Abstract
The rise of antibiotic-resistant strains of bacterial pathogens has necessitated the development of new therapeutics. Antimicrobial peptides (AMPs) are a class of compounds with potentially attractive therapeutic properties, including the ability to target specific groups of bacteria. In nature, AMPs exhibit remarkable structural and functional diversity, which may be further enhanced through genetic engineering, high-throughput screening, and chemical modification strategies. In this review, we discuss the molecular mechanisms underlying AMP selectivity and highlight recent computational and experimental efforts to design selectively targeting AMPs. While there has been an extensive effort to find broadly active and highly potent AMPs, it remains challenging to design targeting peptides to discriminate between different bacteria on the basis of physicochemical properties. We also review approaches for measuring AMP activity, point out the challenges faced in assaying for selectivity, and discuss the potential for increasing AMP diversity through chemical modifications.
Keywords: antimicrobial peptides, cell selectivity, microbiota, machine learning, high-throughput screening, peptide conjugates
1. INTRODUCTION
Antimicrobial peptides (AMPs) are produced by almost all forms of life for defense and communication. Within the human body, they are crucial for maintaining homeostasis between host and commensal microorganisms. Major functions of AMPs include modulation of the host immune system (1) and bacterial growth inhibition through the physical or metabolic disruption of cells (2). AMP activity has been studied extensively in the context of the gut microbiota, one of the largest and most complex ecosystems in the human body. The gut microbial community comprises hundreds of microbial species that contribute to important physiological functions, such as digestion and immune system development (3). A significant perturbation to the structure of the gut microbiota, or dysbiosis, can lead to chronic gastrointestinal diseases and disorders and has been linked with various immunological and neurological disorders (4). Early life dysbiosis is associated with developmental disorders and metabolic diseases later in life (5). One contributing factor is the use of antibiotics, most of which lack targeting specificity and can kill host-associated commensal bacteria along with disease-causing pathogens. For this reason, there has been intense interest in developing chemical or biological agents that can selectively inhibit the growth of specific (e.g., pathogenic) bacteria while preserving normal microbial flora. Another type of microbial community of therapeutic relevance is biofilms, which are microbial aggregates that can form on host tissues or on the surface of medical implants. AMPs have many diverse mechanisms of action that can allow them to target different properties of biofilms. Membrane-penetrating peptides may be advantageous for treating biofilms as they can potentially kill bacteria in different metabolic stages, including persister cells with low metabolic activity that are characteristic of biofilms (5a).
In this regard, AMPs are an attractive class of molecules to develop as engineered therapeutics against infections. AMPs produced by host cells have been mechanistically linked to alterations in the composition of the host’s gut microbiota (6) and demonstrated to have targeting specificity toward particular groups of microorganisms (2). For example, intestinal epithelial cells secrete short cationic AMPs, such as defensins and cathelicidins, that provide defense against pathogens and regulate gut microbial community composition. Reduced defensin production has been associated with inflammatory bowel disease and infection by pathogens such as Clostridioides difficile and Helicobacter pylori (6). In addition to AMPs produced by host cells, microbial cell–derived AMPs can also significantly shape community composition (7). Bacteriocins, ribosomally synthesized peptides produced by bacteria, represent one such class of AMPs that can target specific bacterial groups, often species that are closely related to the bacteria that produce the AMP and thus compete for nutrients.
Engineering AMPs capable of selectively targeting pathogenic or other invading strains of bacteria could enable new, safe ways to intervene in the microbiota to profoundly benefit human health. In recent years, computational methods, particularly machine learning tools, have been developed to identify AMPs on the basis of sequence information and to classify AMPs into broad categories on the basis of their activity spectrum, e.g., their activity against prokaryotic versus eukaryotic cells. While some of the machine learning models can accurately discriminate AMPs from non-AMPs on the basis of the peptides’ sequence, these models have provided limited insights into the mechanisms underlying AMP selectivity and design rules linking AMP sequence and function. There remains a significant need to elucidate sequence motifs or other sequence-driven features that confer targeting efficacy and selectivity against individual bacterial species (or even strains).
Understanding the connections between the primary sequence of an AMP and its targeting mechanism(s) can guide engineering efforts to modify natural AMPs or design synthetic AMPs to confer enhanced selectivity or altogether new capabilities. For example, AMPs can be modified by introducing noncanonical amino acids (ncAAs) or utilizing chemical modifications to expand their structural and chemical diversity and enable conjugation with existing antibiotics. Antibiotic–peptide conjugates are being extensively explored as a new class of drugs with enhanced therapeutic potential due to their improved selectivity profiles. Here, we review the current understanding of AMP mechanisms of action with regard to targeting specificity, summarize different approaches to designing AMPs, and discuss AMP diversification strategies that may be applicable to engineering the next generation of targeting AMPs. While antimicrobials with selective targeting capabilities are not limited to peptides (8, 9), this review focuses on studies that utilize peptides as targeting components to improve selectivity. In the remainder of this review, we define selectivity as the ability of an AMP to inhibit the growth of its target bacterial species relative to other bacterial species.
2. AMP CHARACTERISTICS AND MECHANISMS OF ACTION
AMPs are structurally diverse and exert their antimicrobial effects on a wide range of extracellular and intracellular targets through many different mechanisms. They can adopt several common conformations—α-helical, β-sheet, or random coil—which can change on the basis of their interactions with components of the cell membrane (10). The composition of the bacterial cell membrane is an important determinant of AMP selectivity (11–14). Different genera of gram-positive bacteria—e.g., Staphylococcus, Enterococcus, Bacillus, and Clostridium—have varying compositions of peptidoglycan, phosphatidylglycerol, and phosphatidylethanolamine. Negatively charged lipoteichoic acids and polysaccharides on the cell surfaces of gram-positive bacteria and gram-negative bacteria, respectively, can bind positively charged AMPs to reduce the AMPs’ penetration of the cell membrane or interaction with other cellular components (11). Thus, these cell wall or membrane components can directly impact AMP activity toward particular species or strains of bacteria. AMPs that bind to specific lipids on the cell surface or target intracellular proteins could also have targeting specificity in a microbial community (11). For example, nisin is a well-characterized bacteriocin known to generally target gram-positive bacteria by binding to lipid II, a component of the bacterial cell wall that is more accessible in gram-positive cells (15).
Natural AMPs have been shown to exhibit selectivity toward gram-positive or gram-negative bacteria. Insect defensins are generally known to be more active against gram-positive bacteria (16). An example is defensin-like peptide 4 (DLP4), an insect defensin that is primarily active against gram-positive bacteria (several Staphylococcus aureus strains and Bacillus subtilis) but not gram-negative bacteria (Escherichia coli, Enterobacter aerogenes, and Pseudomonas aeruginosa) (17). In contrast, proline-rich and glycine-rich insect AMPs are generally more potent against gram-negative bacteria (18). Pyrrhocoricin, a proline-rich AMP active against E. coli but not S. aureus, was shown to bind to a fragment of the E. coli heat shock protein DnaK but not its homologous counterpart from S. aureus. In vitro assays showed that pyrrhocoricin diminished the ATPase and protein folding activities of DnaK, suggesting that the mechanism of selectivity for this peptide is species-specific recognition and disruption of an intracellular target.
Due to the diversity of AMP targeting mechanisms, whether on the cell surface or on intracellular targets, there is tremendous potential in designing AMPs that can selectively interact with specific microorganisms. AMPs that target nucleic acids, proteases, and protein synthesis machinery have been discovered and characterized (19). It is important to note that AMP activity is not limited to a single targeting mechanism; many membrane-disruptive cationic AMPs have secondary intracellular mechanisms of action (20). Currently, there is limited understanding of how AMP structure impacts targeting ability; improving this understanding could lead to the development of new classes of antimicrobial agents having minimal off-target effects. One promising approach to elucidating the connection between AMP structure and selectivity is to study the structural patterns of AMPs having known selectivity. This has been enabled by the increasing number of studies on AMPs, the expansion of databases cataloging AMPs, and the development of new methodologies for synthesizing and characterizing these compounds.
3. APPROACHES FOR ENGINEERING AMP SELECTIVITY
Broadly, methods for engineering or discovering AMPs can be grouped into three approaches: rational design, computational design, and high-throughput screening (Figure 1) (21). We highlight notable studies utilizing one or more of these approaches to discover novel targeting AMPs or improve the targeting efficacy of existing AMPs (Table 1). According to these studies, an order of magnitude difference in values of antimicrobial activity between target and nontarget species could be considered selective enough to proceed to more complex models of antimicrobial susceptibility testing, such as community models. Though these studies should try to test as many species in monoculture as possible, a more rigorous test for determining selectivity is by using a community culture within a physiological context.
Figure 1.
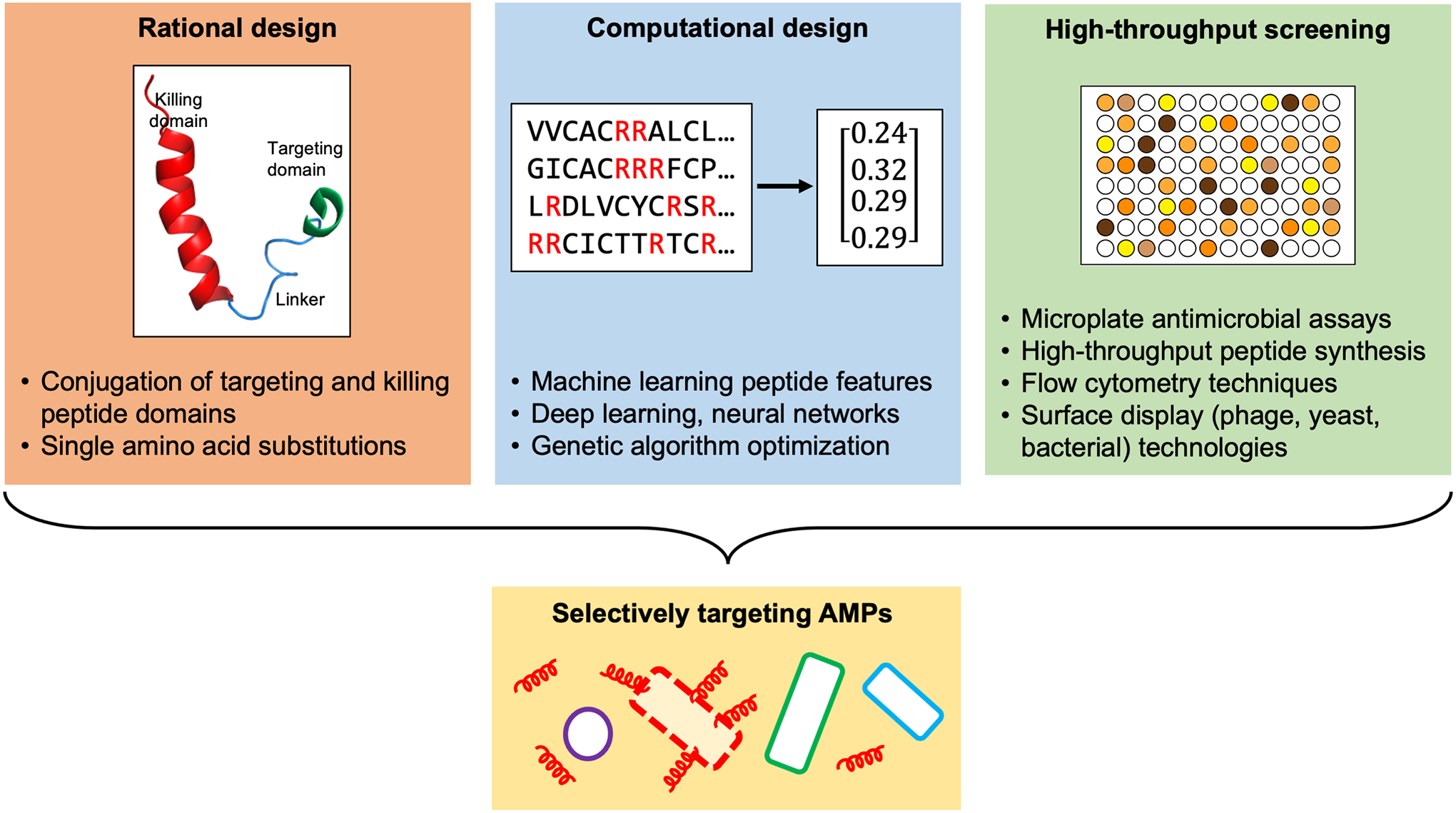
Current approaches for designing new antimicrobial peptides (AMPs) or optimizing existing ones for increased potency or improved selectivity, with examples of rational design, computational design, and high-throughput screening.
Table 1.
Selected examples of targeted AMPs generated using rational design, computational design, or high-throughput screening
| Peptide(s) | Type/characteristics | Discovery/design methodology | Target | Reference |
|---|---|---|---|---|
| cCF10-C4 | Hybrid AMP | Fusion (by a GGG linker) of Enterococcus faecalis–specific peptide pheromone cCF10 to C4, a broad-spectrum AMP modified to have a lower positive charge | E. faecalis; at least eightfold less active on Staphylococcus aureus, Staphylococcus epidermidis, Salmonella typhimurium, Salmonella pullorum, Escherichia coli | 24 |
| Peptide 13 | STAMP, hybrid AMP | Fusion of weakly targeting AMP to targeting AMP previously discovered through phage display; peptides were modified with charged residues at the N or C terminus to alter physicochemical properties | E. coli; at least eightfold less active on Staphylococcus, Pseudomonas, Lactobacillus strains | 75 |
| C16G2 | STAMP | Fusion of two peptides: killing and targeting moieties; targeting domain is a bacterial pheromone; killing domain is derived from broad-spectrum AMP novispirin G10 | Streptococcus mutans in the oral microbiota | 22, 23 |
| RI16-T9W | Amphipathic, helical | Point mutation (T9W) to RI16, a fragment sequence of the cathelicidin PMAP-36; rational design to disrupt sites important for amphipathicity | Pseudomonas aeruginosa | 26 |
| Peptide 3 | Peptide fusion | Fusion of acetyltransferase inhibitor peptide to cell penetrating peptide; targets lipid A biosynthesis | Order of magnitude decrease in MICs for panel of gram-negative bacteria; twofold increase in MIC for S. aureus | 28 |
| Guavanin 2 | Arginine rich, α-helical | Genetic algorithm optimization of a guava peptide; ratio between hydrophobic moment and α-helix propensity was used in fitness function | Selective toward gram-negative species: E. coli, Acinetobacter baumannii (6.25 μM MIC); less active against S. aureus, Streptococcus pyogenes, Listeria ivanovii, E. faecalis (≥50 μM MIC) | 34 |
| SP1, SP2, SP3, SP4 | Short, charged | DBAASP prediction algorithm based on clustering of 9 peptide physiochemical features; modified AMPs included D-enantiomers | Designed to target gram-negative bacteria; varying activities toward E. coli, P. aeruginosa, A. baumannii, Enterococcus cloacae, Klebsiella pneumoniae (≤0.125 – ≥32 μg/ml MIC); not tested on gram-positive bacteria | 37 |
| NN2_0018, NN2_0022, NN2_0024, NN2_0027, NN2_0029, NN2_0035, NN2_0039, NN2_0046, NN2_0050, NN2_0055 | Short, charged | LSTM language model trained on MIC data against E. coli; top 10 sequences synthesized and characterized | Designed to target E. coli; varying activities (0.25 – ≥ 128 μg/ml MIC) toward 30 different gram-positive and gram-negative cultures | 38 |
| PA2-GNU7 | Hybrid AMP, helical | Phage display high-throughput screening and conjugation to GNU7, a broad-spectrum AMP with a GGG linker; the peptides individually or combined separately were less effective than the conjugated peptide | Selectively active on P. aeruginosa in mixed cultures with either E. coli or S. typhimurium | 58 |
| Peptides P1–P18 | Random sequence | Bacterial high-throughput self-screening of surface displayed peptide libraries | Varying activities toward E. coli, P. aeruginosa, A. baumannii (≤2 – >128 μM MIC) | 48 |
| KAM5, KAM8 | Disulfide-cyclized | Phage display high-throughput screening of peptides containing 2-acetylphenylboronic acid moieties | KAM5: S. aureus, no binding to E. coli or Bacillus subtilis; KAM8: A. baumannii, no binding to E. coli or S. aureus | 49 |
| Apidaecin and its analogs | Short, proline-rich | Alanine substitution screening; peptides synthesized using SPOT synthesis | Single amino acid substitutions to apidaecin resulted in varying activities toward P. aeruginosa, E. coli, S. aureus (0.63 – >125 μg/ml MIC) | 44 |
Abbreviations: AMP, antimicrobial peptide; CSP, competence-stimulating peptide; DBAASP, Database of Antimicrobial Activity and Structure of Peptides; LSTM, long short-term memory; MIC, minimum inhibitory concentration; STAMP, specifically targeted antimicrobial peptide.
3.1. Rational Design
A common rational design strategy involves the fusion of targeting and killing peptides. This approach utilizes the relatively high specificity of targeting peptides to narrow the spectrum of killing peptides. A notable example of a synthetic AMP with a narrow activity spectrum is C16G2, a Streptococcus mutans–targeting peptide designed by fusing a Streptococcus competence-stimulating peptide with a fragment of novispirin G10, a broad-spectrum AMP. The fused peptide was demonstrated to have selectivity for S. mutans in mixed community cultures with minimal impact on other species (22, 23). Another example of peptide fusion is cCF10-C6, which targets Enterococcus faecalis. This peptide was designed by fusing the E. faecalis–specific pheromone cCF10 with peptide C6, a fragment of a previously designed broad-spectrum AMP. Modification of this peptide’s net charge by replacing a cationic lysine residue with glutamate further improved specificity of the fusion peptide, allowing it to inhibit E. faecalis at a significantly lower concentration compared with other species in the test panel (24). A targeting peptide can also be used to reduce the antimicrobial activity of an AMP against nontarget species to modulate selectivity. For example, Choudhury et al. (25) fused a Staphylococcus-targeting peptide previously identified from a phage library screen to the broad-spectrum AMPs plectasin and eurosin. This study used minimum inhibitory concentration (MIC) assays to show that, compared with either plectasin or eurosin alone, the fused peptide had significantly reduced antimicrobial activity (more than tenfold) against off-target species while retaining the ability to inhibit the intended Staphylococcus targets.
Beyond fusion strategies, even single amino acid substitutions can have drastic effects on specificity. Zhu et al. (26) showed that a T9W substitution significantly increased the specificity of peptide RI16, a truncated form of porcine myeloid antibacterial peptide, for P. aeruginosa. The result was a greater than tenfold decrease in MIC value for P. aeruginosa compared with E. coli, Salmonella typhimurium, S. aureus, and Staphylococcus epidermidis strains. In rational approaches, amino acid substitutions are typically used to increase AMP amphipathicity, a measure of hydrophobic and hydrophilic structural organization, as this characteristic is an important determinant of antimicrobial activity (27).
Another strategy for engineering selectively active AMPs is to target unique metabolic pathways, proteins, or other types of molecules in a cell. Postma & Liskamp (28) fused a previously discovered E. coli acetyltransferase inhibitor peptide that targets lipid A biosynthesis to a cell-penetrating peptide and found that the fusion increased antimicrobial activity toward a panel of seven gram-negative bacterial species while decreasing activity toward the two gram-positive Staphylococcus strains tested. Muhle & Tam (29) designed peptides to target gram-negative bacteria that mimic the binding sites of lipopolysaccharides, a cell wall component generally unique to gram-negative bacteria. Several of the designed peptides were highly active against E. coli, with submicromolar MIC values, while activity against S. aureus was not detected at concentrations greater than 500 μM. Targeting peptides can also be designed to have indirect antimicrobial effects. Idso et al. (30) discovered a peptide ligand that specifically binds to an epitope of a highly expressed protein, type 3 Fimbrial Shaft (MrkA), on the surface of Klebsiella pneumoniae. The ligand was shown to bind specifically to K. pneumoniae and not E. coli or S. typhimurium. Conjugation of the ligand with a 2,4-dinitrophenyl (DNP) moiety enabled anti-DNP antibodies to specifically bind to the target cell surface and consequently recruit cell-killing macrophages (30).
3.2. Computational Design
Computational models can offer insight into the characteristics of AMPs that contribute to target recognition and activity. Various machine learning models for classifying and designing AMPs have been developed, including neural networks and support vector machine models (31, 32). Genetic algorithms have also been implemented, for example, to optimize a 13-mer AMP against E. coli through iterative rounds of in vitro testing (33) and to design a unique AMP starting from Pg-AMP1, a natural AMP found in guava seeds (34). In the latter study, guavanin 2, the most potent guava AMP identified from the in vitro screen, was further characterized through MIC assays. Interestingly, guavanin 2, which was computationally optimized for antimicrobial activity but not selectivity, was shown to be selective toward gram-negative bacteria among the panel of bacterial species tested in the study. The study proposed that this result could be due to the attraction of the positively charged guavanin 2 peptide by negatively charged phospholipid head groups on gram-negative bacteria. However, it is unclear why the peptide is less active toward gram-positive bacteria, which contain negatively charged teichoic acids on the cell surface. This underscores the challenge of elucidating the molecular mechanisms of selectivity while highlighting the potential for an improved mechanistic understanding to aid computational design of targeting AMPs.
Computational models that can discriminate AMPs from non-AMPs with high accuracy have been developed. In principle, the discriminatory peptide features could be further investigated to understand how AMP primary structure impacts activity. The computational classification models typically incorporate sequence features related to charge and hydrophobicity; however, the models differ with respect to their training datasets and algorithms (35). There are many different approaches for calculating the physicochemical properties of amino acids and encoding the properties into features such as composition, transition, and distribution, as well as pseudoamino acid composition (36). The encoded features are used to train machine learning algorithms to recognize peptide sequence patterns characteristic of AMPs that are distinct from those of non-AMPs. The sequence patterns learned from the classification models have been successfully used for de novo design of AMPs, some of which have shown high specificity for a particular species within a tested panel of bacteria (34, 37, 38).
Despite the progress achieved in the aforementioned studies, there remain hurdles in realizing the potential for using computational design to enhance AMP specificity. One of the main limitations is the availability of experimental activity data needed to train the models. In the Database of Antimicrobial Activity and Structure of Peptides (DBAASP), one of the largest publicly accessible AMP databases, the top 6 test species—E. coli, S. aureus, P. aeruginosa, Candida albicans, B. subtilis, and S. epidermidis—account for approximately 52% of the MIC data. This illustrates the data sparsity problem for designing AMPs against species that are not commonly tested. To address this limitation, recent efforts have attempted to computationally estimate the specificity of an AMP toward particular groups of bacteria. Veltri et al. (39) developed a genetic algorithm–based scheme to generate sequence features, using a fitness function that tracks the occurrence of AMP-associated sequence features. The feature space spanned by the generated sequences was then reduced with a correlation-based filter to remove redundant features. Finally, logistic regression was used to build a model classifying peptides into AMPs and non-AMPs on the basis of their predicted antimicrobial activity. The authors used this method to further classify AMPs as gram-positive targeting, gram-negative targeting, or both, and found good agreement between their results and the peptide classification information in the second version of the Antimicrobial Peptide Database.
Quantitative structure–activity relationship (QSAR) models are yet another approach toward designing selective AMPs. In the context of AMPs, QSAR models can relate a set of calculated peptide sequence features (predictor variables) to experimentally measured antimicrobial activity values (response variables). Majumder et al. (40) trained a QSAR model to predict the activity of AMPs against Acinetobacter baumannii by correlating a set of eight peptide physicochemical features (e.g., charge, hydrophobic moment) with all the MIC data available for 75 AMPs tested against the same ATCC 19606 strain. While the model could theoretically predict the activity of any AMP against A. baumannii, additional experimental validation of the predicted AMP activities is required to assess the accuracy of the model. QSAR models based on physicochemical features have also been extended to predict the activities of both natural peptides and peptides with ncAAs (41).
It is important to note that there is no standardized way to classify a peptide as exclusively gram-positive targeting or gram-negative targeting. Current classifications are based on MIC data for a panel of selected microorganisms against which the AMP of interest has been tested. Typically, the panel comprises a pathogen or other species of interest plus a handful of related species or model organisms such as E. coli or B. subtilis. In an attempt to address the sparsity of available experimental data, Gull & Minhas (42) developed AMP0, a machine learning model that predicts an AMP’s efficacy toward a target species on the basis of the species’ genome or its phylogenetic relation to other species against which the AMP’s efficacy is known. The aforementioned DBAASP also provides a tool that predicts whether an AMP may be active against particular microorganisms. The DBAASP tool was developed by training a machine learning model on key physicochemical peptide features identified through a clustering analysis (37).
3.3. Screening-Based Approaches
A conventional plate-based culture of commensal bacteria on solid media can be an effective screening format for identifying novel, highly selective AMPs. For example, thuricin CD, a two-component C. difficile–targeting AMP produced by Bacillus thuringiensis, was discovered by screening fecal isolates for inhibitory activity, which were indicated by large inhibition zones, on agar plates with C. difficile overlay cultures. Subsequent agar diffusion assays showed that thuricin CD inhibited species in the Clostridioides and Bacillus genera, while having limited activity against a large panel of commensal bacteria (43).
The throughput and coverage of screening-based approaches have benefited from advances in chemical synthesis, and molecular cloning technologies have made it possible to rapidly generate and test peptides with throughputs that are orders of magnitude higher than what was previously possible. For example, Lopez-Perez et al. (44) used SPOT synthesis, a method of synthesizing peptide arrays on membrane supports, to generate hundred-member peptide libraries and evaluate their activity against P. aeruginosa in a high-throughput format. In addition, a full substitution analysis was performed on apidaecin, a proline-rich AMP, by substituting each amino acid at every position of the peptide with all of the other natural amino acids. Recently developed flow-based peptide synthesis technology can achieve throughputs that are orders of magnitude higher than those achieved in conventional solution-based chemical synthesis (45). Albin & Pentelute (46) applied this technology to efficiently synthesize a diverse panel of 43 human AMPs.
The use of genetically encoded peptide libraries in screening has yielded unique, synthetic AMPs. Ritter et al. (47) designed a library of 960 microcin J25 mutants to screen for mutants with specific antimicrobial activity against pathogenic Salmonella species. The library was cloned into an E. coli production strain and screened for activity against a panel of bacteria (two pathogenic Salmonella species and two commensal E. coli strains) using an agar diffusion assay. Ritter et al. (47) discovered a mutant with a single amino acid mutation (I13T) that significantly reduced activity toward commensal E. coli strains relative to the Salmonella species. Tucker et al. (48) developed a peptide screening methodology, called Surface Localized Antimicrobial Display, for generating novel, active AMPs targeted toward E. coli. By expressing peptide libraries on the target cell surface and sequencing the population after iterative rounds of cell culturing, sequences of AMPs interacting with the target cell’s surface can be inferred by calculating the depletion of their respective DNA sequences from the initial population. This method was successfully used to screen a library of 800,000 20-mer peptides to discover AMPs that were active against E. coli. One current limitation to this method is that the target bacterial species must be amenable to genetic manipulation and surface display. Nevertheless, this is a promising technology that provides an efficient method for discovering novel AMPs, particularly against genetically tractable species.
Phage display offers another potentially useful screening method for AMP discovery. McCarthy et al. (49) modified phage-displayed peptides with 2-acetylphenylboronic-acid moieties to covalently target surface imines on bacterial cells. By conjugating the cell-surface-targeting peptide identified from phage display to eosin, a phototoxin, the researchers achieved selective and inducible killing of S. aureus without a significant effect on E. coli or B. subtilis. The screen was also applied to a lipooligosaccharide-deficient mutant of A. baumannii, which identified peptide sequences that targeted the mutant strain but not the wild-type, demonstrating strain-specific killing activity. This study underscores the importance of cell-surface components to AMP activity, raising the possibility that cell-surface-binding peptides can be utilized in combination with chemical conjugation strategies to generate unique, potentially selective antimicrobial molecules.
Fluorescence-activated cell sorting (FACS) is another high-throughput method that can be applied to AMP screening. Scanlon et al. (50) applied FACS on hydrogel droplets of a target microbial species coincubated with an AMP expression host. Briefly, E. coli expressing a Staphylococcus metagenomic library was coincubated with S. aureus in an agarose mixture, which was subsequently fractionated into small droplets by a microfluidic device. Antimicrobial activity in the droplets was assessed using a viability dye. Droplets containing E. coli–expressing AMPs with activity against S. aureus were sorted and sequenced to identify relevant genes. As a proof of concept, the screen was used to identify the bacteriolytic enzyme lysostaphin from the metagenomic library. This FACS workflow could be adapted to screen other compounds that can be expressed and secreted from technically amenable hosts and have antimicrobial activity, including AMPs.
3.4. Current Challenges in Engineering AMPs
The methodological advances described above have enabled substantial progress in improving the activity of AMPs against specific bacteria and in some cases also enhancing their selectivity. However, challenges remain in designing AMPs that can selectively bind or inhibit the target species with minimal off-target effects. This is especially important for applications where AMPs are to be used as therapeutics against pathogens that colonize host-associated microbial communities. AMPs active against a broad spectrum of bacteria could deplete species that are critical for community resilience and host health in addition to fending off the pathogen. Lacticin 3147 is one such broad-spectrum AMP; it is active against pathogenic C. difficile in an in vitro fecal culture but also significantly depletes the population of Lactobacillus and Bifidobacterium, commensal species known to be beneficial for human health (51). In the development of computational models, differences in the way MICs are determined across studies can introduce confounding variability into training data. The use of different media, bacterial strains, and growth conditions (e.g., oxygenation) can lead to discrepancies in MIC determination for the same peptide and species. Experimental data on AMP activity is available only for a very small number of bacterial species. Moreover, the panel of species tested for growth inhibition or loss of viability by an AMP varies from study to study. As absence of evidence does not equate to evidence of absence, developing computational models to learn which peptide features confer targeting specificity can present a major challenge. Along this vein, screening-based approaches have also typically tested peptide libraries against a small number of laboratory strains, rather than against broad, representative panels of the bacterial species that AMPs will likely encounter in therapeutic applications involving host-associated microbiota.
4. CHARACTERIZATION OF ENGINEERED PEPTIDES
Many different types of assays for evaluating antimicrobial activity have been developed. The most commonly used types are summarized in Figure 2. While efforts have been made to standardize antimicrobial activity testing methods, there remain challenges in choosing the appropriate test conditions (52). For example, the composition of the medium and the type of culture vessel used can affect the MIC values obtained in a susceptibility assay. The susceptibility of bacteria to an AMP might be underreported in certain microtiter plates due to adsorption of the AMP on the plastic surface. Peptide precipitation in the test media can also cause variations in the measured MIC values, especially for cationic AMPs. These adsorption and precipitation problems can be mitigated by adding bovine serum albumin to the medium and using polypropylene plates that have lower affinity for protein binding compared with conventional polystyrene plates (53). As short AMPs are usually amphipathic, it is important to consider potential interactions with medium components and culture vessel surfaces.
Figure 2.
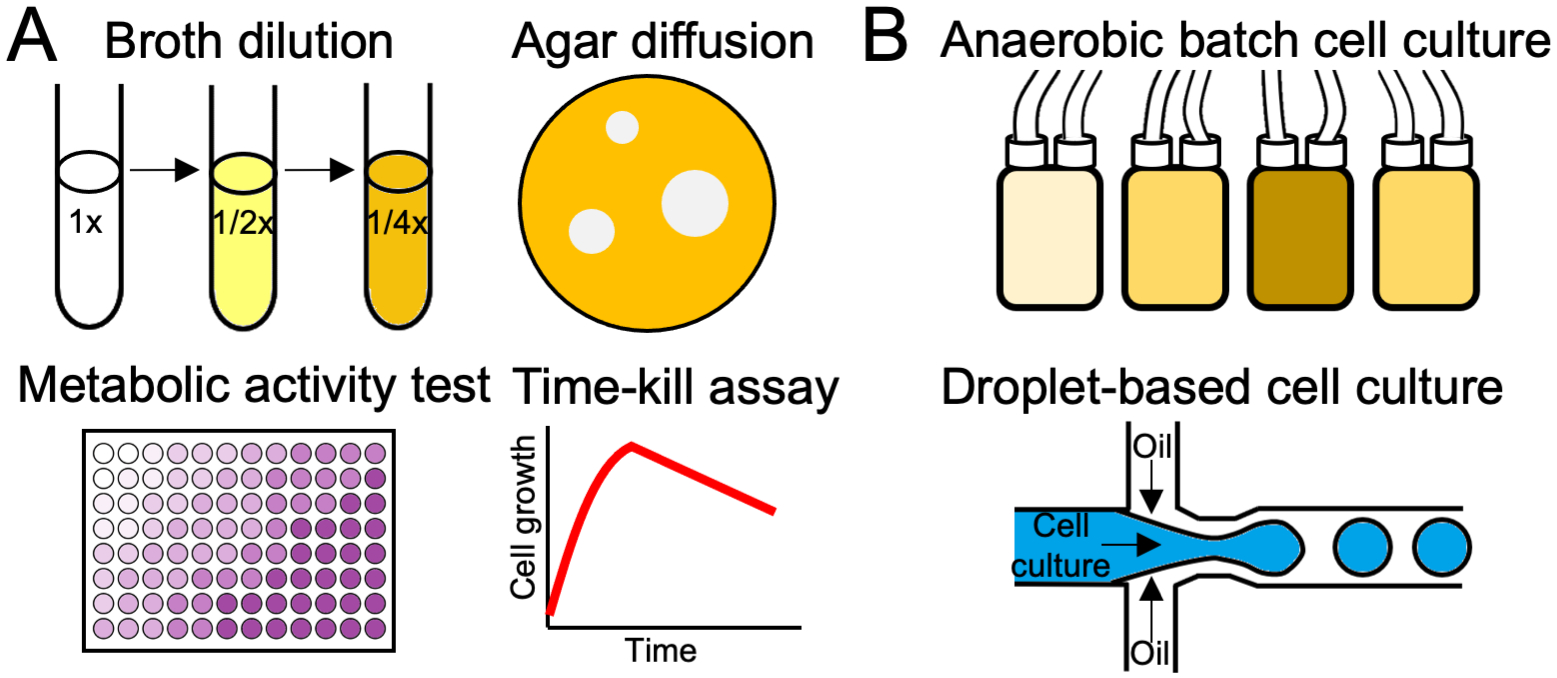
Characterization assays for testing activity or specificity of antimicrobial compounds. (a) Commonly used assays for measuring antimicrobial peptide (AMP) activity against individual species or strains in monoculture. (i) Broth dilution. Serial dilutions of an antimicrobial compound in liquid media are inoculated with the target bacteria. (ii) Agar diffusion. Varying doses of the antimicrobial compound are placed on agar plates inoculated with a bacterial lawn. Antimicrobial activity is measured by the zone of clearance. (iii) Metabolic activity test. A colorimetric or fluorogenic assay is used to assess metabolic activity of the target bacteria by measuring enzyme reduction of a substrate. (iv) Time-kill assay. An assay determines antimicrobial activity over time to measure rate of killing by the test compound. (b) Anaerobic culture systems designed to simulate microbiota within the human body, used to assay the AMP’s effect on microbial diversity. (i) Anaerobic batch cell culture. Small volume bioreactors are used to cultivate microbiota samples. (ii) Droplet-based cell culture. Droplet emulsions are generated using a microfluidic device to segregate a mixed microbial community and isolate low-abundance species.
When developing AMPs as therapeutic agents that target pathogens, it is critical to assess their inhibitory activity under physiologically relevant conditions (53a). For example, the selectivity of AMPs targeting intestinal pathogens such as C. difficile should be tested under conditions compatible with anaerobic commensal bacteria. In this regard, high-throughput methods for the cultivation of anaerobic bacteria, both in monoculture and in mixed community, can facilitate the development of antimicrobials. Auchtung et al. (54) used mini-bioreactor arrays to simultaneously culture 48 fecal microbiota and demonstrated that the communities remained stable one week after inoculation. Droplet-based culture technologies, such as the microfluidic platform developed by Watterson et al. (55), could potentially facilitate screening of rare bacterial species. Both methods offer unique advantages unavailable to conventional culture systems. The mini-bioreactor can support long-term experiments to dynamically capture taxonomic shifts caused by antimicrobial treatment and may be more representative of the gut microbiota because the system allows nutrient flow, cross-feeding, and communication between multiple species. Droplet-based methods can provide insights into the direct impact of antimicrobials on individual species while still maintaining a limited form of interspecies interaction. These methods can be especially useful if the species of interest is at a low abundance in the intact community.
MIC values are usually determined in monocultures, which have the drawback that they may not reflect in vivo growth conditions for some species of interest. Naturally occurring host-associated bacterial communities are often diverse, and cell–cell interactions may play a role in the activity and selectivity of antimicrobial compounds. Complex interactions between microbial species in a community may modulate AMP activity (Figure 3). While an AMP may target one species, it can have subinhibitory effects on other species in the community, triggering defense mechanisms such as changes in gene expression and modifications of the cell wall (56). Adamowicz et al. (57) found that in a cross-feeding coculture system consisting of E. coli, Salmonella enterica, and Methylobacterium extorquens, bacterial species that had a high tolerance to an antibiotic were generally inhibited at lower concentrations of the antibiotic compared with their respective monocultures. On the other hand, MIC determinations in cocultures of multiple species may be confounded by interactions (e.g., competition or mutualism) among species. A notable example is the measured antimicrobial activity of PA2-GNU7, a hybrid peptide designed to selectively kill P. aeruginosa. Despite having little difference in activity toward P. aeruginosa, E. coli, or S. typhimurium monocultures (range of 1–8 μM in MIC), the peptide was shown to selectively kill P. aeruginosa in mixed cultures with either E. coli or S. typhimurium, with an order of magnitude difference in species viability (58). A further confounding factor could be the presence of proteolytic enzymes produced by one species that act to degrade AMPs produced by another species or AMPs added exogenously (59). In this regard, these systems may not be appropriate for determining the direct effects of an antimicrobial on a particular species. Rather, community cultures may be more useful in validating the specificity of targeting peptides to provide a more accurate prediction of their performance in in vivo settings. For example, Guo et al. (22) used a mixed coculture model to show that C16G2 is a targeting AMP capable of significantly decreasing the relative abundance of S. mutans within an oral microbiota. By monitoring the taxonomic shifts induced by an AMP in coculture systems, researchers can study the effects of eliminating specific bacterial groups.
Figure 3.
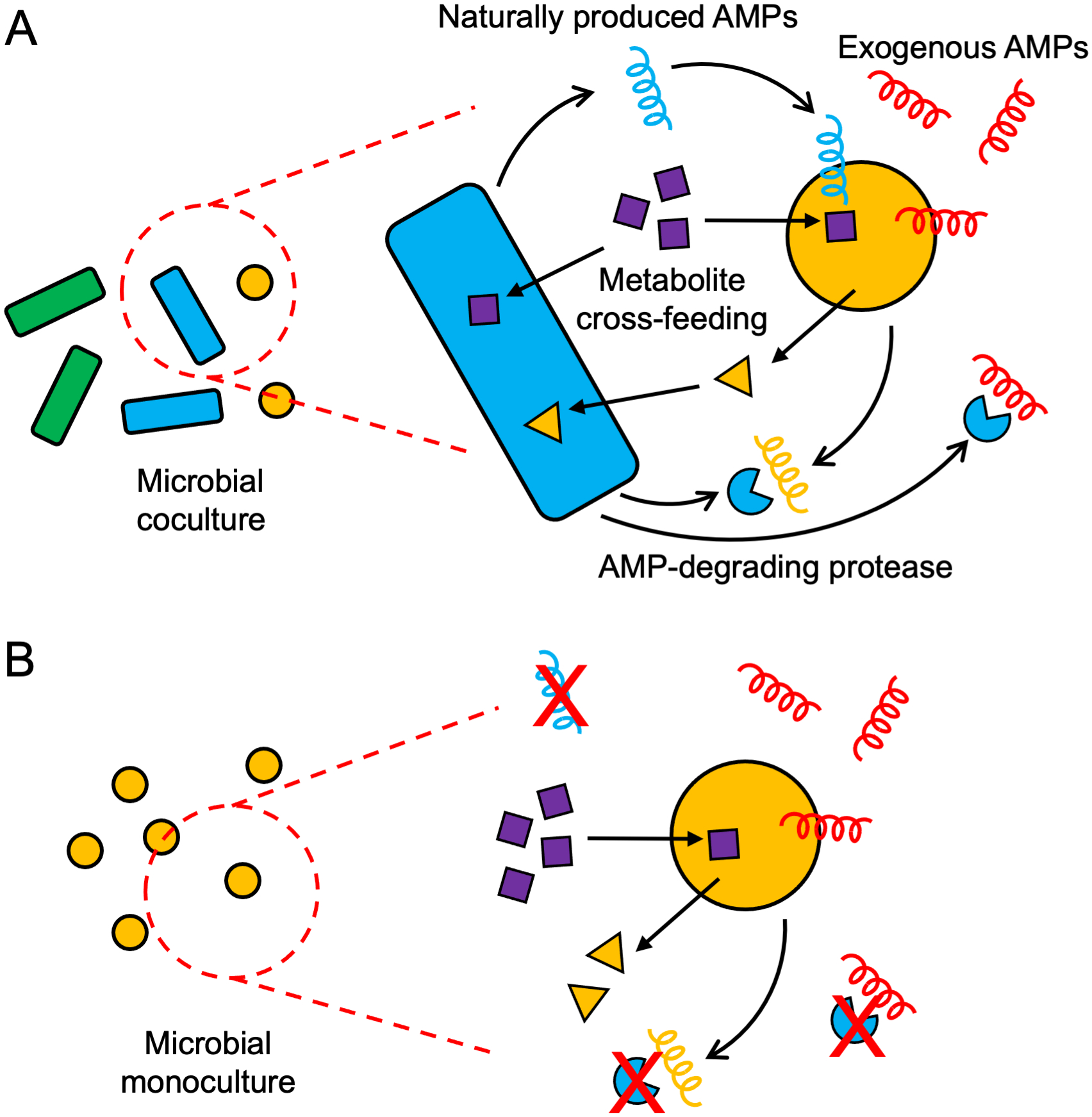
Factors affecting minimum inhibitory concentration (MIC) determination in coculture and monoculture systems. (a) Complex interactions between two microbial species in a coculture system may affect antimicrobial peptide (AMP) sensitivity. (b) A cell in a microbial monoculture misses many of the factors affecting AMP sensitivity that are present in a microbial community.
While there is no standard criterion for translation from preclinical to clinical trials based on MIC values, many AMPs in preclinical development have MIC values on the order of 1 μg/ml or lower toward a target organism as measured using in vitro assays (59a). This is comparable with the range of MIC values obtained for in vitro assays of traditional antibiotics that are considered effective (59b). Most efficacy endpoints defined by standards for antimicrobial susceptibility testing require 100% growth inhibition in an in vitro assay. However, there are unique cases where a certain percentage of growth inhibition or a defined morphological change in the target species is required. MIC values obtained using in vitro assays are dependent on assay conditions and may not necessarily translate to in vivo assays (53a). Though many AMPs in development are meeting MIC standards in vitro, there are additional challenges for therapeutic applications such as protease susceptibility and toxicity. Regarding protease susceptibility, proteases present in serum have been shown to degrade AMPs. For example, cytosolic proteases of red blood cells were directly linked to proteolysis of linear AMPs (59c). A recent study by Starr et al. (59d) attempted to address these issues by screening for AMPs in the presence of red blood cells and serum to mimic physiologically relevant conditions and improve the probability of discovering clinically relevant AMPs. Several nonribosomally synthesized peptides have been approved for clinical use, but no ribosomally synthesized AMPs have been approved yet, though many are undergoing different phases of clinical trials. The main causes for the failure of AMPs progressing through clinical trials are toxicity, poor translation from in vitro to in vivo models, and failure to demonstrate improved activity or selectivity compared with traditional antibiotics (59a). Further chemical modifications to AMPs can improve their therapeutic properties.
5. EXPANDING AMP DIVERSITY
Antimicrobial peptides can be chemically modified to greatly expand their functional diversity and targeting potential (Figure 4). Many AMPs are modified posttranslationally in naturally occurring processes, such as glycosylation and lipidation (60). Judicious posttranslational modification of AMPs in the laboratory can drastically alter AMP properties (61). ncAAs provide additional routes to AMP diversification either during chemical peptide synthesis or by ribosomal incorporation with orthogonal translation machinery (62). Insights into the physicochemical properties of chemically diverse peptides that affect activity and selectivity have the potential to guide the design of targeting AMPs.
Figure 4.
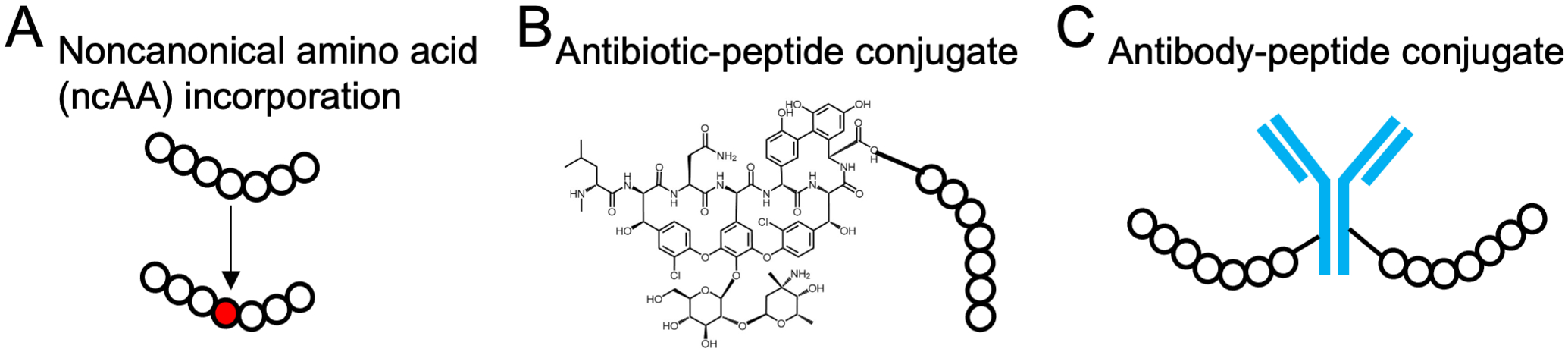
Examples of modified antimicrobial peptides (AMPs). (a) Noncanonical amino acid (ncAA) incorporation. ncAAs may be incorporated through chemical synthesis or genetic code manipulation. (b) Antibiotic–peptide conjugate. (c) Antibody–peptide conjugate.
5.1. Synthetic Modifications
Synthetic modifications to AMPs, which can be accomplished via chemical peptide synthesis or via genetic code manipulation in cells or cell-free systems, can lead to AMPs exhibiting distinct properties such as improved stability or altered selectivity (63, 64). Chemical synthesis strategies have enabled the generation of synthetic structural analogs of natural AMPs, such as selenocysteine-substituted caenopore-5, as well as synthetic forms of naturally posttranslationally modified AMPs, such as glycocin F (64). Vishnepolsky et al. (37) stabilized synthetic AMPs against protease degradation by synthesizing them with D-amino acids, which resulted in AMPs that are completely resistant to proteinase K and α-chymotrypsin digestion while retaining antimicrobial activity toward E. coli. Hicks et al. (65) used chemical synthesis to synthesize 36 peptides with octahydroindolecarboxylic acid and tetrahydroisoquinolinecarboxylic acid residues incorporated into the backbone structure. The authors observed a wide range of MIC values (from 6.25 to > 100 μg/ml) for the peptides against a panel of gram-positive and gram-negative bacteria.
As an alternative to chemical peptide synthesis, genetically encoding ncAAs provides another means to generate AMPs with altered, and potentially augmented, properties (62). Initial work in this area has mainly demonstrated the feasibility of producing bioactive AMPs containing ncAAs. For example, a study by Piscotta et al. (66) used stop codon suppression to incorporate four different ncAAs with different physicochemical properties—meta-substituted trifluoro-, bromo-, chloro-, and nitro-phenylalanine derivatives—into different positions within the peptide microcin J25. Serial dilution assays revealed that the ncAA-containing microcin J25 variants exhibited a wide range of antimicrobial activities toward Salmonella newport, with some substituted clones exhibiting potencies equivalent to the wild-type construct. This demonstrates that ncAA-substituted AMPs retain their activity toward a particular species. In another recent study, Bartholomae et al. (67) used stop codon suppression to generate bioactive nisin variants containing Boc-L-lysine substitutions in both E. coli and Lactococcus lactis, confirming the feasibility of producing ncAA-substituted AMPs in multiple organisms. With physicochemical properties that extend beyond those found in the canonical amino acids, ncAAs can be used to perform protein medicinal chemistry in search of AMPs that exhibit improved therapeutic properties such as enhanced target affinity and specificity (68). In addition, incorporating ncAAs into peptides can provide bioorthogonal chemical handles that enable further modifications, such as conjugation with small molecules, other peptides, or antibodies.
5.2. AMP Conjugates
Selective chemical functionalization of peptides allows AMPs to be covalently linked to many types of small molecules and drugs, including antibodies, antibiotics, lipids, organometallic groups, and other AMPs (69). AMP conjugates have employed broad-spectrum membrane-active AMPs to complement the targeting function of conjugated moieties. Touti et al. (70) designed various antibody–AMP conjugates to target lipopolysaccharide glycans on E. coli and found a conjugate that preferentially killed E. coli in mixed cultures with P. aeruginosa and K. pneumoniae. Peptide nucleic acids, designed to bind to unique mRNA sequences in target species of bacteria, were used to selectively inhibit B. subtilis, E. coli, K. pneumoniae, and S. typhimurium in both monocultures and mixed cultures (71).
AMP conjugates have also been designed to increase the potency of the conjugated moiety toward a species that has developed resistance. For example, Blaskovich et al. (72) conjugated vancomycin to bacterial membrane-targeting peptide sequences modified with lipophilic membrane-penetrating functional groups and demonstrated improved antimicrobial activity of the antibiotic–peptide conjugate against a panel of antibiotic-resistant bacteria. Another study showed that various aliphatic and aromatic linkers within vancomycin–AMP conjugates altered antimicrobial activity toward a panel of gram-positive and gram-negative bacteria (73). A bacteriolysin–AMP conjugate was demonstrated to improve bactericidal activity toward antibiotic-resistant A. baumannii (74). Future design of AMP conjugates incorporating targeting peptides instead of broad-spectrum AMPs can potentially improve species selectivity.
6. CONCLUSIONS AND PERSPECTIVES
AMPs are a highly diverse group of compounds investigated as potential alternatives to antibiotics. Current efforts to discover and design AMPs include combinations of rational and computational design to optimize desired peptide physicochemical properties. High-throughput peptide synthesis and screening technologies have accelerated progress in this field. While there has been increasing success in designing and optimizing AMPs to be highly potent, engineering AMPs to selectively target a particular species of bacteria remains a key challenge. A major bottleneck is the dearth of experimental testing data, as most AMPs cataloged in large databases have been tested only on a small number of bacterial species. Another factor contributing to the data limitation is the variability in assay methodologies across different studies. There is an unmet need for the research community to develop standardized methods for obtaining MIC values, including representative panels of bacterial species that can serve as controls for off-target killing. Additionally, MIC values obtained from monoculture assays may not be representative of AMP activity in microbial communities. Addressing this concern will require the development of culture platforms capable of supporting stable cocultures of multiple species. Despite these limitations, there have been significant advances in the discovery of selectively targeting AMPs, notably from screening peptide libraries. High-throughput assays can considerably increase the rate of AMP discovery and expand the amount of antimicrobial activity data available for uncovering the underlying associations between peptide sequence or structure, in vivo stability, toxicity toward mammalian cells, and antimicrobial activity against a target bacterial species. Additionally, there is exciting potential to greatly expand the functionality of AMPs by incorporating ncAAs and chemical modifications into AMP design. Altogether, these strategies can further the development of targeted antimicrobial peptides to address the need for new therapeutics to combat microbial-associated diseases and disorders.
ACKNOWLEDGMENTS
This work was supported in part by grants from the National Center for Complementary and Integrative Health to A.J. and K.L. (RO1-AT-010282) and from the Ray Nesbitt Chair Endowment to A.J.
Footnotes
DISCLOSURE STATEMENT
The authors are not aware of any affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this review.
LITERATURE CITED
- 1.Mookherjee N, Anderson MA, Haagsman HP, Davidson DJ. 2020. Antimicrobial host defence peptides: functions and clinical potential. Nat. Rev. Drug Discov 19:311–32 [DOI] [PubMed] [Google Scholar]
- 2.Cotter PD, Ross RP, Hill C. 2013. Bacteriocins—a viable alternative to antibiotics? Nat. Rev. Microbiol 11:95–105 [DOI] [PubMed] [Google Scholar]
- 3.Selber-Hnatiw S, Rukundo B, Ahmadi M, Akoubi H, Al-Bizri H, et al. 2017. Human gut microbiota: toward an ecology of disease. Front. Microbiol 8:1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Levy M, Kolodziejczyk AA, Thaiss CA, Elinav E. 2017. Dysbiosis and the immune system. Nat. Rev. Immunol 17:219–32 [DOI] [PubMed] [Google Scholar]
- 5.Arrieta MC, Stiemsma LT, Amenyogbe N, Brown EM, Finlay B. 2014. The intestinal microbiome in early life: health and disease. Front. Immunol 5:427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5a.Batoni G, Maisetta G, Esin S. 2016. Antimicrobial peptides and their interaction with biofilms of medically relevant bacteria. Biochim. Biophys. Acta Biomembr 1858:1044–60 [DOI] [PubMed] [Google Scholar]
- 6.Ostaff MJ, Stange EF, Wehkamp J. 2013. Antimicrobial peptides and gut microbiota in homeostasis and pathology. EMBO Mol. Med 5:1465–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Garcia-Gutierrez E, Mayer MJ, Cotter PD, Narbad A. 2019. Gut microbiota as a source of novel antimicrobials. Gut Microbes 10:1–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de la Fuente-Nunez C, Torres MD, Mojica FJ, Lu TK. 2017. Next-generation precision antimicrobials: towards personalized treatment of infectious diseases. Curr. Opin. Microbiol 37:95–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dias C, Rauter AP. 2019. Membrane-targeting antibiotics: recent developments outside the peptide space. Future Med. Chem 11(3):211–28 [DOI] [PubMed] [Google Scholar]
- 10.Mahlapuu M, Hakansson J, Ringstad L, Bjorn C. 2016. Antimicrobial peptides: an emerging category of therapeutic agents. Front. Cell. Infect. Microbiol 6:194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Malanovic N, Lohner K. 2016. Antimicrobial peptides targeting Gram-positive bacteria. Pharmaceuticals 9(3):59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Epand RM, Epand RF. 2011. Bacterial membrane lipids in the action of antimicrobial agents. J. Pept. Sci 17:298–305 [DOI] [PubMed] [Google Scholar]
- 13.Hollmann A, Martinez M, Maturana P, Semorile LC, Maffia PC. 2018. Antimicrobial peptides: interaction with model and biological membranes and synergism with chemical antibiotics. Front. Chem 6:204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Munch D, Sahl HG. 2015. Structural variations of the cell wall precursor lipid II in Gram-positive bacteria—impact on binding and efficacy of antimicrobial peptides. Biochim. Biophys. Acta Biomembr 1848:3062–71 [DOI] [PubMed] [Google Scholar]
- 15.Scherer KM, Spille JH, Sahl HG, Grein F, Kubitscheck U. 2015. The lantibiotic nisin induces lipid II aggregation, causing membrane instability and vesicle budding. Biophys. J 108:1114–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mylonakis E, Podsiadlowski L, Muhammed M, Vilcinskas A. 2016. Diversity, evolution and medical applications of insect antimicrobial peptides. Philos. Trans. R. Soc. B 371:20150290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Park SI, Kim JW, Yoe SM. 2015. Purification and characterization of a novel antibacterial peptide from black soldier fly (Hermetia illucens) larvae. Dev. Comp. Immunol 52:98–106 [DOI] [PubMed] [Google Scholar]
- 18.Otvos L Jr. 2002. The short proline-rich antibacterial peptide family. Cell. Mol. Life. Sci 59:1138–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Le C-F, Fang C-M, Sekaran SD. 2017. Intracellular targeting mechanisms by antimicrobial peptides. Antimicrob. Agents Chemother 61(4):e02340–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hale JDF, Hancock REW. 2007. Alternative mechanisms of action of cationic antimicrobial peptides on bacteria. Expert Rev. Anti-Infect. Ther 5:951–59 [DOI] [PubMed] [Google Scholar]
- 21.Torres MDT, Sothiselvam S, Lu TK, de la Fuente-Nunez C. 2019. Peptide design principles for antimicrobial applications. J. Mol. Biol 431:3547–67 [DOI] [PubMed] [Google Scholar]
- 22.Guo L, McLean JS, Yang Y, Eckert R, Kaplan CW, et al. 2015. Precision-guided antimicrobial peptide as a targeted modulator of human microbial ecology. PNAS 112:7569–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kaplan CW, Sim JH, Shah KR, Kolesnikova-Kaplan A, Shi W, Eckert R. 2011. Selective membrane disruption: mode of action of C16G2, a specifically targeted antimicrobial peptide. Antimicrob. Agents Chemother 55:3446–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xu L, Shao C, Li G, Shan A, Chou S, et al. 2020. Conversion of broad-spectrum antimicrobial peptides into species-specific antimicrobials capable of precisely targeting pathogenic bacteria. Sci. Rep 10:944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Choudhury A, Islam SMA, Ghidey MR, Kearney CM. 2020. Repurposing a drug targeting peptide for targeting antimicrobial peptides against Staphylococcus. Biotechnol. Lett 42:287–94 [DOI] [PubMed] [Google Scholar]
- 26.Zhu X, Ma Z, Wang J, Chou S, Shan A. 2014. Importance of tryptophan in transforming an amphipathic peptide into a Pseudomonas aeruginosa-targeted antimicrobial peptide. PLOS ONE 9:e114605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Edwards IA, Elliott AG, Kavanagh AM, Zuegg J, Blaskovich MA, Cooper MA. 2016. Contribution of amphipathicity and hydrophobicity to the antimicrobial activity and cytotoxicity of β-hairpin peptides. ACS Infect. Dis 2:442–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Postma TM, Liskamp RMJ. 2016. Triple-targeting Gram-negative selective antimicrobial peptides capable of disrupting the cell membrane and lipid A biosynthesis. RSC Adv 6:65418–21 [Google Scholar]
- 29.Muhle SA, Tam JP. 2001. Design of Gram-negative selective antimicrobial peptides. Biochemistry 40:5777–85 [DOI] [PubMed] [Google Scholar]
- 30.Idso MN, Akhade AS, Arrieta-Ortiz ML, Lai BT, Srinivas V, et al. 2020. Antibody-recruiting protein-catalyzed capture agents to combat antibiotic-resistant bacteria. Chem. Sci 11:3054–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Veltri D, Kamath U, Shehu A. 2018. Deep learning improves antimicrobial peptide recognition. Bioinformatics 34:2740–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee EY, Fulan BM, Wong GC, Ferguson AL. 2016. Mapping membrane activity in undiscovered peptide sequence space using machine learning. PNAS 113:13588–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yoshida M, Hinkley T, Tsuda S, Abul-Haija YM, McBurney RT, et al. 2018. Using evolutionary algorithms and machine learning to explore sequence space for the discovery of antimicrobial peptides. Chemistry 4:533–43 [Google Scholar]
- 34.Porto WF, Irazazabal L, Alves ESF, Ribeiro SM, Matos CO, et al. 2018. In silico optimization of a guava antimicrobial peptide enables combinatorial exploration for peptide design. Nat. Commun 9:1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gabere MN, Noble WS. 2017. Empirical comparison of web-based antimicrobial peptide prediction tools. Bioinformatics 33:1921–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Spanig S, Heider D. 2019. Encodings and models for antimicrobial peptide classification for multi-resistant pathogens. BioData Min 12:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vishnepolsky B, Zaalishvili G, Karapetian M, Nasrashvili T, Kuljanishvili N, et al. 2019. De novo design and in vitro testing of antimicrobial peptides against gram-negative bacteria. Pharmaceuticals 12:82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nagarajan D, Nagarajan T, Roy N, Kulkarni O, Ravichandran S, et al. 2018. Computational antimicrobial peptide design and evaluation against multidrug-resistant clinical isolates of bacteria. J. Biol. Chem 293:3492–509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Veltri D, Kamath U, Shehu A. 2017. Improving recognition of antimicrobial peptides and target selectivity through machine learning and genetic programming. IEEE/ACM Trans. Comput. Biol. Bioinform 14:300–13 [DOI] [PubMed] [Google Scholar]
- 40.Majumder A, Biswal MR, Prakash MK. 2019. Computational screening of antimicrobial peptides for Acinetobacter baumannii. PLOS ONE 14:e0219693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Maccari G, Di Luca M, Nifosi R, Cardarelli F, Signore G, et al. 2013. Antimicrobial peptides design by evolutionary multiobjective optimization. PLOS Comput. Biol 9:e1003212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gull S, Minhas F. 2020. AMP0: species-specific prediction of anti-microbial peptides using zero and few shot learning. IEEE/ACM Trans. Comput. Biol. Bioinform In press. 10.1109/TCBB.2020.2999399 [DOI] [PubMed] [Google Scholar]
- 43.Rea MC, Sit CS, Clayton E, O’Connor PM, Whittal RM, et al. 2010. Thuricin CD, a posttranslationally modified bacteriocin with a narrow spectrum of activity against Clostridium difficile. PNAS 107:9352–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lopez-Perez PM, Grimsey E, Bourne L, Mikut R, Hilpert K. 2017. Screening and optimizing antimicrobial peptides by using spot-synthesis. Front. Chem 5:25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mijalis AJ, Thomas DA 3rd, Simon MD, Adamo A, Beaumont R, et al. 2017. A fully automated flow-based approach for accelerated peptide synthesis. Nat. Chem. Biol 13:464–66 [DOI] [PubMed] [Google Scholar]
- 46.Albin JS, Pentelute BL. 2020. Efficient flow synthesis of human antimicrobial peptides. Aust. J. Chem 73:380–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ritter SC, Yang ML, Kaznessis YN, Hackel BJ. 2018. Multispecies activity screening of microcin J25 mutants yields antimicrobials with increased specificity toward pathogenic Salmonella species relative to human commensal Escherichia coli. Biotechnol. Bioeng 115:2394–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tucker AT, Leonard SP, DuBois CD, Knauf GA, Cunningham AL, et al. 2018. Discovery of next-generation antimicrobials through bacterial self-screening of surface-displayed peptide libraries. Cell 172:618–28.e13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.McCarthy KA, Kelly MA, Li K, Cambray S, Hosseini AS, et al. 2018. Phage display of dynamic covalent binding motifs enables facile development of targeted antibiotics. J. Am. Chem. Soc 140:6137–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Scanlon TC, Dostal SM, Griswold KE. 2014. A high-throughput screen for antibiotic drug discovery. Biotechnol. Bioeng 111:232–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rea MC, Clayton E, O’Connor PM, Shanahan F, Kiely B, et al. 2007. Antimicrobial activity of lacticin 3147 against clinical Clostridium difficile strains. J. Med. Microbiol 56:940–46 [DOI] [PubMed] [Google Scholar]
- 52.Balouiri M, Sadiki M, Ibnsouda SK. 2016. Methods for in vitro evaluating antimicrobial activity: a review. J. Pharm. Anal 6:71–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wiegand I, Hilpert K, Hancock RE. 2008. Agar and broth dilution methods to determine the minimal inhibitory concentration (MIC) of antimicrobial substances. Nat. Protoc 3:163–75 [DOI] [PubMed] [Google Scholar]
- 53a.Mercer DK, Torres MDT, Duay SS, Lovie E, Simpson L, et al. 2020. Antimicrobial susceptibility testing of antimicrobial peptides to better predict efficacy. Front. Cell. Infect. Microbiol 10:326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Auchtung JM, Robinson CD, Britton RA. 2015. Cultivation of stable, reproducible microbial communities from different fecal donors using minibioreactor arrays (MBRAs). Microbiome 3:42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Watterson WJ, Tanyeri M, Watson AR, Cham CM, Shan Y, et al. 2020. Droplet-based high-throughput cultivation for accurate screening of antibiotic resistant gut microbes. eLife 9: e56998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Vasilchenko AS, Rogozhin EA. 2019. Sub-inhibitory effects of antimicrobial peptides. Front. Microbiol 10:1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Adamowicz EM, Flynn J, Hunter RC, Harcombe WR. 2018. Cross-feeding modulates antibiotic tolerance in bacterial communities. ISME J 12:2723–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kim H, Jang JH, Kim SC, Cho JH. 2020. Development of a novel hybrid antimicrobial peptide for targeted killing of Pseudomonas aeruginosa. Eur. J. Med. Chem 185:111814. [DOI] [PubMed] [Google Scholar]
- 59.Sun Z, Zhong J, Liang X, Liu J, Chen X, Huan L. 2009. Novel mechanism for nisin resistance via proteolytic degradation of nisin by the nisin resistance protein NSR. Antimicrob. Agents Chemother 53:1964–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59a.Browne K, Chakraborty S, Chen R, Willcox MD, Black DS, et al. 2020. A new era of antibiotics: the clinical potential of antimicrobial peptides. Int. J. Mol. Sci 21:7047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59b.Ebbensgaard A, Mordhorst H, Overgaard MT, Nielsen CG, Aarestrup FM, Hansen EB. 2015. Comparative evaluation of the antimicrobial activity of different antimicrobial peptides against a range of pathogenic bacteria. PLOS One 10:e0144611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59c.Starr CG, Wimley WC. 2017. Antimicrobial peptides are degraded by the cytosolic proteases of human erythrocytes. Biochim. Biophys. Acta Biomembr 1859:2319–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59d.Starr CG, Ghimire J, Guha S, Hoffmann JP, Wang Y, et al. 2020. Synthetic molecular evolution of host cell-compatible, antimicrobial peptides effective against drug-resistant, biofilm-forming bacteria. PNAS 117:8437–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Koehbach J, Craik DJ. 2019. The vast structural diversity of antimicrobial peptides. Trends Pharmacol. Sci 40:517–28 [DOI] [PubMed] [Google Scholar]
- 61.Wang G 2012. Post-translational modifications of natural antimicrobial peptides and strategies for peptide engineering. Curr. Biotechnol 1:72–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chin JW. 2017. Expanding and reprogramming the genetic code. Nature 550:53–60 [DOI] [PubMed] [Google Scholar]
- 63.Baumann T, Nickling JH, Bartholomae M, Buivydas A, Kuipers OP, Budisa N. 2017. Prospects of in vivo incorporation of non-canonical amino acids for the chemical diversification of antimicrobial peptides. Front. Microbiol 8:124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Li FF, Brimble MA. 2019. Using chemical synthesis to optimise antimicrobial peptides in the fight against antimicrobial resistance. Pure Appl. Chem 91:181–98 [Google Scholar]
- 65.Hicks RP, Bhonsle JB, Venugopal D, Koser BW, Magill AJ. 2007. De novo design of selective antibiotic peptides by incorporation of unnatural amino acids. J. Med. Chem 50:3026–36 [DOI] [PubMed] [Google Scholar]
- 66.Piscotta FJ, Tharp JM, Liu WR, Link AJ. 2015. Expanding the chemical diversity of lasso peptide MccJ25 with genetically encoded noncanonical amino acids. Chem. Commun 51:409–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bartholomae M, Baumann T, Nickling JH, Peterhoff D, Wagner R, et al. 2018. Expanding the genetic code of Lactococcus lactis and Escherichia coli to incorporate non-canonical amino acids for production of modified lantibiotics. Front. Microbiol 9:657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Rezhdo A, Islam M, Huang M, Van Deventer JA. 2019. Future prospects for noncanonical amino acids in biological therapeutics. Curr. Opin. Biotechnol 60:168–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Reinhardt A, Neundorf I. 2016. Design and application of antimicrobial peptide conjugates. Int. J. Mol. Sci 17(5):701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Touti F, Lautrette G, Johnson KD, Delaney JC, Wollacott A, et al. 2018. Antibody-bactericidal macrocyclic peptide conjugates to target Gram-negative bacteria. ChemBioChem 19:2039–44 [DOI] [PubMed] [Google Scholar]
- 71.Mondhe M, Chessher A, Goh S, Good L, Stach JE. 2014. Species-selective killing of bacteria by antimicrobial peptide-PNAs. PLOS ONE 9:e89082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Blaskovich MAT, Hansford KA, Gong Y, Butler MS, Muldoon C, et al. 2018. Protein-inspired antibiotics active against vancomycin- and daptomycin-resistant bacteria. Nat. Commun 9:22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Mishra NM, Briers Y, Lamberigts C, Steenackers H, Robijns S, et al. 2015. Evaluation of the antibacterial and antibiofilm activities of novel CRAMP-vancomycin conjugates with diverse linkers. Org. Biomol. Chem 13:7477–86 [DOI] [PubMed] [Google Scholar]
- 74.Defraine V, Schuermans J, Grymonprez B, Govers SK, Aertsen A, et al. 2016. Efficacy of Artilysin Art-175 against resistant and persistent Acinetobacter baumannii. Antimicrob. Agents Chemother 60:3480–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Tan P, Lai ZH, Zhu YJ, Shao CX, Akhtar MU, et al. 2020. Multiple strategy optimization of specifically targeted antimicrobial peptide based on structure–activity relationships to enhance bactericidal efficiency. ACS Biomater. Sci. Eng 6:398–414 [DOI] [PubMed] [Google Scholar]


