Abbreviations
- 7‐AAD
7‐Aminoactinomycin D
- AKT
protein kinase B
- ANOVA
analysis of variance
- BET
bromodomain and extra‐terminal motif
- BETi
BET inhibitor
- BrdU
5‐bromo‐2'‐deoxyuridine
- CI
Combination Index
- CNV
Copy Number Variation
- E2F
E2 factor (E2F) family of transcription factors
- EGFR
epidermal growth factor receptor
- ERBB
Erb‐B receptor tyrosine kinases (EGFR family)
- FDR
False Discovery Rate
- GI50
Half‐maximal growth inhibitory (GI50) concentration
- GSEA
gene set enrichment analysis
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- MAPK
Mitogen‐activated protein kinase
- MEK
MAPK/ERK kinase
- mRNA
messenger ribonucleic acid
- mTOR
mechanistic target of rapamycin
- mTORi
mTOR inhibitor
- MYC
myelocytomatosis oncogene
- PDAC
pancreatic ductal adenocarcinoma
- PI3K
Phosphoinositide 3‐kinase
- PLK1
Serine/threonine‐protein kinase PLK1
- qPCR
quantitative Polymerase Chain Reaction
- RNA
ribonucleic acid
- RNA‐seq
RNA‐sequencing
- SNP
single‐nucleotide polymorphism
- ZIP
Zero interaction potency
1.
Dear Editor,
Pancreatic ductal adenocarcinoma (PDAC) remains a significant health problem with an increase in the incidence and a five‐year survival rate of only 10% [1]. The Phosphoinositide 3‐kinase‐protein kinase‐B‐mechanistic target of rapamycin (PI3K‐AKT‐mTOR) pathway is a driver pathway in PDAC and an important therapeutic target [2]. We [3] and others [4, 5, 6] have demonstrated that the mTOR kinase is a therapeutic target in PDAC, and rationally designed mTOR inhibitor (mTORi)‐based combination therapies are emerging [2]. However, clinical success has not been satisfactory so far [2] due to tumor adaption, resistance, and lack of predictive biomarkers. This implicates the need to decipher resistance mechanisms, develop rationally defined combination therapies, and find reliable biomarkers. Therefore, we aimed to understand the molecular underpinnings of mTORi resistance and treated 20 well‐characterized (transcriptomics, single nucleotide polymorphisms [SNPs], copy number variations [CNVs]) murine PDAC cell lines [7] with the potent mTORi INK128 (Sapanisertib) to determine the half‐maximal growth inhibitory concentration (GI50) (Figure 1A). The detailed Methods of this study can be found in the Supplementary Methods.
FIGURE 1.
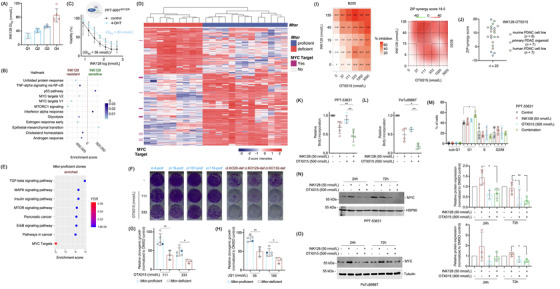
MYC confers mTORi resistance. (A) Heterogeneity in responsiveness to mTORi. Twenty murine PDAC cell lines were treated with a seven‐point dilution of the mTORi INK128 (1000, 500, 100, 50, 10, 5, 1 nmol/L) and analyzed by Thiazolyl Blue Tetrazolium Bromide (MTT) assay after 72 hours. Half‐maximal growth inhibitory concentration (GI50) values were determined and separated into quartiles. (B) MYC and mTOR signatures are enriched in mTORi resistant cells. RNA‐seq data from the most sensitive cells from A (Q1) were compared to the more resistant cells (Q2, Q3 and Q4) by gene set enrichment analysis (GSEA) using the GeneTrail 3.0 platform. The enrichment score and the q values are depicted. (C) Conditional activation of MYC renders cells resistant to mTORi. The murine PPT‐9091MYCER cell line, which has been transduced with a MYCER fusion protein was used. Cells were treated with INK‐128 in the presence or absence of 4‐Hydroxytamoxifen (600 nmol/L) for 72 hours as indicated. Afterward, MTT viability assays were performed. Experiments were performed in n = 3 independent biological experiments and the GI50 values were depicted. (D) Heatmap displaying a hierarchical cluster with the intensities of phosphopeptides (rows) showing statistically significant abundance (two‐sample t‐test, FDR < 0.05, S0 = 1) between Mtor‐proficient and ‐deficient clonal cell lines. Columns display the biological replicates for the individual clones and rows the phosphopeptides intensities (z‐scored, imputed). Phosphopeptides from MYC target proteins (GSEA HALLMARK MYC TARGETS) are indicated in dark pink on the left side. (E) Enriched KEGG pathways in Mtor‐proficient clones, enrichment factor and P value from a Fisher's exact test corrected according to Benjamini‐Hochberg (FDR) performed with each cluster compared to the total phosphoproteome. In addition, the GSEA HALLMARK MYC TARGETS signature is depicted. (F‐H) Reduced clonogenic growth in Mtor‐deficient clones upon BETi treatment. 2,000 cells per well of the Mtor‐proficient clones (n = 4) and the parental cell line PPT4‐ZH363‐Mtor∆E3/lox or 3,000 cells per well of the Mtor‐deficient clones (n = 3) were seeded in 12‐well plates and treated in duplicates with (F) OTX015 (111 and 333 nmol/L) on the following day. Dimethyl sulfoxide (DMSO) was used as vehicle control. (G) Quantification of F. (H) Clonogenic assay as described in (F) was performed using JQ1 (55 and 165 nmol/L). The quantification is depicted. (G) and (H) Unpaired t‐test: *P < 0.05, **P < 0.01. The blue dot marks the parental cell line. (I) Human PDAC organoids (n = 7) were treated with a combination matrix with 4 dilutions of the mTORi INK128 (5, 15, 45, 135 nmol/L) and 5 dilutions of the BETi OTX015 (37, 111, 333, 1000, 3000 nmol/L). After 72 hours viability was measured using CellTiter‐Glo assays. The % inhibition for each combination is shown on the left‐sided panel and the calculated overall ZIP score on the right‐sided panel. (J) Synergistic mode of action between INK‐128 and OTX015 in a subset of murine and human PDAC cells. Overview of ZIP scores for the combination of INK128 with OTX015 in a panel of conventional human PDAC cell lines (n = 7) (data based on MTT), murine PDAC cell lines (n = 8) (data based on clonogenic growth assays) and human PDAC organoids (n = 7) (data based on CellTiter‐ Glo assays). (K) and (L) BrdU Incorporation is reduced upon combination of mTORi and BETi. The murine PDAC cell line PPT‐53631 (K) and the human PDAC cell line PaTu8988T (L) were seeded out in white 96‐well plates and treated in triplicates on the following day with 50 nmol/L INK128, 500 nmol/L OTX015 or the combination thereof. BrdU (10 μmol/L) was added after 24 hours for an additional 2 hours. BrdU incorporation was assessed by a chemiluminescent BrdU Cell Proliferation ELISA Kit and luminescence values were normalized to untreated controls (n = 4–5). One‐way ANOVA with correction for multiple testing according to Tukey: *P < 0.05, **P < 0.01. (M) Combination of mTORi and BETi induces G1 growth arrest. The cell cycle profile of PPT‐53631 after 24 hours treatment with 50 nmol/L INK‐128, 500 nmol/L OTX015 or the combination of the two was determined by 7AAD‐BrdU flow cytometric analysis. Shown is the fraction of cells in sub‐G1, G1, S and G2/M phases of the cell cycle (n = 4). One‐way ANOVA with correction for multiple testing according to Tukey: *P < 0.05. (N) Representative Western Blot and quantification showing MYC protein expression is reduced upon combined treatment with INK‐128 and OTX015 in PPT‐53631. The MYC protein expression in the murine PDAC cell line PPT‐53631 after treatment with 50 nmol/L INK‐128, 500 nmol/L OTX015 or the combination thereof for 24 and 72 hours was determined by immunoblot. MYC protein expression was normalized to loading control and is depicted as relative protein expression normalized to DMSO treated controls. One‐way ANOVA with correction for multiple testing according to Tukey: *P < 0.05, **P < 0.01. (O) Representative Western Blot and quantification showing MYC protein expression is reduced upon combined treatment with INK‐128 and OTX015 in PaTu8988T. The MYC protein expression in the human PDAC cell line PaTuT8988T after treatment with 50 nmol/L INK128, 500 nmol/L OTX015 or the combination thereof for 24 and 72 hours was determined by immunoblot. MYC protein expression was normalized to loading control and is depicted as relative protein expression normalized to DMSO treated controls. One‐way ANOVA with correction for multiple testing according to Tukey: *P < 0.05. Abbreviations: 24h: 24 hours, 72h: 72 hours, ANOVA: analysis of variance, BET: bromodomain and extra‐terminal motif, BETi: BET inhibitor, BrdU: 5‐bromo‐2'‐deoxyuridine, cl: clone, def: (Mtor) deficient, DMSO: dimethyl sulfoxide, ELISA: enzyme‐linked immunosorbent assay, FDR: False Discovery Rate, GI50: Half‐maximal growth inhibitory concentration, GSEA: gene set enrichment analysis, h: hours, KEGG: Kyoto Encyclopedia of Genes and Genomes, mTOR: mechanistic Target of Rapamycin, mTORi: mTOR inhibitor, MTT: Thiazolyl Blue Tetrazolium Bromide, MYC: myelocytomatosis oncogene, MYCER: MYC–estrogen receptor (ER) fusion gene, PDAC: pancreatic ductal adenocarcinoma, prof: Mtor‐proficient, def: Mtor‐deficient, q: q value, RNA: ribonucleic acid, RNA‐seq: RNA‐sequencing, ZIP: zero‐interaction potency
Resistant cell lines showed enrichment of myelocytomatosis oncogene (MYC) signatures (Figure 1B). Similar signatures were enriched in human INK128‐resistant PDAC cell lines (Supplementary Figure S1A and B). Data of a CRISPR/Cas‐drop out screening of PDAC lines showed a significant correlation between mTOR and MYC gene effect scores (Supplementary Figure S1C), demonstrating that some PDACs were co‐addicted to MYC and mTOR. In PPT‐9091MYCER cells, a conditional MYC gain‐of‐function model [8], activation of MYC led to a doubling of the INK128 GI50 value (Figure 1C), corroborating that MYC confers mTORi resistance, which is consistent with a recent report [9]. However, considering the heterogeneity of human cancers and complexity of the MYC network, additional pathways might also contribute to mTORi resistance.
Next, we generated a murine PDAC cell line that allows for the Cre‐mediated deletion of exon 3 of the Mtor gene, called PPT4‐ZH363‐Mtor ∆E3/lox [3]. We used Mtor‐proficient (n = 4) and ‐deficient (n = 3) single‐cell clones to find vulnerabilities associated with genetic mTOR inhibition (Supplementary Figure S2A, B). Downstream signaling, as measured by investigating the phosphorylation of the mTOR target Eukaryotic translation initiation factor 4E‐binding protein 1 (4E‐BP1), was distinctly reduced in the deficient clones (Supplementary Figure S2B). Stable Mtor knockouts showed increased phosphorylation of AKT (Supplementary Figure S2B and C). Functionally, inactivation of Mtor was associated with reduced proliferation (Supplementary Figure S2D). Analysis of transcriptomes demonstrated depletion of mTOR signatures in deficient clones (Supplementary Figure S2E). Further proteomics analyses revealed that proteins corresponding to MYC target genes (Supplementary Figure S2F) and metabolic pathways were the main downregulated signatures (Supplementary Figure S2G). As measured by phospho‐proteomics, signaling by Erb‐B receptor tyrosine kinases/epidermal growth factor receptors (ERBB/EGFR), Insulin, Mitogen‐activated protein kinase (MAPK), and, as expected, mTOR were inhibited in deficient clones and therefore, vice versa enriched in proficient clones. (Figure 1D and E, Supplementary Table S1). Phosphorylation of proteins corresponding to MYC target genes were also downregulated upon Mtor deletion with borderline significance (FDR = 0.06) (Figure 1D and E).
Since we and others have demonstrated synergism of mTORi with MAPK/ERK kinase (MEK) and AKT inhibitors [3], we tested such inhibitors. The efficacy of MEKi at a dose of 5.5 nmol/L (Supplementary Figure S2H) and AKTi (Supplementary Figure S2I) was higher in Mtor‐knockout clones, underscoring the value of the model to define vulnerabilities associated with genetic inhibition of mTOR.
Direct and indirect modes to inhibit MYC have been described [10]. MYC was efficiently blocked by inhibitors of bromodomain and extra‐terminal motif (BET) proteins (Supplementary Figure S3A). We used the BET inhibitors OTX015 and JQ1, and the dual Serine/threonine‐protein kinase PLK1/BET inhibitor BI2536 to evaluate the potential synergism between BETi and mTORi. We observed a strong significant reduction of clonogenic growth induced by all BETi in Mtor‐deficient clones (Figure 1F‐H, Supplementary Figure S3B). Note that the parental cell line, marked as blue dots (Figure 1G and H), responded like the proficient clones. We interpreted from the data of the genetic model that blocking mTOR signaling and MYC might be synergistic in PDAC cells. To directly test the synergism, we used parental PPT4‐ZH363‐Mtor ∆E3/lox cells and observed, indeed, a synergistic reduction of clonogenic growth upon combined INK128 and JQ1 treatment (Supplementary Figure S3C). We extended this finding to a larger panel of murine and human PDAC models using the BET degrader ARV771 and the BET inhibitors OTX015 or JQ1. All inhibitors increased the INK128 sensitivity (Supplementary Figure S3D and E). We calculated the combination index (CI) in 23 human and murine PDAC cell lines using different dose combinations of INK128 and OTX015, and observed values below 1 for most combinations, demonstrating a synergistic effect (Supplementary Figure S3F). Cell lines with the lowest CI values displayed enriched MYC signatures (Supplementary Figure S3G), pointing to the possibility to stratify for combination therapy responders. In addition, we used different assays, treatment periods, and dose matrices via the Synergy Finder platform (https://synergyfinder.fimm.fi/) to calculate a zero‐interaction potency (ZIP) score. High ZIP scores, which indicate synergism of both inhibitors, were found in a proportion of established, two‐dimensional human PDAC cell lines (Supplementary Figure S3H), primary murine PDAC cells (Supplementary Figure S3I), and primary human, three‐dimensional organoid PDAC models (Figure 1I and J). These data demonstrate the existence of a PDAC subtype sensitive for the combination of an mTOR and BET inhibitor across models and species (Figure 1J).
Using live‐cell imaging over a period of 114 hours, we observed a profound growth arrest in the INK128 and OTX015 combination (Supplementary Figure S4A). Consistently, BrdU incorporation was synergistically reduced by the combination therapy in PPT‐53631 (Figure 1K) and PaTu8988T cells (Figure 1L). 7AAD‐BrdU flow cytometric analysis demonstrated that cells treated with the combination therapy were arrested in the G1‐phase of the cell cycle (Figure 1M), a finding corroborated by flow cytometric analysis of propidium iodide‐stained cells (Supplementary Figure S4B and C).
To define the contribution of MYC to growth inhibition, we investigated MYC expression over time. In INK128 treated cells, MYC was maintained with even a trend of increased expression (Figure 1N and O). In contrast, the INK128 and OTX015 combination reduced MYC expression in cells compared to the INK128 monotherapy (Figure 1N and O), pointing to an explanation of the synergistic growth defect. We hypothesized that a more profound inhibition of the mTOR kinase would increase the need of the cells to restore MYC expression. Therefore, we repeated the kinetic analysis using an increased dose of INK128, which was approximately 6–7 fold over the GI50 values. Indeed, at this dose, INK128 increased MYC expression, especially after 72 hours of treatment (Supplementary Figure S4D and E). To different extents, compared to the OTX015 monotherapy, MYC expression was less downregulated by the INK128 and OTX015 combination treatment (Supplementary Figure S4D and E). We analyzed RNA‐seq data in the high‐dose setting. Even if the results were not identical in murine PPT‐53631 and human PaTu‐8988T cells, the combination of INK128 and OTX015 had a profound impact on pro‐proliferative transcriptional networks (Supplementary Figure S4F). This was also evident in mRNA expression profiles of INK128‐treated cells compared to those treated with the combination of OTX015 and INK128. Pro‐proliferative networks, including E2F and MYC, were distinctly inhibited by the combination treatment (Supplementary Figure S4G and H). These results are thus in agreement with the arrest in the G1 phase of the cell cycle and the reduced BrdU incorporation. Furthermore, the partial block in the transcriptional output of E2F and MYC might explain the growth defect also in the INK128 high dose setting, even though MYC protein expression remains. Molecular details of the mechanism that are responsible for the described regulatory circuits remain to be deciphered. Furthermore, heterogeneity of PDACs might impact the circuits activated by the BET and mTOR inhibitor combination therapy.
In summary, we developed a concept to enhance the anti‐tumor activity of mTORi in a subtype of PDACs. Biomarker‐stratified mTORi‐based combination therapies seem promising for further pre‐clinical and clinical development.
FUNDING STATEMENT
This work was supported by the Deutsche Forschungsgemeinschaft (DFG): SFB824 C9 to D.S. and G.S.; SCHN 959/3‐2 to G.S.; SFB1321 (Project‐ID 329628492) P13 to G.S.; SFB1321 (Project‐ID 329628492) P11 to D.S and M.S.R.; SFB1321 S01 and S02 to G.S., M.R., D.S., and R.R; SCHN 959/6‐1 to G.S.; RE 3723/4‐1 to M.R., SFB1361 (Project‐ID ID 393547839) to O.H.K; DFG KR2291‐9‐1/12‐1/14‐1 to O.H.K., Wilhelm‐Sander‐Stiftung (2017.048.2 to G.S. and 2019.086.1 to G.S. and O.H.K.); Deutsche Krebshilfe (70113760 to G.S.; Max Eder Program 111273 to M.R.) and Brigitte und Dr. Konstanze Wegener‐Stiftung (Projekt 65) to O.H.K. This research project/publication was funded by LMU Munich‘s Institutional Strategy LMU excellent within the framework of the German Excellence Initiative to M.S.R.
CONFLICT OF INTEREST
The authors declare no competing interests.
ETHICS APPROVAL AND PATIENT CONSENT STATEMENT
The primary human PDAC cellular organoid models were established and analyzed in accordance with the declaration of Helsinki. The study was approved by the local ethical committee TUM, Klinikum rechts der Isar (Project 207/15, 1946/07, 330/19S), and written informed consent from the patients for research use was obtained prior to the investigation.
PERMISSION TO REPRODUCE MATERIAL FROM OTHER SOURCES
Not applicable.
CLINICAL TRIAL REGISTRATION
Not applicable.
AUTHORS' CONTRIBUTION
Conceptualization and design of the study: C.S., Z.H., K.A. and G.S. Data collection and/or analysis and interpretation were performed by: C.S., Z.H., K.A., M.W., S.K., F.O., L.K., C.S., R.Ö., O.H.K., R.R., M.R., M.S.R., D.S. and G.S. The manuscript was drafted by: C.S., Z.H., K.A. and G.S. All authors revised the manuscript for important intellectual content and approved the final version submitted for publication.
Supporting information
Supporting information
Supporting information
Supporting information
Supporting information
Supporting information
Supporting information
ACKNOWLEDGMENTS
We thank all the patients providing tumor tissue and the Hubrecht Institute for providing engineered cell lines. We thank all colleagues for providing vectors via the Addgene platform.
Christian Schneeweis, Zonera Hassan, and Katja Ascherl equally contributing first authors
DATA AVAILABILITY STATEMENT
Proteome and phosphoproteome data: ProteomeXchange Consortium via the PRIDE partner repository: PXD027779; RNA‐seq via ENA: PRJEB43040 and PRJEB47050.
REFERENCES
- 1. Hessmann E, Schneider G, Meeting 1st Virtual Göttingen‐Munich‐Marburg Pancreatic Cancer. New insights into pancreatic cancer: notes from a virtual meeting. Gastroenterology. 2021;161(3):785–91. [DOI] [PubMed] [Google Scholar]
- 2. Conway JR, Herrmann D, Evans TJ, Morton JP, Timpson P. Combating pancreatic cancer with PI3K pathway inhibitors in the era of personalised medicine. Gut. 2019;68(4):742–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Hassan Z, Schneeweis C, Wirth M, Veltkamp C, Dantes Z, Feuerecker B, et al. MTOR inhibitor‐based combination therapies for pancreatic cancer. Brit J Cancer. 2018;118(3):366–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Brown WS, McDonald PC, Nemirovsky O, Awrey S, Chafe SC, Schaeffer DF, et al. Overcoming Adaptive Resistance to KRAS and MEK Inhibitors by Co‐targeting mTORC1/2 Complexes in Pancreatic Cancer. Cell Reports Medicine. 2020;1(8):100131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Driscoll DR, Karim SA, Sano M, Gay DM, Jacob W, Yu J, et al. mTORC2 Signaling Drives the Development and Progression of Pancreatic Cancer. Cancer Res. 2016;76(23):6911–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Morran DC, Wu J, Jamieson NB, Mrowinska A, Kalna G, Karim SA, et al. Targeting mTOR dependency in pancreatic cancer. Gut. 2014;63(9):1481–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Mueller S, Engleitner T, Maresch R, Zukowska M, Lange S, Kaltenbacher T, et al. Evolutionary routes and KRAS dosage define pancreatic cancer phenotypes. Nature. 2018;554(7690):62–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lankes K, Hassan ZZ, Doffo MJ, Schneeweis C, Lier S, Öllinger R, et al. Targeting the ubiquitin‐proteasome system in a pancreatic cancer subtype with hyperactive MYC. Mol Oncol. 2020;14(12):3048–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Allen‐Petersen BL, Risom T, Feng Z, Wang Z, Jenny ZP, Thoma MC, et al. Activation of PP2A and inhibition of mTOR synergistically reduce MYC signaling and decrease tumor growth in pancreatic ductal adenocarcinoma. Cancer Res. 2018;79(1):209–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wirth M, Mahboobi S, Krämer OH, Schneider G. Concepts to Target MYC in Pancreatic Cancer. Mol Cancer Ther. 2016;15:1792–8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting information
Supporting information
Supporting information
Supporting information
Supporting information
Supporting information
Data Availability Statement
Proteome and phosphoproteome data: ProteomeXchange Consortium via the PRIDE partner repository: PXD027779; RNA‐seq via ENA: PRJEB43040 and PRJEB47050.


