Abstract
Background:
Lipopeptides are potential microbial metabolites that are abandoned with broad spectrum biopharmaceutical properties ranging from antimicrobial, antiviral and anticancer, etc. Clinical studies are not much explored beyond the experimental methods to understand drug mechanisms on target proteins at the molecular level for large molecules. Due to the less available studies on potential target proteins of lipopeptide based drugs, their potential inhibitory role for more obvious treatment on disease have not been explored in the direction of lead optimization. However, Computational approaches need to be utilized to explore drug discovery aspects on lipopeptide based drugs, which are time saving and cost-effective techniques.
Methods:
Here a ligand-based drug discovery approach is coupled with reverse pharmacophore-mapping for the prediction of potential targets for antiviral (SARS-nCoV-2) and anticancer lipopeptides. Web-based servers PharmMapper and Swiss Target Prediction are used for the identification of target proteins for lipopeptides surfactin and iturin produced by Bacillus subtilis.
Results:
The studies have given the insight to treat the diseases with next-generation large molecule therapeutics. Results also indicate the affinity for Angiotensin-Converting Enzymes (ACE) and proteases as the potential viral targets for these categories of peptide therapeutics. A target protein for the Human Papilloma Virus (HPV) has also been mapped.
Conclusion:
The work will further help in exploring computer-aided drug designing of novel compounds with greater efficiency where the structure of the target proteins and lead compounds are known.
Keywords: Antiviral agents, Bacillus subtilis, Drug discovery, Ligands, Lipopeptides, Peptide hydrolases
Introduction
The focus of the current trends in computer-aided drug discovery is to understand the disease mechanism, and further target identification takes place. On the edge of personalized medicine, the system is based upon the molecular basis of fundamentals in drug designing 1. The interplay mentioned above can be addressed by imparting the cheminformatics tools in high throughput format. Cancer is a life-threatening disease which is one of the severe health problem globally. The increasing drug resistance has urged for the search of novel anticancer agents 2. However, steadily increasing drug resistance in the treatment of infectious disease posed a severe problem in antimicrobial therapy and necessitates continuing research on different classes of drug derivatives 3.
Lipopeptides are known as anti-microbial, anti-tumor, anti-inflammatory, anti-hypertensive, anti-parasitic, and anti-cancer compounds 4. The lipopeptides exhibit various biological activities due to the presence of lipid and peptide moiety. Moreover, numerous therapeutically important medicines like peptide therapeutics contain a heterocyclic nucleus. Lipopeptides are amphiphilic compounds of natural origin and found with various therapeutic properties. Surfactin and Iturin lipopeptides produced by Bacillus subtilis (B. subtilis) are reported with antiviral and anticancer properties 5–8. The studies on lipopeptides have been explored for antiviral activities against Corona virus (SARS-nCoV-2) 9. Large molecule therapeutics are found with higher target specificity as compared to small molecules and has the potential to inhibit target proteins from multiple sites figure 1. Hence exploring the drugs with higher selectivity help in subsiding side effects and non-specific cytotoxicity of small-molecule inhibitors in cancer treatment 10,11.
Figure 1.
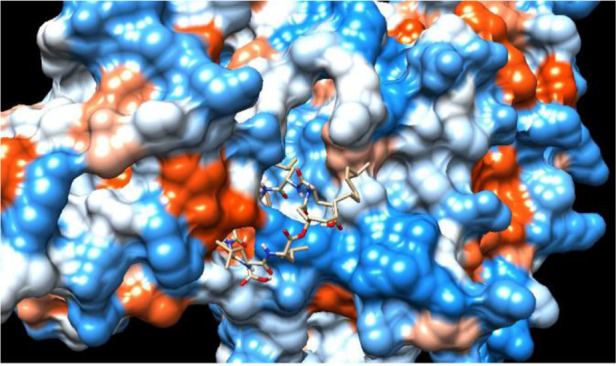
Molecular docking interaction: cyclic peptide drug docked with target protein (electrostatic surface interaction). Ability of cyclic peptide to inhibit target from multiple sites.
The first and subsequent second wave of COVID-19 pandemic due to the outbreak of SARS-nCov-2 (South East Respiratory Syndrome-Novel Corona Virus-2) is concurring the world leading to the global emergency to seek potential treatment options. The current management of the disease through drug repurposing has been an efficient way to combat the corona virus infection. Various target proteins and potential drug options are being explored to defeat the viral pathogenicity 12. Lipopeptides have been known as potential antiviral compounds and such therapeutic options are also being researched 13,14. The need of the hour is to first map the viral targets with the available potential therapeutics in an order to understand their mechanism of action for the novel drug discovery and drug repurposing 12,15. Such studies will facilitate to further accelerate the drug discovery pipeline to combat future pandemics through the discovery of next generation therapeutic options such as peptides and lipopeptides.
There is an urgent need to explore such a new class of drugs where the drug resistance is threatening the world with a slow pipeline of drugs instead of having fledged with increased knowledge and technology-driven facilities 10. The struggle starts with the identification of potential targets for large molecule therapeutics 16. Here a drug discovery approach is coupled with pharmacophore-based virtual screening for the effective prediction of potential targets for lipopeptides. The studies have not been explored in the direction of lead optimization of natural compounds to increase the target specificity of the drug. Though, target identification is the primary step towards novel drug discovery. In silico methods, facilitate the cost-effective ways for target identification and lead optimization in reduced time and chemical exposure 7. Structure-based pharmacophore mapping is a useful technique when insufficient information on ligand molecule is available and therapeutic activity (induce or block) for a particular disease is experimentally proved. It is used to explore information about the receptor site. It gives more indepth insight for the receptor-ligand interaction at the molecular level 17.
In view of the above mentioned facts, compounds derived from synthesis are directed towards the proteomic approach extensively for the identification of potential binding proteins. These techniques are based upon comparative studies on protein expression profiling concerning the presence and absence of a given molecule for a particular cell or tissue. Such methods are not much successful in the discovery of target proteins because these are time consuming and laborious 18. In the case of large molecule natural compounds, the task is even more challenging to execute and incurs huge wastage of chemicals and time 19. Therefore, to bring such natural compounds into the drug discovery pipeline, computational approaches followed by advanced synthetic techniques lead to more significant benefits to improve the health and well-being with the help of next generation drugs 6,20. In the current work in-silico method for target profiling is implemented for the identification of target proteins responsible for cancer and viral infections such as SARS-nCov-2 against anticancer and antiviral lipopeptides surfactin and iturin using web-based servers.
Materials and Methods
Pharmacophore Mapping using PharmMapper
Reverse pharmacophore mapping approach is used with the help of a web-based server PharmMapper. The studies are conducted for identification of target proteins for lipopeptide compounds with anticancer and antiviral properties 4,21. The techniques have been previously utilized for the target proteins for essential oils of Cardomom and bis-pyrimidine compounds 22. The PharmMapper server utilizes the reverse pharmacophore mapping to identify the potential targets for a given query compound. It utilizes an in-built database of pharmacophore models of 23,236 annotated proteins from targetBank, BindingDB, DrugBank, and Potential Drug Target Database (PDTD) with 51,431 ligandabble and 16,159 druggable pharmacophore models. The server compares the query compound with models of the in-built database.
According to the similarity of the pharmacophore of the query compound with identified pharmacophore of the target proteins, the results are provided in the form of Zscore. Alongside the importance and indications of the identified protein in the disease are also provided 23. Here the potential anticancer peptidolipidic compounds of B. subtilis are submitted as the query compound for possible target identification 24. The selection of target proteins is done based on their importance in the cancer disease. The selected lipopeptides are reported with anticancer properties in literature.
The query compounds were submitted to PharmMapper server (http://59.78.96.61/pharmmapper). The PharmMapper server compares the pharmacophore of the query compound with an in-built database of pharmacophore models. It predicts 300 target proteins based on their fitness score and pharmacophoric features. The target protein was ranked as per the fitness score and importance and indication of the protein. Generally, the top 10 proteins with a fitness score of more than 5 were considered to identify the probable target proteins of the query compound 18.
Swiss Target Prediction for target identification
Swiss Target prediction is a web-based server developed by the Swiss Institute of Bioinformatics. This tool is used for the prediction of potential targets of a compound for which targets with Protein ID has not been explored. The website facilitates the estimation of probable targets of a query compound or a bioactive compound. The server has an in-built library of 370,000 known active compounds and more than 3000 target proteins from various species. In the updated version of the server, a large dataset on drug-protein interaction has been made available, which makes it a unique source of information. It is based upon a knowledge-based development approach for the identification of novel targets or secondary targets for uncharacterized compounds or known compounds, respectively. The server accurately predicts the targets for bioactive compounds based on two-dimensional and three-dimensional (2D and 3D) measures of similarity respective to the known compounds 25–27.
Lipopeptides are known as potential anti-cancer, anti-viral compounds. However, the approach towards the identification of the target protein has not been explored. Hence, computational methods for pharmacophore mapping are utilised, which facilitate the identification of potential targets for lipopeptide-based drugs.
Selection and retrieval of lipopeptides
Lipopeptides with potent anticancer properties have been identified and screened through the rigorous literature survey. Surfactin and iturin have been chosen as the most prominent anticancer lipopeptides. The Pub-Chem compound database is utilized to get the compound information. The canonical SMILES annotation of the query compounds were used to submit the molecules to the server and can be used to draw the compound into two-dimensional structure. The information were submitted into the servers in the form of canonical SMILES for query compounds.
Pharmacophore mapping
The two-dimensional structures of the above-mentioned lipopeptides were drawn using the information of canonical SMILES from the PubChem compound database., The 2D sketcher tool of Maestro suite (Schrodinger software package) was used to draw the structures. The server utilizes the sdf and mol2 (mole file formats/extensions for saving chemical structures) file formats for the submission of the query compound for pharmacophore mapping. Further, the individual structures were submitted to the software PharmMapper, which is a free web-based tool for the identification of target proteins based upon the structural complementarity with the submitted drugs. Results obtained through Pharmacophore mapping were further subjected to the structure prediction of the given proteins. Software is capable of providing a minimum of 10–300 target proteins based on their binding vicinity in which the drug should be able to dock and inhibit the target protein. The traditional sequence alignment methodology followed by structure prediction, and further data mining approaches were used for obtaining the three-dimensional structures of predicted proteins.
Sequence alignment & structure prediction
This step was performed using the retrieval of the FASTA sequence of each protein given in the mapping results. The FASTA sequence has further proceeded for sequence alignment using the Basic Local Alignment Search Tool abbreviated as BLAST, a web-based algorithm provided by the National Centre for Bio-technology Information (NCBI). On the basis of alignment score and keeping the query coverage in consideration, the higher similarity structures were retrieved from the Protein DataBank (PDB). The PDB structures were used as a template for the homology modeling of the FASTA sequence of the target proteins predicted through PharmMapper. Some of the FASTA sequences were traced with the 100% similarity in the sequence alignment. However, the experimental structures are available in the protein databank. Hence the structures can be retrieved for further studies.
Target identification
In silico target identification for drug molecules is the primary step in drug discovery, which includes various algorithms for the identification of genes and proteins. The availability of 3D structure of the target protein enables the identification of the best binding mode for understanding the interaction. Pharmacophore is a spatial arrangement of functional groups, is an essential core of the molecule which interacts with the receptor target molecule is the alternative method for deciphering molecular interaction.
Multiple sequence alignment using T-Coffee server
As per pharmacophore mapping results, the mapped protein with PDB ID 2R5K major capsid protein L1 of HPV (Human Papilloma Virus) (Table 1) was further subjected to multiple sequence alignment with SARS-nCoV-2 nucleocapsid protein. The FASTA sequences of both the proteins were retrieved from NCBI and submitted to the T-Coffee server, which uses Clustal_W algorithm for multiple sequence alignment. Multiple sequence alignment is done to have an insight into the percentage similarity index between these two sequences. Sequence similarity searching helps in the identification of feasibility of drug repurposing for a given compound, as established antiviral drugs are re-purposed to treat infection of SARS-nCoV-2 which is responsible for current COVID-19 pandemic.
Table 1.
PharmMapper report for Surfactin: Depicting Hemoglobin subunit, multidrug-resistant operon, Major capsid viral protein and Histones as potential target class
| PharmaMapper rank | PDB ID | Target name | Normalized fit score |
|---|---|---|---|
| +1 | 4YU4 | None | 0.9988 |
| +2 | 3CKI | ADAM 17 | 0.9972 |
| +3 | 1CG8 | Hemoglobin subunit alpha | 0.9957 |
| +4 | 2HC5 | Uncharacterized protein yvyC | 0.9957 |
| +5 | 1M15 | Phosphoenolpyruvate phosphomutase | 0.9944 |
| +6 | 1NVM | 4-hydroxy-2-oxovalerate aldolase | 0.9925 |
| +7 | 1HI5 | Non-secretory ribonuclease | 0.9908 |
| +8 | 2OQC | Uncharacterized protein yxel | 0.9774 |
| +9 | 3ECH | Multidrug resistance operon repressor | 0.9722 |
| +10 | 1FB1 | CREB-binding protein | 0.9079 |
| +72 | 2R5K | Major capsid protein L1 | 0.6073 |
| +73 | 2CY3 | Cytochrome C3–13 kDa | 0.6067 |
| 2NQB | Histone | 0.6002 |
FASTA sequences of Major Capsid protein L1 of HPV:
>pdb|2R5H|D Chain D, Late Major Capsid Protein L1
AVVSTDEYVARTNIYYHAG-
TSRLLAVGHPYFPIKKPNNN-
KILVPKVSGLQYRVFRIHLPDPNKFGFPDTS
FYNPDTQRLVWACVGVEVGRGQPLGVGIS-
GHPLLNKLDDTENASAYAANAGVDNRE-
CISMDYKQTQLCLI
GCKPPIGEHWGKGSPCTQVAVQPGDCP-
PLELINTVIQDGDMVDTGFGAMDFT-
TLQANKSEVPLDICTSIC
KYPDYIKMVSEPYGDSLFFYL-
RREQMFVRHLFNRAGTVGEN-
VPDDLYIKGSGSTANLASSNYFPTPSGSM
VTSDAQIFNKPY
WLQRAQGHNNGICWGNQLFVTVVDTTRST
NMSLCAAISTSETTYKNTNFKEYLRHGEE
YDLQFIFQLCKITLTADVMTYIHSMNSTILED-
WNGGSGGEDPLKKYTFWEVNLKEKFSAD-
LDQFPLGRKF
LLQL
FASTA sequence of Nucleocapsid phosphoprotein of SARS-nCoV-2:
>QHR84456.1 nucleocapsid phosphoprotein [Severe acute respiratory syndrome coronavirus 2]
MSDNGPQNQRNAPRITFGG
PSDSTGSNQNGERSGARSKQRRPQGLPNNTASW
FTALTQHGKEDLKFPRGQ
GVPINTNSSPDDQIGYYRRATRRIRGG-
DGKMKDLSPRWYFYYLGTGPEAGLPY
GANKDGIIWVATEGALN
TPKDHIGTRNPANNAAI-
VLQLPQGTTLPKGFYAEGSRGGSQASSRSSSRSR
NSSRNSTPGSSRGTSPARM
AGNGGDAALALLLLDRLNQLESKMS
GKGQQQQGQTVTKKSAAEASKKPRQKRTAT
KAYNVTQAFGRRGPE
QTQGNFGDQELIRQGTDYKHWPQIAQFAPSAS-
AFFGMSRIGMEVTPSGTWLTYTGAI-
KLDDKDPNFKDQV
ILLNKHIDAYKTFPPTEPKKDKKKKADETQAL-
PQRQKKQQTVTLL
PAADLDDFSKQLQQSMSSADSTQA
Results and Discussion
Prediction of target proteins for anticancer activities of lipopeptides
PharmMapper and Swiss Target Prediction predict the target proteins for a ligand molecule on the basis of the vicinity of the binding cavity of the target proteins, which can probably be inhibited through the query ligand molecule. The query ligand was used here is lipo-peptide surfactin (Figure 2) for which the possible target proteins have been predicted and given in the table 1. According to the PharmMapper report depicted in (Table 1), hemoglobin subunits and membrane proteins have been predicted, which are associated with the hemolytic and surface active properties of surfactin, respectively 28.
Figure 2.
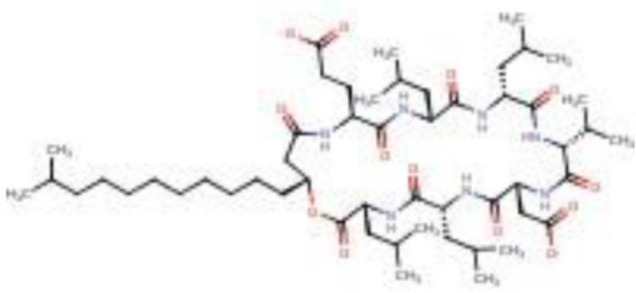
2D structure of cyclic lipopeptide–surfactin (query compound).
As per the PharmMapper results depicted in the (Table 1), top 10 ranked protein IDs are considered as the most significant target proteins. Resulting protein with PDB ID 4YU4 is a hemoglobin subunit that significantly proved to be associated with the hemolytic properties of the surfactin. As per the experimental studies, the hemolytic property is a hinderence of using surfactin as a potential anticancer compound. This was never been explored at the molecular level to understand the factors involved in the hemolytic property of the lipopeptide-based compounds 29. Another protein with PDB ID predicted is 3CK1, a membrane protein which suggests the potential membrane affinity of the compound to lead its pore-forming activity as an antimicrobial compound. As FDA approved lipopeptide daptomycin is successfully used to treat the infections of multidrug-resistant bacteria 30. To explore the anti-cancer activity of compounds, it is necessary to understand the interaction of the ligand at the target site. This study will give an insight into the lead optimization of the surfactin molecule to identify novel analogs with low or non-haemolytic property and higher affinity towards the target proteins for cancer and viral diseases. The studies would be a stepping stone towards the discovery of next-generation anti-cancer and anti-viral compounds 24,31,32.
As per Swiss Target Prediction report in figure 3 and table 2, major proteins predicted are from the apoptotic pathway, which is having significant role in the cancer progression. Inhibition of such a signaling pathway plays a major role in cell death, which has been explained in the mechanism of action of anticancer lipopeptides that they induce the cell cycle arrest and leads to cell death 33. The predicted proteins can be further explored with the help of structural bioinformatics and computer-aided drug designing studies. The work can be extended further for lead optimization studies for a given class of drugs to design a molecule with better efficiency and specificity to inhibit a particular target protein 7,34.
Figure 3.
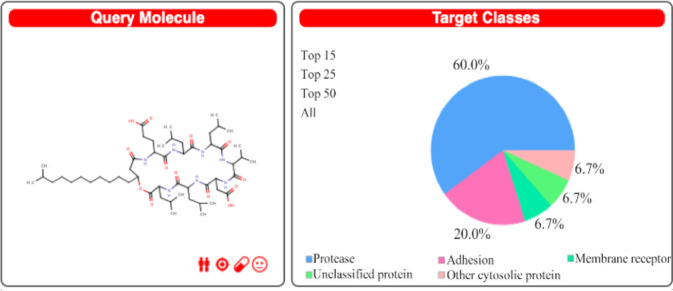
Swiss Target prediction query structure for Surfactin and prediction report with percentage affinity with class of target protein.
Table 2.
Surfactin Swiss Target Prediction report depicting Caspases, Proteases and Membrane receptor proteins as potential targets classes and deciphering Angiotensin Converting enzyme (ACE2) as potential target against SARS-nCoV-2 under top 50 targets aligned
| Target | Common name | Uniprot ID | ChEMBL ID | Target class |
|---|---|---|---|---|
| Caspase-2 | CASP2 | P42575 | CHEMBL4884 | Protease |
| Caspase-3 | CASP3 | P42574 | CHEMBL2334 | Protease |
| Caspase-8 | CASP8 | Q14790 | CHEMBL3776 | Protease |
| Caspase-1 | CASP1 | P29466 | CHEMBL4801 | Protease |
| Proteasome macropain subunit MB1 | PSMB5 | P28074 | CHEMBL4662 | Protease |
| P-selectin | SELP | P16109 | CHEMBL5378 | Adhesion |
| Integrin alpha-V/beta-3 | ITGAV | P06756 | CHEMBL1907598 | Membrane receptor |
| ITGB3 | P05106 | |||
| Caspase-7 | CASP7 | P55210 | CHEMBL3468 | Protease |
| Cathepsin D | CTSD | P07339 | CHEMBL2581 | Protease |
| Thrombin | F2 | P00734 | CHEMBL204 | Protease |
| Selectin E | SELE | P16581 | CHEMBL3890 | Adhesion |
| Leucocyte adhesion molecule-1 | SELL | P14151 | CHEMBL4198 | Adhesion |
| Angiotensin-converting enzyme | ACE | P12821 | CHEMBL1808 | Protease |
The Swiss Target Prediction report in figure 3 is depicting the surfactin structure as a query compound, and a pie chart is depicting the percentage mapping with the target class. The results show surfactin as a potential ligand for major proteases (60%), adhesins, and membrane receptors. Lipopeptides being amphiphilic compounds are found with membrane affinity properties, which can be proved with these results 35,36. Tables 2 and 3 suggest the Caspase proteins as the potential class of target proteins for surfactin. Caspases have a significant role in cancer progression 37. Being a potential target of peptide therapeutics, this aspect can be explored in conjunction with inhibitory/catalytic activity to regularize the class of caspase proteins in cancer management 10,37.
Table 3.
SwissTarget Prediction report for Surfactin
| Target | Uniprot ID | Gene code | ChEMBL ID | Probability | #sim. cmpds (3D/2D) | Target class |
|---|---|---|---|---|---|---|
| Caspase-3 subunit p12 | P42574 | CASP3 | CHEMBL2334 |

|
3/112 | Cysteine protease |
| Caspase-7 subunit p20 | P55210 | CASP7 | CHEMBL3468 |

|
3/111 | Cysteine protease |
| Caspase-6 subunit p18 (by homology) | P55212 | CASP6 | CHEMBL3308 |

|
3/112 | Cysteine protease |
| Caspase-1 subunit p20 | P29466 | CASP1 | CHEMBL4801 |

|
6/147 | Cysteine protease |
| Caspase-4 (by homology) | P49662 | CASP4 | CHEMBL2226 |

|
6/147 | Cysteine protease |
| Caspase-5 subunit p10 (by homology) | P51878 | CASP5 | CHEMBL3131 |

|
6/147 | Cysteine protease |
| Inactive caspase-12 (by homology) | Q6UXS9 | CASP12 |

|
6/147 | Cysteine protease | |
| Caspase-8 subunit p18 | Q14790 | CASP8 | CHEMBL3776 |

|
3/67 | Cysteine protease |
| Complex | P06756/P05106 | ITGAV/ITGB3 | CHEMBL1907598 |

|
13/87 | Membrane receptor |
| Complex | P08514/P05106 | ITGA2B/ITGB3 | CHEMBL2093869 |

|
1/138 | Membrane receptor |
| Renin | P00797 | REN | CHEMBL286 |

|
5/459 | Aspartic protease |
| Cathepsin D | P07339 | CTSD | CHEMBL2581 |

|
5/454 | Aspartic protease |
| Napsin-A (by homology) | O96009 | NAPSA |

|
5/454 | Aspartic protease | |
| Uncharacterized protein (by homology) | H7C469 |

|
1/88 | Aspartic protease | ||
| Caspase-2 subunit p18 | P42575 | CASP2 | CHEMBL4884 |

|
0/18 | Cysteine protease |
Swiss Target Prediction report generated against lipopeptide iturin reveals the G-Protein Coupled Receptors (GPCR) as the most prominent class of targets for iturin (Figure 4 and Table 4). Drugs targeting GPCR’s are used to treat many diseases. More than 40% FDA approved drugs are aimed to target GPCR’s and their related pathways. Now, GPCR’s are being used as biomarkers for early diagnosis of cancer, as they play critical role in activating and regulating signaling pathways associated to cancer 38. Hence, iturin would be a promising compound to be explored to develop as potential anticancer drug 38.
Figure 4.
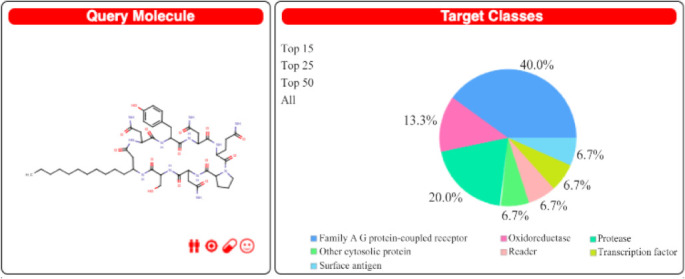
Iturin as query molecule for Swiss Target Prediction with prediction report in percentage for target class.
Table 4.
Swiss Target Report (prediction of GPCR’s, Caspases and ACE as targets for Iturin) and predicted Cyclin dependent kinases (CDK’s) through mapping against Iturin
| Target | Common name | Uniprot ID | ChEMBL ID | Target class |
|---|---|---|---|---|
| Acetylcholine receptor protein delta chain | CHRND | Q07001 | CHEMBL3011 | Ligand-gated ion channel |
| Acidic mammalian chitinase | CHIA | Q9BZP6 | CHEMBL1293197 | Enzyme |
| Acyl-CoA: Dihydroxyacetonephosphateacyltransferase | GNPAT | O15228 | CHEMBL4494 | Enzyme |
| Angiotensin-converting enzyme | ACE | P12821 | CHEMBL1808 | Protease |
| Apetite regulating hormone | GHRL | Q9UBU3 | CHEMBL1921664 | Unclassified protein |
| Baculoviral IAP repeat-containing protein 3 | BIRC3 | Q13489 | CHEMBL5335 | Enzyme |
| Beta secretase 2 | BACE2 | Q9Y5Z0 | CHEMBL2525 | Protease |
| Beta secretase 1 | BACE1 | P56817 | CHEMBL4822 | Protease |
| C-X-C chemokine receptor type 7 | ACKR3 | P25106 | CHEMBL2010631 | Family A G-protein coupled receptor |
| Calcitonin gene-related peptide type 1 receptor | CALCRL | Q16602 | CHEMBL3798 | Family B G-protein coupled receptor |
| Carboxypeptidase B2 isoform A | CPB2 | Q96IY4 | CHEMBL3419 | Protease |
| Carboxypeptidase N catalytic subunit | CPN1 | P15169 | CHEMBL4713 | Protease |
| Caspase-1 | CASP1 | P29466 | CHEMBL4801 | Protease |
| Caspase-2 | CASP2 | P42575 | CHEMBL4884 | Protease |
| Cyclin-dependent kinase 4/cycline D1 | CCND1 | P24385 | CHEMBL1907601 | Kinase |
| CDK4 | P11802 | |||
| CDK2/Cyclin A CCNA2 | CDK2 | P20248 | CHEMBL3038469 | Kinase |
| P24941 | ||||
| Histone deacetylase 8 | HDAC8 | Q9BY41 | CHEMBL3192 | Eraser |
| Chymotrypsin C | CTRC | Q99895 | CHEMBL2386 | Protease |
| Signal transducer and activator of transcription 6 | STAT6 | P42226 | CHEMBL5401 | Transcription factor |
| Melanocortin receptor 5 | MC5R | P33032 | CHEMBL4608 | Family A G-protein coupled receptor |
| E3 SUMO-protein ligase | CBX4 | O00257 | CHEMBL3232685 | Enzyme |
| Ceramide glucosyltransferase | UGCG | Q16739 | CHEMBL2063 | Family A G-protein coupled receptor |
| Neurokine 1 receptor | TACR1 | P25103 | CHEMBL249 | Family A G-protein coupled receptor |
| Cholecystokin B receptor | CCKBR | P32239 | CHEMBL298 | Family A G-protein coupled receptor |
| Interleukin-1 beta | IL1B | P01584 | CHEMBL1909490 | Secreted protein |
| Proteinase-activated receptor 1 | F2R | P25116 | CHEMBL3974 | Family A G-protein coupled receptor |
Prediction of viral targets for antiviral activity of lipopeptides
Pharmacophore mapping studies suggested various viral targets along with the anticancer studies. The studies contain significant findings for the identification of next-generation antiviral drugs to combat the diseases like COVID-19. As per table 1 multidrug resistance operon repressor is found as a predicted target, which suggests the proven antibiotic activity against multidrug resistance strains 6. Table 4 depicts the Angiotensin-Converting Enzyme (ACE) as one of the potential targets of iturin. The affinity of a compound towards ACE2 is significant as Human-ACE2 is a potential target for SARS-nCov-2 Spike-glycoprotein. Inhibition of such interaction plays a vital role in the treatment of Corona virus disease (COVID-19) 9. The findings from table 1 generated through PharmMapper results for surfactin, suggested the affinity towards major capsid protein of Human Papilloma virus (PDB ID: 2R5K). Clustal_W analysis results gave approximately 21% similarity with SARS-nCoV-2 nucleocapsid protein. The results suggest the drug repurposing opportunity with the identification of a similar category of drugs with a high affinity towards SARS-nCoV-2 viral proteins 15,39. Table 4 is also suggesting affinity towards a viral protein Baculoviral IAP repeat-containing protein 40.
Conclusion
Synergistically conducted studies on target protein identification of microbial peptidolipidic compounds give more promising and reliable results regarding the target class of compounds. The experimental studies were touched at the level of mechanistic action but could not lead towards optimization and novel analog designing, which required the structural and molecular information of drug and target protein. Through the in silico studies, the multitargeting efficiency of lipopep-tide compounds is depicted, which is not feasible with a small molecule inhibitor. This suggests the higher specificity and efficiency in binding with the target site. In the current studies, antiviral and anticancer properties of lipopeptide compounds have given various horizons for future perspectives in developing next-generation novel compounds. It is the need of the hour to explore novel compounds to combat future infectious diseases like COVID-19 pandemic and deadly cancer diseases. The study concludes various classes of potential viral targets and cancer target proteins to facilitate the drug discovery process with the possible targets and ligands to study the interaction and discover the promising drug candidates. Utilization of microbial bi-products is much feasible than producing peptide therapeutics in the laboratory, which incurs huge chemical exposure and a financial burden as well with the wastage of chemicals, time and money. Rather computational prediction eases out the journey to some extent. It gives a better understanding of the drug-target interaction at the molecular level. The research can be further explored with molecular dynamics studies for a more in-depth understanding of lead optimization and exploring these compounds with more potency and least side effects.
Acknowledgement
We are thankful to the National Institute of Technology Raipur and Chhattisgarh Council of Science and Technology (CCOST) (Project number 2487/CCOST/MRP/2016, Raipur dated 25.01.2016), India for providing the necessary facilities to prepare the manuscript and permission to publish it.
Footnotes
Conflict of Interest
The authors declare no conflict of interest.
Ethical Approval
This article does not contain any studies with human participants or animals performed by any of the authors.
References
- 1.Bruno A, Costantino G, Sartori L, Radi M. The in silico drug discovery toolbox: applications in lead discovery and optimization. Curr Med Chem 2019;26(21):3838–73. [DOI] [PubMed] [Google Scholar]
- 2.Sauna ZE, Lagassé HAD, Alexaki A, Simhadri VL, Katagiri NH, Jankowski W, et al. Recent advances in (therapeutic protein) drug development. F1000Res 2017;6: 113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Neu HC. The crisis in antibiotic resistance. Science 1992;257(5073):1064–73. [DOI] [PubMed] [Google Scholar]
- 4.Meena KR, Kanwar SS. Lipopeptides as the Antifungal and antibacterial agents: applications in food safety and therapeutics. Biomed Res Int 2015;2015:473050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Caulier S, Nannan C, Gillis A, Licciardi F, Bragard C, Mahillon J. Overview of the antimicrobial compounds produced by members of the Bacillus subtilis group. Front Microbiol 2019;10:302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cochrane SA, Vederas JC. Lipopeptides from Bacillus and Paenibacillus spp.: A gold mine of antibiotic candidates. Med Res Rev 2016;36(1):4–31. [DOI] [PubMed] [Google Scholar]
- 7.Jujjavarapu SE, Dhagat S, Yadav M. Computer-Aided Design of Antimicrobial Lipopeptides As Prospective Drug Candidates. CRC Press LLC; 2019. 146 p. [Google Scholar]
- 8.Pham JV, Yilma MA, Feliz A, Majid MT, Maffetone N, Walker JR, et al. A review of the microbial production of bioactive natural products and biologics. Front Microbiol 2019;10:1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xia S, Liu M, Wang C, Xu W, Lan Q, Feng S, et al. Inhibition of SARS-CoV-2 (previously 2019-nCoV) infection by a highly potent pan-coronavirus fusion inhibitor targeting its spike protein that harbors a high capacity to mediate membrane fusion. Cell Res 2020;30(4): 343–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lau JL, Dunn MK. Therapeutic peptides: Historical perspectives, current development trends, and future directions. Bioorganic Med Chem 2018;26(10):2700–7. [DOI] [PubMed] [Google Scholar]
- 11.Craik DJ, Fairlie DP, Liras S, Price D. The future of peptide-based drugs. Chem Biol Drug Des 2013;81:136–47. [DOI] [PubMed] [Google Scholar]
- 12.Yadav M, Dhagat S, Eswari JS. Emerging strategies on in silico drug development against COVID-19: challenges and opportunities. Eur J Pharm Sci 2020;155: 105522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chowdhury T, Baindara P, Mandal SM. LPD-12: a promising lipopeptide to control COVID-19. Int J Anti-microb Agents 2021;57(1):106218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.de Vries RD, Schmitz KS, Bovier FT, Predella C, Khao J, Noack D, et al. Intranasal fusion inhibitory lipopeptide prevents direct-contact SARS-CoV-2 transmission in ferrets. Science 2021;371(6536):1379–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chartier M, Najmanovich R. Detection of binding site molecular interaction field similarities. J Chem Inf Model 2015;55(8):1600–15. [DOI] [PubMed] [Google Scholar]
- 16.Ciemny M, Kurcinski M, Kamel K, Kolinski A, Alam N, Schueler-Furman O, et al. Protein–peptide docking: opportunities and challenges. Drug Discov Today 2018;23 (8):1530–7. [DOI] [PubMed] [Google Scholar]
- 17.Liu X, Ouyang S, Yu B, Liu Y, Huang K, Gong J, et al. PharmMapper server: a web server for potential drug target identification using pharmacophore mapping approach. Nucleic Acids Res 2010;38 (Web Server issue): W609–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Meshram RJ, Baladhye VB, Gacche RN, Karale BK, Gaikar RB. Pharmacophore mapping approach for drug target identification: a chemical synthesis and in silico study on novel thiadiazole compounds. J Clin Diagn Res 2017;11(5):KF01–KF08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mojsoska B, Jenssen H. Peptides and peptidomimetics for antimicrobial drug design. Pharmaceuticals (Basel) 2015;8(3):366–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sable R, Parajuli P, Jois S. Peptides, peptidomimetics, and polypeptides from marine sources: A wealth of natural sources for pharmaceutical applications. Mar Drugs 2017;15(4):124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kumar S, Singh J, Narasimhan B, Shah SAA, Lim SM, Ramasamy K, et al. Reverse pharmacophore mapping and molecular docking studies for discovery of GTPase HRas as promising drug target for bis-pyrimidine derivatives. Chem Cent J 2018;12(1):106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bhattacharjee B, Chatterjee J. Identification of proapoptopic, anti-inflammatory, anti-proliferative, anti-invasive and anti-angiogenic targets of essential oils in cardamom by dual reverse virtual screening and binding pose analysis. Asian Pac J Cancer Prev 2013;14(6):3735–42. [DOI] [PubMed] [Google Scholar]
- 23.Meshram RJ, Baladhye VB, Gacche RN, Karale BK, Gaikar RB. Pharmacophore mapping approach for drug target identification: A chemical synthesis and in silico study on novel thiadiazole compounds. J Clin Diagn Res 2017;11(5):KF01–KF08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Seydlová G, Svobodová J. Review of surfactin chemical properties and the potential biomedical applications. Cent Eur J Med 2008;123–33. [Google Scholar]
- 25.SwissTargetPrediction: A Web Server for Target Prediction of Bioactive Small Molecules - PubMed [Internet]. [cited 2020 Jul 10]. Available from: https://pubmed.ncbi.nlm.nih.gov/24792161/ [DOI] [PMC free article] [PubMed]
- 26.Gfeller D, Grosdidier A, Wirth M, Daina A, Michielin O, Zoete V. SwissTargetPrediction: A web server for target prediction of bioactive small molecules. Nucleic Acids Res 2014;42(Web Server issue):W32–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Daina A, Michielin O, Zoete V. SwissTargetPrediction: updated data and new features for efficient prediction of protein targets of small molecules. Nucleic Acids Res 2019;47(W1):W357–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Inès M, Dhouha G. Lipopeptide surfactants: Production, recovery and pore forming capacity. Peptides 2015. Sep; 71:100–12. [DOI] [PubMed] [Google Scholar]
- 29.Kracht M, Rokos H, Özel M, Kowall M, Pauli G, Vater J. Antiviral and hemolytic activities of surfactin isoforms and their methyl ester derivatives. J Antibiot (Tokyo) 1999;52(7):613–9. [DOI] [PubMed] [Google Scholar]
- 30.Kopp F, Marahiel MA. Macrocyclization strategies in polyketide and nonribosomal peptide biosynthesis. Nat Prod Rep 2007;24(4):735–49. [DOI] [PubMed] [Google Scholar]
- 31.Lavecchia A, Giovanni C. Virtual screening strategies in drug discovery: A critical review. Curr Med Chem 2013; 20(23):2839–60. [DOI] [PubMed] [Google Scholar]
- 32.Rishton GM. Nonleadlikeness and leadlikeness in biochemical screening. Drug Discov Today 2003;8(2):86–96. [DOI] [PubMed] [Google Scholar]
- 33.Wu YS, Ngai SC, Goh BH, Chan KG, Lee LH, Chuah LH. Anticancer activities of surfactin potential application of nanotechnology assisted surfactin delivery. Front Pharmacol 2017;8:761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Katsila T, Spyroulias GA, Patrinos GP, Matsoukas MT. Computational approaches in target identification and drug discovery. Comput Struct Biotechnol J 2016;14: 177–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Maget-Dana R, Thimon L, Peypoux F, Ptak M. Surfactin/iturin A interactions may explain the synergistic effect of surfactin on the biological properties of iturin A. Biochimie 1992;74(12):1047–51. [DOI] [PubMed] [Google Scholar]
- 36.Maget-Dana R, Peypoux F. Iturins, a special class of pore-forming lipopeptides: biological and physicochemical properties. Toxicology 1994;87(1–3):151–74. [DOI] [PubMed] [Google Scholar]
- 37.Clark A, MacKenzie S. Targeting cell death in tumors by activating caspases. Curr Cancer Drug Targets 2008;8 (2):98–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yu S, Sun L, Jiao Y, Lee LTO. The role of G protein-coupled receptor kinases in cancer. Int J Biol Sci 2018; 14(2):189–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Demirbaş A, Eker H, Elmas ÖF, Ulutaş Demirbaş G, Atasoy M, Türsen Ü, et al. COVID-19 and human papillomavirus: Paradoxical immunity. J Cosmet Derma-tol 2021;20(7):2001–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang J. New strategy for COVID-19 vaccination: targeting the receptor-binding domain of the SARS-CoV-2 spike protein Cell Mol Immunol 2021. Feb;18(2):243–44. [DOI] [PMC free article] [PubMed] [Google Scholar]


