ABSTRACT
β-Lactamase-mediated resistance to ceftazidime-avibactam (CZA) is a serious limitation in the treatment of Gram-negative bacteria harboring Klebsiella pneumoniae carbapenemase (KPC). Herein, the basis of susceptibility to carbapenems and resistance to ceftazidime (CAZ) and CZA of the D179Y variant of KPC-2 and -3 was explored. First, we determined that resistance to CZA in a laboratory strain of Escherichia coli DH10B was not due to increased expression levels of the variant enzymes, as demonstrated by reverse transcription PCR (RT-PCR). Using timed mass spectrometry, the D179Y variant formed prolonged acyl-enzyme complexes with imipenem (IMI) and meropenem (MEM) in KPC-2 and KPC-3, which could be detected up to 24 h, suggesting that IMI and MEM act as covalent β-lactamase inhibitors more than as substrates for D179Y KPC-2 and -3. This prolonged acyl-enzyme complex of IMI and MEM by D179Y variants was not observed with wild-type (WT) KPCs. CAZ was studied and the D179Y variants also formed acyl-enzyme complexes (1 to 2 h). Thermal denaturation and differential scanning fluorimetry showed that the tyrosine substitution at position 179 destabilized the KPC β-lactamases (KPC-2/3 melting temperature [Tm] of 54 to 55°C versus D179Y Tm of 47.5 to 51°C), and the D179Y protein was 3% disordered compared to KPC-2 at 318 K. Heteronuclear 1H/15N-heteronuclear single quantum coherence (HSQC) nuclear magnetic resonance (NMR) spectroscopy also revealed that the D179Y variant, compared to KPC-2, is partially disordered. Based upon these observations, we discuss the impact of disordering of the Ω loop as a consequence of the D179Y substitution. These conformational changes and disorder in the overall structure as a result of D179Y contribute to this unanticipated phenotype.
KEYWORDS: Klebsiella pneumoniae, D179Y, ceftazidime-avibactam resistance, antibiotic resistance, KPC, KPC D179Y, beta-lactamases
INTRODUCTION
The emergence of multidrug-resistant (MDR) Klebsiella pneumoniae-producing KPC β-lactamases has produced a significant challenge to our current armamentarium of antimicrobial therapeutics. In an effort to combat these organisms, alternative therapies were developed, such as the β-lactam-β-lactamase inhibitor combinations ceftazidime-avibactam (CZA), imipenem/relebactam (IMI/REL), and meropenem-vaborbactam (MEM/VAB). CZA was a welcomed addition, as the diazabicyclooctane (DBO) inhibitor avibactam (AVI) rendered many clinical K. pneumoniae isolates possessing K. pneumoniae carbapenemase 2 (KPC-2) and KPC-3 β-lactamases susceptible to ceftazidime (CAZ). The same is true for other combinations.
Early clues as to the importance of the Ω loop in KPC-2 came in the studies of site-directed mutagenesis at the R164 site. Here, Levitt et al. demonstrated that many substitutions at this position resulted in increased CAZ resistance, accompanied by carbapenem susceptibility (1). Equally significant were early studies from 2015 that predicted the rise of the CZA-resistant KPC variants. Winkler et al. performed site-directed mutagenesis of KPC-2 in an isogenic lab Escherichia coli background generating D179A, D179Q, and D179N variants, all of which were CZA resistant (2). Through kinetic analysis, it was concluded that AVI was able to inactivate both KPC-2 and D179N; both were rapidly acylated, but the koff rate was slow. Additionally, D179N exhibited a high affinity for CAZ, and this “burst” may inhibit AVI binding due to preferentially fast binding of CAZ. In the same year, Livermore et al. demonstrated that CZA resistance could be selected for by serial passage of K. pneumoniae in the presence of CZA. These isolates also demonstrated carbapenem susceptibility. Despite being previously performed under laboratory conditions, this was the first report of CZA resistance and carbapenem susceptibility in clinical isolates attributed to the D179Y substitution in a KPC-3 background (3).
Shields et al., in 2016, provided the first case reports of CZA resistance in renal transplant patients (4). In subsequent analyses, Shields et al. determined via conjugation of the native plasmid carrying the KPC-3 D179Y variant into a laboratory strain of E. coli, and via direct cloning of the D179Y variant, that this resistance was attributable to the D179Y substitution in a KPC-3 background. Similarly, they observed CZA resistance concomitant with carbapenem (MEM and ertapenem) susceptibility (5). Even more worrisome was the fact that after serial passage of the D179Y KPC-3 variant in MEM, they were able to select for isolates that were both CZA (16 to 128 μg/mL) and MEM (64 μg/mL) resistant. This came about in three ways: (i) reversion of the D179Y variant to wild-type (WT) KPC-3 combined with an insertion sequence into OmpK36, (ii) conversion to KPC-3 T243A, and (iii) conversion to KPC-3 T243A and a mutation in OmpK36 F362D (6).
In 2017, Compain and Arthur studied D179Y KPC-2 in laboratory strains of E. coli and via kinetic analysis of purified protein (7). This was one of the first studies of D179Y in the KPC-2 background. They serially passaged isolates possessing KPC-2 in CZA and were able to select CZA-resistant variants (32 μg/mL) with simultaneous carbapenem susceptibility (MEM and IMI) confirming the above observations. Kinetic analysis of KPC-2 and the D179Y KPC-2 variant showed that for D179Y there was a drastic reduction in the efficacy of AVI inhibition and residual CAZ hydrolysis (greatly lowered Km offset by a large decrease in kcat) that allows for CZA resistance (7). This was in contrast to what was reported for the D179N in the earlier studies (2). Compain and Arthur also reported a slower carbamylation rate for AVI in the D179Y variant.
At the same time, Barnes et al. performed site-saturation mutagenesis at site 179 in a KPC-2 background (8). Similar to the Compain study, the D179N and D179Y variants demonstrated CZA resistance (16 and 64 μg/mL, respectively) and carbapenem susceptibility was observed for the D179Y variant. Relative to KPC-2, the D179N variant had increased affinity for CAZ and IMI but decreased hydrolysis. Mass spectrometry indicated that the D179N variant trapped CAZ at <15 min and was able to hydrolyze it, whereas IMI was trapped at >15 min. Modeling suggested that in the D179N KPC-2 variant, the H-bonding network between R164 and D179 is disrupted, thereby altering the position of S70 and posing a challenge to acylation. The main conclusion from the study was that D179N preferentially bound CAZ relative to AVI, whereas KPC-2 preferentially bound AVI (8). Also in 2017, Zhang et al. reported CZA resistance mediated by an unusual TIPY insertion in penicillin-binding protein 3 (PBP3) of E. coli (9).
Two later studies done in 2020 furthered the kinetic analysis of the D179Y substitution in both KPC-2 and KPC-3 backgrounds (10, 11). Shapiro et al. and Tsivkovski et al. both recapitulated the CZA resistance in both P. aeruginosa and laboratory strains of E. coli, respectively, and reaffirmed the findings above that the D179Y substitution greatly increases the affinity of CAZ (large decrease in Km) (10, 11). Subsequently, Tsivkovski et al. showed that this gain in affinity was somewhat offset by a lowered kcat. They noted that the D179Y KPC-2 variant had a 20-fold increase in 50% inhibitory concentration (IC50) for AVI inhibition of CAZ hydrolysis and that it hydrolyzed CAZ with a 10-fold higher efficiency (11). Shapiro et al. came to a slightly different conclusion with regard to the CAZ hydrolysis and AVI inhibition. They demonstrated that in the KPC-3 background, the D179Y decrease in Km was offset by a decrease in kcat and that a net higher catalytic efficiency was not obtained for CAZ (10). However, the double mutant of D179Y and T243M in KPC-3 was in qualitative agreement with those of Tsivkovski and Compain. In addition, Shapiro et al. did not show differences in inhibition by AVI between KPC-3 and the D179Y KPC-3 variant. In the clinic, other D179 variants in K. pneumoniae isolates are reported and confer CZA resistance and carbapenem susceptibility. They include KPC-57 (D179V KPC-3), KPC-78 (D179A KPC-3), and GenBank accession no. NG_071204.1 (12, 13).
What is the structural basis for this unusual phenotype? In attempts to answer this question and understand the evolution of this complex substrate specificity, the catalytic mechanisms of susceptibility to IMI/MEM and the resistance to CAZ/CZA of the D179Y variants in KPC-2 and KPC-3 were explored. Our investigations using nuclear magnetic resonance (NMR) and differential scanning fluorimetry suggest that the conformational dynamics and protein disorder underlie the novel properties of the D179Y variants of KPC-2 and KPC-3. This appreciation of dynamic protein changes can inform future structural studies in order to understand novel catalytic properties and design more effective β-lactams and β-lactam inhibitors.
RESULTS AND DISCUSSION
MICs and immunoblots.
Previously reported MICs and immunoblots for analysis of steady-state expression levels (8) were repeated for consistency and are presented in Table S1 and Fig. S1. The KPC-2 D179Y and KPC-3 D179Y variants expressed in E. coli DH10B were reconfirmed to be resistant to CAZ and CZA and susceptible to IMI and MEM.
The major immunogenic epitopes of KPC-2 β-lactamase were mapped previously, and one antibody-binding region consisted of the following amino acid sequence: A172IPGDARDTSSPR184. Therefore, differences observed in expression levels between D179Y KPC-2 and D179Y KPC-3 and their WT counterparts via immunoblot analysis may be due to differential antibody binding, as equal amounts of purified WT and D179Y variant β-lactamases were assayed, and antibody binding differences were noted (Fig. S1). Thus, reverse transcription PCR (RT-PCR) was utilized to circumvent this problem.
RT-PCR.
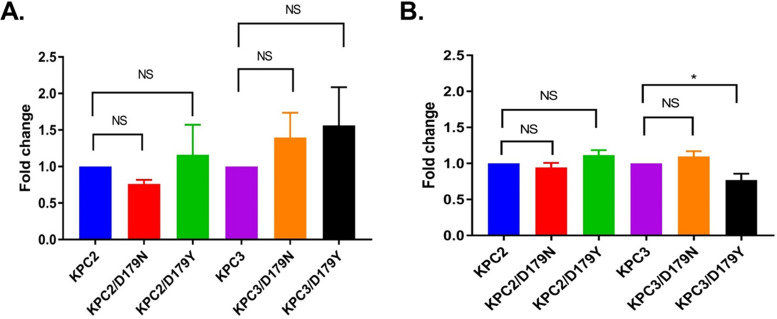
RT-PCR was undertaken because previous epitope mapping revealed that D179 was in a major immunogenic epitope of KPC-2, the β-lactamase the antibody was raised against (14). Results from the RT-PCR demonstrated that mRNA levels of blaKPC-D179N or blaKPC-D179Y (in both a KPC-2 and a KPC-3 background), in either the absence (Fig. 1A) or presence of ceftazidime (32 μg/mL) (Fig. 1B), did not differ significantly from those of their WT counterparts. We conclude that differences in observed MICs were not a result of large disparities in expression between the D179Y variants and the WT enzymes.
FIG 1.
Expression of KPC-2 and KPC-3 did not differ significantly from that of D179N and D179Y variants. RT-PCR data for gene expression by E. coli DH10-β in (A) LB medium and (B) LB medium supplemented with ceftazidime (32 μg/mL) are shown. Expression of KPC-2 and KPC-3 was set at 1.0 (normalized to 16S rRNA), and relative expression of variant genes is presented as the mean ± standard deviations. NS, nonsignificant, P > 0.05. *, KPC3/D179Y expression (0.77 ± 0.09 [P = 0.01]) was not considered significantly decreased since the difference relative to that of KPC3 was ≤2-fold (1.3-fold).
Thermal stability, NMR spectroscopy, and steady-state kinetics.
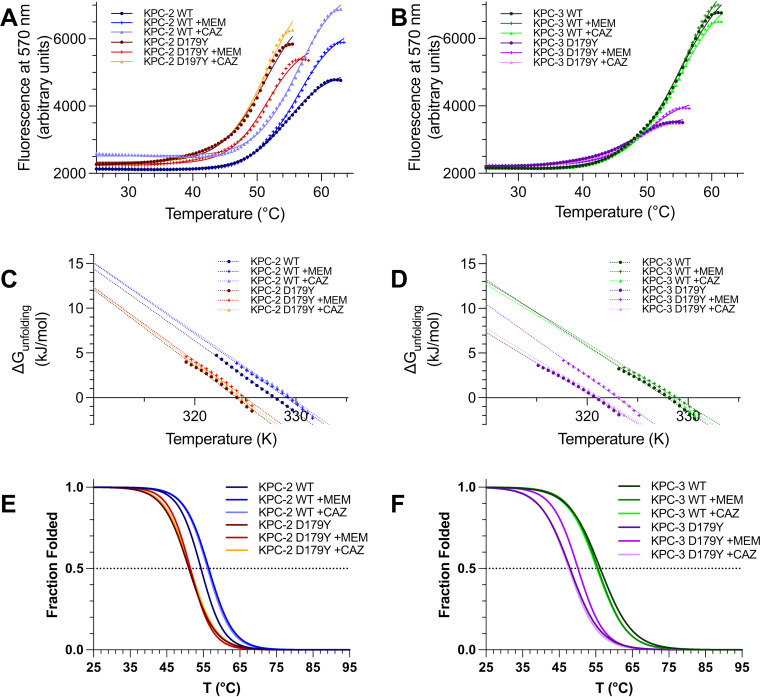
Figure 2 represents our investigations assessing thermal denaturation measured by differential scanning fluorimetry (DSF). KPC-2 and KPC-3 exhibit equal stability, as evidenced by similar melting temperatures (Tm) and similar biological standard state Gibbs energy of unfolding (ΔuG⨁) values for the WT enzymes by DSF (Table 1). In contrast, the D179Y variants of KPC-2 and KPC-3 are less stable than the WT enzymes as evidenced by both Tm and ΔuG⨁.
FIG 2.
Thermal denaturation by differential scanning fluorimetry shows that KPC-2 D179Y (A) and KPC-3 D179Y (B) exhibit lower melting temperatures (Tm) than the corresponding WT enzyme. An analysis to extract the Gibbs free energy of unfolding (ΔuG) confirms that KPC-2 D179Y (C) and KPC-3 D179Y (D) are thermodynamically less stable than the corresponding WT enzymes. The addition of MEM significantly stabilizes KPC-3 D179Y, as evidenced by increases in Tm and ΔuG ⨁. (E) The fraction of folded protein, determined by a sigmoidal Boltzmann fit to differential scanning fluorimetry data, is provided for WT (KPC-2 WT) and D179Y KPC-2 (KPC-2 D179Y) in the apo form, or in the presence of MEM or CAZ. (F) The fraction of folded protein, determined by a sigmoidal Boltzmann fit to differential scanning fluorimetry data, is provide for WT (KPC-3 WT) and D179Y KPC-3 (KPC-3 D179Y) in the apo form, or in the presence of MEM or CAZ.
TABLE 1.
Thermal stability studies by differential scanning fluorimetry
| β-Lactamase |
Tm(°C) |
ΔTm(°C)a |
ΔuG ⨁ (kJ/mol)b |
|||
|---|---|---|---|---|---|---|
| Apo enzyme | +MEM | +CAZ | Apo enzyme | +MEM | +CAZ | |
| KPC-2 | 54.7 ± 0.2 | +1.5 ± 0.3 | +1.6 ± 1.2 | 14.3 | 14.9 | 14.9 |
| D179YKPC-2 | 51.2 ± 0.6 | +0.4 ± 0.2 | +0.4 ± 1.0 | 11.9 | 12.3 | 12.2 |
| KPC-3 | 55.1 ± 1.0 | +0.7 ± 0.4 | −0.2 ± 0.3 | 13.2 | 12.9 | 12.6 |
| D179YKPC-3 | 47.5 ± 0.5 | +2.2 ± 0.3 | +0.5 ± 0.4 | 7.2 | 10.3 | 7.7 |
Change in Tm relative to the apo enzyme.
The value of ΔuG ⨁ is the value of ΔuG at 310.15 K.
In addition to the differences in thermal stability for the apo enzymes using these techniques, the addition of β-lactam substrates also affects thermostability. When examining Tm values, calculating ΔuG⨁ as a thermodynamic measure of stability shows that MEM stabilizes the D179Y variants of KPC-2 and KPC-3, and in the case of D179Y KPC-3, the stabilization is approximately 3 kJ/mol, potentially due to the formation of an additional hydrogen bond between the variant enzyme and MEM.
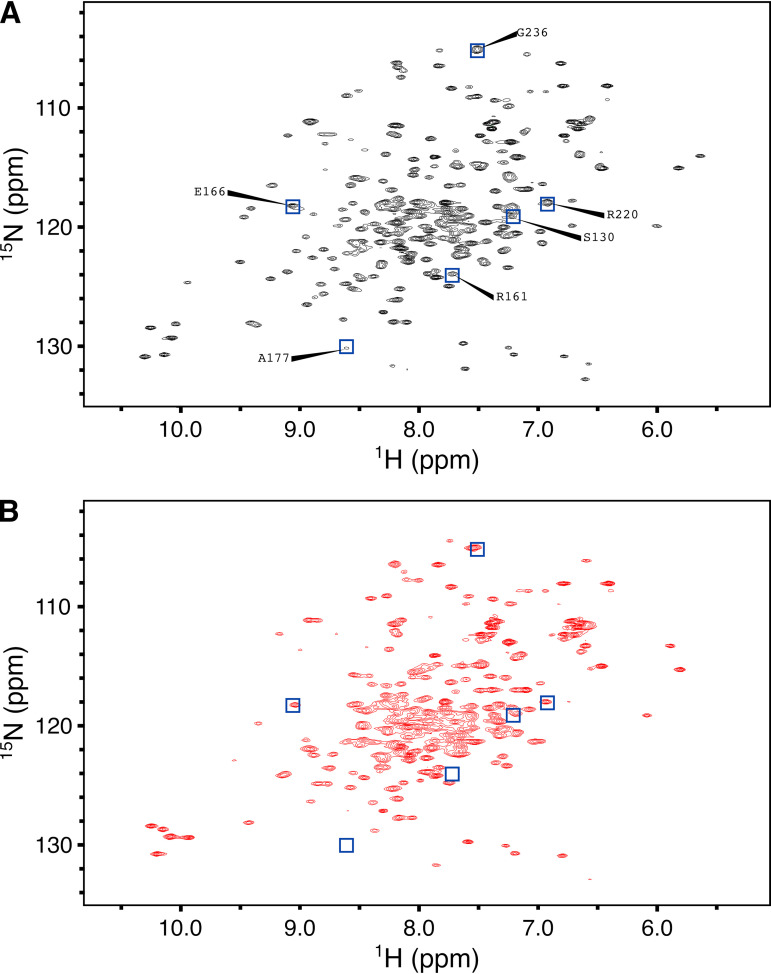
Heteronuclear 1H/15N-HSQC NMR spectra of WT KPC-2 and D179Y KPC-2 also suggest decreased stability of the D179Y mutant (Fig. 3). The HSQC spectra of WT KPC-2, acquired at 308 K, exhibit good chemical shift dispersion, well-defined resonances, and reasonably uniform peak intensities characteristic of a well-folded protein (Fig. 3A). In fact, the spectral quality of WT KPC-2 at 308 K enabled backbone resonance assignment (15). We have added resonance assignments for a select group of residues in Fig. 3A. These resonances correspond to residues near the active site and Ω loop and for which the resonances have sufficient dispersion to enable identification.
FIG 3.
NMR spectra at 308 K of WT KPC-2 (A) and D179Y KPC-2 (B) are markedly different. Poorer chemical shift dispersion and broadening of resonances for D179Y KPC-2 (B) compared to those for WT KPC-2 (A) suggest that D179Y KPC-2 is partially disordered at 308 K. Blue squares with labels indicate resonance assignments from WT KPC-2 for a selected subset of residues near the active site or Ω loop (A). The corresponding positions in the D179Y KPC-2 spectra are also shown as blue squares (B).
In contrast, the HSQC spectra of D179Y KPC-2, acquired at 308 K (Fig. 3B), feature a collapse of chemical shifts for many resonances, broadening of the bulk of resonances, and increased variability in the intensity of resonances, all indicative of NMR spectra for a partially disordered protein (16, 17). Boxes in panel B are the locations in spectral space identical to those in panel A and allow for comparisons of similarities and differences between locations or presence of resonances between the WT spectrum and the D179Y mutant spectrum. Comparing the WT and D179Y constructs at 308 K, fitting of DSF data to a Boltzmann sigmoid (Fig. 2E and F) suggests that the WT protein is >99.9% folded at 308 K, while the D179Y construct is 3% disordered at 308 K, a fraction disordered not reached by WT KPC-2 until 318 K.
As was previously established, WT KPC-2 and KPC-3 are able to hydrolyze MEM (kcat ≈ 5 s−1) and IMI (kcat ≈ 22 s−1) (Fig. S2) (7, 8). Under these steady-state conditions, the D179Y variants do not appreciably hydrolyze MEM or IMI. Progress curves show that the D179Y variant of KPC-2 and KPC-3 poorly hydrolyzes CAZ.
Timed mass spectrometry.
Experiments using electrospray ionization timed mass spectrometry (ESI-MS) were undertaken to investigate if the WT KPC-2 or KPC-3 and the D179Y variants form prolonged (24 h) acyl-complexes with MEM and IMI (Tables 2 and 3 and Fig. S3A and B). Here, 10 μg of each β-lactamase with and without IMI or MEM (1:1 molar ratio) and CAZ (1:25 molar ratio) was incubated for the time indicated and analyzed using the quadrupole time-of-flight (QTOF) mass spectrometer.
TABLE 2.
Mass spectrometry analysis using MEM (atomic mass units [AMU])
| β-Lactamase or antibiotic (1:1 ratio) | Mass spectrometry result (AMU) |
||
|---|---|---|---|
| Apo | +MEM 5 min | +MEM 24 h | |
| MEM | 383 ± 3 | ||
| KPC-2 | 28,319 ± 3 | 28,319 ± 3 | 28,319 ± 3 |
| D179YKPC-2 | 28,366 ± 3 | 28,750 ± 3 | 28,366 ± 3 (90%), 28,750 ± 3 (10%) |
| KPC-3 | 28,344 ± 3 | 28,344 ± 3 | 28,344 ± 3 |
| D179YKPC-3 | 28,393 ± 3 | 28,393 ± 3 (25%), 28,777 ± 3 (75%) | 28,393 ± 3 (90%), 28,777 ± 3 (10%) |
TABLE 3.
Mass spectrometry analysis using IMI (atomic mass units)
| β-Lactamase or antibiotic (1:1 ratio) | Mass spectrometry result (AMU) |
||||
|---|---|---|---|---|---|
| Apo | +IMI 1 h | +IMI 2 h | +IMI 4 h | +IMI 24 h | |
| IMI | 299 ± 3 | ||||
| KPC-2 | 28,319 ± 3 | 28,319 ± 3 | 28,319 ± 3 | 28,319 ± 3 | 28,319 ± 3 |
| D179YKPC-2 | 28,366 ± 3 | 28,665 ± 3 | 28,665 ± 3 | 28,665 ± 3 | 28,366 ± 3 (66%), 28,665 ± 3 (34%) |
| KPC-3 | 28,344 ± 3 | 28,344 ± 3 | 28,344 ± 3 | 28,344 ± 3 | 28,344 ± 3 |
| D179YKPC-3 | 28,393 ± 3 | 28,692 ± 3 | 28,692 ± 3 | 28,393 ± 3 (20%), 28,692 ± 3 (80%) | 28,393 ± 3 (95%), 28,692 ± 3 (5%) |
We show that, unlike the WT enzymes, D179Y variants of KPC-2 and KPC-3 form acyl-enzymes with IMI for at least 24 h (Table 3 and Fig. S3B). Unlike the reaction between CAZ and KPC-2 and KPC-3, wherein acyl-enzymes are not observed (suggesting rapid turnover), D179Y variants also form acyl-enzymes with CAZ but not for prolonged periods of time (Table 4 and Fig. S3C). Following the temporal turnover of CAZ by the D179Y substitution in KPC-2 and KPC-3, we estimate the kcat of CAZ to be 0.014 s−1 and 0.007 s−1 for the D179Y variants of KPC-2 and KPC-3, respectively. The acyl-enzyme complexes of KPC-2 and KPC-3 with AVI were formed in the first 10 s of incubation, and for the KPC-2 D179Y variant, the complex was formed after 1 min incubation (Table 5 and Fig. S3D).
TABLE 4.
Mass spectrometry analysis using CAZ (atomic mass units)
| β-Lactamase or antibiotic (1:25 ratio) | Mass spectrometry result (AMU) |
||||
|---|---|---|---|---|---|
| Apo | +CAZ 15 min | +CAZ 30 min | +CAZ 1 h | +CAZ 2 h | |
| CAZ | 546a ± 3 | ||||
| KPC-2 | 28,319 ± 3 | 28,319 ± 3 | 28,319 ± 3 | 28,319 ± 3 | 28,319 ± 3 |
| D179YKPC-2 | 28,366 ± 3 | 28,831 ± 3 | 28,831 ± 3 | 28,366 ± 3 | 28,366 ± 3 |
| KPC-3 | 28,344 ± 3 | 28,344 ± 3 | 28,344 ± 3 | 28,344 ± 3 | 28,344 ± 3 |
| D179YKPC-3 | 28,393 ± 3 | 28,857 ± 3 | 28,857 ± 3 | 28,857 ± 3 | 28,393 ± 3 |
Difference in measured wt due to loss of pyridine ring.
TABLE 5.
Mass spectrometry analysis using AVI (atomic mass units)
| β-Lactamase or antibiotic (1:2 ratio) | Mass spectrometry result (AMU) |
||||
|---|---|---|---|---|---|
| Apo | +AVI 10 sec | +AVI 20 sec | +AVI 30 sec | +AVI 1 min | |
| AVI | 265 ± 3 | ||||
| KPC-2 | 28,319 ± 3 | 28,584 ± 3 | 28,584 ± 3 | 28,584 ± 3 | 28,584 ± 3 |
| D179YKPC-2 | 28,366 ± 3 | 28,366 ± 3 (55%), 28,631 ± 3 (45%) | 28,366 ± 3 (33%), 28,631 ± 3 (67%) | 28,366 ± 3 (30%), 28,631 ± 3 (70%) | 28,366 ± 3 (25%), 28,631 ± 3 (75%) |
| KPC-3 | 28,344 ± 3 | 28,609 ± 3 | 28,609 ± 3 | 28,609 ± 3 | 28,609 ± 3 |
| D179YKPC-3 | 28,393 ± 3 | 28,393 ± 3 (60%), 28,658 ± 3 (40%) | 28,393 ± 3 (50%), 28,658 ± 3 (50%) | 28,393 ± 3 (25%), 28,658 ± 3 (75%) | 28,393 ± 3 (10%), 28,658 ± 3 (90%) |
Molecular modeling and docking.
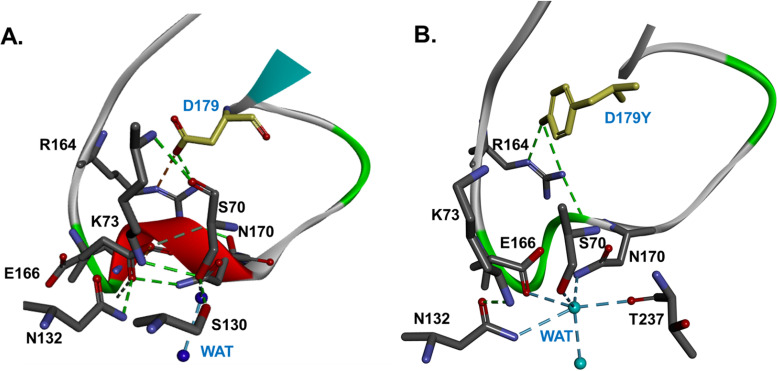
How do we explain these observations? Superimposition of KPC-2 versus KPC-3, both with the D179Y substitutions, shows that the two β-lactamases have similar overall structures (Fig. 4). As expected, differences in the Ω loop are generated by the D179Y substitution. As we will discuss below, KPC-2 and KPC-3 versus D179Y models initially suggest a high degree of similarity. However, NMR and subsequent molecular dynamic simulation and docking analysis show significant structural changes localized in the Ω loop, which we show accounts for the disorder observed and different behavior among the KPC WT enzymes and variants.
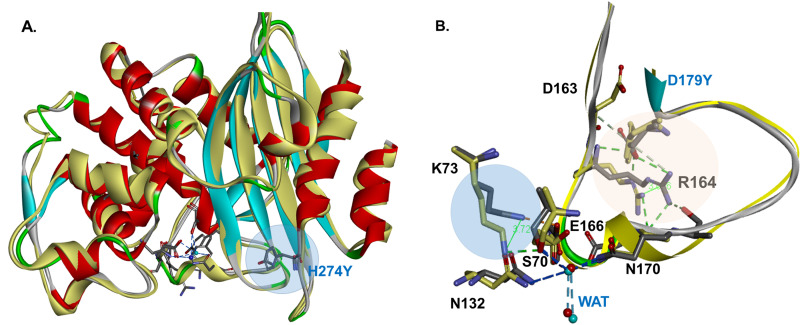
FIG 4.
Superimposition of KPC-2 versus KPC-3, both with the D179Y substitutions, shows that the 2 enzymes have similar overall structures (A). However, the D179Y substitution (yellow) disorders the Ω-loop in KPC-2 and KPC-3 (B). The most significant change is the disruption of the ionic bond between E166 and K73, present in KPC-2 WT but not the variant, and the movement of K73 away from E166 (≈3.7 Å) and toward S70, to an H-bond distance (1.6 Å). R164 is shifting with ∼3 Å, and the ionic bond with D179 present in WT is replaced by steric interactions of Y179 with D163 in the variant (B). The catalytic water is displaced from E166-N170, and with the K73 moving away from E166, toward S70 and S130, the electrostatics of the D179Y active site are significantly changed.
We previously modeled the D179N mutant of KPC-2 (8). The first observation from modeling the D179Y variant is that this substitution disorders the Ω-loop in KPC-2 and KPC-3 (Fig. 4), and the loop becomes larger and more flexible in the D179Y variants. Importantly, E166 and N170 are also likely repositioned (Fig. 5). Additionally, the ionic bond between E166 and K73 present in KPC-2 and KPC-3 may be disrupted. As a consequence of these changes, this model also suggests that K73 is shifted by 3.7 Å in the D179Y variant. With the protonated K73 moving away from E166 and toward S70, the electrostatics in the active site in the D179Y variants are likely significantly changed (Fig. 5).
FIG 5.
KPC-2 (A) versus D179Y (B) models during 500 ps molecular dynamic simulation (MDS). The substitution at position D179 is changing the shape of the loop leading to major interactions being disrupted (an ionic bond D179-R164), with new interactions between Y179-R164 being created. During the KPC-2 MDS, the K73 residue maintains the ionic bond with E166 and is moving away from S70 (from 2.6 to 4 Å). The K73 in the variant is making H-bonds with N132, and the catalytic water is better positioned between E166-N170-S70 in the D179Y variant versus KPC-2 as well (B).
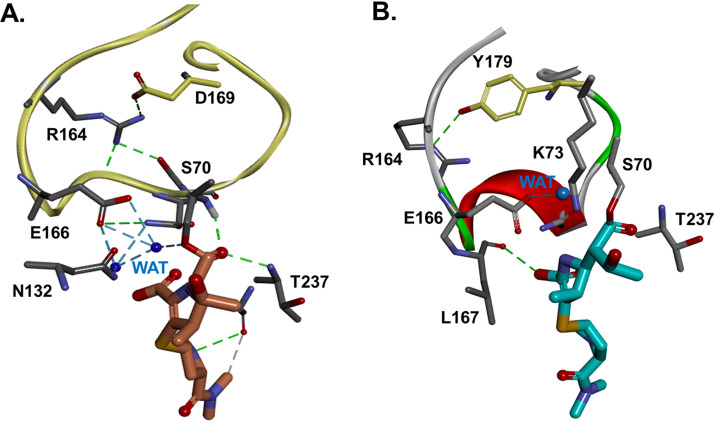
Why is susceptibility to MEM observed? In a further analysis of the WT KPC-2 and MEM complex (Fig. 6), the water molecule is positioned in close proximity to E166-S70 and between E166 and N170. MEM is perfectly positioned in the active site, ready for deacylation. In the D179Y complex with MEM, the water molecule is shifted from the E166-N170-S70 triad. This position, due to changes in the conformation of the Ω loop, may be impeding the MEM deacylation in the D179Y variants. It should be noted that these structural interpretations are based on modeling, NMR, and timed mass spectroscopy data. As a result of this “disorder of the active site,” our analysis suggests that MEM and IMI act as covalent β-lactamase inhibitors for D179Y variants of KPC-2 and KPC-3.
FIG 6.
Molecular models of KPC-2 (A) versus D179Y (B) as acyl-enzymes, with MEM bound. The acyl MEM is perfectly positioned into the active site of KPC-2, ready for deacylation. The catalytic water (WAT) is placed between E166 and N170, makes H-bonds with E166-S70, and is readily available for proton transfer. A second WAT is in the proximity of, and ready to form H-bonds with, N132 and E166.
What is the mechanistic basis of the increased CZA MICs? CAZ seems to be very unique with regard to the D179Y mediated resistance. What has clearly been shown in previous studies is that the D179Y substitution in both KPC-2 and KPC-3 backgrounds definitely increases the affinity of CAZ for the active site (decreases the Km). This points to a key role for the combination of the dimethyl-carboxyl group of the R1 side chain and the piperadine R2 group. Barnes et al. also noted a possible role of the carboxyl moiety of CAZ in resistance (8). Shapiro et al. maintained that docking of CAZ into the active site of KPC-3 caused a “flip of the R1 group” that also involves the carboxyl moiety (10).
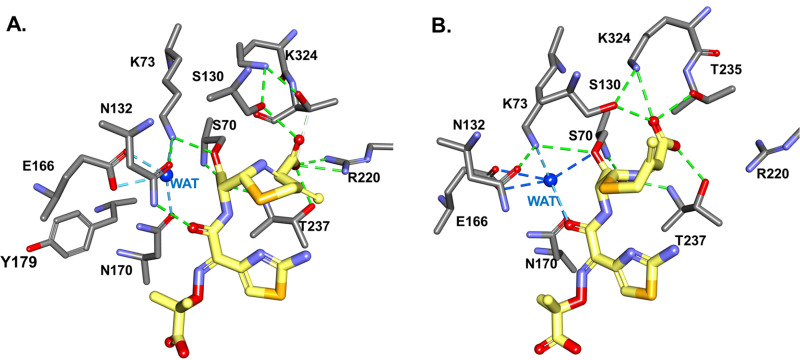
In our analysis, when CAZ was docked into the active site of KPC-2 D179Y variant as an acyl-enzyme complex (in the first part of the MDS simulation), the deacylation water present in the active site did not make H-bonds with S70 (Fig. 7). The H-bond distance between WAT and K73 was preserved for most of the MDS simulation (Fig. S4), and the WAT molecule made/broke bonds with S70. The overall active site of D179Y showed a decrease in the volume of the active site cavity and changes in the electrostatic potential (Fig. S5).
FIG 7.
MDS of CAZ docked into active site of KPC-2 D179Y variant as acyl-enzyme complex, with snap shots at different times: t = 0 ps (A) and t = 200 ps (B). We follow the conformational changes and catalytic water movement during the 500 ps MDS. Deacylation water, present in the active site at the beginning of MDS simulation, does not make H-bonds with S70 (A). The H-bond distance between WAT and K73 is preserved for most of the MDS, and the WAT molecule makes/breaks bonds with S70. This supports the hypothesis that K73 can act as a general base by activating the WAT in the hydrolysis of CAZ.
Conclusions and future considerations.
Herein, we attempted to reconcile an important clinical observation that undermines the effectiveness of CZA in treating KPC-K. pneumoniae infections and tried to understand the biochemical properties that support the paradoxical microbiological data.
In the WT KPC-2 and KPC-3, IMI is hydrolyzed rapidly, making it inactive and thereby conferring a resistant phenotype for IMI. In the case of the D179Y variants, IMI is “trapped” in the active site for a very long period of time, acting primarily as an inhibitor rather than as a substrate. This, coupled with the observation that the D179Y variant proteins are less stable, potentially accumulating less in steady state, suggests that a functional carbapenemase is not present to hydrolyze IMI or MEM. The IMI or MEM that is still free can inactivate the PBPs, hence bringing about a susceptible phenotype in the D179Y variants.
From the molecular modeling, we observed that the catalytic water molecule is shifted from E166-N170-S70 triad, making the D179Y variants deacylation challenged. This is all likely due to the disordered Ω loop. What causes the slower acyl complex formation with AVI in the D179Y variants? Different conformational states may be present compared to those in the WT, again explained by the disordered Ω loop.
When we analyze all published reports, we see that there is always a higher affinity for CAZ in the D179Y variants (decreased Km) than in the WT KPC counterpart. We demonstrate that higher affinity for CAZ comes with a concomitant decrease in both kcat and overall catalytic efficiency in the D179 variants. We also demonstrate for the D179Y variants that a prolonged acyl-enzyme complex with CAZ is formed. However, hydrolysis does still take place. Alternatively stated, there is enough β-lactamase present to more avidly bind, and slowly turnover, CAZ to functionally increase the MIC.
What causes the decreased Km, kcat, and kcat/Km for CAZ in the D179Y variants? From the NMR data, the observed increase in disorder for the protein, likely in the Ω loop, allows CAZ to fit better in the active site, thus lowering the Km for the D179Y variants. But also, this disorder leads to the decreased likelihood of having the active site residues of E166 and N170 and the important water molecule in the proper position to facilitate the reaction and thus the lower kcat and overall lowered kcat/Km.
Separately for AVI, the acyl-enzyme complex is formed more slowly for the D179Y variants than for WT KPC as seen by mass spectrometry. This is also supported by the work of others showing an increase in the IC50 for AVI inhibition of CAZ hydrolysis in the D179Y variant. In the WT KPC-2 and KPC-3, an acyl complex with AVI is formed faster, and they have a lower affinity for CAZ. Alternatively, the D179Y variants form an acyl complex with AVI at a slower rate and have a higher affinity for CAZ. Therefore, the D179Y variants preferentially bind CAZ, and even though it forms a prolonged acyl-enzyme complex, enough hydrolysis still proceeds that we observe a CZA-resistant phenotype.
The D179 substitution across the Ω loop has been observed previously and subjected to intense investigation for many years in class A enzymes. Many of the crystal structures (e.g., TEM, PC1) show significant disorder when there are substitutions at the D179 site. In SHV, with a substitution at the amino acid 164 position, there was observed ligand-dependent disorder (18). In the R164 variants that were studied, significant motion of E166 was not observed in the absence of ligand. However, D179 is located deeper in the Ω loop than R164, forming hydrogen bonds with the backbone nitrogen of R164 and D163, in addition to the salt bridge with the Nε and NH1 atoms of R164. As stated previously, due to this buried location and the increased number of interactions compared to those with R164, we postulated that when D179 is replaced, the Ω loop could be greatly disordered even in the apo state, perhaps similar to what was observed previously for a D179 variant of a distantly related PC1 β-lactamase. As a result, we posit the rearrangement of E166 in the catalytic mechanism. The loss of the salt bridge results in a protein that may be partially disordered across the neck of the Ω loop. Consequently, different conformational states may be present compared with WT. MEM and CAZ binding to the D179 variant stabilizes the protein and orders the local structure, but the protein is still in a different conformational state than the WT binding of the same substrates and the electrostatics of the active site can change.
We conclude that IMI, MEM, and CAZ are “trapped,” but the deacylation (half of the reaction) can still proceed slowly with CAZ perhaps because of the Lys73 that has shifted toward S70 or a repositioning of N170. The lower rate of carbamylation by AVI of the D179Y variants may also contribute to the elevated MICs of CZA (7). We propose that this paradoxical phenotype of CZA resistance accompanied by carbapenem susceptibility in the KPC D179Y variants is brought about by an interplay of all the above factors. In this case, molecular dynamics and NMR have proven to be very useful approaches to understanding structural and dynamic properties as they have been in select cases with class A and B β-lactamases (19–24). Knowing how conformational dynamics affect catalytic properties may affect future strategies in drug design.
MATERIALS AND METHODS
Plasmid constructs.
blaKPC-2 pBR322, blaKPC-2 D179Y pBR322, blaKPC-3 pBR322, and blaKPC-3 D179Y pBR322 were obtained as previously described via site-saturation mutagenesis and subsequent screening by sequencing of the constructs (8, 25). Initially, the E. coli containing blaKPC-2 in pBR322-catI vector was a gift from Fred Tenover (previously of the Centers for Disease Control and Prevention, Atlanta, GA) and was used as the template for the site-saturation mutagenesis experiments.
RT-PCR.
E. coli strains expressing KPC-2, KPC-3, and respective D179N and D179Y variants were cultured overnight in Luria broth (LB) medium. Bacterial cultures were diluted 1:1,000 in fresh LB and grown to mid-exponential phase (optical density at 600 nm [OD600] of ∼0.25) at 37°C. At this point, strains in LB without or with ceftazidime (32 μg/mL) were incubated at 37°C for 30 min and then centrifuged (5,000 × g) at 4°C. Cells were harvested and total mRNA was extracted using the PureLink RNA minikit (Ambion, ThermoFisher) according to the manufacturer’s instructions. Extracted RNA was treated with RNase-free DNase I (Ambion, Thermo Fisher). cDNA was synthesized with the Verso 1-step qRT-PCR system (Fisher Scientific). To confirm the absence of genomic DNA, negative-control reactions were performed with RNA and all reagents except for the reverse transcriptase (no-RT controls). Quantitative PCR (qPCR) was performed on an ABI 7900 fast real-time PCR system (Life Technologies) using the one-step SYBR green kit (Fisher Scientific); 3 μL of cDNA (diluted 1:10) was mixed with 5 nM forward and reverse primers (KPC: CGTGACGGAAAGCTTACAAA, AGCCAATCAACAAACTGCTG; 16S rRNA: GTTAATACCTTTGCTCATTGA, ACCAGGGTATCTAATCCTGTT). Amplification was undertaken in 20 μL final volume using the following protocol: denaturation at 95°C for 12 min, followed by 40 cycles consisting of 15 s at 95°C, 35 s at 60°C. Expression levels were presented in threshold cycle (CT) units, representing the first cycle in which signal above a preset threshold was detected. Experiments were conducted in triplicate. Expression of KPC-2 and KPC-3 was set at 1.0 (normalized to 16S rRNA), and relative expression of variant genes was presented as means ± standard deviations. Differences in expression of D179N and D179Y variants compared to that of respective controls were assessed using one-way analysis of variance (ANOVA) and Sidak’s multiple-comparison test. Significance was defined as ≥2-fold difference in expression and a P value of <0.05.
Steady-state kinetics.
Steady-state kinetic parameters were determined by using an Agilent 8453 diode array spectrophotometer at room temperature. Each assay was performed in 10 mM phosphate-buffered saline (PBS) at pH 7.4. MEM (150 μM) or IMI (150 μM) was incubated with KPC-2 (0.1 μM), KPC-3 (0.1 μM), KPC-2 D179Y (0.5 μM), or KPC-3 D179Y (0.5 μM), and hydrolysis was measured for 6 min. CAZ (50 μM) was incubated with KPC-2 (0.5 μM), KPC-3 (0.5 μM), KPC-2 D179Y (0.5 μM), or KPC-3 D179Y (0.5 μM), and hydrolysis was measured for 6 min.
Mass spectrometry.
Ten micrograms of β-lactamase were incubated with the substrate at a molar ratio of 1:1 (IMI, MEM) and 1:25 (CAZ) in sterile 10 mM phosphate-buffered saline (PBS) at pH 7.4 for a total reaction volume of 20 μL for the times indicated in the figures. Reactions were quenched with 1% acetonitrile and 0.1% formic acid. Samples were analyzed using QTOF Waters Synapt-G2-Si and a Waters Acquity ultrapressure liquid chromatography (UPLC) BEH C18 column (1.7-μm pore size; 2.1 by 50 mm). MassLynxV4.1 was used to deconvolute protein peaks. The tune settings for each data run were as follows: capillary voltage at 3.5 kV, sampling cone at 35, source offset at 35, source temperature of 100°C, desolvation temperature of 500°C, cone gas at 100 L/h, desolvation gas at 800 L/h, and nebulizer bar at 6.0. Mobile phase A was 0.1% formic acid–water. Mobile phase B was 0.1% formic acid–acetonitrile. The mass accuracy for this system is ±5 Da.
Thermal denaturation.
Thermal denaturation experiments by differential scanning fluorimetry were collected using a Bio-Rad CFX96 RT-PCR instrument. All protein samples were prepared as 25 μL aliquots in 96-well Frame Star PCR plates covered with clear seal film. Samples contained 10 μM protein in PBS buffer (pH 7.4) with 5× SYPRO orange dye (Thermo Fisher Scientific), diluted from a 5,000× SYPRO orange stock. Samples indicated as “+MEM” or “+CAZ” contained 200 μM MEM or CAZ, respectively. Fluorescence data were collected using the “HEX” channel for the CFX96 with excitation at 515 to 535 nm and emission at 570 nm with temperature increasing from 25°C to 95°C in 0.5°C increments and a 5-s equilibration at each temperature step. Melting temperature (Tm) values were calculated by fitting fluorescence data to a Boltzmann sigmoidal curve in Prism (version 9.0.2, GraphPad, La Jolla, CA).
NMR spectroscopy.
KPC-2 samples for NMR spectroscopy were prepared in 20 mM phosphate, 50 mM NaCl, and 3% glycerol at pH 5.9. A protease inhibitor cocktail, containing 5 mM ethylenediaminetetraacetic acid, 0.2% sodium azide, and 1 μM 4-(2-aminoethyl)benzenesulfonyl fluoride, was added to each sample. 1H/15N-HSQC spectra were collected at 308 K on a Bruker Avance 600 MHz spectrometer. Spectra were processed with NMRPipe (26) and visualized in Sparky (27).
Molecular modeling and molecular docking.
A structural representation of D179Y variant of KPC-2 β-lactamase was generated using the crystal coordinates of KPC-2 (PDB: 2OV5) and the build and edit protocol of BIOVIA Discovery Studio 2020 Client (Dassault Systèmes, San Diego, CA) molecular modeling software as described previously (1, 25). The crystallographic water molecules were maintained during modeling. The KPC-2 β-lactamase structure and the variant model were solvated and minimized to a root mean square (RMS) of 0.03 Å using conjugate gradient method. To test for stability and possible conformational changes, molecular dynamics simulation (MDS) was conducted for 500 ps on the apo enzymes.
Acyl-meropenem was constructed using the Fragment Builder tools and minimized using a Standard Dynamics Cascade protocol of DS 2020. The acylated meropenem was automatically docked into the active site of KPC-2 and the D179Y variant using CDOCKER module of DS 2020. To obtain acyl-enzyme complexes, the most favorable pose of meropenem demonstrating anticipated active site contacts (such as a short distance [2 to 3 Å] between Ser70: O and C7 of MEM) was chosen. MDS was conducted for 550 ps on KPC-2 and D179Y apo enzymes and as acyl-ceftazidime complexes. The trajectories generated were saved every 2 ps and analyzed.
ACKNOWLEDGMENTS
This study was supported in part by funds and facilities provided by the Cleveland Department of Veterans Affairs, Award Number 1I01BX001974 to R.A.B. from the Biomedical Laboratory Research & Development Service of the VA Office of Research and Development, and the Geriatric Research Education and Clinical Center VISN 10. In addition, research reported in this publication was supported by the National Institute of Allergy and Infectious Diseases of the National Institutes of Health (NIH) under Award Number R21AI142049 to C.J.C., and a grant (MISP 57394) provided by Merck Sharp & Dohme, Corp., Kenilworth, NJ to R.A.B. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH or the Department of Veterans Affairs.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Levitt PS, Papp-Wallace KM, Taracila MA, Hujer AM, Winkler ML, Smith KM, Xu Y, Harris ME, Bonomo RA. 2012. Exploring the role of a conserved class A residue in the omega-loop of KPC-2 beta-lactamase: a mechanism for ceftazidime hydrolysis. J Biol Chem 287:31783–31793. 10.1074/jbc.M112.348540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Winkler ML, Papp-Wallace KM, Bonomo RA. 2015. Activity of ceftazidime/avibactam against isogenic strains of Escherichia coli containing KPC and SHV beta-lactamases with single amino acid substitutions in the omega-loop. J Antimicrob Chemother 70:2279–2286. 10.1093/jac/dkv094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Livermore DM, Warner M, Jamrozy D, Mushtaq S, Nichols WW, Mustafa N, Woodford N. 2015. In vitro selection of ceftazidime-avibactam resistance in Enterobacteriaceae with KPC-3 carbapenemase. Antimicrob Agents Chemother 59:5324–5330. 10.1128/AAC.00678-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shields RK, Potoski BA, Haidar G, Hao B, Doi Y, Chen L, Press EG, Kreiswirth BN, Clancy CJ, Nguyen MH. 2016. Clinical outcomes, drug toxicity, and emergence of ceftazidime-avibactam resistance among patients treated for carbapenem-resistant Enterobacteriaceae infections. Clin Infect Dis 63:1615–1618. 10.1093/cid/ciw636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shields RK, Chen L, Cheng S, Chavda KD, Press EG, Snyder A, Pandey R, Doi Y, Kreiswirth BN, Nguyen MH, Clancy CJ. 2017. Emergence of ceftazidime-avibactam resistance due to plasmid-borne blaKPC-3 mutations during treatment of carbapenem-resistant Klebsiella pneumoniae infections. Antimicrob Agents Chemother 61:e02097-16. 10.1128/AAC.02097-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shields RK, Nguyen MH, Press EG, Chen L, Kreiswirth BN, Clancy CJ. 2017. In vitro selection of meropenem resistance among ceftazidime-avibactam-resistant, meropenem-susceptible Klebsiella pneumoniae isolates with variant KPC-3 carbapenemases. Antimicrob Agents Chemother 61:e00079-17. 10.1128/AAC.00079-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Compain F, Arthur M. 2017. Impaired inhibition by avibactam and resistance to the ceftazidime-avibactam combination due to the D(179)Y substitution in the KPC-2 beta-lactamase. Antimicrob Agents Chemother 61:e00451-17. 10.1128/AAC.00451-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barnes MD, Winkler ML, Taracila MA, Page MG, Desarbre E, Kreiswirth BN, Shields RK, Nguyen MH, Clancy C, Spellberg B, Papp-Wallace KM, Bonomo RA. 2017. Klebsiella pneumoniae carbapenemase-2 (KPC-2), substitutions at ambler position Asp179, and resistance to ceftazidime-avibactam: unique antibiotic-resistant phenotypes emerge from beta-lactamase protein engineering. mBio 8:e00528-17. 10.1128/mBio.00528-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang Y, Kashikar A, Brown CA, Denys G, Bush K. 2017. Unusual Escherichia coli PBP 3 insertion sequence identified from a collection of carbapenem-resistant Enterobacteriaceae tested in vitro with a combination of ceftazidime-, ceftaroline-, or aztreonam-avibactam. Antimicrob Agents Chemother 61:e00389-17. 10.1128/AAC.00389-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shapiro AB, Moussa SH, Carter NM, Gao N, Miller AA. 2021. Ceftazidime-avibactam resistance mutations V240G, D179Y, and D179Y/T243M in KPC-3 beta-lactamase do not alter cefpodoxime-ETX1317 susceptibility. ACS Infect Dis 7:79–87. 10.1021/acsinfecdis.0c00575. [DOI] [PubMed] [Google Scholar]
- 11.Tsivkovski R, Lomovskaya O. 2020. Potency of vaborbactam is less affected than that of avibactam in strains producing KPC-2 mutations that confer resistance to ceftazidime-avibactam. Antimicrob Agents Chemother 64:e01936-19. 10.1128/AAC.01936-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Galani I, Karaiskos I, Angelidis E, Papoutsaki V, Galani L, Souli M, Antoniadou A, Giamarellou H. 2021. Emergence of ceftazidime-avibactam resistance through distinct genomic adaptations in KPC-2-producing Klebsiella pneumoniae of sequence type 39 during treatment. Eur J Clin Microbiol Infect Dis 40:219–224. 10.1007/s10096-020-04000-9. [DOI] [PubMed] [Google Scholar]
- 13.Venditti C, Butera O, Meledandri M, Balice MP, Cocciolillo GC, Fontana C, D'Arezzo S, De Giuli C, Antonini M, Capone A, Messina F, Nisii C, Di Caro A. 2021. Molecular analysis of clinical isolates of ceftazidime-avibactam-resistant Klebsiella pneumoniae. Clin Microbiol Infect 27:1040.e1–1040.e6. 10.1016/j.cmi.2021.03.001. [DOI] [PubMed] [Google Scholar]
- 14.Papp-Wallace KM, Taracila M, Hornick JM, Hujer AM, Hujer KM, Distler AM, Endimiani A, Bonomo RA. 2010. Substrate selectivity and a novel role in inhibitor discrimination by residue 237 in the KPC-2 beta-lactamase. Antimicrob Agents Chemother 54:2867–2877. 10.1128/AAC.00197-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.VanPelt J, Shurina BA, Ramelot TA, Bonomo RA, Page RC. 2019. (1)H, (13)C, and (15)N backbone resonance assignments for KPC-2, a class A serine-beta-lactamase. Biomol NMR Assign 13:139–142. 10.1007/s12104-018-9866-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dyson HJ, Wright PE. 2001. Nuclear magnetic resonance methods for elucidation of structure and dynamics in disordered states. Methods Enzymol 339:258–270. 10.1016/s0076-6879(01)39317-5. [DOI] [PubMed] [Google Scholar]
- 17.Dyson HJ, Wright PE. 2004. Unfolded proteins and protein folding studied by NMR. Chem Rev 104:3607–3622. 10.1021/cr030403s. [DOI] [PubMed] [Google Scholar]
- 18.Sampson JM, Ke W, Bethel CR, Pagadala SR, Nottingham MD, Bonomo RA, Buynak JD, van den Akker F. 2011. Ligand-dependent disorder of the omega loop observed in extended-spectrum SHV-type beta-lactamase. Antimicrob Agents Chemother 55:2303–2309. 10.1128/AAC.01360-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Elings W, Chikunova A, van Zanten DB, Drenth R, Ahmad MUD, Blok AJ, Timmer M, Perrakis A, Ubbink M. 2021. Two beta-lactamase variants with reduced clavulanic acid inhibition display different millisecond dynamics. Antimicrob Agents Chemother 65:e0262820. 10.1128/AAC.02628-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gonzalez MM, Abriata LA, Tomatis PE, Vila AJ. 2016. Optimization of conformational dynamics in an epistatic evolutionary trajectory. Mol Biol Evol 33:1768–1776. 10.1093/molbev/msw052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guo H, Cheng K, Gao Y, Bai W, Wu C, He W, Li C, Li Z. 2020. A novel potent metal-binding NDM-1 inhibitor was identified by fragment virtual, SPR and NMR screening. Bioorg Med Chem 28:115437. 10.1016/j.bmc.2020.115437. [DOI] [PubMed] [Google Scholar]
- 22.Llarrull LI, Tioni MF, Kowalski J, Bennett B, Vila AJ. 2007. Evidence for a dinuclear active site in the metallo-beta-lactamase BcII with substoichiometric Co(II). A new model for metal uptake. J Biol Chem 282:30586–30595. 10.1074/jbc.M704613200. [DOI] [PubMed] [Google Scholar]
- 23.Rydzik AM, Brem J, Chandler SA, Benesch JLP, Claridge TDW, Schofield CJ. 2020. Monitoring protein-metal binding by (19)F NMR - a case study with the New Delhi metallo-beta-lactamase 1. RSC Med Chem 11:387–391. 10.1039/c9md00416e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Softley CA, Zak KM, Bostock MJ, Fino R, Zhou RX, Kolonko M, Mejdi-Nitiu R, Meyer H, Sattler M, Popowicz GM. 2020. Structure and molecular recognition mechanism of IMP-13 metallo-beta-lactamase. Antimicrob Agents Chemother 64:e00123-20. 10.1128/AAC.00123-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Papp-Wallace KM, Taracila MA, Smith KM, Xu Y, Bonomo RA. 2012. Understanding the molecular determinants of substrate and inhibitor specificities in the carbapenemase KPC-2: exploring the roles of Arg220 and Glu276. Antimicrob Agents Chemother 56:4428–4438. 10.1128/AAC.05769-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. 1995. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR 6:277–293. 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 27.Lee W, Tonelli M, Markley JL. 2015. NMRFAM-SPARKY: enhanced software for biomolecular NMR spectroscopy. Bioinformatics 31:1325–1327. 10.1093/bioinformatics/btu830. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1, Fig. S1 to S5, and supplemental materials and methods. Download aac.02124-21-s0001.pdf, PDF file, 0.8 MB (832.5KB, pdf)