ABSTRACT
Proteolysis is essential throughout life, and as more proteases are characterized, our understanding of the roles they play continues to expand. Among other things, proteases are critical for protein turnover and quality control, the activation or inactivation of some enzymes, and they are integral components of signal transduction pathways. This review focuses on a family of proteases in bacteria known as the carboxyl-terminal processing proteases, or CTPs. Members of this family occur in all domains of life. In bacteria, CTPs have emerged as important enzymes that have been implicated in critical processes including regulation, stress response, peptidoglycan remodeling, and virulence. Here, we provide an overview of the roles that CTPs play in diverse bacterial species, and some of the underlying mechanisms. We also describe the structures of some bacterial CTPs, and their adaptor proteins, which have revealed striking differences in arrangements and mechanisms of action. Finally, we discuss what little is known about the distinguishing features of CTP substrates and cleavage sites, and speculate about how CTP activities might be regulated in the bacterial cell. Compared with many other proteases, the study of bacterial CTPs is still in its infancy, but it has now become clear that they affect fundamental processes in many different species. This is a protease family with broad significance, and one that holds the promise of more high impact discoveries to come.
KEYWORDS: carboxyl-terminal processing protease, cell envelope, stress response, gene regulation, cell wall hydrolase
INTRODUCTION
Bacterial cells contain many proteases, belonging to different families, and are associated with a wide range of functions. One widely conserved family is the carboxyl-terminal processing proteases (CTPs), which are assigned as the S41A family of serine proteases in the MEROPS peptidase database (1). Two of their distinguishing features are a serine-lysine catalytic dyad, and a PDZ domain that is thought to be important for substrate recognition. Another common feature is that all bacterial CTPs are known or predicted to be located outside of the cytoplasm. This fits with the fact that they are ATP-independent proteases. However, there are also some differences between them. The MEROPS database assigns some CTPs into subgroups based on homology and domain organization. For example, Escherichia coli Prc/Tsp, was assigned to the C-terminal processing peptidase-1 group. Members of this group are larger than some other CTPs and have additional domains at their N- and C-terminal ends.
Some bacterial CTPs have been identified only by association with a phenotype, but how the CTP affects the phenotype has not been determined in many cases (Table 1). The number of CTP substrates that meet the gold standard of confirmation by both in vivo and in vitro assays is low. Nevertheless, in recent years some bacterial CTPs have been characterized in detail and their substrates identified (Table 1). The picture emerging is one in which CTPs play diverse roles, including protein processing, signal transduction, and quality control. In Gram-negative bacteria, an exciting role has emerged in the degradation of cell wall hydrolases, and it is likely to be a widely conserved phenomenon (2, 3).
TABLE 1.
Bacterial C-terminal processing proteases in the literature
| Species and CTPsa | Notable null phenotypeb | Known or proposed substrates(s) | Selected references |
|---|---|---|---|
| Acinetobacter baumannii | |||
| Ctp WP_000776302.1 | Reduced virulence | None | (23, 24) |
| Bacillus subtilis | |||
| CtpA WP_003231186.1 | DNA damage sensitivity | YneA | (41, 42) |
| CtpB WP_003228041.1 | Sporulation defect | SpoIVFA | (39, 40, 74–76) |
| Bartonella bacilliformis | |||
| CtpA WP_005766244.1 | Not tested | None | (77) |
| Bordetella Bronchiseptica | |||
| CtpA WP_033446625.1 | Defective FHA processing | FhaBc | (27) |
| Borrelia burgdorferi | |||
| CtpA WP_002656708.1 | Reduced growth rate | P13c, BB0323c, OspCc | (29, 30) |
| Brucella suis | |||
| CtpA WP_004690508.1 | Reduced virulence | None | (20) |
| Burkholderia mallei | |||
| CtpA WP_004198008.1 | Reduced virulence | None | (21) |
| Burkholderia pseudomallei | |||
| Prc WP_004534131.1 | Meropenem sensitivity | None | (22) |
| Chlamydia trachomatis | |||
| CT441 WP_010725198.1 | Not tested | NF-κB p65 subunit (host protein)c,d | (31, 32, 35) |
| Escherichia coli K-12 | |||
| Prc WP_032199908.1 | Thermosensitive at low osmolarity | FtsI, MepM, MepS, MltG, MltBc, DigHc | (6, 9, 50, 62–64, 67, 78) |
| Legionella pneumophila | |||
| Tsp WP_010946247.1 | Thermal stress sensitivity | None | (15, 79) |
| Paenibacillus lautus | |||
| CtpA WP_015737691.1 | Not tested | None | (80) |
| Pseudomonas aeruginosa | |||
| CtpA WP_003096067.1 | Reduced virulence | MepM, PA1048, PA1198, PA1199, PA4404 | (3, 68, 69) |
| Prc WP_003091593.1 | Suppresses mucoid phenotype | MucA22c, FoxRc, FiuRc | (47, 55, 59, 81) |
| Pseudomonas putida | |||
| Prc WP_164721827.1 | Altered cell surface signaling | IutYc, PP_0161 (FoxR)c, PP_0351 (FiuR)c | (47, 49) |
| Staphylococcus aureus | |||
| CtpA WP_000342125.1 | Reduced virulence | SosAc | (43, 82) |
| Synechocystis sp. PCC 6803 | |||
| CtpA WP_010873417.1 | Photosynthesis defect | D1 | (12, 14) |
| Vibrio cholerae | |||
| Tsp WP_000650135.1 | Altered TcpP cleavage | TcpPc | (46) |
| Xanthomonas campestris | |||
| Prc WP_014509001.1 | Osmostress sensitivity | VgrS | (44, 45) |
Listed alphabetically by genus. The name of each protease as first reported in the literature is followed by a National Center for Biotechnology Information RefSeq non-redundant protein identification number.
In some cases, multiple phenotypes have been reported for protease null strains (see references) but only one of the notable phenotypes is listed.
Not confirmed to be directly cleaved/degraded by the purified protease in vitro.
EARLY DISCOVERIES OF BACTERIAL CTPS
The first bacterial CTP was reported in E. coli. The transpeptidase Penicillin-Binding Protein 3 (PBP3 or FtsI) is made as a precursor (4). A processing event was found to remove a short C-terminal region from PBP3, and an E. coli strain was identified with a mutation that prevented it (5). The mutation was named prc, to reflect the defect in processing the C-terminus of PBP3 (5). Subsequent work identified the prc gene, which encoded a periplasmic protein with no homology to any known proteins at the time (6). The Prc-dependent cleavage site in PBP3 was also identified, and later work confirmed direct cleavage of PBP3 by Prc in vitro (7, 8).
Another group was interested in understanding why C-terminal amino acids affected protein half-lives in E. coli (9). Wild type λ repressor has a half-life of over 10 h, whereas a variant with the five C-terminal polar amino acids replaced by nonpolar residues has a half-life of only 15 min (10). An E. coli protein that rapidly degraded the mutant λ repressor in vitro, but not the wild type, was purified and named Tsp for Tail-specific protease (9). Tsp and Prc are the same protein, and both names persist in the literature. However, it was apparent that Tsp/Prc was not responsible for degrading λ repressor in vivo, because Tsp/Prc is periplasmic whereas λ repressor is in the cytoplasm (11). Even so, Prc might degrade cell envelope proteins with aberrant C termini, as described later.
Shortly after the discovery of Prc, a CTP involved in the functioning of photosystem II (PSII) was identified in the cyanobacterium Synechocystis sp. PCC 6803 (12). This species is used as a model to investigate the PSII complex, two components of which make up the reaction core, the D1 and D2 proteins. In most oxygenic photosynthetic organisms, the D1 protein has a short C-terminal extension that must be removed to make it functional (13). To find proteins crucial for PSII function, spontaneous photosynthesis-deficient Synechocystis sp. PCC 6803 mutants were isolated (12). One mutation introduced a premature stop codon into a gene that was named ctpA (carboxyl-terminal processing protease) (12). The CtpA protein had a predicted signal sequence to direct it across the thylakoid membrane, and a mature portion that was 28% identical and 50% similar to E. coli Prc (12). Further characterization found that CtpA was responsible for processing the D1 protein into its mature form by removing the C-terminal extension (14).
Since these early discoveries, several other CTPs have been identified in bacteria (Table 1). For example, the Legionella pneumophila Tsp protein is important for survival under thermal stress and inside amoebae (15). Rhizobium leguminosarum CtpA is important for desiccation tolerance, biofilm formation, and cell envelope integrity (16–19). Brucella suis and Burkholderia mallei CTP-defective mutants have disrupted envelopes and reduced virulence, and a Burkholderia pseudomallei CTP mutant has increased sensitivity to meropenem (20–22). An Acinetobacter baumannii ctp null mutant has reduced motility, cell envelope integrity, and virulence, along with increased autolysis (23, 24). These and other examples show that CTPs are important for stress tolerance, cell envelope functions, and virulence, but the substrates of these CTPs are unknown. However, there are CTPs for which much more is known, some of which are described in the following sections.
LIVING UP TO THEIR NAME: CTPS PROPOSED TO PROCESS C-TERMINAL REGIONS
One role played by some CTPs is to process proteins by removing C-terminal regions. Two examples came from the earliest discoveries outlined above: processing of PBP3 (FtsI) and protein D1. The importance of CtpA-dependent processing for D1 function is clear, and it occurs widely in oxygenic photosynthetic organisms (25). In contrast, the significance of PBP3 processing by Prc in E. coli is unclear, although it has been implicated in the filamentous phenotype displayed by both prc and ftsI mutants (5). Three other interesting examples of proposed CTP-dependent processing in bacteria are described below.
A complex example of C-terminal processing occurs in the Gram-negative pathogen Bordetella bronchiseptica. An important virulence factor is the filamentous hemagglutinin (FHA), which is required for adhesion, manipulating cytokine signaling, and for persistence in the host. FHA is synthesized as a massive ∼ 3,700 amino acid protein FhaB, of which the C-terminal ∼ 1,200 amino acids are a prodomain. Removal of this prodomain involves three different proteases, and controls the timing of FHA maturation and release. The N-terminal region of FhaB is exported across the outer membrane, with the C-terminal prodomain remaining in the periplasm. DegP initiates prodomain degradation by removing the extreme C-terminus (ECT) in response to an unknown inducing signal (26). ECT removal by DegP generates a new C-terminus that is recognized by CtpA, which removes most of the remaining prodomain (27). Finally, the remainder of the prodomain is removed by a third protease, SphB1, which results in release of mature FHA from the cell surface (28).
Another CTP named CtpA was identified by searching for proteins with potential C-terminal processing capabilities in the etiological agent of Lyme disease, Borrelia burgdorferi (29). Proteomic analysis identified potential CtpA substrates, all of which were predicted to be outer membrane proteins (29). One was the surface exposed antigen P13, an outer membrane channel-forming protein, which was shown to have its C-terminal region processed by CtpA (29, 30). In addition, CtpA was also shown to remove the C-terminus of the surface exposed lipoprotein OspC, a virulence factor required for the establishment of mammalian infection (30). The authors speculated that C-terminal processing of B. burgdorferi surface-exposed lipoproteins by CtpA might be part of a quality control mechanism (30). This is an interesting idea, which is discussed later.
A questionable example of proposed C-terminal processing by a CTP in the literature comes from the obligate intracellular pathogen Chlamydia trachomatis. The C. trachomatis CTP named CT441 was detected in middle and late stages of infection, which corelated with the cleavage of the host cell p65 subunit of NF-κB (31, 32). A putative cleavage site within p65 was identified, suggesting that CT441 removes its C-terminal region (31, 32). Mutation of the p65 cleavage site, or of the CT441 serine/lysine catalytic dyad, prevented p65 cleavage. However, these studies did not demonstrate direct cleavage of p65 by purified CT441, and the cleavage of p65 by CT441, or in fact by any chlamydial protease, has since been called into question (33, 34). Later, a yeast two-hybrid experiment identified the host protein SRAP1, a co-activator of estrogen receptor α, as an interaction partner of the PDZ domain of CT441, and SRAP1 also interacted with CT441 in vitro (35). Although the authors did not find evidence that CT441 cleaved SRAP1, they did report that CT441 interferes with SRAP1 function in mammalian cells (35). However, that conclusion replied on the use of artificial expression constructs in cultured cells, which does not closely mimic a chlamydial infection. Another study found that CT441 cleaved SRAP1 in vitro, but did not find any evidence that it did so in vivo during the intracellular developmental cycle of C. trachomatis (36). There is also no evidence that CT441 is exported from C. trachomatis cells, which would be required for it to access the host proteins p65 and/or SRAP1. Therefore, although CT441 was proposed to process the C-terminus of p65, its role and the identity of its substrates await unequivocal confirmation.
FLIPPING THE SWITCH: CTPS THAT PARTICIPATE IN REGULATORY PATHWAYS
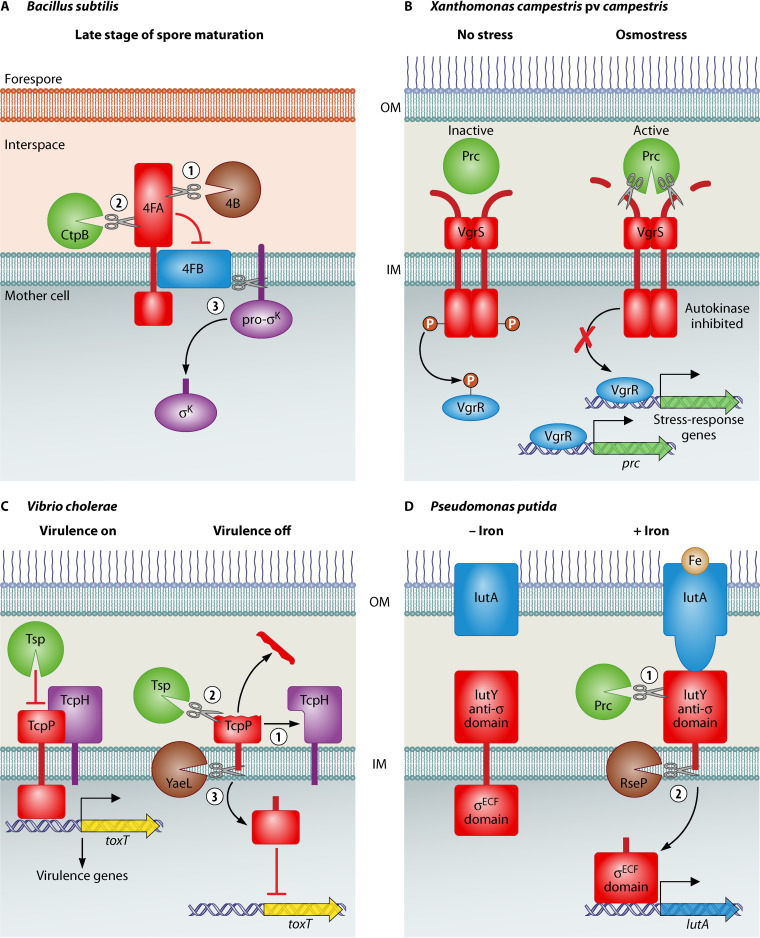
CTPs have also been implicated in regulatory pathways. A well characterized example occurs during sporulation in Bacillus subtilis. Sporulation is a complex process that has been reviewed by others (37). It begins with the formation of a septum close to one pole that gives rise to a sporangium made up of the smaller forespore and the larger mother cell. The mother cell engulfs the forespore so that it is enclosed in the mother cell cytoplasm. The final steps of spore development are controlled by the mother cell specific sigma factor σK, which is produced as the inactive membrane-associated pro-σK. Release of σK requires pro-σK to be cleaved by the membrane-embedded protease SpoIVFB (4FB), which in turn is inhibited by the transmembrane protein, SpoIVFA (4FA; [38]). The N-terminus of 4FA is in the mother cell cytoplasm, whereas the C-terminus is in the interspace separating mother cell and forespore (Fig. 1A). This sets up a system for forespore signaling to be transmitted across the membrane to the mother cell cytoplasm. The inhibition of 4FB by 4FA is relieved by the sequential action of two proteases on the 4FA C-terminus in the interspace, the second of which is a CTP named CtpB (Fig. 1A). The first protease removes the folded C-terminus of 4FA, which protects it from CtpB. The truncated 4FA C-terminus interacts with and activates CtpB, which removes more of the C-terminal segment of 4FA, including the part that inhibits 4FB (39). The uninhibited 4FB then cleaves the membrane-associated pro-σκ, which releases active σK into the mother cell cytoplasm (Fig. 1A). Intriguingly, ctpB expression is controlled by both mother cell and forespore specific sigma factors, and CtpB is produced in both cells (40). However, it is the forespore-specific CtpB that is important for σK activation and the completion of sporulation. Nevertheless, if ctpB is overexpressed in the mother cell only, it triggers sporulation (40). This suggests that CtpB could play additional roles in the mother cell involving unidentified substrates, although this also might be an artifact of ctpB overexpression causing cleavage of non-physiological substrates.
FIG 1.
CTPs involved in regulation. (A) In the final steps of B. subtilis spore development, the SpoIVB protease (4B) removes the C-terminus of SpoIVFA (4FA), which then allows CtpB to remove the 4FA domain that inhibits the SpoIVFB (4FB) protease. 4FB then cleaves pro-sK, releasing active sK into the mother cell cytoplasm. (B) In X. campestris pv. campestris, osmostress activates Prc to cleave the N-terminal periplasmic domain of VgrS. This reduces phosphotransfer to VgrR, which alters its global promoter binding profile, allowing it to induce expression of stress-response genes, and of prc itself, providing a positive feedback loop. (C) In conditions that do not favor V. cholerae virulence gene expression, TcpH dissociates from TcpP, allowing Tsp to cleave close to the C-terminus of TcpP. The truncated TcpP is then further degraded by the YaeL protease. This sequential cleavage of TcpP by Tsp and YaeL inactivates TcpP, so that toxT and the virulence genes controlled by ToxT, are no longer expressed. (D) In P. putida iron-loaded aerobactin (brown circle) binds to the IutA receptor. This causes the IutA signaling domain to interact with IutY, triggering IutY cleavage by Prc. The transmembrane domain of the truncated IutY is then cleaved by RseP (an ortholog of V. cholerae YaeL), which releases the σECF domain into the cytoplasm to induce iutA expression.
B. subtilis has a second CTP called CtpA (41). CtpA was discovered before CtpB but 2 decades passed before anyone identified a role for CtpA, which is controlling the process of DNA damage checkpoint recovery (42). CtpA, and an unrelated protease YlbL, are membrane anchored proteins that degrade the extracellular portion of the transmembrane cell division inhibitor YneA. When DNA damage occurs, yneA expression increases, and the amount of YneA protein overwhelms the proteases and allows it to enforce a checkpoint, which delays cell division. Once DNA repair is complete, yneA expression decreases, the YibL and CtpA proteases clear YneA from the cell, and cell division proceeds (42). An analogous phenomenon has also been reported in Staphylococcus aureus, involving degradation of a cell division inhibitor by CtpA (43).
The plant pathogen Xanthomonas campestris pv. campestris provides another example of a regulatory role, albeit one in which the protease does not live up to the CTP name. A prc deletion mutant had reduced virulence in cabbage, was more susceptible to antibiotics, and had decreased survival under osmostress conditions (44, 45). The reduced osmostress tolerance was attributed to the role played by Prc in regulating an osmostress response (44). Osmostress activates Prc to cleave the N-terminal periplasmic domain of the histidine kinase sensor protein VgrS (Fig. 1B). This reduces VgrS autophosphorylation, which in turn reduces transfer of phosphate to its response regulator partner, VgrR. The reduced phosphorylation of VgrR alters its DNA-binding affinity and changes its global promoter binding profile. Some VgrR-bound promoters control genes important for osmostress-tolerance, and a low level of VgrS-VgrR phosphorylation is optimal to promote bacterial resistance to osmostress (44) (Fig. 1B). The inhibition of VgrS autokinase activity by Prc proteolysis is an effective mechanism to achieve this. VgrR also controls prc expression directly, providing a positive feedback loop (44).
In Vibrio cholerae, a Prc homolog using the alternate Tsp name has been implicated in shutting off the regulatory cascade controlling production of the virulence factors cholera toxin and the toxin-co-regulated pilus (46). TcpP is a transmembrane protein with a C-terminal domain in the periplasm and an N-terminal cytoplasmic DNA binding domain that induces the expression of toxT, encoding a transcriptional activator of virulence factors (Fig. 1C). In conditions favoring virulence factor expression, another transmembrane protein, TcpH, binds to TcpP and protects it from proteolysis. In non-inducing conditions, TcpH dissociates from TcpP, and Tsp is activated to cleave a small C-terminal region from TcpP. This generates a truncated TcpP protein that is a substrate for the inner membrane zinc metalloprotease YaeL, which further degrades TcpP (Fig. 1C). The sequential cleavage of TcpP by Tsp and YaeL inactivates TcpP, so that toxT and the genes encoding cholera toxin and the toxin-co-regulated pilus are no longer expressed (46).
Prc orthologs have been implicated as site-1 proteases in cell surface signaling (CSS) systems of Pseudomonas species. In CSS systems, a ligand, which is often a siderophore, binds to a receptor protein in the outer membrane. The receptor interacts with a transmembrane anti-sigma factor that extends from the periplasm into the cytoplasm, where it controls the activity of a sigma factor of the extracytoplasmic function class (σECF). However, a Pseudomonas putida CCS system, which responds to the siderophore aerobactin, has a hybrid protein called IutY with both transmembrane anti-sigma factor and cytoplasmic σECF domains (47). Genetic experiments led to a model in which extracellular iron-loaded aerobactin binds to the outer membrane receptor protein IutA, which then interacts with IutY and triggers cleavage of the IutY C-terminal periplasmic domain by Prc (47, 48) (Fig. 1D). The truncated IutY is now a substrate for RseP, an ortholog of V. cholerae YaeL, which cleaves the transmembrane domain of IutY and releases the σECF domain into the cytoplasm (47). Prc was also implicated in regulating classical CSS systems in which the sigma and antisigma factors are separate proteins. These were the ferrioxamine (Fox) and ferrichrome (Fiu) responsive systems in both P. putida and P. aeruginosa (47, 49). Therefore, it was suggested that Prc is involved in the control of multiple CSS systems in Pseudomonas species.
The examples above suggest that E. coli Prc orthologs are site 1 proteases in regulated intramembrane proteolysis cascades in V. cholerae, P. putida, and P. aeruginosa, which suggests that it might be a common Prc role (Fig. 1B to D). However, in all cases the involvement of Prc was deduced from genetics and Prc was not shown to cleave the regulatory proteins directly (46–49) (Table 1). These genetic experiments strongly support the involvement of Prc, but leave open the possibility that Prc itself does not cleave the regulatory proteins. For example, if changes in Prc activity causes envelope stress, that might affect the ability of other proteases to cleave the regulatory proteins. On balance though, the case for direct involvement of Prc in these proteolytic cascades is compelling.
DEALING WITH THE TRASH: CTPS IMPLICATED IN PROTEIN QUALITY CONTROL
Some CTPs have been implicated in protein quality control within the bacterial cell envelope. When translation is stalled on the ribosome in E. coli, the 11-amino acid SsrA tag (AANDENYALAA) is added to the C-terminus in a co-translational mechanism (50, 51). This tag marks the mistranslated protein for proteolytic degradation, and it is a widely conserved phenomenon in bacteria (52, 53). In the E. coli cytoplasm, at least four different proteases degrade proteins with SsrA tags (reviewed in [53]). However, data suggests that Prc degrades SsrA-tagged proteins in the periplasm. This was demonstrated with an engineered derivative of cytochrome b562 with an SsrA-tag added to its C-terminus. The tagged cytochrome had a half-life below 5 min in a prc+ strain, but over 1 h in a prc null mutant (50). However, the significance of Prc in degrading naturally SsrA-tagged proteins in vivo is unclear (50). Intriguingly though, in Streptomyces coelicolor and Streptomyces lividans, the gene encoding Prc is located immediately upstream of the two genes required to SsrA-tag proteins (54). When this observation is combined with the data from E. coli, it supports the possibility of a functional link between Prc and SsrA-tagged proteins.
Another Prc might also degrade proteins with aberrant C termini. Pseudomonas aeruginosa has an apparent ortholog of E. coli Prc, which has been named Prc, but also AlgO due to its impact on production of the polysaccharide alginate (55). A critical control point in the regulation of alginate biosynthesis is the inhibition of the AlgU/T sigma factor by the anti-sigma factor MucA (reviewed in [56]). During lung colonization in individuals suffering from cystic fibrosis, P. aeruginosa mutants arise that constitutively produce alginate. These mucoid strains frequently have mutations in the mucA gene, the most common of which is the mucA22 frameshift allele (57, 58). This mutation truncates MucA and adds three arginines to its C-terminus, destabilizing the anti-sigma factor and causing constitutive alginate production. Screens for mutations that suppress the mucA22 phenotype isolated null mutations in prc (55, 59). It was suggested that Prc degrades MucA22, due to its aberrant C-terminus, in conditions that are normally unfavorable for alginate biosynthesis (55, 59, 60). While this is the most likely explanation, Prc has not been shown to degrade MucA22 directly, which leaves open the possibility of an indirect effect. Nevertheless, if Prc does degrade MucA22, it would be another example of protein quality control, and raise the possibility that Prc might degrade other proteins with aberrant C termini.
A third and somewhat different implication of a CTP playing a role in quality control arose from a study of the C-terminal processing of B. burgdorferi surface-exposed lipoproteins by CtpA (30). In the proposed model, low level C-terminal processing of these lipoproteins by CtpA, before the mature protein is inserted into the outer membrane, might be a quality control system for surface lipoprotein translocation. The authors speculated that detection of the released C-terminal peptides might report that proper translocation of surface lipoproteins is occurring (30). Disruption of proper translocation might increase the level of CtpA-dependent processing, raising the level of the C-terminal peptides and triggering a stress response to mitigate the problem. This is an intriguing model, although it is highly speculative and in need of experimental interrogation.
The studies above outline the current support for an involvement of CTPs in bacterial cell envelope protein quality control. It is a particularly attractive hypothesis that C-terminal processing proteases like Prc might target proteins with aberrant C termini. However, the evidence is limited and more work is needed in order to understand how significant this is, and how widespread it might be.
AN EMERGING ROLE IN CELL WALL HYDROLASE DEGRADATION AND THE DISCOVERY OF CTP ADAPTOR PROTEINS
Almost all bacteria have a peptidoglycan cell wall, which determines cell shape and provides resilience against turgor pressure. Peptidoglycan is made up of glycan strands with attached stem peptides. The peptides of parallel glycan chains cross-link via peptide bonds to form a mesh like sacculus surrounding the cell. In addition to enzymes for peptidoglycan synthesis, a number of hydrolases cleave the various bonds in the polymer to facilitate cell growth and division. These hydrolases must be controlled, but the mechanisms by which this is achieved have only begun to emerge recently. Two different CTPs, in two different species, have been found to constrain the activities of some cell wall hydrolases by degrading them. These studies also led to the discovery of CTP adaptor proteins, which are needed for the degradation of some CTP substrates (2, 3).
The first discovery came from E. coli, where the cell wall cross-link hydrolase MepS is more abundant in exponential phase than stationary phase (2). A genetic screen identified two null mutations that increased and equalized MepS abundance at all growth stages (2). One was a prc null mutation, suggesting that Prc degraded MepS. Indeed, a mepS (formerly spr) null mutation was identified originally because it suppressed the thermosensitivity of a prc null mutant at low osmolarity (61). Subsequent experiments confirmed that Prc degrades MepS directly (2). Studies in the last couple of years have also shown that E. coli Prc degrades another cross-link hydrolase, MepM, and the lytic transglycosylase MltG, which cleaves the glycan chains of peptidoglycan (62, 63). The peptidoglycan hydrolases MltB and DigH have also been proposed as likely Prc substrates, although they have not yet been confirmed to be cleaved directly by Prc (64).
As mentioned above, a genetic screen identified two null mutations that stabilized MepS, and one of these affected prc. The second mutation affected nlpI, encoding an outer membrane lipoprotein with tetratricopetide repeat (TPR) motifs (2). In addition to stabilizing MepS, nlpI and prc null mutants also share other common phenotypes (6, 65). It was originally proposed that Prc processes the C-terminus of NlpI, but this was not supported by further investigation (2, 66). Instead, NlpI is an adaptor required for Prc-dependent degradation of MepS in vivo and in vitro (2). It achieves this by binding to Prc and MepS independently, acting as a scaffold to bring them together (2, 67). However, NlpI inhibits the processing of PBP3 by Prc in vitro (7). It has also been suggested that NlpI might not be required for Prc to degrade the cell wall hydrolase substrates MepM and MltG in vivo (62, 63). However, this is unclear for MepM, because although a nlpI null mutation did not cause MepM accumulation in vivo, NlpI massively accelerated Prc-dependent degradation of MepM in vitro (63). Regardless, all these observations suggest the possibility that NlpI might be a substrate-specific adaptor protein.
A similar phenomenon was uncovered in P. aeruginosa, which unlike E. coli, has two CTPs. One is an apparent ortholog of E. coli Prc, but remarkably, it is the other one that has been found to work with an adaptor protein to degrade cell wall hydrolases. This CTP, named CtpA, was found to be important for type III secretion system function and virulence (68). CtpA interaction partners and substrates were identified by biochemical and genetic approaches (3). This identified four CtpA substrates, all of which are predicted cell wall cross-link hydrolases. Two are members of the LytM/M23 peptidase family (MepM and PA4404) and two are members of the NlpC/P60 peptidase family (PA1198 and PA1199, which are homologs of E. coli MepS). Therefore, CtpA degrades proteins that are homologous to the E. coli Prc substrates MepS and MepM. The study also identified an adaptor protein that was named LbcA and was required for CtpA to degrade all four substrates in vivo and in vitro (3). Like E.coli NlpI, LbcA is an outer membrane lipoprotein with TPR motifs. However, LbcA is approximately twice the size of NlpI and their primary sequences are not similar. A more recent study identified a fifth CtpA substrate that is also degraded in an LbcA-dependent manner (69). This substrate, PA1048, is not a predicted cell wall hydrolase, but it has an OmpA-like C-terminal region predicted to bind to peptidoglycan non-covalently. In P. aeruginosa no LbcA-independent CtpA substrate has been identified so far.
The similarities between the E. coli NlpI-Prc and P. aeruginosa LbcA-CtpA systems are obvious. Both consist of a CTP in complex with an outer membrane lipoprotein adaptor, and both degrade cell wall hydrolases. In both cases, the adaptor protein acts as a scaffold that binds to the protease and substrate independently (2, 69). However, while these two systems are similar, they are not orthologs. Prc and CtpA have been assigned to different CTP subgroups, and NlpI and LbcA are not homologous. The differences between them are further emphasized by structural analysis (see below). Therefore, two different CTPs interact with two different adaptor proteins, in two evolutionary divergent species, to achieve the same function. This raises the possibility that this is a widely conserved mechanism used to constrain bacterial cell wall hydrolases. In fact, many CTP-defective mutants of Gram-negative species have been reported to have defective cell envelopes and other phenotypes that might be explained by accumulating cell wall hydrolases (Table 1).
STRUCTURES OF CTPS AND ADAPTOR PROTEINS
The structures of some bacterial CTPs and adaptor proteins have been solved, which has provided valuable insights into their mechanisms of action. However, these studies have also uncovered striking differences, especially in their stoichiometric arrangements.
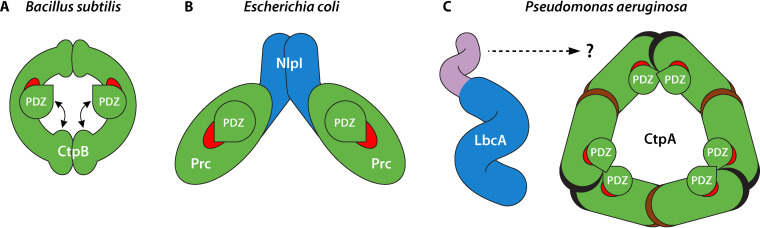
B. subtilis CtpB has a PDZ domain that adopts a similar fold as other PDZ domains, two protease subdomains (cap and core), and dimerization motifs at its N- and C-terminal ends (39). The dimerization domains interact to assemble a ring-like dimer (Fig. 2A). A narrow tunnel between the protease cap and core subdomains is wide enough for an unfolded substrate to enter. The PDZ domain is the most flexible part of the protein and its positioning controls access to the protease tunnel. CtpB switches between inactive and active forms, with the PDZ domain blocking the protease tunnel in the former (Fig. 2A). Once the PDZ domain interacts with a substrate it locks into the active position, allowing the substrate to access the protease tunnel for cleavage to occur. This inhibitory role of the PDZ domain is demonstrated by the fact that removal of the PDZ domain from CtpB yields a constitutively active protease (39).
FIG 2.
Contrasting arrangements of bacterial CTPs. (A) B. subtilis CtpB forms a ring-like dimer via N-terminal to N-terminal and C-terminal to C-terminal interactions. The positioning of the PDZ domain controls access to a narrow protease tunnel (red). CtpB switches between inactive and active forms, defined by whether or not the PDZ domain blocks the protease tunnel. (B) E. coli Prc is a monomer that forms a self-compartmentalized bowl-like structure, with part of the protease domain comprising a vault (red), and a lid-like PDZ domain. Upon interaction with a substrate, the PDZ domain shifts position causing a productive repositioning of the active site residues (not shown). The adaptor protein NlpI is a dimer, with each subunit interacting with one Prc monomer to form the tetrameric arrangement shown. NlpI is required for Prc to degrade MepS, but not for the degradation of any other Prc substrates in vivo. (C) P. aeruginosa CtpA forms a ring-like hexamer, arranged as a trimer of dimers mediated by N-terminal to N-terminal (black) and C-terminal to C-terminal (brown) interactions. The LbcA adaptor protein is a monomer with 11 TPR motifs forming a spiral structure (blue), and a separate N-terminal helical domain (purple) that is required for interaction with CtpA. LbcA is required for CtpA to degrade all five of its known substrates. The CtpA hexamer is inactive in the absence of LbcA because the active site residues (red) are misaligned (not shown). Studies to understand exactly how LbcA interacts with and activates CtpA are in progress.
The next bacterial CTP structure reported was CT441 from C. trachomatis (36). It has a novel N-terminal domain (NTD) that appears to be important for CT441 folding and/or solubility. This NTD is also important for a chaperone activity of CT441 that was uncovered during the structural analysis, and might play a role in protein quality control (36). The NTD is connected to the PDZ domain by a highly flexible region, which prevented the PDZ domain from being defined in the crystal structure. The PDZ domain is followed by a C-terminal domain that mediates the formation of homodimers and contains the active site. However, although CT441 formed dimers in the crystal, it purified as a monomer, meaning that the physiological significance of the dimers is uncertain. The authors speculated that dynamic dimerization might play a role in vivo, perhaps affecting the activation status of the protease. They also hypothesized that movement of the PDZ domain might be important for protease activation (36). However, unlike the PDZ domain of CtpB, the CT441 PDZ domain was required for activity and so it is not only an inhibitor.
E. coli Prc is a monomer that forms a self-compartmentalized bowl-like structure, with a lid-like PDZ domain (67) (Fig. 2B). The PDZ domain of Prc is not an inhibitor, but instead it functions as an activator that is required for protease activity (7, 67). In the inactive state, the catalytic residues of Prc are misaligned. However, when a substrate interacts with the PDZ domain, it changes its position, and this movement triggers a structural change that aligns the active site residues correctly (7). Therefore, in both CtpB and Prc, the PDZ domain plays an important regulatory role, but those roles are different. The structure of E. coli Prc was solved in complex with its NlpI adaptor protein (67). NlpI is a dimer with each subunit of the dimer interacting with one Prc monomer to assemble a 2:2 tetramer (Fig. 2B). NlpI has four TPR motifs, which are all on the outside edge of the dimer. NlpI interacts with Prc mainly via TPR2, whereas TPR1 was implicated in binding to the MepS substrate. Once MepS is bound to NlpI, it is thought that the PDZ domain of Prc interacts with the MepS C-terminus, prompting the activating shift to occur in the Prc catalytic residues. Rotational movement of the substrate-bound PDZ domain might drive translocation of the MepS polypeptide through the protease active site (67). This lever-like mechanism could explain how Prc completely degrades MepS.
The P. aeruginosa LbcA-CtpA system is functionally analogous to the E. coli NlpI-Prc system, but their protease and adaptor protein components are quite different. Structural analysis has revealed that the differences between these systems are even more striking, and that CtpA itself is very different to the other CTPs described above. CtpA consists of a PDZ domain, two protease subdomains (cap and core), and dimerization motifs at its N- and C-terminal ends (70). The N-terminal dimerization domain of CtpA is similar to that of B. subtilis CtpB, but the C-terminal dimerization domain is different. The N termini of two CtpA molecules dimerize in the same way as CtpB. However, instead of N-N and C-C interactions forming a ring-like dimer, the six C-terminal domains of three CtpA dimers interact to form a triangular trimer-of-dimers in the crystal (Fig. 2C). This oligomeric state is supported by the estimated mass of a hexamer from the gel filtration profile of CtpA (70). The CtpA hexamer is inactive because the catalytic residues are beyond hydrogen-bonding distance. This is consistent with the observation that CtpA alone was inactive in substrate degradation in vivo and in vitro (3). Therefore, unlike the B. subtilis CtpB dimer, which fluctuates between active and inactive forms and can be activated by a protein substrate, the CtpA hexamer is locked in an inactive conformation and requires LbcA for activation. The LbcA adaptor is a monomer, in contrast to the dimeric NlpI adaptor of E. coli Prc (67, 70). LbcA contains only α-helices and connecting loops, with 22 of these α-helices comprising the 11 TPR motifs. These 11 TPRs form a spiral, which has the potential to wrap around a substrate for delivery to CtpA (70) (Fig. 2C). Outside of the TPR spiral are four additional N-terminal α-helices that form an extension partially capping the spiral ring. The first of these helices is essential for binding to CtpA (70). Ongoing studies are characterizing the LbcA-CtpA complex. Preliminary indications suggest that up to three LbcA molecules might bind to one CtpA hexamer to assemble a giant, active protease complex (Hao-Chi Hsu, Andrew J. Darwin and Huilin Li, unpublished data).
Although few bacterial CTP structures have been solved, a common theme is that the PDZ domain is a mobile regulatory component, although exactly how it achieves this function differs. The PDZ domain of B. subtilis CtpB is an inhibitor that blocks access to the proteolytic site. In contrast, the PDZ domain of E. coli Prc is a substrate-responsive activator that promotes reconfiguration of the active site residues. These studies have also revealed some other contrasts. For example, E. coli Prc and C. trachomatis CT441 are monomers in solution, whereas B. subtilis CtpB is a dimer, and P. aeruginosa CtpA is a hexamer. Also, although both E. coli Prc and P. aeruginosa CtpA partner with adaptors to degrade cell wall hydrolases, their macromolecular arrangements are completely different (Fig. 2).
FINDING THEIR TARGET: CTP SUBSTRATE AND CLEAVAGE SITE RECOGNITION
What features allow CTPs to recognize their substrates and cleavage sites? Very few studies have examined this, and no clear consensus has emerged. Early attention focused on the importance of the substrate C-terminus, and suggested that non-polar residues might be required for recognition by E. coli Prc. Prc degrades λ repressor when the non-polar sequence WVAAA is added to its C-terminus, but does not degrade the wild-type protein that has RSEYE at its C-terminus (9). The C-terminal SsrA tag that also renders λ repressor susceptible to cleavage by Prc ends with YALAA, which is similar to WVAAA. In addition, changing the final two non-polar amino acids of the SsrA tag from AA to DD protected it from Prc (71). Additional analysis of λ and Arc repressors with altered C termini supported a requirement for non-polar residues at the C-terminus, as well as a free α-carboxyl group (72). However, a free α-carboxyl group cannot be a universal requirement, because X. campestris Prc cleaves the N-terminus of VgrS, and is separated from its C-terminus by the cytoplasmic membrane (44).
It was suggested that E. coli Prc prefers substrates that are not stably folded (72). This might be a key finding, because it is possible that a disordered tail is the critical factor for recognition by a CTP, rather than a non-polar tail. In support of this, the E. coli Prc substrates MltG, MepM, and FtsI have polar residues at or very close to their C termini. The C-terminal tails of P. aeruginosa CtpA substrates, which are critical for degradation, also contain polar residues (73).
CTP cleavage sites have also been investigated, and are usually located some distance from the terminus of the substrate. Prc cleaves PBP3 (FtsI) at a Val-Ile bond (8). Systematic analysis suggested that E. coli Prc prefers small, uncharged residues, or residues with aliphatic side chains, on the N-terminal side of the cleavage site (72). There was a similar preference on the C-terminal side, with the exception that no preference for small residues was found (72). CTP cleaved bonds have also been identified in other species. B. burgdorferi CtpA cleaves an Ala-Leu bond in P13 and an Ala-Glu bond in OspC (30). X. campestris Prc cleaves an Ala-Gln bond in VgrS (44). These findings support a preference for a small amino acid on the N-terminal side of the cleavage site. It is also interesting to consider cleavage site preference when substrates are completely degraded. Perhaps only the first cleavage site has sequence specificity, with subsequent cleavages driven by movement of the substrate through a protease tunnel (67).
KEEPING THINGS UNDER CONTROL: IS CTP ACTIVITY REGULATED?
Another aspect of CTPs that is not clear is whether or not their activities are regulated, and if so, how. An obvious mechanism is to change expression of the CTP-encoding gene. B. subtilis ctpB expression is controlled by mother cell and forespore specific sigma factors, and X. campestris prc expression is directly controlled by VgrR (40, 44). However, there is also evidence that proteolytic activity might be controlled by other mechanisms. The E. coli Prc substrate MepS is degraded more in the stationary growth phase than in exponential phase, whereas MltG appears to be degraded at the same rate regardless of growth phase (2, 62). This suggests a substrate-specific mechanism of regulation. Interestingly, MepS degradation requires the NlpI adaptor protein, whereas MltG degradation does not. NlpI binds to MepS directly and might sequester some of it away from Prc. A more interesting possibility is that NlpI regulates Prc activity when in complex with it, perhaps in response to a growth-rate dependent signal such as the rate of peptidoglycan synthesis. Recently, increased intracellular aromatic amino acid concentration was suggested to affect the level of MepS in a Prc-dependent manner (63). The authors noted that aromatic amino acids are high-cost amino acids, and speculated that their presence could reduce the burden of amino acid biosynthesis and increase the biosynthesis of peptidoglycan precursors (63). Increased peptidoglycan biosynthesis would also require increased levels of hydrolases, such as MepS. Regardless of the mechanism, the growth-rate dependent control of MepS degradation by the NlpI-Prc complex is intriguing. Something similar might also occur in P. aeruginosa, where the levels of CtpA and its LbcA adaptor protein appear constant, regardless of growth phase, but the levels of the substrates are not (3). Therefore, adaptor-dependent control of CTP activity might emerge as a conserved phenomenon.
CONCLUDING REMARKS
CTPs regulate developmental processes, stress responses, and virulence; they are implicated in protein quality control; and they degrade cell wall hydrolases. Even so, there is much we don’t know. Beyond how they recognize substrates, and how their activities are controlled, many other areas also need more investigation. We must identify more CTP substrates, and when possible, confirm them with direct in vitro assays. Substrate identification will help to explain the various ctp null mutant phenotypes (Table 1). Increasing the known substrates should also reveal common themes spanning bacterial genera. For example, do Prc-like CTPs participate broadly in regulated intramembrane proteolysis cascades? Is degradation of cell wall hydrolases a widely conserved phenomenon? CTP adaptor proteins have come to light in the last few years, with the discovery of E. coli NlpI and P. aeruginosa LbcA. It is fascinating that these adaptor proteins share no significant sequence similarity. However, the true significance of this requires more CTP-adaptor systems to be identified and characterized. Are these adaptor proteins limited to Gram negative bacteria, or do Gram positive CTPs have adaptors as well? Perhaps it is also time to reconsider the name of these proteases now that C-terminal processing is clearly not their only role. Some degrade their substrates completely, and X. campestris Prc processes the N-terminus of its substrate. Regardless, the links of these proteases to stress responses, gene regulation, the cell wall, and virulence, means that their continued study provides the potential for broad impacts on our understanding of fundamental processes in all bacterial cells.
ACKNOWLEDGMENTS
CTP research in our laboratory is supported by the National Institute of Allergy and Infectious Diseases (NIAID) of the National Institutes of Health, under Award Numbers R01AI136901 and R21AI51097 to A.J.D., and award number T32AI007180 also supported A.G.S. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
We thank Patrick Lane of ScEYEnce Studios for graphical enhancement of the figures.
Contributor Information
Andrew J. Darwin, Email: andrew.darwin@med.nyu.edu.
Conrad W. Mullineaux, Queen Mary University of London
REFERENCES
- 1.Rawlings ND, Barrett AJ, Bateman A. 2010. MEROPS: the peptidase database. Nucleic Acids Res 38:D227–33. 10.1093/nar/gkp971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Singh SK, Parveen S, SaiSree L, Reddy M. 2015. Regulated proteolysis of a cross-link-specific peptidoglycan hydrolase contributes to bacterial morphogenesis. Proc Natl Acad Sci USA 112:10956–10961. 10.1073/pnas.1507760112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Srivastava D, Seo J, Rimal B, Kim SJ, Zhen S, Darwin AJ. 2018. A proteolytic complex targets multiple cell wall hydrolases in Pseudomonas aeruginosa. mBio 9. 10.1128/mBio.00972-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nakamura M, Maruyama IN, Soma M, Kato J, Suzuki H, Horota Y. 1983. On the process of cellular division in Escherichia coli: nucleotide sequence of the gene for penicillin-binding protein 3. Mol Gen Genet 191:1–9. 10.1007/BF00330881. [DOI] [PubMed] [Google Scholar]
- 5.Hara H, Nishimura Y, Kato J, Suzuki H, Nagasawa H, Suzuki A, Hirota Y. 1989. Genetic analyses of processing involving C-terminal cleavage in penicillin-binding protein 3 of Escherichia coli. J Bacteriol 171:5882–5889. 10.1128/jb.171.11.5882-5889.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hara H, Yamamoto Y, Higashitani A, Suzuki H, Nishimura Y. 1991. Cloning, mapping, and characterization of the Escherichia coli prc gene, which is involved in C-terminal processing of penicillin-binding protein 3. J Bacteriol 173:4799–4813. 10.1128/jb.173.15.4799-4813.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chueh CK, Som N, Ke LC, Ho MR, Reddy M, Chang CI. 2019. Structural basis for the differential regulatory roles of the PDZ domain in C-terminal processing proteases. mBio 10. 10.1128/mBio.01129-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nagasawa H, Sakagami Y, Suzuki A, Suzuki H, Hara H, Hirota Y. 1989. Determination of the cleavage site involved in C-terminal processing of penicillin-binding protein 3 of Escherichia coli. J Bacteriol 171:5890–5893. 10.1128/jb.171.11.5890-5893.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Silber KR, Keiler KC, Sauer RT. 1992. Tsp: a tail-specific protease that selectively degrades proteins with nonpolar C termini. Proc Natl Acad Sci USA 89:295–299. 10.1073/pnas.89.1.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Parsell DA, Silber KR, Sauer RT. 1990. Carboxy-terminal determinants of intracellular protein degradation. Genes Dev 4:277–286. 10.1101/gad.4.2.277. [DOI] [PubMed] [Google Scholar]
- 11.Silber KR, Sauer RT. 1994. Deletion of the prc (tsp) gene provides evidence for additional tail-specific proteolytic activity in Escherichia coli K-12. Mol Gen Genet 242:237–240. 10.1007/BF00391018. [DOI] [PubMed] [Google Scholar]
- 12.Shestakov SV, Anbudurai PR, Stanbekova GE, Gadzhiev A, Lind LK, Pakrasi HB. 1994. Molecular cloning and characterization of the ctpA gene encoding a carboxyl-terminal processing protease. Analysis of a spontaneous photosystem II-deficient mutant strain of the cyanobacterium Synechocystis sp. PCC 6803. J Biol Chem 269:19354–19359. 10.1016/S0021-9258(17)32175-0. [DOI] [PubMed] [Google Scholar]
- 13.Marder JB, Goloubinoff P, Edelman M. 1984. Molecular architecture of the rapidly metabolized 32-kilodalton protein of photosystem II. Indications for COOH-terminal processing of a chloroplast membrane polypeptide. J Biol Chem 259:3900–3908. 10.1016/S0021-9258(17)43182-6. [DOI] [PubMed] [Google Scholar]
- 14.Anbudurai PR, Mor TS, Ohad I, Shestakov SV, Pakrasi HB. 1994. The ctpA gene encodes the C-terminal processing protease for the D1 protein of the photosystem II reaction center complex. Proc Natl Acad Sci USA 91:8082–8086. 10.1073/pnas.91.17.8082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Saoud J, Mani T, Faucher SP. 2021. The tail-specific protease is important for Legionella pneumophila to survive thermal stress in water and inside amoebae. Appl Environ Microbiol 87. 10.1128/AEM.02975-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dong J, Signo KSL, Vanderlinde EM, Yost CK, Dahms TES. 2011. Atomic force microscopy of a ctpA mutant in Rhizobium leguminosarum reveals surface defects linking CtpA function to biofilm formation. Microbiology (Reading) 157:3049–3058. 10.1099/mic.0.051045-0. [DOI] [PubMed] [Google Scholar]
- 17.Gilbert KB, Vanderlinde EM, Yost CK. 2007. Mutagenesis of the carboxy terminal protease CtpA decreases desiccation tolerance in Rhizobium leguminosarum. FEMS Microbiol Lett 272:65–74. 10.1111/j.1574-6968.2007.00735.x. [DOI] [PubMed] [Google Scholar]
- 18.Jun D, Idem U, Dahms TES. 2020. Altered envelope structure and nanomechanical properties of a C-terminal protease A-deficient Rhizobium leguminosarum. Microorganisms 8:1421. 10.3390/microorganisms8091421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jun D, Minic Z, Bhat SV, Vanderlinde EM, Yost CK, Babu M, Dahms TES. 2017. Metabolic adaptation of a C-terminal protease A-deficient Rhizobium leguminosarum in response to loss of nutrient transport. Front Microbiol 8:2617. 10.3389/fmicb.2017.02617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bandara AB, Sriranganathan N, Schurig GG, Boyle SM. 2005. Carboxyl-terminal protease regulates Brucella suis morphology in culture and persistence in macrophages and mice. J Bacteriol 187:5767–5775. 10.1128/JB.187.16.5767-5775.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bandara AB, DeShazer D, Inzana TJ, Sriranganathan N, Schurig GG, Boyle SM. 2008. A disruption of ctpA encoding carboxy-terminal protease attenuates Burkholderia mallei and induces partial protection in CD1 mice. Microb Pathog 45:207–216. 10.1016/j.micpath.2008.05.005. [DOI] [PubMed] [Google Scholar]
- 22.Held K, Gasper J, Morgan S, Siehnel R, Singh P, Manoil C. 2018. Determinants of extreme beta-lactam tolerance in the Burkholderia pseudomallei complex. Antimicrob Agents Chemother 62. 10.1128/AAC.00068-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Roy R, You RI, Chang CH, Yang CY, Lin NT. 2021. Carboxy-terminal processing protease controls production of outer membrane vesicles and biofilm in Acinetobacter baumannii. Microorganisms 9:1336. 10.3390/microorganisms9061336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Roy R, You RI, Lin MD, Lin NT. 2020. Mutation of the carboxy-terminal processing protease in Acinetobacter baumannii affects motility, leads to loss of membrane integrity, and reduces virulence. Pathogens 9:322. 10.3390/pathogens9050322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chang W, Li C, Cui Z, Li W, Song H, Chang H, Fu W, Wang C, Huang T, Luo Y, Shan Y, Wang Y, Wang F, Xu M, Fu A. 2021. Diverged early from CtpB and CtpC, CtpA has evolved to process D1 precursor in oxygenic photosynthetic organisms. Front Plant Sci 12:676036. 10.3389/fpls.2021.676036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Johnson RM, Nash ZM, Dedloff MR, Shook JC, Cotter PA. 2021. DegP initiates regulated processing of filamentous hemagglutinin in Bordetella bronchiseptica. mBio 12:e0146521. 10.1128/mBio.01465-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nash ZM, Cotter PA. 2019. Regulated, sequential processing by multiple proteases is required for proper maturation and release of Bordetella filamentous hemagglutinin. Mol Microbiol 112:820–836. 10.1111/mmi.14318. [DOI] [PubMed] [Google Scholar]
- 28.Coutte L, Antoine R, Drobecq H, Locht C, Jacob-Dubuisson F. 2001. Subtilisin-like autotransporter serves as maturation protease in a bacterial secretion pathway. EMBO J 20:5040–5048. 10.1093/emboj/20.18.5040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ostberg Y, Carroll JA, Pinne M, Krum JG, Rosa P, Bergstrom S. 2004. Pleiotropic effects of inactivating a carboxyl-terminal protease, CtpA, in Borrelia burgdorferi. J Bacteriol 186:2074–2084. 10.1128/JB.186.7.2074-2084.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kumru OS, Bunikis I, Sorokina I, Bergstrom S, Zuckert WR. 2011. Specificity and role of the Borrelia burgdorferi CtpA protease in outer membrane protein processing. J Bacteriol 193:5759–5765. 10.1128/JB.05622-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lad SP, Yang G, Scott DA, Wang G, Nair P, Mathison J, Reddy VS, Li E. 2007. Chlamydial CT441 is a PDZ domain-containing tail-specific protease that interferes with the NF-kappaB pathway of immune response. J Bacteriol 189:6619–6625. 10.1128/JB.00429-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lad SP, Li J, da Silva Correia J, Pan Q, Gadwal S, Ulevitch RJ, Li E. 2007. Cleavage of p65/RelA of the NF-kappaB pathway by Chlamydia. Proc Natl Acad Sci USA 104:2933–2938. 10.1073/pnas.0608393104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Christian J, Vier J, Paschen SA, Hacker G. 2010. Cleavage of the NF-kappaB family protein p65/RelA by the chlamydial protease-like activity factor (CPAF) impairs proinflammatory signaling in cells infected with Chlamydiae. J Biol Chem 285:41320–41327. 10.1074/jbc.M110.152280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen AL, Johnson KA, Lee JK, Sutterlin C, Tan M. 2012. CPAF: a Chlamydial protease in search of an authentic substrate. PLoS Pathog 8:e1002842. 10.1371/journal.ppat.1002842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Borth N, Massier J, Franke C, Sachse K, Saluz HP, Hanel F. 2010. Chlamydial protease CT441 interacts with SRAP1 co-activator of estrogen receptor alpha and partially alleviates its co-activation activity. J Steroid Biochem Mol Biol 119:89–95. 10.1016/j.jsbmb.2010.01.004. [DOI] [PubMed] [Google Scholar]
- 36.Kohlmann F, Shima K, Hilgenfeld R, Solbach W, Rupp J, Hansen G. 2015. Structural basis of the proteolytic and chaperone activity of Chlamydia trachomatis CT441. J Bacteriol 197:211–218. 10.1128/JB.02140-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tan IS, Ramamurthi KS. 2014. Spore formation in Bacillus subtilis. Environ Microbiol Rep 6:212–225. 10.1111/1758-2229.12130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cutting S, Roels S, Losick R. 1991. Sporulation operon spoIVF and the characterization of mutations that uncouple mother-cell from forespore gene expression in Bacillus subtilis. J Mol Biol 221:1237–1256. 10.1016/0022-2836(91)90931-u. [DOI] [PubMed] [Google Scholar]
- 39.Mastny M, Heuck A, Kurzbauer R, Heiduk A, Boisguerin P, Volkmer R, Ehrmann M, Rodrigues CD, Rudner DZ, Clausen T. 2013. CtpB assembles a gated protease tunnel regulating cell-cell signaling during spore formation in Bacillus subtilis. Cell 155:647–658. 10.1016/j.cell.2013.09.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Campo N, Rudner DZ. 2007. SpoIVB and CtpB are both forespore signals in the activation of the sporulation transcription factor sigmaK in Bacillus subtilis. J Bacteriol 189:6021–6027. 10.1128/JB.00399-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Marasco R, Varcamonti M, Ricca E, Sacco M. 1996. A new Bacillus subtilis gene with homology to Escherichia coli prc. Gene 183:149–152. 10.1016/s0378-1119(96)00543-4. [DOI] [PubMed] [Google Scholar]
- 42.Burby PE, Simmons ZW, Schroeder JW, Simmons LA. 2018. Discovery of a dual protease mechanism that promotes DNA damage checkpoint recovery. PLoS Genet 14:e1007512. 10.1371/journal.pgen.1007512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bojer MS, Wacnik K, Kjelgaard P, Gallay C, Bottomley AL, Cohn MT, Lindahl G, Frees D, Veening JW, Foster SJ, Ingmer H. 2019. SosA inhibits cell division in Staphylococcus aureus in response to DNA damage. Mol Microbiol 112:1116–1130. 10.1111/mmi.14350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Deng CY, Zhang H, Wu Y, Ding LL, Pan Y, Sun ST, Li YJ, Wang L, Qian W. 2018. Proteolysis of histidine kinase VgrS inhibits its autophosphorylation and promotes osmostress resistance in Xanthomonas campestris. Nat Commun 9:4791. 10.1038/s41467-018-07228-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liao CT, Liu YF, Chiang YC, Lo HH, Du SC, Hsu PC, Hsiao YM. 2016. Functional characterization and transcriptome analysis reveal multiple roles for prc in the pathogenicity of the black rot pathogen Xanthomonas campestris pv. campestris. Res Microbiol 167:299–312. 10.1016/j.resmic.2016.01.002. [DOI] [PubMed] [Google Scholar]
- 46.Teoh WP, Matson JS, DiRita VJ. 2015. Regulated intramembrane proteolysis of the virulence activator TcpP in Vibrio cholerae is initiated by the tail-specific protease (Tsp). Mol Microbiol 97:822–831. 10.1111/mmi.13069. [DOI] [PubMed] [Google Scholar]
- 47.Bastiaansen KC, Ibanez A, Ramos JL, Bitter W, Llamas MA. 2014. The Prc and RseP proteases control bacterial cell-surface signalling activity. Environ Microbiol 16:2433–2443. 10.1111/1462-2920.12371. [DOI] [PubMed] [Google Scholar]
- 48.Bastiaansen KC, Civantos C, Bitter W, Llamas MA. 2017. New insights into the regulation of cell-surface signaling activity acquired from a mutagenesis screen of the Pseudomonas putida IutY sigma/anti-sigma factor. Front Microbiol 8:747. 10.3389/fmicb.2017.00747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bastiaansen KC, Otero-Asman JR, Luirink J, Bitter W, Llamas MA. 2015. Processing of cell-surface signalling anti-sigma factors prior to signal recognition is a conserved autoproteolytic mechanism that produces two functional domains. Environ Microbiol 17:3263–3277. 10.1111/1462-2920.12776. [DOI] [PubMed] [Google Scholar]
- 50.Keiler KC, Waller PR, Sauer RT. 1996. Role of a peptide tagging system in degradation of proteins synthesized from damaged messenger RNA. Science 271:990–993. 10.1126/science.271.5251.990. [DOI] [PubMed] [Google Scholar]
- 51.Tu GF, Reid GE, Zhang JG, Moritz RL, Simpson RJ. 1995. C-terminal extension of truncated recombinant proteins in Escherichia coli with a 10Sa RNA decapeptide. J Biol Chem 270:9322–9326. 10.1074/jbc.270.16.9322. [DOI] [PubMed] [Google Scholar]
- 52.Hudson CM, Williams KP. 2015. The tmRNA website. Nucleic Acids Res 43:D138–40. 10.1093/nar/gku1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fritze J, Zhang M, Luo Q, Lu X. 2020. An overview of the bacterial SsrA system modulating intracellular protein levels and activities. Appl Microbiol Biotechnol 104:5229–5241. 10.1007/s00253-020-10623-x. [DOI] [PubMed] [Google Scholar]
- 54.Braud S, Lavire C, Bellier A, Mazodier P. 2006. Effect of SsrA (tmRNA) tagging system on translational regulation in Streptomyces. Arch Microbiol 184:343–352. 10.1007/s00203-005-0051-y. [DOI] [PubMed] [Google Scholar]
- 55.Sautter R, Ramos D, Schneper L, Ciofu O, Wassermann T, Koh CL, Heydorn A, Hentzer M, Hoiby N, Kharazmi A, Molin S, Devries CA, Ohman DE, Mathee K. 2012. A complex multilevel attack on Pseudomonas aeruginosa algT/U expression and algT/U activity results in the loss of alginate production. Gene 498:242–253. 10.1016/j.gene.2011.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Damron FH, Goldberg JB. 2012. Proteolytic regulation of alginate overproduction in Pseudomonas aeruginosa. Mol Microbiol 84:595–607. 10.1111/j.1365-2958.2012.08049.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Anthony M, Rose B, Pegler MB, Elkins M, Service H, Thamotharampillai K, Watson J, Robinson M, Bye P, Merlino J, Harbour C. 2002. Genetic analysis of Pseudomonas aeruginosa isolates from the sputa of Australian adult cystic fibrosis patients. J Clin Microbiol 40:2772–2778. 10.1128/JCM.40.8.2772-2778.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Boucher JC, Yu H, Mudd MH, Deretic V. 1997. Mucoid Pseudomonas aeruginosa in cystic fibrosis: characterization of muc mutations in clinical isolates and analysis of clearance in a mouse model of respiratory infection. Infect Immun 65:3838–3846. 10.1128/iai.65.9.3838-3846.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Reiling SA, Jansen JA, Henley BJ, Singh S, Chattin C, Chandler M, Rowen DW. 2005. Prc protease promotes mucoidy in mucA mutants of Pseudomonas aeruginosa. Microbiology (Reading) 151:2251–2261. 10.1099/mic.0.27772-0. [DOI] [PubMed] [Google Scholar]
- 60.Delgado C, Florez L, Lollett I, Lopez C, Kangeyan S, Kumari H, Stylianou M, Smiddy RJ, Schneper L, Sautter RT, Smith D, Szatmari G, Mathee K. 2018. Pseudomonas aeruginosa regulated intramembrane proteolysis: protease MucP can overcome mutations in the AlgO periplasmic protease to restore alginate production in nonmucoid revertants. J Bacteriol 200. 10.1128/JB.00215-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hara H, Abe N, Nakakouji M, Nishimura Y, Horiuchi K. 1996. Overproduction of penicillin-binding protein 7 suppresses thermosensitive growth defect at low osmolarity due to an spr mutation of Escherichia coli. Microb Drug Resist 2:63–72. 10.1089/mdr.1996.2.63. [DOI] [PubMed] [Google Scholar]
- 62.Hsu PC, Chen CS, Wang S, Hashimoto M, Huang WC, Teng CH. 2020. Identification of MltG as a Prc protease substrate whose dysregulation contributes to the conditional growth defect of Prc-deficient Escherichia coli. Front Microbiol 11:2000. 10.3389/fmicb.2020.02000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kim YJ, Choi BJ, Park SH, Lee HB, Son JE, Choi U, Chi WJ, Lee CR. 2021. Distinct amino acid availability-dependent regulatory mechanisms of MepS and MepM levels in Escherichia coli. Front Microbiol 12:677739. 10.3389/fmicb.2021.677739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yakhnina AA, Bernhardt TG. 2020. The Tol-Pal system is required for peptidoglycan-cleaving enzymes to complete bacterial cell division. Proc Natl Acad Sci USA 117:6777–6783. 10.1073/pnas.1919267117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ohara M, Wu HC, Sankaran K, Rick PD. 1999. Identification and characterization of a new lipoprotein, NlpI, in Escherichia coli K-12. J Bacteriol 181:4318–4325. 10.1128/JB.181.14.4318-4325.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tadokoro A, Hayashi H, Kishimoto T, Makino Y, Fujisaki S, Nishimura Y. 2004. Interaction of the Escherichia coli lipoprotein NlpI with periplasmic Prc (Tsp) protease. J Biochem 135:185–191. 10.1093/jb/mvh022. [DOI] [PubMed] [Google Scholar]
- 67.Su MY, Som N, Wu CY, Su SC, Kuo YT, Ke LC, Ho MR, Tzeng SR, Teng CH, Mengin-Lecreulx D, Reddy M, Chang CI. 2017. Structural basis of adaptor-mediated protein degradation by the tail-specific PDZ-protease Prc. Nat Commun 8:1516. 10.1038/s41467-017-01697-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Seo J, Darwin AJ. 2013. The Pseudomonas aeruginosa periplasmic protease CtpA can affect systems that impact its ability to mount both acute and chronic infections. Infect Immun 81:4561–4570. 10.1128/IAI.01035-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Chakraborty D, Darwin AJ. 2021. Direct and indirect interactions promote complexes of the lipoprotein LbcA, the CtpA protease and its substrates, and other cell wall proteins in Pseudomonas aeruginosa. J Bacteriol 203. 10.1128/JB.00393-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hsu H-C, Wang M, Kovach A, Darwin AJ, Li H. 2022. Pseudomonas aeruginosa C-terminal processing protease CtpA assembles into a hexameric structure that requires activation by a spiral-shaped lipoprotein binding partner. mBio 10.1128/mbio.03680-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Keiler KC, Sauer RT. 1996. Sequence determinants of C-terminal substrate recognition by the Tsp protease. J Biol Chem 271:2589–2593. 10.1074/jbc.271.5.2589. [DOI] [PubMed] [Google Scholar]
- 72.Keiler KC, Silber KR, Downard KM, Papayannopoulos IA, Biemann K, Sauer RT. 1995. C-terminal specific protein degradation: activity and substrate specificity of the Tsp protease. Protein Sci 4:1507–1515. 10.1002/pro.5560040808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Chung S, Darwin AJ. 2020. The C-terminus of substrates is critical but not sufficient for their degradation by the Pseudomonas aeruginosa CtpA protease. J Bacteriol 202. 10.1128/JB.00174-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Campo N, Rudner DZ. 2006. A branched pathway governing the activation of a developmental transcription factor by regulated intramembrane proteolysis. Mol Cell 23:25–35. 10.1016/j.molcel.2006.05.019. [DOI] [PubMed] [Google Scholar]
- 75.Pan Q, Losick R, Rudner DZ. 2003. A second PDZ-containing serine protease contributes to activation of the sporulation transcription factor sigmaK in Bacillus subtilis. J Bacteriol 185:6051–6056. 10.1128/JB.185.20.6051-6056.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhou R, Kroos L. 2005. Serine proteases from two cell types target different components of a complex that governs regulated intramembrane proteolysis of pro-sigmaK during Bacillus subtilis development. Mol Microbiol 58:835–846. 10.1111/j.1365-2958.2005.04870.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Mitchell SJ, Minnick MF. 1997. A carboxy-terminal processing protease gene is located immediately upstream of the invasion-associated locus from Bartonella bacilliformis. Microbiology (Reading) 143:1221–1233. 10.1099/00221287-143-4-1221. [DOI] [PubMed] [Google Scholar]
- 78.Keiler KC, Sauer RT. 1995. Identification of active site residues of the Tsp protease. J Biol Chem 270:28864–28868. 10.1074/jbc.270.48.28864. [DOI] [PubMed] [Google Scholar]
- 79.Lawrence A, K Nicholls S, H Stansfield S, M Huston W. 2014. Characterization of the tail-specific protease (Tsp) from Legionella. J Gen Appl Microbiol 60:95–100. 10.2323/jgam.60.95. [DOI] [PubMed] [Google Scholar]
- 80.Li Y, Pan Y, She Q, Chen L. 2013. A novel carboxyl-terminal protease derived from Paenibacillus lautus CHN26 exhibiting high activities at multiple sites of substrates. BMC Biotechnol 13:89. 10.1186/1472-6750-13-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wood LF, Leech AJ, Ohman DE. 2006. Cell wall-inhibitory antibiotics activate the alginate biosynthesis operon in Pseudomonas aeruginosa: Roles of sigma (AlgT) and the AlgW and Prc proteases. Mol Microbiol 62:412–426. 10.1111/j.1365-2958.2006.05390.x. [DOI] [PubMed] [Google Scholar]
- 82.Carroll RK, Rivera FE, Cavaco CK, Johnson GM, Martin D, Shaw LN. 2014. The lone S41 family C-terminal processing protease in Staphylococcus aureus is localized to the cell wall and contributes to virulence. Microbiology (Reading) 160:1737–1748. 10.1099/mic.0.079798-0. [DOI] [PMC free article] [PubMed] [Google Scholar]