ABSTRACT
The epidemiology of macrolide resistance in Mycoplasma (Mycoplasmoides) pneumoniae in the United States is incompletely described. Using a PCR assay targeting common mutations associated with macrolide resistance in M. pneumoniae (23S rRNA gene, A2063G/A2064G), the frequency of macrolide resistance was estimated to be 10% based on analysis of 114 samples tested from January 2014 to September 2021 at Mayo Clinic Laboratories. Seasonality data showed the highest rates of M. pneumoniae infection in the fall/early winter.
KEYWORDS: Mycoplasma pneumoniae, antibiotic resistance, macrolides, pneumonia, surveillance studies
TEXT
Mycoplasma (Mycoplasmoides) pneumoniae causes upper respiratory tract infection, alongside an estimated 20% to 40% of community-acquired pneumonia cases in the general population (1, 2). While an estimated 2 million cases of M. pneumoniae infection occur annually in the United States (3), the true extent of this infection is incompletely defined due to mild symptoms in a majority of infections, similarity of M. pneumoniae-associated symptoms to those of other respiratory pathogens (including viruses), and lack of widespread testing, reporting, and surveillance (4). Although historically serologic testing has been the main approach to M. pneumoniae diagnosis, nucleic acid amplification tests are now the preferred tests (5–7).
Macrolides are a common treatment for respiratory tract infections and a recommended treatment for M. pneumoniae. However, macrolide resistance in M. pneumoniae has been increasing for over 2 decades and is reported to be as high as 90% in some areas in Japan and China and 30% in areas in Europe (5). Worldwide studies of the prevalence of macrolide-resistant M. pneumoniae were amalgamated by Waites et al. in a study published in 2017 (5); however, there was limited data on macrolide-resistant M. pneumoniae prevalence in the United States. In 2019, Waites et al. reported macrolide resistance surveillance data between 2015 and 2018 from 8 states; the overall prevalence of macrolide-resistant M. pneumoniae was 8% (27/360 specimens) and was highest in southern and eastern portions of the United States (15 to 21%) (4). Additionally, Lanata et al. reported the prevalence of macrolide resistance in M. pneumoniae as 3% (14/498) based on M. pneumoniae-positive samples collected in central Ohio from 2015 to 2019 (8). The current study aimed to supplement United States prevalence data by assessing M. pneumoniae macrolide resistance in a convenience sample of specimens that tested positive for M. pneumoniae at Mayo Clinic Laboratories in Rochester, Minnesota. In addition, the performance of a PCR assay designed to detect macrolide resistance in M. pneumoniae by targeting the 23S rRNA gene was compared to a previously described M. pneumoniae assay targeting ptsI (9). Finally, seasonality of M. pneumoniae PCR test positivity was assessed.
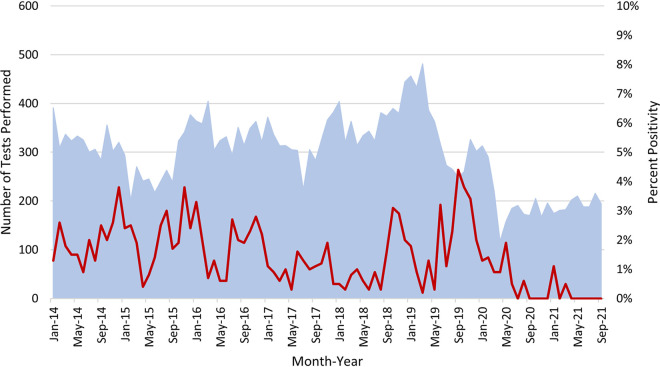
From January 2014 to September 2021, 27,645 patient samples were tested for M. pneumoniae at Mayo Clinic Laboratories using a PCR assay targeting ptsI (9). Overall assay positivity was 2% (410/27,645) (Fig. 1). The highest rates of M. pneumoniae infection generally occurred in the fall/early winter, with peak positivity rates of 3 to 4% between September and January (apart from 2017 to 2018). After April 2020, overall testing and percent positivity rates were lower than those in antecedent times due to the COVID-19 pandemic. Of the 410 positive specimens, 142 (respiratory swabs [n = 92], lower respiratory samples [n = 47], pleural fluids [n = 2], and cerebrospinal fluid [n = 1]) were archived and available for further study.
FIG 1.
M. pneumoniae ptsI PCR tests performed from January 2014 through November 2021 showing total numbers of monthly tests performed (blue background) and monthly percent positivity (red line). There was typically a rise in positivity in late fall/winter, except for COVID-19 pandemic times and, to some extent, the 2017–2018 season. Between February 2017 and August 2018, the overall positivity rate was 1% (52/6,238). Peak annual positivity rates were observed in December 2014 (4%), November 2015 (4%), January 2016 (3%), June 2017 (2%), October 2018 (3%), and September 2019 (4%). Macrolide resistant isolates were found in 2014 (n = 1), 2015 (n = 3), 2016 (n = 3), and 2019 (n = 4), but notably, not all specimens testing positive for M. pneumoniae were assessed for macrolide resistance.
Macrolide resistance in M. pneumoniae corresponds to mutations in the 23S rRNA gene, of which A2063G and A2064G are the most common and confer high-level macrolide resistance (5). A real-time PCR assay using fluorescent resonance energy transfer (FRET) hybridization probes targeting the 23S rRNA gene was designed to detect M. pneumoniae and predict macrolide resistance (by assessing A2063G and A2064G) (Table 1) (10). Specimens were extracted on a MagNA Pure 96 (Roche Diagnostics, Indianapolis, IN) and amplified on a LightCycler 480 (Roche); melting temperature analysis was used to predict macrolide resistance. The wild-type genotype displays a melting temperature of 66.5 ± 2°C, whereas in the presence of A2063G or A2064G, the melting temperature is 60.5 ± 2°C. All PCR-positive samples were subjected to bidirectional Sanger sequencing through reamplification of extracted product for macrolide resistance genotype confirmation. A recovery template was used to monitor for potential PCR inhibition.
TABLE 1.
Primer and probe sequences used for the real-time PCR assay targeting the M. pneumoniae 23S rRNA genea
| Sequence type | Sequence |
|---|---|
| Primers | |
| Forward (F1-Mpn) | 5′-GAAGGAGGTTAGCGCAA-3′ |
| Reverse (R2Mpn) | 5′-TTCTCTACATGATAATGTCC-3′ |
| Probes | |
| FL3 | 5′-CGGGACGGAAAGACCCCGTG-FL-3′ |
| LC3b | 5′-LC610-AGCTTTACTGTAGC+T+TAA+TA+T+TGA-PO4-3′ |
| Recovery template probes | |
| FL | 5′-GGTGCCGTTCACTTCCCGAATAAC-FL-3′ |
| LC | 5′-LC670-CGGATATTTTTGATCTGACCGAAGCG-PO4-3′ |
A 262-bp region of the 23S rRNA gene of M. pneumoniae (121917 to 122178 of GenBank accession number U00089) was targeted using primers and fluorescence resonance energy transfer hybridization probes (set no. 4903; 10× concentration; TIB MolBio, Aldelphia, NJ). Target donor and acceptor probes were synthesized by TIB MolBiol (LCMPRP no. 4903) and labeled with fluorescein and LightCycler Red 610, respectively. A recovery template was added to the master mix to monitor for PCR inhibition. The recovery template was amplified with the same primers used to amplify M. pneumoniae with the amplification region internal to the primers replaced with sequence complementary to the recovery template probes. Recovery template donor and acceptor probes were synthesized by TIB MolBiol and labeled with fluorescein and LightCycler Red 670, respectively.
+, locked nucleic acid.
A previously described assay targeting ptsI (9) was compared to the assay targeting the 23S rRNA gene (Table 2). Notably, 13 previously positive archived samples tested negative by both PCR assays, suggesting sample degradation during prolonged storage. A total of 129 samples tested positive using the ptsI assay, of which only 114 were positive using the 23S rRNA gene assay; all 15 discrepant samples had crossing threshold (Ct) values of >35 cycles via ptsI gene detection. No PCR inhibition was detected.
TABLE 2.
Comparison of results of PCR assays targeting the ptsI and 23S rRNA gene targets
| M. pneumoniae assay parameter |
ptsI gene target |
McNemar’s test P value | |
|---|---|---|---|
| Positive | Negative | ||
| 23S rRNA gene target (no.) | |||
| Positive | 114 | 0 | 0.00006 |
| Negative | 15a | 13 | |
| Total | 129 | 13 | |
| Positive agreement (%) | 88 | ||
| Negative agreement (%) | 100 | ||
Includes 12 respiratory swabs (nasopharyngeal, throat, and nasal), 1 bronchoalveolar lavage fluid sample, 1 pleural/pericardial fluid sample, and 1 sputum sample.
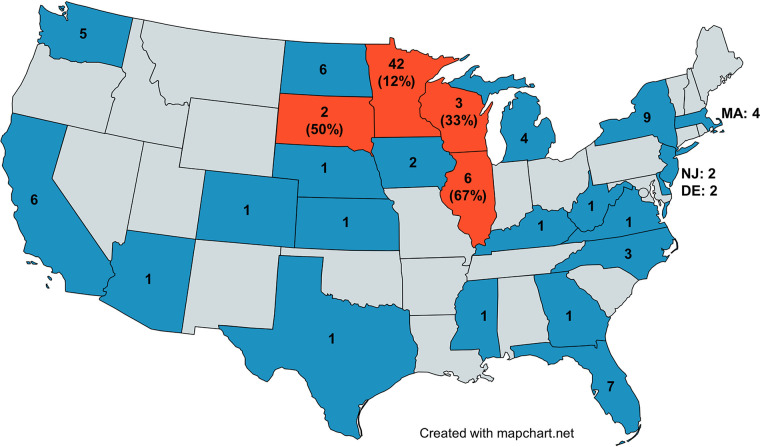
Macrolide resistance prediction for all samples detected by the 23S rRNA gene PCR assay was assessed through melting temperature analysis and confirmed using bidirectional Sanger sequencing. Of the 114 samples tested, 11 (10%) were predicted and confirmed to have A2063G (n = 9) or A2064G (n = 2) mutations. Geographical location of positive samples by state is illustrated in Fig. 2, with demographic data presented in Table 3. All predicted macrolide-resistant specimens were from the Midwest (Minnesota [n = 5], Illinois [n = 4], South Dakota [n = 1], Wisconsin [n = 1]), as were most M. pneumoniae-positive specimens.
FIG 2.
Geographic locations of patients testing positive for M. pneumoniae. Shown is the total number of positive tests by state. States shown in gray had no positive results; states shown in dark blue or orange had positive results. Macrolide-resistant M. pneumoniae was detected in states shown in orange, with the percentage of macrolide-resistant M. pneumoniae listed below the specimen numbers.
TABLE 3.
Patient demographics by sex and age, alongside specimen type, according to macrolide resistance for those testing positive by both the ptsI and 23S rRNA gene targets
| Demographic and population | No. (%) of patients infected with: |
Fisher exact test P value | |
|---|---|---|---|
| Macrolide-resistant M. pneumoniae (n = 11) | Macrolide-susceptible M. pneumoniaea (n = 103) | ||
| Sex | |||
| Female | 5 (45) | 43 (42) | 1.0 |
| Male | 6 (55) | 59 (58) | |
| Age (in yr) | |||
| ≤18 (n = 73) | 6 (55) | 67 (65) | 0.5 |
| ≥19 (n = 41) | 5 (45) | 36 (35) | |
| Mean ± SD (range) age, yr | 25 ± 18 (8–63) | 22 ± 18 (2–78) | 0.3b |
| Specimen type | |||
| Respiratory swabs (nasopharyngeal, nasal, throat) | 6 (8) | 74 (92) | 0.1 |
| Bronchoalveolar lavage fluids/bronchial washings | 2 (13) | 15 (87) | |
| Sputum/tracheal secretions | 2 (8) | 24 (92) | |
| Cerebrospinal fluid | 1 (100) | 0 (0) | |
Sex of one patient was unknown.
Wilcoxon rank sum test.
Despite study limitations of sample selection for convenience (and not being uniformly spread across the United States or time), lack of culture and phenotypic susceptibility testing, and small sample size (many previously positive specimens were unavailable for testing), there are three findings of this study. First, the overall test positivity for M. pneumoniae was highest in the fall/early winter and positivity rates fell during the COVID-19 pandemic, consistent with findings from prior studies (3, 5, 11). Second, although it is possible to accurately detect macrolide resistance in M. pneumoniae using a real-time PCR approach, the described 23S rRNA gene assay is less likely to qualitatively detect M. pneumoniae in clinical specimens than the previously described assay targeting pts1 (Table 2). This suggests that the former should not be used as a standalone assay for M. pneumoniae detection but instead performed on samples testing positive using an assay that more often tends to detect M. pneumoniae. Third, the prevalence of macrolide-resistant M. pneumoniae was 10% overall (and 12% in Minnesota, the state with the largest number of samples tested). If macrolides are used to treat M. pneumoniae, testing for macrolide susceptibility should be considered; in addition, continued surveillance for macrolide-resistant M. pneumoniae should be performed the United States.
ACKNOWLEDGMENTS
We thank the Bacteriology Laboratory at Mayo Clinic, Rochester, MN, for testing and collecting M. pneumoniae-positive patient samples and Matthew Wolf and Seanne Buckwalter for assistance with 23S rRNA gene assay design.
REFERENCES
- 1.Waites KT-R, et al. 2011. Mycoplasma and Ureaplasma, p 970–985. In Versalovic J, Carroll KC, Jorgensen JG, Funke G, Landry ML, Warnock DW (ed), Manual of clinical microbiology, 10th ed. ASM Press, Washington, DC. [Google Scholar]
- 2.Jacobs E, Ehrhardt I, Dumke R. 2015. New insights in the outbreak pattern of Mycoplasma pneumoniae. Int J Med Microbiol 305:705–708. 10.1016/j.ijmm.2015.08.021. [DOI] [PubMed] [Google Scholar]
- 3.Winchell MJ. 2013. Mycoplasma pneumoniae–a national public health perspective. Curr Pediatr Rev 9:324–333. 10.2174/15733963113099990009. [DOI] [Google Scholar]
- 4.Waites KB, Ratliff A, Crabb DM, Xiao L, Qin X, Selvarangan R, Tang Y-W, Zheng X, Bard JD, Hong T, Prichard M, Brooks E, Dallas S, Duffy L, Mixon E, Fowler KB, Atkinson TP, Diekema DJ. 2019. Macrolide-resistant Mycoplasma pneumoniae in the United States as determined from a national surveillance program. J Clin Microbiol 57:e00968-19. 10.1128/JCM.00968-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Waites KB, Xiao L, Liu Y, Balish MF, Atkinson TP. 2017. Mycoplasma pneumoniae from the respiratory tract and beyond. Clin Microbiol Rev 30:747–809. 10.1128/CMR.00114-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Loens K, Ieven M. 2016. Mycoplasma pneumoniae: current knowledge on nucleic acid amplification techniques and serological diagnostics. Front Microbiol 7:448. 10.3389/fmicb.2016.00448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Diaz MH, Winchell JM. 2016. The evolution of advanced molecular diagnostics for the detection and characterization of Mycoplasma pneumoniae. Front Microbiol 7:232–232. 10.3389/fmicb.2016.00232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lanata MM, Wang H, Everhart K, Moore-Clingenpeel M, Ramilo O, Leber A. 2021. Macrolide-resistant Mycoplasma pneumoniae infections in children, Ohio, USA. Emerg Infect Dis 27:1588–1597. 10.3201/eid2706.203206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schmitt BH, Sloan LM, Patel R. 2013. Real-time PCR detection of Mycoplasma pneumoniae in respiratory specimens. Diagn Microbiol Infect Dis 77:202–205. 10.1016/j.diagmicrobio.2013.07.016. [DOI] [PubMed] [Google Scholar]
- 10.Peuchant O, Menard A, Renaudin H, Morozumi M, Ubukata K, Bebear CM, Pereyre S. 2009. Increased macrolide resistance of Mycoplasma pneumoniae in France directly detected in clinical specimens by real-time PCR and melting curve analysis. J Antimicrob Chemother 64:52–58. 10.1093/jac/dkp160. [DOI] [PubMed] [Google Scholar]
- 11.Onozuka D, Hashizume M, Hagihara A. 2009. Impact of weather factors on Mycoplasma pneumoniae pneumonia. Thorax 64:507–511. 10.1136/thx.2008.111237. [DOI] [PubMed] [Google Scholar]