Abstract
The similarities and differences between nervous systems of various species result from developmental constraints and specific adaptations 1–4. Comparative analyses of the prefrontal cortex (PFC), a cerebral cortex region involved in higher-order cognition and complex social behaviors, have identified true and potential human-specific structural and molecular specializations 4–8, such as a PFC enriched anterior-posterior dendritic spine density gradient 5. These changes are likely mediated by divergence in spatiotemporal gene regulation 9–17, which is particularly prominent in the midfetal human cortex 15,18–20. Analyzing human and macaque transcriptomic data 15,20, we identified a transient PFC- and laminar-specific upregulation of cerebellin 2 (CBLN2), a neurexin (NRXN) and glutamate receptor delta GRID/GluD-associated synaptic organizer 21–27, during human midfetal development coinciding with the initiation of synaptogenesis. Moreover, we found that this difference in expression level and laminar distribution of CBLN2 is due to Hominini-specific deletions containing SOX5 binding sites within a retinoic acid-responsive CBLN2 enhancer. In situ genetic humanization of the mouse Cbln2 enhancer drives increased and ectopic laminar Cbln2 expression and promotes PFC dendritic spine formation. These findings suggest a genetic and molecular basis for the disproportionately increased dendritic spines in the Hominini PFC and a developmental mechanism linking dysfunction of the NRXN-GRID-CBLN2 complex to the pathogenesis of neuropsychiatric disorders.
Introduction
Expansion of the PFC in catarrhine primates is associated with increased dendritic complexity and synaptic spines of the pyramidal neurons compared to more posterior cortical association and sensory areas, creating an anterior-posterior gradient of synaptic density 28–30. Moreover, this gradient is exaggerated in humans by the disproportionately high number of spines on pyramidal neurons in the human PFC as compared to the PFC of other analyzed primates 5. Early neocortical synaptogenesis begins during the human midfetal period or equivalent developmental age in other mammals 31, proceeding in an anterior-posterior direction 32 and continues at an accelerated rate well beyond birth 33. In our accompanying study 34, we identified a midfetal PFC-enriched gradient of retinoic acid (RA) concentration and pattern of gene expression in the human and macaque neocortex. Among the identified genes was synaptic organizer, CBLN2, which had the greatest loading of differentially expressed genes representing the anterior/frontal-posterior/temporal axis 35. Members of the CBLN family encode secreted neuronal glycoproteins, which serve as excitatory and inhibitory synaptic organizing molecules 21–27. Thus, we hypothesize that RA-mediated regulation of CBLN2 expression in the developing PFC underlies the disproportionate number of synapses in the human PFC.
Cortical laminar divergence of prefrontal CBLN2 expression
By analyzing RNA-seq data from eleven developing neocortical areas across human and macaque development from the BrainSpan and PsychENCODE projects 15,20, we identified a precocious and transient increase in human (1.9-fold change) and macaque (2.0-fold change) CBLN2 expression in the major areas of the prospective PFC compared to the seven analyzed non-PFC areas during midfetal development (postconception weeks, PCW 13–24) or periods 4–6 according to Kang et al.35 (Fig. 1a, Extended Data Fig. 1a,b). Around the time of late infancy, both human and macaque CBLN2 expression levels are comparable across analyzed neocortical areas, despite modest but statistically significant increased expression in PFC areas compared to non-PFC areas (Human periods 7–10: 1.1-fold change; Macaque periods 7–10: 1.25-fold change Fig. 1a, Extended Data Fig. 1a), which continued at later adult ages (Humans periods 11–14: 1.1-fold change; Macaque periods 11–14: 1.2-fold change; Extended Data Fig. 1a,c). Analysis of the spatiotemporal expression profiles of the paralogs of CBLN2 within each species revealed that CBLN1 and CBLN4 were significantly upregulated in human and macaque PFC, compared to non-PFC areas (Extended Data Fig. 2). While none of the binding partners of the CBLN family members, NRXN1,2,3 and GRID1,2, showed distinct PFC upregulation during midfetal development, all were expressed in the PFC with increasing expression levels during development (Extended Data Fig. 2c).
Fig. 1 |. Phylogenetic, transcriptomic, and regulatory characterization of developmental PFC upregulation of CBLN2.
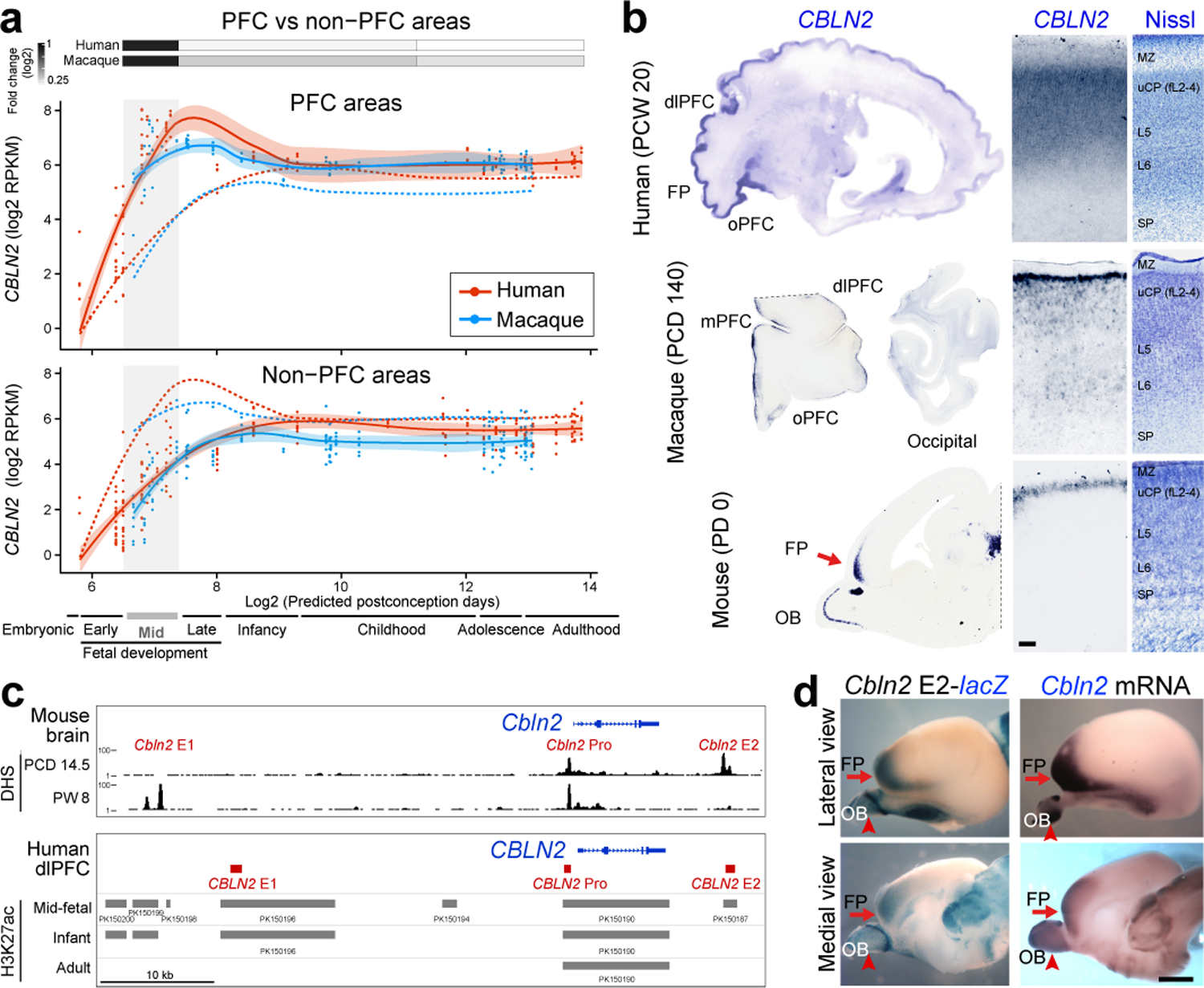
a, CBLN2 spatiotemporal expression in human (red lines) and macaque (blue lines) PFC (solid lines) and non-PFC (dotted lines) areas. Quantification of fold change between PFC and non-PFC expression for periods 4–6 (gray box), 7–10 and 11–14 (two-tailed Mann-Whitney test or two-tailed unpaired t-test P < 1e-4 for all comparisons) shown above plots. For all of the plots, the shading around the lines represents the 95% confidence interval. Developmental periods designed by Kang et al.35 are shown below. b, CBLN2 in situ hybridization on sagittal sections of midfetal human and a neonatal mouse and coronal sections of midfetal macaque prefrontal and occipital lobes. Right panels show laminar CBLN2 expression pattern in PFC and Nissl stain of adjacent section. Human CBLN2 image is from humanbraintranscriptome.org 18,19,37. Experiments were repeated three times for Macaque brain sections and more than five times for mouse brain sections. FP, frontal pole; MZ, marginal zone; SP, subplate; uCP, upper cortical plate. Scale bars, 500 μm. c, DNase 1 hypersensitive sites (DHS) in mouse brain at PCD 14.5 and PW 8 within 50 kb either upstream and downstream of Cbln2 gene. H3K27ac ChIP-seq data from human dlPFC/DFC at mid-fetal age, infancy, and adulthood (bottom panel). Putative cis-regulatory elements were designated as CBLN2 E1, E2, and Pro. d β-Galactosidase activity in transgenic mouse brain carrying mouse Cbln2 E2 conjugated with lacZ reporter at PCD 17. Endogenous Cbln2 expression in age-matched brains by in situ hybridization is shown for comparison. Arrows depict FP; arrowheads, olfactory bulb (OB). Scale bars, 1mm. Experiments were repeated for three brains and two replicates are shown in Extended Data Fig. 3c.
Previous bulk tissue transcriptomic studies have also reported an anterior-posterior gradient of CBLN2 expression in the human midfetal neocortex 18,36,37. We confirmed this finding using in situ hybridization (PCW 21 and 22) and single cell RNA-seq data, showing that CBLN2 was detected broadly in neurons of prospective upper layers (L) 2 to 4 and deeper L5 and 6 of the PFC, but faintly posteriorly (Fig. 1b; Extended Data Fig. 1b,d). We also identified an anterior-posterior gradient of Cbln2 expression in macaque (postconception day, PCD 110, and 140) and mouse (Postnatal day, PD 0) at equivalent developmental ages (Fig. 1b, Extended Fig. 1b, 3a,b). Interestingly, in macaque, Cbln2 expression was mostly observed in L2 corresponding to the prospective upper layers with noticeably weaker expression in the deeper layers (Fig. 1b). In mice, Cbln2 was mostly restricted to the prospective upper layers (Fig. 1b, Extended Data Fig. 3b). Together, our analysis found a progressive extension of CBLN2 expression into the deeper layers of the midfetal PFC in humans compared to macaques, and in macaques compared to mice.
Hominini-specific deletions in RA-responsive CBLN2 enhancer
In order to understand regulatory mechanisms of CBLN2 expression in the midfetal PFC, we analyzed publicly available datasets on the regulatory landscape of developing mouse and human brains generated by the ENCODE and PsychENCODE projects 20,38. Around the mouse Cbln2 locus, we identified three putative cis-regulatory elements marked as DNase 1 hypersensitive sites, which we designated as Cbln2 enhancer 1 (Cbln2 E1, 1452 bp), enhancer 2 (Cbln2 E2, 1005 bp), and promoter (Cbln2 Pro, 316 bp; Fig. 1c, top). The sequences of these putative cis-regulatory elements are moderately conserved between mouse and human (E1, 76.3%; E2, 84.3%; and Pro, 70.0%). Of note, mouse Cbln2 E2 showed DNase 1 hypersensitivity peaks only in PCD 14.5 samples and Cbln2 E1 only in 8 postnatal weeks (PW) samples (Fig. 1c, top). Analysis of genomic sites with differential distribution of H3K27ac, a mark of active enhancers, in the developing and adult human dorsolateral PFC (dlPFC/DFC) indicates that CBLN2 Pro is active at all three analyzed time points (midfetal, early infancy and adult ages), while CBLN2 E1 is active during midfetal age and infancy, and CBLN2 E2 is only active during midfetal age (Fig 1c, bottom).
We next assessed the activity of these putative regulatory regions using transgenic mouse lines in which a lacZ expression cassette was placed downstream of mouse Cbln2 E1, Cbln2 E2, or Cbln2 Pro. While Cbln2 E1-lacZ and Cbln2 Pro-lacZ transgenic lines exhibited no detectable lacZ expression (0 of 10 founders) in the developing cortex between PCD 17 and 18, 50% of Cbln2 E2-lacZ transgenics (3 of 6 founders) showed lacZ expression, with two of three independent lines recapitulating endogenous frontal cortex-enriched Cbln2 expression (Fig. 1d; Extended Data Fig. 3c). These findings indicate that Cbln2 E2 is an enhancer that drives expression in the mouse neonatal frontal cortex.
Given the presence of CBLN2 E2 in human, macaque and mouse, we hypothesized that sequence differences between the orthologs may underlie the species-specific expression pattern of CBLN2. Comparative genomic and phylogenetic analysis identified two separate deletions (122 bp, 20 bp) and insertion (84 bp) in the sequence of the human CBLN2 E2 ortholog as compared to the sequence of the mouse Cbln2 E2 (Fig. 2a). Further comparison across multiple vertebrates revealed that two of the three deleted sequences are jointly absent only in extant members of the Hominini clade (human, common chimpanzee, and bonobo) (Extended Data Fig. 4a,c, 5). The first two of these sequences, which we named Hominini-specific deletion 1 and 2 (HSD1 and 2, Fig. 2a), are highly conserved among other analyzed Haplorhini (gorilla, macaque), Strepsirrhini (lemur), and mouse (Fig. 2a, Extended Data Fig. 4a,c 5). The third inserted sequence (I3) sequence was detected only in mouse and rat of the Muroidea species analyzed (Extended Data Fig. 4a, 5). Interestingly, we did not detect genomic regions orthologous to Cbln2 E2 in analyzed non-placental mammals and non-mammalian chordates, although they do possess the Cbln2 gene (Extended Data Fig. 5).
Fig. 2 |. SOX5 represses CBLN2 through Hominini-specific regulatory deletions.
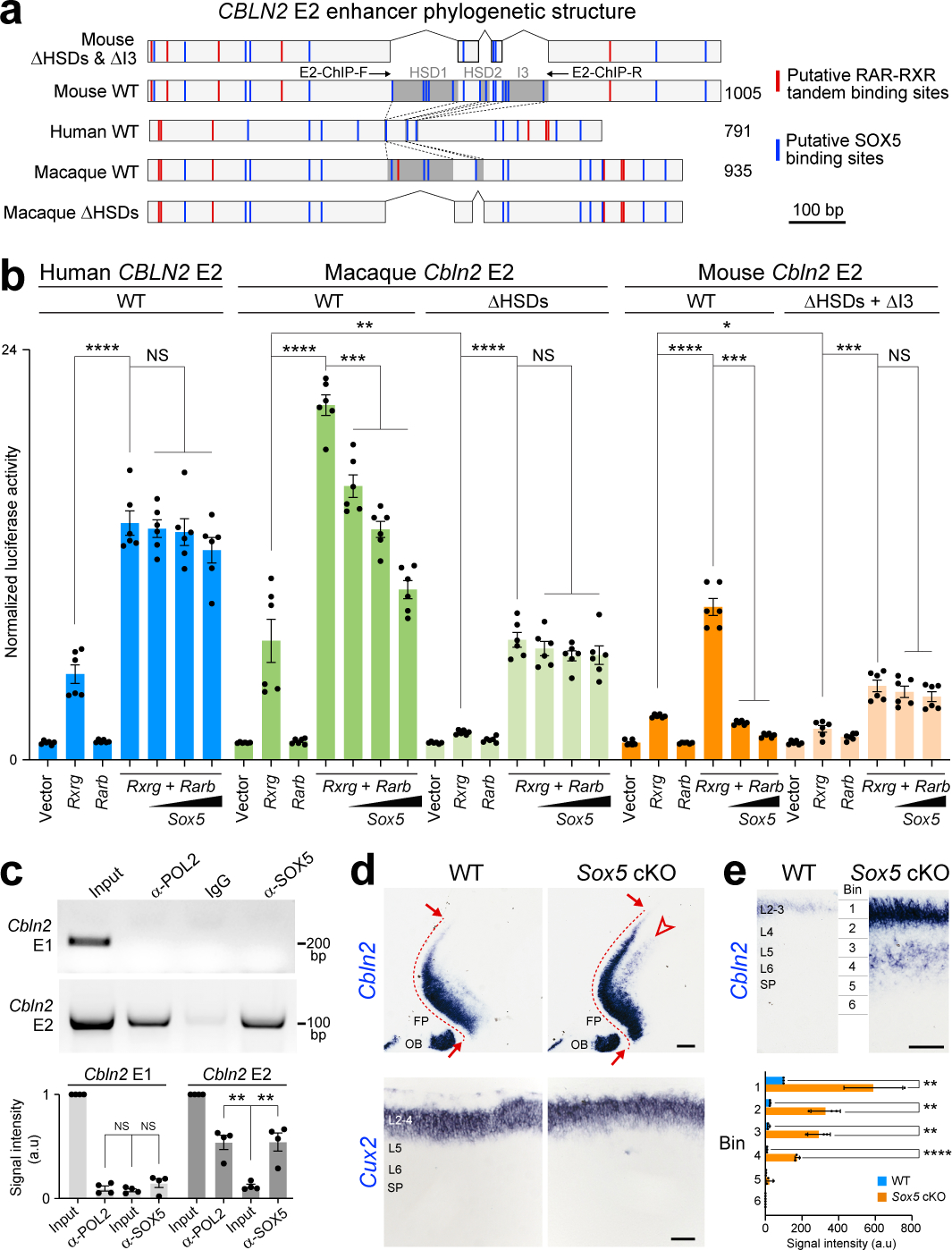
a, Schematics of Cbln2 E2 constructs used for luciferase assays. Shaded dark grey areas in indicate HSD1-2 and I3 (mouse) and HSD1-2 (macaque). All HSDs and I3 are deleted from mouse Cbln2 E2 in ΔHSDs & ΔI3, and all HSDs are deleted from macaque Cbln2 E2 in macaque ΔHSDs. Red and blue lines indicate putative RAR-RXR tandem and SOX5 binding sites, respectively. E2-ChIP-F and R represent ChIP-PCR primers (c) (Table S1). b, Cbln2 E2 luciferase assay in Neuro-2a cells with mouse Rxrg and Rarb and increasing concentrations of Sox5. Two-tailed Student’s t-test; *P = 0.01; **P = 0. 001; ***P = 4e-4 (Macaque), 4e-4 (mouse), 7e-5 (mouse ΔHSDs + ΔI3); ****P < 1e-5, NS, not significant. Error bars: S.E.M.; N = 6 per condition. c, ChIP-PCR assays in PD 0 mouse neocortex using anti-RNA polymerase II (anti-POL2), IgG, and anti-SOX5 and Cbln2 E1 and E2 primers and quantifications of signal intensity below. Two-tailed Student’s t-test; **P = 0.001, 0.003; Error bars: S.E.M.; N = 4 per condition. d, Cbln2 expression in PD 0 Sox5 cKO compared to Sox5 +/+; Emx1-Cre (WT) cortex. N = 3 per genotype. Arrows indicate posterior spread of Cbln2 expression and arrowheads indicate ectopic deep layer expression. FP, frontal pole; OB, olfactory bulb. Scale bars, 200 μm (top); 100 μm (bottom). e, Quantification of cortical Cbln2 signal intensity in six bins spanning from pia to ventricular zone (N = 3). Two-tailed Student’s t-test; ****P = 2e-5 (Bin4); **P = 3e-3 (Bin1), 2e-3 (Bin2), 1e-3 (Bin3). Scale bar, 100 μm.
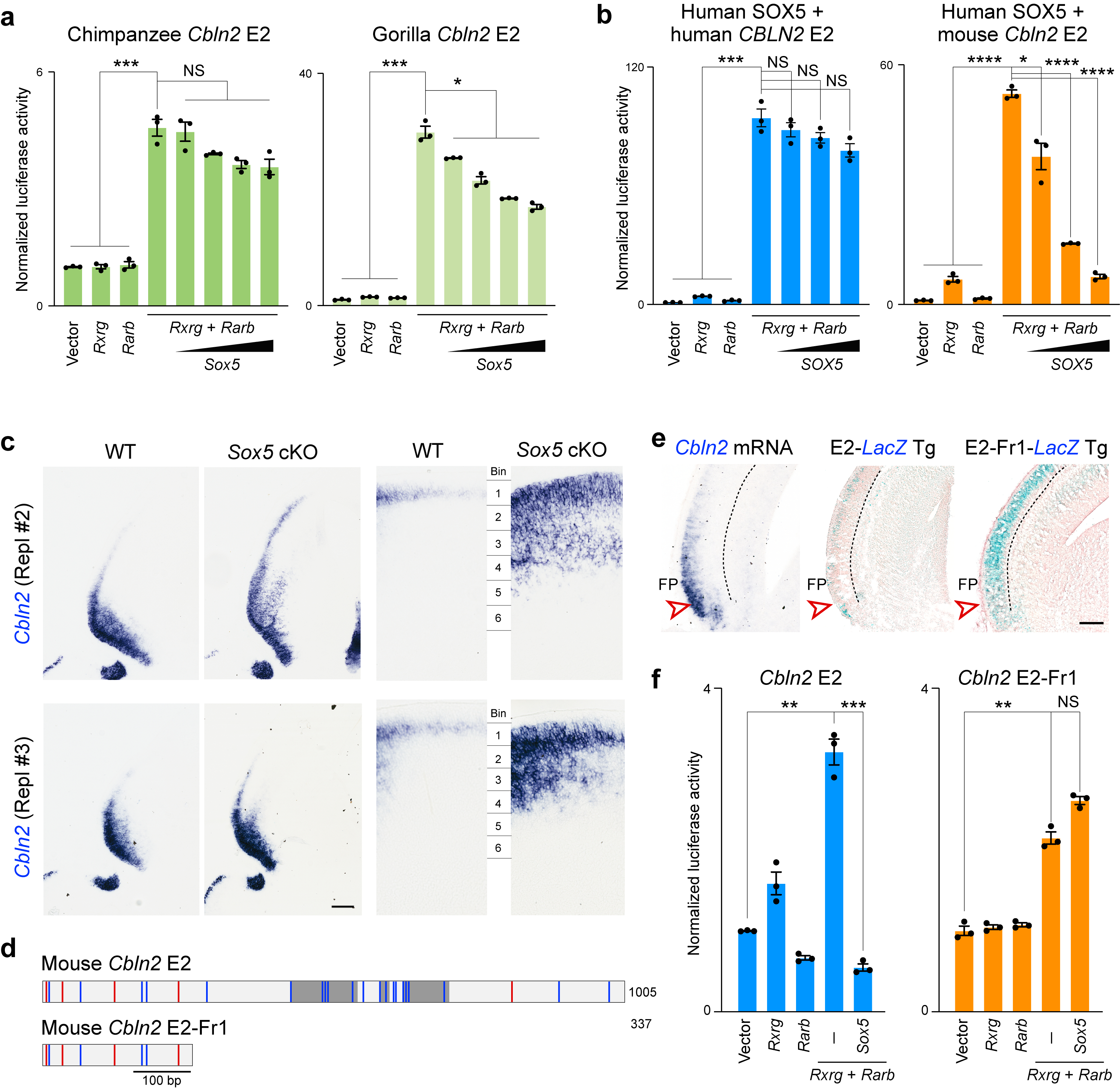
In our accompanying study 34, we found that RA receptors RXRG and RARB are upregulated in the midfetal frontal lobe and that Cbln2 expression was reduced in neonatal Rxrg and Rarb double knockout mice. Thus, we screened for and identified multiple putative RAR-RXR tandem binding sites in CBLN2 E2 (Fig 2a, Extended Data Fig. 4a). Consistent with these findings, we found that overexpression of RXRG and RARB in Neuro2a cell line activated human, chimpanzee, gorilla, macaque, and mouse orthologs of CBLN2 E2 in in vitro luciferase reporter assays more than overexpression of either individually (Fig. 2b; Extended Data Fig. 6a), consistent with previous reports showing that RA receptors exert their effects as heterodimers 39,40.
Hominini-specific repression of CBLN2 E2 enhancer by SOX5
In addition to RA receptors, analysis of Cbln2 E2 also identified multiple conserved putative SOX5 binding sites, including three in HSD1 and a fourth in HSD2 (Fig. 2a, Extended Data Fig. 4a,b), as well as non-conserved sites in mouse HSD1 and 2 orthologs, and in I3 (Fig. 2a). Screening for gained putative human and hominini-specific transcription factor (TF) binding sites identified only a few generic TF binding sites (Extended Data Table 1). Thus, we focused on putative SOX5 binding sites given their loss in Hominini E2 and the role of SOX5 in the specification and development of deep layers cortical projection neurons41,42. We found that mouse Cbln2 E2 activation by RXRG-RARB heterodimer was suppressed by SOX5 in a dose-dependent manner, whereas human and chimpanzee CBLN2 E2 showed no significant suppression by SOX5 at the same doses (Fig. 2b; Extended Data Fig. 6a). Differential suppression of Cbln2 E2 activation was not a consequence of sequence differences between mouse and human SOX5, as activation of mouse Cbln2 E2 was suppressed by both mouse and human SOX5 (Fig. 2b; Extended Data Fig. 6b). Activation of macaque and gorilla Cbln2 E2 showed intermediate suppression of activity at the same doses of SOX5 (Fig. 2b; Extended Data Fig. 6a). Deletion of HSD1, HSD2, and I3 in mouse, or HSD1 and HSD2 in macaque, abolished suppression by SOX5, suggesting that these regions mediate SOX5 suppression (Fig. 2b). Testing for direct in vivo binding using chromatin immunoprecipitation followed by PCR (ChIP-PCR) assays found that endogenous SOX5 bound Cbln2 E2 but not Cbln2 E1 (Fig. 2c).
To understand the functional significance of SOX5 repression of Cbln2 E2 in vivo, we examined Cbln2 cortical expression by creating a cortex-specific Sox5 conditional knockout (cKO; Sox5 loxP/loxP; Emx1-Cre) mouse. As predicted by luciferase assays, the absence of SOX5 led to ectopic expression of Cbln2 in deeper layers of the neonatal frontal cortex (Fig. 2d,e, Extended Data Fig. 6c). There was also an increase and posterior extension of Cbln2 expression in the upper layers (Fig. 2d,e, Extended Data Fig. 6c), likely due to previously described transient Sox5 expression in these layers41. Consistent with these results, the 5’ segment of Cbln2 E2 lacking the two HSD sequences and I3 (Cbln2 E2-Fr1) was not suppressed by SOX5 in luciferase assay (Extended Data Fig. 6d,f). Furthermore, a transgenic mouse line expressing lacZ under the transcriptional control of Cbln2 E2-Fr1, showed deep layer expression of lacZ as compared to Cbln2 E2 (Extended Data Fig. 6d,e). Taken together, these observations suggest SOX5 directly represses Cbln2 expression in the deep layers of a neonatal mouse and midfetal macaque (and likely other non-Hominin primates) frontal cortex by binding HSD sites in Cbln2 E2.
Humanized Cbln2 E2 increases spinogenesis in mouse PFC
To test whether human CBLN2 E2 is sufficient to drive a human-like pattern of Cbln2 expression in the mouse neonatal frontal cortex, we generated a knock-in mouse in which the Cbln2 E2 was replaced in situ (in the orthologous position) with the corresponding human CBLN2 E2 using the CRISPR-Cas9 genome editing technique (Extended Data Fig. 7a–c). The engineered mice carrying homozygous humanized Cbln2 E2 (hCbln2 E2) in both alleles (Homo sapiens;Homo sapiens, Hs;Hs) were viable and fertile, and the expression of the cortical upper layer specific marker, Cux2, and SOX5 were comparable to homozygous wildtype (WT) Cbln2 E2 (Mus musculus;Mus musculus, Mm;Mm) mice (Fig. 3c), indicating that genetically humanized hCbln2 E2 grossly functions as the mouse ortholog. Neocortical Cbln2 expression at PD 0 was increased by 21.7% in hCbln2 E2 (Hs;Hs), as compared to WT by quantitative RT-PCR (Extended Data Fig. 7d). Analysis of expression by in situ hybridization showed that Cbln2 was also transiently upregulated in both prospective upper and deeper layers of the hCbln2 E2 PFC at PCD 18 and PD 1, but not at PCD 16 (Fig. 3a,b; Extended Data Fig. 8). After PD 1, Cbln2 expression extended into the deeper layers of PFC, coinciding with the down-regulation of SOX5 protein expression (Extended Data Fig. 9) in both hCbln2 E2 and WT Cbln2 E2.
Fig. 3 |. In situ genetic humanization of the mouse Cbln2 enhancer drives upregulation and ectopic laminar Cbln2 expression in the neonatal frontal cortex.
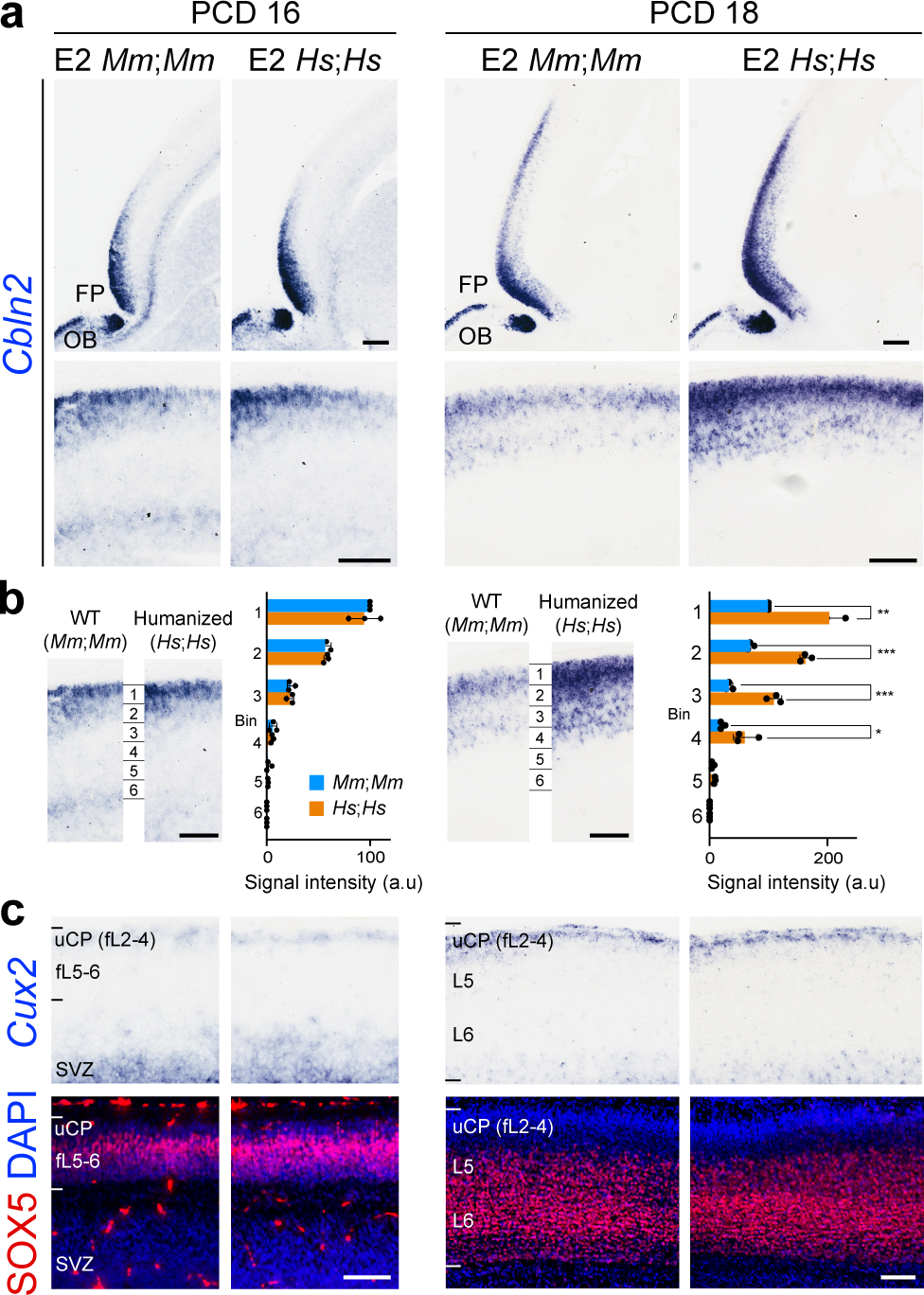
a, Cbln2 expression in the humanized Cbln2 E2 knock-in mouse (hCbln2 E2, Hs;Hs) brain at PCD 16 and 18 compared to WT mouse (Cbln2 E2, Mm;Mm) with higher magnification panels below. FP, frontal pole; OB, olfactory bulb. Scale bars, 200 μm (top); 100 μm (bottom). Experiments were repeated for three brains and two replicates are shown in Extended Data Fig. 8. b, Neocortex was divided into six equal bins spanning from pia to ventricular zone, and Cbln2 signal intensity was quantified for each bin and compared between WT and hCbln2 E2 (Hs;Hs). Two-tailed Student’s t-test; *P = 0.02 (PCD 18), 0.02, 0.04 (PD 1), **P = 1e-3 (PCD18), 1e-3, 3e-3, 3e-3 (PD 1), ***P = 1e-4, 2e-4 (PCD 18); N = 3 per genotype. Scale bars, 100 μm. c, Expression of the upper layer marker, Cux2, and SOX5 in adjacent tissue sections were detected by in situ hybridization and immunostaining, respectively. Scale bars, 100 μm. N = 3.
Members of the CBLN family have been reported to regulate synapse development and function 21–27. Therefore, we hypothesized that a transient increase in Cbln2 expression in the hCbln2 E2 PFC will lead to increased postsynaptic structures and synapse number. We quantified both excitatory and inhibitory postsynaptic density in the medial PFC (mPFC), primary somatosensory area (SSp), and primary visual (VISp) area using immunostaining for postsynaptic proteins PSD-95/DLG4 and gephyrin (GPHN), respectively. In hCbln2 E2 (Hs;Hs) mice, we observed an increase in the density of PSD-95-immunopositive excitatory postsynaptic puncta in both upper (39.5%) and deep layers (47.9%) in the mPFC but not in the SSp and VISp, where Cbln2 expression is not altered, at PD 0 (Extended Data Fig. 10a,b). A similar increase in PSD-95-immunopositive puncta in the mPFC was identified in the Sox5 conditional KO mice (58% for upper layers, 40% for deep layers; Extended Data Fig. 10b). We also observed an increase in the density of GPHN-immunopositive inhibitory postsynaptic puncta number in both upper (24.2%) and deep layers (37.9%) at PD 0 in the hCbln2 E2 PFC but not in the SSp and VISp cortex (Extended Data Fig. 10c,d). In the Sox5 conditional KO mice, anti-GPHN immunostaining revealed a similar increase in the density of inhibitory postsynaptic puncta (27.3% for upper layers, 39.4% for deep layers; Extended Data Fig. 10d). At PD 60, an increase in the density of PSD-95-immunopositive postsynaptic puncta was observed in deep layers of humanized CBLN2 E2 mice (21.4%), and no significant changes were observed in the density of GPHN-immunopositive postsynaptic puncta (Extended Data Fig. 10b,d).
We confirmed the PFC-specific increase in excitatory postsynaptic structures in the hCbln2 E2 (Hs;Hs) mice by quantifying dendritic spines using two different strategies. Using Golgi staining, we identified an increase in mature mushroom shaped spines and long thin spines in both the upper and deep layers of hCbln2 E2 (Hs;Hs) mPFC (Extended Data Fig. 11a) but not in the SSP or VISp (Extended Data Fig. 11b,c). Similarly, we identified an increase in dendritic spines in apical and basal dendrites of both retrogradely labelled upper layer callosally projecting neurons and L5B subcerebrally projecting neurons of the hCbln2 E2 (Hs;Hs) mPFC compared to WT littermates (Fig. 4a,c). To assess whether dendritic spines in the WT and hCbln2 E2 (Hs;Hs) mPFC form synapses, we conducted 3D reconstruction from serial section electron microscopy at PD 60. We found 91.4% of dendritic spines in the hCbln2 E2 mPFC had presynaptic innervation, similar to Cbln2 E2 control (Extended Fig. 11f,g), thus providing evidence against an increase in non-synaptic dendritic spines in hCbln2 E2 (Hs;Hs). Furthermore, we also identified a significant increase overlapping PSD-95/DLG4-immunolabelled postsynaptic and synaptophysin (SYP)-immunolabelled presynaptic puncta in both upper (15.4%) and deep layer (38.6%) of the early postnatal hCbln2 E2 (Hs;Hs) mPFC (Extended Fig. 11d,e). Finally, as increase in dendritic spines and synapse number could be secondary to increased dendritic complexity, which has also been described in human PFC 5, we compared the dendritic arbors in WT Cbln2 E2 compared to hCbln2 E2. We found no significant difference in dendritic complexity in Golgi and retrograde viral tracers labelled dendrites arbors by Sholl Analysis in PFC, SSp or VISp (Fig. 4b; Extended Data Fig. 11a–c).
Fig. 4 |. Increased density of dendritic spines in the mPFC of humanized CBLN2 E2 mice.
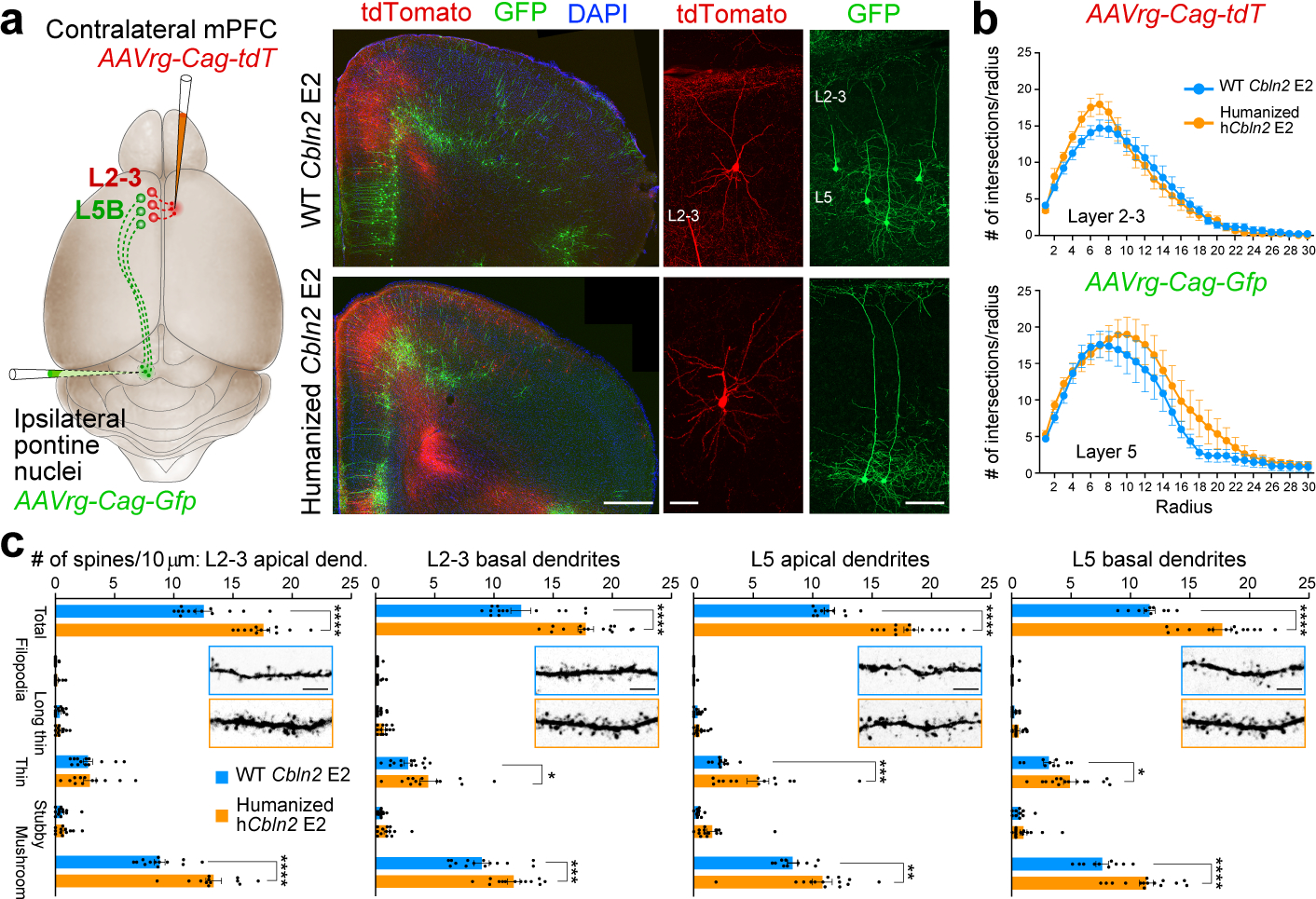
a, Retrogradely labeled callosal L2-3 (red; AAVrg-Cag-tdT) and subcerebral L5B (green; AAVrg-Cag-Gfp) PD 60 pyramidal neurons. Scale bars, 500 μm (left); 20 μm (right). Three brains from each genotype were injected. b, Sholl analysis of retrogradely labelled L2-3 (top) and L5 (below). Two-way ANOVA with Sidak’s multiple comparison method. Error bars: S.E.M.; N = 13 (WT Cbln2 E2), and 15 (hCbln2 E2) L2-3; N = 11 (WT Cbln2 E2), and 15 (hCbln2 E2) L5 (N = 3 brains per genotype). c, Quantification of dendritic spines. Two-way ANOVA with Sidak’s multiple comparison method; *P = 0.03 (L2-3 basal dendrites), 0.03 (L5 basal dendrites); **P = 5e-3, ***P = 4e-4 (L2-3 basal dendrites), 4e-4 (L5 apical dendrites); ****P < 5e-5; Error bars: S.E.M.; N = 13 (WT Cbln2 E2), and 12 (hCbln2 E2) L2-3 apical dendrites; N = 14 (WT Cbln2 E2) and N = 15 (hCbln2 E2) L2-3 basal dendrites; N = 10 (WT Cbln2 E2), and 14 (hCbln2 E2) L5 apical dendrites; N = 10 (WT Cbln2 E2) and N = 15 (hCbln2 E2) L5 basal dendrites (N = 3). Scale bar, 10 μm.
Discussion
In this study, we describe a potential CBLN2-based molecular mechanism and underlying genetic basis for the anterior-posterior gradient of neocortical synaptogenesis and disproportionate increase in dendritic spines within the human PFC. In our accompanying paper 34, we identified that loss of RA signaling in the developing mouse mPFC leads to both reduced expression of Cbln2 and reduced density of excitatory dendritic spines. Regulation of Cbln2 E2 by RXRG and RARB, provides a direct link between these two finding, and may be part of a more complex gene regulatory network regulating development and adaptive changes in PFC connectivity. Furthermore, species-specific regulation of CBLN2 expression is likely only one aspect of phylogenetic differences in connectivity, including described inter-hominin differences 5,6 and accelerated expression of synaptic genes in the developing human neocortex, compared to macaque 15.
While CBLN2 has been implicated in OCD, SCZ, and Tourette’s syndrome 43, it serves as ligands for multiple NRXNs and GRID1, which are ASD- and SCZ-associated proteins, respectively 44,45. Multiple lines of evidence have shown that ASD and SCZ risk genes, converge in time, brain region, cell type and cell compartment with strong evidence for convergence in synapses as well as glutamatergic projection neurons and frontal cortex during human midfetal development 46,47. Consistent with these findings, alterations in the levels of some synaptic proteins and connectivity within the frontal lobe have been described as the possible underlying pathophysiology of both ASD and SCZ 44,48, with dysfunction in the NXRN-GRID trans-synaptic complex, which includes CBLNs, being a putative mechanism and target for interventions.
Methods
Analysis of human and macaque transcriptomic data
Developing human and macaque brain RNA-seq data (counts file) with its metadata information was obtained from BrainSpan (brainspan.org) and PsychENCODE (development.psychencode.org, evolution.psychencode.org) projects 15,20. Timeline of human and macaque development and associated periods were designed by Kang et al. 35. Predicted ages for macaque samples were calculated via the TranscriptomeAge algorithm described in Zhu et al. 15. To perform statistical comparisons, samples from various developmental periods were grouped (periods 4–6, 7–10, and 11–14) and a two-tailed Student’s t-test was used to compare gene expression levels between brain regions and species.
Animals
All experiments using animals were performed in accordance with protocols approved by the Yale University Institutional Animal Care and Use Committee (IACUC). All studies using mice (Mus musculus) and rhesus macaques (Macaca mulatta) were performed in accordance with protocols approved by Yale University’s Institutional Animal Care and Use Committee and NIH guidelines. The animals were housed, and timed-pregnant prenatal and postnatal mouse and monkey brains were obtained in-house at the Yale Animal Resource Center. Mice were reared in group housing less than five mice per cage at 25 °C and 56% humidity in a 12h light:12h dark cycle and provided food and water ad libitum with veterinary care provided by Yale Animal Resource Center. Animals were maintained on the C57BL/6J background. Both sexes were used and randomly assigned for all experiments. The day on which a vaginal plug was observed in mice was designated as PCD 0.5. Mice carrying the floxed Sox5 allele were a kind gift from Véronique Lefebvre 49.
Plasmid construction
For construction of expression vectors used for luciferase assays, full-length cDNAs (Human SOX5, clone ID 30343519; mouse Rarb, clone ID 30608242; mouse Rxrg, clone ID 5707723; all purchased from GE Healthcare) were inserted into pCAGIG vector (Addgene plasmid #11159). For luciferase reporter plasmids, human, chimpanzee, gorilla, macaque, and mouse Cbln2 E2 fragments were PCR-amplified from genomic DNA of individual species and inserted into pGL4.24 vector (Promega, Cat. E8421). The sequences of the PCR primers and synthetic oligonucleotides are listed in Extended Data Table 2.
Postmortem human and macaque tissue
This study was conducted using postmortem human brain specimens or RNA-seq data generated previously 20 from tissue collections at the Department of Neuroscience at Yale School of Medicine, the Brain and Tissue Bank for Developmental Disorders at the University of Maryland, the Clinical Brain Disorders Branch of the National Institute of Mental Health, the Human Fetal Tissue Repository at the Albert Einstein College of Medicine, the Birth Defects Research Laboratory at the University of Washington (R24HD000836), Advanced Bioscience Resources and the Joint MRC–Wellcome Trust Human Developmental Biology Resource (www.hdbr.org; MR/R006237/1). Tissue was collected after obtaining parental or next of kin consent and with approval by the institutional review boards at each institution from which tissue specimens were obtained, the Yale University and the National Institutes of Health. Donated deidentified tissue was handled in accordance with ethical guidelines and regulations for the research use of human brain tissue set forth by the National Institutes of Health (https://oir.nih.gov/sites/default/files/uploads/sourcebook/documents/ethical_conduct/guidelines-biospecimen.pdf) and the WMA Declaration of Helsinki (https://www.wma.net/policies-post/wma-declaration-of-helsinki-ethical-principles-for-medical-research-involving-human-subjects/). All available non-identifying information was recorded for each specimen. No obvious signs of neuropathological alterations were observed in any of the human, macaque or mouse specimens analyzed in this study. The postmortem interval was defined as hours between time of death and time when tissue samples were fresh frozen or started to undergo fixation process.
Transcription factor binding site (TFBS) analysis
The software FIMO from the MEME suite (ver. 4.12.0) was used to identify putative transcription factor binding sites in the human sequence of the CBLN2 E2 enhancer. For this analysis, a set of transcription factor motifs from the JASPAR 2018 dataset (http://jaspar2018.genereg.net/) 50 were used. Then, MAF (Multiple Alignment Format) files were downloaded from the multiz100way table of the UCSC genome browser. An in-house script was used to extract the sequence of each TFBS in each species from the MAF file and identify mutations in core positions of the motif. TFBS were considered conserved in a particular species if they had no mutations in core positions and were considered present in a clade if the TFBS was conserved in at least 25% of the species in the clade. To check for potential loss of TFBS regulatory mechanisms due to the deletions HSD1, HSD2, and I3, we looked for TFBS that are found in these regions and conserved in multiple species that do not have the deletions. To do so, the software FIMO was used to identify instances of transcription factor binding motifs from the JASPAR database. This was performed in two species for each deletion (mouse and macaque for HSD1 and 2, and mouse and rat for I3). Then, the coordinates of putative TFBSs in the regions of interest in macaque and rat were translated to mm10 coordinates using LiftOver (downloaded on May 3rd, 2017) and manually compared to the TFBS identified in the mouse sequence. Instances of the same transcription factor binding motif at the same location in both datasets were considered putatively conserved. SOX5-binding sites and RA receptors binding sites of mouse and human were identified using JASPAR2018 51. The binding sites from other animal species were identified by conservation. RA receptors binding sites include those of RARA::RXRA, RARA::RXRG, RARA, RARB, RARG, RXRA, RXRB, and RXRG.
Generation of transgenic reporter mice
Cbln2 E2 and its related fragments were PCR-amplified from mouse genomic DNA and ligated into pBgn-lacZ 52. Vector was linearized with BglII and KpnI and purified by gel separation followed by phenol/chloroform extraction. Transgene fragment was diluted into microinjection buffer (5 mM Tris-HCl, 0.1 mM EDTA), and injected into the pronucleus of fertilized eggs from B6SJLF1/J mouse purchased from The Jackson Laboratory. Transgenic mice were screened for the presence of transgenes by PCR using the lacZ primer set listed in Extended Data Table 2.
Generation of humanized Cbln2 E2 knock-in mice
The overall strategy for the generation of humanized Cbln2 E2 (hCbln2 E2) follows a previously described protocol 53. The targeting vector carrying hCbln2 E2 mouse was constructed as follows: mouse genomic DNA fragments of 1601 bp (chr18: 86721709–86723309, GRCm38/mm10) and 1909 bp (chr18: 86724521–86726429, GRCm38/mm10) flanking the region containing Cbln2 E2 (chr18: 86723310–86724520, GRCm38/mm10) were PCR-amplified as left and right arm, respectively, using mouse genomic DNA as template. An 852 bp fragment containing human CBLN2 E2 (chr18: 72530473–72531324, GRCh38/hg38) was PCR-amplified using human genomic DNA as a template. These three DNA fragments were ligated into XhoI, HindIII and ClaI sites of the PL451 vector 54. For the construction of the templates of guidance RNA, two sets of top and bottom strand oligomers (see Extended Data Table 2) directing the double strand break at mouse left and right arm were annealed and ligated into BbsI site of pX330-U6-Chimeric_BB-CBh-hSpCas9 vector 55, which was purchased from Addgene (Plasmid #42230). After amplification of insert with T7-tagged primers (see Extended Data Table 2), guidance RNAs were synthesized by T7 RNA polymerase. The coding region of Cas9 was PCR-amplified using pX330-U6-Chimeric_BB-CBh-hSpCas9 as a template and inserted into the pSP64 Poly(A) vector (Promega, Cat. 1241). Vector were digested and linearized with EcoRI. Cas9 mRNA was synthesized by SP6 RNA polymerase. Guidance RNAs and Cas9 mRNA were purified by MEGAclear Transcription Clean-Up Kit (Ambion, Cat. AM1908). The targeting vector, Cas9 mRNA, and two guidance RNAs were mixed at a concentration of (10 ng; 100 ng; 100 ng; 200 ng μl−1) in the microinjection buffer (5mM Tris-HCl pH7.5; 0.1M EDTA) and injected into the pronuclei of fertilized eggs from the B6SJLF1/J mouse strain. Which was purchased from The Jackson Laboratory. The fertilized eggs were then transferred to uterus of female CD-1, purchased from Charles River Laboratories. The first generation (F0) mice with recombined allele were identified by long-distance PCR with a primer set (mP3/mP4) designed outside of targeting vector (Extended Data Table 2, Extended Data Fig. 7a), and confirmed by sequencing. The germ line transmission in F1 generation was confirmed by nested PCR using the primer set of mP3/mP4, followed by hP1/P2 (Extended Data Fig. 7b,c), to exclude the possibility of detecting targeting vector randomly integrated into the genomic DNA. Mice in the following generation were genotyped by PCR with mP1/mP2 and hP1/P2 as indicated in Extended Data Fig. 7.
In situ hybridization
Whole-mount and section in situ hybridization were performed as described previously 56. Antisense digoxigenin (DIG)-labeled RNA probes were synthesized using DIG or Fluorescein RNA Labeling Mix (Roche, Cat. 11277073910). Human and mouse Cbln2 cDNA, and mouse Cux2 cDNA were obtained from GE Healthcare (Clone ID 5727802, 6412317, and 30532644, respectively). Macaque Cbln2 cDNA was synthesized using total RNA from adult macaque dlPFC/DFC region. The PCR fragment was ligated into pCRII vector (ThermoFisher Scientific, Cat. K206001). Tissue sections were obtained from PCW 21 and 22 de-identified postmortem human brains, PCD 110 and 140 postmortem macaque brains, and mouse postmortem brains of various developmental ages. In situ hybridization experiments were repeated using these two sets. Images were taken using Aperio CS2 HR Scanner (Leica; Wetzlar, Germany) and processed by Aperio ImageScope 12.4.3.5008 (Leica).
Immunohistochemistry
Postmortem brains were dissected and fixed with 4% paraformaldehyde overnight at 4°C, followed by 30% sucrose/PBS and embedding in O.C.T. Compound (ThermoFisher Scientific, Cat. 23-730-572). Brains were sectioned at 15–20 μm by cryostat (Leica CM3050S) after they were frozen. For antigen retrieval, sections were treated with R-Buffer AG pH 6.0 (Electron Microscopy Sciences, Cat. 62707–10) for 20 min at 120 °C. The density of excitatory and inhibitory postsynaptic puncta was quantified using immunostaining of PSD-95 (also known as DLG4) 57 and gephyrin (GPHN) 58, respectively. The sources of primary antibodies were anti-PSD-95/DLG4 (1:500; Invitrogen, Cat. 51–6900), anti-GPHN (1:500; Synaptic Systems, Cat. 147011), anti-SYP (1:2000, Sigma-Aldrich, Cat. SAB4200544), anti-SOX5 (1:500; Abcam, Cat. ab94396), and anti-BCL11B/CTIP2 (1:500, Abcam, Cat. ab18465). Secondary antibodies: Alexa Fluor 488-, or 594- conjugated AffiniPure Donkey anti-Rabbit IgG (Jackson ImmunoResearch). For all microscopic analysis, LSM META (Zeiss), and LSM software ZEN2009 were used.
Quantification of postsynaptic and presynaptic puncta marked by immunostaining
For each cortical area, using both the 488 nm channel to detect PSD-95 (also known as DLG4) or gephyrin (GPHN) and the 594 nm channel to detect BCL11B or synaptophysin (SYP), seven serial optical sections at 0.8 μm intervals over a total depth of 5 μm were imaged and the 2nd, 4th, and 6th images were eliminated from further analysis to avoid overlap in counting 59. Area of each image is 0.079 mm2. The number of PSD-95, SYP-, or GPHN-immunopositive puncta on each image was counted using ImageJ (ver.2.0.0-rc-69/1.52p) using the automated Analyze Particles function using a threshold of 985 to 4095, determined based on multiple WT Cbln2 E2 and humanized hCbln2 E2 images. BCL11B-immunonegative cells were considered as L2-4, and BCL11B-immunopositive nuclei were considered as L 5. At least three sections from each animal were selected for counting, and at least 3 animals for each genotype were used.
Golgi staining
A FD Rapid GolgiStain Kit (FD NeuroTechnologies, Cat. PK401) was used to compare neuronal morphologies (~5 basal dendrites/neuron; ~4 neurons per mouse) throughout the prefrontal cortex, S1C, and V1C of WT Cbln2 E2 (N = 3) and humanized hCbln2 E2 (N = 3) mice aged P60. The manufacturer’s protocol was followed, incubating brains in solution A+B for 21 days and solution C for 7 days before sectioning and mounting on TOMO adhesion microscope slides (Matsunami, Cat. TOM-11). The sections were dried overnight, washed and developed by immersing the slides in solution D and solution E mixed in equal amounts for 10 min. The slides were washed twice with distilled water and dehydrated in grades of ethanol and xylene. After cover-slipped with Permount (Fisher Scientific, Cat. 15820100), sections were imaged on a Nikon SDC microscope enabled with brightfield.
Retrograde neuronal tracing with adeno-associated viruses
To differentiate between the callosally and subcerebrally projecting neurons in the mPFC, we performed dual retrograde injections into the contralateral mPFC and the ipsilateral basal pontine nuclei, respectively at PD 30. Briefly, animals were anesthetized by injecting Ketamine/Xylazine solution and head fixed in the stereotactic frame. Thirty minutes prior to surgery, buprenorphine was administered. After lubricating the eyes and shaving the fur, and incision of < 1mm was made. A craniotomy was made with the round 0.5 mm drill bit at the desired co-ordinates (for mPFC: ML ±0.35, AP 1.5, DV 2.5 and for contralateral Pons: ML ± 0.5, AP −4.0 and DV 5.5 from bregma). Using a Hamilton neuros syringe (0.5 ml), we injected 50 nl of AAVrg-Cag-Tdt (Addgene, 59462-AAVrg) into the mPFC and 150 nl of AAVrg-Cag-Gfp (Addgene, 37825-AAVrg) into the pons at P30. To prevent the virus from spreading along the injection tract, the needle was held in place for at least 10 min. After injections, the skin was sutured and animals were returned to the cage. Approximately 3 weeks later (PD 60), the animals were sacrificed and brains were collected. The brains were coronally sectioned on vibratome to obtain 70 μm thick sections. After staining the sections with anti-GFP antibody (1:500, Abcam, Cat. ab13970) and anti-RFP antibodies (1:500, Abcam, Cat. ab124754), the sections were imaged in LSM 800 microscope (ZEISS).
Quantification of dendritic spines and dendritic arbor
The images of entire neurons were acquired at 20X magnification. Nearly 44–50 images/ Z stack were obtained to cover the entire thickness of dendrites. For spine counts, the z stack images were opened in the Reconstruct 1.1.0.0. (Boston, MA, USA) publicly available at http://www.bu.edu/neural/Reconstruct.html 60 and a new series was recreated which enabled us to move across different stacks across z-planes in same image. Whole dendrite was subdivided into segments of 10 μm and number of spines across whole thickness were traced for length and breadth of each spine. Length and breadth ratio was used to determine the spine subtype as described earlier 61,62. After the analysis for each class of spine, standard deviation and p-values are calculated using two-way Anova with Sidak’s multiple comparison method. For Sholl analysis, z-stack images were opened in ImageJ (ver. 2.0.0-rc-69/1.52p) and dendritic arbors were manually traced using the NeuronJ plugin 63. Dendritic complexity was then quantified and plotted using the Sholl Analysis option.
Processing, analysis, and image visualization
For select figure images, slides background was removed and pseudocolored white for improved clarity, as in Fig 1b. To allow robust visualization and analysis, images depicting immunohistochemistry using antibody against PSD-95/DGL4 and GPHN have been inverted and/or pseudocolored, as in Fig. 4.
ChIP-sequencing data analysis
H3K27ac ChIP-seq from the developing human brain was obtained and peaks were called by Li et al. 20 that were converted to the mm10 genome build using LiftOver 64 and visualized using the Integrative Genomics Viewer 65. DNase 1 hypersensitivity annotation was obtained and visualized via the UCSC Genome Browser 66,67.
Luciferase assays
Neuro-2a cells were transfected using Lipofectamine 2000 (ThermoFisher Scientific, Cat. 11668019) with either mouse or human pCAGIG-Sox5, Rxrg, Rarb, or empty pCAGIG, together with one of the pGL4.24 luciferase vectors generated with enhancer sequences as described above. pGL4.73 Renilla luciferase plasmid (Promega, Cat. E6911) was co-transfected to control for transfection efficiency. The luciferase assays were performed 48 h after transfection using the Dual-Luciferase Reporter Assay System (Promega, Cat. E1910) according to the manufacturer’s instructions. Luciferase activity were measured and quantified by GloMax®-Multi Detection System (Promega).
Chromatin immunoprecipitation (ChIP)
The anterior half of cortices from PD 0 mice were dissociated then cross-linked with 1% formaldehyde and processed using EZ-ChIP kit (Sigma-Aldrich, Cat. 17–371). Chromatin fragments bound by endogenous mouse SOX5 were pulled down by anti-SOX5 (Abcam, Cat. ab94396), anti-POL2 antibody (1:1000, Millipore-Sigma, Cat. 05–623, included in the kit), or random IgG control (included in the kit), then detected by PCR using E2-ChIP-F and E2-ChIP-R or E1-ChIP-F, and E1-ChIP-R primers (see Extended Data Table 2).
Quantitative reverse transcription-PCR
Total RNA was isolated using Trizol (Thermo-Fisher Scientific, Cat. 15596026) from freshly dissected neocortices after removal of the olfactory bulb, hippocampus, and striatum. cDNAs were prepared using SuperScript II (Thermo-Fisher Scientific, Cat. 18064022) from more than three independent WT littermates and humanized hCbln2 E2 brains. Quantitative reverse transcription-PCR was performed as described previously 68 using 7000HT Sequence Detection System (Applied Biosystems). At least three biological replicates per transcript were used for every reaction. The copy number of transcripts was normalized against the house keeping TATA-binding protein (TBP) transcript level. For Cbln2 and Tbp primer sets, correlation (R2) was higher than 0.98, and the slope was −3.1 to −3.6 in each standard curve. Primers were designed in a single exon and are listed in Extended Data Table 2.
3D reconstruction of dendrites and denritic spines using electron microscopy
PD 60 WT Cbln2 E2 and humanized hCbln2 E2 mice (N = 3 for each genotype) were intracardially perfused with fixative containing 4% paraformaldehyde and 1% glutaraldehyde in 0.1 M phosphate buffer (pH 7.4). Brains were removed and immersed in same fixative for 3–4 days at 4°C. Coronal 60-mm-thick slices through prefrontal cortex were cut with a vibrotom, post-fixed with 1% OsO4, dehydrated in ethanol, embedded in epoxy resin Durcupan (ACM; Fluka, Buchs, Switzerland) and polymerized on microscope slides. Ultrathin sectioning and electron microscopic investigation were performed as we previously described 69,70 with minor modifications as follow. Segments of the mPFC were dissected and re-embedded into Durcupan blocks. Serial 60-nm-thick sections from mPFC L2-3 were cut by Leica UC7 ultramicrotome, collected on one-slot grids covered with Butvar B-98 films (EMS, Hatfield, PA, USA) and stained with lead citrate. Series of 48–51 consecutive images were photographed with Talos L120C electron microscope (ThermoFisher Scientific, Boston, MA, USA) at 5300 X magnification. For serial imaging, random neuropil segments were chosen although avoiding cell bodies and blood capillaries. 3D reconstructions were performed using the computer program Reconstruct 1.1.0.0. (Boston, MA, USA) publicly available at http://www.bu.edu/neural/Reconstruct.html 60. Sixteen -representable dendrite fragments were traced in the serial images and their innervated spines and non-innervated spine-like membrane protrusions were counted. Percentage of innervated spines among all the protrusions were calculated for each animal.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this paper.
Data availability
None
Extended Data
Extended Data Fig. 1 |. Spatiotemporal expression of CBLN2 in the human and macaque neocortex.
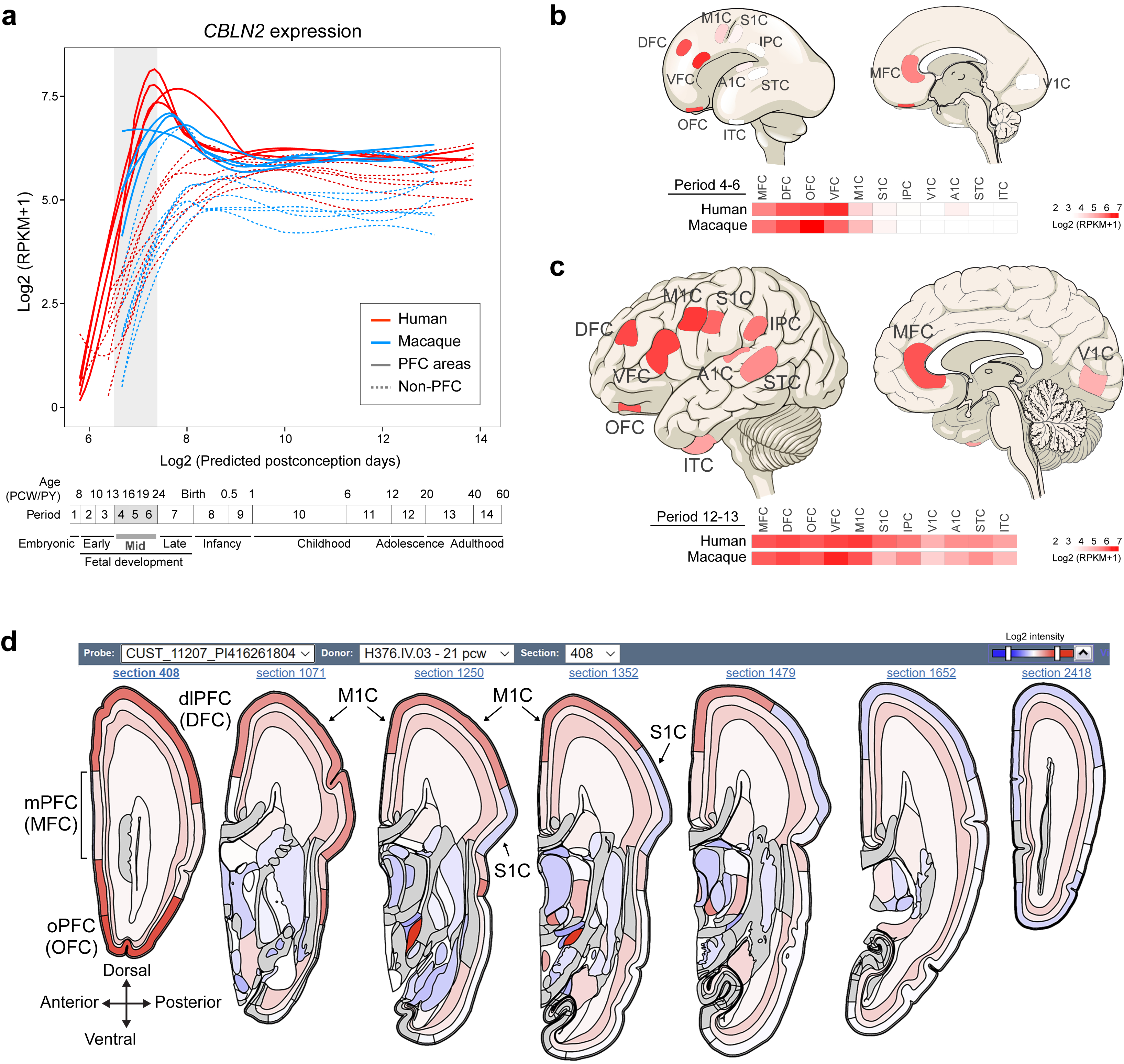
a-c, Spatiotemporal expression of CBLN2 expression in the human and macaque cerebral cortex during development base on BrainSpan (brainspan.org) and PsychENCODE (development.psychencode.org, evolution.psychencode.org) human and macaque RNA-seq data15,20. The RNA-seq data consisted of tissue-level samples comprising the pial surface, marginal zone, cortical plate (layers 2–6) and adjacent subplate zone, of eleven prospective neocortical areas. Red and blue lines indicate human and macaque PFC CBLN2 expression, respectively, and dotted lines represent the non-PFC CBLN2 expression. Vertical grey box demarcates mid-fetal developmental periods. Predicted ages, timeline of human and macaque development and the associated periods are shown below. Visual representation of CBLN2 in human cortex with heatmaps of regional expression in human and macaque below during period 4–6 (b) and period 12–13 (c). Darker reds represent high expression levels. d, Anteroposterior visual representation of human CBLN2 expression at PCW 21 from the BrainSpan human prenatal laser microdissection microarray data (brainspan.org)37.
Extended Data Fig. 2 |. Expression profile of Cbln1,3,4 genes related to dendrite and synapse development and CBLN2 binding partners in macaque and human.
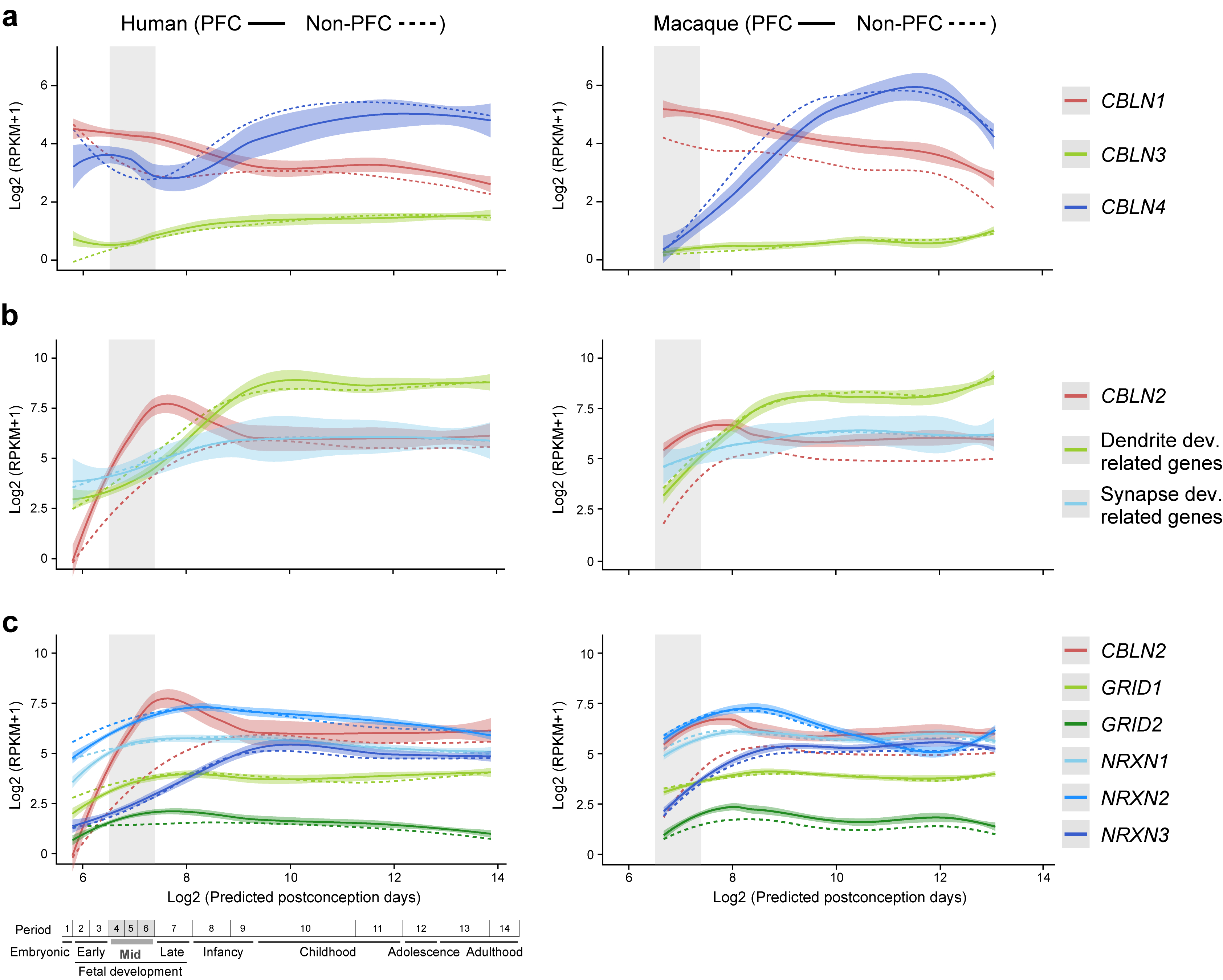
a, Developmental trajectory of CBLN2 expression compared to the expression of key genes related to dendrite development (i.e., MAP1A and CAMK2A) and synapse development genes (i.e., SYP, SYPL1, SYPL2 and SYN1). The lists of genes related to synapse and dendrite development were previously complied and analyzed for their expression trajectories by Kang et al.35 b, Developmental trajectory of CBLN2 expression compared to the expression of genes encoding CBLN2 binding partners (i.e., GRID1, GRID2, NRXN1, NRXN2 and NRXN3). Left and right panels show gene expression in human and macaque, respectively. Gene expression in PFC and non-PFC are indicated by solid and dashed line, respectively. Vertical grey box highlights the mid-fetal developmental periods. Predicted ages, timeline of human and macaque development and the associated periods are shown below. Gene expression values are represented as log2(RPKM+1). For all of these plots, the shading around the lines represents the 95% confidence interval. Abbreviations were as described in Fig. 1 legend.
Extended Data Fig. 3 |. Mouse Cbln2 expression at multiple stages and Cbln2 E2 transgenic lines.
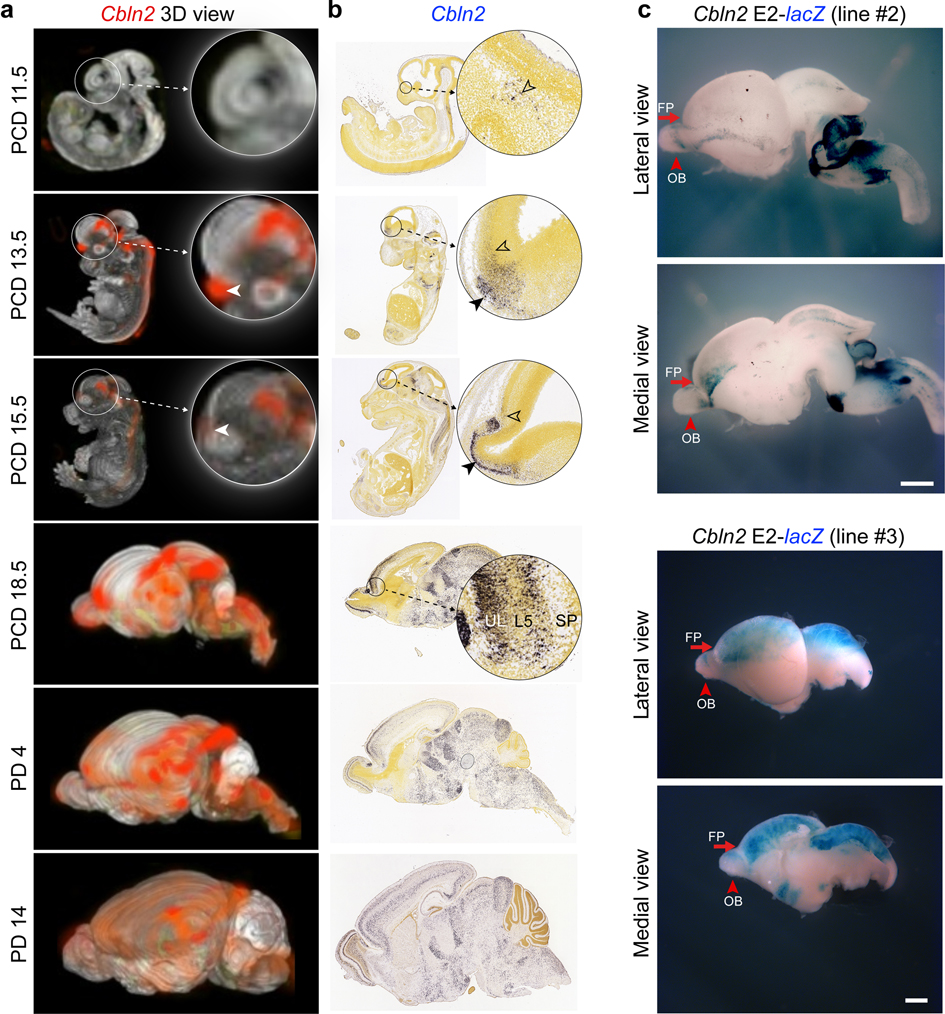
Visualization of Cbln2 expression in wholemount (a) and sagittal sections (b) at PCD 11.5, 13.5, 15.5 18.5 and PD 4, 14 from Allen Brain Atlas developing mouse brain atlas (developingmouse.brain-map.org) 71. Arrowheads highlight early rostral expression, UL, upper layer; L5, layer 5; SP, subplate. c, β-Galactosidase activity in two additional mouse brains from independent transgenic mouse carrying Cbln2 E2 conjugated with lacZ reporter at PCD 17. See Fig. 1c for the third replicate. FP, frontal pole; OB, olfactory bulb. Scale bars, 1 mm.
Extended Data Fig. 4 |. Comparative analysis of CBLN2 E2 deletions across mammals.
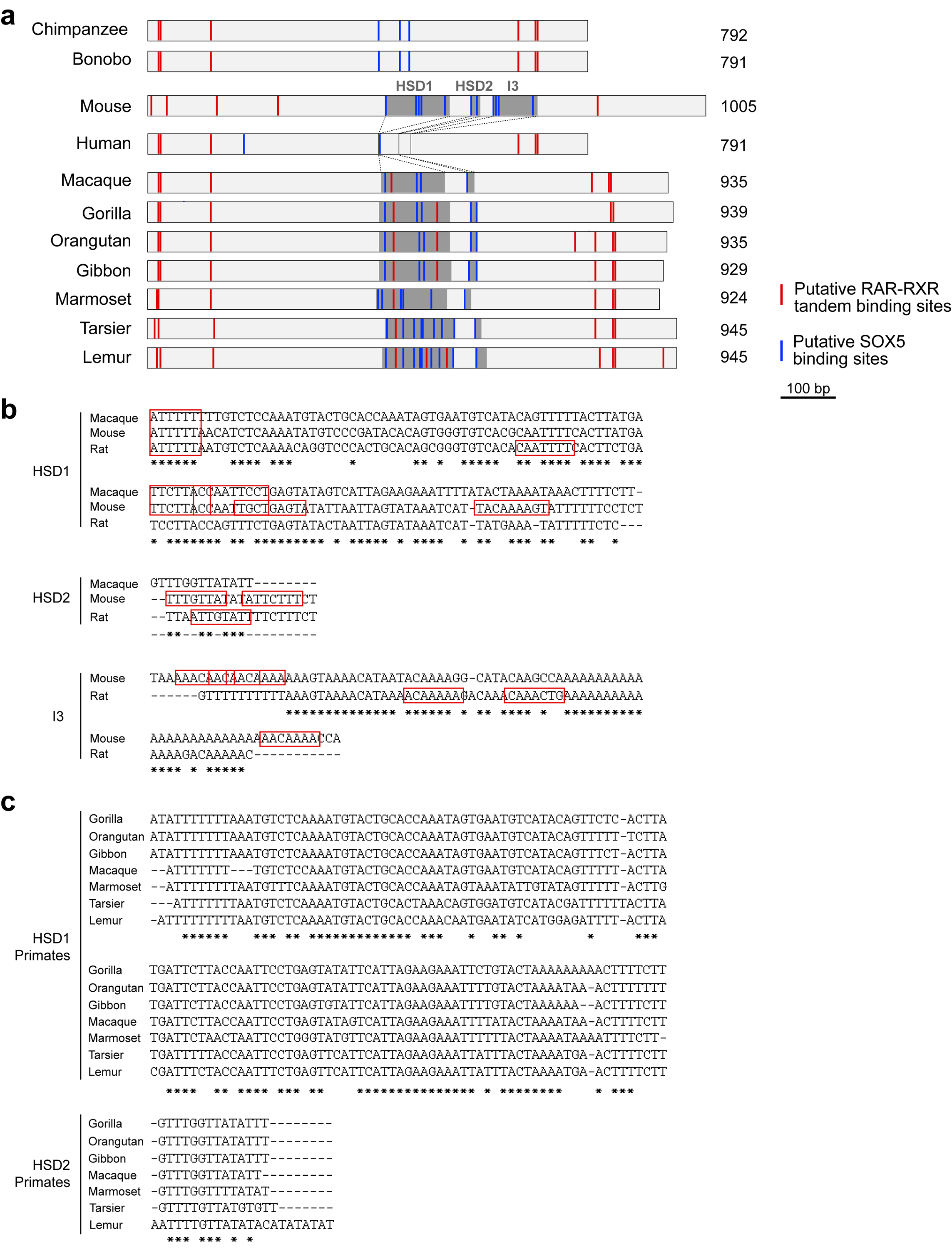
a, Schematic representation of CBLN2 E2 from mouse and primate species including apes (human, common chimpanzee, bonobo, gorilla, orangutan and gibbon), old world monkey (Rhesus macaque), new world monkey (marmoset) and prosimians (tarsier and lemur). HSD1, HSD2 and I3 are shaded. Putative RAR-RXR tandem binding sites indicated as red lines and putative SOX5-binding sites as blue lines. b, Sequence alignments of HSD1, HSD2 and I3 from macaque, mouse, and rat. Putative SOX5 binding sites are shown in red boxes. c, Sequence alignments of HSD1 and HSD2 from primates shown in (a).
Extended Data Fig. 5 |. Conservation of CBLN2 E2 and deleted regions across species.
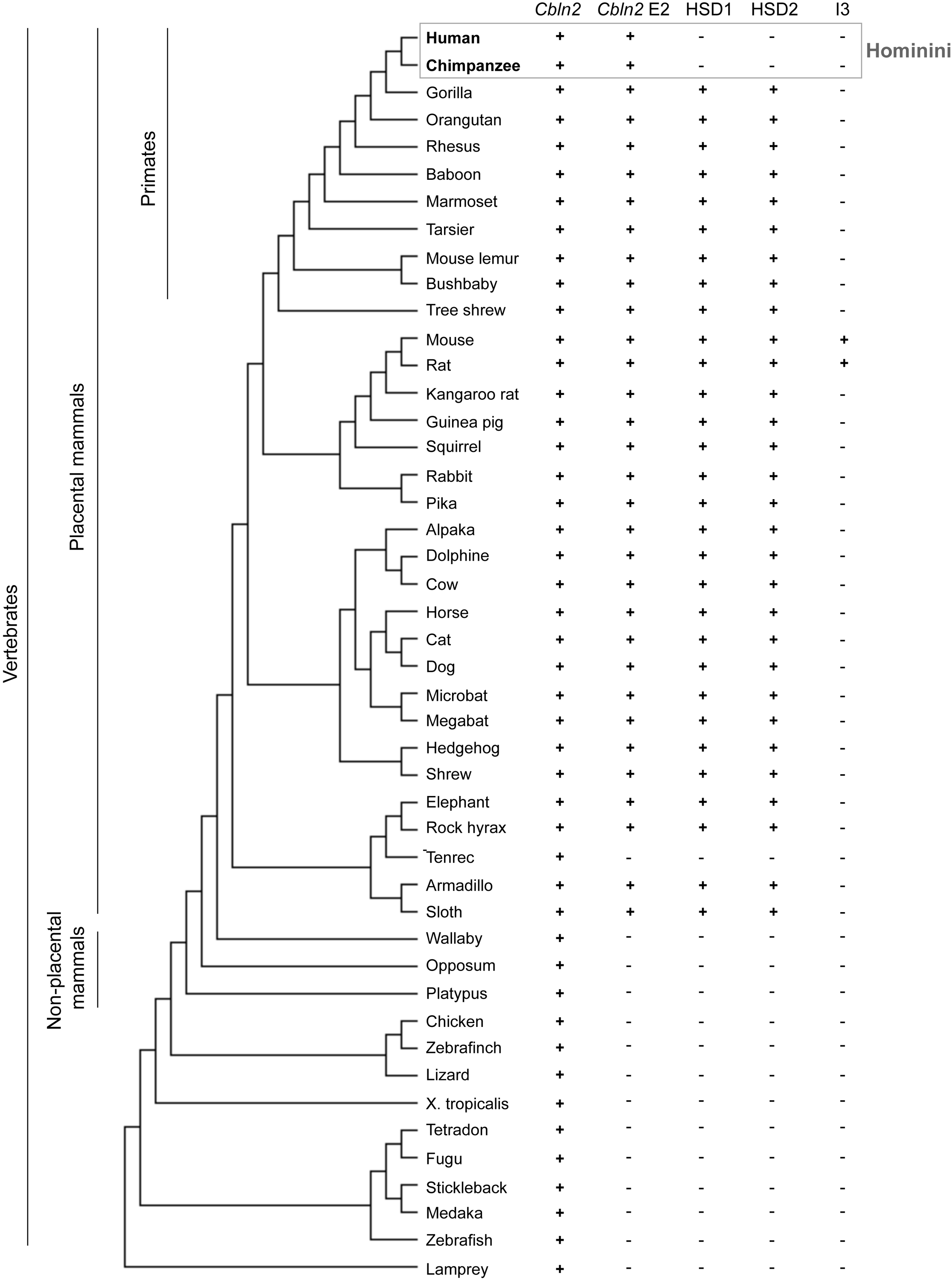
Phylogenetic tree of selected chordates including placental and non-placental mammals with information about presence of the CBLN2/Cbln2 gene and CBLN2 E2/Cbln2 E2. The last three columns describe the presence of HSD1, HSD2 and I3. The mouse E2 sequence was searched in each most updated animal genome browser at UCSC genome browser (as of December 22, 2019). E2 conservation criteria are: 1) identity over 80%; and 2) alignment length over 600 bp compared with mouse E2 (1005 bp).
Extended Data Fig. 6 |. SOX5 directly suppresses RXRG-RARB responsive human, chimpanzee, gorilla and mouse Cbln2 E2 enhancer.
a, Luciferase assay in Neuro-2a cell line from luciferase reporters conjugated to chimpanzee and gorilla wild Cbln2 E2. Two-tailed Student’s t-test; *P = 0.01; ***P = 9e-5 (Chimpanzee), 1e-5 (Gorilla); NS, not significant. Error bars; S.E.M.; N = 3 per condition. b, Overexpression of human SOX5 exerts a similar effect to mouse Sox5 on human and mouse Cbln2 E2 reporters. Two-tailed Student’s t-test; *P = 5e-3; ***P = 4e-5; ****P = 2e-6; 2e-6, 1e-6 (Human Sox5 + mouse Cbln2 E2); NS, not significant; Error bars, S.E.M.; N = 3 per condition. c, Cbln2 expression is upregulated in Sox5 conditional knockout brain at PD 0. Additional two replicates (Repl #2 and 3) not shown in Fig. 2d are shown. Scale bar, 200 μm (left); 100 μm (right). d, Constructs used for luciferase assay and generation of transgenic animals shown in e,f. e, Transgenic mouse brain at PCD 17 carrying Cbln2 E2 (N = 3) or Cbln2 E2 Fr1-lacZ reporters (N = 3). Cbln2 expression in the PFC is indicated by arrowheads. Endogenous Cbln2 expression is also shown for comparison (N = 4). Scale bar, 200 μm. f, Luciferase assay for Cbln2 E2 and Cbln2 E2-Fr1. Two-tailed Student’s t-test; **P = 1e-4 (Cbln2 E2), 2e-4 (Cbln2 E2-Fr1); ***P = 8e-5; NS, not significant; Error bars: S.E.M.; N = 3 per condition.
Extended Data Fig. 7 |. Humanized Cbln2 E2 knock-in mouse shows increased Cbln2 expression in neonatal neocortex.
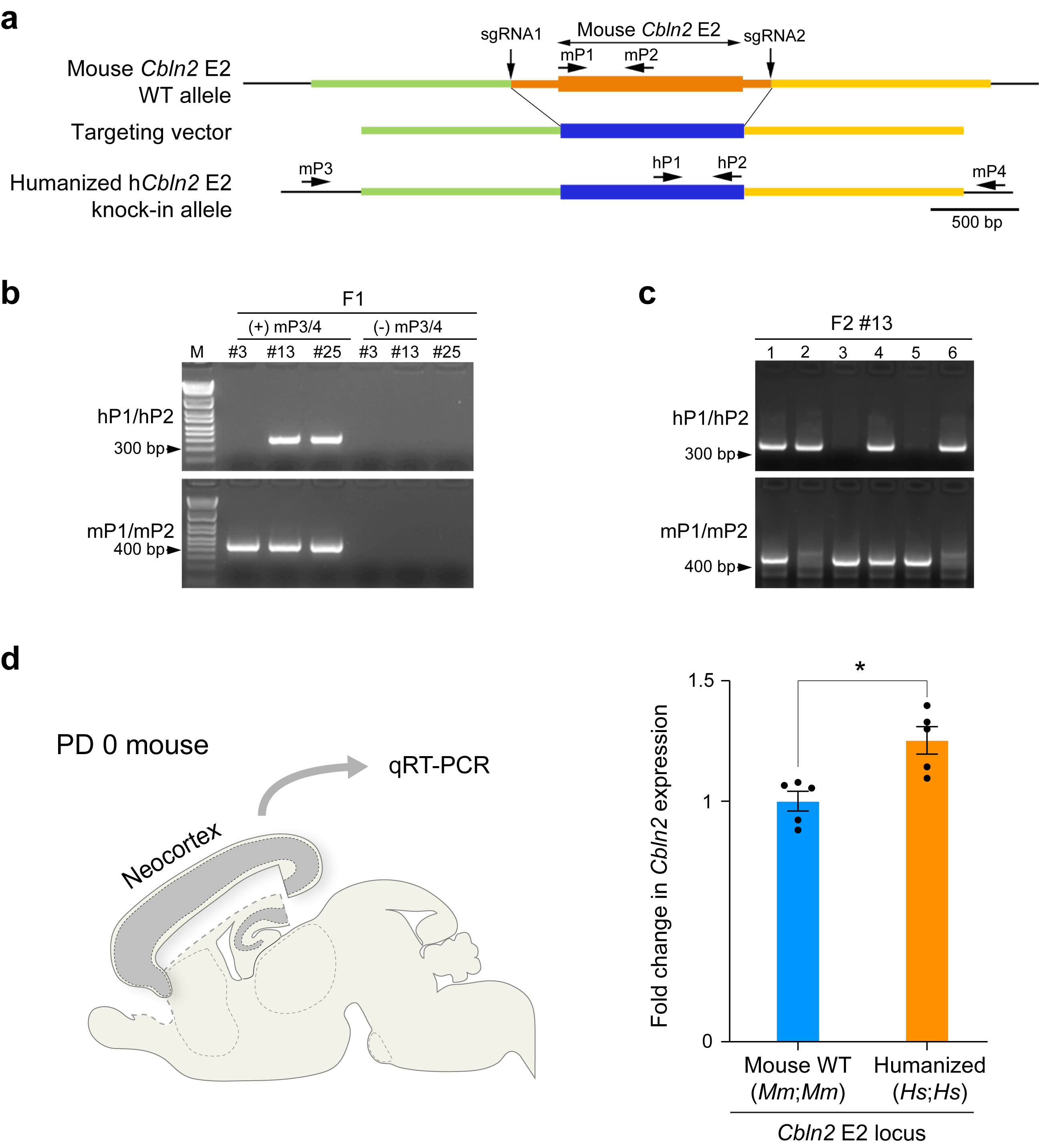
a, Positions of single-guide RNAs (sgRNA1 and 2) to introduce double-strand breaks in the genomic DNA and targeting vector to replace WT mouse Cbln2 E2 region with that of human CBLN2 E2 (humanized hCbln2 E2) are shown. b, Genotyping strategy for F1 mice. Germline transmission in the F1 generation was confirmed by nested PCR using the primer set of mP3/mP4, followed by hP1/P2 as indicated in (a). Two founders #13 and #25 were obtained. c, Mice in the following generation were genotyped by PCR with hP1/P2 and mP1/mP2. An example of genotyping for F2 of line #13 mice is shown (c). d, Comparison of Cbln2 expression between WT Cbln2 E2 (Mm;Mm) and hCbln2 E2 (Hs;Hs) neocortex at PD 0 using quantitative reverse transcription-PCR. RNA was extracted from the neocortex following the removal of hippocampus, olfactory bulb and subpallial regions. Two-tailed Student’s t-test; *P = 0.007; Error bars: S.E.M.; N = 5 per genotype. Genotyping were repeated at least two times in b and c.
Extended Data Fig. 8 |. Additional replicates from Fig. 3.
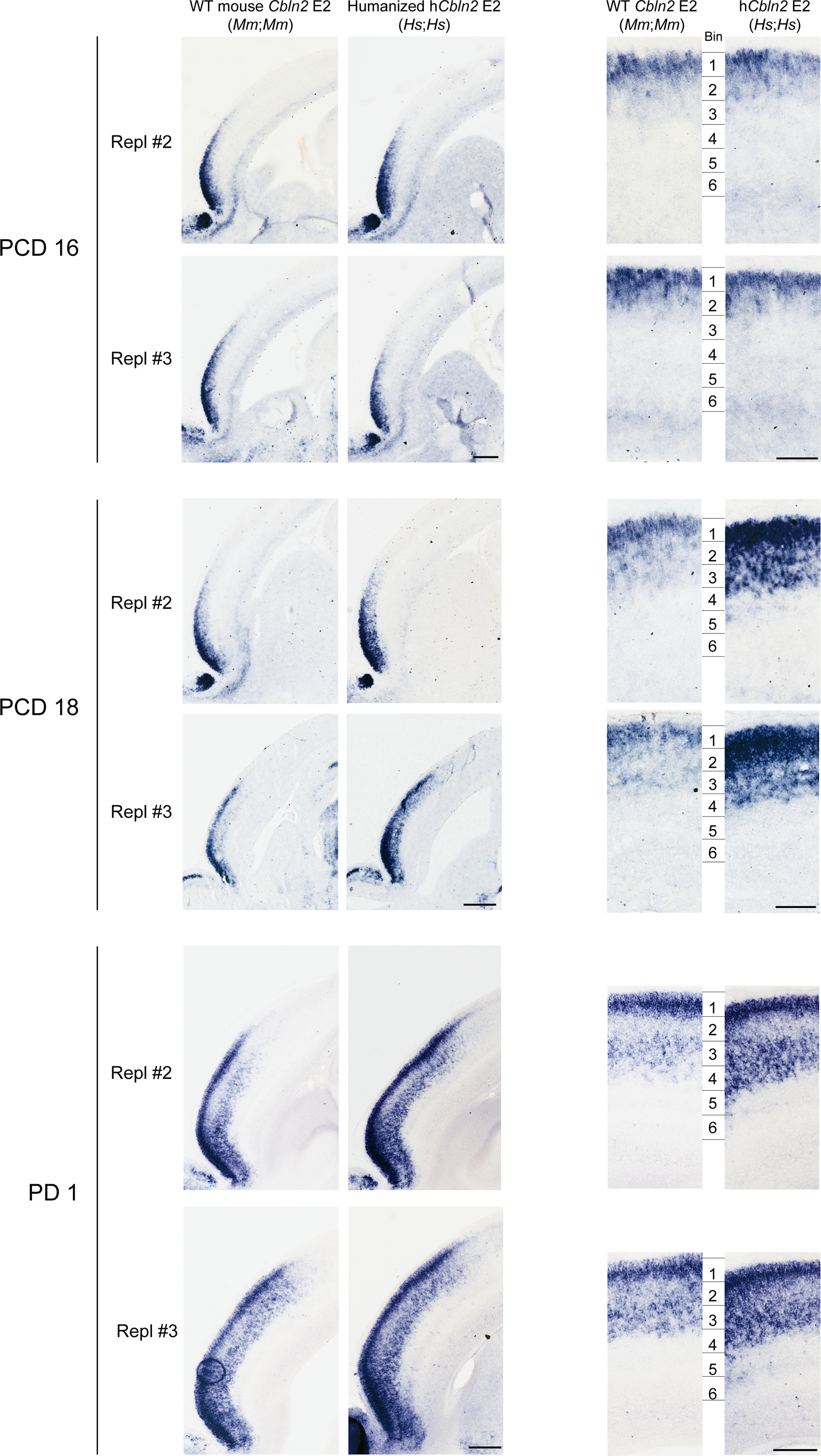
The neocortex of the humanized hCbln2 E2 knock-in prenatal and neonatal mice exhibits upregulated Cbln2 in both upper and deeper layers compared to mice carrying WT Cbln2 E2. Additional two replicates (Repl #2 and 3) not shown in Fig. 3a are shown for PND 16 and PND18. Scale bars, 200 μm (left); 100 μm (right). All analysis and three replicates for PD1. Neocortex was divided into six equal bins spanning from pia to ventricular zone, and Cbln2 signal intensity was quantified for each bin and compared between WT and hCbln2 E2 (Hs;Hs). Two-tailed Student’s t-test; *P = 0.02 (PCD18), 0.02, 0.04 (PD1), **P = 1e-3 (PCD18), 1e-3, 3e-3, 3e-3 (PD1), ***P = 1e-4, 2e-4 (PCD18); N = 3 per genotype. Scale bars, 100 μm. c, Expression of the upper layer marker, Cux2, and SOX5 in adjacent tissue sections were detected by in situ hybridization and immunostaining, respectively. Scale bars, 100 μm. N = 3.
Extended Data Fig. 9 |. Expression of Sox5 in the developing mouse and human neocortex.
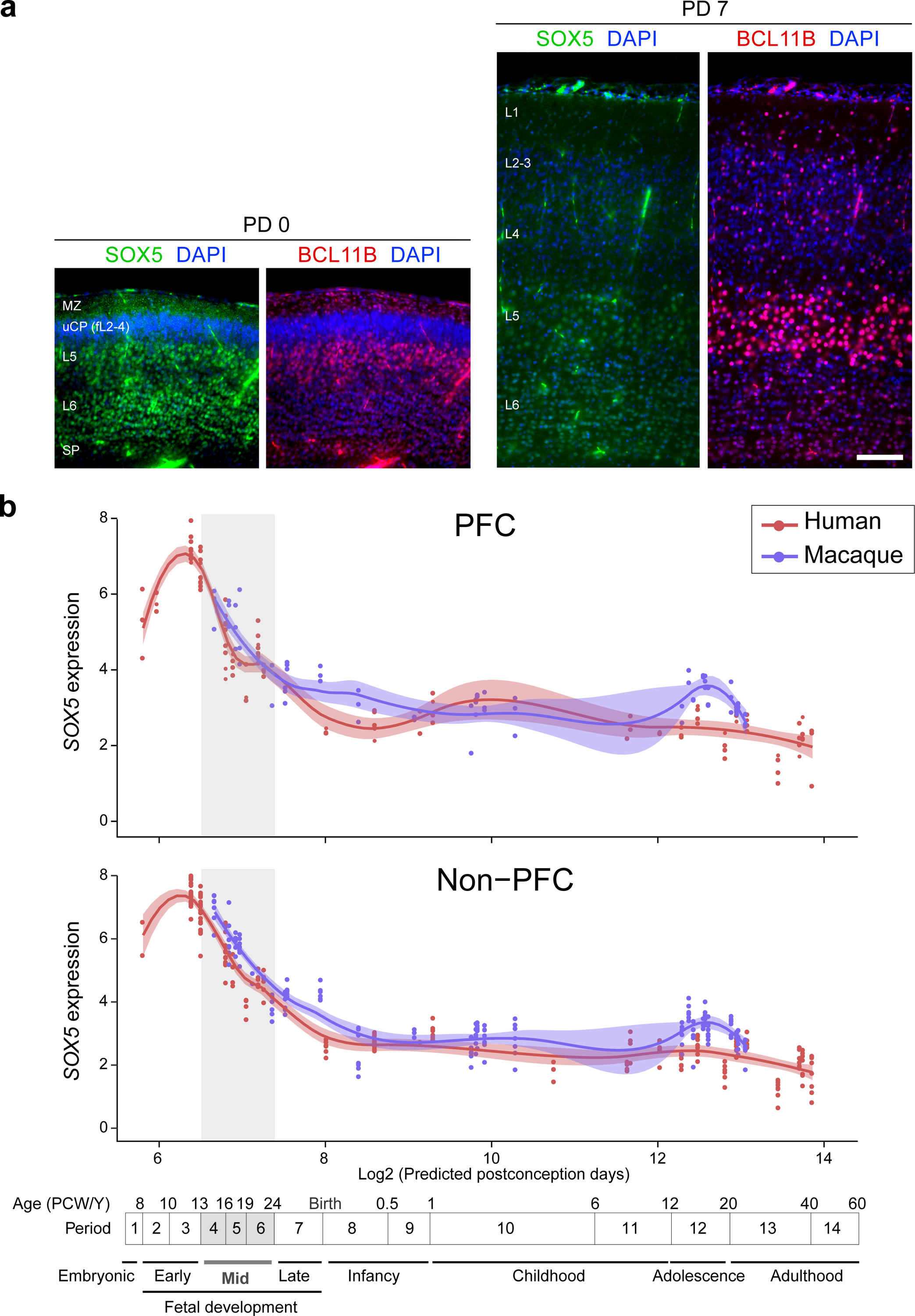
a, Double immunofluorescent staining for SOX5 and BCL11B in PD 0 and PD 7 mouse neocortex. N = 3 per condition. Scale bar, 100 μm. b, SOX5 expression in the PFC and non-PFC areas of the cerebral cortex of human and macaque across development. Red and blue lines indicate human and macaque, respectively. Vertical grey box demarcates mid-fetal developmental periods. For all of those plots, the shading around the lines represents the 95% confidence interval. Predicted ages, timeline of human and macaque development and the associated periods are shown below.
Extended Data Fig. 10 |. Quantification of excitatory and inhibitory postsynaptic puncta.
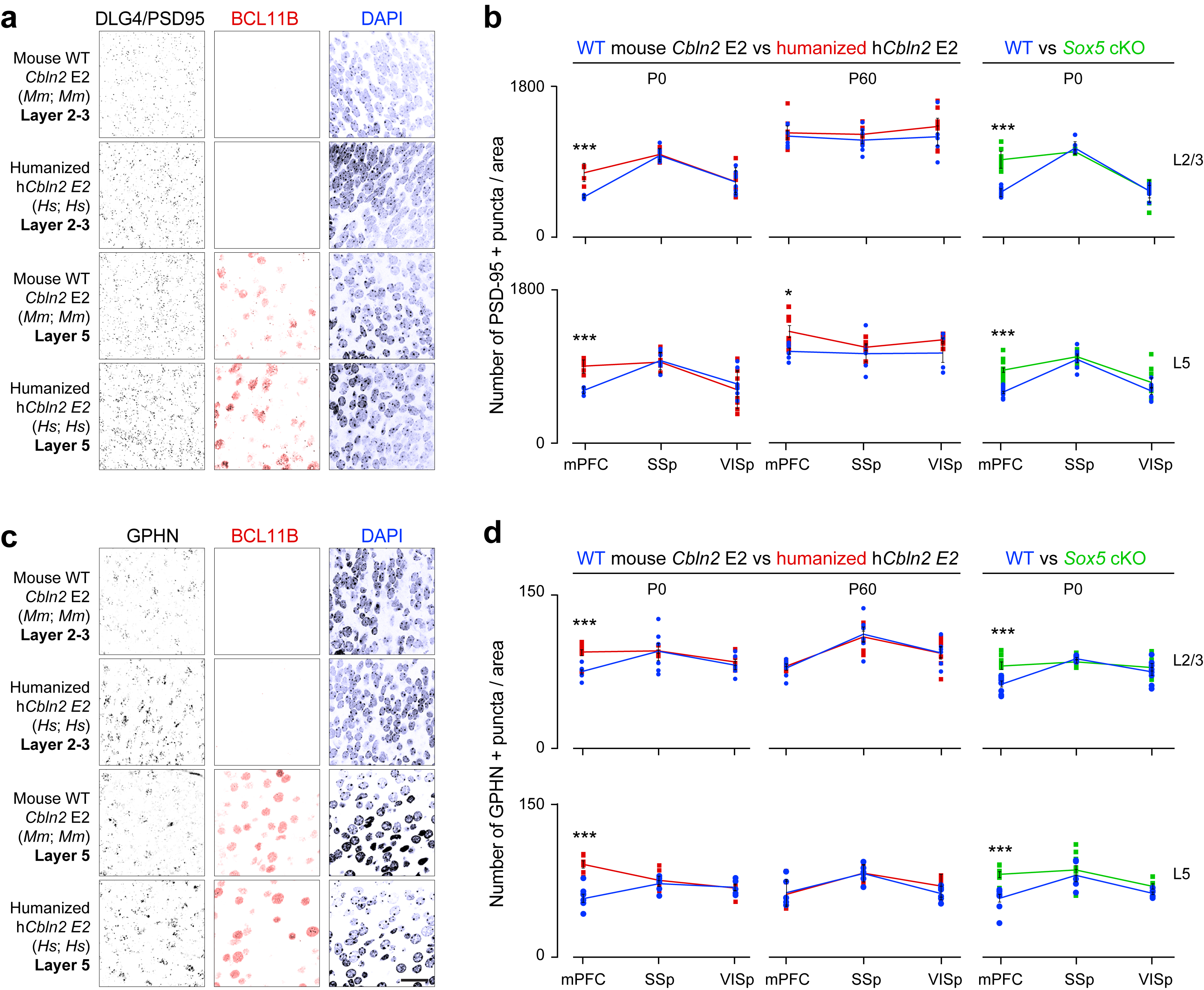
a-d, PD 0 WT and hCbln2 E2 mPFC L2-3 and L5 (BCL11B-immunopositive) immunostained for PSD-95/DLG4 or GPHN. Density of PSD-95/DLG4+ excitatory and GPHN+ inhibitory postsynaptic puncta in PD 0 and PD 60 mPFC, primary somatosensory area (SSp), and primary visual area (VISp) of WT (blue), hCbln2 E2 (orange), and Sox5 cKO mice (green). Two-tailed Student’s test; *P = 0.018; ***P = 4e-4, 1e-4 (b, WT vs. humanized hCbln2 E2 L2-3, and L5, respectively), 1e-4, 7e-5, 4e-4, 2e-4 (d, WT vs. humanized hCbln2 E2 L2-3 and L5, respectively; WT vs. Sox5 cKO L2-3 and L5, respectively); ****P < 1e-5. Error bars: S.E.M.; N = 5 per PD 0 genotype; N = 3 per PD 60 genotype. Scale bar, 25 μm.
Extended Data Fig. 11 |. Quantification of dendritic spines and dendritic complexity using Golgi stain.
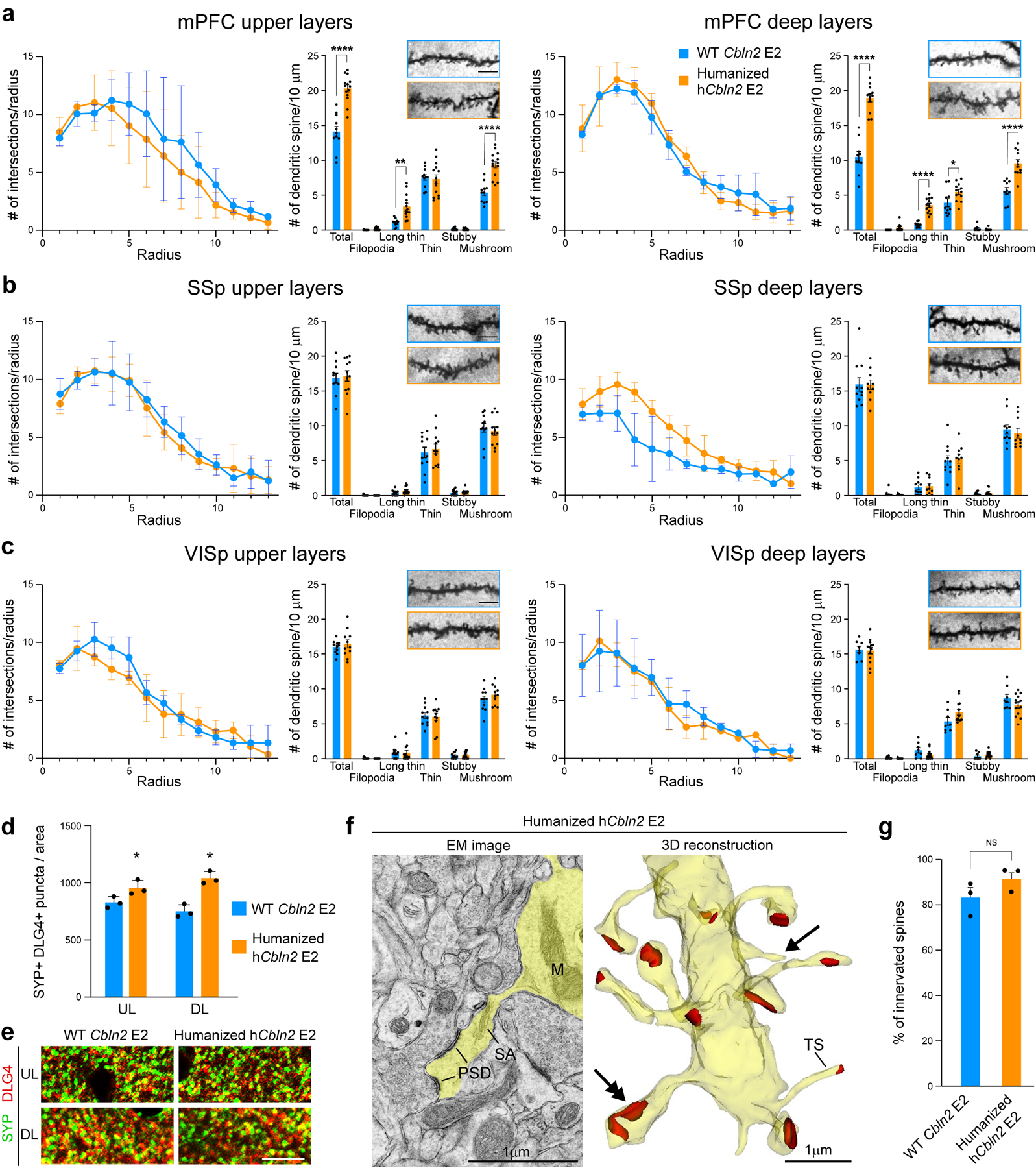
Quantification of Sholl analysis and representative images (inset) of dendritic spines per 10 μm in layer 2–3 and 5 of mPFC (a), SSp (b) and VISp (c) in WT Cbln2 E2 and humanized hCbln2 E2. Two-way ANOVA with Sidak’s multiple comparison method was applied; *P = 0.04; **P = 1e-3, ****P < 0001; Error bars: S.E.M.; N = 16 (WT Cbln2 E2), and 16 (humanized hCbln2 E2) of PD 60 brains (N = 3). Scale bars: 5 mm. d,e. Quantification and representative images of juxtaposed synaptophysin (SYP) and DLG4/PSD-95 immunolabelled puncta in upper (UL) and deep layer (DL) mPFC of WT Cbln2 E2 and humanized hCbln2 E2. Two-tailed Student’s t-test was applied * P = 0.03; Error bars: S.E.M. (N = 3); Scale bar: 10 mm. f, Electron microscopy (EM) image shows dendrite emitting spine with postsynaptic density (PSD) and spine apparatus (SA) (left) in PD 60 humanized hCbln2 E2 mouse. 3D reconstruction (right) of the dendrite with numerous spines showing spine heads and synaptic contacts (red). Double arrow points to the spine shown in EM image. Thin spines without head (TS) and not innervated thin extensions (arrow) are also detected. Abbreviation: M, mitochondria. Sixteen-representable dendrite fragments were traced in the serial images (see Methods). g, Quantification of percentage of innervated spines in mPFC of WT Cbln2 E2 and humanized hCbln2 E2. Two-tailed Student’s t-test was applied. NS, Not significant. Error bars: S.E.M. (N =3)
Extended Data Table 1:
Comparative analysis of the transcription factor binding sites in human CBLN2 E2
| TF | TF_ID | Chromosome | Start(hg38) | End (hg38) | Primate | Euarchontoglires | Laurasiatheria | Afrotheria | Marsupial | Monotreme | Aves | Sarcopterygii | Fish |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| IRF2 | MA0051.1 | chr18 | 72530577 | 72530594 | NO | NO | NO | NO | NO | NO | NO | NO | NO |
| NFATC2 | MA0152.1 | chr18 | 72530837 | 72530843 | NO | NO | NO | NO | NO | NO | NO | NO | NO |
| STAT1::STAT2 | MA0517.1 | chr18 | 72530800 | 72530814 | NO | NO | NO | NO | NO | NO | NO | NO | NO |
| IRF2 | MA0051.1 | chr18 | 72530577 | 72530594 | NO | NO | NO | NO | NO | NO | NO | NO | NO |
| Stat3 | MA0144.1 | chr18 | 72530809 | 72530818 | NO | NO | NO | NO | NO | NO | NO | NO | NO |
| NFATC2 | MA0152.1 | chr18 | 72530837 | 72530843 | NO | NO | NO | NO | NO | NO | NO | NO | NO |
| STAT1::STAT2 | MA0517.1 | chr18 | 72530800 | 72530814 | NO | NO | NO | NO | NO | NO | NO | NO | NO |
| REL | MA0101.1 | chr18 | 72530715 | 72530724 | YES | NO | NO | NO | NO | NO | NO | NO | NO |
| NFKB1 | MA0105.4 | chr18 | 72530712 | 72530724 | YES | NO | NO | NO | NO | NO | NO | NO | NO |
| NFKB1 | MA0105.4 | chr18 | 72530712 | 72530724 | YES | NO | NO | NO | NO | NO | NO | NO | NO |
| NFKB1 | MA0105.4 | chr18 | 72530713 | 72530725 | YES | NO | NO | NO | NO | NO | NO | NO | NO |
| RELA | MA0107.1 | chr18 | 72530715 | 72530724 | YES | NO | NO | NO | NO | NO | NO | NO | NO |
| FOXA1 | MA0148.3 | chr18 | 72530538 | 72530552 | YES | NO | NO | NO | NO | NO | NO | NO | NO |
| FOXP1 | MA0481.1 | chr18 | 72530539 | 72530553 | YES | NO | NO | NO | NO | NO | NO | NO | NO |
| HNF4G | MA0484.1 | chr18 | 72531272 | 72531286 | YES | NO | NO | NO | NO | NO | NO | NO | NO |
| Nkx2-5(var.2) | MA0503.1 | chr18 | 72530508 | 72530518 | YES | NO | NO | NO | NO | NO | NO | NO | NO |
| SREBF1 | MA0595.1 | chr18 | 72530513 | 72530522 | YES | NO | NO | NO | NO | NO | NO | NO | NO |
| NFAT5 | MA0606.1 | chr18 | 72531082 | 72531091 | YES | NO | NO | NO | NO | NO | NO | NO | NO |
| NFATC1 | MA0624.1 | chr18 | 72531082 | 72531091 | YES | NO | NO | NO | NO | NO | NO | NO | NO |
| NFATC3 | MA0625.1 | chr18 | 72531082 | 72531091 | YES | NO | NO | NO | NO | NO | NO | NO | NO |
| GCM1 | MA0646.1 | chr18 | 72530780 | 72530790 | YES | NO | NO | NO | NO | NO | NO | NO | NO |
| NFKB2 | MA0778.1 | chr18 | 72530712 | 72530724 | YES | NO | NO | NO | NO | NO | NO | NO | NO |
| NFKB2 | MA0778.1 | chr18 | 72530712 | 72530724 | YES | NO | NO | NO | NO | NO | NO | NO | NO |
| NFKB2 | MA0778.1 | chr18 | 72530713 | 72530725 | YES | NO | NO | NO | NO | NO | NO | NO | NO |
| FOXC2 | MA0846.1 | chr18 | 72530540 | 72530551 | YES | NO | NO | NO | NO | NO | NO | NO | NO |
| BSX | MA0876.1 | chr18 | 72531261 | 72531268 | YES | NO | NO | NO | NO | NO | NO | NO | NO |
| TFAP2A | MA0003.2 | chr18 | 72531273 | 72531287 | YES | NO | NO | NO | NO | NO | NO | NO | NO |
| NR2F1 | MA0017.1 | chr18 | 72531264 | 72531277 | YES | NO | NO | NO | NO | NO | NO | NO | NO |
| TEAD1 | MA0090.1 | chr18 | 72530683 | 72530694 | YES | NO | NO | NO | NO | NO | NO | NO | NO |
| REL | MA0101.1 | chr18 | 72530715 | 72530724 | YES | NO | NO | NO | NO | NO | NO | NO | NO |
| NFKB1 | MA0105.2 | chr18 | 72530714 | 72530724 | YES | NO | NO | NO | NO | NO | NO | NO | NO |
| NFKB1 | MA0105.3 | chr18 | 72530714 | 72530724 | YES | NO | NO | NO | NO | NO | NO | NO | NO |
| NFKB1 | MA0105.4 | chr18 | 72530712 | 72530724 | YES | NO | NO | NO | NO | NO | NO | NO | NO |
| NFKB1 | MA0105.4 | chr18 | 72530712 | 72530724 | YES | NO | NO | NO | NO | NO | NO | NO | NO |
| NFKB1 | MA0105.4 | chr18 | 72530713 | 72530725 | YES | NO | NO | NO | NO | NO | NO | NO | NO |
| RELA | MA0107.1 | chr18 | 72530715 | 72530724 | YES | NO | NO | NO | NO | NO | NO | NO | NO |
| HNF4A | MA0114.2 | chr18 | 72531271 | 72531285 | YES | NO | NO | NO | NO | NO | NO | NO | NO |
| FOXA1 | MA0148.1 | chr18 | 72530542 | 72530552 | YES | NO | NO | NO | NO | NO | NO | NO | NO |
| FOXA1 | MA0148.2 | chr18 | 72530542 | 72530552 | YES | NO | NO | NO | NO | NO | NO | NO | NO |
| FOXA1 | MA0148.3 | chr18 | 72530538 | 72530552 | YES | NO | NO | NO | NO | NO | NO | NO | NO |
| EBF1 | MA0154.2 | chr18 | 72531274 | 72531284 | YES | NO | NO | NO | NO | NO | NO | NO | NO |
| FOXP1 | MA0481.1 | chr18 | 72530539 | 72530553 | YES | NO | NO | NO | NO | NO | NO | NO | NO |
| HNF4G | MA0484.1 | chr18 | 72531272 | 72531286 | YES | NO | NO | NO | NO | NO | NO | NO | NO |
| Nkx2-5(var.2) | MA0503.1 | chr18 | 72530508 | 72530518 | YES | NO | NO | NO | NO | NO | NO | NO | NO |
| TFAP2C | MA0524.1 | chr18 | 72531273 | 72531287 | YES | NO | NO | NO | NO | NO | NO | NO | NO |
| SREBF1 | MA0595.1 | chr18 | 72530513 | 72530522 | YES | NO | NO | NO | NO | NO | NO | NO | NO |
| NFAT5 | MA0606.1 | chr18 | 72531082 | 72531091 | YES | NO | NO | NO | NO | NO | NO | NO | NO |
| NFATC1 | MA0624.1 | chr18 | 72531082 | 72531091 | YES | NO | NO | NO | NO | NO | NO | NO | NO |
| NFATC3 | MA0625.1 | chr18 | 72531082 | 72531091 | YES | NO | NO | NO | NO | NO | NO | NO | NO |
| GCM1 | MA0646.1 | chr18 | 72530780 | 72530790 | YES | NO | NO | NO | NO | NO | NO | NO | NO |
| NFKB2 | MA0778.1 | chr18 | 72530712 | 72530724 | YES | NO | NO | NO | NO | NO | NO | NO | NO |
| NFKB2 | MA0778.1 | chr18 | 72530712 | 72530724 | YES | NO | NO | NO | NO | NO | NO | NO | NO |
| NFKB2 | MA0778.1 | chr18 | 72530713 | 72530725 | YES | NO | NO | NO | NO | NO | NO | NO | NO |
| FOXC2 | MA0846.1 | chr18 | 72530540 | 72530551 | YES | NO | NO | NO | NO | NO | NO | NO | NO |
| BSX | MA0876.1 | chr18 | 72531261 | 72531268 | YES | NO | NO | NO | NO | NO | NO | NO | NO |
| IRF1 | MA0050.2 | chr18 | 72531209 | 72531229 | YES | YES | NO | NO | NO | NO | NO | NO | NO |
| IRF1 | MA0050.2 | chr18 | 72531209 | 72531229 | YES | YES | NO | NO | NO | NO | NO | NO | NO |
| GATA3 | MA0037.2 | chr18 | 72530872 | 72530879 | YES | YES | YES | NO | NO | NO | NO | NO | NO |
| Stat4 | MA0518.1 | chr18 | 72530982 | 72530995 | YES | YES | YES | NO | NO | NO | NO | NO | NO |
| Hoxa9 | MA0594.1 | chr18 | 72531092 | 72531102 | YES | YES | YES | NO | NO | NO | NO | NO | NO |
| DMRT3 | MA0610.1 | chr18 | 72530878 | 72530888 | YES | YES | YES | NO | NO | NO | NO | NO | NO |
| NFIA | MA0670.1 | chr18 | 72531019 | 72531028 | YES | YES | YES | NO | NO | NO | NO | NO | NO |
| NFIX | MA0671.1 | chr18 | 72531019 | 72531027 | YES | YES | YES | NO | NO | NO | NO | NO | NO |
| VAX2 | MA0723.1 | chr18 | 72530961 | 72530968 | YES | YES | YES | NO | NO | NO | NO | NO | NO |
| GATA3 | MA0037.2 | chr18 | 72530872 | 72530879 | YES | YES | YES | NO | NO | NO | NO | NO | NO |
| FEV | MA0156.1 | chr18 | 72530984 | 72530991 | YES | YES | YES | NO | NO | NO | NO | NO | NO |
| Atoh1 | MA0461.1 | chr18 | 72531171 | 72531178 | YES | YES | YES | NO | NO | NO | NO | NO | NO |
| Stat4 | MA0518.1 | chr18 | 72530982 | 72530995 | YES | YES | YES | NO | NO | NO | NO | NO | NO |
| Hoxa9 | MA0594.1 | chr18 | 72531092 | 72531102 | YES | YES | YES | NO | NO | NO | NO | NO | NO |
| DMRT3 | MA0610.1 | chr18 | 72530878 | 72530888 | YES | YES | YES | NO | NO | NO | NO | NO | NO |
| NFIA | MA0670.1 | chr18 | 72531019 | 72531028 | YES | YES | YES | NO | NO | NO | NO | NO | NO |
| NFIX | MA0671.1 | chr18 | 72531019 | 72531027 | YES | YES | YES | NO | NO | NO | NO | NO | NO |
| VAX2 | MA0723.1 | chr18 | 72530961 | 72530968 | YES | YES | YES | NO | NO | NO | NO | NO | NO |
| Nr2f6(var.2) | MA0728.1 | chr18 | 72530603 | 72530617 | YES | NO | NO | YES | NO | NO | NO | NO | NO |
| CREB3L1 | MA0839.1 | chr18 | 72530971 | 72530984 | YES | NO | NO | YES | NO | NO | NO | NO | NO |
| Nr2f6(var.2) | MA0728.1 | chr18 | 72530603 | 72530617 | YES | NO | NO | YES | NO | NO | NO | NO | NO |
| CREB3L1 | MA0839.1 | chr18 | 72530971 | 72530984 | YES | NO | NO | YES | NO | NO | NO | NO | NO |
| RORA(var.2) | MA0072.1 | chr18 | 72531108 | 72531121 | YES | YES | NO | YES | NO | NO | NO | NO | NO |
| RORA(var.2) | MA0072.1 | chr18 | 72531108 | 72531121 | YES | YES | NO | YES | NO | NO | NO | NO | NO |
| PAX5 | MA0014.2 | chr18 | 72530928 | 72530946 | YES | NO | YES | YES | NO | NO | NO | NO | NO |
| PLAG1 | MA0163.1 | chr18 | 72530895 | 72530908 | YES | NO | YES | YES | NO | NO | NO | NO | NO |
| Gfi1b | MA0483.1 | chr18 | 72530761 | 72530771 | YES | NO | YES | YES | NO | NO | NO | NO | NO |
| POU1F1 | MA0784.1 | chr18 | 72530656 | 72530669 | YES | NO | YES | YES | NO | NO | NO | NO | NO |
| POU2F1 | MA0785.1 | chr18 | 72530658 | 72530669 | YES | NO | YES | YES | NO | NO | NO | NO | NO |
| POU3F1 | MA0786.1 | chr18 | 72530657 | 72530668 | YES | NO | YES | YES | NO | NO | NO | NO | NO |
| POU3F2 | MA0787.1 | chr18 | 72530657 | 72530668 | YES | NO | YES | YES | NO | NO | NO | NO | NO |
| POU3F3 | MA0788.1 | chr18 | 72530657 | 72530669 | YES | NO | YES | YES | NO | NO | NO | NO | NO |
| E2F8 | MA0865.1 | chr18 | 72530901 | 72530912 | YES | NO | YES | YES | NO | NO | NO | NO | NO |
| PAX5 | MA0014.2 | chr18 | 72530928 | 72530946 | YES | NO | YES | YES | NO | NO | NO | NO | NO |
| NR2F1 | MA0017.1 | chr18 | 72530603 | 72530616 | YES | NO | YES | YES | NO | NO | NO | NO | NO |
| PLAG1 | MA0163.1 | chr18 | 72530895 | 72530908 | YES | NO | YES | YES | NO | NO | NO | NO | NO |
| Gfi1b | MA0483.1 | chr18 | 72530761 | 72530771 | YES | NO | YES | YES | NO | NO | NO | NO | NO |
| TFAP2C | MA0524.1 | chr18 | 72530896 | 72530910 | YES | NO | YES | YES | NO | NO | NO | NO | NO |
| POU1F1 | MA0784.1 | chr18 | 72530656 | 72530669 | YES | NO | YES | YES | NO | NO | NO | NO | NO |
| POU2F1 | MA0785.1 | chr18 | 72530658 | 72530669 | YES | NO | YES | YES | NO | NO | NO | NO | NO |
| POU3F1 | MA0786.1 | chr18 | 72530657 | 72530668 | YES | NO | YES | YES | NO | NO | NO | NO | NO |
| POU3F2 | MA0787.1 | chr18 | 72530657 | 72530668 | YES | NO | YES | YES | NO | NO | NO | NO | NO |
| POU3F3 | MA0788.1 | chr18 | 72530657 | 72530669 | YES | NO | YES | YES | NO | NO | NO | NO | NO |
| E2F8 | MA0865.1 | chr18 | 72530901 | 72530912 | YES | NO | YES | YES | NO | NO | NO | NO | NO |
| NR2F1 | MA0017.2 | chr18 | 72530599 | 72530611 | YES | YES | YES | YES | NO | NO | NO | NO | NO |
| CREB1 | MA0018.2 | chr18 | 72530603 | 72530610 | YES | YES | YES | YES | NO | NO | NO | NO | NO |
| Gfi1 | MA0038.1 | chr18 | 72530771 | 72530780 | YES | YES | YES | YES | NO | NO | NO | NO | NO |
| RORA | MA0071.1 | chr18 | 72530603 | 72530612 | YES | YES | YES | YES | NO | NO | NO | NO | NO |
| SOX9 | MA0077.1 | chr18 | 72530577 | 72530585 | YES | YES | YES | YES | NO | NO | NO | NO | NO |
| RELA | MA0107.1 | chr18 | 72530714 | 72530723 | YES | YES | YES | YES | NO | NO | NO | NO | NO |
| RELA | MA0107.1 | chr18 | 72530714 | 72530723 | YES | YES | YES | YES | NO | NO | NO | NO | NO |
| CTCF | MA0139.1 | chr18 | 72530784 | 72530802 | YES | YES | YES | YES | NO | NO | NO | NO | NO |
| Pou5f1::Sox2 | MA0142.1 | chr18 | 72530745 | 72530759 | YES | YES | YES | YES | NO | NO | NO | NO | NO |
| Gfi1b | MA0483.1 | chr18 | 72530772 | 72530782 | YES | YES | YES | YES | NO | NO | NO | NO | NO |
| Gfi1b | MA0483.1 | chr18 | 72531134 | 72531144 | YES | YES | YES | YES | NO | NO | NO | NO | NO |
| Hoxc9 | MA0485.1 | chr18 | 72531091 | 72531103 | YES | YES | YES | YES | NO | NO | NO | NO | NO |
| POU2F2 | MA0507.1 | chr18 | 72530656 | 72530668 | YES | YES | YES | YES | NO | NO | NO | NO | NO |
| POU2F2 | MA0507.1 | chr18 | 72530742 | 72530754 | YES | YES | YES | YES | NO | NO | NO | NO | NO |
| SMAD2::SMAD3::SMAD4 | MA0513.1 | chr18 | 72531140 | 72531152 | YES | YES | YES | YES | NO | NO | NO | NO | NO |
| EMX1 | MA0612.1 | chr18 | 72530960 | 72530969 | YES | YES | YES | YES | NO | NO | NO | NO | NO |
| Pou2f3 | MA0627.1 | chr18 | 72530655 | 72530670 | YES | YES | YES | YES | NO | NO | NO | NO | NO |
| MYF6 | MA0667.1 | chr18 | 72530817 | 72530826 | YES | YES | YES | YES | NO | NO | NO | NO | NO |
| MYF6 | MA0667.1 | chr18 | 72530817 | 72530826 | YES | YES | YES | YES | NO | NO | NO | NO | NO |
| POU1F1 | MA0784.1 | chr18 | 72530742 | 72530755 | YES | YES | YES | YES | NO | NO | NO | NO | NO |
| POU3F4 | MA0789.1 | chr18 | 72530659 | 72530667 | YES | YES | YES | YES | NO | NO | NO | NO | NO |
| POU5F1B | MA0792.1 | chr18 | 72530659 | 72530667 | YES | YES | YES | YES | NO | NO | NO | NO | NO |
| TFAP2B(var.2) | MA0812.1 | chr18 | 72530897 | 72530907 | YES | YES | YES | YES | NO | NO | NO | NO | NO |
| BARX1 | MA0875.1 | chr18 | 72530922 | 72530929 | YES | YES | YES | YES | NO | NO | NO | NO | NO |
| EVX1 | MA0887.1 | chr18 | 72530960 | 72530969 | YES | YES | YES | YES | NO | NO | NO | NO | NO |
| EVX2 | MA0888.1 | chr18 | 72530960 | 72530969 | YES | YES | YES | YES | NO | NO | NO | NO | NO |
| HOXA2 | MA0900.1 | chr18 | 72530960 | 72530969 | YES | YES | YES | YES | NO | NO | NO | NO | NO |
| HOXB2 | MA0902.1 | chr18 | 72530960 | 72530969 | YES | YES | YES | YES | NO | NO | NO | NO | NO |
| HOXB3 | MA0903.1 | chr18 | 72530960 | 72530969 | YES | YES | YES | YES | NO | NO | NO | NO | NO |
| NR2F1 | MA0017.2 | chr18 | 72530599 | 72530611 | YES | YES | YES | YES | NO | NO | NO | NO | NO |
| CREB1 | MA0018.2 | chr18 | 72530603 | 72530610 | YES | YES | YES | YES | NO | NO | NO | NO | NO |
| Gfi1 | MA0038.1 | chr18 | 72530771 | 72530780 | YES | YES | YES | YES | NO | NO | NO | NO | NO |
| RORA | MA0071.1 | chr18 | 72530603 | 72530612 | YES | YES | YES | YES | NO | NO | NO | NO | NO |
| SOX9 | MA0077.1 | chr18 | 72530577 | 72530585 | YES | YES | YES | YES | NO | NO | NO | NO | NO |
| NFKB1 | MA0105.1 | chr18 | 72530714 | 72530723 | YES | YES | YES | YES | NO | NO | NO | NO | NO |
| NFKB1 | MA0105.1 | chr18 | 72530714 | 72530723 | YES | YES | YES | YES | NO | NO | NO | NO | NO |
| NFKB1 | MA0105.2 | chr18 | 72530713 | 72530723 | YES | YES | YES | YES | NO | NO | NO | NO | NO |
| NFKB1 | MA0105.3 | chr18 | 72530713 | 72530723 | YES | YES | YES | YES | NO | NO | NO | NO | NO |
| RELA | MA0107.1 | chr18 | 72530714 | 72530723 | YES | YES | YES | YES | NO | NO | NO | NO | NO |
| RELA | MA0107.1 | chr18 | 72530714 | 72530723 | YES | YES | YES | YES | NO | NO | NO | NO | NO |
| ESR1 | MA0112.2 | chr18 | 72531144 | 72531163 | YES | YES | YES | YES | NO | NO | NO | NO | NO |
| ELF5 | MA0136.1 | chr18 | 72530983 | 72530991 | YES | YES | YES | YES | NO | NO | NO | NO | NO |
| CTCF | MA0139.1 | chr18 | 72530784 | 72530802 | YES | YES | YES | YES | NO | NO | NO | NO | NO |
| Pou5f1::Sox2 | MA0142.1 | chr18 | 72530745 | 72530759 | YES | YES | YES | YES | NO | NO | NO | NO | NO |
| Atoh1 | MA0461.1 | chr18 | 72531105 | 72531112 | YES | YES | YES | YES | NO | NO | NO | NO | NO |
| Gfi1b | MA0483.1 | chr18 | 72530772 | 72530782 | YES | YES | YES | YES | NO | NO | NO | NO | NO |
| Gfi1b | MA0483.1 | chr18 | 72531134 | 72531144 | YES | YES | YES | YES | NO | NO | NO | NO | NO |
| Hoxc9 | MA0485.1 | chr18 | 72531091 | 72531103 | YES | YES | YES | YES | NO | NO | NO | NO | NO |
| POU2F2 | MA0507.1 | chr18 | 72530656 | 72530668 | YES | YES | YES | YES | NO | NO | NO | NO | NO |
| POU2F2 | MA0507.1 | chr18 | 72530742 | 72530754 | YES | YES | YES | YES | NO | NO | NO | NO | NO |
| SMAD2::SMAD3::SMAD4 | MA0513.1 | chr18 | 72531140 | 72531152 | YES | YES | YES | YES | NO | NO | NO | NO | NO |
| EMX1 | MA0612.1 | chr18 | 72530960 | 72530969 | YES | YES | YES | YES | NO | NO | NO | NO | NO |
| Pou2f3 | MA0627.1 | chr18 | 72530655 | 72530670 | YES | YES | YES | YES | NO | NO | NO | NO | NO |
| MYF6 | MA0667.1 | chr18 | 72530817 | 72530826 | YES | YES | YES | YES | NO | NO | NO | NO | NO |
| MYF6 | MA0667.1 | chr18 | 72530817 | 72530826 | YES | YES | YES | YES | NO | NO | NO | NO | NO |
| POU1F1 | MA0784.1 | chr18 | 72530742 | 72530755 | YES | YES | YES | YES | NO | NO | NO | NO | NO |
| POU3F4 | MA0789.1 | chr18 | 72530659 | 72530667 | YES | YES | YES | YES | NO | NO | NO | NO | NO |
| POU5F1B | MA0792.1 | chr18 | 72530659 | 72530667 | YES | YES | YES | YES | NO | NO | NO | NO | NO |
| TFAP2B(var.2) | MA0812.1 | chr18 | 72530897 | 72530907 | YES | YES | YES | YES | NO | NO | NO | NO | NO |
| BARX1 | MA0875.1 | chr18 | 72530922 | 72530929 | YES | YES | YES | YES | NO | NO | NO | NO | NO |
| EVX1 | MA0887.1 | chr18 | 72530960 | 72530969 | YES | YES | YES | YES | NO | NO | NO | NO | NO |
| EVX2 | MA0888.1 | chr18 | 72530960 | 72530969 | YES | YES | YES | YES | NO | NO | NO | NO | NO |
| HOXA2 | MA0900.1 | chr18 | 72530960 | 72530969 | YES | YES | YES | YES | NO | NO | NO | NO | NO |
| HOXB2 | MA0902.1 | chr18 | 72530960 | 72530969 | YES | YES | YES | YES | NO | NO | NO | NO | NO |
| HOXB3 | MA0903.1 | chr18 | 72530960 | 72530969 | YES | YES | YES | YES | NO | NO | NO | NO | NO |
| TFAP2A | MA0003.1 | chr18 | 72530721 | 72530729 | YES | YES | YES | NO | YES | NO | NO | NO | NO |
| Pax4 | MA0068.1 | chr18 | 72530970 | 72530999 | YES | YES | YES | NO | YES | NO | NO | NO | NO |
| TFAP2A | MA0003.3 | chr18 | 72530897 | 72530907 | YES | YES | YES | YES | YES | NO | NO | NO | NO |
| Gfi1 | MA0038.1 | chr18 | 72531133 | 72531142 | YES | YES | YES | YES | YES | NO | NO | NO | NO |
| Foxd3 | MA0041.1 | chr18 | 72530816 | 72530827 | YES | YES | YES | YES | YES | NO | NO | NO | NO |
| TFAP2C | MA0524.2 | chr18 | 72530897 | 72530908 | YES | YES | YES | YES | YES | NO | NO | NO | NO |
| Pou2f3 | MA0627.1 | chr18 | 72530741 | 72530756 | YES | YES | YES | YES | YES | NO | NO | NO | NO |
| POU2F1 | MA0785.1 | chr18 | 72530744 | 72530755 | YES | YES | YES | YES | YES | NO | NO | NO | NO |
| POU3F4 | MA0789.1 | chr18 | 72530745 | 72530753 | YES | YES | YES | YES | YES | NO | NO | NO | NO |
| POU5F1B | MA0792.1 | chr18 | 72530745 | 72530753 | YES | YES | YES | YES | YES | NO | NO | NO | NO |
| TFAP2A(var.2) | MA0810.1 | chr18 | 72530897 | 72530908 | YES | YES | YES | YES | YES | NO | NO | NO | NO |
| TFAP2B | MA0811.1 | chr18 | 72530897 | 72530908 | YES | YES | YES | YES | YES | NO | NO | NO | NO |
| Barhl1 | MA0877.1 | chr18 | 72530921 | 72530930 | YES | YES | YES | YES | YES | NO | NO | NO | NO |
| TFAP2A | MA0003.2 | chr18 | 72530894 | 72530908 | YES | YES | YES | YES | YES | NO | NO | NO | NO |
| TFAP2A | MA0003.3 | chr18 | 72530897 | 72530907 | YES | YES | YES | YES | YES | NO | NO | NO | NO |
| Gfi1 | MA0038.1 | chr18 | 72531133 | 72531142 | YES | YES | YES | YES | YES | NO | NO | NO | NO |
| Foxd3 | MA0041.1 | chr18 | 72530816 | 72530827 | YES | YES | YES | YES | YES | NO | NO | NO | NO |
| Foxa2 | MA0047.1 | chr18 | 72530816 | 72530827 | YES | YES | YES | YES | YES | NO | NO | NO | NO |
| TFAP2C | MA0524.2 | chr18 | 72530897 | 72530908 | YES | YES | YES | YES | YES | NO | NO | NO | NO |
| Pou2f3 | MA0627.1 | chr18 | 72530741 | 72530756 | YES | YES | YES | YES | YES | NO | NO | NO | NO |
| POU2F1 | MA0785.1 | chr18 | 72530744 | 72530755 | YES | YES | YES | YES | YES | NO | NO | NO | NO |
| POU3F4 | MA0789.1 | chr18 | 72530745 | 72530753 | YES | YES | YES | YES | YES | NO | NO | NO | NO |
| POU5F1B | MA0792.1 | chr18 | 72530745 | 72530753 | YES | YES | YES | YES | YES | NO | NO | NO | NO |
| TFAP2A(var.2) | MA0810.1 | chr18 | 72530897 | 72530908 | YES | YES | YES | YES | YES | NO | NO | NO | NO |
| TFAP2B | MA0811.1 | chr18 | 72530897 | 72530908 | YES | YES | YES | YES | YES | NO | NO | NO | NO |
| Barhl1 | MA0877.1 | chr18 | 72530921 | 72530930 | YES | YES | YES | YES | YES | NO | NO | NO | NO |
| TAL1::TCF3 | MA0091.1 | chr18 | 72531153 | 72531164 | YES | YES | YES | NO | NO | NO | NO | YES | NO |
| TAL1::TCF3 | MA0091.1 | chr18 | 72531155 | 72531166 | YES | YES | YES | NO | NO | NO | NO | YES | NO |
| Stat6 | MA0520.1 | chr18 | 72531171 | 72531185 | YES | YES | YES | NO | NO | NO | NO | YES | NO |
| Bhlha15 | MA0607.1 | chr18 | 72531156 | 72531163 | YES | YES | YES | NO | NO | NO | NO | YES | NO |
| TAL1::TCF3 | MA0091.1 | chr18 | 72531153 | 72531164 | YES | YES | YES | NO | NO | NO | NO | YES | NO |
| TAL1::TCF3 | MA0091.1 | chr18 | 72531155 | 72531166 | YES | YES | YES | NO | NO | NO | NO | YES | NO |
| Stat6 | MA0520.1 | chr18 | 72531171 | 72531185 | YES | YES | YES | NO | NO | NO | NO | YES | NO |
| Bhlha15 | MA0607.1 | chr18 | 72531156 | 72531163 | YES | YES | YES | NO | NO | NO | NO | YES | NO |
| Crx | MA0467.1 | chr18 | 72531051 | 72531061 | YES | YES | YES | YES | NO | NO | NO | YES | NO |
| Sox3 | MA0514.1 | chr18 | 72531015 | 72531024 | YES | YES | YES | YES | NO | NO | NO | YES | NO |
| LBX1 | MA0618.1 | chr18 | 72531113 | 72531120 | YES | YES | YES | YES | NO | NO | NO | YES | NO |
| POU6F2 | MA0793.1 | chr18 | 72531111 | 72531120 | YES | YES | YES | YES | NO | NO | NO | YES | NO |
| Dmbx1 | MA0883.1 | chr18 | 72531049 | 72531065 | YES | YES | YES | YES | NO | NO | NO | YES | NO |
| GSC2 | MA0891.1 | chr18 | 72531053 | 72531062 | YES | YES | YES | YES | NO | NO | NO | YES | NO |
| Crx | MA0467.1 | chr18 | 72531051 | 72531061 | YES | YES | YES | YES | NO | NO | NO | YES | NO |
| Sox3 | MA0514.1 | chr18 | 72531015 | 72531024 | YES | YES | YES | YES | NO | NO | NO | YES | NO |
| LBX1 | MA0618.1 | chr18 | 72531113 | 72531120 | YES | YES | YES | YES | NO | NO | NO | YES | NO |
| POU6F2 | MA0793.1 | chr18 | 72531111 | 72531120 | YES | YES | YES | YES | NO | NO | NO | YES | NO |
| Dmbx1 | MA0883.1 | chr18 | 72531049 | 72531065 | YES | YES | YES | YES | NO | NO | NO | YES | NO |
| GSC2 | MA0891.1 | chr18 | 72531053 | 72531062 | YES | YES | YES | YES | NO | NO | NO | YES | NO |
| TAL1::TCF3 | MA0091.1 | chr18 | 72531103 | 72531114 | YES | YES | NO | YES | YES | NO | NO | YES | NO |
| TAL1::TCF3 | MA0091.1 | chr18 | 72531103 | 72531114 | YES | YES | NO | YES | YES | NO | NO | YES | NO |
| TAL1::TCF3 | MA0091.1 | chr18 | 72531101 | 72531112 | YES | YES | YES | YES | YES | NO | NO | YES | NO |
| Atoh1 | MA0461.2 | chr18 | 72531103 | 72531112 | YES | YES | YES | YES | YES | NO | NO | YES | NO |
| mix-a | MA0621.1 | chr18 | 72531112 | 72531122 | YES | YES | YES | YES | YES | NO | NO | YES | NO |
| Neurog1 | MA0623.1 | chr18 | 72531103 | 72531112 | YES | YES | YES | YES | YES | NO | NO | YES | NO |
| Neurog1 | MA0623.1 | chr18 | 72531103 | 72531112 | YES | YES | YES | YES | YES | NO | NO | YES | NO |
| NEUROD2 | MA0668.1 | chr18 | 72531103 | 72531112 | YES | YES | YES | YES | YES | NO | NO | YES | NO |
| NEUROG2 | MA0669.1 | chr18 | 72531103 | 72531112 | YES | YES | YES | YES | YES | NO | NO | YES | NO |
| VAX1 | MA0722.1 | chr18 | 72531113 | 72531120 | YES | YES | YES | YES | YES | NO | NO | YES | NO |
| VSX1 | MA0725.1 | chr18 | 72531113 | 72531120 | YES | YES | YES | YES | YES | NO | NO | YES | NO |
| VSX2 | MA0726.1 | chr18 | 72531113 | 72531120 | YES | YES | YES | YES | YES | NO | NO | YES | NO |
| Dlx1 | MA0879.1 | chr18 | 72531112 | 72531121 | YES | YES | YES | YES | YES | NO | NO | YES | NO |
| EMX2 | MA0886.1 | chr18 | 72531112 | 72531121 | YES | YES | YES | YES | YES | NO | NO | YES | NO |
| Hoxb5 | MA0904.1 | chr18 | 72531108 | 72531123 | YES | YES | YES | YES | YES | NO | NO | YES | NO |
| Hoxd3 | MA0912.1 | chr18 | 72531108 | 72531123 | YES | YES | YES | YES | YES | NO | NO | YES | NO |
| SP1 | MA0079.1 | chr18 | 72530997 | 72531006 | YES | YES | YES | YES | YES | NO | NO | YES | NO |
| TAL1::TCF3 | MA0091.1 | chr18 | 72531101 | 72531112 | YES | YES | YES | YES | YES | NO | NO | YES | NO |
| Atoh1 | MA0461.2 | chr18 | 72531103 | 72531112 | YES | YES | YES | YES | YES | NO | NO | YES | NO |
| mix-a | MA0621.1 | chr18 | 72531112 | 72531122 | YES | YES | YES | YES | YES | NO | NO | YES | NO |
| Neurog1 | MA0623.1 | chr18 | 72531103 | 72531112 | YES | YES | YES | YES | YES | NO | NO | YES | NO |
| Neurog1 | MA0623.1 | chr18 | 72531103 | 72531112 | YES | YES | YES | YES | YES | NO | NO | YES | NO |
| NEUROD2 | MA0668.1 | chr18 | 72531103 | 72531112 | YES | YES | YES | YES | YES | NO | NO | YES | NO |
| NEUROG2 | MA0669.1 | chr18 | 72531103 | 72531112 | YES | YES | YES | YES | YES | NO | NO | YES | NO |
| VAX1 | MA0722.1 | chr18 | 72531113 | 72531120 | YES | YES | YES | YES | YES | NO | NO | YES | NO |
| VSX1 | MA0725.1 | chr18 | 72531113 | 72531120 | YES | YES | YES | YES | YES | NO | NO | YES | NO |
| VSX2 | MA0726.1 | chr18 | 72531113 | 72531120 | YES | YES | YES | YES | YES | NO | NO | YES | NO |
| Dlx1 | MA0879.1 | chr18 | 72531112 | 72531121 | YES | YES | YES | YES | YES | NO | NO | YES | NO |
| EMX2 | MA0886.1 | chr18 | 72531112 | 72531121 | YES | YES | YES | YES | YES | NO | NO | YES | NO |
| Hoxb5 | MA0904.1 | chr18 | 72531108 | 72531123 | YES | YES | YES | YES | YES | NO | NO | YES | NO |
| Hoxd3 | MA0912.1 | chr18 | 72531108 | 72531123 | YES | YES | YES | YES | YES | NO | NO | YES | NO |
NOTE: Yes means BS is conserved (no mutations in core of motif) in at least 25% of the species in that group/clade.
Extended Data Table 2:
List and sequence of relevant sgRNAs and primers
| Oligo name | Use | Sequence |
| mCbln2 sgRNA-L1-top | sgRNA synthesis | caccgTAAGGCGTGTTGCACAAGAT |
| mCbln2 sgRNA-L1-bottom | sgRNA synthesis | aaacATCTTGTGCAACACGCCTTAc |
| mCbln2 sgRNA-R1-top | sgRNA synthesis | caccgATCACAATTTATCTACCGTG |
| mCbln2 sgRNA-R1-bottom | sgRNA synthesis | aaacCACGGTAGATAAATTGTGATc |
| T7- mCbln2-sgRNA-L1 | sgRNA synthesis | gaaattaatacgactcactatagggagaTAAGGCGTGTTGCACAAGAT |
| Primer name | Use | Sequence |
| T7-mCbln2 sgRNA-R1 | sgRNA synthesis | gaaattaatacgactcactatagggagaATCACAATTTATCTACCGTG |
| mP1 | genotyping | GTTGAAATTGATGGTCCAGGTG |
| mP2 | genotyping | AAAATTGCGTGACACCCACT |
| mP3 | genotyping | CAAGCCTTTGGAATGTTGAGCTAC |
| mP4 | genotyping | CCAGAAGTGAAGCCAATGAGAGAA |
| hP1 | genotyping | CCCTAAGGCCTATTACTTT |
| hP2 | genotyping | GAAGGCCTCTCAGCTCATCTTTT |
| LacZ-F | genotyping | ATCCTCTGCATGGTCAGGTC |
| LacZ-R | genotyping | CGTGGCCTGATTCATTCC |
| E2-ChIP-F | ChIP | GTCCCGATACACAGTGGGTG |
| E2-ChIP-R | ChIP | AGCACCATCTATTGGGGAGG |
| E1-ChIP-F | ChIP | TGGGACAAGAGAATGTCAGC |
| E1-ChIP-R | ChIP | GTTACTCAGGACTGAAGCTGAA |
| qPCR-Cbln2-F | quantitative PCR | TGACCCTCAGATGGATTGCAC |
| qPCR-Cbln2-R | quantitative PCR | CTGCTGGGCTCTTGCTTTAAGC |
| qPCR-Tbp-F | quantitative PCR | GTGATGTGAAGTTCCCCATAAGG |
| qPCR-Tbp-R | quantitative PCR | CTACTGAACTGCTGGTGGGGTCA |
Supplementary Material
Acknowledgements
We thank Suxia Bai, Timothy Nottoli and Xiaojun Xing for technical help with generating transgenic mice and genetically humanized mice; Andre Sousa for providing tissue; Belen Lorente-Galdos and Gabriel Santpere for helping with data analysis; Alvaro Duque for using equipment from MacBrainResource (MH113257); Véronique Lefebvre for providing floxed Sox5 mice; and the members of the Sestan laboratory for comments. This work was supported by the National Institutes of Health (HG010898, MH106874, MH106934, MH110926, MH116488), and the Simons Foundation (N.S.). Additional support was provided by the National Institutes of Health T32 fellowship (MH18268) (K.P.), the National Science Foundation Graduate Research Fellowship Program (S.K.M.), the Kavli Foundation and the James S. McDonnell Foundation (N.S.).
Footnotes
Competing Financial Interests
The authors declare no competing financial interests.
References
- 1.Finlay BL & Darlington RB Linked regularities in the development and evolution of mammalian brains. Science 268, 1578–1584 (1995). [DOI] [PubMed] [Google Scholar]
- 2.Barton RA & Harvey PH Mosaic evolution of brain structure in mammals. Nature 405, 1055–1058 (2000). [DOI] [PubMed] [Google Scholar]
- 3.Krubitzer L & Kaas J The evolution of the neocortex in mammals: how is phenotypic diversity generated? Curr. Opin. Neurobiol 15, 444–453 (2005). [DOI] [PubMed] [Google Scholar]
- 4.Passingham RE & Wise SP The Neurobiology of the Prefrontal Cortex: Anatomy, Evolution, and The Origin of Insight. (Oxford University Press, 2015). [Google Scholar]
- 5.Elston GN et al. Specializations of the granular prefrontal cortex of primates: implications for cognitive processing. Anat. Rec. A Discov. Mol. Cell. Evol. Biol 288, 26–35 (2006). [DOI] [PubMed] [Google Scholar]
- 6.Semendeferi K et al. Spatial organization of neurons in the frontal pole sets humans apart from great apes. Cereb. Cortex 21, 1485–1497 (2011). [DOI] [PubMed] [Google Scholar]
- 7.Kwan KY et al. Species-dependent posttranscriptional regulation of NOS1 by FMRP in the developing cerebral cortex. Cell 149, 899–911 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gabi M et al. No relative expansion of the number of prefrontal neurons in primate and human evolution. Proc. Natl. Acad. Sci. U. S. A 113, 9617–9622 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Caceres M et al. Elevated gene expression levels distinguish human from non-human primate brains. Proc. Natl. Acad. Sci. U. S. A 100, 13030–13035 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Khaitovich P et al. Regional patterns of gene expression in human and chimpanzee brains. Genome Res. 14, 1462–1473 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Uddin M et al. Sister grouping of chimpanzees and humans as revealed by genome-wide phylogenetic analysis of brain gene expression profiles. Proc. Natl. Acad. Sci. U. S. A 101, 2957–2962 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Konopka G et al. Human-specific transcriptional networks in the brain. Neuron 75, 601–617 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bauernfeind AL et al. Evolutionary divergence of gene and protein expression in the brains of humans and chimpanzees. Genome Biol Evol 7, 2276–2288 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sousa AMM et al. Molecular and cellular reorganization of neural circuits in the human lineage. Science 358, 1027–1032 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhu Y et al. Spatiotemporal transcriptomic divergence across human and macaque brain development. Science 362, eaat8077 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pollen AA et al. Establishing cerebral organoids as models of human-specific brain evolution. Cell 176, 743–756 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kanton S et al. Organoid single-cell genomic atlas uncovers human-specific features of brain development. Nature 574, 418–422 (2019). [DOI] [PubMed] [Google Scholar]
- 18.Johnson MB et al. Functional and evolutionary insights into human brain development through global transcriptome analysis. Neuron 62, 494–509 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pletikos M et al. Temporal specification and bilaterality of human neocortical topographic gene expression. Neuron 81, 321–332 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li M et al. Integrative functional genomic analysis of human brain development and neuropsychiatric risks. Science 362, eaat7615 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Urade Y et al. Precerebellin is a cerebellum-specific protein with similarity to the globular domain of complement C1q B chain. Proc. Natl. Acad. Sci. U. S. A 88, 1069–1073 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hirai H et al. Cbln1 is essential for synaptic integrity and plasticity in the cerebellum. Nat. Neurosci 8, 1534–1541 (2005). [DOI] [PubMed] [Google Scholar]
- 23.Uemura T et al. Trans-synaptic interaction of GluRdelta2 and Neurexin through Cbln1 mediates synapse formation in the cerebellum. Cell 141, 1068–1079 (2010). [DOI] [PubMed] [Google Scholar]
- 24.Matsuda K et al. Cbln1 is a ligand for an orphan glutamate receptor delta2, a bidirectional synapse organizer. Science 328, 363–368 (2010). [DOI] [PubMed] [Google Scholar]
- 25.Yasumura M et al. Glutamate receptor delta1 induces preferentially inhibitory presynaptic differentiation of cortical neurons by interacting with neurexins through cerebellin precursor protein subtypes. J. Neurochem 121, 705–716 (2012). [DOI] [PubMed] [Google Scholar]
- 26.Wei P et al. The Cbln family of proteins interact with multiple signaling pathways. J. Neurochem 121, 717–729 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Seigneur E & Sudhof TC Genetic ablation of all cerebellins reveals synapse organizer functions in multiple regions throughout the brain. J. Neurosci 38, 4774–4790 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Elston GN Pyramidal cells of the frontal lobe: all the more spinous to think with. J. Neurosci 20, RC95 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jacobs B et al. Regional dendritic and spine variation in human cerebral cortex: a quantitative golgi study. Cereb. Cortex 11, 558–571 (2001). [DOI] [PubMed] [Google Scholar]
- 30.Bianchi S et al. Synaptogenesis and development of pyramidal neuron dendritic morphology in the chimpanzee neocortex resembles humans. Proc. Natl. Acad. Sci. U. S. A 110, 10395–10401 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Molliver ME et al. The development of synapses in cerebral cortex of the human fetus. Brain Res. 50, 403–407 (1973). [DOI] [PubMed] [Google Scholar]
- 32.Voigt T et al. Synaptophysin immunohistochemistry reveals inside-out pattern of early synaptogenesis in ferret cerebral cortex. J. Comp. Neurol 330, 48–64 (1993). [DOI] [PubMed] [Google Scholar]
- 33.Rakic P et al. Concurrent overproduction of synapses in diverse regions of the primate cerebral cortex. Science 232, 232–235 (1986). [DOI] [PubMed] [Google Scholar]
- 34.Shibata M et al. Regulation of prefrontal patterning, connectivity and synaptogenesis by retinoic acid. Accompanying paper.
- 35.Kang HJ et al. Spatiotemporal transcriptome of the human brain. Nature 478, 483–489 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lambert N et al. Genes expressed in specific areas of the human fetal cerebral cortex display distinct patterns of evolution. PLoS One 6, e17753 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Miller JA et al. Transcriptional landscape of the prenatal human brain. Nature 508, 199–206 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.ENCODE Project Consortium. et al. An integrated encyclopedia of DNA elements in the human genome. Nature 489, 57–74 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chiang MY et al. An essential role for retinoid receptors RARbeta and RXRgamma in long-term potentiation and depression. Neuron 21, 1353–1361 (1998). [DOI] [PubMed] [Google Scholar]
- 40.Krezel W et al. Impaired locomotion and dopamine signaling in retinoid receptor mutant mice. Science 279, 863–867 (1998). [DOI] [PubMed] [Google Scholar]
- 41.Kwan KY et al. SOX5 postmitotically regulates migration, postmigratory differentiation, and projections of subplate and deep-layer neocortical neurons. Proc. Natl. Acad. Sci. U. S. A 105, 16021–16026 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shim S et al. Cis-regulatory control of corticospinal system development and evolution. Nature 486, 74–79 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Clarke RA & Eapen V Balance within the neurexin trans-synaptic connexus stabilizes behavioral control. Front. Hum. Neurosci 8, 52 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.State MW & Sestan N The emerging biology of autism spectrum disorders. Science 337, 1301–1303 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sudhof TC Synaptic neurexin complexes: A molecular code for the logic of neural circuits. Cell 171, 745–769 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Willsey AJ et al. Coexpression networks implicate human midfetal deep cortical projection neurons in the pathogenesis of autism. Cell 155, 997–1007 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gulsuner S et al. Spatial and temporal mapping of de novo mutations in schizophrenia to a fetal prefrontal cortical network. Cell 154, 518–529 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lewis DA & Mirnics K Transcriptome alterations in schizophrenia: disturbing the functional architecture of the dorsolateral prefrontal cortex. Prog. Brain Res 158, 141–152 (2006). [DOI] [PubMed] [Google Scholar]
Extended References
- 49.Dy P, Han Y & Lefebvre V Generation of mice harboring a Sox5 conditional null allele. Genesis 46, 294–299 (2008). [DOI] [PubMed] [Google Scholar]
- 50.Mathelier A et al. JASPAR 2016: a major expansion and update of the open-access database of transcription factor binding profiles. Nucleic Acids Res. 44, D110–D115 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Khan A et al. JASPAR 2018: Update of the open-access database of transcription factor binding profiles and its web framework. Nucleic Acids Res 46, D260–D266 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shim S et al. Regulation of EphA8 gene expression by TALE homeobox transcription factors during development of the mesencephalon. Mol. Cell Biol 27, 1614–1630 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang H et al. One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell 153, 910–918 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liu P et al. A highly efficient recombineering-based method for generating conditional knockout mutations. Genome Res 13, 476–484 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cong L et al. Multiplex genome engineering using CRISPR/Cas systems. Science 339, 819–823 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wilkinson DG & Nieto MA Detection of messenger RNA by in situ hybridization to tissue sections and whole mounts. Methods Enzymol. 225, 361–373 (1993). [DOI] [PubMed] [Google Scholar]
- 57.Hunt CA et al. PSD-95 is associated with the postsynaptic density and not with the presynaptic membrane at forebrain synapses. J. Neurosci 16, 1380–1388 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Essrich C et al. Postsynaptic clustering of major GABAA receptor subtypes requires the gamma 2 subunit and gephyrin. Nat. Neurosci 1, 563–571 (1998). [DOI] [PubMed] [Google Scholar]
- 59.Ippolito DM & Eroglu C Quantifying synapses: an immunocytochemistry-based assay to quantify synapse number. J. Vis. Exp 45, 2270 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Fiala JC Reconstruct: A free editor for serial section microscopy. J. Microsc 218, 52–61 (2005). [DOI] [PubMed] [Google Scholar]
- 61.Risher WC et al. Rapid golgi analysis method for efficient and unbiased classification of dendritic spines. PLoS One 9, (2014) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kaur N et al. Neural stem cells direct axon guidance via their radial fiber scaffold. Neuron 107, 1197–1211 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Meijering E et al. Design and validation of a tool for neurite tracing and analysis in fluorescence microscopy images. Cytom. Part A 58, 167–176 (2004). [DOI] [PubMed] [Google Scholar]
- 64.Hinrichs AS et al. The UCSC Genome Browser database: update 2006. Nucleic Acids Res. 34, D590–598 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Robinson JT et al. Integrative genomics viewer. Nat. Biotechnol 29, 24–26 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kent WJ et al. The human genome browser at UCSC. Genome Res. 12, 996–1006 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Rosenbloom KR et al. ENCODE data in the UCSC Genome Browser: year 5 update. Nucleic Acids Res. 41, D56–63 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Shibata M et al. MicroRNA-9 regulates neurogenesis in mouse telencephalon by targeting multiple transcription factors. J. Neurosci 31, 3407–3422 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Morozov YM, Ayoub AE & Rakic P Translocation of synaptically connected interneurons across the dentate gyrus of the early postnatal rat hippocampus. J. Neurosci 26, 5017–5027 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Morozov YM, Mackie K & Rakic P Cannabinoid type 1 receptor is undetectable in rodent and primate cerebral neural stem cells but participates in radial neuronal migration. Int. J. Mol. Sci 21, 1–19 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Thompson CL et al. A high-resolution spatiotemporal atlas of gene expression of the developing mouse brain. Neuron 83, 309–323 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
None