ABSTRACT
Neonatal sepsis is an underrecognized burden on health care systems throughout the world. Antimicrobial drug resistance (AMR) is increasingly prevalent and compromises the use of currently recommended first-line agents. The development of new antimicrobial agents for neonates and children is mandated by regulatory agencies. However, there remains uncertainty about suitable development pathways, especially because of the propensity of premature babies to develop meningoencephalitis as a complication of neonatal sepsis and difficulties studying this disease in clinical settings. We developed a new platform and approach to accelerate the development of antimicrobial agents for neonatal bacterial meningoencephalitis using Pseudomonas aeruginosa as the challenge organism. We defined the pharmacodynamics of meropenem and tobramycin in these models. The percentage of partitioning of meropenem and tobramycin into the cerebrospinal fluid was comparable at 14.3 and 13.7%, respectively. Despite this similarity, there were striking differences in their pharmacodynamics. Meropenem resulted in bactericidal activity in both the cerebrospinal fluid and cerebrum, whereas tobramycin had minimal antibacterial activity. A hollow fiber infection model (HFIM) using neonatal CSF concentration time profiles yielded pharmacodynamics comparable to those observed in the rabbit model. These new experimental models can be used to estimate the pharmacodynamics of currently licensed agents and those in development and their potential efficacy for neonatal bacterial meningoencephalitis.
KEYWORDS: AMR, antimicrobial resistance, drug development, mathematical modeling, meningoencephalitis, meropenem, neonates, pharmacodynamics, pharmacokinetics, tobramycin
INTRODUCTION
Neonatal sepsis is a leading global cause of morbidity and mortality. Infections caused by drug-resistant pathogens are increasingly the norm and compromise therapeutic responses to first-line antimicrobial agents (1). Clinical studies to establish the safety and efficacy of new antimicrobial agents in neonates are notoriously difficult to perform (2). This leads to slow and often failed development of new antibiotics in this special population. New model systems, development pathways, and strategies are urgently required to provide clarity for all stakeholders, including parents, clinicians, sponsors, and regulatory authorities.
There have been concerted efforts to develop new antimicrobial agents to meet the challenge of antimicrobial drug resistance (AMR) via several “push and pull” incentives (3). Legislation in the United States and European Union requires sponsors developing new antimicrobial agents to design drug development programs for children and neonates at an early stage (4, 5). The principal strategy for antimicrobial drug development in children and adolescents is to design regimens that match the drug exposure that has been demonstrated to be effective in adults (6). Unfortunately, this approach may not be suitable for neonates because of important differences in the pathogenesis and clinical manifestations of sepsis in neonates compared with children and adults (7). A deep understanding of the adequacy (or otherwise) of an antimicrobial drug and candidate regimen to treat neonatal meningoencephalitis is critical for assessment of the clinical utility of a new drug/regimen for expanded use in neonates.
Here, we address these issues by developing and characterizing new experimental models that are mimics of neonatal meningoencephalitis and can be used for neonatal antimicrobial drug development. We modified and extended existing rabbit models of bacterial meningitis (8) to estimate the pharmacodynamics of antibacterial agents for neonatal meningoencephalitis. Furthermore, we developed a hollow fiber infection model using a microbiological medium that is similar to neonatal cerebrospinal fluid (CSF) and simulated the concentration-time profile within CSF. We quantified the pharmacodynamics of meropenem and tobramycin in these models and bridged the results to neonates. We considered the implications of these models and experimental results for neonatal drug development in an era of increasing antimicrobial resistance.
RESULTS
Challenge strain, MICs, and mutational frequency of resistance.
The challenge strain for all experiments was Pseudomonas aeruginosa ATCC 27853. The range of meropenem MICs estimated from five independently conducted experiments using broth microdilution with EUCAST methodology was 0.5 to 1 mg/L (mode, 0.5 mg/L) and 0.5 to 1 mg/L (mode, 1 mg/L) for meropenem and tobramycin, respectively. The frequency of mutants able to grow on drug-containing agar at 4 times the MIC was 2.4 × 10−7 and 1.3 × 10−6 for meropenem and tobramycin, respectively.
Development of an experimental rabbit model of neonatal bacterial meningoencephalitis.
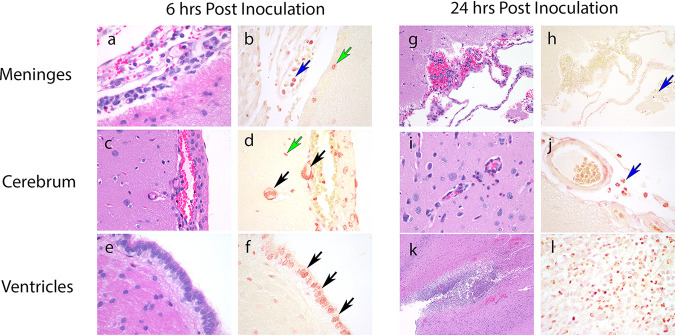
Our original intention was to develop an experimental model that enabled serial sampling of the CSF to define the pharmacokinetics and pharmacodynamics at the effect site. However, preliminary experiments showed this was not possible because repeated anesthesia was poorly tolerated, leading to unacceptable rates of mortality. Hence, a destructive design was employed where each rabbit contributed plasma samples and a single terminal set of observations from each subcompartment. Inoculation of Pseudomonas aeruginosa into the cistern of immunocompetent rabbits resulted in a rapidly progressive and fulminant meningoencephalitis that was universally lethal beyond 30-h. A florid meningoencephalitis was observed macroscopically with a rapid and progressive ventriculitis and meningoencephalitis evident on histopathological sections (Fig. 1). A Gram stain showed Gram-negative bacilli concentrated intracellularly within mononuclear cells. Extracellular bacilli were visible but difficult to distinguish from the brisk inflammatory infiltrate.
FIG 1.
Representative histopathological sections stained with hematoxylin and eosin and Gram stain obtained from untreated rabbits at 6 and 24 h postinoculation. (a and b) At 6 h postinoculation in the meninges there is minimal perivascular hemorrhage, minimal inflammation with intracellular bacteria seen within circulating monocytes (blue arrow), and subpial cerebral microglia (green arrow). (c and d) In the cerebrum there is minimal hemorrhage with no discernible inflammation. There are Gram-negative bacilli in the microglia in the subpial cerebral parenchyma (green arrows) and within blood vessel walls (black arrows). (e and f) In the ventricles there are multiple Gram-negative bacilli within the ependymal lining cells (black arrows). (g and h) At 24 h in the meninges there is mild perivascular hemorrhage, mild heterophilic inflammation and Gram-negative bacilli within circulating and perivascular monocytes (blue arrows). (i and j) There are similar findings in the cerebrum. (k and l) In the ventricles there is acute periventricular hemorrhage, inflammation, and abscess formation with intracellular Gram-negative bacilli visible within intravascular monocycles, microglia, and ependyma. Original magnifications: ×60 (a, b, d, e, f, j, l), ×40 (c and i), ×20 (g and h), ×4 (k).
A delay of 6 h in the initiation of antimicrobial therapy enabled 24 h of antimicrobial therapy to be administered before the onset of mortality in vehicle-treated controls. Infection was well established at this time point (Fig. 1). These well-characterized experimental conditions provided a clinically relevant model system (i.e., severe rapidly lethal infection) but also enabled sufficient time for the elucidation of the pharmacodynamics of meropenem and tobramycin. The regimens of both meropenem and tobramycin resulted in clinically relevant drug exposures (quantified in terms of the area under the curve [AUC]) resulting from currently recommended neonatal regimens.
Pharmacokinetics of meropenem and tobramycin in the central nervous system.
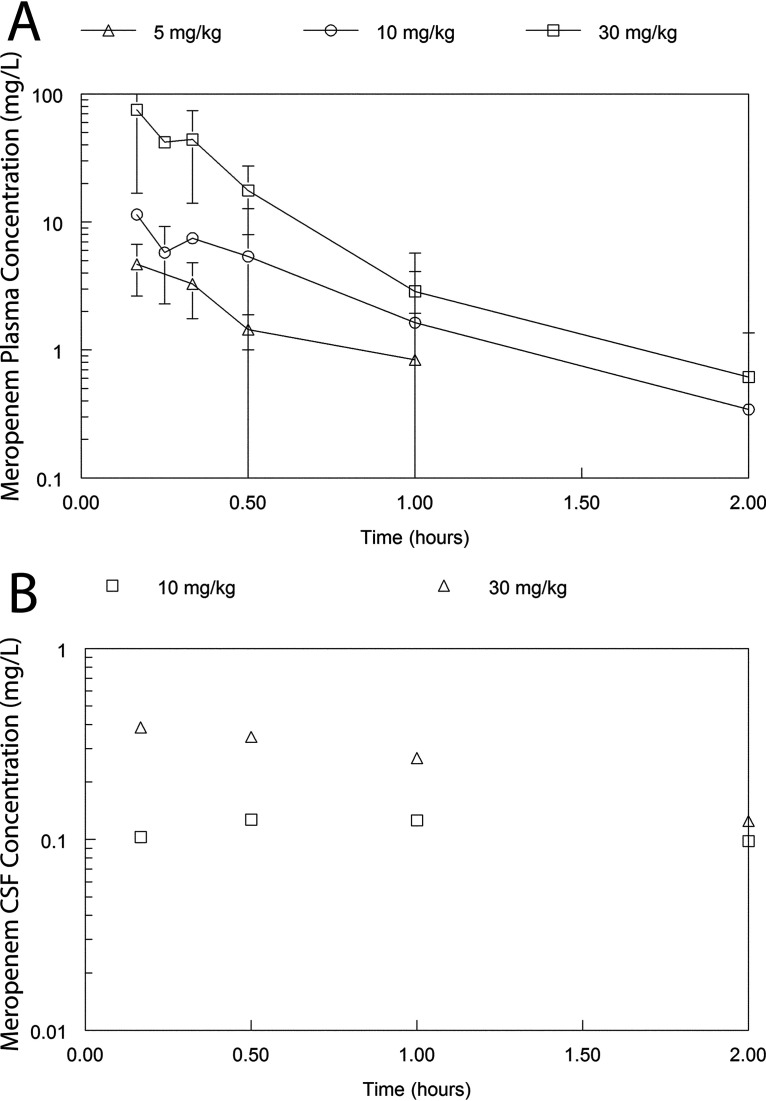
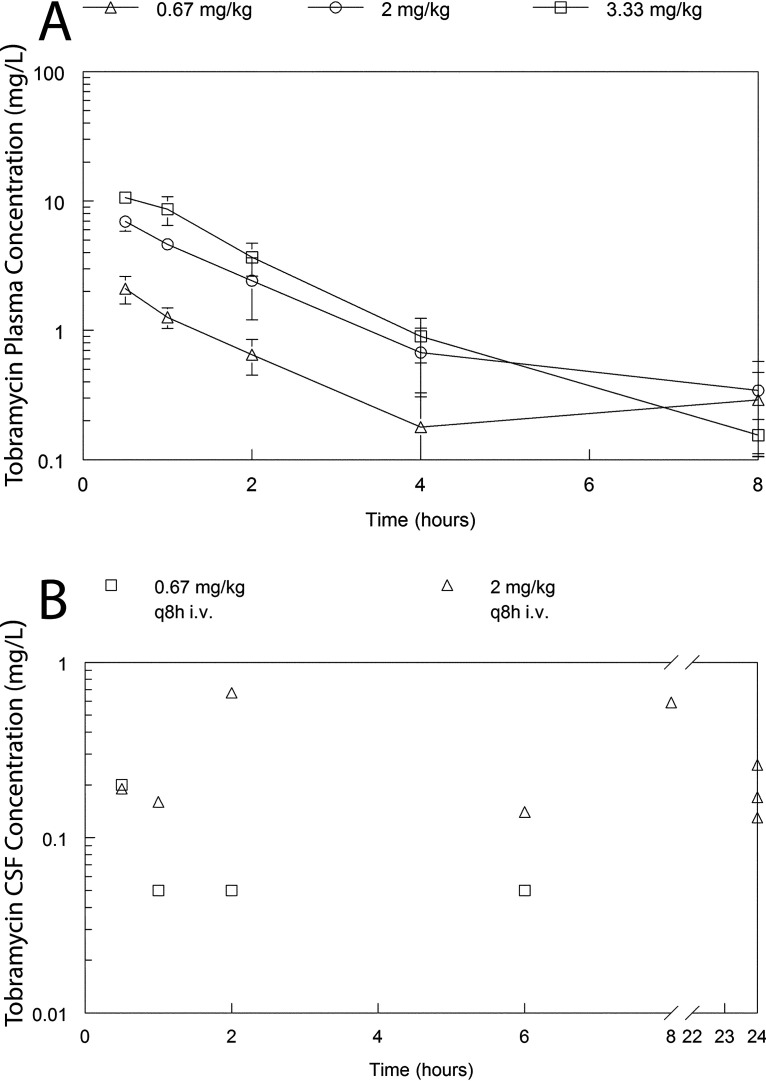
A satellite pharmacokinetics (PK) study was used to define partitioning of meropenem and tobramycin into the CSF (Fig. 2 and 3). The PK of meropenem at 5, 10, and 30 mg/kg body weight every 8 h (q8h) administered intravenously (i.v.) was well described using a linear structural PK model (see “Pharmacokinetic-Pharmacodynamic Modeling” for details). The parameter estimates for the population PK model fitted to the PK data for meropenem and tobramycin are summarized in Tables 1 and 2, respectively. Meropenem readily partitioned into the cerebrum and CSF with an AUCCSF:AUCplasma of 14.3% and was calculated from the Bayesian posterior estimates of those rabbits that had a CSF sample measured. Similarly, the partition ratio of tobramycin was 13.7%. There was a degree of hysteresis for both drugs, with the concentration-time profile in the CSF being much flatter than that observed in plasma. These estimates reflect an estimate obtained from the combined data set from the duration of the study and an assumption that the estimates of drug transfer coefficients are invariant (the rate and extent of drug partitioning may conceivably vary with time as the degree of inflammation changes).
FIG 2.
Concentration-time profiles of meropenem in the plasma and CSF of rabbits receiving 5, 10, and 30 mg/kg as a single dose. Panel A shows the plasma PK from all rabbits (n = 36). Panel B (n = 8) shows the CSF concentrations in a dedicated satellite experiment to estimate the percentage partitioning of meropenem from plasma into the CSF. In panel A, the data are the mean ± the standard deviation. In panel B, each observation is obtained from a single rabbit.
FIG 3.
Concentration-time profiles of tobramycin in the plasma and CSF of rabbits receiving 0.67, 2, and 3.33 mg/kg as a single dose. Panel A shows the plasma PK from all rabbits (n = 36). Panel B shows the CSF concentrations from a dedicated satellite experiment to estimate the percentage of partitioning of tobramycin from plasma into the CSF. In Panel A, the data are the mean ± the standard deviation. In Panel B, each data point represents an observation from a single animal (n = 12).
TABLE 1.
Population PK-PD parameters for meropenema
| Parameterb (units) | Mean | Median | SD |
|---|---|---|---|
| Clearance (L/h) | 3.409 | 3.388 | 0.919 |
| Vol (L) | 1.212 | 1.045 | 0.811 |
| K12 (h−1) | 3.059 | 1.694 | 3.716 |
| K21 (h−1) | 3.347 | 3.113 | 4.469 |
| K13 (h−1) | 0.076 | 0.082 | 0.026 |
| K31 (h−1) | 0.728 | 0.738 | 0.283 |
| Vol CSF (L) | 2.577 | 2.123 | 1.565 |
| Kgmax (log10 CFU/mL/h) | 0.387 | 0.284 | 0.307 |
| popmax (CFU/mL) | 3,352,281 | 144,355 | 4,406,617 |
| kkmax (log10 CFU/mL/h) | 0.859 | 0.860 | 0.117 |
| Hk | 8.866 | 4.517 | 6.678 |
| C50k (mg/L) | 0.075 | 0.079 | 0.021 |
| Initial condition (CFU/mL) | 721 | 739 | 271 |
The population PK-PD parameters for meropenem are from the rabbit model.
Parameters are defined in Materials and Methods.
TABLE 2.
Population PK-PD parameters for tobramycina
| Parametera (units) | Mean | Median | SD |
|---|---|---|---|
| Clearance (L/h) | 0.384 | 0.396 | 0.103 |
| Vol (L) | 0.477 | 0.409 | 0.185 |
| K12 (h−1) | 5.339 | 1.063 | 6.780 |
| K21 (h−1) | 17.882 | 20.726 | 11.632 |
| K13 (h−1) | 0.088 | 0.100 | 0.017 |
| K31 (h−1) | 0.167 | 0.129 | 0.174 |
| Vol CSF (L) | 1.627 | 1.169 | 1.128 |
| Kgmax (log10 CFU/mL/h) | 0.631 | 0.625 | 0.074 |
| popmax (CFU/mL) | 6,841,660.084 | 117,373.215 | 1,2054,144.467 |
| kkmax (log10 CFU/mL/h) | 0.598 | 0.682 | 0.258 |
| Hk | 6.955 | 1.447 | 7.848 |
| C50k (mg/L) | 0.070 | 0.083 | 0.029 |
| Initial condition (CFU/mL) | 252.286 | 140.687 | 202.572 |
The population PK-PD parameters for tobramycin are from the rabbit model.
Parameters are defined in Methods.
Pharmacodynamics of meropenem and tobramycin.
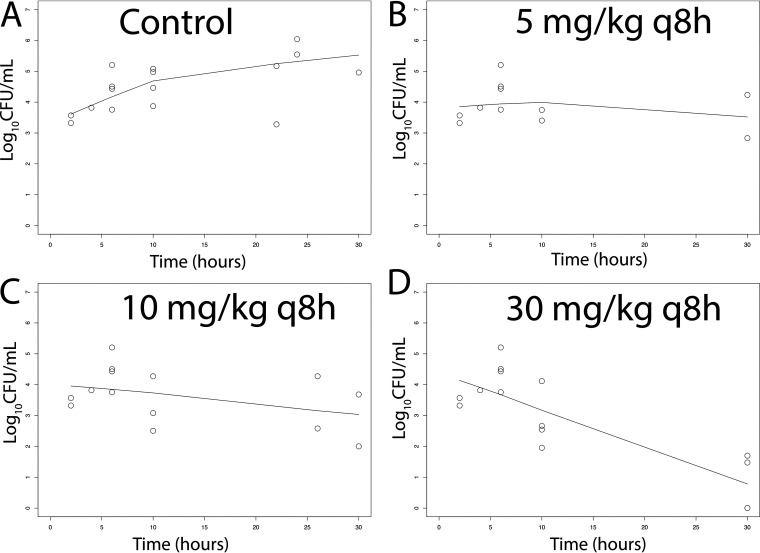
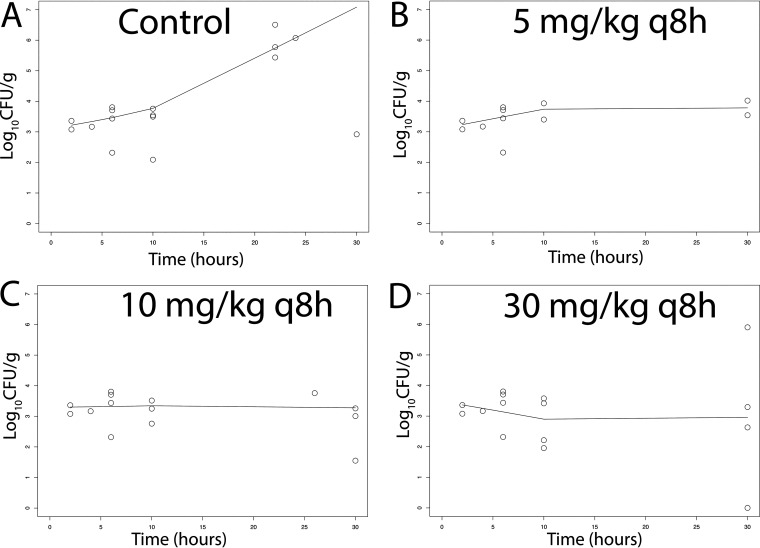
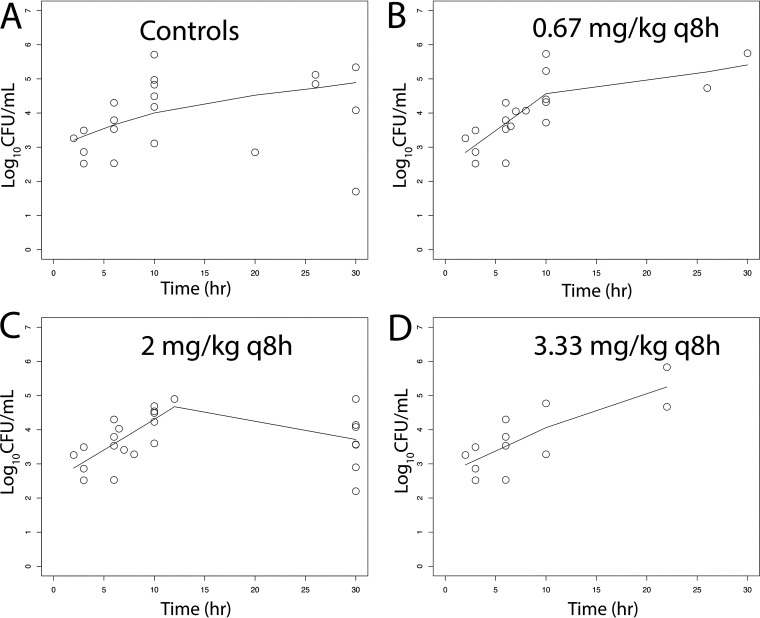
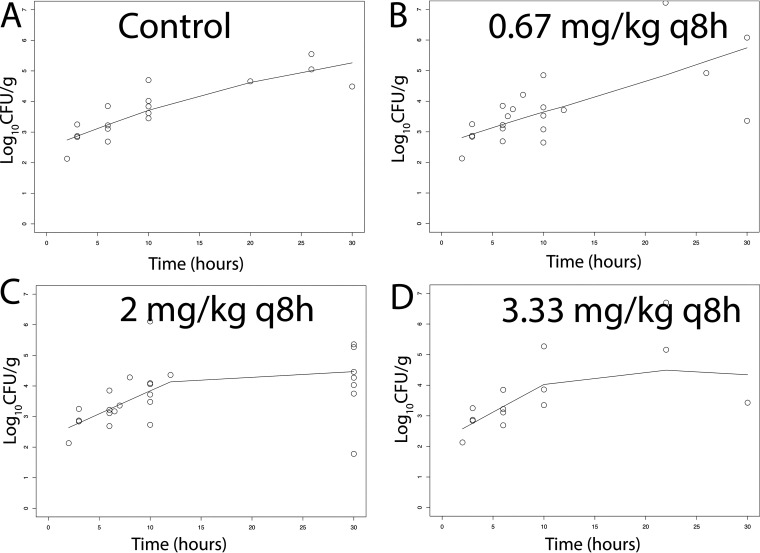
There was a dose-dependent response of meropenem at 5 to 30 mg/kg q8h i.v. in terms of the antibacterial activity in both CSF and the cerebrum. A total of 51 rabbits and multiple experiments were used to define these relationships (including controls) (Fig. 4 and 5). Sterilization was not achieved in any rabbits, although a regimen of 30 mg/kg q8h i.v. induced a progressive decline in bacterial burden in both the CSF and cerebrum throughout the experimental period. The pharmacodynamics in the cerebrum and CSF were comparable. In contrast, there was no obvious dose-exposure-response relationship for tobramycin (n = 55; multiple experiments) despite clinically relevant drug exposures in both plasma and CSF being achieved and fractional partitioning into CSF comparable to that observed with meropenem (Fig. 6 and 7).
FIG 4.
(A–D) Pharmacodynamics of meropenem at 5, 10, and 30 mg/kg q8h i.v. as assessed using the bacterial burden in the CSF of rabbits infected with P. aeruginosa ATCC 27853. Treatment was initiated 6 h postinoculation at time = 0. The solid black lines in each panel are locally weighted scatterplot smoothing (LOWESS) regression lines fitted to the pooled data from each cohort of rabbits.
FIG 5.
(A–D) Pharmacodynamics of meropenem at 5, 10, and 30 mg/kg q8h i.v. as assessed using the bacterial burden in the cerebrum of rabbits infected with P. aeruginosa ATCC 27853. Treatment was initiated 6 h postinoculation at time = 0. The solid black lines in each panel are LOWESS regression lines fitted to the pooled data from each cohort of rabbits.
FIG 6.
(A–D) Pharmacodynamics of tobramycin at 0.67, 2, and 3.33 mg/kg q8h i.v. as assessed using the bacterial burden in the CSF of rabbits infected with P. aeruginosa ATCC 27853. Treatment was initiated 6 h postinoculation at time = 0. The solid black lines in each panel are LOWESS regression lines fitted to the pooled data from each cohort of rabbits. There was no obvious dose exposure response relationship.
FIG 7.
(A–D) Pharmacodynamics of tobramycin at 0.67, 2, and 3.33 mg/kg q8h i.v. as assessed using the bacterial burden in the cerebrum of rabbits infected with P. aeruginosa ATCC 27853. Treatment was initiated 6 h postinoculation at time = 0. The solid black lines in each panel are LOWESS regression lines fitted to the pooled data from each cohort of rabbits. There was no obvious dose exposure response relationship.
Pharmacokinetic-pharmacodynamic modeling.
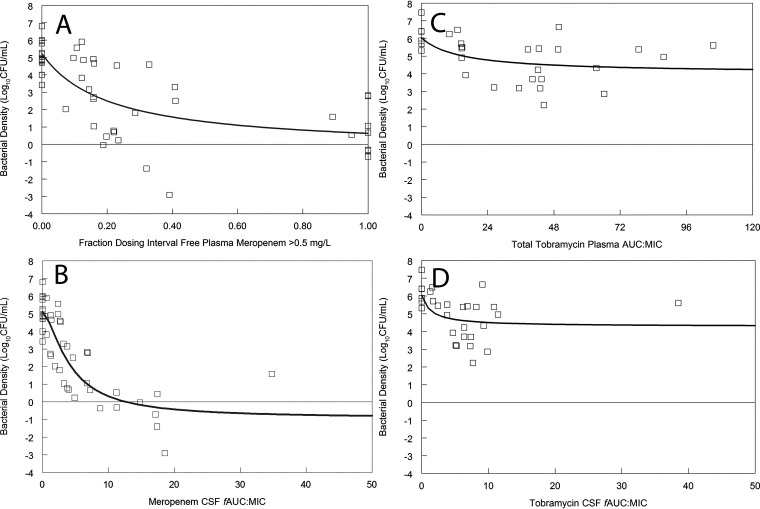
A PK-pharmacodynamic (PD) mathematical model was used to estimate the bacterial burden at 30 h postinoculation (rabbits had been serially sacrificed to define the temporal changes of drug in plasma and CNS along with changes in bacterial burden with time). Several integrative measures of drug exposure were explored, including fT > MIC in plasma and AUC:MIC in both plasma and CSF. We did not use fT > MIC in CSF for meropenem because concentrations were never greater than the MIC of 0.5 mg/L. A regression of various measures of drug exposure and the antibacterial effect is shown in Fig. 8.
FIG 8.
(A to D) The pharmacodynamics of meropenem (A and B) and tobramycin (C and D) in the rabbit model of pseudomonal meningoencephalitis. For meropenem, the pharmacodynamics are quantified using systemic drug exposure (i.e., fT > MIC in plasma; panel A) and drug exposure within the CSF (i.e., AUC:MIC; panel B). For tobramycin, systemic and effect site drug exposures are shown in panels C and D, respectively. Meropenem exhibits significant bactericidal activity in CSF. In contrast, the antibacterial effect of tobramycin is minimal.
Hollow fiber infection model.
Preliminary experiments using Mueller-Hinton broth medium revealed pharmacodynamic effects that were not consistent with broad clinical experiences with meropenem. Bacterial killing was only observed with CSF concentrations (modeled in the HFIM) of 10 mg/L, which is an order of magnitude higher than concentrations achieved in human neonatal CSF (circa0.5 to 1 mg/L) and in the rabbit model of meningoencephalitis. Hence, we used artificial CSF as a mimic of human CSF (9) (Fig. 9).
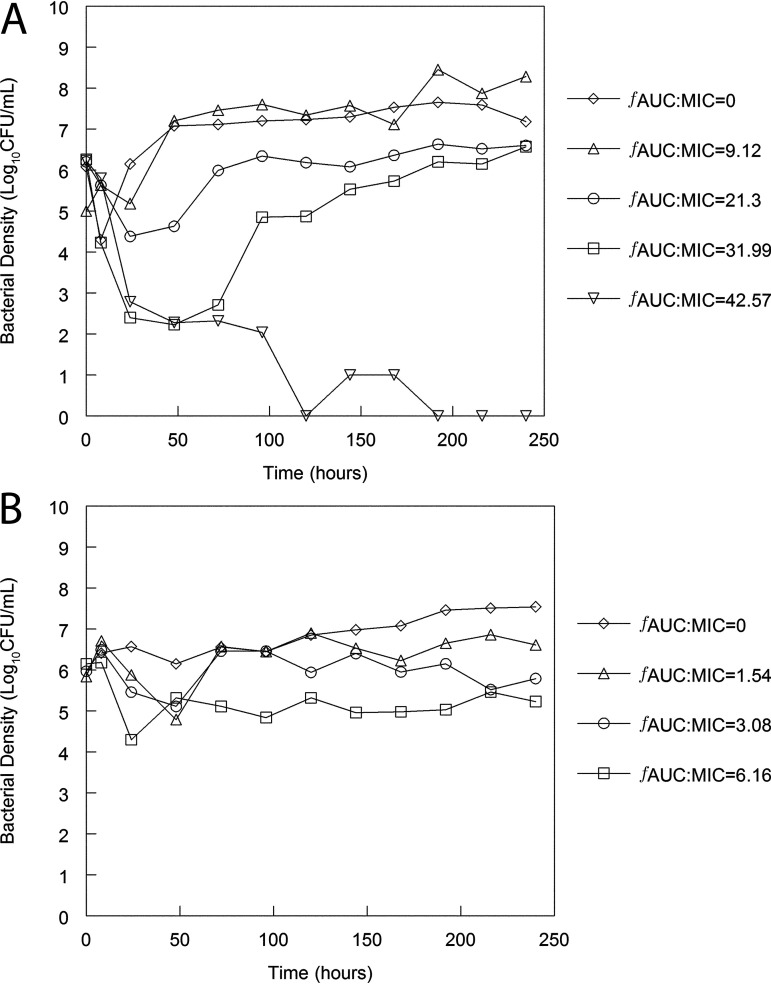
FIG 9.
Pharmacodynamics of meropenem and tobramycin in a hollow fiber infection model with P. aeruginosa ATCC 27853 as the challenge strain. Artificial CSF (aCSF) medium was used as the medium. (A and B) Neonatal CSF concentration time profiles of meropenem (A) and tobramycin (B) were used, and the AUC:MIC values quantify drug exposure in the CSF. An exposure response relationship is evident for meropenem. In contrast, bacterial killing induced by tobramycin is minimal.
Human neonatal-like concentration-time profiles for meropenem and tobramycin were recapitulated in the HFIM. For meropenem, an AUC:MIC of 42.57 was required to achieve sustained bacterial killing. For tobramycin, an AUC:MIC of 6.16 resulted in a reduced bacterial density of only 1 to 2 log10CFU/mL compared with controls. The pharmacodynamics of both meropenem and tobramycin was well aligned with that of the rabbit model of meningoencephalitis shown in Fig. 8.
DISCUSSION
The development of new antimicrobial agents for neonates is mandated by regulatory processes designed to provide new treatment options to a special population with otherwise limited therapeutic options. However, unlike those for adults, the development pathways remain poorly defined. Differences in the pharmacokinetics related to size (allometry) and development of function (ontogeny) are relatively well understood and can be used to scale regimens to match plasma drug exposures that have been demonstrated to be effective in adults (6). However, the pharmacodynamics of a drug-pathogen combination in an adult and neonate are not necessarily comparable because the pathogenesis and disease manifestations are different. Most importantly, neonatal bloodstream infection (regardless of source) may result in meningoencephalitis because of an immature blood-brain barrier, and innate and adaptive immunological defenses are immature (10). Hence, an understanding of the likelihood of a given drug and regimen being effective against both bloodstream and central nervous system infection is paramount.
While it may be tempting to simply conceive the whole problem of antimicrobial drug development and regimen identification for neonatal sepsis as one based on partitioning of drug from plasma into the CSF, this is not necessarily correct. At best, such an assessment is of limited relevance and at worst may be completely misleading. The current study demonstrates that antimicrobial agents may partition into the CSF, but this does not in any way guarantee clinically useful antimicrobial activity in that compartment. In this regard, our results suggest tobramycin should not be used as a monotherapy for meningoencephalitis. The potential utility of an antimicrobial agent for neonatal meningoencephalitis should be initially focused on a pharmacodynamic assessment of the drug-pathogen combination in the central nervous system with subsequent PK-PD bridging studies to identify a potential regimen for premature neonates.
The design of clinical studies for the treatment of neonatal sepsis is possible using primary endpoints such as all-cause 28-day mortality. However, the clinical assessment of a new antimicrobial drug/regimen for the treatment of neonatal meningoencephalitis is more difficult for several reasons. First, there is often significant diagnostic uncertainty for CNS infection with an inability to enroll babies with suspected CNS infection into clinical studies for licensing; second, neonatal meningoencephalitis is associated with longer-term neurodevelopmental abnormalities that manifest well after the inciting event, leading to difficulties linking drug exposure with the ultimate clinical response; and finally, there are significant practical obstacles for enrolling babies with meningoencephalitis into clinical trials powered for inferential testing. For example, a recent study of micafungin versus amphotericin B deoxycholate was terminated early because of slow enrollment (2). These limitations have led the U.S. FDA to consider using laboratory animal data and PK-PD analyses to guide therapeutic decisions in those circumstances where clinical data are unobtainable (7). The FDA has also included conclusions from laboratory animal models in the micafungin license to help guide clinical decision making (11).
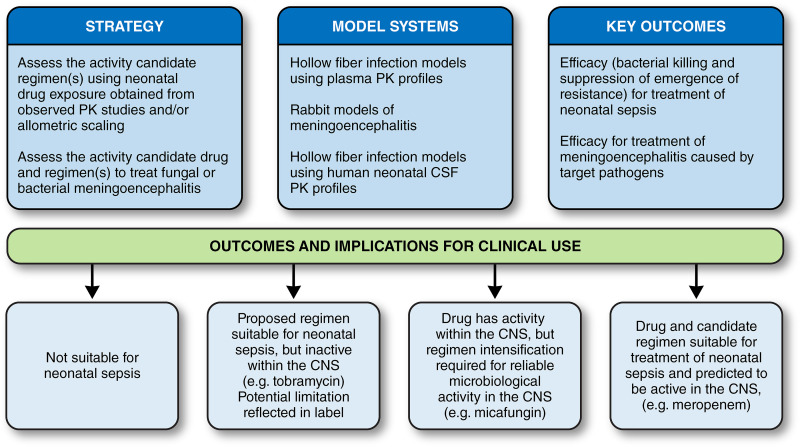
A suite of laboratory models is now available to identify new antimicrobial regimens for neonatal sepsis (10, 12–14). These consist of a series of rabbit models of neonatal sepsis and meningoencephalitis and hollow fiber infection models that enable plasma and CSF concentration time profiles to be simulated. These model systems have strengths and limitations and should be viewed as complementary rather than hierarchical. The principal advantage of the rabbit models is that they are mimics of clinical disease and have anatomical barriers that are ultimately critical determinants of the therapeutic efficacy and pharmacodynamics of any antimicrobial agent; their principal limitation is that they require extensive infrastructure and highly trained personnel to ensure that studies are conducted safely and efficiently. The rabbit model enables infection to be established in the meninges/CSF and cerebrum; these subcompartments are anatomically and pharmacologically distinct and likely to differentially contribute to the clinical manifestations of neonatal meningoencephalitis where both the meninges and cerebrum are involved. HFIM enable human PK (at any effect site) to be simulated and are especially valuable to study the pharmacodynamics of the emergence of resistance because of the ability to use a high-density inoculum. The absence of immunological effectors, the potential binding of drug to plastic, and the inability to replicate multiple clinically relevant anatomic subcompartments are potential limitations. The combination of model systems enables the pharmacodynamics of candidate regimens to be examined from a variety of perspectives, leading to a conclusion as to the likely overall efficacy for treatment of neonatal sepsis (Fig. 9).
How can data from these new model systems be integrated? The use of current models and PK-PD bridging pathways (Fig. 10) enables the utility of old and new antimicrobials for neonatal sepsis to be classified in one of four ways: (i) the drug/regimen is predicted to not be appropriate for neonates (i.e., the regimen causes submaximal antimicrobial effect and/or leads to the rapid emergence of resistance); (ii) the drug/regimen is potentially effective for bloodstream infection but is not active within the CNS (e.g., tobramycin); (iii) the drug/regimen is potentially effective for bloodstream infection and is active within the CNS but only with regimen intensification (i.e., an increased dose and/or altered schedule of administration to optimize the pharmacodynamic target) if that is possible (e.g., micafungin, anidulafungin [7, 10, 12]); and (iv) the drug/regimen is effective for both bloodstream and CNS infection at a standard regimen (e.g., meropenem). Such a classification provides guidance to drug developers, regulatory authorities, and clinicians using a new antibiotic for the treatment of neonatal sepsis.
FIG 10.
Overall strategy model and systems for the development of antimicrobial agents for neonates.
The principal limitation of this study is the requirement to use a destructive design with a relatively limited number of rabbits; while this was the only feasible experimental approach, it does mean strong experimental design is paramount to maximize information and minimize potential bias. More work is required to increase the number of model systems available to develop new antimicrobials. This may include models with other pathogens that cause neonatal sepsis (e.g., Escherichia coli, Klebsiella spp., Acinetobacter baumannii) as well as organisms with resistance mechanisms that are increasingly prevalent throughout the world (e.g., KPC-, OXA-, and NDM-producing pathogens). The performance of the current standard-of-care (e.g., agents recommended by the WHO or similar organizations) can be quantified in these systems and used to benchmark the performance of new antimicrobial agents for neonatal sepsis. There is an ability to study alternative approaches such as combination therapy. Despite the limitations and the relative complexities of our approach, we describe a viable path for the development of new agents for a special population that urgently requires new treatment options but for which well-established clinical development pathways currently do not exist.
MATERIALS AND METHODS
Organism, antimicrobial susceptibility testing, and mutational frequency.
Pseudomonas aeruginosa ATCC 27853 was used as the challenge strain throughout. The MICs of meropenem and tobramycin were estimated using European Committee on Antimicrobial Susceptibility Testing (EUCAST) methodology (15) in five independently conducted experiments. The modal value used for the pharmacodynamic calculations. Pure meropenem and tobramycin (Sigma-Aldrich, UK) were used for all in vitro susceptibility testing.
The mutational frequency of resistance was determined by suspending a sweep of approximately 5 colonies grown overnight in 30 mL Mueller-Hinton broth (Sigma-Aldrich, UK), which was then placed on an orbital shaker at 37°C for 24 h. At 24 h, a 1:1,000 dilution was performed from each inoculum into fresh Mueller-Hinton broth and placed back onto the orbital shaker at 37°C for a further 24 h. Each inoculum was then plated onto drug-free agar and either meropenem- or tobramycin-containing Mueller-Hinton agar (Sigma-Aldrich) at concentrations of 3 or 4 mg/L, respectively, and incubated at 37°C for 24 to 48 h. The mutational frequency was then calculated as the ratio of colonies that grew on drug-containing agar divided by the number of colonies that grew on drug-free Mueller-Hinton agar plates (Oxoid, UK).
Rabbit model of neonatal meningoencephalitis.
All laboratory animal experiments were conducted under UK Home Office project license PAC022930 and approved by the University of Liverpool Animal Welfare Ethics Review Board. A nonneutropenic rabbit model of neonatal meningoencephalitis was developed. Male New Zealand white (NZW) rabbits (Envigo, UK) weighing 2.08 to 3.78 kg at the time of experimentation were used for all experiments and used to characterize dose-exposure-response relationships of meropenem and tobramycin.
A target inoculum of 1 × 105 CFU/mL Pseudomonas aeruginosa ATCC 27853 suspended in phosphate-buffered saline (PBS) was prepared and verified with quantitative culture. Rabbits were inoculated while anesthetized (medetomidine [Vetoquinol, UK], 0.2 mg/kg intramuscularly [i.m.], plus ketamine [Chanelle Pharma, UK], 15 mg/kg i.m.) by injecting 300 μL of the 1 × 105 CFU/mL suspension intrathecally. Animals were serially sacrificed at planned time points unless they exhibited prespecified humane endpoints that mandated immediate sacrifice to minimize suffering.
Postsacrifice, tissue samples were collected for PK-PD analysis. Brain tissue was removed from the skull and divided into quarters. One-quarter was placed in formalin for histopathological analysis, one-quarter was snap-frozen in liquid nitrogen for matrix-assisted laser desorption ionization (MALDI) imaging (analysis pending), one-quarter was placed into sealed labeled bags for PK bioanalysis, and one-quarter was placed into culture tubes with 2 mL PBS before being weighed and homogenized. Brain homogenate was serially diluted with 100 μL plated onto Mueller-Hinton agar and incubated overnight at 37°C before estimating CFU/g brain tissue at each of the time points.
Histopathology.
The progression of infection at a microscopic level was assessed by placing the right cerebral hemisphere from each rabbit in 10% neutral buffered saline. Tissues were embedded in paraffin and sectioned using standard protocols. Sections were stained with hematoxylin and eosin and Gram stain with employment of uninfected rabbits as controls.
In vivo PK-PD studies.
Treatment with meropenem (Venus Pharma GmbH, Germany) or tobramycin (Flynn Pharma, Ireland) was initiated 6 h postinoculation. Meropenem dosages of 5, 10, and 30 mg/kg/dose or tobramycin dosages of 0.67, 2, and 3.33 mg/kg were administered q8h as an i.v. push via the marginal ear vein. Blood samples of approximately 0.5 mL were taken from the opposite ear of each rabbit for PK analyses. Samples were collected predose (0 h) and at 10 min, 15 min, 20 min, 30 min, 1 h, 2 h, and 4 h or 30 min, 1 h, 2 h, 4 h, and 8 h for meropenem and tobramycin, respectively. Whole blood was placed into Eppendorf tubes containing approximately 10 μL heparin (Wockhardt, UK) and centrifuged. Plasma was eluted and stored at –80°C for a maximum of 9 months subsequent to bioanalysis.
A specific satellite pharmacokinetic study to estimate the percentage of partitioning of meropenem and tobramycin was conducted. This study was performed in infected rabbits using the same experimental design as the other pharmacokinetic and pharmacodynamic studies. Rabbits received either 10 (n = 4) or 30 mg/kg/dose (n = 8) meropenem or 0.67 (n = 4) or 2 mg/kg/dose (n = 8) tobramycin. One animal was sacrificed for each dose time point, and a blood sample and CSF sample were collected for PK and PD analysis. CSF samples were collected 10 min, 30 min, 1 h, and 2 h postdose for meropenem and 30 min, 1 h, 2 h, and 6 h postdose for tobramycin.
Hollow fiber infection model of neonatal meningitis.
A hollow fiber infection model (HFIM) was used, simulating the typical neonatal PK profile. The PD response and emergence of resistance of P. aeruginosa ATCC 27853 to a range using human neonatal in CSF concentration-time profiles were determined. An inoculum of 1 × 105 CFU was prepared and injected into the extra capillary space of each hollow fiber cartridge. Preliminary studies were performed with Mueller-Hinton medium, but bacterial killing was not observed with concentration time profiles corresponding to the currently recommended neonatal meropenem regimen of 20 mg/kg q8h. We therefore used artificial CSF (aCSF [9]), which is a mimic of human CSF. The HFIM was incubated in ambient air at 37°C. Serial samples were obtained for pharmacokinetic and pharmacodynamic analyses.
Neonatal CSF concentration time profiles were obtained from the NeoMero study (16) and a small study (n = 6 infants) led by Duke University (data not shown). A range of concentrations for both drugs was studied with a target Cmax for meropenem and tobramycin ranging from 0.1 to 1 mg/L and 0.03 to 0.3 mg/L, respectively. The half-life for meropenem and tobramycin was 6 h and 6.54 h, respectively. Both agents were administered immediately postinoculation. The experimental duration was 10 days. Experiments were conducted once for each drug. Pharmacodynamic samples (approximately 1 mL) were taken from each cartridge at 0 h (predose) and regularly thereafter and plated onto Mueller-Hinton agar.
Bioanalytical assay for meropenem.
Samples of rabbit plasma and rabbit CSF were prepared using solid-phase extraction. An internal standard ([2H6] meropenem [Alsachim, France]) in acetonitrile (5 mg/L; Fisher Scientific UK), of which 250 μL was added to a 96-well protein precipitation plate (Phenomenex, Cheshire, UK). Then, 50 μL of sample, blanks, and calibrators in the range of 0.1 to 50 mg/L and quality controls (0.75, 7.5, and 37.5 mg/L) was mixed with the internal standard, and the plate was placed on an orbital shaker for 2 min. Liquid was drawn through the protein precipitation plate into a collection plate using a positive pressure manifold. Water and 0.1% formic acid (750 μL) were added to each well. The plate was sealed and placed onto an orbital shaker for 5 min before being transferred to the autosampler for analysis by liquid chromatography-tandem mass spectrometry (LC-MS-MS). A similar approach was used for aCSF with adjustment for the lower dynamic range.
LC-MS-MS analysis was performed using an Agilent 1290 Infinity high-performance liquid chromatography (HPLC) instrument coupled to an Agilent 6420 triple quadrupole mass spectrometer fitted with an electrospray source. The LC-MS system was controlled using Agilent MassHunter data acquisition software (ver. B.06.00). Analytes were injected (2 μL) onto a Phenomenex Kinetex C18 100 Å column (2.1 mm by 50 mm, 2.6 μm, 40°C) and separated over a 5.5-min. gradient using a mixture of solvents A and B. Solvent A was LC-MS-grade water with 0.1% (vol/vol) formic acid. Solvent B was HPLC-grade acetonitrile with 0.1% (vol/vol) formic acid. Separations were performed by applying a linear gradient of 2% to 98% solvent B over 4 min at 0.6 mL/min followed by an equilibration step (1.5 min at 2% solvent B).
The mass spectrometer was operated in positive ion mode using a multiple-reaction monitoring (MRM) method. Following an optimization process, the following mass transitions and collision energies (Ce) were used for the analysis: 384.16 > 141.2 (Ce 12 eV) and 390.2 > 147.1 (Ce 16 eV). The mass spectrometer conditions were as follows: capillary voltage of 3.5 kV, fragmentor voltage of 100 V, source gas temperature of 350°C, and gas flow of 11 L/min. The resultant data were processed using Agilent Mass Hunter quantitative analysis (ver. B.05.02).
The lower limit of quantification (LLQ) was 0.1 and 0.01 mg/L for rabbit plasma and aCSF, respectively. The inter- and intraday %CV was <15% and the analyte was stable under all conditions described above.
Bioanalytical assays for tobramycin.
Tobramycin (Alsachim, France) was extracted from rabbit plasma and rabbit CSF using protein precipitation. The internal standard, [13C6] tobramycin (Alsachim, France) was prepared in acetonitrile with 0.1% trifluoroacetic acid (25 mg/L; Fisher Scientific UK). Samples, blanks, and calibrators were added and vortexed for 2 min. Subsequently, water with 0.1% (vol/vol) trifluoroacetic acid (200 μL) was added to each sample. Samples were vortexed for 30 s before being centrifuged for 5 min. Supernatant (150 μL) was added to a 96-well plate before being transferred to the autosampler for analysis by LC-MS-MS. Tobramycin-containing samples from aCSF were prepared using solid-phase extraction.
LC-MS-MS analysis was carried out using an Agilent 1290 Infinity HPLC coupled to an Agilent 6420 triple quadrupole mass spectrometer fitted with an electrospray source. The LC-MS system was controlled using Agilent MassHunter data acquisition software (ver. B.06.00). Analytes were injected (5 μL) onto a Waters HSS T3 100-Å device (2.1 mm by 100 mm, 1.8 μm, 40°C) and separated over a 3.5-min. gradient using a mixture of solvents A and B. Solvent A was LC-MS-grade water with 0.1% (vol/vol) trifluoroacetic acid. Solvent B was LC-MS-grade methanol with 0.1% (vol/vol) trifluoroacetic acid. Separations were performed by applying a linear gradient of 5% to 95% solvent B over 2 min at 0.2 mL/min followed by an equilibration step (1.5 min at 5% solvent B).
The mass spectrometer was operated in positive ion mode using a multiple reaction monitoring (MRM) method. Following an optimization process, the following mass transitions and collision energies were used for the analysis: 468.2 > 324.2 (Ce 10 eV) and 474.2 > 330.3 (Ce 10 eV). The mass spectrometer conditions were as follows: capillary voltage of 3.5 kV, fragmentor voltage of 100 V, source gas temperature of 350°C, and gas flow of 11 L/min. The resultant data were processed using Agilent Mass Hunter quantitative analysis (ver. B.05.02). The limit of assay quantification was 0.1 mg/L with acceptable assay variance.
PK-PD mathematical modeling.
A PK-PD model was fitted to the PK and PD data in a stepwise process using the nonparametric population PK program Pmetrics (17). The PK was initially solved for all rabbits before proceeding to the pharmacodynamic fitting. This was necessary to ensure that a stable solution could be obtained given imbalance in design (i.e., some rabbits had a complete PK data set but only a single terminal estimate of bacterial density in the CSF). A mathematical model was only fitted to the pharmacodynamic data from the CSF.
A three-compartment pharmacokinetic model was fitted to the plasma and CSF data. The structural model took the form:
| (1) |
| (2) |
| (3) |
With the output equations
| (4) |
| (5) |
where XP(1), XP(2), and XP(3) represent the rate of change of mass (mg) of the respective drug in compartments 1, 2, and 3, which represent the central (i.e., bloodstream), peripheral (i.e., rest of the body), and CSF, respectively. RATEIV(1) is the time-delimited zero-order i.v. injection of meropenem or tobramycin into the lateral ear vein; CL is the first-order clearance of drug from the central compartment; V is the volume of the central compartment, and K12, K21, K13, K31 represent the various intercompartmental first-order rate constants. The second output equation contains a term for the volume of the CSF (V_csf).
The fit of the model to the data was assessed by a visual inspection and linear regression of the observed-predicted values before and after the Bayesian step. The Bayesian estimates for each rabbit were resupplied in the fitting of the pharmacodynamic model to the pharmacodynamic data from the CSF (i.e., they were fixed for each rabbit).
The pharmacodynamic structural model took the form
| (6a) |
| (6b) |
With a single output equation,
| (7) |
where XP(4) is the rate of change of bacterial density (CFU/mL) in the CSF, Kgmax is the maximum rate of bacterial growth in the CSF, popmax is the theoretical maximum bacterial density in the CSF, kkmax is the maximum rate of drug-induced bacterial killing, and Hk is the slope function. C50k is the CSF drug concentration where the rate of bacterial killing is half maximal. The bacterial density in CSF immediately postinoculation (initial condition) was estimated as a parameter but is not shown in the differential equations. Drug concentrations in the CSF represented by X(3)/V_csf are explicitly linked to the antibacterial effect. The pharmacodynamics represents a balance between capacity-limited bacterial growth (equation 6a) and drug-induced bacterial killing (equation 6b).
The combined Bayesian posterior estimates for the PK and PD parameters from each rabbit were obtained. The SIM module in ADAPT 5 (18) was used to estimate drug exposure in plasma and CSF as well as several pharmacodynamic endpoints that were of potential relevance. These included the area under the log10 CFU/mL versus time curve and the predicted bacterial density at the end of the experiment (note that this was not necessarily matched by an observation because of the destructive design).
ACKNOWLEDGMENTS
This work was funded by the U.S. Food and Drug Administration (FDA), contract number HHSF223201610082C.
William Hope holds or has recently held research grants with UKRI, National Institutes of Health, National Institute of Health Research, F2G, Spero Therapeutics, Antabio, Pfizer, Bugworks, Phico Therapeutics, BioVersys, GARDP, and NAEJA-RGM. He is (or has recently been) a consultant for Appili Therapeutics, F2G, Spero Therapeutics, NAEJA-RGM, Centauri, Pfizer, Phico Therapeutics, and VenatoRx. He is a member of the Specialist Advisory Committee for GARDP and is the specialty national colead for infectious diseases for the National Institute of Health Research (NIHR). The authors thank Patrick Lane for preparing Figure 10.
REFERENCES
- 1.Thomson KM, Dyer C, Liu F, Sands K, Portal E, Carvalho MJ, Barrell M, Boostrom I, Dunachie S, Farzana R, Ferreira A, Frayne F, Hassan B, Jones E, Jones L, Mathias J, Milton R, Rees J, Chan GJ, Bekele D, Mahlet A, Basu S, Nandy RK, Saha B, Iregbu K, Modibbo F, Uwaezuoke S, Zahra R, Shirazi H, Syed NU, Mazarati J-B, Rucogoza A, Gaju L, Mehtar S, Bulabula ANH, Whitelaw A, van Hasselt JGC, Walsh TR, BARNARDS Group. 2021. Effects of antibiotic resistance, drug target attainment, bacterial pathogenicity and virulence, and antibiotic access and affordability on outcomes in neonatal sepsis: an international microbiology and drug evaluation prospective substudy (BARNARDS). Lancet Infect Dis S1473-3099:e00050‐5. 10.1016/S1473-3099(21)00050-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Benjamin DK, Kaufman DA, Hope WW, Smith PB, Arrieta A, Manzoni P, Kovanda LL, Lademacher C, Isaacson B, Jednachowski D, Wu C, Kaibara A, Walsh TJT. 2018. A phase 3 study of micafungin versus amphotericin B deoxycholate in infants with invasive candidiasis. Pediatr Infect Dis J 37:992–998. 10.1097/INF.0000000000001996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Årdal C, Røttingen J-A, Opalska A, Van Hengel AJ, Larsen J. 2017. Pull Incentives for antibacterial drug development: an analysis by the Transatlantic Task Force on Antimicrobial Resistance. Clin Infect Dis 65:1378–1382. 10.1093/cid/cix526. [DOI] [PubMed] [Google Scholar]
- 4.Agency EM. 2006. European Community. Regulation (EC) No 1901/2006 of the European Parliament and of the Council of medicinal products for paediatric use, amended by Regulation (EC) No 1902/2006. https://eur-lex.europa.eu/legal-content/EN/TXT/PDF/?uri=CELEX:32006R1902&from=EN.
- 5.Pediatric study plans: content of and process for submitting initial pediatric study plans and amended initial pediatric study plans; guidance for industry. 2020. https://www.fda.gov/regulatory-information/search-fda-guidance-documents/pediatric-study-plans-content-and-process-submitting-initial-pediatric-study-plans-and-amended.
- 6.Lestner JM, Smith PB, Cohen-Wolkowiez M, D KB Jr, Hope WW. 2013. Antifungal agents and therapy for infants and children with invasive fungal infections: a pharmacological perspective. Br J Clin Pharmacol 75:1381–1395. 10.1111/bcp.12025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Taormina G, Gopinath R, Moore J, Yasinskaya Y, Colangelo P, Reynolds K, Nambiar S. 2021. A Regulatory review approach for evaluation of micafungin for treatment of neonatal candidiasis. Clin Infect Dis 73:2335–2340. 10.1093/cid/ciab025. [DOI] [PubMed] [Google Scholar]
- 8.Stucki A, Acosta F, Cottagnoud M, Cottagnoud P. 2013. Efficacy of Ceftaroline Fosamil against Escherichia coli and Klebsiella pneumoniae strains in a rabbit meningitis model. Antimicrob Agents Chemother 57:5808–5810. 10.1128/AAC.00285-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Preparation of Artificial CSF. ALZET osmotic pumps. https://www.alzet.com/guide-to-use/preparation-of-artificial-csf/.
- 10.Hope WW, Mickiene D, Petraitis V, Petraitiene R, Kelaher AM, Hughes JE, Cotton MP, Bacher J, Keirns JJ, Buell D, Heresi G, D KB, Jr, Groll AH, Drusano GL, Walsh TJ. 2008. The pharmacokinetics and pharmacodynamics of micafungin in experimental hematogenous Candida meningoencephalitis: implications for echinocandin therapy in neonates. J Infect Dis 197:163–171. 10.1086/524063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Food and Drug Administration. 2019. Supplemental new drug application for Mycamine for treatment of invasive candidiasis in pediatric patients less than 4 months of age. https://www.accessdata.fda.gov/drugsatfda_docs/label/2019/021506s023lbl.pdf.
- 12.Warn PA, Livermore J, Howard S, Felton TW, Sharp A, Gregson L, Goodwin J, Petraitiene R, Petraitis V, Cohen-Wolkowiez M, Walsh TJ, Benjamin DK Jr, Hope WW. 2012. Anidulafungin for neonatal hematogenous Candida meningoencephalitis: identification of candidate regimens for humans using a translational pharmacological approach. Antimicrob Agents Chemother 56:708–714. 10.1128/AAC.05826-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ramos-Martín V, Johnson A, Livermore J, McEntee L, Goodwin J, Whalley S, Docobo-Pérez F, Felton TW, Zhao W, Jacqz-Aigrain E, Sharland M, Turner MA, Hope WW. 2016. Pharmacodynamics of vancomycin for CoNS infection: experimental basis for optimal use of vancomycin in neonates. J Antimicrob Chemother 71:992–1002. 10.1093/jac/dkv451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Darlow CA, Docobo-Perez F, Farrington N, Johnson A, McEntee L, Unsworth J, Jimenez-Valverde A, Gastine S, Kolamunnage-Dona R, de Costa RMA, Ellis S, Franceschi F, Standing JF, Sharland M, Neely M, Piddock L, Das S, Hope W. 2021. Amikacin combined with fosfomycin for treatment of neonatal sepsis in the setting of highly prevalent antimicrobial resistance. Antimicrob Agents Chemother 65:e0029321. 10.1128/AAC.00293-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.European Committee for Antimicrobial Susceptibility Testing (EUCAST) of the European Society of Clinical Microbiology and Infectious Diseases (ESCMID). 2003. Determination of minimum inhibitory concentrations (MICs) of antibacterial agents by agar dilution. Clin Microbiol Infect 9:P509–P515. [DOI] [PubMed] [Google Scholar]
- 16.Germovsek E, Lutsar I, Kipper K, Karlsson MO, Planche T, Chazallon C, Meyer L, Trafojer UMT, Metsvaht T, Fournier I, Sharland M, Heath P, Standing JF, NeoMero Consortium. 2018. Plasma and CSF pharmacokinetics of meropenem in neonates and young infants: results from the NeoMero studies. J Antimicrob Chemother 73:1908–1916. 10.1093/jac/dky128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Neely MN, van Guilder MG, Yamada WM, Schumitzky A, Jelliffe RW. 2012. Accurate detection of outliers and subpopulations with Pmetrics, a nonparametric and parametric pharmacometric modeling and simulation package for R. Ther Drug Monit 34:467–476. 10.1097/FTD.0b013e31825c4ba6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.D’Argenio DZ, Schumitzky A, Wang X. 2009. ADAPT 5 user’s guide: pharmacokinetic/pharmacodynamic systems analysis software. Biomedical Simulations Resource, Los Angeles, CA. [Google Scholar]