ABSTRACT
Flagellum-mediated bacterial motility is important for bacteria to take up nutrients, adapt to environmental changes, and establish infection. The twin-arginine translocation system (Tat) is an important protein export system, playing a critical role in bacterial physiology and pathogenesis. It has been observed for a long time that the Tat system is critical for bacterial motility. However, the underlying mechanism remains unrevealed. In this study, a comparative transcriptomics analysis was performed with extraintestinal pathogenic Escherichia coli (ExPEC), which identified a considerable number of genes differentially expressed when the Tat system was disrupted. Among them, a large proportion of flagellar biosynthesis genes showed downregulation, indicating that transcription regulation plays an important role in mediating the motility defects. We further identified three Tat substrate proteins, MdoD, AmiA, and AmiC, that were responsible for the nonmotile phenotype. The Rcs system was deleted in the Δtat, the ΔmdoD, and the ΔamiAΔamiC strains, which restored the motility of ΔmdoD and partially restored the motility of Δtat and ΔamiAΔamiC. The flagella were also observed in all of the ΔtatΔrcsDB, ΔmdoDΔrcsDB, and ΔamiAΔamiCΔrcsDB strains, but not in the Δtat, ΔmdoD, and ΔamiAΔamiC strains, by using transmission electron microscopy. Quantitative reverse transcription-PCR data revealed that the regulons of the Rcs system displayed differential expression in the tat mutant, indicating that the Rcs signaling was activated. Our results suggest that the Rcs system plays an important role in mediating the motility defects of the tat mutant of ExPEC.
IMPORTANCE The Tat system is an important protein export system critical for bacterial physiology and pathogenesis. It has been observed for a long time that the Tat system is critical for bacterial motility. However, the underlying mechanism remains unrevealed. In this study, we combine transcriptomics analysis and bacterial genetics, which reveal that transcription regulation plays an important role in mediating the motility defects of the tat mutant of extraintestinal pathogenic Escherichia coli. The Tat substrate proteins responsible for the motility defects are identified. We further show that the Rcs system contributes to the motility suppression. We for the first time reveal the link between the Tat system and bacterial motility, which is important for understanding the physiological functions of the Tat system.
KEYWORDS: twin-arginine translocation system, motility, flagella, Rcs system, transcription regulation, extraintestinal pathogenic Escherichia coli
INTRODUCTION
Bacteria utilize various protein transport and secretion systems to deliver proteins that are originally synthesized in the cytoplasm to the subcellular locations beyond the cytoplasm to exert their destined physiological functions (1–3). In many bacteria, a unique protein export system, the twin-arginine translocation (Tat) system, exists to transport a variety of proteins across the cytoplasmic membrane (4). One of the distinguishing features of the Tat system is that it transports fully folded proteins (5). It was originally discovered as a transport system that was closely related to the export of cofactor-containing proteins (6). However, its pleiotropic roles in bacterial physiology have been revealed subsequently (4, 7). A large proportion of them are related to anaerobic respiration, including dehydrogenases, formate dehydrogenases, dimethyl sulfoxide (DMSO) reductases, trimethylamine N-oxide (TMAO) reductase, nitrate reductase, etc. (8). Moreover, the Tat system has been demonstrated to play important roles in the pathogenesis of some important bacterial pathogens, such as pathogenic Escherichia coli, Salmonella, Pseudomonas aeruginosa, and Yersinia pseudotuberculosis (9). In some bacteria, the Tat system is even reported to be essential for viability (10, 11).
Bacterial flagella compose a complex machine that promotes cellular motility, which is important in nutrient acquisition, environmental adaption, and pathogenesis (12–14). The biogenesis of bacterial flagella is under sophisticated transcriptional control. In E. coli, the flhDC operon, which is also known as class I genes, encodes the master regulator that initiates the transcription hierarchy. FlhDC activates the expression of class II genes that encode important regulatory proteins as well as structural components of flagella. Class III genes encode proteins needed in late flagellar assembly, including flagellin FliC. Precise temporal control of flagellar gene expression ensures the production of flagella when needed (15, 16). Apart from this regulatory hierarchy, the expression of bacterial flagellar genes has been reported to be regulated in response to external signals. The Rcs system is an important signal transduction system involved in bacterial envelope stress response and virulence regulation (13). It is composed of three core components, RcsC, RcsD, and RcsB, in which RcsC is a transmembrane histidine kinase that is autophosphorylated when the system is activated and passes the phosphate to RcsD then to RcsB (17). RcsB serves as a response regulator to regulate the expression of downstream genes (17). The Rcs signaling system has been revealed to exert an important regulatory function in controlling bacterial motility as a negative regulator of flagellar biosynthesis (18, 19). In addition, the Rcs system has been reported to be involved in the expression of a variety of genes involved in capsule formation, biofilm formation, and type VI secretion in response to stresses such as envelope stresses, outer membrane damage, and peptidoglycan perturbation (13, 17, 20). Another regulation system involved in the regulation of flagellar expression is the CpxAR two-component system, which is composed of the sensor histidine kinase CpxA and the response regulator CpxR. It can be activated in response to bacterial adhesion to abiotic surfaces, leading to downregulation of fliC expression in enterohemorrhagic E. coli (14). Moreover, appropriate expression of flagellin at different stages of infection has been recently revealed as a new strategy of bacterial pathogenesis (21).
Extraintestinal pathogenic E. coli (ExPEC) is an important bacterial pathogen causing a variety of diseases, including urinary tract infection, septicemia, meningitis, and even death in both humans and farm animals (22–24). Our previous study showed that the Tat system is critical for the virulence of extraintestinal pathogenic E. coli (ExPEC) and revealed that the disruption of the Tat system caused a significant decrease in bacterial motility as reported in other studies (25–27). However, so far, the underlying mechanism of how the Tat system is involved in the regulation of bacterial motility has not been revealed. In this study, we found that in the ExPEC PCN033 strain, deletion of the Tat system caused drastic downregulated expression of flagellar biosynthesis genes. The Tat substrate proteins were further identified that were responsible for the motility defects. We further showed that the Rcs system was a key player that mediates the regulation of motility by the Tat system. Our study for the first time unraveled the link between the Tat system and bacterial motility.
RESULTS
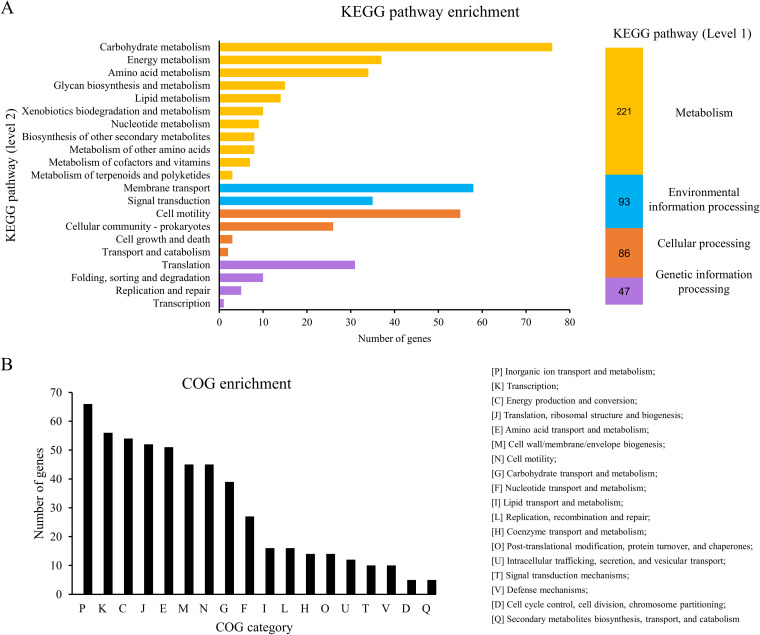
Deletion of the Tat system causes significant alteration in gene expression.
The Tat system has been reported to be involved in a variety of bacterial physiological processes (8). To investigate the influence of the Tat system on gene expression of ExPEC, the transcriptome profiles were compared between ExPEC PCN033 strain (wild type [WT]) and the Tat system-deleted strain (Δtat) by using mRNA sequencing (mRNA-seq). A total of 720 genes were differentially expressed in the Δtat compared with the WT strain [|log2(fold change)| > 1, P < 0.05], among which 508 genes were upregulated and 212 genes were downregulated, indicating an extensive influence on gene expression due to deletion of the Tat system (see Table S1 in the supplemental material). The KEGG database was then used to perform pathway enrichment analysis. As shown in Fig. 1A, metabolism represented the most enriched level 1 KEGG pathways, in which carbohydrate metabolism, membrane transport, and cell motility were the most enriched level 2 KEGG pathways among the differentially expressed genes. Clusters of orthologous groups (COG) enrichment analysis revealed that the differentially expressed genes belonged to a variety of functional categories (Fig. 1B). Specifically, in the Δtat strain, we found that the genes involved in the biosynthesis of polysialic acid, including the kpsMT and kpsFEDUCS operons, and the genes involved in the biosynthesis of colonic acid, including manB and manC, were significantly upregulated (Table 1), indicating that the capsule biosynthesis-related pathway was activated when the Tat system was disrupted. Several stress response genes, including ydeO (28), yciH (29), ydeP (30), elaB (31), yhcN (32), yggT (33), and bhsA (34), were also upregulated in the Δtat strain (Table 1). Moreover, we noticed that 12 genes belonging to the toxin-antitoxin systems were differentially expressed, among which 6 genes were upregulated and 6 were downregulated (Table 1). These data suggest that deletion of the Tat system caused substantial changes in gene expression.
FIG 1.
Differentially expressed genes (Δtat versus WT). (A) KEGG pathway enrichment of the differentially expressed genes. The differentially expressed genes were analyzed by using the BlastKOALA tool (https://www.kegg.jp/blastkoala/) to assign K numbers which were then used for pathway enrichment by using KEGG Mapper (https://www.genome.jp/kegg/mapper/search.html). The numbers inside the vertical bars are the total numbers of genes belonging to each level 2 KEGG pathway. (B) COG enrichment of the differentially expressed genes. COG enrichment was performed with the EggNOG v5.0 database (http://eggnog5.embl.de/#/app/home).
TABLE 1.
Partial differentially expressed genes in Δtat
| No. | Gene ID | Fold change (Δtat/WT) | P value | FDR | Gene name | Description | Function |
|---|---|---|---|---|---|---|---|
| 1 | PPECC33_RS16300 | 115.360 | 3.15E-33 | 3.17E-31 | kpsT | ABC transporter ATP-binding protein | Capsular polysaccharide biosynthesis |
| 2 | PPECC33_RS16295 | 40.224 | 4.71E-70 | 3.48E-67 | HAD-IA family hydrolase | ||
| 3 | PPECC33_RS16305 | 36.504 | 5.56E-40 | 8.21E-38 | kpsM | ABC transporter permease | |
| 4 | PPECC33_RS16290 | 31.779 | 2.63E-49 | 7.46E-47 | Hypothetical protein | ||
| 5 | PPECC33_RS16280 | 17.388 | 1.96E-20 | 1.06E-18 | Hypothetical protein | ||
| 6 | PPECC33_RS16275 | 16.912 | 5.47E-35 | 6.21E-33 | Nucleotidyltransferase family protein | ||
| 7 | PPECC33_RS16245 | 16.912 | 2.53E-42 | 4.66E-40 | kpsF | KpsF/GutQ family sugar-phosphate isomerase | |
| 8 | PPECC33_RS16285 | 11.472 | 3.79E-29 | 3.11E-27 | Glycosyltransferase family 2 protein | ||
| 9 | PPECC33_RS16250 | 11.314 | 1.69E-30 | 1.47E-28 | kpsE | Capsule polysaccharide transporter | |
| 10 | PPECC33_RS16255 | 10.056 | 3.08E-30 | 2.62E-28 | kpsD | Polysaccharide biosynthesis/export family protein | |
| 11 | PPECC33_RS16270 | 8.754 | 9.30E-19 | 4.48E-17 | kpsS | Capsular biosynthesis protein | |
| 12 | PPECC33_RS16265 | 7.945 | 4.89E-31 | 4.42E-29 | kpsC | Capsular polysaccharide biosynthesis protein | |
| 13 | PPECC33_RS16260 | 6.364 | 4.14E-20 | 2.21E-18 | kpsU | 3-Deoxy-manno-octulosonate cytidylyltransferase | |
| 14 | PPECC33_RS10985 | 2.346 | 0.000169 | 0.000959 | wzy | O11 family O-antigen polymerase | |
| 15 | PPECC33_RS10950 | 2.293 | 6.53E-07 | 6.77E-06 | manC | Mannose-1-phosphate guanylyltransferase/mannose-6-phosphate isomerase | |
| 16 | PPECC33_RS10940 | 2.219 | 9.50E-08 | 1.19E-06 | cpsG | Colanic acid biosynthesis phosphomannomutase CpsG | |
| 17 | PPECC33_RS10945 | 2.071 | 1.23E-05 | 9.69E-05 | wcaE | Glycosyltransferase | |
| 18 | PPECC33_RS08185 | 17.509 | 9.76E-06 | 7.82E-05 | ydeO | Acid stress response transcriptional regulator YdeO | Stress response |
| 19 | PPECC33_RS07220 | 5.315 | 0.000105 | 0.00064 | yciH | Stress response translation initiation inhibitor YciH | |
| 20 | PPECC33_RS08195 | 3.837 | 2.65E-07 | 3.01E-06 | ydeP | Acid resistance putative oxidoreductase YdeP | |
| 21 | PPECC33_RS12175 | 2.621 | 1.99E-09 | 3.33E-08 | elaB | Stress response protein ElaB | |
| 22 | PPECC33_RS17710 | 2.549 | 3.38E-12 | 8.26E-11 | yhcN | Peroxide/acid stress response protein YhcN | |
| 23 | PPECC33_RS16005 | 2.445 | 1.14E-06 | 1.12E-05 | yggT | Osmotic shock tolerance protein YggT | |
| 24 | PPECC33_RS05995 | 2.219 | 1.08E-05 | 8.59E-05 | bhsA | Multiple stress resistance protein BhsA | |
| 25 | PPECC33_RS22830 | 105.420 | 1.41E-09 | 2.47E-08 | ghoT | Type V toxin-antitoxin system toxin GhoT | Toxin-antitoxin system |
| 26 | PPECC33_RS22825 | 22.785 | 3.52E-06 | 3.12E-05 | ghoS | Type V toxin-antitoxin system endoribonuclease antitoxin GhoS | |
| 27 | PPECC33_RS01555 | 5.657 | 0.00019 | 0.001056 | yafO | Type II toxin-antitoxin system mRNA interferase toxin YafO | |
| 28 | PPECC33_RS03050 | 3.655 | 0.009247 | 0.028521 | hokE | Type I toxin-antitoxin system toxin HokE | |
| 29 | PPECC33_RS01550 | 2.395 | 0.004016 | 0.014354 | yafN | Type I toxin-antitoxin system antitoxin YafN | |
| 30 | PPECC33_RS24555 | 2.238 | 0.002638 | 0.010115 | lsoB | Type II toxin-antitoxin system antitoxin LsoB | |
| 31 | PPECC33_RS07745 | 0.363 | 1.37E-08 | 2.03E-07 | hokB | Type I toxin-antitoxin system toxin HokB | |
| 32 | PPECC33_RS24945 | 0.409 | 8.06E-07 | 8.20E-06 | Type I toxin-antitoxin system Hok family toxin | ||
| 33 | PPECC33_RS06820 | 0.432 | 6.95E-06 | 5.78E-05 | hokD | Type I toxin-antitoxin system toxin HokD | |
| 34 | PPECC33_RS00080 | 0.473 | 5.55E-07 | 5.87E-06 | mokC | Type I toxin-antitoxin system toxin MokC | |
| 35 | PPECC33_RS00280 | 0.480 | 0.001035 | 0.004596 | Type II toxin-antitoxin system CcdA family antitoxin | ||
| 36 | PPECC33_RS27840 | 0.490 | 0.01337 | 0.038627 | Type I toxin-antitoxin system toxin Ldr family protein |
Deletion of the Tat system disturbs the expression of flagellar biosynthesis genes.
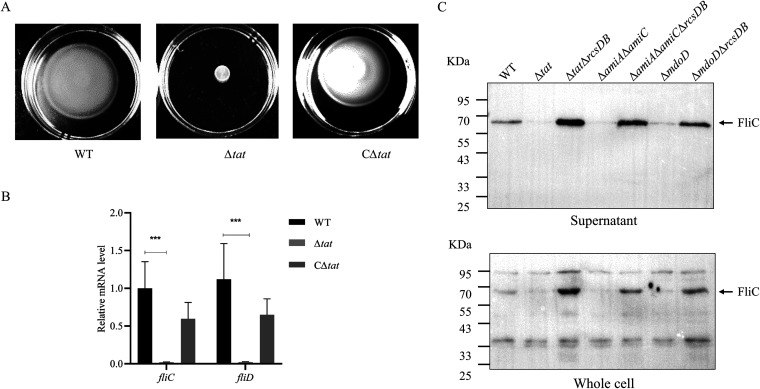
As we reported previously, the deletion of the Tat system in the ExPEC PCN033 strain led to severe defects in motility (Fig. 2A and reference 25). Also, according to the results of the above KEGG pathway enrichment analysis, cell motility is one of the most influenced pathways in the Δtat strain. Among the first 30 genes with the most downregulated expression, 11 were directly related to bacterial flagellar biosynthesis, including fliC and fliD, which encode the flagellin and the flagellar cap protein, respectively (Table 2). Real-time quantitative PCR (RT-qPCR) was performed, which confirmed that the expression levels fliC and fliD were significantly lower in the Δtat strain than in the WT strain (Fig. 2B). Results of Western blot analysis using anti-FliC antibody also showed a significantly lower level of FliC production in the Δtat strain (Fig. 2C). Therefore, the above-described results suggest that disruption of the Tat system interfered with the expression of flagellar biosynthesis genes.
FIG 2.
Disruption of the Tat system interferes with the expression of the flagellar biosynthesis genes. (A) In the swimming assay, 5 μL of cells of each indicated strain at the mid-log phase were spotted onto the swimming plate (tryptone, 10 g/L; yeast, 5 g/L; NaCl, 10 g/L; agar, 0.2%), which was dried at room temperature for 5 min followed by incubation at 37°C for 7 h. The CΔtat strain is the Δtat strain harboring a plasmid expressing tatABC in trans. The assay was performed in triplicate, and the data shown are representative. (B) RT-qPCR analysis. Total RNA was extracted from cells of each indicated strain grown to the mid-log phase. Genomic DNA was removed, and cDNA was synthesized, which was used as the template for qPCR with a QuantStudio 6 Flex fluorescence quantitative PCR instrument. The expression level of the tested gene was calculated using the 2−ΔΔCT method and normalized to the housekeeping gene gapA. Data are displayed as the geometric mean ± standard error of the mean (SEM). Statistical significance between the two groups was analyzed using Student’s t test. ***, P < 0.001. (C) Analysis of flagellin production. Cells of each indicated strain were grown to the mid-log phase in LB at 37°C with shaking. The culture was centrifuged, and the same amount of culture supernatant was collected and concentrated by ultracentrifugation. The same number of bacterial cells was harvested, resuspended in PBS, and lysed by sonication, and these were the whole-cell samples. Samples were normalized and subjected to SDS-PAGE followed by Western blotting using anti-FliC antibody (cat. no. ab93713; Abcam).
TABLE 2.
The first 30 genes with the most downregulated expression in Δtat
| No. | Gene ID | Fold change (Δtat/WT) | P value | FDR | Gene namea | Description |
|---|---|---|---|---|---|---|
| 1 | PPECC33_RS21010 | 0.001 | 1.0E-147 | 4.6E-144 | tatA | Sec-independent protein translocase subunit TatA |
| 2 | PPECC33_RS21015 | 0.001 | 9.10E-66 | 4.48E-63 | tatB | Sec-independent protein translocase subunit TatB |
| 3 | PPECC33_RS21020 | 0.002 | 4.21E-28 | 3.27E-26 | tatC | Sec-independent protein translocase subunit TatC |
| 4 | PPECC33_RS22680 | 0.011 | 2.7E-106 | 6.0E-103 | yjcZ | YjcZ-like family protein |
| 5 | PPECC33_RS10255 | 0.024 | 2.31E-72 | 2.05E-69 | tap | Methyl-accepting chemotaxis protein IV |
| 6 | PPECC33_RS10440 | 0.025 | 1.48E-39 | 2.12E-37 | fliC* | Flagellin FliC |
| 7 | PPECC33_RS10260 | 0.027 | 1.73E-41 | 2.85E-39 | tar | Methyl-accepting chemotaxis protein II |
| 8 | PPECC33_RS06330 | 0.033 | 3.33E-83 | 4.91E-80 | ycgR* | Flagellar brake protein |
| 9 | PPECC33_RS12795 | 0.033 | 1.38E-48 | 3.59E-46 | flxA | FlxA-like family protein |
| 10 | PPECC33_RS10270 | 0.036 | 3.73E-43 | 8.26E-41 | cheA | Chemotaxis protein CheA |
| 11 | PPECC33_RS10245 | 0.040 | 3.04E-58 | 1.12E-55 | cheB | Protein-glutamate methylesterase/protein glutamine deamidase |
| 12 | PPECC33_RS10445 | 0.045 | 5.80E-38 | 8.03E-36 | fliD* | Flagellar filament capping protein FliD |
| 13 | PPECC33_RS10275 | 0.046 | 3.14E-66 | 1.74E-63 | motB* | Flagellar motor protein MotB |
| 14 | PPECC33_RS10240 | 0.049 | 6.55E-53 | 2.23E-50 | cheY | Chemotaxis response regulator CheY |
| 15 | PPECC33_RS10235 | 0.051 | 2.01E-78 | 2.23E-75 | cheZ | Protein phosphatase CheZ |
| 16 | PPECC33_RS10250 | 0.051 | 4.46E-63 | 1.97E-60 | cheR | Protein-glutamate O-methyltransferase CheR |
| 17 | PPECC33_RS05845 | 0.054 | 2.70E-49 | 7.46E-47 | flgK* | Flagellar hook-associated protein FlgK |
| 18 | PPECC33_RS24055 | 0.054 | 8.65E-53 | 2.74E-50 | yjiH | Nucleoside recognition pore and gate family inner membrane transporter |
| 19 | PPECC33_RS05790 | 0.058 | 1.33E-59 | 5.35E-57 | flgM* | Anti-sigma-28 factor FlgM |
| 20 | PPECC33_RS10280 | 0.058 | 1.01E-37 | 1.36E-35 | motA* | Flagellar motor stator protein MotA |
| 21 | PPECC33_RS10265 | 0.061 | 3.95E-42 | 7.00E-40 | cheW | Chemotaxis protein CheW |
| 22 | PPECC33_RS05850 | 0.066 | 8.01E-42 | 1.36E-39 | flgL* | Flagellar hook-associated protein FlgL |
| 23 | PPECC33_RS20080 | 0.068 | 2.30E-33 | 2.37E-31 | uhpT | Hexose-6-phosphate:phosphate antiporter |
| 24 | PPECC33_RS19225 | 0.073 | 2.41E-40 | 3.82E-38 | pdeH | Cyclic-guanylate-specific phosphodiesterase |
| 25 | PPECC33_RS07750 | 0.075 | 1.63E-69 | 1.03E-66 | trg | Methyl-accepting chemotaxis protein Trg |
| 26 | PPECC33_RS10455 | 0.090 | 3.90E-11 | 8.23E-10 | fliT* | Flagella biosynthesis regulatory protein FliT |
| 27 | PPECC33_RS10450 | 0.108 | 5.03E-24 | 3.28E-22 | fliS* | Flagellar export chaperone FliS |
| 28 | PPECC33_RS05785 | 0.118 | 1.25E-20 | 6.93E-19 | flgN* | Flagellr biosynthesis chaperone FlgN |
| 29 | PPECC33_RS03935 | 0.135 | 1.45E-42 | 2.80E-40 | modA | Molybdate ABC transporter substrate-binding protein |
| 30 | PPECC33_RS07110 | 0.137 | 5.51E-20 | 2.91E-18 | trpE | Anthranilate synthase component I |
Genes marked with an asterisk (*) are related to flagellar biosynthesis.
Tat substrates MdoD, AmiA, and AmiC are responsible for motility defects.
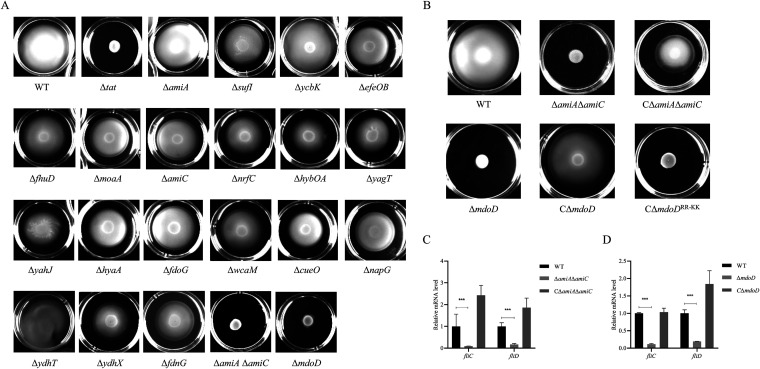
To determine which Tat substrate was responsible for the loss of motility of the ExPEC Δtat strain, a motility assay using soft agar was performed with the 20 Tat substrate-related mutant strains, which were constructed in our previous study (25). AmiA and AmiC are both Tat substrates and have overlapping functions. Therefore, a double deletion mutant, ΔamiAΔamiC, was constructed and tested (35). As shown in Fig. 3, apart from the Δtat and ΔamiAΔamiC strains, which have been previously shown as motility defective (25), ΔmdoD also showed significantly compromised motility. In trans expression of MdoD in ΔmdoD from the pHSG396-Apra-MdoD plasmid and AmiA and AmiC in ΔamiAΔamiC from the pHSG396-Apra-AmiA/AmiC plasmid restored motility (Fig. 3B). As MdoD was a putative Tat substrate and was reported for the first time to be related to bacterial motility, we tested whether the motility phenotype of ΔmdoD depends on the twin-arginine motif in the N-terminal Tat signal peptide. MdoDRR-KK in which the characteristic twin-arginine (R3 and R4) in its signal peptide was replaced with twin-lysine was expressed from the pHSG396-Apra-MdoDRR-KK plasmid in the ΔmdoD strain. It was shown that producing the wild-type MdoD in ΔmdoD restored the motility, while the strain producing MdoDRR-KK was still defective in motility. Our data suggest that it is the Tat substrate proteins MdoD, AmiA, and AmiC that are responsible for the motility defects of the tat mutant.
FIG 3.
Identification of Tat substrate proteins responsible for loss of motility. (A) For the swimming assay with the Tat substrate-related mutants, 5 μL of cells of each indicated strain grown to the mid-log phase was spotted onto the swimming plate (tryptone, 10 g/L; yeast, 5 g/L; NaCl, 10 g/L; agar, 0.2%) and was dried at room temperature for 5 min followed by incubation at 37°C for 7 h. The assay was performed in triplicate, and the data shown are representative. (B) Swimming assay with ΔamiAΔamiC, ΔmdoD, and the strains in trans expressing the corresponding proteins. The CΔmdoD and CΔmdoDRR-KK strains are the ΔmdoD strain encompassing a plasmid encoding the wild type and the mutated MdoD in which the twin-arginine in the signal peptide is replaced with a twin-lysine. CΔamiAΔamiC is the ΔamiAΔamiC strain in trans expressing AmiA and AmiC. (C) and (D) RT-qPCR analysis. Total RNA was extracted from cells of each indicated strain grown to the mid-log phase. Genomic DNA was removed, and cDNA was synthesized and was used as the template for qPCR with a QuantStudio 6 Flex fluorescence quantitative PCR instrument. The expression level of the tested gene was calculated using the 2−ΔΔCT method and normalized to the housekeeping gene gapA. Data are displayed as the geometric mean ± SEM. Statistical significance between the two groups was analyzed using Student’s t test. ***, P < 0.001.
Deletion of rcsDB can partially restore the motility of the tat mutant.
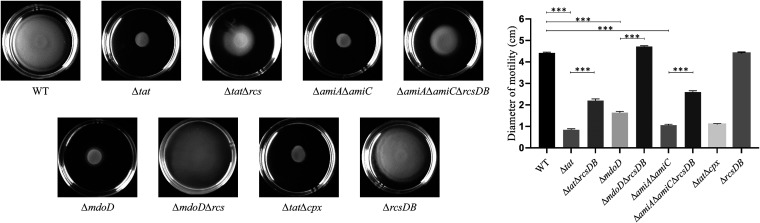
MdoD has been reported as a periplasmic protein involved in glycan biosynthesis, and defects in periplasmic glucan synthesis could induce envelope stress (36–38). AmiA and AmiC are two amidases involved in cell wall remodeling, and their deletion results in cell envelope defects (35). Moreover, several envelope stress response systems, such as the Rcs system and the CpxAR system, have been reported as regulators of bacterial flagella (13, 18, 39). Therefore, a hypothesis was proposed that deletion of the Tat system or the Tat substrates may activate envelope stress response systems which may suppress the expression of flagellar biosynthesis genes. Thus, rcsDB, which encodes the essential components of the Rcs signaling system, was deleted from the Δtat, ΔmdoD, and ΔamiAΔamiC strains, and bacterial motility was tested. In Fig. 4, it is shown that the disruption of the Rcs system completely restored the motility defects of the ΔmdoD strain (P < 0.05) and can partially rescue the motility of the Δtat strain and ΔamiAΔamiC strain (P < 0.05). To test whether the suppressive effect was specific to the Rcs system, another cell envelope stress response system, CpxAR, was deleted in the Δtat strain, which did not rescue the motility phenotype of Δtat (Fig. 4).
FIG 4.
Disruption of the Rcs system restored motility. First, 5 μL of cells of each indicated strain grown the mid-log phase was spotted onto the swimming plate (tryptone, 10 g/L; yeast, 5 g/L; NaCl, 10 g/L; agar, 0.2%) and was dried at room temperature for 5 min followed by incubation at 37°C for 7 h. The assay was performed in 5 replicates for each strain. The representative image is presented on the left. The swimming diameter is measured and shown as the mean ± standard deviation on the right. ***, P < 0.001.
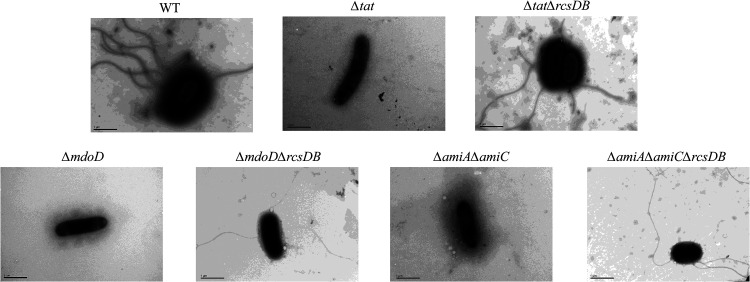
Flagella can be observed in the tat mutant when Rcs was disrupted.
To further confirm that the restored motility phenotype was due to flagellar biosynthesis, transmission electron microscopy analysis was performed to observe bacterial flagella. Consistent with the results of the motility assay, flagella were not seen in Δtat, ΔmodD, or ΔamiAΔamiC but could be observed in the WT, ΔtatΔrcsDB, ΔmodDΔrcsDB, and ΔtatΔrcsDB strains, suggesting that the flagellar biosynthesis was recovered when the Rcs system was deleted (Fig. 5). Western blot analysis also showed that FliC expression and secretion were significantly downregulated in Δtat, ΔmodD, and ΔamiAΔamiC but was restored when Rcs was further disrupted (Fig. 2C). It was noticed that the expression of FliC in the rcsDB-deleted strains was even higher than that in the WT strain. These results suggest that the Rcs system contributes to the disrupted flagellar biosynthesis of the Tat and the Tat substrate mutants.
FIG 5.
Transmission electron microscopy analysis of flagella. Cells of each indicated strain were streaked on an LB agar plate at 37°C for 12 h. A single colony was picked and resuspended in 200 μL of ultrapure water, which was left to stand for 2 h. Then, 10 μL of the bacterial suspension was placed on the grid followed by fixing with 2% phosphotungstic acid staining solution and was imaged using a transmission electron microscope (H-7650; Hitachi, Japan). The scale bar is 1 μm.
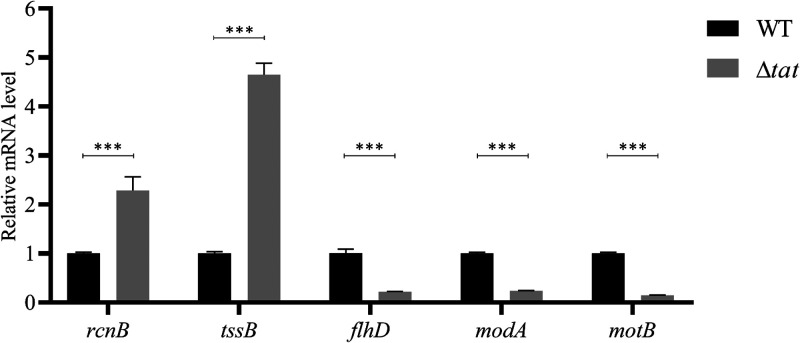
The Rcs system was activated when the Tat system was disrupted.
Previous studies have shown that the Rcs system is the negative regulator of the master transcription regulator of flagellar biosynthesis flhDC (18). The loss of motility could be attributed to activation of the Rcs system in the Δtat strain, which downregulated the expression of the flagellar biosynthesis genes. To verify whether the Rcs system was activated when the Tat system was disrupted, we compared the expression of the genes that have been reported to be regulated by the Rcs system between the WT and the Δtat strain, including flhDC (18), rcnB (40), tssB (20), motB (40), and modA (40). As shown in Fig. 6, the expression of rcnB, and tssB was significantly upregulated in the Δtat strain compared with the WT strain. In comparison, the expression of flhD, modA, and motB was significantly downregulated in the Δtat strain, as they have been shown to be negatively regulated by the Rcs system (40). Thus, the data indicate that the Rcs system was activated when the Tat system was disrupted.
FIG 6.
RT-qPCR analysis of the Rcs regulons. Total RNA was extracted from cells of each indicated strain grown to the mid-log phase. Genomic DNA was removed, and cDNA was synthesized and was used as the template for qPCR with a QuantStudio 6 Flex fluorescence quantitative PCR instrument. The expression level of the tested gene was calculated using the 2−ΔΔCT method and normalized to the housekeeping gene gapA. Data are displayed as the geometric mean ± SEM. Statistical significance between the two groups was analyzed using Student’s t test. ***, P < 0.001.
DISCUSSION
Flagellum-mediated motility is critical for the virulence of several bacterial pathogens in which flagella function at various stages of infection. In enteropathogenic E. coli (EPEC), flagella promote the adhesion of bacteria to epithelial cells (41). In avian pathogenic E. coli (APEC) and Shiga toxin-producing E. coli (STEC), flagella, although not directly involved in adhesion, play important roles in invasion (42–44). Flagella have also been demonstrated to be important for the initial interaction with surfaces, which is critical for the formation of biofilms (45). Moreover, flagella can be recognized by the Toll-like receptor 5, which elicits host immune response, and bacterial pathogens have evolved strategies, such as flagellar expression control, flagellin sequence variations, and flagellin modification, to avoid or evade such immune clearance to establish infection (46). Recently, the Tat system has been reported to play an important role in the pathogenesis of several bacteria (47–49). Interestingly, the compromised motility phenotype has been constantly observed when the Tat system is disrupted in a variety of bacteria, including E. coli (26), Salmonella (27), P. aeruginosa (50), and Yersinia (51). However, so far, the underlying mechanism of how the Tat system is linked to bacterial motility remains unclear.
To understand how the Tat system affects motility, we first compared the mRNA profile between the WT and the Δtat strain. Our data show that a substantial number of genes undergo differential expression, suggesting an important role of the Tat system in the physiology of ExPEC. The results show that the expression of capsular biosynthesis genes and stress response systems is increased in the Δtat strain (Table 1). This is consistent with the findings in a previous report (52). A possible explanation could be that the deletion of the Tat system causes outer membrane defects, which may result in the activation of the stress response systems. Among these envelope stress response systems, the Rcs system has been revealed to positively regulate the expression of capsular polysaccharide biosynthesis (53), and the activation of the Rcs system by the Tat system disruption has been implicated in the previous transcriptomics study (52). In addition, several toxin-antitoxin (TA) systems also showed differential expression in the Δtat strain (Table 1). Among them, ghoT/S showed a drastic upregulation upon Tat deletion, which has been reported to function under stress conditions by reducing metabolism, a possible survival strategy when confronting stresses (54). However, how the Tat system influences these TA systems needs further investigation. The downregulated genes include a substantial amount of flagellar biosynthesis genes, which corresponds with the compromised motility phenotype. This is consistent with a previous study showing that the deletion of the Tat system abolished the synthesis of flagellin in E. coli O157:H7 (26). However, it was not the case in E. coli MC4100, a commensal K-12 strain (52). Whether this difference is attributed to pathotypes remains unclear.
In E. coli, over 30 proteins have been predicted or verified to be transported by the Tat system (8). However, none of them has been revealed to be directly involved in flagellar biosynthesis. To reveal the link between the Tat system and flagellar biosynthesis, we perform a motility phenotype screening with the Tat substrate-related mutants, which identified three Tat substrate proteins, AmiA, AmiC, and MdoD, that are responsible for the loss of motility of the Δtat strain. Deletion of the Tat-dependent amidases AmiA and AmiC simultaneously has been previously shown to cause motility defects in both E. coli and Salmonella (25, 55). We for the first time report that another Tat substrate, MdoD (also known as OpgD), is related to bacterial motility. MdoD is involved in osmoregulated periplasmic glucan (OPG) biosynthesis in which deletion of MdoD affects the glucose backbone structures of periplasmic glucans (56). It has also been reported previously that defects in periplasmic glucan synthesis cause motility defects (36). The osmoregulated periplasmic glucans play important roles in coping with environmental stresses (37, 57). Defects in periplasmic glucans have been reported to induce envelope stress (58, 59). There are several envelope stress response systems that exist in E. coli to cope with envelope stresses (60), among which the Rcs system has been reported to be a negative regulator of flhDC, the master regulator of bacterial flagellar biosynthesis (13, 18). Our result that the disruption of the Rcs system restores the motility of the ΔmdoD strain further demonstrates an important role of Rcs in mediating the motility defects.
It should be noted that the motility was only partially restored for ΔtatΔrcsDB and ΔamiAΔamiCΔrcsDB, suggesting that other factors than the Rcs system may also be involved in the disruption of the motility. AmiA and AmiC are two Tat-exported N-acetylmuramoyl-l-alanine amidases functioning in septum cleavage during cell division (61–63). Moreover, the inactivation of AmiA and AmiC is reported to compromise outer membrane integrity (35). Therefore, peptidoglycan synthesis and outer membrane integrity may be the other factors affecting the motility of the tat mutant.
Conclusions. In conclusion, we show that disruption of the Tat system abolishes motility of extraintestinal pathogenic E. coli and interferes with the expression of a large number of genes, among which, genes involved in flagellar biosynthesis are significantly affected. Tat substrate proteins MdoD, AmiA, and AmiC are identified that are related to the loss of motility. The Rcs system is then demonstrated to play an important role in mediating the loss of motility of the tat mutant. Our study for the first time reveals the underlying link between the Tat system and bacterial motility.
MATERIALS AND METHODS
Strains and plasmid construction.
The strains and plasmids used in this study are all listed in Table 3. All E. coli strains except E. coli χ7213 were grown in lysogeny broth (LB) medium or on LB agar plates at 37°C. E. coli strain χ7213 was cultured in lysogeny broth (LB) medium supplemented with 50 μg/mL of diaminopimelic acid (DAP). The DNA sequences of the primers used in this study are listed in Table S2. The gene deletion strains were constructed according to our previous report (25). Briefly, the pRE112 plasmid containing the homologous arms flanking the target gene to be deleted was used to transform E. coli χ7213 competent cells, which was used as the donor strain for transconjugation. The single exchanged strain was then confirmed by PCR after transconjugation and was incubated on 10% sucrose-containing LB to counterselect the double exchanged strain. The mutant strain was then verified by PCR. To in trans express the Tat substrate proteins in the mutant strains, pHSG396-Apra derivative plasmids were used in which the DNA fragment containing the corresponding coding sequence following a constitutive promoter was inserted into the multiple cloning site. The pHSG396-Apra plasmid was constructed by inserting an apramycin resistance cassette at the NcoI site within the chloramphenicol resistance cassette.
TABLE 3.
Bacterial strains and plasmids used in this study
| Name | Description | Reference or source |
|---|---|---|
| Strains | ||
| ExPEC PCN033 | A virulent clinical strain isolated from diseased pig, wild type | 72 |
| E. coli χ7213 | A diaminopimelic acid autotrophic E. coli strain used for transconjugation | 73 |
| Δtat | As ExPEC PCN033, tatABC deleted | 25 |
| CΔtat | Δtat strain expressing tatABC from a plasmid | |
| ΔmodD | As ExPEC PCN033, modD deleted | |
| ΔamiA | As ExPEC PCN033, amiA deleted | |
| ΔamiC | As ExPEC PCN033, amiC deleted | |
| ΔefeOB | As ExPEC PCN033, efeOB deleted | |
| ΔsufI | As ExPEC PCN033, sufI deleted | |
| ΔmepK | As ExPEC PCN033, mepK (previously known as ycbK) deleted | |
| ΔfhuD | As ExPEC PCN033, fhuD deleted | |
| ΔmoaA | As ExPEC PCN033, moaA deleted | |
| ΔnrfC | As ExPEC PCN033, nrfC deleted | |
| ΔhybOA | As ExPEC PCN033, hybOA deleted | |
| ΔyagT | As ExPEC PCN033, yagT deleted | |
| ΔyahJ | As ExPEC PCN033, yahJ deleted | |
| ΔhyaA | As ExPEC PCN033, hyaA deleted | |
| ΔfdoG | As ExPEC PCN033, fdoG deleted | |
| ΔwacM | As ExPEC PCN033, wcaM deleted | |
| ΔcueO | As ExPEC PCN033, cueO deleted | |
| ΔnapG | As ExPEC PCN033, napG deleted | |
| ΔydhT | As ExPEC PCN033, ydhT deleted | |
| ΔydhX | As ExPEC PCN033, ydhX deleted | |
| ΔfdnG | As ExPEC PCN033, fdnG deleted | |
| ΔamiAΔamiC | As ExPEC PCN033, amiA and amiC deleted | |
| ΔrcsDB | As ExPEC PCN033, rcsDB deleted | This study |
| ΔtatΔrcsDB | As Δtat, rcsDB deleted | This study |
| ΔmdoDΔrcsDB | As ΔmdoD, rcsDB deleted | This study |
| ΔamiAΔamiCΔrcsDB | As ΔamiAΔamiC, rcsDB deleted | |
| ΔtatΔcpxAR | As Δtat, cpxAR deleted | This study |
| Plasmids | ||
| pRE112 | The plasmid used in mutant construction | Lab stock |
| pHSG396-Apra | The plasmid used for in trans protein expression in ExPEC | This study |
| pHSG396-Apra-MdoD | As pHSG396-Apra, a DNA fragment containing a tat promoter followed by the coding sequence of mdoD followed by insertion between XbaI and EcoRI | This study |
| pHSG396-Apra-MdoDRR-KK | As pHSG396-Apra-MdoD, expressing the variant MdoD, where the R3 and R4 residues were both replaced with a lysine | This study |
| pHSG396-Apra-RcsDB | As pHSG396-Apra, a DNA fragment containing a tat promoter followed by the coding sequence of rcsDB inserted between XbaI and EcoRI | This study |
| pHSG396-Apra-AmiA/AmiC | As pHSG396-Apra, a DNA fragment containing the coding sequence of rcsDB followed by a tat promoter inserted between kpnI and SacI | This study |
| pRE112-rcsDB | As pRE112, a DNA fragment containing the upstream 1,000 bp and downstream 1,000 bp of rcsDB inserted between kpnI and SacI | This study |
| pRE112-cpxAR | As pRE112, a DNA fragment containing the upstream 1,000 bp and downstream 1,000 bp of cpxAR inserted between kpnI and SacI | This study |
mRNA-seq.
The ExPEC PCN033 and Δtat strains were subcultured from overnight-grown cultures into fresh LB and grown to the mid-log phase at 37°C with shaking. The cells were harvested and washed with normal saline followed by total RNA extraction using a HiPure bacterial RNA kit (Magen, China) according to the manufacturer’s instructions. RNA quantitation was performed using a Qubit 3.0 device (Thermo Fisher Scientific, Massachusetts, USA). Whole mRNA-seq libraries were constructed by Guangdong Magigene Biotechnology Co., Ltd. (Guangzhou, China), using NEBNext Ultra directional RNA library prep kit for Illumina (New England Biolabs, Massachusetts, USA) following the manufacturer’s recommendations. Briefly, the bacterial 16S rRNA in the total RNA samples were depleted using a Ribo-off rRNA depletion kit (bacteria) (Vazyme, Nanjing, China). Fragmentation was carried out using the RNA first-strand synthesis module (New England Biolabs). The first-strand cDNA was synthesized using a random hexamer primer and M-MuLV reverse transcriptase (New England Biolabs). When synthesizing the second strand of cDNA, a chain-specific library was constructed by replacing dTTP with dUTP to greatly improve the accuracy of the results. The remaining overhangs were converted into blunt ends via exonuclease/polymerase activities. After adenylation of the 3′ ends of DNA fragments, a NEBNext adaptor with a hairpin loop structure was ligated to prepare for hybridization. To select cDNA fragments of preferentially 150 to ∼200 bp in length, the fragments were selected with SpeedBead magnetic carboxylate modified particles (Global Life Sciences Solutions Operations, Buckinghamshire, UK). Then, PCR was performed with Phusion high-fidelity DNA polymerase, universal PCR primers, and index (X) primer. Lastly, PCR products were purified with SpeedBead magnetic carboxylate modified particles, and library insert size was assessed on the Qsep400 high-throughput nucleic acid protein analysis system (Houze Biological Technology Co., Hangzhou, China). The clustering of the index-coded samples was performed on a cBot cluster generation system. After cluster generation, the library was sequenced on an Illumina NovaSeq 6000 platform, and 150-bp paired-end reads were generated.
mRNA-seq data analysis.
The raw RNA-seq data in FASTQ format were processed using Trimmomatic (v0.36) to acquire the clean data (clean reads) (64). Clean reads were mapped to NCBI Rfam databases to remove the rRNA sequences with Bowtie 2 (v2.33) (65). The remaining mRNA sequences were mapped to the reference genome with HISAT 2 (v2.1.0) (66). HTSeq-count (v0.9.1) was used to obtain the read count and function information of each gene according to the result of the mapping (67). To compare the expression level of genes, the fragments per kilobase per million (FPKM) of each gene was calculated. Differentially expressed genes were analyzed using edgeR (v3.16.5) (68). The resulting P value was adjusted using Benjamini and Hochberg’s approach for controlling the false-discovery rate. Genes with an FDR of ≤0.05 and |log2(fold change)| of ≥1 were taken as differentially expressed genes. To do KEGG pathway enrichment, the differentially expressed genes were assigned a K number using the BlastKOALA tool (69). The K numbers were then used for pathway enrichment by using KEGG Mapper (70). The COG enrichment was performed with the EggNOG v5.0 database (71).
Motility assay.
E. coli cells were subcultured 1:100 from overnight-grown cultures in LB and grown at 37°C with shaking to the mid-log phase. Then, 5 μL of the bacterial culture was spotted onto the swimming plate (tryptone, 10 g/L; yeast, 5 g/L; NaCl, 10 g/L; agar, 0.2%), which was dried at room temperature for 5 min followed by incubation at 37°C for 7 h.
Transmission electronic microscopy.
Cells of each indicated strain were streaked on an LB agar plate at 37°C for 12 h. A single colony was picked and resuspended in 200 μL of ultrapure water which was left to stand for 2 h. Then, 10 μL of the bacterial suspension was placed on the grid followed by fix with 2% phosphotungstic acid staining solution and was imaged using a transmission electron microscope (H-7650; Hitachi, Japan).
Real-time quantitative PCR.
Total RNA was extracted using the RNA isolation kit (Tianmo Biotech, China). The purity and degradation of the RNA were analyzed by measuring the absorbance and agarose gel electrophoresis. HiScript Q select RT SuperMix (+gDNA wiper) (Vazyme, China) was used to remove residual genomic DNA and synthesize (cDNA according to the manufacturer’s instructions. The cDNA was used as the template for qPCR using the TB Green premix Ex Taq II kit (TaKaRa Biomedical Technology, Beijing, China) according to the manufacturer’s instructions with a QuantStudio 6 Flex fluorescence quantitative PCR instrument. The expression level of the tested gene was calculated using the 2−ΔΔCT method and normalized to the housekeeping gene gapA.
Western blotting.
To detect flagellin production, cells of each indicated strain were subcultured 1:100 from overnight-grown cultures into LB and grown to the mid-log phase at 37°C with shaking. The culture was centrifuged, and the same amount of culture supernatant was collected and concentrated by ultracentrifugation. The same number of bacterial cells were harvested, resuspended in phosphate-buffered saline (PBS), and lysed by sonication, and these were the whole-cell samples. The samples were normalized and separated on 10% SDS-PAGE and were then transferred to polyvinylidene difluoride (PVDF) membranes and probed with anti-FliC antibody (catalog [cat.] no. ab93713; Abcam, Shanghai, China).
Data availability.
Sequence data have been submitted to the NCBI GEO database, and the accession number is GSE181969.
ACKNOWLEDGMENTS
This work was supported by the National Key Research and Development Program of China (2021YFD1800401) and the National Natural Science Foundation of China (31802211).
We declare no conflict of interest.
Footnotes
Supplemental material is available online only.
Contributor Information
Qi Huang, Email: qhuang@mail.hzau.edu.cn.
Mohamed Y. El-Naggar, University of Southern California
REFERENCES
- 1.Tseng TT, Tyler BM, Setubal JC. 2009. Protein secretion systems in bacterial-host associations, and their description in the Gene Ontology. BMC Microbiol 9:S2. 10.1186/1471-2180-9-S1-S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Costa TR, Felisberto-Rodrigues C, Meir A, Prevost MS, Redzej A, Trokter M, Waksman G. 2015. Secretion systems in Gram-negative bacteria: structural and mechanistic insights. Nat Rev Microbiol 13:343–359. 10.1038/nrmicro3456. [DOI] [PubMed] [Google Scholar]
- 3.Rapisarda C, Tassinari M, Gubellini F, Fronzes R. 2018. Using cryo-EM to investigate bacterial secretion systems. Annu Rev Microbiol 72:231–254. 10.1146/annurev-micro-090817-062702. [DOI] [PubMed] [Google Scholar]
- 4.Palmer T, Berks BC. 2012. The twin-arginine translocation (Tat) protein export pathway. Nat Rev Microbiol 10:483–496. 10.1038/nrmicro2814. [DOI] [PubMed] [Google Scholar]
- 5.DeLisa MP, Tullman D, Georgiou G. 2003. Folding quality control in the export of proteins by the bacterial twin-arginine translocation pathway. Proc Natl Acad Sci USA 100:6115–6120. 10.1073/pnas.0937838100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Berks BC. 1996. A common export pathway for proteins binding complex redox cofactors? Mol Microbiol 22:393–404. 10.1046/j.1365-2958.1996.00114.x. [DOI] [PubMed] [Google Scholar]
- 7.Berks BC, Palmer T, Sargent F. 2003. The Tat protein translocation pathway and its role in microbial physiology. Adv Microb Physiol 47:187–254. 10.1016/s0065-2911(03)47004-5. [DOI] [PubMed] [Google Scholar]
- 8.Palmer T, Sargent F, Berks BC. 2010. The Tat protein export pathway. EcoSal Plus 4. 10.1128/ecosalplus.4.3.2. [DOI] [PubMed] [Google Scholar]
- 9.De Buck E, Lammertyn E, Anne J. 2008. The importance of the twin-arginine translocation pathway for bacterial virulence. Trends Microbiol 16:442–453. 10.1016/j.tim.2008.06.004. [DOI] [PubMed] [Google Scholar]
- 10.Benoit SL, Maier RJ. 2014. Twin-arginine translocation system in Helicobacter pylori: TatC, but not TatB, is essential for viability. mBio 5:e01016-13. 10.1128/mBio.01016-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McDonough JA, Hacker KE, Flores AR, Pavelka MS, Jr, Braunstein M. 2005. The twin-arginine translocation pathway of Mycobacterium smegmatis is functional and required for the export of mycobacterial beta-lactamases. J Bacteriol 187:7667–7679. 10.1128/JB.187.22.7667-7679.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Josenhans C, Suerbaum S. 2002. The role of motility as a virulence factor in bacteria. Int J Med Microbiol 291:605–614. 10.1078/1438-4221-00173. [DOI] [PubMed] [Google Scholar]
- 13.Meng J, Young G, Chen J. 2021. The Rcs system in Enterobacteriaceae: envelope stress responses and virulence regulation. Front Microbiol 12:627104. 10.3389/fmicb.2021.627104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shimizu T, Ichimura K, Noda M. 2016. The surface sensor NlpE of enterohemorrhagic Escherichia coli contributes to regulation of the type iii secretion system and flagella by the Cpx response to adhesion. Infect Immun 84:537–549. 10.1128/IAI.00881-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Osterman IA, Dikhtyar YY, Bogdanov AA, Dontsova OA, Sergiev PV. 2015. Regulation of flagellar gene expression in bacteria. Biochemistry (Mosc) 80:1447–1456. 10.1134/S000629791511005X. [DOI] [PubMed] [Google Scholar]
- 16.Fitzgerald DM, Bonocora RP, Wade JT. 2014. Comprehensive mapping of the Escherichia coli flagellar regulatory network. PLoS Genet 10:e1004649. 10.1371/journal.pgen.1004649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wall E, Majdalani N, Gottesman S. 2018. The complex Rcs regulatory cascade. Annu Rev Microbiol 72:111–139. 10.1146/annurev-micro-090817-062640. [DOI] [PubMed] [Google Scholar]
- 18.Francez-Charlot A, Laugel B, Van Gemert A, Dubarry N, Wiorowski F, Castanie-Cornet MP, Gutierrez C, Cam K. 2003. RcsCDB His-Asp phosphorelay system negatively regulates the flhDC operon in Escherichia coli. Mol Microbiol 49:823–832. 10.1046/j.1365-2958.2003.03601.x. [DOI] [PubMed] [Google Scholar]
- 19.Spöring I, Felgner S, Preuße M, Eckweiler D, Rohde M, Häussler S, Weiss S, Erhardt M. 2018. Regulation of flagellum biosynthesis in response to cell envelope stress in Salmonella enterica serovar Typhimurium. mBio 9:e00736-17. 10.1128/mBio.00736-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hu L, Yu F, Liu M, Chen J, Zong B, Zhang Y, Chen T, Wang C, Zhang T, Zhang J, Zhu Y, Wang X, Chen H, Tan C. 2021. RcsB-dependent regulation of type VI secretion system in porcine extra-intestinal pathogenic Escherichia coli. Gene 768:145289. 10.1016/j.gene.2020.145289. [DOI] [PubMed] [Google Scholar]
- 21.Jozwick AKS, LaPatra SE, Graf J, Welch TJ. 2019. Flagellar regulation mediated by the Rcs pathway is required for virulence in the fish pathogen Yersinia ruckeri. Fish Shellfish Immunol 91:306–314. 10.1016/j.fsi.2019.05.036. [DOI] [PubMed] [Google Scholar]
- 22.Biran D, Ron EZ. 2018. Extraintestinal pathogenic Escherichia coli. Curr Top Microbiol Immunol 416:149–161. 10.1007/82_2018_108. [DOI] [PubMed] [Google Scholar]
- 23.Liu C, Zheng H, Yang M, Xu Z, Wang X, Wei L, Tang B, Liu F, Zhang Y, Ding Y, Tang X, Wu B, Johnson TJ, Chen H, Tan C. 2015. Genome analysis and in vivo virulence of porcine extraintestinal pathogenic Escherichia coli strain PCN033. BMC Genomics 16:717. 10.1186/s12864-015-1890-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vila J, Saez-Lopez E, Johnson JR, Romling U, Dobrindt U, Canton R, Giske CG, Naas T, Carattoli A, Martinez-Medina M, Bosch J, Retamar P, Rodriguez-Bano J, Baquero F, Soto SM. 2016. Escherichia coli: an old friend with new tidings. FEMS Microbiol Rev 40:437–463. 10.1093/femsre/fuw005. [DOI] [PubMed] [Google Scholar]
- 25.Liu J, Yin F, Liu T, Li S, Tan C, Li L, Zhou R, Huang Q. 2020. The Tat system and its dependent cell division proteins are critical for virulence of extra-intestinal pathogenic Escherichia coli. Virulence 11:1279–1292. 10.1080/21505594.2020.1817709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pradel N, Ye C, Livrelli V, Xu J, Joly B, Wu LF. 2003. Contribution of the twin arginine translocation system to the virulence of enterohemorrhagic Escherichia coli O157:H7. Infect Immun 71:4908–4916. 10.1128/IAI.71.9.4908-4916.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Reynolds MM, Bogomolnaya L, Guo J, Aldrich L, Bokhari D, Santiviago CA, McClelland M, Andrews-Polymenis H. 2011. Abrogation of the twin arginine transport system in Salmonella enterica serovar Typhimurium leads to colonization defects during infection. PLoS One 6:e15800. 10.1371/journal.pone.0015800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yamanaka Y, Oshima T, Ishihama A, Yamamoto K. 2014. Characterization of the YdeO regulon in Escherichia coli. PLoS One 9:e111962. 10.1371/journal.pone.0111962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Osterman IA, Evfratov SA, Dzama MM, Pletnev PI, Kovalchuk SI, Butenko IO, Pobeguts OV, Golovina AY, Govorun VM, Bogdanov AA, Sergiev PV, Dontsova OA. 2015. A bacterial homolog YciH of eukaryotic translation initiation factor eIF1 regulates stress-related gene expression and is unlikely to be involved in translation initiation fidelity. RNA Biol 12:966–971. 10.1080/15476286.2015.1069464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Masuda N, Church GM. 2003. Regulatory network of acid resistance genes in Escherichia coli. Mol Microbiol 48:699–712. 10.1046/j.1365-2958.2003.03477.x. [DOI] [PubMed] [Google Scholar]
- 31.Guo Y, Li Y, Zhan W, Wood TK, Wang X. 2019. Resistance to oxidative stress by inner membrane protein ElaB is regulated by OxyR and RpoS. Microb Biotechnol 12:392–404. 10.1111/1751-7915.13369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee J, Hiibel SR, Reardon KF, Wood TK. 2010. Identification of stress-related proteins in Escherichia coli using the pollutant cis-dichloroethylene. J Appl Microbiol 108:2088–2102. 10.1111/j.1365-2672.2009.04611.x. [DOI] [PubMed] [Google Scholar]
- 33.Ito T, Uozumi N, Nakamura T, Takayama S, Matsuda N, Aiba H, Hemmi H, Yoshimura T. 2009. The implication of YggT of Escherichia coli in osmotic regulation. Biosci Biotechnol Biochem 73:2698–2704. 10.1271/bbb.90558. [DOI] [PubMed] [Google Scholar]
- 34.Chen Y, Reinhardt M, Neris N, Kerns L, Mansell TJ, Jarboe LR. 2018. Lessons in membrane engineering for octanoic acid production from environmental Escherichia coli isolates. Appl Environ Microbiol 84:e01285-18. 10.1128/AEM.01285-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ize B, Stanley NR, Buchanan G, Palmer T. 2003. Role of the Escherichia coli Tat pathway in outer membrane integrity. Mol Microbiol 48:1183–1193. 10.1046/j.1365-2958.2003.03504.x. [DOI] [PubMed] [Google Scholar]
- 36.Kannan P, Dharne M, Smith A, Karns J, Bhagwat AA. 2009. Motility revertants of opgGH mutants of Salmonella enterica serovar Typhimurium remain defective in mice virulence. Curr Microbiol 59:641–645. 10.1007/s00284-009-9486-8. [DOI] [PubMed] [Google Scholar]
- 37.Bontemps-Gallo S, Bohin JP, Lacroix JM. 2017. Osmoregulated periplasmic glucans. EcoSal Plus 7. 10.1128/ecosalplus.ESP-0001-2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Meng J, Huang C, Huang X, Liu D, Han B, Chen J. 2020. Osmoregulated periplasmic glucans transmit external signals through Rcs phosphorelay pathway in Yersinia enterocolitica. Front Microbiol 11:122. 10.3389/fmicb.2020.00122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.De Wulf P, McGuire AM, Liu X, Lin EC. 2002. Genome-wide profiling of promoter recognition by the two-component response regulator CpxR-P in Escherichia coli. J Biol Chem 277:26652–26661. 10.1074/jbc.M203487200. [DOI] [PubMed] [Google Scholar]
- 40.Huesa J, Giner-Lamia J, Pucciarelli MG, Paredes-Martínez F, García-del Portillo F, Marina A, Casino P. 2021. Structure-based analyses of Salmonella RcsB variants unravel new features of the Rcs regulon. Nucleic Acids Res 49:2357–2374. 10.1093/nar/gkab060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Girón JA, Torres AG, Freer E, Kaper JB. 2002. The flagella of enteropathogenic Escherichia coli mediate adherence to epithelial cells. Mol Microbiol 44:361–379. 10.1046/j.1365-2958.2002.02899.x. [DOI] [PubMed] [Google Scholar]
- 42.La Ragione RM, Sayers AR, Woodward MJ. 2000. The role of fimbriae and flagella in the colonization, invasion and persistence of Escherichia coli O78:K80 in the day-old-chick model. Epidemiol Infect 124:351–363. 10.1017/S0950268899004045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.LA Ragione RM, Cooley WA, Woodward MJ. 2000. The role of fimbriae and flagella in the adherence of avian strains of Escherichia coli O78:K80 to tissue culture cells and tracheal and gut explants. J Med Microbiol 49:327–338. 10.1099/0022-1317-49-4-327. [DOI] [PubMed] [Google Scholar]
- 44.Best A, La Ragione RM, Sayers AR, Woodward MJ. 2005. Role for flagella but not intimin in the persistent infection of the gastrointestinal tissues of specific-pathogen-free chicks by Shiga toxin-negative Escherichia coli O157:H7. Infect Immun 73:1836–1846. 10.1128/IAI.73.3.1836-1846.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pratt LA, Kolter R. 1998. Genetic analysis of Escherichia coli biofilm formation: roles of flagella, motility, chemotaxis and type I pili. Mol Microbiol 30:285–293. 10.1046/j.1365-2958.1998.01061.x. [DOI] [PubMed] [Google Scholar]
- 46.Rossez Y, Wolfson EB, Holmes A, Gally DL, Holden NJ. 2015. Bacterial flagella: twist and stick, or dodge across the kingdoms. PLoS Pathog 11:e1004483. 10.1371/journal.ppat.1004483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yan X, Hu S, Yang Y, Xu D, Li H, Liu W, He X, Li G, Cai W, Bu Z. 2020. The twin-arginine translocation system is important for stress resistance and virulence of Brucella melitensis. Infect Immun 88:e00389-20. 10.1128/IAI.00389-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Urrutia ÍM, Sabag A, Valenzuela C, Labra B, Álvarez SA, Santiviago CA. 2018. Contribution of the twin-arginine translocation system to the intracellular survival of Salmonella Typhimurium in Dictyostelium discoideum. Front Microbiol 9:3001. 10.3389/fmicb.2018.03001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Avican U, Doruk T, Ostberg Y, Fahlgren A, Forsberg A. 2017. The Tat substrate SufI is critical for the ability of Yersinia pseudotuberculosis to cause systemic infection. Infect Immun 85:e00867-16. 10.1128/IAI.00867-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ochsner UA, Snyder A, Vasil AI, Vasil ML. 2002. Effects of the twin-arginine translocase on secretion of virulence factors, stress response, and pathogenesis. Proc Natl Acad Sci USA 99:8312–8317. 10.1073/pnas.082238299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lavander M, Ericsson SK, Bröms JE, Forsberg A. 2007. Twin arginine translocation in Yersinia. Adv Exp Med Biol 603:258–267. 10.1007/978-0-387-72124-8_23. [DOI] [PubMed] [Google Scholar]
- 52.Ize B, Porcelli I, Lucchini S, Hinton JC, Berks BC, Palmer T. 2004. Novel phenotypes of Escherichia coli tat mutants revealed by global gene expression and phenotypic analysis. J Biol Chem 279:47543–47554. 10.1074/jbc.M406910200. [DOI] [PubMed] [Google Scholar]
- 53.Navasa N, Rodríguez-Aparicio L, Ferrero M, Monteagudo-Mera A, Martínez-Blanco H. 2013. Polysialic and colanic acids metabolism in Escherichia coli K92 is regulated by RcsA and RcsB. Biosci Rep 33:e00038. 10.1042/BSR20130018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cheng HY, Soo VW, Islam S, McAnulty MJ, Benedik MJ, Wood TK. 2014. Toxin GhoT of the GhoT/GhoS toxin/antitoxin system damages the cell membrane to reduce adenosine triphosphate and to reduce growth under stress. Environ Microbiol 16:1741–1754. 10.1111/1462-2920.12373. [DOI] [PubMed] [Google Scholar]
- 55.Fujimoto M, Goto R, Hirota R, Ito M, Haneda T, Okada N, Miki T. 2018. Tat-exported peptidoglycan amidase-dependent cell division contributes to Salmonella Typhimurium fitness in the inflamed gut. PLoS Pathog 14:e1007391. 10.1371/journal.ppat.1007391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lequette Y, Odberg-Ferragut C, Bohin JP, Lacroix JM. 2004. Identification of mdoD, an mdoG paralog which encodes a twin-arginine-dependent periplasmic protein that controls osmoregulated periplasmic glucan backbone structures. J Bacteriol 186:3695–3702. 10.1128/JB.186.12.3695-3702.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bontemps-Gallo S, Lacroix JM. 2015. New insights into the biological role of the osmoregulated periplasmic glucans in pathogenic and symbiotic bacteria. Environ Microbiol Rep 7:690–697. 10.1111/1758-2229.12325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bhagwat AA, Young L, Smith AD, Bhagwat M. 2017. Transcriptomic analysis of the swarm motility phenotype of Salmonella enterica serovar Typhimurium mutant defective in periplasmic glucan synthesis. Curr Microbiol 74:1005–1014. 10.1007/s00284-017-1267-1. [DOI] [PubMed] [Google Scholar]
- 59.Amar A, Pezzoni M, Pizarro RA, Costa CS. 2018. New envelope stress factors involved in σ(E) activation and conditional lethality of rpoE mutations in Salmonella enterica. Microbiology (Reading) 164:1293–1307. 10.1099/mic.0.000701. [DOI] [PubMed] [Google Scholar]
- 60.Mitchell AM, Silhavy TJ. 2019. Envelope stress responses: balancing damage repair and toxicity. Nat Rev Microbiol 17:417–428. 10.1038/s41579-019-0199-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Heidrich C, Templin MF, Ursinus A, Merdanovic M, Berger J, Schwarz H, de Pedro MA, Holtje JV. 2001. Involvement of N-acetylmuramyl-l-alanine amidases in cell separation and antibiotic-induced autolysis of Escherichia coli. Mol Microbiol 41:167–178. 10.1046/j.1365-2958.2001.02499.x. [DOI] [PubMed] [Google Scholar]
- 62.Vermassen A, Leroy S, Talon R, Provot C, Popowska M, Desvaux M. 2019. Cell wall hydrolases in bacteria: insight on the diversity of cell wall amidases, glycosidases and peptidases toward peptidoglycan. Front Microbiol 10:331. 10.3389/fmicb.2019.00331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Vollmer W, Joris B, Charlier P, Foster S. 2008. Bacterial peptidoglycan (murein) hydrolases. FEMS Microbiol Rev 32:259–286. 10.1111/j.1574-6976.2007.00099.x. [DOI] [PubMed] [Google Scholar]
- 64.Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30:2114–2120. 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods 9:357–359. 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kim D, Langmead B, Salzberg SL. 2015. HISAT: a fast spliced aligner with low memory requirements. Nat Methods 12:357–360. 10.1038/nmeth.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Anders S, Pyl PT, Huber W. 2015. HTSeq: a Python framework to work with high-throughput sequencing data. Bioinformatics 31:166–169. 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Robinson MD, McCarthy DJ, Smyth GK. 2010. edgeR: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26:139–140. 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kanehisa M, Sato Y, Morishima K. 2016. BlastKOALA and GhostKOALA: KEGG tools for functional characterization of genome and metagenome sequences. J Mol Biol 428:726–731. 10.1016/j.jmb.2015.11.006. [DOI] [PubMed] [Google Scholar]
- 70.Kanehisa M, Sato Y. 2020. KEGG Mapper for inferring cellular functions from protein sequences. Protein Sci 29:28–35. 10.1002/pro.3711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Huerta-Cepas J, Szklarczyk D, Heller D, Hernández-Plaza A, Forslund SK, Cook H, Mende DR, Letunic I, Rattei T, Jensen LJ, von Mering C, Bork P. 2019. eggNOG 5.0: a hierarchical, functionally and phylogenetically annotated orthology resource based on 5090 organisms and 2502 viruses. Nucleic Acids Res 47:D309–D314. 10.1093/nar/gky1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tang X, Tan C, Zhang X, Zhao Z, Xia X, Wu B, Guo A, Zhou R, Chen H. 2011. Antimicrobial resistances of extraintestinal pathogenic Escherichia coli isolates from swine in China. Microb Pathog 50:207–212. 10.1016/j.micpath.2011.01.004. [DOI] [PubMed] [Google Scholar]
- 73.Roland K, Curtiss R, III, Sizemore D. 1999. Construction and evaluation of a delta cya delta crp Salmonella Typhimurium strain expressing avian pathogenic Escherichia coli O78 LPS as a vaccine to prevent airsacculitis in chickens. Avian Dis 43:429–441. 10.2307/1592640. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Download jb.00612-21-s0001.pdf, PDF file, 0.1 MB (110.6KB, pdf)
Table S2. Download jb.00612-21-s0002.xls, XLS file, 0.1 MB (114.4KB, xls)
Data Availability Statement
Sequence data have been submitted to the NCBI GEO database, and the accession number is GSE181969.