ABSTRACT
As an opportunistic fungal pathogen, Candida albicans is a major cause of superficial and systemic infections in immunocompromised patients. The increasing rate of azole resistance in C. albicans has brought further challenges to clinical therapy. In this study, we collected five isogenic C. albicans strains recovered over discrete intervals from an HIV-infected patient who suffered 2-year recurrent oropharyngeal candidiasis. Azole resistance was known from the clinical history to have developed gradually in this patient, and this was confirmed by MIC assays of each strain. Proteomic techniques can be used to investigate more comprehensively how resistance develops in pathogenic fungi over time. Our study is the first to use tandem mass tag (TMT) labeling combined with liquid chromatography-tandem mass spectrometry (LC-MS/MS) technology to investigate the acquired resistance mechanisms of serial C. albicans isolates at the proteomic level. A total of 4,029 proteins have been identified, of which 3,766 have been quantified. Compared with Ca1, bioinformatics analysis showed that differentially expressed proteins were mainly associated with aspects such as the downregulation of glycolysis/gluconeogenesis, pyruvate metabolism, fatty acid degradation, and oxidative stress response proteins in all four subsequent strains but, remarkably, the activation of amino acid metabolism in Ca8 and Ca14 and increased protection against osmotic stress or excessive copper toxicity, upregulation of respiratory chain activity, and suppression of iron transport in Ca17. By tracing proteomic alterations in this set of isogenic resistance isolates, we acquire mechanistic insight into the steps involved in the acquisition of azole resistance in C. albicans.
KEYWORDS: Candida albicans, quantitative proteomics, acquired azole resistance
INTRODUCTION
Candida albicans is a major fungal pathogen in humans, causing mucosal, cutaneous, and life-threatening systemic infections (1–3). Studies have shown that systemic infections by C. albicans have a crude mortality rate of ∼40% despite antifungal interventions (4–6). The annual attributable cost of candidemia in the United States alone is $1.7 billion (7, 8). Azoles, in particular fluconazole, are the most common antifungal drugs used to treat and prevent candidiasis. However, fluconazole is fungistatic rather than fungicidal, and the treatment therefore invites the development of acquired resistance in the presence of this agent (9, 10). Given its clinical and economic impact, the Centers for Disease Control and Prevention (CDC) added C. albicans to the list of pathogens that pose a potential drug resistance threat (11). Understanding the mechanisms of fluconazole resistance is a critical part of managing our limited antifungal options and maintaining fluconazole as a possible option for the treatment of many candidiasis cases (9).
Several genes involved in ergosterol biosynthesis pathways have been noted as mechanisms of azole resistance, and other mechanisms such as drug efflux pumps, ploidy changes, and loss of heterozygosity (LOH) were also revealed by genome-wide gene expression profile or transcriptional analyses (3, 12–16). However, data gathered from genomic or transcriptomic analyses do not necessarily reflect actual protein functions in the cells (17–19). In a previous study, a gel-based proteomics analysis was employed, which identified 17 proteins with differential expression in a matched set of two C. albicans isolates, namely, an initially susceptible one and a finally resistant counterpart (20). However, this gel-based technique easily misses hydrophobic proteins (20); in addition, due to poor reproducibility, the limited dynamic range, and the incompatibility of proteins separated by electrophoresis with mass spectrometry (MS), it is difficult to make quantitative comparisons between samples on different gels (21). Tandem mass tag (TMT) peptide labeling combined with liquid chromatography-tandem MS (LC-MS/MS) quantitative proteomics can detect more proteins with higher resolution than current conventional methods (22, 23). In this study, we use this quantitative proteomics technique to identify differentially expressed proteins (DEPs) among the posttreatment isolates against the initial isolate (Ca1) collected prior to antifungal therapy. We believe that an improved understanding of global protein changes in C. albicans during the process of acquired drug resistance will help further the development of strategies against this medically important pathogenic fungus.
RESULTS
Global proteomic comparison between Ca1 and serial strains (Ca2, -8, -14, and 17) in C. albicans.
In total, 4,029 proteins were identified by proteomics analysis, of which 3,766 proteins were quantified (see Data Set S1 in the supplemental material). Quality control validations of mass spectrum data and three biological replicates were performed for four comparison groups (Q1 [Ca2 versus Ca1], Q2 [Ca8 versus Ca1], Q3 [Ca14 versus Ca1], and Q4 [Ca17 versus Ca1]), as shown in Fig. S1 and S2. All quantifiable proteins with upregulated (>1.5-fold) and downregulated (<0.667-fold) expression levels are listed in Data Set S2 (P < 0.05). As shown in Table 1, Q2 induced 201 DEPs (63 upregulated and 138 downregulated), which was more than 3 times that of Q3 (35 upregulated and 25 downregulated), 2 times that of Q4 (32 upregulated and 68 downregulated), and 1.8 times that of Q1 (23 upregulated and 86 downregulated). The Q2 group revealed the most upregulated and downregulated proteins. The protein expression levels of common determinants of azole resistance (TAC1, MRR1, UPC2, CDR1, ERG11, ERG3, and HSP90) in these five strains are listed in Table 2. Of these, only CDR1p is significantly upregulated in Q4, according to the proteomics results that we have observed so far.
TABLE 1.
Statistics of differentially expressed proteins
| Group | No. of upregulated DEPs (>1.5-fold) | No. of downregulated DEPs (<0.667-fold) |
|---|---|---|
| Q1 | 23 | 86 |
| Q2 | 63 | 138 |
| Q3 | 35 | 25 |
| Q4 | 32 | 68 |
TABLE 2.
Protein expression levels of common determinants of azole resistance
| UniProt protein accession no. | Gene name | Expression level |
Ca2/Ca1 ratio | Ca8/Ca1 ratio | Ca14/Ca1 ratio | Ca17/Ca1 ratio | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| Ca1 | Ca2 | Ca8 | Ca14 | Ca17 | ||||||
| A0A1D8PN96 | TAC1 | 0.993 | 0.892 | 0.920 | 1.005 | 1.200 | 0.898 | 0.927 | 1.013 | 1.209 |
| Q5A4G2 | MRR1 | 0.993 | 0.892 | 1.059 | 1.087 | 0.956 | 0.898 | 1.066 | 1.095 | 0.963 |
| Q59QC7 | UPC2 | 0.952 | 1.035 | 0.991 | 1.074 | 0.969 | 1.088 | 1.041 | 1.128 | 1.018 |
| Q5ANA3 | CDR1 | 0.555 | 0.547 | 0.571 | 0.664 | 2.570 | 0.986 | 1.028 | 1.196 | 4.627 |
| P10613 | ERG11 | 0.916 | 0.933 | 0.948 | 1.096 | 1.128 | 1.019 | 1.036 | 1.197 | 1.232 |
| Q59VG6 | ERG3 | 1.210 | 1.018 | 0.886 | 0.974 | 0.927 | 0.841 | 0.732 | 0.805 | 0.766 |
| P46598 | HSP90 | 1.027 | 1.096 | 1.007 | 0.923 | 0.945 | 1.068 | 0.981 | 0.899 | 0.921 |
Functional characterization of differentially expressed proteins.
To characterize the functions and subcellular locations of the differentially expressed proteins in serial isolates previously exposed to fluconazole, bioinformatic analyses of Gene Ontology (GO) and subcellular functional annotations were carried out. Based on the GO analysis tool, the biological functions affected by DEPs were classified into the cellular component, molecular function, and biological process. The mainly affected biological functions of DEPs among strains (Ca2, -8, -14, and 17) versus Ca1 are summarized in Table 3. The results showed that the DEPs are mainly involved in the membrane, cell, organelle, catalytic activity, binding, transporter activity, metabolic process, single-organism process, and cellular process. These DEPs were classified according to their subcellular locations (Fig. 1). The subcellular distribution of DEPs indicates that C. albicans resistance to fluconazole mainly affects the cytoplasm, nucleus, plasma membrane (PM), and mitochondria. For Q1, 30%, 29%, and 15% of DEPs are located in the nucleus, cytoplasm, and plasma membrane, respectively. For Q2, 30%, 29%, and 13% of DEPs are located in the cytoplasm, nucleus, and mitochondria, respectively. For Q3, 38% of DEPs are located in the cytoplasm, 25% are located in the nucleus, and 11% are located in the mitochondria. For Q4, 33% of DEPs are located in the cytoplasm, 25% are located in the nucleus, and 16% are located in the plasma membrane.
TABLE 3.
Gene Ontology distribution of differentially expressed proteins
| GO term level 1 | GO term level 2 | No. of proteins (%) |
|||
|---|---|---|---|---|---|
| Q1 | Q2 | Q3 | Q4 | ||
| Biological process | Metabolic process | 45 (34) | 87 (31) | 30 (36) | 36 (28) |
| Single-organism process | 30 (23) | 69 (24) | 28 (33) | 32 (25) | |
| Cellular process | 20 (15) | 53 (19) | 16 (19) | 21 (16) | |
| Cellular component | Membrane | 21 (38) | 46 (36) | 13 (46) | 26 (37) |
| Cell | 17 (31) | 42 (32) | 10 (36) | 25 (35) | |
| Organelle | 6 (11) | 18 (14) | 1 (4) | 9 (13) | |
| Molecular function | Catalytic activity | 51 (59) | 101 (57) | 31 (58) | 43 (50) |
| Binding | 23 (27) | 59 (33) | 19 (36) | 28 (33) | |
| Transporter activity | 6 (7) | 11 (6) | 2 (4) | 9 (11) | |
FIG 1.
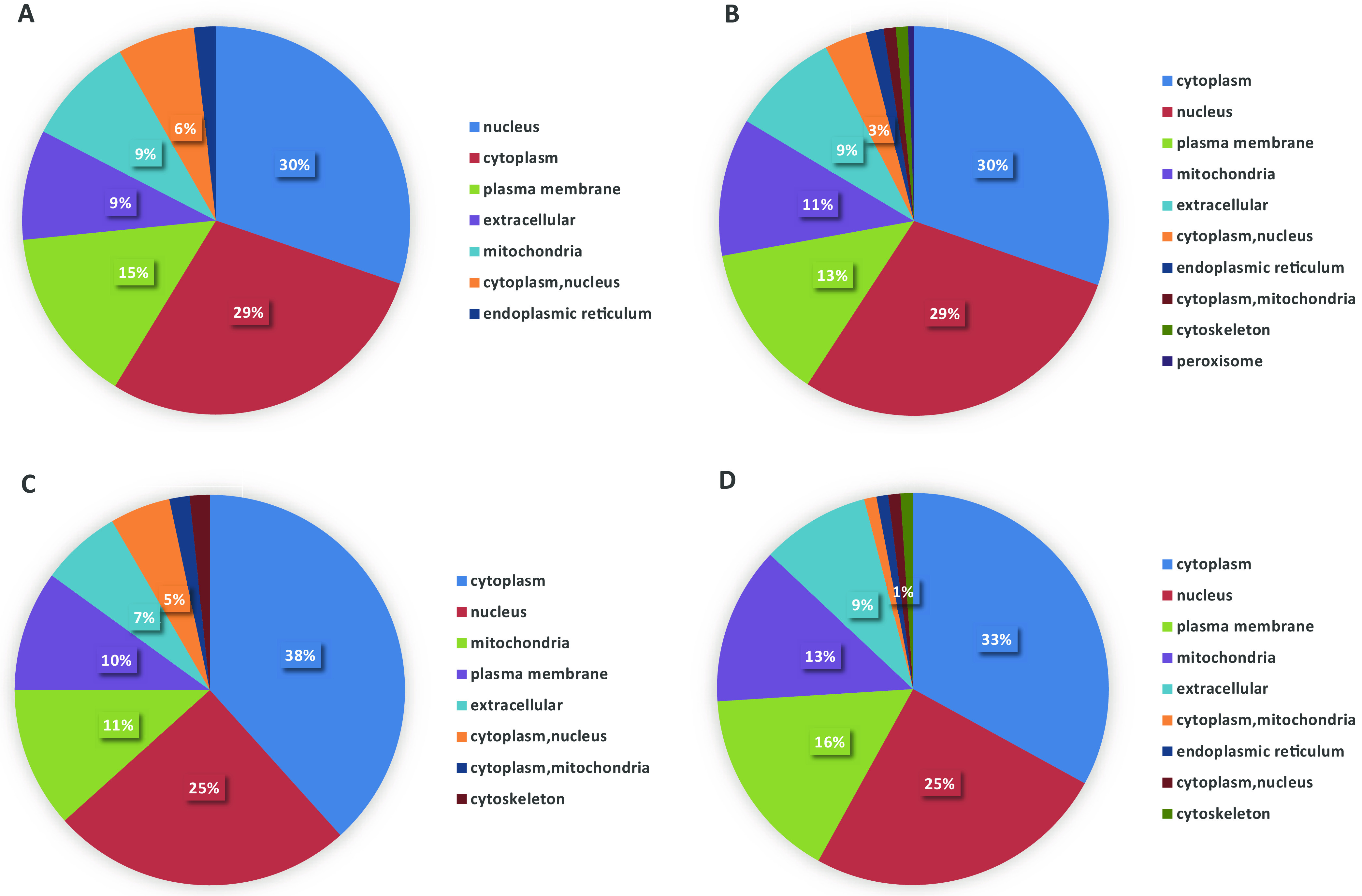
Subcellular locations of differentially expressed proteins. (A) Q1; (B) Q2; (C) Q3; (D) Q4.
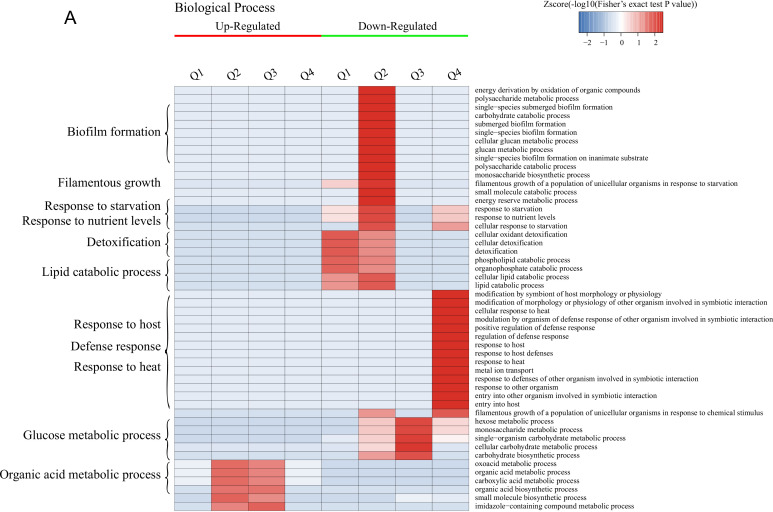
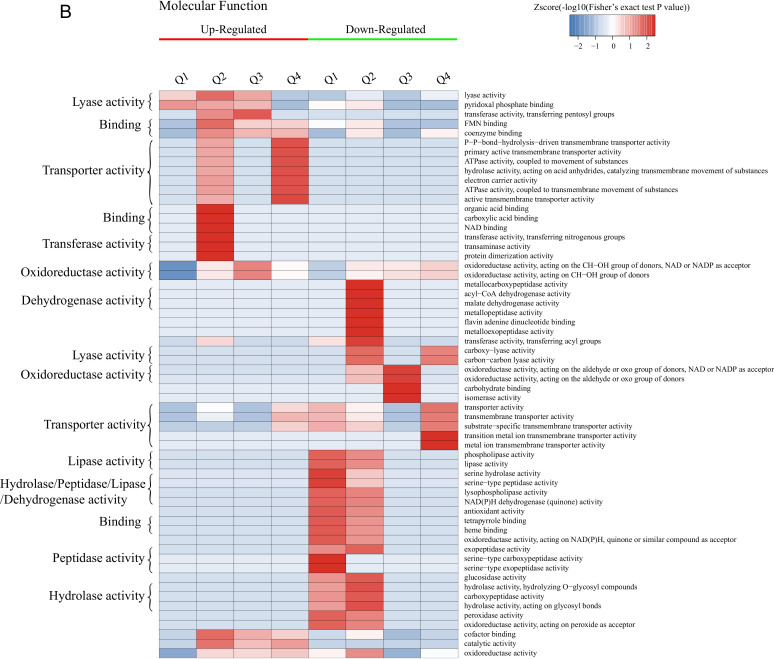
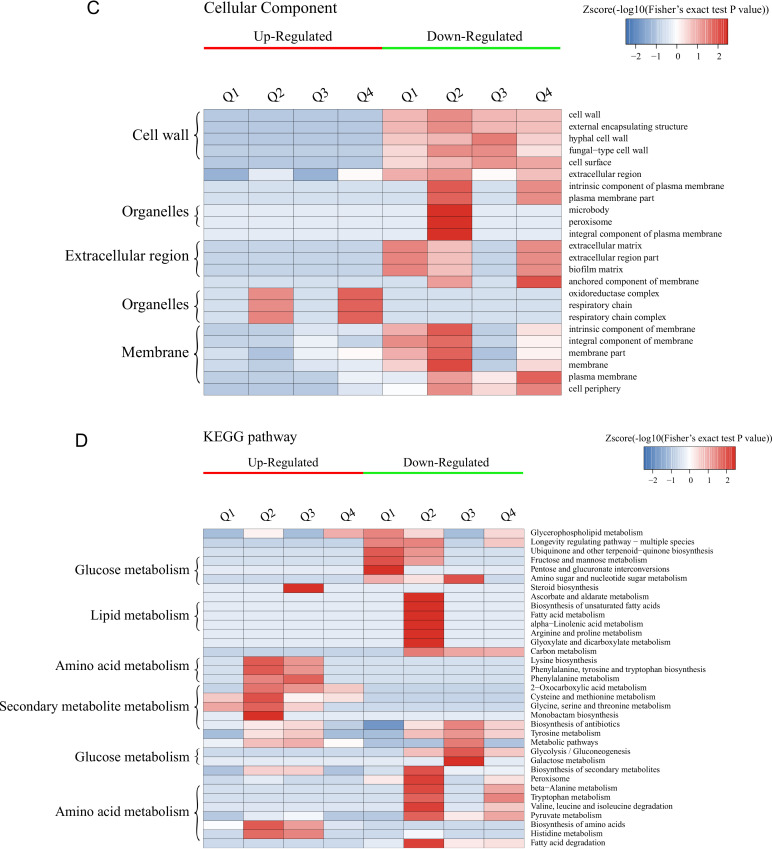
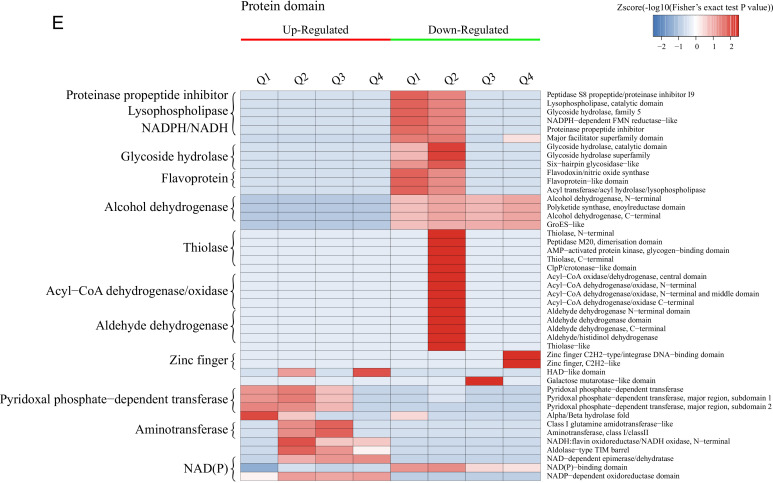
Enrichment-based clustering analysis of the DEPs.
GO enrichment-based clustering analysis of the DEPs was performed to classify the biological processes, molecular functions, and cellular components (Fig. 2A to C and Fig. S3). The abundant upregulated proteins are related to pyridoxal phosphate binding and lyase activity in Q1; imidazole-containing compound metabolic process, histidine biosynthetic/metabolic process, cellular amino acid biosynthetic/metabolic process, and carboxylic acid metabolic/biosynthetic process in Q2 and Q3; cofactor binding, catalytic activity, and oxidoreductase activity in Q2, Q3, and Q4; and flavin mononucleotide (FMN) binding, electron carrier activity, ATPase activity coupled to transmembrane movement of substances, hydrolase activity acting on acid anhydrides catalyzing the transmembrane movement of substances, and the response to toxic substance or chemicals in Q4. The downregulated proteins are mainly involved in peroxidase activity, lipid catabolic process, cellular oxidant detoxification, and the response to oxidative stress in Q1 and Q2; carbohydrate metabolic process and single-organism metabolic process in Q1, Q2, and Q3; galactose metabolic process, hexose metabolic process, monosaccharide metabolic process, and cellular carbohydrate metabolic/biosynthetic process in Q3; and iron ion transmembrane transport, high-affinity iron ion transmembrane transport, and metal ion transmembrane transporter activity in Q4.
FIG 2.
Hierarchical clustering analysis was conducted for the differentially expressed proteins according to biological process (A)-, molecular function (B)-, cellular component (C)-, KEGG pathway (D)-, and protein domain (E)-based enrichment. The P values were transformed into Z-scores for hierarchical clustering analysis. The Z-score is shown in the color key, and red represents significant enrichments.
The results of clustering analysis based on Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment are shown in Fig. 2D and Fig. S4. For all groups, the upregulated DEPs are significantly enriched in the biosynthesis of amino acids and antibiotics, while the downregulated DEPs are mainly related to secondary metabolite biosynthesis and fatty acid degradation. For each group, the upregulated DEPs were mainly associated with monobactam biosynthesis in Q2, steroid biosynthesis in Q3, and glycerophospholipid metabolism in Q4, while the glycerophospholipid metabolism pathway was significantly downregulated for Q1. Furthermore, the upregulated DEPs are enriched in glycine, serine, and threonine metabolism for Q1, Q2, and Q3; cysteine and methionine metabolism in Q1, Q2, and Q4; histidine, phenylalanine, and tyrosine metabolism and the biosynthesis of lysine, phenylalanine, tyrosine, and tryptophan in Q2 and Q3; and 2-oxocarboxylic acid metabolism in Q2, Q3, and Q4. In terms of the downregulated DEPs, these proteins are mainly significantly involved in the peroxisome and the longevity-regulating pathway in multiple species in Q1, Q2, and Q4; pyruvate metabolism, glycolysis/gluconeogenesis, the biosynthesis of antibiotics, and tyrosine metabolism in Q2, Q3, and Q4; and tryptophan metabolism, valine, leucine, and isoleucine degradation, and beta-alanine metabolism in Q4.
The DEPs in all cases were also examined by clustering analysis based on protein domain enrichment analysis (Fig. 2E and Fig. S5). For all four groups, DEPs related to the NADP-dependent oxidoreductase domain are upregulated, while other domains are downregulated, including the NAD(P)-binding domain, the polyketide synthase and enoyl reductase domain, the alcohol dehydrogenase N-terminal domain, the alcohol dehydrogenase C-terminal domain, and the GroES-like domain. Moreover, DEPs involved in pyridoxal phosphate-dependent transferase are upregulated in Q1, Q2, and Q3, coupled with the upregulation of the aldolase-type TIM barrel, NAD-dependent epimerase/dehydratase, NADH:flavin oxidoreductase/NADH oxidase, and N-terminal NAD(P)-binding domains in Q2, Q3, and Q4. The downregulated proteins consist of the glycoside hydrolase superfamily, acyl transferase/acyl hydrolase/lysophospholipase, and major facilitator superfamily domains in Q1 and Q2. For each group, the upregulated DEPs are mainly enriched in the alpha/beta-hydrolase fold in Q1, class I glutamine amidotransferase-like and aminotransferase class I/class II in Q2 and Q3, and the HAD-like domain in Q4. However, the downregulated DEPs are mainly related to the flavoprotein-like domain and flavodoxin/nitric oxide synthase in Q1; the acyl-CoA dehydrogenase/oxidase, thiolase, and thiolase-like domains in Q2; the galactose mutarotase-like domain in Q3; and the zinc finger Cys2His2 (C2H2)-type/integrase DNA-binding domain in Q4.
Dynamic cluster analysis of protein expression patterns.
A total of 1,734 proteins were divided into 12 clusters using Mfuzz analysis, and proteins in the same cluster represent similar expression transformation trends (Fig. 3). Here, we mainly describe clusters showing a consistent or opposite acquired azole resistance tendency. Our analyses indicated that proteins (n = 148) in cluster 8, including Ifu5p, Grp2p, Cdr1p, Gpx1p, Pdr16p, Ifd3p, Ifd6p, and Gdh3p, have increased expression in isolates after previous fluconazole exposure. On the contrary, in cluster 2, 148 proteins, including Ald5p, Ino1p, and Pck1p, show a continuous decrease. Moreover, proteins (n = 104) in cluster 4, including orf19.7166p, His7p, His1p, Gpd1p, Yhb1p, Aro8p, Atf1p, and Rhr2p, are also increased in Ca1, Ca2, and Ca8 but slightly decreased in Ca14 and Ca17.
FIG 3.
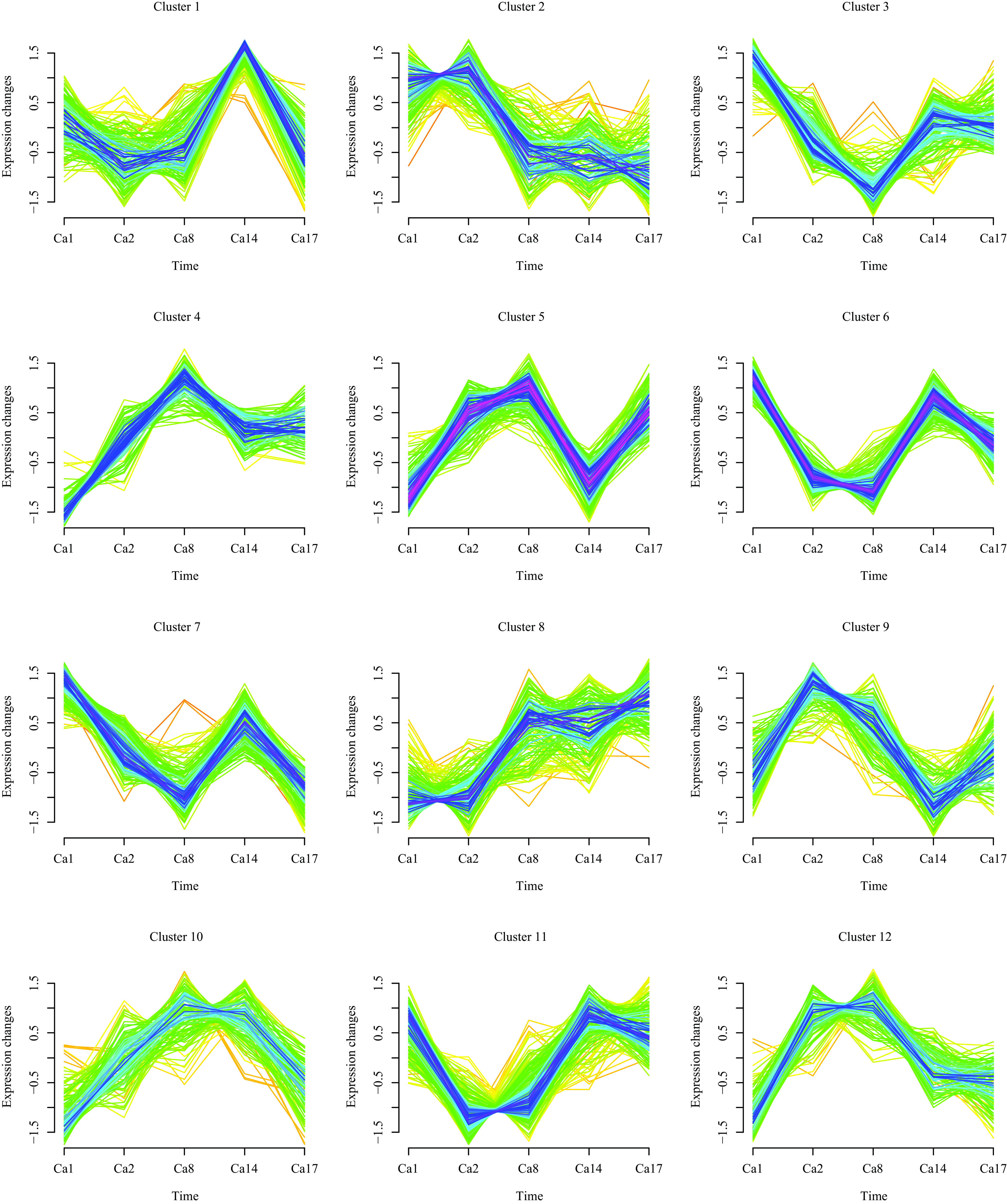
Dynamic clustering of differentially expressed proteins.
DISCUSSION
Drug resistance in fungi is often gradual and incremental by nature, and various resistance strategies may come into being and then fade away over the course of antifungal treatment (16, 24–26). The centerpiece of our study is a series of five isogenic isolates that showed progressively increasing MIC values within a time span of 2 years. It is well known that proteins are the most relevant elements associated with the physiology of microbes (27). Using TMT-labeling and LC-MS/MS methods, we first identify the dynamics of proteomic adaptation at the different stages of the evolution of drug resistance.
Proteome responses in isolate Ca2.
The key factors in isolate Ca2 (MIC = 1 μg · mL−1) are proteins with lyase activity, pyridoxal phosphate binding, and alpha/beta-hydrolase, which include two uncharacterized proteins encoded by orf19.4612 and orf19.7166. Recently, the latter protein has been identified as a Tac1p target that was upregulated in an azole-resistant strain that overexpressed MDR1 (1, 28). Pyridoxal-5′-phosphate (PLP) acts as a cofactor for more than 160 different catalytic functions, and PLP-dependent enzymes are widely involved in cellular processes, principally the biosynthesis of amino acids and amino-acid-derived metabolites (29, 30). KEGG pathway analysis showed that the abundances of proteins involved in glycine, serine, and threonine metabolism and cysteine and methionine metabolism were increased in Ca2. Glycine is the precursor of l-serine and, eventually, cysteine (31). Current evidence indicates that changes in cysteine biosynthesis affect the susceptibility of yeast to various antifungal agents. While glycine is decreased in fluconazole-treated C. albicans cells, it is probably mediated in part by oxidative damage owing to the reduced synthesis of intracellular glutathione (32, 33). Moreover, methionine metabolism is also associated with antifungal efficiency. Methionine is a precursor of S-adenosyl methionine (SAM) (34) that is involved in a number of chemically diverse reactions, such as methylation in ergosterol biosynthesis and aminopropyl donation in polyamine synthesis (35). Increased spermidine and spermine, two predominant polyamines, in eukaryotes can protect C. albicans cells from antifungal killing, which may be related to the attenuated accumulation of reactive oxygen species (ROS) (36, 37). The downregulated proteins related to the peroxisome, fatty acid degradation, and the longevity-regulating pathway are markedly inhibited in Ca2. The peroxisome plays a crucial role in the metabolic activities of eukaryotic cells, including the β-oxidation of fatty acids and the detoxification of hydrogen peroxide (38). Moreover, we observe that the flavoprotein-like domain and the flavodoxin/nitric oxide synthase domain, namely, Pst1p and Pst2p, involved in the oxidative stress response, are also decreased in Ca2. In C. albicans, flavodoxin-like proteins (FLPs) are critical for survival in the host, as a quadruple mutant lacking all four FLPs (pst1Δ pst2Δ pst3Δ ycp4Δ) was more susceptible to various oxidants and became avirulent in a mouse model of systemic candidiasis under conditions where infection with wild-type C. albicans was fatal (39). However, the NADP-dependent oxidoreductase domain is enriched in upregulated DEPs. The coenzyme NADPH/NADP+ transports electrons in NADP-dependent oxidoreductase-catalyzed reactions (40), and the reduced form, NADPH, is required for maintaining redox balance under oxidative stress (31, 41). Moreover, the ERG11 enzyme from C. albicans has been shown to need NADPH-P450 reductase for its functional activity that catalyzes the oxidative removal reaction of the 14α-methyl group from the lanostane frame (42).
Proteome responses in two isolates, Ca8 and Ca14.
In two isolates, Ca8 (MIC = 8 μg · mL−1) and Ca14 (MIC = 32 μg · mL−1), other reduced activities appear, including the lipid catabolic process, cellular oxidant detoxification, and the response to oxidative stress. In addition, carbohydrate metabolic activities and glycolysis/gluconeogenesis are decreased. The ability of the fungus to efficiently assimilate nutrients available within host niches is a crucial attribute for any microbial pathogen (43–45). C. albicans is flexible in metabolism and can adapt to the microenvironment of different anatomical sites. While physiologically relevant sugars, including glucose, fructose, and galactose, exist only at low levels and are even absent in many host niches, other nonfermentable carbon sources, such as amino acids and organic acids, are necessary to support the growth and metabolism of yeast in vivo (46). Thus, we presume that when the metabolic activity of C. albicans cells is disrupted by fluconazole, globally downregulated central carbon metabolism may trigger transformation to a growth form that needs less energy (46). Similarly, the biosynthesis of lysine and aromatic amino acids (phenylalanine, tyrosine, and tryptophan) and histidine, phenylalanine, and tyrosine metabolism are also enhanced in Ca8 and Ca14. The utilization of amino acids as nitrogen sources depends on the respective transaminases in human-pathogenic fungal cells such as upregulated Aro8p noted in this study. This protein is undoubtedly the most versatile aminotransferase in C. albicans, which participates in the degradation of histidine, lysine, and aromatic amino acids as well as in lysine, phenylalanine, and tyrosine biosynthesis (47). Remarkably, enzymes for histidine biosynthesis (His1p, His4p, His5p, and His7p) are upregulated. Indeed, some evidence indicates that His4p and His7p could be potential antifungal targets, as the biosynthetic steps are absent in mammalian hosts (48, 49). In addition to energy sources, amino acids are also major building blocks for proteins and important intermediates in some metabolic pathways (50). For example, the intermediate chorismic acid and the product tryptophan of the aromatic amino acid biosynthesis pathway are precursors for ubiquinone and nicotinamide, respectively, which largely participate in the respiratory chain and cellular redox potential (35).
We note that sterol biosynthesis in Ca14, but without sterol uptake control protein 2 (Upc2p), is upregulated. In this case, while fluconazole targets membrane sterol biosynthesis, intact Upc2p activates the transcriptional factor responsible for ergosterol biosynthesis and restores appropriate sterol levels in order to counter the effect of the azole (51, 52). Moreover, it is known that any imbalance of plasma membrane (PM) lipid constituents such as ergosterol and sphingolipids (SLs) contributes to the development of drug resistance in yeast cells (53–55). Any change in ergosterol biosynthesis by the disruption of ERG genes or a change in the SL composition by the disruption of its biosynthetic genes results in the improper localization of CaCdr1p (belonging to the ABC drug efflux transporters) within lipid rafts (56–58).
Proteome responses in isolate Ca17.
Fluconazole resistance development, fatty acid degradation, pyruvate metabolism, and glycolysis/gluconeogenesis continue to be suppressed, but amino acid metabolism is less disturbed in isolate Ca17 (MIC > 64 μg · mL−1). For energy metabolism and stress responses, electron carrier activities in mitochondria, ATPase activity coupled to the transmembrane movement of substances, and the response to toxic substances are all upregulated. Despite its well-known role as a main energy generator, the mitochondrion is crucial for many secondary processes such as cellular growth, apoptosis, virulence, and, most importantly here, the activation of drug efflux pumps causing enhanced azole resistance (59–61). Analysis of sterol synthesis mutants of Saccharomyces cerevisiae revealed that respiratory development is influenced by the sterol composition (62). Moreover, a previous study showed that ergosterol biosynthesis plays an essential role in maintaining the mitochondrial morphology of S. cerevisiae (63). In addition, the glycerophospholipid metabolism pathway is significantly enhanced, notably orf19.7166 and glycerol-3-phosphate dehydrogenase (Gpd1p). Lipids are viewed as the most adaptable molecules in response to environmental changes and are therefore considered the most suitable targets for stress adaptation (64), such as glycerol against hyperosmotic pressure (65). Accordingly, the haloacid dehalogenase (HAD)-like domain was significantly enriched with upregulated proteins in Ca17, including glycerol-1-phosphatase (Rhr2p) related to glycerol biosynthesis and a P1-type ATPase copper transporter (Crp1p) involved in copper detoxification (66, 67). However, Aqy1p, a water channel in the plasma membrane (68), was downregulated in this study. These results are consistent with the fact that C. albicans cells often secrete glycerin and arabitol when they are confronted with osmotic, temperature, and oxidative stresses (69, 70).
It is well known that iron availability affects various cellular processes such as mitochondrial respiration, electron transport, DNA synthesis and repair, oxygen transport, and other central metabolic pathways (58, 71). ERG11 and ERG5, two key enzymes in ergosterol biosynthesis, are heme-containing enzymes, and iron is essential for heme metabolism (72). Therefore, disrupting iron metabolism can affect the level of ergosterol in yeast and thus affect its sensitivity to azoles. High iron availability has recently been shown to increase β-1,3-glucan levels, which in turn increase the resistance of C. albicans to antifungal drugs (73). In seeming contradiction to the needs of higher mitochondrial respiration, iron transmembrane transporters are downregulated. This could be part of the stress adaptation to restrict iron availability since iron can be toxic by catalyzing the production of reactive free oxygen radicals that severely damage cellular components (74, 75). Along with the suppression of glycolysis and amino acid metabolism, zinc finger Cys2His2(C2H2)-type/integrase DNA-binding domain transcription factors such as Stp2p and the pH response transcription factor Rim101p/PacC are downregulated in Ca17. In C. albicans, Stp2p is required to utilize and catabolize amino acids as carbon sources and produces ammonia extrusion, resulting in an increased environmental pH (76, 77). As the central mediators of alkaline adaptation, the Rim101p/PacC transcription factors are activated by a proteolytic cleavage event at neutral-to-alkaline pH (76). Zinc finger proteins (ZFPs) are classified into various families according to zinc-binding motifs (78, 79). The C2H2 family consists of hundreds of ZFPs that are found in eukaryotes ranging from yeast to humans (79). In contrast, members of the zinc cluster protein family [or Zn(II)2Cys6 (Zn2C6) proteins] are strictly fungal. For example, gain-of-function (GOF) mutations in the transcription factors TAC1, MRR1, and UPC2 are thought to account for the majority of fluconazole resistance in C. albicans (80). However, no significant up- or downregulation of Zn2C6 proteins was observed in all four groups, suggesting that they may not have played a major role in the serial isolates.
Conclusions.
In this study, for the first time, we performed a comprehensive proteomic analysis of serial C. albicans isolates representing the acquisition of fluconazole resistance. As a consequence of the development of fluconazole resistance, carbohydrate and lipid catabolism are all significantly suppressed in Ca2, -8, -14, and 17 compared with isolate Ca1. However, the metabolisms of certain essential amino acids that cannot be synthesized by mammals are upregulated in Ca8 and Ca14. While oxidative stress and peroxidase activity were downregulated in all four subsequent isolates, resistance to osmotic pressure and copper and iron toxicity were upregulated in Ca17, along with mitochondrial respiration that may provide energy for drug efflux pumps. Obviously, the changes in energy metabolism and stress responses under fluconazole represent an adaptation strategy for C. albicans survival in the host, but they may also lead to new strategies for the prevention and treatment of resistant C. albicans infections by directly attacking or even reversing such an adaptation.
MATERIALS AND METHODS
Strains and culture.
Strains Ca1, Ca2, Ca8, Ca14, and Ca17 were obtained from T. C. White (School of Biological Sciences, University of Missouri at Kansas City, Kansas City, MO, USA). All five of these strains were retrieved from an AIDS patient who suffered from recurrent oropharyngeal candidiasis over a period of 2 years (81). The sensitivity of each strain to fluconazole and the established drug resistance mechanism are shown in Table 4. In this series, fluconazole resistance was considered to be gradually acquired in vivo and was related to the doses given to the patient. Moreover, molecular analyses confirmed that these isolates represent a single C. albicans strain and that the resistance levels were stable over 600 generations (81, 82). Five C. albicans isolates were grown at 28°C overnight in yeast extract-peptone-dextrose (YPD) medium with constant shaking at 220 rpm and used as seed cultures. Next, 10 mL of the seed culture was inoculated into 100 mL of fresh YPD liquid medium and then cultured at 28°C for 4 h (220 rpm) until the optical density at 600 nm (OD600) reached 0.8. The cultured cells were centrifuged at 6,000 rpm for 10 min (4°C) and washed twice with cold phosphate-buffered saline (PBS). Each treatment was performed in triplicate.
TABLE 4.
Antifungal profiles of serial C. albicans isolates
| Strain | Fluconazole dose (mg·day−1) | Previously reported MICa (μg · mL−1) | MIC in our studyb (μg · mL−1) | Previously described resistance mechanism(s) | Reference(s) |
|---|---|---|---|---|---|
| Ca1 | 100 | 0.25 | 0.5 | ||
| Ca2 | 100 | 1 | 2 | GOF mutations in MRR1; transient LOH events occurred on chromosome R; overexpression of MDR1 | 83 – 85 |
| Ca8 | 100 | 8 | 8 | Transient ploidy changes | 84 |
| Ca14 | 400 | 32 | 32 | Overexpression of ERG11; loss of allelic variation in ERG11; R467K mutation in ERG11 | 85, 86 |
| Ca17 | 800 | >64 | 128 | GOF mutations in MRR1, TAC1, and UPC2; mitotic recombination on chromosome 5 left arm; overexpression of ERG11, MDR1, CDR1, and CDR2; loss of allelic variation in ERG11; R467K mutation in ERG11 | 12, 13, 83–87 |
Protein extraction and digestion.
Samples were ground with liquid nitrogen into a cell powder and then transferred to a 5-mL centrifuge tube. After that, 4 volumes of lysis buffer (8 M urea, 1% Triton X-100, 10 mM dithiothreitol, and 1% protease inhibitor cocktail) were added to each cell powder. Next, samples were sonicated three times on ice using a high-intensity ultrasonic processor (Scientz). The remaining debris was removed by centrifugation at 20,000 × g for 10 min (4°C). Finally, the protein was precipitated with cold 20% trichloroacetic acid (TCA) for 2 h (−20°C). After centrifugation at 12,000 × g for 10 min (4°C), the supernatant was discarded. The remaining precipitate was washed 3 times with cold acetone. The protein was redissolved in 8 M urea, and the protein concentration was measured with a bicinchoninic acid (BCA) kit according to the manufacturer’s instructions.
For digestion, the protein solution was reduced at 56°C for 30 min with 5 mM dithiothreitol and alkylated for 15 min with 11 mM iodoacetamide at room temperature in the dark. The protein sample was then diluted by adding 100 mM tetraethylammonium bromide (TEAB) to a urea concentration of <2 M. Finally, trypsin was added at a trypsin-to-protein mass ratio of 1:50 for the first digestion overnight and at a trypsin-to-protein mass ratio of 1:100 for a second 4-h digestion.
TMT labeling.
After the trypsin was digested, the peptide was desalted by using a Strata X C18 SPE column (Phenomenex) and vacuum dried. The peptide was reconstituted in 0.5 M TEAB and processed according to the manufacturer’s protocol for the TMT-6plex kit. In brief, 1 U of TMT reagent was thawed and reconstituted in acetonitrile. The peptide mixtures were then incubated at room temperature for 2 h, pooled, desalted, and dried by vacuum centrifugation.
HPLC fractionation.
The tryptic peptides were fractionated into fractions by high-pH reverse-phase high-performance liquid chromatography (HPLC) using an Agilent 300Extend C18 column (5-μm particles, 4.6-mm internal diameter [ID], and 250-mm length). In short, peptides were first separated into 60 fractions with a gradient of 8 to 32% acetonitrile (pH 9.0) over 60 min. The peptides were then combined into 18 fractions and dried by vacuum centrifugation.
LC-MS/MS analysis.
The tryptic peptides were dissolved in 0.1% formic acid (solvent A), directly loaded onto a homemade reversed-phase analytical column (15-cm length and 75-μm ID). The gradient was comprised of an increase from 7% to 25% solvent B (0.1% formic acid in 90% acetonitrile) over 24 min and an increase from 25% to 40% solvent B in 8 min, with climbing to 80% in 4 min and then holding at 80% for the last 4 min, all at a constant flow rate of 350 nL/min, on an EASY-nLC 1000 ultraperformance liquid chromatography (UPLC) system. The peptides were subjected to an nanospray ion (NSI) source followed by tandem mass spectrometry (MS/MS) in a Q Exactive Plus system (Thermo) coupled online to the UPLC system. The electrospray voltage applied was 2.0 kV. For full scans, the m/z scan range was 350 to 1,800, and intact peptides were detected in the Orbitrap instrument at a resolution of 70,000. Peptides were then selected for MS/MS using the normalized collision energy (NCE) setting at 20, and the fragments were detected in the Orbitrap instrument at a resolution of 17,500. A data-dependent procedure that alternated between 1 MS scan and 20 MS/MS scans with 30.0 s of dynamic exclusion was used. The automatic gain control (AGC) was set at 5E4. The fixed first mass was set at 100 m/z.
Database search.
The resulting MS/MS data were processed using the MaxQuant search engine (v.1.5.2.8). Tandem mass spectra were searched against the UniProt Candida albicans database (17,719 sequences) concatenated with a reverse decoy database. Trypsin/P was specified as the cleavage enzyme, allowing up to 2 missed cleavages. The mass tolerances for precursor ions were set at 20 ppm in the first search and 5 ppm in the main search, and the mass tolerance for fragment ions was set at 0.02 Da. Carbamidomethyl on Cys was specified as a fixed modification, and oxidation on Met was specified as a variable modification. The false discovery rate (FDR) was adjusted to <1%, and the minimum score for peptides was set at >40.
Bioinformatics analyses. (i) Annotation methods.
Gene Ontology (GO) proteome annotation was derived from the UniProt-GOA database (http://www.ebi.ac.uk/GOA/) based on three categories: biological process, cellular component, and molecular function. wolfpsort soft was used to predict subcellular localization.
(ii) Functional enrichment analysis.
Two-tailed Fisher’s exact test was employed to test the enrichment of the differentially expressed proteins against all identified proteins. The GO annotations, Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways, and protein domains with a corrected P value of <0.05 were considered significant.
(iii) Enrichment-based clustering.
Enrichment-based clustering analysis was performed using the heatmap.2 function of the gplots R package.
(iv) Dynamic cluster analysis.
For dynamic cluster analysis, the relative expression quantitative data between the different comparison groups were combined according to the protein identification, to form a protein expression level data matrix. Next, the obtained data matrix was transformed by log2 and then normalized so that the expression level of each protein between different comparison groups was a group of numbers with 0 as the mean value and 1 as the standard deviation. Finally, the Mfuzz toolset was used on the normalized data set to perform cluster analysis. Analysis parameters included the following: the cluster number, “c,” was 4, and clustering ambiguity, “m,” was 2.
Data availability.
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD031774.
ACKNOWLEDGMENTS
This study was supported by funds of the National Natural Science Foundation of China (grant no. 81972949 to W.L., grant no. 81573059 and 82073455 to X.L., and grant no. 81903229 to X.Z.), the CAMS Innovation Fund for Medical Sciences (CIFMS) (grant no. 2016-I2M-3-021 to W.L. and X.L.), the basic scientific research fund projects of the Chinese Academy of Medical Sciences (grant no. 2020-PT310-005 to W.L.), the PUMC Youth Fund and Fundamental Research Funds for the Central Universities (grant no. 2017310033 to X.Z.), and the Nanjing Incubation Program for National Clinical Research Center (grant no. 2019060001 to W.L.).
Conceptualization, X.L. and W.L. Data curation, N.S., X.Z., and D.L. Formal analysis, N.S., X.Z., and D.L. Funding acquisition, X.Z., X.L., and W.L. Investigation, N.S., X.Z., D.L., X.L., and W.L. Methodology, N.S. and X.Z. Project administration, X.L. and W.L. Resources, X.Z., X.L., and W.L. Software, N.S. and X.Z. Supervision, X.L. and W.L. Validation, N.S., X.Z., and D.L. Visualization, N.S., X.Z., and D.L. Writing – original draft, N.S. Writing – review and editing, N.S., X.Z., D.L., X.L., and W.L.
We declare no competing interests.
Footnotes
Supplemental material is available online only.
Contributor Information
Xiaofang Li, Email: lxf3568@163.com.
Weida Liu, Email: liuwd@pumcderm.cams.cn.
REFERENCES
- 1.Liu TT, Znaidi S, Barker KS, Xu L, Homayouni R, Saidane S, Morschhäuser J, Nantel A, Raymond M, Rogers PD. 2007. Genome-wide expression and location analyses of the Candida albicans Tac1p regulon. Eukaryot Cell 6:2122–2138. 10.1128/EC.00327-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Prasath KG, Sethupathy S, Pandian SK. 2019. Proteomic analysis uncovers the modulation of ergosterol, sphingolipid and oxidative stress pathway by myristic acid impeding biofilm and virulence in Candida albicans. J Proteomics 208:103503. 10.1016/j.jprot.2019.103503. [DOI] [PubMed] [Google Scholar]
- 3.Zhou Y, Liao M, Zhu C, Hu Y, Tong T, Peng X, Li M, Feng M, Cheng L, Ren B, Zhou X. 2018. ERG3 and ERG11 genes are critical for the pathogenesis of Candida albicans during the oral mucosal infection. Int J Oral Sci 10:9. 10.1038/s41368-018-0013-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pfaller MA, Diekema DJ, Turnidge JD, Castanheira M, Jones RN. 2019. Twenty years of the SENTRY antifungal surveillance program: results for Candida species from 1997-2016. Open Forum Infect Dis 6:S79–S94. 10.1093/ofid/ofy358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pfaller MA, Diekema DJ. 2007. Epidemiology of invasive candidiasis: a persistent public health problem. Clin Microbiol Rev 20:133–163. 10.1128/CMR.00029-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brown GD, Denning DW, Gow NAR, Levitz SM, Netea MG, White TC. 2012. Hidden killers: human fungal infections. Sci Transl Med 4:165rv13. 10.1126/scitranslmed.3004404. [DOI] [PubMed] [Google Scholar]
- 7.Zaoutis TE, Argon J, Chu J, Berlin JA, Walsh TJ, Feudtner C. 2005. The epidemiology and attributable outcomes of candidemia in adults and children hospitalized in the United States: a propensity analysis. Clin Infect Dis 41:1232–1239. 10.1086/496922. [DOI] [PubMed] [Google Scholar]
- 8.Wilson LS, Reyes CM, Stolpman M, Speckman J, Allen K, Beney J. 2002. The direct cost and incidence of systemic fungal infections. Value Health 5:26–34. 10.1046/j.1524-4733.2002.51108.x. [DOI] [PubMed] [Google Scholar]
- 9.Berkow EL, Lockhart SR. 2017. Fluconazole resistance in Candida species: a current perspective. Infect Drug Resist 10:237–245. 10.2147/IDR.S118892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Allen D, Wilson D, Drew R, Perfect J. 2015. Azole antifungals: 35 years of invasive fungal infection management. Expert Rev Anti Infect Ther 13:787–798. 10.1586/14787210.2015.1032939. [DOI] [PubMed] [Google Scholar]
- 11.Fisher MC, Hawkins NJ, Sanglard D, Gurr SJ. 2018. Worldwide emergence of resistance to antifungal drugs challenges human health and food security. Science 360:739–742. 10.1126/science.aap7999. [DOI] [PubMed] [Google Scholar]
- 12.Lyons CN, White TC. 2000. Transcriptional analyses of antifungal drug resistance in Candida albicans. Antimicrob Agents Chemother 44:2296–2303. 10.1128/AAC.44.9.2296-2303.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rogers PD, Barker KS. 2003. Genome-wide expression profile analysis reveals coordinately regulated genes associated with stepwise acquisition of azole resistance in Candida albicans clinical isolates. Antimicrob Agents Chemother 47:1220–1227. 10.1128/AAC.47.4.1220-1227.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mazu TK, Bricker BA, Flores-Rozas H, Ablordeppey SY. 2016. The mechanistic targets of antifungal agents: an overview. Mini Rev Med Chem 16:555–578. 10.2174/1389557516666160118112103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bhattacharya S, Sae-Tia S, Fries BC. 2020. Candidiasis and mechanisms of antifungal resistance. Antibiotics (Basel) 9:312. 10.3390/antibiotics9060312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee Y, Puumala E, Robbins N, Cowen LE. 2021. Antifungal drug resistance: molecular mechanisms in Candida albicans and beyond. Chem Rev 121:3390–3411. 10.1021/acs.chemrev.0c00199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Penesyan A, Kumar SS, Kamath K, Shathili AM, Venkatakrishnan V, Krisp C, Packer NH, Molloy MP, Paulsen IT. 2015. Genetically and phenotypically distinct Pseudomonas aeruginosa cystic fibrosis isolates share a core proteomic signature. PLoS One 10:e0138527. 10.1371/journal.pone.0138527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tian Q, Stepaniants SB, Mao M, Weng L, Feetham MC, Doyle MJ, Yi EC, Dai H, Thorsson V, Eng J, Goodlett D, Berger JP, Gunter B, Linseley PS, Stoughton RB, Aebersold R, Collins SJ, Hanlon WA, Hood LE. 2004. Integrated genomic and proteomic analyses of gene expression in mammalian cells. Mol Cell Proteomics 3:960–969. 10.1074/mcp.M400055-MCP200. [DOI] [PubMed] [Google Scholar]
- 19.Baas T, Baskin CR, Diamond DL, García-Sastre A, Bielefeldt-Ohmann H, Tumpey TM, Thomas MJ, Carter VS, Teal TH, Van Hoeven N, Proll S, Jacobs JM, Caldwell ZR, Gritsenko MA, Hukkanen RR, Camp DG, Smith RD, Katze MG. 2006. Integrated molecular signature of disease: analysis of influenza virus-infected macaques through functional genomics and proteomics. J Virol 80:10813–10828. 10.1128/JVI.00851-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hooshdaran MZ, Barker KS, Hilliard GM, Kusch H, Morschhäuser J, Rogers PD. 2004. Proteomic analysis of azole resistance in Candida albicans clinical isolates. Antimicrob Agents Chemother 48:2733–2735. 10.1128/AAC.48.7.2733-2735.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thompson A, Schäfer J, Kuhn K, Kienle S, Schwarz J, Schmidt G, Neumann T, Johnstone R, Mohammed AKA, Hamon C. 2003. Tandem mass tags: a novel quantification strategy for comparative analysis of complex protein mixtures by MS/MS. Anal Chem 75:1895–1904. 10.1021/ac0262560. [DOI] [PubMed] [Google Scholar]
- 22.Peng C, Andersen B, Arshid S, Larsen MR, Albergaria H, Lametsch R, Arneborg N. 2019. Proteomics insights into the responses of Saccharomyces cerevisiae during mixed-culture alcoholic fermentation with Lachancea thermotolerans. FEMS Microbiol Ecol 95:fiz126. 10.1093/femsec/fiz126. [DOI] [PubMed] [Google Scholar]
- 23.Lietzén N, Ohman T, Rintahaka J, Julkunen I, Aittokallio T, Matikainen S, Nyman TA. 2011. Quantitative subcellular proteome and secretome profiling of influenza A virus-infected human primary macrophages. PLoS Pathog 7:e1001340. 10.1371/journal.ppat.1001340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Prasad R, Singh A. 2013. Lipids of Candida albicans and their role in multidrug resistance. Curr Genet 59:243–250. 10.1007/s00294-013-0402-1. [DOI] [PubMed] [Google Scholar]
- 25.Berman J, Krysan DJ. 2020. Drug resistance and tolerance in fungi. Nat Rev Microbiol 18:319–331. 10.1038/s41579-019-0322-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nishimoto AT, Sharma C, Rogers PD. 2020. Molecular and genetic basis of azole antifungal resistance in the opportunistic pathogenic fungus Candida albicans. J Antimicrob Chemother 75:257–270. 10.1093/jac/dkz400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Coumans JVF, Harvey J, Backhouse D, Poljak A, Raftery MJ, Nehl D, Katz ME, Pereg L. 2011. Proteomic assessment of host-associated microevolution in the fungus Thielaviopsis basicola. Environ Microbiol 13:576–588. 10.1111/j.1462-2920.2010.02358.x. [DOI] [PubMed] [Google Scholar]
- 28.Karababa M, Coste AT, Rognon B, Bille J, Sanglard D. 2004. Comparison of gene expression profiles of Candida albicans azole-resistant clinical isolates and laboratory strains exposed to drugs inducing multidrug transporters. Antimicrob Agents Chemother 48:3064–3079. 10.1128/AAC.48.8.3064-3079.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Eliot AC, Kirsch JF. 2004. Pyridoxal phosphate enzymes: mechanistic, structural, and evolutionary considerations. Annu Rev Biochem 73:383–415. 10.1146/annurev.biochem.73.011303.074021. [DOI] [PubMed] [Google Scholar]
- 30.Ahmed S, DeBerardinis RJ, Ni M, Afroze B. 2020. Vitamin B6-dependent epilepsy due to pyridoxal phosphate-binding protein (PLPBP) defect—first case report from Pakistan and review of literature. Ann Med Surg (Lond) 60:721–727. 10.1016/j.amsu.2020.11.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Katragkou A, Alexander EL, Eoh H, Raheem SK, Roilides E, Walsh TJ. 2016. Effects of fluconazole on the metabolomic profile of Candida albicans. J Antimicrob Chemother 71:635–640. 10.1093/jac/dkv381. [DOI] [PubMed] [Google Scholar]
- 32.Zhu J, Krom BP, Sanglard D, Intapa C, Dawson CC, Peters BM, Shirtliff ME, Jabra-Rizk MA. 2011. Farnesol-induced apoptosis in Candida albicans is mediated by Cdr1-p extrusion and depletion of intracellular glutathione. PLoS One 6:e28830. 10.1371/journal.pone.0028830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kim HS, Fay JC. 2007. Genetic variation in the cysteine biosynthesis pathway causes sensitivity to pharmacological compounds. Proc Natl Acad Sci USA 104:19387–19391. 10.1073/pnas.0708194104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Thomas D, Surdin-Kerjan Y. 1997. Metabolism of sulfur amino acids in Saccharomyces cerevisiae. Microbiol Mol Biol Rev 61:503–532. 10.1128/mmbr.61.4.503-532.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.De Backer MD, Ilyina T, Ma XJ, Vandoninck S, Luyten WH, Vanden Bossche H. 2001. Genomic profiling of the response of Candida albicans to itraconazole treatment using a DNA microarray. Antimicrob Agents Chemother 45:1660–1670. 10.1128/AAC.45.6.1660-1670.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Igarashi K, Kashiwagi K. 2010. Modulation of cellular function by polyamines. Int J Biochem Cell Biol 42:39–51. 10.1016/j.biocel.2009.07.009. [DOI] [PubMed] [Google Scholar]
- 37.Cao Y, Zhu Z, Chen X, Yao X, Zhao L, Wang H, Yan L, Wu H, Chai Y, Jiang Y. 2013. Effect of amphotericin B on the metabolic profiles of Candida albicans. J Proteome Res 12:2921–2932. 10.1021/pr4002178. [DOI] [PubMed] [Google Scholar]
- 38.Chen Y, Yu Q, Wang H, Dong Y, Jia C, Zhang B, Xiao C, Zhang B, Xing L, Li M. 2016. The malfunction of peroxisome has an impact on the oxidative stress sensitivity in Candida albicans. Fungal Genet Biol 95:1–12. 10.1016/j.fgb.2016.07.010. [DOI] [PubMed] [Google Scholar]
- 39.Li L, Naseem S, Sharma S, Konopka JB. 2015. Flavodoxin-like proteins protect Candida albicans from oxidative stress and promote virulence. PLoS Pathog 11:e1005147. 10.1371/journal.ppat.1005147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.You C, Huang R, Wei X, Zhu Z, Zhang Y-HP. 2017. Protein engineering of oxidoreductases utilizing nicotinamide-based coenzymes, with applications in synthetic biology. Synth Syst Biotechnol 2:208–218. 10.1016/j.synbio.2017.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stincone A, Prigione A, Cramer T, Wamelink MMC, Campbell K, Cheung E, Olin-Sandoval V, Grüning N-M, Krüger A, Tauqeer Alam M, Keller MA, Breitenbach M, Brindle KM, Rabinowitz JD, Ralser M. 2015. The return of metabolism: biochemistry and physiology of the pentose phosphate pathway. Biol Rev Camb Philos Soc 90:927–963. 10.1111/brv.12140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Park H-G, Lim Y-R, Eun C-Y, Han S, Han J-S, Cho KS, Chun Y-J, Kim D. 2010. Candida albicans NADPH-P450 reductase: expression, purification, and characterization of recombinant protein. Biochem Biophys Res Commun 396:534–538. 10.1016/j.bbrc.2010.04.138. [DOI] [PubMed] [Google Scholar]
- 43.Miramón P, Lorenz MC. 2017. A feast for Candida: metabolic plasticity confers an edge for virulence. PLoS Pathog 13:e1006144. 10.1371/journal.ppat.1006144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ene IV, Adya AK, Wehmeier S, Brand AC, MacCallum DM, Gow NAR, Brown AJP. 2012. Host carbon sources modulate cell wall architecture, drug resistance and virulence in a fungal pathogen. Cell Microbiol 14:1319–1335. 10.1111/j.1462-5822.2012.01813.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wilson D, Thewes S, Zakikhany K, Fradin C, Albrecht A, Almeida R, Brunke S, Grosse K, Martin R, Mayer F, Leonhardt I, Schild L, Seider K, Skibbe M, Slesiona S, Waechtler B, Jacobsen I, Hube B. 2009. Identifying infection-associated genes of Candida albicans in the postgenomic era. FEMS Yeast Res 9:688–700. 10.1111/j.1567-1364.2009.00524.x. [DOI] [PubMed] [Google Scholar]
- 46.Han T, Cannon RD, Villas-Bôas SG. 2012. Metabolome analysis during the morphological transition of Candida albicans. Metabolomics 8:1204–1217. 10.1007/s11306-012-0416-6. [DOI] [Google Scholar]
- 47.Rząd K, Milewski S, Gabriel I. 2018. Versatility of putative aromatic aminotransferases from Candida albicans. Fungal Genet Biol 110:26–37. 10.1016/j.fgb.2017.11.009. [DOI] [PubMed] [Google Scholar]
- 48.Jastrzębowska K, Gabriel I. 2015. Inhibitors of amino acids biosynthesis as antifungal agents. Amino Acids 47:227–249. 10.1007/s00726-014-1873-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Alcazar-Fuoli L. 2016. Amino acid biosynthetic pathways as antifungal targets for fungal infections. Virulence 7:376–378. 10.1080/21505594.2016.1169360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schrevens S, Van Zeebroeck G, Riedelberger M, Tournu H, Kuchler K, Van Dijck P. 2018. Methionine is required for cAMP-PKA-mediated morphogenesis and virulence of Candida albicans. Mol Microbiol 108:258–275. 10.1111/mmi.13933. [DOI] [PubMed] [Google Scholar]
- 51.MacPherson S, Akache B, Weber S, De Deken X, Raymond M, Turcotte B. 2005. Candida albicans zinc cluster protein Upc2p confers resistance to antifungal drugs and is an activator of ergosterol biosynthetic genes. Antimicrob Agents Chemother 49:1745–1752. 10.1128/AAC.49.5.1745-1752.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hoot SJ, Smith AR, Brown RP, White TC. 2011. An A643V amino acid substitution in Upc2p contributes to azole resistance in well-characterized clinical isolates of Candida albicans. Antimicrob Agents Chemother 55:940–942. 10.1128/AAC.00995-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Singh A, Prasad R. 2011. Comparative lipidomics of azole sensitive and resistant clinical isolates of Candida albicans reveals unexpected diversity in molecular lipid imprints. PLoS One 6:e19266. 10.1371/journal.pone.0019266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mukhopadhyay K, Prasad T, Saini P, Pucadyil TJ, Chattopadhyay A, Prasad R. 2004. Membrane sphingolipid-ergosterol interactions are important determinants of multidrug resistance in Candida albicans. Antimicrob Agents Chemother 48:1778–1787. 10.1128/AAC.48.5.1778-1787.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Singh A, Mahto KK, Prasad R. 2013. Lipidomics and in vitro azole resistance in Candida albicans. OMICS 17:84–93. 10.1089/omi.2012.0075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kundu D, Hameed S, Fatima Z, Pasrija R. 2020. Phospholipid biosynthesis disruption renders the yeast cells sensitive to antifungals. Folia Microbiol (Praha) 65:121–131. 10.1007/s12223-019-00713-3. [DOI] [PubMed] [Google Scholar]
- 57.Pasrija R, Panwar SL, Prasad R. 2008. Multidrug transporters CaCdr1p and CaMdr1p of Candida albicans display different lipid specificities: both ergosterol and sphingolipids are essential for targeting of CaCdr1p to membrane rafts. Antimicrob Agents Chemother 52:694–704. 10.1128/AAC.00861-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Thomas E, Roman E, Claypool S, Manzoor N, Pla J, Panwar SL. 2013. Mitochondria influence CDR1 efflux pump activity, Hog1-mediated oxidative stress pathway, iron homeostasis, and ergosterol levels in Candida albicans. Antimicrob Agents Chemother 57:5580–5599. 10.1128/AAC.00889-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shingu-Vazquez M, Traven A. 2011. Mitochondria and fungal pathogenesis: drug tolerance, virulence, and potential for antifungal therapy. Eukaryot Cell 10:1376–1383. 10.1128/EC.05184-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Guo H, Xie SM, Li SX, Song YJ, Lv XL, Zhang H. 2014. Synergistic mechanism for tetrandrine on fluconazole against Candida albicans through the mitochondrial aerobic respiratory metabolism pathway. J Med Microbiol 63:988–996. 10.1099/jmm.0.073890-0. [DOI] [PubMed] [Google Scholar]
- 61.Dröse S, Brandt U. 2012. Molecular mechanisms of superoxide production by the mitochondrial respiratory chain. Adv Exp Med Biol 748:145–169. 10.1007/978-1-4614-3573-0_6. [DOI] [PubMed] [Google Scholar]
- 62.Parks LW, Smith SJ, Crowley JH. 1995. Biochemical and physiological effects of sterol alterations in yeast—a review. Lipids 30:227–230. 10.1007/BF02537825. [DOI] [PubMed] [Google Scholar]
- 63.Altmann K, Westermann B. 2005. Role of essential genes in mitochondrial morphogenesis in Saccharomyces cerevisiae. Mol Biol Cell 16:5410–5417. 10.1091/mbc.e05-07-0678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Russell NJ, Evans RI, ter Steeg PF, Hellemons J, Verheul A, Abee T. 1995. Membranes as a target for stress adaptation. Int J Food Microbiol 28:255–261. 10.1016/0168-1605(95)00061-5. [DOI] [PubMed] [Google Scholar]
- 65.Hohmann S. 2002. Osmotic stress signaling and osmoadaptation in yeasts. Microbiol Mol Biol Rev 66:300–372. 10.1128/MMBR.66.2.300-372.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Desai JV, Cheng S, Ying T, Nguyen MH, Clancy CJ, Lanni F, Mitchell AP. 2015. Coordination of Candida albicans invasion and infection functions by phosphoglycerol phosphatase Rhr2. Pathogens 4:573–589. 10.3390/pathogens4030573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Schwartz JA, Olarte KT, Michalek JL, Jandu GS, Michel SLJ, Bruno VM. 2013. Regulation of copper toxicity by Candida albicans GPA2. Eukaryot Cell 12:954–961. 10.1128/EC.00344-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Srikantha T, Daniels KJ, Pujol C, Kim E, Soll DR. 2013. Identification of genes upregulated by the transcription factor Bcr1 that are involved in impermeability, impenetrability, and drug resistance of Candida albicans a/α biofilms. Eukaryot Cell 12:875–888. 10.1128/EC.00071-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sánchez-Fresneda R, Guirao-Abad JP, Argüelles A, González-Párraga P, Valentín E, Argüelles J-C. 2013. Specific stress-induced storage of trehalose, glycerol and D-arabitol in response to oxidative and osmotic stress in Candida albicans. Biochem Biophys Res Commun 430:1334–1339. 10.1016/j.bbrc.2012.10.118. [DOI] [PubMed] [Google Scholar]
- 70.Li L, Liao Z, Yang Y, Lv L, Cao Y, Zhu Z. 2018. Metabolomic profiling for the identification of potential biomarkers involved in a laboratory azole resistance in Candida albicans. PLoS One 13:e0192328. 10.1371/journal.pone.0192328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Drakesmith H, Prentice A. 2008. Viral infection and iron metabolism. Nat Rev Microbiol 6:541–552. 10.1038/nrmicro1930. [DOI] [PubMed] [Google Scholar]
- 72.Song J, Zhou J, Zhang L, Li R. 2020. Mitochondria-mediated azole drug resistance and fungal pathogenicity: opportunities for therapeutic development. Microorganisms 8:1574. 10.3390/microorganisms8101574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tripathi A, Liverani E, Tsygankov AY, Puri S. 2020. Iron alters the cell wall composition and intracellular lactate to affect Candida albicans susceptibility to antifungals and host immune response. J Biol Chem 295:10032–10044. 10.1074/jbc.RA120.013413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Mamouei Z, Zeng G, Wang Y-M, Wang Y. 2017. Candida albicans possess a highly versatile and dynamic high-affinity iron transport system important for its commensal-pathogenic lifestyle. Mol Microbiol 106:986–998. 10.1111/mmi.13864. [DOI] [PubMed] [Google Scholar]
- 75.Philpott CC. 2006. Iron uptake in fungi: a system for every source. Biochim Biophys Acta 1763:636–645. 10.1016/j.bbamcr.2006.05.008. [DOI] [PubMed] [Google Scholar]
- 76.Vylkova S, Carman AJ, Danhof HA, Collette JR, Zhou H, Lorenz MC. 2011. The fungal pathogen Candida albicans autoinduces hyphal morphogenesis by raising extracellular pH. mBio 2:e00055-11. 10.1128/mBio.00055-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Martínez P, Ljungdahl PO. 2005. Divergence of Stp1 and Stp2 transcription factors in Candida albicans places virulence factors required for proper nutrient acquisition under amino acid control. Mol Cell Biol 25:9435–9446. 10.1128/MCB.25.21.9435-9446.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.MacPherson S, Larochelle M, Turcotte B. 2006. A fungal family of transcriptional regulators: the zinc cluster proteins. Microbiol Mol Biol Rev 70:583–604. 10.1128/MMBR.00015-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Nishimoto AT, Zhang Q, Hazlett B, Morschhäuser J, Rogers PD. 2019. Contribution of clinically derived mutations in the gene encoding the zinc cluster transcription factor Mrr2 to fluconazole antifungal resistance and CDR1 expression in Candida albicans. Antimicrob Agents Chemother 63:e00078-19. 10.1128/AAC.00078-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Schillig R, Morschhäuser J. 2013. Analysis of a fungus-specific transcription factor family, the Candida albicans zinc cluster proteins, by artificial activation. Mol Microbiol 89:1003–1017. 10.1111/mmi.12327. [DOI] [PubMed] [Google Scholar]
- 81.White T, Pfaller M, Rinaldi M, Smith J, Redding S. 1997. Stable azole drug resistance associated with a substrain of Candida albicans from an HIV-infected patient. Oral Dis 3:S102–S109. 10.1111/j.1601-0825.1997.tb00336.x. [DOI] [PubMed] [Google Scholar]
- 82.Redding S, Smith J, Farinacci G, Rinaldi M, Fothergill A, Rhine-Chalberg J, Pfaller M. 1994. Resistance of Candida albicans to fluconazole during treatment of oropharyngeal candidiasis in a patient with AIDS: documentation by in vitro susceptibility testing and DNA subtype analysis. Clin Infect Dis 18:240–242. 10.1093/clinids/18.2.240. [DOI] [PubMed] [Google Scholar]
- 83.Popp C, Hampe IAI, Hertlein T, Ohlsen K, Rogers PD, Morschhäuser J. 2017. Competitive fitness of fluconazole-resistant clinical Candida albicans strains. Antimicrob Agents Chemother 61:e00584-17. 10.1128/AAC.00584-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ford CB, Funt JM, Abbey D, Issi L, Guiducci C, Martinez DA, Delorey T, Li BY, White TC, Cuomo C, Rao RP, Berman J, Thompson DA, Regev A. 2015. The evolution of drug resistance in clinical isolates of Candida albicans. Elife 4:e00662. 10.7554/eLife.00662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.White TC. 1997. Increased mRNA levels of ERG16, CDR, and MDR1 correlate with increases in azole resistance in Candida albicans isolates from a patient infected with human immunodeficiency virus. Antimicrob Agents Chemother 41:1482–1487. 10.1128/AAC.41.7.1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.White TC. 1997. The presence of an R467K amino acid substitution and loss of allelic variation correlate with an azole-resistant lanosterol 14 alpha demethylase in Candida albicans. Antimicrob Agents Chemother 41:1488–1494. 10.1128/AAC.41.7.1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Rogers PD, Barker KS. 2002. Evaluation of differential gene expression in fluconazole-susceptible and -resistant isolates of Candida albicans by cDNA microarray analysis. Antimicrob Agents Chemother 46:3412–3417. 10.1128/AAC.46.11.3412-3417.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Pfaller MA, Grant C, Morthland V, Rhine-Chalberg J. I994. Comparative evaluation of alternative methods for broth dilution susceptibility testing of fluconazole against Candida albicans. J Clin Microbiol 32:506–509. 10.1128/jcm.32.2.506-509.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Clinical and Laboratory Standards Institute. 2017. Reference method for broth dilution antifungal susceptibility testing of yeasts, 4th ed. Document M27-E4d. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental figures. Download aac.02105-21-s0001.pdf, PDF file, 3.0 MB (3.1MB, pdf)
Supplemental Data Set S1. Download aac.02105-21-s0002.xlsx, XLSX file, 1.6 MB (1.6MB, xlsx)
Supplemental Data Set S2. Download aac.02105-21-s0003.xlsx, XLSX file, 0.1 MB (148.5KB, xlsx)
Data Availability Statement
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD031774.