ABSTRACT
Hospital environments are excellent reservoirs for the opportunistic pathogen Acinetobacter baumannii in part because it is exceptionally tolerant to desiccation. We found that relative to other A. baumannii strains, the virulent strain AB5075 was strikingly desiccation resistant at 2% relative humidity (RH), suggesting that it is a good model for studies of the functional basis of this trait. Consistent with results from other A. baumannii strains at 40% RH, we found the global posttranscriptional regulator CsrA to be critically important for desiccation tolerance of AB5075 at 2% RH. Proteomics experiments identified proteins that were differentially present in wild-type and csrA mutant cells. Subsequent analysis of mutants in genes encoding some of these proteins revealed six genes that were required for wild-type levels of desiccation tolerance. These include genes for catalase, a universal stress protein, a hypothetical protein, and a biofilm-associated protein. Two genes of unknown function had very strong desiccation phenotypes, with one of the two genes predicting an intrinsically disordered protein (IDP) that binds to DNA. Intrinsically disordered proteins are widespread in eukaryotes but less so in prokaryotes. Our results suggest there are new mechanisms underlying desiccation tolerance in bacteria and identify several key functions involved.
IMPORTANCE Acinetobacter baumannii is found in terrestrial environments but can cause nosocomial infections in very sick patients. A factor that contributes to the prevalence of A. baumannii in hospital settings is that it is intrinsically resistant to dry conditions. Here, we established the virulent strain A. baumannii AB5075 as a model for studies of desiccation tolerance at very low relative humidity. Our results show that this trait depends on two proteins of unknown function, one of which is predicted to be an intrinsically disordered protein. This category of protein is critical for the small animals named tardigrades to survive desiccation. Our results suggest that A. baumannii may have novel strategies to survive desiccation that have not previously been seen in bacteria.
KEYWORDS: Acinetobacter baumannii, desiccation, CsrA, intrinsically disordered proteins
INTRODUCTION
Hospital-acquired infections are an important health care concern and economic burden (1, 2), and environmental persistence plays a critical role in the transmission of bacteria that cause these infections (3–6). One such bacterium is Acinetobacter baumannii, an opportunistic pathogen that infects very sick patients. It is responsible for about 2% of nosocomial infections in the United States and Europe, and the frequencies are higher in the rest of the world. A. baumannii is especially problematic because on a global basis, about 45% of isolates are multidrug resistant (7). A factor that contributes to the prevalence of A. baumannii in hospital settings is desiccation tolerance. A. baumannii can survive in a desiccated state on inanimate dry surfaces for days to several months (8–10). These surfaces include materials that are often encountered in the hospital, such as polyvinyl chloride, rubber, and stainless steel (11).
When desiccated, bacteria must respond to diverse stresses that include accumulation of reactive oxygen species, loss of cytoplasmic volume, and loss of cell membrane integrity (12, 13). Proteomics analysis of A. baumannii showed that desiccated cells have higher levels of proteins involved in protein stabilization, antimicrobial resistance, and reactive oxygen species detoxification (14). Attributes of A. baumannii that are associated with desiccation tolerance include biofilm formation (15, 16) and protein aggregation (17). LpxMAB-dependent acetylation of lipid A is essential for survival of A. baumannii ATCC 17978 at 40% relative humidity (RH) (18), and a recA mutant of ATCC 17978, defective in DNA repair, had a pleiotropic phenotype, including a defect in desiccation tolerance (19). katE encoding catalase also contributes to desiccation tolerance (20). Despite these observations, the number of genes identified in A. baumannii that are specifically involved in desiccation tolerance is small. This could be because A. baumannii cells have evolved modified cell structures that are both essential for viability and important for desiccation tolerance. The genetic basis for this would be difficult to uncover in mutant screens. It is also possible that the conditions of desiccation used, typically 30% RH in studies to date, were not sufficiently severe to allow identification of some desiccation tolerance genes. Finally, different strains of A. baumannii differ in desiccation tolerance (21), and no one strain has been developed as a model for desiccation studies.
As part of studies of the GacSA two-component regulatory system and noncoding RNAs in A. baumannii strain AB5075, we constructed a deletion mutant of the global posttranscriptional regulator gene csrA and ascertained that it had a strong desiccation phenotype. Originally isolated from an infected wound of a patient in the United States military health care system, strain AB5075 is multidrug resistant and is highly virulent in an animal model (22). A comprehensive ordered mutant library of AB5075 is available that has two to three sequence-verified transposon (Tn) insertions in each gene and is called the three-allele library (23). We targeted csrA for mutation because its activity is frequently regulated by noncoding RNAs (ncRNAs) controlled by GacSA in other Gammaproteobacteria (24). As we were completing a study of CsrA and its role in desiccation tolerance in AB5075, Farrow et al. reported the construction of ΔcsrA mutants of A. baumannii strains ATCC 17961 and AB09-003 and showed that CsrA in these strains was also important for desiccation tolerance (25).
Here, we extend the work reported by Farrow et al. by developing an assay that can be conveniently used to measure desiccation tolerance at 2% or 30% RH. We used proteomics to identify several new functions that may be controlled by CsrA in A. baumannii at the posttranscriptional level and confirmed some of these. We also identified six CsrA-controlled proteins that confer desiccation tolerance on AB5075 at 2% RH. Several of these have unusual predicted properties that suggest new dimensions of desiccation tolerance.
RESULTS
Characterization of strain AB5075 desiccation tolerance.
Previous studies have shown that A. baumannii can survive in a desiccated state for days to several months (8–11, 20). For these and other desiccation studies, investigators have dried cells at different rates on a variety of different surfaces (21) and usually incubated cells at either 30% RH or in room air, which varied between 25 and 61% RH in one study (20). The recommended RH range in hospitals is between 20 and 60%. We thought it would be useful to the community to have a desiccation assay that is simple and easily reproducible. As described in Materials and Methods, we developed a desiccation assay in which bacterial cells are filtered and dried on polycarbonate membranes and then incubated in sealed plastic containers at 30% RH or 2% RH. After various periods of incubation, cells are washed from filters, and viable cell numbers are determined.
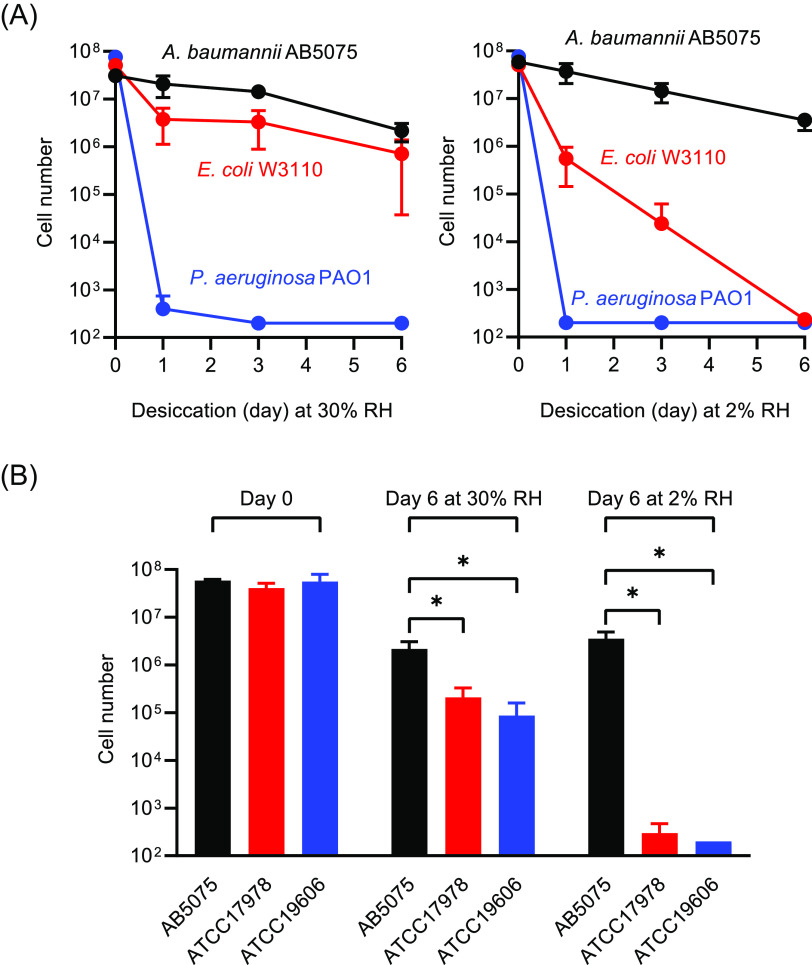
As shown in Fig. 1A, A. baumannii strain AB5075 and Escherichia coli strain W3110, both survived desiccation at 30% RH quite well and better than Pseudomonas aeruginosa strain PAO1. However, at 2% RH, A. baumannii survived far better than either E. coli or P. aeruginosa. As has been reported (20, 26), we found that A. baumannii stationary-phase cells were much more tolerant to desiccation than actively growing cells (Fig. S1 in the supplemental material), and therefore, we routinely used stationary-phase cells in our desiccation assays. We compared the desiccation tolerance of AB5075 to two additional frequently used laboratory strains of A. baumannii, ATCC 17978 and ATCC 19606. We found that AB5075 was strikingly more resistant than the others at 2% RH and significantly more resistant at 30% RH (Fig. 1B). These findings suggested that strain AB5075 could be useful for identifying genes that protect cells in extremely dry conditions. RHs in the Arabian Peninsula, a part of the world where A. baumannii is frequently isolated (27), are often as low as 2 to 5%.
FIG 1.
Desiccation tolerance of A. baumannii. (A) Comparison of A. baumannii AB5075, E. coli W3110, and P. aeruginosa PAO1 at 30% RH or 2% RH. (B) Desiccation tolerance of A. baumannii strains AB5075, ATCC 17978, and ATCC 19606 after 0 days (control) and 6 days of desiccation at 30% RH or 2% RH. Cell numbers represent the total number of viable cells recovered from each membrane. Data are the average of three biological replicates, and standard deviations are shown as error bars. Statistical significance was calculated using a t test: *, P < 0.05. The detection limit is 200 cells.
CsrA is critical for desiccation tolerance of AB5075.
csrA mutants do not have the same phenotypes in all A. baumannii strains (28), but in agreement with others (25, 28) we found that an AB5075 ΔcsrA strain grew as the wild type in defined medium; in our case, M9 minimal medium with 10 mM succinate as a sole carbon source (M9-succinate) (Fig. 2A and Fig. 3B), but poorly on TY agar and had an elongated cell morphology when grown in TY broth (Fig. 2A). The ΔcsrA strain was also defective in growth on other nutrient-rich media, including LB broth, nutrient broth, and tryptic soy broth (TSB). When desiccated at 2% RH after growth in M9-succinate to stationary phase, the AB5075 ΔcsrA mutant lost almost all viability over 6 days (Fig. 2B). The desiccation phenotype was complemented by expressing csrA in trans from a plasmid. ΔcsrA mutant cells incubated for 6 days after being filtered and resuspended in phosphate-buffered saline (PBS) remained fully viable (Fig. 2B). Consistent with this finding, Farrow et al. reported that ΔcsrA mutants of strains ATCC 17961 and AB09-003 did not survive 14 days drying at 40% RH as well as their wild-type (WT) parents (25).
FIG 2.
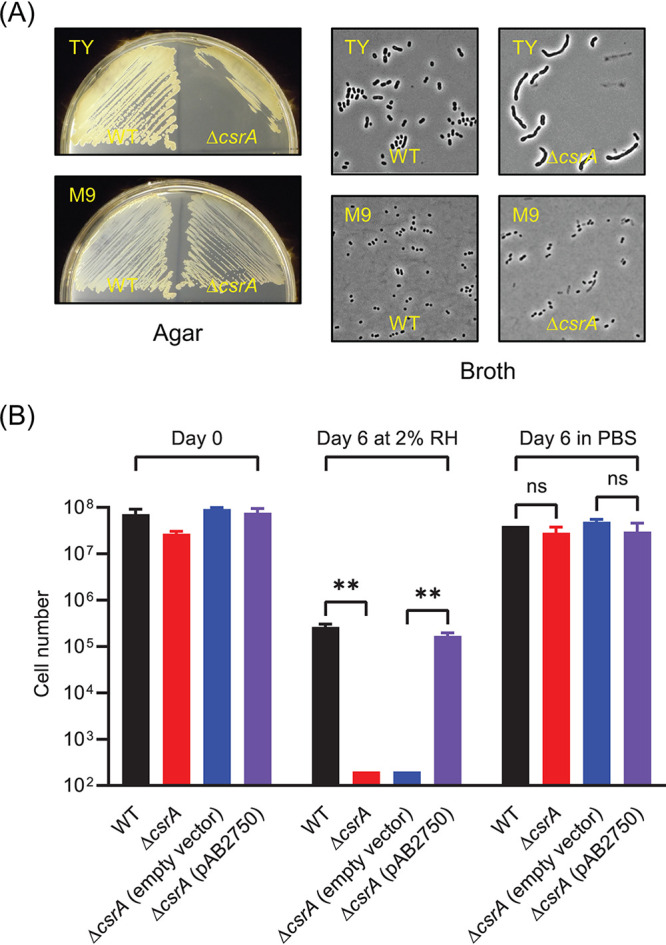
Growth and desiccation tolerance of a ΔcsrA mutant. (A) Comparison of growth of wild type (WT) and a ΔcsrA mutant on TY and M9-succinate agar and of morphologies of cells grown in TY and M9-succinate broth. (B) Desiccation tolerance of WT, ΔcsrA mutant, ΔcsrA mutant with empty vector, and ΔcsrA mutant expressing csrA in trans (pAB2750) at day 0 and at day 6 of desiccation at 2% RH or at 6 days in PBS. The cell number represents the total number of viable cells recovered from each membrane. The data are the average of three biological replicates, and standard deviations are shown as error bars. Statistical significance was calculated using a t test as follows: ns , not significant; **, P < 0.005. The detection limit is 200 cells.
FIG 3.
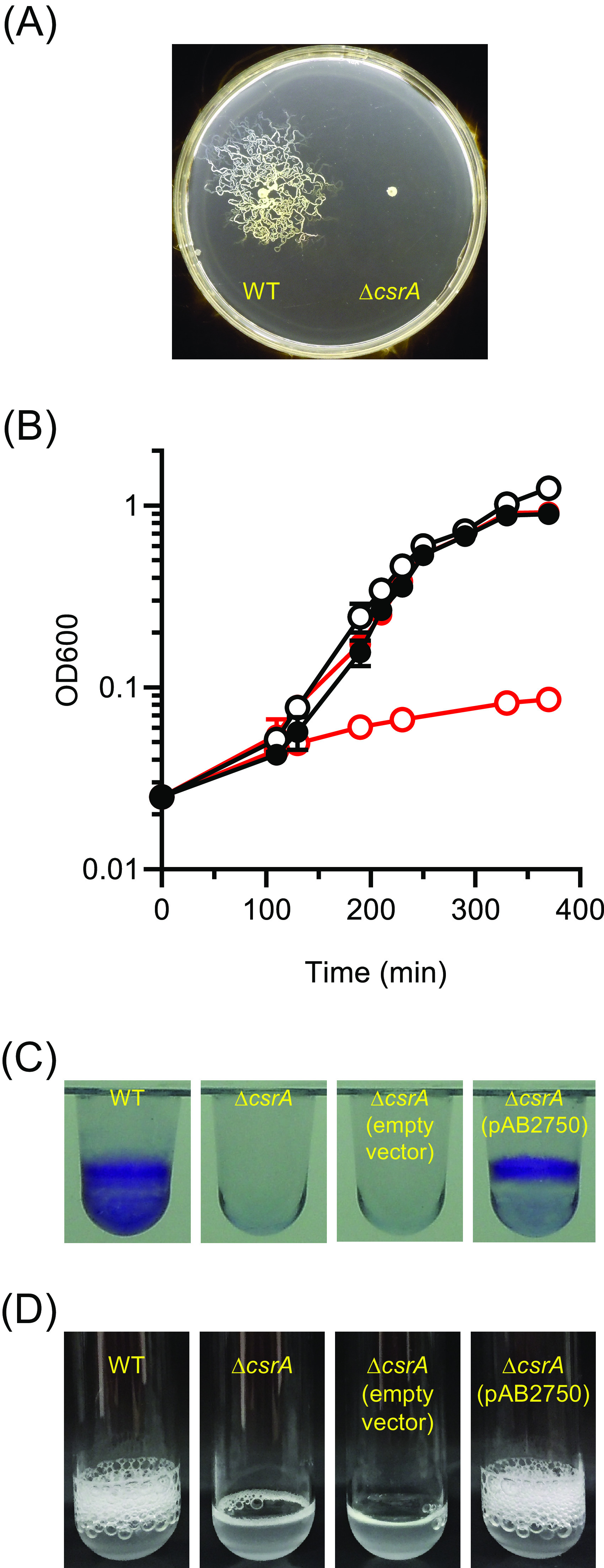
Phenotypes of the ΔcsrA mutant. (A) Twitching motility. (B) Growth of wild type (black symbols) or the ΔcsrA mutant (red symbols) in M9-succinate in the absence (closed symbols) or presence (open symbols) of 0.5% ethanol. The data shown are representative of each strain and condition. (C) Crystal violet staining of biofilms from wild type, ΔcsrA mutant, ΔcsrA mutant with empty vector, and ΔcsrA mutant with pAB2750. (D) Catalase activity of wild type, ΔcsrA mutant, ΔcsrA mutant with empty vector, and ΔcsrA mutant with pAB2750.
Cell processes affected by CsrA in A. baumannii AB5075.
To identify genes whose translation might be regulated by CsrA, we compared the proteomes of wild-type and ΔcsrA cells (Table S1A). There were 97 proteins present at higher levels in the ΔcsrA mutant than the wild type (ratio of ΔcsrA to WT spectral counts ≥ 2.5) (Table S1B). Among these were proteins for type IV pilus assembly, synthesis of the siderophore ferric acinetobactin, and a glutamate/aspartate transporter. The ΔcsrA mutant also had elevated levels of enzymes for the catabolism of hydroxycinnamates, phenylacetate, and quinate. Levels of an alcohol dehydrogenase (ABUW_1621) and an aldehyde dehydrogenase (ABUW_1624) were also elevated. The ΔcsrA mutant was defective in pilus-mediated twitching motility as assessed by movement across a soft agar plate, possibly because an overabundance of pili inhibited twitching (Fig. 3A). The ΔcsrA mutant also had a severe growth defect when grown on succinate in the presence of ethanol, indicating that it is extremely sensitive to this disinfectant (Fig. 3B). One possible explanation for this is that the mutant metabolized ethanol to form toxic acetaldehyde to levels that slowed growth.
There were 106 proteins present in smaller amounts in the ΔcsrA mutant than the wild type (ratio of WT to ΔcsrA spectral counts ≥ 2.5) (Table S1C). A large proportion of these (39%) are annotated as hypothetical proteins. Several membrane proteins and proteins annotated as involved in β-lactam antibiotic resistance (ABUW_1194, ABUW_2619, and ABUW_3497), trehalose synthesis (ABUW_3123), and possibly biofilm formation (ABUW_0916) were in lower abundance in the ΔcsrA mutant than the wild type. As reported previously by Farrow et al., ΔcsrA mutants of strains ATCC 179612 and AB09-003 did not form biofilms (25). We found this to also be the case for strain AB5075, and this phenotype was complemented by expressing the csrA gene in trans (Fig. 3C). The ΔcsrA proteome profile also suggested that CsrA is involved in promoting the expression of proteins involved in oxidative stress, including peroxidase (ABUW_0628) and catalase (katE, ABUW_2436). Indeed, the ΔcsrA mutant lacked detectable catalase activity (Fig. 3D).
Genes important for desiccation tolerance in A. baumannii AB5075.
We took advantage of the three-allele transposon library to test how important some of the gene transcripts that are likely to be controlled by CsrA were for desiccation tolerance (Table 1 and Table S2). We tested two different mutant alleles (transposon insertions in different positions of the gene) for each gene. A. baumannii AB5075 produces opaque and translucent colony variants that interconvert at high frequency and reflect changes in the thickness of capsular exopolysaccharide (29). AB5075 cells with decreased capsule production are about 2.5-fold more sensitive to desiccation (30). Here, we used only opaque colonies of AB5075 and its mutant derivatives in our desiccation assays. Cell cultures in stationary phase were plated at the time of each desiccation assay to confirm that all colonies were opaque.
TABLE 1.
Genes that contribute to desiccation tolerance of A. baumannii AB5075
| Gene | Gene name | Gene annotation | Loss of viability at day 6 relative to WTa | Protein abundance, ratio WT/ΔcsrAb |
|---|---|---|---|---|
| AB5075 (WT) | 1.0 | |||
| ABUW_0916 | Biofilm-associated protein | 1.5 | 2.6 | |
| ABUW_2433 | KGG domain-containing protein | 125 | 4.3 | |
| ABUW_2436 | katE | Catalase | 7.5 | 3.2 |
| ABUW_2437 | Heme oxygenase-like protein | 150 | 10.5 | |
| ABUW_2639 | Universal stress protein family | 6.0 | 4.4 | |
| ABUW_2724 | Hypothetical protein | 4.0 | 2.4 | |
| ABUW_2750 | csrA | Carbon storage regulator | 480 | 8.5 |
Ratio of the viability the wild type at day 6 of desiccation at 2% RH to the viability of each mutant at day 6 of desiccation at 2% RH. Viability was determined as colony-forming units. The viability of the WT is set at 1. All relative losses of viability have P values of <0.05 as computed in Table S2 in the supplemental material. All mutants except the ΔcsrA mutant (grown in M9-succinate) were grown in TSB broth to the stationary phase of growth prior to being desiccated. To compute the relative loss of viability of the csrA mutant, WT was grown in M9-succinate.
Data represent protein abundance as spectral counts.
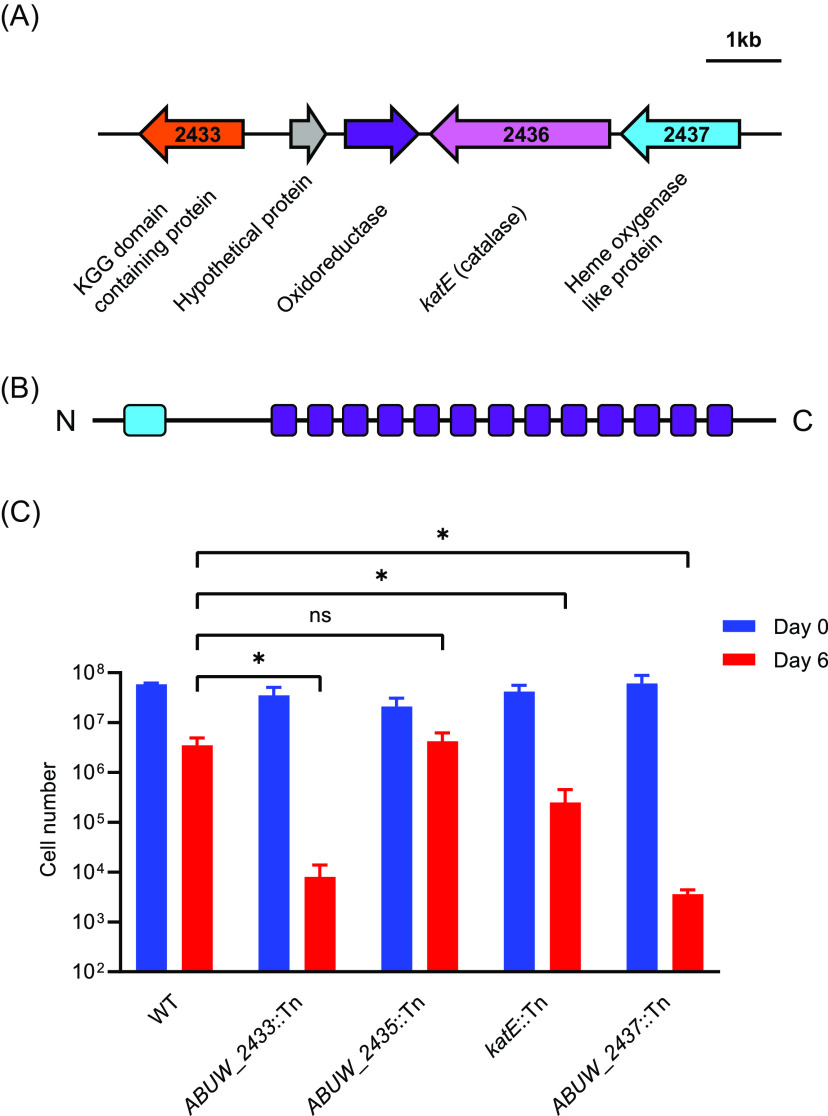
We found that katE, ABUW_2639, and ABUW_2724 mutants were about 5-fold more sensitive to desiccation than the wild type (Table 1; Table S2). ABUW_2639 is annotated as belonging to universal stress protein A family. It and katE have been shown to protect A. baumannii ATCC 17987 from hydrogen peroxide toxicity (31). ABUW_2724 is annotated as a 114-amino-acid hypothetical protein that has a prokaryotic lipoprotein lipid attachment site. We also identified ABUW_0916, encoding a biofilm-associated protein as having a very mild desiccation phenotype. As noted above, biofilms have previously been shown to be important for desiccation tolerance of A. baumannii (15, 16).
ABUW_2433 and ABUW_2437 mutants were greater than 100-fold more sensitive to desiccation than the wild type (Table 1; Fig. 4), and these phenotypes could be partially complemented (Fig. S2). The ABUW_2433 protein has 411 amino acids and is annotated as a KGG domain-containing protein. The full-length protein is predicted to be intrinsically unstructured when queried with the IUPred3 tool (https://iupred.elte.hu/) (32). The single KGG domain (PF10685) comprises 10 amino acids in the N terminus of the protein. The remainder of the protein has a series of AT_hook DNA binding motifs (SM00384). ABUW_2437 is annotated as an iron-containing redox enzyme or a heme-oxygenase-like protein (Fig. 4). The predicted ABUW_2437 transcript has traits characteristic of a target of CsrA posttranscriptional regulation. The DNA sequence predicts a relatively long 5′ untranslated region (316 bp), and there is a predicted CsrA binding motif (GGA) in the ribosome binding site of the transcript.
FIG 4.
A. baumannii genes important for desiccation tolerance. (A) Gene cluster. (B) Diagram of ABUW_2433 (411 amino acids) showing predicted KGG domain (light blue) and AT-hook domains (purple). The full-length protein is predicted to be intrinsically disordered. (C) Desiccation tolerance of mutants at 2% RH. The cell number represents the total number of viable cells recovered from each membrane. The data are the average of three biological replicates, and standard deviations are shown as error bars. Statistical significance was calculated using a t test as follows: ns , not significant; *, P < 0.05. The detection limit is 200 cells.
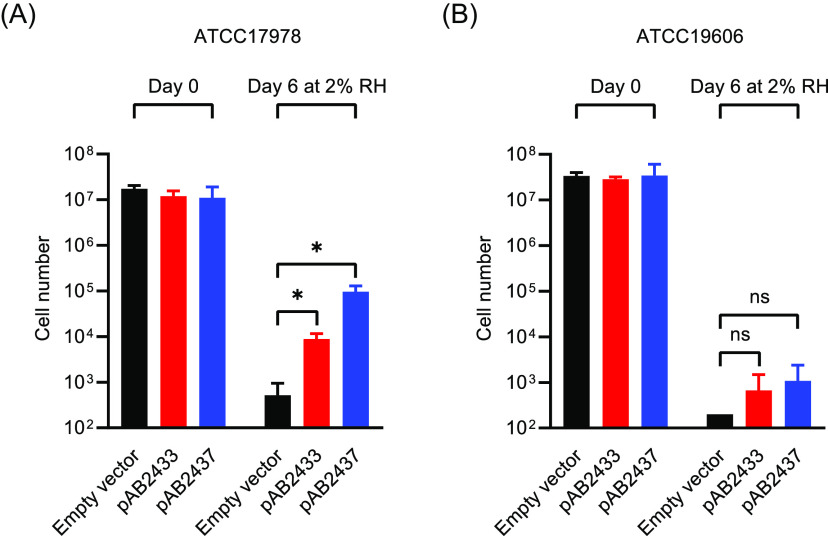
We wondered if ABUW_2433 and ABUW_2437 might play a role in promoting desiccation tolerance of the two A. baumannii strains, ATCC 17978 and ATCC 19606, that do not survive well at 2% RH (Fig. 1B). ATCC 19606 has the gene region shown in Fig. 4 intact, but the gene that is homologous to ABUW_2433, encoding the KGG domain-containing protein, is annotated as a pseudogene. ATCC 17978 appears to be missing a gene homologous to ABUW_2433. However, it has contiguous katE and iron-containing redox protein genes (A1S_1386 and A1S_1385). Expression of the two AB5075 genes in trans improved the survival of ATCC 17978 at 2% RH (Fig. 5), but the changes in viability that were observed for ATCC 19606 were not statistically significant.
FIG 5.
Effect of expression of ABUW_2433 (red bars) or ABUW_2337 (blue bars) genes in trans on desiccation tolerance of A. baumannii ATCC 17978 (A) and ATCC 19606 (B) after 0 days (control) and 6 days of desiccation at 2% RH. Empty vector (black bars) was used as a control. The data are the average of three biological replicates, and standard deviations are shown as error bars. Statistical significance was calculated using a t test as follows: ns , not significant; *, P < 0.05. The detection limit is 200 cells.
DISCUSSION
Depletion of water during desiccation leads to loss of membrane integrity, and accompanying disruption of aerobic respiration results in the generation of reactive oxygen species, including hydrogen peroxide (33). Therefore, it makes sense that katE, encoding catalase, contributes to desiccation tolerance. Previous proteomics analyses of A. baumannii showed that proteins involved in redox defense, including catalase, alkyl peroxidase reductases, and superoxide dismutase, were elevated in stationary-phase cells (34), which is consistent with the observation made by many that stationary-phase cells survive desiccation much better than exponentially growing cells.
Since the desiccation-tolerance genes ABUW_2433 and ABUW_2437 are near or adjacent to katE on the genome and all are likely to be controlled by CsrA, it seemed important to consider that they might somehow mediate oxidative stress tolerance even though the amino acid sequences of the encoded proteins do not have motifs typically associated with reactive oxygen species detoxification. However, in work not shown here, we found that ABUW_2433::Tn and ABUW_2437::Tn mutants were not more sensitive than the wild type to hydrogen peroxide or paraquat, both powerful oxidizing agents. In addition, a study that looked at effects of hydrogen peroxide exposure on gene expression in A. baumannii found that katE, but not ABUW_2433 or ABUW_2437, was expressed at elevated levels, and neither of these genes is part of the OxyR regulon that controls the response to oxidative stress in A. baumannii (35). Thus, we do not think that either of these proteins, which have the greatest defects in desiccation found to date, is likely to function by protecting cells against oxidative stress.
The KGG motif found in ABUW_2433 is also present in some bacterial stress-induced proteins (36, 37) but, to date, has not been directly implicated in protecting against stress. In future work, it will be of interest to determine if this domain contributes to desiccation tolerance phenotype of ABUW_2433. The predicted physical properties of ABUW_2433 provide suggestions as to how it may function. It is an IDP that is highly hydrophilic, with 27% positively charged amino acids residues and 31% negatively charged residues and is predicted to assume a dynamic collapsed or extended conformation, likely depending on its context (Robetta PFRMAT TS prediction; https://robetta.bakerlab.org/). ABUW_2433 has 13 repeated AT-hook DNA binding motifs that extend across about 70% of the protein. This motif preferentially binds to AT-rich sequences in the minor groove of DNA. AT-hook DNA binding motifs are found primarily in eukaryotic proteins, many of which have roles in transcriptional regulation (38, 39). Only 8.5% of annotated AT-hook DNA binding motifs are found in bacteria, and about half of these are found in Gammaproteobacteria, the group to which A. baumannii belongs. We hypothesize that ABUW_2433 binds to A. baumannii DNA to protect it from desiccation-induced damage. IDPs are critical for the microscopic animals called tardigrades to survive desiccation. When desiccated, some of these proteins vitrify and probably trap desiccation-sensitive molecules in a noncrystalline amorphous matrix, thereby protecting them from denaturation or other forms of destruction (40, 41). In plant seeds, an intrinsically disordered regulatory protein has been shown to phase separate upon seed rehydration, allowing it to act as a water sensor (42). IDPs or proteins with IDP domains are much less common in prokaryotes than in eukaryotes, but drawing from work with eukaryotes, they are proposed to play a central role in cellular processes in bacteria that may depend on the formation of molecular condensates (43). It is possible that this is important for the viability of desiccated A. baumannii.
We found that A. baumannii AB5075 survived desiccation for 6 days at 2% RH much better than two other A. baumannii strains that we tested. However, it is important to note that most studies of desiccation tolerance have been carried out at about 30% RH or in room air, and the emphasis has been on the number of days or months that a particular strain remains viable when desiccated. When Farrow et al. (20) tested the survival of several strains that were dried and incubated at an RH of 25 to 61% (mean, 46%), they found AB5075 to have an average survival time of 90 days, whereas strains ATCC 19606 and ATCC 17978 had average survival times of 3 and 34 days, respectively. Even though AB5075 is tolerant to desiccation over months at a mean RH of 46% and for days at 2% RH, we cannot necessarily conclude that the same sets of genes are needed for desiccation tolerance under these two different conditions. For example, Farrow et al. (20) found that the response-regulator protein BmfR was important for desiccation tolerance of ATCC 17978 in long-term desiccation assays, whereas we did not observe a role for bmfR in protecting AB5075 from desiccation stress in shorter-term incubations at 2% RH (Table S2 in the supplemental material).
In conclusion, here, we established a robust assay for desiccation tolerance of a highly virulent strain of A. baumannii and identified two genes, ABUW_2433 and ABUW_2437, that are extremely important for desiccation tolerance. Outside of the Acinetobacter genus, ABUW_2437 has homologs in Pseudomonas stutzeri (40% amino acid identity), but ABUW_2433 does not appear to be present in bacteria other than Acinetobacter. This, and the unusual predicted physical properties of ABUW_2433 as an IDP and possible DNA binding protein, suggest that there is a novel physiological basis for desiccation tolerance in A. baumannii that remains to be explored.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Strains used in this study are listed in Table S3A. Strain AB5075 (AB5075-UW) was used as a wild type (23), and individual transposon mutants were obtained from the Manoil lab comprehensive ordered transposon mutant library at the University of Washington (23). The Tn derivative used to construct this library has an outwardly facing promoter at its 3′ end and thus does not have polar effects on downstream genes. All strains except for the ΔcsrA mutant were routinely grown and maintained in TY medium (10 g tryptone, 5 g yeast extract, and 8 g NaCl in 1,000 mL distilled water) or BBL trypticase soy broth (TSB) medium at 37°C unless otherwise stated. The ΔcsrA mutant was grown in M9 minimal medium with 10 mM succinate as a sole carbon source.
Desiccation assay.
Strains from a frozen stock (−80°C) were streaked onto TY plates and incubated at 37°C. Colonies (three to five) were picked and inoculated into 2 mL of TSB, and cultures were grown overnight at 37°C with a shaking speed of 200 rpm. Overnight cultures were diluted to yield an initial optical density at 600 nm (OD600) of 0.025 in 10 mL TSB in a 50-mL Erlenmeyer flask. Cultures were grown at 37°C with a shaking speed of 200 rpm to mid-log phase (OD600, 0.4 to 0.6) or stationary phase (24 h after inoculation). Cells were harvested by centrifugation and washed twice with Dulbecco's phosphate-buffered saline (DPBS; Gibco), and cell density was adjusted to an OD600 of 1 (about 5 × 108 cells/mL) with DPBS. Cell suspensions (2 spots of 50 μL each per membrane) were filtered onto 0.4-μm Whatman nucleopore polycarbonate track-etched membranes (25 mm diameter) that had been placed in Nalgene analytical filter units, and the membranes were then placed into 15-mL uncapped centrifuge tubes. To obtain the day 0 viable cell number, 1 mL of DPBS was immediately added to one tube and incubated for 5 min at room temperature (24 ± 2°C) on a rotary shaker. Viable cell numbers were determined by plating on TSB agar. For desiccation, tubes with membranes were placed in Snapware containers (2.3 by 6.3 by 8.4 in.) that contained Drierite in the lids of 50-mL centrifuge tubes (×4, 7.5 g of Drierite desiccant in each lid) or saturated calcium chloride hexahydrate solution in a 5-mL beaker (×8) to yield the RHs of 2% or 30% (±2), respectively. The Snapware containers were incubated at room temperature. Digital hygrometers (VWR International Ltd.; catalog no. 35519-045) were placed in each container to monitor the RH. At desired time points, tubes containing membranes were removed from the containers, and 1 mL of DPBS was added to each tube and incubated for 5 min at room temperature on a rotary shaker. Viable cell counts were determined on TSB agar. The wild type and ΔcsrA mutant desiccation data presented are for three biological replicates performed on different days (Table 1 and Table S2 in the supplemental material). We did not perform technical replicates. For Tn mutants that we tested from the three-allele library, we tested two different mutant alleles (Tn insertions in two different locations) for each gene (Table S3). Data are presented for one assay of one allele and two biological replicates of the second allele. Assays were performed on different days. The choice of allele for a given gene was random. Specific allele data are available on request. P values for the differences in viabilities of WT and mutants after 6 days of desiccation are presented in Table S2.
Construction of the ΔcsrA mutant.
In-frame deletion of the csrA (ABUW_2750) gene was generated by overlap extension PCR as described (44). PCR primers are listed in Table S3B. PCR product was cloned into mobilizable suicide vector pEX2-TetRA and transformed into E. coli NEB 10-beta (New England Biolabs). The sequence-verified deletion construct was transformed into E. coli S17-1 and further mobilized into A. baumannii strain AB5075 by conjugation on TY agar. Single recombinant conjugants were first selected on an M9-succinate plate containing 20 μg/mL tetracycline (Tc), and Tc-resistant colonies were further plated onto an M9-succinate plate containing 5% sucrose. Sucrose-resistant and Tc-sensitive colonies were screened by colony PCR and sequencing to validate the expected chromosomal in-frame deletion of the csrA gene.
To complement the ΔcsrA mutant, the full-length csrA gene plus the 15 bp upstream that contains the putative ribosome binding site were PCR amplified and cloned into pMMB67EH-TetRA. The construct was transformed into E. coli NEB 10-beta (New England Biolabs). The sequence-verified construct was transformed into E. coli S17-1 and further mobilized into the ΔcsrA mutant by conjugation on M9-succinate plates containing 20 μg/mL Tc. As a negative control, empty vector pMMB67EH-TetRA was used. The same procedure was used to clone ABUW_2433 and ABUW_2437 for complementation experiments.
Phenotypic characterization of the ΔcsrA mutant.
M9-succinate was used in all experiments. For twitching assays, an overnight culture (16 to 18 h) was diluted to yield OD600 of 0.5, and 2 μL of sample was spotted onto the freshly prepared M9-succinate plate and incubated at 37°C for 24 h. For biofilm assays, an overnight culture was inoculated into 100 μL of M9-succinate in Costar vinyl 96 well U-bottom plates (initial OD600, 0.05), and the plates were sealed with Breath-Easy sealing membranes. After incubation at room temperature for 48 h, culture was removed, the plate was rinsed with tap water twice, and 150 μL of 0.1% crystal violet solution was added to each well. After incubating at room temperature for 15 min, crystal violet solution was removed, the plate was rinsed with tap water 5 times, and the plate was dried at room temperature. For catalase assays, cells were harvested at an OD600 of 0.5, supernatant was removed, and cells were resuspended in DPBS to yield 10 mg wet cell/100 μL DPBS. We placed 100 μL of cell suspensions in 13- by 100-mm borosilicate glass tubes. Then, 100 μL of 1% Triton X-100 and 100 μL of 30% hydrogen peroxide were added, mixed thoroughly, and incubated for 15 min at room temperature (45). Tubes were monitored for production of bubbles, which is indicative of catalase activity.
Label-free protein quantification.
Since the ΔcsrA mutant had a severe growth defect on TY medium, both the wild type and ΔcsrA mutant were grown in M9-succinate. Two biological sample replicates were prepared for each strain. Cells from each culture were harvested at OD600 of 0.5 by centrifugation and washed twice with DPBS, and cells were stored at −80°C before further analysis. Cells were lysed in buffer containing 4% SDS, 100 mM Tris, pH 8.0, and 10 mM dithiothreitol (DTT) by heating at 95°C for 5 min. After cooling to room temperature, the lysates were sonicated with ultrasonication probe on ice to shear DNA. Total protein concentration was determined by the bicinchoninic acid (BCA) assay (Thermo Pierce, Rockford, IL). Five hundred micrograms of each protein lysate were reduced and alkylated and diluted in 8-M urea solution, and the SDS was removed with a 3-kDa-molecular-weight-cutoff filter. After buffer exchange, the protein lysates were digested with trypsin (Promega, Madison, WI) at 37°C overnight, and the digested samples were desalted with 1-cc C18 Sep-Pak solid-phase extraction cartridges (Waters, Milford, MA). The eluted samples were vacuum dried and resuspended in 0.1% formic acid. Reverse-phase nano-liquid chromatography-mass spectrometry (LCMS) analysis of the protein digests was carried out with a Thermo Easy-nLC coupled to a Thermo Q-Exactive Plus Orbitrap mass spectrometer. Triplicate top 20 data-dependent acquisition runs were acquired for each sample, and 1 μg of protein digest was loaded for each run. The peptides were separated by a 50-cm by 75-μm I.D. C8 column (5 μm diameter, 100-Å-pore-size C8 MichromMagic beads) with a 90-min 10% to 30% B gradient (solvent A, 0.1% formic acid in water; solvent B, 0.1% formic acid in acetonitrile; flow rate, 300 nL/min). The MS data acquisition parameters were set as follows: full MS scan resolution, 70K, maximum ion injection time, 100 mS, AGC target, 106; scan range of 400 to 2,000 m/z; MS/MS scan resolution, 17.5K; maximum ion injection time, 100 mS; AGC target, 54; isolation window, 1.6 m/z; HCD NCE 35 scan range of 200 to 2,000 m/z; loop count, 20; intensity threshold, 53; underfill ratio, and 1%; dynamic exclusion, 10 s. High-resolution MS/MS spectra were searched against a target-decoy proteome database of strain AB5075 (a total of 7,678 sequences) downloaded from UniProt (17 October 2017) using Comet (version 2015.02 rev. 1) (46) with the following parameters: precursor peptide mass tolerance, 20 ppm, allowing for −1, 0, +1, +2, or +3 13C offsets; fragment ion mass tolerance, 0.02 Da; static modification, carbamidomethylation of cysteine (57.0215 Da); and variable modification, methionine oxidation (15.9949 Da). The search results were further processed by PeptideProphet (47) for probability assignment to each peptide-spectrum match and ProteinProphet (48) for protein inference and protein probability modeling. The output pepXML files from three technical replicates were grouped for subsequent spectral counting analysis using Abacus (49). The pepXML and protXML files for each sample and combined ProteinProphet file from all samples were parsed into Abacus for spectral counting of each protein. The following filters were applied for extracting spectral counts from MS/MS data sets: (i) the minimum PeptideProphet score that is the best peptide match of a protein must have maxIniProbTH of 0.99; (ii) the minimum PeptideProphet score a peptide must have to be even considered by Abacus is iniProbTH of 0.50; and (iii) the minimum ProteinProphet score a protein group must have in the COMBINED file is minCombinedFilePw of 0.90. Spectral counts for 1,616 proteins were reported across four sample groups (two strains and two biological replicates) with estimated protein false-discovery rate of 1.94%. The protein expression fold changes between wild-type AB5075 and the ΔcsrA mutant were computed from adjusted spectral counts output from Abacus.
Data availability.
The mass spectrometry proteomics data have been deposited to ProteomeXchange Consortium via the PRIDE partner repository with the data set identifier PXD028391.
ACKNOWLEDGMENTS
We thank Hemantha Don Kulasekara and Samuel Miller (University of Washington) for sharing the pEX2-TetRA and pMMB67EH-TetRA vectors. We thank Indranil Biswas for alerting us to the possibility that intrinsically disordered proteins could be involved in desiccation tolerance in A. baumannii.
This work was supported by the Functional Genomics Program, National Institute of Allergy and Infectious Diseases, under grant 1U19AI107775-01.
Footnotes
[This article was published on 14 March 2022. A Table 1 boxhead was updated in the current version, posted on 28 March 2022.]
Supplemental material is available online only.
Contributor Information
Caroline S. Harwood, Email: csh5@uw.edu.
Michael Y. Galperin, NCBI, NLM, National Institutes of Health
REFERENCES
- 1.Magill SS, Edwards JR, Bamberg W, Beldavs ZG, Dumyati G, Kainer MA, Lynfield R, Maloney M, McAllister-Hollod L, Nadle J, Ray SM, Thompson DL, Wilson LE, Fridkin SK, Emerging Infections Program Healthcare-Associated Infections and Antimicrobial Use Prevalence Survey Team. 2014. Multistate point-prevalence survey of health care-associated infections. N Engl J Med 370:1198–1208. 10.1056/NEJMoa1306801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Marchetti A, Rossiter R. 2013. Economic burden of healthcare-associated infection in US acute care hospitals: societal perspective. J Med Econ 16:1399–1404. 10.3111/13696998.2013.842922. [DOI] [PubMed] [Google Scholar]
- 3.Datta R, Platt R, Yokoe DS, Huang SS. 2011. Environmental cleaning intervention and risk of acquiring multidrug-resistant organisms from prior room occupants. Arch Intern Med 171:491–494. 10.1001/archinternmed.2011.64. [DOI] [PubMed] [Google Scholar]
- 4.Huang SS, Datta R, Platt R. 2006. Risk of acquiring antibiotic-resistant bacteria from prior room occupants. Arch Intern Med 166:1945–1951. 10.1001/archinte.166.18.1945. [DOI] [PubMed] [Google Scholar]
- 5.Munoz-Price LS, Namias N, Cleary T, Fajardo-Aquino Y, Depascale D, Arheart KL, Rivera JI, Doi Y. 2013. Acinetobacter baumannii: association between environmental contamination of patient rooms and occupant status. Infect Control Hosp Epidemiol 34:517–520. 10.1086/670209. [DOI] [PubMed] [Google Scholar]
- 6.Thom KA, Johnson JK, Lee MS, Harris AD. 2011. Environmental contamination because of multidrug-resistant Acinetobacter baumannii surrounding colonized or infected patients. Am J Infect Control 39:711–715. 10.1016/j.ajic.2010.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Harding CM, Hennon SW, Feldman MF. 2018. Uncovering the mechanisms of Acinetobacter baumannii virulence. Nat Rev Microbiol 16:91–102. 10.1038/nrmicro.2017.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Giannouli M, Antunes LC, Marchetti V, Triassi M, Visca P, Zarrilli R. 2013. Virulence-related traits of epidemic Acinetobacter baumannii strains belonging to the international clonal lineages I-III and to the emerging genotypes ST25 and ST78. BMC Infect Dis 13:282. 10.1186/1471-2334-13-282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jawad A, Seifert H, Snelling AM, Heritage J, Hawkey PM. 1998. Survival of Acinetobacter baumannii on dry surfaces: comparison of outbreak and sporadic Isolates. J Clin Microbiol 36:1938–1941. 10.1128/JCM.36.7.1938-1941.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Antunes LCS, Imperi F, Carattoli A, Visca P. 2011. Deciphering the multifactorial nature of Acinetobacter baumannii pathogenicity. PLoS One 6:e22674. 10.1371/journal.pone.0022674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wendt C, Dietze B, Dietz E, Rüden H. 1997. Survival of Acinetobacter baumannii on dry surfaces: comparison of outbreak and sporadic isolates. J Clin Microbiol 35:1394–1397. 10.1128/jcm.35.6.1394-1397.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lebre PH, Maayer PD, Cowan DA. 2017. Xerotolerant bacteria: surviving through a dry spell. Nat Rev Microbiol 15:285–296. 10.1038/nrmicro.2017.16. [DOI] [PubMed] [Google Scholar]
- 13.Potts M. 1994. Desiccation tolerance of prokaryotes. Microbiol Rev 58:755–805. 10.1128/mr.58.4.755-805.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gayoso CM, Mateos J, Méndez JA, Fernández-Puente P, Rumbo C, Tomás M, de Ilarduya ÓM, Bou G. 2014. Molecular mechanisms involved in the response to desiccation stress and persistence in Acinetobacter baumannii. J Proteome Res 13:460–476. 10.1021/pr400603f. [DOI] [PubMed] [Google Scholar]
- 15.Espinal P, Marti S, Vila J. 2012. Effect of biofilm formation on the survival of Acinetobacter baumannii on dry surfaces. J Hosp Infect 80:56–60. 10.1016/j.jhin.2011.08.013. [DOI] [PubMed] [Google Scholar]
- 16.Orsinger-Jacobsen SJ, Patel SS, Vellozzi EM, Gialanella P, Nimrichter L, Miranda K, Martinez LR. 2013. Use of a stainless steel washer platform to study Acinetobacter baumannii adhesion and biofilm formation on abiotic surfaces. Microbiology (Reading) 159:2594–2604. 10.1099/mic.0.068825-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang X, Cole CG, DuPai CD, Davies BW. 2020. Protein aggregation is associated with Acinetobacter baumannii desiccation tolerance. Microorg 8:343. 10.3390/microorganisms8030343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Boll JM, Tucker AT, Klein DR, Beltran AM, Brodbelt JS, Davies BW, Trent MS. 2015. Reinforcing lipid A acylation on the cell surface of Acinetobacter baumannii promotes cationic antimicrobial peptide resistance and desiccation survival. mBio 6:e00478-15. 10.1128/mBio.00478-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Aranda J, Bardina C, Beceiro A, Rumbo S, Cabral MP, Barbé J, Bou G. 2011. Acinetobacter baumannii RecA protein in repair of DNA damage, antimicrobial resistance, general stress response, and virulence. J Bacteriol 193:3740–3747. 10.1128/JB.00389-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Farrow JM, Wells G, Pesci EC. 2018. Desiccation tolerance in Acinetobacter baumannii is mediated by the two-component response regulator BfmR. PLoS One 13:e0205638. 10.1371/journal.pone.0205638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zeidler S, Müller V. 2019. Coping with low water activities and osmotic stress in Acinetobacter baumannii: significance, current status and perspectives. Environ Microbiol 21:2212–2230. 10.1111/1462-2920.14565. [DOI] [PubMed] [Google Scholar]
- 22.Jacobs AC, Thompson MG, Black CC, Kessler JL, Clark LP, McQueary CN, Gancz HY, Corey BW, Moon JK, Si Y, Owen MT, Hallock JD, Kwak YI, Summers A, Li CZ, Rasko DA, Penwell WF, Honnold CL, Wise MC, Waterman PE, Lesho EP, Stewart RL, Actis LA, Palys TJ, Craft DW, Zurawski DV. 2014. AB5075, a highly virulent isolate of Acinetobacter baumannii, as a model strain for the evaluation of pathogenesis and antimicrobial treatments. mBio 5:e01076-14. 10.1128/mBio.01076-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gallagher LA, Ramage E, Weiss EJ, Radey M, Hayden HS, Held KG, Huse HK, Zurawski DV, Brittnacher MJ, Manoil C. 2015. Resources for genetic and genomic analysis of emerging pathogen Acinetobacter baumannii. J Bacteriol 197:2027–2035. 10.1128/JB.00131-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lapouge K, Schubert M, Allain FH-T, Haas D. 2008. Gac/Rsm signal transduction pathway of gamma-proteobacteria: from RNA recognition to regulation of social behaviour. Mol Microbiol 67:241–253. 10.1111/j.1365-2958.2007.06042.x. [DOI] [PubMed] [Google Scholar]
- 25.Farrow JM, Wells G, Palethorpe S, Adams MD, Pesci EC. 2020. CsrA supports both environmental persistence and host-associated growth of Acinetobacter baumannii. Infect Immun 88:e00259-20. 10.1128/IAI.00259-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zeidler S, Müller V. 2019. The role of compatible solutes in desiccation resistance of Acinetobacter baumannii. Microbiologyopen 8:e00740. 10.1002/mbo3.740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Borgio JF, Rasdan AS, Sonbol B, Alhamid G, Almandil NB, AbdulAzeez S. 2021. Emerging status of multidrug-resistant bacteria and fungi in the Arabian Peninsula. Biology 10:1144. 10.3390/biology10111144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hubloher JJ, Schabacker K, Müller V, Averhoff B. 2021. CsrA coordinates compatible solute synthesis in Acinetobacter baumannii and facilitates growth in human urine. Microbiol Spectr 9:e01296-21. 10.1128/Spectrum.01296-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chin CY, Tipton KA, Farokhyfar M, Burd EM, Weiss DS, Rather PN. 2018. A high-frequency phenotypic switch links bacterial virulence and environmental survival in Acinetobacter baumannii. Nat Microbiol 3:563–569. 10.1038/s41564-018-0151-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tipton KA, Chin C-Y, Farokhyfar M, Weiss DS, Rather PN. 2018. Role of capsule in resistance to disinfectants, host antimicrobials, and desiccation in Acinetobacter baumannii. Antimicrob Agents Chemother 62:e01188-18. 10.1128/AAC.01188-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Elhosseiny NM, Amin MA, Yassin AS, Attia AS. 2015. Acinetobacter baumannii universal stress protein A plays a pivotal role in stress response and is essential for pneumonia and sepsis pathogenesis. Int J Med Microbiol 305:114–123. 10.1016/j.ijmm.2014.11.008. [DOI] [PubMed] [Google Scholar]
- 32.Erdős G, Pajkos M, Dosztányi Z. 2021. IUPred3: prediction of protein disorder enhanced with unambiguous experimental annotation and visualization of evolutionary conservation. Nucleic Acids Res 49:W297–W303. 10.1093/nar/gkab408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.França MB, Panek AD, Eleutherio ECA. 2007. Oxidative stress and its effects during dehydration. Comp Biochem Physiol A Mol Integr Physiol 146:621–631. 10.1016/j.cbpa.2006.02.030. [DOI] [PubMed] [Google Scholar]
- 34.Soares NC, Cabral MP, Gayoso C, Mallo S, Rodriguez-Velo P, Fernández-Moreira E, Bou G. 2010. Associating growth-phase-related changes in the proteome of Acinetobacter baumannii with increased resistance to oxidative stress. J Proteome Res 9:1951–1964. 10.1021/pr901116r. [DOI] [PubMed] [Google Scholar]
- 35.Juttukonda LJ, Green ER, Lonergan ZR, Heffern MC, Chang CJ, Skaar EP. 2019. Acinetobacter baumannii OxyR regulates the transcriptional response to hydrogen peroxide. Infect Immun 87:e00413-18. 10.1128/IAI.00413-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Asteri I-A, Boutou E, Anastasiou R, Pot B, Vorgias CE, Tsakalidou E, Papadimitriou K. 2011. In silico evidence for the horizontal transfer of gsiB, a σ(B)-regulated gene in gram-positive bacteria, to lactic acid bacteria. Appl Environ Microbiol 77:3526–3531. 10.1128/AEM.02569-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Robbe-Saule V, Lopes MD, Kolb A, Norel F. 2007. Physiological effects of Crl in Salmonella are modulated by sigmaS level and promoter specificity. J Bacteriol 189:2976–2987. 10.1128/JB.01919-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Aravind L, Landsman D. 1998. AT-hook motifs identified in a wide variety of DNA-binding proteins. Nucleic Acids Res 26:4413–4421. 10.1093/nar/26.19.4413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Su L, Deng Z, Leng F. 2020. The mammalian high mobility group protein AT-Hook 2 (HMGA2): biochemical and biophysical properties, and its association with adipogenesis. Int J Mol Sci 21:3710. 10.3390/ijms21103710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Boothby TC, Tapia H, Brozena AH, Piszkiewicz S, Smith AE, Giovannini I, Rebecchi L, Pielak GJ, Koshland D, Goldstein B. 2017. Tardigrades use intrinsically disordered proteins to survive desiccation. Mol Cell 65:975–984.e5. 10.1016/j.molcel.2017.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hesgrove C, Boothby TC. 2020. The biology of tardigrade disordered proteins in extreme stress tolerance. Cell Commun Signal 18:178. 10.1186/s12964-020-00670-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dorone Y, Boeynaems S, Flores E, Jin B, Hateley S, Bossi F, Lazarus E, Pennington JG, Michiels E, Decker MD, Vints K, Baatsen P, Bassel GW, Otegui MS, Holehouse AS, Exposito-Alonso M, Sukenik S, Gitler AD, Rhee SY. 2021. A prion-like protein regulator of seed germination undergoes hydration-dependent phase separation. Cell 184:4284–4298.e27. 10.1016/j.cell.2021.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cohan MC, Pappu RV. 2020. Making the case for disordered proteins and biomolecular condensates in bacteria. Trends Biochem Sci 45:668–680. 10.1016/j.tibs.2020.04.011. [DOI] [PubMed] [Google Scholar]
- 44.Horton RM, Ho SN, Pullen JK, Hunt HD, Cai Z, Pease LR. 1993. Gene splicing by overlap extension. Methods Enzymol 217:270–279. 10.1016/0076-6879(93)17067-f. [DOI] [PubMed] [Google Scholar]
- 45.Iwase T, Tajima A, Sugimoto S, Okuda K, Hironaka I, Kamata Y, Takada K, Mizunoe Y. 2013. A simple assay for measuring catalase activity: a visual approach. Sci Rep 3:3081. 10.1038/srep03081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Eng JK, Jahan TA, Hoopmann MR. 2013. Comet: an open‐source MS/MS sequence database search tool. Proteomics 13:22–24. 10.1002/pmic.201200439. [DOI] [PubMed] [Google Scholar]
- 47.Keller A, Nesvizhskii AI, Kolker E, Aebersold R. 2002. Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal Chem 74:5383–5392. 10.1021/ac025747h. [DOI] [PubMed] [Google Scholar]
- 48.Nesvizhskii AI, Keller A, Kolker E, Aebersold R. 2003. A statistical model for identifying proteins by tandem mass spectrometry. Anal Chem 75:4646–4658. 10.1021/ac0341261. [DOI] [PubMed] [Google Scholar]
- 49.Fermin D, Basrur V, Yocum AK, Nesvizhskii AI. 2011. Abacus: a computational tool for extracting and pre‐processing spectral count data for label‐free quantitative proteomic analysis. Proteomics 11:1340–1345. 10.1002/pmic.201000650. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Download jb.00479-21-s0001.xlsx, XLSX file, 0.3 MB (349.3KB, xlsx)
Table S2. Download jb.00479-21-s0002.xlsx, XLSX file, 0.01 MB (14.8KB, xlsx)
Table S3. Download jb.00479-21-s0003.xlsx, XLSX file, 0.01 MB (15.4KB, xlsx)
supplemental material. Download jb.00479-21-s0004.pdf, PDF file, 0.03 MB (35.1KB, pdf)
Data Availability Statement
The mass spectrometry proteomics data have been deposited to ProteomeXchange Consortium via the PRIDE partner repository with the data set identifier PXD028391.