Abstract
The prefrontal cortex (PFC) and its connections with the mediodorsal thalamus (MD) are crucial for cognitive flexibility and working memory 1 and are thought to be altered in disorders such as autism 2,3 and schizophrenia 4,5. While developmental mechanisms governing regional patterning of the cerebral cortex have been characterized in rodents 6-9, the mechanisms underlying the development of PFC-MD connectivity and the lateral expansion of PFC with distinct granular layer 4 in primates 10,11 remain elusive. Here we report an anterior/frontal-to-posterior/temporal, PFC-enriched, gradient of retinoic acid (RA), a signaling molecule regulating neural development and function 13-15, and genes regulated by RA in the early- and midfetal human and macaque neocortex. We observed multiple potential RA sources, including primate-specific expression and cortical expansion of RA synthesizing enzymes compared to mouse. Furthermore, RA signaling is largely confined to the prospective PFC by CYP26B1, an RA-catabolizing enzyme, upregulated in the prospective motor cortex. Gene deletion in mice revealed that RA signaling through RXRG and RARB, and CYP26B1-dependent catabolism are required for proper molecular patterning of prefrontal and motor areas, development of PFC-MD connectivity, intra-PFC dendritic spinogenesis, and expression of the layer 4 marker RORB. Together, these findings reveal a critical role for RA signaling in PFC development and, potentially, its evolutionary expansion.
Introduction
The PFC reaches its greatest complexity in anthropoid primates (monkeys and apes), which appear to uniquely have many prefrontal areas covering the entire anterior two-thirds of the frontal lobe and a well-defined granular layer 4 10,11. Our previous analyses revealed that the transcriptomic differences between neocortical areas are most prominent during primate midfetal development 16,17, a crucial period for neuronal specification and the initial assembly of neocortical neural circuits 18. Thus, we hypothesized that the molecular processes governing the development and evolutionary diversification of the PFC could be revealed by differential regional gene expression analysis of the midfetal primate neocortex.
Enrichment of RA-related genes in midfetal frontal cortex
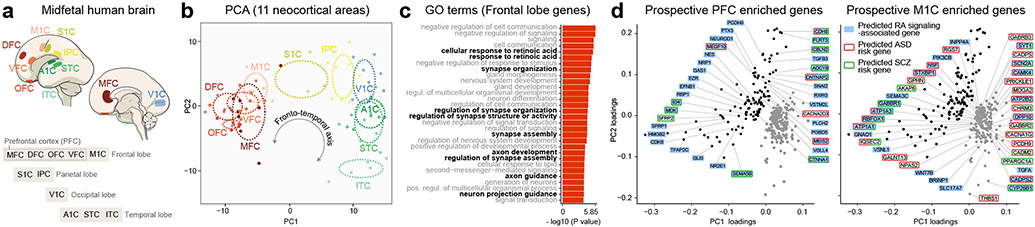
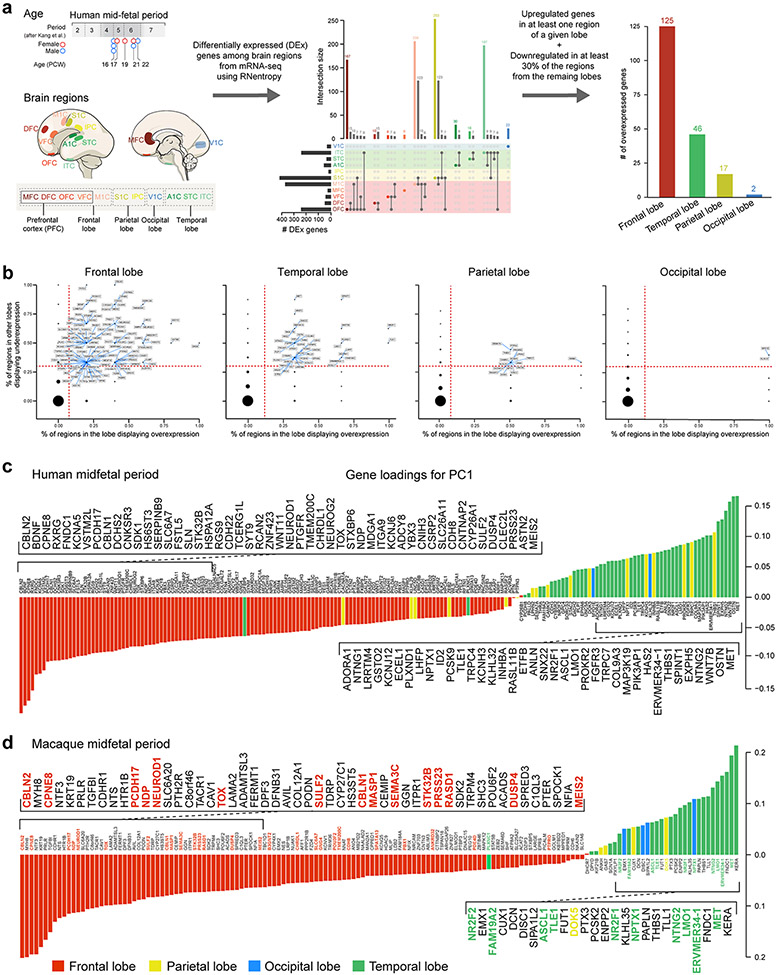
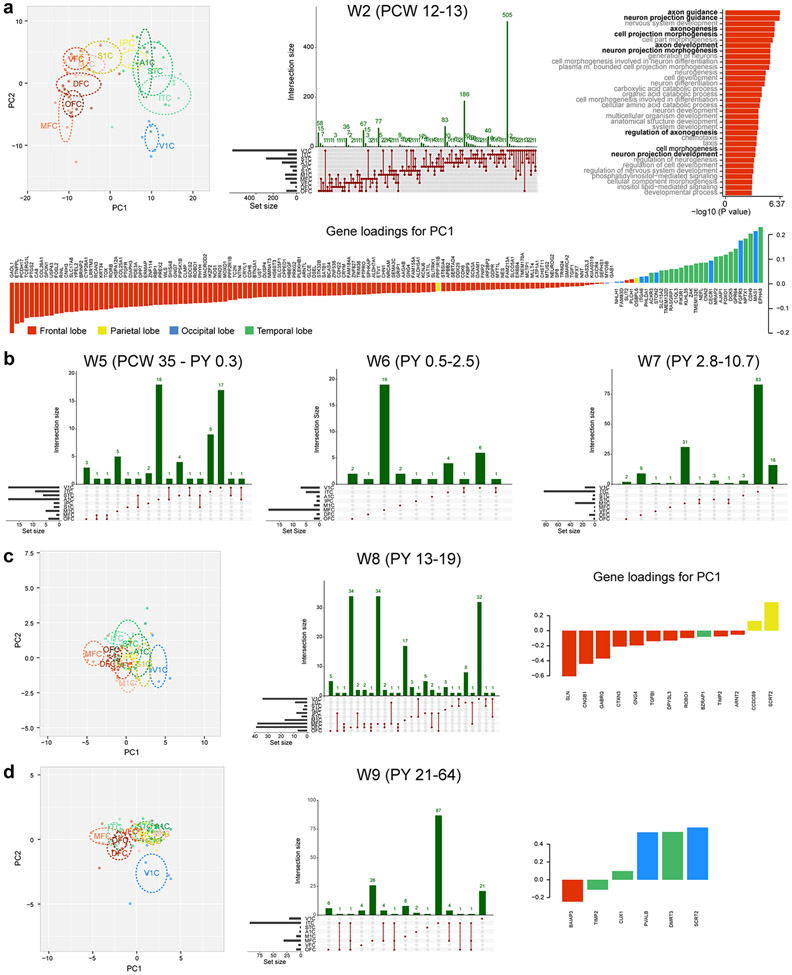
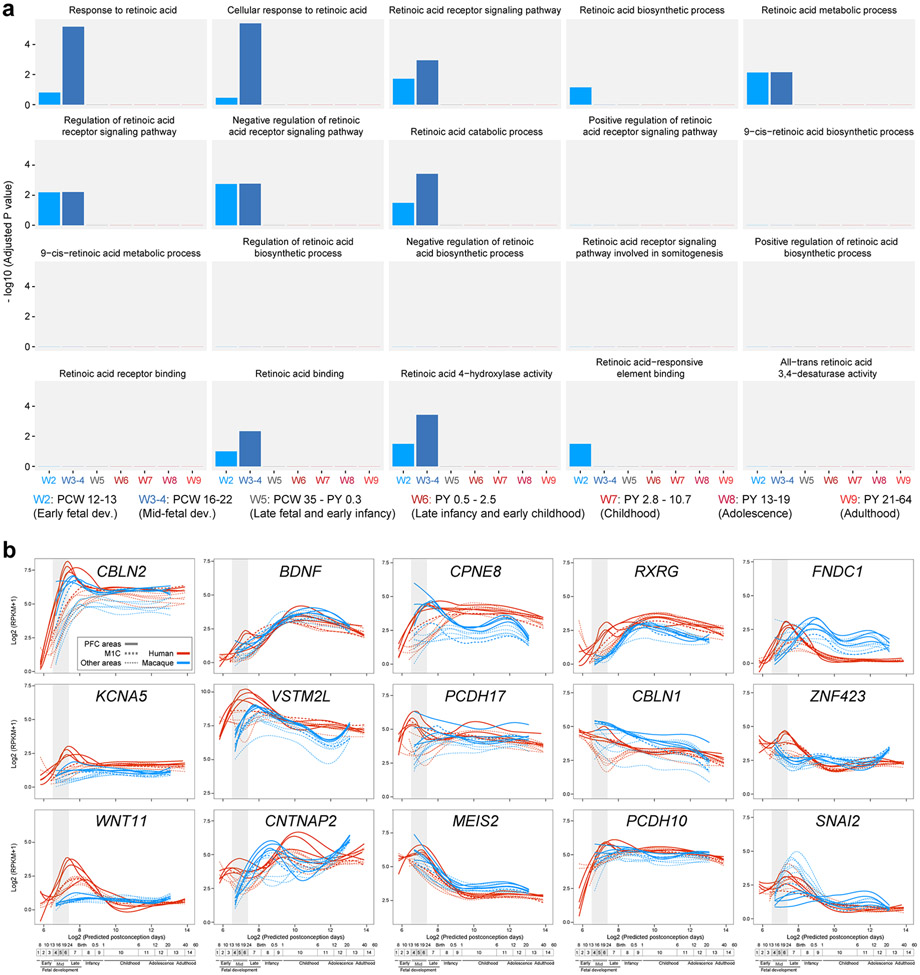
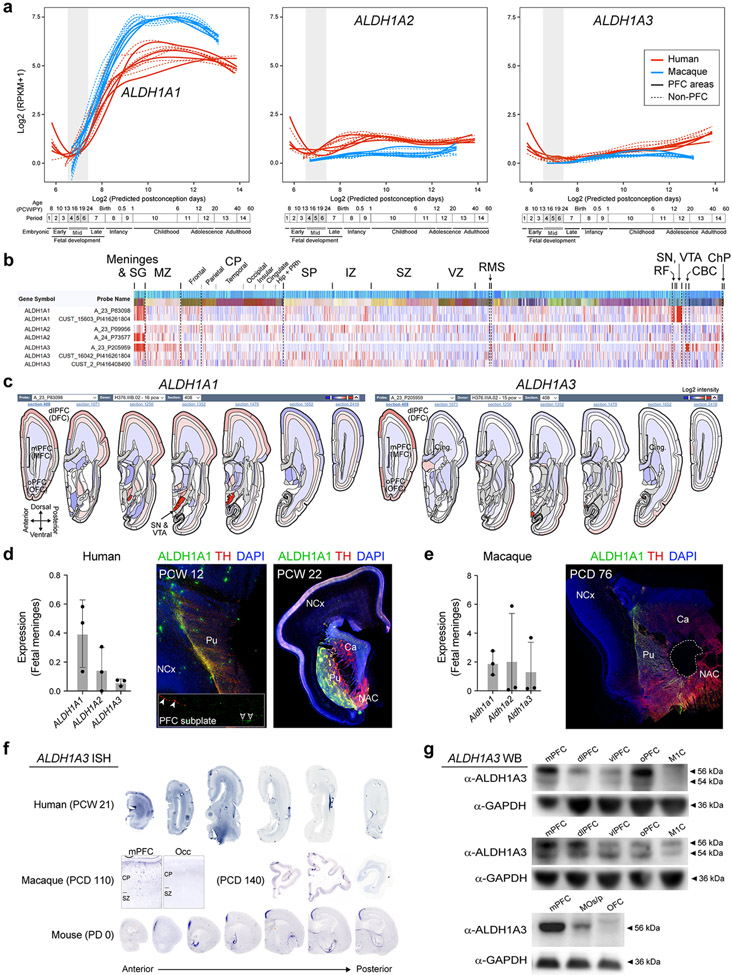
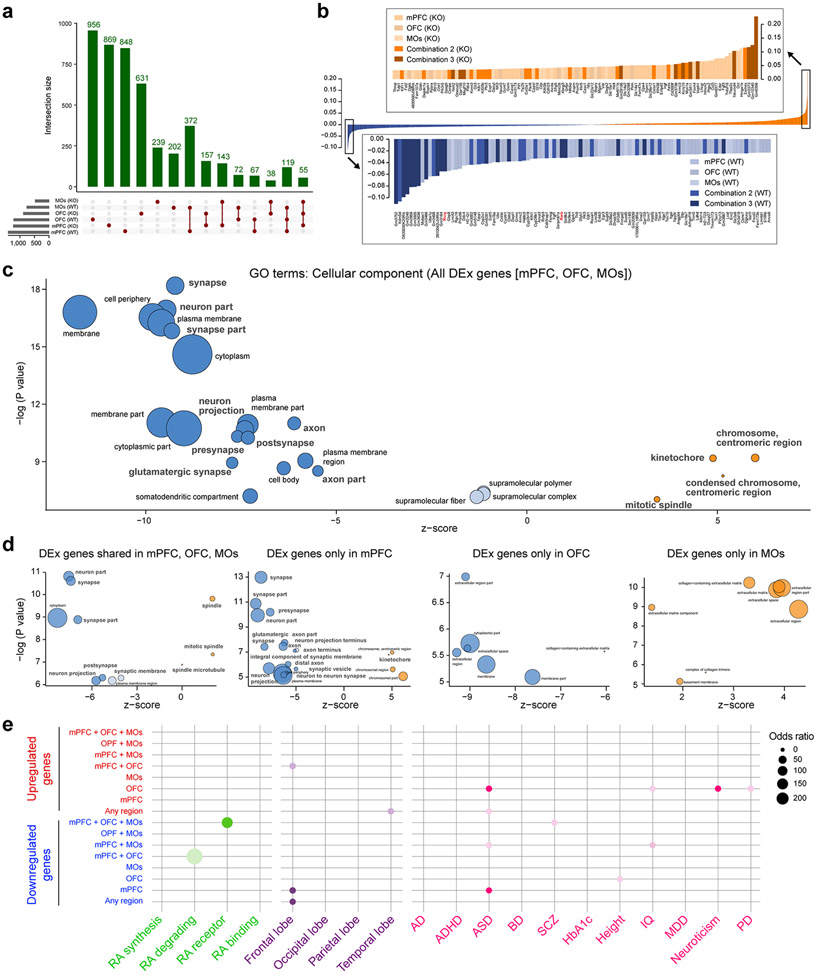
Using human BrainSpan RNA-seq data 17, we screened for genes that are differentially upregulated in the midfetal frontal lobe. The midfetal data consisted of tissue-level samples ranging in age from post-conception weeks (PCW) 16 to 22, including four prospective PFC areas (medial, mPFC/MFC; orbital, oPFC/OFC; dorsolateral, dlPFC/DFC; and ventrolateral, vlPFC/VFC) and the primary motor cortex (M1C). Gene expression in these frontal areas was compared to areas within the parietal (primary sensory cortex, S1C; and inferior parietal cortex, IPC), occipital (primary visual cortex, V1C), and temporal lobes (primary auditory cortex, A1C; superior temporal cortex, STC; and inferior temporal cortex, ITC) (Fig. 1a; Extended Data Fig. 1a). We identified 190 protein-coding genes that are specifically upregulated, using stringent criteria, in at least one area, within a lobe in comparison with areas from other lobes including 125 in the frontal areas, which were able to differentiate the four brain lobes and most areas within them, as observed by principal component analysis (PCA) (Fig. 1b; Extended Data Fig. 1a,b). Moreover, the first principal component (PC1), which accounts for the highest variability present in the data, corresponded to the anterior-posterior/frontal-temporal axis (Fig. 1b). Gene Ontology (GO) analysis of the frontally upregulated genes identified an enrichment of genes associated with categories such as “response to retinoic acid”, “synapse organization/assemble” and “axon development/guidance’ (Fig. 1c, d; Extended Data Tables 1 and 2), suggesting that these genes may play a role in frontal lobe patterning and circuit development. Many of the same frontal lobe enriched and RA-related genes, were upregulated in the early and midfetal human and Macaca mulatta frontal cortex using PsychENCODE dataset 19 (Extended Data Fig. 1c,d). We also observed prominent frontal enrichment of many of the same genes during the early fetal period (Extended Data Fig. 1,2,3). Closer analysis of the spatiotemporal profile of the fetal frontally-upregulated genes revealed largely transient nature of the enrichment, with mainly conserved and some divergent expression patterns between human and macaque (Extended Data Fig. 1).
Fig. 1 ∣. Predicted RA signaling-associated genes are upregulated in human midfetal prospective frontal areas.
a, Diagram of the analyzed eleven human midfetal areas from the four lobes. b, PCA of genes specifically enriched in at least one area within a lobe. Ellipses are centered on the mean of the points of a given area and the size of the axes corresponds to their standard deviation on each component. c, GO terms associated with frontally upregulated genes and their unadjusted p-value d, PC loadings with labeled genes being upregulated in PFC (black dots) and M1C (gray dots). Genes highlighted in blue are associated with RA signaling and genes with red or green outlines are predicted ASD or SCZ risk genes, respectively. For reproducibility information, see Methods.
Analyzing only the RNA-seq data encompassing the five midfetal frontal areas 17, predicted RA signaling-associated genes, such as CBLN2, RXRG, CDH8, MEIS2, and RBP1 were among the genes upregulated in the PFC compared to the M1C (Fig. 1d; Extended Data Table 2; see our accompanying study 20), while the RA degrading enzyme CYP26B1 was upregulated in M1C (Fig. 1d), consistent with our previous microarray-based findings 21. We also identified multiple predicted autism spectrum disorder (ASD) (Group 1-3 gene.sfari.org) and schizophrenia (SCZ; Score of 3 or higher from szdb.org)) risk genes, upregulated in the midfetal frontal cortex (Fig. 1d; Extended Data Table 2)
PFC-enriched gradient of RA in the midfetal cortex
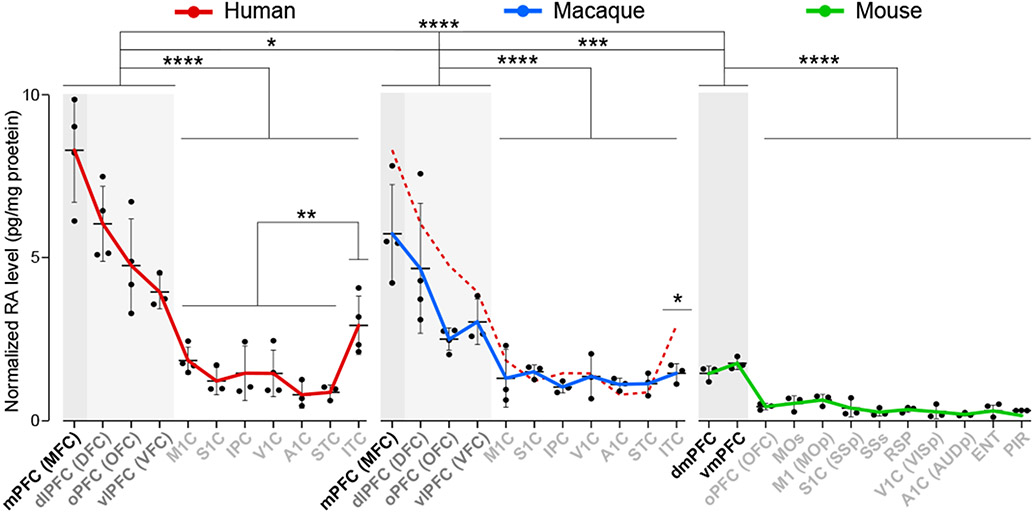
RA is a diffusible biologically active derivative of vitamin A implicated in neural tube patterning, neurogenesis, cell differentiation, and synaptic function 12,13,22-27. Moreover, alterations in RA signaling have been implicated in the pathophysiology of ASD 28-30 and SCZ 14,31,32. Given the enrichment of RA-related genes among the upregulated fetal frontal genes, we assessed RA concentrations in different areas of the midfetal human and macaque, and neonatal mouse neocortex (approximately equivalent developmental ages), using an enzyme-linked immunosorbent assay (ELISA). We revealed a PFC-enriched anterior-posterior gradient of RA concentration in midfetal human and macaque neocortex, with homologous mPFC exhibiting the highest concentration within each species (Fig. 2). Overall, RA concentrations were significantly higher in prospective primate PFC areas compared to more posterior areas. Comparison across the three species, identified higher concentrations of RA in both the human mPFC and all four PFC areas overall as compared to the other two species (Fig. 2), as well as in macaque compared to mouse (Fig. 2). Interestingly, ITC, an association area within the temporal lobe thought to exhibit unique features and connectivity in humans 33, had a higher RA concentration among non-frontal areas in humans (Fig. 2) with a two-fold increase in RA concentration in human compared to macaque.
Fig. 2 ∣. PFC-enriched antero-posterior gradient of RA.
a, RA concentrations in human (PCW 16, 18, 19, 21) and macaque (Four PCD 110 brains), and mouse (Four PD 1 brains) (N = 3-4 for each areal sample) cortical areas. One-way ANOVA with post-hoc Dunnett’s adjustment or two-tailed unpaired t-test: ****P < 1e-4, ***P=0.0005, (Macaque PFC vs. Mouse PFC), **P = 0.005 (Human non-PFC minus ITC vs. Human ITC) *P = 0.01 (Human PFC vs. Macaque PFC), P = 0.04 (Human ITC vs. Macaque ITC); Errors bars: S.E.M. Dashed red line in macaque graph represent human RA concentrations. Single assay was done for each brain sample.
Expanded cortical expression of RA synthesizing enzymes in primates
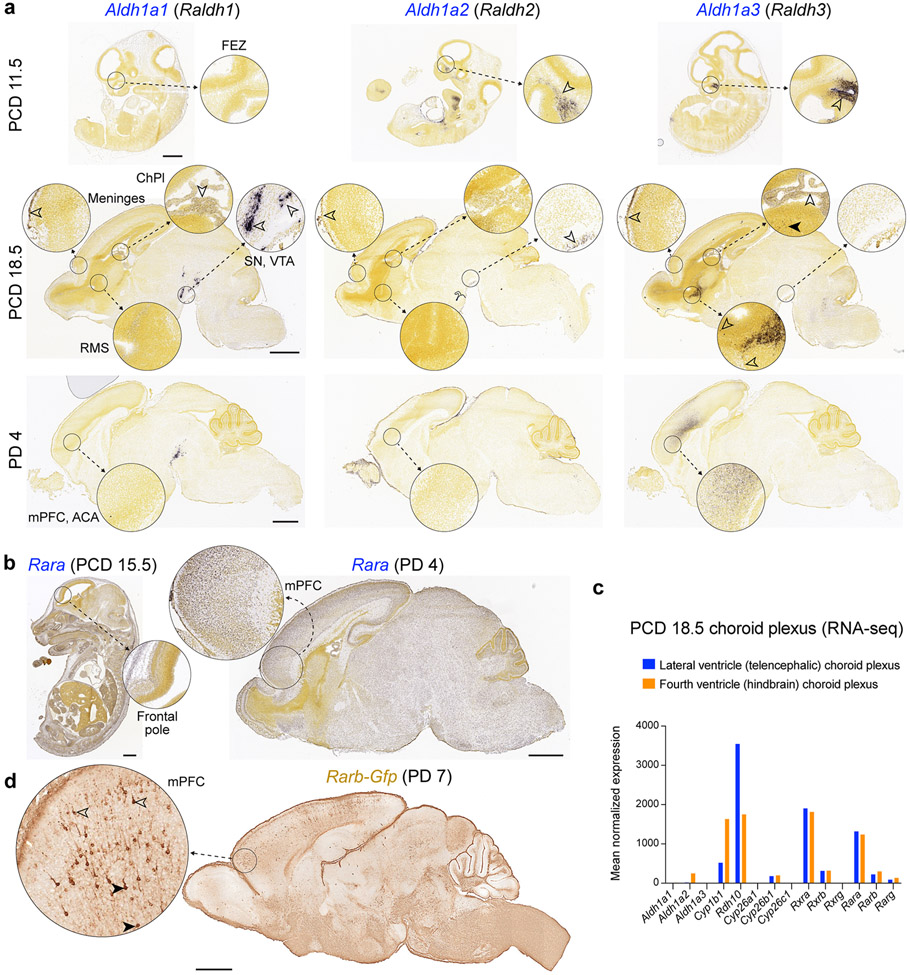
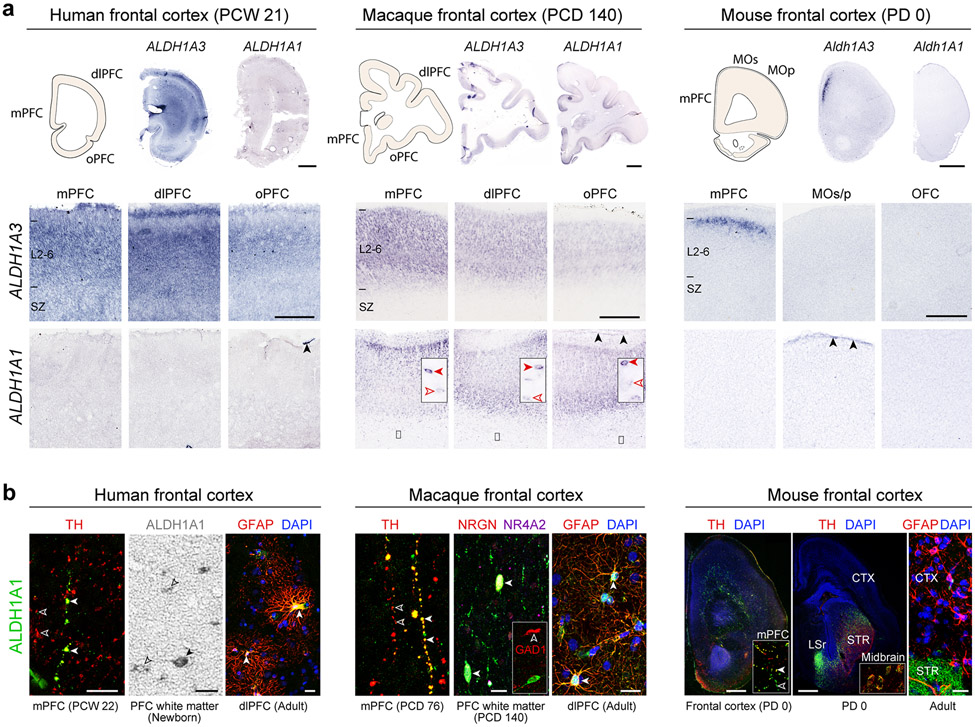
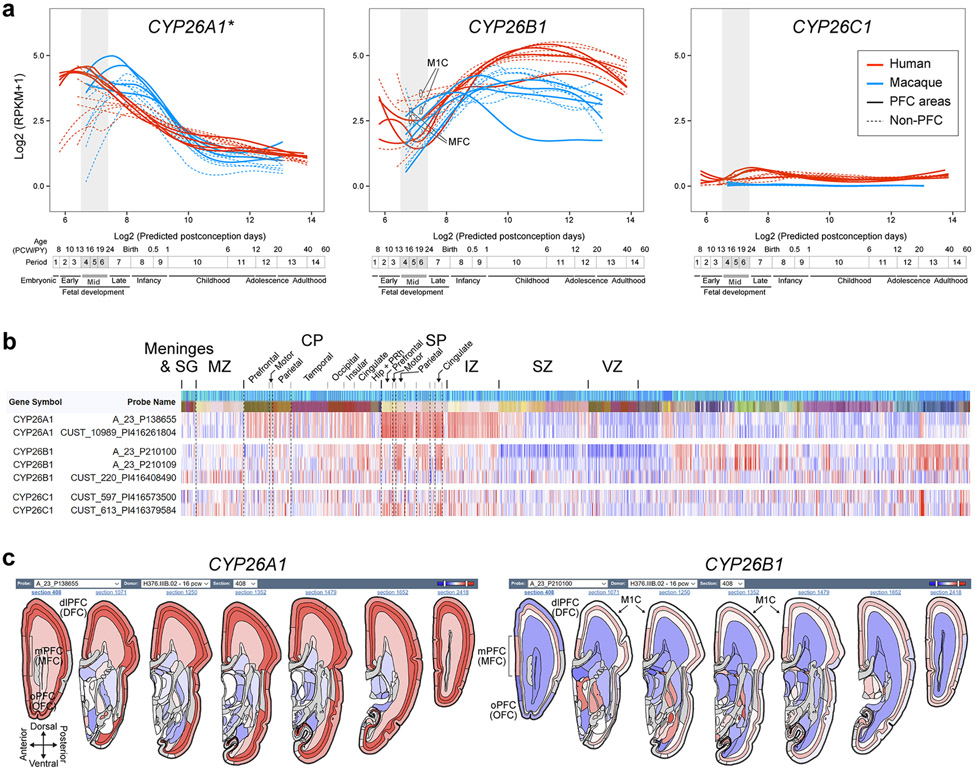
The observed enrichment and primate-specific lateral expansion of RA levels in the fetal PFC led us to systematically examine the spatio-temporal expression of a RA synthesizing enzymes, ALDH1A1-3, and degrading enzymes, CYP26A1, CYP26B1 and CYP26C1 27,28 in human, macaque and mouse (For full descriptions, please see supplemental results). Briefly, we observed multiple potential fetal cortical RA sources, including conserved expression of ALDH1A1 in meninges and midbrain axons, and ALDH1A3 in the mPFC 26. We also, observed a primate-specific expression of ALDH1A1 in astrocytes and frontal subplate neurons, and lateral frontal expression of ALDH1A1 and ALDH1A3 (Extended Data Fig. 6). These primate-specific expression patterns were complementary to the increase and lateral extension in RA in the mid- and late-fetal primate PFC and the lateral extension of granular PFC in anthropoid primates 10,11.
Developmental PFC RA signaling mediated by RXRG and RARB
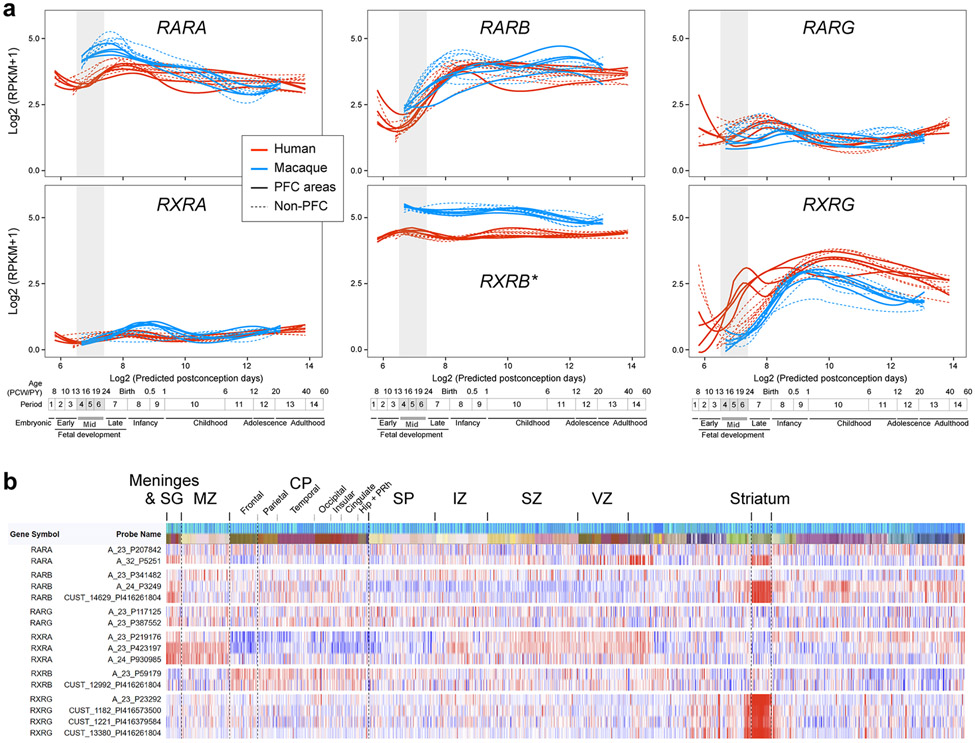
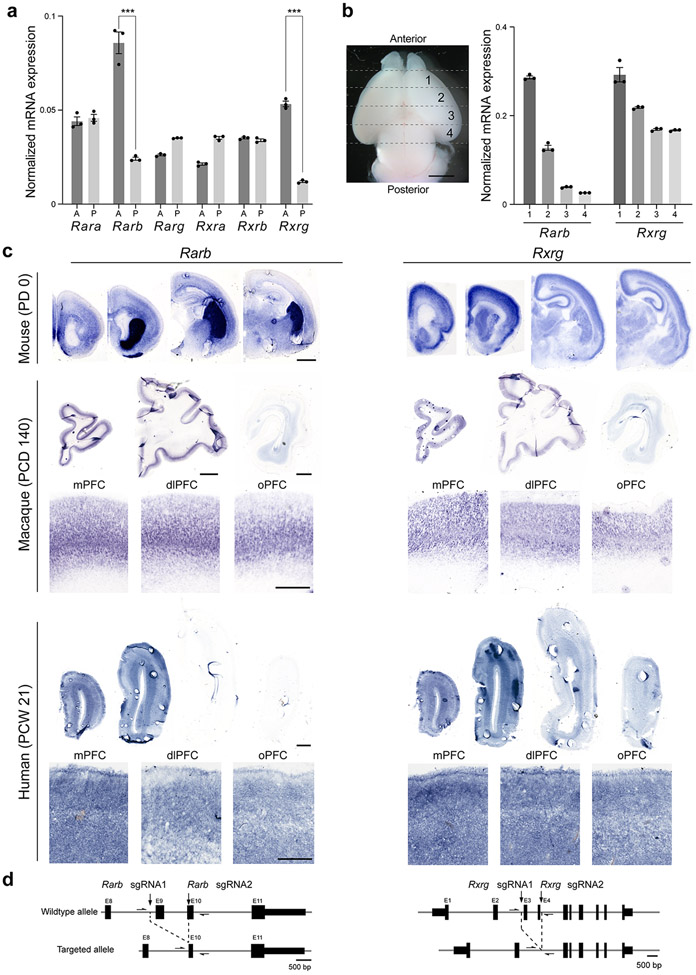
Given the midfetal frontal cortical upregulation of both RA and RA synthesizing enzymes, we further assessed the expression of RA-dependent receptors and RA-responsive downstream genes in the developing human, macaque and mouse cortex. In addition to RXRG, which is upregulated in the human midfetal PFC 21 (Extended Data Fig 1c), multiple genes encoding RA receptors were also detected in the developing human, macaque and mouse cortex or cultured primary human midfetal cortical neurons (Extended Fig. 5,8,9,15b). Of these, only RARB and RXRG exhibited an obvious higher anterior to lower posterior gradient of expression (Extended Data Fig. 8,9a).
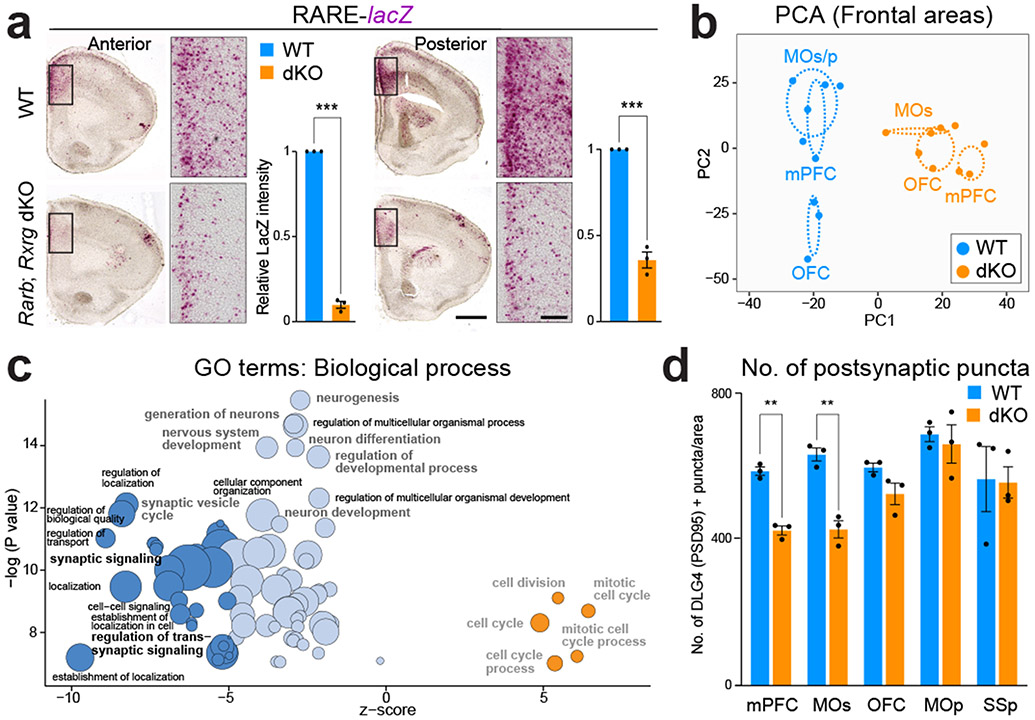
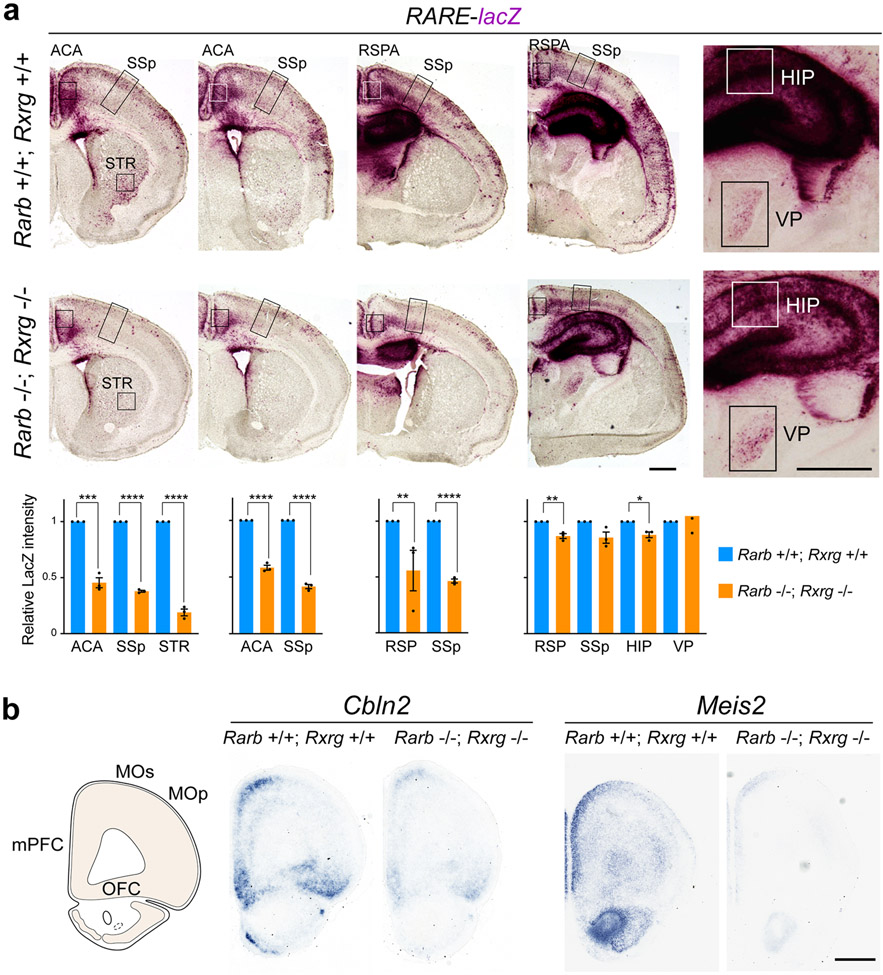
The RARB-RXRG heterodimer has been previously shown to mediate RA signaling in the adult mouse cortex and striatum, and be required for learning, locomotion and dopamine signaling 34,35. To assess whether RARB-RXRG are required for RA signaling activity in the developing mouse frontal cortex, we generated constitutive Rarb and Rxrg double knockout (dKO) mice (Extended Data Fig. 9d), which, consistent with previous findings 34,35, are viable. Using RARE-lacZ reporter line 36 where lacZ is under the transcriptional control of a RA response element, we identified a significant reduction of RA signaling in the PD0 dKO mPFC compared to control mice (Fig. 3a, Extended Data Fig. 10a). In addition, we found a less extensive reduction of RA signaling in the anterior cingulate area (ACA) and retrosplenial area (Extended Data Fig. 10a). Furthermore, expression of frontally enriched RA regulated genes, Cbln2 20 and Meis2, which are induced by RA in human and chimpanzee cerebral organoids and repressed by RAR antagonists in human cortical neurons (Fig. 1d, Extended Data Fig. 11b,15d), was reduced in the dKO mice (Extended Data Fig. 10b). Together, these findings indicate that RARB and RXRG mediate RA signaling in the developing mouse mPFC.
Fig. 3 ∣. Reduced RA signaling in the PFC of mice lacking Rxrg and Rarb leads to downregulation of genes involved in synapse and axon development.
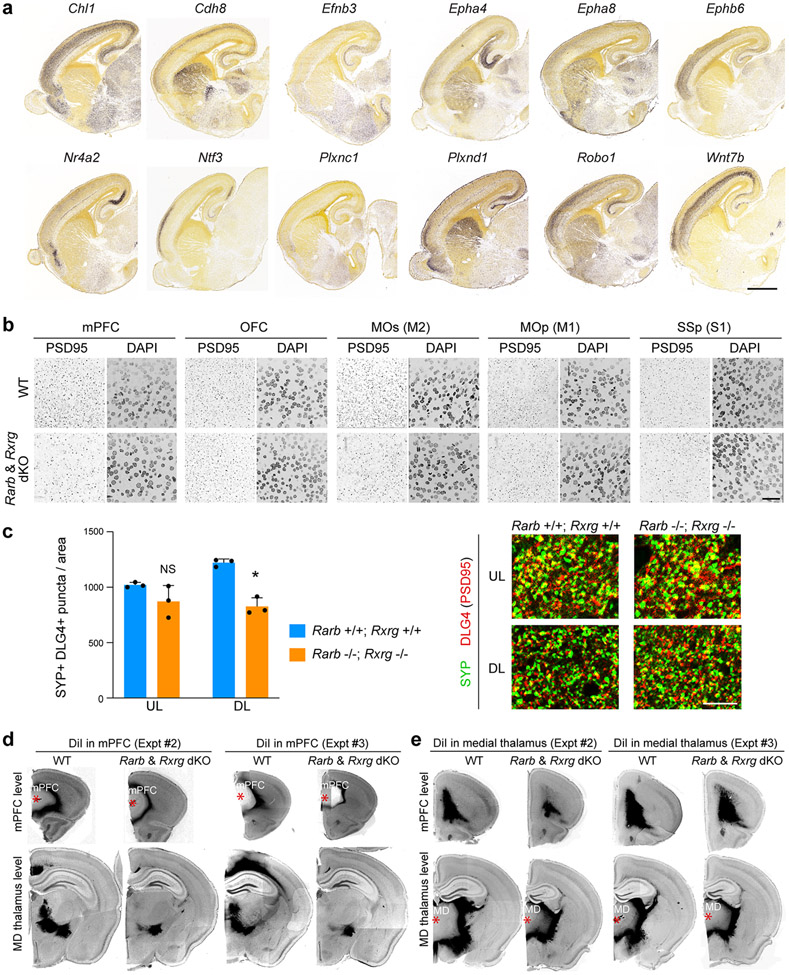
a, β-Galactosidase histochemical staining in Rxrg +/+;Rarb +/+; (WT); RARE-lacZ (Blue) and Rxrg −/−;Rarb −/− (dKO); RARE-lacZ (orange) brains at PD 0. Two-tailed Student’s t-test: WT vs. dKO: ***P =1e-6; 3e-4, Errors bars: S.E.M.; N = 3 per genotype; Scale bars: 200 μm; 50 μm (inset). b, First, two principal components calculated from the expression of DEx protein-coding genes between WT and dKO littermates in at least one of the three frontal cortex areas (mPFC, MOs, and OFC). c, GO terms associated with DEx genes showing their z-score and unadjusted p values. Z-score represents the proportion of upregulated versus downregulated genes in the dKO compared to WT associated to each GO term. Dark blue: z-score < −5; light blue: z-score −5,0; orange: z-score >0. Size of the bubbles are proportional to the total number of DEx genes associated to the given GO term. d, Quantification of excitatory synapses marked by DLG4/PSD95 in the mPFC, MOs, OFC, MOp, and SSp regions from PD 0 WT and dKO mice brains. Two-tailed Student’s t-test: **P = 6e-4; 2e-3; Errors bars; S.E.M.; N = 3 per genotype. For reproducibility information, see Methods.
RXRG and RARB regulate PFC connectivity
To understand the functional significance of RA signaling through the RARB-RXRG heterodimer in the developing cortex, we performed RNA-seq analysis of different regions/areas of the PD 0 mouse frontal cortex (mPFC; secondary motor cortex (MOs) and the adjacent parts of the primary motor cortex (MOp); and OFC) microdissected from dKO and wild-type (WT) littermates. We identified 4768 differentially expressed (DEx) protein-coding genes between the two genotypes in at least one of the areas with the highest number of DEx genes in the mPFC (Extended Data Fig. 12a; Extended Data Table 3). PCA based on the expression of these DEx genes separated the WT and dKO along PC1, with the mPFC showing the greatest distance between the WT and the dKO, further supporting the notion that the mPFC was most affected by the reduction of RA signaling (Fig. 3b).
The GO enrichment analysis revealed that terms associated with genes that are overexpressed in the WT frontal cortex compared to the dKO frontal cortex were highly related to the process of synaptogenesis and cellular components related to synapses and axons, whereas the ones overexpressed in the dKO are related to the regulation of the cell cycle (Fig. 3c; Extended Data Fig. 12c,d). In addition, when analyzing DEx genes in different regions/areas of the frontal cortex, only genes overexpressed exclusively in the mPFC were associated with the cellular components, axons and synapses (Extended Data Fig. 12c,d). Several of the genes downregulated in the dKO related to axon guidance and synapse development displayed an anterior-enrichment in WT neonatal mouse cortex (Extended Data Fig. 13a; Extended Data Table 4). We also observed a significant enrichment of homologous genes specifically upregulated in the human midfetal frontal lobe among the genes that were downregulated in the dKO mice frontal cortex and mPFC exclusively (Extended Data Fig. 12b,e). Overall, these results suggest a possible role for RA signaling in the regulation of synaptogenesis and axon development, specifically in the mPFC.
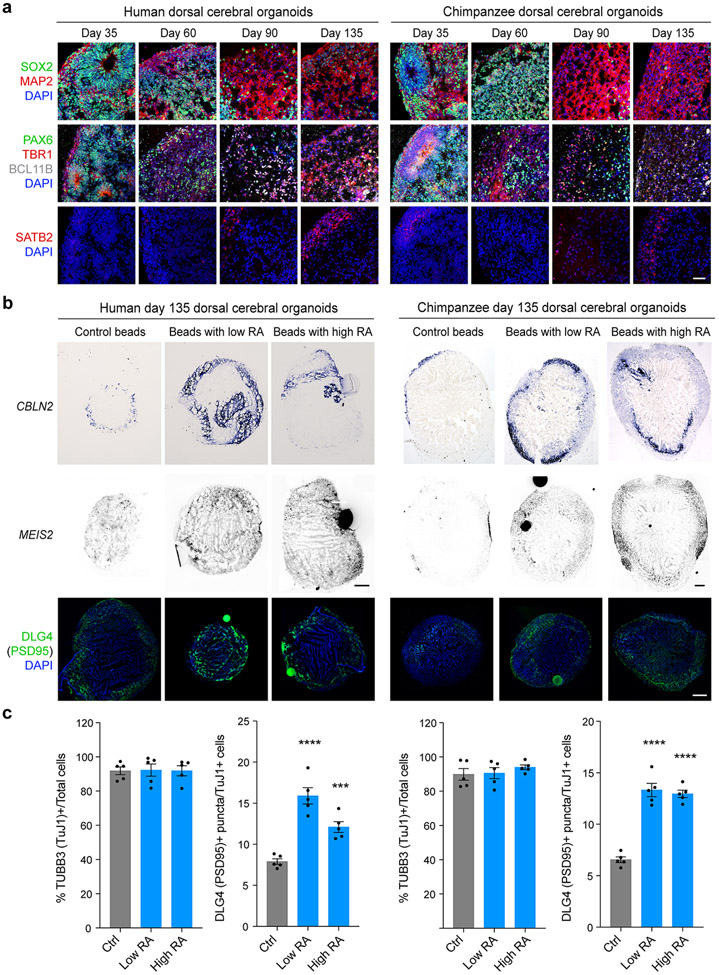
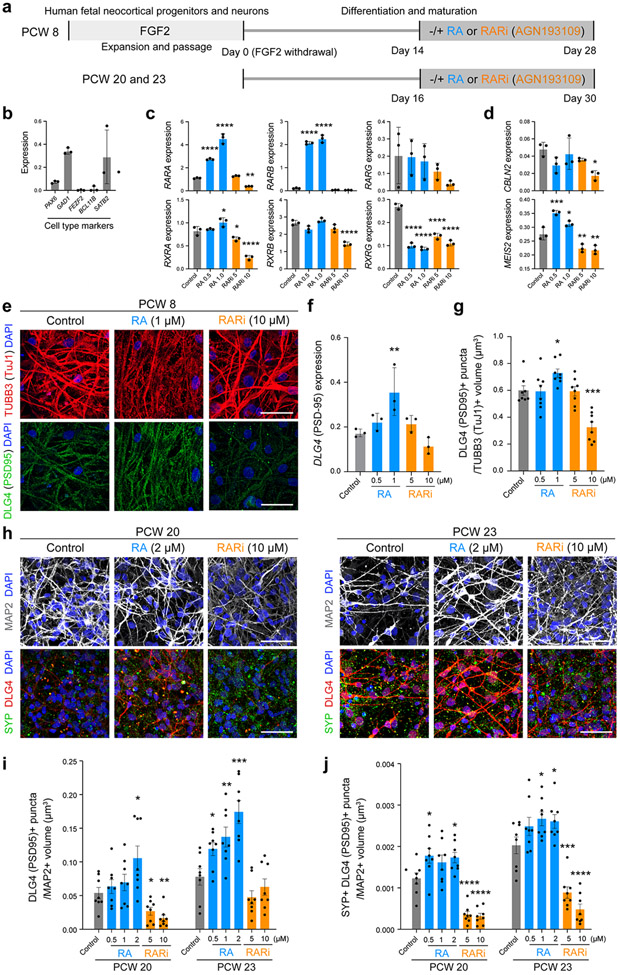
We analyzed the role of RA in synaptogenesis by quantifying synapse number in multiple regions of the mouse PD 0 cortex between dKO mice and WT littermate controls. We identified a significant reduction of DLG4/PSD95-positive excitatory postsynaptic densities in the mPFC and MOs, but not in the OFC, primary motor area (MOp) and primary somatosensory area (SSp) (Fig. 3d, Extended Data Fig. 13b). Similarly, we identified a significant decrease of Synaptophysin (SYP) and PSD95 co-immunolabelled puncta in deep layers of the mPFC (Extended Data Fig 13c) and total and mushroom dendritic spines in mPFC neurons labelled by retrograde viral tracer AAVrg-Cag-Gfp (Extended Data Fig. 14e). Of note, there was no difference observed in dendritic complexity of mPFC upper layers neurons (Extended Data Fig. 14e). In addition, treatment of human primary cortical neurons with RA induced expression of DLG4/PSD95 mRNA, and DLG4/PSD95 and SYP co-immunolabelled synaptic puncta while applying RA receptor inhibitor AGN193109 had the opposite effect (Extended Data Fig. 15e-j). Similarly, placement of RA-soaked beads with both human and chimpanzee cerebral organoids induced DLG4/PSD95 expression (Extended Data Fig. 11b,c). We further assessed the role of RA signaling in regulating synaptic organizer CBLN2 and local connectivity in our accompanying study20.
Rxrg and Rarb are required for development of mPFC-MD thalamus connectivity
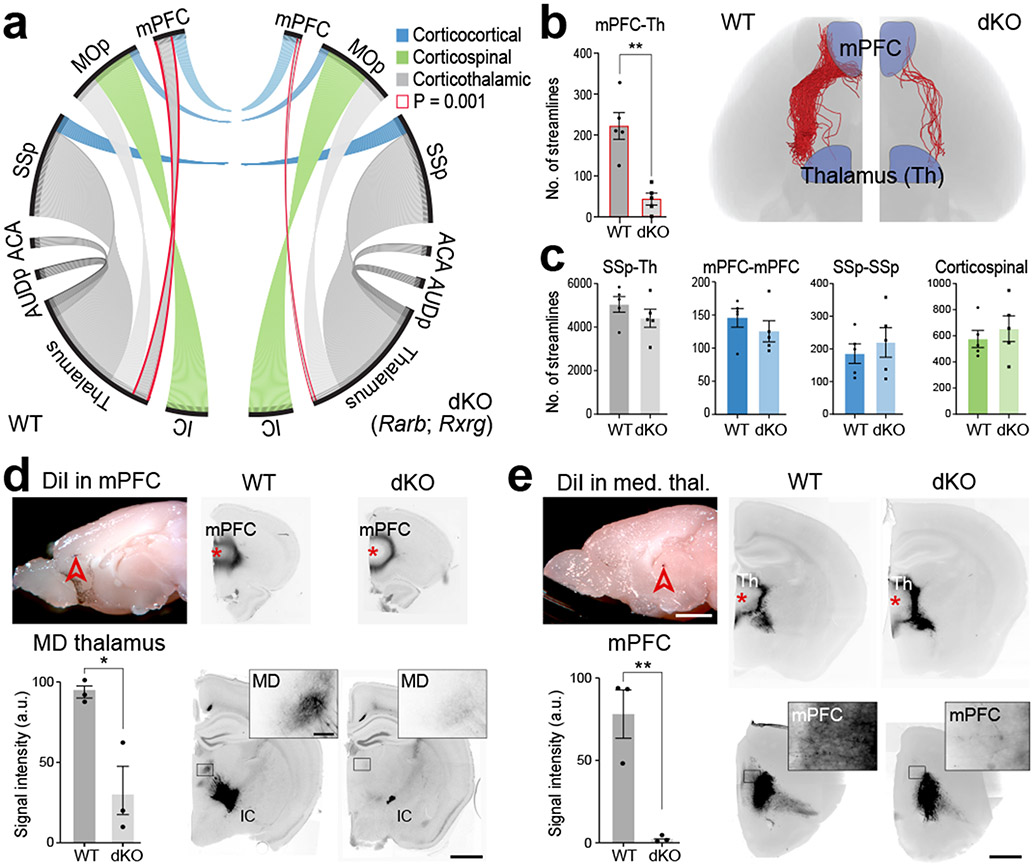
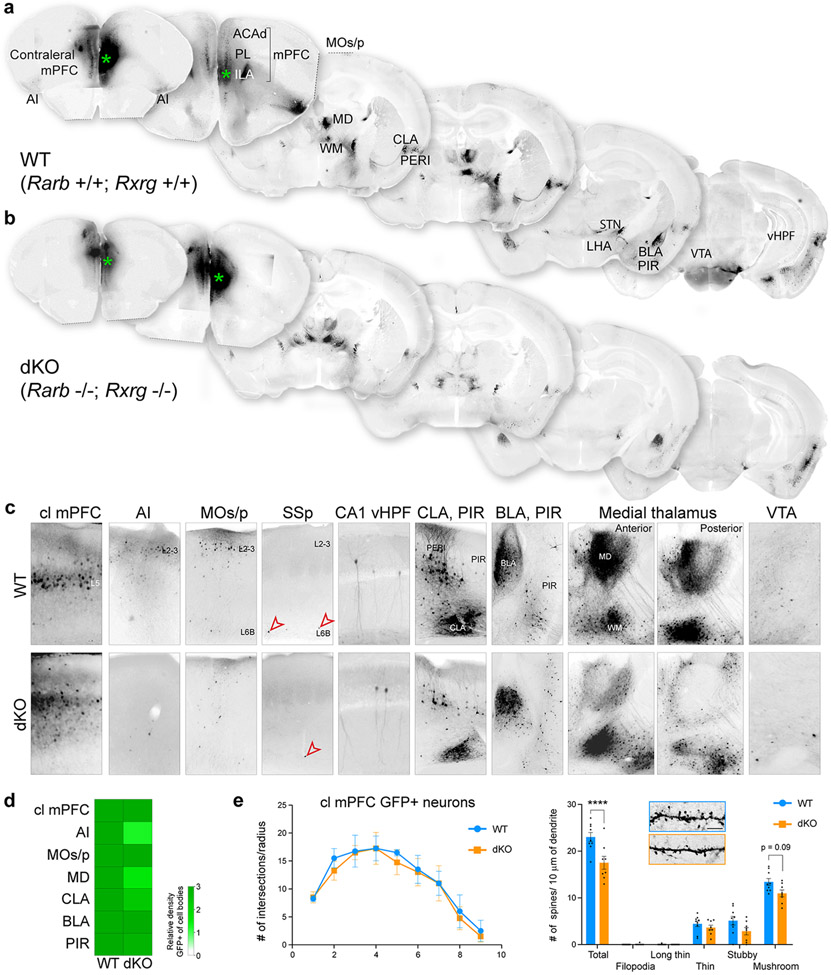
We investigated the role of RA signaling on long-range connections from the mPFC using Diffusion Tensor Imaging (DTI). We identified a reduction in long-range connections between the mPFC and thalamus in dKO compared to WT mice at PD 5 (Fig. 4a,b). There was no difference in connections between the left and right mPFC at PD 5 (Fig. 4a,c). Due to limitations of the technique and developmental stage, we were unable to study the connections between the mPFC and striatum, or basolateral amygdala (BLA) using DTI. Anterograde lipophilic axon tracing experiments of either mPFC and MD at PD 21 similarly identified reciprocal reduction of MD-PFC connectivity, as well as a reduction of fibers in the internal capsule (Fig. 4d,e; Extended Data Fig. 13d). To more fully explore mPFC connectivity in dKO, we used a retrograde viral tracer to study inputs into the PD 30 mPFC. We identified reduced inputs from the MD and anterior insula in dKO while identifying no obvious changes in inputs from the contralateral mPFC, MOs/p, internal capsule, claustrum, piriform cortex, amygdala and ventral hippocampus (Extended Data Fig. 14a-d). Finally, we assessed for changes in dopaminergic innervation of the mPFC and found no significant difference between WT and dKO (Extended Data Fig. 16f).
Fig. 4 ∣. Altered mPFC-MD connectivity in mice lacking Rarb and Rxrg.
a, Representation of number (#) of streamlines generated as a connectivity measurement between the cortical areas, thalamus and internal capsule (IC) at PD 5 using DTI. b, Visualization and quantification of streamlines between mPFC and thalamus in WT and dKO brains. Two-tailed unpaired t-test: **P = 1e-3; N = 5 per genotype. Errors bars: S.E.M. c, Quantification of select corticothalamic, corticocortical and corticospinal streamlines. Two-tailed unpaired t-test; P = 0.2 (SSp-Th), 0.3 (mPFC-mPFC), 0.5 (SSp-SSp), 0.5 (Corticospinal); Errors bars: S.E.M; N = 5. d, e, DiI placement in the mPFC and medial thalamus in WT and dKO brain at PD 21 with labelled processes in the MD (inset), and mPFC (inset). Two-tailed Student’s t-test: *P = 9e-3, **P = 3e-3; N = 3 per genotype; Errors bars: S.E.M.; Scale bars: 1 mm; 100 μm (inset).
While there is reduced RA activity in the outer shell of the striatum in the dKO brain (Extended Data Fig. 10a), we found that fibers traverse through the medial aspect of the striatum, suggesting that alterations in RA signaling in the lateral striatum do not affect the guidance of reciprocal mPFC-MD connectivity. RA signaling has been previously implicated in thalamic development 37, thus we assessed whether other thalamocortical connections were altered in the dKO mice at PD 5 using DTI. There was no difference in thalamocortical connectivity with the MOp, primary auditory area (AUDp), or SSp (Fig. 4a,c; Extended Data Fig. 16b) or formation of barrel fields in the SSp (Extended Data Fig. 16c). Given that Aldh1a3 is expressed in the anterior cingulate area (ACA) 26, we also examined whether thalamocortical innervation was altered in the ACA and found no difference in the number of streamlines (Fig. 4a; Extended Data Fig. 16b). The corticospinal tract (CST) and connections across the corpus callosum between left and right mPFC, SSp and MOp in the dKO showed no difference in the number of streamlines compared to control (Fig. 4a,c, Extended Data Fig. 16b). Furthermore, scalar indexes, which describe the microstructural integrity of white matter, were similar between WT and dKO mice in the corpus callosum, anterior commissure and internal capsule (Extended Data Fig. 16a). In addition, the width of the CST at PD 30 was slightly increased in the dKO mice (Extended Data Fig. 16d). The reduction of connections between the mPFC and thalamus was not due to cell death in the mPFC (Extended Data Fig. 16e). In summary, deletion of Rxrg and Rarb leads to reduction of RA signaling specifically in mPFC, as well as selective reduction of reciprocal mPFC-MD connectivity.
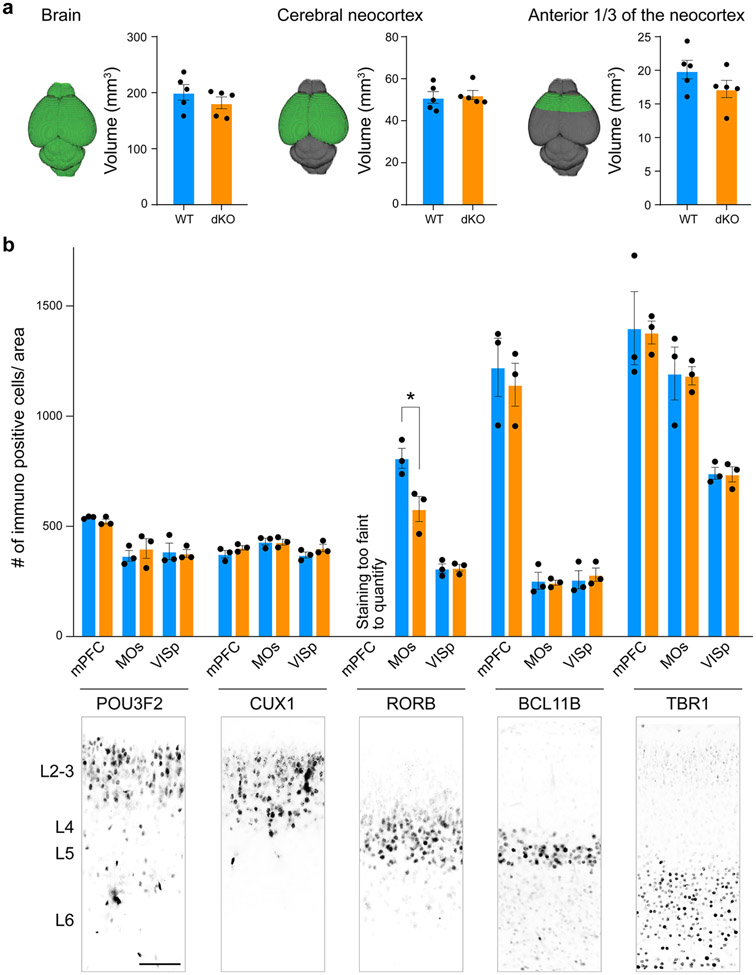
RA signaling has been previously described to be involved in the regulation of proliferation, cell cycle timing and cortical laminar development 22-24, which could underlie our findings. Thus, we assessed if the constitutive deletion of RXRG and RARB lead to changes in cortical size and laminar organization. The volume of the brain, cortex and frontal cortex at PD 5 were not significantly different when measured by MRI (Extended Data Fig. 17a). We also quantified the number of neurons using the upper layer markers CUX1 and POU3F2/BRN2, layer 4 marker RORB, and deep layer markers BCLL11B/CTIP2 and TBR1 and found no difference between WT and dKO in the mPFC, motor cortex or V1 (Extended Data Fig. 17b). We did find a modest reduction in RORB labeled neurons in the motor cortex (Extended Data Fig. 17b). Of note, the mouse mPFC lacks a granular layer 4 and had minimal RORB labeled cells.
Ectopic RA signaling expands medial thalamic connectivity and layer 4 identity
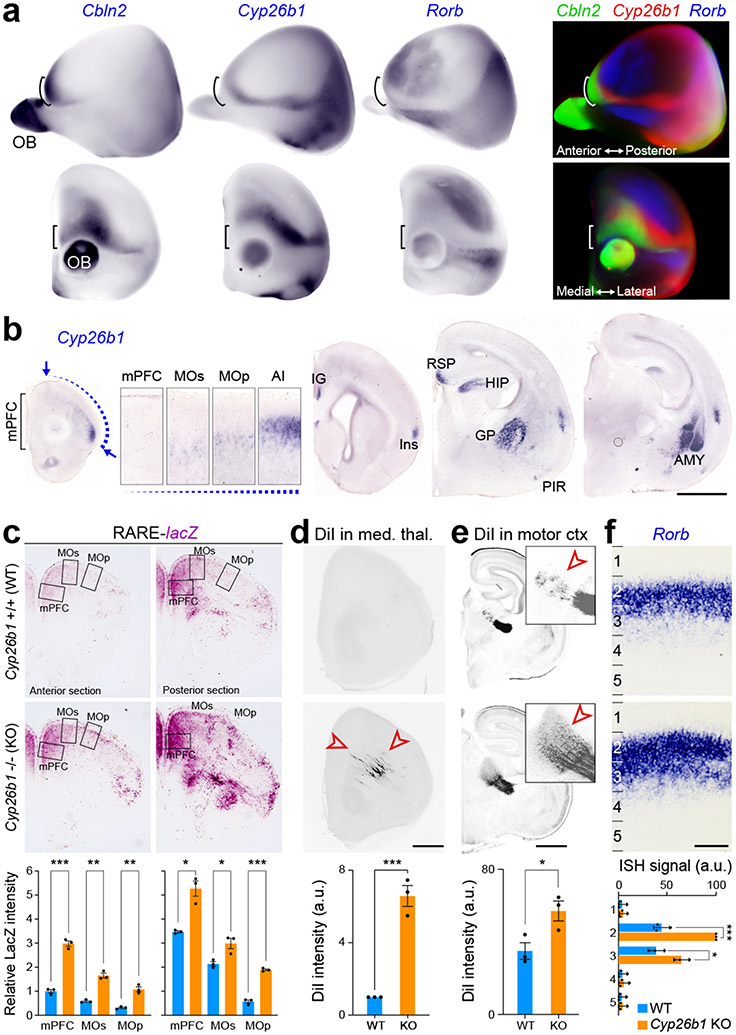
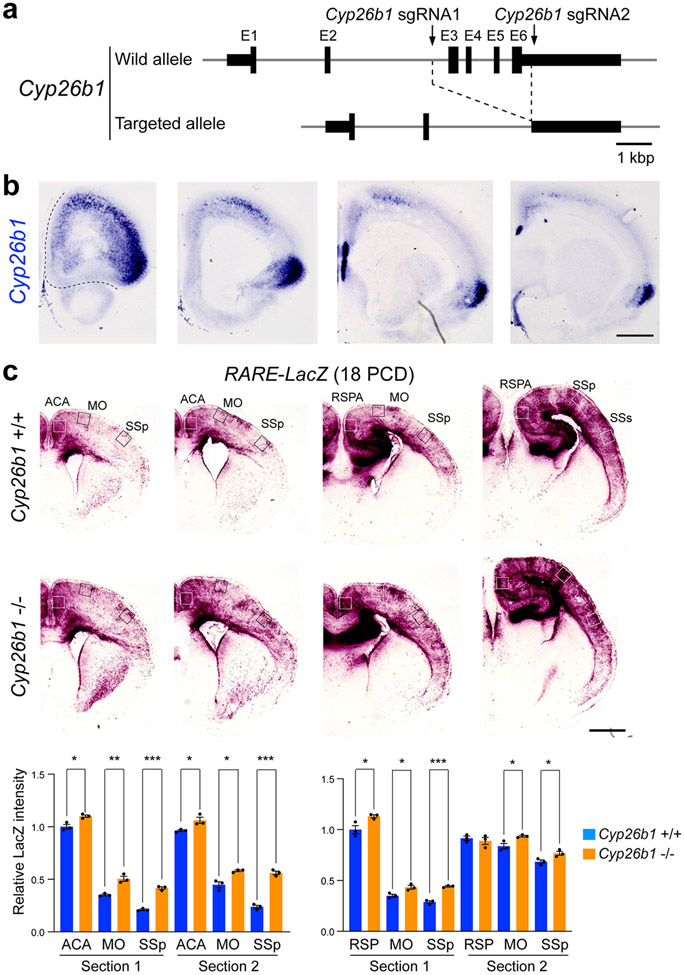
MD innervation of the primate frontal cortex is expanded laterally compared to rodents, with the primate PFC areas having a more prominent layer 4 10,11. Thus, we investigated whether the expansion of RA signaling is sufficient to increase and laterally extend MD innervation and alter layer 4 development in the mouse neonatal frontal cortex by genetically deleting the RA-degrading enzyme CYP26B1 in mice (Extended Data Fig. 18a). Consistent with previous findings in humans 21 and mice 24, whole-mount and serial tissue section in situ hybridization revealed that Cyp26b1 is upregulated in the prospective motor cortex (MOs/MOp) and anterior insula compared to the mPFC and OFC, defined by high expression of Cbln2, and SSp, defined by Rorb (Fig. 5a,b; Extended Data Fig. 18b). While mice with constitutive deletion of Cyp26b1 died perinatally, it has been previously shown that genetic deletion of Cyp26b1 in mice resulted in expansion of RA signaling in multiple organs prenatally 38 and postnatally in the mouse mPFC 24. To investigate the possible role of CYP26B1 in restricting RA signaling to the mouse medial frontal cortex, we generated Cyp26b1 KO mice that also harbored the RARE-lacZ transgene. The lack of Cyp26b1 resulted in spreading of RA signaling dorsolaterally toward the MOs and MOp regions of the RARE-lacZ reporter mouse line at postconceptional day (PCD) 18 (Fig. 5c). This increase was not robustly observed in more posterior regions (Extended Data Fig. 18c).
Fig. 5 ∣. Increased RA signaling promotes mPFC-medial thalamic connectivity.
a, Lateral (top) and coronal (bottom) views of whole-mount in situ hybridization (ISH) of PD 0 mouse cortex with pseudo-color merge of Cbln2, Cyp26b1 and Rorb. b, Expression of Cyp26b1 in the PD 0 mouse brain using ISH. Cyp26b1 expression displays a gradient from the insula (AI) to MOs (insets). Arrows indicate Cyp26b1 expression at the insula and MOs. Scale bars, 200 μm.. c, β-Galactosidase histochemical staining of the mPFC of Cyp26b1+/+ (WT); RARE-lacZ and Cyp26b1−/− (KO); RARE-lacZ mouse brains at PCD 18. Relative signal intensity was quantified in the boxed area compared anterior mPFC expression (mPFC, MOs and MOp). Two-tailed Student’s t-test: WT vs. Cyp26b1 KO: ***P = 1e-4; 6e-5, **P = 4e-4; 1e-3, *P = 4e-3; 2e-2; N = 3 per genotype; Errors bars: S.E.M. d,e, Representative images and quantification of DiI labeling in PCD 18 frontal MOp/s (d) and MD thalamus (e) after DiI was placed in the MD thalamus or mPFC of WT and Cyp26b1 KO brains. Two-tailed Student’s t-test: ***P = 4e-4; *P = 0.03; N = 3 per genotype and condition. Scale bars: 200 μm. f, Representative image and quantification of expression of Rorb in WT and KO mice in PCD 18 primordial motor cortex. Two-tailed Student’s t-test: WT vs. Cyp26b1 KO: ***P = 2e-4, *P = 0.01; N = 3 per genotype; Errors bars: S.E.M. AMY, amygdala; GP, Globus pallidus; HIP, Hippocampus; IN, insula; IG, Indusium griseum; OB, Olfactory Bulb; RSP, Retrosplenial cortex.
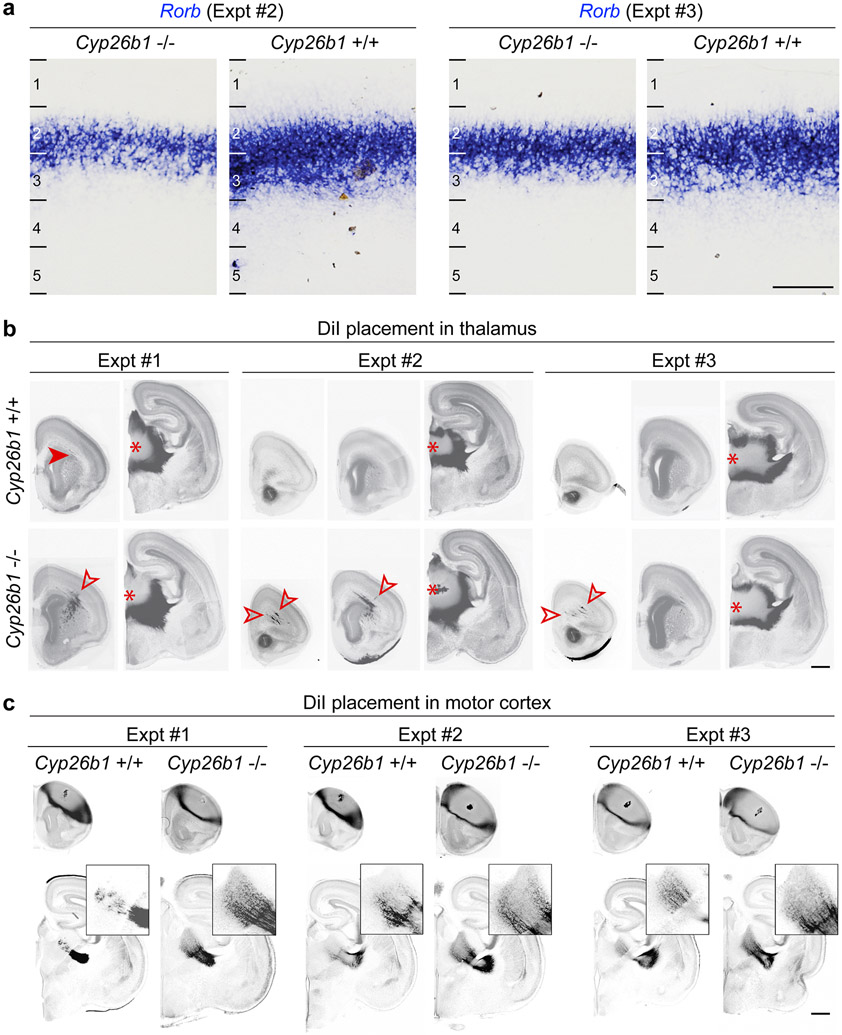
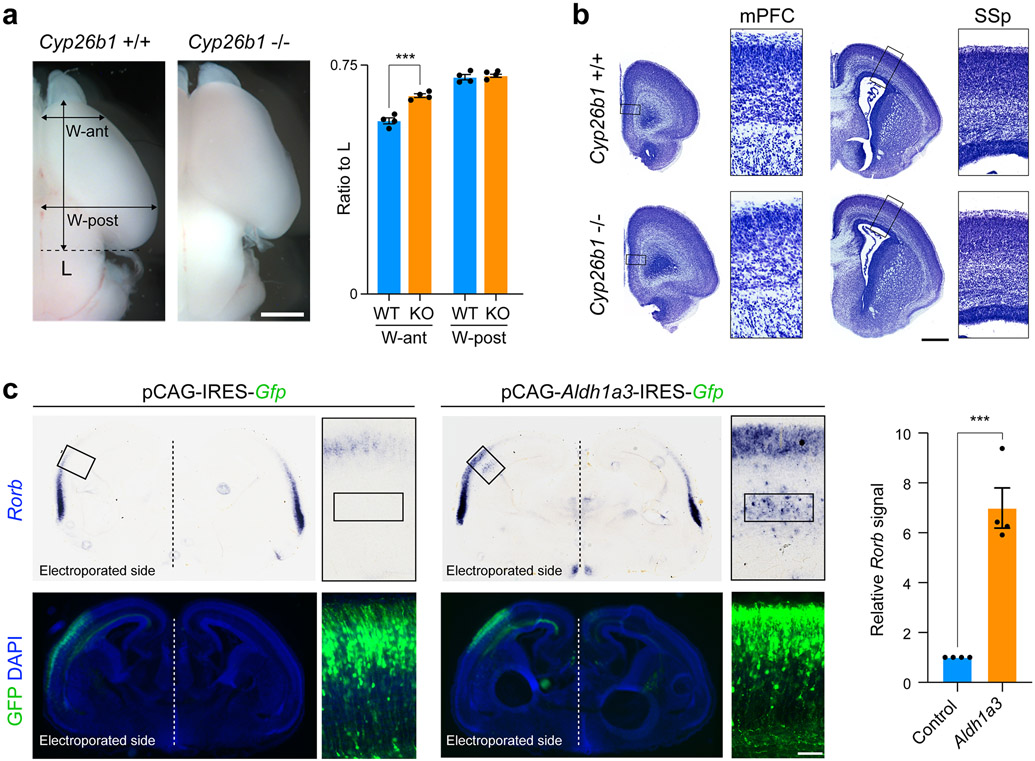
Next, we used Cyp26b1 KO mice to study the effects of expansion of RA signaling into the dorsolateral frontal cortex by placing DiI into the medial thalamus of fixed postmortem WT and KO brains, harvested at PCD 18 due to perinatal lethality of the KO mice. Histological analysis revealed that WT littermates had occasional thalamocortical axons within the medial and dorsolateral frontal white matter and cortex at this age. In contrast, KO mice showed precocious and robust innervation of both the medial and dorsolateral frontal cortex by the axons originating from the medial thalamus (Fig. 5d, Extended Data Fig. 19b). When DiI was placed in motor cortex, KO mice showed increased signal in the developing thalamus (Fig. 5e; Extended Data Fig. 19c). We also observed moderately enlarged frontal cortex with grossly normal cytoarchitecture of the cortical wall and cortical plate analyzed areas of the Cyp26b1 KO cortex (Extended Data Fig. 20a, b).
Furthermore, we observed an upregulation and expansion in laminar expression of layer 4 marker Rorb, in the motor cortex of neonatal Cyp26b1 KO mice (Fig. 5f; Extended Data Fig. 19a) and after misexpression of Aldh1a3 in the dorsolateral fronto-parietal cortex using in utero electroporation (Extended Data Fig. 20c). In summary, we identified that ectopic RA signaling in the perinatal mouse frontal cortex leads to expansion of medial thalamocortical innervation as well as the ectopic regional and laminar expansion of Rorb, expression both characteristics of the lateral granular PFC in anthropoid primates10,11.
Conclusions
We showed that RA signaling is required for proper prefrontal gene expression, spinogenesis, and long-range connectivity. We propose that the early/midfetal cortical expansion of RA signaling underlies the lateral expansion of PFC areas and MD innervation in primates. As the expansion of the prefrontal and temporal association areas have been proposed to be one of the evolutionary underpinnings of complex cognition 9-11, it will be important to explore whether RA signaling has a broader role in developmental specification and expansion of association areas, as well as disorders affecting cognition. Please see supplementary material for expanded discussion.
Methods
Analysis of human and macaque transcriptomic data
Developing human and macaque brain RNA-seq data (counts file) with its metadata information was obtained from BrainSpan (brainspan.org) and PsychENCODE (development.psychencode.org/#; evolution.psychencode.org/#) projects 17,19. Timeline of human and macaque development and associated periods were designed by Kang et al.39.
For human midfetal periods 4-6, a total of 73 mRNA samples corresponding to eleven prospective neocortical areas, comprising of the pial surface, marginal zone, cortical plate (layers 2-6) and adjacent subplate zone, of eleven prospective neocortical areas, from windows 3 and 4 (PCW 16-22) were considered for analyses (Extended Data Fig. 1). The human neocortical areas under study are orbital (oPFC/OFC), dorsolateral (dlPFC/DFC), ventrolateral (vlPFC/VFC), medial (mPFC/MFC) prefrontal cortex, and primary motor cortex (M1C) from the frontal lobe; primary somatosensory cortex (S1C) and posterior inferior parietal cortex (IPC) from the parietal lobe; primary auditory cortex (A1C), posterior superior temporal cortex (STC), and inferior temporal cortex (ITC) from the temporal lobe; and primary visual cortex (V1C) from the occipital lobe. A TMM normalization procedure was applied (function normalizeCounts from tweeDEseq package in R) to the expression of 15724 protein-coding genes that show sufficiently large counts (determined with function filterByExpr from edgeR package in R). To identify genes that are upregulated in a given brain lobe, we first applied RNentropy 40, available as a package in R, to determine which genes are differentially expressed among the eleven neocortical areas. Then, we considered a gene to be specifically overexpressed in a given lobe if i) there is at least one area in this lobe where the gene is significantly upregulated, ii) the gene is not upregulated in any area of the other lobes, and iii) the gene is under-expressed in at least 30% of the areas from the remaining lobes. Similarly, we identified genes that are specifically upregulated in the PFC compared to the M1C, or vice versa, by first running RNentropy pairwise comparisons between M1C and each of the prefrontal areas independently. Then, a gene was considered to be upregulated in PFC if i) it is upregulated in a prefrontal area in at least one of the comparisons, ii) it is not upregulated in M1C in any of the comparisons, and iii) it is under-expressed in M1C in at least three of the comparisons. A gene was considered to be upregulated in M1C if i) it is overexpressed in MOp in at least three of the comparisons, ii) it is not upregulated in any PFC area, and iii) it is under-expressed in a prefrontal area in at least one of the comparisons. Principal component analyses were performed using the prcomp function in R by centering the log2-transformed expression data of the selected genes. Significant GO terms were obtained via goana function from the limma package in R, reporting the ones with at least 10 genes in the background and at least 5 in the dataset. Frontal lobe, PFC and M1C upregulated genes were characterized for association with retinoic acid (RA) signaling, autism spectrum disorder (ASD) and schizophrenia (SCZ) using the following criteria. Association with RA signaling was defined by both dysregulation in PD 0 Rarb/Rxrg double knockout frontal cortex RNA-sequencing dataset and literature review identifying association with RA signaling. Association with ASD was based on https://gene.sfari.org/database/human-gene/. Association with SCZ was based on a total score of 3 or higher on http://www.szdb.org/gene_rank.php. Information is provided in Extended Data Table 3.
Predicted ages for macaque samples were calculated via the TranscriptomeAge algorithm described in Zhu et al.19. To perform statistical comparisons, samples from various developmental periods were grouped (periods 4-6, 7-10, and 11-14) and a two-tailed Student's t-test was used to compare gene expression levels between brain regions and species.
Animals
All studies using mice (Mus musculus) and rhesus macaques (Macaca mulatta) were performed in accordance with protocols approved by Yale University’s Institutional Animal Care and Use Committee and NIH guidelines. The animals were housed, and timed-pregnant prenatal and postnatal mouse and monkey brains were obtained in-house at the Yale Animal Resource Center.
Mice were reared in group housing less than five mice per cage at 25 °C and 56% humidity in a 12h light:12h dark cycle and provided food and water ad libitum with veterinary care provided by Yale Animal Resource Center. Both sexes were used and randomly assigned for all experiments. Animals were maintained on the C57BL/6J background. The day on which a vaginal plug was observed was designated as PCD 0.5 in mice. For timed pregnancies in monkey, females were housed with males for 3 days and the middle day was designated after observation of a vaginal plug and subsequent pregnancy as PCD 1. By this method, the estimated age of a monkey fetus has a maximal variation of ±1 day in its 165-day gestation. All monkeys were tested negative for herpes B virus and tuberculosis. RARE-lacZ (Tg(RARE-Hspa1b/lacZ)12Jrt) mice and timed pregnant CD-1 mice for in utero electroporation were purchased from Jackson Laboratory and Charles River Laboratories, respectively.
Postmortem human and macaque tissue
This study was conducted using postmortem human brain specimens or RNA-seq data generated previously 17 from tissue collections at the Department of Neuroscience at Yale School of Medicine, the Brain and Tissue Bank for Developmental Disorders at the University of Maryland, the Clinical Brain Disorders Branch of the National Institute of Mental Health, the Human Fetal Tissue Repository at the Albert Einstein College of Medicine, the Birth Defects Research Laboratory at the University of Washington (R24HD000836), Advanced Bioscience Resources and the Joint MRC–Wellcome Trust Human Developmental Biology Resource (www.hdbr.org; MR/R006237/1). Tissue was collected after obtaining parental or next of kin consent and with approval by the institutional review boards at each institution from which tissue specimens were obtained, the Yale University and the National Institutes of Health. Donated deidentified tissue was handled in accordance with ethical guidelines and regulations for the research use of human brain tissue set forth by the National Institutes of Health (https://oir.nih.gov/sites/default/files/uploads/sourcebook/documents/ethical_conduct/guidelines-biospecimen.pdf) and the WMA Declaration of Helsinki (https://www.wma.net/policies-post/wma-declaration-of-helsinki-ethical-principles-for-medical-research-involving-human-subjects/). All available non-identifying information was recorded for each specimen. No obvious signs of neuropathological alterations were observed in any of the human, macaque or mouse specimens analyzed in this study. The postmortem interval was defined as hours between time of death and time when tissue samples were fresh frozen or started to undergo fixation process.
Generation of Rarb, Rxrg and Cyp26b1 knockout mice using CRISPR-Cas9 gene editing technique
The overall strategy for the generation of Rxrg and Rarb KO mice follows a previously described protocol using CRISPR-Cas9 genome editing technique 41. For the construction of the templates of guidance RNAs, two sets of top and bottom strand oligomers (see Extended Data Table 4) directing the double-strand break at targeting sites were annealed and ligated into BbsI site of pX330-U6-Chimeric_BB-CBh-hSpCas9 vector 42, which was purchased from Addgene (Plasmid #42230). After amplification of insert with T7-tagged primers (see Extended Data Table 5), guidance RNAs were synthesized by T7 RNA polymerase. The coding region of Cas9 was PCR amplified using pX330-U6-Chimeric_BB-CBh-hSpCas9 as a template and inserted into the pSP64 Poly(A) vector (Promega, Cat. P1241). Vectors were digested and linearized with EcoRI. Cas9 mRNA was synthesized by SP6 RNA polymerase. Guidance RNAs and Cas9 mRNA were purified by MEGAclear Transcription Clean-Up Kit (Ambion, AM1908). Cas9 mRNA and two guidance RNAs were mixed at a concentration of 10 ng or 100 ng/μl in the microinjection buffer (5 mMTris-HCl pH7.5; 0.1 M EDTA) and injected into the pronuclei of fertilized eggs from B6SJLF1/J mouse strain purchased from The Jackson Laboratory. The fertilized eggs were then transferred to uterus of female CD-1 mouse strain, purchased from Charles River Laboratories. The first generation (F0) mice with recombined alleles were identified by PCR with two primer sets designed outside and inside of targeted area (Extended Data Table 5; Extended Data Fig. 9d), confirmed by genome DNA sequencing. The germline transmission in F1 generation was confirmed by the same sets of PCR primers. For generation of Rxrg KO mice, a pair of guidance RNAs flanking whole exon 3 and 4 were designed to delete a large part of DNA binding domain (Extended Data Fig. 8d) 43. For generation of Rarb KO mice, a pair of guidance RNAs were designed to delete the whole of exon 9 and a part of exon 10 (Extended Data Fig. 9d). As a result, α-helical sheets of H4 to H8 in the ligand binding domain were deleted, and a frameshift occurred in the rest of C-terminal region, which results in total abolition of receptor activity 43. For generation of Cyp26b1 KO mice, a pair of guidance RNAs were designed to delete the whole of exon 3 and 6, as described previously, to abolish enzymatic activity (Extended Data Fig. 18a) 44. All primer sequences are listed in Extended Data Table 5.
Human primary neocortical cultures and differentiation
Fresh tissues from prenatal human brain specimens (PCW 8-HSB#765, sex not determined; PCW 20-HSB#781, female; PCW 23-HSB#784, male) were maintained in ice-cold Hibernate-E (ThermoFisher Scientific, Cat. A1247601) and processed within 12 to 18 hours. Primary cortical neural stem cells from PCW 8 cortical tissue were isolated from the dissected neocortical proliferative zones (i.e., ventricular zone and subventricular zone). Primary cortical neural progenitors and neurons from PCW 20 or 23 cortical tissue were isolated from dissected neocortical walls, including cortical plate zones. Briefly, the tissue was mechanically separated into small pieces, incubated with 2 mg/ml papain (Brainbits, PAP) for 20 min, and gently triturated to a single cell suspension with 0.1 mg/ml DNase I (STEMCELL Technologies, Cat. 07900) in HBSS (ThermoFisher Scientific, Cat. 88284). Cortical neural stem cell expansion and differentiation were performed as previously described 45. Cells were plated onto poly-L-ornithine/laminin-coated wells at a density of 2 × 105 cells in 24 well plate (IBIDI) and cultured with DMEM/F12 supplemented with 1x N2 (ThermoFisher Scientific, Cat. 17502048), 1x B27 (ThermoFisher Scientific, Cat. 17504044), 10 ng FGF2 (R&D Systems, Cat. 3718-FB) and 1% penicillin/streptomycin (ThermoFisher Scientific, Cat. 15140122). The expanded cortical stem cells were replated onto poly-L-ornithine/laminin-coated wells at a density of 1 × 105 cells in a 24 well plate, and differentiation to neurons was induced 2 days after plating by FGF2 withdrawal. The isolated cortical neural progenitors and neurons were plated onto poly-L-ornithine/laminin-coated wells at a density of 2 × 105 cells in 24 well plate (IBIDI) and cultured without FGF2. On day 4 after the FGF2 withdrawal of cortical neural stem cell culture and day 2 of cortical progenitor and neuron culture, the medium was replaced with Neurobasal medium (ThermoFisher Scientific, Cat. 21103049) containing 1x N2 (ThermoFisher Scientific, Cat. 17502048), 1x B27 (ThermoFisher Scientific, Cat. 17504044), 10 ng/ml BDNF (Abcam, Cat. 9794), 10 ng/ml GDNF (R&D Systems, Cat. 212-GD) and 1% penicillin/streptomycin (Fisher Scientific, Cat. 15140148). After neural differentiation and further maturation (Extended Data Fig. 15a), cortical neurons were exposed to varying doses of all-trans retinoic acid (Sigma-Aldrich, Cat. R2625-50MG) or pan-retinoic acid receptor antagonist AGN193109 (Tocris, Cat. 5758) for another 14 days. See Extended Data Fig. 15a for summary.
Human and chimpanzee cerebral organoid culture
For maintenance of human (cell line: HSB311 #36 46,47) and chimpanzee (cell line: 3651D 48), induced pluripotent stem cells were dissociated to single cells with Accutase (ThermoFisher Scientific, Cat. 00-4555-56), plated at a density of 1 X 105 cells/cm2 in a Matrigel (BD)-coated 6 well plates (Falcon) with mTeSR1 (STEMCELL Technologies, Cat. 85850) containing 5 μM Y27632, ROCK inhibitor (Sigma-Aldrich, Cat. SCM075). ROCK inhibitor was removed at 24 hours after plating, and cells were cultured for another 4 days before the next passage. Cerebral organoids were generated by directed differentiation protocol as previously described 45-47. Human and chimpanzee induced pluripotent stem cells were dissociated into single cells using Accutase (ThermoFisher Scientific, Cat. 00-4555-56). Neural induction was directed by dual SMAD and WNT inhibition using neural induction medium supplemented with 100 nM LDN193189 (STEMCELL Technologies, Cat. 72147), 10 μM SB431542 (Selleck Chemicals, Cat. S1067) and 2 μM XAV939 (Sigma-Aldrich, Cat. X3004-5MG). The dissociated cells were reconstituted with the neural induction medium and plated at 10,000 cells per well in a 96 well v-bottom ultra-low-attachment plate (Sumitomo Bakelite). To increase the cell survival and aggregate formation, 10 μM Y-27632 (Sigma-Aldrich, Cat. SCM075) was added for the first one day. After 10 days of stationary culture, organoids were transferred to 6-well ultra-low-attachment plate (Millipore Sigma) and kept onto an orbital shaker rotating at speed of 90 rpm/min to enhance the nutrient and gas exchanges. From day 18, organoids were cultured with neural differentiation media supplemented with 1x CD lipid concentrate (ThermoFisher Scientific, Cat. 11905031), 5 ug/ml heparin (STEMCELL Technologies, Cat. 07980), 20 ng/ml BDNF (Abcam, Cat. 9794), 20 ng/ml GDNF (R&D Systems, Cat. 212-GD), 200 μM cAMP (Sigma-Aldrich, Cat. 20-198), and 200 μM ascorbic acid (Sigma-Aldrich, Cat. A92902) for further neuronal maturation (Extended Data Fig. 11a). On day 133, an RA-soaked bead was attached to each cerebral organoid by embedding with and solidifying the growth factor-reduced Matrigel (Corning. Cat. 354230) and further cultured for 48 hours before collection.
Preparation of RA-soaked bead
AG1-X2 Resin (150 or 200 μm in diameter, Bio Rad, Cat. 140-1231) were added with 1 M formic acid for 1 hour and washed with distilled H2O (5 x 5 min). Beads were completely dried in 37°C incubator overnight. Dried beads were soaked in either 1 mg/ml or 5 mg/ml of all-trans-retinoic acid (Sigma-Aldrich, Cat. R2625-50MG) dissolved in DMSO (Sigma-Aldrich, Cat. D8418) for 1 hour and then washed twice in DMEM (Gibco) before being placed onto human or chimpanzee organoids.
In situ hybridization
Whole-mount and section in situ hybridization was performed as described previously 49. Antisense digoxigenin (DIG)-labeled RNA probes were synthesized using DIG RNA Labeling Mix (Roche, Cat. 11277073910). Human and mouse ALDH1A1 (Clone ID 2988388 and 6477503, respectively), ALDH1A3 (Clone ID 6208628 and 6515355, respectively), RXRG (Clone ID 4635470 and 5707723, respectively), RARB (Clone ID 30341884 and 30608242, respectively), and mouse Rorb (Clone ID 5358124), Cbln2 (Clone ID 6412317), Cyp26b1 (Clone ID 6400154). cDNAs were purchased from GE Healthcare for template preparation. Mouse Meis2 DNA was a gift from John Rubenstein. For macaque in situ hybridization, human probes were used because of high similarity between human and macaque transcripts (97.7% identity in ALDH1A1; 95.4% identity in ALDH1A3; 97.7% in RXRG; 98.8% in RARB). Sections were obtained from PCW 21, 22 human brains and PCD 110, 140 macaque brains. In situ hybridization were repeated using these 2 sets. Images were taken using Aperio CS2 HR Scanner (Leica; Wetzlar, Germany) and processed by Aperio ImageScope (ver. 12.4.3.5008, Leica). In Fig. 5a, Cbln2, Cyp26b1, and Rorb expression data were merged by converting color whole-mount in situ hybridization into black-and-white images, then merged as separate RGB channels. Images were aligned manually by N.S. In Fig. 5f, and Extended Data Fig. 19a the cortical plate was divided into 5 equal bins and Rorb intensity in each bin was quantified using ImageJ (ver.2.0.0-rc-69/1.52p).
Enzyme-linked immunosorbent assay (ELISA)
Eleven neocortical areas were dissected from four fresh frozen postmortem human midfetal brains (PCW 16, 18, 18 and 19) and four fresh frozen macaque brains (all four PCD 110) as described in Zhu et al. 2018 19. Twelve neocortical areas were dissected from three fresh mouse brains at P1, based on Paxinos, 2007 50 and the Allen Mouse Brain Atlas (mouse.brain-map.org/static/atlas) 51. Each human and macaque brain sample was further microdissected into three pieces and weighed. Each piece was independently homogenized using a Dounce homogenizer in three to four volumes of homogenizing buffer (isopropanol: ethanol=2:1; 1 mg/ml butylated hydroxytoluene), followed by centrifugation at 10K rpm for 10 minutes at 4 °C. Supernatant was used for both determination of protein concentration by Pierce BCA Protein Assay Kit (Thermo-Fisher Scientific, Cat. 23227), and RA concentrations using ELISA colorimetric detection Kit according to the manufacturer's instructions (MyBioSource, Cat. MBS705877). This kit could not distinguish all-trans-retinol, all-trans-retinal or. all-trans-retinoic acid). Thus, the concentration of RA was concentration of all forms.
Quantitative reverse transcription-PCR
Total RNA was isolated from freshly microdissected cortices after removal of the olfactory bulb and striatum (Extended Data Fig. 9a,b) using Trizol (Thermo-Fisher Scientific, Cat. 15596026). cDNAs were prepared using SuperScript II (Thermo-Fisher Scientific, Cat. 18064022) from three independent WT cerebral hemispheres. Quantitative reverse transcription (RT)-PCR was performed as described previously 52 using 7000HT Sequence Detection System (Applied Biosystems). At least three replicates per transcript were used for every reaction. The copy number of transcripts was normalized against the house keeping TATA-binding protein (TBP) transcript level. For Rxra,b,g, Rara,b,c and Tbp primer sets, correlation (R2) was higher than 0.98, and the slope was −3.1 to −3.6 in each standard curve. Primers to detect the expression of the genes above were designed in a single exon. Primer sequences are listed in Extended Data Table 5.
β-Galactosidase histochemical staining
Brains were dissected from PD 0 RARE-lacZ mouse pups and drop-fixed in 4% paraformaldehyde for 2 hours at 4 °C, followed by embedding in O.C.T. Compound (ThermoFisher Scientific, Cat. 23-730-572). Brains were sectioned at 20 μm by cryostat (Leica CM3050S) after they were frozen. β-Galactosidase staining followed the protocol described by Kokubu et al 53. We used Red-gal (Sigma-Aldrich, Cat. RES1364C-A102X) for the chromogenic reaction. Intensity of β-Galactosidase staining was quantified using ImageJ (ver.2.0.0-rc-69/1.52p).
Nissl staining
Postmortem brains were immersion fixed in 4% paraformaldehyde overnight at 4°C, followed by embedding in OCT. Brains were sectioned at 15-20 μm by cryostat (Leica, CM3050S) after they were frozen. After PBS wash, sections were dehydrated using increasing concentration of ethanol, followed by Cresyl violet, wash, and second ethanol dehydration. Sections were cover-slipped with Permount (Fisher Scientific, Cat. 15820100).
Immunohistochemistry
Human (PCW 12, PCW 16, PCW 19, PCW 20, PCW 21, PCW 22, PCW 24, newborn, and adult [42, 47 and 79 years old; PMI 5-15 hours]), macaque (PCD 40, PCD 76, PCD 114, PCD 140, adult [4.5, 8 and 11 years old; PMI under 1 hour]), and mouse (PD 0 and adult [4 months]) brain tissue samples, the cultured human cortical neurons, and the cerebral organoids were fixed in 4% paraformaldehyde at 4°C. Brain tissues and cerebral organoids were frozen and embedded in O.C.T. compound (ThermoFisher Scientific, Cat. 23-730-572). Embedded Samples were sectioned at 15-20 μm for mouse brain and cerebral organoids, and 50 μm for macaque and human brain by cryostat (Leica, CM3050S). Tissue sections were mounted and dried overnight.
For immunohistochemistry of mouse brain in Extended Data Fig. 16d,e,f, sections were treated with or without R-Buffer AG pH 6.0 (Electron Microscopy Sciences, Cat. 62707-10) for antigen retrieval, followed by washing in PBS (3 × 5 min) and incubation in blocking buffer (5% (v/v) normal donkey serum (Jackson ImmunoResearch Laboratories, Cat. 017-000-121), 1% (w/v) bovine serum albumin, (Sigma-Aldrich, Cat. A9647-100G), and 0.3% (v/v) Triton X-100 in PBS) for 1 hour at room temperature. Sections were incubated for 24-48 hours at 4°C with diluted primary antibodies in blocking buffer. Sections were then washed in PBS (3 × 5 min) incubated with fluorescent secondary antibodies for 2 hours at room temperature. After washing in PBS (3 × 5 min), Tissue was cover-slipped with Vectashield (Vector laboratories, Cat. H-1000). For immunohistochemistry of human, macaque, and mouse brain sections in Extended Data Fig. 4d,e; 7b antigen retrieval was performed using R-Buffer A pH 6.0 (Electron Microscopy Sciences, Cat. 62706-10) for postnatal tissue or R-Buffer AG pH 6.0 (Electron Microscopy Sciences, Cat. 62707-10) for prenatal tissue. Sections were incubated in 1% hydrogen peroxide/PBS to quench endogenous peroxidase activity. Sections were washed in PBS (3 × 10 min) and incubated in blocking solution containing 5% (v/v) normal donkey serum (Jackson ImmunoResearch Laboratories, Cat. 017-000-121), 1% (w/v) bovine serum albumin (Sigma-Aldrich, Cat. A9647-100G), and 0.3% (v/v) Triton X-100 in PBS for 1 h at room temperature. Primary antibodies were diluted in blocking solution and incubated with tissues sections overnight at 4°C. Sections were washed with PBS (3 × 10 min) prior to being incubated with the appropriate biotinylated or fluorescent secondary antibodies (Jackson ImmunoResearch Labs) for 1.5 h at room temperature. All secondary antibodies were raised in donkey and diluted at 1:250 in blocking solution. Sections were subsequently washed in PBS and incubated with avidin-biotin-peroxidase complex (Vectastain ABC Elite kit; Vector Laboratories) for 1 h at room temperature. Finally, sections were washed in PBS (3 × 15 min) and signals were developed using a DAB Peroxidase (HRP) Substrate Kit (Vector Laboratories, Cat. NC9276270) according to the manufacturer’s protocol. Following washes in PBS, sections were dried, dehydrated, and cover-slipped with Permount (Fisher Scientific, Cat. 15820100). For immunofluorescence, sections were washed in PBS with 0.3% (v/v) Triton X-100 and treated with the Autofluorescence Eliminator Reagent (Millipore, Cat. 2160) according to manufacture instructions, and cover-slipped with Vectashield (Vector laboratories, Cat. H-1000). For immunocytochemistry shown in Extended Data Fig. 15, the fixed primary cortical neurons were washed in PBS (3 × 15 min) and incubated with 0.3% (v/v) Triton X-100 and 10% (v/v) donkey serum (Jackson ImmunoResearch Laboratories, Cat. 017-000-121) in PBS for 40 min. For immunohistochemistry shown in Extended Data Fig. 11a,b, the cerebral organoid sections were washed in PBS (3 × 15 min) and incubated with 0.5% (v/v) Triton X-100 and 10% (v/v) donkey serum (Jackson ImmunoResearch Laboratories, Cat. 017-000-121) in PBS for 2 h. Primary cortical neurons and the organoid sections were incubated with primary antibodies in 10% (v/v) donkey serum at 4°C overnight. Samples were then washed in PBS (3 × 5 min), and incubated with fluorescent secondary antibodies for 2 hours at room temperature in 10% (v/v) donkey serum. All sections and tissues processed for immunofluorescence were cover-slipped with Vectashield (Vector laboratories, Cat. H-1000), after washing in PBS (3 × 5 min).
The sources of primary antibodies were anti-ALDH1A1 (1:500, Abcam, Cat. 52492), anti-BCL11B/CTIP2 (1:500, Abcam, Cat. ab18465), anti-cleaved capase3 (1:500; Cell Signaling, Cat. 9611), anti-CUX1 (1:500, Santa Cruz Biotechnology, Cat. sc-13024), anti-GFAP (1:2000, Sigma-Aldrich, Cat. G3893), anti-NR4A2 (1:100, R&D systems, Cat. AF2156), anti-GAD1 (1:50, R&D systems, Cat. AF2086), anti-NRGN (1:50, R&D systems, Cat. AF7947), anti-PAX6 (1:200; R&D Systems, Cat. AF8150), anti-POU3F2/BRN2 (1:500, Sigma-Aldrich, Cat. SAB2702086), anti-DLG4/PSD95 (1:500; Invitrogen, Cat. 51-6900), anti-SYP (1:2000, Sigma-Aldrich, Cat. SAB4200544), anti-L1CAM (1:500; Millipore-Sigma, Cat. ABT143), anti-RORB (1:500, Novus Biologicals, Cat. NBP1-82532), anti-SATB2 (1:200, Abcam, Cat. ab92446), anti-SOX2 (1:200, R&D Systems Cat. MAB2018), anti-TBR1 (1:200, Abcam, Cat. ab31940), anti-TH (1:1000, Immunostar, Cat. 22941), and anti-TUBB3 (1:400; TuJ1 clone, Abcam, Cat. 18207). Secondary antibodies: Alexa Fluor 488-, 594-, or 647-conjugated AffiniPure Donkey anti-IgG (1: 200; Jackson ImmunoResearch). For all microscopic analysis, images were acquired using Aperio CS2 HR Scanner (Leica; Wetzlar, Germany), ApoTome.2 microscope (Zeiss), LSM510 META (Zeiss), LSM800, or LSB880 confocal microscope (Zeiss) and assembled in Aperio ImageScope 12.4.3.5008 (Leica), Zeiss ZEN2009, ImageJ (ver. 2.0.0-rc-69/1.52p), Adobe Photoshop (ver. 12.0 ×64), and Adobe Illustrator (ver. 23.1.1).
Western blotting.
Five prospective neocortical areas (mPFC, dlPFC, vlPFC, oPFC, and M1C) were dissected from fresh frozen brains of human and macaque, and three neocortical areas (mPFC, Mos/p, and OFC) from fresh mouse brains. Dissected tissues were lysed by sonication in RIPA buffer with protease inhibitors (Sigma-Aldrich, Cat. 11836153001). Following centrifugation at 10,000 xg, supernatant protein concentrations were quantified using Pierce BCA Protein Assay Kit (Thermo-Fisher Scientific, Cat. 23227). Supernatants were mixed with 4x Laemmli Sample Buffer (Bio Rad, Cat. 1610747), boiled, and electrophoresed on NuPage Bis-Tris gels (ThermoFisher Scientific), followed by protein transfer to PVDF membranes (Bio Rad, Cat. 1620174). Blotted membranes were incubated in blocking buffer (TBS/5% milk) for 1 hour and then transferred to blocking buffer with primary antibodies diluted at 1:1000 at 4°C overnight. Membranes were then washed in TBS/ 0.1% Tween20 (3 × 5 min), and incubated with secondary antibodies conjugated with HRP. Secondary antibodies were diluted at 1:1000. After washing with TBS/ 0.1% Tween20 (3 × 5 min), protein bands were visualized using SuperSignal West Pico Chemiluminescent Substrate (ThermoFisher Scientific, Cat. 34580). The sources of primary antibodies were anti-ALDH1A3 (1:2000, Novus Biologicals, Cat. NBP2-15339), and anti-GAPDH (1:5000, Abcam, Cat. 9485). Of note, two bands were identified using anti-ALDH1A3 antibody, a 56 kDa band likely ALDH1A3 and a 54 kDa band likely ALDH1A1.
Droplet digital PCR for gene expression quantitation
An aliquot of the total RNA that was extracted from meninges (human: PCW 19, 19, and 20; macaque: PCD 80, 80, and 110) or primary neural cultures were used for secondary validation through ddPCR analysis. 200 ng of total RNA was used for cDNA synthesis using SuperScript III First-strand synthesis Supermix (ThermoFisher Scientific, Cat. 18018400) and subsequently diluted with nuclease-free water. ddPCR was carried out using the Bio-Rad QX100 system. After each PCR reaction mixture consisting of ddPCR master mix and custom primers/probe set was partitioned into 15,000–20,000 droplets, parallel PCR amplification was carried out. Endpoint PCR signals were quantified and Poisson statistics was applied to yield target copy number quantification of the sample. Two-color PCR reaction was utilized for the normalization of gene expression by the housekeeping gene TBP. All pre-designed assays used in validation can be found in Extended Data Table 5. For each region, the gene expression was compared between species by one-way ANOVA followed by Dunnet’s multi comparison test.
Quantification of postsynaptic and presynaptic puncta marked by immunostaining
For each region of both WT and dKO mice, using the 488 nm or 594 nm channels to detect either synaptophysin (SYP) immunolabelling, DLG4/PSD95 immunolabelling and DAPI nuclear counterstain, seven serial optical sections at 0.8 μm intervals over a total depth of 5 μm were imaged and the 2nd, 4th, and 6th images were eliminated from further analysis to avoid overlap in counting 54. Area of each image is 0.079 mm2. The number of SYP and PSD95 immunolabelled puncta on each image was counted using ImageJ (ver. 2.0.0-rc-69/1.52p) using the automated Analyze Particles function using a threshold of 985 to 4095, determined based on multiple WT and dKO images. At least two sections from each animal were selected for counting, and at least 3 animals for each genotype were used. The total number and volume of SYP and DLG4/PSD95 immunolabelled puncta and TuJ1 (anti-TUBB3 antibody) or MAP2 in cultured neurons were analyzed with 20 z-stack (0.49 μm intervals) images per each 8 field for each condition and the total number of DLG4/PSD95 immunolabelled puncta and TuJ1 immunolabelled neurons in cerebral organoids were analyzed with 5 z-stack images using Volocity (ver. 6.3.1) and Spotfire (ver. 11.2.0) software.
Retrograde neuronal tracing with adeno-associated viruses
Briefly, WT (N = 4) and Rarb;Rxrg dKO (N = 4) animals were anesthetized by injecting Ketamine/Xylazine solution and head fixed in the stereotactic frame. Animals were injected with Burenorphine 30 mins prior to the surgery. After lubricating the eyes and shaving the fur, and incision of < 1mm was made. A craniotomy was made with the round 0.5 mm drill bit at the desired co-ordinates (mPFC: ML ±0.35, AP 1.5, DV 2.5 from bregma). Using a 0.5 μl Hamilton neuros syringe, we inject 50 nl of AAVrg-Cag-Gfp (Addgene, #37825-AAVrg) into the mPFC. To prevent the virus from spreading along the injecting tract, the needle was held in place for at least 10 min. After injections, the skin was sutured and animals were returned to the cage. Post-surgery, animals were injected with Meloxicam for 48 hours. Three weeks later, the animals were sacrificed and brains were collected. The brains were coronally sectioned on vibratome to obtain 70 μm thick sections. After staining the sections with anti-GFP antibody (1:500, Abcam, Cat. ab13970) and DAPI the sections were imaged in LSM 800 microscope (ZEISS). Intensity and density of labelling was quantified on a scale of 0 to 3 with 0 being no labelling, 1 being weak or sparse cellular labelling (<10%), 2 being strong and less than 50% of cell labelled, and 3 being dense labelling. This discrete approach of quantification was used due to variability in injection site.
Quantification of dendritic spines and arborization from viral tracing sections
The contralateral mPFC to the injection site was used for this analysis; WT (N = 4) and Rarb;Rxrg dKO (N=4). For Sholl analysis, the images of entire neurons were acquired at 20X magnification. For spine counts, Z-stack images across entire dendritic thickness with 29-33 images/ stack were obtained. The Z-stack images were opened in the Reconstruct (ver. 1.1.0.0, Boston, MA, USA) publicly available at https://www.bu.edu/neural/Reconstruct.html 55 and a new series was recreated which enabled us to move across different stacks across z-planes in same image. Whole dendrite was subdivided into segments of 10 μm and number of spines across whole thickness were traced for length and breadth of each spine. After tracing, the length and breadth of the spines their ratio was used to determine the spine subtype as described earlier 56. After the analysis for each class of spine, standard deviation and p-values are calculated using two-way ANOVA with Sidak’s multiple comparison method. For Sholl analysis, Z-stack images were opened in ImageJ (ver. 2.0.0-rc-69/1.52p) and dendritic arbors were manually traced using the NeuronJ plugin 57. Dendritic complexity was then quantified and plotted using the Sholl Analysis option.
Plasmid construction
For construction of expression vectors, full-length cDNAs (mouse Aldh1a3, Clone ID 6515355, purchased from GE Healthcare) were inserted into pCAGIG vector (pCAGIG was obtained from Addgene (Plasmid #11159).
In utero electroporation
In utero electroporation was performed as previously described 52. Plasmid DNA (4 μg/μl) was injected into the lateral ventricle of embryonic mice at E13.5-E14.5 and transferred into the cells of the ventricular zone by electroporation (five 50-ms pulses of 40V at 950-ms intervals). Brains were dissected at PD 0. Brains and tissue sections of electroporated animals were analyzed for GFP expression after fixation with 4% paraformaldehyde for 12 hours at 4 °C.
Mouse RNA-seq data generation and analysis
Postmortem mouse brains were dissected at PD 0 in ice-cold sterile PBS, fresh frozen, and stored at −80 °C. Brains were incubated in RNAlater-Ice at −20 °C for 12-16 hours prior to further dissection. mPFC, MOs, and OFC were microdissected based on Paxinos, 2007 50 and the Allen Mouse Brain Atlas (mouse.brain-map.org/static/atlas) 51 and RNA was isolated using RNeasy Plus Micro kit with additional on-column DNAase step (Qiagen, Cat. 74134). RNA quality and amount were quantified using High Sensitivity RNA Screen Tape Assay (Agilent, Cat. 5067-5579), and concentration was standardized to 10 ng/μl. SMART-seq v4 Ultra Low Input Kit (Takara Bio, Cat. 634890) was used to create cDNA, and concentration was quantified using Quant-iT PicoGreen dsDNA Assay Kit (ThermoFisher Scientific, Cat. P11496). Nextera XT DNA library Preparation Kit (Illumina, Cat. FC-131-1024) were used to create cDNA libraries for sequencing. Libraries were normalized and sequenced at the Yale Center for Genomic Analysis (YGCA) using the NovaSeq with 100 bp paired end reads. Reads from each library were mapped against the mouse assembly GRCm38 using STAR v.2.6.0a (gtf and fasta files downloaded from Ensembl version 94; parameters: --readFilesIn $j1 $j2 --outSAMattributes All --outFilterMultimapNmax 1 --outSAMstrandField intronMotif --outFilterIntronMotifs RemoveNoncanonical --quantMode TranscriptomeSAM --outFilterMismatchNoverLmax 0.1 --alignSJoverhangMin 8 --alignSJDBoverhangMin 1 --outSAMunmapped Within --outFilterType BySJout). Counts were obtained using featureCounts v1.6.2 with -p parameter.
To compare the gene expression patterns of three WT vs three Rarb/Rxrg dKO mice, a TMM a procedure was applied (function normalizeCounts from tweeDEseq package in R) to the expression of 15085 protein-coding genes that show sufficiently large counts (determined with function filterByExpr from edgeR package in R). We assessed DEx genes in each brain region (mPFC, OFC, and MOs) running RNentropy independently among WT and dKO mice per region. Genes overexpressed in a given condition are those that are both significantly upregulated in that condition and significantly downregulated in the opposite condition according to RNentropy. The same criterion was applied for identification of downregulated genes. Genes with an inconsistent pattern of expression between regions were excluded. Principal component analyses were performed using the prcomp function in R by centering the log2-transformed expression data of the selected genes. Significant GO terms were obtained via goana function from the limma package in R and plotted using function GOBubble from GOplot package in R. Fisher test enrichments calculated for RA related genes (RA synthesis: Rdh10, Rdh5, Aldh1a1/Raldh1, Aldh1a2/Raldh2, Aldh1a3/Raldh3, Adh1, Adh5, Adh7; RA degradation: Cyp26a1, Cyp26b1, Cyp26c1: RA receptors: Rara, Rarb, Rarg, Rxra, Rxrb, Rxrg; RA binding: Ttr, Rlbp1, Rbp1, Rbp2, Rbp3, Rbp4, Fabp5), genes overexpressed in individual lobes of the midfetal human cortex based on Fig. 1, and neuropsychiatric disease related genes (downloaded from Li et al. 30) in up- and downregulated genes. Genes associated with the GO terms: “axon guidance”, “axon guidance receptor”, “axon development”, and “ephrin” were manually screened for anterior to posterior gradient using developingmouse.brain-map.org 58, gensat.org 59 and Elsen et al.60. Mouse RNA-seq data were deposited into the NCBI GEO database (Accession number GSE142851).
Diffusion-weighted magnetic resonance imaging and tractography
Five PD 5 postmortem dKO homozygotes and five WT C57BL/6 mouse brains were drop fixed in 4% paraformaldehyde solution in 0.1 M PBS for 48 hours. They were subsequently transferred to 0.1 M PBS and just before imaging to Fomblin (Sigma-Aldrich, Cat. 317926). The diffusion-weighted images were acquired on a Bruker BioSpin 9.4 T MRI (Bruker GmbH, Ettlingen, Germany) using a standard 3D Stejskal-Tanner spin-echo sequence with 30 different angles of diffusion sensitization at b value of 1000 s/mm2 and the following parameters: repetition time=2000ms; echo time=25.616ms; diffusion encoding duration = 4ms. The in-plane resolution was 0.11mm and slice thickness was 0.22mm. Overall scanning time was ~24hours.
Image processing and tractography
Cerebral cortical regions of interest (ROI) and thalamus were manually defined according to Paxinos, 2007 50 and the Allen Mouse Brain Atlas (mouse.brain-map.org/static/atlas) 51 by D.A. and K.P. without prior knowledge of the experimental groups. Image preprocessing was done with Advanced Normalization Tools (ANTs, ver. 2.2.0.0.dev297-gf23cb). The reconstruction of axonal pathways was performed with MRtrix3 61 software (ver. 3.0.0-65-g91788533) using constrained spherical deconvolution 62 and probabilistic tracking (iFOD2) with FOD amplitude cut-off of 0.1. The thalamus was used as a seeding point and each cortical ROI was used as a termination mask. To evaluate the integrity of the major white matter tracts between the groups, both internal capsules, anterior commissure and corpus callosum were manually delineated according to Paxinos 2007 50 and the Allen Mouse Brain Atlas (mouse.brain-map.org/static/atlas) 51 by D.A. and K.P. without prior knowledge of the experimental groups. Values of the fractional anisotropy (FA), apparent diffusion coefficient (ADC), radial (RD) and axial (AD) diffusivity were calculated using underlying scalar maps derived by MRtrix3.
Anterograde tracing of axons
For anterograde tracing of axons between mPFC and thalamus, brains were collected at either PCD 18 or P21, and fixed overnight in 4% paraformaldehyde at 4 °C. Brains were then hemidissected. A crystal of 1,1#-dioctadecyl-3,3,3#,3# tetra-methyl-indocarbocyanine perchlorate (DiI, Sigma-Aldrich, Cat. 24364) was inserted either into the mPFC, MD nucleus of the thalamus, or medial thalamus under the stereomicroscope. The size of the crystal is ~200 μm. Brains were then placed in 1% paraformaldehyde in PBS and left for 14 days at 37 °C. Following DiI diffusion, the brains were sectioned coronally on a vibrating microtome (Leica) at 80 μm thickness and stained with DAPI. Sections were mounted onto glass and immediately sealed in VECTASHIELD Hardset Antifade Mounting Medium (VECTOR Laboratories, Cat. H-1400). Slides were analyzed under ApoTome.2 microscope (Zeiss) and intensity was quantified using ImageJ (ver.2.0.0-rc-69/1.52p).
Processing, analysis, and image visualization
To allow robust visualization and analysis, images depicting DiI tracing or immunohistochemistry using antibody against PSD95/DGL4 have been inverted and/or pseudo colored, as in Fig. 4, 5. In addition, background was removed for in situ hybridization experiments and images were pseudocolored and superimposed in Fig. 5 and Extended Data Fig. 4, 9,10, using Adobe Photoshop.
Extended Data
Extended Data Fig. 1 ∣. Workflow for analysis of midfetal RNA-seq data and genes upregulated in individual lobes.
a, Human developmental neocortical RNA-seq dataset and workflow for analysis to identify genes upregulated in each cortical lobe. b, Genes upregulated in the frontal, temporal, parietal and occipital lobes in comparison to the other lobes. We identified 190 protein-coding genes that are specifically upregulated, using stringent criteria, in at least one area within a lobe in comparison with areas from other lobes: 125 in the frontal lobe, 46 in the temporal lobe, 17 in the parietal lobe, and 2 in the occipital lobe. The X-axis represents proportion of putative areas in the frontal lobe in which the gene is significantly upregulated according to RNentropy. The Y-axis represents proportion of areas in the other lobes in which the gene is significantly downregulated according to RNentropy. Upregulated genes, the ones delimited by dashed red lines, are labeled. c,d, Gene loadings of PC1 from PCA of protein-coding genes that are specifically enriched in one of the four lobes of the midfetal human (c) and macaque (d) cortex. Colors represent the cortical lobe where the gene was found to be specifically upregulated. For reproducibility information, see Methods.
Extended Data Fig. 2 ∣. Analysis of gene upregulated in individual cortical lobes during other developmental periods.
a, Analysis of upregulated genes during window 2 (early fetal period, PCW 12-13 specimens in the BrainSpan dataset 17). See Fig. 1a for further explanation. b, Number of upregulated genes during late prenatal, and early postnatal stages in individual lobes are significantly reduced compared to earlier stages. c, d, Analysis of upregulated genes during window 8 (postnatal year, PY 13-19) and W9 (PY 21-64). See Fig. 1a for further explanation. For reproducibility information, see Methods.
Extended Data Fig. 3 ∣. Extended RA specific GO analysis and spatiotemporal expression of select genes upregulated in the midfetal frontal lobe.
a, Analysis for statistically significant enrichment of upregulated genes during developmental and adult stages for GO terms associated with RA. X-axis represents windows analyzed, which are defined at the bottom of the figure. b, Spatiotemporal expression of select genes upregulated in the midfetal frontal lobe from Fig. 1 in sixteen neocortical areas across human (red) and macaque (blue) development using BrainSpan (brainspan.org) and PsychENCODE (evolution.psychencode.org) RNA-seq data 17,19. Thick full lines represent four PFC areas, thick dotted line represents the primary motor cortex (M1C) and thin dotted lines represent the other non-frontal neocortical areas. Vertical gray box demarcates midfetal periods analyzed in Fig. 1. Timeline of human and macaque development and the associated periods designed by Kang et al. 39 shown below. Predicted ages were calculated using the TranscriptomeAge algorithm 19, which aligns our earliest macaque samples (PCD 60) with human early midfetal samples. Distinct global patterns of spatiotemporal expression were observed. For example, precocious expression in the frontal lobe/PFC followed by broad expression in all eleven neocortical areas (e.g. CBLN2, BDNF), transient enrichment in the frontal lobe/PFC (e.g. WNT11, PCDH17) and downregulation in non-PFC areas during midfetal development (e.g. MEIS2).
Extended Data Fig. 4 ∣. Expression of RA synthesizing enzymes in the developing human and macaque cortex.
a, Spatiotemporal expression of genes encoding RA synthesizing enzymes, ALDH1A1, 2 and 3, in eleven neocortical areas of human and macaque during prenatal and postnatal development using BrainSpan (brainspan.org) and PsychENCODE (evolution.psychencode.org) RNA-seq data 17,19. Red and blue lines indicate human and macaque, respectively, and dotted lines represent the non-PFC expression in the PFC plot. Vertical gray box demarcates midfetal developmental periods. Predicted ages, timeline of human and macaque development, and the associated periods are shown below 19,63. b, Heatmap of normalized (z-score) microarray signals computed for genes encoding RA synthesizing enzymes from the BrainSpan human prenatal laser microdissection microarray data63 (brainspan.org). Left columns represent gene name and specific probe. Each column represents regions of the brain labelled above the heatmaps. Darker reds represent high expression levels. c, Anteroposterior visual representation of human ALDH1A1 and ALDH1A3 expression at PCW 16 and 15 respectively from BrainSpan atlas58. ALDH1A1, 2 and 3 expression in midfetal human (PCW 19, 19, 20) (d, left) and macaque (PCD 80, 80, 110) (e, left) meninges. Immunostaining for ALDH1A1 and TH in human PCW 12 and 22 brains (d, right), and in macaque PCD 76 brain (e, right). White arrowheads, and open arrowheads indicate ALDH1A1+;TH+ and ALDH1A1+;TH− axons in the subplate, respectively. NCX, neocortex; PU, putamen; NAC, nucleus accumbens; CA, CA subfields of hippocampus. Errors bars: S.D. N = 3. f, Anterior to posterior expression of ALDH1A3 mRNA in human (PCW 21), macaque (PCD 140) and mouse (PD 0) brain. N = 2 for human and macaque, N = 3 for mouse. g, Western blot using ALDH1A3 antibody in human (PCW 20), macaque (PCD 114) and mouse (PD 0) frontal cortex areas. In macaque and human, there are two bands likely representing ALDH1A3 (56kDa) and ALDH1A1 (54kDa). Experiments were repeated at least two times for each animal species. See Extended Data Fig. 6a for schemas of frontal areas.
Extended Data Fig. 5 ∣. Expression of genes encoding RA synthesizing enzymes in the developing mouse cortex.
Expression of Aldh1a1, Aldh1a2 and Aldh1a3 at PCD 11.5, 18.5 and PD 4 (a) and Rara at PCD 15.5 and PD 4 (b) from the Allen Developing Mouse Brain Atlas58 (developingmouse.brain-map.org). c, Expression of RA related genes in the PCD 18.5 choroid plexus (CP). Blue bars represent lateral ventricle CP and orange bars represent fourth ventricle CP. Data from Lun et al.64. d, Analysis of Rarb-gfp mouse line from the GENSAT project59 (gensat.org) at PD 7 revealed GFP expression in upper (open arrows) and deep layer (solid arrows) pyramidal neurons in the mPFC at PD 7. Scale bars: 1 mm. FEZ, Frontonasal ectodermal zone; ChPl, Choroid plexus; RMS, Rostral migratory stream; SN, Substantia nigra; VTA, Ventral tegmental area; mPFC, Medial prefrontal cortex; ACA, anterior cingulate area.
Extended Data Fig. 6 ∣. Expression of RA synthesizing enzymes in the developing and adult cortex.
a, Representative images of ALDH1A1 and ALDH1A3 in situ hybridization of human (PCW 21; N = 2), macaque (PCD 110 and 140; N = 1 each), and mouse (PD 0; N = 3) PFC. Red, black, and open arrowheads indicate ALDH1A1-expressing subplate neurons (insets), astrocytes and meninges, respectively. Scale bars: 200 μm (mouse); 2 mm (human and macaque); 100 μm (mouse, lower panel); 500 μm (human and macaque, lower panel). b, ALDH1A1 immunofluorescence (green) and immunohistochemistry (brown). Human: left, solid and open arrowheads indicate TH+;ALDH1A1+ and TH+;ALDH1A1− axons, respectively; middle, solid and open arrowheads indicate subplate neurons and astrocytes, respectively; right, arrowheads indicate GFAP+;ALDH1A1+ astrocytes. Macaque: left, solid and open arrowheads indicate TH+;ALDH1A1+ and TH+;ALDH1A1− axons, respectively; middle, solid and open arrowheads indicate putative excitatory NRGN+;NR4A2+;ALDH1A1+ and inhibitory GAD1+;ALDH1A1− subplate neurons, respectively; right, arrowheads indicate GFAP+;ALDH1A1+ astrocytes. Mouse: left, solid and open arrowheads indicate TH+;ALDH1A1+ and TH+;ALDH1A1− mPFC axons (inset), respectively; middle, TH+ (red) and ALDH1A1+ (green) midbrain neurons and axons in striatum (STR), lateral septal nucleus (LSR) and cortex (CTX); right, GFAP+;ALDH1A1− astrocytes. N = 3. Scale bars: 20 μm (human, macaque, and mouse left and middle panels); 1 mm (mouse right panel). For reproducibility information, see Methods.
Extended Data Fig. 7. ∣. Expression of genes encoding RA degrading enzymes in the developing human and macaque cortex.
a, Expression of RA degrading enzymes in individual regions of the cerebral cortex of human and macaque during development. Red and blue lines indicate human and macaque, respectively, and dotted lines represent the non-PFC expression in the PFC plot and vice versa. Vertical gray box demarcates midfetal developmental periods. Predicted ages, timeline of human and macaque development, and the associated periods are shown below 19,39. * Data for CYP26A1 was not present in Zhu et al.19 and the data analyzed individually. The expression level for human and macaque were not normalized and can’t be directly compared. b, Heatmap of normalized (z-score) microarray signals computed for genes encoding RA degrading enzymes from the BrainSpan human prenatal laser microdissection microarray data63 (brainspan.org). Left column represents gene and specific probe. Rows represent regions of the brain. Darker reds represent high expression levels. c, Anteroposterior visual representation of human CYP26A1 and CYP26B1 expression at PCW 15 respectively from the BrainSpan atlas.
Extended Data Fig. 8 ∣. Expression of genes encoding RA receptors in the developing human and macaque cortex.
a, Expression of genes encoding RA receptors in individual regions of the cerebral cortex of human and macaque during prenatal and postnatal development. Red and blue lines indicate human and macaque, respectively, and dotted lines represent the non-PFC expression in the PFC plot and vice versa. Vertical gray box demarcates midfetal developmental periods. Predicted ages, timeline of human and macaque development, and the associated periods are shown below 19,39. * Data for RXRB was not present in Zhu et al 19 and the data were analyzed individually. The expression level for human and macaque were not normalized and cannot be compared. b, Heatmap of normalized (z-score) microarray signals computed for genes encoding RA receptors from the BrainSpan human prenatal laser microdissection microarray data63 (brainspan.org). Left column represents gene and specific probe. Rows represent regions of the brain. Darker reds represent high expression levels.
Extended Data Fig. 9 ∣. RARB and RXRB are expressed in a moderate anterior to posterior gradient in the developing neocortex.
a, Quantitative PCR analysis of Rara, b, g and Rxra, b, g transcripts in the anterior and posterior half of mouse cortex at PD 0. Two-tailed Student’s t-test: ***P = 4e-4, 1e-4; N = 3 per condition; Errors bars: S.E.M. b, Quantitative PCR analysis of Rarb and Rxrg transcripts in four sections dissected out of the cortical plate in anterior-posterior direction. Both genes showed an anterior-posterior gradient in expression level. Two-tailed Student’s t-test; N = 3 per condition; Errors bars: S.E.M. c, Expression of Rarb and Rxrg in mouse (PD 0), macaque (PCD 140) and human (PCW 21) brains by in situ hybridization. Higher magnification images of the regions of anterior cortex. Rarb and Rxrg transcripts are upregulated in the anterior part of the cortex in all three. Scale bars, 200 μm (mouse); 2 mm (human); 500 μm (human, higher magnification). N = 2 for human and macaque, N = 3 for mouse. d, Strategies for the generation of Rarb and Rxrg KO mice using CRISPR-Cas9 technique 41.
Extended Data Fig. 10 ∣. RA signal in the neonatal mouse forebrain.
a, β-Galactosidase histochemical staining of more posterior regions of Rarb+/+; Rxrg+/+ (WT); RARE-lacZ and Rarb−/−; Rxrg−/− (dKO); RARE-lacZ mouse brains at PD 0. Signal intensity in the boxed area (ACA, SSp, RSPA, STR, HIP and VP) was quantified. Note the reduced activity in anteromedial structures including ACA and RSPA (RSP). There is also reduced expression in HIP and lateral STR, but not the thalamus. Two-tailed Student’s t-test; *P = 9e-3, **P = 7e-4; 3e-3 (from left), ***P = 2e-4, ****P = 2e-7; 1e-5; 4e-5; 5e-6; 6e-6 (from left). N = 3 per genotype: Errors bars: S.E.M.; Scale bars, 200 μm. b, Cbln2 and Meis2 expression in PD 0 WT and dKO mutant brain by in situ hybridization at PD 0. Note that Cbln2 and Meis2 expression in mPFC was decreased in dKO. Scale bar, 200 μm. N = 3 per genotype. ACA, Anterior cingulate area; CP, caudoputamen; HIP, Hippocampus; RSPA/RSP, Retrosplenial area; STR, striatum; VP, Ventroposterior thalamus.
Extended Data Fig. 11 ∣. RA regulates CBLN2, MEIS2, and DLG4/PSD95 expression in human and chimpanzee cerebral organoids.
a, Expression of cortical neural stem/progenitor markers (PAX6 and SOX2), cortical cell type-specific markers (BCL11B, SATB2 and TBR1) and a pan-neuronal marker (MAP2) across the differentiation times show the dorsal cortical identity of the organoids derived from human and chimpanzee induced pluripotent stem cells. Scale bars, 50 μm. Each experiment used with 3-5 replicate organoids per times and conditions. b, In situ hybridization for CBLN2 and MEIS2, immunostaining for PSD95/DLG4, and DAPI nucleic acid (nuclear) staining in human and chimpanzee day 135 organoids exposed to low or high concentration RA-soaked bead for 48 hours. Scale bars, 500 μm. One experiment has done with 3-5 replicate organoids per times and conditions. c, Proportion of TuJ1 immuno-positive neurons in day 135 cerebral organoids was similar across conditions. The ratio of total number of PSD95+ synaptic puncta to total number of TuJ1+ cells in the day 135 cerebral organoids was significantly increased in RA-soaked bead applied conditions. Two-tailed t-test, compared to the condition without bead (Control, Ctrl), Errors bars: S.E.M. ***P = 5e-4, ****P< 1e-4. N=5 (multiple sections for each organoid) per condition.
Extended Data Fig. 12 ∣. Additional analysis of RNA-seq experiments.
a, Number of upregulated genes between PD 0 WT and dKO mice per region and phenotype, as well as combinations of regions and phenotypes. b, Gene loadings of the first principal component from PCA in Fig. 3. Colors represent the frontal cortex region where the gene was found to be upregulated. c, Cellular component GO terms associated with the total list of 4,768 DEx genes found, showing their z-score and nominal P values. Z-score represents the proportion of upregulated versus downregulated genes in the list of DEx genes associated to each GO term (i.e. z-score = (#up - #down) / sqrt (#all DEx associated to the GO term)). Dark blue: z-score <−5; light blue: z-score (−5,0]; orange: z-score >0. Size of the bubbles are proportional to the total number of DEx genes associated to the given GO term. d, Cellular component GO terms associated with DEx genes found in all three frontal cortex regions, and DEx genes unique to each region (mPFC, OFC, MOs) and their unadjusted p value. e, Enrichment of RA related genes (green), genes upregulated in individual lobes of the midfetal human cortex based on Fig. 1 (purple), and psychiatric disease related genes in up- and downregulated genes (pink). DEx genes are separated by genes that are DEx only in the given region (mPFC, OFC, MOs), genes that are DEx in the two given regions (mPFC + OFC, mPFC + MOs, OFC + MOs), genes that are DEx in all three regions (mPFC + OFC + MOs). Circles plotted for significant enrichments (P value <0.05), in darker color, significance is considering the adjusted p value. Diameter of circle is associated with odds ratio per legend.
Extended Data Fig. 13 ∣. Altered synaptic density and axonal projections in Rxrg and Rarb dKO mice.
a, Example of downregulated genes between PD 0 Rarb+/+; Rxrg+/+ (WT) and Rarb−/−; Rxrg−/− (dKO) that displayed an anterior to posterior gradient in PCD 18 mouse embryo (images are from the Allen Developing Mouse Brain Atlas; developingmouse.brain-map.org/ 58). Scale bar: 1 mm. b, Immunostaining for PSD95/DLG4 in the cortical subregions (mPFC, MOs, OFC, MOp, and SSp) of WT and dKO brain at PD 0. Each region as shown in Fig. 2c. Scale bar: 25 μm. N = 3 per genotype. c, Quantification and representative images of co-localized synaptophysin (SYP) and PSD95 puncta in PD 0 WT and dKO mPFC. Two-tailed Student’s t-test; *P = 0.01; N = 3 per genotype; Errors bars: S.E.M.; Scale bar: 10 μm. d, e, DiI placement in mPFC (c) and medial thalamus (d) with tracing data in WT and dKO brain at P21. Additional two replicates of experiment shown in Fig. 4d,e are shown. Asterisks: DiI crystal placement. Scale bar: 1 mm.
Extended Data Fig. 14 ∣. Analysis of axonal inputs to the mPFC and dendritic spines in Rarb and Rxrg dKO mice.
Anteroposterior series of PD 30 coronal sections of representative WT (a) and dKO (b) with retrograde viral tracer AAVrg-Cag-Gfp injected into mPFC (green asterisk). Sections were inverted and converted to grayscale. Black regions indicate the presence of viral tracer. High magnification representative images of labelled regions (c) and quantification of labelling using 0-3 scale (d) (N = 4 for each genotype; see methods section for more details). e, Quantification of Sholl analysis (Two-way ANOVA with Sidak’s multiple comparison method; P = .440- >0.999); Error bars: S.E.M.) and dendritic spines of deep layer neurons in the contralateral mPFC in WT (blue) and dKO (orange) (N = 4 for each genotype). Two-way ANOVA with Sidak’s multiple comparison method was applied; *P = 0.09; ****P < 1e-4; Error bars: S.E.M. Inset shows representative images of dendritic spines. Scale bar: 5 μm. ACAd, Dorsal anterior cingulate area; AI, Anterior Insula; BLA, Basolateral amygdala; vHPF, ventral hippocampal fields; CLA, claustrum; cl MPFC, Contralateral medial prefrontal cortex; ILA, Infralimbic area; LHA, Lateral habenula; MD, The mediodorsal nucleus of the thalamus; MOs/p, The secondary and primary motor areas. PER, Perirhinal cortex PIR, Piriform cortex PL, Prelimbic area; SSp, Primary somatosensory area; STN, Subthalamic nucleus; VTA, Ventral tegmental area.
Extended Data Fig. 15 ∣. RA regulates CBLN2, MEIS2, and DLG4/PSD95 expression and synaptogenesis in human fetal cortical neurons.
a, Schematics of human cortical neuron differentiation and modulation of RA signaling. From PCW 8 neocortical tissue, cortical neural stem cells were isolated and expanded with FGF2 for 20 days. Neural differentiation was initiated by the mitogen withdrawal. After 14 days of differentiation, cells were treated with varying doses of RA or RA receptor inhibitor (RARi, AGN193109) for another 14 days. From PCW 20 or 23 neocortical tissue, cortical neural progenitors and neurons were isolated and cultured without FGF2. After 16 days of culture, cells were treated with RA or RARi for another 14 days. Gene expression of cell type markers (b), RA receptors (c) RA regulated genes CBLN2 and MEIS2 (d) in PCW 8 cortical cells after 28 days of differentiation, measured by ddPCR. One way ANOVA and multiple comparison, compared to vehicle (0.01% DMSO)-treated control. Errors bars: S.E.M. *P <0.05 (RXRA expression, RA1.0, P = 0.0192, RARi5, P = 0.0277; CBLN2 expression, RARi10, P = 0.0135; MEIS2 expression, RA1.0, P = 0.0456), **P <0.01 (RARA expression, RARi10, P = 0.0039; MEIS2 expression, RARi5, P = 0.0057, PARi10, P = 0.0028), ***P<0.001 (MEIS2 expression, RA0.5, P = 0.0003), ****P<0.0001. N=3 experimental replicates per condition. e, TUBB3/TUJ1 and DLG4/PSD95 expression in RA or RARi-treated PCW 8 cortical cells. Scale bar: 50 μm. f, PSD95 transcript in PCW 8 cortical cells was significantly increased in RA 1 μM condition quantified by ddPCR. Two-tailed t-test, compared to the vehicle treatment. Errors bars: S.E.M. **P = 0.0083. N= 8 fields per condition. g, The ratio of total DLG4/PSD95+ puncta volume to total TUJ1+ volume in PCW 8 cortical cells was increased in the RA condition and reduced in the RARi condition. Two-tailed t-test, compared to the vehicle treatment. Errors bars: S.E.M. *P = 0.0152, ***P = 0.0001. h, MAP2, synaptophysin (SYP) and DLG4/PSD95 expression in RA or RARi-treated PCW 20 and 23 cortical cells. Scale bar: 50 μm. i, The ratio of total PSD95+ puncta volume to total MAP2+ volume in PCW 20 and 23 cortical cells was increased in RA conditions and reduced in RARi conditions. Two-tailed t-test, compared to the vehicle treatment. Errors bars: S.E.M. *P <0.05 (20 pcw-RA2, P = 0.0221; 20 pcw-RARi5, P = 0.0193; 23 pcw-RA0.5, P = 0.0192), **P <0.01 (20 pcw-RARi10, P = 0.0018; 23 pcw-RA1, P = 0.0081), ***P = 0.0004. N = 8 fields per condition. j, The ratio of total number of SYP+ and PSD95+ colocalized synaptic puncta to total MAP2+ volume in PCW 20 and 23 neocortical cells was increased in RA conditions and reduced in RARi conditions. Two-tailed t-test, compared to the vehicle treatment. Errors bars: S.E.M. *P (20 pcw-RA0.5, P = 0.0271; 20 pcw-RA2, P = 0.0159; 23 pcw-RA1, P = 0.0289; 23 pcw-RA2, P = 0.0447), ***P = 0.0004, ****P<0.0001. N= 8 fields per condition.
Extended Data Fig. 16 ∣. Analysis of axonal projections and cell death in Rarb and Rxrg dKO mice.
a, Four scalar indexes which describe microstructural integrity do not differ in the four major white-matter tracts (corpus callosum, anterior commissure, left and right internal capsules) between WT and dKO mice. FA Comparisons: Corpus callosum (two-tailed unpaired t-test, P = 0.4); Anterior cingulate (two tailed mann-whitney test, P = 0.2); Internal Capsule left (two-tailed unpaired t-test, P = 0.07); Internal Capsule right (two-tailed unpaired t test, P = 0.8). ADC Comparisons: Corpus callosum (two-tailed unpaired t-test, p value 0.1); Anterior cingulate (two-tailed unpaired t-test, p value 0.1); Internal Capsule left (two-tailed unpaired t-test, p value 0.9); Internal Capsule right (two-tailed unpaired t-test, P = 0.3). RD Comparisons: Corpus callosum (two-tailed unpaired t-test, P = 0.1); Anterior cingulate (two-tailed unpaired t-test, P = 0.1); Internal Capsule left (two tailed mann-whitney test, P = 0.1); Internal Capsule right (two-tailed unpaired t-test, P = 0.3). AD Comparisons: Corpus callosum (two-tailed unpaired t-test, P = 0.08); Anterior cingulate (two-tailed unpaired t-test, P = 0.1); Internal Capsule left (two-tailed unpaired t-test, P = 0.4); Internal Capsule right (two-tailed unpaired t-test, P = 0.2). N=5; Errors bars: S.E.M. b, Number of streamlines of MOp-thalamus and corticocortical tracts, did not differ between WT and dKO. Two-tailed unpaired t-test for MOp-MOp, MOp-Th, and AUDp-th (P = 0.1, 0.07, and 0.6, respectively). Two-tailed Mann-Whitney test for ACA-Th (P = 0.3). N = 5 per genotype; Errors bars: S.E.M. c, Barrel formation was examined by Nissl staining at PD 5 in WT and dKO. N = 3 per genotype. d, Representative images and quantification of corticospinal tract (CST) width using L1CAM expression at PD 5 in WT and dKO. The width of the corticospinal tract (shown in brackets) is slightly increased in dKO. Two-tailed Student’s t-test: *P = 0.02; N = 5 per genotype; Errors bars: S.E.M. e, Representative images and quantification of of apoptotic cells in the mPFC detected by cleaved caspase3 (actCASP3) between WT and dKO. Two-tailed Student’s t-test: WT vs. dKO; NS; N = 5 per genotype; Errors bars: S.E.M.; Scale bars: 500 μm; 200 μm (b,f); 1 mm (c); 100 μm (d). f, Representative images and quantification of tyrosine hydroxylase (TH) immunolabeled axons in wildtype and dKO mouse frontal cortex at PD 5 (N = 4). Rectangles represent regions analyzed. Clockwise starting from left-most: mPFC, MOs, OFC. Errors bars: S.E.M.
Extended Data Fig. 17 ∣. Analysis of neocortical layers and tissue volume in Rarb and Rxrg dKO mice.
a, Representative images and quantification of total volume of the brain (left), cerebral neocortex (middle), anterior one-third of the neocortex (right) of wildtype (WT) littermates and Rarb;Rxrg dKO at PD 5. Two-tailed unpaired t-test for total volume of the brain and anterior one-third of the neocortex. Two-tailed Mann-Whitney test for volume of cerebral neocortex. P = 0.3, 0.1, 0.6. N = 5. Errors bars: S.E.M. b, Number of immune-positive cells expressing cortical layer markers POU3F2/BRN2, CUX1, RORB, BCL11B/CTIP2, TBR1 in wildtype and dKO mouse mPFC, MOs, and primary visual cortex (VISp) at PD 5 (N = 3). (Unpaired t-test: WT vs. dKO *P = 0.03; Errors bars: S.E.M.). RORB expression in mouse PFC was too faint to quantify. Below each graph is a representative image of each marker. Scale bar: 200 μm
Extended Data Fig. 18 ∣. RA signal in posterior cortical regions of Cyp26b1 KO mice.
a, Strategy for the generation of Cyp26b1−/− (Cyp26b1 KO) mice using CRISPR-Cas9 gene editing technique 41. b, Cyp26b1 expression in PD 0 mice cortex by in situ hybridization. The colorimetric staining was purposefully extended compared to the experiment in Fig. 5a to better visualize low expressing locations. c, β-Galactosidase staining of more posterior regions of control Cyp26b1+/+; RARE-lacZ (Ctrl) and Cyp26b1−/−; RARE-lacZ (KO) mouse brains at PCD 18. Scale bar: 500 μm. Intensity of signal in the boxed areas (ACA, MO, SSp) was quantified. Increase in RA signaling in Cyp26b1 KO brains is less significant in posterior regions. Two-tailed Student’s t-test: Ctrl vs. Cyp26b1 KO: *P = 0.01; 0.04; 9e-3; 7e-3; 6e-3; 0.02; 0.02; **P = 4e-3; 1e-3; 4e-3; 4e-3, N = 3 per genotype; Errors bars: S.E.M.; Scale bars: 200 μm.
Extended Data Fig. 19 ∣. Rorb expression and medial thalamocortical projections in the neonatal Cyp26b1 KO mice.
a, Additional two replicates of Rorb expression in WT and Cyp26b1 KO brains from Fig. 5f are shown. Scale bar: 100 μm. b, DiI was placed in the medial thalamus of WT and Cyp26b1 KO brains, and signal was detected in the PFC. Additional two replicates of experiment in Fig. 5d are shown. N = 3 per genotype and condition. Arrowheads: Thalamocortical innervation of the medial and dorso-lateral frontal cortex. Asterisks: DiI crystals placed. Scale bar: 400 μm. c, DiI was placed in the frontal motor cortex of WT and Cyp26b1 KO brains, and signal was detected in the medial thalamus. Additional two replicates of experiment in Fig. 5e are shown. N = 3 per genotype and condition. Of note, due to restriction related to Covid-19, PD 0 brains were left 2 months in cold room after 3 weeks in 37 degree. Scale bar: 400 μm.
Extended Data Fig. 20 ∣. Ectopic RA signaling leads to enlargement of the lateral region of the frontal cortex and Rorb upregulation.
a, Representative image and quantification of the ratio of brain width at anterior and posterior cortex to total brain length at PCD 18 in Cyp26b1 KO brain compared WT. Two-tailed Student’s t-test: WT vs. Cyp26b1 KO: ***P = 3e-4; N = 4 per genotype; Errors bars: S.E.M. b, Nissl staining reveals that the cortical wall and cortical plate are grossly normal when analyzed in the mPFC and SSp of Cyp26b1 KO. N = 3 per genotype. c, Representative image and quantification of Rorb expression after electroporation of either control pCAG-IRES-Gfp or pCAG-Aldh1a3-IRES-Gfp expression vector plasmid in the dorso-lateral fronto-parietal wall at PCD 14. Brains were dissected out at PD 5. Boxed region represents region of higher magnification to the right. GFP expression as a marker of misexpressing cells are shown in lower panels. Rorb signal intensity in the boxed area in the cortex was quantified. Two-tailed Student’s t-test: ***P = 2e-4; N = 4 per genotype; Errors bars: S.E.M.; Scale bars: 500 μm (a); 200 μm (b); 1 mm (c); 40 μm and 500 μm (d).
Supplementary Material
Extended Data Table 1: Gene ontogeny of midfetal frontal lobe enriched genes
Extended Data Table 2: Annotation of midfetal prospective PFC and M1C enriched genes
Extended Data Table 3: List of differentially expressed genes from wildtype and Rarb;Rxrg dKO PD 0 frontal cortex. obtained using RNentropy72. Briefly, the analysis is done in two steps: i) a global sample specificity test is computed based on their specificity with a background distribution, adjusting their P values with a Benjamini-Hochberg procedure; ii) the expression in each sample of the significant genes that passed the default global threshold in step i) are then compared with the expression in the remaining samples with a local sample specificity test, obtaining unadjusted P values. In this table, those results are summarized per phenotype.
Extended Data Table 4: List and annotation of PD 0 mouse differentially expressed or human PFC enriched genes related to axon development
Extended Data Table 5: List of relevant primers and sgRNA
Acknowledgements
We thank Suxia Bai and Timothy Nottoli for technical help in generation of gene-edited mouse lines; Fahmeed Hyder and Jelena Mihailovic for their assistance with MRI diffusion-weighted sequence design and conducting the MRIs; Alvaro Duque for use of equipment from MacBrainResource (MH113257); John Rubenstein for sharing reagents; Fuchen Li and Marco Onorati for generating human iPSCs used in this study; Yoav Gilad and Bryan Pavlovic for providing chimpanzee iPSCs and the members of the Sestan laboratory for comments. This work was supported by the National Institutes of Health (HG010898, MH106874, MH106934, MH110926, MH116488), and Simons Foundation (N.S.). The project that gave rise to these results received the support of a fellowship from “la Caixa” Foundation (ID 100010434). The fellowship code is LCF/BQ/PI19/11690010. Additional support was provided by the National Institutes of Health T32 fellowship (MH18268) (K.P.), the National Science Foundation Graduate Research Fellowship Program (S.K.M), Ministerio de Ciencia e Innovación, Spain (PID2019-104700GA-I00) (G.S), Kavli Foundation, and the James S. McDonnell Foundation (N.S.).
Footnotes
Competing Financial Interests
The authors declare no competing financial interests.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this paper.
Data availability
The mouse RNA-seq data are available at the NCBI GEO under the accession numbers GSE142851.
References
- 1.Fuster JM The Prefrontal Cortex (Elsevier, 2015). [Google Scholar]
- 2.Amaral DG et al. Neuroanatomy of autism. Trends Neurosci. 31, 137–45 (2008). [DOI] [PubMed] [Google Scholar]
- 3.Willsey AJ et al. Coexpression networks implicate human midfetal deep cortical projection neurons in the pathogenesis of autism. Cell 155, 997–1007 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tan H-Y et al. Dysfunctional and compensatory prefrontal cortical systems, genes and the pathogenesis of schizophrenia. Cereb. Cortex 17, i171–81 (2007). [DOI] [PubMed] [Google Scholar]
- 5.Giraldo-Chica M et al. Prefrontal-thalamic anatomical connectivity and executive cognitive function in schizophrenia. Biol. Psychiatry 83, 509–517 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fukuchi-Shimogori T & Grove EA Neocortex patterning by the secreted signaling molecule FGF8. Science 294, 1071–1074 (2001). [DOI] [PubMed] [Google Scholar]
- 7.Cholfin JA & Rubenstein JLR Frontal cortex subdivision patterning is coordinately regulated by Fgf8, Fgf17, and Emx2. J. Comp. Neurol 509, 144–155 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.O’Leary DD & Sahara S Genetic regulation of arealization of the neocortex. Curr. Opin. Neurobiol 18, 90–100 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Geschwind DH & Rakic P Cortical evolution: Judge the brain by its cover. Neuron 80, 633–647 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Preuss TM Do rats have prefrontal cortex? The rose-woolsey-akert program reconsidered. J. Cogn. Neurosci 7, 1–24 (1995). [DOI] [PubMed] [Google Scholar]
- 11.Petrides M et al. The prefrontal cortex: comparative architectonic organization in the human and the macaque monkey brains. Cortex. 48, 46–57 (2012). [DOI] [PubMed] [Google Scholar]
- 12.Durston AJ et al. Retinoic acid causes an anteroposterior transformation in the developing central nervous system. Nature 340, 140–144 (1989). [DOI] [PubMed] [Google Scholar]
- 13.Marshall H et al. Retinoic acid alters hindbrain Hox code and induces transformation of rhombomeres 2/3 into a 4/5 identity. Nature 360, 737–741 (1992). [DOI] [PubMed] [Google Scholar]
- 14.Lamantia AS Forebrain induction, retinoic acid, and vulnerability to schizophrenia: insights from molecular and genetic analysis in developing mice. Biol. Psychiatry 46, 19–30 (1999). [DOI] [PubMed] [Google Scholar]
- 15.Maden M Retinoic acid in the development, regeneration and maintenance of the nervous system. Nat. Rev. Neurosci 8, 755–65 (2007). [DOI] [PubMed] [Google Scholar]
- 16.Pletikos M et al. Temporal specification and bilaterality of human neocortical topographic gene expression. Neuron 81, 321–32 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li M et al. Integrative functional genomic analysis of human brain development and neuropsychiatric risks. Science 362, eaat7615 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Silbereis JC et al. The cellular and molecular landscapes of the developing human central nervous system. Neuron 89, 248–68 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhu Y et al. Spatiotemporal transcriptomic divergence across human and macaque brain development. Science 362, eaat8077 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shibata M. et al. Accompanying Paper [Google Scholar]
- 21.Johnson MB et al. Functional and evolutionary insights into human brain development through global transcriptome analysis. Neuron 62, 494–509 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Siegenthaler JA et al. Retinoic acid from the meninges regulates cortical neuron generation. Cell 139, 597–609 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Haushalter C et al. Retinoic acid controls early neurogenesis in the developing mouse cerebral cortex. Dev. Biol 430, 129–141 (2017). [DOI] [PubMed] [Google Scholar]
- 24.Larsen R et al. The thalamus regulates retinoic acid signaling and development of parvalbumin interneurons in postnatal mouse prefrontal cortex. eNeuro 6, ENEURO.0018–19.2019 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Aoto J et al. Synaptic signaling by all-trans retinoic acid in homeostatic synaptic plasticity. Neuron 60, 308–320 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Smith D et al. Retinoic acid synthesis for the developing telencephalon. Cereb. Cortex 11, 894–905 (2001). [DOI] [PubMed] [Google Scholar]
- 27.Dmetrichuk JM et al. Retinoic acid induces neurite outgrowth and growth cone turning in invertebrate neurons. Dev. Biol 294, 39–49 (2006). [DOI] [PubMed] [Google Scholar]
- 28.Moreno-Ramos OA et al. Whole-exome sequencing in a south American cohort links ALDH1A3, FOXN1 and retinoic acid regulation pathways to autism spectrum disorders. PLoS One 10, e0135927 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xu X et al. Excessive UBE3A dosage impairs retinoic acid signaling and synaptic plasticity in autism spectrum disorders. Cell Res. 28, 48–68 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kumar S et al. Impaired neurodevelopmental pathways in autism spectrum disorder: a review of signaling mechanisms and crosstalk. J. Neurodev. Disord 11, 10 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Goodman AB Three independent lines of evidence suggest retinoids as causal to schizophrenia. Proc. Natl. Acad. Sci. U. S. A 95, 7240–4 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Reay WR et al. Polygenic disruption of retinoid signalling in schizophrenia and a severe cognitive deficit subtype. Mol. Psychiatry 25, 719–731 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rilling JK Comparative primate neurobiology and the evolution of brain language systems. Current Opinion in Neurobiology 28, 10–14 (2014) [DOI] [PubMed] [Google Scholar]
- 34.Chiang MY et al. An essential role for retinoid receptors RARβ and RXRγ in long-term potentiation and depression. Neuron 21, 1353–1361 (1998). [DOI] [PubMed] [Google Scholar]
- 35.Krezel W et al. Impaired locomotion and dopamine signaling in retinoid receptor mutant mice. Science 279, 863–867 (1998). [DOI] [PubMed] [Google Scholar]
- 36.Rossant J et al. Expression of a retinoic acid response element-hsplacZ transgene defines specific domains of transcriptional activity during mouse embryogenesis. Genes Dev. 5, 1333–1344 (1991). [DOI] [PubMed] [Google Scholar]
- 37.Wilde JJ et al. Diencephalic size is restricted by a novel interplay between GCN5 acetyltransferase activity and retinoic acid signaling. J. Neurosci 37, 2565–2579 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yashiro K et al. Regulation of retinoic acid distribution is required for proximodistal patterning and outgrowth of the developing mouse limb. Dev. Cell 6, 411–422 (2004). [DOI] [PubMed] [Google Scholar]
Extended References
- 39.Kang HJ et al. Spatiotemporal transcriptome of the human brain. Nature 478, 483–489 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zambelli F et al. RNentropy: An entropy-based tool for the detection of significant variation of gene expression across multiple RNA-Seq experiments. Nucleic Acids Res. 46, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang H et al. One-step generation of mice carrying mutations in multiple genes by CRISPR/cas-mediated genome engineering. Cell 153, 910–918 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cong L et al. Multiplex genome engineering using CRISPR/Cas systems. Science 339, 819–823 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Renaud JP et al. Crystal structure of the RAR-γ ligand-binding domain bound to all-trans retinoic acid. Nature 378, 681–689 (1995). [DOI] [PubMed] [Google Scholar]
- 44.MacLean G et al. Apoptotic extinction of germ cells in testes of Cyp26b1 knockout mice. Endocrinology 148, 4560–4567 (2007). [DOI] [PubMed] [Google Scholar]
- 45.Micali N et al. Variation of Human Neural Stem Cells Generating Organizer States In Vitro before Committing to Cortical Excitatory or Inhibitory Neuronal Fates. Cell Rep. 31, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xiang Y et al. Fusion of regionally specified hPSC-derived organoids models human brain development and interneuron migration. Cell Stem Cell 21, 383–398.e7 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pollen AA et al. Establishing cerebral organoids as models of human-specific brain evolution. Cell 176, 743–756 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gallego Romero I et al. A panel of induced pluripotent stem cells from chimpanzees: a resource for comparative functional genomics. Elife 4, (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wilkinson DG & Nieto MA Detection of messenger RNA by in situ hybridization to tissue sections and whole mounts. Methods Enzymol. 225, 361–73 (1993). [DOI] [PubMed] [Google Scholar]
- 50.Paxinos G Atlas of the Developing Mouse Brain: at E17.5, PD 0, and P6. (Academic Press, 2007). [Google Scholar]
- 51.Dong HW The Allen Reference Atlas: A Digital Color Brain Atlas of the C57BL/6J Male Mouse (John Wiley & Sons, 2008). [Google Scholar]
- 52.Shibata M et al. MicroRNA-9 modulates Cajal-Retzius cell differentiation by suppressing Foxg1 expression in mouse medial pallium. J. Neurosci 28, 10415–10421 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kokubu H & Lim J X-gal staining on adult mouse brain sections. Bio Protoc. 4, pii: e1064 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ippolito DM & Eroglu C Quantifying synapses: An immunocytochemistry-based assay to quantify synapse number. J. Vis. Exp 45, 2270 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fiala JC Reconstruct: A free editor for serial section microscopy. J. Microsc 218, 52–61 (2005). [DOI] [PubMed] [Google Scholar]
- 56.Risher WC et al. Rapid golgi analysis method for efficient and unbiased classification of dendritic spines. PLoS One 9, e107591 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Meijering E et al. Design and validation of a tool for neurite tracing and analysis in fluorescence microscopy images. Cytom. Part A 58, 167–176 (2004). [DOI] [PubMed] [Google Scholar]
- 58.Thompson CL et al. A high-resolution spatiotemporal atlas of gene expression of the developing mouse brain. Neuron 83, 309–323 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gong E et al. A gene expression atlas of the central nervous system based on bacterial artificial chromosomes. Nature 425, 917–925 (2003). [DOI] [PubMed] [Google Scholar]
- 60.Elsen GE et al. The protomap is propagated to cortical plate neurons through an Eomes-dependent intermediate map. Proc. Natl. Acad. Sci. U. S. A 110, 4081–4086 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tournier J-D et al. MRtrix: Diffusion tractography in crossing fiber regions. Int. J. Imaging Syst. Technol 22, 53–66 (2012). [Google Scholar]
- 62.Tournier JD et al. Robust determination of the fibre orientation distribution in diffusion MRI: Non-negativity constrained super-resolved spherical deconvolution. Neuroimage 35, 1459–1472 (2007). [DOI] [PubMed] [Google Scholar]
- 63.Miller JA et al. Transcriptional landscape of the prenatal human brain. Nature 508, 199–206 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lun MP et al. Spatially heterogeneous choroid plexus transcriptomes encode positional identity and contribute to regional CSF production. J. Neurosci 35, 4903–4916 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Extended Data Table 1: Gene ontogeny of midfetal frontal lobe enriched genes
Extended Data Table 2: Annotation of midfetal prospective PFC and M1C enriched genes
Extended Data Table 3: List of differentially expressed genes from wildtype and Rarb;Rxrg dKO PD 0 frontal cortex. obtained using RNentropy72. Briefly, the analysis is done in two steps: i) a global sample specificity test is computed based on their specificity with a background distribution, adjusting their P values with a Benjamini-Hochberg procedure; ii) the expression in each sample of the significant genes that passed the default global threshold in step i) are then compared with the expression in the remaining samples with a local sample specificity test, obtaining unadjusted P values. In this table, those results are summarized per phenotype.
Extended Data Table 4: List and annotation of PD 0 mouse differentially expressed or human PFC enriched genes related to axon development
Extended Data Table 5: List of relevant primers and sgRNA
Data Availability Statement
The mouse RNA-seq data are available at the NCBI GEO under the accession numbers GSE142851.