Abstract
The relationships between adaptive evolution, phenotypic plasticity, and canalization remain incompletely understood. Theoretical and empirical studies have made conflicting arguments on whether adaptive evolution may enhance or oppose the plastic response. Gene regulatory traits offer excellent potential to study the relationship between plasticity and adaptation, and they can now be studied at the transcriptomic level. Here, we take advantage of three closely related pairs of natural populations of Drosophila melanogaster from contrasting thermal environments that reflect three separate instances of cold tolerance evolution. We measure the transcriptome-wide plasticity in gene expression levels and alternative splicing (intron usage) between warm and cold laboratory environments. We find that suspected adaptive changes in both gene expression and alternative splicing tend to neutralize the ancestral plastic response. Further, we investigate the hypothesis that adaptive evolution can lead to decanalization of selected gene regulatory traits. We find strong evidence that suspected adaptive gene expression (but not splicing) changes in cold-adapted populations are more vulnerable to the genetic perturbation of inbreeding than putatively neutral changes. We find some evidence that these patterns may reflect a loss of genetic canalization accompanying adaptation, although other processes including hitchhiking recessive deleterious variants may contribute as well. Our findings augment our understanding of genetic and environmental effects on gene regulation in the context of adaptive evolution.
Keywords: adaptive evolution, transcriptomic plasticity, genetic canalization, Drosophila melanogaster
Significance.
It is unclear whether adaptive evolution is concordant or discordant with regulatory plasticity, especially for splicing plasticity that is rarely studied. Here, we analyzed RNA-seq data from three pairs of natural fly populations that represent separate adaptive evolution to cold climate. We found that adaptive evolution is generally discordant with the ancestral plasticity between cold and warm temperatures for gene expression abundance and splicing. We also investigate the hypothesis that adaptation leads to decanalization of the selected traits. By comparing the expression variance between inbred and outbred samples, we found evidence that adaptation may lead to genetic decanalization for expression abundance but not for splicing. Our study reveals the relationship between adaptation, plasticity, and canalization in three instances in nature.
Introduction
For organisms to cope with environmental changes, two important strategies are adaptive evolution and phenotypic plasticity (Meyers and Bull 2002). Phenotypic plasticity is the phenomenon of a single genotype producing different phenotypes under different environmental conditions. Producing different phenotypes from a genotype often requires an intermediate step, such as gene expression change (Ghalambor et al. 2007; Pfennig et al. 2010). It has also been shown in many cases that expression evolution contributes to adaptive evolution (Fraser et al. 2010; Nourmohammad et al. 2017). Studying the interactions between expression plasticity and adaptive expression evolution can generate insights into how these two processes help organisms respond to environmental changes.
Adaptation to a new environment may change gene expression in the same direction as the initial plasticity, which suggests that the initial plasticity is beneficial, shifting the phenotype toward the optimal phenotypic values in the selective environment (Via 1993; Chevin et al. 2010). Alternatively, a plastic response can be deleterious if it shifts the phenotype away from the optimum under environmental perturbation, and adaptive evolution should restore the phenotype toward the ancestral state (Scheiner 1993; von Heckel et al. 2016). The latter scenario predicts a negative relationship between the direction of plastic change and that of evolutionary change, resulting in a “counter-gradient” pattern (Conover and Schultz 1995). Finally, environmentally induced expression change may have little effect on fitness. In that case, evolutionary change is determined by drift, showing no relation with the plasticity in terms of direction. Many recent laboratory and field studies have observed a “counter-gradient” pattern for plasticity and adaptive evolution for gene expression (Levine et al. 2011; Dayan et al. 2015; Ghalambor et al. 2015; Ragland et al. 2015; Huang and Agrawal 2016; Ho and Zhang 2018; Koch and Guillaume 2020; Fischer et al. 2021), while some other studies found that adaptive plasticity is more common (Kenkel and Matz 2017; Mallard et al. 2020; Bittner et al. 2021; Josephs et al. 2021). However, few of them have studied multiple natural population pairs that reflect separate adaptation events. Moreover, the relationship between plasticity and adaptive evolution for other regulatory traits such as splicing is unexplored.
On the contrary, adaptation to a new environment might have a significant consequence on the level of canalization of the selected trait. Canalization refers to the capacity of a trait to maintain a constant phenotype under genetic or environmental perturbation (Waddington 1942; Flatt 2005). This concept has been defined operationally in different ways (Dworkin 2005). Environmental canalization has been described as the opposite of phenotype plasticity (Nijhout and Davidowitz 2003), as indicated by the constancy of a trait across environments. Hence, plasticity can be viewed as a type of environmental decanalization. However, environmental canalization can alternatively be defined based on the trait variation observed among individuals in one environment compared with others (Stearns and Kawecki 1994). Similarly, genetic canalization can be described as the constancy of the phenotype under heritable perturbations, such as mutations (de Visser et al. 2003). It can be measured as the inverse of the variance caused by mutations (Flatt 2005).
Canalization can potentially constrain evolution by reducing phenotypic variation (Charlesworth et al. 1982; Maynard Smith et al. 2020), or it may facilitate evolution by allowing cryptic genetic variation to accumulate (Gibson and Dworkin 2004; Masel 2005; Paaby and Rockman 2014). If a trait is under stabilizing selection in the ancestral environment, its developmental canalization may be favored by selection (Gavrilets and Hastings 1994; Wagner et al. 1997). However, when adaptive evolution shifts a trait to a new optimum, the previous canalization mechanism may be undermined. This possibility, that adaptive evolution could result in decanalization, was hinted at when blowflies with newly evolved insecticide resistance were found to have bristle asymmetry and prolonged development (Clarke and McKenzie 1987; McKenzie and Game 1987) and also when in vitro selection on an enzyme resulted in reduced robustness to both genetic and environmental perturbations (Hayden et al. 2012). A natural example of decanalization of the same trait that evolved adaptively was then provided by Lack et al. (2016b), who found that highland Ethiopian Drosophila melanogaster with derived large wing size had greatly reduced genetic robustness of wing development. One subsequent study found that environmental decanalization may not have occurred for these Ethiopian fly wings (Pesevski and Dworkin 2020), highlighting the potential distinctness of genetic and environmental canalization. A further study showed that when the wing size in the same species was artificially selected, both genetic and environmental decanalization of wing development ensued (Groth et al. 2018). However, long-term lab selection of D. melanogaster for rapid development was suggested to increase canalization of that trait (Ghosh et al. 2019). Whether a decanalizing effect of directional selection exists for a substantial proportion of adaptive gene regulatory traits has not previously been investigated, and this question represents one focus of the present study.
In this study, we take advantage of African and European populations of D. melanogaster that have experienced parallel adaptation to colder climates to study the interaction between adaptation and plasticity as well as canalization in gene expression. Originating from a warm sub-Saharan range (Lachaise et al. 1988; Pool et al. 2012; Sprengelmeyer et al. 2020), populations have independently occupied colder environments at least three times: in higher latitudes of Eurasia (here represented by the France FR population) and in the highlands of Ethiopia (EF population) and South Africa (SD population). Flies from these three locations were paired with genetically similar populations from warm regions: Egypt (EG), Ethiopia lowland (EA), and South Africa lowland (SP), respectively, together representing three different population pairs: Mediterranean (MED), Ethiopian (ETH), and South African (SAF). These cold-adapted populations have evolved separately from the warm-adapted ones for ∼1,000–2,000 years (∼15,000–30,000 generations) (Sprengelmeyer et al. 2020). They show parallel changes in cold tolerance, as measured by recovery after prolonged 4 °C cold exposure (Pool et al. 2017) and greater egg-to-adult survival at 15 °C (Huang et al. 2021). These cold-adapted populations also show a genome-wide excess of parallel allele frequency shifts (Pool et al. 2017).
In our related study (Huang et al. 2021), we identified evolutionary changes in gene expression abundance and alternative splicing (intron usage) between cold- and warm-adapted populations for each pair at a low temperature approximating the derived cold environment. Here, we rear the same crosses from these populations at a warm temperature, approximating the ancestral environment, and analyze transcriptomes of adult females. First, we compare the directions of gene regulatory plasticity between cold and warm environments and the directions of evolutionary changes between cold and warm populations for both expression abundance and intron usage. We find adaptive evolution tends to counter plasticity by restoring the ancestral regulation (neutralizing the ancestral plasticity). Second, we test the hypothesis that adaptively evolved traits representing novel states are more likely to undermine canalization mechanisms that had evolved to buffer the ancestral state. Since we previously analyzed gene regulation from some parental inbred lines of the outbred crosses we otherwise analyze (Huang et al. 2021), we use inbreeding as a form of genetic perturbation (Réaale and Roff 2003; de Visser et al. 2003) to study regulatory canalization in the cold and warm populations and its potential disruption due to adaptive evolution. We use the ratio of regulatory trait variance among inbred samples to that among outbred samples as a potential signal of genetic decanalization. We indeed find transcriptome-wide evidence consistent with adaptive evolution leading to decanalization for expression abundance traits, although other processes such as hitchhiking deleterious variants may contribute to this signal as well.
Results
Adaptive Evolution and Naive Plasticity in Expression and Splicing
We collected RNA-seq data from whole female adult samples representing the three warm/cold population pairs described above, raised at temperatures intended to reflect either an ancestral warmer environment (25 °C) or a derived cooler environment (15 °C). For each of these six populations, the transcriptomes from eight unique outbred crosses were analyzed (fig. 1A). We first performed principal component analysis (PCA) on the normalized expression read counts across samples from different environments and populations. There are signals of temperature environment on transcriptome variation, separating the samples from the cold environment from the respective samples from the warm environment along the PC1 (supplementary fig. S1, Supplementary Material online). Then we characterized the genes/introns that showed consistent plastic expression/splicing changes between 15 and 25 °C rearing environments in warm-adapted populations (P). Formally, we required the direction of plasticity for at least seven out of the eight crosses to be consistent with the average plastic change (see Materials and Methods). We refer to this pattern from warm-adapted populations as ancestral or naive plasticity, hypothetically representing the initial influence of derived cooler environments on expression/splicing prior to any cold adaptation. Across the six populations (three warm/cold-adapted population pairs), 14–65% of all gene expression traits met the consistent plasticity criterion (at least seven out of eight crosses showing the same direction of plastic change), as did 24–36% of intron usage traits (table 1A). The Mediterranean pair had the greatest proportions of consistently plastic traits for both expression and splicing. Across all populations and traits, an average 29.9% of genes/introns show consistent plasticity by our criteria. In light of our expected false-positive rate (7%), we estimate that approximately 77% of the identified genes are true positives for plasticity, which provides a substantially enriched set of plasticity candidates for the transcriptome-wide analyses described below.
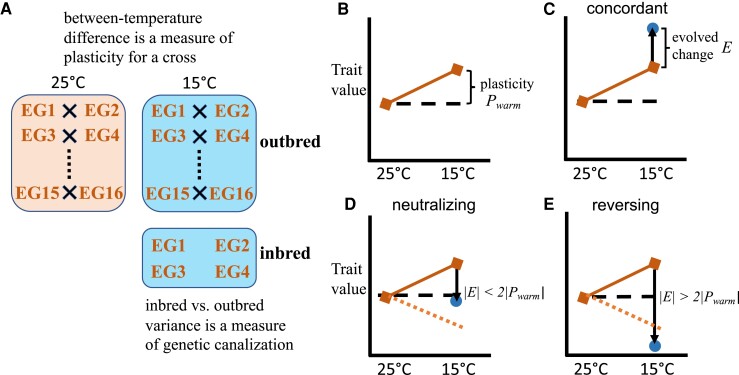
Fig. 1.
Illustrations of the experimental design and different expectations for adaptative changes regarding the naive plasticity. (A) Experimental design is shown for one population (EG, a warm population) as an example. Crosses generated outbred offspring at 15 °C (derived condition) and 25 °C (ancestral condition) to study transcriptome plasticity. Inbred strains were reared at 15 °C to study genetic canalization by comparing the variance of trait value for inbred versus outbred samples at 15 °C. Whole female adults were collected from the offspring. (B–E) Conceptual illustrations of different expectations for evolved changes and naive plasticity. (B) Naive plasticity for a warm population. The difference between trait values at 15 °C versus 25 °C is a measure of plasticity (Pwarm). (C) The evolved change (E, solid arrow) is the difference between trait values between cold (dot) and warm population (square) at 15 °C. If the evolved change is in the same direction as the naive plasticity, the evolved change is regarded as “concordant.” (D) If the evolved change is in the opposite direction as the naive plasticity (E × Pwarm < 0) and moves the trait closer to the warm population’s trait value at 25 °C (|E| < 2|Pwarm|), it is regarded as “neutralizing.” The orange dashed line indicates the level of plastic change but in the opposite direction. (E) If the evolved change moves the trait further from the warm population’s trait value at 25 °C with a magnitude greater than the initial plasticity in the opposite direction (E × Pwarm < 0; |E| < 2|Pwarm|), it is regarded as “reversing.” The “concordant” and “reversing” categories are both regarded as “novelty” for the canalization analysis because they each take the cold-adapted population’s trait value in its home environment farther away from the warm-adapted population’s trait value in its own home environment than naive plasticity would have done on its own.
Table 1.
Numbers of Gene Expression Abundance and Intron Usage Traits Showing Plasticity in Warm- and Cold-Adapted Populations for Each Population Pair
| A | ||||||
|---|---|---|---|---|---|---|
| All | MED | ETH | SAF | |||
| Population | Warm | Cold | Warm | Cold | Warm | Cold |
| Expression abundance | 4,890 (43%) | 7,453 (65%) | 3,286 (28%) | 1,627 (14%) | 2,195 (19%) | 2,303 (20%) |
| Intron usage | 3,219 (32%) | 3,658 (36%) | 2,884 (29%) | 2,358 (24%) | 2,504 (25%) | 2,399 (24%) |
| B | ||||||
|---|---|---|---|---|---|---|
| PST outliers | MED | ETH | SAF | |||
| Population | Warm | Cold | Warm | Cold | Warm | Cold |
| Expression abundance | 138 (41%) | 205 (60%) | 126 (37%) | 93 (27%) | 117 (35%) | 118 (35%) |
| Intron usage | 113 (32%) | 116 (33%) | 77 (22%) | 114 (32%) | 94 (27%) | 78 (22%) |
Panel A shows all the genes/introns, and panel B shows those with elevated expression/splicing differentiation between adult females from cold- and warm-adapted populations at 15 °C (PST outliers identified in Huang et al. 2021). The percentage in parentheses indicates the proportion showing consistent plasticity.
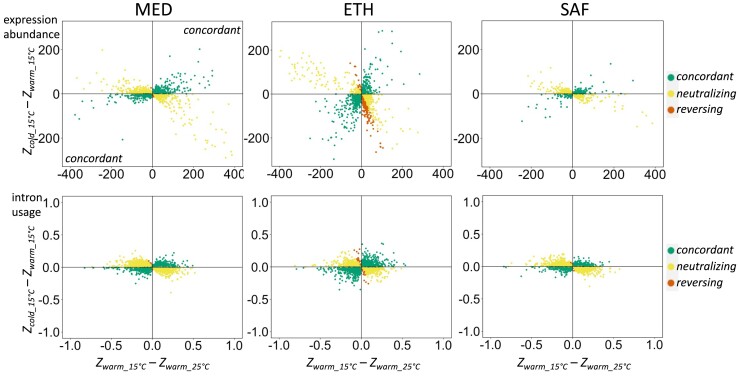
We then set out to assess the general relationship between a warm-adapted population’s naive plasticity and the evolved difference between the populations in the derived environment (fig. 1B–E). Here, we are asking whether evolutionary changes are concordant or discordant with naive plasticity, as opposed to asking how plasticity itself has evolved. We compared the directions of initial plasticity (P) in warm-adapted populations and evolved change (E), which is the difference in expression between cold- and warm-adapted populations at 15 °C. If P and E were in the same direction (i.e., P × E > 0), the evolved change was defined as “concordant” (fig. 1C). Considering all plastic genes for gene expression abundance, the proportion of “concordant” changes was 53% in MED, 55% in ETH, and 39% in SAF. For intron usage, the proportion of “concordant” changes was 41% in MED, 53% in ETH, and 38% in SAF (fig. 2).
Fig. 2.
The plastic responses of the warm-adapted population (Pwarm, x-axis) and the evolved differences in expression between populations in the derived cold environment (E, y-axis) for all genes (upper panel) and introns (lower panel) that showed consistent naive plasticity. Dots located in the lower left and upper right quadrants indicate that the plasticity responses and evolutionary changes are concordant.
We further partitioned those traits where P and E were in opposite directions (“discordant,” P×E < 0). If the evolved change moved expression/splicing farther from ancestral levels than the ancestral plasticity did (the magnitude of evolved change was more than twice that of plasticity, |E| > |2P|), we defined it as “reversing” (fig. 1E). If, instead, the evolved change brought the cold population at 15 °C back closer to the warm population at 25 °C, then we defined the evolved change as “neutralizing” (fig. 1D). These categories may have distinct evolutionary interpretations, since neutralizing changes are consistent with a single trait optimum across thermal environments, whereas for reversing changes, the observation that evolution increased trait differentiation between the populations in their home environments could indicate distinct adaptive optima in cold versus warm environments. We found that among “discordant” changes, “reversing” was a relatively small fraction across population pairs (fig. 2; on average 11% for expression abundance and 1.7% for intron usage). It is worth noting that the Ethiopia pair appears to have a much higher proportion of “reversing” changes (32% for expression and 3.6% for intron usage) than the other two pairs, suggesting distinct evolution in the Ethiopia pair.
To examine whether putative adaptive evolution in expression/splicing in cold populations followed the initial plastic changes, we focused on genes showing elevated population differentiation in gene expression abundance or intron usage between warm- and cold-adapted populations based on Huang et al. (2021) (about 339 outlier genes and 351 outlier intron junctions for each population pair). These candidates for adaptive gene regulatory evolution were identified using a PST outlier approach, focusing on the top 5% of genes/introns for each population pair separately (Huang et al. 2021; see Materials and Methods). Among these outliers (an average of 339 genes per population pair for expression and 351 introns for splicing), the numbers of expression abundance genes also passing the cutoff for plasticity were in the range of 93–205 across population pairs and the numbers of divergent intron usage traits passing the plasticity cutoff were in the range of 77–116 (table 1B).
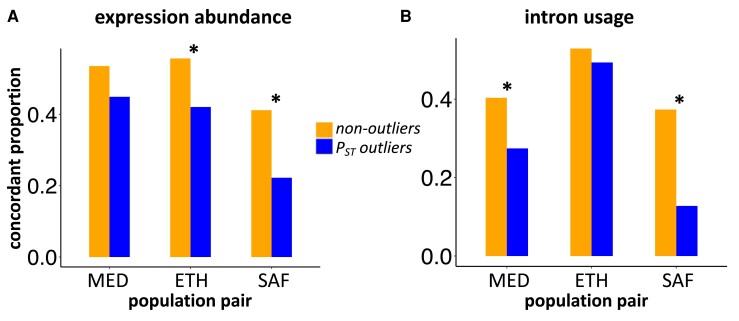
For both expression abundance and intron usage, we observed a general pattern that plastic PST outliers had lower proportions of “concordant” naive expression plasticity than plastic nonoutliers across all population pairs (fig. 3). In other words, putatively adaptively evolved divergence tended to oppose naive plasticity. For expression abundance, we found the difference in the “concordant” proportion was significant for the ETH [χ2 = 8.3, degree of freedom (df) = 1, P = 0.0039] and SAF (χ2 = 15.1, df = 1, P = 9.9e−05) pairs but only marginally significant for the MED pair (χ2 = 3.6, df = 1, P = 0.058). For intron usage, the difference in the “concordant” proportion was significant for the MED (χ2 = 6.7, df = 1, P = 0.0096) and the SAF (χ2 = 21.7, df = 1, P = 3.2e−06) pairs but not the ETH pair (χ2 = 0.24, df = 1, P = 0.62). Mirroring transcriptome-wide patterns, a large majority of PST outliers with “discordant” changes were “neutralizing” rather than “reversing”; the average proportion of “reversing” is 4% for gene expression and 9% for intron usage. Hence, the largest share of putatively adaptive regulatory changes served to mitigate ancestral plasticity.
Fig. 3.
The proportion of genes (A) and introns (B) classified as “concordant” of naive plasticity for PST outliers and nonoutliers. *Proportion is significantly different between PST outliers and nonoutliers (P < 0.01).
Because the same data from the warm-adapted population in the cold environment was used to estimate both plastic and evolutionary changes, it is possible that random deviations in this estimate could yield a bias (Mallard et al. 2018; Ho and Zhang 2019). For example, if this quantity was underestimated, then plasticity would become more negative, but evolution would become more positive. This potential bias toward discordant changes should be more pronounced for nonoutliers in light of their more uncertain direction of evolution (whereas outliers tend to have greater magnitudes of evolution, for which the direction is less likely to be altered by measurement error). This suggestion assumes that outliers reflect true evolution (adaptive or otherwise) and were not generated via measurement error; we note that each population’s extreme high and low values were excluded from PST calculations as a strategy to reduce the influence of measurement error (Huang et al. 2021). In contrast to the above prediction based on bias, we observed above that PST outliers had more discordant changes than nonoutliers (fig. 3).
The potential bias toward discordant changes should only exist if overlapping data are used to estimate plasticity and evolution. Therefore, we repeated the above analysis using nonoverlapping subsets of the data to estimate plasticity and evolution. We randomly split the shared data (the warm-adapted population in 15 °C, Zwarm_15°C), with four samples being used to estimate plasticity (Pwarm = Zwarm_15°C′ − Zwarm_25°C) and the other four being used to estimate evolution (E = Zcold_15°C − Zwarm_15°C′) and identify PST outliers, for all possible subdivisions of the data. The results were qualitatively similar to those reported above (supplementary fig. S3, Supplementary Material online vs. fig. 3). All three population pairs again showed an excess of discordant changes among PST outliers for expression. For splicing, the MED and SAF pairs showed a significant excess of discordant changes among outliers (supplementary fig. S3, Supplementary Material online).
Gene Ontology Functional Enrichment for Plastic Genes/Splicing
Functional analysis [Gene Ontology (GO) enrichment testing] of all plastic genes (regardless of PST outlier status) with expression/splicing showing either concordant, neutralizing or reversing evolved changes is shown in supplementary table S4, Supplementary Material online. For gene expression, we found more significant GO categories for genes showing “neutralizing” changes among three population pairs, compared with “concordant” or “reversing.” While for splicing, most of the significant GO categories were found for those showing “concordant” in the MED pair (supplementary table S4, Supplementary Material online). Furthermore, we considered the candidates for adaptive differentiation between warm- and cold-adapted populations (Huang et al. 2021) and performed similar GO enrichment tests. The power of these analyses may be reduced because only 71–203 adaptation candidates with consistent plasticity were assigned among our three categories (see the above). We ran separate GO enrichment analyses based on whether the potential adaptive evolution is “concordant” or else “neutralizing” the initial plasticity (there were fewer than 10 genes/introns showing “reversing” changes, and therefore, this category was not considered). We only found significant GO terms for the MED pair. For “concordant,” there is one significant term, “lipid particle”; for “neutralizing,” the significant terms are “coenzyme binding,” “cofactor binding,” “ion transmembrane transporter activity,” “magnesium ion binding,” “NAD binding,” and “transferase activity, transferring acyl groups.” Interestingly, “lipid particle” is also found in the GO terms for PST outliers at the adult stage in the MED pair (Huang et al. 2021).
Gene Regulatory Evolution and Genetic Canalization
We then explored how selection history may influence gene regulatory canalization. Our primary interest was to test whether transcriptomic evolution supported the hypothesis that adaptive evolution may be accompanied by a loss of canalization, which was suggested in the case of D. melanogaster wing size evolution in nature and in the laboratory (Lack et al. 2016b; Groth et al. 2018). Here, we leveraged previously collected RNA-seq data (Huang et al. 2021) from a subset of the parental inbred strains of the outbred crosses that all other data and analyses were derived from. Inbreeding reflects a broad genetic perturbation due to the homozygosity of recessive deleterious variants (which tend to be abundant in natural populations due to the limited capacity of natural selection to completely purge them). Comparisons between inbred and outbred data can therefore illuminate the relative strength of genetic canalization between different groups (Réaale and Roff 2003; de Visser et al. 2003). For example, we would predict that for a gene expression abundance trait, a population with relatively lesser canalization for this gene would have greater deviations in expression level in the inbred samples compared with the outbred samples, whereas a population with greater canalization for this gene would have relatively less difference in the variability of inbred and outbred samples (less effect of the perturbation). We therefore calculated, for each relevant regulatory trait in each population, the variance of this trait among inbred (Vinbred) and outbred (Voutbred) samples. Inbred data were available, and hence, these comparisons were made for the colder 15 °C environments only.
Although we had a particular interest in decanalization involving potential adaptive changes showing regulatory novelty, we began by comparing Vinbred and Voutbred across all expression and splicing traits, many of which may be evolving neutrally. For each population, the Vinbred and Voutbred are highly correlated (supplementary fig. S4, Supplementary Material online). Compared between populations, warm-adapted populations had much lower average Voutbred than cold-adapted populations (table 2), in spite of having somewhat higher genetic diversity than their cold-adapted counterparts (Lack et al. 2016a). The reasons for this pattern are unclear; it is possible that the stressed transcriptomic profiles of warm-adapted flies made them relatively more similar. In contrast, warm-adapted populations had higher Vinbred than cold-adapted populations, except for the SAF pair (table 2). Concordantly, we found that warm-adapted populations consistently had more genes and introns showing Vinbred > Voutbred than cold-adapted populations (fig. 4A and B). The much greater increase in Vinbred relative to Voutbred in warm-adapted populations may reflect the relatively greater stress experienced by these flies at 15 °C, compared with those from populations better adapted to such conditions (in other words, the effects of genetic and environmental perturbation may be synergistic). Alternatively, the warm-adapted populations may be expected to hold a somewhat greater number of recessive deleterious variants to be exposed by inbreeding (see Discussion).
Table 2.
Transcriptome-Wide Mean Variance of Expression or Intron Usage among Inbred Strains or Outbred Crosses within a Population, Averaged across all Genes/Intron Usages
| Pair | MED | ETH | SAF | ||||
|---|---|---|---|---|---|---|---|
| Population | Warm | Cold | Warm | Cold | Warm | Cold | |
| Expression abundance | Vinbred | 23,438 | 15,965 | 61,003 | 37,384 | 12,847 | 34,075 |
| Voutbred | 5,132 | 10,412 | 5,083 | 25,574 | 3,125 | 61,581 | |
| Intron usage | Vinbred | 0.0071 | 0.0067 | 0.0113 | 0.0095 | 0.0064 | 0.0073 |
| Voutbred | 0.0039 | 0.0059 | 0.0033 | 0.0049 | 0.0042 | 0.0089 | |
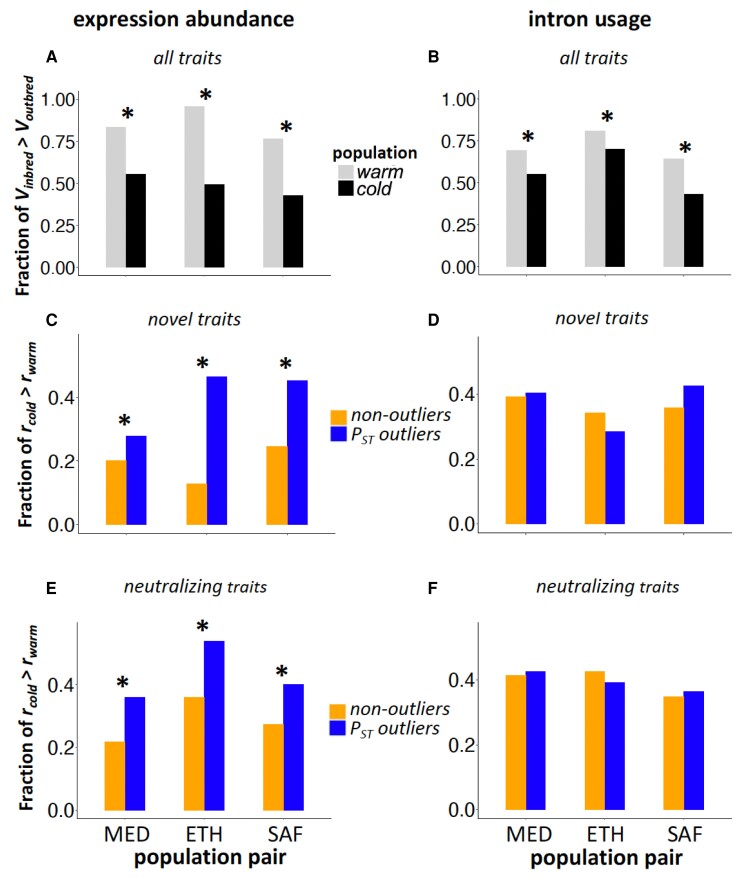
Fig. 4.
Genetic decanalization measured by Vinbred relative to Voutbred (r). (A and B) Transcriptome-wide fraction of regulatory traits for which r > 1 for each warm-adapted and cold-adapted population is shown for expression abundance (A) and intron usage (B). (C–F) Fraction of traits with novel regulatory changes (C and D) or with neutralizing changes (E and F) for which rcold > rwarm for PST outliers and nonoutlier controls is shown for expression abundance (left) and intron usage (right). *Fractions are significantly different (all with P < 0.05).
Regardless of the forces shaping transcriptome-wide patterns of inbred and outbred variance, we were interested in asking whether genes potentially under recent adaptive regulatory evolution (PST outliers) show distinct patterns of inbred versus outbred trait variance consistent with decanalization, compared with other genes. We suggest that larger inbred-to-outbred variance ratios may indicate the trait is more sensitive to the genetic perturbation of inbreeding. Thus, if adaptive evolution has resulted in decanalization, we predict that PST outliers will have elevated ratios of Vinbred to Voutbred in cold-adapted populations compared with their warm-adapted counterparts. However, one alternative explanation for such a result is the hitchhiking of deleterious recessive regulatory variants since any given selected haplotype may have linked harmful variants that increase in frequency alongside the favored variant (Chun and Fay 2011; Good and Desai 2014). Fortunately, these competing hypotheses make contrasting predictions with regard to adaptive changes that produce “novel” states distinct from those observed in ancestral range populations versus those “neutralizing” naive plasticity to maintain ancestral regulatory states. The decanalization hypothesis hinges on adaptation producing new phenotypes that are not fully buffered by ancestral canalization mechanisms. Therefore, this hypothesis would predict that novel adaptive regulatory changes should have greater potential to undermine ancestral buffering mechanisms and be more sensitive to inbreeding disruption than neutralizing changes, and therefore, novel outliers should have greater ratios of inbred versus outbred variance. However, the deleterious hypothesis depends only on the hitchhiking process itself and not the direction of the adaptive regulatory change, and therefore, it predicts no difference between novel and neutralizing changes.
In testing the above predictions, we first classified all expression and splicing traits based on whether their evolution was novel or “neutralizing” in the cold environment. Similar to the plasticity section, we used the two values Pwarm and E to characterize each regulatory trait (supplementary fig. S2, Supplementary Material online). To classify novel changes, we required that the evolved change took the expression value out of the ancestral plasticity range, which included both cases where P × E > 0 (“concordant”) and those where P × E < 0 and |E| > |2P| (“reversing”). The remaining traits showing Pwarm × E < 0 and |E| < |2P| were again classified as neutralizing. Averaged across three population pairs, 37% of all expression abundance and 45% of all intron usage traits were deemed novel.
We then compared novel/neutralizing results from all genes to those from PST outliers. For each of our PST outlier genes showing elevated differentiation between a warm and cold population (Huang et al. 2021) that also showed evidence for regulatory novelty, we therefore tested whether the Vinbred/Voutbred ratio (r) was higher in the cold population (rcold) or the warm population (rwarm). If the directional selection has had important decanalizing effects on the evolved regulatory traits, we would predict that novel PST outliers would be more likely to have a larger rcold than rwarm, compared with nonoutlier genes classified as a novel. For gene expression abundance, we found that the fraction of genes with rcold > rwarm was significantly higher for outliers than the nonoutliers for all three population pairs (fig. 4C; MED pair: 29% vs. 20%, χ2 = 6.7, df = 1, P = 0.0098; ETH pair: 47% vs. 13%, χ2 = 182, df = 1, P < 2.2e−16; SAF pair: 45% vs. 24%, χ2 = 33, df = 1, P = 8.0e−9.). The result is consistent with the hypothesis that directional selection on expression abundance led to decanalization of these same traits.
Alternatively, the above difference between novel outliers and novel nonoutliers is also compatible with the hitchhiking of deleterious variants. Furthermore, a qualitatively similar excess of rcold > rwarm genes was observed for neutralizing expression changes as well (fig. 4E; MED pair: 36% vs. 22%, χ2 = 12, df = 1, P = 0.0005; ETH pair: 54% vs. 36%, χ2 = 13, df = 1, P = 0.0003; SAF pair: 40% vs. 27%, χ2 = 11, df = 1, P = 0.0008). Since the deleterious model predicts a greater probability of rcold > rwarm for all types of outliers, this result for neutralizing changes might indicate a role for this hitchhiking process.
Since the decanalization model does make a stronger prediction of inbreeding disruption for novel outliers, and the deleterious model does not, we conducted a more formal test asking if novel outliers showed a more enhanced rcold > rwarm probability (compared with novel nonoutliers) than neutralizing outliers (compared with neutralizing nonoutliers). We performed a regression GLM analysis to model the above probability as a function of outlier status, novel/neutralizing category, and the magnitude of evolved change |E| standardized by the expression of the warm-adapted population in the cold condition. We included magnitude in this analysis in light of the possibility that under the deleterious hitchhiking model, stronger expression differences were under stronger selection and therefore carried more hitchhiking variants with them (but qualitatively similar results were obtained without the magnitude term). We tested for a significant interaction between outlier status and novel/neutralizing category. The MED and SAF pairs returned nonsignificant results, in different directions, as indicated in figure 4. However, the ETH pair yielded a significant interaction, indicating a greater decanalization signal for a novel than neutralizing outliers (P = 0.0015). Hence, we find some support for the prediction of the decanalization hypothesis that novel states should be more likely to undermine ancestral buffering mechanisms.
In contrast to the results for gene expression, for intron usage, the fraction of genes with rcold > rwarm was not significantly different between outliers and nonoutliers for any population pair or partition of outlier introns. Two pairs showed a qualitatively similar pattern for novel changes (fig. 4D), but just one did so for neutralizing changes (fig. 4F). Hence, there was no clear support for decanalization or deleterious hitchhiking for the putatively adaptive alternative splicing traits examined.
Discussion
Gene regulation can play a key role in mediating complex interactions between genotype, environment, and fitness. In this study, we use parallel cold-adapted natural fly populations to investigate the interplay between gene regulatory evolution, plastic effects of the environment on a trait, and canalization, which may buffer against such effects. Regulatory plasticity may be beneficial if it is aligned with a trait’s fitness–environment gradient (e.g., if a gene contributing to cold tolerance is upregulated under cold conditions). In other cases, environmental stresses may trigger unfavorable shifts in gene regulation.
Here, focusing on outliers that may reflect adaptive regulatory evolution, we observe evidence of adaptive gene expression divergence opposing the initial plasticity for gene expression and splicing across three pairs of natural populations that evolved in parallel (fig. 3). These patterns are consistent with several previous studies, which found similar “counter-gradient” patterns in expression (Levine et al. 2011; Dayan et al. 2015; Ghalambor et al. 2015; Huang and Agrawal 2016; Ho and Zhang 2019; Koch and Guillaume 2020). Unlike previous studies, we also examine splicing evolution and find that it shows a similar pattern, suggesting that this “counter-gradient” result may be general for transcriptomic traits. Further, among the expression/splicing traits showing evolution opposing the initial plasticity, the majority of them are neutralizing the initial plastic response, that is, restoring the expression toward the ancestral state, rather than reversing changes generating novel regulatory states.
There are at least two potential reasons why adaptive evolution may neutralize the initial plasticity. First, the initial plasticity may have represented a maladapted state induced by the perturbation of a cold environment. Therefore, subsequent adaptation to cold will restore the expression/splicing back to the ancestral optimal level. On the other hand, some of the initial plasticity may be beneficial in the short term. The naive populations may respond to a new environment by altering expression/splicing immediately, for example, stress response, which allows them to persist in the environment with certain costs. Once better mechanisms of coping with the environment have evolved, the stress response may no longer be induced. Without stress response activation, regulation for the cold-adapted population in the cold environment could revert toward that observed for the warm-adapted population in the warm environment. From the GO enrichment test (supplementary table S2, Supplementary Material online), we do find significant enrichment for GO terms “response to stimulus” and “response to stress” among the genes that show naive plasticity, although they are not limited to genes showing “neutralizing” changes. The presence of stress response genes among the “neutralizing” category could make sense if warm-adapted flies in the cold environment manifest a stressed transcriptional state, then it could make sense for (less-stressed) cold-adapted flies in that same cold environment to show stress response gene regulation more aligned with that of the warm-adapted population in its home environment. In other cases, novel regulatory states at stress response genes might protect against the challenges of the cold environment.
It is worth noting that, although the neutralizing cases are enriched in PST outliers relative to nonoutliers, ∼20–50% of the outliers show “concordant” changes (fig. 3). For GO enrichment tests for those showing “concordant” plasticity evolution, only the MED pair has significant terms. These significant GO terms in MED are related to the synapse and behavior, which is consistent with other findings that synapse-associated genes are associated with cold tolerance (Mackay et al. 2012; Pool et al. 2017).
Notably, the ETH pair has many more genes/introns showing reversing changes than the other two pairs. The ETH highland population exhibits distinct phenotypic evolution such as darker pigmentation (Bastide et al. 2014), larger body size (Pitchers et al. 2013; Lack et al. 2016b), and reduced reproductive rate (Lack et al. 2016b), raising the possibility that the strong plasticity-reversing expression change in EF may contribute to the unique phenotypic evolution of this high altitude (3,050 m) population. We note that the ETH pair was previously found to have unique regulatory evolution in other respects (Huang et al. 2021): a large majority of its adult (and larval) PST outliers had increased rather than decreased expression, and it showed particular importance of trans-regulatory evolution compared with the other population pairs. Hence, while multi-population comparisons like this one may illuminate evolutionary patterns of general significance, the specific ecological and evolutionary context of each population may importantly influence transcriptome-wide patterns of change.
Plasticity can be reduced by environmental canalization (Debat and David 2001; Liefting et al. 2009), which may or may not share a common basis with genetic canalization (Wagner et al., 1997; Meiklejohn and Hartl 2002; Flatt 2005; Pesevski and Dworkin 2020). Here, we explored whether putatively adaptive regulatory evolution may disrupt genetic canalization using inbreeding as a broad genetic perturbation. We first observed a transcriptome-wide pattern of warm-adapted populations showing greater vulnerability to the genetic perturbation of inbreeding than cold-adapted populations in all pairs for both expression abundance and intron usage (fig. 4A and B). One explanation is that a low temperature is a form of environmental stress to the warm-adapted populations but not so much to the cold-adapted ones and that this stress induces a relatively uniform transcriptomic response. In combination, the environmental perturbation of cold stress may have compromised the ability of warm-adapted organisms to buffer the genetic perturbation of inbreeding and led to a greater reduction in canalization (higher Vinbred/Voutbred ratio) than the less-stressed cold-adapted organisms (Chen et al. 2015). Although this inbreeding–environment interaction has been observed before, the underlying mechanisms are not understood (Kristensen et al. 2006; Reed et al. 2012). Another potential contributor to the apparently greater transcriptome-wide effects of inbreeding on warm-adapted populations is a potentially larger number of recessive deleterious variants. All three of our cold-adapted populations have slightly lower genomic diversity than their warm-adapted counterparts and appear to have gone through mild population bottlenecks after the population pairs diverged (Lack et al. 2016a; Sprengelmeyer et al. 2020). After the initial generations of such a bottleneck, the recessive load is expected to be lower in the bottlenecked population than in the nonbottlenecked counterpart (Kirkpatrick and Jarne 2000), and hence, our warm-adapted populations would be predicted to harbor a greater recessive load than their cold-adapted partners.
Aside from the above transcriptome-wide differences in canalization, adaptation could reduce canalization if selection shifts gene regulation outside of the buffered ancestral range. Previously, evidence of decanalization linked to adaptive evolution in nature was limited to transient developmental stability in insecticide-resistant blowflies (Clarke and McKenzie 1987; McKenzie and Game 1987) and wing abnormalities in large-winged Ethiopian D. melanogaster (Lack et al. 2016b). Here, we find that genes with putatively adaptive expression changes are more vulnerable to inbreeding in all three population pairs (fig. 4C and E). These results are consistent with the adaptation–decanalization hypothesis, but they could also be generated if linked deleterious recessive regulatory variants hitchhiked along with favored haplotypes. We reasoned that only the decanalization model predicts that “novel” regulatory changes should result in greater vulnerability to inbreeding, and we found some evidence in support of this prediction. Specifically, we found that for the ETH population pair, outlier genes with novel regulatory changes were significantly more vulnerable to inbreeding. It is intriguing that decanalization evidence was strongest for the ETH pair, given that wing size evolution in the cold-adapted member of this pair was the morphological inspiration for the adaptation–decanalization hypothesis (Lack et al. 2016b).
Our findings involving large numbers of putatively adaptive expression traits could hint at a much broader relationship between adaptive evolution and decanalization, at least for gene expression. However, variation in the strength of evidence for decanalization among our population pairs suggests that evolutionary context may be an important factor modulating the relationship between adaptation and canalization. Further work is needed to understand this relationship more clearly. For example, to what extent do adaptive changes by themselves break down canalization versus decanalization occurring separately as a prerequisite for adaptive trait change? Also, to the extent that decanalization occurs concurrently with adaptation trait evolution, how often does it occur as a pleiotropic byproduct of the adaptive change itself versus arising from linked deleterious variants?
In contrast, evidence for decanalization was not observed to a significant degree for putatively adaptive intron usage changes (fig. 4D). It is possible that our splicing PST outliers may be less enriched for targets of strong adaptive evolution than our expression outliers. Alternatively, there may be a meaningful difference in how the two traits are buffered. While our plasticity data suggest that expression and splicing are similarly susceptible to environmental plasticity (table 1), it is possible that intron usage traits are less vulnerable to the genetic perturbation of inbreeding, perhaps due to differences in the properties of biological networks governing expression and splicing.
In this study, we leveraged parallel adaptive divergence among natural populations of a model organism to broadly examine the relationship between adaptive evolution, thermal environment, and gene regulation. However, much work remains to be done. Our finding that gene expression evolution tends to neutralize ancestral plasticity is consistent with many, although not all, previous studies (see Introduction) and suggests that many plastic expression changes may be maladaptive in novel environments. For the first time, we also showed a similar pattern for alternative splicing in the context of intron usage. It will therefore be of interest to assess the degree to which a broader range of traits (gene regulatory and otherwise) show counter-gradient patterns of evolution and plasticity. Our investigation of the relationship between adaptive gene regulatory evolution and decanalization particularly calls out for further research in light of this topic’s lack of prior study. It was only relatively recently that an instance of morphological evolution in nature was found to yield decanalization of the same structure’s development (wing size in our highland ETH population; Lack et al. 2016b). We find some evidence that putatively adaptive gene expression traits are linked to (genetic) decanalization on a transcriptome-wide scale, which raises the question of whether a more general role for adaptive evolution in undermining canalization may exist. However, there is a considerable need to investigate this relationship across a broader range of organisms and traits.
Materials and Methods
RNA Sample Collection and Sequencing
As described in previous publications (Lack et al. 2016a; Pool et al. 2016; Huang et al. 2021), we have three D. melanogaster population pairs, each representing cold-adapted and warm-adapted populations from the same region: a Mediterranean pair (France FR and Egypt EG), an Ethiopian pair (EF and EA), and a South African pair (SD and SP). Within each of these six populations, we selected sixteen strains and assigned them to eight crosses. These strains had been inbred for eight generations. To reduce within-cross variance, pairs of strains for a cross were chosen based on minimal overlapping heterozygosity genome-wide. Each cross was conducted concurrently at both 25 °C, the control warm condition, and 15 °C, the derived cold condition (fig. 1A). Twenty virgin females and 20 males were collected from maternal and paternal lines, respectively, for each cross and allowed to mate and lay eggs for a week in half-pint bottles. Each bottle contained standard Drosophila medium (containing molasses, cornmeal, yeast, agar, and antimicrobial agents). To collect samples for RNA-seq, 30 female F1 offspring were collected 4–5 days after eclosion and shock-frozen in liquid nitrogen immediately. To compare the effects of inbred and outbred on expression variation after adaptive evolution, we also reared four inbred lines at 15 °C for each population (fig. 1A) and collected 30 females 4–5 days after eclosion. A temperature of 15 °C represented the derived environment where the cold-adapted populations underwent selection.
For RNA extraction, 30 females for each sample were homogenized using TissueLyser II (Qiagen, Hilden, Germany). Total mRNA was isolated using the Magnetic mRNA Isolation Kit (New England Biolabs, Ipswich, MA, USA) and cleaned up using the RNeasy MinElute Cleanup Kit (Qiagen, Hilden, Germany). Strand-specific libraries were prepared using the NEBNext mRNA Library Prep Reagent Set for Illumina. Libraries were size-selected for approximately 150 bp inserts using AMPureXP beads (Beckman Coulter, CA, USA). The libraries were quantified using Bioanalyzer and manually multiplexed for sequencing. All libraries were sequenced on a HiSeq2500 (V4) with 2 × 75 bp paired-end reads in two flow cells. Numbers of paired-end reads generated for different libraries can be found in supplementary table S1, Supplementary Material online. Data from parental outbred samples, parental inbred samples, and all F1 samples were each largely sequenced on the same flow cell as other samples of the same type, with a minority of samples having an initially low depth of coverage subsequently topped off by further sequencing on the same machine.
Quantifying Gene Expression and Intron Usage Frequency
The paired-end sequence reads from each of the F1 samples were mapped to the transcribed regions annotated in D. melanogaster (release 6, BDGP6.84) using STAR with parameters from the ENCODE3 STAR-RSEM pipeline (Li and Dewey 2011; Dobin et al. 2013). Numbers of mapped read pairs for different samples can be found in supplementary table S2, Supplementary Material online. For gene expression, the numbers of reads mapped to each gene were quantified using RSEM (Li and Dewey 2011). Reads mapped to the rRNA were excluded from the analysis. Supplementary table S3, Supplementary Material online provides the read counts for each gene in each sample. The expression abundance for each gene was the number of reads mapped to the gene per million reads (standardized by total reads mapped to the transcriptome).
To visually describe the transcriptome variation among samples, we first performed PCA for the F1 samples from different temperature conditions across populations. We used DESeq2 (Love et al. 2014) to construct the data object from the matrix of count data output from RSEM and performed variance stabilizing transformation (vst). The top 5,000 genes with the highest variance across samples at the transformed scale were used for PCA with the function prcomp in R.
To quantify intron usage, we used Leafcutter (Li et al. 2018) to estimate the excision frequencies of alternative introns. Leafcutter took the alignment files generated by STAR as input to quantify the usage of each intron. Then Leafcutter formed clusters that contained all overlapping introns that shared a donor or acceptor splice site. The default parameters were used: >50 reads supporting each intron cluster and <500 kb for intron length. The numbers of intron excision events for different clusters in each sample can be found in supplementary table S3, Supplementary Material online. The intron usage frequency is the number of intron excision events divided by the total events per cluster. It is worth noting that Leafcutter only detects exon–exon junction usage and is unable to quantify 5′ and 3′ end usage and intron retention, which may be confounded by differential bias among libraries.
Comparison of Naive Plasticity and Evolutionary Changes
The detailed methods of identifying candidates for adaptive evolution between cold and warm populations can be found in Huang et al. (2021). Briefly, that study used PST statistics to quantify gene regulatory divergence (for both expression abundance and intron usage) between cold- and warm-adapted populations in each pair. That approach was used because the goal was to identify transcriptomic traits with unusually high population differentiation relative to within-population variation (representing candidates for local adaptation), as opposed to simply testing whether any significant population difference in expression/splicing existed at all. The upper 5% of PST quantile was used to identify outliers that are more likely to have been under adaptive evolution. (evaluated separately for each population pair). While this threshold is necessarily arbitrary, in light of our present study’s focus on transcriptome-wide patterns, it is not essential that every PST outlier be a true product of local adaptation. Instead, we simply need a group of genes that is meaningfully enriched for true local adaptation targets and that contains enough such genes to provide reasonable power for the analyses described below. Given that we are studying population pairs that have occupied contrasting environments for tens of thousands of generations (Sprengelmeyer et al. 2020) and the differ in multiple traits (Bastide et al. 2016; Lack et al. 2016b; Pool et al. 2017), it seems reasonable to propose that PST outliers are enriched for targets of local adaptation, and that differences between outliers and nonoutliers are informative regarding the characteristics of genes under adaptive transcriptomic evolution. However, we note that factors such as population differences in organ size could also influence our whole-organism RNA-seq data, as further discussed in Huang et al. (2021).
To quantify plasticity, we used the difference in gene expression between rearing conditions for each cross. For expression abundance, we calculated the change of expression abundance from cold to warm conditions for each of the eight crosses (where expression in both conditions was >0). To identify genes showing naive plasticity in warm-adapted populations, we required consistent expression changes (consistently higher or lower expression for the cross in cold conditions than that in warm conditions) in at least seven out of eight crosses following the direction of average change among the eight crosses. Similarly, for intron usage, we calculated the between-environment differences in intron usage proportion for each of the eight crosses. The criterion to identify plastic splicing in warm-adapted populations was consistent differences in at least seven out of eight crosses showing the same direction as the average difference. In the absence of true plasticity, only 7% of genes should meet this criterion by chance (two-sided binomial calculation). Null simulations in which all 16 values were normally distributed confirmed that the false-positive rate should be close to 7%. While a 7% false-positive rate is slightly higher than the conventional 5%, it represents a reasonable compromise in light of the data available and the goals of our study. Here, our need is to define a set of plasticity-enriched genes of adequate size to offer power for the analyses described below.
To examine whether adaptation to cold environments tends to enhance or oppose the naive expression plasticity (fig. 1B), we first calculated the expression value for every gene in each population at each condition. For expression abundance, the median abundance among eight F1 samples was used as the expression value for a population in a certain condition (Z). For intron usage, the corresponding value was the median of intron usage proportion among the eight F1 samples. To determine whether a regulatory trait evolved to enhance or oppose ancestral plastic regulation (fig. 1C–E), we first examined “naive plasticity” (warm-adapted population in a cold environment, Zwarm_15°C, compared with the warm environment, Zwarm_25°C; fig. 1B). We then asked whether evolution in the cold-adapted population enhanced or opposed the ancestral naive plasticity. Formally, we asked whether the differences in trait values between cold- and warm- adapted populations in the cold environment, E = Zcold_15°C − Zwarm_15°C, were in the same direction as the plastic response of the warm population, Pwarm = Zwarm_15°C − Zwarm_25°C. If E × Pwarm > 0, the evolved regulation was assigned as “concordant” (fig. 1C). If E × Pwarm < 0, the evolved regulation was assigned as “discordant,” and these discordant traits were further partitioned into two categories. We defined as “neutralizing” traits in which the cold population’s “home” value (Zcold_15°C) evolved to be closer to the warm population’s “home” value (Zwarm_25°C) than predicted by the warm population’s plastic response, and, therefore, |E| < 2|Pwarm| (fig. 1D). In contrast, we defined as “reversing” traits in which an evolved change E was both opposing the direction of plasticity P and took Zcold_15°C farther from Zwarm_25°C than predicted by the warm population’s plastic response (|E| > 2|Pwarm|; fig. 1E). For both categories where evolved expression was farther from the warm population’s “home” value range (either concordant or reversing), we defined them as “novel.”
The differences in the “concordant” to “discordant” ratio between PST outliers and nonoutliers were tested based on the chi-squared test in R version 3.3.0. for both expression abundance and splicing. It is worth noting that the test assumes that regulation is independent among genes and introns. However, it is possible that the expression or splicing of different genes is causally related because of the shared regulatory network.
Because the calculations of Pwarm and E included a common value, Zwarm_15°C, any random measurement error on this value may generate an artifact of a negative relationship between Pwarm and E (Mallard et al. 2018; Ho and Zhang 2019). We therefore repeated this analysis while subdividing the data for estimating Zwarm_15°C: four random crosses were used to estimate Pwarm and the other crosses were used to estimate E. We used the latter four crosses of warm-adapted populations at 15 °C for measuring E and the eight crosses of cold-adapted populations at 15 °C to identify PST outliers and nonoutliers. Then, we used the same criteria to categorize the evolutionary changes into “concordant” or “discordant” and repeated the same analysis for all sets of subdivided data (70 sets). Since we used Zwarm_15°C estimated from different data sets to calculate Pwarm and E, the potential artifact of a negative relationship between Pwarm and E should be removed. The fraction of the subdivided data (70 sets) showing an excess of concordant changes in PST outliers versus nonoutliers was the P-value for testing whether PST outliers had a significantly lower concordant proportion than nonoutliers.
GO Enrichment Test for Expression Plasticity
GO enrichment tests were performed using the R package “clusterProfiler” (Yu et al. 2012) based on the fly genome annotation (Carlson 2019). For expression abundance, the gene ID for concordant, neutralizing, or reversing outliers were input as the focal gene list, while all the genes used in studying expression abundance divergence were input as the universe gene list. For intron usage, they were mapped to gene regions to generate the universe gene list. Genes including concordant, neutralizing, or reversing outlier intron junctions were used in the focal gene list. The types of GO terms being tested contained all three subontologies: Biological Process (BP), Cellular Component (CC), and Molecular Function (MF). Selection of over-represented GO terms was based on adjusted P-value < 0.1 using the “BH” FDR method (Benjamini and Hochberg 1995) for each subontology. Based on the description of the GO terms, only the enriched GO terms with distinct functions were reported. If multiple GO terms were over-represented by the same set of candidate genes, only the term annotated with the fewest genes was reported.
Testing the Decanalizing Effect of Adaptation on Gene Expression
We examined the level of canalization for gene expression under inbreeding as a genetic perturbation. For each population, we selected four outbred F1 samples from which we also had data from one of the parental inbred lines. For gene expression abundance, we calculated the variance among four inbred samples and that among four outbred samples for each population. For intron usage, we re-ran Leafcutter based on all inbred samples (24 samples in total for six populations) and all outbred samples (24 samples). Then we calculated the variance for inbred samples (Vinbred) and for outbred samples (Voutbred) for each population. While variance estimates from only four values are expected to be noisy, our focus is exclusively on transcriptome-wide correlations and not conclusions about individual genes. Only those expression abundance or intron usage traits with variance above 0 for both inbred and outbred samples in both populations for each pair were used in this comparison. We first compared the transcriptome-wide fractions showing Vinbred > Voutbred between cold and warm populations for both expression abundance and intron usage. Because the hypothesis is about recent adaptive change to novel states resulting in decanalization, we focused on the set of genes/introns that evolved expression novelty (identified in the previous section). To test the role of selection on canalization, we compared the fractions showing a higher Vinbred to Voutbred ratio (r) in the cold population (rcold) than that in the warm population (rwarm) for PST outliers with that for nonoutlier controls using a chi-squared test. We note that we are only comparing variances between inbred and outbred samples of the same gene from the same population and not between genes that may have different average expression levels. The comparisons between the PST outliers and nonoutliers were made for the “novelty” category and for the “neutralizing” category.
To distinguish between the decanalization model and the deleterious model, we performed a generalized linear model (GLM) regression analysis on expression abundance across genes for each population pair:
where Pr is whether a gene showed rcold > rwarm as opposed to rcold > rwarm. Status is whether a gene was a PST outlier or nonoutlier. Category is whether a gene was classified as “novelty” or “neutralizing.” M is the magnitude of evolved trait change, which was calculated by the absolute value of evolved expression change |E| standardized by the expression level of the warm-adapted population at 15 °C. We tested for a significant interaction between Status and Category variables by comparing the above full model against a reduced model without the interaction term. A likelihood ratio test was used to determine significance.
Supplementary Material
Acknowledgments
The authors thank Colin Dewey for helpful discussions. The authors also thank the anonymous reviewers for their helpful comments on the manuscript. This work was funded by the NSF DEB grant 1754745 to J.E.P. and by the NIH NIGMS grant F32GM106594 to J.B.L.
Supplementary Material
Supplementary data are available at Genome Biology and Evolution online.
Data Availability
The raw RNA-seq reads for samples in the cold condition (15 °C) are available from the Sequence Read Archive (SRA) under the BioProject PRJNA720479, and those from the warm condition (25 °C) are under the BioProject PRJNA758705. See supplementary table S3, Supplementary Material online and relevant codes are available at https://github.com/YuhengHuang87/ExpressionPlasticityCanalization.
Literature Cited
- Bastide H, Lange JD, Lack JB, Yassin A, Pool JE. 2016. A variable genetic architecture of melanic evolution in Drosophila melanogaster. Genetics 204(3):1307–1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastide H, Yassin A, Johanning EJ, Pool JE. 2014. Pigmentation in Drosophila melanogaster reaches its maximum in Ethiopia and correlates most strongly with ultra-violet radiation in sub-Saharan Africa. BMC Evol Biol. 14(1):179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc: Ser B (Methodological) 57(1):289–300. [Google Scholar]
- Bittner NKJ, Mack KL, Nachman MW. 2021. Gene expression plasticity and desert adaptation in house mice. Evolution 75(6):1477–1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson M. 2019. org.Dm.eg.db: Genome wide annotation for Fly. R package version 3.8.2.
- Charlesworth B, Lande R, Slatkin M. 1982. A Neo-Darwinian commentary on macroevolution. Evolution 36(3):474–498. [DOI] [PubMed] [Google Scholar]
- Chen J, Nolte V, Schlötterer C. 2015. Temperature stress mediates decanalization and dominance of gene expression in Drosophila melanogaster. PLOS Genet. 11(2):e1004883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chevin L-M, Lande R, Mace GM. 2010. Adaptation, plasticity, and extinction in a changing environment: towards a predictive theory. PLoS Biol. 8(4):e1000357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chun S, Fay JC. 2011. Evidence for hitchhiking of deleterious mutations within the human genome. PLoS Genet. 7(8):e1002240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke GM, McKenzie JA. 1987. Developmental stability of insecticide resistant phenotypes in blowfly; a result of canalizing natural selection. Nature. 325(6102):345–346. [Google Scholar]
- Conover DO, Schultz ET. 1995. Phenotypic similarity and the evolutionary significance of countergradient variation. Trends Ecol Evol. 10(6):248–252. [DOI] [PubMed] [Google Scholar]
- Dayan DI, Crawford DL, Oleksiak MF. 2015. Phenotypic plasticity in gene expression contributes to divergence of locally adapted populations of Fundulus heteroclitus. Mol Ecol. 24(13):3345–3359. [DOI] [PubMed] [Google Scholar]
- Debat V, David P. 2001. Mapping phenotypes: canalization, plasticity and developmental stability. Trends Ecol Evol. 16(10):555–561. [Google Scholar]
- de Visser J, et al. 2003. Perspective: evolution and detection of genetic robustness. Evolution 57(9):1959–1972. [DOI] [PubMed] [Google Scholar]
- Dobin A, et al. 2013. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29(1):15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dworkin I. 2005. Canalization, cryptic variation, and developmental buffering: a critical examination and analytical perspective. In: Hallgrımsson B, Hall BK, editors. Variation, a central concept in biology. New York: Academic Press. p. 131–158. [Google Scholar]
- Fischer EK, Song Y, Hughes KA, Zhou W, Hoke KL. 2021. Nonparallel transcriptional divergence during parallel adaptation. Mol Ecol. 30(6):1516–1530. [DOI] [PubMed] [Google Scholar]
- Flatt T. 2005. The evolutionary genetics of canalization. Q Rev Biol. 80(3):287–316. [DOI] [PubMed] [Google Scholar]
- Fraser HB, Moses AM, Schadt EE. 2010. Evidence for widespread adaptive evolution of gene expression in budding yeast. Proc Natl Acad Sci U S A. 107(7):2977–2982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavrilets S, Hastings A. 1994. A quantitative-genetic model for selection on developmental noise. Evolution 48(5):1478–1486. [DOI] [PubMed] [Google Scholar]
- Ghalambor Cameron K, et al. 2015. Non-adaptive plasticity potentiates rapid adaptive evolution of gene expression in nature. Nature 525(7569):372–375. [DOI] [PubMed] [Google Scholar]
- Ghalambor CK, McKAY JK, Carroll SP, Reznick DN. 2007. Adaptive versus non-adaptive phenotypic plasticity and the potential for contemporary adaptation in new environments. Funct Ecol. 21(3):394–407. [Google Scholar]
- Ghosh SM, Satish KM, Jayaram M, Joshi A. 2019. Does long-term selection for development time result in canalization: a test using Drosophila melanogaster. Front Ecol Evol. 7:228. [Google Scholar]
- Gibson G, Dworkin I. 2004. Uncovering cryptic genetic variation. Nat Rev Genet. 5(9):681–690. [DOI] [PubMed] [Google Scholar]
- Good BH, Desai MM. 2014. Deleterious passengers in adapting populations. Genetics 198(3):1183–1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groth BR, Huang Y, Monette MJ, Pool JE. 2018. Directional selection reduces developmental canalization against genetic and environmental perturbations in Drosophila wings. Evolution 72(8):1708–1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayden EJ, Weikert C, Wagner A, Lehner B. 2012. Directional selection causes decanalization in a group I ribozyme. PLoS ONE. 7(9):e45351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho W-C, Zhang J. 2018. Evolutionary adaptations to new environments generally reverse plastic phenotypic changes. Nat Commun. 9(1):350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho W-C, Zhang J. 2019. Genetic gene expression changes during environmental adaptations tend to reverse plastic changes even after the correction for statistical nonindependence. Mol Biol Evol. 36(3):604–612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Agrawal AF. 2016. Experimental evolution of gene expression and plasticity in alternative selective regimes. PLOS Genet. 12(9):e1006336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Lack JB, Hoppel GT, Pool JE. 2021. Parallel and population-specific gene regulatory evolution in cold-adapted fly populations. Genetics 218(3):iyab077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Josephs EB, Van Etten ML, Harkess A, Platts A, Baucom RS.. 2021. Adaptive and maladaptive expression plasticity underlying herbicide resistance in an agricultural weed. Evol Lett. 5(4):432–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenkel CD, Matz MV. 2017. Gene expression plasticity as a mechanism of coral adaptation to a variable environment. Nat Ecol Evol. 1(1):14. [DOI] [PubMed] [Google Scholar]
- Kirkpatrick M, Jarne P. 2000. The effects of a bottleneck on inbreeding depression and the genetic load. Am Nat. 155(2):154–167. [DOI] [PubMed] [Google Scholar]
- Koch EL, Guillaume F. 2020. Restoring ancestral phenotypes by reduction of plasticity is a general pattern in gene expression during adaptation to different stressors in Tribolium castaneum. Mol Ecol. 29(20):3938–3953. [DOI] [PubMed] [Google Scholar]
- Kristensen TN, Sørensen P, Pedersen KS, Kruhøffer M, Loeschcke V. 2006. Inbreeding by environmental interactions affect gene expression in Drosophila melanogaster. Genetics 173(3):1329–1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lachaise D, et al. 1988. Historical biogeography of the Drosophila melanogaster species subgroup. In: Hecht MK, Wallace B, Prance GT, editors. Evolutionary biology. Evolutionary biology. Vol. 22. Boston (MA): Springer. [Google Scholar]
- Lack JB, Lange JD, Tang AD, Corbett-Detig RB, Pool JE. 2016a. A thousand fly genomes: an expanded Drosophila genome nexus. Mol Biol Evol. 33(12):3308–3313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lack JB, Monette MJ, Johanning EJ, Sprengelmeyer QD, Pool JE. 2016b. Decanalization of wing development accompanied the evolution of large wings in high-altitude Drosophila. Proc Natl Acad Sci U S A. 113(4):1014–1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine MT, Eckert ML, Begun DJ. 2011. Whole-Genome Expression Plasticity across Tropical and Temperate Drosophila melanogaster Populations from Eastern Australia. Mol Biol Evol. 28(1):249–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li YI, et al. 2018. Annotation-free quantification of RNA splicing using LeafCutter. Nat Genet. 50(1):151–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Dewey CN. 2011. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 12(1):323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liefting M, Hoffmann AA, Ellers J. 2009. Plasticity versus environmental canalization: population differences in thermal responses along a latitudinal gradient in Drosophila serrata. Evolution 63(8):1954–1963. [DOI] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Gen Biol. 15(12):31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackay TFC, et al. (2012). The Drosophila melanogaster genetic reference panel. Nature 482(7384):173–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallard F, Jakšić A, Schlötterer C. 2018. Contesting the evidence for non-adaptive plasticity. Nature 555(7698):E21–E22. [DOI] [PubMed] [Google Scholar]
- Mallard F, Nolte V, Schlötterer C. 2020. The evolution of phenotypic plasticity in response to temperature stress. Genome Biol Evol. 12(12):2429–2440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masel J. 2005. Evolutionary capacitance may be favored by natural selection. Genetics 170(3):1359–1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maynard Smith J, et al. 1985. Developmental constraints and evolution: a perspective from the mountain lake conference on development and evolution. Quart Rev Biol. 60:265–287. [Google Scholar]
- McKenzie JA, Game AY. 1987. Diazinon resistance in Lucilia cuprina; mapping of a fitness modifier. Heredity 59(3):371–381. [Google Scholar]
- Meiklejohn CD, Hartl DL. 2002. A single mode of canalization. Trends Ecol Evol. 17(10):468–473. [Google Scholar]
- Meyers LA, Bull JJ. 2002. Fighting change with change: adaptive variation in an uncertain world. Trends Ecol Evol. 17(12):551–557. [Google Scholar]
- Nijhout HF, Davidowitz G. 2003. Developmental perspectives on phenotypic plasticity, canalization, and fluctuating asymmetry. In: Polak M, editor. Developmental instability: causes and consequences. MIT Press. p. 3–13. [Google Scholar]
- Nourmohammad A, et al. 2017. Adaptive evolution of gene expression in Drosophila. Cell Reports 20(6):1385–1395. [DOI] [PubMed] [Google Scholar]
- Paaby AB, Rockman MV. 2014. Cryptic genetic variation: evolution’s hidden substrate. Nat Rev Genet. 15(4):247–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pesevski M, Dworkin I. 2020. Genetic and environmental canalization are not associated among altitudinally varying populations of Drosophila melanogaster. Evolution 74(8):1755–1771. [DOI] [PubMed] [Google Scholar]
- Pfennig DW, et al. 2010. Phenotypic plasticity’s impacts on diversification and speciation. Trends in Ecology & Evolution 25(8):459–467. [DOI] [PubMed] [Google Scholar]
- Pitchers W, Pool JE, Dworkin I. 2013. Altitudinal clinal variation in wing size and shape in African Drosophila melanogaster: one cline or many? Evolution 67(2):438–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pool JE, et al. 2012. Population genomics of sub-Saharan Drosophila melanogaster: African diversity and non-african admixture. PLOS Genet. 8(12):e1003080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pool JE, Braun DT, Lack JB. 2017). Parallel evolution of cold tolerance within Drosophila melanogaster. Mol Biol Evol. 34(2):349–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ragland GJ, et al. 2015. Differences in performance and transcriptome-wide gene expression associated with Rhagoletis (Diptera: Tephritidae) larvae feeding in alternate host fruit environments. Mol Ecol. 24(11):2759–2776. [DOI] [PubMed] [Google Scholar]
- Réaale D, Roff DA. 2003. Inbreeding, developmental stability, and canalization in the sand cricket Gryllus firmus. Evolution 57(3):597–605. [PubMed] [Google Scholar]
- Reed DH, Fox CW, Enders LS, Kristensen TN. 2012. Inbreeding-stress interactions: evolutionary and conservation consequences: inbreeding-stress interactions. Ann N Y Acad Sci. 1256(1):33–48. [DOI] [PubMed] [Google Scholar]
- Scheiner SM. 1993. Genetics and evolution of phenotypic plasticity. Annu Rev Ecol Syst. 24:35–68. [Google Scholar]
- Sprengelmeyer QD, et al. 2020. Recurrent collection of Drosophila melanogaster from Wild African environments and genomic insights into species history. Mol Biol Evol. 37(3):627–638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stearns SC, Kawecki TJ. 1994. Fitness sensitivity and the canalization of life history traits. Evolution 48(5):1438–1450. [DOI] [PubMed] [Google Scholar]
- Via S. 1993. Adaptive phenotypic plasticity: target or by-product of selection in a variable environment? Am Nat. 142(2):352–365. [DOI] [PubMed] [Google Scholar]
- von Heckel K, Stephan W, Hutter S. 2016. Canalization of gene expression is a major signature of regulatory cold adaptation in temperate Drosophila melanogaster. BMC Genomics 17(1):574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waddington CH. 1942. Canalization of development and the inheritance of acquired characters. Nature. 150(3811):563–565. [Google Scholar]
- Wagner GP, Booth G, Bagheri-Chaichian H. 1997. A population genetic theory of canalization. Evolution 51(2):329–347. [DOI] [PubMed] [Google Scholar]
- Yu G, Wang L-G, Han Y, He Q-Y. 2012. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS: J Integrative Biol. 16(5):284–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw RNA-seq reads for samples in the cold condition (15 °C) are available from the Sequence Read Archive (SRA) under the BioProject PRJNA720479, and those from the warm condition (25 °C) are under the BioProject PRJNA758705. See supplementary table S3, Supplementary Material online and relevant codes are available at https://github.com/YuhengHuang87/ExpressionPlasticityCanalization.