ABSTRACT
The multi-antifungal drug-resistant strain (NUBS21012) of Trichophyton rubrum was isolated from a patient with recurrent tinea corporis. The resistant strain encoded Phe at codon 393 instead of Leu (L393F) in the squalene epoxidase (SQLE) gene. The expression of genes encoding ATP-binding cassette transporter proteins increased in the strain compared to that of other strains. This result provides evidence that ATP-binding cassette transporter proteins are closely associated with azole resistance.
KEYWORDS: deletion mutation, onychomycosis, squalene epoxidase, terbinafine, Trichophyton rubrum, azole, dermatophytes, dermatophytoses
INTRODUCTION
Dermatophytoses are common fungal skin infections in humans (1–3). More than 60% to 70% of the infections are caused by dermatophytes, predominantly Trichophyton rubrum (50%) (3). Terbinafine (TRF) has been used as a treatment for dermatophytosis; however, TRF-resistant strains have recently been isolated from human patients around the world (4–7). Almost all TRF-resistant strains encode F397L or L393F amino acid substitutions in the squalene epoxidase (SQLE) gene (4–7).
In this study, we isolated a TRF-resistant strain of T. rubrum from a patient with recurrent tinea corporis. The patient had been treated with TRF and had been switched to ravuconazole (RVZ) for 1 year and 6 months. However, the treatment did not cure the tinea corporis completely, and the systemic skin lesions progressed to fungal granuloma. We show that a susceptible strain can become resistant to azole compounds after overmedication. Switching from TRF to azoles in a case of TRF resistance can generate a strain resistant to TRF and azoles.
Case report.
A 60-year-old female with Cushing’s syndrome had tinea corporis on the face, back, chest, abdomen, buttocks, crotch, and both thighs. She had dermatophytosis for more than 10 years and experienced chronic and recurrent tinea corporis that was treated with TRF hydrochloride cream. Subsequently, in 2020, she experienced tinea corporis on the back that was treated with oral and topical TRF, but the treatment failed. She was then treated with oral fosravuconazole (F-RVZ) and topical luliconazole (Table 1). The first isolate (strain NUBS21011) was obtained from a tinea corporis lesion on the face on 11 March 2020; a L393F mutation was detected in the SQLE gene of this TRF-resistant isolate (Table 1) (8). In this study, an additional strain (NUBS21012) was later isolated from a recurrent tinea corporis lesion on the face on 6 September 2021 (after 1 year and 6 months of oral F-RVZ treatment) (Table 1).
TABLE 1.
Strains, MICs of antifungal drugs, and mutation sites in the SQLE and TERG genesa
| Strain | Origin | Isolation date | Treatment history (administration periods) | MICs (μg/mL) |
Amino acid substitution in SQLEb | Amino acid substitution in TERGc | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| AMB | GRF | ITZ | VRZ | RVZ | TRF | ||||||
| NUBS21011 | Tinea corporis | 11 March 2020 | Oral and topical TRF (1 yr) | 1 | 0.25 | 0.5 | 0.25 | <0.03 | 32 | L393F | Not detected |
| NUBS21012 | Tinea corporis (fungal granuloma) | 6 September 2021 | Oral fosravuconazole and topical LCZ (1 yr 6 mo) | 0.5 | 8 | 16 | 8 | 16 | 32 | L393F | Y136H |
| N45 | Tinea unguium | 8 June 2020 | Unknown | NT | NT | <0.03 | NT | <0.03 | <0.03 | NT | NT |
| N46 | Tinea corporis | 2 June 2020 | Unknown | NT | NT | <0.03 | NT | <0.03 | <0.03 | NT | NT |
NUBS, Nihon University College of Bioresource Sciences; AMB, amphotericin B; GRF, griseofulvin; TRF, terbinafine; ITZ, itraconazole; VRZ, voriconazole; RVZ, ravuconazole; LCZ, luliconazole; SQLE, squalene epoxidase; TERG, cytochrome P450 51; NT, not tested.
Amino acid sequences encoded by the SQLE gene in T. rubrum CBS 118892 (GenBank accession no. XM_003233797).
Amino acid sequences were compared to those encoded by the Trichophyton rubrum CBS 118892 cytochrome P450 51 (TERG_05717) mRNA, complete coding DNA sequence (CDS) (GenBank accession no. XM_003236932.1).
To assess the susceptibility of the isolates to amphotericin B (AMB), griseofulvin (GRF), TRF, itraconazole (ITZ), voriconazole (VRZ), and RVZ, the broth microdilution assay was performed based on the Clinical and Laboratory Standards Institute (CLSI) M38-A2 guidelines with modifications, as previously described (9, 10). The MICs against NUBS21011 and NUBS21012 are listed in Table 1.
To sequence the hot spot mutations in SQLE of the isolates, primers were prepared based on the conserved sequence of the T. rubrum SQLE (see Table S1 in the supplemental material) (8). PCR amplification and sequencing analysis were conducted as reported previously (8). The SQLE gene of the TRF-resistant strains (NUBS21011 and NUBS21012) encoded the L393F mutation (Table 1).
To mRNA sequence the azole target cytochrome P450 51 genes of the clinical isolates (NUBS21011 and NUBS21012), primers were prepared based on the conserved sequence of the Trichophyton rubrum CBS 118892 cytochrome P450 51 (GenBank accession no XM_003236932.1) (Table S1). The cDNA synthesis process and sequencing analysis were done according to a previous report (8). Sequences of the cytochrome P450 51 genes in NUBS21011 and NUBS21012 were deduced to be 1,584 bp long, and they encoded a protein of 528 amino acids. The homology for the gene of NUBS21011 was 100% identical to that of the T. rubrum CBS 118892 cytochrome P450 51. In addition, the Y136H mutation was identified in the gene of NUBS21012 (Table 1).
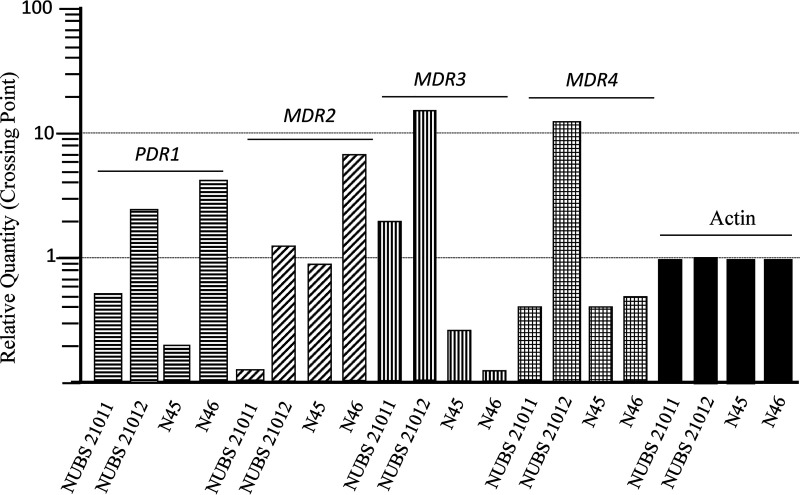
To analyze the expression levels of the ABC transporter family (PDR1, MDR2, MDR3, and MDR4) genes, we performed real-time quantitative PCR (RT-qPCR) as follows. Strains were cultured for 4 days at 28°C in Sabouraud’s dextrose broth (SDB; 1% peptone and 2% dextrose). ITZ exposure was carried out according to the previously reported method of Monod et al., who added ITZ to evaluate the expression of genes in four Trichophyton species (11). All strains were cultured for 3 h at 28°C in SDB with 0.05 μg/mL ITZ. The cDNA synthesis process, RT-qPCR (Thermal Cycler Dice; TaKaRa), and analysis were done as described previously (12). The primer pairs of PDR1, MDR2, MDR3, and MDR4 were used according to previous reports (11, 13). The MDR3 and MDR4 transcript levels were 10 to 100 times higher in NUBS21012 than in the TRF-resistant strain NUBS21011 and antifungal-susceptible strains N45 and N46 (Fig. 1).
FIG 1.
Transcript levels of the actin-encoding gene, PDR1, MDR1, MDR2, MDR3, and MDR4 in TRF-resistant strains (NUBS21011 and NUBS21012) and TRF-susceptible strains (N45 and N46) of T. rubrum. All strains were cultured in SDB containing 0.05 μg/mL ITZ for 3 h at 28°C. Expression levels of all genes were normalized to that of the actin-encoding gene and are expressed as a relative quantity.
It is speculated that the RVZ treatment induced the additional multidrug resistance in the TRF-resistant strain. Therefore, the long-term administration of antifungal agents may lead to the development of multidrug resistance.
Several single-point mutations in the SQLE gene have been identified in TRF-resistant strains of T. rubrum isolated from tinea pedis and tinea unguium around the world, including I121M, V237I, L393F, L393S, F397L, F415S, H440Y, and F484Y in SQLE (4–7). Previously, we performed an epidemiological study of the prevalence of TRF-resistant T. rubrum in Japanese patients (8). All TRF-resistant strains carried the L393F mutation in the SQLE gene (8). Therefore, TRF resistance seems to mainly result from single-point mutations in the SQLE gene.
Resistance to azoles can be linked to the overexpression of genes encoding ABC transporter proteins, including the PDR1, MDR2, MDR3, and MDR4 genes in dermatophytes (11, 12, 14). In this study, the MDR3 and MDR4 transcript levels were 10 to 100 times higher in NUBS21012 than in the TRF-resistant strain (NUBS21011) and antifungal-susceptible strains (N45 and N46). The transcript levels of MDR2 and MDR3 were also increased by the ITZ addition stimulation (11, 13). Our result provides evidence that ABC transporter proteins of MDR3 and MDR4 are closely associated with azole resistance. In addition, the multidrug-resistant strain NUBS21012 was also less susceptive to GRF (MIC, 8 μg/mL) than that of NUBS21011 (MIC, 0.25 μg/mL) (Table 1). MDR3 and MDR4 also might be related to resistance for GRF.
Moreover, Y136H was identified in the P450 51 gene of NUBS21012, but it was not detected in the first isolated strain (NUBS21011). Thus, it has not been reported to demonstrate the gene mutation and azole drug resistance. However, we could not clarify in this study whether this point mutation is related to the drug resistance mechanism.
Data availability.
The sequences determined in this study have been deposited into the GenBank database (GenBank accession no. LC662767 and LC662768).
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Kwon-Chung KJ, Bennett EJ. 1992. Dermatophytoses, p 105–161. In Medical Mycology. Lea & Febiger, Philadelphia, PA. [Google Scholar]
- 2.Reiss E, Shadomy HJ, Lyon I. 2012. Dermatophytosis, p 527–566. In Fundamental medical mycology, Wiley-Blackwell, Hoboken, NJ. [Google Scholar]
- 3.Lipner SR, Scher RK. 2019. Onychomycosis: clinical overview and diagnosis. J Am Acad Dermatol 80:835–851. doi: 10.1016/j.jaad.2018.03.062. [DOI] [PubMed] [Google Scholar]
- 4.Osborne CS, Leitner I, Favre B, Ryder NS. 2005. Amino acid substitution in Trichophyton rubrum squalene epoxidase associated with resistance to terbinafine. Antimicrob Agents Chemother 49:2840–2844. doi: 10.1128/AAC.49.7.2840-2844.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yamada T, Maeda M, Alshahni MM, Tanaka R, Yaguchi T, Bontems O, Salamin K, Fratti M, Monod M. 2017. Terbinafine resistance of Trichophyton clinical isolates caused by specific point mutations in the squalene epoxidase gene. Antimicrob Agents Chemother 61:e00115-17. doi: 10.1128/AAC.00115-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Saunte DML, Hare RK, Jørgensen KM, Jørgensen R, Deleuran M, Zachariae CO, Thomsen SF, Bjørnskov-Halkier L, Kofoed K, Arendrup MC. 2019. Emerging terbinafine resistance in Trichophyton: clinical characteristics, squalene epoxidase gene mutations, and a reliable EUCAST method for detection. Antimicrob Agents Chemother 63:e01126-19. doi: 10.1128/AAC.01126-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Monod M, Méhul B. 2019. Recent findings in onychomycosis and their application for appropriate treatment. J Fungi 5:20. doi: 10.3390/jof5010020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hiruma J, Noguchi H, Hase M, Tokuhisa Y, Shimizu T, Ogawa T, Hiruma M, Harada K, Kano R. 2021. Epidemiological study of terbinafine-resistant dermatophytes isolated from Japanese patients. J Dermatol 48:564–567. doi: 10.1111/1346-8138.15745. [DOI] [PubMed] [Google Scholar]
- 9.Itoi S, Kano R, Hasegawa A, Hasegawa A, Kamata H. 2012. In vitro activities of antifungal agents against clinical isolates of dermatophytes from animals. J Vet Med Sci 74:1067–1069. doi: 10.1292/jvms.12-0057. [DOI] [PubMed] [Google Scholar]
- 10.CLSI. 2008. Reference method for broth dilution antifungal susceptibility testing of filamentous fungi; approved standard, 2nd ed. Document M38-A2. Clinical Laboratory Standards Institute, Philadelphia, PA. [Google Scholar]
- 11.Monod M, Feuermann M, Salamin K, Fratti M, Makino M, Alshahni MM, Makimura K, Yamada T. 2019. Trichophyton rubrum azole resistance mediated by a new ABC transporter, TruMDR3. Antimicrob Agents Chemother 63:e00863. doi: 10.1128/AAC.00863-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kano R. 2021. ATP-binding cassette (ABC) transporter proteins in highly terbinafine-resistant strains of Trichophyton indotineae (Former species name: Trichophyton interdigitale). Med Mycol J 62:21–25. doi: 10.3314/mmj.20-00014. [DOI] [PubMed] [Google Scholar]
- 13.Martins MP, Franceschini AC, Jacob TR, Rossi A, Martinez-Rossi NM. 2016. Compensatory expression of multidrug-resistance genes encoding ABC transporters in dermatophytes. J Med Microbiol 65:605–610. doi: 10.1099/jmm.0.000268. [DOI] [PubMed] [Google Scholar]
- 14.Sacheli R, Hayette MP. 2021. Antifungal resistance in dermatophytes: genetic considerations, clinical presentations and alternative therapies. J Fungi 7:983. doi: 10.3390/jof7110983. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Download aac.02393-21-s0001.pdf, PDF file, 0.08 MB (83.2KB, pdf)
Data Availability Statement
The sequences determined in this study have been deposited into the GenBank database (GenBank accession no. LC662767 and LC662768).