Abstract
Proteins are the essential functional biomolecules profoundly implicated in all aspects of pancreatic tumorigenesis and its progression. While common genomic factors, such as KRAS, TP53, SMAD4, and CDKN2A have been well recognized in association of pancreatic ductal adenocarcinoma (PDAC), our understanding of functional changes at the proteome level merits further investigation. Malignance associated proteome alterations can be attributed to the convoluted outcomes from genetic, epigenetic and environmental factors in initiating and progressing PDAC, and may reflect on changes in protein expressional level, structure, localization, as well as post-translational modifications (PTMs) status. The study of localized or systemic proteome alterations in PDAC, as well as its precursor lesions, such as pancreatic intraepithelial neoplasia (PanIN) and mucinous pancreatic cystic neoplasm, would provide unique perspectives in elucidating functional molecular events underlying PDAC. While efforts have been made, challenges still exist to comprehensively integrate much of the proteomic discovery to the perspectives gained from genomic studies in the context of biomarker discovery. Novel approaches and data from well-defined longitudinal clinical studies and experimental models are needed to facilitate the study of PDAC and precursor lesions for early detection and intervention.
Keywords: Pancreatic cancer, Proteomics, Mass spectrometry, Post-translational modification, Glycosylation
Introduction
In the United States, pancreatic cancer is the third leading cause of cancer death with a 5-year survival rate of 8%, and is predicted to be the second leading cause of cancer death by the year 2030 [1, 2]. Pancreatic ductal adenocarcinoma (PDAC) represents the vast majority of pancreatic cancers. This lethal disease is characterized by its poor prognosis and rapid development of drug resistance; and it is difficult to detect at early stages when treatments are most effective. Genomic studies have revealed common gene mutations in PDAC, including oncogenic activation of KRAS and the frequent inactivation of TP53, SMAD4, and CDKN2A tumor suppressors [3-6]. While KRAS mutations arise early in the natural history of PDAC, mutations in TP53 and SMAD4 are later events in cancer progression with higher frequency in invasive disease [7]. These genomic mutations can profoundly affect protein characteristics, function and interactions at various levels. However, changes in the genome and the transcriptome do not always correlate with proteome alterations at functional level [8]. Cancer cells and the surrounding microenvironment are directly influenced by their functional proteomes and associated biochemical processes. Malignancy-associated proteome alterations can extend beyond protein expressional changes and polymorphisms, and may include numerous post-translational and/or isoform changes that also impact specific signaling pathways and interaction of protein complexes in disease settings [9-11]. Such proteome alterations in cancer are dynamic and closely interplay with a reprogrammed metabolism that is required for tumor growth.
Currently, the knowledge of extended protein networks associated with PDAC tumorigenesis has lagged behind, in part, due to the enormous complexity, dynamic changes and high heterogeneity associated with the expanded proteome. Several PDAC associated protein families are found to be involved in the development and progression of PDAC. Mucins are a heavily glycosylated protein family that are frequently associated with epithelial mucosa. The abnormal expression, glycosylation, localization of various mucin molecules are correlated with PDAC and its progression [12-16]. Carcinoembryonic antigen-related cell adhesion molecules (CEACAMs) are another family of cell surface proteins that are implicated in facilitating tumor progression. CEACAMs 1, 5, and 6 have been linked with PDAC progression [17, 18]. While these two PDAC associated protein families[12-16] have been extensively studied, the investigation of the broader interactive protein changes and their roles in neoplastic progression, metastasis and chemoresistance of PDAC remain uncertain. There has been a knowledge gap in linking genetic, epigenetic and environmental factors to the broader proteome alterations for better understanding the complementary molecular events that are underlying pancreatic tumorigenesis. The study of localized or systemic proteome alterations in PDAC and its precursor lesions, such as pancreatic intraepithelial neoplasia (PanIN) and intraductal papillary mucinous neoplasm (IPMN) / mucinous cystic neoplasm (MCN), would provide unique perspectives at a functional level to facilitate the earlier detection and intervention of this lethal disease.
MS based proteomics
Proteins are the essential functional biomolecules that participate in a vast array of physiological cellular activities, constituting ~50% of the dry mass of a cell [19] and reaching up to 60-80 mg/ml concentration in human plasma. Post-translational modifications (PTMs) of proteins add an additional layer of complexity to the protein, significantly affecting the folding, localization, activation/deactivation or stability of the proteins. Proteomics can deliver dynamic information such as protein turnover, protein interactions and status of PTMs, and thereby provide a real time picture of cellular function under biological conditions [9]. It represents an emerging paradigm in biomedical research in the post genomic era, emphasizing functional changes, which are clinically highly relevant [9, 20, 21]. The advances in mass spectrometry and bioinformatics have enabled interrogation of a vast number of proteins and their PTMs in a complex biological sample at a global scale or using a targeted, highly specific fashion. The technologic advances in proteomic analyses permit reproducible, accurate, information-rich data sets that can be analyzed on their own or integrated with other “omics” data [9, 20].
In PDAC, proteomic efforts have investigated a variety of relevant clinical specimens, including pancreatic tissues, serum/plasma, pancreatic juice, cyst fluid, urine and bile fluid, with research goals spanning from mechanistic studies to elucidate complex pathways implicated in pancreatic tumorigenesis, to the discovery of protein biomarkers for early detection or as therapeutic targets [22-29]. These studies have pioneered the technology, generated important data sets and provided useful guidance for future PDAC study. However, challenges still remain in gaining the depth and breadth of a proteomics analysis to interrogate low abundant proteins with robust quantification, and to dissect the enormous and complex interactions and pathways embedded in the informatics rich data sets. In a proteomic analysis, while sample preparation strategies may vary due to individual study design and specimen type, the overall flow of shotgun proteomic analysis of clinical samples remains similar, as illustrated in Figure 1. Studies can be designed to elicit information with a variety of perspectives, such as protein expressional profiles, polymorphisms, status of PTMs, protein changes in subcellular compartments, and interactions between proteins and/or other biomolecules. Metaproteomics, which studies the proteomes of environmental sources, further extends the applications to explore the composition and functional changes in the gut microbiome, as relevant to pancreatic diseases. With improved sensitivity and resolution, investigation of proteome changes within a single cell is also becoming feasible. The information embedded in proteomic data is complex and enormous. The knowledge needed to comprehensively interpret an information-rich proteomic data set may extend beyond the sequence databases and knowledge bases that are currently available, as many disease-associated protein changes are post translational and/or have not been well defined, highlighting the technical challenges in analyzing proteomic data.
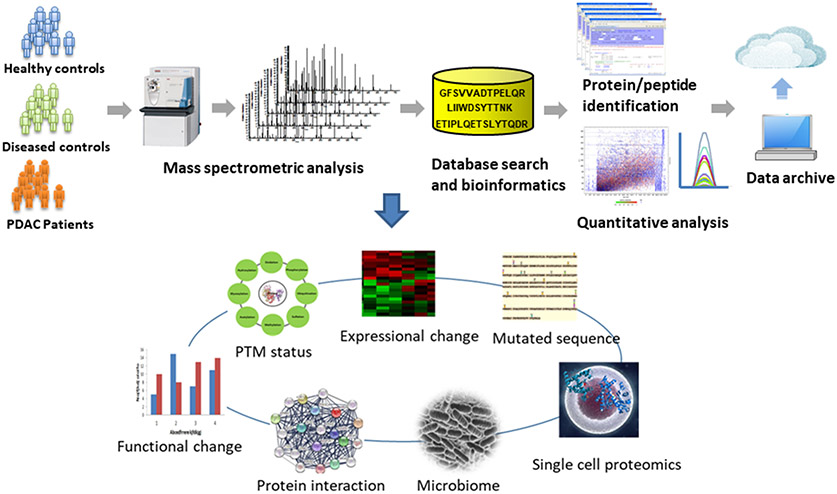
Figure 1.
A schematic illustration of proteomic work flow for comparative study of PDAC vs controls. Regardless study designs and specimen types, the platform typically consists of three modules, including sample preparation, LC MS/MS analysis and bioinformatics for data processing.
Alterations in PDAC proteome
The profound changes in protein expression in PDAC are driven by a variety of complex, multifaceted molecular events implicated in pancreatic tumorigenesis and disease progression. Many of the differential proteins identified in PDAC tissue by proteomics are involved in, not only cancer cells, but also protein-driven interactions between the cancer cells and the tumor stroma to orchestrate PDAC tumor growth, migration, angiogenesis, invasion, metastasis, and immunologic escape (Figure 2). PDAC is associated with enormous stroma, with fibroblasts, vascular endothelial cells, acinar cells and immune cells along with extracellular matrix (ECM). This underscores the importance of the tumor microenvironment in promoting pancreatic cancer progression and drug resistance. In fact, pancreatic cancer stroma may account for 50%-90% of the total volume of a PDAC tumor. The cross-talk between cancer cells and the surrounding microenvironment may induce production and secretion of stimulatory growth factors and cytokines by cancer and stromal cells to recruit vasculature or suppress immune surveillance and promote tumor development [30, 31]. Such malignancy-induced phenomena are mirrored in the proteome alterations in cancer cells and the surrounding stromal cells in pancreatic cancer tissue, including fibroblasts, vascular endothelial cells, immune cells, and acinar cells. In-depth proteomic analysis of fibroblasts, vascular endothelial cells, and acinar cells isolated from tumor tissue has yet to be reported. Nevertheless, analysis of whole tumor tissues could reveal proteome alterations of these cellular and non-cellular components of the tumor microenvironment. For acinar cells, which are unique to pancreas, a number of exocrine pancreas digestive enzymes, including trypsin subtypes, lipase, carboxypeptidase, elastase, phospholipase, and amylase, are down-regulated in pancreatic cancer tissue, reflecting the possible replacement or destruction of acinar cell mass due to cancer [25, 32-34]. Activation of stromal fibroblasts into cancer-associated fibroblasts - a myofibroblast phenotype that promotes invasion of cancer cells, is evidenced by their functional changes driven by an altered proteome [35, 36]. Differential expression of proteins from vascular endothelial cells and immune cells are also observed, including thymocyte differentiation antigen (THY1 or CD90) - a biomarker for molecular imaging of PDAC [37], and galectins which are a family of proteins profoundly implicated in immune response and pancreatic cancer survival [33, 38-45]. The expression of Galectin-1 was inversely correlated with the survival of pancreatic cancer, suggesting that it could be a prognostic marker of PDAC [40, 45].
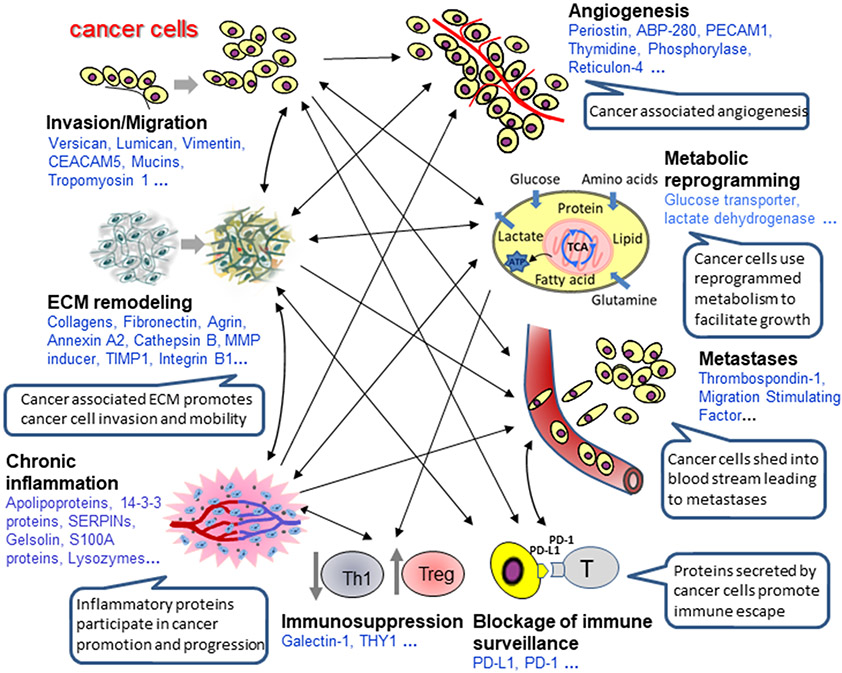
Figure 2.
Differential proteins in PDAC. Differential proteins identified in PDAC tissues are broadly involved in many aspects of tumorigenesis to facilitate cancer progression, including tumor growth, migration, angiogenesis, invasion, metastasis and immunologic escape, through orchestration of protein crosstalk between cancer cells and tumor microenvironment.
Proteins involved in wounding response and inflammatory pathways, such as apolipoproteins, SERPIN proteins, 14-3-3 proteins, S100A proteins, serum amyloid P-component, complements, gelsolin, lysozymes, and alpha-1-acid glycoprotein 1, to name a few, are largely over-expressed in PDAC tissue [25, 32-34]. As an integrated feature of PDAC, proteome alterations in extracellular matrix (ECM) are also involved in facilitating cancer growth by remodeling the ECM to promote invasion and angiogenesis. Many proteins involved in the ECM structure and organization are upregulated in pancreatic cancer tissue, including annexins, collagens, decorin, dermatopontin, EMILINs, fibrillins, fibrinogens, fibronectin, lumican, vitronectin, laminin, myosins, periostin, transgelins, transforming growth factor-beta-induced protein, versican, argins, integrins, cathepsins, and utrophin [25, 32-34], consistent with the persistent activation of pancreatic stellate cells that mediate stromal fibrosis through secretion of ECM proteins [46]. These proteome alterations in PDAC tissue are not only concurrent with the functional changes in PDAC, but strongly correlate with histological observations in pancreatic adenocarcinoma. The knowledge gained could be further assessed to inform the biomarker discovery and therapeutic target development. In addition to THY1 and galectins, several other proteins have been studied as potential PDAC biomarkers. Plectin-1 has been investigated as a molecular imaging marker for detection of primary and metastatic pancreatic cancer [47, 48]. Prolargin and osteoglycin were shown to be associated pancreatic cancer survival, and potential prognostic markers [40]. Gelsolin and TIMP1 were tested in plasma as a composite biomarker in separating the early stage PDAC patients from healthy controls and patients with chronic pancreatitis [49].
The functional alterations in PDAC tumor microenvironment create intense physical, oxidative and nutrient-poor stress for the cancer cells. In response, the PDAC tumors utilize various metabolic reprogramming mechanisms for survival, adaptation, and proliferation. This includes upregulation of proteins that increase glycolysis and biosynthesis, such as glucose transporter GLUT1 and other glycolytic enzymes [50]. One of the biosynthesis pathways, pentose phosphate pathway (PPP) can be upregulated in PDAC tumors to provide sustain increased need of building blocks for ribose synthesis [10, 50]. The uptake of glucose and glutamine by cancer cells may fuel an increased glycan biosynthesis through hexosamine biosynthetic pathway (HBP), leading to the overall elevated level of glycosylation on many proteins in PDAC tissue [51]. PDAC cells are frequently addicted to glutamate for maintaining re-dox balance. Interfering the glutamine pathway of cancer cells might result in reducing cell proliferation and sensitizing chemo-resistant PDAC cells [52]. In addition, PDAC cells could utilize autophagy and micropinocytosis to overcome nutrient deprivation [53, 54]. All of these metabolic and functional changes in cancer cells and tumor environment dynamically interplay with and correspond to the proteome alterations in PDAC as the disease progresses.
PDAC-associated proteome alterations can extend beyond pancreatic tumor tissue. Analysis of relevant bodily fluids, such as blood and pancreatic juice or cyst fluids, reveals proteomic alterations that may represent a systemic or localized changes dependent, in part, on their proximity to the cancer site. The proteomic composition of these bodily fluids are dramatically different in association with their physiologic function. For example, plasma/serum represents the proteome of the circulating system, which includes many functional blood proteins and proteins shed from tissues, while pancreatic juice contains many secreted enzymes, and cyst fluid may include mucins and other tissue-derived proteins. The unique proteome differences originating from the physiological nature of these bodily fluids may determine a unique perspective in developing concentration-based protein biomarkers for bodily fluid detection, i.e. a novel and specific biomarker for a certain bodily fluid. More detailed discussion on the proteomic analysis of bodily fluid could be found in the literature [26].
Aberrant glycosylation in PDAC
Protein glycosylation can broaden the complexity and functionality of proteins and glycosylation changes have been implicated in pancreatic tumorigenesis. The glycan component of a mucin can make up more than 50% of its molecular weight and plays an important role in modulating the functionality of the protein in tumorigenesis, as well as cell-cell interaction within the tumor microenvironment. Among the most frequently occurring PTMs, aberrant glycosylation has been recognized as a hallmark associated with PDAC. In fact, CA 19-9, the current clinical biomarker for PDAC monitoring, is a glycosylation test that detects abnormal changes associated with sialylated Lewis antigen of mucins and other protein carriers (MUC1, MUC5AC and MUC16 are major carriers of CA 19-9) [12, 55-57]. Aberrant glycosylation changes can involve the structural modifications of glycan moieties, as well as the occupancy changes on protein glycosylation sites [58]. Altered glycoforms of MUC1, MUC4 and MUC5AC are observed early in pancreatic cancer progression (PanINs) to late stage metastatic disease [15], including the elevation of fucosylated core structures, fucose and Lewis antigen in the blood of PDAC patients [56].
Glycomic studies have also revealed a number of glycan structure changes associated with PDAC, including increased protein fucosylation and sialylation detected in serum [59], and several hyper-fucosylated glycoproteins, including triacylglycerol lipase and pancreatic α-amylase, observed in cyst fluids from IPMN and MCN [60]. Recent reports also observed the occupancy changes on N-glycosylation of many PDAC associated proteins involved in TGF-β, TNF and NF-kappa-B pathways, including MUC5AC, CEACAM5, insulin-like growth factor binding protein (IGFBP3), and galectin-3-binding protein (LGALS3BP) [51, 61]. In addition, abnormal protein glycosylation can significantly affect the biochemical and mechanical properties of extracellular matrix (ECM), influencing the formation of cancer associated ECM, which promotes tumor cell migration and invasion [62-66]. Disruption of protein glycosylation through inhibition of N-glycosylation or the HBP pathway impacts the signaling cascade affecting expression of receptor tyrosine kinases (RTKs). Inhibition of glycosylation also enhances chemosensitivity of drug-resistant PDAC cells, underscoring the importance of glycosylation in perpetuating cancer survival [52, 67, 68]. While these studies open a window to explore the glycoproteome in PDAC, comprehensive analysis of intact glycopeptides or glycoproteins still remains a technical challenge. Much work remains to dissect on the complex mechanisms underlying the glycosylation events involved in PDAC.
Other protein PTMs in PDAC
In addition to the glycosylation discussed above, several common PTMs are also altered in the PDAC proteomes and contribute to the tumorigenesis, progression and metastasis. These PTMs may include phosphorylation, ubiquitination, sumoylation, and acetylation, playing pivotal roles in essentially all major signaling pathways implicated in PDAC. Three of these pathways are further discussed below (Figure 3).
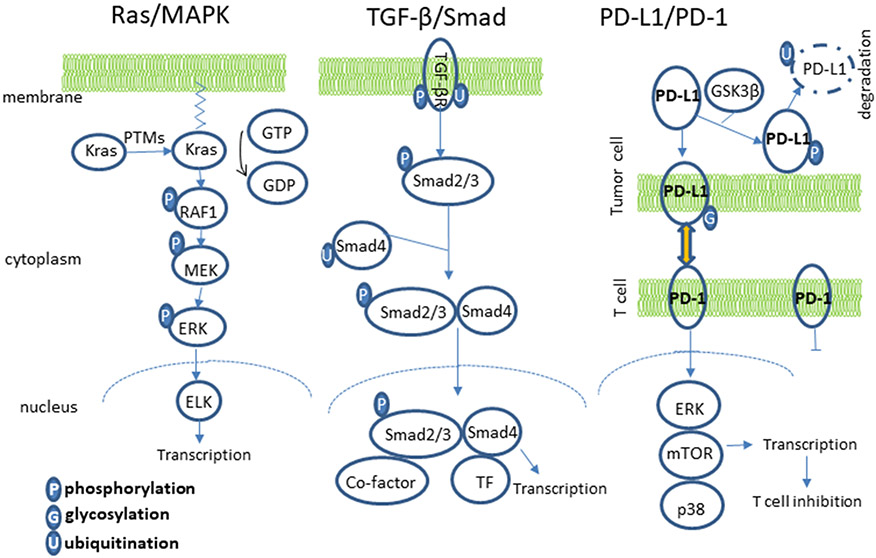
Figure 3.
PTMs in signaling pathways. PTMs play a pivotal role in regulating signaling pathways involved in pancreatic tumorigenesis and metastasis, including RAS/MAPK, TGF-β/SMAD and PD-L1/PD-1.
In the RAS/MAPK pathway, which is activated in over 90% of PDAC, KRAS and other signal mediators are subject to multiple PTMs. Nascent K-RAS proteins are subject to 3 sequential PTMs at the C terminal (farnesylation, proteolysis and methylation) to gain affinity to the plasma membrane, which is required for its activity [69, 70]. The isoform K-Ras4A has additional palmitoylation to gain enhanced membrane affinity [71]. These PTMs are considered constitutional and are usually not affected by RAS activation status or disease condition [69]. However, these PTMs are required for the proper trafficking and localization of RAS into the membrane. Other PTMs of K-RAS, such as phosphorylation or ubiquitination, have been shown to result in membrane disassociation or enhanced K-RAS activity, respectively [69]. Phosphorylation and dephosphorylation of the multiple kinases and mediators in the RAS/MAPK pathway are pivotal in transmitting the extracellular signal to the cell nucleus to activate or modulate gene transcription. Gene mutations or PTMs leading to alterations in the phosphorylation can result in substantial impact in this signaling pathway and affect disease conditions.
Dysregulation of the TGF-β signaling pathway is very common at tumor initiation or during tumor progression of pancreas. It can act as tumor suppressing or tumor promoting, depending on the tumor stage and microenvironment. All signaling mediators in the TGF-β pathway are subject to extensive PTMs [72]. For TGF-β receptors, various PTMs, including phosphorylation and ubiquitination, are critical for the initiation and regulation of the signal transduction into the nucleus. The downstream SMADs are also subject to extensive and stringent regulation by multiple PTMs. Among the various SMADs in this pathway, SMAD4 is a critical common co-factor, which is deactivated in approximately 50% of PDAC. The deactivation of SMAD4 in PDAC are usually the results of gene deletion, frameshift mutation, and single point mutation. Gene deletion and frameshift mutation can result in loss or reduced expression of SMAD4 protein. Most of the SMAD4 single point mutations found in PDAC cause a change in single amino acid that enhances affinity with ubiquitin (E3) ligase, and thus higher protein ubiquitination and accelerated degradation [73], leading to inhibition of transcriptional response of TGF-β.
Disabling immune checkpoints is emerging as a promising approach of immunotherapy for several types of cancer. The key T-cell checkpoint ligand, PD-L1, shows increased positivity in multiple cancers including PDAC [74, 75]. PD-L1 is subject to extensive regulation by PTMs. The stability of PD-L1 is extensively regulated by the ubiquitin/proteasome pathway [76]. Phosphorylation of PD-L1 by GSK3β increases its affinity to ubiquitin (E3) ligase, leading to degradation [77]. N-Linked glycosylation of PD-L1 creates a spatial barrier and disrupts GSK3β and PD-L1 interaction, leading to PD-L1 protein stabilization [77]. N-linked glycosylation of PD-L1 is also required for the proper interaction of PD-L1 and PD-1 [76]. These PTMs directly affect the functionality of PD-L1. Immunosuppression is very prominent in the progression of PDAC. However, recent results using checkpoint blockage have suggested that pancreatic cancer is resistant to this initial immunotherapy approach. Furthermore, the positivity of PD-L1 in PDAC is controversial in the literature, ranging from scarcely expressed to highly expressed in PDAC [74, 75, 78, 79]. The discrepancy on the PD-L1 staining between different studies could be in part attributed to the antibody used and the PTMs of PD-L1. In light of the complex PTMs of PD-L1, targeting PTMs of PD-L1 could represent a novel strategy to improve the effectiveness of immunotherapy for pancreatic cancer.
Proteome at early stages of PDAC
The precursors of pancreatic carcinoma may include PanIN lesions and pancreatic cystic neoplasms with malignant potential, such as IPMNs and MCNs. Both PanIN 3 and IPMN/MCN--with high grade dysplasia (both forms of carcinoma in-situ) show many similar molecular features with PDAC, including changes in the protein profile. Studies have shown that many differentially expressed proteins in PanIN 3 lesions, such as galectin 1, annexin A4 and A5, vimentin and laminin, are also concurrently dysregulated in PDAC tissue, suggesting that the dysregulation at functional level may start early prior to the development of invasive tumor [25, 33]. Oncogene c-MYC was identified as a prominent regulatory protein in the network of dysregulated proteins identified in PanIN 3 tissues. Such an observation from a proteome perspective, while preliminary, supports the pathological and genomic progression model of PanINs to PDAC [80-83].
In the proteomic study of pancreatic malignancy associated with cysts, one study suggested that the detection of protein family members of amylase, mucins, CEACAMs, and S100 proteins in cyst fluid might facilitate the discrimination of pancreatic cyst with malignant potential from benign lesions [84]. These proteins are among the proteins differentially expressed in PDAC. Systematic proteomic investigation of cystic tissue specimens obtained from surgical procedures or endoscopic ultrasound guided fine needle aspiration (EUS-FNA) have not been reported, but would provide useful insights and expand our understanding on IPMN/MCN progression to pancreatic carcinoma. To date, the molecular details on the transition of proteomes and key protein networks at the early stages of human PDAC progression largely remain unclear. One of the major hurdles of clinical proteomic study on PDAC progression are the limited resources of clinically and pathologically well-defined specimens from patients with pre-cancer or early stage disease, as early detection of PDAC still remains a significant clinical challenge.
Confounding proteome changes
The changes in PDAC proteomes are multifaceted and can be convoluted by associated diseases or disease complications such as chronic pancreatitis, jaundice and diabetes [85-89]. Mounting evidence has shown that PDAC and chronic pancreatitis share many similar clinical and molecular features, and such similarities reflect in not only the tissue proteome but also the bodily fluids [34, 87, 88, 90-94]. As inflammation is a critical component of cancer progression, many inflammation-related proteins are concurrently overexpressed in the tissue lesions of both PDAC and chronic pancreatitis [34]. Fibrosis is one of the fundamental histological abnormalities observed in both PDAC and chronic pancreatitis [46, 95, 96]. Persistent activation of pancreatic stellate cells promotes fibrosis and enhances secretion of ECM components. As a consequence, proteins that are associated with ECM and stellate cells are frequently elevated in both PDAC and chronic pancreatitis [34]. In bodily fluids, the proteome and glycoproteome of plasma/serum of PDAC can be confounded by chronic pancreatitis, jaundice and diabetes due to the systemic proteome changes in the circulating system. The proteomes of pancreatic juice and EUS-FNA biopsies can be influenced by the level of obstruction of the main pancreatic ducts due to inflammation, jaundice or other non-cancerous diseases [97, 98]. While these confounding alterations pose a significant challenge in dissecting malignant signals for biomarker development, they may also be components of the complex mechanisms involved in pancreatic tumorigenesis, including inflammation, ECM remodeling, angiogenesis, fibrosis, immune response and diminishing of acinar cells.
Concluding remarks
While the signature mutations of PDAC, including near ubiquitous oncogenic mutations of KRAS and the frequent inactivation of TP53, SMAD4, and CDKN2A tumor suppressors have been well recognized [3-6], there is a need to collect sufficient data to define the cascading proteome alterations and functional drivers in PDAC. Such alterations may include changes in protein expression, amino acid sequence, PTM status, interaction networks and subcellular distribution associated with low- and high-grade dysplasia, at early and late stage PDAC. The discoveries from the emerging field of proteogenomics [99], which integrates genomic and transcriptomic information to enhance proteomic analysis, are expected to facilitate the interface and convergence of current understanding of pancreatic tumorigenesis from genomic and functional perspectives. The development of quantitative spectral library-based platform technology, such as SWATH [100], which affords enormous multiplex capability and records digital archive of a whole proteome for retrospective analysis, carries an exciting technological advance to assist clinical biomarker development. The development of databases and bioinformatics for metaproteomic analysis of human gut microbiome has provided a powerful tool to interrogate the interplay of microbiota with host response at functional level. While exciting perspective has been demonstrated and increasing efforts have been made in exploring proteome alterations associated with PDAC and its progression, significant challenges exist. In addition to technical hurdles, the timeline and precise mechanism of PDAC progression from low-grade precursors (PanINs, mucinous cystic lesions) to invasive cancerous lesion are difficult to assess, and still remain vague. Heterogeneities associated with patients and specimen collection can also confound the analysis and data interpretation. Well-defined clinical longitudinal studies and experimental models are needed to provide well characterized samples for proteome analysis. The study of cancer stem cells and the presence of intra-tumoural heterogeneity in PDAC, which has direct implications for targeted or immunotherapeutic interventions, pose additional challenges for single cell proteomic analysis [101]. The co-existence of these challenges and knowledge gaps define the opportunities in current proteomic study of PDAC.
Acknowledgment
The authors are grateful to Dr. Ruedi Aebersold (ETH Zurich) for his long-term support and collaboration in proteomic study of pancreatic cancer. This work is supported in part with federal fund from the National Institutes of Health under grant R01CA180949. The authors are grateful for the supports from Canary Foundation and Walters Foundation for early detection of pancreatic cancer.
Reference List
- [1].Rahib L, Smith BD, Aizenberg R, Rosenzweig AB, Fleshman JM, and Matrisian LM, Projecting cancer incidence and deaths to 2030: the unexpected burden of thyroid, liver, and pancreas cancers in the United States. Cancer Res. 74 (2014) 2913–2921. [DOI] [PubMed] [Google Scholar]
- [2].Siegel RL, Miller KD, and Jemal A, Cancer statistics, 2019. CA Cancer J. Clin 69 (2019) 7–34. [DOI] [PubMed] [Google Scholar]
- [3].Hezel AF, Kimmelman AC, Stanger BZ, Bardeesy N, and DePinho RA, Genetics and biology of pancreatic ductal adenocarcinoma. Genes Dev. 20 (2006) 1218–1249. [DOI] [PubMed] [Google Scholar]
- [4].Hidalgo M, Pancreatic cancer. N. Engl. J. Med 362 (2010) 1605–1617. [DOI] [PubMed] [Google Scholar]
- [5].Maitra A and Hruban RH, Pancreatic cancer. Annu. Rev. Pathol 3 (2008) 157–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Vincent A, Herman J, Schulick R, Hruban RH, and Goggins M, Pancreatic cancer. Lancet 378 (2011) 607–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Distler M, Aust D, Weitz J, Pilarsky C, and Grutzmann R, Precursor lesions for sporadic pancreatic cancer: PanIN, IPMN, and MCN. Biomed. Res Int 2014 (2014) 474905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Tian Q, Stepaniants SB, Mao M, Weng L, Feetham MC, Doyle MJ, Yi EC, Dai H, Thorsson V, Eng J, Goodlett D, Berger JP, Gunter B, Linseley PS, Stoughton RB, Aebersold R, Collins SJ, Hanlon WA, and Hood LE, Integrated genomic and proteomic analyses of gene expression in Mammalian cells. Mol. Cell Proteomics 3 (2004) 960–969. [DOI] [PubMed] [Google Scholar]
- [9].Aebersold R and Mann M, Mass-spectrometric exploration of proteome structure and function. Nature 537 (2016) 347–355. [DOI] [PubMed] [Google Scholar]
- [10].Campbell SL and Wellen KE, Metabolic Signaling to the Nucleus in Cancer. Mol. Cell 71 (2018) 398–408. [DOI] [PubMed] [Google Scholar]
- [11].Harper JW and Bennett EJ, Proteome complexity and the forces that drive proteome imbalance. Nature 537 (2016) 328–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Hollingsworth MA and Swanson BJ, Mucins in cancer: protection and control of the cell surface. Nat. Rev. Cancer 4 (2004) 45–60. [DOI] [PubMed] [Google Scholar]
- [13].Kaur S, Kumar S, Momi N, Sasson AR, and Batra SK, Mucins in pancreatic cancer and its microenvironment. Nat. Rev. Gastroenterol. Hepatol 10 (2013) 607–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Remmers N, Bailey JM, Mohr AM, and Hollingsworth MA, Molecular pathology of early pancreatic cancer. Cancer Biomark. 9 (2010) 421–440. [DOI] [PubMed] [Google Scholar]
- [15].Remmers N, Anderson JM, Linde EM, DiMaio DJ, Lazenby AJ, Wandall HH, Mandel U, Clausen H, Yu F, and Hollingsworth MA, Aberrant expression of mucin core proteins and o-linked glycans associated with progression of pancreatic cancer. Clin. Cancer Res 19 (2013) 1981–1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Simeone DM, Ji B, Banerjee M, Arumugam T, Li D, Anderson MA, Bamberger AM, Greenson J, Brand RE, Ramachandran V, and Logsdon CD, CEACAM1, a novel serum biomarker for pancreatic cancer. Pancreas 34 (2007) 436–443. [DOI] [PubMed] [Google Scholar]
- [17].Beauchemin N and Arabzadeh A, Carcinoembryonic antigen-related cell adhesion molecules (CEACAMs) in cancer progression and metastasis. Cancer Metastasis Rev. 32 (2013) 643–671. [DOI] [PubMed] [Google Scholar]
- [18].Gebauer F, Wicklein D, Horst J, Sundermann P, Maar H, Streichert T, Tachezy M, Izbicki JR, Bockhorn M, and Schumacher U, Carcinoembryonic antigen-related cell adhesion molecules (CEACAM) 1, 5 and 6 as biomarkers in pancreatic cancer. PLoS. One 9 (2014) e113023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Milo R, What is the total number of protein molecules per cell volume? A call to rethink some published values. Bioessays 35 (2013) 1050–1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Aebersold R and Mann M, Mass spectrometry-based proteomics. Nature 422 (2003) 198–207. [DOI] [PubMed] [Google Scholar]
- [21].Altelaar AF, Munoz J, and Heck AJ, Next-generation proteomics: towards an integrative view of proteome dynamics. Nat. Rev. Genet 14 (2013) 35–48. [DOI] [PubMed] [Google Scholar]
- [22].Aspinall-O'Dea M and Costello E, The pancreatic cancer proteome - recent advances and future promise. Proteomics. Clin. Appl 1 (2007) 1066–1079. [DOI] [PubMed] [Google Scholar]
- [23].Cecconi D, Palmieri M, and Donadelli M, Proteomics in pancreatic cancer research. Proteomics 11 (2011) 816–828. [DOI] [PubMed] [Google Scholar]
- [24].Coleman O, Henry M, McVey G, Clynes M, Moriarty M, and Meleady P, Proteomic strategies in the search for novel pancreatic cancer biomarkers and drug targets: recent advances and clinical impact. Expert. Rev. Proteomics 13 (2016) 383–394. [DOI] [PubMed] [Google Scholar]
- [25].Pan S, Brentnall TA, Kelly K, and Chen R, Tissue proteomics in pancreatic cancer study: discovery, emerging technologies, and challenges. Proteomics 13 (2013) 710–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Pan S, Brentnall TA, and Chen R, Proteomics analysis of bodily fluids in pancreatic cancer. Proteomics 15 (2015) 2705–2715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Pawa N, Wright JM, and Arulampalam TH, Mass spectrometry based proteomic profiling for pancreatic cancer. JOP. 11 (2010) 423–426. [PubMed] [Google Scholar]
- [28].Sun C, Rosendahl AH, Ansari D, and Andersson R, Proteome-based biomarkers in pancreatic cancer. World J Gastroenterol. 17 (2011) 4845–4852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Tonack S, Aspinall-O'Dea M, Neoptolemos JP, and Costello E, Pancreatic cancer: proteomic approaches to a challenging disease. Pancreatology. 9 (2009) 567–576. [DOI] [PubMed] [Google Scholar]
- [30].Brown LF, Guidi AJ, Schnitt SJ, Van De WL, Iruela-Arispe ML, Yeo TK, Tognazzi K, and Dvorak HF, Vascular stroma formation in carcinoma in situ, invasive carcinoma, and metastatic carcinoma of the breast. Clin. Cancer Res 5 (1999) 1041–1056. [PubMed] [Google Scholar]
- [31].Liotta LA and Kohn EC, The microenvironment of the tumour-host interface. Nature 411 (2001) 375–379. [DOI] [PubMed] [Google Scholar]
- [32].Chen R, Yi EC, Donohoe D, Pan S, Eng J, Crispin DA, Lane Z, Goodlett DA, Bronner MP, Aebersold R, and Brentnall TA, Pancreatic Cancer Proteome: the Proteins that Underlie Invasion, Metastasis, and Immunologic Escape. Gastroenterology 129 (2005) 1187–1197. [DOI] [PubMed] [Google Scholar]
- [33].Pan S, Chen R, Reimel BA, Crispin DA, Mirzaei H, Cooke K, Coleman JF, Lane Z, Bronner MP, Goodlett DR, McIntosh MW, Traverso W, Aebersold R, and Brentnall TA, Quantitative proteomics investigation of pancreatic intraepithelial neoplasia. Electrophoresis 30 (2009) 1132–1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Pan S, Chen R, Stevens T, Bronner MP, May D, Tamura Y, McIntosh MW, and Brentnall TA, Proteomics portrait of archival lesions of chronic pancreatitis. PLoS. One 6 (2011) e27574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Brentnall TA, Lai LA, Coleman J, Bronner MP, Pan S, and Chen R, Arousal of cancer-associated stroma: overexpression of palladin activates fibroblasts to promote tumor invasion. PLoS. One 7 (2012) e30219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].von AD, Bhagat TD, Nagrath D, Maitra A, and Verma A, The role of stromal cancer-associated fibroblasts in pancreatic cancer. J. Hematol. Oncol 10 (2017) 76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Foygel K, Wang H, Lutz AM, Chen R, Machtaler S, Pysz M, Lowe A, Tian L, Carrigan T, Brentnall TA, and Willmann JK, Ultrasonic Molecular Imaging of Thy1 for Pancreatic Cancer Detection. Gastroenterology Submitted (2013) 885–894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Belo AI, van der Sar AM, Tefsen B, and van D, I, Galectin-4 Reduces Migration and Metastasis Formation of Pancreatic Cancer Cells. PLoS. One 8 (2013) e65957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Chen R, Pan S, Ottenhof NA, de Wilde RF, Wolfgang CL, Lane Z, Post J, Bronner MP, Willmann JK, Maitra A, and Brentnall TA, Stromal galectin-1 expression is associated with long-term survival in resectable pancreatic ductal adenocarcinoma. Cancer Biol. Ther 13 (2012) 899–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Chen R, Dawson DW, Pan S, Ottenhof NA, de Wilde RF, Wolfgang CL, May DH, Crispin DA, Lai LA, Lay AR, Waghray M, Wang S, McIntosh MW, Simeone DM, Maitra A, and Brentnall TA, Proteins associated with pancreatic cancer survival in patients with resectable pancreatic ductal adenocarcinoma. Laboratory Investigation 95 (2015) 43–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Martinez-Bosch N and Navarro P, Targeting Galectin-1 in pancreatic cancer: immune surveillance on guard. Oncoimmunology. 3 (2014) e952201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Merlin J, Stechly L, de BS, Monte D, Leteurtre E, Van S I, Huet G, and Pigny P, Galectin-3 regulates MUC1 and EGFR cellular distribution and EGFR downstream pathways in pancreatic cancer cells. Oncogene 30 (2011) 2514–2525. [DOI] [PubMed] [Google Scholar]
- [43].Song S, Ji B, Ramachandran V, Wang H, Hafley M, Logsdon C, and Bresalier RS, Overexpressed galectin-3 in pancreatic cancer induces cell proliferation and invasion by binding Ras and activating Ras signaling. PLoS. One 7 (2012) e42699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Tang D, Yuan Z, Xue X, Lu Z, Zhang Y, Wang H, Chen M, An Y, Wei J, Zhu Y, Miao Y, and Jiang K, High expression of Galectin-1 in pancreatic stellate cells plays a role in the development and maintenance of an immunosuppressive microenvironment in pancreatic cancer. Int. J. Cancer 130 (2012) 2337–2348. [DOI] [PubMed] [Google Scholar]
- [45].Tang D, Zhang J, Yuan Z, Gao J, Wang S, Ye N, Li P, Gao S, Miao Y, Wang D, and Jiang K, Pancreatic satellite cells derived galectin-1 increase the progression and less survival of pancreatic ductal adenocarcinoma. PLoS. One 9 (2014) e90476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Shimizu K, Mechanisms of pancreatic fibrosis and applications to the treatment of chronic pancreatitis. J Gastroenterol. 43 (2008) 823–832. [DOI] [PubMed] [Google Scholar]
- [47].Bausch D, Thomas S, Mino-Kenudson M, d.Fernandez, Bauer TW, Williams M, Warshaw AL, Thayer SP, and Kelly KA, Plectin-1 as a novel biomarker for pancreatic cancer. Clin. Cancer Res 17 (2011) 302–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Kelly KA, Bardeesy N, Anbazhagan R, Gurumurthy S, Berger J, Alencar H, DePinho RA, Mahmood U, and Weissleder R, Targeted nanoparticles for imaging incipient pancreatic ductal adenocarcinoma. PLoS. Med 5 (2008) e85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Pan S, Chen R, Brand RE, Hawley S, Tamura Y, Gafken PR, Milless BP, Goodlett DR, Rush J, and Brentnall TA, Multiplex targeted proteomic assay for biomarker detection in plasma: a pancreatic cancer biomarker case study. J. Proteome Res 11 (2012) 1937–1948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Ying H, Kimmelman AC, Lyssiotis CA, Hua S, Chu GC, Fletcher-Sananikone E, Locasale JW, Son J, Zhang H, Coloff JL, Yan H, Wang W, Chen S, Viale A, Zheng H, Paik JH, Lim C, Guimaraes AR, Martin ES, Chang J, Hezel AF, Perry SR, Hu J, Gan B, Xiao Y, Asara JM, Weissleder R, Wang YA, Chin L, Cantley LC, and DePinho RA, Oncogenic Kras maintains pancreatic tumors through regulation of anabolic glucose metabolism. Cell 149 (2012) 656–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Pan S, Chen R, Tamura Y, Crispin DA, Lai LA, May DH, McIntosh MW, Goodlett DR, and Brentnall TA, Quantitative glycoproteomics analysis reveals changes in N-glycosylation level associated with pancreatic ductal adenocarcinoma. J Proteome Res 13 (2014) 1293–1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Chen R, Lai LA, Sullivan Y, Wong M, Wang L, Riddell J, Jung L, Pillarisetty VG, Brentnall TA, and Pan S, Disrupting glutamine metabolic pathways to sensitize gemcitabine-resistant pancreatic cancer. Sci. Rep 7 (2017) 7950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Commisso C, Davidson SM, Soydaner-Azeloglu RG, Parker SJ, Kamphorst JJ, Hackett S, Grabocka E, Nofal M, Drebin JA, Thompson CB, Rabinowitz JD, Metallo CM, Vander Heiden MG, and Bar-Sagi D, Macropinocytosis of protein is an amino acid supply route in Ras-transformed cells. Nature 497 (2013) 633–637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Halbrook CJ and Lyssiotis CA, Employing Metabolism to Improve the Diagnosis and Treatment of Pancreatic Cancer. Cancer Cell 31 (2017) 5–19. [DOI] [PubMed] [Google Scholar]
- [55].Szajda SD, Waszkiewicz N, Chojnowska S, and Zwierz K, Carbohydrate markers of pancreatic cancer. Biochem. Soc. Trans 39 (2011) 340–343. [DOI] [PubMed] [Google Scholar]
- [56].Yue T, Goldstein IJ, Hollingsworth MA, Kaul K, Brand RE, and Haab BB, The prevalence and nature of glycan alterations on specific proteins in pancreatic cancer patients revealed using antibody-lectin sandwich arrays. Mol. Cell Proteomics 8 (2009) 1697–1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Yue T, Partyka K, Maupin KA, Hurley M, Andrews P, Kaul K, Moser AJ, Zeh H, Brand RE, and Haab BB, Identification of blood-protein carriers of the CA 19-9 antigen and characterization of prevalence in pancreatic diseases. Proteomics 11 (2011) 3665–3674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Pan S, Brentnall TA, and Chen R, Glycoproteins and glycoproteomics in pancreatic cancer. World J Gastroenterol. 22 (2016) 9288–9299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Zhao J, Qiu W, Simeone DM, and Lubman DM, N-linked glycosylation profiling of pancreatic cancer serum using capillary liquid phase separation coupled with mass spectrometric analysis. J. Proteome. Res 6 (2007) 1126–1138. [DOI] [PubMed] [Google Scholar]
- [60].Mann BF, Goetz JA, House MG, Schmidt CM, and Novotny MV, Glycomic and proteomic profiling of pancreatic cyst fluids identifies hyperfucosylated lactosamines on the N-linked glycans of overexpressed glycoproteins. Mol. Cell Proteomics 11 (2012) M111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Nigjeh EN, Chen R, Allen-Tamura Y, Brand RE, Brentnall TA, and Pan S, Spectral library-based glycopeptide analysis-detection of circulating galectin-3 binding protein in pancreatic cancer. Proteomics. Clin. Appl 11 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Janik ME, Litynska A, and Vereecken P, Cell migration-the role of integrin glycosylation. Biochim. Biophys. Acta 1800 (2010) 545–555. [DOI] [PubMed] [Google Scholar]
- [63].Leeming DJ, Bay-Jensen AC, Vassiliadis E, Larsen MR, Henriksen K, and Karsdal MA, Post-translational modifications of the extracellular matrix are key events in cancer progression: opportunities for biochemical marker development. Biomarkers 16 (2011) 193–205. [DOI] [PubMed] [Google Scholar]
- [64].Malik R, Lelkes PI, and Cukierman E, Biomechanical and biochemical remodeling of stromal extracellular matrix in cancer. Trends Biotechnol. 33 (2015) 230–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Itano N, Zhuo L, and Kimata K, Impact of the hyaluronan-rich tumor microenvironment on cancer initiation and progression. Cancer Sci. 99 (2008) 1720–1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Provenzano PP and Hingorani SR, Hyaluronan, fluid pressure, and stromal resistance in pancreas cancer. Br. J Cancer 108 (2013) 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Contessa JN, Bhojani MS, Freeze HH, Rehemtulla A, and Lawrence TS, Inhibition of N-linked glycosylation disrupts receptor tyrosine kinase signaling in tumor cells. Cancer Res 68 (2008) 3803–3809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Dwek RA, Butters TD, Platt FM, and Zitzmann N, Targeting glycosylation as a therapeutic approach. Nat. Rev. Drug Discov 1 (2002) 65–75. [DOI] [PubMed] [Google Scholar]
- [69].Ahearn IM, Haigis K, Bar-Sagi D, and Philips MR, Regulating the regulator: post-translational modification of RAS. Nat. Rev. Mol. Cell Biol 13 (2011) 39–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Silvius JR and l'Heureux F, Fluorimetric evaluation of the affinities of isoprenylated peptides for lipid bilayers. Biochemistry 33 (1994) 3014–3022. [DOI] [PubMed] [Google Scholar]
- [71].Laude AJ and Prior IA, Palmitoylation and localisation of RAS isoforms are modulated by the hypervariable linker domain. J. Cell Sci 121 (2008) 421–427. [DOI] [PubMed] [Google Scholar]
- [72].Xu P, Lin X, and Feng XH, Posttranslational Regulation of Smads. Cold Spring Harb. Perspect. Biol 6 (2016) a022087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Wan M, Huang J, Jhala NC, Tytler EM, Yang L, Vickers SM, Tang Y, Lu C, Wang N, and Cao X, SCF(beta-TrCP1) controls Smad4 protein stability in pancreatic cancer cells. Am. J. Pathol 166 (2005) 1379–1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Hou YC, Chao YJ, Hsieh MH, Tung HL, Wang HC, and Shan YS, Low CD8(+) T Cell Infiltration and High PD-L1 Expression Are Associated with Level of CD44(+)/CD133(+) Cancer Stem Cells and Predict an Unfavorable Prognosis in Pancreatic Cancer. Cancers. (Basel) 11 (4) (2019) E541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Lu C, Paschall AV, Shi H, Savage N, Waller JL, Sabbatini ME, Oberlies NH, Pearce C, and Liu K, The MLL1-H3K4me3 Axis-Mediated PD-L1 Expression and Pancreatic Cancer Immune Evasion. J. Natl. Cancer Inst 109 (6) (2017). doi: 10.1093/jnci/djw283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Hsu JM, Li CW, Lai YJ, and Hung MC, Posttranslational Modifications of PD-L1 and Their Applications in Cancer Therapy. Cancer Res. 78 (2018) 6349–6353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Li CW, Lim SO, Xia W, Lee HH, Chan LC, Kuo CW, Khoo KH, Chang SS, Cha JH, Kim T, Hsu JL, Wu Y, Hsu JM, Yamaguchi H, Ding Q, Wang Y, Yao J, Lee CC, Wu HJ, Sahin AA, Allison JP, Yu D, Hortobagyi GN, and Hung MC, Glycosylation and stabilization of programmed death ligand-1 suppresses T-cell activity. Nat. Commun 7 (2016) 12632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Liang X, Sun J, Wu H, Luo Y, Wang L, Lu J, Zhang Z, Guo J, Liang Z, and Liu T, PD-L1 in pancreatic ductal adenocarcinoma: a retrospective analysis of 373 Chinese patients using an in vitro diagnostic assay. Diagn. Pathol 13 (1) (2018) 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Zheng L, PD-L1 Expression in Pancreatic Cancer. J. Natl. Cancer Inst 109 (6) (2017). doi: 10.1093/jnci/djw304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Hruban RH, Wilentz RE, Goggins M, Offerhaus GJ, Yeo CJ, and Kern SE, Pathology of incipient pancreatic cancer. Ann. Oncol 10 Suppl 4 (1999) 9–11. [PubMed] [Google Scholar]
- [81].Hruban RH, Goggins M, Parsons J, and Kern SE, Progression model for pancreatic cancer. Clin. Cancer Res 6 (2000) 2969–2972. [PubMed] [Google Scholar]
- [82].Hruban RH, Wilentz RE, and Kern SE, Genetic progression in the pancreatic ducts. Am. J Pathol 156 (2000) 1821–1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Notta F, Chan-Seng-Yue M, Lemire M, Li Y, Wilson GW, Connor AA, Denroche RE, Liang SB, Brown AM, Kim JC, Wang T, Simpson JT, Beck T, Borgida A, Buchner N, Chadwick D, Hafezi-Bakhtiari S, Dick JE, Heisler L, Hollingsworth MA, Ibrahimov E, Jang GH, Johns J, Jorgensen LG, Law C, Ludkovski O, Lungu I, Ng K, Pasternack D, Petersen GM, Shlush LI, Timms L, Tsao MS, Wilson JM, Yung CK, Zogopoulos G, Bartlett JM, Alexandrov LB, Real FX, Cleary SP, Roehrl MH, McPherson JD, Stein LD, Hudson TJ, Campbell PJ, and Gallinger S, A renewed model of pancreatic cancer evolution based on genomic rearrangement patterns. Nature 538 (2016) 378–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Ke E, Patel BB, Liu T, Li XM, Haluszka O, Hoffman JP, Ehya H, Young NA, Watson JC, Weinberg DS, Nguyen MT, Cohen SJ, Meropol NJ, Litwin S, Tokar JL, and Yeung AT, Proteomic analyses of pancreatic cyst fluids. Pancreas 38 (2009) e33–e42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Brand R, The diagnosis of pancreatic cancer. Cancer J. 7 (2001) 287–297. [PubMed] [Google Scholar]
- [86].Hidalgo L, Repiso A, Romero M, Navajas J, Sanchez-Simon R, Gomez-Rodriguez R, and Carrobles JM, Obstructive jaundice as a complication of a peptic duodenal ulcer mimicking pancreatic cancer. Endoscopy 42 Suppl 2 (2010) E294–E295. [DOI] [PubMed] [Google Scholar]
- [87].Lowenfels AB, Maisonneuve P, Cavallini G, Ammann RW, Lankisch PG, Andersen JR, Dimagno EP, Andren-Sandberg A, and Domellof L, Pancreatitis and the risk of pancreatic cancer. International Pancreatitis Study Group. N. Engl. J. Med 328 (1993) 1433–1437. [DOI] [PubMed] [Google Scholar]
- [88].Malka D, Hammel P, Maire F, Rufat P, Madeira I, Pessione F, Levy P, and Ruszniewski P, Risk of pancreatic adenocarcinoma in chronic pancreatitis. Gut 51 (2002) 849–852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Pannala R, Basu A, Petersen GM, and Chari ST, New-onset diabetes: a potential clue to the early diagnosis of pancreatic cancer. Lancet Oncol. 10 (2009) 88–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Chen R, Pan S, Cooke K, Moyes KW, Bronner MP, Goodlett DR, Aebersold R, and Brentnall TA, Comparison of pancreas juice proteins from cancer versus pancreatitis using quantitative proteomic analysis. Pancreas 34 (2007) 70–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Chen R, Brentnall TA, Pan S, Cooke K, Moyes KW, Lane Z, Crispin DA, Goodlett DR, Aebersold R, and Bronner MP, Quantitative proteomics analysis reveals that proteins differentially expressed in chronic pancreatitis are also frequently involved in pancreatic cancer. Mol. Cell Proteomics. 6 (2007) 1331–1342. [DOI] [PubMed] [Google Scholar]
- [92].Chen R, Crispin DA, Pan S, Hawley S, McIntosh MW, May D, Anton-Culver H, Ziogas A, Bronner MP, and Brentnall TA, Pilot study of blood biomarker candidates for detection of pancreatic cancer. Pancreas 39 (2010) 981–988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Pan S, Chen R, Crispin DA, May D, Stevens T, McIntosh MW, Bronner MP, Ziogas A, Anton-Culver H, and Brentnall TA, Protein alterations associated with pancreatic cancer and chronic pancreatitis found in human plasma using global quantitative proteomics profiling. J. Proteome Res 10 (2011) 2359–2376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Rosty C, Geradts J, Sato N, Wilentz RE, Roberts H, Sohn T, Cameron JL, Yeo CJ, Hruban RH, and Goggins M, p16 Inactivation in pancreatic intraepithelial neoplasias (PanINs) arising in patients with chronic pancreatitis. Am. J. Surg. Pathol 27 (2003) 1495–1501. [DOI] [PubMed] [Google Scholar]
- [95].Leake I, Pancreatic cancer: surprising role for fibrosis. Nat. Rev. Gastroenterol. Hepatol 11 (2014) 396. [DOI] [PubMed] [Google Scholar]
- [96].Tang D, Wu Q, Zhang J, Zhang H, Yuan Z, Xu J, Chong Y, Huang Y, Xiong Q, Wang S, Tian Y, Lu Y, Ge X, Shen W, and Wang D, Galectin-1 expression in activated pancreatic satellite cells promotes fibrosis in chronic pancreatitis/pancreatic cancer via the TGF-beta1/Smad pathway. Oncol. Rep 39 (2018) 1347–1355. [DOI] [PubMed] [Google Scholar]
- [97].Krishna NB, Mehra M, Reddy AV, and Agarwal B, EUS/EUS-FNA for suspected pancreatic cancer: influence of chronic pancreatitis and clinical presentation with or without obstructive jaundice on performance characteristics. Gastrointest. Endosc 70 (2009) 70–79. [DOI] [PubMed] [Google Scholar]
- [98].Zhou L, Lu Z, Yang A, Deng R, Mai C, Sang X, Faber KN, and Lu X, Comparative proteomic analysis of human pancreatic juice: methodological study. Proteomics 7 (2007) 1345–1355. [DOI] [PubMed] [Google Scholar]
- [99].Nesvizhskii AI, Proteogenomics: concepts, applications and computational strategies. Nat. Methods 11 (2014) 1114–1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Aebersold R, Bensimon A, Collins BC, Ludwig C, and Sabido E, Applications and Developments in Targeted Proteomics: From SRM to DIA/SWATH. Proteomics. 16 (2016) 2065–2067. [DOI] [PubMed] [Google Scholar]
- [101].Irish JM, Kotecha N, and Nolan GP, Mapping normal and cancer cell signalling networks: towards single-cell proteomics. Nat. Rev. Cancer 6 (2006) 146–155. [DOI] [PubMed] [Google Scholar]