Abstract
Matrix metalloproteinases (MMPs) are Zn(II) dependent endopeptidases involved in the degradation of collagen. Unbalanced collagen breakdown results in numerous pathological conditions, including cardiovascular and neurodegenerative diseases and tumor growth and invasion. Matrix metalloproteinase-1 (MMP-1) is a member of the MMPs family. The enzyme contains catalytic and structural Zn(II) ions. Despite many studies on the enzyme, there is little known about the synergy between the two Zn(II) metal ions and the enzyme and substrate dynamics in MMP-1 structure-function relationships. We performed a computational study of the MMP-1•triple-helical peptide (THP) enzyme•substrate complex to provide this missing insight. Our results revealed Zn(II) ions' importance in modulating the long-range correlated motions in the MMP-1•THP complex. Overall, our results reveal the importance of the catalytic Zn(II) and the role of the structural Zn(II) ion in preserving the integrity of the enzyme active site and the overall enzyme-substrate complex synergy with the dynamics of the enzyme and the substrate. Notably, both Zn(II) sites participate in diverse networks of long-range correlated motions that involve the CAT and HPX domains and the THP substrate, thus exercising a complex role in the stability and functionality of the MMP-1•THP complex. Both the Zn(II) ions have a distinct impact on the structural stability and dynamics of the MMP-1•THP complex. The study shifts the paradigm from the "local role" of the Zn(II) ions with knowledge about their essential role in the long-range dynamics and stability of the overall enzyme•substrate (ES) complex.
1. INTRODUCTION
Collagen, a major component in the extracellular matrix of connective tissues, is the most abundant protein in the human body [1-9]. It adopts a triple-helical supersecondary structure rich in the amino acids glycine (G) and proline (P) [10]. Essential features of this unique structural protein include the ability to impart tensile strength and thermal stability to various tissues of tendons, ligaments, and skin [11-13]. The degradation of collagen (collagenolysis) is an enzyme-catalyzed process involved in many biological events such as tissue remodeling, wound healing, angiogenesis, and cellular differentiation [14-15]. In turn, poorly regulated collagenolysis contributes to cardiovascular diseases, neurodegenerative diseases, cancer, and arthritis [16-19]. Enzymes catalyzing collagen hydrolysis include matrix metalloproteinases (MMPs). Interstitial (types I-III) collagen is cleaved by collagenolytic MMPs at a specific site into ¾ and ¼ fragments [20-21].
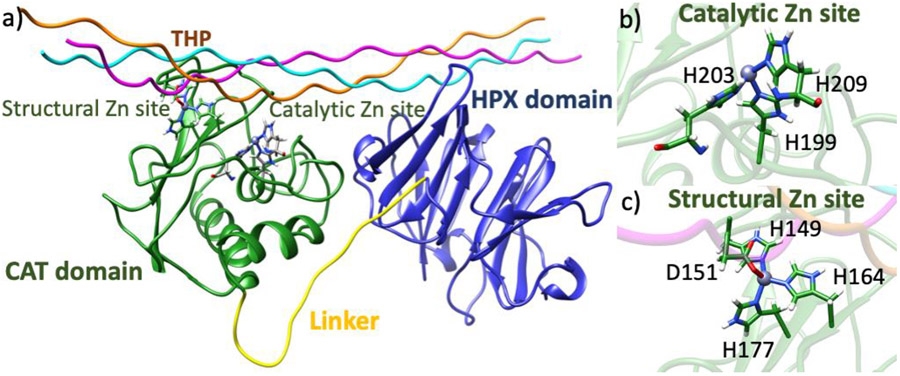
MMPs are multidomain Zn(II) dependent endopeptidases [22]. There are 23 identified human MMPs, the majority of which are characterized by an N-terminal catalytic domain (CAT), C-terminal hemopexin-like (HPX) domain, and a proline-rich linker connecting the domains (Fig. 1a) [23-24]. Effective degradation of collagen fibrils is executed only if interdomain communication is possible via the linker, thus highlighting the necessity of both domains [25-27]. The collagenolytic mechanism of matrix metalloproteinase-1 (MMP-1) has been studied both experimentally and computationally [25-33]. MMP-1 efficiently catalyzes the degradation of interstitial collagen utilizing both the CAT and HPX domains [21]. Allosteric communications between the CAT and HPX domains have a pivotal role in the activity and substrate recognition of the enzyme [25,30,34]. The MMP-1 HPX domain consists of a four-blade β propeller with a Ca(II) ion [28]. The HPX domain interacts with triple-helical substrates via the hydrophobic S10' exosite of blade 1 and residues of blade 2 [26,28]. The interactions of the blade 1 exosite with the triple-helix initiate collagenolysis by orienting the CAT domain for collagen scissile bond hydrolysis through the linker [25,27].
Fig. 1.
a) MMP-1•triple-helical peptide (THP) complex. b) Catalytic site with Zn(II) coordinated to H199, H203, and H209. c) Structural site with Zn(II) coordinated to H149, D151, H164, H177.
The highly conserved CAT domain contains two Zn(II) and three Ca(II) ions [22,35]. One of the two Zn(II) ions is located at the active site of the MMP-1 CAT domain and directly participates in catalysis. The second Zn(II) is also bound within the MMP-1 CAT domain, but outside the active site and at 8-10 Å distance from the catalytic Zn(II). Little is known about the role of the second Zn(II) in catalysis, although its importance has been indicated by studies showing that MMPs possess high affinities for structural Zn(II) [36,37].
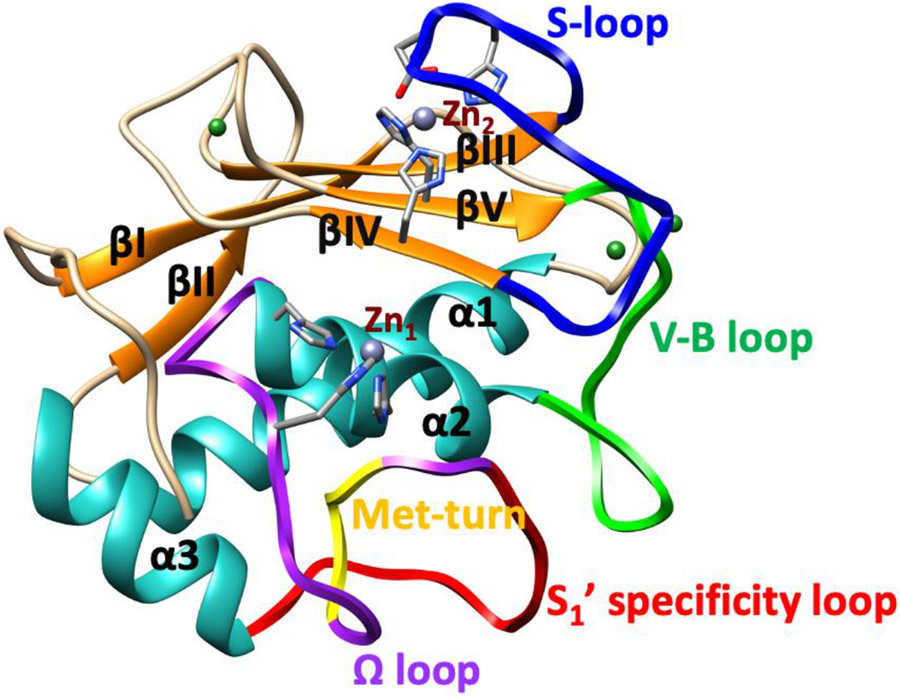
The catalytic Zn(II) is bound by three histidines (H199, H203, H209) in a conserved HEXGHXXGXXH motif (Fig. 1b) [22]. Close to the catalytic Zn(II) site, MMPs possess several other binding sites (subsites) that vary in size, charge, and hydrophobicity, which can interact with the substrate and a variety of inhibitors. Subsites are denoted as S1, S2, S3, Sn if they are on the left side of the catalytic Zn(II) ion and S1', S2', S3', Sn', respectively if they are on the right side of the Zn(II) ion. The interactions with these subsites are important for substrate binding and can be utilized in MMP-selective inhibitor design. The respective interacting substrate residues on the N-terminal side of the scissile bond are designated P1, P2, P3, while the residues of the C-terminal side are designated P1', P2' P3' respectively [38-40]. The carbonyl group of the substrate P1 glycine residue coordinates to the catalytic Zn(II) for instigating hydrolysis [22]. Thus, the catalytic Zn(II) ion plays a key role in the cleavage mechanism, [41-43], and the S1' subsite is essential for substrate selectivity [44]. The enzyme's R195 occludes the S1' subsite (pocket), influencing the size of substrate entering the pocket [45-46]. This pocket contains mainly hydrophobic residues and is comprised of a characteristic Ω loop, which includes the S1' specificity loop linking α-helices 2 and 3 (Fig. 2) [47]. The loop holds hydrophobic methionine M217, part of the highly conserved 1,4-β-turn referred to as the "Met-turn" (Fig. 2). M217 is actively engaged in maintaining the structural integrity of the active site by forming a hydrophobic sheath below the catalytic Zn(II) [48-50].
Fig. 2.
Structure of MMP-1 CAT domain. The secondary structure elements (β-strands (orange) and α helices (light sea green)), the Zn(II) ions (Zn1 & Zn2 (grey)), Ca(II) ions (forest green), S-loop (blue), V-B loop (green), Ω-loop (purple) which includes the S1’ specificity loop (red) and Met-turn (yellow) are depicted.
The structural Zn(II) ion is tetrahedrally coordinated to three histidine residues (H149, H164, H177) and an aspartic acid (D151) (Fig. 1c) [51,52]. The structural Zn(II) preserves the secondary and tertiary structure of the enzyme and is surrounded by three β-strands (βIII, βIV, and βV) [53]. The fifth β-strand connects to α-helix 2 via the V-B loop (183RWTNNFREY191) (Fig. 2), which is integral for the expression of collagenolytic activity [54]. A long S-shaped loop called the S-loop connects the antiparallel β-strands III and IV (Fig. 2) [53,42], stretching into a cleft-sided 'bulge' and continues into the β-strand IV. The bulge segment and the second half of the S-loop form the upper rim of the catalytic site [53]. The S-loop is positioned above the catalytic site and is stabilized by the structural Zn(II) and Ca(II) in the CAT domain [55].
Earlier studies on MMP-2 and MMP-3 reported the impact of metal ions on the back opening of the S1' pocket and the conversion of α-helix to β-sheet structures [41,55]. Subsequent spectroscopic studies of MMP-1 and a series of mutants aimed to probe the role of the Zn(II) ions on the enzyme structure and catalysis [56]. In these studies, MMP-1 mutant forms were designed only to bind the catalytic or structural Zn(II) but not both at the same time. The studies revealed that the structural Zn(II) plays a role in preserving the enzyme's secondary and tertiary structure. The studies also show that its presence influences the coordination number of the catalytic Zn(II). The removal of the catalytic Zn(II) ion influenced the secondary and tertiary structure of the CAT domain and decreased the rate of catalytic turnover. However, a THP substrate was not examined in these studies. Therefore, the effect of the Zn(II) ions on the dynamics of the enzyme•substrate complex between MMP-1 and collagen, a native macromolecular substrate, was not explored. In particular, it is not known how each of the Zn(II) ions influence the dynamics of the local sites, the binding of triple-helical substrates, and crucial interactions of CAT and HPX domains as well as the overall dynamics of the MMP-1•THP complex. Furthermore, the distinct effect of the individual Zn(II) ions on the long-range correlated motions in MMP-1•THP has not been explored.
Interactions between distant regions in an enzyme can have a significant impact on substrate binding, the active site structure, and the interactions stabilizing the transition state of the catalyzed reaction and enabling product release [57,58]. Experimental and computational mutagenesis studies have demonstrated the stimulating effects of such remote interactions on catalysis in MMP-1 [28,32]. However, the role of the Zn(II) ions on these long-range interactions in MMP-1 is entirely unexplored. To understand the remote long-range interactions in MMP-1 and the effect of Zn(II) ions on them, one needs to explore the inherent conformational flexibility of the MMP-1•THP complex. The dynamics will provide an understanding not only about the local flexibility of the substrate binding site, the CAT and HPX domains, and the linker region but crucially on the long-range correlated motions and domain-domain interactions that drive MMP-1 functionality and regulation. Knowledge about the influence of both Zn(II) ions on the long-range dynamics and substrate binding in the MMP-1•THP initial complex is important for understanding the Zn(II) ion's role in MMP-1 catalytic mechanism.
We hypothesize that both of the Zn(II) ions in the MMP-1 CAT domain could directly influence the structure and the dynamics of the CAT domain, THP binding, HPX domain, and ultimately MMP-1 function by introducing perturbations in the active site and conformational changes and long-range correlated motions in the overall structure. We presently performed 1-microsecond Molecular Dynamics (MD) simulations on various forms of the MMP-1•THP complex: the native MMP-1•THP complex (that contains both the catalytic and structural Zn(II)); MMP-1•THP complex without the structural Zn(II); MMP-1•THP complex without the catalytic Zn(II); and MMP-1•THP complex without both Zn(II) ions. MD simulations have allowed us to investigate the effect of the Zn(II) ions on substrate binding and long-range interactions and correlated motions between distal regions of the enzyme.
2. METHODOLOGY
2.1. System Preparation
Initial coordinates for modeling were taken from the X-ray crystallographic structure of MMP-1 complexed with a THP (PBD ID code: 4AUO) [28]. The THP numbering for the MMP-1•THP complexes in the present study was assigned based on the sequence within the triple-helical region of type II collagen, whereby the scissile bond is found at residues G775-L776. The three strands within the THP are referred to as leading (L), middle (M), and trailing (T). This numbering is consistent with our prior MD studies [31]. Missing atoms were added using Swiss PDB Viewer [59]. The active form of the enzyme was prepared by substituting A200 with E200. The protonation states of Zn(II) coordinated histidines were predicted using Propka [60]. Hydrogen atoms were added using the leap module of amber 18. Amber's primary protein model, ff14SB [61] force field, was used for all simulations. The parameters of structural Zn(II) coordinated to three histidines and aspartic acid was taken directly from Zinc Amber Force Field (ZAFF) developed by Peters et al. [62] The parameters for a bonded model of catalytic Zn(II) bound by three histidines were prepared using python-based Metal Centre Parameter Builder MCPB.py v3.0 [63]. Force constants were calculated using the Seminario method [64], and the charges of amino acids were derived from Restricted Electrostatic Potential (RESP) [65] fits.
In addition to the two Zn(II) ions, the crystal structure of the enzyme•substrate complex contained three Ca(II) ions in the CAT domain and one Ca(II) ion in the HPX domain of MMP-1 (Fig. 2). These Ca(II) ions were present in all of the MD simulations (Fig. S2). The systems for MD simulations were as follows: i) the native MMP-1•THP complex (that contains both the catalytic and structural Zn(II) (MMP1 Zn1 Zn2)); ii) MMP-1•THP complex without the structural Zn(II) (MMP1 Zn1); iii) MMP-1•THP complex without the catalytic Zn(II) (MMP1 Zn2) and MMP-1•THP complex without both Zn(II) ions (MMP1 no Zn).
2.2. Molecular Dynamics
MD simulations were carried out on the various system configurations using Amber 18 [66]. The Periodic Boundary conditions were applied to all systems. The enzyme•substrate complex was neutralized by adding Cl− counter ions. The protein was solvated with TIP3P [67] water molecules in a rectangular box at an approximate distance of 10 Å farther from the protein. Energy minimization was performed in 2 steps, with the solvent molecules minimized, restraining the solute particles, followed by the minimization of the entire system. Both steps were performed using the steepest descent in the initial 5000 steps and conjugate gradient in the final 5000 steps. The system was then slowly heated from 0 K to 300 K with solute molecules restrained for 100 ps with a harmonic potential of 4.0 kcal/mol/Å in the NVT canonical ensemble. The temperature was controlled by Langevin dynamics [68] using a 1.0 ps−1 collision frequency. SHAKE Algorithm [69] was applied to constrain all X─H bonds. Heating was followed by equilibrating the system at 300 K without restraint for 3 ns in the NPT ensemble. Pressure set at 1 bar was maintained by Brendson barostat [70]. The production MD was performed in GPU version of Amber 18 code from the equilibrated structure for 1 μs with a time step of 2 fs in NPT ensemble. Long-range electrostatic interactions in the periodic system were evaluated by Particle Mesh Ewald (PME) method [71] with a 10 Å cut- off considering the Vander Waals interactions. Hydrogen bond interactions were analyzed with the CPPTRAJ module of AMBER [72]. Root Mean Square Deviation (RMSD), Root Mean Square Fluctuation (RMSF), and Radius of Gyration (RoG) were performed on the C-α atoms of the enzyme•substrate complex using CPPTRAJ [72]. The RoG is a measure of the compactness of the simulated system and is calculated using the formula:
where M is the total mass of the protein, mi is the mass of the atom i, Rc is the mass center, and ri is the distance between atom i and Rc [73].
Dynamic Cross-Correlation Analysis (DCCA) and Principal Component Analysis (PCA) were done on the C-α atoms of the protein using the final 500 ns MD trajectory with the R package Bio3D [74]. The Dynamic Cross-Correlation Analysis (DCCA) provides information about the collective correlated and anticorrelated motions between remote parts of the simulated system. The cross-correlation between the positions of the ith and jth atoms were represented graphically using the Cij matrix. All the α carbon atoms of the enzyme•substrate complex was included to create the Cij matrix. The elements of the matrix Cij can vary between C(i,j) = −1.0 (for completely anticorrelated motions) to C(i,j) = 1.0 (for completely correlated motions) [75]. Complicated motions can also be analyzed by PCA. PCA reduces the dimensionality of motions by extracting only the most important motions from a few principal components (PCs) [73]. The method uses the eigenvalues of a covariance matrix that describe the covariances of the positions of the C-α atoms. In this study, we calculated PC1 and plotted it on the molecular structure using Bio3D tool.
3. RESULTS AND DISCUSSION
3.1. Overall Dynamics
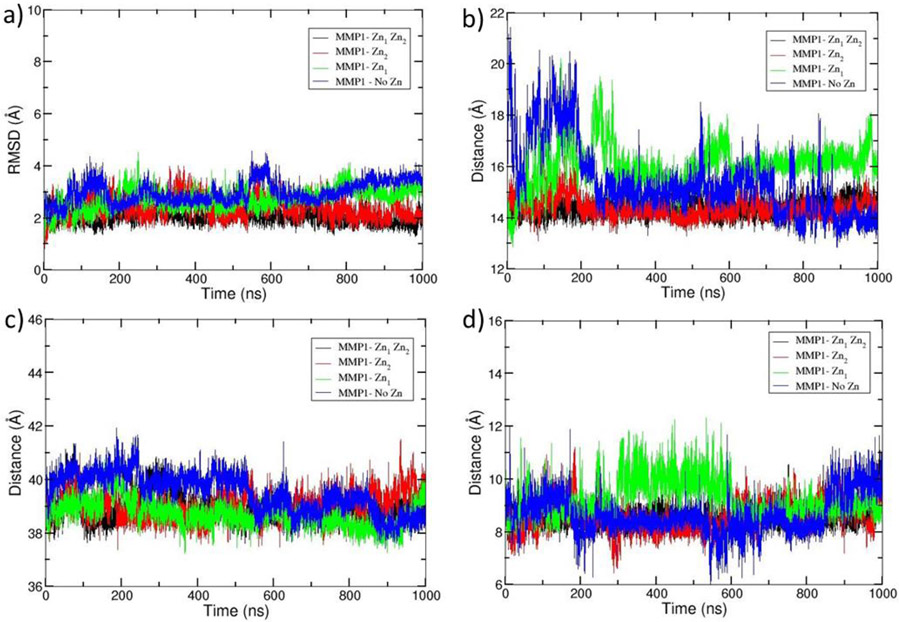
The trajectory of MD simulation was analyzed to gain direct insight into the structural dynamics of the enzyme. The results manifest that all systems equilibrated at around 600 ns with an average RMSD of 2.0, 2.3, 2.7, and 2.9 Å for (MMP1 Zn1 Zn2), (MMP1 Zn2), (MMP1 Zn1), and (MMP1 no Zn), respectively (Fig. 3a). The gradual increase of RMSD values with the elimination of Zn(II) ions indicated an influence on the overall stability. It prompted us to investigate the impact of eliminating the metal ions in the catalytic and structural sites. Analysis of the distance between the center of mass of catalytic Zn(II) and structural Zn(II) coordinated residues revealed that an average distance of 14.39 Å and 14.38 Å exists for (MMP1 Zn1 Zn2) and (MMP1 Zn2), respectively (Fig. 3b), representing stable structures. However, an increase to average distances of 16.04 Å and 15.41 Å were observed with the removal of structural Zn(II) and both Zn(II) ions, respectively, from the enzyme•substrate complex with rapid fluctuations destabilizing the system without Zn(II) ions (Fig. 3b). The examination of average distance between the center of mass of the CAT and HPX domains in the enzyme•substrate complexes showed that the domains are at a distance of 38.94 Å in (MMP1 Zn1 Zn2) (Fig. 3c). Though some fluctuations were observed during the initial period of the trajectory, equilibration was observed at around 400 ns. The removal of each of the Zn(II) ions led to minor changes in the interdomain distance, 38.70 Å for (MMP1 Zn1) and 39.03 Å for (MMP1 Zn2) (Fig 3c), while the simultaneous removal of the both Zn(II) ions had slightly stronger effect (39.52 Å). The stability of the systems was also explored by measuring the distance between the center of mass of the catalytic site to the center of mass of the scissile bond (Fig. 3d). The distance in (MMP1 Zn1 Zn2) was stable throughout the simulation and averaged at 8.52 Å. An average distance of 8.56 Å was observed when the catalytic Zn(II) was removed, but with higher fluctuations. The system reached equilibrium after 600 ns with Zn(II) removed from the structural site and displayed an average distance of 9.23 Å. Large fluctuations were observed when the system was devoid of metal ions, with an averaged distance of 8.64 Å. (Fig. 3d).
Fig. 3.
Comparison of the overall MD properties of the simulated systems. a) RMSD, b) distance between the center of mass of catalytic Zn(II) and structural Zn(II) coordinated residues, c) distance between the center of mass of CAT and HPX domains, and d) distance between the center of mass of the catalytic site and the scissile bond. The simulated systems were MMP-1•THP complex with both the catalytic and structural Zn(II) present (MMP1 Zn1 Zn2) (black), without structural Zn(II) (MMP1 Zn1) (green), without catalytic Zn(II) (MMP1 Zn2) (red), and without both Zn(II) ions (MMP1 no Zn) (blue).
We further explored the effects of the Zn(II) ions on the conformation of the linker region. Radius of Gyration (RoG) of the linker for (MMP1 Zn1 Zn2) was 11.74 Å, which then slightly increased to 11.83 Å for (MMP1 Zn1), 11.88 Å for (MMP1 Zn2), and 11.88 Å for (MMP1 no Zn) (Fig. S4 (e)) indicating that the Zn(II) ions did not appear to have a remarkable influence on the RoG of the linker. The stability of the systems was extensively studied using additional calculations, including RMSDs and RMSFs of the domains, Ca(II) ions, loops, and substrate (Fig. S1, S2, and S3).
3.2. Effect of the Catalytic and the Structural Zn(II) Ions on the Local Hydrogen Bonding Interactions
To explore the effects of the catalytic and structural Zn(II) ions on the local interactions in the respective binding sites, we performed an analysis of the hydrogen-bonding networks.
3.2.1. Hydrogen bonding interactions in the catalytic Zn(II) site
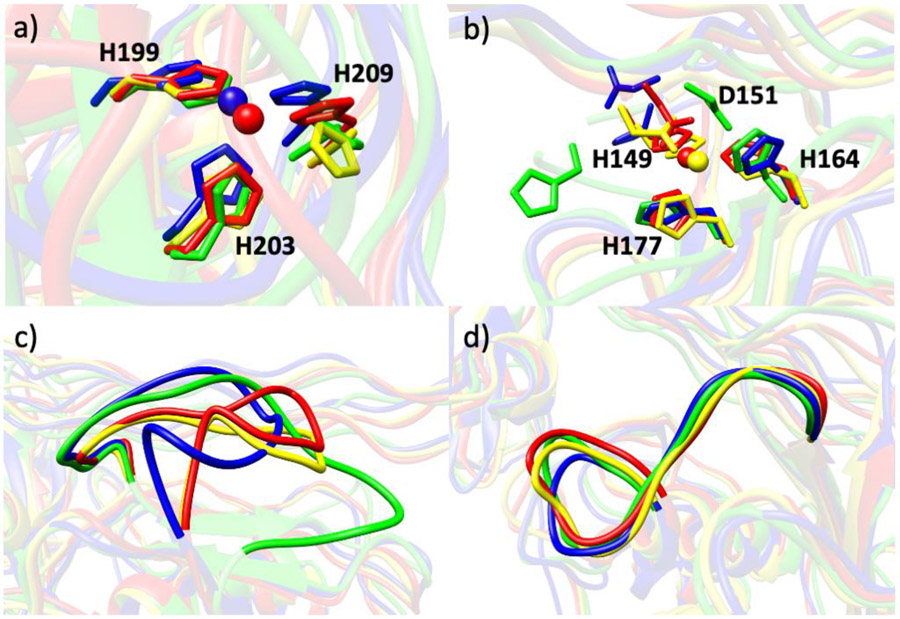
A considerable number of hydrogen bonding interactions were observed involving H203. As introduced earlier, H199, H203, and H209 are the residues bound to the catalytic Zn(II) (Fig. 4a). H203 was stabilized via hydrogen bonding interactions with E200 (37%), L207 (25%), and G206 (35%), respectively, in (MMP1 Zn1 Zn2), where the percentage denotes how often a hydrogen bond is present between the respective atoms during the total simulation time. E200 has a crucial role in the catalytic mechanism [76-78]. H199 interacts with L216 (69%) of the Met-turn, while H209 interacts with S208 (15%) through hydrogen bonding. These interactions may enhance the stability of the active site. With the removal of catalytic Zn(II) from the active site, new hydrogen bonding interactions arose between H199 and R195 (11%) and H199 and H203 (46%). The carboxylate oxygens of E200 were involved in hydrogen bonding with H203 (19%) and H199 (63%). H203 maintained its stabilizing interaction with G206 (26%) and H199 with L216 (12%) similarly to (MMP1 Zn1 Zn2). The H199 and R195 interaction (36%) persisted even with the removal of the structural Zn(II). The system was further investigated by removing the Zn(II) ions from both the catalytic and structural sites. Various new hydrogen bonding interactions were identified. H209 interacted with V82 of the N-terminal loop of the enzyme through a hydrogen bond (19%). The possibility exists that the enzyme complex without Zn(II) ions may result in an increase in fluctuations of the N-terminal loop leading the V82 residue to move closer to the active site. An interaction of E200 with H203 was observed, which was similar to (MMP1 Zn1 Zn2). An additional interaction in the system was the involvement of the E200 carboxylate oxygens with H199 (32% and 18%).
Fig. 4.
The superimposed (a) catalytic site, (b) structural site, (c) S-loop, and (d) V-B loop of (MMP1 Zn1 Zn2) (red), (MMP1 Zn1) (blue), (MMP1 Zn2) (yellow), and (MMP1 no Zn) (green).
3.2.2. Hydrogen bonding interactions in the structural Zn(II) site
We next examined interactions involving the structural Zn(II), coordinated to H149, D151, H164, and H177 (Fig. 4b). Two strong hydrogen bonding interactions existed between the peptide backbone nitrogen and oxygen atoms of H164 and H177 (89% and 58%) in (MMP1 Zn1 Zn2) (Fig. S5). The protonated ND1 of H164 made a hydrogen bond with the side chain carbonyl oxygen of Q774 (58%) of the T strand of the THP, where Q was the hydrogen bond acceptor. The backbone oxygen atom and carboxylate oxygen of S-loop residue D151 formed two hydrogen bonds with the backbone nitrogen of S153 (41% and 16%, respectively) of the same loop. It reduced to a single hydrogen bond interaction when the Zn(II) ion was removed from the catalytic site. The interaction between H164 and Q774 (68%) of the T strand was retained in (MMP1 Zn2) as in (MMP1 Zn1 Zn2). The distortion caused in the site by the removal of structural Zn(II) in (MMP1 Zn1) allowed the Zn(II) bonded residues to be engaged with the neighboring residues via hydrogen bonding. A new hydrogen bond was observed between D151 and R150 (29%). H149 formed a single hydrogen bond with R146 (29%) and two hydrogen bonds with the carboxylate oxygens of D148 of the S-loop (24% and 18%, respectively). V145 of βIII hydrogen-bonded with H177 (16%). Due to a conformational change in the structural site, the side chain of H164 flipped towards Q774, which in turn switched the atoms forming the hydrogen bond between H164 and Q774. In particular, the deprotonated NE2 of H164, which coordinated to the structural Zn(II) in the WT enzyme•substrate complex, becomes properly oriented to interact with the peptide backbone nitrogen atom of Q774 (10%). Therefore, Q774 changed its role from a hydrogen bond acceptor in (MMP1 Zn1 Zn2) as well as in (MMP1 Zn2) (Fig. S6) to a hydrogen bond donor in (MMP1 Zn1).
The interaction of H164 with H177 (81%) was maintained as in (MMP1 Zn1 Zn2). The results indicate that eliminating the structural Zn(II) from the enzyme promotes interaction between the S-loop residues and the β-sheets surrounding the structural Zn(II). Simulations of MMP-1 without substrate present (unpublished data) suggested key differences with respect to the MMP-1•THP complex that indicate the substrate influences the interactions in the catalytic Zn(II) site.
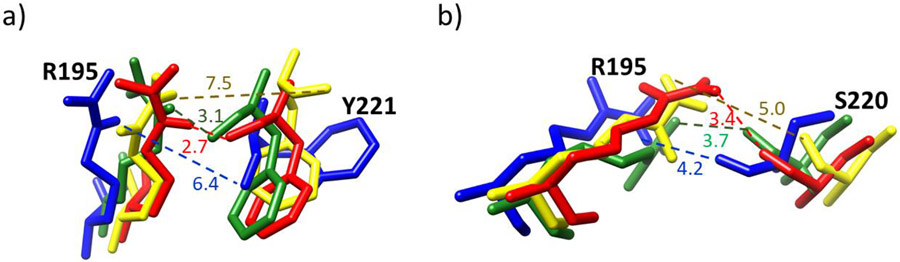
To explore in more detail the role of the structural Zn(II) in MMP-1, a further analysis identified interactions involving two key residues of the CAT domain, R195 and M217. R195 was hydrogen-bonded to Y221 (66%) located at the entrance of the specificity pocket (Fig. 5a and Fig. S7, S9). R195 was also involved in close interaction with S220 (20%) (Fig. 5b and Fig. S7). These interactions of the catalytic site might have a role in positioning the R195 near the pocket. The deformation of the S-loop by the elimination of the structural Zn(II) strongly weakened the interaction of R195 with Y221 and S220 in the CAT domain (Fig. 5 and Fig. S7, S9). The involvement of the structural Zn(II) in substrate selectivity has yet to be examined experimentally, although it was previously observed that removal of the structural Zn(II) reduced the catalytic activity of the MMP-1 CAT domain towards a synthetic substrate [56].
Fig. 5.
a) Superimposed distances (in Angstroms) between donor nitrogen (NE) of R195 with acceptor oxygen (O) of Y221 in (MMP1 Zn1 Zn2) (red), (MMP1 Zn1) (blue), (MMP1 Zn2) (yellow), and (MMP1 no Zn) (green). The R195 – Y221 hydrogen bond is lost when the Zn(II) ion is removed from the structural site. b) Distance between donor nitrogen (NH2) of R195 with acceptor oxygen (O) of S220 in (MMP1 Zn1 Zn2) (red), (MMP1 Zn1) (blue), (MMP1 Zn2) (yellow), and (MMP1 no Zn) (green).
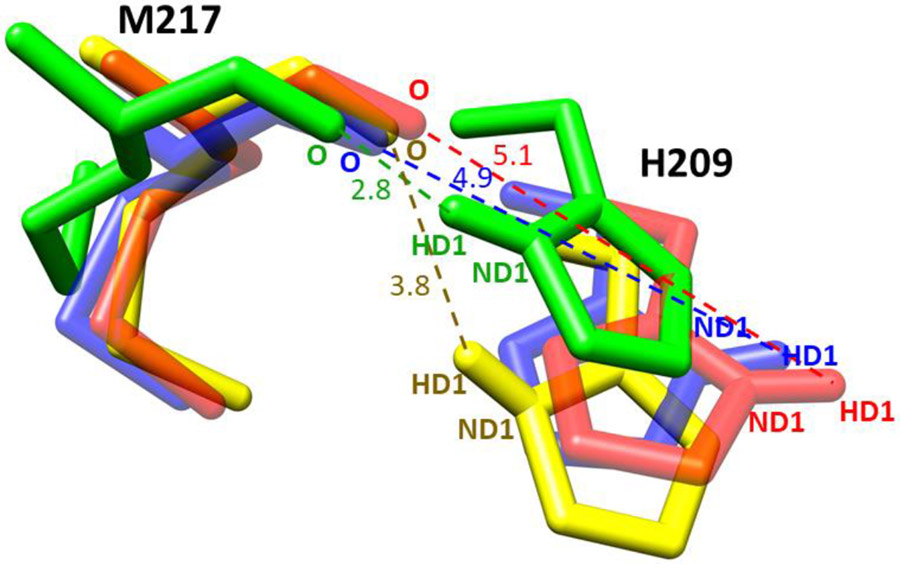
M217, which is a part of the Met-turn residues in the CAT domain, was not involved in hydrogen bonding interaction with any of the Zn(II) coordinating histidines in the active site of (MMP1 Zn1 Zn2) as well as (MMP1 Zn1) (Fig. 6 and Fig. S8). With the removal of the catalytic Zn(II), M217 moved closer to H209 (Fig. S10) and formed a flexible hydrogen bond (22%) when both of the Zn(II) ions were removed from the enzyme•substrate complex. This highlights the importance of both Zn(II) ions in preserving the structure of the catalytic site by maintaining the hydrophobic sheath formed by M217 of the Met-turn below the catalytic site. The results show that the structural integrity of the catalytic site depends not only on the catalytic Zn(II) but also on the structural Zn(II). A prior study of the MMP-1 CAT domain had indicated that catalytic activity was impacted by the structural Zn(II), with the suggestion that partial unfolding had occurred near the catalytic site when the structural Zn(II) was removed [56]. A prior MD study had indicated the importance of the structural Zn(II) in the MMP-2 CAT domain [55]. The polar interactions in the MMP-2 CAT domain Ω-loop were not perturbed by the removal of structural Zn(II), except for the A419-T426 H-bond [55]. The structural Zn(II) was found to restrain the conformation of solvent-exposed loop regions (βIV-βV connection loop and S-loop), allowing interaction with other structural elements [55]. The structural Zn(II) stabilized Y182, H193, and F195 of the S3 binding site, whereas the entrance to the S1' subsite was not impacted by the structural Zn(II) [55]. However, the prior MD study was performed without a THP present. A more recent study indicated that the presence of a THP influences the dynamics of MMP-1 [30].
Fig. 6.
Superimposed distances (in Angstroms) between the acceptor oxygen (O) of M217 and donor nitrogen (ND1) of catalytic site residue H209 in (MMP1 Zn1 Zn2) (red), (MMP1 Zn1) (blue), (MMP1 Zn2) (yellow), and (MMP1 no Zn) (green). Hydrogen bond formation takes place when both of the Zn(II) ions are removed from the system, which may alter the structural orientation of the catalytic site.
The structural Zn(II) together with a Ca(II) ion constrains the S-loop [53]. The S-loop of (MMP1 Zn1 Zn2) is located above the catalytic cleft (Fig. 2). H149 and H164 coordinated to the Zn(II) have a stabilizing π stacking interaction with F155 of the S-loop. W184, which is part of the V-B loop, was stabilized by hydrophobic interaction with L162. R146 of the S-loop formed four hydrogen bonds with D181 and a single hydrogen bond with E180. D181 and E180 are part of the V-B loop. E182 hydrogen-bonded with G159 of the S-loop.
Removal of both Zn(II) ions from the MMP-1•THP complex resulted in D151 forming four hydrogen bonds with R146 (27%, 24%, 22%, and 18%) and three hydrogen bonds with S153 of the S-loop. The hydrogen bond of H164 with the trailing (T) strand of the THP remained the same (10%) as in (MMP1 Zn1). The S-loop became highly flexible with the removal of both Zn(II) ions from the catalytic domain (Fig. 4c). This likely induces both correlated and anticorrelated motions with different residues from the N-terminal loop leading V82 to form a hydrogen bond with H209. The participation of the same loop in forcing the hydrophobic methionine residue to engage in a hydrogen bonding interaction with histidine is suspected.
3.3. Effects of the Catalytic and the Structural Zn(II) Ions on the Long-Range Correlated Motions in the MMP-1•THP Complex
3.3.1. Collective Correlated Motions in the MMP1 Zn1 Zn2 system
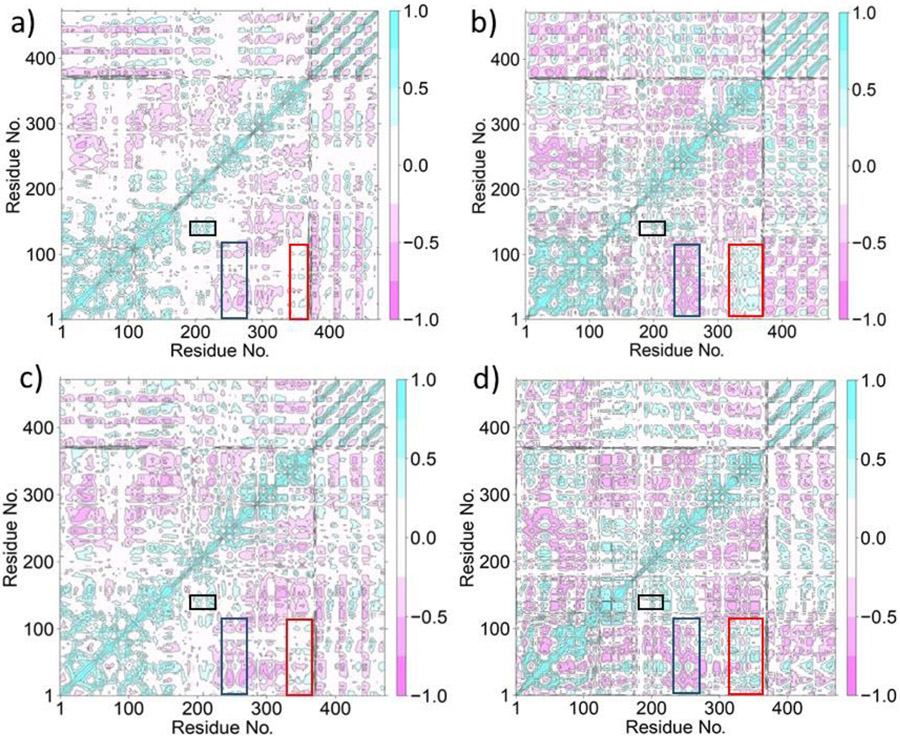
The effects of the catalytic and the structural Zn(II) ions on the long-range collective correlated motions in the MMP-1•THP complex were analyzed using Dynamic Cross-Correlation Analysis (DCCA). DCCA of MMP1 Zn1 Zn2 (Fig. 7a) revealed that the specificity loop showed a correlated motion with residues of the HPX blade 1 region. This correlation supports the finding that the binding of blade 1 residues of the HPX domain with triple-helical substrates facilitates the binding of the CAT domain to the substrate through the linker [26]. Additionally, previous studies have demonstrated an increase in collagenase activity with the mutation of blade 2 residues [28,32]. Residues 302-308 of blade 2 show correlated interactions with the specificity loop, whereas residues 309-347 show no correlation, and residues 349-350 were anticorrelated to the specificity loop. Blade 2 residues (323, 327-333, 337-352) show anti-correlation to the V-B loop (residues 180-184). Allosteric interactions between the HPX and CAT domains, facilitated by the linker and triple-helical substrate, were previously described to result in the opening of the MMP-1 catalytic pocket, as measured by the distance between N171 and T230 [30] (respectively N152 and T211 in our simulations).
Fig. 7.
Dynamic Cross-Correlation Analysis of MMP-1•THP. a) (MMP1 Zn1 Zn2), b) (MMP1 Zn1), c) (MMP1 Zn2), and d) (MMP1 no Zn). The positive correlations (from 0 to 1.0) between residues are shown in cyan color and negative correlations (from 0 to −1.0) are represented in pink color. The boxes in the figure show the following important motions: Correlated motion between the S1' specificity loop and blade 1 residues (black), anticorrelated motion between the blade 2 residues and the CAT domain involving S-loop and V-B loop residues (blue), correlated motion between blade 4 residues and the CAT domain involving S-loop and V-B loop residues (red). Residues 1-162 (crystal structure numbers 81-242) denote the CAT domain, 179-367 (crystal structure numbers 259-447) denote the HPX domain, and 372-472 (crystal structure numbers 963-995) denotes the THP substrate.
The S-loop is maintained by the structural Zn(II), and thus the structural Zn(II) exhibited a strong positive correlation to the loop residues. Importantly, other than the structural Zn(II), S-loop residues demonstrated positive correlation with the catalytic Zn(II). Notably, both the Zn(II) ions were positively correlated with the V-B loop (residues 180-185 and 191-192). The only residue of the V-B loop anticorrelated to the structural Zn(II) was R189. Residues 187-189 of the V-B loop have anti-correlation to residues 149-154 of the S-loop, whereas residues 180-184 of the V-B loop have a positive correlation to the S-loop. The various correlated and anticorrelated motions with the S-loop may be incumbent for the functioning of the V-B loop (Fig. 4d). Mutation of the RWTNNFREY sequence of the V-B loop abated enzymatic activity by 2-fold [54]. Thus, the loop has a crucial role in the collagenolytic activity of MMP-1. The linker of MMP-1 has a glycine residue (G252) which is conserved in all type I collagenases [79]. This residue demonstrated a positive correlation with the catalytic Zn(II) coordinated histidines and the S1' specificity loop. G775 and L776 of the scissile peptide bond showed anticorrelated motions with the structural Zn(II). The correlations involving the THP showed that residues 777-783 of the L strand were involved in anti-correlation with residues 763-770 of the same strand. Correlated motions were coupled with anticorrelated motions within the same strand. This trend was seen in M and T strands as well. From this distinct pattern within the THP, it can be concluded that the region of THP proximal to the CAT domain behaves differently from the region of the strands near the HPX domain. This may facilitate disruption of the triple-helix near the active site, which is needed for efficient collagenolysis, while an intact triple-helix is favorable for binding to the HPX domain [25].
3.3.2. Correlated Motions in MMP1 Zn1
We further delineated the impact of structural Zn(II) on the correlated dynamics of the MMP-1•THP complex (Fig. 7b). The correlated movements of the S1' specificity loop with the S10' exosite were less compared to the native MMP-1 system, but a greater number of residues were involved in correlated motion in comparison to (MMP1 Zn2), discussed in section 3.3.3. Both of the Zn(II) ions, and the catalytic Zn(II) in particular, were essential for maintaining this positive correlation. A new negative correlation was observed with residues 314-322 and 333-338 of HPX blade 2 and the S-loop. A larger segment from blade 4 was involved in a positive correlation to the S-loop and the V-B loop. Consequently, a significant number of intense anticorrelated motions involving blade 2 residues (315-323 and 331-341) with the V-B loop and S-loop residues were noted. Linker residues 243 and 244 expressed negative correlation to the S-loop residues. Anticorrelated movements also arose between residues 223 and 224 of the specificity loop and the S-loop. S-loop residues demonstrated negative correlation to residues 214-217 located near the S1' subsite. Anticorrelated movements in the THP were mainly observed within the middle portion and tail of each strand. The terminal residues of each THP strand did not seem to be involved in negative correlations among themselves. S-loop residues (148-160) displayed no correlation with residues 763-774 of the THP L strand in contrast to the anticorrelated movements in (MMP1 Zn1 Zn2). When the THP residues were not involved in correlation with the loop, they tended to lose their anticorrelating properties with other residues of the same strand. This is likely to affect the interaction of the substrate with the respective CAT and HPX domains, which may ultimately influence catalysis.
3.3.3. Correlated Motions in MMP1 Zn2
DCCA of the system with the catalytic Zn(II) removed show some interesting differences compared to the wild-type enzyme (Fig. 7c). The initial part of the S10' exosite was involved in a positive correlation with the specificity loop in comparison to the (MMP1 Zn1 Zn2) system. Importantly, the structural Zn(II) maintained a strong correlation with S-loop residues. A part of this loop, including residues 148-157, was involved in anticorrelated movements with the βII of the CAT domain. The anticorrelated motions of blade 2 residues with the V-B loop intensified upon removal of the catalytic Zn(II) ion, indicating that the catalytic Zn(II) stabilizes the interaction between the V-B loop and HPX blade 2. The development of intense anticorrelated motions can be a result of an increase in correlated movements of the blade 4 residues of the HPX domain with the S-loop and the V-B loop. Residues 243-252 of the linker expressed a negative correlation with S-loop residues. G252 of the linker retained its positive correlation with the S1' specificity loop [79]. The anticorrelated motions of the V-B loop with the S-loop in (MMP1 Zn1 Zn2) were highly reduced when the system lacked the catalytic Zn(II), suggesting the necessity of the proper orientation of the catalytic site with the presence of Zn(II). The removal of catalytic Zn(II) also induced changes in the N-terminal loop residues (residues 84-87). This loop is proximal to the catalytic cleft. The distortion in the active site region with the removal of the Zn(II) ion might have triggered the loop residues to be involved in anticorrelated movements with the V-B loop. The terminal loop (residues 81-87) was also engaged in anti-correlation with residues 102-110 of the connecting loop of β-sheet I and α-helix 1, and loop residues 88-91 have anti-correlation with residues 262-264 of HPX blade 1. Accordingly, a significant number of random correlations were observed in this system. THP regions that expressed no correlation with the HPX domain in (MMP1 Zn1 Zn2) now exhibited correlated motions. For example, residues 785-792 of the L strand positioned near the HPX domain moved in positive correlation with residues 333-335 of blade 2. The same strand residues 788-792 showed a positive correlation to 318-320 of HPX blade 2. The anticorrelating characteristics within the same THP strand as observed in (MMP1 Zn1 Zn2) were observed in this system, too. The THP M and T strands demonstrate the same trend as the L strand.
3.3.4. Combined Effect of the Catalytic Zn(II) ion and the Structural Zn(II) ion on the Correlated Motions in the MMP-1•THP Complex
The majority of the correlated motions observed when the catalytic Zn(II) and structural Zn(II) were removed individually were observed in the system without any Zn(II) ions (Fig. 7d). A lower segment of the S10' exosite residues were involved in positive correlation with the S1' specificity loop. Anticorrelated movements were observed with the HPX blade 2 residues and the S-loop, similar to the system without the structural Zn(II). The S-loop maintained positive correlation with the V-B loop. However, some anticorrelated movements were observed with V-B loop residues 186-189 and S-loop residues 149-152. Removal of Zn(II) ions mainly affected the N-terminal loop residues of the enzyme. Residues 83-87 of the loop were anticorrelated to residues 94-99 of β-sheet I and 129-133 of β-sheet II. The α-helix 1 exhibited strong anti-correlation to residues 156 and 157 of the S-loop. The most striking difference observed upon removing zinc ions from the system was that the terminal residues of L, M, and T strands of the THP expressed no correlation with residues 93-380 of the CAT and HPX domains. These include the S1'specificity pocket as well as the S10' exosite.
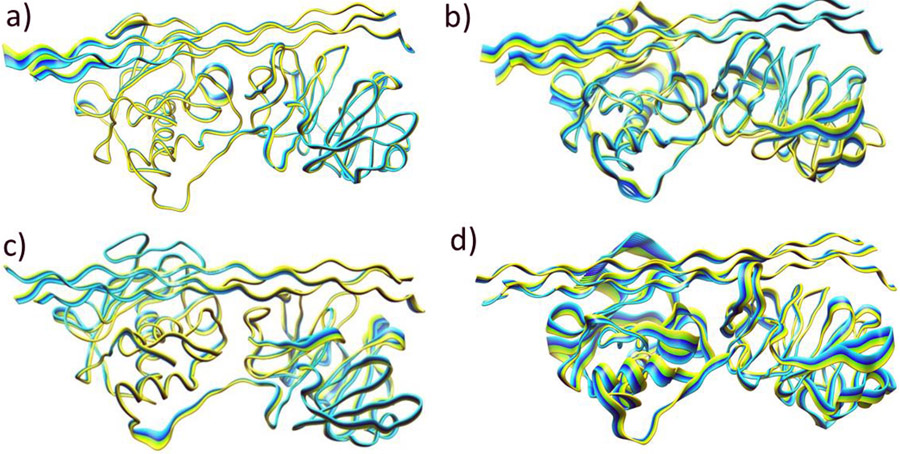
3.3.5. Effect of the Zn(II) Ions on the Main Direction of Movements
The effect of the Zn(II) ions on the main direction of the movements in the MMP-1•THP complex was further explored using PCA on PC1. PCA plots show the main direction of the fluctuations in the residues of the enzyme. Overall, the PCA confirmed and complemented the results from the DCCA. Both the CAT and HPX domains showed minimal movement in the (MMP1 Zn1 Zn2) system (Fig. 8a). The direction of motion of the THP region containing the scissile bond was towards the CAT domain. Flexible motions were observed in the V-B loop. The domains influenced the linker movements, i.e., the part of the linker proximal to the CAT domain moved in the same direction as the domain.
Fig. 8.
Principal Component Analysis (PC1) of MMP-1•THP. a) (MMP1 Zn1 Zn2), b) (MMP1 Zn1), c) (MMP1 Zn2), and d) (MMP1 no Zn). Yellow to blue is the direction of motion of the protein.
Domain motions were rather more substantial for the enzyme•substrate complexes without the structural Zn(II) (Fig. 8b). The CAT domain elements tended to move in different directions, both towards the THP and away from it. In contrast to (MMP1 Zn1 Zn2), flexible motions were observed in HPX domain of (MMP1 Zn1) system. Elimination of Zn(II) from the structural site induced more flexibility to the linker region near the CAT domain and resulted in the movement of linker residues away from the same domain. Induced flexibility in the linker region could affect interdomain communications. The THP strands located near the CAT domain were directed away from the domain, and the THP strands near the HPX domain moved towards the domain. Thus, different regions of the same strand moved in different directions. Motions were also observed with the terminal residues of the THP near the CAT domain.
The HPX domain showed increased flexible motions compared to the CAT domain when the system is without the catalytic Zn(II) (Fig. 8c). As observed in (MMP1 Zn1 Zn2), the CAT domain movements were towards the peptide, and the exosite moved in the direction of the CAT domain. Hence, it is assumed that the catalytic Zn(II) does not induce directional changes in the exosite. However, some regions displayed more flexibility. Linker motions were similar to (MMP1 Zn1 Zn2), but probably because of the change in the catalytic site caused by the elimination of the catalytic Zn(II), linker residues positioned near the catalytic domain were highly flexible. A change in the direction of motion of the S-loop was observed. S-loop residues now moved in the direction of the HPX domain and not the CAT domain.
Elimination of both the metals from MMP-1 made the system highly unstable (Fig. 8d). Intensive motions were observed in the CAT domain and especially in the S-loop. The domain tended to move away from the THP. The S-loop underwent large fluctuations. The HPX domain moved in the direction opposite to that of the CAT domain. Minimal movement was observed in HPX blade 1 and blade 2 residues. The linker remained flexible, and the region positioned near the CAT domain moved in the direction of the HPX domain and vice-versa. Thus, the structural Zn(II) plays a major role in the directional orientation of the linker. Reduced movements were seen in the THP strands, but they showed movement in different directions similar to the (MMP1 Zn1) system.
4. CONCLUSIONS
The studies highlighted the distinct importance of the catalytic and structural Zn(II) ions in a synergy with the enzyme and substrate flexibilities for the overall stability of the MMP-1•THP Complex. The local interactions within the enzyme's binding site unveiled the influence of structural Zn(II) on the catalytic site. To summarize, the results identified R195 and M217 as important residues for conserving the active site's integrity. Removing the catalytic Zn(II) from the active site resulted in new hydrogen bonding interactions involving the coordinated histidines, i.e., H199 was hydrogen-bonded to H203. Lack of Zn(II) in the catalytic site would leave the enzyme inactive as collagen's degradation occurs by the respective peptide bond's cleavage via a tetra-coordinated Zn(II) intermediate [22]. Analysis of the correlated motions in (MMP1 Zn1), (MMP1 Zn2), and (MMP1 no Zn) systems showed they could not completely retain the positive correlation between the S10' subsite and the S1' specificity loop. Thus, both Zn(II) ions are necessary for maintaining communication between the exosite and subsite for effective catalysis. The appearance of anticorrelated motions of the blade 2 residues with the V-B loop and S-loop when the system lacks catalytic Zn(II) may be attributed to a long-range interaction of the V-B and S-loop with the blade 4 residues. Increased flexible motions in the HPX domain, as observed in the PCA, support our finding that the catalytic site might be involved in long-range interactions with the HPX domain's residues. This would be consistent with experimental studies [25,29-30]. Removal of the structural Zn(II) from the CAT domain had a surprising effect on the catalytic site. The structural Zn(II) affected the interactions of R195, which serves as a "gatekeeper" in the MMP-1 S1' subsite and impacts substrate selectivity [45-46]. As was observed in the (MMP1 Zn1) system, the HPX blade 2 residues demonstrated anti-correlation to the V-B and S-loop residues, with the correlations being intense. There was an increased number of correlated motions in the HPX blade 4 residues with the loops. Blade 4 residues may be necessary for the effective functioning of the Zn(II) ions. Since the S-loop is maintained by the structural Zn(II), removing the metal ion from the enzyme would impact the loop interactions. Consequently, the S-loop loses the anticorrelating property with the triple-helical substrate. Instability in the CAT and HPX domain movements was observed in the PCA Analysis. Removing both Zn(II) ions renders the system highly unstable. The THP lost its correlating property with the S1' subsite and the S10' exosite. The impact was also observed in the interactions involving the N-terminal loop. The motions were intense in the CAT domain, and the S-loop became flexible. Even though the structural Zn(II) maintains the S-loop, removing both the Zn(II) ions from the system induced more flexible motions in the S-loop. A minor role of the catalytic Zn(II) in the stability of the S-loop is suspected.
The study asserts the crucial contributions of the catalytic and structural Zn (II) ions on the log-range correlated dynamics involving not only the CAT domain but also the HPX domain and the THP substrate. The results advocate about shifting the paradigm about the local effects of the Zn (II) ions with understanding about their distinct impact in the long-range dynamics and stability of the ES complex. Our study is intended to stimulate the overall interest in investigating the role of Zn(II) ions in metalloenzyme reaction mechanisms and further demonstrates the power of computational molecular physics (CMP) for an in-depth understanding of the interrelationships between enzyme catalytic mechanisms, dynamics and function [80].
Supplementary Material
Acknowledgments.
This research was supported by NIH grant GM132873 (to TKC and GBF).
Footnotes
The authors declare no competing financial interest.
REFERENCES
- 1.Piez KA, Eigner EA, and Lewis MS (1963) The Chromatographic Separation and Amino Acid Composition of the Subunits of Several Collagens. Biochemistry 2:58–66 [Google Scholar]
- 2.Gallop PM, Blumenfield OO, Seifter S (1972) Structure and Metabolism of Connective tissue proteins. Ann Rev Biochem 41:617–672 [DOI] [PubMed] [Google Scholar]
- 3.Miller EJ (1971) Isolation and Characterization of the Cyanogen Bromide Peptides from the α1(II) Chain of Chick Cartilage Collagen. Biochemistry 10:3030–3034 [DOI] [PubMed] [Google Scholar]
- 4.Trelstad RL, Kang AH, Toole BP, Gross J (1972) Collagen Heterogeneity: High resolution separation of native [α1(I)]2α2 and [α1(II)]3 and their component α chains. J Biol Chem 247:6469–6473 [PubMed] [Google Scholar]
- 5.Chung E, Miller EJ (1974) Collagen Polymorphism: Characterization of Molecules with the Chain Composition [α1(III)]3 in Human Tissues. Science 183:1200–1201 [DOI] [PubMed] [Google Scholar]
- 6.Trelstad RL (1974) Human aorta collagens: Evidence for three distinct species. Biochem Biophys Res Commun 57:717–725 [DOI] [PubMed] [Google Scholar]
- 7.Epstein EH, Munderloh NH (1975) Isolation and characterization of CNBr peptides of human (alpha 1 (III)) 3 collagen and tissue distribution of (alpha 1 (I)) 2 alpha 2 and (alpha 1 (III)) 3 collagens. J Biol Chem 250:9304–9312 [PubMed] [Google Scholar]
- 8.Butler WT, Birkendal-Hansen H, Beegle WF, Taylor RE, Chung E (1975) Proteins of the periodontium. Identification of collagens with the [alpha1(I)]2alpha2 and [alpha1(III)]3 structures in bovine periodontal ligament. J Biol Chem 250:8907–8912 [PubMed] [Google Scholar]
- 9.Seyer JM, Hutcheson ET, Kang AH (1978) Chemotactic attraction of human fibroblasts to type I, II, and III collagens and collagen-derived peptides. Proc Natl Acad Sci USA 75:871–875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Van Der Rest M, Garrone R (1991) Collagen Family of Proteins. FASEB J 5:2814–2823 [PubMed] [Google Scholar]
- 11.Nalinanon S, Benjakul S, Kishimura H, Osako K (2011) Type I collagen from the skin of ornate threadfin bream (Nemipterus hexodon): Characteristics and effect of pepsin hydrolysis. Food Chem 125:500–507 [Google Scholar]
- 12.Myllyharju J (2004) Molecular biology and biosynthesis of collagens. In: Massaro EJ and Rogers JM (ed) The Skeleton: Biochemical, Genetic, and Molecular Interactions in Development and Homeostasis, Humana Press, Totowa [Google Scholar]
- 13.Foegeding EA, Lanier TC, Hultin HO (1996) Characteristics of edible muscle tissue. In: Fennema OR (Ed) Food Chemistry Marcel Dekker, Inc; New York, USA [Google Scholar]
- 14.Marneros AG, Olsen BR (2001) The Role of Collagen-Derived Proteolytic Fragments in Angiogenesis. Matrix Biol 20:337–345 [DOI] [PubMed] [Google Scholar]
- 15.Streuli C (1999) Extracellular matrix remodelling and cellular differentiation. Curr Opin Cell Biol 11:634–640 [DOI] [PubMed] [Google Scholar]
- 16.Folgueras AR, Pendás AM, Sánchez LM, López-Otín C (2004) Matrix Metalloproteinases in Cancer: From New Functions to Improved Inhibition Strategies. Int J Dev Biol 48:411–424 [DOI] [PubMed] [Google Scholar]
- 17.Rosenberg GA (2009) Matrix metalloproteinases and their multiple roles in neurodegenerative diseases Lancet Neurol 8:205–216 [DOI] [PubMed] [Google Scholar]
- 18.Spinale FG (2002) Matrix metalloproteinases: regulation and dysregulation in the failing heart. Circ Res 90:520–530 [DOI] [PubMed] [Google Scholar]
- 19.Burrage PS, Mix KS, Brinckerhoff CE (2006) Matrix metalloproteinases: role in arthritis. Front Biosci 11:529–543 [DOI] [PubMed] [Google Scholar]
- 20.Page-McCaw A, Ewald AJ, Werb Z (2007) Matrix metalloproteinases and the regulation of tissue remodelling. Nat Rev Mol Cell Biol 8:221–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fields GB (2013) Interstitial collagen catabolism. J Biol Chem 288:8785–8793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gomis-Ruth FX (2003) Structural aspects of the metzincin clan of metalloendopeptidases. Mol. Biotechnol 24:157–202 [DOI] [PubMed] [Google Scholar]
- 23.Li J, Brick P, O'Hare MC, Skarzynski T, Lloyd LF, Curry VA, Clark IM, Bigg HF, Hazleman BL, Cawston TE, Blow DM (1995) Structure of full-length porcine synovial collagenase reveals a C-terminal domain containing a calcium-linked, four-bladed β-propeller. Structure 3:541–549 [DOI] [PubMed] [Google Scholar]
- 24.Nagase H, Woessner JF (1999) Matrix metalloproteinases. J Biol Chem 274:21491–21494 [DOI] [PubMed] [Google Scholar]
- 25.Bertini I, Fragai M, Luchinat C, Melikian M, Toccafondi M, Lauer JL, Fields GB (2012) Structural Basis for Matrix Metalloproteinase 1-Catalyzed Collagenolysis. J Am Chem Soc 134:2100–2110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Arnold LH, Butt L, Prior SH, Read C, Fields GB, Pickford AR (2011) The interface between catalytic and hemopexin domains in matrix metalloproteinase 1 conceals a collagen binding exosite. J Biol Chem 286:45073–45082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cerofolini L, Fields GB, Fragai M, Geraldes CFGC, Luchinat C, Parigi G, Ravera E, Svergun DI, Teixeira JMC (2013) Examination of Matrix Metalloproteinase-1 in Solution: A Preference for the Pre-Collagenolysis State. J Biol Chem 288:30659–30671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Manka SW, Carafoli F, Visse R, Bihan D, Raynal N, Farndale RW, Murphy G, Enghild JJ, Hohenester E, Nagase H (2012) Structural insights into triple-helical collagen cleavage by matrix metalloproteinase 1. Proc Natl Acad Sci USA 109:12461–12466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lauer-Fields JL, Chalmers MJ, Busby SA, Minond D, Griffin PR, Fields GB (2009) Identification of specific hemopexin-like domain residues that facilitate matrix metalloproteinase collagenolytic activity. J Biol Chem 284:24017–24024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kumar L, Nash A, Harms C, Planas-Iglesias J, Wright D, Klein-Seetharaman J, Sarkar SK (2020) Allosteric communications between domains modulate the activity of matrix metalloprotease-1. Biophys J 119:360–374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Karabencheva-Christova TG, Christov CZ, Fields GB (2018) Conformational dynamics of matrix metalloproteinase-1•Triple-helical peptide complexes. J Phys Chem B 122:5316–5326 [DOI] [PubMed] [Google Scholar]
- 32.Singh W, Fields GB, Christov CZ, Karabencheva-Christova TG (2016) Effects of mutations on structure-function relationships of matrix metalloproteinase-1. Int J Mol Sci 17:1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chung L, Dinakarpandian D, Yoshida N, Lauer-Fields JL, Fields GB, Visse R, Nagase H (2004) Collagenase unwinds triple-helical collagen prior to peptide bond hydrolysis. EMBO J 23:3020–3030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Manka SW, Brew K (2020) Thermodynamic and Mechanistic Insights into Coupled Binding and Unwinding of Collagen by Matrix Metalloproteinase 1. J Mol Biol 432:5985–5993 [DOI] [PubMed] [Google Scholar]
- 35.Terp GE, Cruciani G, Christensen IT, Jørgensen FS (2002) Structural Differences of Matrix Metalloproteinases with Potential Implications for Inhibitor Selectivity Examined by the GRID/CPCA Approach. J Med Chem 45:2675–2684 [DOI] [PubMed] [Google Scholar]
- 36.Bode W, Reinemer P, Huber R, Kleine T, Schnierer S, Tschesche H (1994) The X-ray crystal structure of the catalytic domain of human neutrophil collagenase inhibited by a substrate analogue reveals the essentials for catalysis and specificity. EMBO J 13:1263–1269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Salowe SP, Marcy AI, Cuca GC, Smith CK, Kopka IE, Hagman WK, Hermes JD (1992) Characterization of zinc-binding sites in human stromelysin-1: stoichiometry of the catalytic domain and identification of a cysteine ligand in the proenzyme. Biochemistry 31:4535–4540 [DOI] [PubMed] [Google Scholar]
- 38.Gupta SP (2012) Matrix metalloproteinase inhibitors: specificity of binding and structure-activity relationships. Exper Suppl 103:35–57 [PubMed] [Google Scholar]
- 39.Babine RE, Bender SL (1997) Molecular recognition of protein-ligand complexes: applications to drug design. Chem Rev 97:1359–1472 [DOI] [PubMed] [Google Scholar]
- 40.Abramowi N, Schechter I, Berger A (1967) On the size of the active site in proteases II. Carboxypeptidase-A. Biochem Biophys Res Commun 29:862–867 [DOI] [PubMed] [Google Scholar]
- 41.Wetmore DR, Hardman KD (1996) Roles of the propeptide and metal ions in the folding and stability of the catalytic domain of stromelysin (matrix metalloproteinase 3). Biochemistry 35:6549–6558 [DOI] [PubMed] [Google Scholar]
- 42.Tallant C, Marrero A, Gomis-Rüth FX (2010) Matrix metalloproteinases: Fold and function of their catalytic domains. Biochim Biophys Acta 1803:20–28 [DOI] [PubMed] [Google Scholar]
- 43.Bertini I, Calderone V, Fragai M, Luchinat C, Maletta M, Yeo KJ (2006) Snapshots of the Reaction Mechanism of Matrix Metalloproteinases. Angew Chem Int Ed 45:7952–7955 [DOI] [PubMed] [Google Scholar]
- 44.Aureli L, Gioia M, Cerbara I, Monaco S, Fasciglione GF, Marini S, Ascenzi P, Topai A, Coletta M (2008) Structural bases for substrate and inhibitor recognition by matrix metalloproteinases. Curr Med Chem 15:2192–2222 [DOI] [PubMed] [Google Scholar]
- 45.Borkati N, Winkler FK, Williams DH, D'Arcy A, Broadhurst MJ, Brown PA, Johnson WH, Murray EJ (1994) Structure of the catalytic domain of human fibroblast collagenase complexed with an inhibitor. Struct Biol 1:106–l 10 [DOI] [PubMed] [Google Scholar]
- 46.Lovejoy B, Cleasby A, Hassell AM, Longley K, Luther MA, Weigl D, McGeehan G, McElroy AB, Drewry D, Lambert MH, Jordan SR (1994) Structure of the catalytic domain of fibroblast collagenase complexed with an inhibitor. Science 263:375–377 [DOI] [PubMed] [Google Scholar]
- 47.Lovejoy B, Welch AR, Carr S, Luong C, Broka C, Hendricks RT, Campbell JA, Walker KAM, Martin R, Van Wart H, Browner MF (1999) Crystal structures of MMP-1 and −13 reveal the structural basis for selectivity of collagenase inhibitors. Nat Struct Biol 6:217–221 [DOI] [PubMed] [Google Scholar]
- 48.Bode W, Fernandez-Catalan C, Tschesche H, Grams F, Nagase H, Maskos K (1999) Structural properties of matrix metalloproteinases. Cell Mol Life Sci 55:639–652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bode W, Gomis-Ruth FX, Stockler W (1993) Astacins, serralysins, snake venom and matrix metalloproteinases exhibit identical zinc-binding environments (HEXXHXXGXXH and Met-turn) and topologies and should be grouped into a common family, the 'metzincins'. FEBS Lett 331:134–140 [DOI] [PubMed] [Google Scholar]
- 50.Stöcker W, Grams F, Baumann U, Reinemer P, Gomis-Rüth FX, McKay DB, Bode W (1995) The metzincins—topological and sequential relations between the astacins, adamalysins, serralysins, and matrixins (collagenases) define a superfamily of zinc-peptidases. Protein Sci 4:823–840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Iyer S, Visse R, Nagase H, Acharya KR (2006) Crystal Structure of an Active Form of Human MMP-1. J Mol Biol 362:78–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Massova I, Kotra LP, Mobashery S (1998) Structural insight into the binding motifs for the calcium ion and the non-catalytic zinc in matrix metalloproteases. Bioorg Med Chem Lett 8:853–858 [DOI] [PubMed] [Google Scholar]
- 53.Maskos K (2005) Crystal structures of MMPs in complex with physiological and pharmacological inhibitors. Biochimie 87:249–263 [DOI] [PubMed] [Google Scholar]
- 54.Chung L, Shimokawa K, Dinakarpandian D, Grams F, Fields GB, Nagase H (2000) Identification of the RWTNNFREY-(183-191) region as a critical segment of matrix metalloproteinase 1 for the expression of collagenolytic activity. J Biol Chem 275:29610–29617 [DOI] [PubMed] [Google Scholar]
- 55.Diaz N, Suarez D (2007) Molecular dynamics simulations of matrix metalloproteinase 2: role of the structural metal ions. Biochemistry 46:8943–8952 [DOI] [PubMed] [Google Scholar]
- 56.Yang H, Makaroff K, Paz N, Aitha M, Crowder MW, Tierney DL (2015) Metal ion dependence of the matrix metalloproteinase-1 mechanism. Biochemistry 54:3631–3639 [DOI] [PubMed] [Google Scholar]
- 57.Lewis JC (2019) Beyond the Second Coordination Sphere: Engineering Dirhodium Artificial Metalloenzymes To Enable Protein Control of Transition Metal Catalysis. Acc Chem Res 52:576–584 [DOI] [PubMed] [Google Scholar]
- 58.Lee J, Goodey NM (2011) Catalytic Contributions from Remote Regions of Enzyme Structure. Chem Rev 111:7595–7624 [DOI] [PubMed] [Google Scholar]
- 59.Guex N, Peitsch MC (1997) SWISS-MODEL and the Swiss-Pdb Viewer: an environment for comparative protein modeling. Electrophoresis 18:2714–2723 [DOI] [PubMed] [Google Scholar]
- 60.Bas DC, Rogers DM, Jensen JH (2008) Very fast prediction and rationalization of pKa values for protein–ligand complexes. Proteins 73:765–783 [DOI] [PubMed] [Google Scholar]
- 61.Maier JA, Martinez C, Kasavajhala K, Wickstrom L, Hauser K, Simmerling C (2015) ff14SB: Improving the Accuracy of Protein Side Chain and Backbone Parameters from ff99SB. J Chem Theory Comput 11:3696–3713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Peters MB, Yang Y, Wang B, Füsti-Molnar L, Weaver MN, Merz KM (2010) Structural Survey of Zinc-Containing Proteins and Development of the Zinc AMBER Force Field (ZAFF) J Chem Theory Comput 6:2935–2947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Li P, Merz KM (2016) MCPB. Py: A Python Based Metal Center Parameter Builder. J Chem Inf Model 56:599–604 [DOI] [PubMed] [Google Scholar]
- 64.Seminario JM (1996) Calculation of intramolecular force fields from second-derivative tensors. Int J Quantum Chem 60:1271–1277 [Google Scholar]
- 65.Cieplak P, Cornell WD, Bayly C, Kollman PA (1995) Application of the Multimolecule and Multiconformational RESP Methodology to Biopolymers: Charge Derivation for DNA, RNA, and Proteins. J Comput Chem 16:1357–1377 [Google Scholar]
- 66.Case DA, Ben-Shalom IY, Brozell SR, Cerutti DS, Cheatham T, Cruzeiro VWD, Darden TA, Duke RE, Ghoreishi D, Gilson MK, Gohlke H, Goetz AW, Greene D, Harris R, Homeyer N, Izadi S, Kovalenko A, Kurtzman T, Lee TS, LeGrand S, Li P, Lin C, Liu J, Luchko T, Luo R, Mermelstein DJ, Merz KM, Miao Y, Monard G, Nguyen C, Nguyen H, Omelyan I, Onufriev A, Pan F, Qi R, Roe DR, Roitberg A, Sagui C, Schott-Verdugo S, Shen J, Simmerling CL, Smith J, Salomon-Ferrer R, Swails J, Walker RC, Wang J, Wei H, Wolf RM, Wu X, Xiao L, et al. (2018) AMBER 2018 [Google Scholar]
- 67.Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML (1983) Comparison of simple potential functions for simulating liquid water. J Chem Phys 79:926–935 [Google Scholar]
- 68.Davidchack RL, Handel R, Tretyakov MV (2009) Langevin Thermostat for Rigid Body Dynamics. J Chem Phys 130:234101. [DOI] [PubMed] [Google Scholar]
- 69.Ryckaert JP, Ciccotti G, Berendsen HJ (1977) Numerical Integration of the Cartesian Equations of Motion of a System with Constraints: Molecular Dynamics of n-Alkanes. J Comput Phys 23:327–341 [Google Scholar]
- 70.Berendsen HJC, Postma JPM, van Gunsteren WF, DiNola A, Haak JR (1984) Molecular Dynamics with Coupling to an External Bath. J Chem Phys 81:3684–3690 [Google Scholar]
- 71.Darden T, York D, Pedersen L (1993) Particle Mesh Ewald: An N·log(N) Method for Ewald Sums in Large Systems. J Chem Phys 98:10089–10092 [Google Scholar]
- 72.Roe DR, Cheatham TE (2013) III PTRAJ and CPPTRAJ: Software for Processing and Analysis of Molecular Dynamics Trajectory Data. J Chem Theory Comput 9:3084–3095 [DOI] [PubMed] [Google Scholar]
- 73.Genheden S, Reymer A, Saenz-Méndez P, Eriksson LA (2017). Computational Chemistry and Molecular Modelling Basics. The Royal Society of Chemistry, pp 1–38 [Google Scholar]
- 74.Grant BJ, Rodrigues APC, ElSawy KM, McCammon JA, Caves LSD (2006) Bio3d: An R Package for the Comparative Analysis of Protein Structures. Bioinformatics 22:2695–2696 [DOI] [PubMed] [Google Scholar]
- 75.Arnold GE, Ornstein RL (1997) Molecular dynamics study of time-correlated protein domain motions and molecular flexibility: cytochrome P450BM-3. Biophys J. 73:1147–1159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Cha J, Pedersen MV, and Auld DS (1996) Metal and pH dependence of heptapeptide catalysis by human matrilysin. Biochemistry 35:15831–15838 [DOI] [PubMed] [Google Scholar]
- 77.Cha J, Auld DS (1997) Site-Directed Mutagenesis of the Active Site Glutamate in Human Matrilysin: Investigation of Its Role in Catalysis. Biochemistry 36:16019–16024 [DOI] [PubMed] [Google Scholar]
- 78.Johnson LL, Pavlovsky AG, Johnson AR, Janowicz JA, Man CF, Ortwine DF, Purchase CF II, White AD, Hupe DJ (2000) A rationalization of the acidic pH dependence for stromelysin-1 (matrix metalloproteinase-3) catalysis and inhibition. J Biol Chem 275:11026–11033 [DOI] [PubMed] [Google Scholar]
- 79.Tsukada H, Pourmotabbed T (2002) Unexpected crucial role of residue 272 in substrate specificity of fibroblast collagenase. J Biol Chem 277:27378–27384 [DOI] [PubMed] [Google Scholar]
- 80.Brini E, Simmerling C, Dill K (2020) Protein storytelling through physics. Science 370:3041. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.