Summary
Mass-spectrometry-based proteomics enables quantitative analysis of thousands of human proteins. However, experimental and computational challenges restrict progress in the field. This review summarizes the recent flurry of machine-learning strategies using artificial deep neural networks (or “deep learning”) that have started to break barriers and accelerate progress in the field of shotgun proteomics. Deep learning now accurately predicts physicochemical properties of peptides from their sequence, including tandem mass spectra and retention time. Furthermore, deep learning methods exist for nearly every aspect of the modern proteomics workflow, enabling improved feature selection, peptide identification, and protein inference.
Keywords: proteomics, deep learning, neural networks, bioinformatics, mass spectrometry, peptides, MS/MS, retention time
In this review, Meyer summarizes and contrasts the different machine-learning strategies that use neural networks for (1) prediction of peptide properties from sequence and (2) peptide/protein identification. Limitations and opportunities of these deep learning tools in mass-spectrometry-based proteomics are also discussed.
Introduction
Nearly all cells in an organism share one genome that acts as a library of instructions to produce diverse specialized cells and tissues, each of which have unique and dynamic proteomes (Jiang et al., 2020) that reflect the biological milieu of physiology or disease. Liquid chromatography-tandem mass spectrometry (LC-MS/MS) is currently the most effective method to discover and quantify the human proteome (Aebersold and Mann, 2016), enabling a proteomic depth now comparable with RNA sequencing (Bekker-Jensen et al., 2017). Mass spectrometry-based proteomics is an essential approach for the study of biological systems, and is routinely applied for diverse applications beyond relative or absolute proteome quantification (Xie et al., 2011), including proteome stability measurement (Jarzab et al., 2020; Mateus et al., 2016) and biomarker discovery (Meyer and Schilling, 2017).
Computational methods are required for many parts of the modern proteomics workflow, and the success of computational methods relies on deep understanding of the technical and experimental details (Figure 1). For example, peptides must be identified from their tandem mass spectra, which requires an underlying comprehension of peptide fragmentation. Peptides must also be quantified, usually based on their elution profile, which can be challenging due to missing values, interfering masses, or shifts in chromatographic retention time. Countless software tools exist to facilitate and expedite various proteomic data analysis, including for design of data collection, feature selection, peptide and protein identification and quantification, and biological interpretation (Marx, 2020; Tsiamis et al., 2019). Still, many computational challenges prevent more sensitive and accurate peptide identification and quantification in the field of shotgun proteomics (Schubert et al., 2017; Sinitcyn et al., 2018).
Figure 1.
General proteomics workflow highlighting challenges
Peptides are produced from enzymatic hydrolysis of the isolated proteome and analyzed by liquid chromatography-tandem mass spectrometry (LC-MS/MS). This process involves detecting features and assigning retention times to peptides, detecting precursor peptide charge, and then measuring the fragment ion spectra or tandem mass spectra for a peptide. The collection of tandem mass spectra is then subject to peptide-spectra matching to identify peptides, and the original set of proteins is inferred. Red stars indicate that deep learning tools now facilitate these aspects of the workflow.
Along with the recent explosion of machine learning applications in economic and scientific sectors, machine learning tools have emerged to facilitate proteomic analysis. Machine learning is a subfield of artificial intelligence (Alzubi et al., 2018; Domingos, 2012). Any machine learning model can be thought of as a mathematical function approximator, which learns a relationship that connects input data (X) to output data (y) when the underlying relationship is not known from first principles. For example, machine learning could take many examples of peptide sequences (inputs) and their measured retention times (outputs) to build a model that then predicts retention time for other unmeasured peptides. Thus, the benefit of machine learning is that once we build a model to learn some relationship based on measured examples, we can predict the model output in the future given only the input, which saves the cost of measuring that output.
Among the many types of machine learning models, “deep learning” with artificial neural networks provides highly generalizable function approximation (Figures 2A–2C) (LeCun et al., 2015). A key feature driving the success of deep learning is the ability to automatically learn data representation, which obviates a need for time-consuming feature selection or data engineering. Over the last decade, deep learning has become democratized through the wide availability of cheap graphics processing units (GPUs) and the emergence of public software libraries written in Python (namely TensorFlow [Abadi et al., 2016] and PyTorch [Paszke et al., 2017]). Deep learning models often outperform standard machine learning models for many problems given a large enough set of example data (or “training data”).
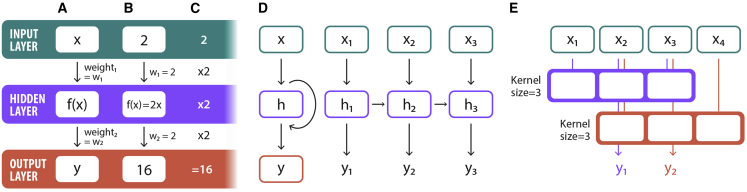
Figure 2.
Basic neural network background
(A and B) Neural networks are simply collections of math operations that transform an input (x) to an output (y). Inputs and outputs are connected to the neuron by weights, which are linear operators that multiply the previous value. The function in the hidden layer can be anything. In the simplest case with one neuron in the hidden layer (A), the input value is multiplied by the first weight, and then the new value x∗weight1 is input to the function in the neuron. The output of that function is multiplied by weight2 to calculate the output. When a neural network is “trained,” inputs are passed forward through the math to compute the output. The value of the output is compared with the true known value of y, and then the weights are updated slightly to make the output value closer to the true value of y. A simple example of this is shown in (B) where weight1 is 2, the function is 2∗x, and weight2 is 2. Note that neuron functions are often not linear (such as a rectified linear unit (ReLU) or sigmoid).
(C) The output y of this neural network is 16 when the input = 2.
(D) A simple recurrent neural network accepts sequence or time series data and adds a connection between the hidden layer and itself across time points, which allows the network to learn interactions between inputs in the series.
(E) A simple one-dimensional convolutional neural network showing how local patterns are summarized by a filter kernel into a new output vector with fewer dimensions.
Within deep learning, there are many types of models that differ primarily in how their neurons are connected. In particular, sequence data are well suited for recurrent neural networks (RNNs) (Rumelhart et al., 1986), including gated recurrent units (GRU) (Chung et al., 2014) or long short-term memory (LSTM) (Hochreiter and Schmidhuber, 1997) (Figure 2D). Convolutional neural networks (CNNs) are another architecture for spatially arranged data (Figure 2E). Originally introduced in 1980 (Fukushima, 1980), CNNs learn to filter local patterns in data, which makes them excellent for image classification (Krizhevsky et al., 2012; Kuo and Huang, 2018). Both mass spectra and peptide sequences have local patterns that make the CNN architecture a logical choice for predicting peptide spectra.
One limitation of deep learning is that usually tens of thousands of example data points are required to effectively train a neural network; however, models trained on a large number of examples for one task can be applied to significantly fewer examples in a new task using a process called transfer learning (Bozinovski, 2020; Lima et al., 2017). Transfer learning can be applied across tasks when the input data have similar structure, and enables transfer of learned data patterns across disparate tasks, such as between object classification and drug classification (Meyer et al., 2019).
Deep learning is increasingly applied to a variety of biomedical research problems (Ching et al., 2018). Recent increases in the speed and sensitivity of proteomic data collection have produced the quantities of data needed for training deep learning models. Deep learning models have emerged to predict peptide properties (Guan et al., 2019) from only a primary sequence, including tandem mass spectra (Gessulat et al., 2019; Tiwary et al., 2019; Zeng et al., 2019; Zhou et al., 2017), ion mobility (Meier et al., 2020), and retention time (Ma et al., 2017, 2018; Moruz and Käll, 2017). Furthermore, deep learning has been applied to improve peptide identification (Demichev et al., 2020; Tran et al., 2017, 2019), protein inference (Kim et al., 2017), and peak detection (Zohora et al., 2019). Other recent reviews cover more generally machine learning in proteomics (Bouwmeester et al., 2020a), machine learning specifically for data-independent acquisition (DDA) experiments (Xu et al., 2020), and more comprehensively all aspects of deep learning in proteomics (Wen et al., 2020a). In comparison, here I summarize neural network approaches for peptide property prediction and peptide/protein identification, provide perspective on how deep learning solves the associated challenges, and contrast the different deep learning strategies. Finally, limitations and opportunities of deep learning tools for mass spectrometry-based proteomics are also discussed.
Methods for predicting peptide properties
Predicting fragment ion intensities
Although the mass of possible peptide fragment ions can easily be predicted from any amino acid sequence, the exact intensities of those fragments depend on the unique chemistry of each peptide sequence (Huang et al., 2005; Tabb et al., 2003). Several reports of deep learning methods to predict fragment ion intensities in peptide MS/MS have emerged dating back to at least 2006 (Table 1). Most of these methods are based on repurposing specialized neural networks originally designed for natural language processing (RNN [Rumelhart et al., 1986] or a specific type of RNN, LSTM [Hochreiter and Schmidhuber, 1997]) (Figure 3). RNNs in particular are uniquely well suited to deal with the sequential nature of peptides, whose properties depend on local and long-range interactions between their amino acids. Fortunately, the common use of higher-energy collisional dissociation (HCD) between Q-TOF and Orbitrap instruments produces overall very similar spectra (Szabó et al., 2021), making predictions from models trained on one data type generally useful to the whole community.
Table 1.
Methods for fragment ion intensity prediction
| Year | Name | Neural network details | Comments | Citations |
|---|---|---|---|---|
| 2005 | PeptideART | feedforward network | engineered peptide feature inputs, outputs of fragment probabilities | Arnold et al. (2005), Li et al. (2011) |
| 2017 | pDeep | bidirectional LSTM, multi-output regression; Keras v1.2.1, TensorFlow 0.12.1 | limited to peptides of up to 20 amino acids | Zhou et al. (2017) |
| 2018 | DeepMatch | bidirectional LSTM, weak supervision | direct integration with peptide spectrum matching algorithm outperforms COMET | Schoenholz et al. (2018) |
| 2018a | Prosit (latin for “of benefit”) | encoder: bidirectional GRU with dropout and attention, parallel encoding of precursor charge and collision energy; decoder: bidirectional GRU with dropout and time-distributed dense; multi-output regression Keras 2.1.1 and TensorFlow 1.4.0 | over half a million training peptides and 21 million MS/MS spectra at multiple collision energies, predicts MS/MS spectra and retention time, integration with database search to decrease FDR, integration with Skyline (MacLean et al., 2010), web tool https://www.proteomicsdb.org/prosit/ | Gessulat et al. (2019) |
| 2019a | DeepMass | encoder: three bidirectional LSTM with 385 units each; decoder: four fully connected dense layers 768 units each; multi-output regression TensorFlow v.1.7.0 |
predicted fragmentation with accuracy similar to repeated measure of the same peptide's fragmentation. Predicted spectra used for DIA data analysis nearly equivalent to spectral libraries | Tiwary et al. (2019) |
| 2019 | pDeep2 | bidirectional LSTM, multi-output regression | original pDeep model adapted to predict spectra of modified peptides using transfer learning | Zeng et al. (2019) |
| 2019a | N/A | encoder: bidirectional LSTM with dropout; iRT model, two dense layers, tanh, single output regression. Charge state distribution model, two dense layers, softmax activation, multi-output regression length 5 for charge 1–5. Spectral prediction model, a time-distributed dense layer with sigmoid activation function, multi-output regression; Keras | predicts retention time, precursor charge state distribution, and fragment ion spectra | Guan et al. (2019) |
| 2019 | MS2CNN | basic CNN architecture, engineered peptide features as input with a CNN kernel size of 4 | better than pDeep for prediction of spectra from +3 charge state peptide precursors | Lin et al., 2019 |
| 2020a | DeepDIA | hybrid CNN and bidirectional LSTM, CNN first extracts features from pairs of amino acids, then LSTM, then dense layer. Multi-output regression of the b/year ions, including water/ammonia losses. Keras 2.2.4 and TensorFlow 1.11 | predicts MS/MS spectra and indexed retention time (iRT). Slightly more protein identifications from DIA analysis of Hela proteome than libraries from DDA or Prosit | Yang et al. (2020) |
| 2020 | N/A | sequence-to-sequence CNN | full-spectrum prediction, not only fragment ions | Liu et al. (2020) |
Abbreviation are as follows: FDR, false discovery rate; N/A, not applicable.
Indicates methods that predict other factors apart from fragment ion spectra.
Figure 3.
Concept of LSTM neural network applied to fragment ion spectra prediction for peptides
Each amino acid in the sequence is converted to a string of ones and zeros unique to that amino acid (called “one-hot encoding”). The encoded sequences are fed into one or more bidirectional LSTM layers. The output from the hidden layers is essentially multi-output regression, where real values are predicted for each possible b and y ions corresponding to the relative abundances of those fragments. Network weights are learned that accurately convert a given sequence in the correct proportions of fragment ions.
The reproducibility of repeated peptide fragmentation spectra for the same peptide can be considered an upper limit benchmark to determine the best possible performance of a spectral prediction algorithm. Early work from Predrag Radivojac's group determined the Pearson correlation between fragment ion intensities across repeated measurement of the same peptide and found overall good agreement ranging from 0.76 to 0.93 depending on the source and charge state of the peptides (Li et al., 2011). The authors developed a two-layer neural network to predict tandem mass spectra named PeptideART (Arnold et al., 2005). PeptideART-predicted spectra with Pearson correlation approaching the reproducibility of measurements across different experiments, which showed the promise of this approach (Table 1).
In 2017, a leap forward in peptide fragment ion intensity prediction was achieved using a bidirectional LSTM (Zhou et al., 2017) trained on proteome tools fragmentation spectra (Zolg et al., 2017). This model, named pDeep, was one of the first spectral prediction tools that could be considered “deep” as opposed to “shallow,” which refers to the use of many network layers (Table 1). pDeep predicts fragmentation by HCD, electron transfer dissociation, and electron transfer higher-energy collisional dissociation with a median Pearson correlation between predicted and observed spectra of over 0.9. At the time of introduction, pDeep greatly outperformed other spectral prediction algorithms, and its performance approached the theoretical upper limit set by technical reproducibility of repeat spectra collection. The authors further found that their network could differentiate isobaric amino acid differences (for example, GG versus N, or isoleucine versus leucine). This seminal paper opened the floodgates for additional deep learning methods of spectral and peptide property prediction.
Two years later, an updated model was published by the same group, pDeep2 (Zeng et al., 2019), which was trained and tested on approximately 8,000,000 peptide-spectra matches (PSMs). The full model was further adapted for prediction of fragmentation spectra from peptides containing 22 post-translational modifications (PTMs) using transfer learning (Table 1). Transfer learning is a process by which a neural network is trained on a large dataset, and then later adapted to another similar but distinct type of prediction by tuning with a smaller training dataset (Bozinovski, 2020; Lima et al., 2017; Meyer et al., 2019). To learn modified spectra, as few as 7,000 PTM PSMs were required. Transfer learning from the full model was critical to enable accurate PTM spectra prediction.
Another approach to peptide sequence prediction called DeepMatch directly incorporated the process of peptide identification (Schoenholz et al., 2018) (Table 1). To achieve this, the authors used a neural network with three parts: (1) an LSTM to predict fragments for a sequence, (2) a fully connected network to represent those fragments, and (3) a scoring system to compare the observed spectra with the predicted fragments for a sequence. Weak supervision was used to train this combined system, which means labels were assigned for each experimental spectrum based on the best scoring peptide (due to a lack of known “gold standard” PSMs). DeepMatch identified significantly more peptides than a traditional peptide identification software Comet (Eng et al., 2013), even after Comet results were refined with Percolator (Käll et al., 2007).
The following spring of 2019 brought a pair of spectra prediction papers: Prosit (Gessulat et al., 2019) and DeepMass (Tiwary et al., 2019). Both models focused on prediction of only b/year fragment ions, and were mostly applied to tryptic peptides. Prosit used a GRU (Chung et al., 2014) with attention (Bahdanau et al., 2016) architecture trained on the ProteomeTools synthetic peptide resource of 550,000 tryptic peptides measured by 21 million tandem mass spectra at various collision energies. Prosit predicts both tandem mass spectra and peptide retention time with high quality, and the quality of predicted spectra exceeded that of experimental measurement in some cases. Prosit was integrated into a database search to lower false discovery rates, and was applied to predict spectra for non-tryptic peptides. DeepMass also produced spectra within the measurement uncertainty of repeated MS/MS events, and the authors performed model analysis to show how distant amino acids interact to influence fragment ion intensity. Furthermore, both Prosit and DeepMass were used to generate spectral libraries for data-independent acquisition (DIA) proteomics experiments for any organisms based only on protein sequences (Table 1).
An obvious application of peptide property prediction is for DIA mass spectrometry (Meyer and Schilling, 2017; Meyer et al., 2017), which provides better analysis depth and more consistent peptide quantification across samples than DDA. One original limitation slowing adoption of DIA was the requirement for a spectral library, including the retention time for each peptide. Usually those libraries are built from separate exhaustive sample fractionation and repeated mass spectrometry analysis by DDA. DeepDIA takes advantage of property prediction to enable streamlined DIA analysis without a need to collect spectral libraries (Yang et al., 2020) (Table 1). DeepDIA identified slightly more proteins from DIA analysis of Hela proteome samples than libraries from Prosit or from DDA. DeepDIA also performed better than DirectDIA, which is a strategy that generates spectral libraries directly from DIA data similar to DIA-Umpire (Tsou et al., 2015).
Most of the above-mentioned spectra prediction algorithms used RNN architectures, but the CNN and variations thereof are also well suited for prediction from ordered peptide sequence data. A model named MS2CNN showed that CNNs predict fragment ion spectra with good accuracy (Lin et al., 2019). Instead of using the CNN directly on the peptide sequence to learn spatial structure of amino acids, the model input was an engineered feature vector (Table 1). Notably, the MS2CNN model was slightly worse at spectral prediction than pDeep for peptides with +2 precursor charge, but slightly better for peptides with +3 precursor charge.
Tandem mass spectra from the most common dissociation, HCD, are primarily composed of sequence informative b and y ions from the N- and C-terminal portions of the peptide, respectively, after fragmentation of the amide bond along the peptide backbone. Therefore, most spectra prediction strategies only predict the abundance of these fragments. However, MS/MS spectra can have significant contributions from other types of ions, such as losses of water or ammonia, or internal ions resulting from multiple peptide backbone cleavages. Liu et al. (2020) built a model that predicts these non-backbone fragments along with the typical backbone fragments. In contrast with other models, they used a sequence-to-sequence CNN architecture. They showed that their model outperformed pDeep, DeepMass, and Prosit for overall spectra prediction, probably due in part to the lack of non-backbone ion prediction by these other methods.
The unique chemistry of a peptide sequence determines its observable properties. Therefore, a neural network model that learns any peptide property captures the fundamental chemistry, and can be applied to predict other peptide properties. As an example of this, Guan et al. (2019) trained peptide property encoding networks with three different decoders for various peptides, including retention time, precursor charge state distribution, and fragmentation pattern in tandem mass spectra. These models were accurate relative to other work, and the added prediction of charge state distribution may be useful for peptide identification tasks.
Predicting retention time and collisional cross-section
In addition to some of the tandem mass spectrum prediction models above that also predict peptide retention time, several other models have been introduced to predict only peptide retention time (Table 2). These models have different strengths and weaknesses, but in general deep learning methods outperform other peptide retention time prediction methods. The success of these diverse neural network architectures can be useful to inform other peptide property prediction tasks.
Table 2.
Methods for prediction of retention time
| Year | Name | Neural network details | Comment | Citation |
|---|---|---|---|---|
| 2003 | N/A | fully connected neural network with 2 hidden layers, 20 inputs and one output | 95% of retention predictions within 10% of the true value | Petritis et al. (2003) |
| 2006 | N/A | fully connected neural network with 16 inputs, 4 hidden neurons, and 1 output | mean prediction error ~5.8% | Shinoda et al. (2006) |
| 2006 | N/A | 1,052 input nodes, 24 hidden nodes, 1 output node | average elution time precision of 1.5% | Petritis et al. (2006) |
| 2017 | DeepRT | feature extraction by LSTM and CNN, retention prediction from bagged ensemble of standard prediction models. Theano (0.9.0 dev1), Keras (1.0.1), and sklearn (0.17.1) | 95% of retention predictions within 28 min versus best benchmark of 45.8 min | Ma et al. (2017) |
| 2018 | DeepRT+ | capsule network (a type of CNN) | 95% of retention predictions within 15.7 min versus DeepRT at 24.7 min or best benchmark of 45.8 min | Ma et al. (2018) |
| 2019 | Prosita (latin for “of benefit”) | encoder: bidirectional GRU with dropout and attention, parallel encoding of precursor charge and collision energy; decoder: bidirectional GRU with dropout and time-distributed dense; multi-output regression Keras 2.1.1 and TensorFlow 1.4.0 | over half a million training peptides and 21 million MS/MS spectra at multiple collision energies, predicts MS/MS spectra and retention time, integration with database search to decrease FDR, integration with Skyline (cite), web tool https://www.proteomicsdb.org/prosit/ | Gessulat et al. (2019) |
| 2019 | DeepMassa | encoder: three bidirectional LSTM with 385 units each; decoder: four fully connected dense layers 768 units each; multi-output regression TensorFlow v.1.7.0 |
predicted fragmentation with accuracy similar to repeated measure of the same peptide's fragmentation. Predicted spectra used for DIA data analysis nearly equivalent to spectral libraries | Tiwary et al. (2019) |
| 2019 | N/A | encoder: bidirectional LSTM with dropout; iRT model, two dense layers, tanh, single output regression. Charge state distribution model, two dense layers, softmax activation, multi-output regression length 5 for charge 1–5. Spectral prediction model, a time-distributed dense layer with sigmoid activation function, multi-output regression; Keras | predicts retention time, precursor charge state distribution, and fragment ion spectra | Guan et al. (2019) |
| 2020 | DeepDIAa | hybrid CNN and bidirectional LSTM, CNN first extracts features from pairs of amino acids, then LSTM, then dense layer. Multi-output regression of the b/year ions, including water/ammonia losses. Keras 2.2.4 and TensorFlow 1.11 | predicts MS/MS spectra and indexed retention time (iRT). Slightly more protein identifications from DIA analysis of Hela proteome than libraries from DDA or Prosit | Yang et al. (2020) |
| 2020 | DeepLC | hybrid network: three CNN input paths (1) one-hot amino acid sequence, (2) amino acid pairs, and (3) amino acid composition. One dense input of peptide features. Inputs concatenated and processed through dense layers | predicts retention time for previously unseen peptide modifications | Bouwmeester et al. (2020b) |
| 2020 | AutoRT | ensemble of 10 best CNN and LSTM, networks returned by transfer learning. Keras 2.2.4 and TensorFlow 1.13.1 | used predicted retention time as a filter to assess identification strategies for mutated peptides | Wen et al. (2020b) |
Abbreviations are as follows: FDR, false discovery rate; N/A, not applicable.
Indicates methods that predict other factors beyond retention time.
Maybe the first example of an artificial neural network applied to predict any peptide property was from Richard Smith's group in 2003 (Petritis et al., 2003). In this early work, a simple feedforward 3-layer neural network was used: the input layer had 20 neurons (one for each amino acid), the hidden layer had 2 neurons, and the output was a single value between 0 and 1 indicating relative peptide retention time. The input layer took counts for each amino acid in the peptide. This simple neural network design allowed determination of relative weights for each amino acid's contribution to the observed elution time. The authors found that 95% of the predicted peptide retention times were within 10% of their true value, and the predicted retention time helped disambiguate isobaric peptides with the same molecular formula. This approach was later improved by adding inputs of several features, including sequence instead of only composition (Petritis et al., 2006). A similar strategy was published for peptides from LysC digestion of the E. coli proteome (Shinoda et al., 2006).
Roughly 15 years later, Siqi Lui's group revisited the use of neural networks for peptide retention time prediction by using LSTM, but also CNNs (Fukushima, 1980). Their first iteration, DeepRT (Ma et al., 2017), used the LSTM and CNN for feature extraction from the peptide sequences followed by an ensemble of more traditional models, predictions from which were averaged by “bagging” (Breiman, 1996). The following year, they introduced DeepRT+ (Ma et al., 2018), which used a type of CNN called a capsule network (Sabour et al., 2017) to make retention time predictions. DeepRT+ trained on data from C18 peptide retention was even able to be retrained to predict retention time of peptides on a completely different stationary phase, such as strong cation exchange (Table 2). At the time each of these models was introduced, they significantly outperformed previous benchmarks set by GPTime (Maboudi Afkham et al., 2016) and Elude (Moruz et al., 2010).
The ability to accurately predict peptide retention time could be used to reveal whether putative peptide identifications are true or false hits. Such filtration of putative PSMs would be especially useful when new data analysis strategies are needed, for example, for identification of peptides resulting from somatic mutation. Wen et al. (2020b) developed a strategy called autoRT based on an ensemble of CNN and LSTM models (Table 2). AutoRT was more accurate than the other retention time prediction models according to median absolute error. AutoRT was useful in assessing various filtration strategies for peptide identification; two-stage false discovery rate calculation and additional validation by the PepQuery algorithm (Wen et al., 2019) were useful in limiting putative peptides with large errors in observed versus predicted retention time.
Much like the proteome tools PTM peptide dataset was used by pDeep2 to predict spectra for modified peptides, a model named DeepLC used the same data to enable prediction retention times for modified peptides (Bouwmeester et al., 2020b) (Table 2). DeepLC was trained and tested using 20 diverse datasets, including reversed-phase, hydrophilic interaction, and strong cation exchange chromatography. As expected, the model performance improved with increasing dataset size. A key difference to the DeepLC strategy is inclusion of one input layer that encodes amino acids by their chemical composition. The authors showed that this allowed them to predict the retention time for modifications that were not included in the training data. The authors also showed a further generalization of this strategy where they encode amino acids as a modified form of glycine. This approach is promising for predicting and evaluation of retention times for modified peptides.
LSTM networks are also extremely accurate at predicting other peptide properties. For example, Meier et al. (2020) recently described a deep learning strategy for accurate prediction of peptide collisional cross-sections (CCSs) as measured on the TimsTOF instrument (Meier et al., 2015). This strategy enabled prediction of peptide CCS values with ∼1% median relative error, and SHAP analysis (Lundberg and Lee, 2017) revealed how amino acids contribute to the observed CCS. As demonstrated for peptide retention time prediction (Wen et al., 2020b), CCS prediction may prove to be a useful filter for the peptide identification process.
Deep learning methods for peptide and protein identification
In addition to peptide property prediction, numerous deep learning strategies have emerged to enable better peptide and protein identification. Deep learning models are effective in detecting LC-MS features (Kantz et al., 2019; Zohora et al., 2019), assessing if spectra are high enough quality to be identified (Ma, 2017), and even predicting which peptides from a protein are likely observable (i.e., proteotypic) (Serrano et al., 2019). Deep neural networks are also effective in de novo peptide sequencing (Tran et al., 2017, 2019), which is essentially the opposite task described for spectrum prediction above. Instead of asking “what is the spectra for this peptide?” they ask “what peptide explains this spectra?” DeepNovo achieves this using both a CNN and LSTM to learn features of MS/MS, fragments, and sequence patterns (Tran et al., 2017) (Table 3). DeepNovo outperformed other de novo sequencing algorithms according to multiple metrics and was able to reconstruct over 97% of antibody sequences with over 97% accuracy without a sequence database.
Table 3.
Methods for protein and peptide identification
| Year | Name | Neural network details | Comment | Citation |
|---|---|---|---|---|
| 2012 | Barista | special type of network or tripartite graph where layers represent proteins, peptides, and spectra | protein inference through integration of protein and peptide identification | Spivak et al. (2012) |
| 2017 | DeepPep | CNN, torch7 framework | predicts peptide probability from binarized protein sequence, protein scored based on change in peptide prediction without each protein | Kim et al. (2017) |
| 2017 | DeepNovo | LSTM/CNN hybrid network built with TensorFlow | application to DDA data. Iteratively predicts one amino acid at each step. Up to 64% better than previous algorithms | Tran et al. (2017) |
| 2018 | DeepMatch | bidirectional LSTM, weak supervision | spectral prediction integrated with peptide identification | Schoenholz et al. (2018) |
| 2019 | DeepNovo | LSTM/CNN hybrid network built with TensorFlow | adapted to DIA data by incorporating the retention time dimension | Tran et al. (2018) |
| 2020 | DIA-NN | ensemble of dense, feedforward classifiers. Implemented with Cranium DNN | operates with or without a user-supplied spectral library | Demichev et al. (2020) |
| 2020 | DeepRescore | uses AutoRT and pDeep2 models | generates new scores derived from comparing observed peptide properties to deep learning-predicted properties. Those scores are input to Percolator | Wen et al. (2020b) |
In a subsequent paper, DeepNovo was adapted to identify peptides from DIA data by incorporating the retention time dimension (Tran et al., 2019). CNNs learned to embed precursor and fragment ion profiles over time, and again LSTM networks decoded the best matching amino acids. Peptides identified by DeepNovo-DIA were highly complementary to those found by two other library-free methods, PECAN (Ting et al., 2017) and DirectDIA. Compared with these other tools, DeepNovo-DIA did not require any database, and enabled discovery of human leukocyte antigen peptides.
DeepMatch (Schoenholz et al., 2018), mentioned in the tandem mass spectra prediction section, also directly incorporates the peptide identification process with their spectral prediction. Another approach related to DeepDIA (Yang et al., 2020) showed that the supplementation of experimental spectral libraries with predicted libraries could enable improved coverage of specific proteins of interest, such as membrane proteins (Lou et al., 2020). Similarly, Searle et al. (2020) generated libraries in silico but showed that empirically correcting the library further improved the number of detectable peptides. All these approaches using predicted spectra and retention time used standard downstream DIA processing tools, e.g., Spectronaut (Bruderer et al., 2015).
The key step in peptide identification is assigning a score that segregates true matches from decoy matches. Approaches, such as PeptideProphet (Keller et al., 2002) and Percolator (Käll et al., 2007), build models from multiple peptide scores or properties to improve the separation of decoys and targets and increase peptide identification. Deep neural networks were recently applied to build such a classifier for discrimination of peptide peaks in DIA. DIA-NN uses multiple interference correction strategies and a simple dense feedforward neural network to improve peptide identification from DIA experiments (Demichev et al., 2020) (Table 3). DIA-NN significantly outperformed other tools for peptide identification from DIA data (Gotti et al., 2020). DIA-NN can operate with or without a spectral library, and is therefore an excellent addition to our proteomic toolkit.
An extension of the idea to use predicted peptide properties, such as retention time, to filter peptide identifications (Wen et al., 2020b) is to rather use predicted peptide properties to re-score peptide identifications. DeepRescore does exactly this using both predicted retention time and predicted tandem mass spectra. Compared with DIA-NN that replaces Percolator to learn the difference between targets and decoys, DeepRescore directly used features derived from comparison with deep learning-based predictions to add scores for input to Percolator. This strategy improved the sensitivity and quality of non-tryptic peptide identifications purified from human leukocyte antigen.
Protein inference has also been tackled with neural networks. An early example was Barista, which combined protein and peptide identification tasks into a single goal of optimizing protein identifications (Spivak et al., 2012) (Table 3). This approach produced up to 34% more protein identifications than competing approaches. More recently, an interesting strategy called DeepPep (Kim et al., 2017) approached protein inference using CNN models trained to predict the probabilities of identified peptides from binarized protein sequences with 1 where peptides match and 0 everywhere else (Table 3). Once trained, the model was used to predict the peptide probabilities by iteratively holding out each of the protein sequence inputs, enabling calculation of each protein's importance in explaining the peptide list. DeepPep performed similar to the five other compared models, and thus provides a new perspective on protein inference.
Discussion and future prospects
For decades, each proteomic experiment has been treated independently of all previous work, requiring de novo re-identification of previously observed peptides. This is partially due to the original data collection strategy for peptide discovery, stochastic DDA. A recent shift has devoted significant energy to reuse previous peptide identifications (Martens and Vizcaíno, 2017) and more comprehensively sample peptides using data collection by DIA. A shortfall of DIA originally was the requirement for library spectra, but ingenious strategies based on co-elution of peptide precursor ions and fragment ions changed this (Tsou et al., 2015), including for PTM discovery (Meyer et al., 2017). These new deep learning tools to predict spectra and retention time now remove nearly all barriers to DIA analysis, and will likely push the field of proteomics further toward DIA analysis (Van Puyvelde et al., 2020).
Although we have seen great progress in recent years in the application of deep learning to proteomics, there are still several limitations to widespread adoption. First, making predictions from trained models is feasible on standard computers, but training new models requires specialized hardware called graphics processing units, or GPUs. Furthermore, different deep learning libraries are used for model generations, namely TensorFlow or PyTorch, and these models are not intercompatible (see “neural network details” columns in Tables 1, 2, and 3). These requirements are compounded by the need for specialist scientists who understand the advanced computational requirements along with the relevant advanced mass spectrometry details. These challenges may limit the adoption of deep learning methods in proteomics but these barriers may be ameliorated with a shift to public cloud resources (Neely, 2021).
Another limitation in the current proteomics deep learning landscape is the lack of unified metrics to use for comparing tools. Currently, each paper has its own comparison between tools using different datasets and different metrics. Machine learning scientists in other fields often create a unified set of tasks to serve as benchmarks by which new strategies can be compared (for example, see ImageNet [Krizhevsky et al., 2012]). A common set of peptides to be used for property prediction tasks for proteomics would benefit the field and enable more direct comparison across various algorithms. This would also enable more scrutiny of the reported results as anyone would more easily be able to verify the performance of an arbitrary algorithm.
A major unexplored opportunity for the application of deep learning with proteomics is for automating the interpretation of large omic datasets. This will require framing experiments as regression or classification problems instead of hypothesis generation experiments. Interpretation of data by deep learning will require the production of thousands of proteome examples, which represents a major barrier given the average throughput of most experiments. However, recent advances have enabled much higher throughput proteome quantification, such as microflow LC (Bian et al., 2020), short gradient LC (Bache et al., 2018), and even the lack of LC altogether with direct infusion shotgun proteome analysis (Meyer et al., 2020).
Another potential source of large volumes of data required for deep learning is from public repositories; there is growing hunger for machine-readable datasets, including detailed metadata about the sample preparation, individual treatments, and data acquisition. Unfortunately, requirements for additional metadata come at a cost of less desire by researchers to use repositories. However, increasing the amount of metadata provided with all studies would open up new possibilities in training neural networks for additional tasks. For example, is this concentration of trypsin associated with observing a specific proteolytic cleavage, or is a common reagent associated with a detrimental artifact? Or across similar sample preparation and data collection parameters, can we learn how gene knockouts influence the proteome more generally? These questions might become tractable if available datasets included very detailed metadata. These questions would also be more tractable if sample preparation and data collection protocols were more standardized across labs and continents.
One fascinating example of deep learning for omic data interpretation completely ignores the peptide and protein identification process and uses DIA data maps as images for sample classification by a CNN (Zhang et al., 2020). Instead of learning from curated identities of proteins or peptide quantities, this work shows that deep learning can directly classify samples into diseased or healthy conditions from the raw data. Notably, benign thyroid nodules were distinguished from papillary thyroid carcinoma with over 91% accuracy, and the model based on the DIA-based image (called DIA tensor) performed better than a model based on quantified proteins. Further work could use model analysis methods to determine which pixels (corresponding to peptide fragments) are responsible for the differential classification.
With the explosion of deep learning tools for mass spectrometry in recent years, it will take time for the dust to settle and reveal those tools that are both easy to use and effective. We have already seen that integration with Skyline software (MacLean et al., 2010) has increased adoption and use of Prosit (Gessulat et al., 2019). It will also take time for experiments to determine the best ways to incorporate these new tools. For example, by optimizing the isolation windows for in-silico-predicted spectral libraries, recent work showed that even an old Q-Exactive could identify over 7,000 human proteins from a single analysis (Doellinger et al., 2020).
In summary, deep learning tools have transformed the field of proteomics over the last few years, and will likely be ingrained in all aspects of proteomics for the foreseeable future.
Acknowledgments
Dasom Hwang illustrated the figures from concept sketches.
Declaration of interests
The authors declare no competing interests.
References
- Abadi M., Barham P., Chen J., Chen Z., Davis A., Dean J., Devin M., Ghemawat S., Irving G., Isard M., et al. TensorFlow: a system for large-scale machine learning. arXiv. 2016 160508695 Cs. [Google Scholar]
- Aebersold R., Mann M. Mass-spectrometric exploration of proteome structure and function. Nature. 2016;537:347–355. doi: 10.1038/nature19949. [DOI] [PubMed] [Google Scholar]
- Alzubi J., Nayyar A., Kumar A. Machine learning from theory to algorithms: an overview. J. Phys. Conf. Ser. 2018;1142:012012. [Google Scholar]
- Arnold, R.J., Jayasankar, N., Aggarwal, D., Tang, H., and Radivojac, P. (2005). A machine learning approach to predicting peptide fragmentation spectra. In Biocomputing 2006, (Maui, Hawaii: World Scientific), pp. 219–230. [PubMed]
- Bache N., Geyer P.E., Bekker-Jensen D.B., Hoerning O., Falkenby L., Treit P.V., Doll S., Paron I., Müller J.B., Meier F., et al. A novel LC system embeds analytes in pre-formed gradients for rapid, ultra-robust proteomics. Mol. Cell. Proteomics. 2018;17:2284–2296. doi: 10.1074/mcp.TIR118.000853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahdanau D., Cho K., Bengio Y. Neural machine translation by jointly learning to align and translate. arXiv. 2016 14090473 Cs Stat. [Google Scholar]
- Bekker-Jensen D.B., Kelstrup C.D., Batth T.S., Larsen S.C., Haldrup C., Bramsen J.B., Sørensen K.D., Høyer S., Ørntoft T.F., Andersen C.L., et al. An optimized shotgun strategy for the rapid generation of comprehensive human proteomes. Cell Syst. 2017;4:587–599.e4. doi: 10.1016/j.cels.2017.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bian Y., Zheng R., Bayer F.P., Wong C., Chang Y.-C., Meng C., Zolg D.P., Reinecke M., Zecha J., Wiechmann S., et al. Robust, reproducible and quantitative analysis of thousands of proteomes by micro-flow LC–MS/MS. Nat. Commun. 2020;11:157. doi: 10.1038/s41467-019-13973-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouwmeester R., Gabriels R., Van Den Bossche T., Martens L., Degroeve S. The age of data-driven proteomics: how machine learning enables novel workflows. Proteomics. 2020;20:1900351. doi: 10.1002/pmic.201900351. [DOI] [PubMed] [Google Scholar]
- Bouwmeester R., Gabriels R., Hulstaert N., Martens L., Degroeve S. DeepLC can predict retention times for peptides that carry as-yet unseen modifications (Bioinformatics) bioRxiv. 2020 doi: 10.1101/2020.03.28.013003. [DOI] [PubMed] [Google Scholar]
- Bozinovski S. Reminder of the first paper on transfer learning in neural networks, 1976. Informatica. 2020;44 doi: 10.31449/inf.v44i3.2828. [DOI] [Google Scholar]
- Breiman L. Bagging predictors. Mach. Learn. 1996;24:123–140. [Google Scholar]
- Bruderer R., Bernhardt O.M., Gandhi T., Miladinović S.M., Cheng L.-Y., Messner S., Ehrenberger T., Zanotelli V., Butscheid Y., Escher C., et al. Extending the limits of quantitative proteome profiling with data-independent acquisition and application to acetaminophen-treated three-dimensional liver microtissues. Mol. Cell. Proteomics. 2015;14:1400–1410. doi: 10.1074/mcp.M114.044305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ching T., Himmelstein D.S., Beaulieu-Jones B.K., Kalinin A.A., Do B.T., Way G.P., Ferrero E., Agapow P.-M., Zietz M., Hoffman M.M., et al. Opportunities and obstacles for deep learning in biology and medicine. J. R. Soc. Interface. 2018;15:20170387. doi: 10.1098/rsif.2017.0387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung J., Gulcehre C., Cho K., Bengio Y. Empirical evaluation of gated recurrent neural networks on sequence modeling. arXiv. 2014 14123555 Cs. [Google Scholar]
- Demichev V., Messner C.B., Vernardis S.I., Lilley K.S., Ralser M. DIA-NN: neural networks and interference correction enable deep proteome coverage in high throughput. Nat. Methods. 2020;17:41–44. doi: 10.1038/s41592-019-0638-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doellinger J., Blumenscheit C., Schneider A., Lasch P. Isolation window optimization of data-independent acquisition using predicted libraries for deep and accurate proteome profiling. Anal. Chem. 2020;92:12185–12192. doi: 10.1021/acs.analchem.0c00994. [DOI] [PubMed] [Google Scholar]
- Domingos P. A few useful things to know about machine learning. Commun. ACM. 2012;55:78–87. [Google Scholar]
- Eng J.K., Jahan T.A., Hoopmann M.R. Comet: an open-source MS/MS sequence database search tool. Proteomics. 2013;13:22–24. doi: 10.1002/pmic.201200439. [DOI] [PubMed] [Google Scholar]
- Fukushima K. Neocognitron: a self-organizing neural network model for a mechanism of pattern recognition unaffected by shift in position. Biol. Cybern. 1980;36:193–202. doi: 10.1007/BF00344251. [DOI] [PubMed] [Google Scholar]
- Gessulat S., Schmidt T., Zolg D.P., Samaras P., Schnatbaum K., Zerweck J., Knaute T., Rechenberger J., Delanghe B., Huhmer A., et al. Prosit: proteome-wide prediction of peptide tandem mass spectra by deep learning. Nat. Methods. 2019;16:509–518. doi: 10.1038/s41592-019-0426-7. [DOI] [PubMed] [Google Scholar]
- Gotti C., Roux-Dalvai F., Joly-Beauparlant C., Leclercq M., Mangnier L., Droit A. Extensive and accurate benchmarking of DIA acquisition methods and software tools using a complex proteomic standard (Bioinformatics) bioRxiv. 2020 doi: 10.1101/2020.11.03.365585. [DOI] [PubMed] [Google Scholar]
- Guan S., Moran M.F., Ma B. Prediction of LC-MS/MS properties of peptides from sequence by deep learning. Mol. Cell. Proteomics. 2019;18:2099–2107. doi: 10.1074/mcp.TIR119.001412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochreiter S., Schmidhuber J. Long short-term memory. Neural. Comput. 1997;9:1735–1780. doi: 10.1162/neco.1997.9.8.1735. [DOI] [PubMed] [Google Scholar]
- Huang Y., Triscari J.M., Tseng G.C., Pasa-Tolic L., Lipton M.S., Smith R.D., Wysocki V.H. Statistical characterization of the charge state and residue dependence of low-energy CID peptide dissociation patterns. Anal. Chem. 2005;77:5800–5813. doi: 10.1021/ac0480949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarzab A., Kurzawa N., Hopf T., Moerch M., Zecha J., Leijten N., Bian Y., Musiol E., Maschberger M., Stoehr G., et al. Meltome atlas—thermal proteome stability across the tree of life. Nat. Methods. 2020;17:495–503. doi: 10.1038/s41592-020-0801-4. [DOI] [PubMed] [Google Scholar]
- Jiang L., Wang M., Lin S., Jian R., Li X., Chan J., Dong G., Fang H., Robinson A.E., Snyder M.P., et al. A quantitative proteome map of the human body. Cell. 2020;183:269–283.e19. doi: 10.1016/j.cell.2020.08.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Käll L., Canterbury J.D., Weston J., Noble W.S., MacCoss M.J. Semi-supervised learning for peptide identification from shotgun proteomics datasets. Nat. Methods. 2007;4:923–925. doi: 10.1038/nmeth1113. [DOI] [PubMed] [Google Scholar]
- Kantz E.D., Tiwari S., Watrous J.D., Cheng S., Jain M. Deep neural networks for classification of LC-MS spectral peaks. Anal. Chem. 2019;91:12407–12413. doi: 10.1021/acs.analchem.9b02983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller A., Nesvizhskii A.I., Kolker E., Aebersold R. Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal. Chem. 2002;74:5383–5392. doi: 10.1021/ac025747h. [DOI] [PubMed] [Google Scholar]
- Kim M., Eetemadi A., Tagkopoulos I. DeepPep: deep proteome inference from peptide profiles. PLoS Comput. Biol. 2017;13:e1005661. doi: 10.1371/journal.pcbi.1005661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krizhevsky A., Sutskever I., Hinton G.E. Advances in Neural Information Processing Systems. International Conference on Neural Information Processing Systems; 2012. Imagenet classification with deep convolutional neural networks; pp. 1097–1105.https://dl.acm.org/doi/10.5555/2999134.2999257 [Google Scholar]
- Kuo P.-H., Huang C.-J. A green energy application in energy management systems by an artificial intelligence-based solar radiation forecasting model. Energies. 2018;11:819. [Google Scholar]
- LeCun Y., Bengio Y., Hinton G. Deep learning. Nature. 2015;521:436. doi: 10.1038/nature14539. [DOI] [PubMed] [Google Scholar]
- Li S., Arnold R.J., Tang H., Radivojac P. On the accuracy and limits of peptide fragmentation spectrum prediction. Anal. Chem. 2011;83:790–796. doi: 10.1021/ac102272r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lima E., Sun X., Dong J., Wang H., Yang Y., Liu L. Learning and transferring convolutional neural network knowledge to ocean front recognition. IEEE Geosci. Remote Sens. Lett. 2017;14:354–358. [Google Scholar]
- Lin Y.-M., Chen C.-T., Chang J.-M. MS2CNN: predicting MS/MS spectrum based on protein sequence using deep convolutional neural networks. BMC Genomics. 2019;20 doi: 10.1186/s12864-019-6297-6. https://bmcgenomics.biomedcentral.com/articles/10.1186/s12864-019-6297-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu K., Li S., Wang L., Ye Y., Tang H. Full-spectrum prediction of peptides tandem mass spectra using deep neural network. Anal. Chem. 2020;92:4275–4283. doi: 10.1021/acs.analchem.9b04867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lou R., Tang P., Ding K., Li S., Tian C., Li Y., Zhao S., Zhang Y., Shui W. Hybrid spectral library combining DIA-MS data and a targeted virtual library substantially deepens the proteome coverage. IScience. 2020;23:100903. doi: 10.1016/j.isci.2020.100903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundberg S.M., Lee S.-I. In: Advances in Neural Information Processing Systems 30. Guyon I., Luxburg U.V., Bengio S., Wallach H., Fergus R., Vishwanathan S., Garnett R., editors. Curran Associates, Inc; 2017. A unified approach to interpreting model predictions; pp. 4765–4774. [Google Scholar]
- Ma C. DeepQuality: mass spectra quality assessment via compressed sensing and deep learning. arXiv. 2017 171011430 Q-Bio. [Google Scholar]
- Ma C., Zhu Z., Ye J., Yang J., Pei J., Xu S., Zhou R., Yu C., Mo F., Wen B., et al. DeepRT: deep learning for peptide retention time prediction in proteomics. arXiv. 2017 170505368 Q-Bio. [Google Scholar]
- Ma C., Ren Y., Yang J., Ren Z., Yang H., Liu S. Improved peptide retention time prediction in liquid chromatography through deep learning. Anal. Chem. 2018;90:10881–10888. doi: 10.1021/acs.analchem.8b02386. [DOI] [PubMed] [Google Scholar]
- Maboudi Afkham H., Qiu X., The M., Käll L. Uncertainty estimation of predictions of peptides’ chromatographic retention times in shotgun proteomics. Bioinformatics. 2016 doi: 10.1093/bioinformatics/btw619. [DOI] [PubMed] [Google Scholar]
- MacLean B., Tomazela D.M., Shulman N., Chambers M., Finney G.L., Frewen B., Kern R., Tabb D.L., Liebler D.C., MacCoss M.J. Skyline: an open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics. 2010;26:966–968. doi: 10.1093/bioinformatics/btq054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martens L., Vizcaíno J.A. A golden age for working with public proteomics data. Trends Biochem. Sci. 2017;42:333–341. doi: 10.1016/j.tibs.2017.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marx V. When computational pipelines go ‘clank’. Nat. Methods. 2020;17:659–662. doi: 10.1038/s41592-020-0886-9. [DOI] [PubMed] [Google Scholar]
- Mateus A., Määttä T.A., Savitski M.M. Thermal proteome profiling: unbiased assessment of protein state through heat-induced stability changes. Proteome Sci. 2016;15:13. doi: 10.1186/s12953-017-0122-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meier F., Beck S., Grassl N., Lubeck M., Park M.A., Raether O., Mann M. Parallel accumulation–serial fragmentation (PASEF): multiplying sequencing speed and sensitivity by synchronized scans in a trapped ion mobility device. J. Proteome Res. 2015;14:5378–5387. doi: 10.1021/acs.jproteome.5b00932. [DOI] [PubMed] [Google Scholar]
- Meyer J.G., Schilling B. Clinical applications of quantitative proteomics using targeted and untargeted data-independent acquisition techniques. Expert Rev. Proteomics. 2017;14:419–429. doi: 10.1080/14789450.2017.1322904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer J.G., Mukkamalla S., Steen H., Nesvizhskii A.I., Gibson B.W., Schilling B. PIQED: automated identification and quantification of protein modifications from DIA-MS data. Nat. Methods. 2017;14:646–647. doi: 10.1038/nmeth.4334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meier F., Köhler N.D., Brunner A.-D., Wanka J.-M.H., Voytik E., Strauss M.T., Theis F.J., Mann M. Deep learning the collisional cross sections of the peptide universe from a million training samples. Syst. Biol. 2020 doi: 10.1038/s41467-021-21352-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer J.G., Liu S., Miller I.J., Coon J.J., Gitter A. Learning drug function from chemical structure with convolutional neural networks and random forests. J. Chem. Inf. Model. 2019;59:4438–4449. doi: 10.1021/acs.jcim.9b00236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer J.G., Niemi N.M., Pagliarini D.J., Coon J.J. Quantitative shotgun proteome analysis by direct infusion. Nat. Methods. 2020;17:1222–1228. doi: 10.1038/s41592-020-00999-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moruz L., Käll L. Peptide retention time prediction. Mass Spectrom. Rev. 2017;36:615–623. doi: 10.1002/mas.21488. [DOI] [PubMed] [Google Scholar]
- Moruz L., Tomazela D., Käll L. Training, selection, and robust calibration of retention time models for targeted proteomics. J. Proteome Res. 2010;9:5209–5216. doi: 10.1021/pr1005058. [DOI] [PubMed] [Google Scholar]
- Neely B.A. Cloudy with a chance of peptides: accessibility, scalability, and reproducibility with cloud-hosted environments. J. Proteome Res. 2021 doi: 10.1021/acs.jproteome.0c00920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paszke A., Gross S., Chintala S., Chanan G., Yang E., DeVito Z., Lin Z., Desmaison A., Antiga L., Lerer A. https://openreview.net/forum?id=BJJsrmfCZ
- Petritis K., Kangas L.J., Ferguson P.L., Anderson G.A., Paša-Tolić L., Lipton M.S., Auberry K.J., Strittmatter E.F., Shen Y., Zhao R., et al. Use of artificial neural networks for the accurate prediction of peptide liquid chromatography elution times in proteome analyses. Anal. Chem. 2003;75:1039–1048. doi: 10.1021/ac0205154. [DOI] [PubMed] [Google Scholar]
- Petritis K., Kangas L.J., Yan B., Monroe M.E., Strittmatter E.F., Qian W.-J., Adkins J.N., Moore R.J., Xu Y., Lipton M.S., et al. Improved peptide elution time prediction for reversed-phase liquid chromatography-MS by incorporating peptide sequence information. Anal. Chem. 2006;78:5026–5039. doi: 10.1021/ac060143p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rumelhart D.E., Hinton G.E., Williams R.J. Learning representations by back-propagating errors. Nature. 1986;323:533–536. [Google Scholar]
- Sabour S., Frosst N., Hinton G.E. Dynamic Routing between Capsules. ArXiv. 2017 171009829 Cs. [Google Scholar]
- Schoenholz S.S., Hackett S., Deming L., Melamud E., Jaitly N., McAllister F., O’Brien J., Dahl G., Bennett B., Dai A.M., et al. Peptide-spectra matching from weak supervision. arXiv. 2018 180806576 Q-Bio Stat. [Google Scholar]
- Schubert O.T., Röst H.L., Collins B.C., Rosenberger G., Aebersold R. Quantitative proteomics: challenges and opportunities in basic and applied research. Nat. Protoc. 2017;12:1289–1294. doi: 10.1038/nprot.2017.040. [DOI] [PubMed] [Google Scholar]
- Searle B.C., Swearingen K.E., Barnes C.A., Schmidt T., Gessulat S., Küster B., Wilhelm M. Generating high quality libraries for DIA MS with empirically corrected peptide predictions. Nat. Commun. 2020;11 doi: 10.1038/s41467-020-15346-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serrano G., Guruceaga E., Segura V. DeepMSPeptide: peptide detectability prediction using deep learning. Bioinformatics. 2019;36:1279–1280. doi: 10.1093/bioinformatics/btz708. [DOI] [PubMed] [Google Scholar]
- Shinoda K., Sugimoto M., Yachie N., Sugiyama N., Masuda T., Robert M., Soga T., Tomita M. Prediction of liquid chromatographic retention times of peptides generated by protease digestion of the Escherichia coli proteome using artificial neural networks. J. Proteome Res. 2006;5:3312–3317. doi: 10.1021/pr0602038. [DOI] [PubMed] [Google Scholar]
- Sinitcyn P., Rudolph J.D., Cox J. Computational methods for understanding mass spectrometry-based shotgun proteomics data. Annu. Rev. Biomed. Data Sci. 2018;1:207–234. [Google Scholar]
- Spivak M., Weston J., Tomazela D., MacCoss M.J., Noble W.S. Direct maximization of protein identifications from tandem mass spectra. Mol. Cell. Proteomics. 2012;11 doi: 10.1074/mcp.M111.012161. M111.012161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szabó D., Schlosser G., Vékey K., Drahos L., Révész Á. Collision energies on QTof and Orbitrap instruments: how to make proteomics measurements comparable? J. Mass Spectrom. 2021;56:e4693. doi: 10.1002/jms.4693. [DOI] [PubMed] [Google Scholar]
- Tabb D.L., Smith L.L., Breci L.A., Wysocki V.H., Lin D., Yates J.R. Statistical characterization of ion trap tandem mass spectra from doubly charged tryptic peptides. Anal Chem. 2003;75:1155–1163. doi: 10.1021/ac026122m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ting Y.S., Egertson J.D., Bollinger J.G., Searle B.C., Payne S.H., Noble W.S., MacCoss M.J. PECAN: library-free peptide detection for data-independent acquisition tandem mass spectrometry data. Nat. Methods. 2017;14:903–908. doi: 10.1038/nmeth.4390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiwary S., Levy R., Gutenbrunner P., Salinas Soto F., Palaniappan K.K., Deming L., Berndl M., Brant A., Cimermancic P., Cox J. High-quality MS/MS spectrum prediction for data-dependent and data-independent acquisition data analysis. Nat. Methods. 2019;16:519–525. doi: 10.1038/s41592-019-0427-6. [DOI] [PubMed] [Google Scholar]
- Tran N.H., Zhang X., Xin L., Shan B., Li M. De novo peptide sequencing by deep learning. Proc. Natl. Acad. Sci. U S A. 2017;114:8247–8252. doi: 10.1073/pnas.1705691114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran N.H., Qiao R., Xin L., Chen X., Liu C., Zhang X., Shan B., Ghodsi A., Li M. Deep learning enables de novo peptide sequencing from data-independent-acquisition mass spectrometry. Nature Methods. 2018;16:63–66. doi: 10.1038/s41592-018-0260-3. [DOI] [PubMed] [Google Scholar]
- Tran N.H., Qiao R., Xin L., Chen X., Liu C., Zhang X., Shan B., Ghodsi A., Li M. Deep learning enables de novo peptide sequencing from data-independent-acquisition mass spectrometry. Nat. Methods. 2019;16:63–66. doi: 10.1038/s41592-018-0260-3. [DOI] [PubMed] [Google Scholar]
- Tsiamis V., Ienasescu H.-I., Gabrielaitis D., Palmblad M., Schwämmle V., Ison J. One thousand and one software for proteomics: tales of the toolmakers of science. J. Proteome Res. 2019;18:3580–3585. doi: 10.1021/acs.jproteome.9b00219. [DOI] [PubMed] [Google Scholar]
- Tsou C.-C., Avtonomov D., Larsen B., Tucholska M., Choi H., Gingras A.-C., Nesvizhskii A.I. DIA-Umpire: comprehensive computational framework for data-independent acquisition proteomics. Nat. Methods. 2015;12:258–264. doi: 10.1038/nmeth.3255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Puyvelde B., Willems S., Gabriels R., Daled S., De Clerck L., Vande Casteele S., Staes A., Impens F., Deforce D., Martens L., et al. Removing the hidden data dependency of DIA with predicted spectral libraries. Proteomics. 2020;20:1900306. doi: 10.1002/pmic.201900306. [DOI] [PubMed] [Google Scholar]
- Wen B., Wang X., Zhang B. PepQuery enables fast, accurate, and convenient proteomic validation of novel genomic alterations. Genome Res. 2019;29:485–493. doi: 10.1101/gr.235028.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen B., Zeng W., Liao Y., Shi Z., Savage S.R., Jiang W., Zhang B. Deep learning in proteomics. Proteomics. 2020;20:1900335. doi: 10.1002/pmic.201900335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen B., Li K., Zhang Y., Zhang B. Cancer neoantigen prioritization through sensitive and reliable proteogenomics analysis. Nat. Commun. 2020;11 doi: 10.1038/s41467-020-15456-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie F., Liu T., Qian W.-J., Petyuk V.A., Smith R.D. Liquid chromatography-mass spectrometry-based quantitative proteomics. J. Biol. Chem. 2011;286:25443–25449. doi: 10.1074/jbc.R110.199703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L.L., Young A., Zhou A., Röst H.L. Machine learning in mass spectrometric analysis of DIA data. Proteomics. 2020;20:1900352. doi: 10.1002/pmic.201900352. [DOI] [PubMed] [Google Scholar]
- Yang Y., Liu X., Shen C., Lin Y., Yang P., Qiao L. In silico spectral libraries by deep learning facilitate data-independent acquisition proteomics. Nat. Commun. 2020;11:146. doi: 10.1038/s41467-019-13866-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng W.-F., Zhou X.-X., Zhou W.-J., Chi H., Zhan J., He S.-M. MS/MS spectrum prediction for modified peptides using pDeep2 trained by transfer learning. Anal. Chem. 2019;91:9724–9731. doi: 10.1021/acs.analchem.9b01262. [DOI] [PubMed] [Google Scholar]
- Zhang F., Yu S., Wu L., Zang Z., Yi X., Zhu J., Lu C., Sun P., Sun Y., Selvarajan S., et al. Phenotype classification using proteome data in a data-independent acquisition tensor format. J. Am. Soc. Mass Spectrom. 2020;31:2296–2304. doi: 10.1021/jasms.0c00254. [DOI] [PubMed] [Google Scholar]
- Zhou X.-X., Zeng W.-F., Chi H., Luo C., Liu C., Zhan J., He S.-M., Zhang Z. pDeep: predicting MS/MS spectra of peptides with deep learning. Anal. Chem. 2017;89:12690–12697. doi: 10.1021/acs.analchem.7b02566. [DOI] [PubMed] [Google Scholar]
- Zohora F.T., Rahman M.Z., Tran N.H., Xin L., Shan B., Li M. DeepIso: a deep learning model for peptide feature detection from LC-MS map. Sci. Rep. 2019;9:17168. doi: 10.1038/s41598-019-52954-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zolg D.P., Wilhelm M., Schnatbaum K., Zerweck J., Knaute T., Delanghe B., Bailey D.J., Gessulat S., Ehrlich H.-C., Weininger M., et al. Building ProteomeTools based on a complete synthetic human proteome. Nat. Methods. 2017;14:259–262. doi: 10.1038/nmeth.4153. [DOI] [PMC free article] [PubMed] [Google Scholar]