Abstract
Due to high selection advances and shortened generation interval, genomic selection (GS) is now an effective animal breeding scheme. In broilers, many studies have compared the accuracy of different GS prediction methods, but few reports have demonstrated phenotypic or genetic changes using GS. In this study, the paternal chicken line B underwent continuous selection for 3 generations. The chicken 55 k SNP chip was used to estimate the genetic parameters and detect genomic response regions by selective sweep analysis. The heritability for body weight (BW), meat production, and abdominal fat traits were ranged from 0.12 to 0.38. A high genetic correlation was found between BW and meat production traits, while a low genetic correlation (<0.1) was found between meat production and abdominal fat traits. Selection resulted in an increase of about 516 g in BW and 140 g in breast muscle weight. Percentage of breast muscle and whole thigh were increased 0.8 to 1.5%. No change was observed in abdominal fat percentage. The genomic estimated breeding value advances was positive for BW and meat production (except whole thigh percentage), while negative for abdominal fat percentage. By selective sweep analysis, 39 common chromosomal regions and 102 protein coding genes were found to be influenced, including MYH1A, MYH1B, and MYH1D of the MYH gene family. Tight junction pathway as well as myosin complex related terms were enriched. This study demonstrates the effective use of GS for improvements in BW and meat production in chicken line B. Further, genomic regions, responsive to intensive genetic selection, were identified to contain genes of the MYH family.
Key words: chicken, genomic selection, body weight, meat production traits, selective sweep
INTRODUCTION
Within the past few decades, intensive genetic selection has been applied to broilers, which has emphasized enhanced body weight (BW) and meat production (Morris and Pollott, 1997). Each is a direct determinant of economic benefit. In 2005, BW at 28 and 42 d of age was increased threefold (28 d: 1,396 g, 42 d: 4,202 g) compared to that (28 d: 316 g, 42 d: 905 g) in 1957 for the Ross broiler (Barbut, 2019). The conventional best linear unbiased prediction (BLUP) method is a widely applied breeding approach to complex traits, such as shank length, egg production, and disease resistance. With the use of high-throughput SNP detection techniques, and the advantages for shorten breeding rotation and improved prediction accuracy, genomic estimated breeding value (GEBV) is preferred and is proved to be more effective in modern farm animal breeding system.
Genomic selection (GS) was first used to estimate GEBV by evaluating the cumulative effect of genome-wide markers, with the assumption that at least one marker would share the same linkage disequilibrium (LD) with the major quantitative trait locus (QTL) of interest (Meuwissen et al., 2001). This approach has been widely used for farm animal breeding of beef cattle (Pollak et al., 2012), dairy cow (Schaeffer, 2006), pig (Ibáñez-Escriche et al., 2014), and chicken (Preisinger, 2012). For chicken, estimates of prediction accuracy and genetic parameters for different models (e.g., PBLUP, GBLUP, and SSGBLUP) have been reported for BW, meat production, and meat quality (Le Bihan-Duval et al., 2008; Chen et al., 2011; R. Liu et al., 2019b; Abdollahi-Arpanahi et al., 2015). However, few reports have evaluated the multigenerational breeding process in chicken populations selected using the GS method. In this study, GS was used to improve BW and meat production in broiler line B for several generations. Phenotypic data were collected from generation 4 (G4) to generation 7 (G7). In this manner, the real effect of GS on this broiler line B was evaluated.
Long-term artificial selection or intensive genetic selection of farm animals results in a genomic signature that influences the specific traits (Kristensen et al., 2015; Berry, 2018). For example, the modern chicken (meat-type and egg-type chicken), established during the domestication and goal-directed breeding, likely has a detectable signature in the genome (Rubin et al., 2010; Ericsson et al., 2014; Qanbari et al., 2015; Gu et al., 2020). By selective sweep analysis, candidate chicken genes have been identified that are associated with pigmentation (e.g., EDN3, RALY, and LGR4), muscularity (e.g., WWP1, DLK1), and reproductive performance (e.g., TSHR) (Ericsson et al., 2014; Qanbari et al., 2015; D. Li et al., 2020a; X. Huang et al., 2020b). Therefore, it's effective to detect the human-driven selection signature in genome using selective sweep method, and unveil the latent genetic mechanism for economic traits Sheng et al. 2015. and Zan et al. 2017 indicated that associated genomic regions and SNPs were detected after divergent selection for BW in chicken. A list of potential selection signatures has been identified among nine distinct broiler breeding lines aimed to various selection purpose (Stainton et al., 2015). Among these, BW and meat production were prominent traits in the breeding system of chicken. Hence, it's essential to explore the chromosomal regions associated with each trait as a consequence of genetic selection.
In current study, this broiler population (n = 4,201) underwent GS program for three generations was used to analyze the phenotypic process and genetic parameter. And the 2 selective sweep methods were used to uncover the latent genomic signature left by GS.
MATERIALS AND METHODS
Population and Breeding Scheme
All experimental protocols related to chickens were conducted in accordance with guidelines established by the Ministry of Science and Technology (Beijing, China). Ethical approval was conferred by the Animal Welfare and Ethics Committee of the Institute of Animal Sciences, Chinese Academy of Agricultural Sciences (IAS-CAAS, Beijing, China) with the reference number: IAS2019-44.
Chickens were fast-growing white-feathered pure line B that had been selected for multiple generations based on BW and meat production (Li et al., 2020b). This chicken line was a synthetic strain generated by Foshan Gaoming Xinguang Agriculture and Animal Husbandry Co., Ltd. (Foshan, Guangdong, China). From G4 to G7, individuals with both genotype and phenotype records were used as the training set. The test set comprised chickens with genotype but not phenotype records. Sires with top GEBV for BW, breast muscle weight and whole thigh weight were selected. The detailed selection scheme is shown in Figure S1. All chickens for each generation were housed in individual cages, with recommended environmental conditions and a corn-soybean meal diet (Table S1). Feed and water were provided ad libitum. A total of 4,399 chickens were included in the study.
Phenotypic Statistics
After a 12-h fasting, G4 to G7, chickens were slaughtered at 42 d (Figure S1). Pectoral muscle and whole thigh were stripped, and the abdominal fat was separated completely. Next, BW, breast muscle weight (BrW), whole thigh weight (ThW), and abdominal fat weight (AbFW) were recorded. The relative weight of BrW, ThW, and AFW was calculated as follows:
Breast muscle percentage (BrP) = BrW / BW × 100%.
Whole thigh percentage (ThP) = ThW / BW × 100%.
Abdominal fat percentage (AbFP) = AbFW / BW × 100%.
After statistical analysis of 7 traits, individuals were eliminated from consideration if the phenotypic index deviated more than 3 standard deviations from the average value. A total of 4,217 chicken were retained in the final analysis.
Genotyping and Quality Control
Genomic DNA was obtained from vein blood by the phenol-chloroform extraction, evaluated by quality control for integrality and concentration. Qualified DNA was genotyped with an accustomed chicken SNP array, which is designed based on the Gallus gallus 5.0 assembly and includes 52,060 SNPs (Liu et al., 2019c). PLINK software (v 1.9) was used for quality control of genotyping data (Purcell et al., 2007). Totally, 16 individuals were eliminated due to a lower call rate (–mind 0.1), and 7,327 SNPs were excluded due to a lower call rate (<90%) or minor allele frequency (<5%), as well as the SNPs located in W chromosome (only 5 SNPs). Then 44,733 SNPs were retained after quality control. Missing alleles were imputed using Beagle 5.1 (Browning et al., 2018). A total of 44,733 SNPs from 4,201 chickens (male: 2,632, female: 1,569) were retained for subsequent genetic parameter estimation.
Estimation of GEBV and Genetic Parameter
In this study, SNPs data was used to produce a genomic relationship matrix (GRM) by the method of VanRaden (2008). GEBV and variance component estimates for phenotype were calculated based on univariate animal model constructed by ASReml v4.1 (Gilmour et al., 2015) in an R environment. The univariate model was defined as follows:
where y is the vector of phenotype, b is the vector of fixed effects, including gender and generation effect, a is the vector of random additive genetic effects, e is the vector of random residual effect. X and Z are design matrices relating observation to the corresponding fixed and random effects, respectively. The variance-covariance structure assumed for random effect was following:
where G is the GRM based on SNP data, I is the identity matrix, σa2 is the additive genetic variance, σe2 is the residual variance. The heritability of phenotypes was calculated by: h2 = σa2/(σa2 + σe2). A bivariate animal model was used to evaluate the genetic correlation (rg) and phenotypic correlation (rp) between any two traits:
where y, b, a, e, X, and Z were same as above equation.
Calculation of LD Decay
The LD decay extent defined by the pair-wise r2 estimation across SNP distance was evaluated in each and all generations using PopLDdecay program (Zhang et al., 2019.). The r2 was defined as the following equation:
where was the frequency of the A1B1 haplotype, and , , and are the frequency of A2B2, A1B2, and A2B1 haplotype, respectively. The , , , and were the frequency of A1, A2, B2, and B2 alleles, respectively. The distance from the initial position (harboring top r2 value) to the end position (r2 = 0.1) was considered as the LD decay distance. The LD decay level was plotted with a window size of 1-Mb in R environment.
Detection of Selective Sweeps
Based on the methods of population differentiation and genomic site polymorphism, we explored the candidate divergent regions by scanning chromosomal regions for high fixation index (Fst) (Weir and Cockerham, 1984) value and obvious distinction in genetic diversity (π ratio in log2 scale). The G4 population was regarded as the genomic original population, G5, G6, and G7 were considered genomic evolutionary populations which were used for selective signature comparison to the G4 population. Considering the LD decay extent, the half of the window size when LD decays to 0.1 (around 150-kb) was set as an appropriate sliding window to detect selection signature (Table S2). Therefore, the Fst and π value were calculated in a 150-kb sliding window with 75-kb steps using VCFtools software (Danecek et al., 2011). The top 5% of Fst (G5 v G4: 0.013; G6 v G4: 0.024; G7 v G4: 0.031) and log2(π ratio) (G5 v G4: −0.18; G6 v G4: −0.24; G7 v G4: −0.27) was defined as a significant threshold, and the regions meeting the significant level of these 2 methods were regarded as a candidate divergent region in each generation. The common divergent regions in 3 generations (G5–G7) were defined as divergent regions. Visualization of these analyses was implemented by ggplot2 packages in R (Gómez-Rubio, 2017).
Gene Annotation, Kyoto Encyclopedia of Genes and Genomes, and Gene Ontology Enrichment Analysis
The candidate selective regions were annotated using biomaRt package in R (Durinck et al., 2009). Protein-coding genes located within and overlapping with the candidate regions were extracted to perform KEGG and GO enrichment analysis with KOBAS 3.0 (Xie et al., 2011). A P-value of 0.05 was regarded as a threshold for significant enrichment.
RESULTS
Estimation of Genetic Parameter
The estimates of heritability, as well as genetic and phenotypic correlations for production performance are shown in Table 1. Heritability for ThP, ThW, and BW had a low trend (0.12–0.17), with a moderate estimate for BrW, BrP, AbFW, and AbFP (0.24–0.38). BrW and ThW were highly correlated with BW (rg: 0.60–0.76, rp: 0.74–0.79). There was a relatively high phenotypic correlation (rp = 0.53) between BrW and ThW, while the genetic correlation was relatively low between them (rg = 0.18). There was a high negative correlation (rg = −0.52) between BrP and ThP. For AbFW, genetic correlation was low for BW, BrW, and ThW (rg: −0.02 to 0.13). As expected, weight and relative weight of abdominal fat had extremely high levels of genetic and phenotypic correlation (rg = 0.98, rp = 0.95). And a same result was also observed for BrW and ThW (rg: 0.56–0.74; rp: 0.67–0.79). There was a low pairwise genetic correlation between AbFW or AbFP and other traits (BW, BrW, BrP, ThW, and ThP), which ranged from −0.02 to −0.23.
Table 1.
Estimates of heritability, phenotypic correlation and genetic correlation for the traits1.
| BW | BrW | ThW | AbFW | BrP | ThP | AbFP | |
|---|---|---|---|---|---|---|---|
| BW | 0.17 ± 0.03 | 0.79 ± 0.01 | 0.74 ± 0.01 | 0.36 ± 0.02 | 0.26 ± 0.02 | 0.01 ± 0.02 | 0.10 ± 0.02 |
| BrW | 0.60 ± 0.06 | 0.24 ± 0.03 | 0.53 ± 0.01 | 0.21 ± 0.02 | 0.79 ± 0.01 | −0.07 ± 0.02 | −0.001 ± 0.02 |
| ThW | 0.76 ± 0.05 | 0.18 ± 0.11 | 0.14 ± 0.02 | 0.26 ± 0.02 | 0.11 ± 0.02 | 0.67 ± 0.01 | 0.07 ± 0.02 |
| AbFW | 0.13 ± 0.09 | −0.06 ± 0.09 | −0.02 ± 0.10 | 0.34 ± 0.03 | −0.02 ± 0.02 | −0.01 ± 0.02 | 0.95 ± 0.002 |
| BrP | 0.07 ± 0.09 | 0.74 ± 0.03 | −0.28 ± 0.10 | −0.15 ± 0.07 | 0.38 ± 0.03 | −0.12 ± 0.02 | −0.09 ± 0.02 |
| ThP | −0.11 ± 0.13 | -0.48 ± 0.10 | 0.56 ± 0.09 | −0.23 ± 0.11 | −0.52 ± 0.09 | 0.12 ± 0.02 | −0.01 ± 0.02 |
| AbFP | −0.06 ± 0.09 | −0.17 ± 0.08 | −0.17 ± 0.10 | 0.98 ± 0.004 | −0.16 ± 0.07 | −0.20 ± 0.10 | 0.37 ± 0.03 |
Bold diagonal is heritability, upper diagonal is phenotypic correlation, and lower diagonal is genetic correlation. Abbreviations: AbFP, abdominal fat percentage; AbFW, abdominal fat weight; BrP, breast muscle percentage; BW, body weight; BrW, breast muscle weight; ThW, whole thigh weight; ThP, whole thigh percentage.
Effect of Genomic Selection
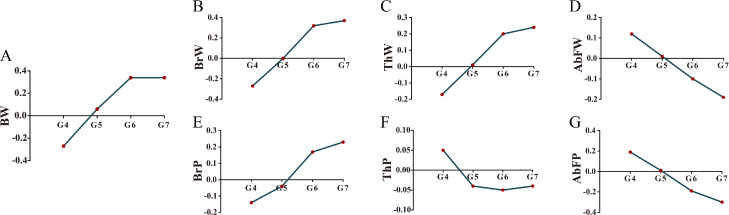
The phenotypic and genetic trends for all traits are presented in Table 2 and Figure 1, respectively. Compared to G4, a fluctuating increment was observed for BW, meat production, and yield. An average increment of 516.2 g for BW was realized. The BW gain in male chickens (573.4 g) was higher than that in female chickens (484.7 g). A similar result was found for the meat production trait, an average of 119.7 to 161.8 g of weight gain was observed for BrW and ThW as well as a 0.5 to 1.8% increment for BrP and ThP. Even though AbFW was elevated by 8.6 g, only 0.1% phenotypic change was found for AbFP. Similar results were found by GEBV estimation for all traits with the exception for ThP (Table S3). Genomic selection for AbFW (from 0.12 to −0.19) and AbFP (from 0.19 to −0.3) revealed a negative genetic process. The GEBV result was shown in Figure 1.
Table 2.
Phenotypic alteration undergoing GS breeding for multigenerations1.
| BW |
BrW |
ThW |
AbFW |
BrP |
ThP |
AbFP |
||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Male | Female | Male | Female | Male | Female | Male | Female | Male | Female | Male | Female | Male | Female | |||||||||||||||
| G4 | 2,001.4 ± 172.0 | 1,794.8 ± 145.2 | 396.8 ± 52.5 | 372.6 ± 48.1 | 452.9 ± 46.1 | 401.7 ± 38.5 | 31 ± 9.3 | 32.6 ± 9.4 | 19.8 ± 1.6 | 20.7 ± 1.6 | 22.6 ± 1.3 | 22.4 ± 1.3 | 1.5 ± 0.5 | 1.8 ± 0.5 | ||||||||||||||
| G5 | 2,498.1 ± 162.1 | 2,332.7 ± 123.4 | 553.2 ± 53.0 | 545.8 ± 47.8 | 576.3 ± 48.9 | 519.9 ± 38.6 | 30.9 ± 10.3 | 40.1 ± 10.1 | 22.1 ± 1.4 | 23.4 ± 1.5 | 23.1 ± 1.1 | 22.3 ± 1.0 | 1.2 ± 0.4 | 1.7 ± 0.4 | ||||||||||||||
| G6 | 2,418.7 ± 166.5 | 2,224.3 ± 134.2 | 504.6 ± 50.1 | 494.6 ± 46.3 | 561.9 ± 47.2 | 486.2 ± 38.5 | 43.2 ± 9.7 | 40.7 ± 9.4 | 20.8 ± 1.2 | 22.2 ± 1.4 | 23.2 ± 0.9 | 21.9 ± 1.1 | 1.8 ± 0.4 | 1.8 ± 0.4 | ||||||||||||||
| G7 | 2,574.8 ± 155.0 | 2,279.5 ± 136.6 | 555.2 ± 52.0 | 500.8 ± 47.4 | 614.7 ± 47.3 | 521.4 ± 39.8 | 39.6 ± 9.6 | 42.9 ± 9.7 | 21.6 ± 1.4 | 22 ± 1.5 | 23.9 ± 1.1 | 22.9 ± 1.1 | 1.5 ± 0.3 | 1.9 ± 0.4 | ||||||||||||||
| Process 2 | 573.4 | 484.7 | 158.4 | 128.2 | 161.8 | 119.7 | 8.6 | 10.3 | 1.8 | 1.3 | 1.3 | 0.5 | 0.0 | 0.1 | ||||||||||||||
| Total 3 | 516.2 | 140.5 | 136.3 | 9.6 | 1.5 | 0.8 | 0.0 | |||||||||||||||||||||
All the phenotype statistics were recorded from generation 4 to generation 7. Abbreviations: AbFP, abdominal fat percentage; AbFW, abdominal fat weight; BW, body weight; BrP, breast muscle percentage; BrW, breast muscle weight; ThW, whole thigh weight; ThP, whole thigh percentage.
The process was a statistics for the average difference of phenotype of male and female chickens, which was calculated by: phenotype(G7) – phenotype(G4).
The total was calculated as same as above, but didn't distinguish the gender.
Figure 1.
The genetic changes in different generations. (A–G) The genetic changes from G4 to G7 for BW, BrW, ThW, AbFW, BrP, ThP, and AbFP, respectively. The vertical axis was shown as the ratio of mean GEBV to standard deviation.
LD Decay Analysis
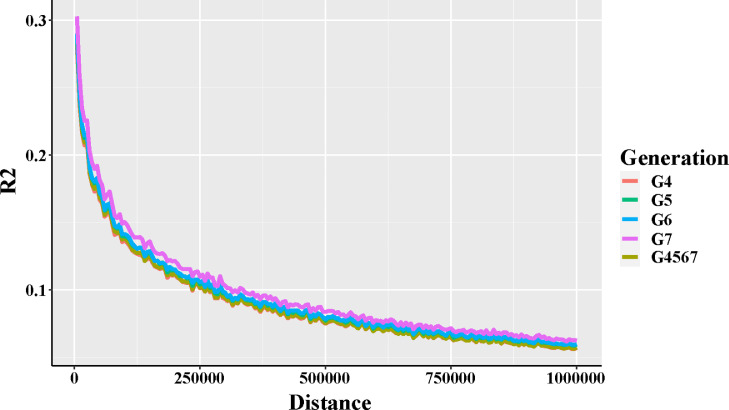
The LD (r2) values for each generation using all SNP-pairs were calculated and plotted along 1-Mb distances (Figure 2). The r2 for overall generation was ranged from 0.05 to 0.28 in genomic distance of 5 to 1,000-kb. The distance of decay (from top to 0.1) was 295-kb in line B, and an increasing trend with expectation was observed at different generations (G4: 290-kb, G5: 330-kb, G6: 335-kb, G7: 390-kb).
Figure 2.
Average LD decay with increased physical distance between paired SNPs in each generation. The different color indicated four generations and merged populations (G4567). The LD decay distance (reduced to 0.1) was gradually increased (G7 > G6 > G5 > G4).
Genome-Wide Selective Sweep Analysis
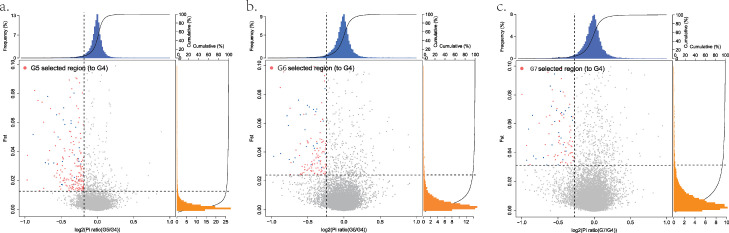
GS was applied for 3 generations to line B in order to improve BW and meat production traits. Theoretically, candidate genomic region affecting BW and meat production traits should be positively selected, retaining a selection signature. We scanned the genome for regions with high Fst values and high differences in genetic diversity (π in log2 scale) in 150-kb sliding windows. A total of 260 regions were detected by comparison of G5 and G4 (Fst > 0.013, log2π < −0.184) (Figure 3A, Table S4), with 406 protein coding genes and 53 long non-coding RNAs (lncRNAs) annotated within those regions (Table S5). By comparison of G6 and G4, 189 candidate regions (Fst > 0.024, log2π < −0.236) (Figure 3B, Table S4), with 290 protein coding genes as well as 43 lncRNAs were found (Table S5). By comparison of G7 and G4, 137 candidate regions (Fst > 0.031, log2π < −0.273) (Figure 3C, Table S4), with 284 protein coding genes as well as 29 lncRNAs were identified (Table S5). These results demonstrated a decrease in the number of divergent regions and an elevated trend of genomic differentiation level in each selected generation.
Figure 3.
Selected regions in each generation compared to G4. (A–C) Selected regions in G5, G6, and G7, respectively. The x-axis represented π in log2 scale, and y-axis represented Fst index. The blue bars indicated the frequency of different π in log2 scale, and the orange bar indicated the frequency of different Fst value. The red dots represented the regions under selection, the blue dots represented the selected regions among three comparisons, while the gray dots represented the unselected regions. The dotted line indicated the top 5% threshold of Fst index and genetic diversity. Some limitations on Fst value and π were shown in figures, the full results was shown as Table S4.
Among the 3 comparisons, 102 protein coding genes and 8 lncRNAs located within 39 common regions were detected, uncovering a latent core function for growth and development with consistent breeding. And a list of candidate SNPs within these regions were detected frequency difference in different generations (Table S5). By annotation and enrichment, genes related to muscle development and movement (e.g., motor activity, myosin complex, and actin filament binding) were identified, including MYH1A, MYH1B, and MYH1D (Table 3 and S6). Further, tight junction may be a latent regulation core (Table 3 and S7).
Table 3.
Enrichment analysis for common genes obtained by selective sweep analysis1.
| Term | Input | Background | P-value | Corrected P-value 2 | Genes | |
|---|---|---|---|---|---|---|
| KEGG pathway | Tight junction | 7 | 139 | 9.73E-06 | 1.24E-04 | MYH1A, MYH1B, MYH1D, etc. |
| Ether lipid metabolism | 2 | 78 | 1.63E-02 | 2.26E-01 | CHPT1, LPCAT2 | |
| Phosphonate and phosphinate metabolism | 1 | 6 | 3.55E-02 | 2.26E-01 | CHPT1 | |
| GO term | Motor activity | 5 | 29 | 7.78E-07 | 2.97E-04 | MYH1A, MYH1B, MYH1D, etc. |
| Myosin complex | 5 | 35 | 1.80E-06 | 3.43E-04 | MYH1A, MYH1B, MYH1D, etc. | |
| Actin filament binding | 5 | 146 | 1.07E-03 | 1.03E-01 | MYH1A, MYH1B, MYH1D, etc. | |
| Regulation of MAPK cascade | 2 | 14 | 3.00E-03 | 1.91E-01 | MMP2, GRB2 | |
| Cellular response to ionizing radiation | 2 | 14 | 3.00E-03 | 1.91E-01 | LIG4, GRB2 | |
| Positive regulation of actin filament polymerization | 2 | 25 | 8.46E-03 | 2.26E-01 | PFN1, GRB2 | |
| Spermatid development | 2 | 32 | 1.32E-02 | 2.26E-01 | HID1, ENSGALG00000012766 | |
| Transforming growth factor beta receptor signaling pathway | 2 | 46 | 2.54E-02 | 2.26E-01 | APPL2, HPGD | |
| Protein import into nucleus | 2 | 48 | 2.74E-02 | 2.26E-01 | NUP85, APPL2 | |
| Cellular protein metabolic process | 1 | 5 | 3.05E-02 | 2.26E-01 | GIGYF2 |
The significant pathways and top 10 GO terms were enriched by the common genes located in the 39 differential regions in genome.
The corrected P-value was calculated using B-H method.
DISCUSSION
Heritability and Correlation Among BW and Meat Production Traits
In current study, estimated heritability based on SNP data for BW and meat production traits was low to medium (0.12–0.38). Although heritability estimates for those traits were relatively low, results were within a normal range previously reported for broilers (Abdollahi‐Arpanahi et al., 2015; Grupioni et al., 2015; Teng et al., 2019). For quality chicken or crossbred populations, a medium and high heritability estimate based on the GBLUP method for BW (0.38), BrW (0.42), BrP (0.34), thigh muscle yield (0.52), AbFW (0.52), and AbFP (0.45) has been reported (Liu et al., 2017; Zhang et al., 2017). Those values are larger than those found for this population, which may be due to differences in genomic marker density and kinship (Norman et al., 2018). Besides that, line B is a high inbred population, which could result in a smaller estimates of additive genetic variance and heritability (Powell et al., 2010). In Abdollahi-Arpanahi et al report, a lower heritability for BW (0.05–0.10) in fast-growing broiler was estimated ( Abdollahi-Arpanahi et al., 2016), which was lower than that in this study.
The estimate of genetic correlation between BW and meat product traits (BrW, ThW) was strong (0.60–0.76). Similarly, a high correlation (0.82–0.86) was calculated using a pedigree-based matrix between BW and muscle weight by Venturini et al. (2014). There was low correlation between BW and BrP (0.07) as well as ThP (−0.11). A medium genetic correlation (0.18) between BrW and ThW was found, with a high negative correlation (−0.52) between BrP and ThP. Similar results were reported by Cruz et al., a high correlation was found among BW, BrW, and ThW (Cruz et al., 2020). Herein, we found AbFW to be genetically correlated with BW (0.13), although the relationship was close to zero between AbFP and BW (−0.06). These results suggested selection for BW was accompanied by an increase in abdominal fat (Li et al., 2021), even though, no change was detected in AbFP. For modern breeding programs, higher meat production and less abdominal fat are preferred in fast-growing broilers. The inclusion of AbFW with negative coefficient in the breeding procedure is therefore reasonable and effective.
Effects of GS in Line B
Compared to traditional breeding schemes for farm animals, GS is beneficial to shorten generation intervals and enhance genetic gain of target traits based on SNP information (Meuwissen et al., 2001). GS has been applied to broiler (e.g., Aviagen, Cobb) selection and breeding to improve growth and meat production (Avendaño et al., 2012; Hidalgo et al., 2021).
Tremendous improvements in BW, BrW, and ThW were obtained after 3 generations using GS, although a fluctuation was observed in G6 generation compared to G5 generation. This phenotypic fluctuation is commonly observed in animal breeding program. In Ullengala et al. (2020) report, the BW and shank length of a synthetic chicken line tend to increase after selection of eight generations, but a negative changes was observed in G6 and G7 compared to G5. Which may be due to limited prediction accuracy and SNP density (Norman et al., 2018) or the different environment conditions. Ullengala et al. reported an average BW gain of 40 g in each generation was acquired after selection based on pedigree information (Ullengala et al., 2020). In current study, an average BW gain of 172 g in each generation was obtained based on the described GS breeding scheme. Which indicated superior breeding progress of GS program as expected (Lillehammer et al., 2013). In accordance with the breeding goal, the breeding scheme had a clear effect on meat production. BrW and ThW were both increased by over 30% (approximately 140 g) after selection of 3 generations. Similar trends were observed for BrP and ThP (0.8–1.5%). AbFW was also increased in G7 compared to G4, but almost no change was found for AbFP (<0.05%).
Similar but smaller increments were found by GEBV estimation for BW and meat production traits, indicating a stably enhanced production performance. This result is consistent with a trend between GEBV and phenotype, also reported previously (Ullengala et al., 2020). The GEBV of AbFW and AbFP exhibited a negative change after selection, indicating that the phenotypic increase of AbFW may be caused by the enhancement for BW, considering genetical correlation between AbFW and BW. Simultaneously, there was no increase in AbFP as evidenced by synchronization enhancement for fat deposition and BW during the breeding process. Additionally, we found that genetic gain of ThW was positively enhanced, while it's decreased for ThP, which may be caused by relatively higher enhancement GEBV for BW than that for ThW. In summary, more than 50% BW gain was due to improved breast and thigh weight. These results demonstrate the excellent breeding effect of this GS program.
LD Decay and Selective Signatures in Genome After GS
For line B, BW and meat production were improved significantly after only 3 generations using GS technology. Likewise, the distance of LD decay was gradually enhanced among different generations (G7 > G6 > G5 > G4). In previous reports, LD extent was elevated with the increase of generation for Baier yellow chicken and Langshan chicken (Zhang et al., 2018.), which demonstrated a result of positive selection of genomic sites. As expected, the nucleotide diversity (π) was gradually decreased from G4 to G7. Both results suggested the increased LD decay distance and decreased nucleotide diversity were caused by the intensive selection using the GS program in multigenerations Wang et al. (2017). have illustrated that the bottlenecking occurred in the aviculture of Yuanbao chicken due to a weaker LD decay and lower nucleotide diversity. Similar, we cannot exclude the bottleneck effect in the GS program, although only a small change was found in LD decay and nucleotide diversity among 4 generations.
Based on selective sweep theory, the frequency of favorable alleles and adjacent sites could be enhanced in a short time under the positive selection, which could result in decreased polymorphism within specific genomic regions (Smith and Haigh, 1974). As Zhou et al. ( 2018) reported, evidence of population differentiation and selected regions can be explored by the combined use of Fst and π. As such, genome-wide candidate selected regions were identified by calculating Fst and π. And only the regions significantly selected in 3 comparisons could be recognized as the selective signatures in genome, which could avoid the extreme bias of allele frequency in each generation. A total of thirty-nine 150-kb regions were found to exhibit genomic differentiation and selective signatures after continuous selection from the starting generation, G4. Relatively small differences in Fst were found, which may be due to the identical origin of the chickens and the restricted number of selection generations. In a previous study, clear genetic divergence was observed in both the selected line and in the natural line during a long-term selection (more than 15 generations), even though each line originated from the same quality chicken population (L. Liu et al., 2019a). In addition, some regions had a relatively high differentiation (Fst > 0.10) as judged by the 3comparisons, indicating a pivotal feature in those chromosomal regions (e.g., chr2: 30225001-30375000, chr4: 43950001-44100000, chr18: 450001-600000).
By gene annotation and enrichment, a list of 102 protein coding genes and 8 lncRNAs were identified within candidate regions. Muscle-associated GO terms, including motor activity, myosin complex, actin filament binding, and myofibril, were most significantly enriched. This result indicated that genes related to muscle development and composition may be responsive to genetic selection (e.g., MYH1A, MYH1B, and MYH1D). Myosin heavy chain is an important components of myosin, and has considerable effect on muscle development (Chakkalakal et al., 2012; Vikne et al., 2012). Liu et al. reported MYH1E and MYH1A to be key target genes of functional lncRNAs, identifying a regulation point for skeletal muscle development in multiple embryonic periods for chicken (Z. Liu et al., 2019d). MYH1B is a marker for fast-type muscle fiber, and the stability of this gene have a regulatory effect on muscle fiber type switching in chicken ( Yu et al., 2021). And Zeng et al. have reported the MYH1B has a crucial role in the development of thigh muscle rather than pectoral muscle in the immunocastration chicken model ( Zeng et al., 2020). Likewise, the MYH gene family has been shown to be similarly important in deer (Jia et al., 2020), pig (Fazarinc et al., 2020), and cattle (Picard and Gagaoua, 2020). Consistent with previous studies, significantly enriched tight junction pathway is crucial to growth rate and myoblast differentiation in chickens (He et al., 2020; Huang et al., 2020a).
CONCLUSIONS
In conclusion, 3 generations of genomic selection in the pure chicken line B resulted in significant phenotypic increments in BW (over 20%) and meat production (over 30%). With the aim of enhancing BW and meat production, slight genomic differentiation was obtained. Candidate genes responsive to selection (e.g., MYH1A, MYH1B, and MYH1D) within genomic differentiation regions were annotated to muscle development and movement. These genes are likely regulation targets that facilitate growth and meat production using this animal breeding system.
ACKNOWLEDGMENTS
The work was supported by the Agricultural Science and Technology Innovation Program (CAAS-ZDRW202005), the National Nonprofit Institute Research Grant (Y2020PT02), the China Agriculture Research System of MOF and MARA (CARS-41), the Agricultural Science and Technology Innovation Program (ASTIP-IAS04; ASTIP-IAS-TS-15).
DISCLOSURES
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
Supplementary material associated with this article can be found in the online version at https://doi.org/10.1016/j.psj.2022.101856.
Appendix. Supplementary materials
REFERENCES
- Abdollahi-Arpanahi R., Morota G., Valente B.D., Kranis A., Rosa G.J., Gianola D. Assessment of bagging GBLUP for whole-genome prediction of broiler chicken traits. J. Anim. Breed Genet. 2015;132:218–228. doi: 10.1111/jbg.12131. [DOI] [PubMed] [Google Scholar]
- Abdollahi-Arpanahi R., Morota G., Valente B.D., Kranis A., Rosa G.J.M., Gianola D. Differential contribution of genomic regions to marked genetic variation and prediction of quantitative traits in broiler chickens. Genet Sel Evol. 2016;48:10. doi: 10.1186/s12711-016-0187-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avendaño S., Watson K., Kranis A. Genomics in poultry breeding—into consolidation phases'. Proceedings XXIV World's Poultry Congress, 5–9 August 2012; Salvador, Bahia, Brazil; 2012. [Google Scholar]
- Barbut S. Recent myopathies in broiler's breast meat fillets. Worlds Poult. Sci. J. 2019;75:559–582. [Google Scholar]
- Berry D.P. Symposium review: breeding a better cow–will she be adaptable? J. Dairy Sci. 2018;101:3665–3685. doi: 10.3168/jds.2017-13309. [DOI] [PubMed] [Google Scholar]
- Browning B.L., Zhou Y., Browning S.R. A one-penny imputed genome from next-generation reference panels. Am. J. Hum. Genet. 2018;103:338–348. doi: 10.1016/j.ajhg.2018.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakkalakal J.V., Kuang S., Buffelli M., Lichtman J.W., Sanes J.R. Mouse transgenic lines that selectively label Type I, Type IIA, and Types IIX+B skeletal muscle fibers. Genesis. 2012;50:50–58. doi: 10.1002/dvg.20794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C.Y., Misztal I., Aguilar I., Tsuruta S., Meuwissen T.H., Aggrey S.E., Wing T., Muir W.M. Genome-wide marker-assisted selection combining all pedigree phenotypic information with genotypic data in one step: an example using broiler chickens. J. Anim. Sci. 2011;89:23–28. doi: 10.2527/jas.2010-3071. [DOI] [PubMed] [Google Scholar]
- Cruz V.A., Grupioni N.V., Mendonça G.G., Venturini G.C., Ledur M.C., Peixoto J.O., Munari D.P. Genetic parameters for performance and carcass traits in a paternal 1 lineage of broiler. Anais Acad. Bras. Ciências. 2020;92:e20180697. doi: 10.1590/0001-3765202020180697. [DOI] [PubMed] [Google Scholar]
- Danecek P., Auton A., Abecasis G., Albers C.A., Banks E., DePristo M.A., Handsaker R.E., Lunter G., Marth G.T., Sherry S.T., McVean G., Durbin R., Genomes Project Analysis G. The variant call format and VCFtools. Bioinformatics. 2011;27:2156–2158. doi: 10.1093/bioinformatics/btr330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durinck S., Spellman P.T., Birney E., Huber W. Mapping identifiers for the integration of genomic datasets with the R/Bioconductor package biomaRt. Nat. Protocols. 2009;4:1184. doi: 10.1038/nprot.2009.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ericsson M., Fallahsharoudi A., Bergquist J., Kushnir M.M., Jensen P. Domestication effects on behavioural and hormonal responses to acute stress in chickens. Physiol. Behav. 2014;133:161–169. doi: 10.1016/j.physbeh.2014.05.024. [DOI] [PubMed] [Google Scholar]
- Fazarinc G., Vrecl M., Poklukar K., Škrlep M., Batorek-Lukač N., Brankovič J., Tomažin U., Čandek-Potokar M. Expression of myosin heavy chain and some energy metabolism-related genes in the longissimus dorsi muscle of krškopolje pigs: effect of the production system. Front. Vet. Sci. 2020;7 doi: 10.3389/fvets.2020.533936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmour A., Gogel B., Cullis B., Welham S., Thompson R. VSN International Ltd; Hemel, Hempstead, UK: 2015. ASReml User Guide Release 4.1 Structural Specification. [Google Scholar]
- Gómez-Rubio V. ggplot2-elegant graphics for data analysis. J. Stat. Software. 2017;77:1–3. [Google Scholar]
- Grupioni N., Cruz V., Stafuzza N., Freitas L., Ramos S., Savegnago R., Peixoto J., Ledur M., Munari D. Phenotypic, genetic and environmental parameters for traits related to femur bone integrity and body weight at 42 days of age in a broiler population. Poult. Sci. 2015;94:2604–2607. doi: 10.3382/ps/pev257. [DOI] [PubMed] [Google Scholar]
- Gu H., Zhu T., Li X., Chen Y., Wang L., Lv X., Yang W., Jia Y., Jiang Z., Qu L. A joint analysis strategy reveals genetic changes associated with artificial selection between egg-type and meat-type ducks. Anim. Genet. 2020;51:890–898. doi: 10.1111/age.13014. [DOI] [PubMed] [Google Scholar]
- He M., Wu P., Chen F., Zhang B., Chen L., Zhang T., Zhang L., Li P., Wang J., Zhang G. Transcriptome analysis of leg muscles in fast and slow growth Bian chickens. Anim. Biotechnol. 2020;31:295–305. doi: 10.1080/10495398.2019.1588129. [DOI] [PubMed] [Google Scholar]
- Hidalgo J., Lourenco D., Tsuruta S., Masuda Y., Breen V., Hawken R., Bermann M., Misztal I. Investigating the persistence of accuracy of genomic predictions over time in broilers. J. Anim. Sci. 2021;99 doi: 10.1093/jas/skab239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H., Liu L., Li C., Liang Z., Huang Z., Wang Q., Li S., Zhao Z. Fat mass- and obesity-associated (FTO) gene promoted myoblast differentiation through the focal adhesion pathway in chicken. 3 Biotech. 2020;10:403. doi: 10.1007/s13205-020-02386-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X., Otecko N.O., Peng M., Weng Z., Li W., Chen J., Zhong M., Zhong F., Jin S., Geng Z., Luo W., He D., Ma C., Han J., Ommeh S.C., Zhang Y., Zhang X., Du B. Genome-wide genetic structure and selection signatures for color in 10 traditional Chinese yellow-feathered chicken breeds. BMC Genomics. 2020;21:316. doi: 10.1186/s12864-020-6736-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibáñez-Escriche N., Forni S., Noguera J.L., Varona L. Genomic information in pig breeding: science meets industry needs. Livestock Sci. 2014;166:94–100. [Google Scholar]
- Jia B., Liu Y., Li Q., Zhang J., Ge C., Wang G., Chen G., Liu D., Yang F. Altered miRNA and mRNA expression in sika deer skeletal muscle with age. Genes (Basel) 2020;11 doi: 10.3390/genes11020172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kristensen T.N., Hoffmann A.A., Pertoldi C., Stronen A.V. What can livestock breeders learn from conservation genetics and vice versa? Front, Genet, 2015;6:38. doi: 10.3389/fgene.2015.00038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Bihan-Duval E., Debut M., Berri C.M., Sellier N., Santé-Lhoutellier V., Jégo Y., Beaumont C. Chicken meat quality: genetic variability and relationship with growth and muscle characteristics. BMC Genet. 2008;9:53. doi: 10.1186/1471-2156-9-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D., Sun G., Zhang M., Cao Y., Zhang C., Fu Y., Li F., Li G., Jiang R., Han R. Breeding history and candidate genes responsible for black skin of Xichuan black-bone chicken. BMC Genomics. 2020;21:1–15. doi: 10.1186/s12864-020-06900-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Liu R., Zheng M., Feng F., Liu D., Guo Y., Zhao G., Wen J. New insights into the associations among feed efficiency, metabolizable efficiency traits and related QTL regions in broiler chickens. J. Anim. Sci. Biotechnol. 2020;11:65. doi: 10.1186/s40104-020-00469-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Zheng M., Zhao G., Wang J., Liu J., Wang S., Feng F., Liu D., Zhu D., Li Q. Identification of QTL regions and candidate genes for growth and feed efficiency in broilers. Genet. Select. Evol. 2021;53:1–17. doi: 10.1186/s12711-021-00608-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lillehammer M., Meuwissen T.H., Sonesson A.K. Genomic selection for two traits in a maternal pig breeding scheme. J. Anim. Sci. 2013;91:3079–3087. doi: 10.2527/jas.2012-5113. [DOI] [PubMed] [Google Scholar]
- Liu L., Cui H., Xing S., Zhao G., Wen J. Effect of divergent selection for intramuscular fat content on muscle lipid metabolism in chickens. Animals (Basel) 2019;10:4. doi: 10.3390/ani10010004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu R., Xing S., Wang J., Zheng M., Cui H., Crooijmans R., Li Q., Zhao G., Wen J. A new chicken 55K SNP genotyping array. BMC Genomics. 2019;20:410. doi: 10.1186/s12864-019-5736-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu R., Zheng M., Wang J., Cui H., Li Q., Liu J., Zhao G., Wen J. Effects of genomic selection for intramuscular fat content in breast muscle in Chinese local chickens. Anim. Genet. 2019;50:87–91. doi: 10.1111/age.12744. [DOI] [PubMed] [Google Scholar]
- Liu T., Luo C., Wang J., Ma J., Shu D., Lund M.S., Su G., Qu H. Assessment of the genomic prediction accuracy for feed efficiency traits in meat-type chickens. PLoS One. 2017;12 doi: 10.1371/journal.pone.0173620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z., Han S., Shen X., Wang Y., Cui C., He H., Chen Y., Zhao J., Li D., Zhu Q., Yin H. The landscape of DNA methylation associated with the transcriptomic network in layers and broilers generates insight into embryonic muscle development in chicken. Int. J. Biol. Sci. 2019;15:1404–1418. doi: 10.7150/ijbs.35073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meuwissen T.H., Hayes B.J., Goddard M.E. Prediction of total genetic value using genome-wide dense marker maps. Genetics. 2001;157:1819–1829. doi: 10.1093/genetics/157.4.1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris A.J., Pollott G.E. Comparison of selection based on phenotype, selection index and best linear unbiased prediction using data from a closed broiler line. Br. Poult. Sci. 1997;38:249–254. doi: 10.1080/00071669708417981. [DOI] [PubMed] [Google Scholar]
- Norman A., Taylor J., Edwards J., Kuchel H. Optimising genomic selection in wheat: effect of marker density, population size and population structure on prediction accuracy. G3: Genes Genomes Genet. 2018;8:2889–2899. doi: 10.1534/g3.118.200311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picard B., Gagaoua M. Muscle fiber properties in cattle and their relationships with meat qualities: an overview. J. Agric. Food Chem. 2020;68:6021–6039. doi: 10.1021/acs.jafc.0c02086. [DOI] [PubMed] [Google Scholar]
- Pollak E., Bennett G., Snelling W., Thallman R., Kuehn L. Genomics and the global beef cattle industry1. Anim. Product. Sci. 2012;52:92–99. [Google Scholar]
- Powell J.E., Visscher P.M., Goddard M.E. Reconciling the analysis of IBD and IBS in complex trait studies. Nat. Rev. Genet. 2010;11:800–805. doi: 10.1038/nrg2865. [DOI] [PubMed] [Google Scholar]
- Preisinger R. Genome-wide selection in poultry. Anim. Product. Sci. 2012;52:121–125. [Google Scholar]
- Purcell S., Neale B., Todd-Brown K., Thomas L., Ferreira M.A., Bender D., Maller J., Sklar P., de Bakker P.I., Daly M.J., Sham P.C. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qanbari S., Seidel M., Strom T.M., Mayer K.F., Preisinger R., Simianer H. Parallel selection revealed by population sequencing in chicken. Genome Biol. Evol. 2015;7:3299–3306. doi: 10.1093/gbe/evv222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin C.J., Zody M.C., Eriksson J., Meadows J.R., Sherwood E., Webster M.T., Jiang L., Ingman M., Sharpe T., Ka S., Hallböök F., Besnier F., Carlborg O., Bed'hom B., Tixier-Boichard M., Jensen P., Siegel P., Lindblad-Toh K., Andersson L. Whole-genome resequencing reveals loci under selection during chicken domestication. Nature. 2010;464:587–591. doi: 10.1038/nature08832. [DOI] [PubMed] [Google Scholar]
- Schaeffer L.R. Strategy for applying genome-wide selection in dairy cattle. J. Anim. Breed. Genet. 2006;123:218–223. doi: 10.1111/j.1439-0388.2006.00595.x. [DOI] [PubMed] [Google Scholar]
- Sheng Z., Pettersson M.E., Honaker C.F., Siegel P.B., Carlborg Ö. Standing genetic variation as a major contributor to adaptation in the Virginia chicken lines selection experiment. Genome Biol. 2015;16:219. doi: 10.1186/s13059-015-0785-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith J.M., Haigh J. The hitch-hiking effect of a favourable gene. Genet. Res. 1974;23:23–35. [PubMed] [Google Scholar]
- Stainton J.J., Haley C.S., Charlesworth B., Kranis A., Watson K., Wiener P. Detecting signatures of selection in nine distinct lines of broiler chickens. Anim. Genet. 2015;46:37–49. doi: 10.1111/age.12252. [DOI] [PubMed] [Google Scholar]
- Teng J., Gao N., Zhang H., Li X., Li J., Zhang H., Zhang X., Zhang Z. Performance of whole genome prediction for growth traits in a crossbred chicken population. Poult. Sci. 2019;98:1968–1975. doi: 10.3382/ps/pey604. [DOI] [PubMed] [Google Scholar]
- Ullengala R., Prince L.L.L., Haunshi S., Paswan C., Chatterjee R. Estimation of breeding value, genetic parameters and maternal effects of economic traits in rural male parent line chicken using pedigree relationships in an animal model. J. Anim. Breed. Genet. 2020;138:418–431. doi: 10.1111/jbg.12531. [DOI] [PubMed] [Google Scholar]
- VanRaden P.M. Efficient methods to compute genomic predictions. J. Dairy Sci. 2008;91:4414–4423. doi: 10.3168/jds.2007-0980. [DOI] [PubMed] [Google Scholar]
- Venturini G., da CRUZ V., Rosa J., Baldi F., El Faro L., Ledur M., Peixoto J.d.O., Munari D. Genetic and phenotypic parameters of carcass and organ traits of broiler chickens. Embrapa Suínos Aves-Artigo Periódico Indexado (ALICE) 2014;13:10294–10300. doi: 10.4238/2014.December.4.24. [DOI] [PubMed] [Google Scholar]
- Vikne H., Gundersen K., Liestøl K., Maelen J., Vøllestad N. Intermuscular relationship of human muscle fiber type proportions: slow leg muscles predict slow neck muscles. Muscle Nerve. 2012;45:527–535. doi: 10.1002/mus.22315. [DOI] [PubMed] [Google Scholar]
- Wang M.S., Otecko N.O., Wang S., Wu D.D., Yang M.M., Xu Y.L., Murphy R.W., Peng M.S., Zhang Y.P. An evolutionary genomic perspective on the breeding of dwarf chickens. Mol. Biol. Evol. 2017;34:3081–3088. doi: 10.1093/molbev/msx227. [DOI] [PubMed] [Google Scholar]
- Weir B.S., Cockerham C.C. Estimating F-statistics for the analysis of population structure. Evolution. 1984;38:1358–1370. doi: 10.1111/j.1558-5646.1984.tb05657.x. [DOI] [PubMed] [Google Scholar]
- Xie C., Mao X., Huang J., Ding Y., Wu J., Dong S., Kong L., Gao G., Li C.-Y., Wei L. KOBAS 2.0: a web server for annotation and identification of enriched pathways and diseases. Nucleic. Acids. Res. 2011;39:W316–W322. doi: 10.1093/nar/gkr483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J.A., Wang Z.J., Yang X., Ma M.T., Li Z.H., Nie Q.H. LncRNA-FKBP1C regulates muscle fiber type switching by affecting the stability of MYH1B. Cell Death Discov. 2021;7:73. doi: 10.1038/s41420-021-00463-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zan Y., Sheng Z., Lillie M., Rönnegård L., Honaker C.F., Siegel P.B., Carlborg Ö. Artificial selection response due to polygenic adaptation from a multilocus, multiallelic genetic architecture. Mol. Biol. Evol. 2017;34:2678–2689. doi: 10.1093/molbev/msx194. [DOI] [PubMed] [Google Scholar]
- Zeng Y.T., Wang C., Zhang Y., Xu L., Zhou G.B., Zeng C.J., Zuo Z.C., Song T.Z., Zhu Q., Yin H.D., Zhang M. Improvac immunocastration affects the development of thigh muscles but not pectoral muscles in male chickens. Poult Sci. 2020;99:5149–5157. doi: 10.1016/j.psj.2020.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C., Dong S.S., Xu J.Y., He W.M., Yang T.L. PopLDdecay: a fast and effective tool for linkage disequilibrium decay analysis based on variant call format files. Bioinformatics. 2019;35:1786–1788. doi: 10.1093/bioinformatics/bty875. [DOI] [PubMed] [Google Scholar]
- Zhang M., Han W., Tang H., Li G., Zhang M., Xu R., Liu Y., Yang T., Li W., Zou J., Wu K. Genomic diversity dynamics in conserved chicken populations are revealed by genome-wide SNPs. BMC Genomics. 2018;19:598. doi: 10.1186/s12864-018-4973-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z., Xu Z.-Q., Luo Y.-Y., Zhang H.-B., Gao N., He J.-L., Ji C.-L., Zhang D.-X., Li J.-Q., Zhang X.-Q. Whole genomic prediction of growth and carcass traits in a Chinese quality chicken population. J. Anim. Sci. 2017;95:72–80. doi: 10.2527/jas.2016.0823. [DOI] [PubMed] [Google Scholar]
- Zhou Z., Li M., Cheng H., Fan W., Yuan Z., Gao Q., Xu Y., Guo Z., Zhang Y., Hu J., Liu H., Liu D., Chen W., Zheng Z., Jiang Y., Wen Z., Liu Y., Chen H., Xie M., Zhang Q., Huang W., Wang W., Hou S., Jiang Y. An intercross population study reveals genes associated with body size and plumage color in ducks. Nat. Commun. 2018;9:2648. doi: 10.1038/s41467-018-04868-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.