ABSTRACT
The body of vertebrate embryos forms by posterior elongation from a terminal growth zone called the tail bud. The tail bud is a source of highly motile cells that eventually constitute the presomitic mesoderm (PSM), a tissue that plays an important role in elongation movements. PSM cells establish an anterior-posterior cell motility gradient that parallels a gradient associated with the degradation of a specific cellular signal (FGF) known to be implicated in cell motility. Here, we combine the electroporation of fluorescent reporters in the PSM with time-lapse imaging in the chicken embryo to quantify cell diffusive movements along the motility gradient. We show that a simple microscopic model for random cell motility induced by FGF activity along with geometric confinement leads to rectified tissue elongation consistent with our observations. A continuum analog of the microscopic model leads to a macroscopic mechano-chemical model for tissue extension that couples FGF activity-induced cell motility and tissue rheology, and is consistent with the experimentally observed speed and extent of elongation. Together, our experimental observations and theoretical models explain how the continuous addition of cells at the tail bud combined with lateral confinement can be converted into oriented movement and drive body elongation.
KEY WORDS: Chick morphogenesis, Embryo elongation, Rectified motility, Tissue expansion
Summary: A theoretical framework for the dynamics of body elongation in vertebrate embryos is provided, along with experimental observations that are consistent with it.
INTRODUCTION
Most vertebrate species exhibit an elongated body axis. This characteristic pattern is established during embryogenesis as the tissues progressively form in an anterior-to-posterior direction. Microsurgical ablation of the posterior presomitic mesoderm (PSM), which contains the precursors of skeletal muscles and the axial skeleton, severely reduces posterior elongation movements, indicating that this tissue plays a major role in the control of posterior elongation of the embryonic axis. Analysis of cell motility in the chicken embryo PSM (Bénazéraf et al., 2010) shows that there is an anterior-posterior gradient in the activity of cells. However, locally, the motility inside the PSM of chicken and zebrafish embryos is manifested by random, undirected, Brownian-like cellular motion (Bénazéraf et al., 2010; Lawton et al., 2013). These random, diffusive movements contrast with the oriented cell intercalation movements controlling elongation of the anterior parts of the embryo. Within the PSM, this motility gradient is downstream of a chemical gradient of the secreted fibroblast growth factor FGF, as shown schematically in Fig. 1A. FGF is known to play an important role in cell motility (Delfini et al., 2005); indeed, increasing FGF concentration in the PSM causes cell motility to increase without any orientation preference (Bénazéraf et al., 2010), reducing the speed of elongation. Earlier qualitative models focused on the role of the motility gradient as a driving force for elongation (Bénazéraf et al., 2010), but how this motility gradient is related to elongation is still unclear.
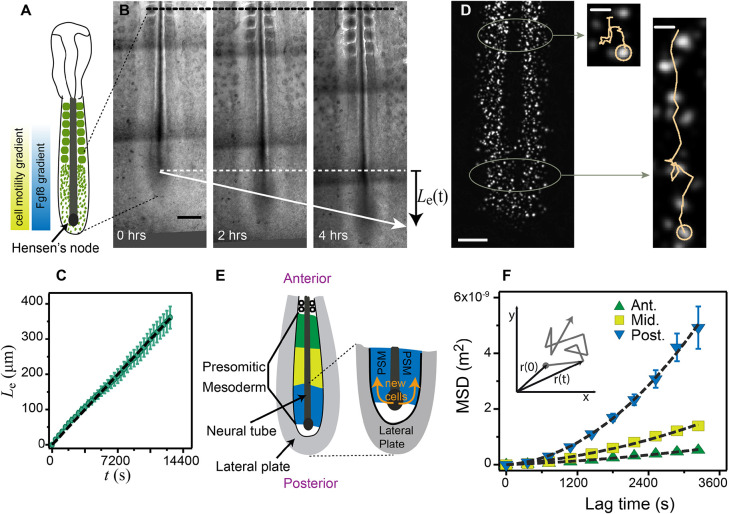
Fig. 1.
Axis elongation and cell diffusion in chicken embryo. (A) Schematic of an embryo at HH stage 10. Cell motility decreases from posterior to anterior, in correlation with a decrease in FGF concentration. A gradient of cell density (green) opposite to the motility gradient is shown in the schematic embryo. (B) Time series of an elongating embryo. The black dashed line shows the reference point for tracking the posterior elongation. Le(t) is the distance over which the Hensen's node advances over time. Scale bar: 200 µm. (C) Elongation of the PSM, as a function of time. The slope gives the average elongation rate 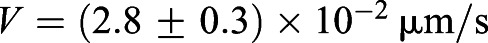 (n=5, mean±s.e.m.). (D) Electroporated cells inside the PSM. Anterior cells advance a shorter distance than the posterior cells for the same duration of time (here 4 h). Scale bars: 200 µm (whole PSM); 25 µm (zoomed tracks). (E) Schematic of the PSM showing the three regions considered for MSD analysis shown in F, as well as the depiction of the neighboring tissues. New stem cells are generated by division of the progenitor cells inside the tail bud (TB), and move into the PSM. The movement of cells in the PSM is limited by the neural tube medially, the somites anteriorly and the lateral plate laterally. (F) Average MSD from five embryos (1000 cells) for the anterior (Ant.), middle (Mid.) and posterior (Post.) regions of the PSM. Dashed lines are fits to Eqn 1. Inset shows a sketch of random motion.
(n=5, mean±s.e.m.). (D) Electroporated cells inside the PSM. Anterior cells advance a shorter distance than the posterior cells for the same duration of time (here 4 h). Scale bars: 200 µm (whole PSM); 25 µm (zoomed tracks). (E) Schematic of the PSM showing the three regions considered for MSD analysis shown in F, as well as the depiction of the neighboring tissues. New stem cells are generated by division of the progenitor cells inside the tail bud (TB), and move into the PSM. The movement of cells in the PSM is limited by the neural tube medially, the somites anteriorly and the lateral plate laterally. (F) Average MSD from five embryos (1000 cells) for the anterior (Ant.), middle (Mid.) and posterior (Post.) regions of the PSM. Dashed lines are fits to Eqn 1. Inset shows a sketch of random motion.
The observation that at a local level cell motility is random and undirected needs to be reconciled with the emergence of global body elongation. As new cells enter the PSM, they are exposed to a high concentration of FGF and become highly motile, but do not move in an oriented manner. Owing to the confinement of motility driven by the presence of relatively immobile and stiff lateral tissues (Bénazéraf et al., 2017), the cells generate an effective mechanical pressure. Thus, a potential mechanism for the rectification that leads to the observed directional cell velocity and body elongation could be this mechanical pressure. Because the expression of FGF is highest at the posterior PSM and decreases away from it, there is a natural reduction in motility anteriorly, consistent with observations. Eventually, the effects of anterior adhesion dominate over motility and this causes cellular condensation into somites.
To quantify these processes, here we use experimental observations to measure the effective diffusivity of cells as well as their advection speed as a function of their location relative to the last formed somite. Our observations suggest a minimal microscopic cellular description of a zone of proliferating cells with high motility, which we use to develop both a quantitative cellular model and an equivalent macroscopic continuum theory, thus providing a theoretical and computational framework for body elongation. These complementary approaches yield simple expressions for the speed and scale of body elongation that are consistent with our experimental measurements, with potential implications for our understanding of outgrowth-driven morphogenesis in other settings in which FGF-driven gradients in motility and contractility drive the extension of the limb bud (Gros et al., 2010) and the gut (Nerurkar et al., 2019).
RESULTS
Experimental observations
Our experiments were carried out with chicken embryos at Hamburger-Hamilton (HH) stages 10-11 (Hamburger and Hamilton, 1992), corresponding to the period when the elongation of the embryo is most substantial (Denans et al., 2015). A time series of an elongating PSM is shown in Fig. 1B (see also Movie 1). To measure the elongation rate, we registered the movement of the embryo with respect to the last-formed somite at the beginning of the experiment (t=0) as depicted by the black dashed line in Fig. 1B, and tracked the advancement of the Hensen's node, Le(t) as a function of time. In Fig. 1C, we see that Le(t) increases linearly with time, with a mean elongation rate V=(2.8±0.3)×10−2 μm/s (averaged from five embryos).
To evaluate the role of cell motility on overall body elongation, we then examined the movement of cells by electroporating the PSM cells with fluorescent reporters that label cell nuclei specifically (Hatakeyama and Shimamura, 2008), as shown in Fig. 1D (see also Movie 1). As a first approximation, the motion of the cells can be considered two-dimensional in the anterior-posterior and medio-lateral directions because the relative dorso-ventral depth of the PSM is small. In the reference frame of the last-formed somite at t=0, during a fixed acquisition time of 4 h, posterior cell trajectories showed a larger net displacement than the anterior ones, consistent with prior experiments (Bénazéraf et al., 2010). To quantify the variations of cell motility along the body axis, we divided the PSM into three regions (anterior, middle and posterior), as shown schematically in Fig. 1E, and obtained the mean square displacement (MSD) of the cells given by < Δr2(t)>, where 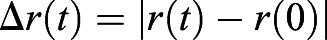 defines the distance that the cell travels in a lag time t, as shown in the inset of Fig. 1F (Qian et al., 1991; Wirtz, 2009).
defines the distance that the cell travels in a lag time t, as shown in the inset of Fig. 1F (Qian et al., 1991; Wirtz, 2009).
Decomposing the motion into a random diffusive term and an oriented drift term, we write the MSD as:
| (1) |
where D is the effective cell diffusivity and v is the local population drift velocity (Qian et al., 1991). This model for cell dynamics is in accordance with previous findings for chicken (Bénazéraf et al., 2010) and zebrafish (Lawton et al., 2013) PSM elongation. Fig. 1F shows the mean MSD curves of the three regions (anterior, middle, posterior) defined in Fig. 1E and the fit to Eqn 1 (black dashed curve). From the fits, we obtain DPost=(3.5±0.7)×10-2 μm2/s, DMid=(2.1±0.4)×10-2 μm2/s, DAnt=(1.4±0.3)×10-2 μm2/s, vPost=(2.0±0.2)×10-2 μm/s, vMid=(1.1±0.1)×10-2 μm/s and vAnt=(0.6±0.1)×10-2 μm/s (mean±s.e.m.). These estimates confirm the presence of a motility gradient of cells along the AP axis. [Note that one might argue that the ‘drift’ v could be the result of a persistent random walk rather than a drift. However, this kind of dynamics will not show a gradual change in the slope of 〈Δr2(t)〉 as is observed here.]
Our observations quantify how the cells move in space and time along the PSM. When new cells are added close to the tail bud (TB), they are highly motile but move in random directions; as they move further away from the zone of proliferation in the TB, on average they gradually slow down and stop moving.
Next, we provide microscopic and macroscopic chemo-mechanical mathematical and computational models based on the observed gradient in motility and the confinement due to the more rigid lateral tissues, and show that they explain the observed rectified cell motility and the resulting posterior body elongation.
Microscopic cellular model
We start with a simple cellular model built to mimic the experimental observations in a two-dimensional setting with cells surrounded by three confining lateral walls and a free boundary corresponding to the TB. Our simplifying assumptions, which are used in both cellular and continuum models, are that the width of the PSM is constant, which is known to be approximately true for the region in which cells are motile (Bénazéraf et al., 2017), and that the cells cannot escape from the PSM owing to the constraints imposed by the somites anteriorly, the lateral plate laterally and the neural plate medially. In the discrete model, we model individual cells as soft, elastic, disks that move randomly in a manner analogous to a Brownian particle, recognizing that the cause of random movement is not related to the temperature of the environment but instead corresponds to the random activity (motility) of the cell (Berthier and Kurchan, 2013; Mallory et al., 2014). The equation of motion for an overdamped cell with coordinate ri(t), assuming that inertial effects are negligible, is:
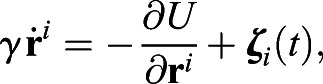 |
(2) |
where γ is the viscous friction coefficient experienced by each cell, 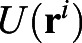 is a potential that prevents cell-cell overlap, and ζi(t) is random force. The viscous friction is a result of the interaction of cells with their environment and with each other. The repulsive interaction between cells of diameter a guarantees that two cells cannot occupy the same position; a simple form that suffices for this is given by:
is a potential that prevents cell-cell overlap, and ζi(t) is random force. The viscous friction is a result of the interaction of cells with their environment and with each other. The repulsive interaction between cells of diameter a guarantees that two cells cannot occupy the same position; a simple form that suffices for this is given by:
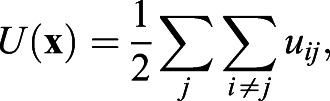 |
(3) |
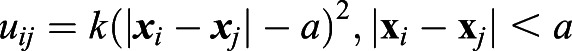 |
(4) |
and
| (5) |
Note that in our simulations the cell-cell excluded volume interactions are conservative and thus collisions are elastic but overdamped. The random force ζi(t) is assumed to have zero-mean and normally distributed with Gaussian statistics such that:
| (6) |
and
| (7) |
where  is the single-cell activity/motility and ζi,α are the x or y components of ζi.
is the single-cell activity/motility and ζi,α are the x or y components of ζi.
We also assume that the microscopic diffusivity of a (Brownian) cell is related to the activity by the relation 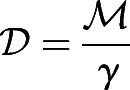 , i.e. that the fluctuation-dissipation theorem is valid and tantamount to assuming that the time scales on which we consider the system are long compared with the time scales for individual cell movements. We simulate the dynamics of the cells following Eqns 1-7 in a domain that has one free boundary at the PSM at which new cells are injected with the same initial activity; at each time step, we turn off the activity (and thus decrease the fraction of motile cells) with a probability which changes exponentially with time, to mimic the gradual decrease in the activity of the cells owing to a reduction in FGF concentration anteriorly, following:
, i.e. that the fluctuation-dissipation theorem is valid and tantamount to assuming that the time scales on which we consider the system are long compared with the time scales for individual cell movements. We simulate the dynamics of the cells following Eqns 1-7 in a domain that has one free boundary at the PSM at which new cells are injected with the same initial activity; at each time step, we turn off the activity (and thus decrease the fraction of motile cells) with a probability which changes exponentially with time, to mimic the gradual decrease in the activity of the cells owing to a reduction in FGF concentration anteriorly, following:
| (8) |
where τ is the slowest time scale associated with kinetics of degradation of FGF, which initiates once the cell enters the TB. We will assume that the initial FGF level and τ are both constant, which is approximately true in the experimental setup for the observed times of 4 h. At the posterior-end corresponding to the TB, we assume that cells move as the body elongates owing to a constant rate at which they are added in the space that is not occupied by other cells (Fig. 2A,B). The movement of the cells from the TB to the PSM region is limited by the available space in the PSM. In our model, if there is no free space, no cells are added, so the rate of adding cells is limited by the motion of the TB, the diffusion and advection of cells away from the TB and the maximal cell packing density ρ0 [the ratio of the area covered by the cells (disks) to the total PSM area]. We do not include cell division in our model, because it has been shown that interfering with cell division in the PSM does not affect the tailbud elongation considerably during the time scale of our analysis (Bénazéraf et al., 2010).
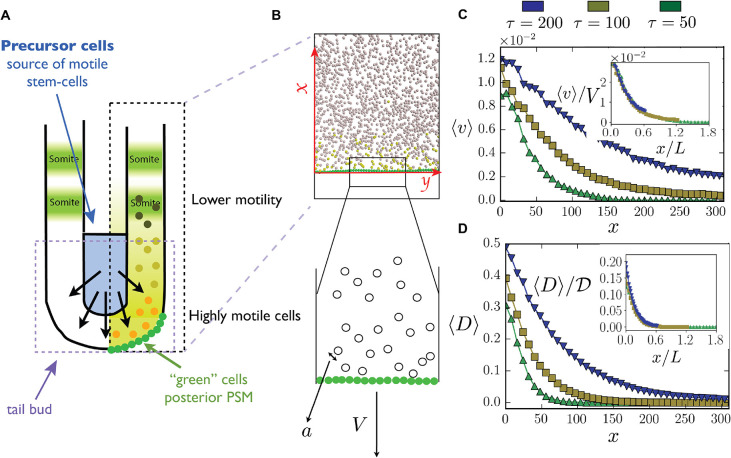
Fig. 2.
Microscopic cell-based simulation. (A,B) Schematic view of the presomitic mesoderm used as a basis for cellular simulation. Yellow and gray spheres represent motile and immotile cells, respectively. The green spheres form a connected wall, which represents the border of the TB, and move in response to pressure applied by the motile cells. The wall velocity is V and the cell diameter is a. (C,D) Velocity and motility profiles for different activity decay rate τ values are calculated as in the experiments, by fitting to Eqn 1. Insets: Scaling x by L, 〈v〉 by V and 〈D〉 by 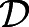 , shows that the curves collapse onto each other, demonstrating that L is the relevant length scale over which cells are motile. Here, we used
, shows that the curves collapse onto each other, demonstrating that L is the relevant length scale over which cells are motile. Here, we used 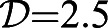 , k=100, a=1 and γ=50 in simulation units. See section 2 in the supplementary information.
, k=100, a=1 and γ=50 in simulation units. See section 2 in the supplementary information.
The TB boundary is modeled as a wall of immobile cells attached to their neighbors by elastic springs with spring constants, kchain=2k, and allowed to move posteriorly as a result of the mechanical pressure exerted by the motile cells anterior to it. Our simulations show that after an initial transient state, the motion of the wall reaches a steady state whereby cells added at a constant rate cause the wall to move at a constant velocity (Movie 2). The quasi-one-dimensional dynamics manifest in the simulations arises because of the impermeable boundary conditions in the direction perpendicular to the direction of elongation.
The fraction of motile cells is much larger near the moving wall, and as one moves anteriorly, this density falls off quickly because of the decrease in FGF activity. Changing FGF activity by changing τ, and thus varying the probability with which each individual cells stops moving (and therefore the total fraction of moving cells), changes the velocity and motility profiles (see Fig. 2C,D).
To understand these numerical results qualitatively, we note that a new cell can be added next to the (moving) boundary (x=a, y∈[0, Ymax]) only when there is a gap of order of the size of one cell a (see Fig. 2B). Thus, our model is a cellular analog of the Brownian ratchet introduced in the context of molecular polymerization (Peskin et al., 1993). In the PSM region where the internal tissue resistance to cell motion dominates any external resistance, as in our simulations, the rate of elongation is limited by the waiting time for a gap to open that allows for the addition of a cell of size a, i.e. 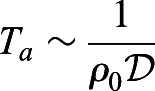 . Here,
. Here, 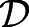 is the activity-driven diffusivity of a single cell (consistent with Eqn 1). In this limit of diffusion-limited elongation (Ta≪τ), the speed of elongation scales as a/Ta, i.e.:
is the activity-driven diffusivity of a single cell (consistent with Eqn 1). In this limit of diffusion-limited elongation (Ta≪τ), the speed of elongation scales as a/Ta, i.e.:
| (9) |
Similarly, the length scale over which the fraction of motile cells falls off exponentially is:
| (10) |
We note that the other limit, i.e. τ≪Ta corresponding to adding cells that are not active, will lead to a jammed state and is not relevant here. In Fig. 2C,D, we show the decay in the speed of the cells and the effective diffusivity as a function of the distance from the TB for different values of τ. In the inset, we see that the rescaled elongation velocity and cell diffusivity using the relations shown in Eqns 9 and 10 as a function of location from the wall is consistent with our simple scaling arguments. However, as can be noticed from the ordinate in the inset of Fig. 2C, the wall velocity in our simulations,  , is a consequence of the collective effect of elongation driven by the addition of multiple cells. To test that this is indeed the case, we varied both cell diffusivity
, is a consequence of the collective effect of elongation driven by the addition of multiple cells. To test that this is indeed the case, we varied both cell diffusivity  and maximal cell packing ρ0 and confirmed that the overall elongation rate
and maximal cell packing ρ0 and confirmed that the overall elongation rate  (see Fig. S3) is consistent with experimental results (Fig. S4).
(see Fig. S3) is consistent with experimental results (Fig. S4).
Macroscopic continuum theory
Although the multicellular model for the dynamics of PSM elongation provides a qualitative and quantitative model by accounting for cell addition at the TB and gradual decay in motility driven by FGF degradation, an effective coarse-grained description of the process would be even more useful. There are a few reasons for this: they crystallize the processes at play in terms of the laws of conservation of mass and momentum of an active fluid, and characterize the parameter dependences in terms of dimensionless parameters and scaling laws, both of which have the feature of being generalizable beyond the specific case of body elongation. With this in mind, we now provide an effective macroscopic one dimensional continuum theory that links the density of motile cells ρ(x, t) and the velocity field v(x, t) of motile cells as a function of location x in a fixed lab frame. A hydrodynamic description of the diffusion, advection and degradation of motile cells can then be written in terms of the equations for mass and momentum balance (in the viscously dominated limit) as:
 |
(11) |
| (12) |
The first equation describes the variations in the density of actively motile cells that also gradually decay at a rate τ, whereas the second equation characterizes how the stresses associated with the active pressure generated by the random motility of the cells causes the tissue to respond and push the TB. Here, η is the viscosity of the PSM and ξ is the viscous friction associated with motion of the elongating PSM relative to the surrounding tissues (endoderm, ectoderm, neural tube and lateral plate). We note that these friction coefficients define a characteristic viscous screening length lξ∼(η/ξ)1/2 that we assume is of the order of a few cell sizes, i.e. lξ∼a.
The equations need a closure relation for the pressure; a minimal relation assumes that the active pressure is proportional to the density of motile cells p∼αρ, consistent with both our microscopic model, with  , and also consistent with earlier simulations of active Brownian particles (Mallory et al., 2014). Although other relations of the form p∼αρq are plausible, we choose to stay with a minimal model here.
, and also consistent with earlier simulations of active Brownian particles (Mallory et al., 2014). Although other relations of the form p∼αρq are plausible, we choose to stay with a minimal model here.
To complete the formulation of the problem, we need to specify some boundary conditions for the free-boundary problem associated with the moving TB assuming the domain of interest to be x∈[s(t), ∞), with the boundary of the TB being s(t). Motile cells enter the domain at a rate proportional to the difference between their local density and the maximum cell density ρ0, so that the cell flux is  , where
, where  is the injection rate of cells from the progenitor zone into the TB. This flux must be balanced by diffusion and advection of cells from the boundary, i.e.
is the injection rate of cells from the progenitor zone into the TB. This flux must be balanced by diffusion and advection of cells from the boundary, i.e.
| (13) |
which is reminiscent of a generalized Stefan-like condition in moving boundary problems in solidification. It is also necessary to satisfy force balance at the moving boundary so that:
| (14) |
where F is the resisting pressure exerted by the tissue ahead of the TB, and  . In the cellular simulation, this force is a result of the dynamic friction between the wall cells and the substrate and thus depends on the velocity; here, we have assumed a more general form. Far from the TB, we assume that due to degradation of FGF, the density and velocity of motile cells vanishes so that:
. In the cellular simulation, this force is a result of the dynamic friction between the wall cells and the substrate and thus depends on the velocity; here, we have assumed a more general form. Far from the TB, we assume that due to degradation of FGF, the density and velocity of motile cells vanishes so that:
| (15) |
Together, Eqns 11 and 12, along with the above boundary conditions (Eqns 13-15), determine the spatiotemporal evolution of the density and velocity fields in the elongating embryo as well as the speed of elongation of the embryo itself.
To understand the dependence of the solution of Eqns 11 and 12 on the problem parameters, we can rewrite the equations in a form that depends on five dimensionless variables: 
Here Π1 is the ratio of the cell size a and the elongation length scale  , Π2 is the ratio of the maximum active stress αρ0 and the external viscous friction ξVL∼ξV2τ, and Π3 is the ratio of the internal viscous stress ηV/L and the active stress αρ0. We note that the parameter
, Π2 is the ratio of the maximum active stress αρ0 and the external viscous friction ξVL∼ξV2τ, and Π3 is the ratio of the internal viscous stress ηV/L and the active stress αρ0. We note that the parameter  , where lξ is the viscous screening length defined ealier (after Eqn 12). Finally, there are two dimensionless parameters associated with the boundary conditions Π4, which is the ratio of the external stress at the PSM boundary F and the active stress αρ0, and Π5, which is the scaled ratio of the internal cell diffusivity
, where lξ is the viscous screening length defined ealier (after Eqn 12). Finally, there are two dimensionless parameters associated with the boundary conditions Π4, which is the ratio of the external stress at the PSM boundary F and the active stress αρ0, and Π5, which is the scaled ratio of the internal cell diffusivity  and the rate of addition of cells at the PSM boundary
and the rate of addition of cells at the PSM boundary  .
.
By rescaling all the velocities by the interface velocity V, all the lengths by elongation length scale L=Vτ, and the density by the maximal close packing density ρ0, the dimensionless form of Eqns 11-15 can be written in terms of the dimensionless variables  ,
,  and
and  as:
as:
| (16) |
and
| (17) |
along with the boundary conditions:
 |
(18) |
| (19) |
and
| (20) |
For parameter values, we assume that  , the typical degradation time scale τ≈1−2×104 s, the viscosity of the PSM η∼104−105 Pa · s and the friction coefficient ξ≈1012−1013 Pa · s/m2 measured using the micropipette aspiration technique (Guevorkian et al., 2010) (see section 1 in the supplementary information). This yields Π1≈0.005−0.08 and Π2Π3=0.03−10.
, the typical degradation time scale τ≈1−2×104 s, the viscosity of the PSM η∼104−105 Pa · s and the friction coefficient ξ≈1012−1013 Pa · s/m2 measured using the micropipette aspiration technique (Guevorkian et al., 2010) (see section 1 in the supplementary information). This yields Π1≈0.005−0.08 and Π2Π3=0.03−10.
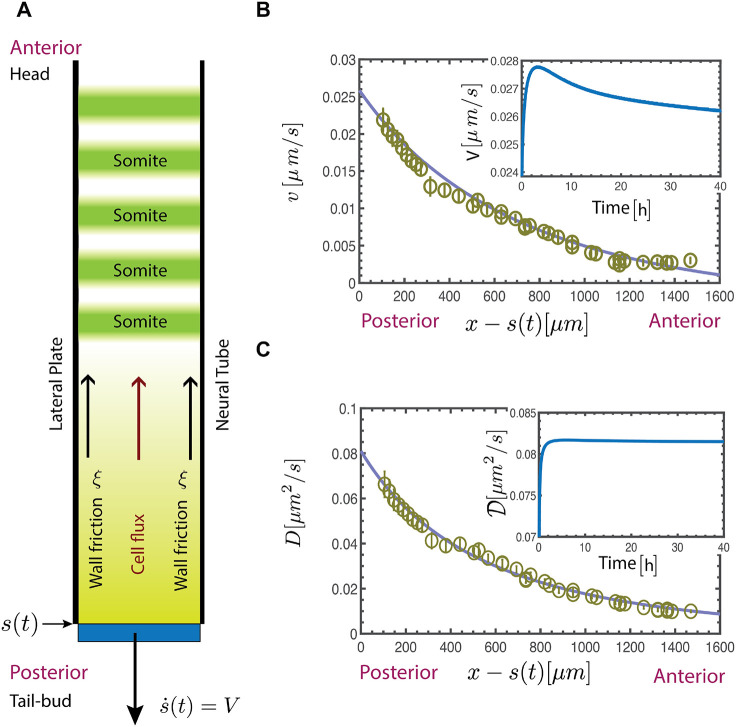
Using the following parameter values Π1=0.01, Π2=2, Π3=1, Π4≈0.005, Π5=0.003, we solve the initial boundary value problem (Eqns 17-21) using a finite difference method. In Fig. 3B,C, we see that our calculated profiles for the velocity v(x) and diffusivity  compare well with the experimentally observed profiles, and are suggestive of a simple exponential law (see also Fig. S5). In Fig. 3B, we show that the system evolves into a steady state following a short transient (insets), with an active zone near the TB that moves at constant speed. Both the cellular motility (Fig. 3B) and diffusivity (Fig. 3C) decay towards the anterior where the FGF degrades. Comparing our numerical results for the moving average of cell velocity 〈v〉 and the rescaled diffusivity
compare well with the experimentally observed profiles, and are suggestive of a simple exponential law (see also Fig. S5). In Fig. 3B, we show that the system evolves into a steady state following a short transient (insets), with an active zone near the TB that moves at constant speed. Both the cellular motility (Fig. 3B) and diffusivity (Fig. 3C) decay towards the anterior where the FGF degrades. Comparing our numerical results for the moving average of cell velocity 〈v〉 and the rescaled diffusivity  with the experimentally determined values as a function of position relative to Hensen's node (see Materials and Methods) we see that the profiles compare well. Furthermore, on converting the results to dimensional values, we can estimate the active stress αρ0≈5−50 Pa, qualitatively similar to measurements on amniote embryos (Zhou et al., 2009; Serwane et al., 2017). From the simulations, we can also estimate the typical length associated with motility decay L∼1200 μm and the length scale lξ∼1800 μm associated with friction, which are of the same order as the experimental estimates, L=Vτ∼260−520 μm and
with the experimentally determined values as a function of position relative to Hensen's node (see Materials and Methods) we see that the profiles compare well. Furthermore, on converting the results to dimensional values, we can estimate the active stress αρ0≈5−50 Pa, qualitatively similar to measurements on amniote embryos (Zhou et al., 2009; Serwane et al., 2017). From the simulations, we can also estimate the typical length associated with motility decay L∼1200 μm and the length scale lξ∼1800 μm associated with friction, which are of the same order as the experimental estimates, L=Vτ∼260−520 μm and  , 3-15 times the cell size.
, 3-15 times the cell size.
Fig. 3.
Macroscopic continuum model. (A) Schematic showing half of the PSM in the neighborhood of the TB, the position of which is s(t). (B,C) Experimentally measured velocity v(x) and diffusivity D(x) profiles (calculated using Eqn 1) as a function of distance from the TB compares well with the results of our continuum theory obtained by solving Eqn 11 and 12 for v(x) and  (continuous lines). The insets show the velocity (B) and the diffusivity (C) at the free boundary of the TB as a function of time; following a short initial transient, we see that the speed and diffusivity reach a steady state.
(continuous lines). The insets show the velocity (B) and the diffusivity (C) at the free boundary of the TB as a function of time; following a short initial transient, we see that the speed and diffusivity reach a steady state.
A sensitivity analysis of our continuum model (see section 3 in the supplementary information) shows that the parameters Π1, the ratio of the cell size to the elongation length scale that arises from activity, and Π3, the ratio of viscous stresses to the active stress, affect the velocity and motility profile significantly. The other parameters, Π2, Π4 and Π5, have a negligible effect on our results over a range relevant for our problem (for the range of parameter values used, see Table S3; for the associated results, see Figs S6-S8). The dependence of the length scales corresponding to the typical motility and velocity profiles as a function of the dimensionless parameters Π1 and Π3 is shown in Fig. S10.
DISCUSSION
In vertebrate embryos, posterior structures are formed sequentially by a combination of cell proliferation and cell motility that together leads to body elongation. Although it has long been observed that the elongation process involves the posterior displacement of the TB, the physical mechanisms underlying this have not been clearly elucidated. Here, we have quantified this process experimentally and theoretically and shown that it occurs as a result of two effects: the addition of motile cells at a boundary (the TB) and confinement both laterally and anteriorly. These two effects lead to the rectification of random cell diffusivity, which generates the forces underlying elongation and the emergence of characteristic velocity and activity/diffusivity length scales. Simple cellular models and a one-dimensional continuum framework capture the essence of the behavior in terms of a pair of fundamental dimensionless parameters that characterize the ratio of the active driving stress to the internal and external viscous stresses, and are consistent with a narrow boundary layer of activity near the TB, in agreement with observations. Natural extensions of the models include adding the effects of dimensionality, such as the lateral growth in the absence of FGF decay (Bénazéraf et al., 2010), two-dimensional compressive stresses that generate vortical flows near the TB (Xiong et al., 2020), and the gradients in viscosity from the anterior to the posterior region (Mongera et al., 2018). Such multidimensional models could also be modified to describe other embryonic outgrowths processes that exhibit graded diffusive behavior of cells downstream of FGF signaling, such as the budding of vertebrate limbs (Gros et al., 2010) and the onset of gut elongation (Nerurkar et al., 2019).
MATERIALS AND METHODS
Chicken embryo preparation and electroporation
Fertilized chicken eggs were obtained from a commercial provider (Les Couvoirs de l'Est, Willgottheim, France) and incubated at 37°C in a humidified incubator. After 24 h, HH stage 4-5 embryos were mounted on filter paper and transferred ventral side up to 35 mm agar/albumen petri dishes for injection (Chapman et al., 2001). Electroporation of the PSM was performed using H2B-mCherry or H2B-Venus nuclear markers as described previously (Bénazéraf et al., 2010). Electroporated embryos were returned to incubator and left to grow to HH stage 10-11 before imaging.
Time-lapse imaging and track analysis
The imaging procedure used here was similar to a previously described procedure (Czirók et al., 2002; Rupp et al., 2003). Briefly, the embryos were transferred to custom-made six-well observation chambers containing agar/albumin gel, and positioned ventral side up. The time-lapse imaging was performed at 37°C using a motorized upright microscope (Leica DMR, Leica Microsystems) with a 10× objective (N.A. 0.3) and a CCD digital camera (QImaging Retiga 1300i) at a rate of 10 frames/h. At each time point, brightfield and fluorescent images of the embryo were taken at three various fields to cover the total length of the PSM. Post-acquisition processing was performed on images as described previously (Czirók et al., 2002; Rupp et al., 2003) to obtain a 2D time series. Cell tracking and trajectory analysis was performed on fluorescent images using custom-made MATLAB (MathWorks) routines. Trajectories were analyzed on 4-h long movies. To account for cell position modification due to tissue drift, we divided our movies to 1 h segments. The location of each cell was defined with respect to the node at the beginning of each 1 h trajectory. For each cell trajectory, the MSD was calculated and adjusted with Eqn 1 to obtain D and v. Further, the moving average filter, Smooth (MATLAB), was used for the plots of D and v as a function of position to obtain Fig. 3.
Discrete cell simulations
The discrete cell simulations were performed by discretizing Eqn 2 using the Euler–Maruyama method and integrating in time. In the initial state, all the particles are arranged in a square lattice (particles are slightly perturbed from the lattice) in a region that has dimensions of 80a×160a. At a given time, the particles can be at x∈[0,Xmax], y∈[0,Ymax], where Xmax is the position of the rigid boundary and Ymax=160a is the lateral width. We use reduced, dimensionless units in which all energies are given in terms of a typical energy w: U*=U/w, all lengths in terms of the typical cell size a, r*=r/a and masses in terms of a mass unit M, m*=m/M. Using these fundamental units, we can also reduce the effective temperature, i.e. the active random motility  , time
, time  , fraction ρ*=Na3/V (V is the volume) and any other physical quantity of interest, where
, fraction ρ*=Na3/V (V is the volume) and any other physical quantity of interest, where  and ρ* are all dimensionless.
and ρ* are all dimensionless.
Supplementary Material
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Conceptualization: O.P., L.M.; Methodology: I.R., K.G., L.M.; Software: I.R., A.G.; Validation: A.G., L.M.; Formal analysis: I.R., K.G., A.G., L.M.; Investigation: I.R., K.G., L.M.; Resources: O.P., L.M.; Data curation: K.G.; Writing - original draft: I.R., K.G., L.M.; Writing - review & editing: A.G., O.P., L.M.; Visualization: I.R., A.G.; Supervision: O.P., L.M.; Project administration: L.M.; Funding acquisition: O.P., L.M.
Funding
This work was partially supported by the French Agence Nationale de la Recherche (ANR-14-CE32-0009-01 to K.G.), the Human Frontier Science Program (RGP0051/2012 to O.P.), and grants and fellowships from the Schlumberger Foundation and the MacArthur Foundation, and the National Institutes of Health (1R01HD097068 to O.P. and L.M.). Deposited in PMC for release after 12 months.
References
- Bénazéraf, B., Beaupeux, M., Tchernookov, M., Wallingford, A., Salisbury, T., Shirtz, A., Shirtz, A., Huss, D., Pourquié, O., François, P.et al. (2017). Multi-scale quantification of tissue behavior during amniote embryo axis elongation. Development 144, 4462-4472. 10.1242/dev.150557 [DOI] [PubMed] [Google Scholar]
- Bénazéraf, B., Francois, F., Baker, R. E., Denans, N., Little, C. and Pourquié, O. (2010). A random cell motility gradient downstream of fgf controls elongation of an amniote embryo. Nature 466, 248-252. 10.1038/nature09151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berthier, L. and Kurchan, J. (2013). Non-equilibrium glass transitions in driven and active matter. Nat. Phys. 9, 310-314. 10.1038/nphys2592 [DOI] [Google Scholar]
- Chapman, S., Collignon, J., Schoenwolf, G. and Lumsden, A. (2001). Improved method for chick whole-embryo culture using a filter paper carrier. Dev. Dyn. 220, 284-289. [DOI] [PubMed] [Google Scholar]
- Czirók, A., Rupp, P., Rongish, B. and Little, C. (2002). Multi-field 3d scanning light microscopy of early embryogenesis. J. Microsc. 206, 209-217. 10.1046/j.1365-2818.2002.01032.x [DOI] [PubMed] [Google Scholar]
- Delfini, M.-C., Dubrulle, J., Malapert, P., Chal, J. and Pourquié, O. (2005). Control of the segmentation process by graded mapk/erk activation in the chick embryo. Proc. Natl. Acad. Sci. U.S.A. 102, 11343-11348. 10.1073/pnas.0502933102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denans, N., Iimura, T. and Pourquié, O. (2015). Hox genes control vertebrate body elongation by collinear wnt repression. Elife 4, e04379. 10.7554/eLife.04379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gros, J., Hu, J. K.-H., Vinegoni, C., Feruglio, P. F., Weissleder, R. and Tabin, C. J. (2010). Wnt5a/jnk and fgf/mapk pathways regulate the cellular events shaping the vertebrate limb bud. Curr. Biol. 20, 1993-2002. 10.1016/j.cub.2010.09.063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guevorkian, K., Colbert, M.-J., Durth, M., Dufour, S. and Brochard-Wyart, F. (2010). Aspiration of biological viscoelastic drops. Phys. Rev. Lett. 104, 218101. 10.1103/PhysRevLett.104.218101 [DOI] [PubMed] [Google Scholar]
- Hamburger, V. and Hamilton, H. (1992). A series of normal stages in the development of the chick embryo. Dev. Dyn. 195, 231-272. 10.1002/aja.1001950404 [DOI] [PubMed] [Google Scholar]
- Hatakeyama, J. and Shimamura, K. (2008). Method for electroporation for the early chick embryo. Dev. Growth Differ. 50, 449-452. 10.1111/j.1440-169X.2008.01040.x [DOI] [PubMed] [Google Scholar]
- Lawton, A. K., Nandi, A., Stulberg, M. J., Dray, N., Sneddon, M. W., Pontius, W., Emonet, T. and Holley, S. A. (2013). Regulated tissue fluidity steers zebrafish body elongation. Development 140, 573-582. 10.1242/dev.090381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallory, S., Šarić, A., Valeriani, C. and Cacciuto, A. (2014). Anomalous thermomechanical properties of a self-propelled colloidal fluid. Physical Review E 89, 052303. 10.1103/PhysRevE.89.052303 [DOI] [PubMed] [Google Scholar]
- Mongera, A., Rowghanian, P., Gustafson, H. J., Shelton, E., Kealhofer, D. A., Carn, E. K., Serwane, F., Lucio, A. A., Giammona, J. and Campàs, O. (2018). A fluid-to-solid jamming transition underlies vertebrate body axis elongation. Nature 561, 401-405. 10.1038/s41586-018-0479-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nerurkar, N. L., Lee, C., Mahadevan, L. and Tabin, C. J. (2019). Molecular control of macroscopic forces drives formation of the vertebrate hindgut. Nature 565, 480-484. 10.1038/s41586-018-0865-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peskin, C. S., Odell, G. M. and Oster, G. F. (1993). Cellular motions and thermal fluctuations: the brownian ratchet. Biophys. J. 65, 316. 10.1016/S0006-3495(93)81035-X [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian, H., Sheetz, M. P. and Elson, E. L. (1991). Single particle tracking. analysis of diffusion and flow in two-dimensional systems. Biophys. J. 60, 910-921. 10.1016/S0006-3495(91)82125-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rupp, P., Rongish, B., Czirók, A. and Little, C. (2003). Culturing of avian embryos for time-lapse imaging. BioTechniques 34, 274-278. 10.2144/03342st01 [DOI] [PubMed] [Google Scholar]
- Serwane, F., Mongera, A., Rowghanian, P., Kealhofer, D. A., Lucio, A. A., Hockenbery, Z. M. and Campàs, O. (2017). In vivo quantification of spatially-varying mechanical properties in developing tissues. Nat. Methods 14, 181. 10.1038/nmeth.4101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirtz, D. (2009). Particle-tracking microrheology of living cells: principles and applications. Annu. Rev. Biophys. 38, 301-326. 10.1146/annurev.biophys.050708.133724 [DOI] [PubMed] [Google Scholar]
- Xiong, F., Ma, W., Bénazéraf, B., Mahadevan, L. and Pourquié, O. (2020). Mechanical coupling coordinates the co-elongation of axial and paraxial tissues in avian embryos. Dev. Cell 55, 354-366. 10.1016/j.devcel.2020.08.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, J., Kim, H. Y. and Davidson, L. A. (2009). Actomyosin stiffens the vertebrate embryo during crucial stages of elongation and neural tube closure. Development 136, 677-688. 10.1242/dev.026211 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.