Summary
Pangenome analysis is fundamental to explore molecular evolution occurring in bacterial populations. Here, we introduce Pagoo, an R framework that enables straightforward handling of pangenome data. The encapsulated nature of Pagoo allows the storage of complex molecular and phenotypic information using an object-oriented approach. This facilitates to go back and forward to the data using a single programming environment and saving any stage of analysis (including the raw data) in a single file, making it sharable and reproducible. Pagoo provides tools to query, subset, compare, visualize, and perform statistical analyses, in concert with other microbial genomics packages available in the R ecosystem. As working examples, we used 1,000 Escherichia coli genomes to show that Pagoo is scalable, and a global dataset of Campylobacter fetus genomes to identify evolutionary patterns and genomic markers of host-adaptation in this pathogen.
Keywords: pangenome analysis, bacterial comparative genomics, bacterial evolution, data visualization, object-oriented programming, R, pangenome reconstruction
Graphical abstract
Highlights
-
•
Pagoo is a software framework to analyze bacterial pangenomes
-
•
It can be used with the output of any pangenome reconstruction software
-
•
Genetic, phenotypic, and other metadata are integrated in a single object or file
-
•
Pagoo is extensible and interacts easily with other microbial genomics packages
Motivation
Despite the extensive availability of software for bacterial pangenome reconstruction, current tools for pangenome data analysis do not provide end-to-end solutions because, once visualizations are generated, users cannot use the same framework to return to the data and perform further comparisons and more complex analyses using standardized methods. To fill this gap, we developed Pagoo, a flexible and extensible but standardized framework that allows performing integrative pangenome analysis in a simple and reproducible way.
Ferrés and Iraola develop a software package in R to integrate and analyze bacterial pangenome data. This allows creating a single programming object that stores all the data, as well as methods to produce standard analyses and visualizations for comparative genomics.
Introduction
The exponentially growing number of diverse bacterial genomes has prompted pangenome reconstruction as a gold standard to uncover molecular evolution of bacterial populations (Tettelin et al., 2005; Vernikos et al., 2015). This is because of the high intraspecific diversity observed in bacterial genomes, which are affected by horizontal gene transfer, variations in effective population size, and constant colonization of new niches. As these forces are influential in determining pangenome size and structure (McInerney et al., 2017), pangenome comparisons can reveal genome evolutionary dynamics associated with important biological processes, such as speciation, host adaptation, pathogenicity, or the acquisition of antimicrobial resistance. Pangenome reconstruction is typically performed from genes annotated in a set of whole-genome sequences. In general, coding sequences of different strains are grouped in orthologous clusters based on different similarity criteria. Then, pangenome data inform about the belonging of each gene encoded in each genome to a certain orthologous cluster. In recent years, several software tools have been developed to reconstruct bacterial pangenomes, such as Roary (Page et al., 2015), panX (Ding et al., 2018), PanOCT (Fouts et al., 2012), PIRATE (Bayliss et al., 2019), PEPPAN (Zhou et al., 2020), or Panaroo (Tonkin-Hill et al., 2020). These tools focus on automation of steps, improvement of clustering algorithms, and optimization of computational costs to process thousands of sequences of increasingly large genomic datasets. Many of these software include data visualization modules along with other specific software that have been developed with this purpose, including Phandango (Hadfield et al., 2018) or PanViz (Pedersen et al., 2017). However, current tools do not provide end-to-end solutions for customized pangenome data analysis, since, once visualizations are generated, users cannot use the same framework to return to the data and perform further comparisons and more complex analyses using standardized methods.
Here, we introduce Pagoo, a pangenome post-processing tool that can take the output produced by pangenome reconstruction software tools providing a standardized framework for its analysis. Pagoo is based on an object-oriented design built on a class system in R (R Core team, 2017), which implements: (1) an integrative data structure for standardized storage of pangenome information, such as orthologous clusters, sequences, annotations, and metadata, in a single object (shareable through a single file); (2) a set of straightforward methods for responsive querying, handling, and subsetting of this data structure; and (3) a set of standard statistics and active visualizations leveraging flexible downstream comparative analyses. Along with extensive documentation, we show how Pagoo interacts with other widely used microbial genomics tools and the R ecosystem for improved analysis of molecular evolution in bacterial populations.
New approaches
Description of the software
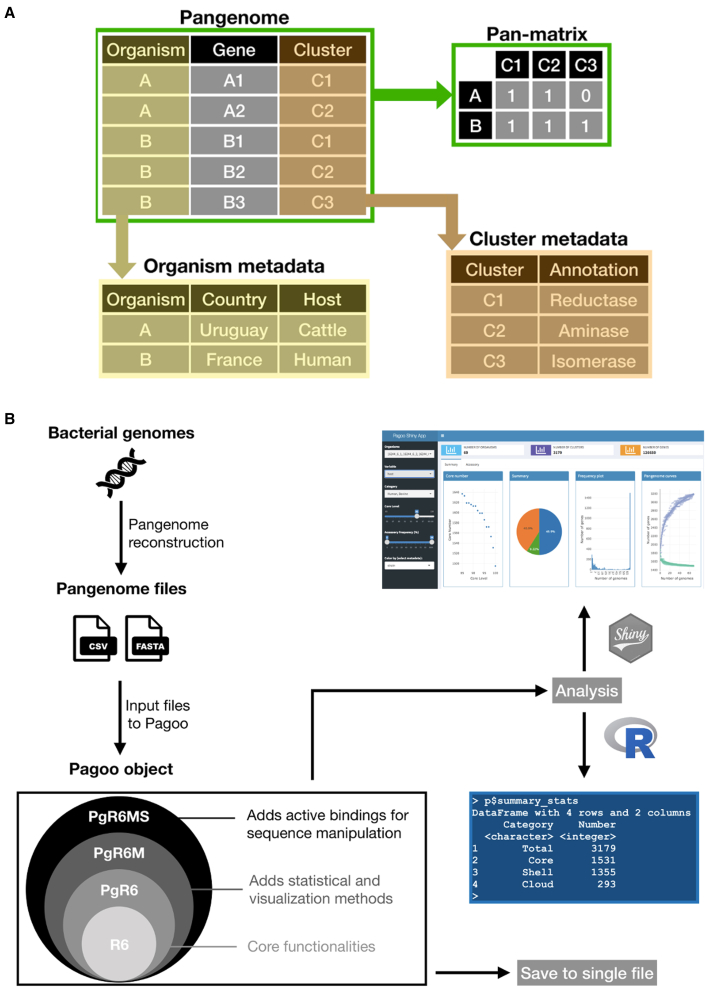
A pangenome can be represented as individual genes that belong to organisms (genomes), which are then assigned to a cluster of orthologous genes. Pagoo stores this as a three-column matrix, with one column identifying an individual gene, the next one identifying the organism that this gene belongs to, and the last one identifying the orthologous cluster that the gene was assigned by the pangenome reconstruction method. Optionally, this matrix can contain additional columns as gene-specific metadata, such as annotations, functional assignments, or genomic coordinates. Orthologous clusters and organisms can also take metadata represented as two different matrices, with the condition that each one must contain a column that correctly maps each observation (cluster or organism) into the former matrix. Gene sequences can also be added to this structure, with the condition that their names must also map to rows in the first matrix (Figure 1A). This relational structure optimizes data storage avoiding duplication, enables flexibility to work with different types of metadata, and facilitates complex querying and analysis.
Figure 1.
Framework and overall design of Pagoo
(A) Example of the relational structure implemented to store, link, and operate over different pangenome data types.
(B) General description of the workflow from assembled genomes to Pagoo analysis. Once pangenome files are created with any available pangenome reconstruction software, these files can be loaded to create the Pagoo object. The specific R6 classes store and manage different data types that can store all the information in a single file or perform comparative analyses using the R console interface or the Pagoo Shiny application.
A salient and unique feature of Pagoo is that these data structures are stored and managed in an encapsulated, object-oriented fashion using the R6 package as backend. In contrast with traditional R programming, the R6 paradigm considers that methods belong to objects rather than to generic functions, so an object contains both the data and embedded methods to analyze it. In this context, the Pagoo object is built on three R6 classes. PgR6 is the most basic class that contains methods and functions for data handling and subsetting. Then, PgR6M inherits all the methods and fields from PgR6 and incorporates statistical methods and visualization tools based on the ggplot2 package (Wickham, 2011). PgR6MS inherits all capabilities from the others and adds methods for manipulation of biological sequences using the Biostrings package (Figure 1B). These classes support the main data types that typically represent a pangenome, providing a synergistic framework to manage both the raw data and methods to perform operations and explore results with customized visualizations. Moreover, any of these classes could be further inherited and easily extended by third-party applications.
Another remarkable feature of Pagoo is that raw data stored in the pangenome object are kept unaltered in the background, while users can query, mutate, or subset the object using active bindings. This allows changing the state of the object without altering the original data. For example, users can temporarily hide certain organisms from the dataset, actively set thresholds that change the definition of core genes, or extract specific information from organisms, genes, clusters, or sequences. Class-specific methods for generic subset operators are also implemented, enabling extraction of relevant fields straight from the object by using standard R subset notation.
Pagoo provides specific methods for generating the pangenome object from scratch by formatting output files from any pangenome reconstruction software. In addition, Pagoo provides specific functions to automatically generate the pangenome object from output files produced by Roary and Panaroo. Other popular pangenome reconstruction software, such as PIRATE and PEPPAN, have functions to transform their outputs into Roary's output format, making them compatible with Pagoo. Then, pangenome can be saved with any changes to the object along with the unaltered original data as a single file. Pagoo has been built and tested in all three major operative systems (Linux, Windows, and Mac). A detailed explanation of each method and operator for data input, saving, and loading, and for specific comparative analyses, is provided in the online user manual (GitHub: https://iferres.github.io/pagoo/). Together, this implementation represents a conceptual advance for pangenome data handling, facilitating reproducibility, and enabling multiple and flexible analyses.
Data analysis and visualization
Pagoo includes statistical and visualization methods. Customized plots and statistical analyses can be generated directly from the pangenome object using active bindings on the console, or by deploying a built-in R-Shiny application. This interactive application is divided into two main components: (1) a general dashboard that interactively displays summary statistics, including number of organisms, orthologous clusters and genes, core and accessory genome sizes, gene frequency barplots, pangenome curves, and scrollable information about core genome clusters and genes (i.e., annotation or any other metadata); and (2) a specific dashboard showing clustering of genomes according to accessory gene distances and principal-component analysis (PCA), genome-specific accessory genome sizes, visualization of gene presence/absence matrix with associated metadata, and information about accessory gene clusters (Figure S1). This interactive application allows responsive exploration of evolutionary trends in bacterial populations, guiding downstream analyses on the console that can be performed over the same pangenome object using methods provided by Pagoo or leveraging the interaction with other R tools.
Creating recipes for more complex analyses
Remarkably, more complex comparative pangenome analyses can be performed by applying concise code recipes. We define recipes as relatively short snippets that pipe pangenome information extracted from the object as input to other R tools. We have developed example recipes (available in the online user manual at GitHub: https://iferres.github.io/pagoo/articles/6-Recipes.html) to build core genome phylogenies, identify population structure, explore genome-wide selective pressures acting over the core genes, and compare individual gene sequences against specific databases. Importantly, the development and implementation of recipes enable full reproducibility of publication-quality figures generated directly from the pangenome object. Despite the R ecosystem currently including dozens of packages that allow performing diverse and complex comparative analyses, it is possible that some specific tasks could be difficult to implement in R. In this sense, Pagoo recipes can be designed to interact with external tools to generate results outside the R session and integrate them into the pangenome object using standard R data input functions.
Results
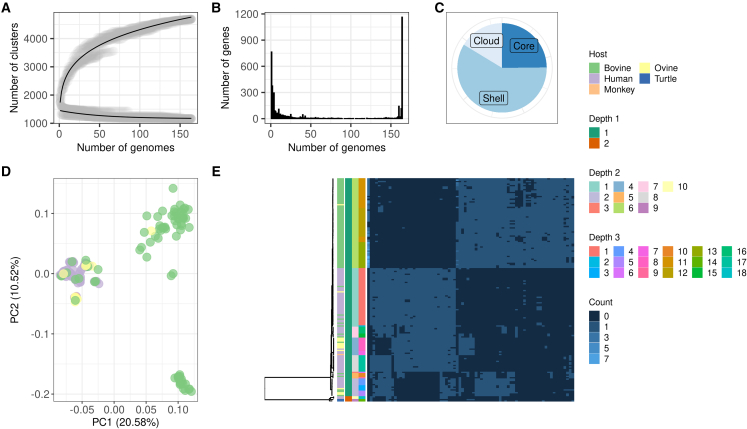
As a working example, we used a previously published study on the genomic evolution of Campylobacter fetus (Iraola et al., 2017), a zoonotic pathogen that presents a strong population structure with different lineages adapted to livestock or humans. In brief, we used Pagoo to analyze a pangenome reconstructed from 164 C. fetus genomes (Figure 2). Using simple and readable R code we were able to recover the main diversity trends reported for this species, such as a marked difference in accessory gene patterns between livestock- and human-adapted lineages. Specifically, we identified a set of 78 highly discriminant accessory genes that can differentiate between these lineages and could be used to develop molecular typing tools (Table S1). In addition, automatic extraction of core genes from the Pagoo object enabled robust phylogenomic analyses in interaction with other R packages, such as DECIPHER (Wright, 2015), for multiple sequence alignment, phangorn (Schliep, 2011) for phylogenetic reconstruction, and Rhierbaps (Tonkin-Hill et al., 2018) for population structure inference. This analysis revealed the same population structure composed by eight main C. fetus lineages as reported in the original study (Iraola et al., 2017). Then, we used Pagoo to compare phylogenies built from every single core gene against the core genome phylogeny. This allowed us to rank the core genes based on their goodness to recover the population structure, mcp4 being the one with the closest distance (Figure S2A). This enables the future development of high-resolution C. fetus typing methods using amplicon sequencing of single core genes. Also, the PCA based on the accessory genes allowed to recover previously observed patterns of variation separating bovine-adapted C. fetus from other hosts, including humans (discrimination given by PC1 in observed in Figure 2D). Using this approach we also detected a separation between bovine-adapted genomes given by PC2 (Figure 2D). Further exploration of this demonstrated a specific group of genomes from Spain with particular accessory gene patterns, suggesting a previously unnoticed geographic structuring of this pathogen.
Figure 2.
Results extracted from the pangenome object
Exploration of the C. fetus pangenome using information directly extracted from the pangenome object and customized esthetics.
(A) Pangenome and core genome curves with gray circles representing different sub-samples at increasing numbers of genomes; the black lines show the fitting to the power law and exponential decay functions, respectively.
(B) The distribution of genes in different subsets of genomes.
(C) The distribution of the pangenome in core genes and accessory genes (shell and cloud genes).
(D) A principal components analysis generated from the gene presence/absence matrix whose first principal component (PC1) clearly distinguishes between two groups of genomes, one of mostly bovine-derived strains (right), and the other with various hosts (left).
(E) A heatmap (right) with gene count (paralogs) abundance (in columns) and per organisms in (rows, associated to a phylogenetic tree [left]) inferred from a concatenated alignment of core genes. Between the tree and the heatmap, the color bars indicate, from left to right, the host associated to each isolate, and three different depth levels of subpopulations inferred by the Rhierbaps package.
An additional dataset consisting of hundreds of multi-drug resistant E. coli genomes (Decano and Downing, 2019) was used to test scalability by measuring the time it takes Pagoo to perform certain operations. First, Pagoo was able to upload the output from Roary, including gene sequences automatically extracted from GFF3 annotation files, and automatically build its relational structure for 500 genomes in ∼20 min. This time does not scale linearly because it depends on the number and size of gene clusters, but was completed in reasonable time (<2 h) for 1,000 genomes. Second, once the Pagoo object is created, information can be queried on the fly in matter of seconds or minutes. For example, one single operation can extract all core genes from 1,000 genomes in ∼2 min. Also, visualizations such as gene distribution plots can be rendered in seconds just using single operations (Figure S2B).
Discussion
The advent of high-throughput sequencing technologies more than 15 years ago pushed microbiology toward the field of comparative genomics, which rapidly transitioned from studies including few to thousands of genomes (Vernikos et al., 2015). This substantially increased the complexity of datasets, requiring new approaches to systematically handle and track different components of interrelated pangenomic data. Pagoo introduces a framework underpinned in a concept that leverages the simplicity of storing all the information in a standardized and reproducible manner in a single, shareable object, as well as providing specific methods to query and analyze it. The implemented classes can be easily inherited and extended, so users can eventually incorporate methods able to cope with any kind of metadata added to the pangenomes, such as genomic coordinates or structural information. Pagoo not only allows to perform basic exploratory analysis through the Shiny application, but also facilitates expert bioinformatic analysis to deliver more complex results through the R console ecosystem working in concert with other population genomic packages. The advantage of using an interpreted language for post-processing of pangenomic data, like R, in contrast to compiled tools that run from the terminal interface, is that users can go back and forward to the data, producing both general and fine-grained analyses in a single environment. Pagoo's encapsulated and object-oriented nature brings simplicity and intuition to the experience of working with these kinds of complex relational data, in combination with the active-binding feature of R6 classes, which opens the possibility of changing the state of the object as a whole and its behavior. On the contrary, the classic functional object-oriented R programming works well for simpler data structures, but falls short and becomes inconvenient when dealing with complex and potentially mutable structures, which pangenomes are. Overall, Pagoo's design aims to improve and facilitate current practices on the analysis of molecular evolution in bacterial populations.
Limitations of the study
Although the R ecosystem includes dozens of packages to perform diverse comparative genomic analyses, it is possible that some specific tasks are difficult to implement in R or are better covered by other pangenome analysis tools. Also, Pagoo has been tested with hundreds to thousands of genomes, but a potential limitation is the time it can take to generate Pagoo objects from extremely big datasets. In particular, Pagoo's Shiny application for dynamic visualization has been designed to work with small to medium size datasets.
STAR★Methods
Key resources table
Resource availability
Lead contact
Further information and requests for resources should be directed to and will be fulfilled by the lead contact, Gregorio Iraola (giraola@pasteur.edu.uy).
Materials availability
This study did not generate new unique reagents.
Method details
Re-analysis of C. fetus dataset
Genome assemblies were obtained from a previously published study by (Iraola et al., 2017). Genome re-annotation was performed with Prokka (Seemann, 2014) and the pangenome was reconstructed using Roary (Page et al., 2015) with default parameters. Assessment of genome quality revealed that 4 assemblies contained an abnormal number of genes (Figure S2C), so they were hidden from the dataset using Pagoo’s ‘drop()’ function. This function is recommended only with exploratory purposes or when the number of hidden organisms is small. Otherwise, it is recommended to reconstruct the pangenome by removing undesired genomes from the beginning. Plots describing summary statistics were generated using Pagoo’s built-in methods and the ggplot2 package (Wickham, 2011).
Core genes were extracted from the pangenome object using Pagoo’s method ‘core_seqs_4_phylo’, were aligned with DECIPHER (Wright, 2015) and a reference phylogeny was reconstructed from concatenated core gene sequences using the phangorn package (Schliep, 2011). The Rhierbaps package (Tonkin-Hill et al., 2018) was used to infer population structure. Each individual core gene was aligned to reconstruct gene-specific phylogenies as described above. The topological distance between the reference phylogeny and gene-specific phylogenies was calculated with the ‘dist.topo’ function from the ape package (Paradis and Schliep, 2019). A tanglegram was plotted to compare the reference phylogeny with the closest topology obtained with single genes.
Scalability assessment using E. coli genomes
To evaluate scalability, annotated GFF3 files from 1,000 E. coli genomes were obtained from (Decano and Downing, 2019). We used subsets of 10, 100, 500 and 1,000 genomes to build pangenomes using Roary (Page et al., 2015) with default parameters. Then, we assessed the time Pagoo takes to complete five different operations using these pangenome datasets (Figure S2B). The evaluated operations were grouped in two categories: (1) loading pangenome data, and (2) querying already loaded pangenome data. In the first category, we evaluated the time it takes to: (1.1) create a Pagoo object using the built-in function ‘roary_2_pagoo()’ providing only the gene presence/absence matrix file, (1.2) create a Pagoo object using the ‘roary_2_pagoo()’ function but also providing the GFF3 files to include sequences into the Pagoo object, and (1.3) create a Pagoo object from already loaded information and sequences into the R session using the ‘pagoo()’ function. In the second category we measured the time it takes to (2.1) retrieve core genome sequences, and (2.2) generate a pangenome frequency plot. Each operation was repeated 10 times.
Quantification and statistical analysis
A Principal Component Analysis (PCA) based on accessory gene presence/absence patterns was generated with Pagoo's method ‘pan_pca()’. Then, those accessory genes which most contributed to discriminate between livestock-associated and human-associated lineages were identified based on the eigenvector of the first component. Genes with a loading value lower than −0.05 and greater than 0.05 were selected (Figure S2D; Table S1).
Acknowledgments
We thank Pablo Fresia, Daniela Costa, and Andrés Parada for insightful comments and suggestions during testing of Pagoo. I.F. is funded by grant ANII-POS_NAC_2018_1_151494 from the Agencia Nacional de Investigación e Innovación (ANII), Uruguay. This work has been partially funded by the G4 Research Groups Program from Institut Pasteur Montevideo and Banco de Seguros del Estado (BSE) of Uruguay.
Author contributions
G.I. and I.F. conceived the idea. I.F. developed the software and performed experiments. G.I. and I.F. wrote the manuscript.
Declaration of interests
The authors declare no competing interests.
Published: September 27, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.crmeth.2021.100085.
Contributor Information
Ignacio Ferrés, Email: iferres@pasteur.edu.uy.
Gregorio Iraola, Email: giraola@pasteur.edu.uy.
Supplemental information
Data and code availability
-
•
Pagoo code has been deposited and is available at the Comprehensive R Archive Network (CRAN). Pagoo’s source code is also available at GitHub. We provide a Singularity (Kurtzer et al., 2017) image with all dependencies needed to fully reproduce analyses using the two working dataset (C. fetus and E. coli), from downloading the data to generating the plots. The Singularity definition file along with the scripts are publicly available at GitHub. The prebuilt Singularity image is hosted at Singularity-hub (Sochat et al., 2017).
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- Bayliss S.C., Thorpe H.A., Coyle N.M., Sheppard S.K., Feil E.J. PIRATE: a fast and scalable pangenomics toolbox for clustering diverged orthologues in bacteria. GigaScience. 2019;8:1–9. doi: 10.1093/gigascience/giz119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decano A.G., Downing T. An Escherichia coli ST131 pangenome atlas reveals population structure and evolution across 4,071 isolates. Sci. Rep. 2019;9:17394. doi: 10.1038/s41598-019-54004-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding W., Baumdicker F., Neher R.A. panX: pan-genome analysis and exploration. Nucleic Acids Res. 2018;46:e5. doi: 10.1093/nar/gkx977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fouts D.E., Brinkac L., Beck E., Inman J., Sutton G. PanOCT: automated clustering of orthologs using conserved gene neighborhood for pan-genomic analysis of bacterial strains and closely related species. Nucleic Acids Res. 2012;40:e172. doi: 10.1093/nar/gks757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadfield J., Croucher N.J., Goater R.J., Abudahab K., Aanensen D.M., Harris S.R. Phandango: an interactive viewer for bacterial population genomics. Bioinformatics. 2018;34:292–293. doi: 10.1093/bioinformatics/btx610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iraola G., Forster S.C., Kumar N., Lehours P., Bekal S., García-Peña F.J., Paolicchi F., Morsella C., Hotzel H., Hsueh P.-R., et al. Distinct Campylobacter fetus lineages adapted as livestock pathogens and human pathobionts in the intestinal microbiota. Nat. Commun. 2017;8:1367. doi: 10.1038/s41467-017-01449-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurtzer G.M., Sochat V., Bauer M.W. Singularity: scientific containers for mobility of compute. PLoS One. 2017;12:e0177459. doi: 10.1371/journal.pone.0177459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McInerney J.O., McNally A., O’Connell M.J. Why prokaryotes have pangenomes. Nat. Microbiol. 2017;2:1–5. doi: 10.1038/nmicrobiol.2017.40. [DOI] [PubMed] [Google Scholar]
- Page A.J., Cummins C.A., Hunt M., Wong V.K., Reuter S., Holden M.T.G., Fookes M., Falush D., Keane J.A., Parkhill J. Roary: rapid large-scale prokaryote pan genome analysis. Bioinformatics. 2015;31:3691–3693. doi: 10.1093/bioinformatics/btv421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paradis E., Schliep K. Ape 5.0: an environment for modern phylogenetics and evolutionary analyses in R. Bioinformatics. 2019;35:526–528. doi: 10.1093/bioinformatics/bty633. [DOI] [PubMed] [Google Scholar]
- Pedersen T.L., Nookaew I., Wayne Ussery D., Månsson M. PanViz: interactive visualization of the structure of functionally annotated pangenomes. Bioinformatics. 2017;33:1081–1082. doi: 10.1093/bioinformatics/btw761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core team (2017). R: A Languague and Environment for Statistical Computing.
- Schliep K.P. phangorn: phylogenetic analysis in R. Bioinformatics. 2011;27:592–593. doi: 10.1093/bioinformatics/btq706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seemann T. Prokka: rapid prokaryotic genome annotation. Bioinformatics. 2014;30:2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- Sochat V.V., Prybol C.J., Kurtzer G.M. Enhancing reproducibility in scientific computing: metrics and registry for singularity containers. PLoS One. 2017;12:e0188511. doi: 10.1371/journal.pone.0188511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tettelin H., Masignani V., Cieslewicz M.J., Donati C., Medini D., Ward N.L., Angiuoli S.V., Crabtree J., Jones A.L., Durkin A.S., et al. Genome analysis of multiple pathogenic isolates of Streptococcus agalactiae: implications for the microbial “pan-genome”. Proc. Natl. Acad. Sci. U.S.A. 2005;102:13950–13955. doi: 10.1073/pnas.0506758102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tonkin-Hill G., Lees J.A., Bentley S.D., Frost S.D.W., Corander J. RhierBAPS: an R implementation of the population clustering algorithm hierBAPS. Wellcome Open Res. 2018;3:93. doi: 10.12688/wellcomeopenres.14694.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tonkin-Hill G., MacAlasdair N., Ruis C., Weimann A., Horesh G., Lees J.A., Gladstone R.A., Lo S., Beaudoin C., Floto R.A., et al. Producing polished prokaryotic pangenomes with the Panaroo pipeline. Genome Biol. 2020;21:180. doi: 10.1186/s13059-020-02090-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vernikos G., Medini D., Riley D.R., Tettelin H. Ten years of pan-genome analyses. Curr. Opin. Microbiol. 2015;23:148–154. doi: 10.1016/j.mib.2014.11.016. [DOI] [PubMed] [Google Scholar]
- Wickham H. ggplot2. WIREs Comput. Stat. 2011;3:180–185. [Google Scholar]
- Wright E.S. DECIPHER: harnessing local sequence context to improve protein multiple sequence alignment. BMC Bioinformatics. 2015;16:322. doi: 10.1186/s12859-015-0749-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z., Charlesworth J., Achtman M. Accurate reconstruction of bacterial pan- and core genomes with PEPPAN. Genome Res. 2020;30:1667–1679. doi: 10.1101/gr.260828.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
Pagoo code has been deposited and is available at the Comprehensive R Archive Network (CRAN). Pagoo’s source code is also available at GitHub. We provide a Singularity (Kurtzer et al., 2017) image with all dependencies needed to fully reproduce analyses using the two working dataset (C. fetus and E. coli), from downloading the data to generating the plots. The Singularity definition file along with the scripts are publicly available at GitHub. The prebuilt Singularity image is hosted at Singularity-hub (Sochat et al., 2017).
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.