Abstract
Visual inspection of the cervix with acetic acid (VIA), though error prone, has long been used for screening women and to guide management for cervical cancer. The automated visual evaluation (AVE) technique, in which deep learning is used to predict precancer based on a digital image of the acetowhitened cervix, has demonstrated its promise as a low-cost method to improve on human performance. However, there are several challenges in moving AVE beyond proof-of-concept and deploying it as a practical adjunct tool in visual screening. One of them is making AVE robust across images captured using different devices. We propose a new deep learning based clustering approach to investigate whether the images taken by three different devices (a common smartphone, a custom smartphone-based handheld device for cervical imaging, and a clinical colposcope equipped with SLR digital camera-based imaging capability) can be well distinguished from each other with respect to the visual appearance/content within their cervix regions. We argue that disparity in visual appearance of a cervix across devices could be a significant confounding factor in training and generalizing AVE performance. Our method consists of four components: cervix region detection, feature extraction, feature encoding, and clustering. Multiple experiments are conducted to demonstrate the effectiveness of each component and compare alternative methods in each component. Our proposed method achieves high clustering accuracy (97%) and significantly outperforms several representative deep clustering methods on our dataset. The high clustering performance indicates the images taken from these three devices are different with respect to visual appearance. Our results and analysis establish a need for developing a method that minimizes such variance among the images acquired from different devices. It also recognizes the need for large number of training images from different sources for robust device-independent AVE performance worldwide.
Keywords: cervical cancer screening, deep clustering, automated visual inspection, transfer learning
I. Introduction
Cervical cancer, a disease that is primarily caused by persistent infection from high risk types of human papillomavirus (HPV), is one of the most common cancers in women worldwide [1]. Its mortality and morbidity are especially high among women in low- and medium- resource countries or regions (LMIC), due to the shortage of medical personnel and the lack of sufficient access to effective screening and treatment programs. VIA (visual inspection with acetic acid), a method involves visually examining the cervix appearance without magnification after application of a weak (3%-5%) acetic acid solution, has served as a low-cost screening method that is commonly employed in LMIC. However, its accuracy is considered inadequate due to high inter- and intra-reader variability [2]. Since clinical colposcopes that are traditionally used to magnify and illuminate the cervix are expensive, medical practitioners and field workers in several screening programs have been equipped with relatively inexpensive and easy-to-use mobile colposcope and imaging devices [3,4]. Such programs mainly use digital images for the training of nurses or practitioners in VIA in combination with remote consultation with experienced colposcopists and, minimally, for documentation of clinical examination. More recently, researchers have employed machine learning techniques to automatically analyze the captured cervical images to identify abnormal cases [5,6]. Toward this, we recently reported on our deep learning-based technique called automatic visual evaluation (AVE) [7]. AVE was evaluated on a large Cervigram® image dataset collected using a now-obsolete film camera with ring flash and fixed focus during a multi-year longitudinal natural history study carried out two decades ago by the National Cancer Institute (NCI) in Guanacaste, Costa Rica [8]. The results obtained on that dataset demonstrated AVE can achieve performance significantly superior to human interpretation of the same images in identifying images with high grade cervical intraepithelial neoplasia (CIN). They suggested that AVE could become a meaningful adjunct screening tool. However, to use AVE in public health, a number of additional practical issues need to be addressed [9,10]. Among these, a primary challenge is acquisition of standardized “good-quality” images of the cervix in health and disease, and to study technical and physiologic influences on AVE decision-making. It is unclear how much the choice of imaging device affects AVE performance. Further, we need to know which imaging factors, such as light source and device ergonomics, affect the ease of capturing good quality images. Understanding these points would not only help balance the variability and consistency impacted by time-cost challenges that are often faced during medical data collecting, but also inform us on how image variety influences AVE. To this end we conducted a study in Nigeria that used three devices to capture images of the cervix of human papillomavirus (HPV)-infected women during screening. The devices were a common smartphone, a custom smartphone-based mobile handheld device for cervical imaging, and a clinical colposcope equipped with SLR digital camera-based imaging capability. In this paper, we report our initial efforts to examine differences in visual characteristics in images captured using these devices. We aim to assess to what extent cervix images taken by one digital device are visually different from the images taken by another device based strictly on their appearance with no knowledge of any abnormality exhibited on them. Specifically, we present an unsupervised approach using only the intrinsic similarity within each data subset and the image source device information is only used in the algorithm evaluation. Our approach comprises several components: cervix extraction, feature extraction, feature encoding, and clustering. We integrate feature extraction, feature encoding and clustering into a single deep learning network following state-of-art clustering methods.
There are a number of deep learning based clustering methods that have been developed in recent years [11–14]. Most of the existing deep clustering methods use an autoencoder architecture and comprise two main components: 1) embedded representation generation, and 2) feature clustering. The training phase of these methods consists of two stages: a pretraining stage and fine-tuning stage. In the pretraining stage, only the autoencoder is trained and the k-means technique is used to cluster the features embedded by the pretrained autoencoder. In the fine-tuning stage, the encoder or autoencoder (where weights are initialized by the pretrained autoencoder) and the clustering layer (where weights are initialized by the k-means cluster centroids) are jointly tuned. Methods are varied based on the architecture of the autoencoder and the loss functions used (viz., clustering loss and reconstruction loss). For a more comprehensive survey or list of deep clustering methods, please refer to [14]. We compare our method with several state of the art deep clustering networks, viz. DEC [11], DCEC [12], and DynAE [13] using our data. One significant difference between above three networks and our network is the way in which features are extracted. In our approach, the images are input to an ImageNet pretrained model before sending to an autoencoder, while in all other networks, the images are input to an autoencoder directly for extracting features. Our approach achieves a high clustering performance (accuracy of ~97%) on our dataset which is significantly superior to the results obtained from the three selected deep clustering methods. We also examine and compare several alternative choices in each component, e.g., features from different ImageNet pretrained classification networks, features encoded with different lengths, conventional or network layer based clustering methods, and different ways of training for clustering networks. Experiments for analyzing the effectiveness of feature encoding and clustering network are also conducted.
The high clustering accuracy indicates that images taken by the three imaging devices are highly distinguishable across devices, which further emphasizes the need of investigating the robustness of AVE across multiple devices. Our work on comparing the differences of devices on AVE performance is ongoing, and we will report that result to the community when it is done. In the following, we first describe the datasets and the imaging devices used in this research in Section 2. Then in Section 3, we present our method and elucidate the algorithms in each component in detail. We evaluate the performance, compare the different choices, and discuss the results in Section 4. Section 5 concludes the paper and provides directions for future work.
II. Datasets and Imaging Devices
In order to evaluate use of new technologies, such as AVE, to serve as reliable adjuncts to VIA, NCI has recently initiated several studies for triage of women testing HPV-positive. Among the several research goals of this effort, one is to evaluate AVE performance and robustness across images acquired using different devices. To this end, three imaging devices were used in the study conducted in Nigeria shown in Fig. 1, viz., Samsung Galaxy® S8 cellphone, MobileODT EVA®, and Zeiss® colposcope with Nikon® camera. The MobileODT EVA device is a hand-held colposcope built around a low-cost Android smartphone. It has additional hardware for magnification and illumination, i.e., optical lens and oriented polarized lighting source, as well as customized software for patient record management and expert consulting/collaboration. The Zeiss® colposcope is a standard device commonly used in colposcopy clinics. To obtain digital images through the Zeiss® colposcope, an adaptor is used to mount a Nikon® D700 SLR camera on the colposcope top.
Fig. 1.
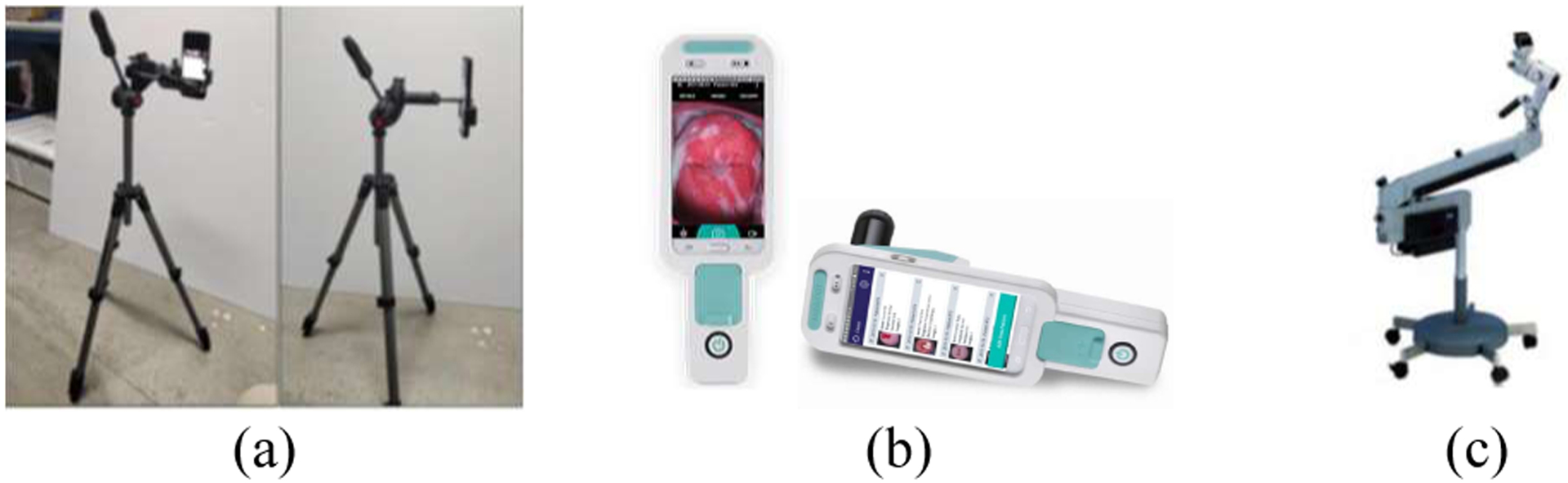
The three imaging devices. (a) Samsung Galaxy S8 on tripod; (b) MobileODT EVA (from its website); (c) Zeiss colposcope (from its website).
The data used in this study was collected on HPV positive women at the colposcopy clinic of the Obafemi Awolowo University Teaching Hospitals Complex (OAUTHC) in Nigeria. The study was approved by National Cancer Institute (NCI) and OAUTHC ethical Institutional Review Boards. The study participants were presented with and provided written consent for the data to be used for research and subsequent machine learning studies. The details of the methods are described in [15]. For each participating woman, multiple images of the cervix were captured using each device. All the images of the same patient were taken by the same provider. The images were taken at least one minute after the application of the acetic acid. Acetic acid could be re-applied during the process if it was deemed necessary by the provider. Judgement as to whether the quality of the images was satisfactory was at the discretion of the care provider, who followed the operational guidelines developed by the team after some testing in a pilot experiment. Examples of these guidelines include: for stability, the S8 cellphone was screwed on a tripod stand, and EVA was mounted on a MobileODT provided stand; the light on the back of the S8 cellphone was kept on for illumination; to avoid motion blur effect of clicking while capturing the image, “Open Camera” application which collects three snaps of the images after one click was used on S8 cellphone, hand-wave feature for image capture was used on EVA, and “Case Air Wi-Fi tethering system” application for image capture was used on colposcope. All of the images and related patient data were recorded and managed using the software application developed by MobileODT.
The dataset used in our analysis consists of patient data collected from December 2018 to November 2019. It contains 988 patients. The total number of images is 13792, among which the number of images from cellphone, EVA, and colposcope are 6113, 3594, and 4085, respectively. The sizes (width × height) of cellphone images, EVA images and colposcope images are around 3000 × 4000 (or 4000 × 3000), 3100 × 4100, and 6000 × 4000, respectively. There is a large visual variety in image content not only due to the differences in the appearance of the cervix related to the woman’s age, parity, and cervix anatomy and condition, but also due to non-disease or non-cervix related factors such as illumination, focus, specular reflection, presence of clinical instruments, and variable zoom and angle to the cervix. Several example images from each device are shown in Fig. 2. Based on the histology results, the numbers of patients having precancer or cancer versus normal or signs of HPV infection are 54 and 934, respectively. Since there are not enough cases (precancer+) in the current dataset for training an AVE model, we aim to use an unsupervised method to investigate the (in)homogeneity of image appearances across these devices.
Fig. 2.
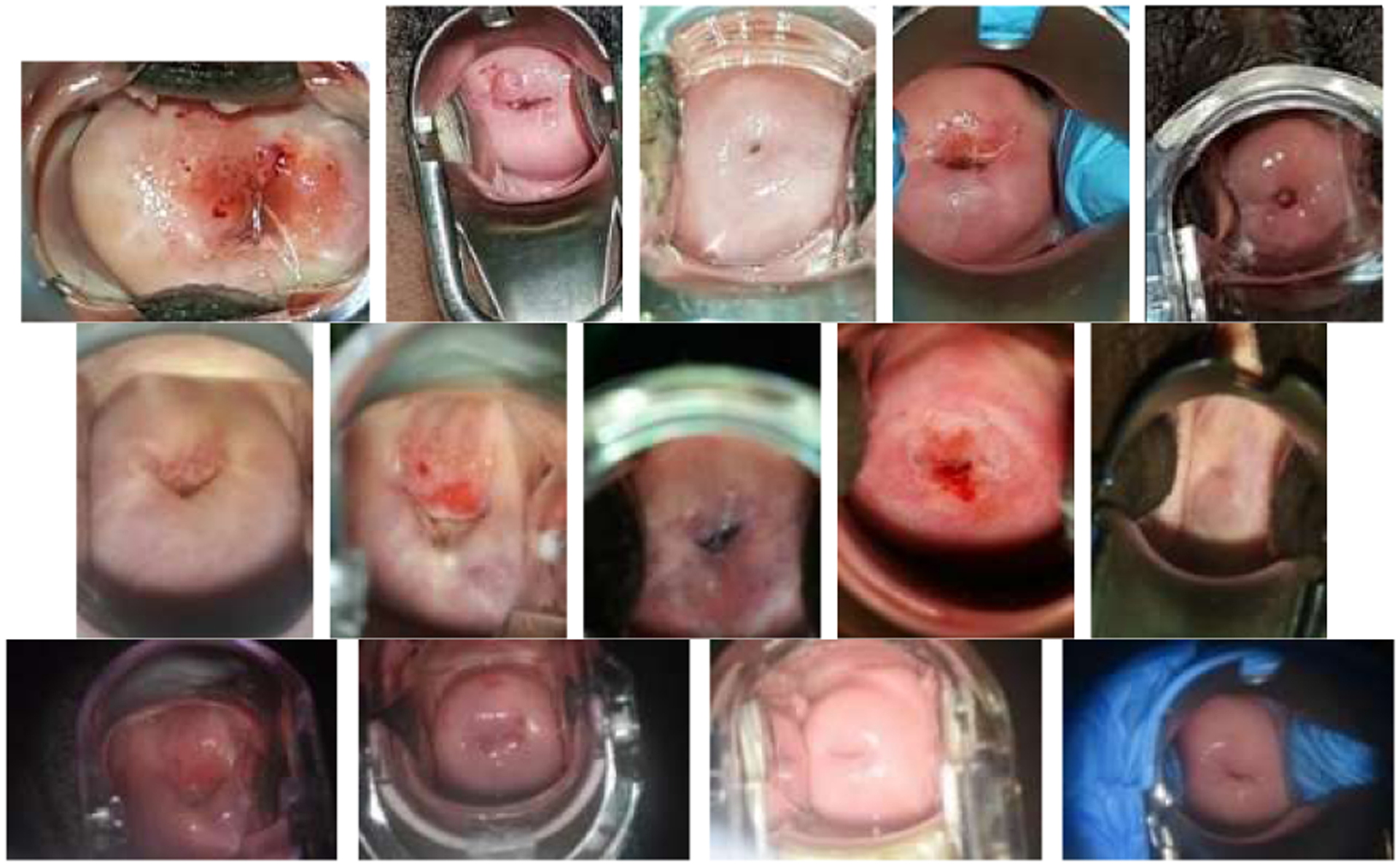
Example images from the three imaging devices (1st row: cellphone; 2nd row: EVA; 3rd row: colposcope).
III. Methods
The proposed method consists of four main components: cervix extraction, feature extraction, feature encoding and clustering, as shown in Fig. 3. Since it is the characteristic of cervix that is of interest, our first step is to localize the cervix region in order to reduce the influence by the area outside the cervix. In previous work, we developed a cervix detector that was based on the object detection deep learning network, Faster RCNN. The cervix detector was trained with around 1600 images in a different set of cervix images (that is, not the Nigeria images used in this current work). Several examples of cervix extraction results for each device are shown in Fig. 4. The cervix region in each image is then cropped out and resized to a uniform size (224 × 224). We use resized cervix images as the input for the subsequent components. Given that we have a limited number of images, we decide to use representative deep learning classification networks that have achieved cornerstone performance on the ImageNet dataset to extract features, in order to take advantage of the technique of transfer learning. To this end, we use and compare two well-known ImageNet pretrained networks: ResNet50 and Vgg16. As we demonstrate later, features obtained from both networks are very effective for our data and contribute significantly to the clustering performance that outperforms recent deep clustering networks, even when we use a simple conventional clustering method. For ResNet50, the features are extracted from the average pooling layer (feature vector length is 2048). For Vgg16, the features are extracted from the first fully connected layer (feature vector length is 4096). As shown in the t-SNE (t-distributed stochastic neighbor embedding) plots in Fig. 5, the features of images of the three devices extracted from both networks could be separated very well by a clustering method. This demonstrates the effectiveness of the technique of transfer learning and the strength of deep learning models trained with a dataset of millions of images (ImageNet). Although the t-SNE plots suggest the ResNet50 and Vgg16 features are good sets of features for separating images from different devices, the feature vector dimensions are relatively high with respect to the number of images in the dataset. This implies that the clustering may encounter difficulties due to the curse of dimensionality problem. So we apply an autoencoder to reduce their dimensionality. Compared to the architectures of the autoencoders used in state-of-art deep clustering networks, our autoencoder has a very simple architecture. It consists of only one hidden layer and the number of nodes in the hidden layer is set to be much less than that of the input layer (e.g., only 100 nodes). The output of the hidden layer, the encoded feature vector, is then sent to be the input of a clustering method. The ablation experiments in Section 4 indicate the encoded features by the autoencoder can improve the clustering performance by a significant margin for both ResNet50 and Vgg16 features.
Fig. 3.
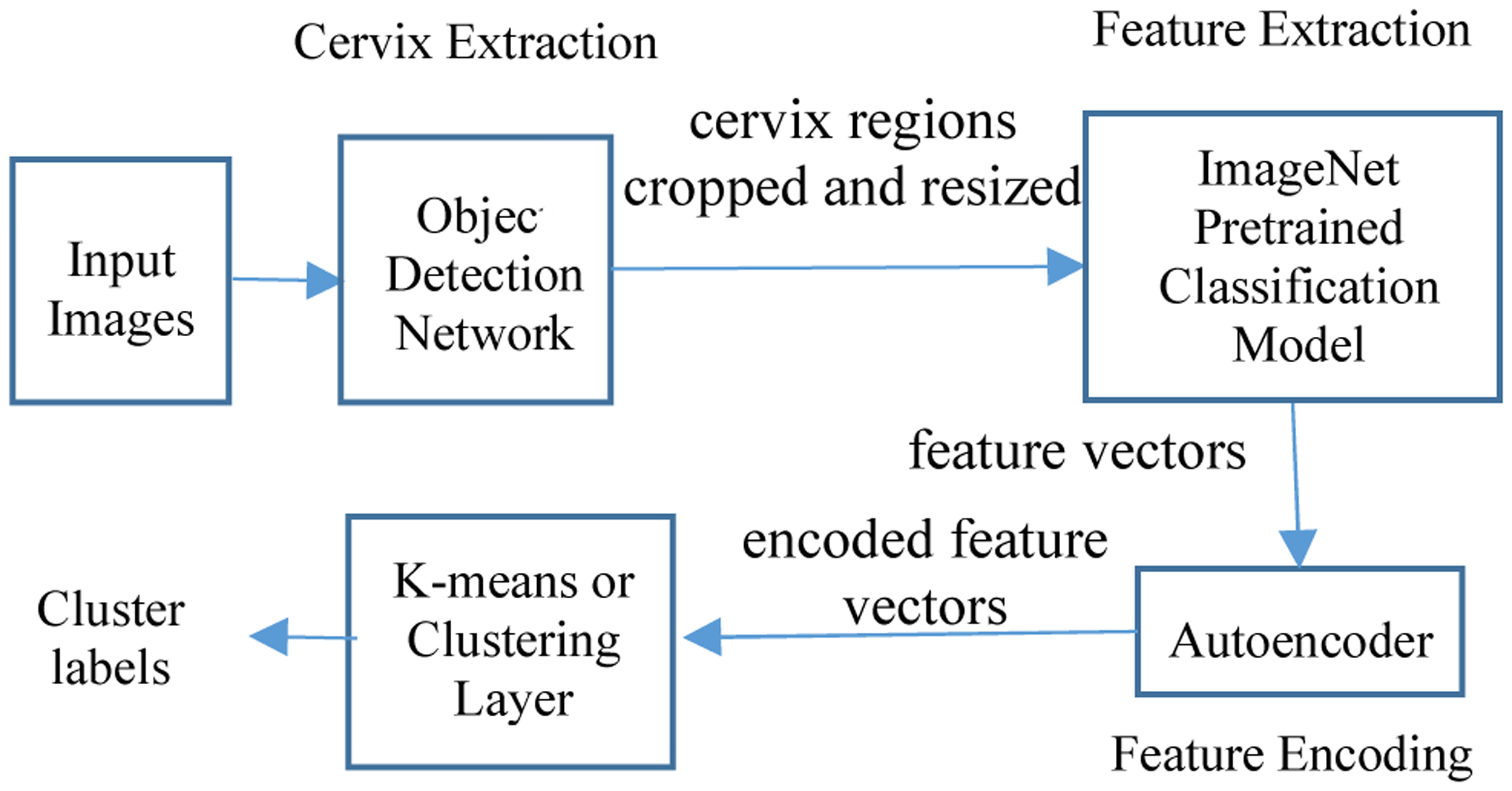
The diagram of the proposed method.
Fig. 4.
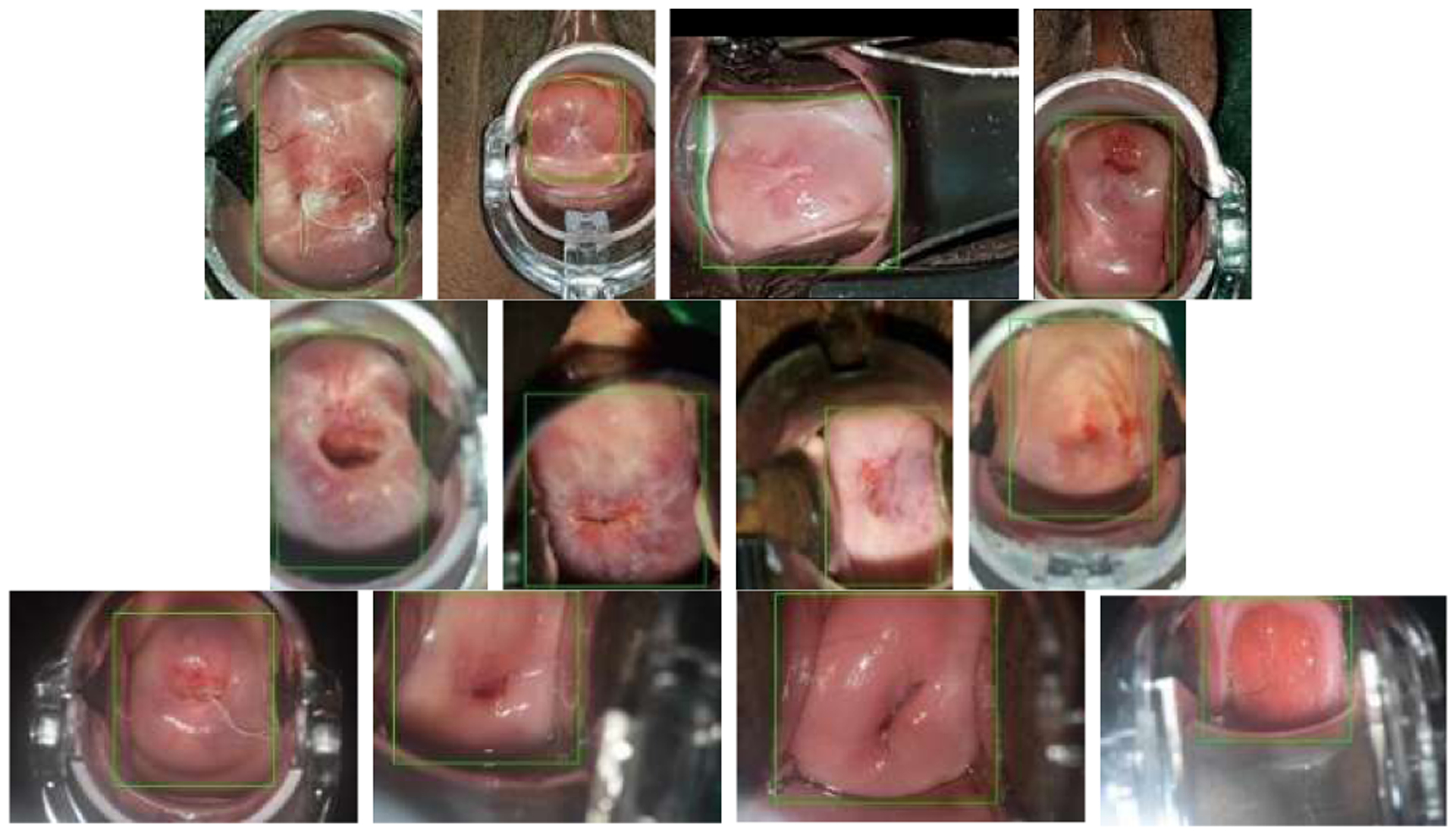
Examples of cervix extraction (1st row: cellphone; 2nd row: EVA; 3rd row: colposcope)
Fig. 5.
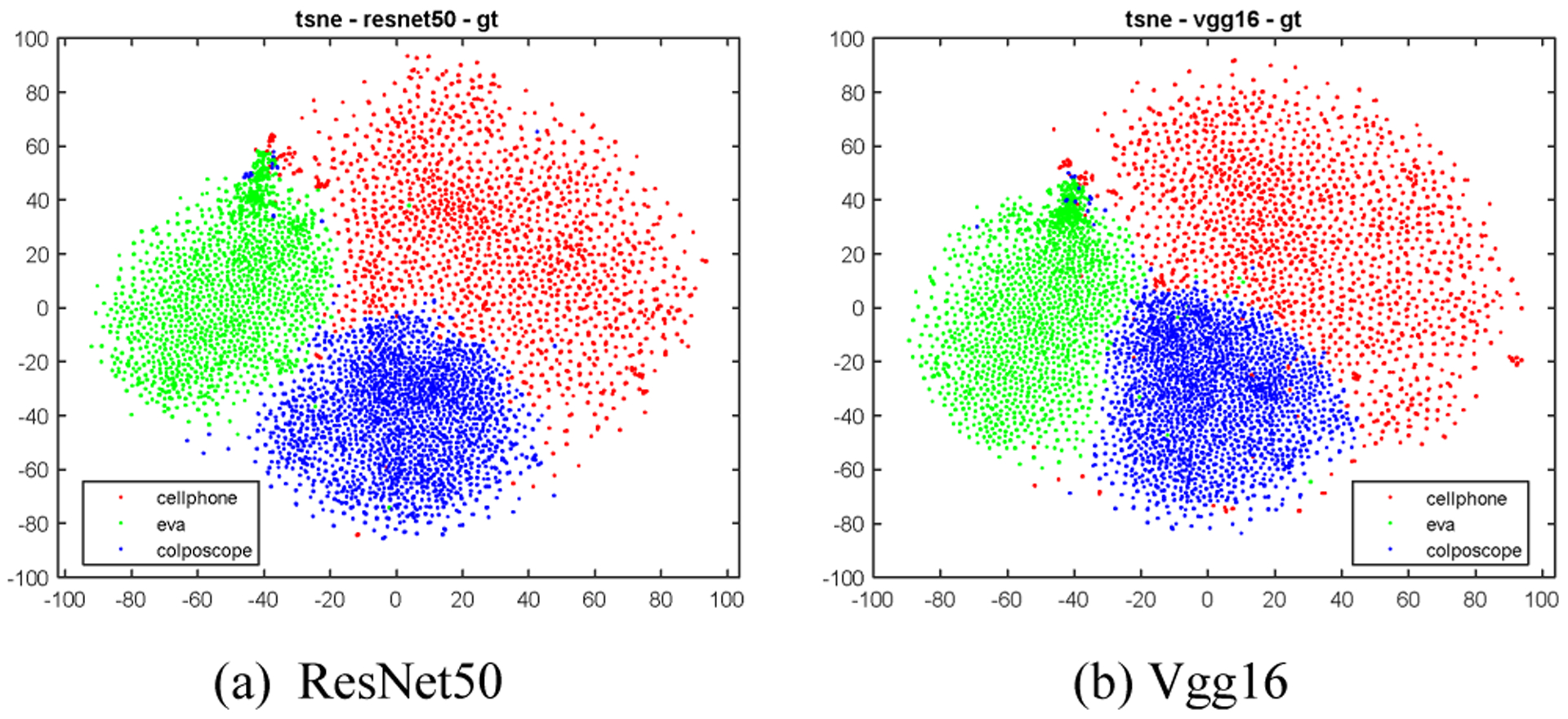
T-SNE plots of ResNet50/Vgg16 features of all the images in the dataset.
The performance of clustering depends on the effectiveness of both the features and the method of clustering. For the clustering, we first apply the k-means method. Specifically, we run k-means with 20 different random centroid seeds and select the best result based on the value of inertia (within-cluster sum-of-squares criterion). We find this simple method achieves very good performance on both the original and the encoded features. We then adopt a clustering layer [11] to see if it can further improve the performance. Different from k-means, it can also be combined with the feature extractor and encoder to make the entire approach a deep neural network that can be trained end-to-end. The clustering layer is connected to the encoded layer of the autoencoder. The cluster centroids , are the trainable parameters (weights) of the clustering layer and the probability of the encoded feature vector of each image belonging to a cluster j (soft assignments qij) is calculated using Student’s t-distribution:
| (1) |
The loss for the clustering layer is a KL divergence loss between the soft assignments qij and the predefined target distribution pij of soft assignments:
| (2) |
| (3) |
To evaluate the performance, two most commonly used metrics are the clustering accuracy (ACC) and the normalized mutual information (NMI). ACC is defined as
| (4) |
where M is a mapping function representing all possible one-to-one mappings between cluster assignments and ground truth labels. ACC finds the best matching between cluster assignments cp and ground truth labels yt using the Hungarian algorithm. NMI is defined as
| (5) |
where I and H denote the mutual information and entropy function respectively.
IV. Experimental Results and Discussion
We carry out several ablation experiments to compare and analyze the effectiveness of the methods used in feature extraction, feature encoding, and clustering. In existing deep clustering literature, the majority of the reported results are on the whole set (such as MNIST-full). Although the methods do not use the ground truth labels explicitly in the network, the parameter tuning process of the training of the clustering network could be influenced unintentionally by the evaluation metrics that are calculated using the ground truth label information. In our experiments, we are careful to not use any supervisory information to guide the parameter tuning process, for example, we use the loss value, intra-cluster distance, and inter-cluster distance as guidance, rather than the performance value in terms of ACC and NMI. Nonetheless, it is desirable not only to examine the clustering capability of the method with its seen data, but also to evaluate the method with unseen data. Therefore, we divide the dataset into a training set and a hold-out set using the ratio of 80/20 for images of each device. The training set is used to train the autoencoder, the clustering methods (k-means, clustering layer), or the combined network of the encoder and clustering layer, and the hold-out set is used to check how the fixed model performs on an unseen set. The algorithm is implemented using Python and Keras, and the experiments are run on a Lambda GPU server with 8 GPU cards.
A. The advantage of using autoencoder
As indicated by the t-SNE plots in Fig. 5, both the ResNet50 and Vgg16 features could be used to distinguish images from different devices very well. However, when applying the k-means to cluster these features, we observe that ResNet50 features outperform Vgg16 features significantly (ACC 0.934 vs 0.901). One reason could be that the dimension of Vgg16 features (4096) is much higher than that of the ResNet50 features (2048) with respect to the size of the dataset. Therefore, as discussed in Section 3, we use an autoencoder with only one hidden layer to reduce the feature length. We set the number of nodes in the hidden layer (also the length of the encoded feature) to be 100. The activation functions used in both the hidden layer and output layer of the autoencoder are the sigmoid function. The loss function of the model is mean squared error and the model is trained using the optimization algorithm Adam with a learning rate of 0.001 and default values for other parameters of Adam. The batch size is 256 for ResNet50 features and is 16 for Vgg16 features. The number of epochs is 1000. The t-SNE plots of the encoded features are given in Fig.6 and they indicate that, like the original features, these encoded features can distinguish images of three devices pretty well. We first apply k-means to cluster the original ResNet50/Vgg16 features and the encoded ResNet50/Vgg16 features. The k-means implementation of the scikit-learn package is used. It runs with 20 different centroid seeds and the best result is selected in terms of inertia. The corresponding ACC and NMI values for each type of features are listed in Table I respectively. As indicated there, the performance of the encoded features is significantly higher than that of the original features for both types of features (e.g., ACC is from 0.911 to 0.951 for Vgg16, and from 0.937 to 0.975 for ResNet50, for hold-out set), while the ResNet50 features outperform the Vgg16 features. The corresponding confusion matrices of the encoded ResNet50 feature are listed in Table II.
Fig. 6.
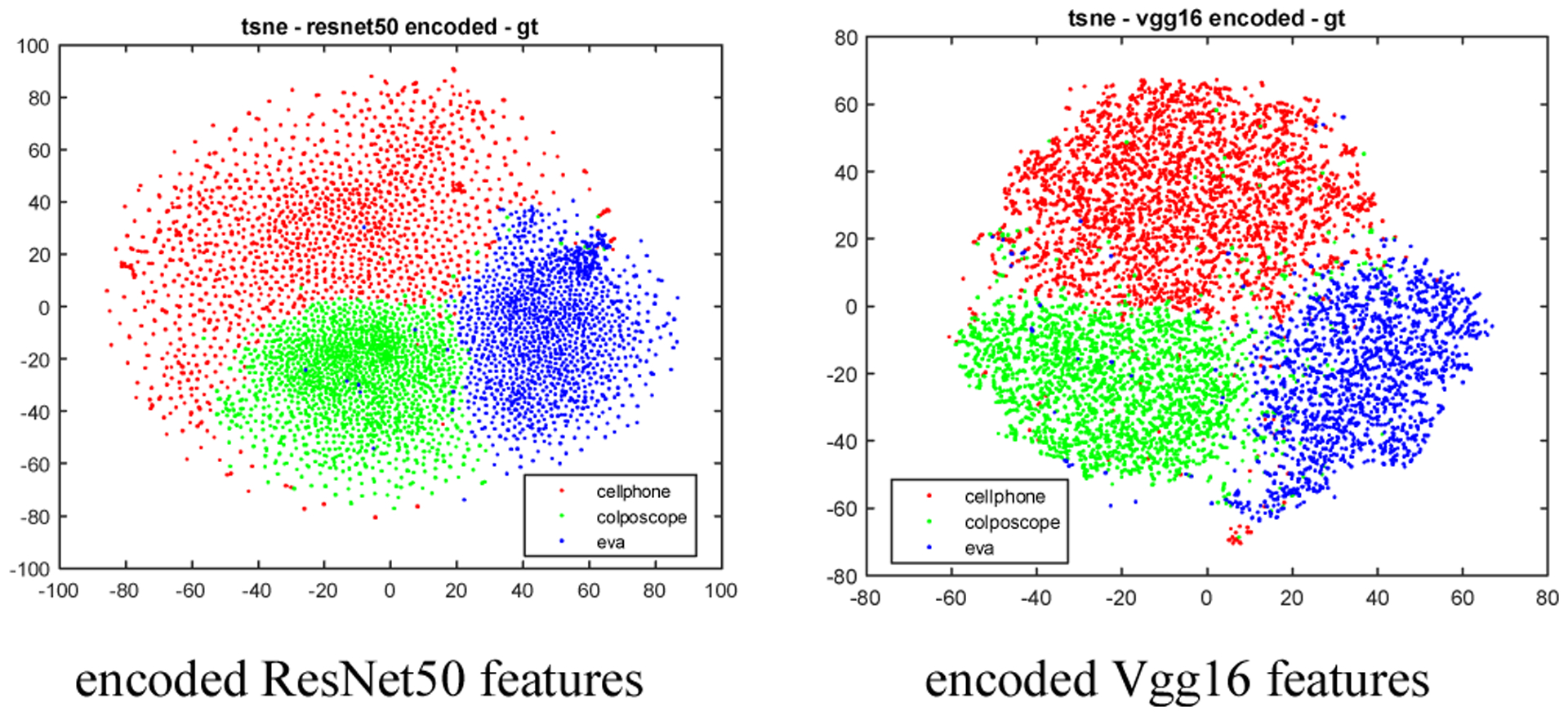
T-SNE plots of the encoded ResNet50 and Vgg16 features (length = 100) of the images in the training set
Table I.
Clustering Performance with or without Autoencoder
| Method | Training set | Hold-out set | ||
|---|---|---|---|---|
| ACC | NMI | ACC | NMI | |
| ResNet50 + k-means | 0.934 | 0.757 | 0.937 | 0.762 |
| ResNet50 + encoder + k-means | 0.977 | 0.889 | 0.975 | 0.882 |
| Vgg16 + k-means | 0.901 | 0.704 | 0.911 | 0.729 |
| Vgg16 + encoder + k-means | 0.958 | 0.818 | 0.951 | 0.793 |
Table II.
Confusion Matrix for “ResNet50 + Encoder + K-means”
| Training set | Hold-out set | ||||||
|---|---|---|---|---|---|---|---|
| Cell | EVA | Colpo | Cell | EVA | Colpo | ||
| Cell | 4713 | 8 | 25 | Cell | 1171 | 0 | 9 |
| EVA | 51 | 2848 | 35 | EVA | 26 | 713 | 9 |
| Colpo | 126 | 19 | 3208 | Colpo | 26 | 6 | 799 |
B. Performance using the clustering layer
As demonstrated in Table I, k-means, a well-known, conventional, simple clustering method, can achieve very good clustering results (ACC above 90% for all features and ACC about 97% for encoded ResNet50 features) for our application. This is most likely because of the strong representativeness of the features we selected and generated. However, it is desirable to use a clustering layer since it can be integrated with the layers for feature extraction and feature encoding, which makes all three components function together as one neural network. We first freeze the encoder layer and only train the clustering layer. We train the clustering layer with the weights (the cluster centroids) initialized randomly or initialized by the results of k-means. K-means doesn’t have tunable hyperparameters once the number of clusters is set. On the contrary, there are multiple hyperparameters for the clustering layer. We use two measures in addition to loss to help choose the values of hyperparameters: inertia, and Dunn index (the ratio of the minimum of inter-cluster distances and maximum of intra-cluster distances). The smaller the value of inertia and the larger the value of Dunn index, the better. We also fine-tune the encoder and the clustering layer jointly. Table III lists the results obtained for the encoded ResNet50 features. As it shows, the performance is slightly better than that of k-means when fine-tuning the encoder and clustering layer jointly (ACC is almost the same and NMI is 2% higher for the hold out set). For jointly fine-tuning the encoder and the clustering layer, the parameters used are Adam optimizer with default parameters (learning rate = 0.001), convergence threshold = 0.001, update interval = 100, batch size = 256, and maximal iteration = 50K. These results indicate that, for our highly representative features, a conventional k-means that has less training time and complexity can achieve a close performance compared to a clustering network.
Table III.
Clustering Performance with Clustering Layer
| Encoded ResNet50 feature | Training set | Hold-out set | ||
|---|---|---|---|---|
| ACC | NMI | ACC | NMI | |
| Train clustering layer only (random init.) | 0.953 | 0.806 | 0.949 | 0.799 |
| Train clustering layer only (k-means init.) | 0.953 | 0.806 | 0.949 | 0.799 |
| Fine-tune encoder and clustering layer jointly (k-means init.) | 0.977 | 0.894 | 0.979 | 0.904 |
C. Comparison with other deep clustering methods
We test three representative deep clustering networks on our dataset: DEC [11], DCEC [12] and a very recent architecture DynAE [13]. They all use an autoencoder and a clustering layer and consist of two stages: 1) the pretraining stage in which the autoencoder is trained for the initialization of the encoder, and 2) the fine-tuning stage in which the autoencoder/encoder and the clustering layer are then jointly fine-tuned. According to the publications cited above, of the two stages, the pretraining stage contributes considerably more to achieving high performance. DEC uses a stacked autoencoder (the encoder network dimensions are set to be d–500–500–2000–10 for all datasets, where d is the input data dimension, e.g. 784 for the MNIST dataset) and all layers are fully connected. In DEC, the encoder and clustering layer are jointly fine-tuned using the KL divergence loss. DCEC uses a convolutional autoencoder (containing several convolutional layers, deconvolutional layers and a fully connected layer in between) to incorporate the spatial relationship of pixels. DCEC keeps the decoder in the network fine-tuning stage and uses a combination of reconstruction loss and clustering loss to fine-tune the encoder. DynAE proposes a new autoencoder called dynamic autoencoder to tackle the trade-off between reconstruction loss and clustering loss. It adopts the same autoencoder architecture as DEC but in the pretraining stage, it uses both data augmentation and an adversarially constrained interpolation. This pretraining method increases the performance by a considerable margin (ACC from 0.861 to 0.971) on the MNIST dataset, while the additional gain of using fine-tuning with dynamic loss function is much less significant (< 0.02). This demonstrates that the performance of a deep clustering network heavily depends on the features as well as the importance of feature learning in the pretraining stage. Although all of these deep clustering networks achieved high performance, especially DynAE, on benchmark datasets like MNIST, none of them obtain comparable performance to our approach on our dataset. We believe performance disparity occurs largely because of the way the features are extracted/learned before the fine-tuning stage (in which the clustering layer is involved). Fig. 7 shows the t-SNE plots of the features obtained in the pretraining stage of DEC, DCEC, and DynAE respectively. Compared to Fig. 5 and Fig. 6, it is evident that the features from the ImageNet pretrained classification models has clear advantage. For these three networks, the images are the direct input of the autoencoder and the autoencoder needs to be trained from scratch with our limited dataset. We could focus on finding a better autoencoder architecture for those deep clustering methods, but with our limited number of images, we have opted to take advantage of transfer learning with a simple conventional autoencoder which has demonstrated high effectiveness and representative capability on our dataset. We would also fine tune our network using different loss functions (like the dynamic loss developed by DynAE) to further improve the current performance. However, our main goal of this work is to analyze the appearance difference between images of different imaging devices, and our simple method has demonstrated that they are well distinguishable across these three devices.
Fig. 7.
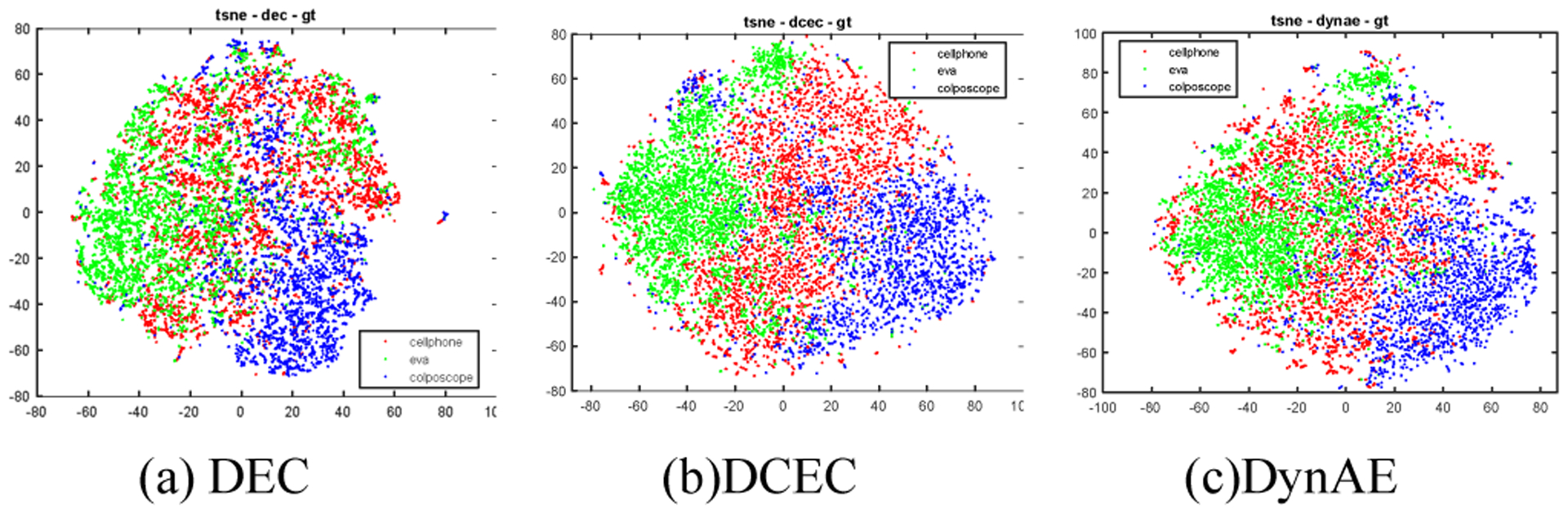
T-SNE plots of embedded features obtained from the autoencoder pretraining stage of existing deep clustering methods.
V. Conclusions
Although automated visual evaluation (AVE) utilizing deep learning technique has demonstrated its promise as an effective adjunct for the screening of cervical cancer on a multi-year population-based dataset, there are multiple critical challenges needed to be addressed to pave the way for real-world deployment, such as image quality control, network explanation and interpretation, and algorithm robustness across imaging devices. In this paper, we report our first step towards the analysis and improvement of AVE for multiple devices. Specifically, we carry out a study in Nigeria to collect cervix images using three representative imaging devices and examine whether the appearance of images taken by different devices are different from each other. We develop an approach to clustering the cervix extracted from the images. Given the limited number of images in our dataset, we propose a simple method for feature extraction and encoding but it still achieves high clustering accuracy because of the very good representativeness of the features. The high clustering performance indicates that images from different devices look differently from each other and can be well distinguished. This emphasizes the need to find the balance between the variance among the images and the number of images to be collected for the training of the AVE algorithm in order to achieve good generalization if different imaging devices are used. It also emphasizes the importance of understanding whether AVE makes the correct classification decision based on the right information, i.e., the fundamental clinical appearance difference that separates the normal from the abnormal, particularly when multiple devices are used.
Acknowledgments
This research was supported by the Intramural Research Program of the National Library of Medicine and the National Cancer Institute (NCI) both part of the National Institutes of Health (NIH). This research was also supported in part by an appointment to the NCI Research Participation Program. This program is administered by the Oak Ridge Institute for Science and Education through an interagency agreement between the U.S. Department of Energy and the NIH. The EVA system devices and data management software were donated by MobileODT. The company had no role in design, analysis, interpretation, and finalization of the manuscript.
The study was partially funded by the Global Good (Seattle, USA).
Contributor Information
Kanan T. Desai, National Cancer Institute, National Institutes of Health, USA
Anabik Pal, National Library of Medicine, National Institutes of Health, Bethesda, USA.
L. Rodney Long, National Library of Medicine, National Institutes of Health, Bethesda, USA.
Mark Schiffman, National Cancer Institute, National Institutes of Health, USA.
Sameer Antani, National Library of Medicine, National Institutes of Health, Bethesda, USA.
References
- [1]. https://www.who.int/health-topics/cervical-cancer#tab=tab_1 .
- [2].Jeronimo J; Massad LS; Castle PE; Wacholder S; Schiffman M Interobserver agreement in the evaluation of digitized cervical images. Obstet. Gynecol 2007, 110, 833–840 [DOI] [PubMed] [Google Scholar]
- [3].Yeates KE; Sleeth J; Hopman W; Ginsburg O; Heus K; Andrews L; et al. Evaluation of a smartphone-based training strategy among health care workers screening for cervical cancer in northern Tanzania: the Kilimanjaro method. J Glob Oncol. 2016. May 4;2(6):356–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Matti R; Gupta V; D’Sa DK, Sebag C.; Peterson CW; Levitz D. Introduction of mobile colposcopy as a primary screening tool for different socioeconomic populations in urban India. Pan Asian J Obs Gyn 2019; 2(1):4–11 [Google Scholar]
- [5].Xu T; Zhang H; Huang X; Zhang S; Metaxas DN Multimodal Deep Learning for Cervical Dysplasia Diagnosis. Medical Image Computing and Computer-Assisted Intervention – MICCAI; 2016. [Google Scholar]
- [6].Fernandes K; Chicco D; Cardoso JS; Fernandes J Supervised deep learning embeddings for the prediction of cervical cancer diagnosis. PeerJ Computer Science 4:e154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Hu L; Bell D; Antani S; Xue Z; Yu K; Horning MP; et al. An observational study of deep learning and automated evaluation of cervical images for cancer screening. J Natl Cancer Inst 2019; 111(9): 923–932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Herrero R; Schiffman M; Bratti C; Hildesheim A; Balmaceda I; Sherman ME; et al. Design and methods of a population-based natural history study of cervical neoplasia in a rural province of Costa Rica: The Guanacaste Project.Rev. Panam. Salud Publica 1997, 15, 362–375. [DOI] [PubMed] [Google Scholar]
- [9].Guo P; Singh S; Xue Z, Long LR; Antani S Deep learning for assessing image focus for automated cervical cancer screening. The IEEE Conference on Biomedical & Health Informatics (BHI), 2019. [Google Scholar]
- [10].Xue Z; Novetsky AP; Einstein MH; et al. A demonstration of automated visual evaluation of cervical images taken with a smartphone camera. Int J Cancer. 2020; 10.1002/ijc.33029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Xie j.; Girshick R; Farhadi A. Unsupervised deep embedding for clustering analysis, In: International Conference on Machine Learning (ICML), 2016 [Google Scholar]
- [12].Guo X; Liu X; Zhu E; Yin J Deep Clustering with Convolutional Autoencoders. International Conference on Neural Information Processing (ICONIP), 2017. [Google Scholar]
- [13].Mrabah N; Khan NM; Ksantini R Deep clustering with a dynamic autoencoder: from reconstruction towards centroids construction. arXiv preprint arXiv:1901.07752, 2020. [DOI] [PubMed] [Google Scholar]
- [14].Min E; Guo X; Liu Q; Zhang G; Cui J; Long J A survey of clustering with deep learning: From the perspective of network architecture. IEEE Access, vol. 6, pp. 39501–39514, 2018. [Google Scholar]
- [15].Desai KT; Ajenifuja KO; Banjo A; et al. Design and feasibility of a novel program of cervical screening in Nigeria: self-sampled HPV testing paired with visual triage. Infect Agents Cancer. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]


