ABSTRACT
Microbial communities occupy diverse niches in nature, and community members routinely exchange a variety of nutrients among themselves. While large-scale metagenomic and metabolomic studies shed some light on these exchanges, the contribution of individual species and the molecular details of specific interactions are difficult to track. In this study, we follow the exchange of vitamin B1 (thiamin) and its intermediates between microbes within synthetic cocultures of Escherichia coli and Vibrio anguillarum. Thiamin contains two moieties, 4-amino-5-hydroxymethyl-2-methylpyrimidine (HMP) and 4-methyl-5-(2-hydroxyethyl)thiazole (THZ), which are synthesized by distinct pathways using enzymes ThiC and ThiG, respectively, and then coupled by ThiE to form thiamin. Even though E. coli ΔthiC, ΔthiE, and ΔthiG mutants are thiamin auxotrophs, we observed that cocultures of ΔthiC-ΔthiE and ΔthiC-ΔthiG mutants are able to grow in a thiamin-deficient medium, whereas the ΔthiE-ΔthiG coculture does not. Further, the exchange of thiamin and its intermediates in V. anguillarum cocultures and in mixed cocultures of V. anguillarum and E. coli revealed that there exist specific patterns for thiamin metabolism and exchange among these microbes. Our findings show that HMP is shared more frequently than THZ, concurrent with previous observations that free HMP and HMP auxotrophy is commonly found in various environments. Furthermore, we observe that the availability of exogenous thiamin in the media affects whether these strains interact with each other or grow independently. These findings collectively underscore the importance of the exchange of essential metabolites as a defining factor in building and modulating synthetic or natural microbial communities.
IMPORTANCE Vitamin B1 (thiamin) is an essential nutrient for cellular metabolism. Microorganisms that are unable to synthesize thiamin either fully or in part exogenously obtain it from their environment or via exchanges with other microbial members in their community. In this study, we created synthetic microbial cocultures that rely on sharing thiamin and its biosynthesis intermediates and observed that some of them are preferentially exchanged. We also observed that the coculture composition is dictated by the production and/or availability of thiamin and its intermediates. Our studies with synthetic cocultures provide the molecular basis for understanding thiamin sharing among microorganisms and lay out broad guidelines for setting up synthetic microbial cocultures by using the exchange of an essential metabolite as their foundation.
KEYWORDS: HMP, nutrient exchange, synthetic microbial coculture, thiamin, thiazole, vitamin B1
INTRODUCTION
Microorganisms inhabit diverse natural habitats and ecosystems and are engaged in a multitude of interactions, including sharing and competing for essential nutrients. Microbial communities or consortia are shaped via these positive and/or negative interactions and have their own unique metabolic network that is defined by the spatial distribution, physiology, and availability of nutrients among the microbial participants (1–3). The exchange of biomolecules such as sugars, nucleobases, amino acids, vitamins, electron acceptors, fermentation by-products, and metal-chelating siderophores is found to occur between members of natural and synthetic microbial consortia (1, 4–8). Some members of a microbial consortia may stop synthesizing a metabolite that is readily available in their environment and eventually become auxotrophic for that nutrient (8, 9). Auxotrophy is beneficial for an individual organism, as it allows for the reduction in metabolic burden and/or genome size (10, 11). For example, in experiments with Escherichia coli, about 13% of mutants auxotrophic for vitamins, amino acids, and nucleotides show a higher fitness than the wild-type strain when the missing nutrient is provided exogenously in sufficient quantities in the growth medium (12). Another study shows that the coevolution of a coculture of the sulfate-reducing bacterium Desulfovibrio vulgaris and the archaeon Methanococcus maripaludis over 103 generations leads to loss-of-function mutations in the sulfur-reducing genes in D. vulgaris. Further, deleting these genes increases the yield of the corresponding D. vulgaris strains in coculture with M. maripaludis by an optical density at 600 nm (OD600) of ∼0.05 in comparison to the wild-type strain (13). Conversely, microorganisms that produce a metabolite to share or exchange with their fellow community dwellers (commensalism or mutualism, respectively) secure a position of prominence as they become indispensable for the consortium (14). Ruminococcus bromii, a starch-degrading bacterium associated with the human gut, converts starch to sugars that are substrates for other gut bacteria, thus playing the role of a keystone species (15). Thus, auxotrophy and metabolite sharing are important features for the formation and sustenance of microbial communities, and synthetic communities developed for biotechnological applications or for studying microbe-microbe interactions are often designed using these principles.
Among the commonly shared metabolites in microbial communities are vitamins (1, 16, 17). The activated forms of vitamins play an indispensable role as cofactors for numerous enzymes in primary metabolism across all domains of life. Several metagenomic analyses reveal that the water-soluble B vitamins are readily exchanged in marine microbial communities, the human gut microbiota, and communities associated with insects and other hosts (17–20). Of these, vitamin B1 (thiamin) is an important member of the B vitamins that plays an essential role in carbohydrate, amino acid, and lipid metabolism by assisting enzymes in conducting “impossible” decarboxylations (21). In the human gut microbiome, B1 auxotrophy appears to be most widespread at both the genus and family level, indicating that it is a commonly shared metabolite (17).
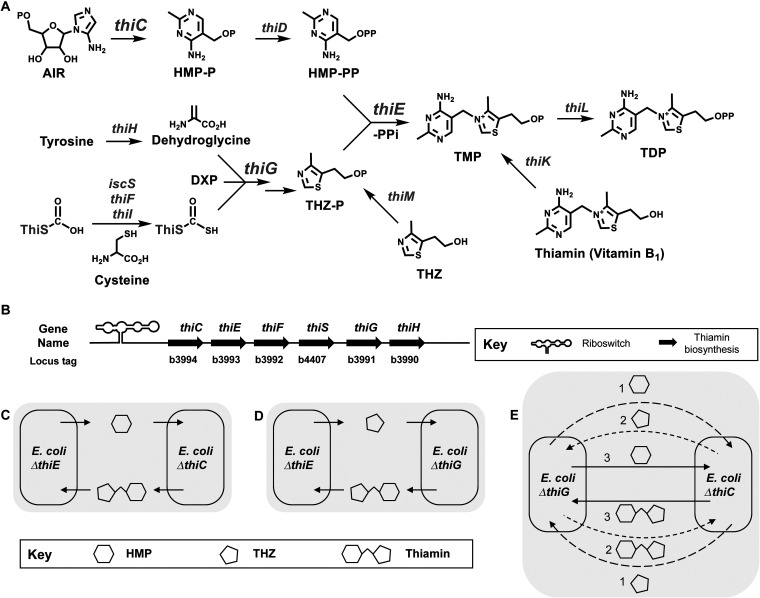
Even though most bacteria, plants, and eukaryotes such as fungi are capable of thiamin biosynthesis, many other organisms are unable to produce it and instead acquire it from their surroundings or their diet. The structure of thiamin consists of a 5-membered 4-methyl-5-(2-hydroxyethyl)thiazole ring (THZ) and a 6-membered 4-amino-5-hydroxymethyl-2-methylpyrimidine ring (HMP), which are synthesized in the natural world in their phosphorylated forms, THZ-P and HMP-P, respectively, in two distinct branches of the thiamin biosynthesis pathway (Fig. 1A) (22–26). The HMP-P is further phosphorylated to HMP-PP, following which the diphosphate is displaced by an attack via the THZ-P ring nitrogen to form a methylene bridge between the two rings to yield thiamin monophosphate (TMP) (Fig. 1A) (27). A final phosphorylation of TMP yields thiamin diphosphate (TDP), which is used as a cofactor by enzymes for cellular metabolism (28, 29). In some bacteria and yeast, thiamin is pyrophosphorylated directly to form TDP with the help of thiamin pyrophosphokinase (30, 31).
FIG 1.
Overview of the thiamin biosynthesis pathway in E. coli strain K-12 substrain MG1655. (A) Major steps in the thiamin biosynthesis pathway are depicted. All phosphate groups (-PO32−) are indicated as P. The 4-amino-5-hydroxy-2-methylpyrimidine phosphate (HMP-P) ring is formed by rearrangement of its precursor AIR by the enzyme ThiC. 4-Methyl-5-(2-hydroxyethyl)thiazole (THZ) is formed by the enzyme ThiG. The sulfur in the THZ ring is transferred via enzymes IscS, ThiI, and ThiF to form a thiocarboxylate moiety on the C terminus of the enzyme ThiS. The 1-deoxyxylulose-5-phosphate is synthesized by Dxs, and the ThiH enzyme converts tyrosine to dehydroglycine. ThiD, ThiM, and ThiK act as kinases for HMP-P, THZ, and thiamin, respectively. The enzyme ThiE attaches the HMP-PP and the THZ-P rings together in the final steps of the pathway to form thiamin monophosphate (TMP), and ThiL phosphorylates it to form the active form of the cofactor, thiamin pyrophosphate/diphosphate (TDP). Abbreviations: AIR, 5′-phosphoribosyl-5-aminoimidazole; DXP, 1-deoxyxylulose-5-phosphate; HMP-P, 4-amino-5-hydroxymethyl-2-methylpyrimidine phosphate; HMP-PP, 4-amino-5-hydroxymethyl-2-methylpyrimidine diphosphate; THZ-P, 4-methyl-5-(2-hydroxyethyl)thiazole phosphate; THZ, 4-methyl-5-(2-hydroxyethyl)thiazole. Gene names are shown in gray. (B) Arrangement of the genes involved in the de novo biosynthesis pathway for thiamin in E. coli K-12 MG1655. (C to E) We hypothesize that the coculture of EcΔthiC-EcΔthiE strains can survive by exchanging HMP and thiamin (C), the coculture of EcΔthiE-EcΔthiG strains can survive by exchanging THZ and thiamin (D), and the coculture of EcΔthiC-EcΔthiG strains can survive in three possible scenarios where metabolites are exchanged in the following pairs (E): exchange of HMP and THZ (1), exchange of THZ and thiamin (2), and exchange of HMP and thiamin (3).
Thiamin and its intermediates THZ and HMP are stable under physiological conditions and are salvaged from the environment by organisms for producing thiamin. Metagenomic analysis of the human gut microbiome reveals that the thiamin biosynthesis and salvage pathways display the largest variety of intermediates and noncanonical metabolic precursors (17). Recent findings implicate HMP as an important metabolite in shaping marine algal and bacterial consortia (32). Additionally, studies show that there exist thiamin auxotrophs that lack thiamin transporters but instead contain putative transporters for the uptake of HMP and/or THZ, which permit the salvage of these intermediates to produce thiamin (16, 33, 34). Examples of HMP and thiamin transporters and their uptake have been reported widely in literature (16, 35–37). On the other hand, information on THZ uptake and exchange is limited to only a handful of studies that predict a THZ transporter and show the uptake of the precursor carboxythiazole (16, 35, 36, 38, 39).
The modular nature of thiamin biosynthesis, where HMP and THZ are found to be independently synthesized and salvaged, makes this pathway a unique candidate for studying metabolic cross talk within microbial cocultures. To experimentally validate some of these findings, we require a simple model system whose members are engaged in thiamin, THZ, and HMP exchange. Such a system will allow us to (i) understand the molecular principles of thiamin biosynthesis occurring beyond an individual organism within a community and (ii) establish the design principles of building synthetic communities sustained by thiamin biosynthesis and uptake with diverse biotechnological applications.
This study involves a series of thiamin-dependent synthetic cocultures using E. coli, a Gram-negative bacterium that is capable of de novo thiamin synthesis and salvage and that is a member of several environmental and enteric microbial communities. In E. coli, the formation of the HMP-P ring is catalyzed by the enzyme ThiC, the THZ-P ring is synthesized by a host of enzymes including ThiG, and subsequently, these rings are coupled together by ThiE to form TMP. Also, no known transporters and salvage enzymes of HMP or THZ or their analogues are found in E. coli, and only one known transporter, ThiBPQ, exists to facilitate thiamin transport (32, 35, 36, 39, 40). We generated E. coli strain K-12 substrain MG1655 thiamin biosynthesis mutants—the ΔthiC, ΔthiE and ΔthiG mutants—which are impaired in de novo thiamin biosynthesis and thus are thiamin auxotrophs. We then set up pairwise synthetic cocultures of these three E. coli mutants to study their growth over short time periods. We also analyzed the exchange of thiamin, THZ, and HMP at a molecular level and its effect on the coculture composition. Further, we studied similar cocultures of another gammaproteobacterium, Vibrio anguillarum, and finally, mixed cocultures of E. coli and V. anguillarum to understand the extent to which our findings on thiamin metabolism within the E. coli cocultures hold true for other bacteria.
A unique property of the thiamin-based synthetic cocultures we have devised is that these are reliant on the exchange of precursors and intermediates within a single metabolic pathway, in comparison to other synthetic coculture studies in literature which involve exchange of molecules derived from two or more metabolic pathways (1, 4, 6). The advantages of a coculture system which is based on the biosynthesis of a single metabolite are the following: (i) the growth conditions of individual strains are similar, as they are auxotrophs for the same metabolite, (ii) the regulation of the biosynthesis and uptake of individual intermediates along the pathway can be studied, and (iii) coupling the results we observe from our system with genetic data from isolates and metagenomes has the potential to improve predictions and hypotheses of B1-related auxotrophy and metabolite exchange in natural systems.
Our results indicate that the intermediates in a single biochemical pathway can be exchanged with different efficiencies. In addition, we show that the exchange of nutrients in the thiamin biosynthesis pathway can also modulate the ratios of the organisms that are engaged in the cross talk.
RESULTS
E. coli thiamin biosynthesis auxotrophic mutants show concentration-dependent increases in growth when supplemented with thiamin or its biosynthesis intermediates.
E. coli is capable of producing TDP de novo and also contains genes to salvage thiamin from its environment (Fig. 1A). All the major genes for thiamin synthesis and salvage are found in three operons: (i) thiCEFSGH, which conducts de novo thiamin biosynthesis (Fig. 1B), (ii) thiMD, which codes for kinases in the salvage pathway, and (iii) thiBPQ, which codes for an ABC-type thiamin transporter (25) (see Fig. S1A in the supplemental material). All three operons are regulated by TDP-dependent riboswitches (41).
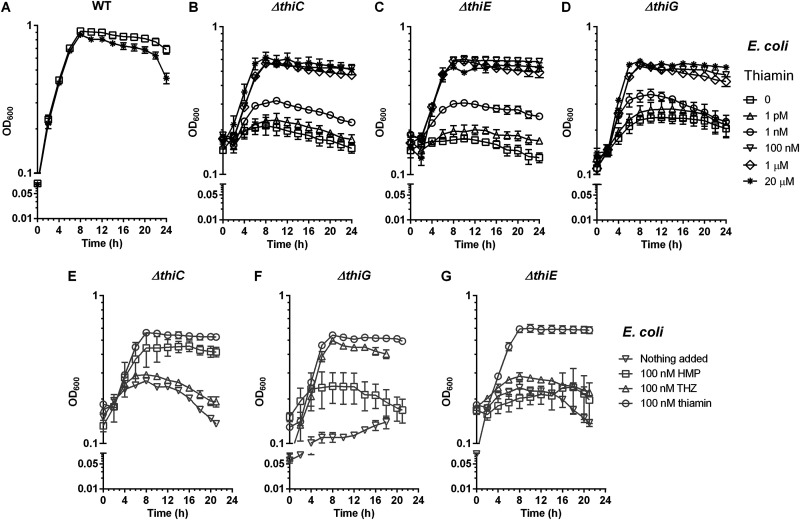
To begin our studies, we created three knockout E. coli K-12 MG1655 strains, EcΔthiC, EcΔthiE, and EcΔthiG (referred to as the thi mutant strains), and noted that all three strains grew without any growth disadvantage in a nutrient-rich medium (Fig. S2A). Next, we tested their growth in a thiamin-deficient minimal medium containing M9 salts with glucose and NH4Cl as the carbon and nitrogen sources, respectively. We expected that they would require exogenously added thiamin for growth in this medium, but to our surprise, all three strains survived well in the first passage (P1) from the nutrient-rich to the minimal medium (Fig. S2C). Analysis of the M9 medium without any cells as a control showed the absence of thiamin and its phosphorylated analogues, indicating that thiamin is indeed carried over by the cells (Fig. S3). A second passage (P2) of the mutants in the thiamin-deficient minimal medium showed significantly lesser growth than that of the wild-type strain, and the third passage (P3) showed no growth, indicating that the thi mutant strains were indeed thiamin auxotrophs (Fig. 2A to D, no thiamin added trace; see also Fig. S2E). Our results match similar observations in the literature, which note that thiamin stored inside the cells during their growth in rich medium is carried over into a few generations of cell growth (42, 43). For all future experiments, the P2 cells were used, as this allowed us to have some cells from the controls for thiamin quantitation experiments while yet showing a sufficient difference in optical density (OD600) between the single-culture and coculture growth experiments.
FIG 2.
Supplementation of the thiamin mutants of E. coli K-12 MG1655 with thiamin, HMP, and THZ in M9 medium. (A to D) Growth phenotype of the wild-type strain (A), the EcΔthiC strain (B), the EcΔthiE strain (C), and the EcΔthiG strain (D). The following symbols in panels A to D represent the indicated concentrations of thiamin: □, nothing added; ○, 1 pM; Δ, 1 nM; ∇, 100 nM; ◊, 1 μM; *, 20 μM. (E to G) Supplementation of the EcΔthiC mutant (E), the EcΔthiG mutant (F), and the EcΔthiE mutant (G). Symbols in panels E to G: ∇, no HMP/THZ; □, 100 nM HMP; Δ, 100 nM THZ; ○, 100 nM thiamin. Means ± standard errors of the means of results from three independent experiments are plotted.
Next, to determine the minimum thiamin concentration required by the thi mutant strains, we tested their growth in minimal medium supplemented with thiamin concentrations ranging from 0 to 20 μM. We found that while these strains show low growth with up to 1 nM thiamin, they are able to achieve an OD600 of ∼0.6 with 100 nM, 1 μM, and 20 μM thiamin (Fig. 2B to D). This shows that thiamin is the growth-limiting nutrient for the thi mutants. To ensure that thiamin is not limiting in our assays, all further experiments were conducted with 20 μM thiamin unless otherwise stated. We further complemented each knockout strain with a plasmid containing the deleted gene and confirmed that growth can be restored in these strains in minimal medium in the absence of thiamin, as also observed in previous studies (Fig. S4) (44–46).
We expect the EcΔthiC and EcΔthiG strains to be impaired in the biosynthesis of the intermediates HMP and THZ, respectively, while the EcΔthiE mutant should not be able to link them together to synthesize thiamin. To test this, we fed HMP and THZ to the thi mutants in various concentrations ranging from 0 to 1 μM. The EcΔthiC strain survived only when supplemented with HMP, not with THZ (Fig. 2E; Fig. S5), as did the EcΔthiG strain when supplemented with THZ but not with HMP (Fig. 2F; Fig. S5), and the EcΔthiE strain was unable to survive with either HMP or THZ alone (Fig. 2G; Fig. S5). This confirms that the metabolic phenotypes of the thi mutants are correlated with their genotypes.
Specific cocultures of the thiamin biosynthesis mutants grow in minimal medium with no exogenously added thiamin.
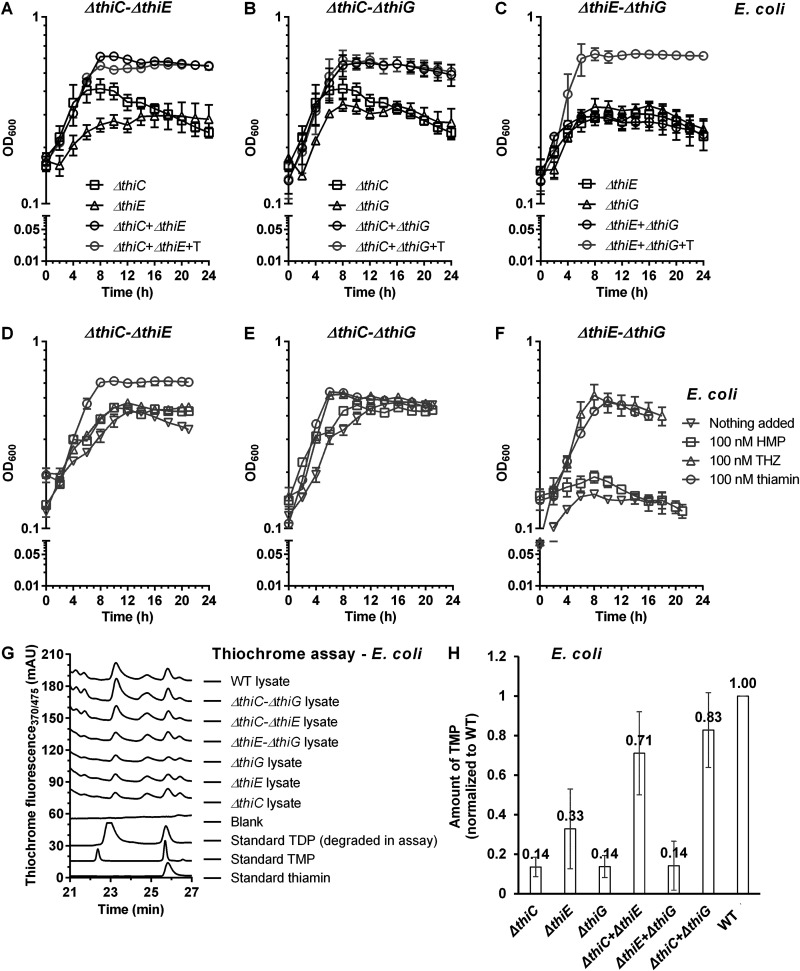
Next, we constructed three pairwise cocultures, EcΔthiC-EcΔthiE (Ec-CE), EcΔthiC-EcΔthiG (Ec-CG), and EcΔthiE-EcΔthiG (Ec-EG), and studied their growth in thiamin-deficient minimal medium. We hypothesized that if the thi mutant strains can share thiamin biosynthesis intermediates among themselves and produce thiamin, the cocultures will survive, as opposed to the single cultures, which are auxotrophic and perish under similar growth conditions. We started these cocultures with 9:1, 1:1, and 1:9 ratios of the two strains within the coculture and observed their growth over a period of 24 h. We observed that both the Ec-CE and the Ec-CG co-cultures showed increased survival at a 1:9 ratio of the Ec deltathiC to Ec deltathiE and Ec deltathiC to Ec deltathiG strains, respectively, compared to the individual pure strains (Fig. 3A and B, data for the 9:1 and 1:1 ratios not shown). On the other hand, the Ec-EG coculture showed no difference in growth in comparison to its individual pure cultures (Fig. 3C).
FIG 3.
Thiamin biosynthesis mutants of E. coli K-12 MG1655 grow in pairwise cocultures in thiamin-deficient M9 medium. (A to C) Coculture of the EcΔthiC-EcΔthiE strains (A), the EcΔthiE-EcΔthiG strains (B), and the EcΔthiC-EcΔthiG strains (C). (D to F) Supplementation of the ΔthiC-ΔthiE coculture (D), the ΔthiE-ΔthiG coculture (E), and the ΔthiC-ΔthiG coculture (F). Symbols in panels D to F represent the following: ∇, no HMP/THZ; □, 100 nM HMP; Δ, 100 nM THZ; ○, 100 nM thiamin. (G) HPLC of thiochrome assay samples to detect thiamin from coculture lysates. (H) Amount of thiochrome monophosphate (TMP), normalized to that in the WT strain, detected in lysates of monocultures or cocultures of the thiamin biosynthesis mutants grown in P2. Means ± standard errors of the means of results from three independent experiments are plotted.
There are several possibilities of exchange of thiamin and its intermediates that account for the survival of the Ec-CE and Ec-CG cocultures (Fig. 1C to E). The EcΔthiC strain cannot synthesize HMP, but if it can acquire it from its environment, it can combine the HMP with the THZ it synthesizes to form thiamin. Alternately, it can acquire thiamin directly from its environment. Similarly, the EcΔthiG strain cannot synthesize THZ but it can grow if it acquires THZ or thiamin from its surrounding. On the other hand, the EcΔthiE strain can synthesize both the HMP and the THZ intermediates but is unable to combine them to form thiamin and needs to acquire it from its growth medium. The growth observed in the Ec-CE coculture can be explained only if the EcΔthiE strain supplemented the EcΔthiC strain with HMP and the EcΔthiC strain in return supplemented the EcΔthiE strain with thiamin (Fig. 1C and 3A). This indicates that both HMP and thiamin are likely being exchanged in the medium. Along similar lines, the Ec-EG coculture would grow if the EcΔthiE strain supplemented the EcΔthiG strain with THZ and the EcΔthiG strain, in return, supplemented the EcΔthiE strain with thiamin (Fig. 1D and 3C). Since the Ec-EG coculture does not grow, and we know that THZ and thiamin are salvaged by the E. coli cells based on our feeding studies and that thiamin is also exchanged as per the results of the Ec-CE coculture, this result suggests that THZ is not released by EcΔthiE mutant at sufficient concentrations and thus does not support the growth of the EcΔthiG mutant. The absence of any annotated THZ transporters in E. coli also supports this hypothesis. Interestingly, the Ec-CG coculture shows a higher OD600 than the individual pure cultures grown in thiamin-deficient medium (Fig. 3B). The Ec-CG coculture can grow in three scenarios: (i) the EcΔthiC strain and the EcΔthiG strain provided the other with THZ and HMP, respectively, (ii) the EcΔthiC strain provided the EcΔthiG strain with THZ and the EcΔthiG strain synthesized thiamin and provided it back to the EcΔthiC strain, or (iii) the EcΔthiG strain provided the EcΔthiC strain with HMP and the EcΔthiC strain synthesized thiamin and provided it back to the EcΔthiG strain (Fig. 1E). Since the data for the Ec-EG coculture indicates that THZ is likely not being exchanged, only the third possibility remains for the Ec-CG coculture, that is, HMP and thiamin are exchanged among the thiamin biosynthesis mutants. Incidentally, several reports in literature note the exchange or release of HMP among microbial communities, confirming our observation (33, 47).
Next, we compared carbon sources to understand whether these results hold true across different growth conditions. In addition to glucose, we chose pyruvate and succinate, since their utilization as carbon sources via the Kreb’s cycle is linked to thiamin availability. Similar to what we observed with glucose, the Ec-CE and Ec-CG cocultures showed growth in pyruvate and succinate minimal media without thiamin while the Ec-EG did not, and the growth of the Ec-CG coculture was highest among the three (Fig. S6). Since glucose, pyruvate, and succinate metabolism in cells require thiamin-utilizing enzymes, our growth studies imply that the Ec-CE and Ec-CG cocultures are able to synthesize thiamin. The Ec-CE coculture showed lower growth in the presence of pyruvate and succinate than in the presence of glucose, and thus we continued with glucose as the carbon source for all further experiments.
Analysis of the cocultures demonstrates that the exchange of HMP and thiamin aids their survival.
To probe the growth patterns observed for the Ec-CE, Ec-CG, and Ec-EG cocultures, we conducted a supplementation study with a range of HMP and THZ concentrations (Fig. 3D to F; Fig. S5). We observed that while the Ec-CE and Ec-CG cocultures each show growth without or with supplementation with both molecules, the Ec-EG coculture survives only when fed with THZ, not with HMP (Fig. 3F). Interestingly, the EcΔthiG mutant can survive with 1 nM THZ, whereas the EcΔthiC mutant requires 100 nM HMP to survive (Fig. S5A, I, and K). This indicates that E. coli differs in its ability to either acquire and/or utilize the thiamin biosynthesis intermediates THZ and HMP. This result also sheds light on one of our preliminary observations that when the Ec-CE and Ec-CG cocultures were started at a total OD600 of 0.01 in the P2 passage instead of 0.1, they were unable to survive (data not shown). We attribute this to the lack of an adequate pool of thiamin intermediates at the start that would allow the coculture strains to begin dividing and cooperating, thus ensuring their survival.
De novo biosynthesis of thiamin occurs within cocultures.
To verify that the growth of the cocultures is due to the de novo biosynthesis of thiamin, we analyzed the lysates of the cells grown in thiamin-deficient medium from the second passage, P2, for single cultures and cocultures for the presence of thiamin and its phosphorylated versions TMP and TDP. To do so, we used the thiochrome assay, which employs an oxidation reaction under alkaline conditions to generate a fluorescent derivative of thiamin (23). First, we noted that under the thiochrome assay conditions we used, the standard thiochrome diphosphate formed is unstable and undergoes dephosphorylation as demonstrated by high-performance liquid chromatography (HPLC) analysis (Fig. 3G). Next, we analyzed the lysates of the Ec-CE cocultures and the Ec-CG cocultures and noted that the levels of thiochrome monophosphate in them were significantly higher than those in their respective single cultures when measured at 24 h and similar to those in the wild-type E. coli cell lysate (Fig. 3G and H). In contrast, the amounts of thiochrome monophosphate detected from the lysates of the Ec-EG cocultures and their respective single cultures were similar and were significantly lower than those of the wild-type lysate (Fig. 3G and H). This implies that thiamin is synthesized de novo in the Ec-CE and Ec-CG cocultures. Further, liquid chromatography-tandem mass spectrometry (LC-MS/MS) analysis of these samples confirmed the presence of thiamin in the lysates of the cocultures (Fig. S7). Taken together, these results show that the Ec-CE and the Ec-CG cocultures grow due to de novo thiamin synthesis, whereas the Ec-EG cocultures do not survive, as they are unable to produce thiamin.
Vibrio anguillarum thiamin mutants follow a similar pattern of exchange as E. coli.
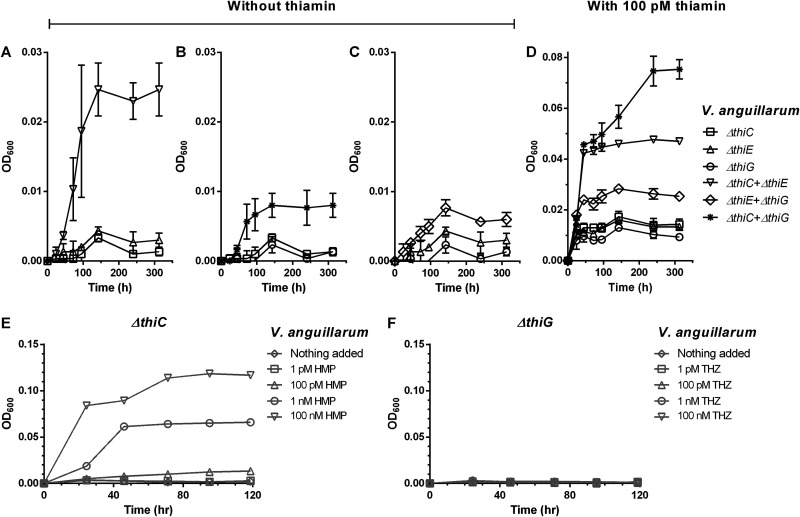
In order to determine whether the pattern of exchange of thiamin biosynthesis intermediates observed in E. coli is conserved across other microbes, we analyzed another gammaproteobacterium, Vibrio anguillarum strain PF430-3, which is capable of de novo thiamin biosynthesis and salvage (Fig. S1B). Similar to the previous experiment, V. anguillarum ΔthiC, ΔthiE, and ΔthiG mutant strains were grown individually and in pairwise cocultures in thiamin-deficient M9 medium (Fig. 4).
FIG 4.
Thiamin biosynthesis mutants of V. anguillarum strain PF430-3 grow in pairwise cocultures in thiamin-deficient M9 medium. (A to C) Cocultures of the VaΔthiC-VaΔthiE strains (A), the VaΔthiC-VaΔthiG strains (B), and the VaΔthiE-VaΔthiG strains (C) without thiamin supplementation. (D) Cocultures of the VaΔthiC, VaΔthiE, and VaΔthiG strains supplemented with 100 pM thiamin. (E) HMP supplementation of the VaΔthiC mutant. (F) THZ supplementation of the VaΔthiG mutant. The following symbols in panels E and F represent the indicated concentrations of HMP and THZ, respectively: ◊, nothing added; □, 1 pM; Δ, 100 pM; ○, 1 nM; ∇, 100 nM. Means ± standard deviations of results from three independent experiments are plotted.
Since V. anguillarum and E. coli are both gammaproteobacteria containing the same set of thiamin genes except the kinase gene thiM, we expect V. anguillarum to show a similar pattern of exchange (Fig. S1). The V. anguillarum ΔthiC, ΔthiE, and ΔthiG single cultures showed no background growth in the P1 passage and a concentration-dependent increase in growth starting with nanomolar concentrations of supplemented thiamin (Fig. S8). For the cocultures grown without supplemented thiamin, we observed that the V. anguillarum CE (Va-CE) coculture showed significant growth, followed by the Va-CG coculture, while the Va-EG coculture showed background growth similar to what we observed in E. coli (Fig. 4A to C). When supplemented with 100 pM thiamin, the Va-CE, Va-CG, and Va-EG cocultures grew significantly better than the single cultures, reiterating that the cocultures were likely producing thiamin (Fig. 4D). Interestingly, even though the V. anguillarum ΔthiC strain shows a concentration-dependent increase in growth when supplemented with HMP similar to its E. coli counterpart, the VaΔthiG strain does not grow with exogenously added THZ (Fig. 4E and F). This indicates that unlike the E. coli ΔthiG mutant, whose growth can be complemented by thiamin and THZ, the V. anguillarum ΔthiG mutant can be complemented only by thiamin. This may be attributed to the absence of the thiazole kinase gene thiM in V. anguillarum, annotated as a salvage enzyme that phosphorylates THZ to produce THZ-P for incorporation in thiamin biosynthesis in E. coli and other organisms (Fig. 1; Fig. S1) (48).
V. anguillarum and E. coli thiamin mutants exchange thiamin and its biosynthesis intermediates among themselves.
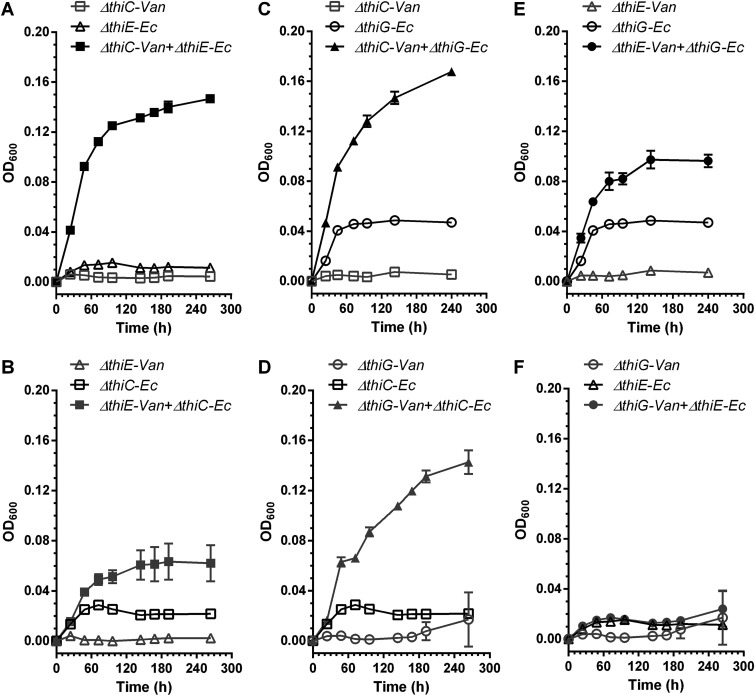
Finally, to test whether the pattern of exchange that we observe occurs between different species, mixed cocultures of the thiamin biosynthesis mutants of E. coli and V. anguillarum in a pairwise manner were studied (Fig. 1C, D, and E describe all possible combinations of exchange). We observed that the V. anguillarum ΔthiC-E. coli ΔthiE (VaC-EcE) mixed cocultures grow in the absence of thiamin (Fig. 5A). This observation, along with the results obtained so far, shows that HMP is provided by EcE to VaC cells, which synthesize thiamin and provide it back to the EcE cells, thus leading to the growth of the coculture. The VaE-EcC, VaC-EcG, and VaG-EcC mixed cocultures also survived in the absence of thiamin as expected, further corroborating the exchange of HMP and thiamin (Fig. 5B to D). We noted that the final OD600 achieved by the VaC-EcE and VaC-EcG cocultures is greater than that of their corresponding reverse combinations, VaE-EcC and VaG-EcC cocultures, respectively. Since V. anguillarum has lesser growth in the thiamin-deficient medium than E. coli, it is likely that the amount of HMP provided by the VaE or the VaG strain to the EcC strain is lesser, leading to less growth of VaE-EcC and VaG-EcC cocultures.
FIG 5.
Mixed-species cocultures of the thiamin biosynthesis mutants of V. anguillarum PF430-3 and E. coli K-12 MG1655 in thiamin-deficient M9 medium. Mixed cocultures of the VaΔthiC-EcΔthiE strains (A), the VaΔthiE-EcΔthiC strains (B), the VaΔthiC-EcΔthiG strains (C), the VaΔthiG-EcΔthiC strains (D), the VaΔthiE-EcΔthiG strains (E), and the VaΔthiG-EcΔthiE strains (F) without thiamin supplementation. All empty gray and black symbols represent single cultures of V. anguillarum and E. coli, respectively. All filled symbols represent the cocultures, with gray and black colors representing the corresponding reverse combinations. Means ± standard deviations of results from three independent experiments are plotted.
Surprisingly, we observed that the VaE-EcG coculture also grew significantly better than the individual strains without thiamin supplementation (Fig. 5E), even though its overall growth was lower than that of the VaC-EcG coculture. This result is in contrast to what was observed in the Va-EG or Ec-EG cocultures, which did not grow beyond the background level. This indicates that THZ synthesized by the V. anguillarum ΔthiE strain is available in the medium at a concentration that allows E. coli ΔthiG to grow and produce thiamin and share it in return with V. anguillarum ΔthiE. The reverse combination coculture VaG-EcE does not survive in the absence of thiamin. This may be attributed to V. anguillarum lacking ThiM as mentioned previously, which restricts its ability to salvage THZ (Fig. 4F and 5F). All mixed cocultures of V. anguillarum and E. coli were able to survive with 100 pM of supplemented thiamin, as expected (Fig. S9).
To summarize, the CE and CG cocultures of E. coli or V. anguillarum strains can survive in the absence of externally supplemented thiamin, whereas the EG cocultures cannot. These experiments confirm that HMP and thiamin are shared within the synthetic cocultures but that THZ may not be as easily shared. Further, results obtained from the mixed cocultures of E. coli and V. anguillarum strains suggest that even though THZ is picked up when present at higher concentrations, it might not be readily shared among microorganisms, as the concentrations produced are too low to be salvaged.
The ratio of the individual strains within the coculture is determined by the exchange of thiamin and its biosynthesis intermediates.
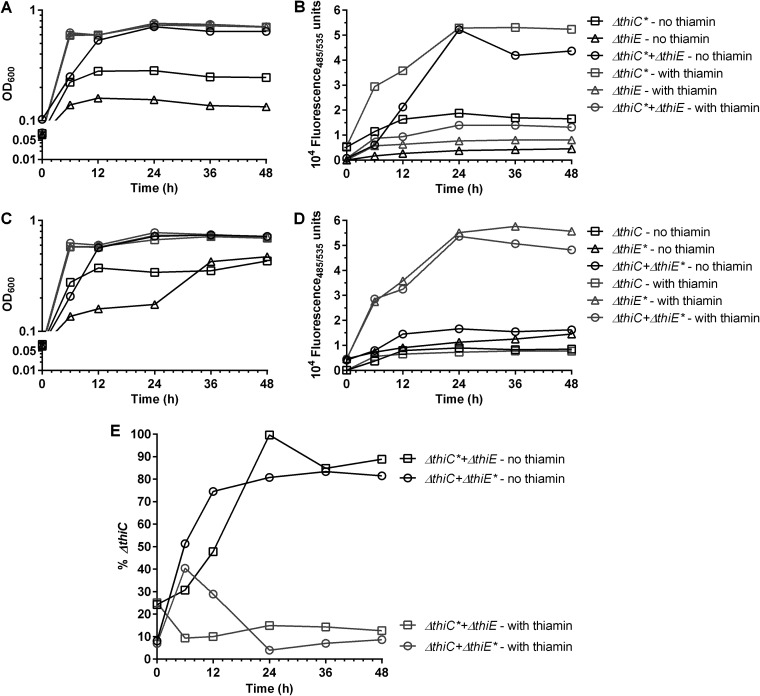
Our observations of the coculture experiments thus far are based on the total OD600 of the cocultures. To understand and quantify the contribution of each individual strain, we created the thi mutants fluorescently labeled with green fluorescent protein (GFP) and set up the pairwise Ec-CE and Ec-CG cocultures in thiamin-deficient minimal medium, where one of the strains in each coculture was fluorescently labeled (Fig. S2B and D). This approach allows us to quantify the amount of each strain in the coculture by using two parameters, i.e., (i) the total OD600 and (ii) the fluorescence of the coculture, which indicates the growth of the GFP-tagged strain. Briefly, we generated a standard curve of fluorescence versus OD600 for each individual strain, after which the cocultures (GFP strain indicated with an asterisk)—Ec-C*E and Ec-CE* with the controls Ec-C*E* and Ec-CE and a similar set for the Ec-CG cocultures—were set up. We then noted the increase in the OD600 and fluorescence values over time and mapped the fluorescence signal of the coculture to the standard curve of the corresponding GFP-tagged strain, allowing us to quantify its OD600 in the coculture (Fig. S10). The remaining untagged strain numbers were then calculated by subtracting this number from the total OD600, eventually yielding the ratios of the two strains over the course of the coculture growth.
Our experiments and subsequent calculations showed that the quantities of the strains in the cocultures change over a period of 24 h when no thiamin is exogenously provided (Fig. 6; Fig. S11). The OD600 of the Ec-C*E coculture increases over time as expected (Fig. 6A). The fluorescence of the coculture also increased, indicating that the quantity of the GFP-marked EcΔthiC* strain increased over time (Fig. 6B). Next, we observed that for the Ec-CE* coculture in the absence of thiamin, the fluorescence did not increase even though the OD600 value increased over time, reiterating the result that the EcΔthiC strain increased in numbers in the coculture (Fig. 6C and D).
FIG 6.
Growth phenotypes and fluorescence of the monocultures and cocultures of the thiamin mutant strains. The strains containing the GFPmut2 cassette are marked with an asterisk. Black symbols, without thiamin; gray symbols, with thiamin. (A and B) OD600 (A) and fluorescence (B) of EcΔthiC*-EcΔthiE cocultures. (C and D) OD600 (C) and fluorescence (D) of EcΔthiC-EcΔthiE* cocultures. (E) Percentage of EcΔthiC cells in the EcΔthiC*-EcΔthiE cocultures and the EcΔthiC-EcΔthiE* cocultures. Average values of results from two independent experiments are plotted.
Interestingly, in the presence of thiamin, even though the OD600 of the Ec-C*E coculture increased over time, and the fluorescence increase in the EcΔthiC* single culture cells was proportional to its growth as expected, the total fluorescence of the Ec-C*E coculture increased very slightly over time (Fig. 6A and B). This indicates that the ratio of the individual strains remains constant over time with respect to the starting ratio. Also, both the OD600 and the fluorescence of the Ec-CE* coculture and the EcΔthiE* single culture increased over time, confirming that the numbers of the two participating strains do not deviate in the coculture in the presence of thiamin (Fig. 6C and D).
Upon quantifying the Ec-C*E and Ec-CE* coculture results, we found that in the absence of thiamin, the percentage of the EcΔthiC cells in the cocultures increased over time to attain an average ratio of ∼8:2 of EcΔthiC to EcΔthiE cells at 24 h (Fig. 6E). Also, the presence of GFP does not alter the final ratios of the strains in the cocultures, as illustrated by the Ec-C*E and Ec-CE* cocultures showing similar ratios. Comparable ratios were obtained when the Ec-C*G cocultures were similarly analyzed (Fig. S9B). When the cocultures were further transferred at the end of 24 h of growth to a fresh thiamin-deficient M9 medium in passage P3, the new ratios held constant over a period of 24 h (Fig. S11A and C). Additionally, even after the continued growth of the P2 cocultures for another ∼24 h, the ratios attained stayed constant (Fig. 6E; Fig. S11B). We hypothesize that this change in the ratio of the two strains results from the exchange of HMP and thiamin, which equilibrates after ∼24 h and subsequently stabilizes. However, in the presence of exogenously added thiamin, the exchange is no longer necessary and hence the ratios of the two strains remain mostly unaltered.
DISCUSSION
Thiamin, an essential nutrient for living organisms, assists enzymes in executing key decarboxylation reactions in primary metabolism. Several studies based on metagenomic analyses predict that thiamin and its building blocks HMP and THZ can be salvaged by both thiamin auxotrophs and prototrophs (16, 17, 47). In this study, we investigated the mechanism of thiamin synthesis and exchange within microbial cocultures through a molecular lens.
It has been reported that secondary transporters such as PnuT, which facilitate bidirectional transport of thiamin, are found more often in prototrophs, whereas the ABC family primary transporters such as ThiT, which promote thiamin uptake, are found more often in auxotrophs (16, 17). It has also been observed for both marine and gut microbial communities that some organisms in the community might be auxotrophic for the biosynthesis of both THZ and HMP, whereas certain others in the same community can produce both these intermediates but lack the ability to combine them to form thiamin (16, 17). These observations reiterate that thiamin sharing is common among microorganisms.
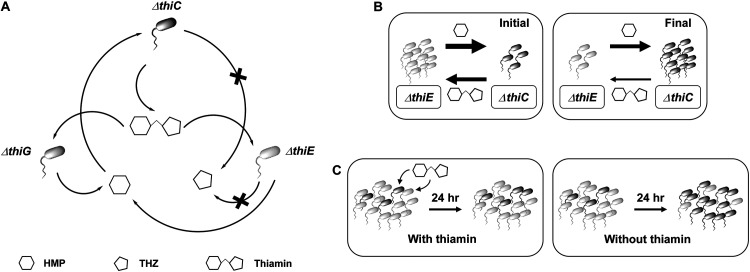
To better understand the specifics of the exchange of thiamin and its intermediates in a community, we created synthetic cocultures with bacterial strains with defined thiamin auxotrophy patterns. Our results from the E. coli and V. anguillarum cocultures as well as their mixed cocultures suggest that thiamin and HMP are commonly exchanged among microorganisms, whereas the exchange of THZ may occur less frequently and under specific conditions (Fig. 7A). These results corroborate previous observations, where (i) marine organisms Synechococcus sp. strain WH8102 and Dunaliella tertiolecta have been shown to release HMP in their growth medium, and thiamin and HMP concentrations in seawater show day-night variation suggesting changes in their release and consumption, (ii) the haptophyte Emiliania huxleyi has been shown to utilize HMP and its analogue more efficiently than thiamin, and (iii) most aquatic microbes are pyrimidine (thiamin) auxotrophs lacking thiC, and other prevalent taxa require intact thiamin (17, 32, 33, 47). Our results show that the Ec-EG and Va-EG cocultures do not grow, and we attribute this to the inability of THZ to be shared (illustrated in the schematic shown in Fig. 7A and 1D). However, the the mixed cocultures of VaC-EcG and VaE-EcG cocultures both show growth, which may be a result of the exchange of THZ between these organisms (Fig. 5C and E and 1D and E). Of these, the growth of the VaE-EcG coculture was surprising and unexpected based on our previous results, and we reason that there is only one possibility for how these two thiamin auxotroph strains may support one another’s growth—V. anguillarum ΔthiE supplies THZ to E. coli ΔthiG, which produces thiamin and in turn returns it to V. anguillarum ΔthiE, enabling it to grow and the coculture to be sustained over 12 days (∼300 h) (Fig. 5E and 1D). In the VaC-EcG coculture, there are three possibilities, as illustrated in the schematic in Fig. 1E showing the potential exchanges, briefly, (i) VaΔthiC → THZ → EcΔthiG, EcΔthiG → thiamin → VaΔthiC, (ii) VaΔthiC → THZ → EcΔthiG, EcΔthiG →HMP → VaΔthiC, or (iii) EcΔthiG → HMP → VaΔthiC, VaΔthiC → thiamin → EcΔthiG. Based on the observation that the VaE-EcG coculture is able to grow, it opens up the possibility for any of these to occur. However, as the OD600 of the VaC-EcG coculture is significantly higher than that of the VaE-EcG coculture, it is likely that the two cocultures have different patterns of exchanges (Fig. 5C and E). Based on this observation, we hypothesize that the VaC-EcG coculture may follow possibilities (ii) and (iii), and this needs to be investigated further.
FIG 7.
Proposed model for the exchange of thiamin biosynthesis intermediates in the cocultures and effects of the exchange on the coculture dynamics. (A) Proposed molecular exchanges among the thiamin mutants. (B) Model for exchanges in the coculture of the ΔthiC-ΔthiE strains without thiamin at the initial and final stages (after 24 h) of coculture. The thickness of the arrows is proportional to the amounts of the respective nutrients being released. (C) Ratios of the two strains in the coculture differ based on the presence or absence of thiamin. Black cells, ΔthiC strain; gray cells, ΔthiE strain.
The unexpected growth of the VaE-EcG mixed coculture in thiamin-deficient medium might be explained based on some of the characteristics of the coculture inhabitants. We hypothesize that the cells of VaΔthiE lyse owing to the longer incubation time of ∼300 h, as opposed to EcΔthiE cells in the EcE-EcG cocultures, which are grown for only 24 h. This results in the release of THZ in the medium, a sufficient amount of which then accumulates and is salvaged by the EcΔthiG cells, and thus the VaE-EcG coculture survives. But had this been the case, the Va-EG coculture, which showed no growth for ∼300 h, should have also survived (Fig. 4C). We hypothesize that this inability to grow is because unlike E. coli, which harbors the thiazole kinase ThiM, V. anguillarum lacks this enzyme and is hence unable to convert exogenous THZ to THZ-P, which is subsequently routed into thiamin biosynthesis (Fig. 1A). This hypothesis is further supported by two additional observations: (i) the VaΔthiG strain does not grow when supplemented with up to 100 nM THZ (Fig. 4F), and (ii) the VaG-EcE coculture is unable to survive in thiamin-deficient medium (Fig. 5F). It is also possible that instead of or alongside lysis, V. anguillarum exports THZ into the medium using yet-unannotated transporters, which enables EcΔthiG to grow in the VaE-EcG coculture.
When calculating the ratios of the two strains in the coculture, we noted that the EcΔthiC strain increases in the coculture over time and the ratios of EcΔthiC to EcΔthiG and EcΔthiC to EcΔthiE finally stabilize at ∼8:2. The role of the EcΔthiG or EcΔthiE strains in both cocultures is to provide HMP, whereas that of EcΔthiC is to produce thiazole and subsequently thiamin. Let us take the instance of the Ec-CE coculture. The EcΔthiE strain (present in a higher amount at the start) produces HMP and supplies it to the EcΔthiC strain. The EcΔthiC strain produces THZ, combines it with the HMP that it obtains, and provides EcΔthiE with thiamin in return. However, since producing THZ appears to be the rate-determining step in thiamin biosynthesis, the numbers of the EcΔthiC strain need to be higher to fulfill the thiamin requirement of the coculture (49). An alternate explanation may be that as the thiamin-producing EcΔthiC strain grows and replicates, it will require more thiamin, and hence, it needs a small but continuous supply of HMP. Thus, just enough thiamin to aid the survival of some EcΔthiE cells to ensure the availability of HMP is released from the EcΔthiC strain in the coculture (Fig. 7B). In conclusion, our results strongly suggest that the strain that produces thiamin or both thiamin and THZ numerically dominates the coculture once a cross talk has been established.
We also observed that when the cocultures of the E. coli thi mutants are supplemented with thiamin, the ratios of the two strains in the cocultures do not deviate much from the starting ratios (Fig. 7C). This suggests that when a nutrient is available in plenty in a community of auxotrophs, they may not interact with each other. But when the nutrient is unavailable or scarce, a cross talk that allows for the microorganisms to share it may evolve, which includes enhancing or limiting certain interactions and microbial populations. This subsequently shapes the community composition and relative abundance of its members, and these principles may be utilized to design synthetic microbial cocultures. Indeed, the seasonal blooms of marine microorganisms which either produce or utilize thiamin alter the concentrations of thiamin biosynthesis intermediates in seawater, and when the microbial numbers are low, the overall concentrations of the intermediates remain at an equilibrium (50). Such changes in the community composition have also been reported earlier for synthetic cocultures based on their differential abilities of nutrient exchange or uptake (4, 5, 45).
Finally, we hypothesize that the reason for HMP being exchanged more readily than THZ among auxotrophs is that the biosynthesis of THZ involves more steps and intermediates than the biosynthesis of HMP and is also perhaps metabolically more expensive (49). THZ is assembled by thiG (or THI4 in eukaryotes) using three intermediates, ThiS-thiocarboxylate, dehydroglycine, and 1-deoxyxylulose-5-phosphate (DXP), each of which has intriguing properties. The sulfur which is installed in the THZ ring is transferred from cysteine onto the carboxylate end of ThiS, a small protein synthesized solely for this purpose, to form ThiS-thiocarboxylate (51). Dehydroglycine is unstable under aqueous conditions and needs to be protected in the active site of the enzyme that synthesizes it (52). Finally, DXP, an isoprenoid biosynthesis intermediate, requires thiamin for its own biosynthesis. Also, THI4, the eukaryotic homolog of ThiG, is a single turnover enzyme that itself provides the S atom of the THZ ring that it synthesizes (53). In contrast, HMP is synthesized by a single-step rearrangement of the substrate aminoimidazole ribotide (AIR) by ThiC, an intermediate common to purine, vitamin B1, and vitamin B12 biosynthesis pathways (24, 54, 55). Thus, the biosynthesis of HMP may have a lower metabolic expense than that of THZ, making it easier and less costly in a competitive sense for organisms to share HMP rather than THZ. Interestingly, one study reports that the ratio of the thiC gene to the total number of thiG and thi4 genes in marine microbes is in the 0.06 to 0.28 range, always less than 1 (47). Congruently, another study reported higher concentrations of HMP than thiamin in surface waters of the Sargasso Sea and that the abundance of thiC genes was lesser than that of the thiG genes at depths ranging from 0 to 80 m (33). Even beyond marine ecosystems, there is a propensity for HMP exchange within the human gut microbiome (HGM) as well, wherein out of the 2,228 reference genomes studied, 199 were HMP auxotrophs, whereas only 114 were THZ auxotrophs (17). These studies, taken together with our observations, point to HMP and possibly other pyrimidine intermediates as key nutrients in determining the dynamics of nutrient exchange and subsequently microbial abundance (32, 40, 50).
Our results indicate that the rules of exchange of thiamin and its intermediates are broadly similar across organisms, and variations may be predicted based on growth conditions and the genome sequences of the interacting species. Variation in thiamin-related genotypes is evident among natural populations, but characterizing the behavior of model organisms as done here is expected to aid predictions of exchange among natural communities (33, 47). We also observe temporal changes in the ratios of the thi mutants in our cocultures based on the ability of the strains to either make B1 or a B1 biosynthesis intermediate, or use exogenously added B1. Our findings inform the physiology of single microbial members with regard to thiamin metabolism within the context of a microbial community. Finally, our study highlights the nature of interdependencies that arise from relying on acquiring essential metabolites from the environment or from fellow community members.
Conclusions.
In this study, we designed a unique coculture system based on the exchange of intermediates derived from a single metabolic pathway, i.e., vitamin B1 and its biosynthesis intermediates. We conclude that the sharing of vitamin B1 and its intermediates is modulated by their availability and the presence of biosynthesis, salvage, and transporter proteins in cells. Exchange forms the basis of building an interacting community of microbes but may also be a feasible mechanism to halt interactions or limit the success of portions of a community, e.g., provision of thiamin rather than HMP to prevent dominance of pyrimidine auxotrophs. Finally, our investigations at the molecular level underscore the specific role of metabolite exchange in determining, stabilizing, and sustaining the collective metabolism and composition of our microbial cocultures.
MATERIALS AND METHODS
Chemicals and reagents.
All the chemicals used were obtained either from TCI, HiMedia, or Sigma unless otherwise specified. The enzymes used were obtained from TaKaRa.
Strains and plasmids.
The E. coli K-12 MG1655 strain containing pKD46 and the plasmids pKD3 and pProEX-Hta were a gift from Nishad Matange at IISER Pune, India. The plasmids pCA24N-EcthiC, pCA24N-EcthiG, and pCA24N-EccobT were obtained from the ASKA collection hosted at IISER Pune, India. The E. coli KL-16 strain harboring the GFPmut2-kanR cassette was a gift from Deepa Agashe at NCBS, Bangalore, India.
Generating single-gene knockouts in E. coli.
All the single-knockout mutants of E. coli K-12 MG1655 used in the study were generated using recombination by the λ Red recombineering system (42, 56). The primer sequences used for generating the gene knockouts and for their verification are listed in Table S1 in the supplemental material. For generating the strains marked with GFP, we flipped out the kanr cassette from the thi mutants of E. coli K-12 MG1655. We then cloned and inserted the GFPmut2::kanR cassette from the E. coli KL-16 strain into the thi mutants, after the aidB gene, in the reverse orientation with respect to the aidB gene. This gave us the following E. coli mutants: thiC aidB1633::GFPmut2-kanR (EcΔthiC*), thiE aidB1633::GFPmut2-kanR (EcΔthiE*), and thiG aidB1633::GFPmut2-kanR (EcΔthiG*). The GFPmut2-kanR insertions were carried out using the same λ Red recombineering system mentioned above.
Primary culture setup (LB and P1 cultures) for E. coli.
E. coli K-12 MG1655 wild type (WT) and ΔthiC, ΔthiE, and ΔthiG mutants were grown in LB aerobically at 37°C, 180 rpm, for 6 to 8 h. The cultures were centrifuged at 6,500 rpm for 1 min, and the pellets were washed three times with 1× M9 salts by resuspending them using a vortex for each wash. This step was used to make sure that the cells do not carry over any residual nutrients from LB. These cells were used to start passage 1 (P1) cultures (first subcultures in minimal medium) in M9 medium containing NH4Cl, glucose, and inosine (50 μM), in 4 mL of medium in 25-mL test tubes, at a starting OD600 of 0.05 and were incubated aerobically at 37°C, 180 rpm, for 16 to 18 h. Cells grown in P1 were centrifuged at 6,500 rpm for 1 min, and the pellets were washed three times with 1× M9 salts. For the thi mutant rescue experiments, the mutants with or without pProEx-Hta or pCA24N plasmids harboring the genes mentioned were grown similarly in P1, supplemented with or without thiamin (20 μM).
Pairwise coculture setup of E. coli mutants in P2.
E. coli cells washed after P1 were used to start their cocultures in P2 (second subcultures in minimal salts medium with composition as described above) at a starting OD600 of 0.1, in a 96-well plate with a lid, with 200 μL medium in each well, and were incubated aerobically at 37°C, with ∼240 rpm orbital shaking, for 24 to 96 h, with OD600 reading and fluorescence reading at excitation/emission values of 485/535 after every shaking cycle of ∼300 s, with the upper lid at a temperature 2°C higher than 37°C (in EnSight) to avoid condensation, inside a plate reader (either Tecan or EnSight, respectively). Alternately, the cells were grown in 25-mL test tubes with 4 mL medium each at 37°C, 180 rpm, in a shaker incubator. The media used were supplemented with various nutrients as and when required at the concentrations mentioned for both the thiamin requirement and the HMP and THZ feeding studies. The two different mutants used for cocultures were inoculated either singly as a control or in their coculture at ratios of 1:9, 1:1, or 9:1. Both the single cultures and the cocultures were inoculated at a starting OD600 of 0.1; that is, e.g., for the cocultures with the 1:9 ratio, the two mutants were mixed at a starting OD600 of 0.01 and 0.09 of the individual mutants, respectively. When the alternate carbon sources were used, glucose (final concentration in the medium, 22.2 mM) was replaced with 33.3 mM Na succinate or 44.4 mM Na pyruvate, to keep the amount of carbon fed the same for all media.
V. anguillarum cultures and experiments.
Vibrio anguillarum PF430-3 (57, 58) wild type and ΔthiC, ΔthiE, and ΔthiG mutants (47) were used in experiments. All were reisolated from cryopreserved stock using marine broth agar plates and liquid medium (59) with estuarine surface water from the ModMon Neuse River Estuary monitoring station 180 (60) as the base medium. Cells from liquid ZoBell cultures in late exponential or early stationary phase were washed and centrifuged (9,000 × g, 3 min) three times with 1× M9 medium without thiamin and then resuspended in M9 medium without thiamin. Absorbance at 600 nm (i.e., optical density [OD]) was measured using a spectrophotometer (Genesys 30; Thermo Scientific). Based on the OD of resuspended cell cultures, 0.001 OD of washed cells was added (final density) at the start of each experiment.
Cocultures of PF430-3 strains were started by adding 0.001 OD (final concentration) of each strain to M9 medium. Cultures were grown in clear sterile polystyrene tubes and incubated in the dark at 20°C with daily homogenization by repetitive inversion. Thiamin hydrochloride, HMP, and THZ used in experiments were purchased from Fisher Scientific, TCI, and Alfa Aesar, respectively, at ≥98% HPLC purity. Fresh solutions of vitamins were prepared under reduced light in a laminar flow hood with autoclaved MilliQ water as the diluent. Solutions were kept on ice while the experiments were being set up.
E. coli JW5549 ΔthiG761::kan (Keio Collection) was reisolated as described for PF430-3 but using M9 as the base for ZoBell solid and liquid media. Cells were washed and resuspended in M9 medium without B1 as for PF430-3. Cocultures of V. anguillarum and E. coli thi mutants were initiated by adding 0.001 OD of each cell type to M9 medium without amended B1. Growth conditions were the same as for PF430-3.
Additional methods pertaining to medium and cloning techniques used, the thiochrome assay, and fluorescence versus OD600 correlation can be found in the supplemental material.
ACKNOWLEDGMENTS
We are grateful to the labs of Deepa Agashe and Nishad Matange for providing specific E. coli strains and plasmids used in our research. We thank Gayathri Pananghat, Manjula Reddy, and Nishad Matange for insightful discussions. We thank the PerkinElmer-IISER Pune Centre of Excellence, the IISER Pune Biological Mass spectrometry facility, and the labs of Sandanaraj Britto and Thomas Pucadyil and their members for help with instrumentation. We also thank Sandeep Krishna, Shashi Thutupalli, Ateek Shah, Yamini Mathur, and Yashwant Kumar for their critical input. We acknowledge Gina Welsing, Mir Nasir Ahmed, Rituparna Ghosh, and Saswata Nayak for their contributions to the project during their lab rotations and short-term research stints.
R.R.M.S. is supported by fellowships from IISER Pune and the Council for Scientific and Industrial Research (CSIR), India–NET. The research is supported by funds provided by the Ministry of Science and Technology, Government of India Department of Biotechnology (DBT), Ramalingaswami Re-Entry Fellowship BT/RLF/Re-Entry/12/2014 to A.B.H. and is in part based upon work supported by the U.S. National Science Foundation under grant no. OCE-2049388 to R.W.P.
Footnotes
Supplemental material is available online only.
Contributor Information
Ryan W. Paerl, Email: rpaerl@ncsu.edu.
Amrita B. Hazra, Email: amrita@iiserpune.ac.in.
Elizabeth Anne Shank, University of Massachusetts Medical School.
REFERENCES
- 1.Zengler K, Zaramela LS. 2018. The social network of microorganisms—how auxotrophies shape complex communities. Nat Rev Microbiol 16:383–390. doi: 10.1038/s41579-018-0004-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.D'Souza G, Shitut S, Preussger D, Yousif G, Waschina S, Kost C. 2018. Ecology and evolution of metabolic cross-feeding interactions in bacteria. Nat Prod Rep 35:455–488. doi: 10.1039/c8np00009c. [DOI] [PubMed] [Google Scholar]
- 3.Douglas AE. 2020. The microbial exometabolome: ecological resource and architect of microbial communities. Philos Trans R Soc Lond B Biol Sci 375:20190250. doi: 10.1098/rstb.2019.0250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shou W, Ram S, Vilar JMG. 2007. Synthetic cooperation in engineered yeast populations. Proc Natl Acad Sci USA 104:1877–1882. doi: 10.1073/pnas.0610575104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cooper MB, Kazamia E, Helliwell KE, Kudahl UJ, Sayer A, Wheeler GL, Smith AG. 2019. Cross-exchange of B-vitamins underpins a mutualistic interaction between Ostreococcus tauri and Dinoroseobacter shibae. ISME J 13:334–345. doi: 10.1038/s41396-018-0274-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pande S, Shitut S, Freund L, Westermann M, Bertels F, Colesie C, Bischofs IB, Kost C. 2015. Metabolic cross-feeding via intercellular nanotubes among bacteria. Nat Commun 6:6238. doi: 10.1038/ncomms7238. [DOI] [PubMed] [Google Scholar]
- 7.Wang P-H, Correia K, Ho H, Venayak N, Nemr K, Flick R, Mahadevan R, Edwards EA. 2019. An interspecies malate-pyruvate shuttle reconciles redox imbalance in an anaerobic microbial community. ISME J 13:1042–1055. doi: 10.1038/s41396-018-0333-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kazamia E, Helliwell KE, Purton S, Smith AG. 2016. How mutualisms arise in phytoplankton communities: building eco‐evolutionary principles for aquatic microbes. Ecol Lett 19:810–822. doi: 10.1111/ele.12615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Helliwell KE, Wheeler GL, Leptos KC, Goldstein RE, Smith AG. 2011. Insights into the evolution of vitamin B12 auxotrophy from sequenced algal genomes. Mol Biol Evol 28:2921–2933. doi: 10.1093/molbev/msr124. [DOI] [PubMed] [Google Scholar]
- 10.Giovannoni SJ, Tripp HJ, Givan S, Podar M, Vergin KL, Baptista D, Bibbs L, Eads J, Richardson TH, Noordewier M, Rappe MS, Short JM, Carrington JC, Mathur EJ. 2005. Genome streamlining in a cosmopolitan oceanic bacterium. Science 309:1242–1245. doi: 10.1126/science.1114057. [DOI] [PubMed] [Google Scholar]
- 11.Helliwell KE, Wheeler GL, Smith AG. 2013. Widespread decay of vitamin-related pathways: coincidence or consequence? Trends Genet 29:469–478. doi: 10.1016/j.tig.2013.03.003. [DOI] [PubMed] [Google Scholar]
- 12.D'Souza G, Waschina S, Pande S, Bohl K, Kaleta C, Kost C. 2014. Less is more: selective advantages can explain the prevalent loss of biosynthetic genes in bacteria. Evolution 68:2559–2570. doi: 10.1111/evo.12468. [DOI] [PubMed] [Google Scholar]
- 13.Hillesland KL, Lim S, Flowers JJ, Turkarslan S, Pinel N, Zane GM, Elliott N, Qin Y, Wu L, Baliga NS, Zhou J, Wall JD, Stahl DA. 2014. Erosion of functional independence early in the evolution of a microbial mutualism. Proc Natl Acad Sci USA 111:14822–14827. doi: 10.1073/pnas.1407986111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Banerjee S, Schlaeppi K, van der Heijden MGA. 2018. Keystone taxa as drivers of microbiome structure and functioning. Nat Rev Microbiol 16:567–576. doi: 10.1038/s41579-018-0024-1. [DOI] [PubMed] [Google Scholar]
- 15.Ze X, Duncan SH, Louis P, Flint HJ. 2012. Ruminococcus bromii is a keystone species for the degradation of resistant starch in the human colon. ISME J 6:1535–1543. doi: 10.1038/ismej.2012.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Romine MF, Rodionov DA, Maezato Y, Osterman AL, Nelson WC. 2017. Underlying mechanisms for syntrophic metabolism of essential enzyme cofactors in microbial communities. ISME J 11:1434–1446. doi: 10.1038/ismej.2017.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rodionov DA, Arzamasov AA, Khoroshkin MS, Iablokov SN, Leyn SA, Peterson SN, Novichkov PS, Osterman AL. 2019. Micronutrient requirements and sharing capabilities of the human gut microbiome. Front Microbiol 10:1316. doi: 10.3389/fmicb.2019.01316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sañudo-Wilhelmy SA, Gómez-Consarnau L, Suffridge C, Webb EA. 2014. The role of B vitamins in marine biogeochemistry. Annu Rev Mar Sci 6:339–367. doi: 10.1146/annurev-marine-120710-100912. [DOI] [PubMed] [Google Scholar]
- 19.Sharma V, Rodionov DA, Leyn SA, Tran D, Iablokov SN, Ding H, Peterson DA, Osterman AL, Peterson SN. 2019. B-vitamin sharing promotes stability of gut microbial communities. Front Microbiol 10:1485. doi: 10.3389/fmicb.2019.01485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rodionova IA, Li X, Plymale AE, Motamedchaboki K, Konopka AE, Romine MF, Fredrickson JK, Osterman AL, Rodionov DA. 2015. Genomic distribution of B-vitamin auxotrophy and uptake transporters in environmental bacteria from the Chloroflexi phylum. Environ Microbiol Rep 7:204–210. doi: 10.1111/1758-2229.12227. [DOI] [PubMed] [Google Scholar]
- 21.Soderberg T. 2016. Organic chemistry with a biological emphasis, vol II. Chemistry Publications, University of Minnesota, Morris, MN. [Google Scholar]
- 22.Hazra A, Chatterjee A, Begley TP. 2009. Biosynthesis of the thiamin thiazole in Bacillus subtilis: identification of the product of the thiazole synthase-catalyzed reaction. J Am Chem Soc 131:3225–3229. doi: 10.1021/ja806752h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hazra AB, Han Y, Chatterjee A, Zhang Y, Lai R, Ealick SE, Begley TP. 2011. A missing enzyme in thiamin thiazole biosynthesis: identification of TenI as a thiazole tautomerase. J Am Chem Soc 133:9311–9319. doi: 10.1021/ja1110514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chatterjee A, Hazra AB, Abdelwahed S, Hilmey DG, Begley TP. 2010. A radical dance in thiamine biosynthesis: mechanistic analysis of the bacterial hydroxymethylpyrimidine phosphate synthase. Angew Chem Int Ed Engl 49:8653–8656. doi: 10.1002/anie.201003419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Begley TP, Downs DM, Ealick SE, McLafferty FW, Van Loon APGM, Taylor S, Campobasso N, Chiu H-J, Kinsland C, Reddick JJ, Xi J. 1999. Thiamin biosynthesis in prokaryotes. Arch Microbiol 171:293–300. doi: 10.1007/s002030050713. [DOI] [PubMed] [Google Scholar]
- 26.Jurgenson CT, Begley TP, Ealick SE. 2009. The structural and biochemical foundations of thiamin biosynthesis. Annu Rev Biochem 78:569–603. doi: 10.1146/annurev.biochem.78.072407.102340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Peapus DH, Chiu HJ, Campobasso N, Reddick JJ, Begley TP, Ealick SE. 2001. Structural characterization of the enzyme-substrate, enzyme-intermediate, and enzyme-product complexes of thiamin phosphate synthase. Biochemistry 40:10103–10114. doi: 10.1021/bi0104726. [DOI] [PubMed] [Google Scholar]
- 28.Nakayama H, Hayashi R. 1972. Biosynthesis of thiamine pyrophosphate in Escherichia coli. J Bacteriol 109:936–938. doi: 10.1128/jb.109.2.936-938.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Imamura N, Nakayama H. 1982. thiK and thiL loci of Escherichia coli. J Bacteriol 151:708–717. doi: 10.1128/jb.151.2.708-717.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schyns G, Potot S, Geng Y, Barbosa TM, Henriques A, Perkins JB. 2005. Isolation and characterization of new thiamine-deregulated mutants of Bacillus subtilis. J Bacteriol 187:8127–8136. doi: 10.1128/JB.187.23.8127-8136.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nosaka K, Kaneko Y, Nishimura H, Iwashima A. 1993. Isolation and characterization of a thiamin pyrophosphokinase gene, THI80, from Saccharomyces cerevisiae. J Biol Chem 268:17440–17447. doi: 10.1016/S0021-9258(19)85354-1. [DOI] [PubMed] [Google Scholar]
- 32.Gutowska MA, Shome B, Sudek S, Mcrose DL, Hamilton M, Giovannoni SJ, Begley TP, Worden AZ. 2017. Globally important haptophyte algae use exogenous pyrimidine compounds more efficiently than thiamin. mBio 8:e01459-17. doi: 10.1128/mBio.01459-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Carini P, Campbell EO, Morré J, Sañudo-Wilhelmy SA, Thrash JC, Bennett SE, Temperton B, Begley T, Giovannoni SJ. 2014. Discovery of a SAR11 growth requirement for thiamin’s pyrimidine precursor and its distribution in the Sargasso Sea. ISME J 8:1727–1738. doi: 10.1038/ismej.2014.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Paerl RW, Bouget FY, Lozano JC, Vergé V, Schatt P, Allen EE, Palenik B, Azam F. 2017. Use of plankton-derived vitamin B1 precursors, especially thiazole-related precursor, by key marine picoeukaryotic phytoplankton. ISME J 11:753–765. doi: 10.1038/ismej.2016.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rodionov DA, Vitreschak AG, Mironov AA, Gelfand MS. 2002. Comparative genomics of thiamin biosynthesis in procaryotes. New genes and regulatory mechanisms. J Biol Chem 277:48949–48959. doi: 10.1074/jbc.M208965200. [DOI] [PubMed] [Google Scholar]
- 36.Jaehme M, Slotboom DJ. 2015. Diversity of membrane transport proteins for vitamins in bacteria and archaea. Biochim Biophys Acta 1850:565–576. doi: 10.1016/j.bbagen.2014.05.006. [DOI] [PubMed] [Google Scholar]
- 37.Belda E, Moya A, Bentley S, Silva FJ. 2010. Mobile genetic element proliferation and gene inactivation impact over the genome structure and metabolic capabilities of Sodalis glossinidius, the secondary endosymbiont of tsetse flies. BMC Genomics 11:449. doi: 10.1186/1471-2164-11-449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Paerl RW, Bertrand EM, Rowland E, Schatt P, Mehiri M, Niehaus TD, Hanson AD, Riemann L, Bouget FY. 2018. Carboxythiazole is a key microbial nutrient currency and critical component of thiamin biosynthesis. Sci Rep 8:5940. doi: 10.1038/s41598-018-24321-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rodionov DA, Hebbeln P, Eudes A, Beek J, Rodionova IA, Erkens GB, Slotboom DJ, Gelfand MS, Osterman AL, Hanson AD, Eitinger T. 2009. A novel class of modular transporters for vitamins in prokaryotes. J Bacteriol 191:42–51. doi: 10.1128/JB.01208-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jenkins AH, Schyns G, Potot S, Sun G, Begley TP. 2007. A new thiamin salvage pathway. Nat Chem Biol 3:492–497. doi: 10.1038/nchembio.2007.13. [DOI] [PubMed] [Google Scholar]
- 41.Miranda-Ríos J, Navarro M, Soberón M. 2001. A conserved RNA structure (thi box) is involved in regulation of thiamin biosynthetic gene expression in bacteria. Proc Natl Acad Sci USA 98:9736–9741. doi: 10.1073/pnas.161168098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL, Mori H. 2006. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol Syst Biol 2:2006.0008. doi: 10.1038/msb4100050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kim J, Copley SD. 2007. Why metabolic enzymes are essential or nonessential for growth of Escherichia coli K12 on glucose. Biochemistry 46:12501–12511. doi: 10.1021/bi7014629. [DOI] [PubMed] [Google Scholar]
- 44.Bazurto JV, Farley KR, Downs DM. 2016. An unexpected route to an essential cofactor: Escherichia coli relies on threonine for thiamine biosynthesis. mBio 7:e01840-15. doi: 10.1128/mBio.01840-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Costliow ZA, Degnan PH. 2017. Thiamine acquisition strategies impact metabolism and competition in the gut microbe Bacteroides thetaiotaomicron. mSystems 2:e00116-17. doi: 10.1128/mSystems.00116-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vander Horn PB, Backstrom AD, Stewart V, Begley TP. 1993. Structural genes for thiamine biosynthetic enzymes (thiCEFGH) in Escherichia coli K-12. J Bacteriol 175:982–992. doi: 10.1128/jb.175.4.982-992.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Paerl RW, Sundh J, Tan D, Svenningsen SL, Hylander S, Pinhassi J, Andersson AF, Riemann L. 2018. Prevalent reliance of bacterioplankton on exogenous vitamin B1 and precursor availability. Proc Natl Acad Sci USA 115:E10447–E10456. doi: 10.1073/pnas.1806425115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mizote T, Nakayama H. 1989. The thiM locus and its relation to phosphorylation of hydroxyethylthiazole in Escherichia coli. J Bacteriol 171:3228–3232. doi: 10.1128/jb.171.6.3228-3232.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Leonardi R, Roach PL. 2004. Thiamine biosynthesis in Escherichia coli—in Vitro reconstitution of the thiazole synthase activity. J Biol Chem 279:17054–17062. doi: 10.1074/jbc.M312714200. [DOI] [PubMed] [Google Scholar]
- 50.Suffridge CP, Bolaños LM, Bergauer K, Worden AZ, Morré J, Behrenfeld MJ, Giovannoni SJ. 2020. Exploring vitamin B1 cycling and its connections to the microbial community in the North Atlantic Ocean. Front Mar Sci 7:606342. doi: 10.3389/fmars.2020.606342. [DOI] [Google Scholar]
- 51.Taylor SV, Kelleher NL, Kinsland C, Chiu HJ, Costello CA, Backstrom AD, McLafferty FW, Begley TP. 1998. Thiamin biosynthesis in Escherichia coli. Identification of this thiocarboxylate as the immediate sulfur donor in the thiazole formation. J Biol Chem 273:16555–16560. doi: 10.1074/jbc.273.26.16555. [DOI] [PubMed] [Google Scholar]
- 52.Challand MR, Martins FT, Roach PL. 2010. Catalytic activity of the anaerobic tyrosine lyase required for thiamine biosynthesis in Escherichia coli. J Biol Chem 285:5240–5248. doi: 10.1074/jbc.M109.056606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chatterjee A, Abeydeera ND, Bale S, Pai PJ, Dorrestein PC, Russell DH, Ealick SE, Begley TP. 2011. Saccharomyces cerevisiae THI4p is a suicide thiamine thiazole synthase. Nature 478:542–546. doi: 10.1038/nature10503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang Y, Morar M, Ealick SE. 2008. Structural biology of the purine biosynthetic pathway. Cell Mol Life Sci 65:3699–3724. doi: 10.1007/s00018-008-8295-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mehta AP, Abdelwahed SH, Fenwick MK, Hazra AB, Taga ME, Zhang Y, Ealick SE, Begley TP. 2015. Anaerobic 5-hydroxybenzimidazole formation from aminoimidazole ribotide: an unanticipated intersection of thiamin and vitamin B12 biosynthesis. J Am Chem Soc 137:10444–10447. doi: 10.1021/jacs.5b03576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci USA 97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tan D, Lo Svenningsen S, Middelboe M. 2015. Quorum sensing determines the choice of antiphage defense strategy in Vibrio anguillarum. mBio 6:e00627-15. doi: 10.1128/mBio.00627-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Silva-Rubio A, Avendaño-Herrera R, Jaureguiberry B, Toranzo AE, Magariños B. 2008. First description of serotype O3 in Vibrio anguillarum strains isolated from salmonids in Chile. J Fish Dis 31:235–239. doi: 10.1111/j.1365-2761.2007.00878.x. [DOI] [PubMed] [Google Scholar]
- 59.ZoBell CE. 1941. Studies on marine bacteria. I. The cultural requirements of heterotrophic aerobes. J Mar Res IV:42–75. [Google Scholar]
- 60.Paerl HW, Hall NS, Hounshell AG, Luettich RA, Jr, Rossignol KL, Osburn CL, Bales J. 2019. Recent increase in catastrophic tropical cyclone flooding in coastal North Carolina, USA: long-term observations suggest a regime shift. Sci Rep 9:10620. doi: 10.1038/s41598-019-46928-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental file 1. Download jb.00503-21-s0001.pdf, PDF file, 1.1 MB (1.1MB, pdf)