Abstract
Background
Vascular smooth muscle cell (SMC) apoptosis is involved in major cardiovascular diseases. Smad2 is a transcription factor implicated in aortic aneurysm. The molecular mediators of Smad2-driven SMC apoptosis are not well defined. Here we have identified a Smad2-directed mechanism involving MET and FAS, both encoding cell membrane signaling receptors.
Methods and results
Guided by microarray analysis in human primary aortic SMCs, loss/gain-of-function (siRNA/overexpression) indicated that Smad2 negatively and positively regulated, respectively, the gene expression of Met which was identified herein as anti-apoptotic and that of Fas, a known pro-apoptotic factor. While co-immunoprecipitation suggested a physical association of Smad2 with p53, chromatin immunoprecipitation followed by quantitative PCR revealed their co-occupancy in the same region of the MET promoter. Activating p53 with nutlin3a further potentiated the suppression of MET promoter-dependent luciferase activity and the exacerbation of SMC apoptosis that were caused by Smad2 overexpression. These results indicated that Smad2 in SMCs repressed the transcription of MET by cooperating with p53, and that Smad2 also activated FAS, a target gene of its transcription factor activity.
Conclusions
Our study suggests a pro-apoptotic mechanism in human SMCs, whereby Smad2 negatively and positively regulates MET and FAS, genes encoding anti-apoptotic and pro-apoptotic factors, respectively.
Keywords: Smad2, p53, MET, FAS, Apoptosis, Smooth muscle cell
Highlights
-
•
Smad2 inhibits anti-apoptotic MET expression in human vascular smooth muscle cells.
-
•
Smad2 and p53 co-occupy the promoter of MET repressing its transcription.
-
•
Smad2 activates FAS, which encodes a pro-apoptotic factor.
-
•
Results identify novel pro-apoptotic regulations in human smooth muscle cells.
Introduction
Vascular smooth muscle cell (SMC) is the principal cell type that constitutes the strength and contractility of the artery wall. Moreover, these cells form a signaling hub, sensing microenvironmental perturbation and in response, undergoing cell state transitions. As such, dysregulated SMC homeostasis is the basis of major vascular diseases – for example, excessive apoptosis contributes to aortic aneurysms [1,2].
SMC apoptosis is a highly regulated program involving complex interplay of extracellular signal, cell membrane receptors, and intracellular responses. TGFβ signaling is regarded as critical in aortic aneurysms [3]. Smad2 and Smad3 represent major effector proteins of TGFβ signaling [4]. Via cytosol-to-nucleus translocation, these two transcriptional factors transmit extracellular signals to intracellular reactions to gene expression [5,6]. Whereas Smad3 in SMCs is reportedly pro- or anti-proliferative in restenosis or atherosclerosis [[7], [8], [9], [10]], Smad2 was linked to SMC apoptosis and aortic aneurysm [[11], [12], [13]]. This complexity of disparate Smad2/3 functions underscores the importance to delineate the Smad2-specific regulatory mechanisms that underlie SMC apoptosis, whereas current knowledge is limited [[14], [15], [16], [17]].
To quest for Smad2 downstream signaling mediators, in this study we performed microarray after Smad2 silencing in human primary aortic SMCs (AoSMC). Consistent with the reported Smad2 function in SMC apoptosis [13], FAS (a.k.a CD95) was prominently down-regulated. FAS encodes a cell membrane receptor with a pro-apoptotic role well-documented in SMCs [18,19]. However, a Smad2 regulation of FAS in SMCs was not previously reported. More interestingly, MET (encoding Met), another top-ranked gene, was implicated herein as repressed by Smad2. Met is a proto-oncoprotein known to be pro-proliferative in multiple malignancies [20]. Whether it plays a role in SMC apoptosis was not clear. Given the background knowledge presented above, it was expectable that Smad2 as a transcription activator may turn on the pro-apoptotic gene FAS. However, Smad2-mediated repression of MET transcription was somewhat surprising.
We further found that the traditionally regarded activator of transcription, Smad2, partnered with p53, a master inhibitory transcription factor that inactivates gene expression. The two transcription factors formed a complex binding to the same site of MET promoter thereby blocking its transcription. Our data also showed that Met inhibited AoSMC apoptosis. Therefore, this study uncovered a Smad2-directed negative and positive regulation of MET and FAS, genes encoding anti- and pro-apoptotic factors, respectively. Our results thus provide new insight for better understanding the complex Smad2 regulations of apoptosis in AoSMCs.
Results
Smad2 negatively regulates the transcription of Met, which is found here to be anti-apoptotic in human SMCs
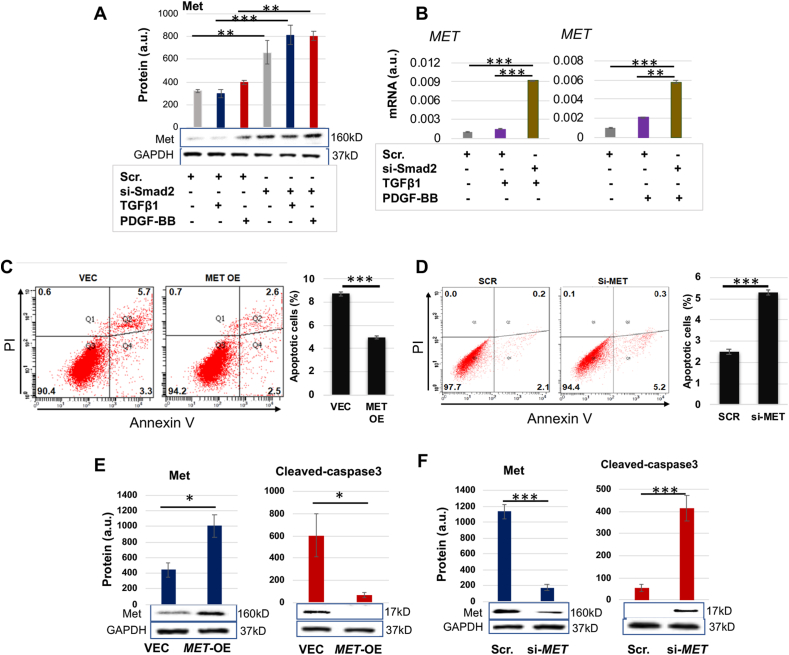
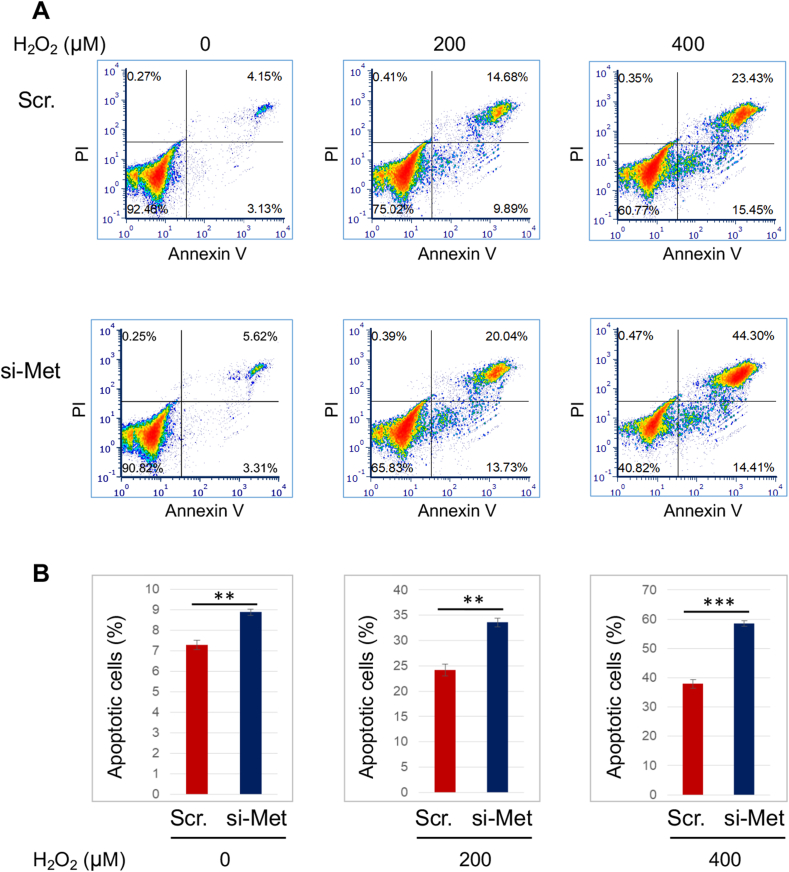
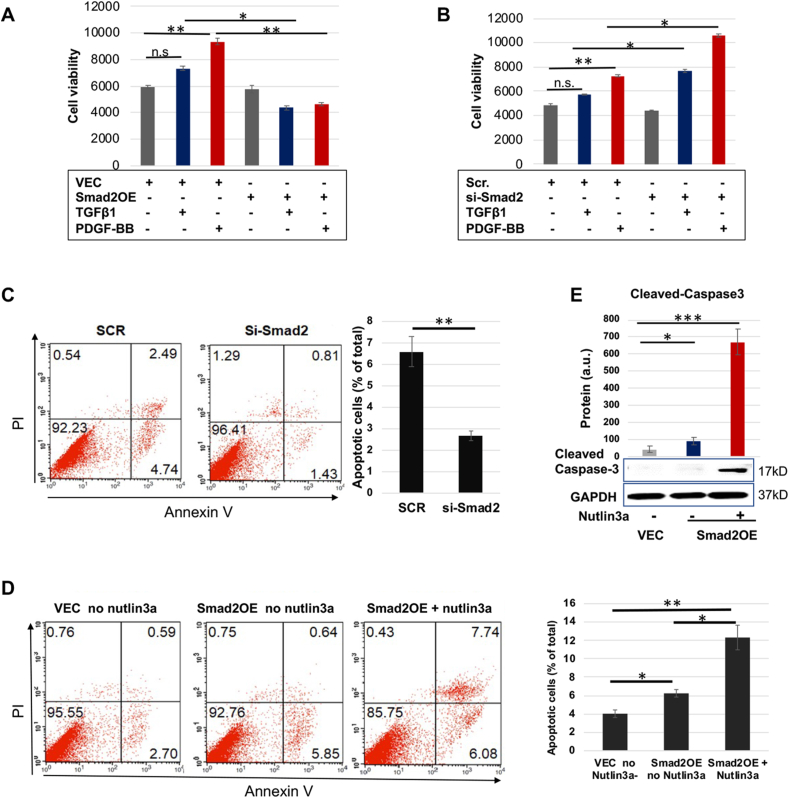
As a top-ranked gene that was upregulated due to Smad2 knockdown, MET caught our interest in the microarray analysis (Fig. S1). Met is expressed in SMCs and known as a pro-mitotic cell surface signaling receptor [20], yet its role in SMC apoptosis was not previously delineated. We thus first verified that Smad2-specific silencing with siRNA markedly increased MET expression at both protein (Fig. 1A) and mRNA levels (Fig. 1B) in the presence of TGFβ1 (or PDGF-BB). We included PDGF-BB as an extra condition in consideration of its reported effect of indirectly activating Smad2 [21]. We then determined the role of Met in AoSMC apoptosis through FACS sorting of apoptotic cells and Western blotting of cleaved Caspase3. SMCs are relatively resistant to death. The ratio of apoptotic cells versus total cells is generally small, and a change in apoptotic SMC number would be masked if a comparison were made between total cell numbers. We thus used percent apoptotic cells (versus total SMCs) as a parameter for comparison rather than using total cells which include mostly non-apoptotic cells and only a small portion of apoptotic cells. As shown in Fig. 1C, 8.71% of the total AoSMC population was detected as apoptotic in the control of empty vector, and MET overexpression reduced this percentage to 4.96% (a 43% reduction, see black bars of averaged values). MET silencing (Fig. 1D), on the other hand, increased percent apoptotic cells to 5.3% from 2.5% (scrambled siRNA control). In accordance, MET overexpression and silencing substantially decreased and increased cleaved caspase3 protein, respectively (Fig. 1E and F). To further confirm the anti-apoptotic role of Met observed here in AoSMCs, we next applied H2O2 which aggressively induces SMC apoptosis [22,23]. As shown in Fig. 2, treatment with H2O2 exacerbated apoptosis in a concentration-dependent manner in AoSMCs transfected either with scrambled siRNA or MET-specific siRNA. Of note, more severe apoptosis occurred in AoSMCs with (vs without) MET silencing. For example, after 16 h treatment with 0.4 mM H2O2, 58.71% (44.30% + 14.41%, late and early phases) of total cells were detected as apoptotic when MET was silenced, whereas without MET silencing only 38.88% (23.43% + 15.45%) apoptotic cells were detected.
Fig. 1.
Met is anti-apoptotic in AoSMCs and negatively regulated by Smad2.
A and B. Smad2 loss-of-function increases MET expression at protein and mRNA levels (Western blots in A and qRT-PCR analyses in B).
C and D. Met gain- and loss-of-function respectively inhibits and enhances AoSMC apoptosis (FACS assay). Black bars represent averaged values based on three independent repeat experiments (mean ± SD, n = 3). The apoptotic cells include Q2 and Q4, late- and early-phase apoptotic populations, respectively. The Flow Jo software was used.
E and F. Met gain- and loss-of-function respectively inhibits and enhances AoSMC apoptosis (cleaved caspase3 assay).
Human primary aortic SMCs (AoSMCs) were transfected with scrambled siRNA (Scr), Smad2-specific siRNA, empty vector (VEC), or overexpression plasmid (OE) for 12 h in basal medium (no FBS). The cells were cultured for another 12 h in fresh basal medium (no transfection reagents) to recover, and then treated with solvent or 10 ng/ml TGFβ1 or 50 ng/ml PDGF-BB for 20 h before harvest for Western blot and qRT-PCR analyses.
Quantification: Densitometry of Western blots (similar ECL exposure) from independent repeat experiments was normalized (to GAPDH) and then averaged to calculate mean ± SEM (n = 3 independent experiments). Readings of triplicate qRT-PCR reactions were normalized (to GAPDH) and averaged to calculate mean ± SD (n = 3 repeats). a.u., arbitrary unit.
Statistics: One-way ANOVA followed by Bonferroni post-hoc test in A and B; Student's t-test in C–F; ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001.
Fig. 2.
MET silencing increases H2O2-induced apoptotic AoSMCs.
AoSMCs were transfected with scrambled siRNA (Scr) or MET-specific siRNA for 12 h in full medium, and then cultured in fresh medium until sub-confluency. The culture was continued in the presence of indicated concentrations of H2O2 for another 16 h, and then used for FACS assay (A), and the FCS Express 6 software was used. The assay was performed in a different time than that in Fig. 1, Fig. 5. Quantification (B): Percent apoptotic cells vs total cells; mean ± SD; paired Student's t-test, ∗∗P < 0.01, ∗∗∗P < 0.001.
Taken together, we not only identified a Smad2-specific negative regulation of MET, but also elucidated an anti-apoptotic role of Met in AoSMCs.
Smad2 and p53 co-immunoprecipitate (co-IP) with each other and co-repress MET transcription
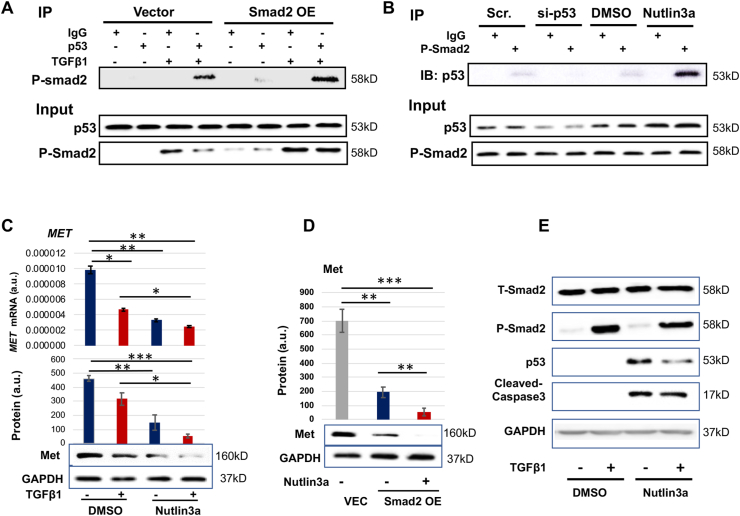
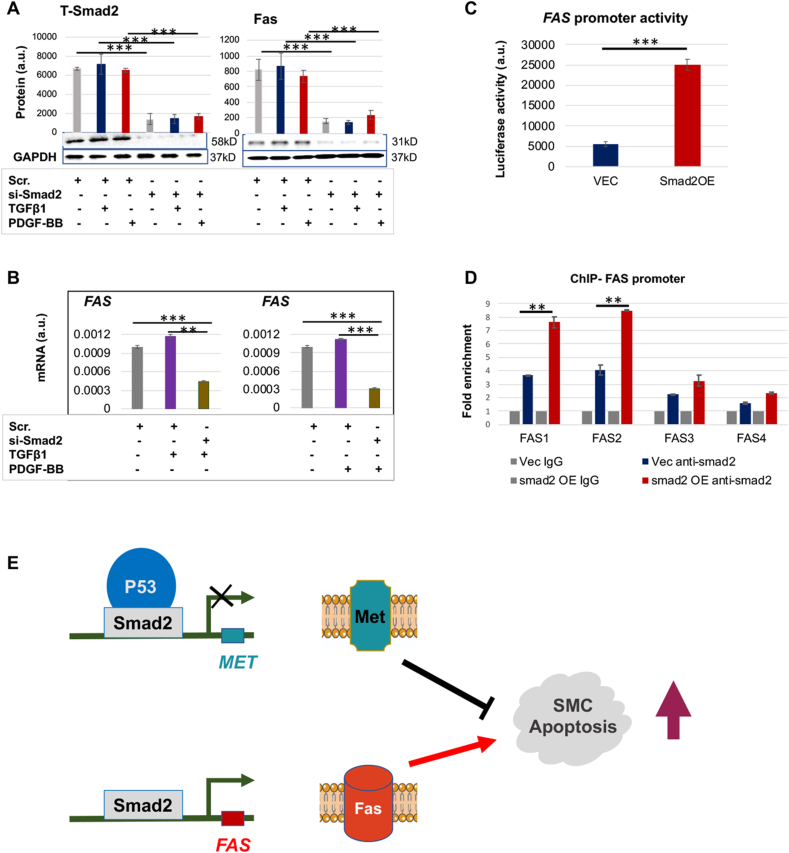
Smad2 is well known for transcriptional activation. As such, its inhibitory effect on MET expression appeared provocative against the familiar view. Interestingly, using a software (LASAGNA-Search 2.0) to predict transcription factor binding sites, we noticed that Smad2 and p53 may share the same binding region (−209 bp from the transcription start site) on the MET promoter. We thus inferred that Smad2 and p53 may function as a complex in regulating MET transcription. In order to test this hypothesis, we performed co-IP experiments using an antibody for p53 to detect co-IP'ed Smad2 and also an antibody for P-Smad2 to detect co-IP'ed p53. The data indicated that while P-Smad2 (activated form) co-IP'ed specifically with p53 against the IgG background (Fig. 3A), p53 also co-IP'ed specifically with P-Smad2 (Fig. 3B). Further indicating functional specificity of the observed co-IP, while Smad2 overexpression increased the P-Smad2 co-IP with p53, activating p53 (with nutlin3a) enhanced its co-IP with P-Smad2 (Fig. 3A and B). Importantly, robust P-Smad2/p53 co-IP occurred in the presence of TGFβ1 which stimulates Smad2 phosphorylation and its nuclear translocation and hence function. Indicative of a functional outcome of this P-Smad2/p53 association, while using TGFβ1 to activate Smad2 reduced MET expression (mRNA and protein), activating p53 with nutlin3a further significantly potentiated this effect (Fig. 3C). The Smad2-specificity of this effect was confirmed in Fig. 3D via Smad2 overexpression. An alternative scenario is that p53 may potentiate the Smad2 function by regulating its expression level. However, p53 activation did not increase Smad2, neither its total protein nor phosphorylated form (Fig. 3E). Taken together, these data indicate that Smad2 and p53 interact with each other or function in the same protein complex regulating MET expression in AoSMCs.
Fig. 3.
Smad2 cooperates with p53 in regulating MET expression.
A and B. Co-IP between P-Smad2 and p53. C and D. Activating p53 potentiates MET transcription inhibition in a Smad2-dependent manner. E. Activating p53 does not reduce Smad2.
Human primary aortic SMCs (AoSMCs) were cultured, transfected, treated, and assayed as described for Fig. 1. Quantification was performed as described for Fig. 1; mean ± SD for mRNA levels in C (n = 3 repeats) and mean ± SEM for Western blots, n = 3 independent repeat experiments. Statistics: One-way ANOVA/Bonferroni post-hoc test; ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001.
Smad2 and p53 co-occupy the same MET promoter region
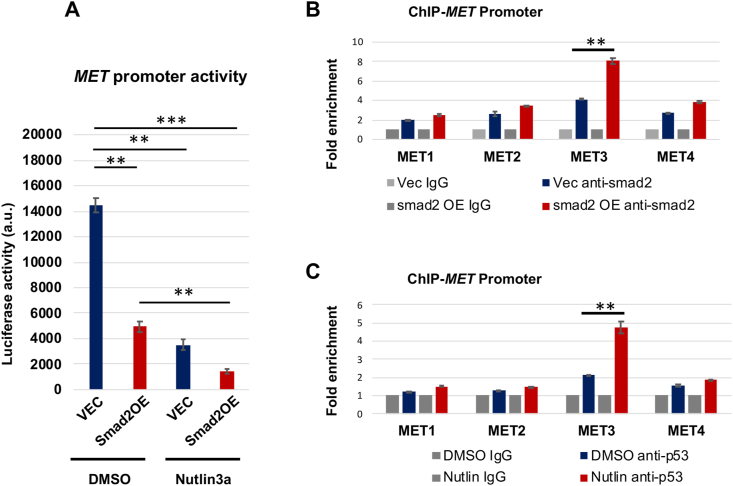
Given the above result of Smad2/p53 cooperativity in regulating MET expression, we next determined whether they functioned by directly binding to the MET promoter. We first performed MET promoter activity assay using plasmids containing the luciferase gene placed downstream of the MET promoter. As shown in Fig. 4A, while Smad2 overexpression significantly reduced MET promoter activity, p53 activation further enhanced this effect. We then used chromatin immunoprecipitation (ChIP)-qPCR to determine Smad2/p53's physical binding to 4 different MET promoter regions: 1100 (MET1), −900 (MET2), −200 (MET3), and +50 (MET4). These regions were chosen based on software prediction. Interestingly, only one (MET3) GAGGCAGACAGACACGTGCTGGGGCGG of the 4 DNA regions exhibited significantly more co-IP with the transcription factors as a result of Smad2 overexpression or p53 activation (Fig. 4B and C). These results suggest that Smad2 and p53 bind to the same region in the MET promoter thereby cooperatively regulating MET transcription.
Fig. 4.
Smad2 and p53 enrich in the same region of MET promoter.
A. p53 activation with nutlin3a enhances the Smad2 function of inhibiting MET promoter activity (luciferase assay).
B and C. Smad2 and p53 co-occupy in the same MET promoter region (ChIP-qPCR).
Human primary aortic SMCs (AoSMCs) culture, transfection, and data quantification were performed as described for Fig. 1. Cells were treated with nutlin3a for 2 h before harvest. Mean ± SD, n = 3 repeats. Statistics: One-way ANOVA/Bonferroni post-hoc test; ∗∗P < 0.01, ∗∗∗P < 0.001.
p53 activation further potentiates AoSMC apoptosis that is enhanced by Smad2 overexpression
Up to this point, we had identified an anti-apoptotic role of Met in AoSMCs and Smad2's negative transcriptional regulation of MET. We had also found p53/Smad2 co-occupancy in the same MET promoter region and their cooperativity that potentiated the Smad2 regulatory function. Collectively, these new lines of information logically suggested that p53 may potentiate the pathophysiological role of Smad2 in promoting AoSMC apoptosis. To test this proposition, we first verified the role of Smad2 in AoSMC apoptosis. As shown in Fig. 5 (A and B), Smad2 silencing promoted, and its overexpression inhibited AoSMC viability, suggesting a pro-apoptotic role of Smad2. However, reduced cell viability could result from attenuated proliferation or aggravated apoptosis or both. In support of a positive role for Smad2 in AoSMC apoptosis, FACS data showed that percent apoptotic AoSMCs dropped from 6.57% to 2.67% after Smad2 silencing (Fig. 5C). Consistently, percent apoptotic cells rose from 4.1% to 6.3% after Smad2 overexpression (Fig. 5D). More interestingly, activation of p53 by nutlin3a further increased percent apoptotic cells to 12.3% (Fig. 5D). This result was also confirmed by Western blot analysis of cleaved caspase3 (Fig. 5E).
Fig. 5.
p53 activation potentiates Smad2's pro-apoptotic function.
A and B. AoSMC viability assay (CellTiter-Glo).
C and D. Apoptosis assay (FACS). Black bars represent averaged values based on three independent repeat experiments (mean ± SD, n = 3). The apoptotic cells include Q2 and Q4, late- and early-phase apoptotic populations, respectively. The Flow Jo software was used.
E. Apoptosis assay (cleaved caspase3).
Prior to assays, human primary aortic SMCs (AoSMCs) were cultured, transfected, and cytokine-treated, as described for Fig. 1. Quantification was performed as described for Fig. 1; mean ± SD, n = 3 repeats.
Statistics: One-way ANOVA/Bonferroni post-hoc test; Student's t-test was performed in C; ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001; n.s. not significant.
Smad2 as a transcription factor positively regulates FAS transcription
We also looked into the positive role of Smad2 as a transcription activator. Among the top-ranked genes whose expression was reduced by Smad2 siRNA (microarray, Fig. S1), FAS was the only one with an established pro-apoptotic role but not known for regulation by Smad2 in SMCs. We therefore verified this microarray result through quantitative real time PCR (qRT-PCR) and Western blot analysis for mRNA and protein levels of FAS. Indeed, Smad2 knockdown effectively abated FAS expression at both protein (Fig. 6A) and mRNA levels (Fig. 6B). We then asked whether Smad2 regulated FAS transcription directly. Luciferase assay (Fig. 6C) indicated that Smad2-specific overexpression boosted FAS promoter activity by 5-fold. Furthermore, ChIP assay (Fig. 6D) suggested that the Smad2 binding to the FAS promoter around the FAS1 (−1450bp) and FAS2 (−1400bp) regions was significantly enhanced by Smad2 overexpression as compared to the empty vector control (VEC). These results indicate that in SMCs the transcription factor Smad2 positively regulates the transcription of the pro-apoptotic gene FAS, likely through binding to its promoter.
Fig. 6.
Smad2 positively regulates AoSMC expression of FAS.
Human primary aortic SMCs (AoSMCs) were transfected with scrambled siRNA (Scr), Smad2-specific siRNA, empty vector (VEC), or Smad2 overexpression plasmid (Smad2-OE) for 12 h in basal medium (no FBS). The cells were cultured for another 12 h in fresh basal medium (no Lipofectamine) to recover, and then treated with solvent or 10 ng/ml TGFβ1 or 50 ng/ml PDGF-BB for 20 h before harvest for Western blot and qRT-PCR analyses.
A and B. Smad2 loss-of-function reduces FAS expression at protein and mRNA levels (Western and qRT-PCR analyses). C. Smad2 gain-of-function enhances FAS promoter activity (luciferase assay). D. Smad2 enrichment at FAS promoter regions (ChIP-qPCR).
Quantification: Densitometry of Western blots (similar ECL exposure) from independent repeat experiments was normalized (to GAPDH) and then averaged to calculate mean ± SEM, n = 3 independent experiments. Readings of triplicate qRT-PCR reactions were normalized (to GAPDH) and averaged to calculate mean ± SD (n = 3 repeats). a.u., arbitrary unit.
Statistics: One-way ANOVA/Bonferroni post-hoc test; Student's t-test was performed in C; ∗∗P < 0.01, ∗∗∗P < 0.001.
E. Schematic working model of Smad2 regulations of MET and FAS that promote SMC apoptosis. While Smad2 partnering with P53 represses the transcription of MET, it also activates the transcription of FAS. The sum of decreased Met (anti-apoptotic) and increased Fas (pro-apoptotic) prompts SMC apoptosis.
Immunohistochemistry for the expression of Smad2, Fas, and Met in aneurysmal rat arteries
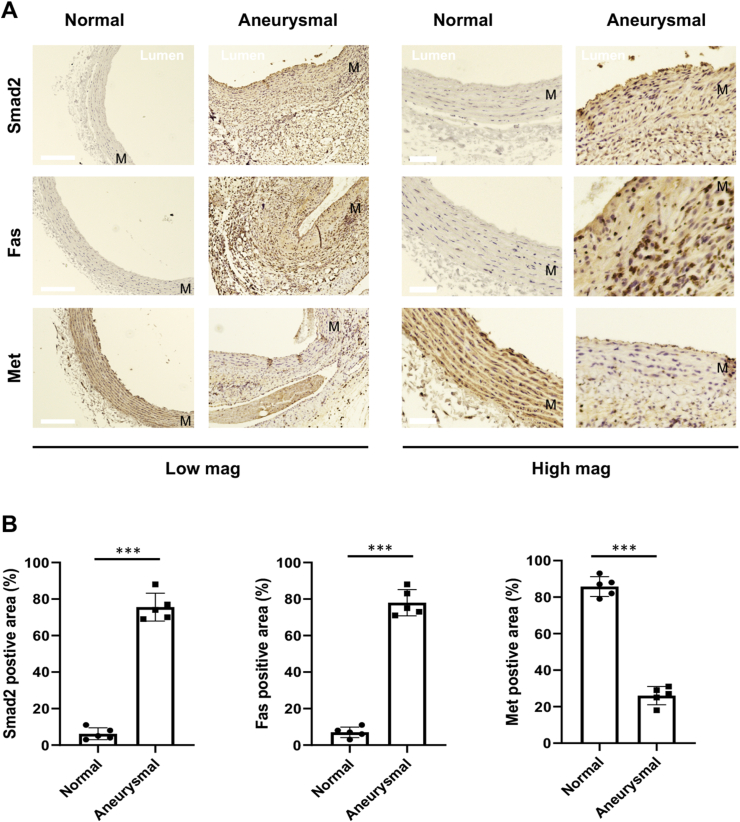
Since a pro-aneurysmal role has been reported for Smad2 [12], which was found here as an important regulator of MET and FAS in AoSMCs in vitro, we finally determined the expression of these players in vivo in aneurysmal arteries where SMC apoptosis is a major event [24]. We performed elastase infusion in rat abdominal aortic arteries which often induces marked aneurysm 7 days after the infusion [25]. Immunohistochemistry on aorta cross sections indicated that Smad2 increased in aneurysmal aortic arteries versus non-aneurysmal arteries, in agreement with previous reports [11,15]. More interestingly, whereas Fas increased, Met decreased in elastase-treated arteries as compared to normal artery controls, in particular in the medial layer where SMCs reside (Fig. 7, Fig. S2). This in vivo observation was consistent with our in vitro data that indicated Smad2-directed, respective (positive and negative) regulations of FAS and MET expression in AoSMCs that underwent apoptosis.
Fig. 7.
Immunohistochemistry of Smad2, Fas, and Met in aneurysmal arteries. The elastase-induced model of rat abdominal aortic aneurysm was performed as described in Methods. The elastase-infused aorta segments or non-infused normal controls were collected at 7 days post infusion, and artery cross-sections were prepared for immunostaining.
A. Representative immunostained artery cross-sections. Note different colors: blue-stained nuclei and brown-colored staining of Smad2, Fas, and Met. More stained sections from multiple animals are presented in Fig. S2. M = medial layer. Scale bar: 100 μm for low-mag pictures and 20 μm for high-mag pictures.
B. Quantification. Percent immunostained area (vs the total imaged artery tissue area) was measured using Image-J. The averaged data values (from 4 sections of each animal) were used to calculate mean ± SEM (n = 5 rats) for each animal group. Statistics: Unpaired Student t-test; ∗∗∗P < 0.001. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
Discussion
In this study we made an interesting finding that Smad2, generally perceived as an activating transcription factor [26], repressed the transcription of MET. Namely, Smad2 co-opted p53's function of transcriptional repression to inhibit MET transcription. This Smad2 negative regulation of MET in SMCs was not previously reported. We also found that Met was anti-apoptotic in AoSMCs, a function that could not be simply extrapolated from its known pro-proliferative role because apoptosis and mitosis are disparate cellular programs. On the other hand, Smad2 activated the transcription of FAS, a well-documented pro-apoptotic gene [18]. With these results, we are able to interpret the Smad2 pro-apoptotic mechanism from a bipartite perspective – Smad2 turns on pro-apoptotic FAS while turning off anti-apoptotic MET – actions orchestrated for aggravating AoSMC apoptosis. Given a reportedly pro-apoptotic role for Smad2 in SMCs with a link to the potentially lethal aortic aneurysms [[11], [12], [13],17], our findings are significant for deciphering Smad2's molecular regulations that underlie AoSMC apoptosis.
We dissected the Smad2/p53 partnership based on different layers of evidence. First, co-IP data indicated that Smad2 and p53 were interacting or in the same protein complex. Second, they bound to the same region of the MET promoter (−209 bp with respect to TSS), as revealed by ChIP-qPCR mapping of different MET promoter sequences. Third, activating p53 enhanced the Smad2-specific inhibitory effect on MET promoter-dependent transcriptional activity and MET gene expression. Fourth, activating p53 did not lead to an increase of P-Smad2 or its total protein, ruling out this alternative scenario. Lastly yet importantly, the functional outcomes of Smad2 overexpression, p53 activation, and/or MET silencing were all consistently verified as exacerbated AoSMC apoptosis. These results hence collectively suggest that when activated in the AoSMC apoptotic program, Smad2, though commonly known as a transcription activator, repressed MET by recruiting p53. Both binding to the same MET promoter site, Smad2 and p53 may form a complex. Their occupancy at MET could thereby be stabilized for the repression of MET expression. In accordance with an anti-apoptotic role for Met in AoSMCs, an involvement of Met in pro-proliferative pathways (such as ERK, STAT3, and AKT, Fig. S3) was also observed here in AoSMCs, consistent with the Met signaling pathways previously reported with cancer cells [27,28]. Of note, a p53/Smad2 partnership was also found in an earlier report [29]. However, these two factors were found to regulate CDKN1A through transcriptional activation in Xenopus embryos, a mechanism apparently different from that of MET repression observed herein. Therefore, although the role of p53 in SMCs appears controversial and context dependent [12,30], the results from our experimental setting suggest that p53 and Smad2 cooperatively repress MET and promote SMC apoptosis. On the other hand, with a focus on Smad2 and p53, our data cannot rule out participation of other factors in this MET-regulatory complex.
By contrast, FAS was positively regulated by Smad2, a result that can be readily interpreted based on the known transcription-activating function of Smad2 [31]. Indeed, loss- and gain-of-function and ChIP-qPCR experiments supported the role for Smad2 as a transcription factor directly binding to the FAS promoter activating its transcription. Since no p53-binding sequences were found (through software) in the FAS promoter, we do not expect the Smad2/p53 cooperativity is applicable for FAS. An early study showed TGFβ1-dependent Fas induction and subsequent apoptosis in the human gastric SNU-620 carcinoma cell line [32]. However, this study delineated that Smad3 but not Smad2 was responsible for the TGFβ1-induced Fas upregulation. Therefore, the discrepancy between our result and this early report is interesting which highlights the cell type and context dependent nature of the TGFβ/Smad signaling and its biological manifestation [6,33].
In summary, our results collectively unravel a regulatory mechanism whereby Smad2 represses MET while activating FAS transcription. Likely through this two-pronged regulation, Smad2 could effectively execute its pro-apoptotic function (Fig. 6E). Particularly interesting, a Smad2/p53 partnership enables an activation-to-repression transition of Smad2's function in gene expression.
Conclusions
We have identified a previously unrecognized pro-apoptotic regulation in human SMCs, whereby the same transcription factor (Smad2) negatively and positively regulates the transcription of anti-apoptotic and pro-apoptotic factors (Met and Fas), respectively and concurrently. This regulation manifests functionally as Smad2-prompted SMC apoptosis. Our findings could contribute to better understanding of the molecular events that occur in aortic aneurysms [34], a degenerative vascular disease featuring SMC apoptosis with no medical therapy available [24]. Further dissection of Smad2-dominated SMC apoptotic mechanisms is needed to evaluate the utility of its targeting toward translational medicine.
Materials and methods
Materials
Human aortic smooth muscle cells (AoSMCs, CC-2571), smooth muscle cell basal medium (SmBM, CC-3181), and SmBM plus Single Quots of supplements (CC-3182) were purchased from Lonza (Walkersville, MD). Recombinant Human TGFβ1 was purchased from Thermo Fisher Scientific (PHG9214). Recombinant Human PDGF-BB (520BB050) was from R&D Systems (Minneapolis, M). Cell Titer-Glo 2.0 Assay kit was purchased from Promega (G9242). Scrambled control and smad2-or MET-specific siRNA were from Thermo Fisher Scientific (Waltham, MA. Scrambled: AM4635; SMAD2:4427037, ID-S8397; MET: 4427038, ID-s8700). Opti-MEM I Reduced Serum Medium, Lipofectamine3000 and Lipofectamine RNAiMAX Transfection Reagent were from Thermo Fisher Scientific (31985062, L3000008 and 13778150). Pierce Fast Western Blot Kit was from Thermo Fisher Scientific (35050). Dead Cell Apoptosis Kit with Annexin V Alexa Fluor 488 & Propidium Iodide (PI) was from Thermo Fisher Scientific (V13241). Nutlin 3a was purchased from Millipore Sigma (SML0580).
Human aortic smooth muscle cell (AoSMC) culture and transfection
Human AoSMCs were cultured in SmBM with supplements (full medium) in a humidified incubator with 5% CO2 at 37 °C and used at passage 5–7, as we previously reported [35]. For Smad2 plasmid (Addgene, 11734) transfection, AoSMCs were cultured in full medium till 80–90% confluence and changed to basal medium (0% FBS) 2 h before transfection. The cells were transfected with the Smad2 plasmid using Lipofectamine3000 (following the manufacturer's instruction) for 12 h and cultured with fresh basal medium (no Lipofectamine) for another 24 h. PDGF-BB (50 ng/ml) or TGFβ1 (10 ng/ml) was then added and 12 h after treatment cells were harvested for assays. Transfection with siRNAs and treatment with PDGF-BB or TGFβ1 followed the same procedures and conditions except that the RNAi Max transfection reagent was used (following manufacturer's protocol) at 70–80% AoSMC confluency. For MET plasmid (Addgene, 37560), lentivirus was packaged using LentiX 293T cell line (TaKaRa, 632180) and transduced into AoSMCs. Cell apoptosis or Western blot analysis were performed at 48 h post infection.
Microarray and analysis
We followed the method we used in our recent publication [35]. Briefly, human SMCs were transfected with scrambled siRNA or si-Smad2 in basal medium for 24 h, then stimulated with TGFβ (10 ng/ml) for 12 h. The cells were collected for total RNA isolation using TRIzol reagent following the manufacturer's instruction (Thermo Fisher Scientific, Cat#15596026). Microarray (100 ng/sample) was then performed using Affymetrix Microarrays (Human Transcriptome Array 2.0). The data were analyzed using Transcriptome Analysis Console.
Quantitative real-time PCR (qRT-PCR)
As we previously described [35], total RNA was extracted from cell lysates using the TRIzol reagent (Thermo-Fisher Scientific, Cat#15596026) and used for cDNA synthesis with the High-Capacity cDNA Reverse Transcription kit (Thermo Fisher Scientific, Cat# 4368814). In each 20 μl reaction, 10 ng of cDNA was amplified through quantitative real-time PCR using PowerUp SYBR Green Master Mix (Thermo Fisher Scientific, A25778), and mRNA expression was determined using 7500 Fast Real-Time PCR System (Applied Biosystems, Carlsbad, CA). Levels of mRNAs were normalized to glyceraldehyde 3-phosphate dehydrogenase (GADPH) using the ΔΔCt method. qRT-PCR was done in triplicate reactions. The primers used are listed in Table S1.
Western blot analysis
We did the procedures similar to that in our recent report [35]. Briefly, cells were lysed in RIPA buffer (50 mM Tris, 150 mM NaCl, 1% Nonidet P-40 and 0.1% sodium dodecyl sulfate) containing Halt Protease and Phosphatase Inhibitor Cocktail (Thermo Fisher Scientific, Cat#78440). Protein concentration was determined using a Pierce BCA Protein Assay kit (Thermo Fisher Scientific, Cat#23227). Whole-cell lysates were mixed with Laemmli loading buffer, boiled at 95 °C for 5 min. Proteins were separated by 12% SDS–PAGE and transferred to a PVDF membrane. Immunoblotting was carried out using specific primary antibodies and Fast Western Optimized HRP Reagent. In order to detect multiple proteins, the whole blot was cut into strips prior to hybridization with different antibodies (for the original blots, please see Supplemental Materials). Specific protein bands on blots were illuminated using ECL detection reagents and recorded with an Azure C600 imager (Azure Biosystems, Dublin, CA). Band intensity was quantified using the ImageJ 64 software (https://imagej.nih.gov/ij/). Densitometry data were normalized to GAPDH (loading control). The information for the antibodies is presented in Table S2.
Cell apoptosis assay
After 24 h of starvation, transfected AoSMCs were treated for 24 h with DMSO control or Nutlin3a (final 5 nM). Harvested cells were washed in cold PBS and then resuspended in annexin-binding buffer (106 cells/mL). FITC annexin V (5 μL) and propidium iodide (PI) working solution (1 μL) were added (to each 100 μL cell suspension). After incubation at room temperature, annexin-binding buffer (400 μL) was added to each 100 μL of cell suspension. Analysis of apoptotic cells followed our recent publication [35] using BD FACS Calibur (BD Biosciences, NJ, USA) and the Flow Jo or FCS Express 6 software (specified in figure legends).
Cell viability assay
For cell viability assay, 5000 AoSMCs were seeded in each well of the 96-well white plate. Then cells were transfected with smad2 plasmid or empty vector, and scrambled siRNA or si-Smad2. Following transfection and TGFβ1 or PDGF-BB treatment (or solvent control), AoSMCs were washed once with PBS and then 50 μl of PBS plus 50 μl of CellTiter-Glo reagent was added in each well. Cell viability was analyzed using the Flexstation 3 plate reader to read 96-well plates.
Co-immunoprecipitation (co-IP)
Co-immunoprecipitation was performed following the instruction of Pierce Crosslink Immunoprecipitation Kit (Thermo Scientific, 26147). In brief, AoSMCs were transfected with empty vector (VEC) or smad2 overexpression plasmid (Addgene, 11734), or AoSMCs were starved and treated with Nutlin 3a for 24 h. Post transfected or treated AoSMCs were lysed on ice for 30min in IP Lysis/Wash Buffer (included in kit) containing Halt protease inhibitor cocktail (Thermo Fisher Scientific, 87785). Cell lysate were centrifuged at 12000 rpm for 10min at 4 °C. During centrifugation, we performed binding of antibodies to protein A/G plus agarose and crosslinking the bound antibodies following the instruction. After centrifugation, the pre-cleared lysate was added to the antibody-crosslinked resin in the column, and incubated with gentle end-over-end mixing for overnight at 4 °C. The proteins were finally eluted from the column and subjected to SDS-PAGE and Western blot analysis.
Luciferase assay for the constructs containing MET promoter or FAS promoter
The 1.7 kb promoter region of MET (−1500 to +200) was cloned from human AoSMC genomic DNA. It was then subcloned into a promoter reporter vector (EMPTY_PROM, Switchgear Genomics, S790005). Similarly, 2.3 kb FAS gene promoter (with 5′ UTR) (−2000 to 300) was amplified and subcloned into the EMPTY_PROM (Switchgear Genomics, S790005). The primers for cloning are presented in Table S3. Correct sequences of the clones were verified through sequencing. For luciferase activity assay, we followed the manufacturer’ instruction using the Lightswitch luciferase assay reagent (Switchgear Genomics, LS010). We first transfected cells with a luciferase assay plasmid, by seeding 5000 AoSMCs/well in 96-well plates and culturing for 24 h (no Lipofectamine). We then transfected the cells with an empty vector (VEC) or Smad2-overexpressing plasmid (Smad2-OE) for 24 h. For FAS promoter activity assay, the cell culture medium was changed to fresh full medium for a 24-h incubation before assay. For MET promoter activity assay, the cell culture medium was changed to fresh basal medium and the cells were cultured for 12 h, and then treated with Nutlin 3a (5 ng/ml) for 24 h. The luciferase activity assay was performed by adding 100 μl Assay Solution per well. The cells were incubated for 30 min at room temperature before reading in Luminometer System (Applied Biosystems, Foster City, CA). Luciferase activity reading was normalized to cell number, and duplicate plates were used.
Chromatin immunoprecipitation (ChIP) assay
We used the ChIP assay method we recently reported [35]. ChIP was performed using the Pierce Magnetic ChIP kit (Thermo Fisher Scientific, 26157). Briefly, AoSMCs, transfected with VEC or Smad2-OE (and treated with 10 ng/ml TGFβ1 for 2 h), or treated with Nutlin3a (5 nM for 24 h) were cross-linked with 1% formaldehyde for 10min at room temperature. Cross-linking reactions were stopped by the addition of a 1/10 volume of 10X glycine and incubated at room temperature for 5 min. The cells were washed with ice-cold PBS, collected, and then nuclei were extracted after cell lysis. Micrococcal nuclease was added to the nuclei suspension to digest the DNA for 15 min at 37 °C, and MNase Stop Solution was added to stop the reaction. The nuclei were recovered and re-suspended in IP Dilution Buffer, and then sonicated (Four 5-s pulses at 20 Watts for 1 × 106 cells) to break the nuclear membrane. Chromatin extracts containing DNA fragments with an average size of 500 base pairs were immunoprecipitated overnight at 4 °C using an anti-Smad2 antibody (Abcam, ab71109), anti-p53 (ab1101), or IgG control (included in kit). ChIP-grade Protein A/G Magnetic beads were added and incubated for 4 h at 4 °C. RNAse A and Proteinase K were used to digest RNA and protein. The purified DNA was used for qPCR using Applied Biosystems 7500 Fast Real-Time PCR System (Applied Biosystems, Carlsbad, CA) and PowerUp SYBR Green Master Mix (Thermo Fisher Scientific, A25778). The primers designed to amplify selected regions of the MET or FAS promoter are listed in Table S4.
Elastase induced abdominal aortic aneurysm (AAA) in rats
All animal procedures conformed to the NIH Guide for the Care and Use of Laboratory Animals and were in compliance with the Institutional Animal Care and Use Committee at University of Virginia. All surgeries were performed under isoflurane anesthesia (through inhaling, flow rate 2 ml/min), and all efforts were made to minimize suffering. Animals were euthanized in a chamber gradually filled with CO2. Male Sprague-Dawley rats of 300–350 g were used (from Charles River). The animals were kept under standard conditions before surgery. AAA was induced using the elastase method detailed in our recent report [25]. In brief, in an anesthetized rat, a 10-mm segment of the infrarenal aorta was dissected after mid-line laparotomy. From the right saphenous artery, a PE-10 tube was introduced into the infrarenal aorta, and the proximal and distal parts of the isolated aorta were temporarily ligated. Type-I porcine pancreatic elastase (Sigma-Aldrich) of total 2.7 units in 0.27 ml PBS was continuously infused over 0.9 h into the aorta through the PE-10 tube connected to a syringe pump. The ligation and PE-10 tube were then removed, the laparotomy was sutured, and the animal was kept on a warm pad for recovery.
Immunohistochemistry
Animals were euthanized at 7 days after elastase infusion, and infused and non-infused aortas were collected, fixed, paraffin-embedded, and then cut to prepare 5-μm sections. We performed essentially the same immunohistochemistry procedures as included in our recent report [35]. Briefly, slides were deparaffinized and rehydrated, and then subjected to antigen retrieval in citrate buffer at 80 °C in a high-pressure cooker. A primary antibody (see antibodies listed in Table S2) and ImmPRESS HRP (horse radish peroxidase) Anti-Rabbit IgG Polymer Detection Kit (Vector Laboratories, MP-7451-15) were used to visualize the protein of interest. Images were taken under an EVOS microscope (Thermo Fisher Scientific).
Statistical analysis
Data are generally presented as mean ± standard deviation (SD) except for Western blots (specified in figure legends) which were repeated in three independent experiments performed in different days. After determination of data normal distribution, differences between two groups were analyzed by Student's t-test for independent samples; for comparison between more than two sets of experimental conditions, we applied one-way analysis of variance (ANOVA) followed by Bonferroni post-hoc test. P values < 0.05 were considered as statistically significant. Significance in all figures is indicated as follows: ∗ p < 0.05, ∗∗ p < 0.01, ∗∗∗P < 0.001.
Ethical approval and consent to participate
The research was performed with institutional approvals including biosafety and chemical protocols.
Consent for publication
All authors agreed on the publication.
Availability of supporting data
Supporting data is included as supplemental materials.
Funding
This work was supported by NIH grants R01HL-068673 (to K.C.K.) and R01HL129785 (to K.C.K. and L.-W.G.), and R01HL133665 (to L.-W.G.), and Overseas Research Fellowships, The Uehara Memorial Foundation (to T.S.).
Authors' contributions
X.X. designed and performed in vitro experiments and did immunohistochemistry, T.S. performed the surgery of elastase-induced aneurysm model and prepared artery samples. X.X. and L-W.G. analyzed data and wrote the manuscript. L-W.G. and K.C.K. critically reviewed the manuscript.
Declaration of competing interest
The authors have declared no competing interests and no industry or financial relationship to disclose.
Acknowledgements
None.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.athplu.2021.08.005.
Contributor Information
Lian-Wang Guo, Email: lg8zr@virginia.edu.
K. Craig Kent, Email: CK8AQ@hscmail.mcc.virginia.edu.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Clement M., Chappell J., Raffort J., Lareyre F., Vandestienne M., Taylor A.L., Finigan A., Harrison J., Bennett M.R., Bruneval P., Taleb S., Jorgensen H.F., Mallat Z. Vascular smooth muscle cell plasticity and autophagy in dissecting aortic aneurysms. Arterioscler Thromb Vasc Biol. 2019;39:1149–1159. doi: 10.1161/ATVBAHA.118.311727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shankman L.S., Gomez D., Cherepanova O.A., Salmon M., Alencar G.F., Haskins R.M., Swiatlowska P., Newman A.A., Greene E.S., Straub A.C., Isakson B., Randolph G.J., Owens G.K. KLF4-dependent phenotypic modulation of smooth muscle cells has a key role in atherosclerotic plaque pathogenesis. Nat Med. 2015;21:628–637. doi: 10.1038/nm.3866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Golledge J. Abdominal aortic aneurysm: update on pathogenesis and medical treatments. Nat Rev Cardiol. 2018;16:225–242. doi: 10.1038/s41569-018-0114-9. [DOI] [PubMed] [Google Scholar]
- 4.Schepers D., Tortora G., Morisaki H., MacCarrick G., Lindsay M., Liang D., Mehta S.G., Hague J., Verhagen J., van de Laar I., Wessels M., Detisch Y., van Haelst M., Baas A., Lichtenbelt K., Braun K., van der Linde D., Roos-Hesselink J., McGillivray G., Meester J., Maystadt I., Coucke P., El-Khoury E., Parkash S., Diness B., Risom L., Scurr I., Hilhorst-Hofstee Y., Morisaki T., Richer J., Desir J., Kempers M., Rideout A.L., Horne G., Bennett C., Rahikkala E., Vandeweyer G., Alaerts M., Verstraeten A., Dietz H., Van Laer L., Loeys B. A mutation update on the LDS-associated genes TGFB2/3 and SMAD2/3. Hum Mutat. 2018;39:621–634. doi: 10.1002/humu.23407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shi X., DiRenzo D., Guo L.W., Franco S.R., Wang B., Seedial S., Kent K.C. TGF-beta/Smad3 stimulates stem cell/developmental gene expression and vascular smooth muscle cell de-differentiation. PloS One. 2014;9 doi: 10.1371/journal.pone.0093995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.David C.J., Massague J. Contextual determinants of TGFbeta action in development, immunity and cancer. Nat Rev Mol Cell Biol. 2018;19:419–435. doi: 10.1038/s41580-018-0007-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shi X., Guo L.W., Seedial S., Takayama T., Wang B., Zhang M., Franco S.R., Si Y., Chaudhary M.A., Liu B., Kent K.C. Local CXCR4 upregulation in the injured arterial wall contributes to intimal hyperplasia. Stem Cell. 2016;34:2744–2757. doi: 10.1002/stem.2442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.DiRenzo D.M., Chaudhary M.A., Shi X., Franco S.R., Zent J., Wang K., Guo L.W., Kent K.C. A crosstalk between TGF-beta/Smad3 and Wnt/beta-catenin pathways promotes vascular smooth muscle cell proliferation. Cell Signal. 2016;28:498–505. doi: 10.1016/j.cellsig.2016.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shi X., Guo L.W., Seedial S.M., Si Y., Wang B., Takayama T., Suwanabol P.A., Ghosh S., DiRenzo D., Liu B., Kent K.C. TGF-beta/Smad3 inhibit vascular smooth muscle cell apoptosis through an autocrine signaling mechanism involving VEGF-A. Cell Death Dis. 2014;5:e1317. doi: 10.1038/cddis.2014.282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen P.Y., Qin L., Li G., Tellides G., Simons M. Smooth muscle FGF/TGFbeta cross talk regulates atherosclerosis progression. EMBO Mol Med. 2016;8:712–728. doi: 10.15252/emmm.201506181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gomez D., Al Haj Zen A., Borges L.F., Philippe M., Gutierrez P.S., Jondeau G., Michel J.B., Vranckx R. Syndromic and non-syndromic aneurysms of the human ascending aorta share activation of the Smad2 pathway. J Pathol. 2009;218:131–142. doi: 10.1002/path.2516. [DOI] [PubMed] [Google Scholar]
- 12.Gomez D., Kessler K., Michel J.B., Vranckx R. Modifications of chromatin dynamics control Smad2 pathway activation in aneurysmal smooth muscle cells. Circ Res. 2013;113:881–890. doi: 10.1161/CIRCRESAHA.113.301989. [DOI] [PubMed] [Google Scholar]
- 13.Redondo S., Ruiz E., Santos-Gallego C.G., Padilla E., Tejerina T. Pioglitazone induces vascular smooth muscle cell apoptosis through a peroxisome proliferator-activated receptor-gamma, transforming growth factor-beta1, and a Smad2-dependent mechanism. Diabetes. 2005;54:811–817. doi: 10.2337/diabetes.54.3.811. [DOI] [PubMed] [Google Scholar]
- 14.Rodriguez-Vita J., Sanchez-Lopez E., Esteban V., Ruperez M., Egido J., Ruiz-Ortega M. Angiotensin II activates the Smad pathway in vascular smooth muscle cells by a transforming growth factor-beta-independent mechanism. Circulation. 2005;111:2509–2517. doi: 10.1161/01.CIR.0000165133.84978.E2. [DOI] [PubMed] [Google Scholar]
- 15.Gomez D., Kessler K., Borges L.F., Richard B., Touat Z., Ollivier V., Mansilla S., Bouton M.C., Alkoder S., Nataf P., Jandrot-Perrus M., Jondeau G., Vranckx R., Michel J.B. Smad2-dependent protease nexin-1 overexpression differentiates chronic aneurysms from acute dissections of human ascending aorta. Arterioscler Thromb Vasc Biol. 2013;33:2222–2232. doi: 10.1161/ATVBAHA.113.301327. [DOI] [PubMed] [Google Scholar]
- 16.Michel J.B., Jondeau G., Milewicz D.M. From genetics to response to injury: vascular smooth muscle cells in aneurysms and dissections of the ascending aorta. Cardiovasc Res. 2018;114:578–589. doi: 10.1093/cvr/cvy006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tashima Y., He H., Cui J.Z., Pedroza A.J., Nakamura K., Yokoyama N., Iosef C., Burdon G., Koyano T., Yamaguchi A., Fischbein M.P. Androgens accentuate TGF-beta dependent erk/smad activation during thoracic aortic aneurysm formation in marfan syndrome male mice. J Am Heart Assoc. 2020;9 doi: 10.1161/JAHA.119.015773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chan S.W., Hegyi L., Scott S., Cary N.R., Weissberg P.L., Bennett M.R. Sensitivity to Fas-mediated apoptosis is determined below receptor level in human vascular smooth muscle cells. Circ Res. 2000;86:1038–1046. doi: 10.1161/01.res.86.10.1038. [DOI] [PubMed] [Google Scholar]
- 19.Geng Y.J., Henderson L.E., Levesque E.B., Muszynski M., Libby P. Fas is expressed in human atherosclerotic intima and promotes apoptosis of cytokine-primed human vascular smooth muscle cells. Arterioscler Thromb Vasc Biol. 1997;17:2200–2208. doi: 10.1161/01.atv.17.10.2200. [DOI] [PubMed] [Google Scholar]
- 20.Finisguerra V., Prenen H., Mazzone M. Preclinical and clinical evaluation of MET functions in cancer cells and in the tumor stroma. Oncogene. 2016;35:5457–5467. doi: 10.1038/onc.2016.36. [DOI] [PubMed] [Google Scholar]
- 21.Porsch H., Mehic M., Olofsson B., Heldin P., Heldin C.H. Platelet-derived growth factor beta-receptor, transforming growth factor beta type I receptor, and CD44 protein modulate each other's signaling and stability. J Biol Chem. 2014;289:19747–19757. doi: 10.1074/jbc.M114.547273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shaw R.L., Norton C.E., Segal S.S. Apoptosis in resistance arteries induced by hydrogen peroxide: greater resilience of endothelium versus smooth muscle. Am J Physiol Heart Circ Physiol. 2021;320:H1625–H1633. doi: 10.1152/ajpheart.00956.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen B.Y., Huang C.C., Lv X.F., Zheng H.Q., Zhang Y.J., Sun L., Wang G.L., Ma M.M., Guan Y.Y. SGK1 mediates the hypotonic protective effect against H2O2-induced apoptosis of rat basilar artery smooth muscle cells by inhibiting the FOXO3a/Bim signaling pathway. Acta Pharmacol Sin. 2020;41:1073–1084. doi: 10.1038/s41401-020-0357-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lu H., Daugherty A. Aortic aneurysms. Arterioscler Thromb Vasc Biol. 2017;37:e59–e65. doi: 10.1161/ATVBAHA.117.309578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shirasu T., Koyama H., Miura Y., Hoshina K., Kataoka K., Watanabe T. Nanoparticles effectively target rapamycin delivery to sites of experimental aortic aneurysm in rats. PloS One. 2016;11 doi: 10.1371/journal.pone.0157813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Saadat S., Noureddini M., Mahjoubin-Tehran M., Nazemi S., Shojaie L., Aschner M., Maleki B., Abbasi-Kolli M., Rajabi Moghadam H., Alani B., Mirzaei H. Pivotal role of TGF-beta/smad signaling in cardiac fibrosis: non-coding RNAs as effectual players. Front Cardiovasc Med. 2020;7:588347. doi: 10.3389/fcvm.2020.588347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang Y., Xia M., Jin K., Wang S., Wei H., Fan C., Wu Y., Li X., Li X., Li G., Zeng Z., Xiong W. Function of the c-Met receptor tyrosine kinase in carcinogenesis and associated therapeutic opportunities. Mol Canc. 2018;17:45. doi: 10.1186/s12943-018-0796-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang H., Rao B., Lou J., Li J., Liu Z., Li A., Cui G., Ren Z., Yu Z. The function of the HGF/c-Met Axis in hepatocellular carcinoma. Front Cell Dev Biol. 2020;8:55. doi: 10.3389/fcell.2020.00055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cordenonsi M., Dupont S., Maretto S., Insinga A., Imbriano C., Piccolo S. Links between tumor suppressors: p53 is required for TGF-beta gene responses by cooperating with Smads. Cell. 2003;113:301–314. doi: 10.1016/s0092-8674(03)00308-8. [DOI] [PubMed] [Google Scholar]
- 30.Mercer J., Figg N., Stoneman V., Braganza D., Bennett M.R. Endogenous p53 protects vascular smooth muscle cells from apoptosis and reduces atherosclerosis in ApoE knockout mice. Circ Res. 2005;96:667–674. doi: 10.1161/01.RES.0000161069.15577.ca. [DOI] [PubMed] [Google Scholar]
- 31.Coda D.M., Gaarenstroom T., East P., Patel H., Miller D.S., Lobley A., Matthews N., Stewart A., Hill C.S. Distinct modes of SMAD2 chromatin binding and remodeling shape the transcriptional response to NODAL/Activin signaling. eLife. 2017;6 doi: 10.7554/eLife.22474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim S.G., Jong H.S., Kim T.Y., Lee J.W., Kim N.K., Hong S.H., Bang Y.J. Transforming growth factor-beta 1 induces apoptosis through Fas ligand-independent activation of the Fas death pathway in human gastric SNU-620 carcinoma cells. Mol Biol Cell. 2004;15:420–434. doi: 10.1091/mbc.E03-04-0201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Aragon E., Wang Q., Zou Y., Morgani S.M., Ruiz L., Kaczmarska Z., Su J., Torner C., Tian L., Hu J., Shu W., Agrawal S., Gomes T., Marquez J.A., Hadjantonakis A.K., Macias M.J., Massague J. Structural basis for distinct roles of SMAD2 and SMAD3 in FOXH1 pioneer-directed TGF-beta signaling. Genes Dev. 2019;33:1506–1524. doi: 10.1101/gad.330837.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dai X., Shen J., Annam N.P., Jiang H., Levi E., Schworer C.M., Tromp G., Arora A., Higgins M., Wang X.F., Yang M., Li H.J., Zhang K., Kuivaniemi H., Li L. SMAD3 deficiency promotes vessel wall remodeling, collagen fiber reorganization and leukocyte infiltration in an inflammatory abdominal aortic aneurysm mouse model. Sci Rep. 2015;5:10180. doi: 10.1038/srep10180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xie X., Urabe G., Marcho L., Stratton M., Guo L.W., Kent C.K. ALDH1A3 regulations of matricellular proteins promote vascular smooth muscle cell proliferation. iScience. 2019;19:872–882. doi: 10.1016/j.isci.2019.08.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Supporting data is included as supplemental materials.