Summary
Neuronal birthdate is one of the major determinants of neuronal phenotypes. However, most birthdating methods are retrospective in nature, allowing very little experimental access to the classified neuronal subsets. Here, we introduce four neurogenic tagging mouse lines, which can assign CreER-loxP recombination to neuron subsets that share the same differentiation timing in living animals and enable various experimental manipulations of the classified subsets. We constructed a brain atlas of the neurogenic tagging mouse lines (NeuroGT), which includes holistic image data of the loxP-recombined neurons and their processes across the entire brain that were tagged on each single day during the neurodevelopmental period. This image database, which is open to the public, offers investigators the opportunity to find specific neurogenic tagging driver lines and the stages of tagging appropriate for their own research purposes.
Keywords: neurogenic tagging, transgenic mouse, CreER-loxP system, NeuroGT, neuronal birthdate
Graphical abstract
Highlights
-
•
We generate four CreER mouse drivers to tag neurons on the basis of time of neurogenesis
-
•
The drivers enable birthdate-based classification and manipulation of neurons
-
•
We develop NeuroGT, an image atlas of tagged neurons across the entire brain
-
•
Through NeuroGT, researchers can find driver lines suitable for their own research
Motivation
Neuronal birthdates are commonly used to classify neurons. However, the current nucleotide-based birthdating method is retrospective in nature and does not allow the manipulation of classified neurons. To overcome this limitation, we developed neurogenic tagging mouse lines that can induce CreER-loxP recombination in neurons with similar birthdates, and the recombination tag can be subsequently used for experimental manipulation of the classified neuron subsets. To encourage the use of this resource, we launched the NeuroGT database, which contains image data of the tagged neurons across the entire brain.
Hirata et al. develop neurogenic tagging CreER driver mice and the NeuroGT database to showcase the mouse lines. The resource can be used to classify neurons on the basis of their generation timing and manipulate the classified neuron subsets for research purposes.
Introduction
During development, neurons are generated over a protracted time window. Mounting evidence shows that neurogenic timing, which is often referred to as the neuronal birthdate, has immense impacts on neuronal phenotypes. In the neocortex, the birthdate determines layer positioning, connection patterns, and molecular and physiological properties of neurons (McConnell, 1989; Lodato and Arlotta, 2015). The chronological specification of neuronal fates is not a unique property of the neocortex but rather a generally conserved strategy in various nervous systems for generating neuronal diversity (Suzuki and Hirata, 2013). Even without affecting the intrinsic molecular differences, the birth timing itself could cause differential neuronal phenotypes because neurons with different birthdates are influenced by a changing environment (Hirata and Iwai, 2019). Because of its intimate relationships with various neuronal phenotypes, neuronal birthdating has been a reliable standard for the classification of neurons in various nervous systems for over half a century (Bayer and Altman, 1987; Govindan et al., 2018).
Traditionally, neuronal birthdating has been performed by using nucleotide analogs, such as radioactive thymidine and, more recently, bromodeoxyuridine (BrdU) or 5-ethyynyl uridine (EdU), which are incorporated into the DNA during the S phase to mark the final round of the cell cycle. Although these techniques have substantially improved, they are still descriptive histological techniques to determine neuronal birthdates in sacrificed animals. Recently, we developed a neurogenic tagging method by which tamoxifen (TM)-dependent CreER (Feil et al., 1997) recombination was induced in olfactory bulb neurons in a birthdate-dependent manner (Hirata et al., 2019). Given that this technique is based on irreversible loxP recombination, the tag can be used later to explore the birthdate-classified neuronal subset in various ways.
This neurogenic tagging method uses driver mouse lines in which CreER is expressed only transiently within a short time window immediately after neuronal fates are committed (Figure 1A). Consequently, a single administration of TM at a certain developmental stage induces the recombination of loxP sequences only in the cells that express CreER. In the previous study, we used the enhancer of the neurog2 gene to achieve the transient expression of CreER in a bacterial artificial chromosome (BAC) transgenic mouse. Although the biological principle underlying this method differs from that of nucleotide-based birthdating, the CreER driver that we developed achieved birthdate-dependent neuron tagging in the brain regions such as the olfactory bulb (Hirata et al., 2019) and cerebellum (Tran-Anh et al., 2020; Zhang et al., 2020), mirroring the endogenous expression patterns of neurog2.
Figure 1.
Neurogenic tagging and the driver mouse lines
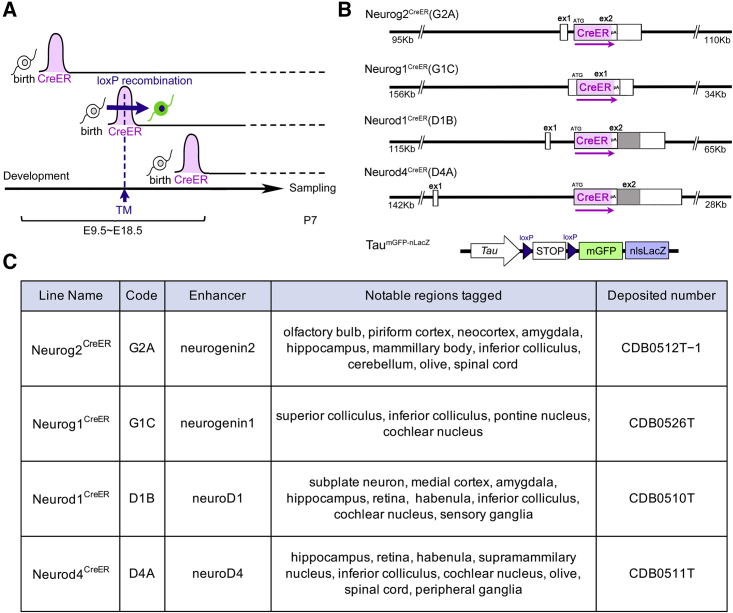
(A) In neurogenic tagging mouse lines, CreER is transiently expressed within a short neurogenic time window in various neurons. TM injection on a single day during E9.5–E18.5 induces loxP recombination only in cells expressing CreER. The mouse embryos were raised up to P7, and the brains were sampled for visualization of tagged neurons.
(B) The BAC genomic constructs recombined with the CreER cassette that were used to generate the four neurogenic tagging driver lines. Boxes show the exons, in which the shaded part represents originally protein-coding sequences and the white part untranslated sequences. The approximate lengths of the genome upstream and downstream of the CreER cassette are indicated under each construct. The bottom panel shows a schematic of the gene structure of the TaumGFP-nLacZ reporter. Both reporter proteins, mGFP and nls-βGAL, were used to visualize tagged neurons.
(C) Four neurogenic tagging mouse lines were characterized in this study. The mice can be obtained from RIKEN BDR (http://www2.clst.riken.jp/arg/TG%20mutant%20mice%20list.html).
To cover other brain regions, we developed three neurogenic tagging driver lines by using the neuronal differentiation genes neurog1, neurod1, and neurod4 (Figures 1B and 1C), all of which are basic-helix-loop-helix transcription factors that are transiently expressed during the maturation phase of neurons (Kim et al., 2011; Sudarov et al., 2011; Aprea et al., 2014). Together with the previous line developed by using neurog2, the collection of driver lines covers most of the mouse nervous systems. Our aim is to contribute this resource to the scientific community. To encourage the use of this resource, we have developed a brain atlas of neurogenic tagging mouse lines (NeuroGT). This database contains section images of the entire brain visualized for tagged neurons as well as their processes; these images were obtained from postnatal mice that received a single TM injection on each day during the neurogenetic period. Researchers interested in particular brain regions can find appropriate mouse lines and tagging stages for their research purposes.
Results
NeuroGT and neurogenic tagging driver mice
The neurogenic tagging drivers Neurog2CreER(G2A), Neurog1CreER(G1C), Neurod1CreER(D1B), and Neurod4CreER(D4A) were developed as described in STAR Methods and are deposited at the RIKEN Center for Biosystems Dynamics Research (RIKEN BDR) (Figures 1B and 1C). To showcase the tagged neuron images comprehensively, each neurogenic tagging line was crossed with TaumGFP-nLacZ mouse (Hippenmeyer et al., 2005), which is a global neuronal Cre reporter that expresses dual nucleus- and membrane-localized reporters under a constitutive neuronal promoter after the excision of the loxP-STOP-loxP cassette (Figure 1B). Staged pregnant mice were then intraperitoneally injected with TM only once during the gestation stages at embryonic day 9.5 (E9.5)–E18.5 (hereafter called TM9.5–TM18.5, referring to TM injection stages), and the tagged offspring were delivered and raised up to postnatal day 7 (P7).
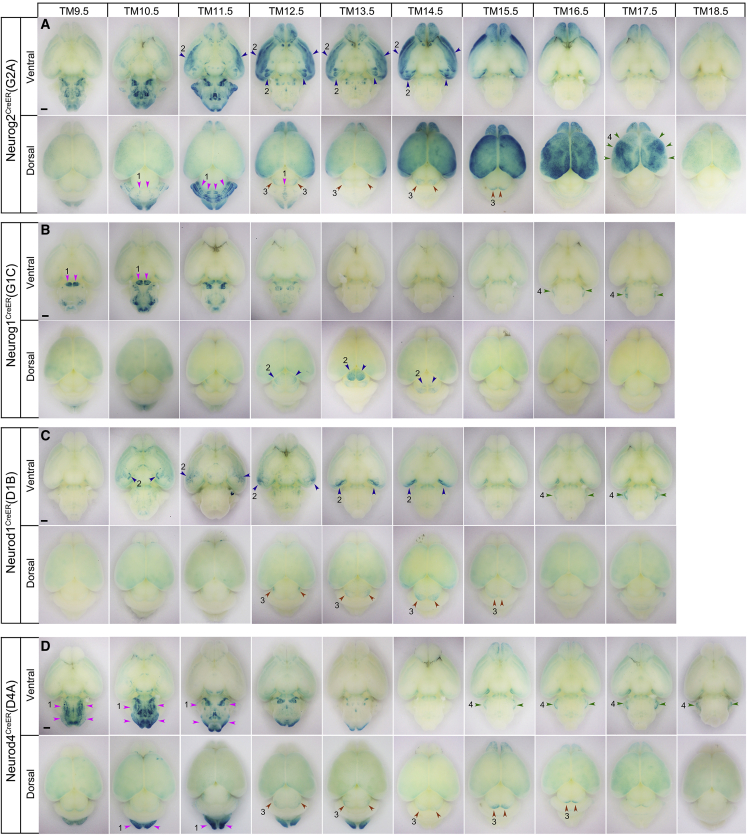
Figure 2 shows X-gal-stained images of whole-mount brains tagged by the four drivers at different TM stages. The tagged neurons stained blue by the nucleus-localized (nls)-βGAL reporter were distributed in different brain regions depending on the TM injection stages and on the driver lines. Some brain regions were tagged by multiple drivers, whereas others were tagged specifically only by a single driver (Figure 2 and Table 1). From the overall external appearance, a neurogenic wave was observed to migrate from posterior regions, such as the hindbrain, to the more anterior regions according to the TM injection stages (Figure 2). In the brain regions tagged by multiple drivers, the spatiotemporal patterns of tagging were basically similar (Table 1), indicating that all the driver lines capture a similar neurogenetic time window in neurons.
Figure 2.
Neurogenic-tagged neurons in whole-mount brains
Ventral and dorsal views of P7 brains tagged by Neurog2CreER(G2A) (A), Neurog1CreER(G1C) (B), Neurod1CreER(D1B) (C), and Neurod4CreER(D4A) (D) drivers. The TM injection stages are indicated at the top. Magenta arrowheads 1 show cerebellar Purkinje stripes in (A), pontine nuclei in (B), and olive nuclei and other structures in the medulla in (D). Blue arrowheads 2 show the olfactory cortex and amygdala nuclei in (A) and (C), and the superior colliculus in (B). Brown arrowheads 3 show the inferior colliculus in (A), (C), and (D). Green arrowheads 4 show the area-specific distribution of superficial cortical neurons in (A), and cochlear nuclei in (B), (C), and (D). Scale bars, 1 mm.
Table 1.
Comparison of neurogenic-tagged stages identified in current study and the birthdates determined in previous papers
| Region | Neurogenic-tagged stage | Driver line | Tagged territory | Birthdate reported | Method | Reference |
|---|---|---|---|---|---|---|
| Olfactory bulb | TM11.5–TM17.5 | Neurog2CreER(G2A) | from deep to surface | E10.5–E17.5 | [3H]thymidine | Hinds, 1968 |
| Neurod4CreER(D4A) | ||||||
| Piriform cortex | TM11.5–TM14.5 | Neurog2CreER(G2A) | E12.5 (peak) | BrdU | Sarma et al., 2011 | |
| Amygdala | TM12.5–TM15.5 | Neurog2CreER(G2A) | distinct nuclei | E11.5–E15.5 | [3H]thymidine | McConnell and Angevine, 1983 |
| Neurod1CreER(D1B) | ||||||
| Neocortex | TM12.5–TM17.5 | Neurog2CreER(G2A) | from deep to surface | E11.5–E17.5 | [3H]thymidine | Caviness, 1982 |
| Subplate neuron (neocortex) | TM11.5–TM12.5 | Neurod1CreER(D1B) | E11.5–E12.5 | BrdU | Hoerder-Suabedissen and Molnar, 2013 | |
| Hippocampus pyramidal neuron | TM13.5–TM17.5 | Neurog2CreER(G2A) | from deep to surface | E12.5–E16.5 | [3H]thymidine | Caviness, 1973 |
| Neurod1CreER(D1B) | ||||||
| Neurod4CreER(D4A) | ||||||
| Habenula | TM11.5–TM16.5 | Neurog2CreER(G2A) | from lateral to medial | E11.5–E16.5 | [3H]thymidine | Angevine, 1970 |
| Neurod1CreER(D1B) | ||||||
| Neurod4CreER(D4A) | ||||||
| Mammillary body | TM11.5–TM16.5 | Neurog2CreER(G2A) | from lateral to medial | E9.5–E13.5 | BrdU | Szabo et al., 2015 |
| Neurog1CreER(G1C) | ||||||
| Supramammillary nucleus | TM10.5–TM16.5 | Neurod4CreER(D4A) | E10.5–E14.5 | [3H]thymidine | Shimada and Nakamura, 1973 | |
| Superior colliculus | TM11.5–TM14.5 | Neurog1CreER(G1C) | E11.5–E14.5 | [3H]thymidine | Edwards et al., 1986 | |
| Inferior colliculus | TM12.5–TM16.5 | Neurog2CreER(G2A) | from lateral to medial | E12.5–E15.5 | [3H]thymidine | Zeng et al., 2009 |
| Neurog1CreER(G1C) | ||||||
| Neurod1CreER(D1B) | ||||||
| Neurod4CreER(D4A) | ||||||
| Cerebellum Purkinje cell | TM10.5–TM12.5 | Neurog2CreER(G2A) | sagittal stripes | E10.5–E12.5 | BrdU | Hashimoto and Mikoshiba, 2003 |
| Pontine nucleus | TM9.5–TM11.5 | Neurog1CreER(G1C) | E12.5–E14.5 | BrdU | Kawauchi et al., 2006 | |
| Trapezoid body | TM11.5–TM12.5 | Neurog2CreER(G2A) | E11.5–E12.5 | [3H]thymidine | Pierce, 1973 | |
| Neurog1CreER(G1C) | ||||||
| Neurod4CreER(D4A) | ||||||
| Inferior olive | TM10.5–TM12.5 | Neurog2CreER(G2A) | E9.5–E11.5 | [3H]thymidine | Pierce, 1973 | |
| Neurod4CreER(D4A) | ||||||
| Cochlear nucleus | TM15.5–TM17.5 | Neurog1CreER(G1C) | E14.5–E18.5 (second wave) | BrdU | Shepard et al., 2019 | |
| Neurod1CreER(D1B) | ||||||
| Neurod4CreER(D4A) | ||||||
| Spinal cord | TM9.5–TM13.5 | Neurog2CreER(G2A) | from ventral to dorsal | E9.5–E13.5 | [3H]thymidine | Nornes and Carry, 1978 |
| Neurod4CreER(D4A) | ||||||
| Dorsal root ganglion | TM9.5–TM12.5 | Neurod1CreER(D1B) | E10.5–E13.5 | [3H]thymidine | Lawson and Biscoe, 1979 | |
| Neurod4CreER(D4A) |
Representative brain regions are tagged by the neurogenic tagging driver lines. The TM stages for tagging are compared with the birthdates determined by using nucleotide analogs in mice. The reported birthdates are adjusted to set the day of mating at E0.5.
To expose the internal structures, we collected and coronally sectioned brains tagged at different TM stages by individual drivers. The interspaced serial sections were antibody stained with diaminobenzidine (DAB) for either the nls-βGAL or the membrane-localized mGFP reporter. All these sections were converted into high-resolution digital images (e.g., Figures 6B–6D) and subgrouped into datasets according to three categories, namely the driver line, TM stage, and stained reporter (nls-βGAL or mGFP). Each dataset defined by the combination of the three categories contained 142–180 interspaced section images, which were aligned along the entire anterocaudal axis of the brain (approximately 1.2 mm in length). This coverage is sufficiently high to identify even a small nucleus or structure in the brain (Table 1).
Figure 6.
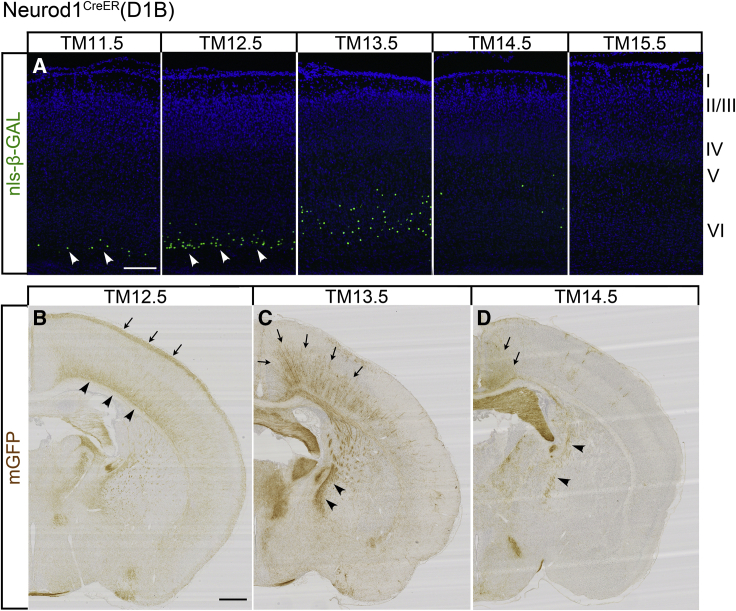
Cortex and telencephalic hemispheres tagged by Neurod1CreER(D1B)
(A) P7 cortical sections stained for nls-βGAL. Subplate neurons in the sublayer VIb are labeled at TM11.5–TM12.5 (arrowheads). Later TM injections label only a small number of neurons in the layers V and VI. Scale bar, 200 μm.
(B–D) mGFP staining of telencephalic sections prepared from P7 mice tagged at TM12.5 (B), TM13.5 (C), and TM14.5 (D). In (B), the labeling at the bottom (arrowheads) and the top (arrows) of the cortical plate corresponds to the typical positions of basal dendrites and axons of surviving subplate neurons. During later TM stages (C and D), the tagged neurons are concentrated on the medial cortical areas (arrows), and their axons selectively project to the medial part of the striatum (arrowheads). Scale bar, 500 μm.
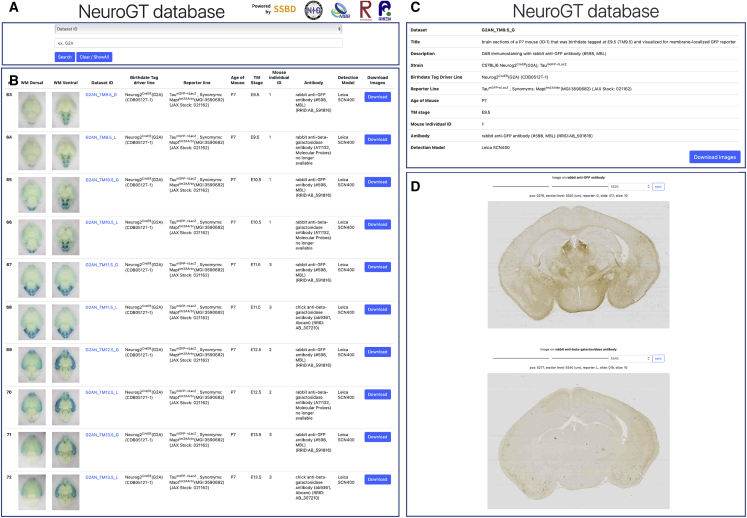
All datasets of the high-resolution section images (835 GB of data for 13,538 images in 84 datasets including TM-negative controls) and whole-mount images shown in Figure 2 are available in the NeuroGT database (https://ssbd.riken.jp/neurogt/). Users can search for these datasets from a web browser by using terms that match the meta-information stored in the database, such as their identifier, driver name, TM stage, or reporter used for staining (Figure 3A). The search results are displayed as a list of links to the dataset page (Figure 3B). To enable this search and visualization, development of the NeuroGT was based on the SSBD (Systems Science of Biological Dynamics) database (Tohsato et al., 2016). The dataset pages at the next stage allow users to download the high-resolution section images (Figure 3C) and interactively view their thumbnails together with the meta-information. The thumbnail images of nls-βGAL- and mGFP-stained sections are stacked separately in order along the anteroposterior axis (Figure 3D) and can be viewed sequentially by dragging the slider (Video S1). The search function for anatomical regions will be implemented in the future. Through NeuroGT, researchers can identify the drivers useful for their research and obtain them from RIKEN BDR (http://www2.clst.riken.jp/arg/TG%20mutant%20mice%20list.html).
Figure 3.
Screenshots from NeuroGT database
(A) Input window for keyword search.
(B) An example of a search result, which is returned as links to individual dataset pages with whole-mount brain images and selected meta-information.
(C) Meta-information of high-resolution section images and a button that allows users to download the images.
(D) Interactive viewer of the image thumbnails. The images of nls-βGAL and mGFP are stacked separately. By dragging the slider, coronal sections can be viewed sequentially along the anteroposterior axis. The sync button automatically matches the section level of the two reporter images. See also Video S1.
In the interactive viewer of thumbnail images, coronal section images stained either for mGFP or nls-βGAL reporter can be sequentially seen along the anteroposterior brain axis by dragging the slider. The sync button at each thumbnail stack matches the section level of the other reporter stack.
Table 1 summarizes the brain regions that were tagged by individual drivers. When the tagged stages were compared with the reported birthdates determined in mice by using nucleotide analogs, the ranges were fairly consistent (Table 1). The exceptions might be the pontine nucleus and dorsal root ganglion. Considering the short time lag often observed between the final S phase and the expression of CreER (Florio et al., 2012; Toma et al., 2014; Hirata et al., 2019; see also this paper), the TM stages for labeling these regions slightly preceded the birthdates determined with nucleotide analogs, suggesting that the loxP recombination might have occurred in pre-mitotic neuronal progenitors in these regions. Table 1 lists only limited brain regions from areas containing extensive tagged neurons. The NeuroGT database can be used to access and visualize a specific brain region of interest.
The following sections describe the features of each driver line, focusing on specific regions that can be effectively tagged by a particular line.
Neurog2CreER(G2A) driver using the neurog2 enhancer
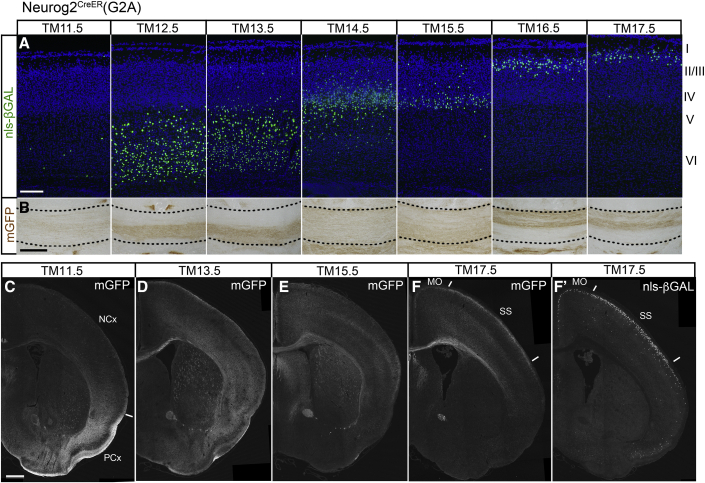
The Neurog2CreER(G2A) line tags the most extensive brain regions. Specifically, at TM10.5–TM12.5, sagittal stripes in the cerebellum were labeled (magenta arrowheads 1 in Figure 2A), reflecting birthdate-dependent generation of cerebellar Purkinje neurons (Hashimoto and Mikoshiba, 2003; Namba et al., 2011). TM injection between TM11.5 and TM17.5 heavily labeled olfactory bulb neurons, as reported previously (Hirata et al., 2019), and neurons in the central olfactory areas and the amygdala (blue arrowheads 2 in Figure 2A). The dense labeling of the piriform cortex at TM11.5–TM12.5 demarcated a sharp border from the neocortex that was labeled by later TM injections, as will be described in the next paragraph (see also Figure 4C). In the inferior colliculus of the midbrain, a neurogenic gradient from the lateral to the mediocaudal was marked at TM12.5–TM16.5 (brown arrowheads 3 in Figure 2A) as previously reported (Altman and Bayer, 1981). Labeling of the inferior colliculus was also observed in the same spatiotemporal pattern by other neurogenic tagging drivers (brown arrowheads 3 in Figures 2C and 2D; Table 1).
Figure 4.
Cortex and telencephalic hemispheres tagged by Neurog2CreER(G2A)
(A) P7 cortical sections stained for nls-βGAL. The tagged neurons with green-stained nuclei are arranged inside-out across the cortical layers according to the TM injection stages shown at the top.
(B) P7 corpus callosum (between dotted lines) stained for mGFP. Note that axons tagged at TM12.5–TM13.5 and TM16.5–TM17.5 course through the ventral and dorsal parts of the callosum, respectively. See also Figure S1.
(C–F) mGFP staining of telencephalic sections from P7 mice tagged at TM11.5 (C), TM13.5 (D), TM15.5 (E), and TM17.5 (F). Different fiber systems and neurons are visualized depending on the stage of TM injection. (F′) A section stained for nls-βGAL from the same mouse as that in (F). The white line in (C) indicates the boundary between the neocortex (NCx) and the piriform cortex (PCx). In (F) and (F′), tagged neurons are more abundant in the somatosensory area (SS) than in the motor area (MO).
Scale bars, 200 μm (A and B) and 500 μm (C–F).
In cross-sections of the neocortex, the inside-out birth-order arrangement of neurons was clearly visible with the nls-βGAL reporter (Figure 4A). As expected from the neurog2 expression, only excitatory projection neurons, but not GABAergic inhibitory neurons, were tagged in the neocortex (Figures S1A–S1C). The mGFP reporter prominently labeled neuronal fibers, including axons and dendrites (Figures 4B–4F). In a close-up of the corpus callosum (Figure 4B), long axons projecting from the tagged neocortical neurons were visualized. Interestingly, the axons of the early-born (TM12.5–TM13.5) and late-born (TM16.5–TM17.5) cortical neurons formed dorsoventrally segregated fascicles within the callosum, presenting a feature that would have been difficult to recognize by nucleotide-based birthdating. Previous studies have reported the dorsoventral segregation of callosal axons from the medial and lateral cortical areas (Piper et al., 2009; Nishikimi et al., 2011; Zhou et al., 2013). The present axon compartments formed by the early-born and late-born callosal neurons appeared to be different from those based on the distinct cortical areas (Figures S1D–S1G), adding complexity to the organization of the corpus callosum.
Another unique feature of the neocortex was the patchy areal distribution of superficial neurons in the layer II/III tagged around the late TM17.5 stage (green arrowheads 4 in Figure 2A). Somatosensory and visual areas contained packed X-gal-labeled neurons, whereas frontal, motor, and medial cingulate areas contained less abundant labeled neurons. The biased areal distribution of these superficial neurons was also confirmed in neocortical sections (Figures 4F and 4F′). A previous nucleotide-based neuronal birthdating study only reported a neurogenetic gradient in the superficial layers (Bayer and Altman, 1991). Perhaps the discrete mosaic distribution of superficial neurons was more readily recognizable in a whole-mount cortical representation by using neurogenic tagging.
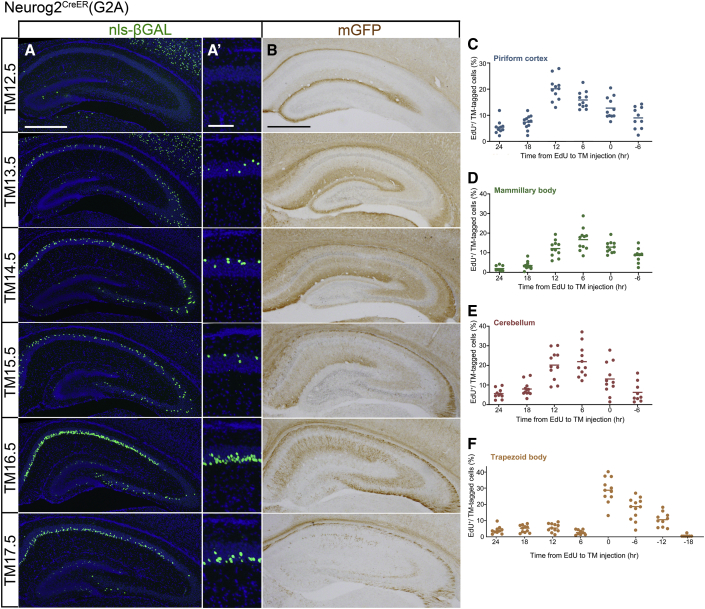
In the hippocampus, pyramidal neurons in the CA1–CA3 regions were labeled with embryonic TM injections (Figure 5A). Interestingly, deep and superficial neurons in the CA1 layer were separately labeled by different TM injection times (Figure 5A′). They appear to be the neuronal subsets in the radial axis that have recently attracted much attention (Slomianka et al., 2011; Soltesz and Losonczy, 2018). The membrane-localized mGFP reporter, on the other hand, visualized the laminar organization of the hippocampus (Forster et al., 2006), where laminar-specific afferent axons from other brain regions and local projections of hippocampal proper neurons were differentially labeled depending on the TM stage (Figure 5B).
Figure 5.
Hippocampus and time course of neurons tagged by Neurog2CreER(G2A)
(A and B) nls-βGAL (A, A′) and mGFP (B) staining of hippocampal sections prepared from P7 mice that were tagged at different stages indicated on the left. (A′) High magnification of the CA1 pyramidal layer in (A). Neurons in the deep and superficial sublayers are labeled at TM13.5–TM15.5 and TM16.5–TM17.5, respectively. In (B), afferent axons such as the perforant path are prominently labeled at early TM12.5–TM13.5, whereas dendrites and efferent axons of hippocampal pyramidal neurons are labeled at late TM16.5–TM17.5. Scale bars, 500 μm (A and B) and 100 μm (A′).
(C–F) The proportion of EdU and nls-βGAL double-positive neurons in the piriform cortex layer II (C), mammillary body (D), midline compartment of the cerebellum (E), and the trapezoid body (F). TM was injected at the fixed TM12.5 stage, and EdU was injected at the indicated time before the TM injection. The dots show the values for individual mice, and the horizontal lines show the means. Each value was calculated from 127–254 (C), 86–395 (D), 101–331 (E) and 113–366 (F) nls-βGAL-positive neurons.
To characterize the timing of individual neurons that underwent TM-induced recombination in relation to their last cell cycle, we conducted a double injection of TM at the fixed TM12.5 stage and of EdU at a certain time point either before or after TM injection (Figures 5C–5F). Although the time courses for the emergence of double-positive neurons for EdU and the nls-βGAL reporter varied across brain regions, in several brain domains such as the piriform cortex (Figure 5C), mammillary body (Figure 5D), and cerebellum (Figure 5E), TM-induced recombination was most prevalent in neurons 6–12 h after the final DNA synthesis. This time course was similar to that previously observed in olfactory bulb neurons (Hirata et al., 2019). There were, however, a few exceptions; for example, neurons in the trapezoid body were most frequently double labeled when EdU and TM were co-injected (Figure 5F), suggesting that TM-induced recombination occurs in the progenitor stage before the final DNA synthesis. Nonetheless, a peak of double-positive neurons was detected in a specific time interval between EdU and TM injections, implying that the TM-induced recombination marks the neurons only within a short differentiation time window (Figure 5F).
Neurog1CreER(G1C) driver using the neurog1 enhancer
The characteristic regions tagged by the Neurog1CreER(G1C) driver were the pontine nucleus (magenta arrowheads 1 in Figure 2B) and the superior colliculus in the midbrain (blue arrowheads 2 in Figure 2B), both of which were not labeled in whole-mount brain preparations by the other three drivers (Figures 2A–2D and Table 1).
In sections of the superior colliculus, neurons in the superficial sensory layers that receive retinal axons were labeled at TM12.5–TM13.5, and neurons in the deep motor layers were labeled sparsely for a more protracted TM11.5–TM14.5 (Figures S2A and S2B). This spatiotemporal pattern of labeling resembles the reported pattern of neurogenesis in the superior colliculus (Edwards et al., 1986), where superficial and deep-layer neurons are generated as distinct compartments following different time courses. Scattered neurons in the deep motor layers were also tagged by the other drivers at the same TM11.5–TM14.5 stages (Figures S2C and S2D), although they were invisible in the whole-mount preparations (Figure 2). The observation that the superficial layer neurons were only tagged by the Neurog1CreER(G1C) driver seems to be consistent with the idea that the superficial sensory layers and the deep motor layers of the superior colliculus are populated with neuronal populations of distinct origins.
Neurod1CreER(D1B) driver using the neurod1 enhancer
The Neurod1CreER(D1B) driver tagged only several externally visible structures in the whole-mount brains; some olfactory areas and the amygdala were significantly labeled at TM10.5–TM14.5 (blue arrowheads 2 in Figure 2C). The labeled amygdala nuclei overlapped with, but were distinct from, those labeled by the Neurog2CreER(G2A) driver (Figure 2A). Neurons in the inferior colliculus (brown arrowheads 3 in Figures 2A, 2C, and 2D) and the cochlear nucleus (green arrowheads 4 in Figures 2B–2D) were labeled following the same spatiotemporal pattern shared by the other drivers (Table 1). The late labeling of the cochlear nucleus after TM15.5 appeared to correspond with the second wave of neurogenesis in the dorsal cochlear nucleus reported recently (Shepard et al., 2019).
In cross-sections of the neocortex, a tangential band of neurons was labeled at the border between the neocortex and white matter at TM11.5–TM12.5 (Figure 6A). These cells appeared to be surviving subplate neurons in layer VIb (Friedlander and Torres-Reveron, 2009). The mGFP reporter selectively visualized their characteristic dense basal dendrites at the bottom of the cortical plate and widespread axons in layer 1 (Figure 6B; Clancy and Cauller, 1999). Tagging at TM13.5–TM14.5 labeled only a few cortical neurons scattered in layers VI and V (Figure 6A). These neurons were mainly positioned in the medial cortex (Figures 6C and 6D), and their axons were observed to selectively project to the medial part of the striatum (Figures 6C and 6D), exhibiting a unique feature reported for neurons in the medial prefrontal area (Gerfen et al., 2013; Mailly et al., 2013). Later-stage TM injections did not label neurons in the upper layers (Figure 6A), showing a clear contrast to the inside-out pattern throughout the cortical plate induced by the Neurog2CreER(G2A) driver (Figure 4A).
Neurod4CreER(D4A) driver using the neurod4 enhancer
The Neurod4CreER(D4A) driver abundantly tagged neurons in the caudal parts of the brain such as the medulla and the spinal cord (magenta arrowheads 1 in Figure 2D). The spatial pattern and time course of labeling in these caudal regions resembled those of Neurog2CreER(G2A) (Figure 2A and Table 1), consistent with the idea that neurod4 is a downstream effector of neurog2 (Seo et al., 2007; Masserdotti et al., 2015). However, there were differences between the two drivers; for example, the neocortex and cerebellar Purkinje cells were not significantly labeled by the Neurod4CreER(D4A) driver (Figure 2D and Table 1).
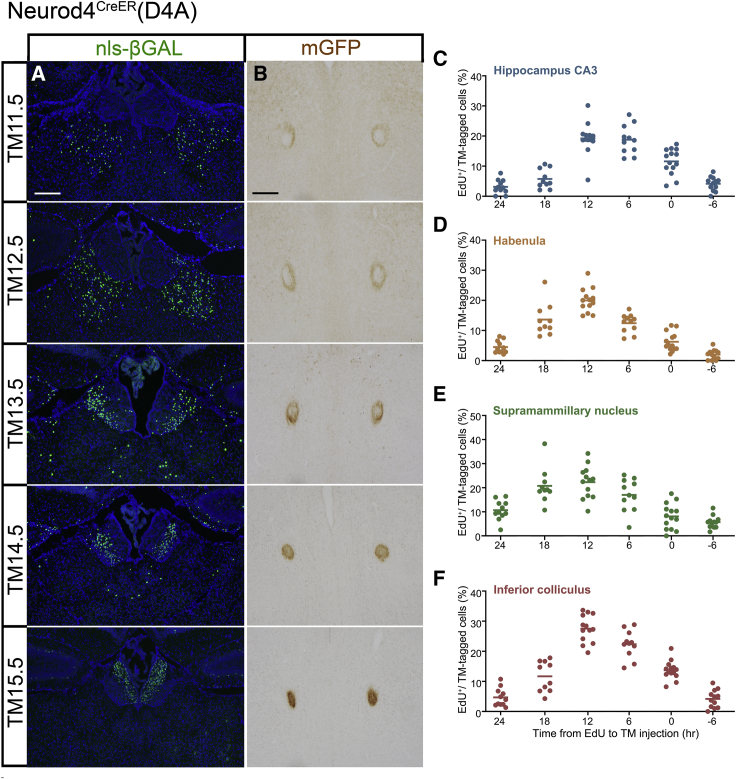
This driver notably visualized long projecting axons in several brain regions. For example, mGFP-labeled optic axons from the retinal ganglion cells tagged at TM13.5–TM14.5 were targeted at the superficial layers of the superior colliculus (Figure S2D). The habenular neurons in the thalamus were generated in a lateral-to-medial gradient (Figure 7A) (Angevine, 1970; Aizawa et al., 2007), whereas their axons exhibited a periphery-to-center arrangement within the fasciculus retroflexus, which is the axon tract formed by habenular neurons (Figure 7B).
Figure 7.
Habenula and time course of neurons tagged by Neurod4CreER(D4A)
(A and B) nls-βGAL staining of the habenula nucleus (A) and mGFP staining of the fasciculus retroflexus (B) obtained from P7 mice that were tagged at different stages indicated on the left. Depending on the TM stage, habenular neurons are labeled from lateral to medial (A). Their axons are labeled from the periphery to the center of the fasciculus retroflexus (B). Scale bar, 200 μm.
(C–F) The proportion of EdU and nls-βGAL double-positive neurons in the hippocampus CA3 (C), habenula (D), supramammillary nucleus (E), and inferior colliculus (F). TM was injected at the fixed E14.5 stage, and EdU was injected at the indicated time before TM injection. The dots show the values of individual mice, and the horizontal lines show the means. Each value was calculated from 20–153 (C), 71–257 (D), 43–173 (E), and 52–199 (F) nls-βGAL-positive neurons.
Using this driver, we also characterized the cell-cycle timing of neurons that underwent TM-induced recombination. TM injection at the fixed E14.5 stage labeled neurons in the hippocampal CA3 region (Figure 7C), habenula (Figure 7D), supramammillary nucleus (Figure 7E), and inferior colliculus (Figure 7F). The time-spaced injection of EdU indicated that neurons were most susceptible to TM-induced recombination at 6–12 h after the final DNA synthesis. Although the time course of double labeling seemed slightly delayed compared with that of Neurog2CreER(G2A) (Figures 5C–5F), such a comparison might not be meaningful, as different neurons and TM stages were involved and we could not quantitatively determine the time lag in the gene expression cascade from neurog2 to neurod4.
Discussion
Neurogenic tagging resource
In the past few decades, various important discoveries have been made through neuronal birthdating (Bayer and Altman, 1987; Govindan et al., 2018). The neurogenic tagging resource presented in this study resolves some of the limitations of the previous method and forges a link between this traditional histological classification and the new experimental approaches and applications. Specifically, neurogenic tagging can be easily combined with various molecular genetic tools such as optogenetics and chemogenetics (Deisseroth et al., 2006; Alexander et al., 2009), which enable the functional manipulation of birthdate-classified neuronal subsets. We hope that the unique ideas of researchers will lead to important discoveries using this resource, given that the neuronal birthdate is so fundamental to organizing the neural circuitry.
This neurogenic tagging method uses the expression timing of neuronal differentiation genes; therefore, the underlying biological principle is significantly different from that of the traditional method using nucleotide analogs. The four neuronal differentiation genes exploited in our resources were selected on the basis of their seemingly transient expression during the maturation phase of neurons as shown in a previous study (Mattar et al., 2008) and databases (Allen Developmental Mouse Brain Atlas, https://developingmouse.brain-map.org; GENSAT: Gene Expression Nervous System Atlas, http://www.gensat.org/index.html). As expected, the neurons underwent loxP recombination 6–12 h after the final DNA synthesis in different parts of the nervous system (Figures 5C–5E and 7C–7F), and recapitulated the patterns consistent with those seen in birthdating analyses using nucleotide analogs (Table 1). In some exceptional cases, such as the trapezoid body (Figure 5F), TM-induced recombination seemed to occur within a short time window during the progenitor stage before the final DNA synthesis. In the pontine nucleus and the dorsal root ganglion (Table 1), the earlier tagging stages, compared with the previously determined birthdates, also suggest that TM-induced recombination occurs pre-mitotically in these regions. Thus, neurogenic tagging using neuronal differentiation genes appears to mark the timing of neuron commitment rather than cell-cycle exit (Hirata et al., 2019). Recently, increasing evidence has indicated that neuron commitment and cell-cycle exit are dissociable processes (Imayoshi et al., 2013; Hardwick and Philpott, 2014; Oberst et al., 2019). A prime example is the cortical basal progenitor, which is the fate-restricted neural progenitor that undergoes additional cell divisions but only generates neurons (Lodato and Arlotta, 2015; Hevner, 2019). Our ongoing study shows that these cortical basal progenitors are indeed tagged by one of the neurogenic tagging driver lines.
During development, the neurod1 gene is widely expressed in many nervous systems (Miyata et al., 1999; Mattar et al., 2008), and a short enhancer element of this gene is commonly used to study neuronal differentiation in vivo and in vitro (Guerrier et al., 2009). Somewhat contradictory to the general use of this gene, the Neurod1CreER(D1B) driver tagged only a restricted subset of neurons (Figures 2C and 6). This might be because we obtained only one mouse line that exhibited significant CreER activity after the intensive production of transgenic mice by using the neurod1 gene. Regardless, the unique characteristics of neuronal tagging by the Neurod1CreER(D1B) driver shown in this study and NeuroGT demonstrate the usefulness of this driver line for some specific purposes.
The present collection of four neurogenic tagging drivers does not fully cover all brain regions. For example, the majority of neurons in the striatum and hypothalamus were unlabeled by any of the drivers. To complement the current collection a promising candidate is the Ascl1 gene, which is expressed in the basal plate of the neural tube in a manner complementary to that of neuronal differentiation genes used in this study (Ma et al., 1997; Wilkinson et al., 2013). Furthermore, the mice that express CreER under the Ascl1 gene enhancer have been reported to apparently show birthdate-dependent recombination (Tg(Ascl1-cre/Esr1∗)1Jejo, MGI: 3767428, Battiste et al., 2007; Ascl1tm1.1(cre/ERT2)Jejo, MGI: 4452601, Kim et al., 2008; Sudarov et al., 2011). In the future, we hope to include detailed image data of neurons tagged by Ascl1CreER mice in the NeuroGT database.
To date, multiple CreER-expressing mice under the control of neuronal differentiation genes have been generated by several groups (Neurog2tm1(cre/Esr1∗)And, MGI: 2652037, Zirlinger et al., 2002; Tg(Neurog1-cre/ERT2)1Good/J, MGI: 3807088, Koundakjian et al., 2007; Neurog2iCreERT2, Winpenny et al., 2011; Neurog2tm1(icre/ERT2∗)Ggc, MGI: 5431772, Florio et al., 2012; Tg(Neurod1-cre/ERT2)M1Fcal, MGI: 5582823, Aprea et al., 2014). Although these mice have been primarily used for cell lineage analyses related to the manipulated genes, they are theoretically applicable to birthdate-based classification and manipulation of neurons. Our Nerog2CreER(G2A) driver mouse generated by BAC transgenesis has a greater CreER activity than that under the endogenous locus in Neurog2-CreER knockin mice (Zirlinger et al., 2002). However, suitable drivers differ depending on the research purpose. It is important to note that information in the NeuroGT database can be useful even for developing a research design that does not use our mouse lines.
Features highlighted by the present neurogenic tagging analyses
Our original motivation for developing the neurogenic tagging method was to visualize axon projections of birthdate-classified neuron subsets in the olfactory bulb in adulthood. This method was indeed effective in revealing the birthdate-dependent olfactory axon trajectories (Hirata et al., 2019). It has also been applied to examine birthdate-dependent compartments in the cerebellum (Tran-Anh et al., 2020; Zhang et al., 2020). The present analyses using the multiple neurogenic tagging drivers added interesting findings, three of which are discussed below.
Corpus callosum
Within the corpus callosum, early-born (TM12.5–TM13.5) and late-born (TM16.5–TM17.5) cortical neurons formed segregated axon bundles (Figures 4B and S1D–S1G), consistent with the current knowledge that the callosal projections are formed by a subset of layers II/III and V neurons in the neocortex (Fame et al., 2011). The segregated construction of the callosum by pioneering and following axons has been proposed (Koester and O'Leary, 1994; Rash and Richards, 2001). Although the previous studies have considered the time lag of axon arrivals from the medial and lateral cortical areas (Piper et al., 2009; Nishikimi et al., 2011; Zhou et al., 2013), the present analysis indicates that the time lag between the early-born and late-born callosal neurons in different layers also underlies the organization of this axon bundle, which is the largest in placental mammals (Suarez et al., 2014).
Fasciculus retroflexus
Neurogenic tagging revealed a periphery-to-center axon topology within the fasciculus retroflexus (Figures 7A and 7B). A previous axon-tracing analysis reported that the medial and lateral habenula nuclei project their axons into the core and shell of this tract, respectively (Herkenham and Nauta, 1979). To explain this organization, the present study provides a hint: late-growing axons might penetrate the center of the bundle pre-formed by early-growing axons. In general, for axon tracts positioned on the brain surface, late-growing axons are sequentially added to the pial superficial space, displacing the pre-existing axons deeper (Walsh et al., 1983; Walsh and Guillery, 1985; Inaki et al., 2004; Yamatani et al., 2004). Although little is known about the development of internally located axon tracts, new axons are found to grow in the center of pre-formed axon bundle in the mushroom body of Drosophila (Kurusu et al., 2002). Within the optic nerve, which is an isolated axon bundle, newly growing axons are positioned near the center (Walsh, 1986).
Hippocampus pyramidal neurons
Although hippocampal neurons in the single pyramidal layer have been regarded as a homogeneous population, recent studies indicate that hippocampal pyramidal neurons are in fact heterogeneous along the radial axis (Slomianka et al., 2011; Soltesz and Losonczy, 2018). The deep and superficial subsets in the layer express different genes, make different connections, and are expected to have distinct functions in different behavioral contexts (Mizuseki et al., 2011; Lee et al., 2014; Danielson et al., 2016). These subsets are assumed to differentiate at different times in an inside-out pattern (Slomianka et al., 2011; Soltesz and Losonczy, 2018). The present neurogenic tagging analysis confirmed this assumption and allowed effective classification of these neuronal subsets (Figures 5A and 5A′). An advantage of this neurogenic tagging is that this classification can now be connected to functional tests. We hope that the neurogenic tagging resource will benefit ingenious ideas that will lead to future scientific discoveries.
Limitations of the study
Neurogenic tagging is different from nucleotide-based neuronal birthdating. The timing of recombination in the cell cycle differs among neurons and appears to take place prior to the final mitosis in some neurons. It is important to consider this difference when the neurogenic tagging approach is taken, as this difference might be beneficial in some cases. In the NeuroGT database, the anatomical ontology of tagged brain regions is not yet available but will be included in the future to enable searching by anatomical regions.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Chicken anti-β-GAL | Abcam | Cat# ab9361; RRID: AB_307210 |
| Rabbit anti-GFP | MBL International | Cat# 598; RRID: AB_591816 |
| Chicken anti-GFP antibody | Abcam | Cat# ab13970; RRID: AB_300798 |
| Rat monoclonal anti-CTIP2 antibody | Abcam | Cat# ab51502; RRID: AB_882455 |
| Mouse monoclonal anti-SATB2 antibody | Abcam | Cat# ab51502; RRID: AB_882455 |
| Rabbit anti-GABA antibody | Sigma-Aldrich | Cat# A2052; RRID: AB_477652 |
| Rabbit anti-NRP1 antibody | Kawakami et al., 1996 | |
| Donkey biotin-SP-conjugated anti-rabbit IgG | Jackson Immunoresearch | Cat# 711-065-152; RRID:AB_2340593 |
| Donkey Alexa488-conjugated anti-rabbit IgG | Life Technologies | Cat# A-21206; RRID: AB_141708 |
| Donkey Cy3-conjugated anti-rabbit IgG | Jackson Immunoresearch | Cat# 711-165-152; RRID: AB_2307443 |
| Donkey Alexa488-conjugated anti-chicken IgY | Jackson Immunoresearch | Cat# 703-545-155; RRID: AB_2340375 |
| Donkey biotin-SP-conjugated anti-chicken IgY | Jackson Immunoresearch | Cat# 703-065-155; RRID: AB_2313596 |
| Donkey Cy3-conjugated anti-rat IgG | Jackson Immunoresearch | Cat# 712-166-153; RRID: AB_2340669 |
| Donkey Cy3- conjugated anti-mouse IgG | Jackson Immunoresearch | Cat# 715-165-150; RRID: AB_2340813 |
| Chemicals, peptides, and recombinant proteins | ||
| Tamoxifen | Sigma-Aldrich | T5648; CAS: 10540-29-1 |
| Corn oil | Sigma-Aldrich | C8267; CAS: 8001-30-7 |
| Progesterone | Fujifilm Wako | 161-14531; CAS: 57-83-0 |
| 5-ethynyl-2'-deoxyuridine (EdU) | Tokyo Chemical Industry | E1057; CAS: 61135-33-9 |
| Alexa555 azide triethylammonium salt | Thermo Fisher Scientific | A20012 |
| X-gal | Fujifilm Wako | 029-15043; CAS: 7240-90-6 |
| Elite ABC kit | Vectastain | PK-6100; RRID: AB_2336819 |
| DAB | Dojindo | 347-00904; CAS: 7411-49-5 |
| Deposited data | ||
| NeuroGT database | This paper | https://ssbd.riken.jp/neurogt/ |
| Raw images for NeuroGT database and all figures, and counts of EdU-positive and -negative neurons | This paper | SSBD:repository: https://doi.org/10.24631/ssbd.repos.2021.03.001 |
| Experimental models: organisms/strains | ||
| Mouse: C57BL/6 | Japan SLC | RRID: MGI:5295404 |
| Mouse: ICR | Japan SLC | RRID: MGI:5462094 |
| Mouse: Neurog2CreER(G2A); C57BL/6-Tg(Neurog2-cre/ERT2)G2ATahi |
Hirata et al., 2019 | Accession # CDB0512T−1: http://www2.clst.riken.jp/arg/TG%20mutant%20mice%20list.html |
| Mouse: Neurog1CreER(G1C); C57BL/6-Tg(Neurog1-cre/ERT2)G1CTahi |
This paper | Accession # CDB0526T: http://www2.clst.riken.jp/arg/TG%20mutant%20mice%20list.html |
| Mouse: Neurod1CreER(D1B); C57BL/6-Tg(Neurod1-cre/ERT2)D1BTahi |
This paper | Accession #: CDB0510T: http://www2.clst.riken.jp/arg/TG%20mutant%20mice%20list.html |
| Mouse: Neurod4CreER(D4A); C57BL/6-Tg(Neurod4-cre/ERT2) D4ATahi |
This paper | Accession # CDB0511T: http://www2.clst.riken.jp/arg/TG%20mutant%20mice%20list.html |
| Mouse: TaumGFP-nLacZ; 129P2-Mapttm2Arbr |
Hippenmeyer et al., 2005 | RRID: IMSR_JAX:021162 |
| Mouse: ROSA26R-Cre; Gt(ROSA)26Sortm1Sor |
Soriano, 1999 | MGI: 1861932 |
| Oligonucleotides | ||
| Cre primer1: 5’-TAAAGATATCTCACGTA CTGACGGTG-3’ |
Hirata et al., 2019 | N/A |
| Cre primer2: 5’-TCTCTGACCAGAG TCATCCTTAGC-3’ |
Hirata et al., 2019 | N/A |
| Recombinant DNA | ||
| BAC clone RP24-347K19 | BACPAC Resource Center | RP24-347K19 |
| BAC clone RP23-280C11 | BACPAC Resource Center | RP23-280C11 |
| BAC clone RP23-55O18 | BACPAC Resource Center | RP23-55O18 |
| CreERT2 | Feil et al., 1997 | N/A |
| p23loxZeo | Dr. Junji Takeda, Osaka University | N/A |
| p24loxZeo | Dr. Junji Takeda, Osaka University | N/A |
| Software and algorithms | ||
| Cell Sens Standard (ver.1.16) | Olympus | N/A |
| Fluoview FV-ASW (ver 4.02) | Olympus | N/A |
| SCN400 Client (version 2.2.0.3789) | Leica | N/A |
| Photoshop CC 2021 (22.2.0) | Adobe | N/A |
| Fiji / ImageJ (2.0.0) | Schindelin et al., 2012 | RRID: SCR_002285 |
| Microsoft Excel (Mac 2019, 16.46) | Microsoft | N/A |
| GraphPad Prism 8 | GraphPad | N/A |
| Other | ||
| Slide scanner (SCN400) | Leica | N/A |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to Tatsumi Hirata (tathirat@nig.ac.jp).
Materials availability
The neurogenic tagging mouse lines are deposited at RIKEN BDR (http://www2.clst.riken.jp/arg/TG%20mutant%20mice%20list.html) and provided with a completed materials transfer agreement. The accession numbers are as follows: Neurog2CreER(G2A) driver, CDB0512T−1; Neurog1CreER(G1C) driver, CDB0526T; Neurod1CreER(D1B) driver CDB0510T; Neurod4CreER(D4A) driver, CDB0511T. All other reagents generated in this study are available from the Lead Contact.
Data and code availability
All images acquired in this study are available in the NeuroGT database at https://ssbd.riken.jp/neurogt/. Raw images and unprocessed data related to this study and NeuroGT are archived in SSBD:repository (https://doi.org/10.24631/ssbd.repos.2021.03.001).
Experimental model and subject details
Mice
C57BL/6 wild-type mice (RRID: MGI: 5295404) and outbred ICR foster mothers (RRID: MGI: 5462094) were purchased from Japan SLC Inc. The TaumGFP-nLacZ reporter mice, officially known as 129P2-Mapttm2Arbr (RRID: IMSR_JAX: 021162, Hippenmeyer et al., 2005), with a CD-1 mixed background were provided by Dr. Silvia Arber at Friedrich Miescher Institute for Biomedical Research and backcrossed with C57BL/6 wild-type mice for at least four generations before use in this study. All mice were maintained in the animal facility of the National Institute of Genetics or RIKEN BDR. They were group housed in conventional cages under controlled conditions (temperature 23 ± 2°C, humidity 50 ± 10%, 12 h light-dark cycle), and food and water were provided ad libitum. All procedures for the care and treatment of mice were approved by the Institutional Animal Committees and carried out according to their guidelines.
Heterozygous neurogenic tagging driver mice were mated with homozygous reporter mice. The day on which a vaginal plug was detected and the day of birth were designated as embryonic day 0.5 (E0.5) and postnatal day 0 (P0), respectively. TM treatment was performed by intraperitoneally injecting a staged pregnant mouse with 250 μL of corn oil (C8267, CAS#8001-30-7, Sigma-Aldrich) containing 9 mM tamoxifen (T5648, CAS#10540-29-1, Sigma-Aldrich) and 5 mM progesterone (161-14531, CAS# 57-83-0, Fujifilm Wako). As TM often delays delivery, when pups were not born by E19.5, they were collected by caesarian delivery and given to ICR foster mothers. Brain sampling was performed indiscriminately once pups grew up to the appropriate age, and only brains of the desired genotype were later selected by PCR genotyping of the reserved tissues for CreER internal sequences. At the time of sampling, the sex of the mice was still obscure and therefore undetermined; for the Nerog2CreER(G2A) driver, it is highly likely that only male mice were sampled because the transgene seems to be located on the Y chromosome (Hirata et al., 2019).
Methods details
Generation of neurogenic tagging mouse lines
The Neurog2CreER(G2A), officially named C57BL/6-Tg(Neurog2-cre/ERT2)G2ATahi, (accession No. CDB0512T−1: http://www2.clst.riken.jp/arg/TG%20mutant%20mice%20list.html) was established as described previously (Hirata et al., 2019).
The Neurog1CreER(G1C) line, officially named C57BL/6-Tg(Neurog1-cre/ERT2)G1CTahi, was generated using the genomic BAC clone RP24-347K19 encoding the mouse neurog1, which was obtained from the BACPAC Resource Center (Children’s Hospital Oakland Research Institute, Oakland, CA). The entire coding sequence of exon1 was replaced by CreER(T2) (Feil et al., 1997) (gifted by Dr. Pierre Chambon), and the loxP site in the vector backbone (pTARBAC) was deleted by replacement with the zeocin resistance gene in the p24loxZeo (gifted by Dr. Junji Takeda). For the Neurod1CreER(D1B) line, officially named C57BL/6-Tg(Neurod1-cre/ERT2)D1BTahi, the CreER(T2) coding sequence was inserted at the first ATG sequence of exon2 in the BAC clone RP23-280C11 encoding the mouse neurod1 (BACPAC Resource Center). For the Neurod4CreER(D4A) line, officially named C57BL/6-Tg(Neurod4-cre/ERT2)D4ATahi, the CreER(T2) coding sequence was inserted at the first ATG sequence of exon2 in the BAC clone RP23-55O18 encoding the mouse neurod4 (BACPAC Resource Center), and the loxP site in the vector backbone (pBACe3.6) was deleted by replacement with the zeocin resistance gene in the p23loxZeo (gifted by Dr. Junji Takeda). Except for the above-mentioned wild-type loxP sequences, the BAC vector backbone sequences, including a few genes, were unmodified.
The constructed BAC recombinants were injected into fertilized eggs with a C57BL/6 background, and the tail genomic DNA of the resulting mice was assayed by PCR for the integration of the transgene. Transgene containment was determined by PCR using internal Cre recombinase primers 5’-TAAAGATATCTCACGTACTGACGGTG-3’ and 5’-TCTCTGACCAGAGTCATCCTTAGC-3’, resulting in the amplification of 300-bp fragments. In total, eleven, two, and five mice were found to have random integrations of the transgenes for neurog1, neurod1, and neurod4, respectively, including multiple BAC constructs for each gene. These mouse lines were assayed for CreER activity by crossing them with ROSA26R Cre reporter mice, officially named Gt(ROSA)26Sortm1Sor (RRID: MGI: 1861932, Soriano, 1999). After crossing, pregnant mice were injected with TM solution at E12.5 or E14.5. Embryos were dissected from the dam at E18.5-E19.5, and their isolated brains were whole-mount stained with X-gal (5-bromo-4-chloro-3-indoyl-f3-D-galactopyranoside, 029-15043, CAS#7240-90-6, Fujifilm Wako) as will be described later. Through this screen, Neurog1CreER(G1C), Neurod1CreER(D1B), and Neurod4CreER(D4A) were identified by the highest recombination rate among the transgenic mice for each gene.
Histochemistry
The mice were anesthetized and transcardially perfused with 4% paraformaldehyde (PFA)/phosphate-buffered saline (PBS). For whole-mount visualization of the nls-β-GAL signals, the brains were immediately dissected and immersed in PBS containing 1 mg/mL of X-gal, 5 mM K3[Fe(CN)6], 5 mM K4[Fe(CN)6], 2 mM MgCl2, and 1% Tween 20 for 2.5 hours at 37°C (Saga et al., 1992). To prepare cryosections, the dissected brains were further fixed with 4% PFA/PBS for 8–24 hours, immersed in 30% sucrose in PBS, and frozen in a 2:1 mixture of OCT compound (Sakura Finetek) and 30% sucrose/PBS. Coronal sections of 20-μm thickness were cut on a cryostat, and mounted on MAS-coated glass slides (Matsunami Glass).
The sections were washed with 10 mM Tris-HCl (pH 7.4), 130 mM NaCl and 0.1% Tween 20 (TBST) and incubated with rabbit anti-GFP antibody (1:1000, #598, MBL International Cat# 598, RRID: AB_591816) diluted in PBS containing 0.1% Tween 20 and 0.5% blocking reagent (FP1020, PerkinElmer) overnight at 4°C. After treatment with 0.7% H2O2 in methanol for 5 min at 4°C, antibody labeling was detected using donkey biotin-SP-conjugated anti-rabbit IgG antibody (1:1000, Jackson Immunoresearch Cat# 711-065-152. RRID: AB_2340593), amplified with the Elite ABC kit (1:300, PK-6100, Vectastain, RRID: AB_2336819), and visualized in TBST containing 0.13 mg/mL 3,3’-diaminobenzidine tetrahydrochloride (DAB, 347-00904, CAS#7411-49-5, Dojindo) and 0.03% H2O2. In some specimens, the binding of anti-GFP antibodies was fluorescently visualized with donkey Alexa488-conjugated anti-rabbit IgG (1:1000, Life Technologies Cat# A-21206, RRID: AB_141708). For antibody staining of nls-β-GAL, the sections were heat-treated in an antigen retrieval solution (HistoVT One, Nacalai Tesque) by autoclaving at 105°C for 2 min and incubated with chicken anti-βGAL antibody (1:2000, Abcam Cat# ab9361, RRID: AB_307210) diluted in PBS containing 0.1% Tween 20 and 0.5% blocking reagent overnight at 4°C. The bound antibodies were detected with donkey Alexa488-conjugated anti-chicken IgY (1:1000, Jackson Immunoresearch Cat# 703-545-155, RRID: AB_2340375). In NeuroGT database, the binding of anti-βGAL antibodies was enzymatically visualized with DAB. For that, the sections were treated with 0.7% H2O2/ methanol for 5 min at 4°C after antigen retrieval and incubated with the anti-βGAL primary antibody, donkey biotin-SP-conjugated anti-chicken IgY (1:1000, Jackson Immunoresearch Cat# 703-065-155, RRID: AB_2313596), and the Elite ABC kit, as described above.
For the double immunostaining of cortical neurons (Figures S1A and S1B), antigen-retrieved sections were incubated with chicken anti-βGAL antibody together with rat monoclonal anti-CTIP2 (1:1000, Abcam, Cat# ab18465, RRID: AB_2064130) or mouse monoclonal anti-SATB2 (1:1000, Abcam, Cat# ab51502, RRID: AB_882455) antibody, and then stained with donkey Alexa488-conjugated anti-chicken IgY and donkey Cy3-conjugated anti-rat IgG (1:1000, Jackson Immunoresearch Cat# 712-166-153, RRID:AB_2340669) or donkey Cy3-conjugated anti-mouse IgG (1:1000, Jackson Immunoresearch Cat# 715-165-150, RRID:AB_2340813) antibodies. In Figure S1C, to minimize cross-reactions of the anti-rabbit secondary antibody, sections were first stained with rabbit anti-GABA (1:2000, Sigma-Aldrich, Cat# A2052, RRID: AB_477652) and donkey Cy3-conjugated anti-rabbit IgG (1:1000, Jackson Immunoresearch Cat# 711-165-152, RRID: AB_2307443) antibodies, and then visualized for βGAL as described. Rabbit anti-NRP1 antibody was prepared and used as previously described (1:2000, Kawakami et al., 1996; Sato et al., 1998). In Figures S1E and S1G, sections were immunostained for mGFP with a chicken anti-GFP (1:1000; Abcam Cat# ab13970, RRID: AB_300798) and anti-chicken IgY antibodies, after staining for NRP1 as described above. The fluorescent samples were counterstained with DAPI (4',6-diamidino-2-phenylindole, 045-30361, CAS#28718-90-3, Fujifilm Wako).
Image acquisition
Whole-mount brain images (Figure 2) were captured with a digital camera (EOS 6D, Canon) mounted on a dissection microscope (SZ61, Olympus). The sections (Figures 4A, 4B, 5A, 5B, 6A, 7A, 7B, S2A, and S2C) were imaged with a CCD camera (Olympus DP71) attached to a conventional fluorescent microscope (Zeiss Axioplan2) using Cell Sens Standard software. Wide tile images were constructed from multiple images using the Photomerge tool in Photoshop software (Adobe). In some cases (Figures 4C–4F and S1), fluorescent images were captured with an inverted confocal microscope (Olympus IX81 FV1000) using the Fluoview software (FV-ASW ver4.02). Wide images were automatically constructed via multi-area time-lapse processing with a built-in mosaic imaging tool. The transmission images in Figures 6B–6D, S2B, and S2D were cropped from the digital images prepared for the NeuroGT database, for which the imaging procedures are explained in the following section. All the obtained images were rotated and cropped, and the brightness and contrast were non-linearly and equally adjusted across the entire image using Photoshop software (Adobe).
EdU incorporation assay
A thymidine analog, 5-ethynyl-2'-deoxyuridine (EdU, Tokyo Chemical Industry) was intraperitoneally injected into a pregnant mouse (50 mg/kg body weight) at the indicated time points before or after TM injection. The brains were dissected from the mice at P0, and coronal sections of 16-μm thickness were prepared as explained above. The sections were antigen-retrieved as described above and immunostained with chicken anti-βGAL and donkey Alexa488-conjugated anti-chicken IgY antibodies. Subsequently, the incorporated EdU was detected in a solution of 0.1 M Tris (pH 7.6), 2 mM CuSO4, 3 μM Alexa555 azide triethylammonium salt (A20012, Thermo Fisher Scientific), and 10 mM ascorbic acid for 40 min at room temperature. The numbers of neurons labeled for βGAL and those doubly labeled for βGal and EdU were manually counted for each mouse with a 40× objective lens (Plan-Apochromat) under a fluorescent microscope (Zeiss Axioplan2) with 38 HE (ex470/40, em525/50) and 15 (ex549/12, em590) filter sets. The exact numbers of animals and neurons used for quantification are shown in the figures and legends. The scattered plots (Figures 5C–5F and 7C–7F) were generated using the Prism8 software (GraphPad).
Construction of the NeuroGT database
The brains were collected from P7 mice that had been tagged on a single day during E9.5–E18.5 using each neurogenic tagging mouse driver. Whole brains were cut into serial coronal sections (20-μm thick) from anterior to posterior, and each section was pasted on a batch of 10 glass slides in rotation. From the batch, three interspaced slides were antibody stained for the mGFP reporter, and three others were antibody stained for the nls-βGAL reporter. The antibody reactions were enzymatically visualized using DAB, as described above. Slide-fixed sections were dehydrated and mounted using Entellan New (Merck). The staining procedures and antibodies were slightly modified for the samples, and all these details are provided as meta-information in the NeuroGT database.
Images of the glass slides containing the immunostained multiple sections were acquired with a slide scanner (SCN400, Leica) using its designated software (Leica SCN400 Client Version 2.2.0.3789). The contrast and brightness were adjusted automatically. The images acquired using a 20× objective lens were extracted and separated into individual images of single sections using the Fiji distribution package of ImageJ (RRID: SCR_002285; Schindelin et al., 2012). The images of individual sections were aligned along the antero-posterior axis of the brain by rotating at 180° if needed.
The NeuroGT database was established as a web-based system using the Django framework with the PostgreSQL database on a Linux container with the Nginx server. An interactive image viewer was constructed using JavaScript. To encourage the operation, the database was organized as SSBD (Tohsato et al., 2016) with common meta-information such as organism name and contact information of the corresponding author in a unified format. The meta-information specific to NeuroGT, including driver name, TM stage and reporter, has also been provided.
Quantification and statistical analysis
The exact numbers of animals and neurons used for quantification are shown in Figures 5C–5F and 7C–7F. The proportions of tagged neurons containing EdU were calculated using Excel for Mac (Microsoft) and the raw counting data were archived in SSBD:repository (https://doi.org/10.24631/ssbd.repos.2021.03.001). The scattered plots and means were generated using the GraphPad Prism 8. In this study, the purpose of quantification was to show a trend of the continuous developmental process but not to differentiate a specific developmental stage from the others; thus, dichotomous significance testing was not performed.
Additional resources
All images acquired in this study are available in the NeuroGT database at https://ssbd.riken.jp/neurogt/. Raw images and other data are available in SSBD:repository (https://doi.org/10.24631/ssbd.repos.2021.03.001).
Acknowledgments
The authors thank Yash Hiromi and Yan Zhu for their comments on the manuscript, Yumiko Hatanaka and Hidenori Aizawa for providing helpful information, and Pierre Chambon for CreERT2, Junji Takeda for p23loxZeo and p24loxZeo, and Silvia Arber for TaumGFP-nLacZ mice. This research was supported by MEXT/JSPS KAKENHI grants (19HP7002 and 20H03345) to T.H., Platform for Advanced Bioimaging Support (JP16H06280) and ROIS Challenging Exploratory Research Projects for the Future Grant awarded to T.H., and JSPS KAKENHI grant (JP18H05412) to S.O.
Author contributions
T.H. conceived the research, performed the experiments, analyzed the data, and wrote the manuscript with help from other authors. G.S. and H.K. established the mouse lines. S.O. and T.F. converted the images of neurogenic-tagged serial sections into high-resolution digital data. Y.T., H.I., and S.O. processed the image data and constructed the NeuroGT database.
Declaration of interests
The authors declare no competing interests.
Published: May 21, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.crmeth.2021.100012.
Supplemental information
References
- Aizawa H., Goto M., Sato T., Okamoto H. Temporally regulated asymmetric neurogenesis causes left-right difference in the zebrafish habenular structures. Dev. Cell. 2007;12:87–98. doi: 10.1016/j.devcel.2006.10.004. [DOI] [PubMed] [Google Scholar]
- Alexander G.M., Rogan S.C., Abbas A.I., Armbruster B.N., Pei Y., Allen J.A., Nonneman R.J., Hartmann J., Moy S.S., Nicolelis M.A., et al. Remote control of neuronal activity in transgenic mice expressing evolved G protein-coupled receptors. Neuron. 2009;63:27–39. doi: 10.1016/j.neuron.2009.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altman J., Bayer S.A. Time of origin of neurons of the rat inferior colliculus and the relations between cytogenesis and tonotopic order in the auditory pathway. Exp. Brain Res. 1981;42:411–423. doi: 10.1007/BF00237506. [DOI] [PubMed] [Google Scholar]
- Angevine J.B., Jr. Time of neuron origin in the diencephalon of the mouse. An autoradiographic study. J. Comp. Neurol. 1970;139:129–187. doi: 10.1002/cne.901390202. [DOI] [PubMed] [Google Scholar]
- Aprea J., Nonaka-Kinoshita M., Calegari F. Generation and characterization of Neurod1-CreER(T2) mouse lines for the study of embryonic and adult neurogenesis. Genesis. 2014;52:870–878. doi: 10.1002/dvg.22797. [DOI] [PubMed] [Google Scholar]
- Battiste J., Helms A.W., Kim E.J., Savage T.K., Lagace D.C., Mandyam C.D., Eisch A.J., Miyoshi G., Johnson J.E. Ascl1 defines sequentially generated lineage-restricted neuronal and oligodendrocyte precursor cells in the spinal cord. Development. 2007;134:285–293. doi: 10.1242/dev.02727. [DOI] [PubMed] [Google Scholar]
- Bayer S.A., Altman J. Directions in neurogenetic gradients and patterns of anatomical connections in the telencephalon. Prog. Neurobiol. 1987;29:57–106. doi: 10.1016/0301-0082(87)90015-3. [DOI] [PubMed] [Google Scholar]
- Bayer S.A., Altman J. Raven Press; 1991. Neocortical Development. [Google Scholar]
- Caviness V.S., Jr. Time of neuron origin in the hippocampus and dentate gyrus of normal and reeler mutant mice: an autoradiographic analysis. J. Comp. Neurol. 1973;151:113–120. doi: 10.1002/cne.901510203. [DOI] [PubMed] [Google Scholar]
- Caviness V.S., Jr. Neocortical histogenesis in normal and reeler mice: a developmental study based upon [3H]thymidine autoradiography. Brain Res. 1982;256:293–302. doi: 10.1016/0165-3806(82)90141-9. [DOI] [PubMed] [Google Scholar]
- Clancy B., Cauller L.J. Widespread projections from subgriseal neurons (layer VII) to layer I in adult rat cortex. J. Comp. Neurol. 1999;407:275–286. [PubMed] [Google Scholar]
- Danielson N.B., Zaremba J.D., Kaifosh P., Bowler J., Ladow M., Losonczy A. Sublayer-specific coding dynamics during spatial navigation and learning in hippocampal area CA1. Neuron. 2016;91:652–665. doi: 10.1016/j.neuron.2016.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deisseroth K., Feng G., Majewska A.K., Miesenbock G., Ting A., Schnitzer M.J. Next-generation optical technologies for illuminating genetically targeted brain circuits. J. Neurosci. 2006;26:10380–10386. doi: 10.1523/JNEUROSCI.3863-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards M.A., Caviness V.S., Jr., Schneider G.E. Development of cell and fiber lamination in the mouse superior colliculus. J. Comp. Neurol. 1986;248:395–409. doi: 10.1002/cne.902480308. [DOI] [PubMed] [Google Scholar]
- Fame R.M., MacDonald J.L., Macklis J.D. Development, specification, and diversity of callosal projection neurons. Trends Neurosci. 2011;34:41–50. doi: 10.1016/j.tins.2010.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feil R., Wagner J., Metzger D., Chambon P. Regulation of Cre recombinase activity by mutated estrogen receptor ligand-binding domains. Biochem. Biophys. Res. Commun. 1997;237:752–757. doi: 10.1006/bbrc.1997.7124. [DOI] [PubMed] [Google Scholar]
- Florio M., Leto K., Muzio L., Tinterri A., Badaloni A., Croci L., Zordan P., Barili V., Albieri I., Guillemot F., et al. Neurogenin 2 regulates progenitor cell-cycle progression and Purkinje cell dendritogenesis in cerebellar development. Development. 2012;139:2308–2320. doi: 10.1242/dev.075861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forster E., Zhao S., Frotscher M. Laminating the hippocampus. Nat. Rev. Neurosci. 2006;7:259–267. doi: 10.1038/nrn1882. [DOI] [PubMed] [Google Scholar]
- Friedlander M.J., Torres-Reveron J. The changing roles of neurons in the cortical subplate. Front Neuroanat. 2009;3:15. doi: 10.3389/neuro.05.015.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerfen C.R., Paletzki R., Heintz N. GENSAT BAC cre-recombinase driver lines to study the functional organization of cerebral cortical and basal ganglia circuits. Neuron. 2013;80:1368–1383. doi: 10.1016/j.neuron.2013.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govindan S., Oberst P., Jabaudon D. In vivo pulse labeling of isochronic cohorts of cells in the central nervous system using FlashTag. Nat. Protoc. 2018;13:2297–2311. doi: 10.1038/s41596-018-0038-1. [DOI] [PubMed] [Google Scholar]
- Guerrier S., Coutinho-Budd J., Sassa T., Gresset A., Jordan N.V., Chen K., Jin W.L., Frost A., Polleux F. The F-BAR domain of srGAP2 induces membrane protrusions required for neuronal migration and morphogenesis. Cell. 2009;138:990–1004. doi: 10.1016/j.cell.2009.06.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardwick L.J., Philpott A. Nervous decision-making: to divide or differentiate. Trends Genet. 2014;30:254–261. doi: 10.1016/j.tig.2014.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto M., Mikoshiba K. Mediolateral compartmentalization of the cerebellum is determined on the "birth date" of Purkinje cells. J. Neurosci. 2003;23:11342–11351. doi: 10.1523/JNEUROSCI.23-36-11342.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herkenham M., Nauta W.J. Efferent connections of the habenular nuclei in the rat. J. Comp. Neurol. 1979;187:19–47. doi: 10.1002/cne.901870103. [DOI] [PubMed] [Google Scholar]
- Hevner R.F. Intermediate progenitors and Tbr2 in cortical development. J. Anat. 2019;235:616–625. doi: 10.1111/joa.12939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinds J.W. Autoradiographic study of histogenesis in the mouse olfactory bulb. I. Time of origin of neurons and neuroglia. J. Comp. Neurol. 1968;134:287–304. doi: 10.1002/cne.901340304. [DOI] [PubMed] [Google Scholar]
- Hippenmeyer S., Vrieseling E., Sigrist M., Portmann T., Laengle C., Ladle D.R., Arber S. A developmental switch in the response of DRG neurons to ETS transcription factor signaling. PLoS Biol. 2005;3:e159. doi: 10.1371/journal.pbio.0030159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirata T., Iwai L. Timing matters: a strategy for neurons to make diverse connections. Neurosci. Res. 2019;138:79–83. doi: 10.1016/j.neures.2018.09.006. [DOI] [PubMed] [Google Scholar]
- Hirata T., Shioi G., Abe T., Kiyonari H., Kato S., Kobayashi K., Mori K., Kawasaki T. A novel birthdate-labeling method reveals segregated parallel projections of mitral and external tufted cells in the main olfactory system. eNeuro. 2019;6 doi: 10.1523/ENEURO.0234-19.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoerder-Suabedissen A., Molnar Z. Molecular diversity of early-born subplate neurons. Cereb. Cortex. 2013;23:1473–1483. doi: 10.1093/cercor/bhs137. [DOI] [PubMed] [Google Scholar]
- Imayoshi I., Isomura A., Harima Y., Kawaguchi K., Kori H., Miyachi H., Fujiwara T., Ishidate F., Kageyama R. Oscillatory control of factors determining multipotency and fate in mouse neural progenitors. Science. 2013;342:1203–1208. doi: 10.1126/science.1242366. [DOI] [PubMed] [Google Scholar]
- Inaki K., Nishimura S., Nakashiba T., Itohara S., Yoshihara Y. Laminar organization of the developing lateral olfactory tract revealed by differential expression of cell recognition molecules. J. Comp. Neurol. 2004;479:243–256. doi: 10.1002/cne.20270. [DOI] [PubMed] [Google Scholar]
- Kawakami A., Kitsukawa T., Takagi S., Fujisawa H. Developmentally regulated expression of a cell surface protein, neuropilin, in the mouse nervous system. J. Neurobiol. 1996;29:1–17. doi: 10.1002/(SICI)1097-4695(199601)29:1<1::AID-NEU1>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- Kawauchi D., Taniguchi H., Watanabe H., Saito T., Murakami F. Direct visualization of nucleogenesis by precerebellar neurons: involvement of ventricle-directed, radial fibre-associated migration. Development. 2006;133:1113–1123. doi: 10.1242/dev.02283. [DOI] [PubMed] [Google Scholar]
- Kim E.J., Battiste J., Nakagawa Y., Johnson J.E. Ascl1 (Mash1) lineage cells contribute to discrete cell populations in CNS architecture. Mol. Cell Neurosci. 2008;38:595–606. doi: 10.1016/j.mcn.2008.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim E.J., Hori K., Wyckoff A., Dickel L.K., Koundakjian E.J., Goodrich L.V., Johnson J.E. Spatiotemporal fate map of neurogenin1 (Neurog1) lineages in the mouse central nervous system. J. Comp. Neurol. 2011;519:1355–1370. doi: 10.1002/cne.22574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koester S.E., O'Leary D.D. Axons of early generated neurons in cingulate cortex pioneer the corpus callosum. J. Neurosci. 1994;14:6608–6620. doi: 10.1523/JNEUROSCI.14-11-06608.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koundakjian E.J., Appler J.L., Goodrich L.V. Auditory neurons make stereotyped wiring decisions before maturation of their targets. J. Neurosci. 2007;27:14078–14088. doi: 10.1523/JNEUROSCI.3765-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurusu M., Awasaki T., Masuda-Nakagawa L.M., Kawauchi H., Ito K., Furukubo-Tokunaga K. Embryonic and larval development of the Drosophila mushroom bodies: concentric layer subdivisions and the role of fasciclin II. Development. 2002;129:409–419. doi: 10.1242/dev.129.2.409. [DOI] [PubMed] [Google Scholar]
- Lawson S.N., Biscoe T.J. Development of mouse dorsal root ganglia: an autoradiographic and quantitative study. J. Neurocytol. 1979;8:265–274. doi: 10.1007/BF01236122. [DOI] [PubMed] [Google Scholar]
- Lee S.H., Marchionni I., Bezaire M., Varga C., Danielson N., Lovett-Barron M., Losonczy A., Soltesz I. Parvalbumin-positive basket cells differentiate among hippocampal pyramidal cells. Neuron. 2014;82:1129–1144. doi: 10.1016/j.neuron.2014.03.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lodato S., Arlotta P. Generating neuronal diversity in the mammalian cerebral cortex. Annu. Rev. Cell Dev. Biol. 2015;31:699–720. doi: 10.1146/annurev-cellbio-100814-125353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Q., Sommer L., Cserjesi P., Anderson D.J. Mash1 and neurogenin1 expression patterns define complementary domains of neuroepithelium in the developing CNS and are correlated with regions expressing notch ligands. J. Neurosci. 1997;17:3644–3652. doi: 10.1523/JNEUROSCI.17-10-03644.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mailly P., Aliane V., Groenewegen H.J., Haber S.N., Deniau J.M. The rat prefrontostriatal system analyzed in 3D: evidence for multiple interacting functional units. J. Neurosci. 2013;33:5718–5727. doi: 10.1523/JNEUROSCI.5248-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masserdotti G., Gillotin S., Sutor B., Drechsel D., Irmler M., Jorgensen H.F., Sass S., Theis F.J., Beckers J., Berninger B., et al. Transcriptional mechanisms of proneural factors and REST in regulating neuronal reprogramming of astrocytes. Cell Stem Cell. 2015;17:74–88. doi: 10.1016/j.stem.2015.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattar P., Langevin L.M., Markham K., Klenin N., Shivji S., Zinyk D., Schuurmans C. Basic helix-loop-helix transcription factors cooperate to specify a cortical projection neuron identity. Mol. Cell Biol. 2008;28:1456–1469. doi: 10.1128/MCB.01510-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McConnell J., Angevine J.B., Jr. Time of neuron origin in the amygdaloid complex of the mouse. Brain Res. 1983;272:150–156. doi: 10.1016/0006-8993(83)90372-4. [DOI] [PubMed] [Google Scholar]
- McConnell S.K. The determination of neuronal fate in the cerebral cortex. Trends Neurosci. 1989;12:342–349. doi: 10.1016/0166-2236(89)90041-6. [DOI] [PubMed] [Google Scholar]
- Miyata T., Maeda T., Lee J.E. NeuroD is required for differentiation of the granule cells in the cerebellum and hippocampus. Genes Dev. 1999;13:1647–1652. doi: 10.1101/gad.13.13.1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuseki K., Diba K., Pastalkova E., Buzsaki G. Hippocampal CA1 pyramidal cells form functionally distinct sublayers. Nat. Neurosci. 2011;14:1174–1181. doi: 10.1038/nn.2894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Namba K., Sugihara I., Hashimoto M. Close correlation between the birth date of Purkinje cells and the longitudinal compartmentalization of the mouse adult cerebellum. J. Comp. Neurol. 2011;519:2594–2614. doi: 10.1002/cne.22640. [DOI] [PubMed] [Google Scholar]
- Nishikimi M., Oishi K., Tabata H., Torii K., Nakajima K. Segregation and pathfinding of callosal axons through EphA3 signaling. J. Neurosci. 2011;31:16251–16260. doi: 10.1523/JNEUROSCI.3303-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nornes H.O., Carry M. Neurogenesis in spinal cord of mouse: an autoradiographic analysis. Brain Res. 1978;159:1–6. doi: 10.1016/0006-8993(78)90105-1. [DOI] [PubMed] [Google Scholar]
- Oberst P., Fievre S., Baumann N., Concetti C., Bartolini G., Jabaudon D. Temporal plasticity of apical progenitors in the developing mouse neocortex. Nature. 2019;573:370–374. doi: 10.1038/s41586-019-1515-6. [DOI] [PubMed] [Google Scholar]
- Pierce E.T. Time of origin of neurons in the brain stem of the mouse. Prog. Brain Res. 1973;40:53–65. doi: 10.1016/S0079-6123(08)60679-2. [DOI] [PubMed] [Google Scholar]
- Piper M., Plachez C., Zalucki O., Fothergill T., Goudreau G., Erzurumlu R., Gu C., Richards L.J. Neuropilin 1-Sema signaling regulates crossing of cingulate pioneering axons during development of the corpus callosum. Cereb. Cortex. 2009;19(Suppl 1) doi: 10.1093/cercor/bhp027. i11–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rash B.G., Richards L.J. A role for cingulate pioneering axons in the development of the corpus callosum. J. Comp. Neurol. 2001;434:147–157. doi: 10.1002/cne.1170. [DOI] [PubMed] [Google Scholar]
- Saga Y., Yagi T., Ikawa Y., Sakakura T., Aizawa S. Mice develop normally without tenascin. Genes Dev. 1992;6:1821–1831. doi: 10.1101/gad.6.10.1821. [DOI] [PubMed] [Google Scholar]
- Sarma A.A., Richard M.B., Greer C.A. Developmental dynamics of piriform cortex. Cereb. Cortex. 2011;21:1231–1245. doi: 10.1093/cercor/bhq199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato Y., Hirata T., Ogawa M., Fujisawa H. Requirement for early-generated neurons recognized by monoclonal antibody lot1 in the formation of lateral olfactory tract. J. Neurosci. 1998;18:7800–7810. doi: 10.1523/JNEUROSCI.18-19-07800.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B., et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo S., Lim J.W., Yellajoshyula D., Chang L.W., Kroll K.L. Neurogenin and NeuroD direct transcriptional targets and their regulatory enhancers. EMBO J. 2007;26:5093–5108. doi: 10.1038/sj.emboj.7601923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shepard A.R., Scheffel J.L., Yu W.M. Relationships between neuronal birthdates and tonotopic positions in the mouse cochlear nucleus. J. Comp. Neurol. 2019;527:999–1011. doi: 10.1002/cne.24575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada M., Nakamura T. Time of neuron origin in mouse hypothalamic nuclei. Exp. Neurol. 1973;41:163–173. doi: 10.1016/0014-4886(73)90187-8. [DOI] [PubMed] [Google Scholar]
- Slomianka L., Amrein I., Knuesel I., Sorensen J.C., Wolfer D.P. Hippocampal pyramidal cells: the reemergence of cortical lamination. Brain Struct. Funct. 2011;216:301–317. doi: 10.1007/s00429-011-0322-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soltesz I., Losonczy A. CA1 pyramidal cell diversity enabling parallel information processing in the hippocampus. Nat. Neurosci. 2018;21:484–493. doi: 10.1038/s41593-018-0118-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soriano P. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat. Genet. 1999;21:70–71. doi: 10.1038/5007. [DOI] [PubMed] [Google Scholar]
- Suarez R., Gobius I., Richards L.J. Evolution and development of interhemispheric connections in the vertebrate forebrain. Front. Hum. Neurosci. 2014;8:497. doi: 10.3389/fnhum.2014.00497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudarov A., Turnbull R.K., Kim E.J., Lebel-Potter M., Guillemot F., Joyner A.L. Ascl1 genetics reveals insights into cerebellum local circuit assembly. J. Neurosci. 2011;31:11055–11069. doi: 10.1523/JNEUROSCI.0479-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki I.K., Hirata T. Neocortical neurogenesis is not really "neo": a new evolutionary model derived from a comparative study of chick pallial development. Dev. Growth Differ. 2013;55:173–187. doi: 10.1111/dgd.12020. [DOI] [PubMed] [Google Scholar]
- Szabo N.E., Haddad-Tovolli R., Zhou X., Alvarez-Bolado G. Cadherins mediate sequential roles through a hierarchy of mechanisms in the developing mammillary body. Front. Neuroanat. 2015;9:29. doi: 10.3389/fnana.2015.00029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tohsato Y., Ho K.H.L., Kyoda K., Onami S. SSBD: a database of quantitative data of spatiotemporal dynamics of biological phenomena. Bioinformatics. 2016;32:3471–3479. doi: 10.1093/bioinformatics/btw417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toma K., Kumamoto T., Hanashima C. The timing of upper-layer neurogenesis is conferred by sequential derepression and negative feedback from deep-layer neurons. J. Neurosci. 2014;34:13259–13276. doi: 10.1523/JNEUROSCI.2334-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran-Anh K., Zhang J., Nguyen-Minh V.T., Fujita H., Hirata T., Sugihara I. Common origin of the cerebellar dual somatotopic areas revealed by tracking embryonic Purkinje cell clusters with birthdate tagging. eNeuro. 2020;7 doi: 10.1523/ENEURO.0251-20.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh C. Age-related fiber order in the ferret's optic nerve and optic chiasm. J. Neurosci. 1986;6:1635–1642. doi: 10.1523/JNEUROSCI.06-06-01635.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh C., Guillery R.W. Age-related fiber order in the optic tract of the ferret. J. Neurosci. 1985;5:3061–3069. doi: 10.1523/JNEUROSCI.05-11-03061.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh C., Polley E.H., Hickey T.L., Guillery R.W. Generation of cat retinal ganglion cells in relation to central pathways. Nature. 1983;302:611–614. doi: 10.1038/302611a0. [DOI] [PubMed] [Google Scholar]
- Wilkinson G., Dennis D., Schuurmans C. Proneural genes in neocortical development. Neuroscience. 2013;253:256–273. doi: 10.1016/j.neuroscience.2013.08.029. [DOI] [PubMed] [Google Scholar]
- Winpenny E., Lebel-Potter M., Fernandez M.E., Brill M.S., Gotz M., Guillemot F., Raineteau O. Sequential generation of olfactory bulb glutamatergic neurons by Neurog2-expressing precursor cells. Neural Dev. 2011;6:12. doi: 10.1186/1749-8104-6-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamatani H., Sato Y., Fujisawa H., Hirata T. Chronotopic organization of olfactory bulb axons in the lateral olfactory tract. J. Comp. Neurol. 2004;475:247–260. doi: 10.1002/cne.20155. [DOI] [PubMed] [Google Scholar]
- Zeng S.J., Lin Y.T., Tian C.P., Song K.J., Zhang X.W., Zuo M.X. Evolutionary significance of delayed neurogenesis in the core versus shell auditory areas of Mus musculus. J. Comp. Neurol. 2009;515:600–613. doi: 10.1002/cne.22076. [DOI] [PubMed] [Google Scholar]
- Zhang J., Tran-Anh K., Hirata T., Sugihara I. Striped distribution pattern of Purkinje cells of different birthdates in the mouse cerebellar cortex studied with the Neurog2-CreER transgenic line. Neuroscience. 2020;462:122–140. doi: 10.1016/j.neuroscience.2020.07.028. [DOI] [PubMed] [Google Scholar]
- Zhou J., Wen Y., She L., Sui Y.N., Liu L., Richards L.J., Poo M.M. Axon position within the corpus callosum determines contralateral cortical projection. Proc. Natl. Acad. Sci. U S A. 2013;110:E2714–E2723. doi: 10.1073/pnas.1310233110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zirlinger M., Lo L., McMahon J., McMahon A.P., Anderson D.J. Transient expression of the bHLH factor neurogenin-2 marks a subpopulation of neural crest cells biased for a sensory but not a neuronal fate. Proc. Natl. Acad. Sci. U S A. 2002;99:8084–8089. doi: 10.1073/pnas.122231199. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
In the interactive viewer of thumbnail images, coronal section images stained either for mGFP or nls-βGAL reporter can be sequentially seen along the anteroposterior brain axis by dragging the slider. The sync button at each thumbnail stack matches the section level of the other reporter stack.
Data Availability Statement
All images acquired in this study are available in the NeuroGT database at https://ssbd.riken.jp/neurogt/. Raw images and unprocessed data related to this study and NeuroGT are archived in SSBD:repository (https://doi.org/10.24631/ssbd.repos.2021.03.001).