plot of single-cell RNA-sequencing datasets MTAB-5530 and E-MTAB-4617 from zebrafish adult whole kidney marrow and spleen showing expression of zferv1a (ENSDARG00000110878) in multiple haematopoietic lineages. P=1.95×10−11 for CD4+ versus the other clusters and P=1.97×10−45 for T cells versus the other clusters (Mann–Whitney with paired Wilcoxon test). HSPCs, haematopoietic stem and progenitor cells.
Modulation of zferv1a expression
To investigate whether zferv1a can affect the development of the tissues where it is expressed, we performed an overexpression assay. We injected a vector containing zferv1a alongside the same vector containing the eGFP cassette to use as a positive control. Each plasmid was injected in equimolar amount or GFP alone and the expression of zferv1a and GFP was measured by qPCR. Overexpression of GFP resulted in a high level of GFP transcript detected by qPCR, whereas no expression of zferv1a was detected (Fig. S6). We then checked for induction of zferv1a (ENSDARG00000110878) in our datasets of larvae stimulated with type I interferon or with chikungunya (CHIKV) virus (Levraud et al., 2019; NCBI BioProject accession number: PRJNA531581). No expression was detected in the dataset of type I interferon-injected larvae (3 dpf). Expression was detected in the dataset of CHIKV-infected fish (4 dpf). However, although CHIKV infection triggers a very strong interferon response, with 1000-fold induction of some interferon-stimulated genes (ISGs), there was only a 1.7-fold increase for zferv1a, which was not statistically significant.
DISCUSSION
We describe here a novel reporter line in zebrafish that allows the expression of an endogenous retrovirus to be followed in real time in vivo. Our results validate and refine previously published expression data of zferv in the thymus and specifically in the lymphoid lineage. We identified a close relative to zferv, here named zferv1a, and showed transcriptional activity mostly restricted to T lymphocytes, as previously reported (Frazer et al., 2012; Shen and Steiner, 2004), but also observed in neurons.
Stability of the engineered-ERV transgene
Using a transgenic approach, we imaged in vivo in real time the behaviour of an ERV for the first time. Transgenesis entails random insertion of the transgene in the genome, which in some cases can disrupt neighbouring gene expression or induce silencing of the transgene (Akitake et al., 2011). Our initial ERV transgene Tg(ltr5:GFP:ltr3)sh500 was engineered to resemble the ERV structure and keep both LTR sequences. However, this line started to show signs of genetic instability and silencing from the F2 generation. It is possible that the location of the insertion triggered silencing of the transgene; however, we suspect that because of the similarity to a retrovirus genome, the transgene was targeted by epigenetic mechanisms specific to ERV. Indeed, transcriptional repression of ERV has been extensively studied and key histone and DNA methylation modifications identified (Maksakova et al., 2008; Rowe et al., 2010; Turelli et al., 2014), which could be targeting the LTR promoter we used. Previous work on zferv identified amplification of the zferv locus initially in T cells from an acute lymphoblastic leukaemia zebrafish model and, although at a lesser rate, also in normal T cells (Frazer et al., 2012). We suspect that genomic amplification of the initial transgene ltr5:GFP:ltr3 would be facilitated in the initial sh500 transgenic line as the transgene would be subjected to the transposase activity of other functional ERVs. This, in turn, might have activated targeting and silencing of the transgene within a generation. Either or both of those mechanisms might explain why we still observed some mild (neuron-specific) silencing phenotypes in the second transgenic line using a less-recognisable ERV structure, with the 3′LTR sequence replaced by a normal polyA tail in Tg(ltr5:GFP:pA)sh533.
The role of ERVs in T lymphocytes
Despite stability issues of the transgene, we consistently observed the signature expression of zferv in lymphocytes, as already identified in zebrafish transcriptional studies by others (Frazer et al., 2012; Shen and Steiner, 2004). The role of ERV expression in T cells is still under investigation. Each step of T-cell differentiation is accompanied by a sequence of wide transcriptional changes that allows (1) commitment to the T-cell lineage, (2) T-cell receptor (TCR) rearrangement and (3) positive or negative selection (Mingueneau et al., 2013). It is possible therefore that transcriptional de-repression of neighbouring genes allows ERV to be expressed during those stages, which would explain such a strong expression during those early phases of T-cell development.
Whether ERV expression is merely a side effect of neighbouring gene activation or plays a crucial role during T-cell development is an important question to answer. ERVs do confer advantages to our adaptive immune system, with important implications for infectious and cancer therapeutic approaches. T cells that have been presented with ERV peptides during their initial thymic selection have a higher sensitivity to exogenous retrovirus infection, conferring an advantage in fighting HIV (Young et al., 2012). Furthermore, very recently T-cell populations were found with the ability to recognise the antigen of multiple HERVs in patients with haematological cancers, with HERV-T cell less present in healthy donors (Saini et al., 2020). This finding might lead to selective therapies using HERV as a target to kill tumour cells.
Relevance of neuronal expression of ERV for human studies
Here, we provide evidence for a member of the ERV family being expressed in the healthy developing brain of a vertebrate. Reporter expression suggests that this neuronal expression might be more easily silenced than in T cells; the relevance of this finding for endogenous zferv expression, and its physiological significance, remain to be investigated. The role of endogenous retrovirus expression in developing neurons deserves further investigation as the potential role of ERVs in neurodegenerative and neurological diseases is a subject under intense study. The implications of aberrant presence of ERVs in the brain in human neurological diseases are actively being investigated, with the human HERV-K found to be upregulated in cortical neurons of amyotrophic lateral sclerosis (ALS) and MS patients (Li et al., 2015; Mameli et al., 2007; Perron et al., 1997), with currently active clinical trials targeting ERVs with anti-retroviral therapies for ALS, MS and the autoinflammatory disease AGS (Gold et al., 2018, 2019; Rice et al., 2018).
Our finding in embryos and adults suggests a role for ERVs in young neurons, potentially highlighting a role for ERVs in neuron development and maturation. There is evidence that other types of retroelements are expressed in neurons where they might function as regulators of progenitor differentiation. Indeed, brain expression of other retroelements has previously been reported, mainly focusing on the long interspersed nuclear element L1 family of non-LTR elements, which makes up 17% of all genomic DNA, constituting the largest percentage of the human genome of all retroelements (Scott et al., 1987). In the brain, L1 elements are de-repressed to allow neuronal precursor cells to differentiate and contribute to individual somatic mosaicism in human (Coufal et al., 2009) and mouse (Muotri et al., 2005). The latest study also used an engineered reporter approach by tagging GFP to the retroelement L1 (Muotri et al., 2005). Although L1 elements are not related to the ERV that is the subject of our study, it was recently discovered that additional groups of retroelements are enriched in specific regions of the human brain, in particular, LTR elements in the cerebellum (Bogu et al., 2019 preprint). With the advances in single-cell sequencing, it is possible that expression of different groups of ERVs, specifically human ERVs, will also be identified in the brain to begin a complete understanding of the presence and role of ERVs in brain development and diseases (Evans and Erwin, 2020).
Conclusions and future work
The zebrafish has been successfully used to model human neurodegenerative and neurodevelopmental disorders. Early time points of brain development, which are particularly difficult to study in mammals, are studied with ease in the zebrafish because of its transparency. As such, zebrafish models of neurodevelopmental autoinflammatory disorders (Hamilton et al., 2020; Haud et al., 2011; Kasher et al., 2015) or neurodegenerative diseases, such as MS (Kulkarni et al., 2017) or ALS (Da Costa et al., 2014), could be combined with our zferv1a reporter line to elucidate the role of ERVs in the development of these diseases.
This study describes the first zebrafish reporter line for an ERV, providing a tool for investigating how certain pathological states activate ERV expression in specific cell lineages, using a real-time in vivo approach.
MATERIALS AND METHODS
Zebrafish husbandry
We used the following transgenic lines: Tg(mpeg1:mCherryCAAX)sh378 to label the membrane of macrophages and microglia (Bojarczuk et al., 2016), Tg(lck:mCherry) to label T cells (Langenau et al., 2004) and NK cells (Hernández et al., 2018), Tg(CD4:mCherry) to label CD4+ T cells and some macrophage/dendritic cells (Dee et al., 2016) and the Tg(lyz:nfsb-mCherry)sh260 for neutrophils (Buchan et al., 2019).
All zebrafish were raised in the Bateson Centre at the University of Sheffield in UK Home Office-approved aquaria and maintained following standard protocols (Nüsslein-Volhard and Dham, 2002). Tanks were maintained at 28°C with a continuous re-circulating water supply and a daily light/dark cycle of 14 h/10 h. All procedures were performed on embryos younger than 5.2 dpf, which were therefore outside of the Animals (Scientific Procedures) Act, to standards set under AWERB (Animal Welfare and Ethical Review Bodies) and UK Home Office-approved protocols.
Identification of zferv genomes
We used the published full zferv nucleotide sequence (AF503912) as a query for a nucleotide BLAST search on zebrafish genome assembly on the Ensembl website (http://www.ensembl.org/Danio_rerio/Tools/Blast?db=core), using the most recent reference genome (GRCz11, July 2017), choosing the ‘BLAT’ option favouring highly similar sequences, and disabling the filter for species-specific repeats. We selected only hits with homology found in both the gag/pol and the env regions. Subsequent sequence alignments of LTRs, repetitive elements and open reading frames, and annotations were performed using DNA Strider 2.0.
zferv1a full genome cloning
The full zferv1a sequence was subcloned from fosmid 1930h03 (Sanger Center, Cambridge, UK) predicted to contain a ∼37 kb of chromosome 4 encompassing the sequence, according the annotations of the zv9 version of the zebrafish genome (Ensembl.org). The sequence, plus ∼100 bases of flanking genomic sequence on each side, was amplified using primers 5′-CCCTGCTCATTCAACACCATAC-3′ and 5′-CCCGTCTGTGAATTACCAAGC-3′, and cloned into PCR4-TOPO (Thermo Fisher).
RNA in situ hybridisation
The env1a probe was made from the plasmid containing the full zferv1a genome with the following primers: BamH1_env1a_fwd, TGGGGATCCATGAATAAAATAAACAAATTGG, and SmaI_env1a_rev, CCACCCGGGCACCATATCCAATAGTTCCTCC, generating a 1935 bp (env1a) fragment. The pol1a probe was made from the same plasmid using BamH1_pol1a_fwd, TGGGGATCCGGCAGCAGACGCCGCTGCTA, and Xba1_pol1a_rev, TGGTCTAGACCTCAGGCTCCTCAGTGTCT, generating a 1295 bp fragment. The env1b probe was made from cDNA using BamH1_env1b_fwd, TGGGGATCCatgaATATAAACAAATTGGTGG, and SmaI_env1b_rev, CCACCCGGGCAGAACGCTATAGTCAGTGCTC, generating a 635 bp (env1a) fragment. Fragments were subsequently cloned into Zero Blunt™ TOPO™ vector (Invitrogen) and reverse transcribed into RNA using the DIG labelling kit (Roche) (SP6 enzyme for antisense probe and T7 for sense probe). Whole-mount in situ hybridisation on wild-type nacre zebrafish was performed to identify the spatial and temporal pattern of zferv1a expression using the envelope (env) probe env1a and the reverse transcriptase probe pol1a and from zferv1b using the envelope probe env1b. We fixed wild-type embryos from the nacre strain at different stages and followed an in situ hybridisation protocol as previously described (Thisse and Thisse, 2008).
Generation of zferv1a and lck:mCherry transgenic lines
LTR5 and LTR3 regions of zferv1a were cloned into p5E and p3E Gateway entry vectors, respectively, using the following primers: BamH1_LTR3′_fwd, TGTGGATCCTCTCTTTCGAGATCAAGAGAGGG; Xho1_LTR3′_rev, ACACTCGAGGCTGCACCTTGTTGGAAAATGG; kpn1_LTR5′_fwd, TGTGGTACCCCTCTCTCTTGTAAAGGTTGAGGG; and sacII_LTR5′_rev ACACCGCGGTTTAATAATGGTTTTGTCTCCC.
An LR reaction was performed into the pDestCRY-CFP vector, which was injected into one-cell-stage zebrafish embryos to create the following transgenic lines: Tg(ltr5:GFP:ltr3)sh500 and Tg(ltr5:GFP:pA)sh533, subsequently called Tg(zferv1a:GFP)sh533.
The previously described lck promoter (Langenau et al., 2004) was used to create the transgene lck:mCherry in the pDestCRYmCherry vector by Gateway recombination. Stable lines were created by injecting these constructs with Tol2 mRNA in one-cell-stage embryos and selecting founders using the red eye marker.
Tyrosinase CRISPR/Cas9 crispant generation
Synthetic SygRNA (crRNA and tracrRNA) (Merck) in combination with Cas9 nuclease protein (Merck) was used for gene editing. Transactivating RNAs (tracrRNA) and gene-specific CRISPR RNAs (crRNA) were resuspended to a concentration of 20 µM in nuclease-free water containing 10 mM Tris-HCl pH 8. SygRNA complexes were assembled on ice immediately before use using a 1:1:1 ratio of crRNA:tracrRNA:Cas9 protein. Gene-specific crRNAs were designed using the online tool CHOPCHOP (http://chopchop.cbu.uib.no/). Tyrosinase crRNA tyr: GGACTGGAGGACTTCTGGGG(AGG) (Isles et al., 2021).
Flow cytometry analysis
The head kidney of three adult Tg(zferv1a:GFP)sh533 and three wild-type nacre fish were dissected and collected into cold Leibovitz 15 media supplemented with 20% foetal bovine serum and 0.5 mM EDTA. The tissue was pipetted up and down to create a single-cell suspension and kept on ice. Negative gate was set up using cells from nacre animals with the Blue530 detector on an FACSARIA IIu flow cytometry machine and all analysis carried out using the FlowJo software. Gating was performed as described in Fig. S5 using the GFP-positive population back-gated onto the different cell type populations: monocytes, progenitors, T cells and erythrocytes.
Colocalisation imaging and analysis
Tg(zferv1a:GFP)sh533 was crossed to Tg(lck:mCherry), Tg(CD4:mCherry), Tg(lyz:nfsb-mCherry)sh260 and Tg(mpeg1:mCherry:CAAX)sh378, and double-positive animals were selected and imaged at 5 dpf using the Perkin Elmer Spinning Disk system for 3 h. Whole z-stacks were imported into Fiji to perform colocalisation analysis using the Colocalisation Threshold Plugin. Whole z-stacks at three different time points during the time lapse in four different larvae were used with the same threshold values to determine the colocalisation coefficient by Pearson's correlation.
Single-cell RNA-sequencing dataset analysis
Expression of zferv1a (ENSDARG00000110878) in different haematopoietic lineages was obtained from single-cell RNA-sequencing databases of seven different transgenic lines each labelling a different lineage (Athanasiadis et al., 2017). Single-cell RNA-sequencing raw counts files of zebrafish blood cells were obtained from datasets available on Array Express (https://www.ebi.ac.uk/arrayexpress/; accession codes: E-MTAB-5530 and E-MTAB-4617). The data were then processed and analysed using the Seurat software package for R (Hao et al., 2021; Satija et al., 2015). The data were selected to have a minimum of 200 genes in each cell and then merged and normalised. The Seurat wrapper fastMNN was used to correct for batch effects and uniform manifold approximation and projection (UMAP) analysis was performed on the resulting combined dataset, which resulted in nine clusters. The function ‘FindAllMarkers’ was used to find the differentially expressed genes in each cluster and these were compared with marker genes published in the studies to assign cluster identity. Expression distributions across clusters were obtained for ENSDARG00000110878. Differential expression of CD4+ cells and T cells versus all cells was determined (average log2 fold change=1.98, adjusted P-value=5.31e−81).
Statistical differential expression was performed in Seurat between each cluster with Mann–Whitney test followed by a Kruskal–Wallis multiple testing correction.
zferv1a overexpression
The plasmid containing the full zferv1a gene was injected in equimolar concentration alongside a plasmid containing GFP into one-cell-stage zebrafish embryos, using embryos injected with GFP only as control. RNA was extracted from 30 hours post-fertilisation GFP-positive embryos from both groups using Trizol and cDNA was synthesised as previously described (Hamilton et al., 2020). qPCR was performed to measure the level of expression of GFP and zferv1a normalised to the ef1a reference gene using the following primers: egfpforward, CCATCTTCTTCAAGGACGAC; egfpreverse, CGTTGTGGCTGTTGTAGTTG; ZFERVpol-1S, GCTAGGACATCCCATTGTGT; ZFERVpol-2A, GGGAATGTGTTCTGGTGTCT; Ef1a-1S, GCTGATCGTTGGAGTCAACA; Ef1a-2A, ACAGACTTGACCTCAGTGGT.
Statistical analyses
All statistical analyses were performed in GraphPad Prism where data was entered using a grouped table for ANOVA analysis (more than two samples or two variables). Sample distribution was assessed using frequency of distribution analysis. n (experimental unit) number for each experiment is stated in figure legends. P-values are indicated in legends and asterisks used for graphs with multiple comparisons: *P<0.05, **P<0.01, ***P<0.001, ****P<0.0001. Following the recommendation of the American Statistical Association, we do not associate a specific P-value with significance (Wasserstein et al., 2019).
Supplementary Material
Acknowledgements
We thank the flow cytometry facility at the University of Sheffield, The Light Microscopy Facility and the Zebrafish facility staff at The Bateson Centre for their technical help. We thank Adam Hurlstone (The University of Manchester) for supplying the Tg(lck:mCherry) transgenic line, Ana Cvejic for the immune single-cell sequencing dataset and Angela Ciuffi from the Institute of Microbiology (IMUL), Lausanne University Hospital and University of Lausanne for supervising A.D.-J. Imaging was carried out in the Wolfson Light Microscopy Facility, supported by an MRC grant (G0700091) and a Wellcome Trust grant (GR077544AIA).
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Conceptualization: S.A.R., N.H.; Methodology: J.-P.L., N.H.; Validation: N.H.; Formal analysis: E.V.C., N.H.; Investigation: A.C., H.M.I., J.-P.L., N.H.; Resources: A.D.-J., S.A.R., N.H.; Data curation: H.A.R., E.V.C., J.J.P., E.G.C., J.-P.L., N.H.; Writing - original draft: N.H.; Writing - review & editing: H.A.R., E.V.C., H.M.I., S.A.R., J.-P.L., N.H.; Visualization: J.-P.L., N.H.; Supervision: S.A.R., N.H.; Project administration: S.A.R., N.H.; Funding acquisition: S.A.R., N.H.
Funding
This work was supported by a European Leukodystrophy Association fellowship (Association Européenne contre les Leucodystrophies; ELA 2016-012F4 to N.H.), a Medical Research Council Programme Grant (MR/M004864/1 to S.A.R.), a studentship from the MRC Discovery Medicine North (DiMeN) Doctoral Training Partnership (MR/N013840/1) and a Sir Jules Thorn PhD scholarship to H.A.R., and an Agence Nationale de la Recherche grant (ANR-16-CE20-0002-03 to J.-P.L.). Open Access funding provided by The University of Sheffield Bateson Centre. Deposited in PMC for immediate release.
References
- Ahmad, S., Mu, X., Yang, F., Greenwald, E., Park, J. W., Jacob, E., Zhang, C.-Z. and Hur, S. (2018). Breaching self-tolerance to Alu duplex RNA underlies MDA5-mediated inflammation. Cell 172, 797-810.e13. 10.1016/j.cell.2017.12.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akitake, C. M., Macurak, M., Halpern, M. E. and Goll, M. G. (2011). Transgenerational analysis of transcriptional silencing in zebrafish. Dev. Biol. 352, 191-201. 10.1016/j.ydbio.2011.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Athanasiadis, E. I., Botthof, J. G., Andres, H., Ferreira, L., Lio, P. and Cvejic, A. (2017). Single-cell RNA-sequencing uncovers transcriptional states and fate decisions in haematopoiesis. Nat. Commun. 8, 2045. 10.1038/s41467-017-02305-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baudino, L., Yoshinobu, K., Morito, N., Kikuchi, S., Fossati-Jimack, L., Morley, B. J., Vyse, T. J., Hirose, S., Jørgensen, T. N., Tucker, R. M.et al. (2014). Dissection of genetic mechanisms governing the expression of serum retroviral Gp70 implicated in murine lupus nephritis. J. Immunol. 181, 2846-2854. 10.4049/jimmunol.181.4.2846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benitez-Guijarro, M., Lopez-Ruiz, C., Tarnauskaitė, Ž., Murina, O., Mohammad, M. M., Williams, T. C., Fluteau, A., Sanchez, L., Vilar-Astasio, R., Garcia–Canadas, M.et al. (2018). RNase H2, mutated in Aicardi–Goutières syndrome, promotes LINE–1 retrotransposition. EMBO J. 37, e98506. 10.15252/embj.201798506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bermejo, A. V., Ragonnaud, E., Daradoumis, J. and Holst, P. (2020). Cancer associated endogenous retroviruses: ideal immune targets for adenovirus-based immunotherapy. Int. J. Mol. Sci. 21, 1-21. 10.3390/ijms21144843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogu, G. K., Reverter, F., Marti-Renom, M. A., Snyder, M. P. and Guigó, R. (2019). Atlas of transcriptionally active transposable elements in human adult tissues. BioRxiv. 10.1101/714212 [DOI] [Google Scholar]
- Bojarczuk, A., Miller, K. A., Hotham, R., Lewis, A., Ogryzko, N. V., Kamuyango, A. A., Frost, H., Gibson, R. H., Stillman, E., May, R. C.et al. (2016). Cryptococcus neoformans intracellular proliferation and capsule size determines early macrophage control of infection. Sci. Rep. 6, 21489. 10.1038/srep21489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourque, G., Burns, K. H., Gehring, M., Gorbunova, V., Seluanov, A., Hammell, M., Imbeault, M., Izsvák, Z., Levin, H. L., Macfarlan, T. S.et al. (2018). Ten things you should know about transposable elements. Genome Biol. 19, 1-12. 10.1186/s13059-018-1577-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchan, K. D., Prajsnar, T. K., Ogryzko, N. V., de Jong, N. W. M., van Gent, M., Kolata, J., Foster, S. J., van Strijp, J. A. G. and Renshaw, S. A. (2019). A transgenic zebrafish line for in vivo visualisation of neutrophil myeloperoxidase. PLoS ONE 14, e0215592. 10.1101/456541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiappinelli, K. B., Strissel, P. L., Desrichard, A., Li, H., Henke, C., Akman, B., Hein, A., Rote, N. S., Cope, L. M., Snyder, A.et al. (2015). Inhibiting DNA methylation causes an interferon response in cancer via DsRNA including endogenous retroviruses. Cell 162, 974-986. 10.1016/j.cell.2015.07.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung, H., Calis, J. J. A., Wu, X., Sun, T., Yu, Y., Sarbanes, S. L., Thi, V. L. D., Shilvock, A. R., Hoffmann, H.-H., Rosenberg, B. R.et al. (2018). Human ADAR1 prevents endogenous RNA from triggering translational shutdown. Cell 172, 811-824.e14. 10.1016/j.cell.2017.12.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuong, E. B. (2018). The placenta goes viral: retroviruses control gene expression in pregnancy. PLoS Biol. 16, e3000028. 10.1371/journal.pbio.3000028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuong, E. B., Elde, N. C. and Feschotte, C. (2016). Regulatory evolution of innate immunity through co-option of endogenous retroviruses. Science 351, 1083. 10.1126/science.aad5497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coufal, N. G., Garcia-Perez, J. L., Peng, G. E., Yeo, G. W., Mu, Y., Lovci, M. T., Morell, M., O'Shea, K. S., Moran, J. V. and Gage, F. H. (2009). L1 retrotransposition in human neural progenitor cells. Nature 460, 1127-1131. 10.1038/nature08248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crow, Y. J., Chase, D. S., Schmidt, J. L., Szynkiewicz, M., Forte, G. M. A., Gornall, H. L., Oojageer, A., Anderson, B., Pizzino, A., Helman, G.et al. (2015). Characterization of human disease phenotypes associated with mutations in TREX1, RNASEH2A, RNASEH2B, RNASEH2C, SAMHD1, ADAR, and IFIH1. American Journal of Medical Genetics Part A, 167A, 296-312. 10.1002/ajmg.a.36887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Da Costa, M. M. J., Allen, C. E., Higginbottom, A., Ramesh, T., Shaw, P. J. and McDermott, C. J. (2014). A new zebrafish model produced by TILLING of SOD1-related amyotrophic lateral sclerosis replicates key features of the disease and represents a tool for in vivo therapeutic screening. Dis. Model. Mech. 7, 73-81. 10.1242/dmm.012013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dee, C. T., Nagaraju, R. T., Athanasiadis, E. I., Gray, C., Fernandez del Ama, L., Johnston, S. A., Secombes, C. J., Cvejic, A. and Hurlstone, A. F. L. (2016). CD4-transgenic zebrafish reveal tissue-resident Th2- and regulatory T cell–like populations and diverse mononuclear phagocytes. J. Immunol. 197, 3520-3530. 10.4049/jimmunol.1600959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans, T. A. and Erwin, J. A. (2020). Retroelement-derived RNA and its role in the brain. Semin. Cell Dev. Biol. 114, 68-80. 10.1016/j.semcdb.2020.11.001 [DOI] [PubMed] [Google Scholar]
- Feschotte, C. (2008). Transposable elements and the evolution of regulatory networks. Nat. Rev. Genet. 9, 397-405. 10.1038/nrg2337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frazer, J. K., Batchelor, L. A., Bradley, D. F., Brown, K. H., Dobrinski, K. P., Lee, C. and Trede, N. S. (2012). Genomic amplification of an endogenous retrovirus in zebrafish T-cell malignancies. Adv. Hematol. 2012, 627920. 10.1155/2012/627920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Göke, J., Lu, X., Chan, Y.-S., Ng, H.-H., Ly, L.-H., Sachs, F. and Szczerbinska, I. (2015). Dynamic transcription of distinct classes of endogenous retroviral elements marks specific populations of early human embryonic cells. Cell Stem Cell 16, 135-141. 10.1016/j.stem.2015.01.005 [DOI] [PubMed] [Google Scholar]
- Gold, J., Marta, M., Meier, U. C., Christensen, T., Miller, D., Altmann, D., Holden, D., Bianchi, L., Adiutori, R., MacManus, D.et al. (2018). A phase II baseline versus treatment study to determine the efficacy of raltegravir (isentress) in preventing progression of relapsing remitting multiple sclerosis as determined by gadolinium-enhanced MRI: the INSPIRE Study. Multiple Sclerosis Related Disorders 24, 123-128. 10.1016/j.msard.2018.06.002 [DOI] [PubMed] [Google Scholar]
- Gold, J., Rowe, D. B., Kiernan, M. C., Vucic, S., Mathers, S., van Eijk, R. P. A., Nath, A., Garcia Montojo, M., Norato, G., Santamaria, U. A.et al. (2019). Safety and tolerability of triumeq in amyotrophic lateral sclerosis: the lighthouse trial. Amyotrophic Lateral Sclerosis Frontotemp. Degener. 20, 595-604. 10.1080/21678421.2019.1632899 [DOI] [PubMed] [Google Scholar]
- Hamilton, N., Rutherford, H. A., Petts, J. J., Isles, H. M., Weber, T., Henneke, M., Gärtner, J., Renshaw, S. A. and Dunning, M. J. (2020). The failure of microglia to digest developmental apoptotic cells contributes to the pathology of RNASET2-deficient leukoencephalopathy. Glia 68, 1531-1545. 10.1002/glia.23829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao, Y., Hao, S., Andersen-Nissen, E., Mauck, W. M., Zheng, S., Butler, A., Lee, M. J., Wilk, A. J., Darby, C. and Zager, M.et al. (2021). Integrated analysis of multimodal single-cell data. Cell 184, 3573-3587.e29. 10.1016/j.cell.2021.04.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haud, N., Kara, F., Diekmann, S., Henneke, M., Willer, J. R., Hillwig, M. S., Gregg, R. G., Macintosh, G. C., Gärtner, J., Alia, A.et al. (2011). Rnaset2 mutant zebrafish model familial cystic leukoencephalopathy and reveal a role for RNase T2 in degrading ribosomal RNA. Proc. Natl. Acad. Sci. USA 108, 1099-1103. 10.1073/pnas.1009811107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernández, P. P., Strzelecka, P. M., Athanasiadis, E. I., Hall, D., Robalo, A. F., Collins, C. M., Boudinot, P., Levraud, J.-P. and Cvejic, A. (2018). Single-cell transcriptional analysis reveals ILC-like cells in zebrafish. Sci. Immunol. 3, eaau5265. 10.1126/sciimmunol.aau5265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isles, H. M., Loynes, C. A., Alasmari, S., Kon, F. C., Henry, K. M., Kadochnikova, A., Hales, J., Muir, C. F., Keightley, M.-C., Kadirkamanathan, V.et al. (2021). Pioneer neutrophils release chromatin within in vivo swarms. eLife 10, e68755. 10.7554/eLife.68755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston, J. B., Silva, C., Holden, J., Warren, K. G., Clark, A. W. and Power, C. (2001). Monocyte activation and differentiation augment human endogenous retrovirus expression: implications for inflammatory brain diseases. Ann. Neurol. 50, 434-442. 10.1002/ana.1131 [DOI] [PubMed] [Google Scholar]
- Kasher, P. R., Jenkinson, E. M., Briolat, V., Gent, D., Morrissey, C., Zeef, L. A. H., Rice, G. I., Levraud, J.-P. and Crow, Y. J. (2015). Characterization of Samhd1 morphant zebrafish recapitulates features of the human type I interferonopathy Aicardi-Goutières Syndrome. J. Immunol. 194, 2819-2825. 10.4049/jimmunol.1403157 [DOI] [PubMed] [Google Scholar]
- Kissa, K., Murayama, E., Zapata, A., Cortés, A., Perret, E., Machu, C. and Herbomel, P. (2008). Live imaging of emerging hematopoietic stem cells and early thymus colonization. Blood 111, 1147-1156. 10.1182/blood-2007-07-099499 [DOI] [PubMed] [Google Scholar]
- Knerr, I., Huppertz, B., Weigel, C., Dötsch, J., Wich, C., Schild, R. L., Beckmann, M. W. and Rascher, W. (2004). Endogenous retroviral syncytin: compilation of experimental research on syncytin and its possible role in normal and disturbed human placentogenesis. Mol. Hum. Reprod. 10, 581-588. 10.1093/molehr/gah070 [DOI] [PubMed] [Google Scholar]
- Kulkarni, P., Yellanki, S., Medishetti, R., Sriram, D., Saxena, U. and Yogeeswari, P. (2017). Novel zebrafish EAE model: a quick in vivo screen for multiple sclerosis. Multiple Sclerosis Related Disorders 11, 32-39. 10.1016/j.msard.2016.11.010 [DOI] [PubMed] [Google Scholar]
- Kunarso, G., Chia, N.-Y., Jeyakani, J., Hwang, C., Lu, X., Chan, Y.-S., Ng, H.-H. and Bourque, G. (2010). Transposable elements have rewired the core regulatory network of human embryonic stem cells. Nat. Genet. 42, 631-634. 10.1038/ng.600 [DOI] [PubMed] [Google Scholar]
- Lander, E. S., Linton, L. M., Birren, B., Nusbaum, C., Zody, M. C., Baldwin, J., Devon, K., Dewar, K., Doyle, M., Fitzhugh, W.et al. (2001). Correction: Initial sequencing and analysis of the human genome. Nature 412, 565-566. 10.1038/35087627 [DOI] [PubMed] [Google Scholar]
- Langenau, D. M., Ferrando, A. A., Traver, D., Kutok, J. L., Hezel, J.-P. D., Kanki, J. P., Zon, L. I., Look, A. T. and Trede, N. S. (2004). In vivo tracking of T cell development, ablation, and engraftment in transgenic zebrafish. Proc. Natl Acad. Sci. USA 101, 7369-7374. 10.1073/pnas.0402248101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levraud, J.-P., Jouneau, L., Briolat, V., Laghi, V. and Boudinot, P. (2019). IFN-stimulated genes in Zebrafish and humans define an ancient arsenal of antiviral immunity. J. Immunol. 203, 3361-3373. 10.4049/jimmunol.1900804 [DOI] [PubMed] [Google Scholar]
- Li, W., Lee, M. H., Henderson, L., Tyagi, R., Bachani, M., Steiner, J., Campanac, E., Hoffman, D. A., Von Geldern, G., Johnson, K.et al. (2015). Human endogenous retrovirus-K contributes to motor neuron disease. Sci. Transl. Med. 307. 10.1126/scitranslmed.aac8201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, P., Du, J., Goodier, J. L., Hou, J., Kang, J., Kazazian, H. H., Zhao, K. and Yu, X.-F. (2017). Aicardi-Goutières syndrome protein TREX1 suppresses L1 and maintains genome integrity through exonuclease-independent ORF1p depletion. Nucleic Acids Res. 45, 4619-4631. 10.1093/nar/gkx178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lokossou, A. G., Toudic, C., Nguyen, P. T., Elisseeff, X., Vargas, A., Rassart, É., Lafond, J., Leduc, L., Bourgault, S., Gilbert, C.et al. (2020). Endogenous retrovirus-encoded Syncytin-2 contributes to exosome-mediated immunosuppression of T cells. Biol. Reprod. 102, 185-198. 10.1093/biolre/ioz124 [DOI] [PubMed] [Google Scholar]
- Maksakova, I. A., Mager, D. L. and Reiss, D. (2008). Endogenous retroviruses - keeping active endogenous retroviral-like elements in check: the epigenetic perspective. Cell. Mol. Life Sci. 65, 3329-3347. 10.1007/s00018-008-8494-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mameli, G., Astone, V., Arru, G., Marconi, S., Lovato, L., Serra, C., Sotgiu, S., Bonetti, B. and Dolei, A. (2007). Brains and peripheral blood mononuclear cells of Multiple Sclerosis (MS) Patients Hyperexpress MS-Associated Retrovirus/HERV-W endogenous retrovirus, but not human Herpesvirus 6. J. Gen. Virol. 88, 264-274. 10.1099/vir.0.81890-0 [DOI] [PubMed] [Google Scholar]
- Mandl, J. N., Monteiro, J. P., Vrisekoop, N. and Germain, R. N. (2013). T cell-positive selection uses self-ligand binding strength to optimize repertoire recognition of foreign antigens. Cell 38, 263-274. 10.1016/j.immuni.2012.09.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi, S., Lee, X., Li, X.-, Veldman, G. M., Finnerty, H., Racie, L., LaVallie, E., Tang, X.-Y., Edouard, P., Howes, S.et al. (2000). Syncytin is a captive retroviral envelope protein involved in human placental morphogenesis. Nature 403, 785-789. 10.1038/35001608 [DOI] [PubMed] [Google Scholar]
- Mingueneau, M., Kreslavsky, T., Gray, D., Heng, T., Cruse, R., Ericson, J., Bendall, S., Spitzer, M. H., Nolan, G. P., Kobayashi, K.et al. (2013). The transcriptional landscape of Αβ T cell differentiation. Nat. Immunol. 14, 619-632. 10.1038/ni.2590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muotri, A. R., Chu, V. T., Marchetto, M. C. N., Deng, W., Moran, J. V. and Gage, F. H. (2005). Somatic mosaicism in neuronal precursor cells mediated by L1 retrotransposition. Nature 435, 903-910. 10.1038/nature03663 [DOI] [PubMed] [Google Scholar]
- Nüsslein-Volhard, C. and Dham, R. (2002). Zebrafish: A Practical Approach. New York: Oxford University Press. [Google Scholar]
- Okahara, G., Matsubara, S., Oda, T., Sugimoto, J., Jinno, Y. and Kanaya, F. (2004). Expression analyses of Human Endogenous Retroviruses (HERVs): tissue-specific and developmental stage-dependent expression of HERVs. Genomics 84, 982-990. 10.1016/j.ygeno.2004.09.004 [DOI] [PubMed] [Google Scholar]
- Perron, H., Garson, J. A., Bedin, F., Beseme, F., Paranhos-Baccala, G., Komurian-Pradel, F., Mallet, F., Tuke, P. W., Voisset, C., Blond, J. L.et al. (1997). Molecular identification of a novel retrovirus repeatedly isolated from patients with multiple sclerosis. Proc. Natl. Acad. Sci. U.S.A. 94, 7583-7588. 10.1073/pnas.94.14.7583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pi, W., Yang, Z., Wang, J., Ruan, L., Yu, X., Ling, J., Krantz, S., Isales, C., Conway, S. J., Lin, S.et al. (2004). The LTR enhancer of ERV-9 human endogenous retrovirus is active in oocytes and progenitor cells in transgenic zebrafish and humans. Proc. Natl Acad. Sci. USA 101, 805-810. 10.1073/pnas.0307698100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renshaw, S. A. and Trede, N. S. (2012). A model 450 million years in the making: zebrafish and vertebrate immunity. Dis. Model. Mech. 5, 38-47. 10.1242/dmm.007138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renshaw, S. A., Loynes, C. A., Trushell, D. M. I., Elworthy, S., Ingham, P. W. and Whyte, M. K. B. (2006). A transgenic zebrafish model of neutrophilic inflammation. Blood 108, 3976-3978. 10.1182/blood-2006-05-024075 [DOI] [PubMed] [Google Scholar]
- Rice, G. I., Meyzer, C., Bouazza, N., Hully, M., Boddaert, N., Semeraro, M., Zeef, L. A. H., Rozenberg, F., Bondet, V., Duffy, D.et al. (2018). Reverse-transcriptase inhibitors in the Aicardi – Goutières Syndrome. N. Engl. J. Med. 6), 2275-2277. 10.1056/NEJMc1810983 [DOI] [PubMed] [Google Scholar]
- Rowe, H. M., Jakobsson, J., Mesnard, D., Rougemont, J., Reynard, S., Aktas, T., Maillard, P. V., Layard-Liesching, H., Verp, S., Marquis, J.et al. (2010). KAP1 controls endogenous retroviruses in embryonic stem cells. Nature 463, 237-240. 10.1038/nature08674 [DOI] [PubMed] [Google Scholar]
- Saini, S. K., Due Ørskov, A., Bjerregaard, A.-M., Unnikrishnan, A., Holmberg-Thydén, S., Borch, A., Jensen, K. V., Anande, G., Bentzen, A. K., Marquard, A. M.et al. (2020). Human endogenous retroviruses form a reservoir of T cell targets in hematological cancers. Nat. Commun. 11, 1-14. 10.1038/s41467-020-19464-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satija, R., Farrell, J. A., Gennert, D., Schier, A. F. and Regev, A. (2015). Spatial reconstruction of single-cell gene expression data. Nat. Biotechnol. 33, 495-502. 10.1038/nbt.3192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott, A. F., Schmeckpeper, B. J., Abdelrazik, M., Comey, C. T., O'Hara, B., Rossiter, J. P., Cooley, T., Heath, P., Smith, K. D. and Margolet, L. (1987). Origin of the human L1 elements: proposed progenitor genes deduced from a consensus DNA sequence. Genomics 1, 113-125. 10.1016/0888-7543(87)90003-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen, C.-H. and Steiner, L. A. (2004). Genome structure and thymic expression of an endogenous retrovirus in zebrafish. J. Virol. 78, 899-911. 10.1128/JVI.78.2.899-911.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiroma, T., Sugimoto, J., Oda, T., Jinno, Y. and Kanaya, F. (2001). Search for active endogenous retroviruses: identification and characterization of a HERV-E gene that is expressed in the pancreas and thyroid. J. Hum. Genet. 46, 619-625. 10.1007/s100380170012 [DOI] [PubMed] [Google Scholar]
- Tavakolian, S., Goudarzi, H. and Faghihloo, E. (2019). Evaluating the expression level of HERV-K Env, Np9, Rec and Gag in breast tissue. Infectious Agents Cancer 14, 1-5. 10.1186/s13027-019-0260-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thisse, C. and Thisse, B. (2008). High-resolution in situ hybridization to whole-mount zebrafish embryos. Nat. Protoc. 3, 59-69. 10.1038/nprot.2007.514 [DOI] [PubMed] [Google Scholar]
- Thomas, C. A., Tejwani, L., Trujillo, C. A., Negraes, P. D., Herai, R. H., Mesci, P., Macia, A., Crow, Y. J. and Muotri, A. R. (2017). Modeling of TREX1-dependent autoimmune disease using human stem cells highlights L1 accumulation as a source of neuroinflammation. Cell Stem Cell 21, 319-331.e8. 10.1016/j.stem.2017.07.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trede, N. S., Langenau, D. M., Traver, D., Look, A. T. and Zon, L. I. (2004). The use of zebrafish to understand immunity. Immunity 20, 367-379. 10.1016/S1074-7613(04)00084-6 [DOI] [PubMed] [Google Scholar]
- Treger, R. S., Pope, S. D., Kong, Y., Tokuyama, M., Taura, M. and Iwasaki, A. (2019). The lupus susceptibility locus Sgp3 encodes the suppressor of endogenous retrovirus expression SNERV. Immunity 50, 334-347.e9. 10.1016/j.immuni.2018.12.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turelli, P., Castro-Diaz, N., Marzetta, F., Kapopoulou, A., Raclot, C., Duc, J., Tieng, V., Quenneville, S. and Trono, D. (2014). Interplay of TRIM28 and DNA methylation in controlling human endogenous retroelements. Genome Res. 24, 1260-1270. 10.1101/gr.172833.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasserstein, R. L., Schirm, A. L. and Lazar, N. A. (2019). Moving to a world beyond ‘p<0.05’. Am. Stat. 73 Suppl. 1, 1-19. 10.1080/00031305.2019.1583913 [DOI] [Google Scholar]
- Young, G. R., Ploquin, M. J.-Y., Eksmond, U., Wadwa, M., Stoye, J. P. and Kassiotis, G. (2012). Negative selection by an endogenous retrovirus promotes a higher-avidity CD4+ T cell response to retroviral infection. PLoS Pathog. 8, e1002709. 10.1371/journal.ppat.1002709 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.