Abstract
Objective
Abnormal liver function and liver injury related to COVID-19 during hospitalization has received widespread attention. However, the long-term observation of patients’ liver functions after discharge has not been investigated. This study intends to analyze the abnormal liver function in patients one year after they are discharged.
Methods
Serum liver function tests were analyzed for the first time immediately after hospitalization (T1), before discharge (T2), a median of 14.0 (14.0, 15.0) days after discharge (T3) and 1 year (356.0 (347.8, 367.0) days) after discharge (T4). Patients with at least one serum parameter (ALT, AST, ALP, GGT and TB) exceeding the upper limit of reference range were defined as having abnormal liver function.
Results
For the 118 COVID-19 patients with a median follow-up time of 376.0 (71.5, 385.3) days from onset to the end of the follow-up after discharge, the proportion with abnormal liver function in T1, T2, T3 and T4 were 32.2%, 45.8%, 54.8% and 28.8%, respectively. The proportion of patients with at least once abnormal liver function detected from T1 to T2, T1 to T3, T1 to T4 was 60.2%, 77.4% and 88.9%, respectively. From T1 to T4, the ALT, AST, GGT and BMI at admission were significantly higher in the patients with persistently abnormal liver function than in the patients with persistently normal liver function. Abnormal liver function was mainly manifested in the elevation of GGT and TB levels. Multivariate logistics regression analysis showed that age and gender-adjusted ALT (odds ratio [OR]=2.041, 95% confidence interval [CI]: 1.170–3.561, P=0.012) at admission was a risk factor for abnormal liver function in the T4 stage.
Conclusion
Abnormal liver function in patients with COVID-19 can persist from admission to one year after discharge, and therefore, the long-term dynamic monitoring of liver function in patients with COVID-19 is necessary.
Keywords: COVID-19, serum, liver function, follow-up
Introduction
COVID-19 is a global disease and is considered to be a multi-organ disease with complex clinical manifestations.1 Acute sequelae of COVID-19 are also a focus of attention today with more and more reports showing organ damage in COVID-19 patients after discharge.2 Liver injury or abnormalities were reported in COVID-19 patients during and after hospitalization.3 A Chinese study showed that 76.3% of 417 COVID-19 patients had abnormal liver function test results and 21.5% had liver injury during hospitalization.3 A large retrospective cohort study showed that abnormal aspartate aminotransferase (AST) and direct bilirubin levels at admission were independent predictors of COVID-19-related mortality. The study also emphasized the need to monitor liver function, especially AST and direct bilirubin levels in COVID-19 inpatients.4 Liver enzyme abnormalities correlate with disease severity and liver infection with severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) directly leads to liver damage in patients with COVID-19. Therefore, the clearance of virus from the liver and the long-term prognosis for COVID-19 patients need to be monitored.5 COVID-19 disease severity was an independent risk factor for liver injury, while elevated levels of AST and total bilirubin (TB) on admission indicated an increased risk of death.6 The alanine aminotransferase (ALT), alkaline phosphatase (ALP) and γ-glutamyl transpeptidase (GGT) of patients remained abnormally higher for 14 days after discharge.7 A follow-up study showed that liver function improved in patients with COVID-19 3–6 months after discharge.8 However, the outcome for the patient’s liver function 1 year after discharge has not yet been discussed. Here, we aim to present temporal trends in liver function outcomes from admission to 12 months after discharge in a cohort of hospitalized patients with COVID-19-induced pneumonia.
Patients and Methods
Patients
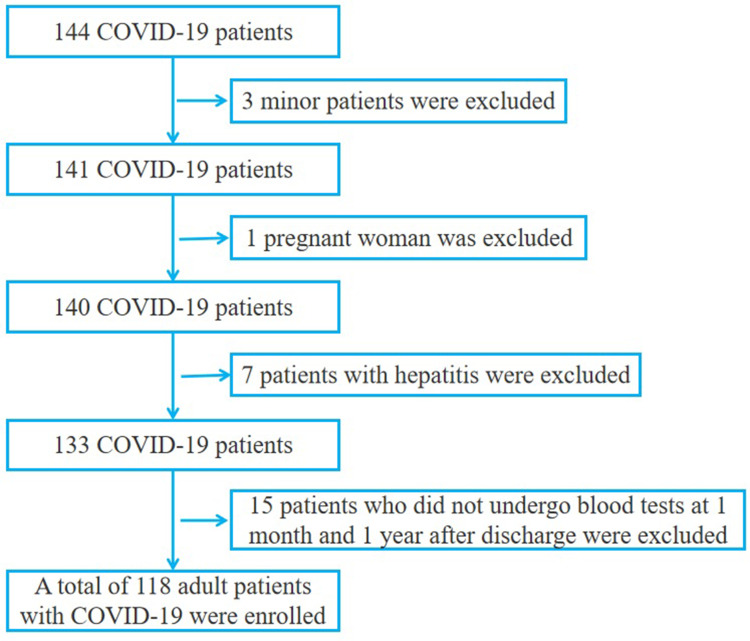
From January 16, 2020 to April 1, 2020, a total of 144 patients with confirmed COVID-19 were placed in wards of Taizhou hospital in Zhejiang province, which is affiliated with Wenzhou Medical University. These patients were diagnosed with COVID-19 according to the Chinese Government Diagnosis and Treatment Guidelines (5th trial version).9 The criteria for severe and non-severe COVID-19 were also categorized according to the aforementioned guidelines.9,10 The inclusion and exclusion criteria are shown in Figure 1. Firstly, 3 minors and 1 pregnant woman were excluded from the study. Then patients with diseases that affect the results of liver function tests conducted upon admission to hospital, such as viral hepatitis (hepatitis B and hepatitis C), autoimmune hepatitis, alcoholic liver disease, non-alcoholic fatty liver disease, biliary obstruction, transmissible hemochromatosis and hepatolenticular degeneration. In addition, patients that had been prescribed anti-tuberculosis drugs (isoniazid), anti-fungal drugs (azole drugs) and anti-epileptic drugs were excluded. Finally, 6 patients with hepatitis B and 1 patient with hepatitis C were excluded. Then 15 patients who did not undergo blood tests at 1 month and 1 year after discharge were excluded. In the end, 118 patients over 18 years old were included in the study. There was no significant difference in the proportion of severe cases, male sex ratio and mean age between the excluded 15 patients and the included 118 patients. The follow-up period was conducted from January 16, 2020 to February 20, 2021. The 118 COVID-19 patients had a median follow-up of 376.0 (71.5, 385.3) days from onset to the end of the follow-up after discharge, with the longest follow-up time being 397 days. The median time from onset to admission was 5.0 (3.0, 9.0) days; the median time from admission to discharge was 21.0 (13.0, 27.3) days, and the median follow-up time after discharge was 340.5 (41.0, 360.0). Four stages were defined as follows: T1: the first blood test after admission, with the median time from onset to blood collection being 7.0 (4.0, 10.0) days (N=118); T2: the blood test before discharge (N=118); T3: the first blood test after discharge, with a median follow-up of 14.0 (14.0, 15.0) days after discharge (N=115); T4: one year after discharge, with a median follow-up of 356.0 (347.8, 367.0) days after discharge (N=66). 53 patients did not participate in the follow-up during T4. The proportion of severe cases, male sex ratio and average age of these 53 patients were similar to the 66 patients who participated in the follow-up, and the difference was not statistically significant. This study was approved by the Medical Ethics Committee of Taizhou Hospital in Zhejiang Province affiliated with Wenzhou Medical University (#K20200209).
Figure 1.
Included and excluded COVID-19 patients.
Methods
The T1 and T2 blood samples were collected during hospitalization, while the T3 and T4 blood samples were collected during the outpatient follow-up. All samples were collected in the morning from patients who fasted. Serum samples were centrifuged at 3500 rpm for 5 min and biochemical parameters were performed with an AU5800 (OLYMPUS, Japan). EDTA-K2 anticoagulant samples were measured using a Sysmex 2100D routine hematology analyzer (Sysmex, Japan) for routine blood examinations. EDTA-K2 anticoagulant samples were measured using a FACSCanto II flow cytometer (BD, USA) for cytokines. Normal reference range for adult liver function: ALT: male: 9–50 (U/L), female: 7–40 (U/L); AST: male: 15–40 (U/L), female: 13–35 (U/L); ALP: male: 45–125 (U/L), female: 35–135 (U/L); GGT: male: 10–60 (U/L), female: 7–45 (U/L); TB: male and female: 3.4–20.5 (μmol/L). Based on previously published articles,11 anything beyond the upper limit reference range for liver function was defined as abnormal liver function.
Statistical Analysis
The Mann–Whitney U-test was used to detect the differences between non-severe and severe patient laboratory indicators in the T1 stage, abnormal distribution was reported as a median (interquartile range [IQR]). Categorical variables were reported as numbers (percentages) in the T1 stage, and the differences between categorical variables were compared using the chi-squared test. Logistics regression analysis was used to evaluate the likelihood of T1 test results being risk factors for liver function abnormalities in stages T3 and T4. We treated laboratory parameters as categorical variables in the logistics regression analysis according to quartiles. Variables with P <0.1 in univariate logistics regression analysis were included in the multifactor logistics regression analysis. Odds ratios (OR) and 95% confidence intervals (CI) for each parameter were also calculated. A P value <0.05 was considered as statistically significant. All statistical analyses were conducted using SPSS (v. 24.0).
Results
This analysis included 118 COVID-19 patients (48.8 ± 13.9 years) consisting of 63 males (47 non-severe patients and 16 severe patients) and 55 females (40 non-severe patients and 15 severe patients) (Table 1). The age of severe COVID-19 patients was significantly higher than that of non-severe patients in both males (P=0.007) and females (P=0.005). The clinical information and laboratory parameters of the patients at first admission (T1) are shown in Table 1. For male patients, serum ALT and AST levels were significantly higher in severe COVID-19 compared with non-severe COVID-19 patients (P=0.013, P=0.001), while ALP serum levels in non-severe COVID-19 patients were significantly higher than those in severe patients (P=0.032). Serum AST and GGT were significantly higher in female patients with severe COVID-19 than in non-severe COVID-19 patients (P=0.029, P=0.009). Male and female patients with severe COVID-19 had a lower ALB level than those with non-severe COVID-19 (P=0.018, P=0.028). The neutrophil count and neutrophil ratio were significantly higher in male patients with severe COVID-19 than in non-severe COVID-19 patients (P<0.001, P<0.001). The lymphocyte count, lymphocyte ratio and monocyte ratio were significantly lower in male patients with severe COVID-19 than in non-severe COVID-19 patients (P<0.001, P<0.001, P=0.002). The lymphocyte count was significantly lower in female patients with severe COVID-19 than in non-severe COVID-19 patients (P=0.001). The levels of CRP and IL-6 in male (P=0.011, P=0.024) and female (P=0.002, P=0.041) patients with severe COVID-19 were significantly higher than those in non-severe COVID-19 patients. The IL-10 level in females with severe COVID-19 were significantly higher than those in non-severe COVID-19 patients (P=0.019). A significantly higher proportion of female patients with severe COVID-19 had fever symptoms compared to non-severe patients (P=0.028). Oxygen inhalation and antibiotic therapy in severe male (P=0.009, P=0.020) and female (P=0.024, P=0.003) patients was significantly higher than that in non-severe patients. Male and female patients with severe COVID-19 had a significantly higher proportion of individuals undergoing immunoglobulin and methylprednisolone therapy than non-severe patients (all P<0.001).
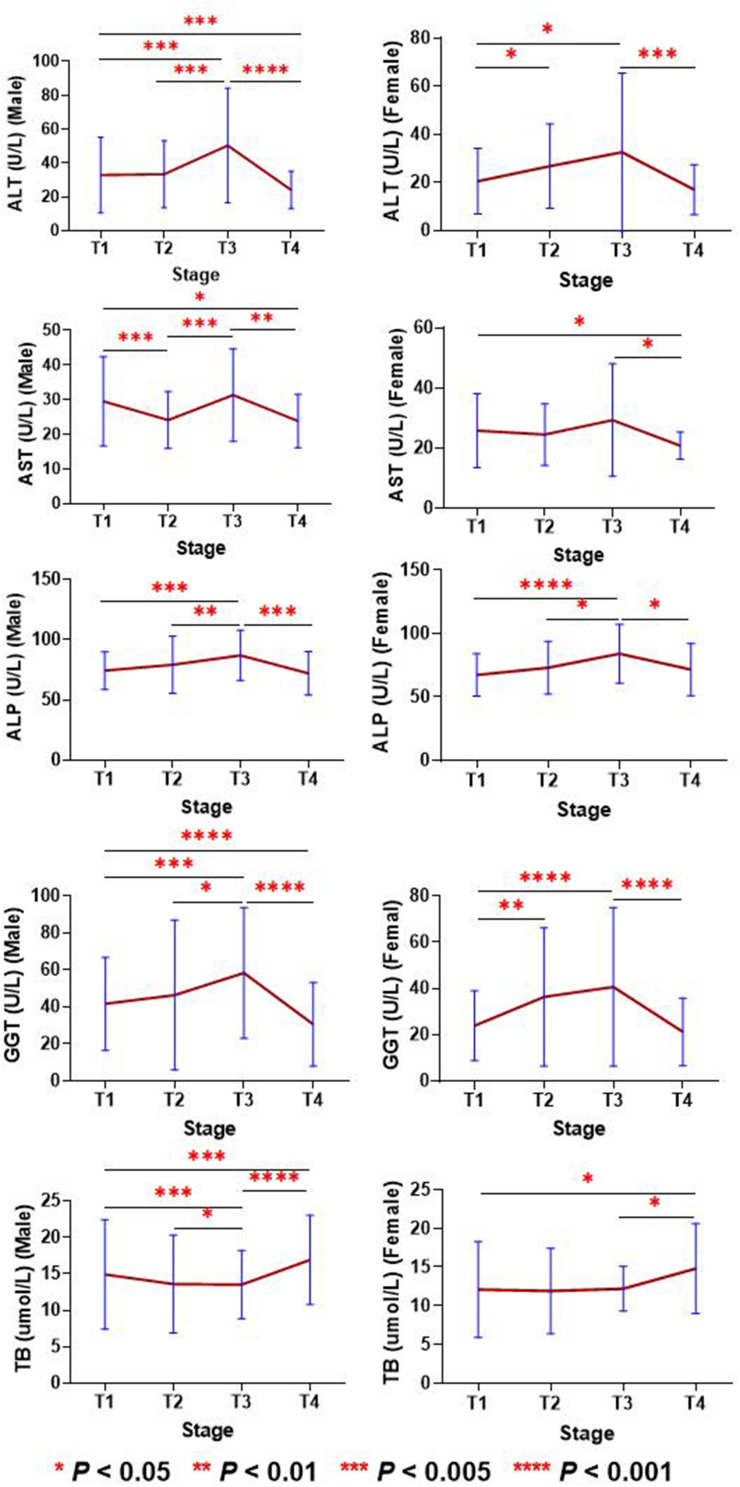
2. The changes in liver function parameters at the serum level from stage T1 to T4 are shown in Figure 2. The serum levels of ALT, ALP and GGT in males and females increased from T1 to T3, and then fell back to the T1 level or lower in T4. In both males and females, AST decreased in the T2 stage, increased in the T3 stage and continued to decrease in the T4 stage to values lower than that of the T1 stage. The TB levels in T2 and T3 were lower in males and females than in T1, then rose higher in T4 to levels higher than in T1. At the end, we observed that ALT, AST, ALP and GGT of both females and males showed a decreasing trend in the T4 stage, while TB in the T4 stage showed an increasing trend.
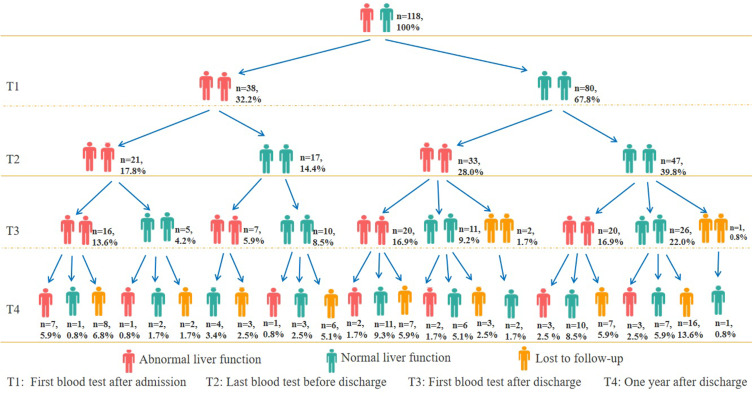
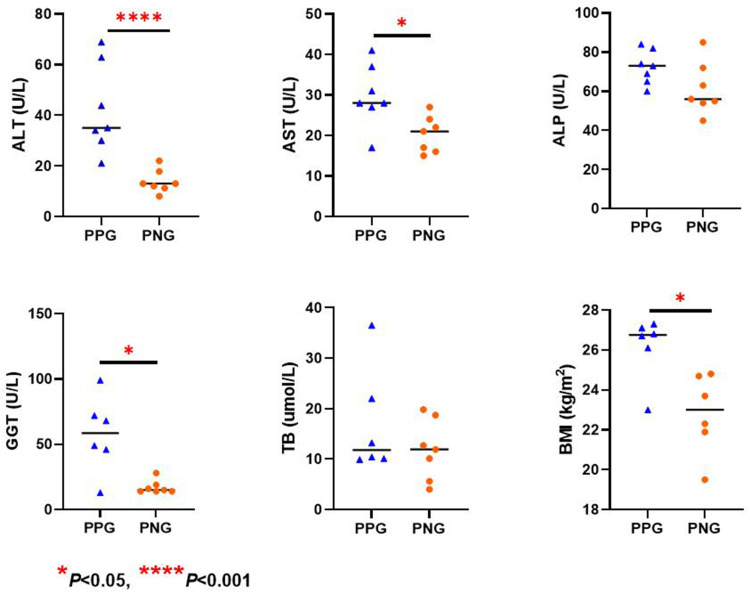
3. The proportion of patients with abnormal liver function in T1, T2, T3 and T4 was 32.2% (38/118), 45.8% (54/118), 54.8% (63/115) and 28.8% (19/66), respectively (Figure 3). The proportion of patients with at least once instance of abnormal liver function detected from T1 to T2, T1 to T3 and T1 to T4 was 60.2% (71/118), 77.4% (89/115) and 88.9% (56/63), respectively. The proportion with abnormal liver function in T3 was significantly higher than that in T1 (P<0.001) and T4 (P<0.001). The proportion of patients with abnormal liver function in T2 was similar to that in T3 (P=0.19), while the proportion of patients with abnormal liver function in T4 was similar to that in T1 (P=0.74). Seven patients (persistently positive group, PPG) with COVID-19 had persistently abnormal liver function from stage T1 to T4. Seven patients (persistently negative group, PNG) with COVID-19 had persistently normal liver function from stage T1 to T4. We found that ALT, AST, GGT and BMI at admission were significantly higher in the PPG than in the PNG (Figure 4). A total of 23 patients with COVID-19 had normal liver function before discharge, but had abnormal liver function at stage T3 and/or T4 (Figure 3).
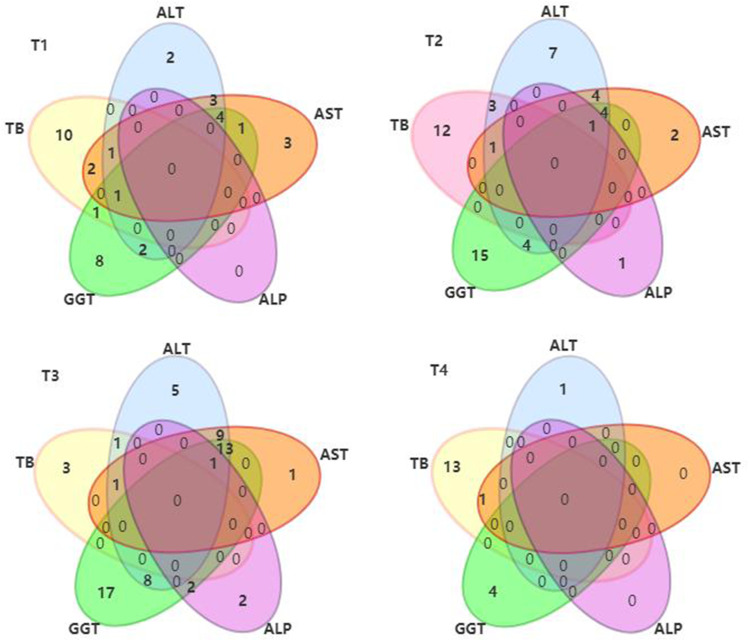
4. As shown in Figure 5, abnormal liver function in T1 was mainly due to an abnormally elevated TB level alone (N=10, 8.5%), followed by an abnormally elevated GGT level alone (N=8, 6.8%). In T2, an abnormal increase of GGT alone was dominant (N=15, 12.7%), followed by an abnormal increase of TB alone (N=12, 10.2%). The abnormal liver function of T3 was mainly due to the GGT level alone (N=17, 14.8%), followed by an abnormal increase of ALT, AST and GGT combined (N=13, 11.3%). T4 was dominated by an abnormal increase of TB alone (N=13, 19.7%).
5. The association of clinical parameters at first admission with abnormal liver function in the T3 and T4 stage is shown in Table 2. Multivariate logistics regression analysis showed that BMI (OR=1.649, 95% CI: 1.059–2.568, P=0.027), GGT (OR=1.983, 95% CI: 1.282–3.066, P=0.002) and percentage of CD8+ T cells (OR=1.727, 95% CI: 1.118–2.667, P=0.014) were risk factors for abnormal liver function in T3 when adjusted for age and sex. IL-2 (OR=0.636, 95% CI: 0.423–0.955, P=0.029) was found to be a protective factor for abnormal liver function in T3 after age and gender calibration. Multivariate logistics regression analysis showed that age and sex-adjusted ALT (OR=2.041, 95% CI: 1.170–3.561, P=0.012) was a risk factor for stage T4 liver dysfunction.
Table 1.
Demographics and Baseline Characteristics of COVID-19 Patients
| Variables | Total (N=118) | Male (N=63) | Female (N=55) | ||||
|---|---|---|---|---|---|---|---|
| Non-Severe (N=47) | Severe (N=16) | P value | Non-Severe (N=40) | Severe (N=15) | P value | ||
| Age (year) | 47.0 (39.0–57.0) | 43.0 (36.0–54.0) | 54.5 (47.0–68.0) | 0.007 | 45.5 (39.3–54.8) | 62.0 (52.0–65.0) | 0.005 |
| BMI (kg/m2) | 24.5 (21.9–26.7) | 24.9 (22.7–27.4) | 26.1 (24.8–28.1) | 0.127 | 22.5 (20.8–25.1) | 24.5 (22.3–27.1) | 0.050 |
| ALT (U/L) | 21.0 (14.0–34.3) | 26.0 (18.0–37.0) | 36.0 (24.0–54.5) | 0.013 | 15.5 (11.0–23.5) | 18.0 (12.0–29.0) | 0.448 |
| AST (U/L) | 24.5 (20.0–31.0) | 24.0 (20.0–30.0) | 36.5 (27.0–57.8) | 0.001 | 21.5 (18.3–26.8) | 28.0 (22.0–30.0) | 0.029 |
| ALP (U/L) | 71.0 (59.8–82.0) | 75.0 (67.0–87.0) | 69.5 (62.5–75.0) | 0.032 | 71.0 (54.0–83.8) | 61.0 (50.0–69.0) | 0.130 |
| GGT (U/L) | 25.5 (17.0–42.5) | 30.0 (23.0–47.0) | 48.0 (26.0–63.5) | 0.127 | 16.5 (13.3–22.8) | 24.0 (18.0–37.0) | 0.009 |
| TB (μmol/L) | 12.0 (8.4–16.6) | 13.1 (8.6–17.0) | 15.3 (11.4–22.3) | 0.146 | 11.3 (7.0–16.2) | 9.9 (6.3–15.5) | 0.597 |
| TP (g/L) | 69.0 (66.5–74.2) | 70.1 (66.6–74.6) | 68.3 (62.8–76.9) | 0.748 | 68.1 (66.0–74.2) | 69.0 (63.3–72.4) | 0.813 |
| ALB (g/L) | 39.6 (37.1–42.1) | 40.9 (37.6–44.5) | 37.4 (35.0–41.0) | 0.018 | 39.4 (37.6–42.1) | 38.0 (33.0–39.9) | 0.028 |
| Crea (μmol/L) | 75.0 (66.0–88.0) | 85.0 (76.5–92.0) | 87.0 (72.8–94.5) | 0.923 | 65.5 (60.0–70.8) | 67.0 (62.0–75.0) | 0.173 |
| BUN (mmol/L) | 4.0 (3.2–5.2) | 4.4 (3.8–5.1) | 4.6 (4.0–8.2) | 0.215 | 3.4 (2.7–4.1) | 4.0 (3.1–5.6) | 0.148 |
| UA (μmol/L) | 259.0 (213.5–327.0) | 317.0 (237.8–369.3) | 255. 5 (190.3–352.0) | 0.195 | 237.5 (208.5–263.8) | 257.0 (165.0–295.0) | 0.584 |
| GLU (mmol/L) | 6.7 (5.5−8.6) | 5.9 (5.2–7.9) | 7.8 (6.9–14.0) | 0.025 | 6.7 (5.6–8.9) | 7.4 (5.5–8.3) | 0.895 |
| TG (mmol/L) | 1.3 (1.0–2.1) | 1.6 (1.1–2.1) | 1.3 (0.9–2.4) | 0.479 | 1.1 (0.9–1.9) | 1.2 (0.9–1.6) | 0.828 |
| TC (mmol/L) | 3.9 (3.4–4.6) | 3.8 (3.4–4.4) | 4.1 (3.7–4.9) | 0.169 | 3.7 (3.4–4.5) | 3.9 (3.2–4.2) | 0.865 |
| HDL-C (mmol/L) | 1.0 (0.9−1.2) | 1.0 (0.8–1.1) | 1.0 (0.9–1.1) | 0.961 | 1.1 (0.9–1.3) | 1.1 (0.9–1.2) | 0.467 |
| LDL-C (mmol/L) | 2.5 (2.2–3.1) | 2.6 (2.3–3.1) | 2.9 (2.3–3.5) | 0.198 | 2.5 (2.2–3.2) | 2.3 (1.9–2.9) | 0.199 |
| IgG (g/L) | 12.6 (10.9–14.3) | 12.2 (10.5–13.8) | 12.2 (10.0–15.9) | 0.903 | 13.3 (11.3–14.6) | 12.2 (10.7–17.5) | 0.586 |
| IgA (g/L) | 2.3 (1.8–3.0) | 2.3 (1.9–3.2) | 2.5 (2.0–3.1) | 0.910 | 2.3 (1.7–3.1) | 2.2 (1.3–2.8) | 0.234 |
| IgM (g/L) | 1.0 (0.8−1.4) | 0.8 (0.7–1.1) | 0.8 (0.5–1.0) | 0.481 | 1.4 (0.9–1.7) | 1.1 (1.0–1.3) | 0.308 |
| C3 (g/L) | 1.3 (1.2–1.5) | 1.4 (1.3–1.6) | 1.5 (1.2–1.6) | 0.958 | 1.2 (1.1−1.4) | 1.2 (1.1 −1.4) | 0.824 |
| C4 (g/L) | 0.4 (0.3–0.5) | 0.4 (0.3–0.5) | 0.4 (0.3–0.5) | 0.508 | 0.3 (0.3–0.4) | 0.4 (0.3–0.5) | 0.074 |
| C1Q (mg/L) | 229.0 (201.5–250.0) | 226.0 (199.0–243.3) | 220.1 (182.8–243.6) | 0.460 | 228.3 (205.5–258.3) | 242.0 (211.3–278.4) | 0.492 |
| ASO (KIU/L) | 54.6 (26.2–102.3) | 56.0 (32.7–114.5) | 57.4 (26.4–129.7) | 0.965 | 30.9 (19.9–83.3) | 61.3 (39.6–146.2) | 0.063 |
| CRP (mg/L) | 10.1 (3.3–26.0) | 9.2 (4.3–22.2) | 25.3 (12.0–56.4) | 0.011 | 4.3 (1.1–14.0) | 21.3 (9.7–37.6) | 0.002 |
| WBC (×109/L) | 5.1 (4.4–6.8) | 5.0 (4.5–6.4) | 6.8 (5.7–8.6) | 0.002 | 5.0 (4.2–6.6) | 4.4 (3.8–6.2) | 0.199 |
| RBC (×1012/L) | 4.5 (4.2–4.9) | 4.9 (4.5–5.2) | 4.8 (4.2–5.3) | 0.868 | 4.3 (4.2–4.5) | 4.4 (4.1–4.5) | 0.379 |
| Neutrophil count (×109/L) | 3.4 (2.6–5.0) | 3.3 (2.4–4.5) | 5.5 (4.6–7.4) | <0.001 | 3.3 (2.6–4.5) | 3.1 (2.4–5.0) | 0.828 |
| Lymphocyte count (×109/L) | 1.2 (0.8–1.6) | 1.3 (1.0–1.6) | 0.7 (0.4–0.9) | <0.001 | 1.4 (1.0–1.7) | 0.9 (0.6–1.0) | 0.001 |
| Monocyte coun (×109/L) | 0.4 (0.3–0.5) | 0.5 (0.4–0.6) | 0.4 (0.3–0.6) | 0.402 | 0.4 (0.3–0.5) | 0.3 (0.2–0.5) | 0.325 |
| Neutrophil ratio (%) | 67.2 (58.4–76.4) | 65.4 (56.9–71.0) | 82.7 (72.8–89.8) | <0.001 | 65.0 (56.6–71.4) | 71.2 (60.5–81.9) | 0.112 |
| Lymphocyte ratio (%) | 23.5 (16.0–29.5) | 24.5 (20.1–28.1) | 10.2 (6.4–15.5) | <0.001 | 26.4 (21.1–34.2) | 21.2 (11.1–28.5) | 0.089 |
| Monocyte ratio (%) | 7.8 (5.8–10.4) | 9.3 (6.9–10.7) | 5.3 (3.5–7.8) | 0.002 | 7.2 (6.2–9.0) | 7.6 (5.1–11.3) | 0.835 |
| HB (g/L) | 135.0 (126.8–149.3) | 147.0 (135.0–157.0) | 147.0 (126.3–157.5) | 0.458 | 129.0 (123.3–135.0) | 130.0 (123.0–137.0) | 0.880 |
| PLT (×109/L) | 203.0 (165.8–247.0) | 204.0 (166.0–241.0) | 182.5 (133.5–253.5) | 0.463 | 215.0 (188.3–259.3) | 175.0 (132.0−247.0) | 0.052 |
| IL-2 (pg/mL) | 1.4 (0.9–1.8) | 1.4 (0.9–1.8) | 1.0 (0.6–1.4) | 0.077 | 1.4 (1.1–2.1) | 1.3 (0.9–2.0) | 0.524 |
| IL-4 (pg/mL) | 1.5 (0.9–2.1) | 1.7 (0.9−2.2) | 1.2 (0.7–1.9) | 0.231 | 1.4 (0.9–2.1) | 1.4 (1.1–2.3) | 0.481 |
| IL-6 (pg/mL) | 7.4 (3.3–17.5) | 6.7 (3.1–15.6) | 16.6 (6.0–40.0) | 0.024 | 5.3 (2.7–11.9) | 10.9 (5.1–32.6) | 0.041 |
| IL-10 (pg/mL) | 3.6 (2.6–5.7) | 3.6 (2.4–5.3) | 3.9 (3.1–8.5) | 0.081 | 3.2 (2.3–4.7) | 4.4 (3.4–8.2) | 0.019 |
| TNF-α (pg/mL) | 1.2 (0.6–1.9) | 1.2 (0.6–2.0) | 1.3 (0.6–2.0) | 0.935 | 1.1 (0.5–1.8) | 1.1 (0.5–1.6) | 0.828 |
| IFN-γ (pg/mL) | 1.9 (1.2–2.8) | 2.0 (1.4–3.0) | 1.5 (1.0–2.4) | 0.380 | 2.0 (1.1–3.1) | 2.1 (1.0–2.8) | 0.931 |
| Percentage of CD4+ T cells (%) | 29.9 (23.4–35.9) | 29.3 (23.6–36.4) | 23.6 (16.5–33.3) | 0.053 | 31.6 (26.7–36.6) | 28.0 (25.6–35.6) | 0.199 |
| Percentage of CD8+ T cells (%) | 23.5 (19.4–29.6) | 25.7 (20.9–33.5) | 21.2 (14.4–26.2) | 0.023 | 23.4 (20.2–28.5) | 25.9 (16.2–30.3) | 0.938 |
| Percentage of CD3+ T cells (%) | 57.3 (49.7–66.7) | 58.4 (53.5–66.9) | 44.5 (41.3–56.0) | <0.001 | 61.1 (52.0–68.0) | 54.1 (45.0–59.0) | 0.107 |
| Symptoms - no. (%) | |||||||
| Fever | 86 (72.9) | 34 (72.3) | 15 (93.8) | 0.152 | 23 (57.5) | 14 (93.3) | 0.028 |
| Cough | 58 (49.2) | 21 (44.7) | 9 (56.3) | 0.424 | 21 (52.5) | 7 (46.7) | 0.700 |
| Expectoration | 23 (19.5) | 8 (17.0) | 3 (18.8) | 1.000 | 9 (22.5) | 3 (20.0) | 1.000 |
| Comorbidity- no. (%) | |||||||
| Hypertension | 21 (17.8) | 9 (19.1) | 6 (37.5) | 0.251 | 4 (10.0) | 2 (13.3) | 0.660 |
| Diabetes | 11 (9.3) | 4 (8.5) | 3 (18.8) | 0.506 | 3 (7.5) | 1 (6.7) | 1.000 |
| Treatment - no. (%) | |||||||
| Oxygen inhalation | 88 (74.6) | 29 (61.7) | 16 (100.0) | 0.009 | 28 (70.0) | 15 (100.0) | 0.024 |
| Antibiotics | 35 (29.7) | 10 (21.3) | 9 (56.3) | 0.020 | 7 (17.5) | 9 (60.0) | 0.003 |
| Antiviral drug | 118 (100.0) | 47 (100.0) | 16 (100) | 40 (100.0) | 15 (100.0) | ||
| Immunoglobulin | 34 (28.8) | 2 (4.3) | 15 (93.8) | <0.001 | 4 (10.0) | 13 (86.7) | <0.001 |
| Methylprednisoloe | 35 (29.7) | 3 (6.4) | 15 (93.8) | <0.001 | 4 (10.0) | 13 (86.7) | <0.001 |
| Chinese medicine | 118 (100.0) | 47 (100.0) | 16 (100.0) | 40 (100.0) | 15 (100.0) | ||
Abbreviations: no. (%), number; ALT, Alanine aminotransferase; AST, Aspartate aminotransferase; ALP, Alkaline phosphatase; GGT, γ-glutamyl transpeptidase; TB, Total bilirubin; TP, Total protein; ALB, Albumin; Crea, Creatinine; BUN, Blood urea nitrogen; UA, Uric acid; GLU, Glucose; TG, Triglyceride; TC, Total cholesterol; HDL-C, High density lipoprotein cholesterol; LDL-C, Low density lipoprotein cholesterol; IgG, Immunoglobulin G; IgA, Immunoglobulin A; IgM, Immunoglobulin M; C3, Complement C3; C4, Complement C4; C1q, Complement C1q; ASO, Anti streptolysin O; CRP, C-reactive protein; WBC, White blood cell count; N, Neutrophil count; L, Lymphocyte count; M, Monocyte count; NR, Neutrophil ratio; LR, Lymphocyte ratio; MR, Monocyte ratio; RBC, Red blood cell count; Hb, Hemoglobin; PLT, Platelet; IL-2, Interleukin-2; IL-4, Interleukin-4; IL-6, Interleukin-6; IL-10, Interleukin-10; TNF-α, Tumor necrosis factor-α; IFN-γ, Interferon-γ.
Figure 2.
Changes in liver function parameters from stage T1 to T4 in patients with COVID-19. *P value < 0.05; **P value < 0.01; ***P value < 0.005; ****P value < 0.001.
Figure 3.
Dynamic changes in abnormal liver function for 118 patients with COVID-19 from stage T1 to T4.
Figure 4.
Comparison of liver function indicators at admission in COVID-19 patients with persistent abnormal liver function (N=7) and persistent normal liver function (N=7). *P value < 0.05; ****P value < 0.001.
Figure 5.
The combination of ALT, AST, ALP, GGT and TB exceeded the reference range in COVID-19 positive individuals with abnormal liver function.
Table 2.
Association of First Admission Clinical Parameters with Abnormal Liver Function of T3 and T4 Stage
| Variables | T3 Stage | T4 Stage | ||||||
|---|---|---|---|---|---|---|---|---|
| Univariate★ OR (95% CI) | P values | Multivariate★ OR (95% CI) | P values | Univariate★ OR (95% CI) | P values | Multivariate★ OR (95% CI) | P values | |
| BMI (kg/m2) | 1.730 (1.176, 2.546) | 0.005 | 1.649 (1.059, 2.568) | 0.027 | 0.967 (0.578, 1.617) | 0.898 | ||
| ALT (U/L) | 1.785 (1.205, 2.645) | 0.004 | 1.349 (0.791, 2.300) | 0.272 | 1.807 (0.979, 3.334) | 0.058 | 2.041 (1.170, 3.561) | 0.012 |
| AST (U/L) | 1.358 (0.949, 1.943) | 0.094 | 0.866 (0.482, 1.557) | 0.631 | 1.511 (0.899, 2.542) | 0.119 | ||
| ALP (U/L) | 1.156 (0.818, 1.635) | 0.411 | 1.201 (0.715, 2.019) | 0.489 | ||||
| GGT (U/L) | 1.827 (1.224, 2.729) | 0.003 | 1.983 (1.282, 3.066) | 0.002 | 1.091 (0.634, 1.879) | 0.752 | ||
| TB (μmol/L) | 1.095 (0.776, 1.545) | 0.696 | 1.115 (0.674, 1.844) | 0.672 | ||||
| TP (g/L) | 1.073 (0.758, 1.518) | 0.692 | 1.057 (0.644, 1.737) | 0.826 | ||||
| ALB (g/L) | 1.250 (0.856, 1.827) | 0.249 | 1.352 (0.780, 2.343) | 0.283 | ||||
| Crea (μmol/L) | 0.875 (0.557, 1.374) | 0.562 | 1.008 (0.499, 2.036) | 0.982 | ||||
| BUN (mmol/L) | 0.872 (0.601, 1.267) | 0.474 | 0.729 (0.416, 1.278) | 0.270 | ||||
| UA (μmol/L) | 1.048 (0.727, 1.510) | 0.802 | 0.936 (0.557, 1.575) | 0.805 | ||||
| GLU (mmol/L) | 0.927 (0.648, 1.326) | 0.679 | 1.084 (0.659, 1.783) | 0.751 | ||||
| TG (mmol/L) | 1.108 (0.780, 1.574) | 0.566 | 0.588 (0.337, 1.025) | 0.061 | 0.585 (0.336, 1.019) | 0.059 | ||
| TC (mmol/L) | 1.030 (0.730, 1.454) | 0.864 | 1.324 (0.803, 2.813) | 0.272 | ||||
| HDL-C (mmol/L) | 0.874 (0.618, 1.236) | 0.446 | 1.189 (0.725, 1.950) | 0.493 | ||||
| LDL-C (mmol/L) | 1.023 (0.724, 1.445) | 0.899 | 1.296 (0.788, 2.132) | 0.307 | ||||
| IgG (g/L) | 0.896 (0.627, 1.280) | 0.547 | 0.779 (0.461, 1.318) | 0.352 | ||||
| IgA (g/L) | 0.852 (0.595, 1.219) | 0.380 | 0.937 (0.544, 1.616) | 0.815 | ||||
| IgM (g/L) | 0.832 (0.559, 1.238) | 0.364 | 1.018 (0.536, 1.931) | 0.957 | ||||
| C3 (g/L) | 1.363 (0.937, 1.981) | 0.105 | 1.042 (0.625, 1.738) | 0.873 | ||||
| C4 (g/L) | 1.155 (0.806, 1.655) | 0.433 | 1.020 (0.607, 1.714) | 0.941 | ||||
| C1Q (mg/L) | 0.968 (0.675, 1.389) | 0.860 | 1.120 (0.653, 1.920) | 0.681 | ||||
| ASO (KIU/L) | 1.110 (0.773, 1.594) | 0.570 | 0.851 (0.497, 1.455) | 0.555 | ||||
| CRP (mg/L) | 1.034 (0.720, 1.484) | 0.858 | 1.172 (0.683, 2.010) | 0.565 | ||||
| WBC (×109/L) | 0.977 (0.694, 1.375) | 0.893 | 0.753 (0.455, 1.248) | 0.271 | ||||
| Neutrophil count (×109/L) | 0.953 (0.674, 1.349) | 0.787 | 0.756 (0.453, 1.261) | 0.284 | ||||
| Lymphocyte count (×109/L) | 1.213 (0.849, 1.733) | 0.288 | 0.757 (0.446, 1.283) | 0.300 | ||||
| Monocyte coun (×109/L) | 0.985 (0.693, 1.400) | 0.932 | 1.334 (0.802, 2.219) | 0.267 | ||||
| Neutrophil ratio (%) | 0.823 (0.578, 1.173) | 0.282 | 1.009 (0.606, 1.681) | 0.973 | ||||
| Lymphocyte ratio (%) | 1.192 (0.831, 1.709) | 0.340 | 0.937 (0.548, 1.604) | 0.813 | ||||
| Monocyte ratio (%) | 1.080 (0.760, 1.536) | 0.667 | 1.504 (0.900, 2.612) | 0.119 | ||||
| RBC (×1012/L) | 1.344 (0.879, 2.056) | 0.172 | 1.013 (0.567, 1.810) | 0.966 | ||||
| HB (g/L) | 1.428 (0.916, 2.228) | 0.116 | 1.023 (0.565, 1.854) | 0.939 | ||||
| PLT (×109/L) | 0.850 (0.600, 1.202) | 0.358 | 0.867 (0.525, 1.433) | 0.578 | ||||
| IL-2 (pg/mL) | 0.716 (0.501, 1.024) | 0.067 | 0.636 (0.423, 0.955) | 0.029 | 1.103 (0.662, 1.837) | 0.706 | ||
| IL-4 (pg/mL) | 0.832 (0.590, 1.172) | 0.292 | 0.998 (0.597, 1.666) | 0.993 | ||||
| IL-6 (pg/mL) | 1.231 (0.865, 1.752) | 0.249 | 1.103 (0.662, 1.839) | 0.706 | ||||
| IL-10 (pg/mL) | 1.038 (0.736, 1.465) | 0.830 | 1.147 (0.697, 1.887) | 0.589 | ||||
| TNF-α (pg/mL) | 0.777 (0.547, 1.104) | 0.159 | 1.115 (0.668, 1.861) | 0.678 | ||||
| IFN-γ (pg/mL) | 0.867 (0.612, 1.228) | 0.421 | 0.924 (0.561, 1.522) | 0.757 | ||||
| Percentage of CD4+ T cells (%) | 1.027 (0.726, 1.453) | 0.879 | 1.330 (0.800, 2.210) | 0.271 | ||||
| Percentage of CD8+ T cells (%) | 1.472 (1.023, 2.118) | 0.037 | 1.727 (1.118, 2.667) | 0.014 | 1.032 (0.610, 1.745) | 0.906 | ||
| Percentage of CD3+ T cells (%) | 1.147 (0.804, 1.635) | 0.449 | 1.342 (0.783, 2.297) | 0.284 | ||||
Abbreviations: ALT, Alanine aminotransferase; AST, Aspartate aminotransferase; ALP, Alkaline phosphatase; GGT, γ-glutamyl transpeptidase; TB, Total bilirubin; TP, Total protein; ALB, Albumin; Crea, Creatinine; BUN, Blood urea nitrogen; UA, Uric acid; GLU, Glucose; TG, Triglyceride; TC, Total cholesterol; HDL-C, High density lipoprotein cholesterol; LDL-C, Low density lipoprotein cholesterol; IgG, Immunoglobulin G; IgA, Immunoglobulin A; IgM, Immunoglobulin M; C3, Complement C3; C4, Complement C4; C1q, Complement C1q; ASO, Anti streptolysin O; CRP, C-reactive protein; WBC, White blood cell count; N, Neutrophil count; L, Lymphocyte count; M, Monocyte count; NR, Neutrophil ratio; LR, Lymphocyte ratio; MR, Monocyte ratio; RBC, Red blood cell count; Hb, Hemoglobin; PLT, Platelet; IL-2, Interleukin-2; IL-4, Interleukin-4; IL-6, Interleukin-6; IL-10, Interleukin-10; TNF-α, Tumor necrosis factor-α; IFN-γ, Interferon-γ; ★, Adjusted by age and gender. OR, odds ratio; CI, confidence interval.
Discussion
Abnormalities of liver enzymes are common in patients with COVID-19.5 One study showed that liver injury and abnormal liver function test results during hospitalization were as high as 21.5% and 76.3%, respectively.3 Another study showed that 58% of patients had abnormal liver function upon admission, and revealed that abnormal liver function at admission was an independent predictor of ICU transfer or death.11 In our study, 32.2% of patients had abnormal liver function when they were admitted to hospital (stage T1). At this time, the patients were still in the early stage of the disease process, and did not receive treatment. The damage of patients’ liver function may have been caused by the SARS-CoV-2 virus. Studies have shown that the spike protein of SARS-CoV-2 binds to the ACE2 receptor of bile duct cells, resulting in decreased function and hepatobiliary injury.12 As the disease progressed, the proportion of abnormal liver function up to discharge (stage T2) was 45.8%, which was probably due to liver damage being caused by the SARS-CoV-2 virus, drugs and cytokine storm syndrome. Studies have shown that using many medications to treat the SARS-CoV-2 virus, such as antiviral drugs, acetaminophen, steroids, corticosteroids, immunomodulators, antibiotics and antipyretics, may cause hepatotoxicity, resulting in liver dysfunction.13,14 One study showed that abnormal liver function may be related to hepatocyte injury caused by ischemia and hypoxia, and a systemic immune response caused by cytokine storm syndrome.15 We currently know that two consecutive negative SARS-CoV-2 RNA tests in nasopharyngeal swab samples were determined as the hospital’s discharge standard.16 In this study, the proportion of abnormal liver function one month after discharge (T3) was the highest (54.8%), which shows that although the patients reached the discharge standard and damage caused by the virus had not taken place, liver damage caused by the treatment process still persisted. It is therefore suggested that great attention be paid to the monitoring and protection of liver function after discharge. A study showed the abnormal values of ALT, GGT and ALP remained abnormally elevated for 14 days after discharge.7 This result is similar to the one found in our study. We found that the proportion of patients with abnormal liver function continued to be 28.8% one year after discharge (T4). In particular, 7 patients with COVID-19 continued to have abnormal liver function from stage T1 to stage T4. This result suggests that abnormal liver function can occur in patients for a long time after discharge, and it is therefore necessary to pay attention to long-term liver function after discharge. Although the proportion of abnormal liver function in the T1, T2, T3 and T4 stages was 32.2%, 45.8%, 54.8% and 28.8%, respectively, the proportion was lower or close to 50%. However, only 7 patients were negative throughout the entire process. We observed that the proportion of patients with abnormal liver function detected at least once from T1 to T2, T1 to T3 and T1 to T4 was 60.2%, 77.4% and 88.9%, respectively. This reveals that the population with abnormal liver function in each stage is different. Some patients have the alternation of normal and abnormal liver function, indicating that on the one hand, the liver is damaged and on the other hand, it has strong self-healing function.17 However, the proportion of patients with abnormal liver function detected at least once from T1 to T4 is amazing, therefore, long-term dynamic monitoring of patients’ liver function status is necessary.
SARS-CoV-2 is the virus that causes COVID-19.18 The hypothesized mechanism is through the host angiotensin-converting enzyme 2 (ACE2) receptor, which is present in lung tissue.19 There has also been direct evidence of liver infection with the SARS-CoV-2 virus, which directly causes liver damage in patients with COVID-19.5 The ACE2 receptor has been expressed in hepatic bile duct cells.20 COVID-19 patients infected with SARS-CoV-2 with an impaired liver function exhibit ACE2 receptor overexpression and cytokine storm overexpression, which ultimately exacerbates liver function impairment and mortality.21 In our study, individual patients with liver function impairment were mainly distinguished by elevated GGT and/or TB. GGT is a standard liver enzyme test which reflects the involvement of the biliary tract.22 GGT and TB levels are commonly monitored as indicators of the trend and degree of cholestasis.23 Our results further verified that liver dysfunction in COVID-19 patients was mainly caused by biliary tract injury, which was consistent with many previous reports. One study shows that cholangiopathy is a late complication of severe COVID-19, which may lead to progressive biliary tract injury and liver failure.24 An in vitro experiment showed that SARS-CoV-2 infection disrupted bile acid transport by regulating the expression of tight junction formation and bile acid transport related genes. This study supports the view that liver injury in patients suffering from COVID-19 may be caused by direct bile duct cell injury and bile acid accumulation caused by viral infection.25
A cytokine storm is characteristic of COVID-19.26 Significant release of proinflammatory cytokines and changes in the number and function of lymphocytes are related symptoms.27 We found that the percentage of CD8+ T cells in the first blood test (T1) was a risk factor for stage T3 liver dysfunction. Studies have shown lymphocyte count is a risk factor associated with prolonged shedding time of SARS-CoV-2.28 Another paper has also shown that the CD8+ T cell count is a risk factor for an increased duration of positive SARS-CoV-2.29 IL-2, an important factor in the occurrence of COVID-19-induced cytokine storm, was identified in this study as a protective factor for abnormal liver function in T3. One study found that there was higher IL-2 induction levels in asymptomatic or mild patients than in moderate or severe patients. It was thus proposed that IL-2 had a determining effect on the severity of COVID-19.30 The decrease of IL-2 concentration in the plasma of patients with COVID-19 is an early warning sign of patients deteriorating due to the disease. Moderate supplementation of IL-2 can improve the immune disorder of patients with severe COVID-19 and reduce their mortality.31 Our study shows that BMI is a risk factor for T3 stage abnormal liver function after discharge from hospital. Studies have shown that the severity of SARS-CoV-2 disease increased with an increase in BMI and that obesity was a risk factor for the severity of SARS-CoV-2.32 One article showed that mortality was strongly associated with BMI.33 Another article suggests that the indexes of liver function in obese patients were significantly abnormal, and that the antibody titer of COVID-19 in obese patients was negatively correlated with BMI.34 Our research shows that GGT is a risk factor for stage T3 liver function abnormality. Studies have shown that GGT is associated with the severity of SARS-CoV-2 infection.35 Elevated GGT levels are more common in severe patients, and GGT is positively correlated with a prolonged hospital stay and disease severity.36 Our research shows that ALT were risk factors for abnormal liver function in the T4 stage. A meta-analysis showed that elevated ALT was significantly associated with a poor outcome of the disease.37 COVID-19 patients with elevated levels of ALT or AST had significantly more severe symptoms, and their admission rate to ICU was higher.38 The relationship between ALT and abnormal liver function after 1 year in our study suggests that it can be used as a long-term monitoring indicator for abnormal liver function.
In conclusion, our study showed that hospitalized COVID-19 patients had abnormal liver function that persisted for up to 1 year after discharge, with ALT being a risk factor for abnormal liver function after 1 year. The long-term dynamic monitoring of liver function in patients with COVID-19 is necessary.
Ethical Approval and Informed Consent
Informed consent was obtained from the patients included in this study. This research did not affect the patients’ health and privacy. All procedures performed in the studies involving human participants accorded with the ethical standards of the Medical Ethics Committee of Taizhou Hospital in Zhejiang Province affiliated with Wenzhou Medical University, and with the 1964 Helsinki Declaration and its later amendments, or other comparable ethical standards.
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Gupta A, Madhavan MV, Sehgal K, et al. Extrapulmonary manifestations of COVID-19. Nat Med. 2020;26:1017–1032. doi: 10.1038/s41591-020-0968-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Al-Aly Z, Xie Y, Bowe B. High-dimensional characterization of post-acute sequelae of COVID-19. Nature. 2021;594:259–264. doi: 10.1038/s41586-021-03553-9 [DOI] [PubMed] [Google Scholar]
- 3.Cai Q, Huang D, Yu H, et al. COVID-19: abnormal liver function tests. J Hepatol. 2020;73:566–574. doi: 10.1016/j.jhep.2020.04.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ding ZY, Li GX, Chen L, et al. Association of liver abnormalities with in-hospital mortality in patients with COVID-19. J Hepatol. 2021;74:1295–1302. doi: 10.1016/j.jhep.2020.12.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang Y, Liu S, Liu H, et al. SARS-CoV-2 infection of the liver directly contributes to hepatic impairment in patients with COVID-19. J Hepatol. 2020;73:807–816. doi: 10.1016/j.jhep.2020.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang SS, Dong L, Wang GM, et al. Progressive liver injury and increased mortality risk in COVID-19 patients: a retrospective cohort study in China. World J Gastroenterol. 2021;27:835–853. doi: 10.3748/wjg.v27.i9.835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.An YW, Song S, Li WX, et al. Liver function recovery of COVID-19 patients after discharge, a follow-up study. Int J Med Sci. 2021;18:176–186. doi: 10.7150/ijms.50691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li G, Du L, Cao X, et al. Follow-up study on serum cholesterol profiles and potential sequelae in recovered COVID-19 patients. BMC Infect Dis. 2021;21:299. doi: 10.1186/s12879-021-05984-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.National Health Commission of the PRC. Diagnosis and treatment protocol for COVID-19 (trial version 5); 2020. Available from: http://www.nhc.gov.cn/jkj/s3577/202002/a5d6f7b8c48c451c87dba14889b30147.shtml. Accessed April 1, 2022.
- 10.Shen B, Yi X, Sun Y, et al. Proteomic and metabolomic characterization of COVID-19 patient sera. Cell. 2020;182:59–72. doi: 10.1016/j.cell.2020.05.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Piano S, Dalbeni A, Vettore E, et al. Abnormal liver function tests predict transfer to intensive care unit and death in COVID-19. Liver Int. 2020;40:2394–2406. doi: 10.1111/liv.14565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lozano-Sepulveda SA, Galan-Huerta K, Martínez-Acuña N, et al. SARS-CoV-2 another kind of liver aggressor, how does it do that? Ann Hepatol. 2020;19:592–596. doi: 10.1016/j.aohep.2020.08.062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Boeckmans J, Rodrigues RM, Demuyser T, et al. COVID-19 and drug-induced liver injury: a problem of plenty or a petty point? Arch Toxicol. 2020;94:1367–1369. doi: 10.1007/s00204-020-02734-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Marjot T, Webb GJ, Barritt AS, et al. COVID-19 and liver disease: mechanistic and clinical perspectives. Nat Rev Gastroenterol Hepatol. 2021;18:348–364. doi: 10.1038/s41575-021-00426-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li X, Zhang ZC, Zhang PL. Severe COVID-19 patients with liver injury: a seven-case series. Eur Rev Med Pharmacol Sci. 2020;24:7855–7860. doi: 10.26355/eurrev_202007_22290 [DOI] [PubMed] [Google Scholar]
- 16.Han H, Luo Q, Mo F, et al. SARS-CoV-2 RNA more readily detected in induced sputum than in throat swabs of convalescent COVID-19 patients. Lancet Infect Dis. 2020;20:655–656. doi: 10.1016/S1473-3099(20)30174-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang R, Zhang X, Gracia-Sancho J, et al. Liver regeneration: cellular origin and molecular mechanisms. Liver Int. 2022. doi: 10.1111/liv.15174 [DOI] [PubMed] [Google Scholar]
- 18.Yan R, Zhang Y, Li Y, et al. Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science. 2020;367:1444–1448. doi: 10.1126/science.abb2762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang S, Qiu Z, Hou Y, et al. AXL is a candidate receptor for SARS-CoV-2 that promotes infection of pulmonary and bronchial epithelial cells. Cell Res. 2021;31:126–140. doi: 10.1038/s41422-020-00460-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jothimani D, Venugopal R, Abedin MF, et al. COVID-19 and the liver. J Hepatol. 2020;73:1231–1240. doi: 10.1016/j.jhep.2020.06.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ali FEM, Mohammedsaleh ZM, Ali MM, et al. Impact of cytokine storm and systemic inflammation on liver impairment patients infected by SARS-CoV-2: prospective therapeutic challenges. World J Gastroenterol. 2021;27:1531–1552. doi: 10.3748/wjg.v27.i15.1531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Neuman MG, Malnick S, Chertin L. Gamma glutamyl transferase - an underestimated marker for cardiovascular disease and the metabolic syndrome. J Pharm Pharm Sci. 2020;23:65–74. doi: 10.18433/jpps30923 [DOI] [PubMed] [Google Scholar]
- 23.Cutrin JC, Cantino D, Biasi F, et al. Reperfusion damage to the bile canaliculi in transplanted human liver. Hepatology. 1996;24:1053–1057. doi: 10.1002/hep.510240512 [DOI] [PubMed] [Google Scholar]
- 24.Faruqui S, Okoli FC, Olsen SK, et al. Cholangiopathy after severe COVID-19: clinical features and prognostic implications. Am J Gastroenterol. 2021;116:1414–1425. doi: 10.14309/ajg.0000000000001264 [DOI] [PubMed] [Google Scholar]
- 25.Zhao B, Ni C, Gao R, et al. Recapitulation of SARS-CoV-2 infection and cholangiocyte damage with human liver ductal organoids. Protein Cell. 2020;11:771–775. doi: 10.1007/s13238-020-00718-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Meng QF, Tian R, Long H, et al. Capturing cytokines with advanced materials: a potential strategy to tackle COVID-19 cytokine storm. Adv Mater. 2021;33:e2100012. doi: 10.1002/adma.202100012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang L, Liu S, Liu J, et al. COVID-19: immunopathogenesis and immunotherapeutics. Signal Transduct Target Ther. 2020;5:128. doi: 10.1038/s41392-020-00243-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shi D, Wu W, Wang Q, et al. Clinical characteristics and factors associated with long-term viral excretion in patients with severe acute respiratory syndrome coronavirus 2 Infection: a Single-Center 28-Day Study. J Infect Dis. 2020;222:910–918. doi: 10.1093/infdis/jiaa388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lin A, He ZB, Zhang S, et al. Early risk factors for the duration of severe acute respiratory syndrome coronavirus 2 viral positivity in patients with coronavirus disease 2019. Clin Infect Dis. 2020;71:2061–2065. doi: 10.1093/cid/ciaa490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tjan LH, Furukawa K, Nagano T, et al. Early differences in cytokine production by severity of coronavirus disease 2019. J Infect Dis. 2021;223:1145–1149. doi: 10.1093/infdis/jiab005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shi H, Wang W, Yin J, et al. The inhibition of IL-2/IL-2R gives rise to CD8 + T cell and lymphocyte decrease through JAK1-STAT5 in critical patients with COVID-19 pneumonia. Cell Death Dis. 2020;11:429. doi: 10.1038/s41419-020-2636-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li Y, Shang Y, Yang Y, et al. Factors associated with a positive severe acute respiratory syndrome coronavirus 2 testing in suspected cases presenting with pneumonia: a retrospective cohort study in a single medical center. Respiration. 2020;99:739–747. doi: 10.1159/000508398 [DOI] [PubMed] [Google Scholar]
- 33.Mani VR, Kalabin A, Valdivieso SC, et al. New York inner city hospital COVID-19 experience and current data: retrospective analysis at the epicenter of the American coronavirus outbreak. J Med Internet Res. 2020;22:e20548. doi: 10.2196/20548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shang L, Wang L, Zhou F, et al. Long-term effects of obesity on COVID-19 patients discharged from hospital. Immun Inflamm Dis. 2021;9:1678–1685. doi: 10.1002/iid3.522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Weber S, Hellmuth JC, Scherer C, et al. Liver function test abnormalities at hospital admission are associated with severe course of SARS-CoV-2 infection: a prospective cohort study. Gut. 2021;70:1925–1932. doi: 10.1136/gutjnl-2020-323800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu J, Yu C, Yang Q, et al. The clinical implication of gamma-glutamyl transpeptidase in COVID-19. Liver Res. 2021;5:209–216. doi: 10.1016/j.livres.2021.09.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sharma A, Jaiswal P, Kerakhan Y, et al. Liver disease and outcomes among COVID-19 hospitalized patients - A systematic review and meta-analysis. Ann Hepatol. 2021;21:100273. doi: 10.1016/j.aohep.2020.10.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chaibi S, Boussier J, Hajj WE, et al. Liver function test abnormalities are associated with a poorer prognosis in Covid-19 patients: results of a French cohort. Clin Res Hepatol Gastroenterol. 2021;45:101556. doi: 10.1016/j.clinre.2020.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]