Abstract
The human gut microbiome is crucial for human health and disease but exhibits extensive individual-level strain variation. Distinct strains encode and express different functions. The resulting emergent properties therefore differentially affect human health and disease in a personalized manner. Pryszlak et al. have made strides in tackling the challenge of genome-based microbiome screening which will ultimately yield strain-level understanding of the functional roles played by the human gut microbiome.
The human gut microbiome is crucial for human health and disease but exhibits extensive individual-level strain variation. Distinct strains encode and express different functions. The resulting emergent properties might therefore differentially affect human health and disease in a personalized manner. Pryszlak et al. have made strides in tackling the challenge of genome-based microbiome screening which will ultimately yield strain-level understanding of the functional roles played by the human gut microbiome.
Main text
The composition and the potential role of the human microbiome in modulating host health and disease is well established (Heintz-Buschart and Wilmes, 2018). The gut microbiome alone is composed of thousands of different microbial species, and alterations from the norm (dysbiosis) have been linked to numerous chronic diseases. Additionally, the composition of the gut microbiome is crucial in how the human body metabolizes drugs and which essential functions such as fermentation of indigestible food components as well as the synthesis of essential vitamins it contributes. Importantly, the microbiome exhibits individual-level strain variation (Schloissnig et al., 2013), whereby each strain may be functionally relevant (Figure 1). Strain-conferred functional complements are established right after birth (Wampach et al., 2018) and may dictate health trajectories throughout life. Consequently, given the inherent functional relevance of strains (as established in classical infection biology), it is of utmost importance to develop methods that allow the resolution of an individual’s microbiome at the strain-level, which further allows an in-depth assessment of their respective functions.
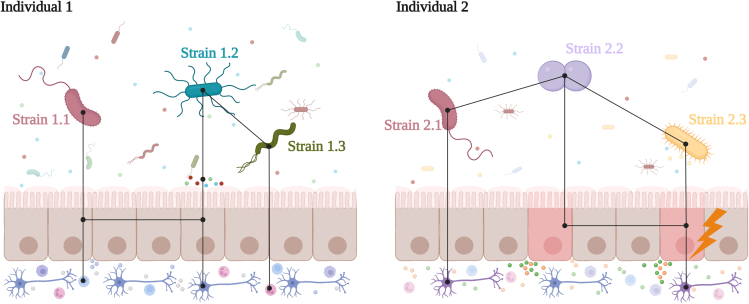
Figure 1.
Representation of individual-level strain variation in two human individuals
Distinct strains have distinct effects on host cells as well as on the functional networks that are established between microbial and host cells. The emergent functional properties may be influenced by early colonization and host genetics, which in turn may trigger critical disease processes over a lifetime. Figure created with Biorender (https://biorender.com/).
In addition to the role that microbial strains may play in triggering diseases, the current and next frontier of medicine with regard to human diseases is treatments tailored to each individual (Petrosino, 2018). To account for the significant degree of interpersonal variation, personalized screening will become increasingly relevant to precisely study the genotype and phenotype of an individual along with other essential endogenous factors such as the immune system and the microbiome. Individual-level variation is relevant in this context, not least in relation to hypothesis-driven investigations of the mechanisms underlying disease processes triggered by specific strains. However, the development of personalized microbiome screening methodologies has so far been challenging. Specifically, significant challenges are associated with culturing individual strains as well as amplifying their genomes.
To study the essential functions of the gut microbiome, omics methodologies are used to gain crucial insights into the complex interrelationships between the microbiome and the host. This has been and is currently addressed through mechanistic studies including via in vitro and ex vivo models (Shah et al., 2016), interventions (Cani et al., 2021), and animal models (Kostic et al., 2013). Current microbiome studies predominantly focus on reconstructing microbial genomes from sequence data into either metagenomically assembled genomes (MAGs) and/or by single amplified genomes (SAGs) (Arikawa et al., 2021). While MAGs allow insights into the abundance of different taxa, SAGs provide essential information regarding uncultivated and low-abundance strains. However, both methods have limitations. Specifically, MAGs are prone to chimeric assemblies, whereas MAG and SAG recovery is in general influenced by sequencing depth. Moreover, the untargeted and cost-prohibitive nature of these methods do not allow for recovering complete genomes of specific microbes and/or their respective gene complements (Franzosa et al., 2015). Thus, while the individual strain-level information is critical for the extensive understanding of the microbial functional repertoire that contributes to human physiology, much is still unknown. Therefore, microbiome-oriented studies need a comprehensive toolset to enable new and targeted discoveries.
Here we preview an improved protocol that allows the characterization of complex microbial communities such as the gut microbiome not only in a targeted fashion, but also to gain crucial insights at the functional level. The principle behind this methodology centers on microfluidic systems (Colin et al., 2015), whereby droplets containing microbial cells of interest may be screened. Pryszlak et al. have leveraged the nuances and strengths of culture-independent methods with a targeted hypothesis-driven approach (Pryszlak et al., 2021). Based on fecal samples, they demonstrate instantaneous targeting and sorting of bacterial cells via their droplet-based microfluidic method. Their methodology allows for the preparation of fecal samples for whole genome sequencing and for the subsequent characterization of individual bacterial cells, bypassing traditional and limiting culturing methods. The preparation of fecal samples includes the estimation of the absolute cell count, which, in turn, allows for the efficient encapsulation of single cells. Based on this principle, the cell counts of fecal samples were obtained by flow cytometry across samples of varying diversity. Aside from infant stool samples whose absolute cell counts were lower, all other samples tested exhibited a similar range of absolute cell counts.
Pryszlak et al. further introduce a targeted approach for single cells by which any microbial strain of interest can be enriched inside single droplets. This is further complemented by the recovery of target strains without the need for a reference database by leveraging the marker gene sequences previously established through the metagenomic operations taxonomic unit (mOTU) methodology (Milanese et al., 2019). Based on the marker gene information, primer pairs and probes were designed directly from the shotgun metagenomic sequencing data. Crucially, the approach may be applied to samples of varied origins, including but not limited to sediments, marine, freshwater, etc. To further enrich the cells and improve genome recovery and completion, each microbial cell was subsequently sorted into monodispersed droplets. The marker genes within each droplet were thereafter amplified by PCR, resulting in each droplet containing amplified DNA of an individual microbial cell. The individual cells were further detected by the fluorescent 6-FAM signal from cleaved TaqMan probes, followed by cell sorting to enrich for bacteria of interest.
The taxa comprising the human microbiome follow a typical rank abundance distribution ranging from highly abundant taxa themselves comprised of strains to more lowly abundant taxa. The methodology by Pryszlak et al. importantly allows for the culture-independent characterization of low abundance taxa which were typically undersampled in previous microbiome studies (Gibbons et al., 2017). Interestingly, an additional outcome of the methodology established by Pryszlak et al. is the removal of artifactual residual amplicon sequences obtained as a byproduct of the PCR reaction. The developed workflow utilizes biotinylated PCR primers for amplicon removal. In this process, a chemical handle is incorporated into the amplicons, allowing them to be selectively bound to magnetic beads. Amplicon removal is based on the principle of size selection using magnetic streptavidin beads to efficiently remove smaller amplicon fragments. This process leaves unbound genomic DNA in the supernatant, here the low-abundance target genome, which can further be amplified to obtain high-quality, pure DNA. This procedure is essential to identify low-abundance species and to gain clearer insights into the underlying complexity of the composition of microbial species from each individual sample.
In order to obtain high-quality genomes, a specific number of droplets/cells are required. As a proof of concept, Pryszlak et al. therefore demonstrated that Bacillus subtilis, a bacterial strain not resident in the gut and spiked into the fecal samples in different concentrations (1:50, 1:100, 1:250), was recovered at the expected numbers. By targeting and encapsulating this strain alone, positive droplets, as expected, were observed via fluorescence-activated single-cell droplet sorting and sequencing. With this approach, it was demonstrated that the lower limit of detection for positive cells was up to a concentration of 0.4% for samples spiked with B. subtilis. Aiming to obtain a high-quality genome of a target species, Pryszlak et al. further assessed the number of B. subtilis cells required for reconstructing a high-quality genome. The results showed that the obtained B. subtilis genome had a relatively even genomic coverage across all the samples tested. Furthermore, it was determined that ∼4,000 targeted and sorted cells are required for cell enrichment allowing for sufficient coverage of the target genome. This method complements but crucially also augments the widely used SAG methodology (Labonté et al., 2015), which is not only an untargeted approach but also only results in low to average genome quality.
The methodology outlined by Pryszlak et al. allows testing of hypotheses by leveraging in silico information for the targeted enrichment and sequencing of microbial strains. It is also of interest to further assess the applications of the described method toward the enrichment of previously unknown and uncharacterized taxa. Furthermore, the method may shed light on the “rare” taxa conundrum (Jousset et al., 2017), such as highlighting their role in human health and physiology despite their limited abundance. Although questions remain regarding the time, efficiency of the method, and capital investment required for undertaking such screenings, the applications and pros outweigh the limitations and the unanswered queries. Furthermore, future studies may employ and take advantage of this novel technique to identify and characterize an individual’s microbiota, leading to truly personalized microbiome screening modalities.
Potential applications of this methodology include, but are not limited to, understanding the effects of the microbiome from a bottom-up approach, i.e., single cell/genome resolution. Going forward, the method described by Pryszlak et al. will not only allow for screening the microbiome, but potentially also host cells, including susceptible human cell populations relevant from a disease point of view. Therefore, the described methodological foundation will enable personalized medicine approaches in future, e.g., by disentangling and modeling the complex interactions between the human system and the microbiome on an individual basis. Importantly, this will further help unravel mechanisms underlying an altered gut microbiome in the context of the triggering and sustaining of disease processes. Such insights will in turn allow the steering of microbiome toward specific endpoints, either through tailored therapies or lifestyle changes.
As highlighted in this preview, the individual-level variations may affect the hypothesis-driven assessment of the role of the microbiome in human diseases. Furthermore, we are often unaware about all the functions of a cell including those of microbial and human origin. Thus it remains unclear as to how different cell types interact with each other, especially given the inherent individual-level variation also apparent in the gut microbiome (Heintz-Buschart and Wilmes, 2018). This individual-level variation needs to be connected to individual-derived susceptible cell populations from the host to get a better understanding of microbiome-driven pathogenesis. To allow for the assessment of individual-level variations, microfluidic methods, in general, provide a suitable avenue with respect to modeling personalized approaches. Collectively, microfluidic devices allow analysis at spatial scales, which are especially relevant from a host-microbiome point of view. Leveraging the use of gut-on-chip co-culture, e.g., the human microbial crosstalk (HuMiX) model (Shah et al., 2016), with the isolation and cultivation modalities developed by Pryszlak et al., we might be able to screen relevant human cell types obtained from individual patients along with the corresponding microbiome members. Such approaches would allow individual-resolved mechanistic insights into host-microbiome crosstalk. Furthermore, it will allow us to enrich in previously unknown host-modulatory taxa that have been reported to influence human health while simultaneously mitigating the challenges of culturing. Overall, the new methodology developed by Pryszlak et al. advances the microbiome field by facilitating future insights into the “dark matter” of the human microbiome—including unknown species and functions, as well as the relevance of individual-level variation in the microbiome in relation to their impact on sustaining health or triggering disease.
Acknowledgments
This project has received funding from the European Research Council (ERC) under the European Union’s Horizon 2020 research and innovation program (grant agreement 863664).
Declaration of interests
P.W. declares being listed as an inventor on patents PCT/EP2013/056607, PCT/EP2016/062024, PCT/US2017/061602, and PCT/EP2019/081424.
References
- Arikawa K., Ide K., Kogawa M., Saeki T., Yoda T., Endoh T., Matsuhashi A., Takeyama H., Hosokawa M. Recovery of strain-resolved genomes from human microbiome through an integration framework of single-cell genomics and metagenomics. Microbiome. 2021;9:202. doi: 10.1186/s40168-021-01152-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cani P.D., de Hase E.M., van Hul M. Gut microbiota and host metabolism: From proof of concept to therapeutic intervention. Microorganisms. 2021;9 doi: 10.3390/microorganisms9061302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colin P.Y., Kintses B., Gielen F., Miton C.M., Fischer G., Mohamed M.F., Hyvönen M., Morgavi D.P., Janssen D.B., Hollfelder F. Ultrahigh-throughput discovery of promiscuous enzymes by picodroplet functional metagenomics. Nat. Commun. 2015;6:10008. doi: 10.1038/ncomms10008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franzosa E.A., Huang K., Meadow J.F., Gevers D., Lemon K.P., Bohannan B.J.M., Huttenhower C. Identifying personal microbiomes using metagenomic codes. Proc. Natl. Acad. Sci. USA. 2015;112:E2930–E2938. doi: 10.1073/pnas.1423854112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbons S.M., Kearney S.M., Smillie C.S., Alm E.J. Two dynamic regimes in the human gut microbiome. PLoS Comput. Biol. 2017;13:e1005364. doi: 10.1371/journal.pcbi.1005364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heintz-Buschart A., Wilmes P. Human Gut Microbiome: Function Matters. Trends in Microbiology. 2018;26:563–574. doi: 10.1016/j.tim.2017.11.002. [DOI] [PubMed] [Google Scholar]
- Jousset A., Bienhold C., Chatzinotas A., Gallien L., Gobet A., Kurm V., Küsel K., Rillig M.C., Rivett D.W., Salles J.F., et al. Where less may be more: How the rare biosphere pulls ecosystems strings. ISME Journal. 2017;11:853–862. doi: 10.1038/ismej.2016.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kostic A.D., Howitt M.R., Garrett W.S. Exploring host-microbiota interactions in animal models and humans. Genes Dev. 2013;27:701–718. doi: 10.1101/gad.212522.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labonté J.M., Swan B.K., Poulos B., Luo H., Koren S., Hallam S.J., Sullivan M.B., Woyke T., Wommack K.E., Stepanauskas R. Single-cell genomics-based analysis of virus-host interactions in marine surface bacterioplankton. ISME J. 2015;9:2386–2399. doi: 10.1038/ismej.2015.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milanese A., Mende D.R., Paoli L., Salazar G., Ruscheweyh H.J., Cuenca M., Hingamp P., Alves R., Costea P.I., Coelho L.P., et al. Microbial abundance, activity and population genomic profiling with mOTUs2. Nat. Commun. 2019;10:1014. doi: 10.1038/s41467-019-08844-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrosino J.F. The microbiome in precision medicine: The way forward. Genome Medicine. 2018;10 doi: 10.1186/s13073-018-0525-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pryszlak A., Wenzel T., Seitz K.W., Hildebrand F., Kartal E., Raffaele Cosenza M., Benes V., Merten C. Enrichment of gut microbiome strains for cultivation-free genome sequencing using droplet microfluidics. Cell Reports Methods. 2021;2:100137-1–100137-12. doi: 10.1016/j.crmeth.2021.100137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schloissnig S., Arumugam M., Sunagawa S., Mitreva M., Tap J., Zhu A., Waller A., Mende D.R., Kultima J.R., Martin J., et al. Genomic variation landscape of the human gut microbiome. Nature. 2013;493:45–50. doi: 10.1038/nature11711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah P., Fritz J.V., Glaab E., Desai M.S., Greenhalgh K., Frachet A., Niegowska M., Estes M., Jäger C., Seguin-Devaux C., et al. A microfluidics-based in vitro model of the gastrointestinal human-microbe interface. Nat. Commun. 2016;7:11535. doi: 10.1038/ncomms11535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wampach L., Heintz-Buschart A., Fritz J.V., Ramiro-Garcia J., Habier J., Herold M., Narayanasamy S., Kaysen A., Hogan A.H., Bindl L., et al. Birth mode is associated with earliest strain-conferred gut microbiome functions and immunostimulatory potential. Nat. Commun. 2018;9:5091. doi: 10.1038/s41467-018-07631-x. [DOI] [PMC free article] [PubMed] [Google Scholar]