Abstract
As essential regulators of mitochondrial quality control, mitochondrial dynamics and mitophagy play key roles in maintenance of metabolic health and cellular homeostasis. Here we show that knockdown of the membrane-inserted scaffolding and structural protein caveolin-1 (Cav-1) and expression of tyrosine 14 phospho-defective Cav-1 mutant (Y14F), as opposed to phospho-mimicking Y14D, altered mitochondrial morphology, and increased mitochondrial matrix mixing, mitochondrial fusion and fission dynamics as well as mitophagy in MDA-MB-231 triple negative breast cancer cells. Further, we found that interaction of Cav-1 with mitochondrial fusion/fission machinery Mitofusin 2 (Mfn2) and Dynamin related protein 1 (Drp1) was enhanced by Y14D mutant indicating Cav-1 Y14 phosphorylation prevented Mfn2 and Drp1 translocation to mitochondria. Moreover, limiting mitochondrial recruitment of Mfn2 diminished formation of the PINK1/Mfn2/Parkin complex required for initiation of mitophagy resulting in accumulation of damaged mitochondria and ROS (mtROS). Thus, these studies indicate that phospho-Cav-1 may be an important switch mechanism in cancer cell survival which could lead to novel strategies for complementing cancer therapies.
Keywords: Cav-1, Mitochondrial dynamics, Mitophagy, mtROS, Mitofusin 2, Dynamin-related protein 1
1. Introduction
Caveolin-1 (Cav-1), a primary structural protein of caveolae and key regulator of cellular signaling, has also been shown to regulate mitochondrial function and cellular energy metabolism [[1], [2], [3]]. In brown adipose tissue, reduction in Cav-1 expression lead to morphological alterations of mitochondria (C) and Cav-1 null mice show evidence of a switch to glycolysis and mitochondrial dysfunction in white adipose tissue [4]. Cav-1 also directly regulates the assembly of the mitochondrial respiratory chain. In fibroblasts, depletion of Cav-1 leads to inhibition of electron transport chain (ETC) complex I, complex IV, and complex V subunits [5]. Evidence of spatial and biochemical cooperation between Cav-1 and mitochondria indicate that Cav-1 positive vesicles identified by immunogold labeling using electron microscopy can be found around double membrane structures with crista protruding into the matrix. This result was confirmed by immunoblotting, with both the α and β isoforms of Cav-1 observed in the mitochondrial fraction [6]. Cav-1 was also enriched in the mitochondria associated membrane (MAM) compartment, where membranes from the endoplasmic reticulum (ER) and mitochondria are tethered by trypsin-sensitive filaments [7]. Cav-1 knockout mice exhibited a significant reduction in mitochondria and ER contacts in the liver [8] whereas in cancer cells, Cav-1 abrogated mitochondria-ER interactions associated with ER stress [9]. Thus, it is becoming increasingly clear that Cav-1 is an important regulator of mitochondrial structure and function, however, the role of Cav-1 in regulating mitochondrial dynamics and turnover has not been reported.
Mitochondria are highly dynamic organelles due in large part to membrane fusion and fission events which regulate mitochondrial health, turnover and function [10]. Several pathological conditions including cancer, cardiovascular disease, neurodegenerative disease, and diabetes are associated with dysregulated mitochondrial dynamics [[11], [12], [13]]. A critical balance of mitochondrial fusion and fission events must be achieved to maintain mitochondrial homeostasis [14]. Mitochondrial damage promotes mitochondrial reactive oxygen species (mtROS) production and the release of immunogenic and apoptogenic factors [15]. Mitochondrial autophagy (mitophagy) is a selective mechanism for elimination of damaged mitochondria, reduces mtROS generation and excessive inflammation, and can promote cell survival [[16], [17], [18], [19]]. Recent studies have demonstrated a negative regulatory role of Cav-1 in global cellular autophagy. For example, increased autophagy was detected in adipocytes derived from Cav-1 deficient mice [20]. Cav-1 deficiency was also shown to promote autophagosome-lysosome fusion and lysosomal function in breast cancer cells [21]. In addition, Cav-1 was found to associate with the ATG5-ATG12 complex where it played a role in suppressing autophagosome formation in lung epithelium [22] and aortic endothelium [23]. However, the role of Cav-1 in mitophagy per se has not yet been investigated. Here, we show that phosphorylated Cav-1 negatively regulates the initiation of mitophagy resulting in accumulation of damaged mitochondria, mtROS production, and cytoplasmic mtDNA accumulation in breast cancer cells.
2. Results
2.1. Mitochondrial morphology and damage control are altered by Cav-1 depletion
Mitochondrial exist in a variety of shapes and morphological states which affect functional parameters ranging from ATP and ROS generation to the regulation of cell motility and cell death [[24], [25], [26]]. Here, we silenced Cav-1 in the triple negative breast cancer cell line MDA-MB-231 using Cav-1-specific siRNA. After 48–72 h, the fluorescent mitochondrial dye tetramethyl rhodamine methylester (TMRM) was used to stain mitochondria in live cells which we visualized by confocal microscopy and quantified using Image J and Imaris software. Over 30,000 individual mitochondria were measured and classified into different groups based on length. In the absence of Cav-1, we observed a 2-fold increase in the number of fragmented mitochondria denoted by < 15 pixels in length, whereas the relative abundance of elongated mitochondria in the intermediate group (30–100 pixels) was 32% less in Cav-1 depleted cells (Fig. 1A). The average length of all 30,000 individual mitochondria analyzed decreased significantly by 15% following Cav-1 gene silencing (Fig. 1B). Therefore, Cav-1 may play a role in regulating mitochondrial morphology in breast cancer cells.
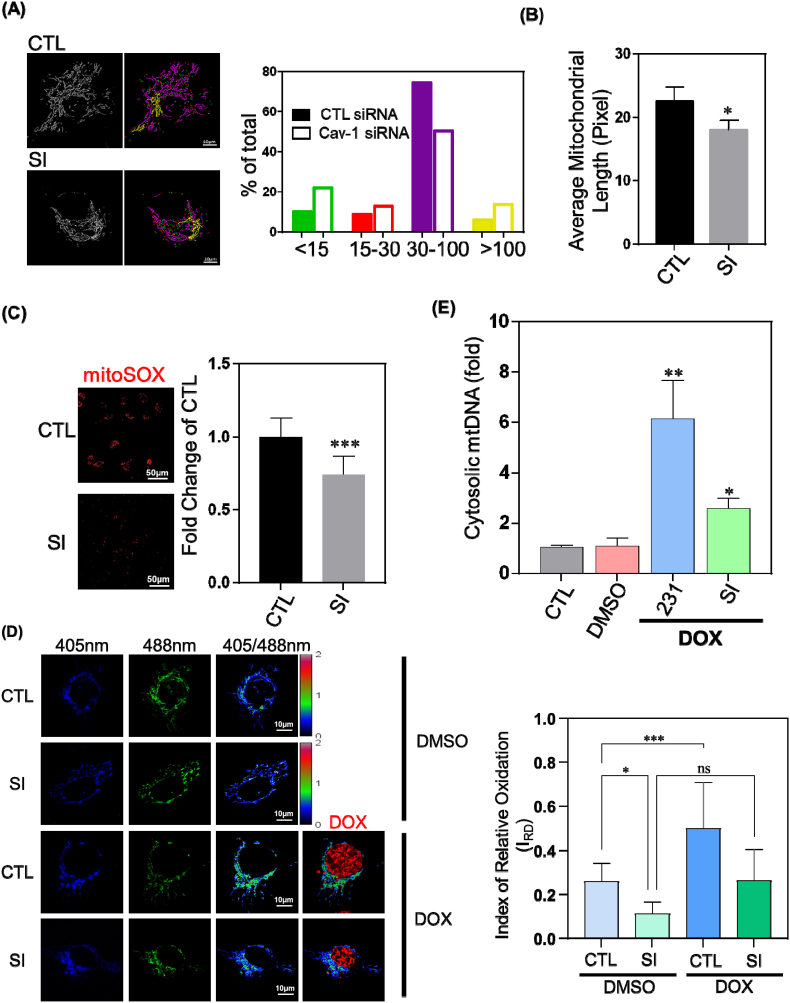
Fig. 1.
Caveolin-1 Regulates Mitochondrial Shape and Susceptibility to Damage. MDA-MB-231 cells treated with control siRNA (CTL) or Cav-1 siRNA (SI) were stained with 50 nM TMRM for 30 min or 5 μM MitoSox for 10 min at 37 °C and visualized by confocal microscopy.
(A) Over 30,000 individual mitochondria were analyzed with an ImageJ plugin and categorized into different length groups. Quantification revealed that subpopulations with the shortest and longest length both increased, whereas the number of mitochondria in the intermediate group decreased in Cav-1 depleted cells.
(B) Average length was calculated by dividing total length by the number of mitochondria. Mitochondrial length after Cav-1 knockdown was reduced by 15% compared to control. Data are mean ± SEM, n ≥ 15; *p < 0.05 vs control by t-test.
(C) Mitochondrial ROS was measured by fluorescence intensity of MitoSox. Depletion of Cav-1 reduced mitochondrial ROS by 26% as compared to control siRNA group. Data are mean ± SEM, n = 10; ***p < 0.001 vs control by t-test.
(D) Cells infected by retrovirus encoding mito-roGFP2-ORP1 were treated with 10 μM doxorubicin for 4 h and then visualized by confocal microscopy. The fluorescence ratio at 510 nm peak emission from excitation at 405 nm and 488 nm reflects mitochondrial oxidative status. Index of relative oxidation (IRO) was calculated using equation IRO = 1-(R-RH2O2)/(RDTT- RH2O2). Cells with low Cav-1 expression exhibited reduced level of mitochondrial oxidation even after challenged by doxorubicin. Data are mean ± SEM, n ≥ 10; *p < 0.05, ***p < 0.001 by ANOVA.
(E) After treating with 10 μM doxorubicin for 24 h, cytosolic fractions were collected and analyzed by RT-PCR. Bar graph summarizes the fold change of mtDNA in the cytosol normalized to control. Cav-1 inhibition diminished mitochondrial damage induced by doxorubicin by over 50%. Data are mean ± SEM, n = 3–5; *p < 0.05, **p < 0.01 vs control by ANOVA.
Mitochondrial damage leads to accumulation of mtDNA [27] in the cytosol and enhanced mtROS production [28], thus mtROS and cytosolic mtDNA level reflect the extent of mitochondrial damage. Using the live-cell mitochondria-targeted fluorogenic dye MitoSOX Red, whose fluorescence increases upon oxidation by superoxide and binding to nucleic acid [29], we found that Cav-1 depletion using siRNA reduced mitoSOX fluorescence, suggesting there may be less mtROS generation (Fig. 1C). To clarify whether Cav1 siRNA decreased mtROS production, mitochondria-targeted (mt) redox biosensor mt-roGFP2-Orp1 was used to generate a specific ratiometric index of mitochondrial redox status [30,31]. mt-roGFP2-Orp1 is a well-established biosensor whose fluorescence directly and quantifiably relates to mitochondrial ROS levels. To verify the mitochondrial targeting property of the biosensor, we co-labeled cells expressing mt-roGFP2-Orp1 with TMRM. The colocalization of biosensor and TMRM confirmed mitochondrial targeting (Fig. S1A). mt-roGFP2-Orp1 has two excitation peaks (∼400 nm and ∼480 nm) and a maximum absorption at 510 nm. Oxidation increases fluorescence at 400 nm excitation whereas reduction increases fluorescence at 480 nm excitation. Therefore, the ratio of fluorescence emitted from alternating 405 nm and 488 nm excitation reflects the redox status of the biosensor's environment [32], which in the case of mt-roGPF2-Orp1 is the mitochondria.
To establish the fluorescence range of mt-roGPF2-Orp1, cells were treated with 3 mM DTT or 5 mM H2O2 for 20 min to obtain min-max readings of fully reduced (normalized to 0) or oxidized (normalized to 1) mitochondria (Fig. S1B). Consistent with the results observed with mitoSOX, Cav-1 knockdown significantly reduced mtROS production. Upon challenge with the chemotherapeutic drug doxorubicin to induce mitochondrial damage and mimic cancer chemotherapy, as shown in Fig. 1D, mtROS production dramatically increased in cells treated with control siRNA whereas the increase was not significant in Cav-1 depleted cells suggesting Cav-1 depletion suppresses mtROS production. We also assessed accumulation of cytosolic mtDNA following exposure to doxorubicin and observed less cytosolic mtDNA in Cav-1 depleted breast cancer cells suggesting cells with reduced Cav-1 expression have a greater capability of repairing mitochondrial damage (Fig. 1E).
2.2. Mitochondrial dynamics are reduced by Cav-1 expression
To explore the cause of altered mitochondrial shape and reduced mitochondrial damage in Cav-1 depleted cells, we first assessed mitochondrial dynamics which strongly impact mitochondrial shape and quality control functions. The frequency of mitochondrial fission and fusion events is a direct indicator of mitochondrial dynamics. Therefore, we conducted time-lapse recordings of 3-dimensional (3D) segments of mitochondria for 200s by live-cell confocal microscopy in control, Cav-1 siRNA treated (SI), and Cav-1 adenovirus (AD) infected MDA-MB-231 cells following siRNA mediated depletion (rescue protocol). We then measured the number of fusion and fission events per minute. As shown in Fig. 2A, the number of fusion and fission events increased following Cav-1 depletion. We also tracked the kinetics of mitochondria movement (Fig. 2B) and observed enhanced mitochondrial velocity in Cav-1 depleted cells. Taken together, these data indicate that reduction in Cav-1 expression in breast cancer cells increases mitochondrial fission/fusion dynamics.
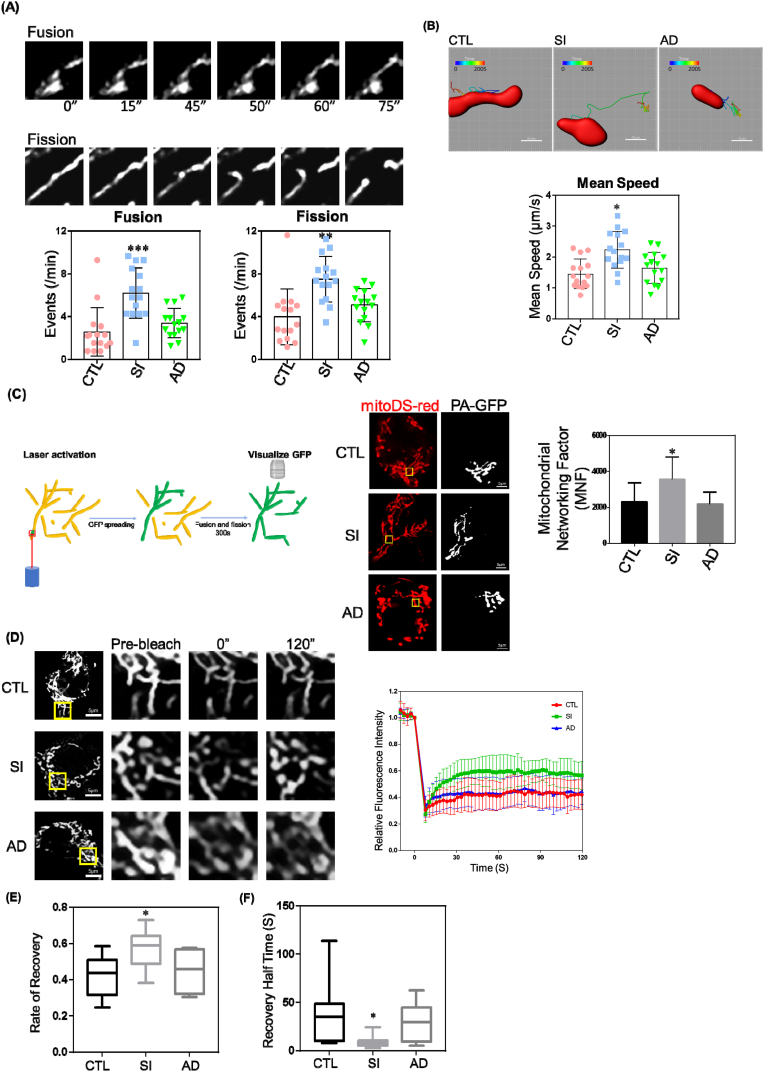
Fig. 2.
Depletion of Cav-1 Increases Mitochondrial Dynamics. MDA-MB-231 cells treated with control siRNA (CTL), Cav-1 siRNA (SI), or Cav-1 adenovirus (AD) to rescue Cav-1 expression for 48–72 h were stained with 50 nM TMRM for 30 min at 37 °C and visualized by confocal microscopy.
(A) Mitochondrial fusion and fission events per minute were increased in Cav-1 depleted cells (SI) as compared to control and Ad-Cav-1 rescue group.
(B) Mean speed of mitochondrial movement was quantified by Imaris software. The motility of mitochondria in Cav-1 depleted cells increased significantly. Data are mean ± SEM, n = 15; *p < 0.05, **p < 0.01, ***p < 0.001 vs control by ANOVA.
(C) CTL, SI and AD cells were co-transfected with mito-Ds-Red and mito-Photoactive (PA)-GFP for 48 h. Live cells were visualized by confocal microscopy. Mitochondrial Networking Factor (MNF) which represents the area of photoactivated GFP spreading was calculated using ImageJ. The loss of Cav-1 enhanced MNF demonstrating wider spreading of activated GFP which was inhibited by rescuing Cav-1 expression.
(D) CTL, SI and AD cells were transfected with mito-Ds-Red for 48 h. Live cells were visualized by confocal microscopy. Fluorescence recovery after photobleaching (FRAP) performed on cells after Cav-1 knockdown ± Ad-Cav-1 rescue were transfected with mito-DS-Red. Mean fluorescence recovery curves after photobleaching region of interest (ROI) normalized to the last prebleach image.
(E) Quantification of fluorescence recovery rate revealed greater fluorescent signal recovery in Cav-1 depleted cells.
(F) Quantification of recovery half time revealed faster fluorescent signal recovery in Cav-1 depleted cells. Data are mean ± SEM, n ≥ 10; *p < 0.05, **p < 0.01, ***p < 0.001 vs control by ANOVA. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
Continuous fission and fusion of mitochondria enables matrix mixing and this exchange ensures mitochondrial DNA stability and maintenance of mitochondrial health [28]; vice versa, the degree of matrix mixing appropriately reflects mitochondrial fusion and fission dynamics. Therefore, we assessed the level of mitochondrial matrix mixing by calculating mitochondrial networking factor (MNF), an index of photoactivated GFP (PA-GFP) dispersion within the mitochondrial network [33]. In this experiment, PA-GFP was activated in a small region of the mitochondrial network which was then tracked as it spread-out from the photoactivated region and mixed with the matrix of connected regions over 5 min. We calculated the area of PA-GFP fluorescence and defined this as the MNF. Mito-Ds-Red (used to visualize all mitochondria) and mito-PA-GFP (activated by a focused laser) were co-transfected into control and Cav-1 siRNA treated cells with or without Cav-1 adenovirus to rescue expression in MDA-MB-231 cells. As shown in Fig. 2C, MNF increased by nearly 35% in cells depleted of Cav-1 (SI). Moreover, rescue of Cav-1 expression with Cav-1 adenovirus (AD) after siRNA depletion prevented the increase in MNF.
Next, we used fluorescence recovery after photobleaching (FRAP) to study the velocity of protein recovery in the mitochondria. After photobleaching, the fluorescence signal within the bleached region was quenched and recovery of the fluorescence signal was achieved by diffusion of “in-network” GFP into the bleached site. The degree and rate of fluorescence recovery represents matrix protein motility, and mitochondria-associated protein motility relates to mitochondrial fusion and fission dynamics [34]. Cells depleted of Cav-1 had a faster and more intense fluorescence recovery, which was abrogated by rescue of Cav-1 expression (Fig. 2D–F). Elevated MNF and increased FRAP following knockdown of Cav-1 suggests that Cav-1 is a negative regulator of mitochondrial dynamics and network formation.
2.3. Cav-1 deficiency induces mitophagy
While studying mitochondrial morphological changes, we noticed a loss of mitochondrial mass in Cav-1 depleted MDA-MB-231 cells (Fig. 3A) suggesting a reduction in Cav-1 expression may promote mitophagy. Mitophagy is the selectively autophagic process of degrading damaged mitochondria. Thus, we analyzed the expression of autophagosome maker LC3-II. Consistently, LC3-II expression was elevated after Cav-1 downregulation and restored by Cav-1 rescue (Fig. S2) indicating that Cav-1 negatively regulates autophagy.
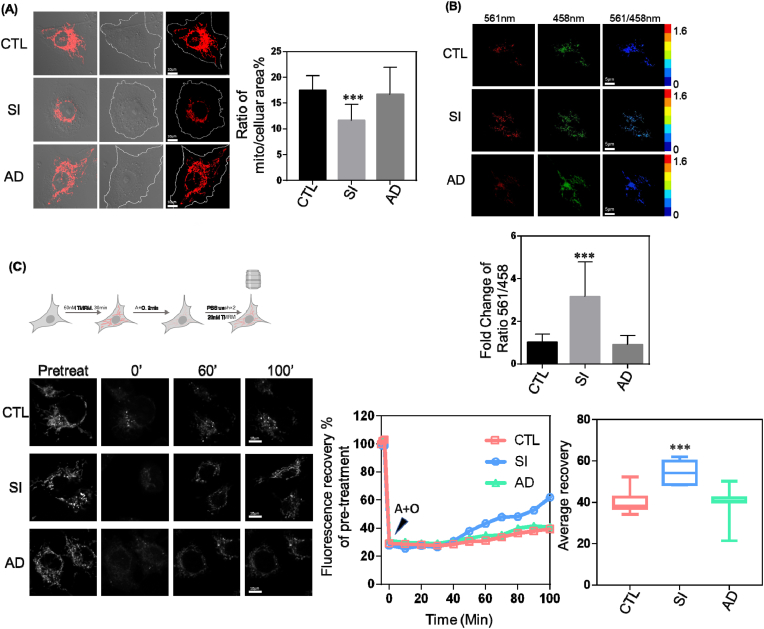
Fig. 3.
Mitophagy is Enhanced by Reduced Cav-1 Expression. MDA-MB-231 cells treated with control siRNA (CTL), Cav-1 siRNA (SI), or Cav-1 adenovirus (AD) to rescue Cav-1 expression for 48–72 h were stained with 50 nM TMRM for 30 min at 37 °C and visualized by confocal microscopy.
(A) Quantification of TMRM revealed a 35% reduction in mitochondrial mass in Cav-1 siRNA treated cells.
(B) CTL, SI and AD cells transfected with mito-Keima for 48 h were visualized by confocal microscopy. The mito-Keima signal was captured by alternating 458 nm and 561 nm excitation and emitted fluorescence expressed as a ratio of 561/458 was used to reflect mitophagy. Bar graph depicts the fold change in 561/458 fluorescence relative to control. Cav-1 SI induced a 3-fold increase in the 561/458 fluorescence ratio compared with control and Cav-1 reconstituted cells.
(C) The fluorescence signal of TMRM recovered by 60% after 4 μM antimycin and 10 μM oligomycin insult as compared to 40% recovery in control and Ad-Cav-1 rescued cells. Data are mean ± SEM, n ≥ 10; ***p < 0.001 vs control by ANOVA.
To directly visualize mitophagy, we used mito-Keima, a pH-dependent fluorescent reporter obtained from coral which allows for the rapid determination and quantitative assessment of mitochondria engulfment by the autophagosome, as this results in a change in emission maxima from 458 nm (neutral pH) to 561 nm (acidic) [[35], [36], [37]]. As shown in Fig. 3B, the 561/458 nm ratio was about three times higher in Cav-1 depleted cells which was fully reversed by reconstitution of Cav-1 expression with Ad-Cav-1 suggesting augmentation of mitophagy upon Cav-1 knockdown.
TMRM is a cell-permeant fluorescent dye which is readily sequestered and fluoresces brightly in healthy mitochondria with normal mitochondrial membrane potential (MMP) but becomes dim when mitochondria are damaged and MMP decreases. Therefore, we monitored TMRM fluorescence recovery after insult to live cells to assess mitochondrial repair processes. Since mitophagy removes damaged mitochondria and thereby promotes healthy mitochondrial networks, cells with active mitophagy should recover faster after induction of mitochondrial damage. In this study, cells labeled with 50 nM TMRM were treated with 4 μM antimycin and 10 μM oligomycin for 2 min to induce mitochondrial damage and then washed twice with PBS and incubated in complete media containing 20 nM TMRM while imaging. After antimycin and oligomycin treatment, the fluorescent signal in all three groups (control, Cav-1 knockdown, and rescue) went down. About 30 min later, the TMRM signal of Cav-1 knockdown cells started increasing while the other two groups exhibited minor changes. After 100 min, the fluorescence signal in the Cav-1 knockdown group had recovered to 60% of the starting value which was significantly higher than control and Cav-1 reconstituted cells (Fig. 3C). Therefore, these results suggest that Cav-1 is a critical regulator of mitophagy and mitochondrial damage control.
2.4. Cav-1 regulation of mitochondrial dynamics via interaction with key regulators of mitochondrial fission and fusion events
To determine the underlying molecular mechanism by which Cav-1 regulates mitochondrial morphological dynamics and mitophagy, we examined Cav-1 interactions with the primary regulators of mitochondrial fusion and fission, namely Mitofusin 2 (Mfn2) and Dynamin-related protein 1 (Drp1) [11]. Mitochondria and other membrane fractions were isolated for determination of Drp1 and Mfn2 protein levels in each fraction by Western blotting. Quantification of Western blots indicated that the level of Mfn2 increased by 60% and Drp1 by 30% in the mitochondria fraction of Cav-1 depleted cells, while the amount of both proteins present in the membrane fraction significantly decreased (Fig. 4A). Upon rescue of Cav-1 expression, redistribution of Mfn2 and Drp-1 was observed with recovery of the original distribution of both Mfn2 and Drp-1 between the mitochondria and membrane fractions. We also observed that both Mfn2 and Drp-1 co-immunoprecipitated with Cav-1 (Fig. S3) suggesting a potential close interaction. To further verify close interaction between Cav-1 and either Mfn2 or Drp-1, we used Fluorescence Resonance Energy Transfer (FRET) to detect fluorescently-tagged molecules within a proximity of 10-100 Å. Mfn2-YFP, Drp1-YFP and Cav1-CFP were transfected into HEK cells which have very low endogenous Cav-1 expression. In each experiment, the fluorescence intensity of CFP was recorded before and after YFP photobleaching in fixed samples to measure FRET efficiency. As shown in Fig. 4B, there was 25% and 16% FRET efficiency for Mfn2-YFP/Cav-1-CFP and Drp1-YFP/Cav-1-CFP pairs respectively, suggesting a close interaction between Cav-1 with Mfn2 and Drp-1. Therefore, Cav-1 closely interacts with and regulates the function of Mfn2 and Drp1 fission and fusion machinery proteins.
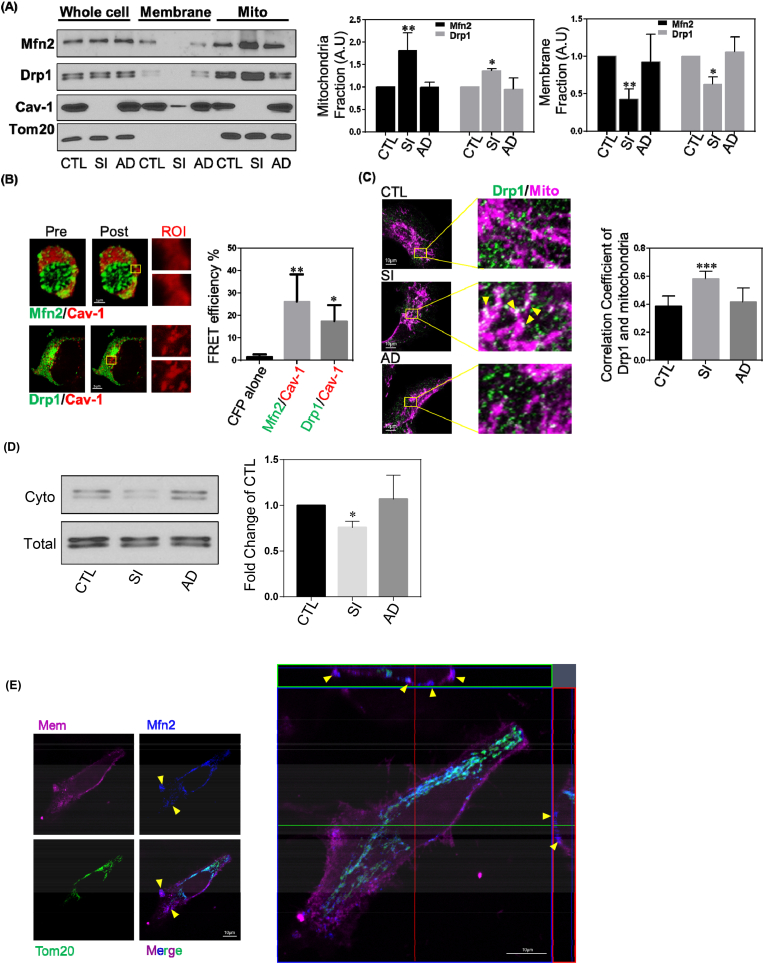
Fig. 4.
Cav-1 Binds to Mfn2 and Drp1.
(A) Representative images of Western blots of MDA-MB-231 cells treated with Cav-1 siRNA with of without rescue with Cav-1 adenovirus. Membrane and mitochondria fractions were isolated and relative band intensities (densitometry) were determined by ImageJ. Bar graph depicts fold-change in Mfn2 and Drp1 levels in the whole cell lysates, membrane and mitochondrial fractions. Without a change in whole cell expression, Cav-1 depletion increased Mfn2 and Drp1 levels in the mitochondrial fraction. Data are mean ± SEM, n ≥ 3; **p < 0.01, *p < 0.05 vs control by ANOVA.
(B) HEK cells co-transfected with Cav-1-CFP and Mfn2-YFP or Drp1-YFP were fixed and imaged by confocal microscopy. FRET intensity was calculated from CFP signal induced by 458 nm excitation after photobleaching YFP. Representative images depicting increase in CFP intensity after photobleaching region of interest. Bar graph summary of FRET efficiency of Mfn2-YFP and Drp1-YFP with Cav-1-CFP. Data are the mean ± SEM, n = 6; **p < 0.01, *p < 0.05 vs CFP alone by ANOVA.
(C) MDA-MB-231 cells treated with control or Cav-1 siRNA, with and without rescue with Cav-1 adenovirus, were fixed and immunostained for Tom20 and Drp1. Quantification of Pearson's correlation coefficient of co-localized Drp1 and mitochondria labels are from over 50 cells. Data are mean ± SEM, n ≥ 10; *p < 0.05, ***p < 0.001 vs control by ANOVA.
(D) Cytosolic fractions versus total cell lysates were used for Drp1 Western blot. In Cav-1 repleted cells, the cytosolic Drp1 level decreased. Data are mean ± SEM, n ≥ 3; *p < 0.05 vs control by ANOVA
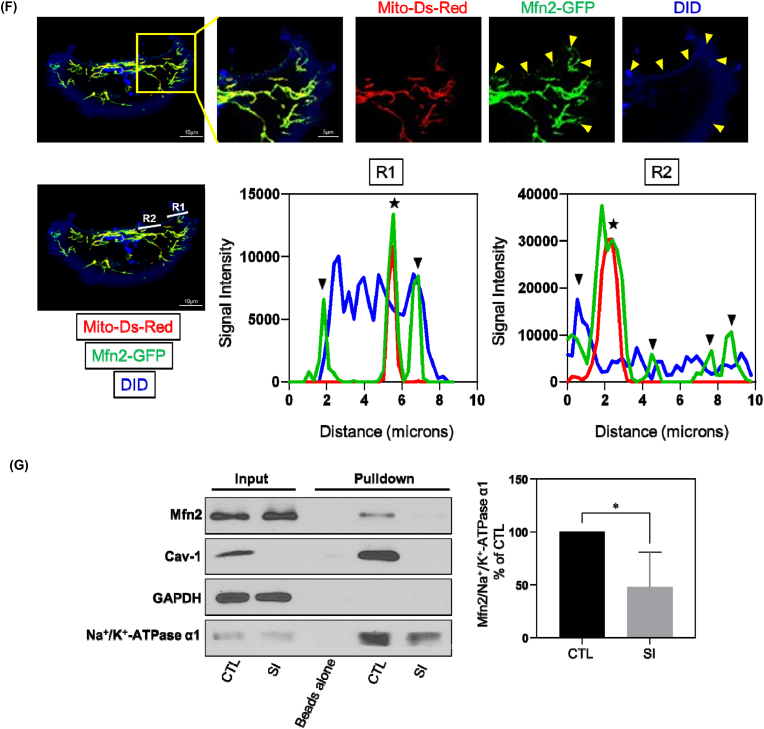
(E) MDA-MB-231 cells stained with MemBrite® Fix 568 were washed and fixed with 4% PFA for 20 min at RT, and then immunostained for Mfn2 and Tom20. 35 optical sections of 0.15 μm were used to reconstruct 3D images. The yellow arrow heads represent non-mitochondrial Mfn2 colocalizing with plasma membrane fluorescent label. The orthogonal view revealed spatial correlation of Mfn2 and plasma membrane fluorescence.
(F) MDA-MB-231 cells transfected with Mfn2-GFP and mito-Ds-Red were stained with DID and visualized by confocal microscopy. The yellow arrow heads in the representative image denote non-mitochondrial Mfn2 colocalizing with the plasma membrane. The white lines, Region1 (R1) and Region2 (R2), indicated the line scan of Mfn2-GFP, mito-Ds-Red and DID fluorescence was analyzed using ImageJ, respectively. The black arrow heads signify non-mitochondrial Mfn2-GFP colocalizing with plasma membrane and stars signify Mfn2-GFP colocalizing with mitochondria.
(G) MDA-MB-231 cells treated with control or Cav-1 siRNA were incubated with EZ-Link™ Sulfo–NHS–SS-Biotin for 45 min on ice and then lysed. The plasma membrane proteins labeled with biotin were captured by Streptavidin-coupled Dynabeads. Samples were then subjected to Western Blot analysis. GAPDH, a cytoplasmic maker, was not found in biotin surface membrane pulldown fraction suggesting there's no cytoplasmic contamination. Bar graph depicts fold-change of Mfn2 detected in biotin labeled plasma membrane fraction relative to membrane inserted Na+/K+-ATPase α1 subunit which revealed a decrease in plasma membrane-localized Mfn2 after Cav-1 depletion. Data are mean ± SEM, n = 4; *p < 0.05 vs control by Student's t-test. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
Furthermore, as shown in Fig. 4C, immunostaining of Drp-1 in Cav-1 siRNA treated cells revealed greater colocalization of Drp-1 and mitochondrial membrane protein Tom20 than in control siRNA treated cells. Since Drp-1 is a cytosolic protein [38], we assessed its level in the cytosol and found a reduction in cytoplasmic Drp-1 levels in Cav-1 depleted cells (Fig. 4D). Taken together, these data strongly suggest that Cav-1 impedes Drp-1 translocation from the cytosol to the mitochondrial membrane.
Mfn2 localized at the mitochondrial outer membrane is a primary mediator of mitochondrial fusion events. Moreover, Mfn2 is also found at the plasma membrane where it serves a non-canonical function as a stabilizer of endothelial adherens junctions (AJs) [39]. To investigate if Mfn2 localizes to the plasma membrane in breast cancer MDA-MB-231 cells, we labeled cells with the plasma membrane dye MemBrite® Fix, which accumulates in the plasma membrane and withstands fixation. From 3-D reconstructions of Z-stack images, in addition to endogenous Mfn2 immunostaining colocalized with mitochondria label Tom20, Mfn2 was also observed at the plasma membrane (Fig. 4E). Additionally, we also co-transfected GFP-tagged Mfn2 and mito-Ds-Red, and then stained cells with lipophilic tracker DID, a fluorescent dye retained in the lipid bilayer of membranes. As shown in Fig. 4F, colocalization of Mfn2-GFP and DID was also observed. Therefore, Mfn2 localizes to the plasma membrane in MDA-MB-231 breast cancer cells.
Next, we evaluated whether Cav-1 regulates the ability of Mfn2 at the plasma membrane to translocate to the mitochondrial membrane. We conducted biotin-streptavidin pulldown of plasma membrane proteins in control and Cav-1 siRNA treated cells. EZ-Link™ Sulfo–NHS–SS-Biotin, which labels cell surface primary amines, was applied to cell cultures which were then lysed and incubated with streptavidin-coupled Dynabeads to capture biotin labeled plasma membrane proteins. Western blot analysis revealed a decrease in the level of Mfn2 in plasma membrane fractions after Cav-1 knockdown (Fig. 4G). In combination with the observation of increased mitochondrial Mfn2 after Cav-1 depletion, these data support the conclusion that Cav-1 negatively regulates Mfn2 trafficking from the plasma membrane to mitochondria.
To determine if Cav-1 regulates mitochondrial dynamics through associating with Mfn2 and Drp1, we simultaneously down-regulated the expression of Cav-1 and either Mfn2 or Drp1 (Fig. S4A), and then measured the morphology and dynamics of mitochondria. Compared to Mfn2 depleted cells, the average mitochondrial length of cells treated with both Mfn2 and Cav-1 siRNA was significantly reduced which indicates a greater level of mitochondrial fission. Similarly, a higher level of mitochondrial fusion was found in Drp1 and Cav-1 double-depleted cells (Fig. S4B) suggesting Cav-1 regulates both mitochondrial fusion and fission events by sequestering Mfn2 and Drp1. We further quantified the number of mitochondrial fusion and fission events per minute after double knockdown (Fig. S4C). These results show that fusion is increased in Cav-1 depleted cells in the absence of Drp1, and likewise, fission was increased in Cav-1 and Mfn2 double-depleted cells. Additionally, fusion dynamics did not change upon Cav-1 and Mfn2 double knockdown compared to Mfn2 knockdown alone suggesting the role of Cav-1 in regulation of fusion is through Mfn2. Analysis of fission dynamics similarly showed no change upon Cav-1 and Drp1 double knockdown compared to Drp1 depletion alone which implies Cav-1 regulates fission by sequestering Drp1.
2.5. Cav-1 regulates mitochondrial dynamics in a phosphorylation-dependent manner
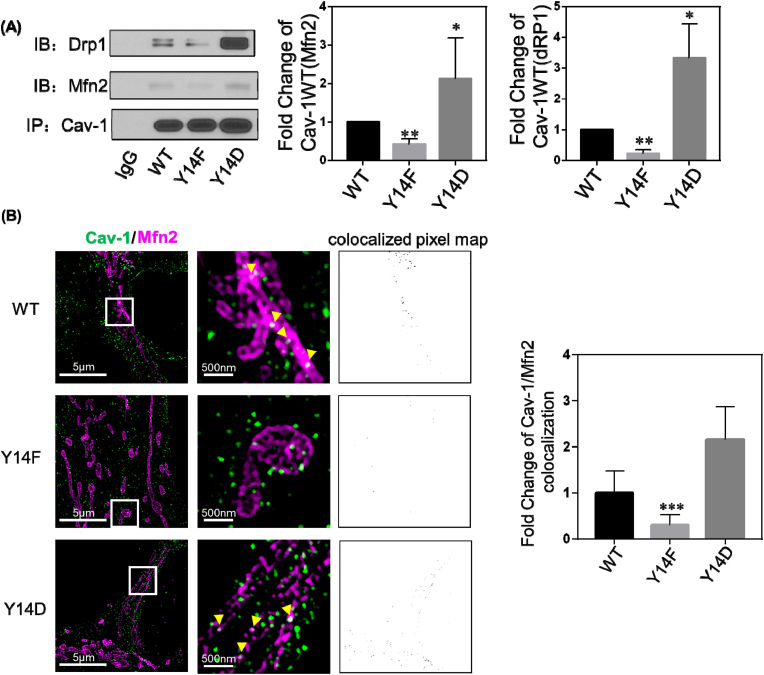
Cav-1 binds to multiple signaling molecules through its scaffolding domain (CSD), and phosphorylation of Cav-1 on tyrosine-14 has been shown to facilitate the binding of various cellular proteins to the CSD [40]. We therefore investigated whether interactions between Cav-1 and Mfn2/Drp1 are phosphorylation-dependent. WT, phospho-mimicking Y14D, and phospho-defective Y14F Cav1 mutants were expressed in MCF-7 cells in which endogenous Cav-1 expression is very low, and then interaction between Cav-1 and Mfn-2 and Drp-1 were assessed by immunoprecipitation and Western blotting. As shown in Fig. 5A, co-IP of Mfn-2 and Drp-1 with Cav-1 was enhanced by the phospho-mimicking Y14D mutant and reduced by phospho-defective Y14F mutant indicative of a phosphorylation-dependent interaction.
Fig. 5.
Cav-1 Phosphorylation Status Affects its Association with Mfn2 and Drp1.
(A) MCF-7 cells transfected with Cav-1 wildtype (WT), Y14F, and Y14D mutants were lysed after 48 h and immunoprecipitated with anti-Cav-1 antibody. The pellets were washed and blotted for Cav-1, Drp1, and Mfn2. Bar graph depicts fold-change in interaction with Cav-1. Data are mean ± SEM, n = 3; *p < 0.05 vs wildtype by ANOVA.
(B) Cav-1 depleted MDA231 cells were co-transfected with Cav-1 WT, Y14F, Y14D and Mfn2-myc cDNA, fixed, and immunostained for Cav-1 and myc. Images were acquired with a DeltaVision OMX 3D-SIM system. Bar graph summarizing colocalization of Cav-1 and Mfn2 indicates inhibition of colocalization in Y14F mutant expressing cells. Data are mean ± SEM, n = 10; ***p < 0.001 vs WT by ANOVA.
Three-dimensional structured illumination microscopy (3D‐SIM) was next used to visualize Cav-1 mutants and Mfn2. 3D‐SIM super-resolution of a predetermined illumination pattern can reveal fine structural details with a resolution of approximately 50 nm in the x–y plane and 125 nm in the z plane [41]. By analyzing 3D - SIM images of two fluorophores, we found that the phospho-defective Y14F mutant exhibited significantly reduced association with Mfn2, and that the opposite was true for phospho-mimicking Y14D mutant as compared to WT-Cav-1 (Fig. 5B). This result is consistent with co-immunoprecipitation data and re-iterates the critical role of Cav-1 Y14 phosphorylation in negative regulation of mitochondrial fusion protein Mfn2.
To assess whether Cav-1 binding to Mfn2 and Drp-1 affects mitochondrial function, we next assessed mitophagy using mito-Keima. As shown in Fig. S5A, the phosphor-defective Y14F Cav-1 mutant reduced the 561/458 nm ratio of mito-Keima, whereas phospho-mimicking Y14D-Cav-1 dramatically increased mitophagy indicating the negative regulation of Cav-1 on mitophagy may be via phosphorylation-dependent sequestration of Mfn2. Mitochondrial dynamics were also evaluated in cells expressing the Y14 Cav-1 phospho-mutants. Cells transfected with Y14F exhibited greater frequency of fusion and fission events (Fig. S5B), velocity of mitochondrial movement (Fig. S5C), and mitochondrial protein mobility as measured by FRAP (Figs. S5D–E) as compared to cells expressing WT and Y14D Cav-1 mutant. Therefore, these data further indicate that Cav-1 directly associates with mitochondrial fusion and fission machinery Mfn2 and Drp1 in a phosphorylation dependent manner and that this association negatively regulates mitophagy and mitochondrial fission/fusion and trafficking dynamics.
2.6. Inhibition of PINK1/Parkin mitophagy pathway by Cav-1/Mfn2 interaction
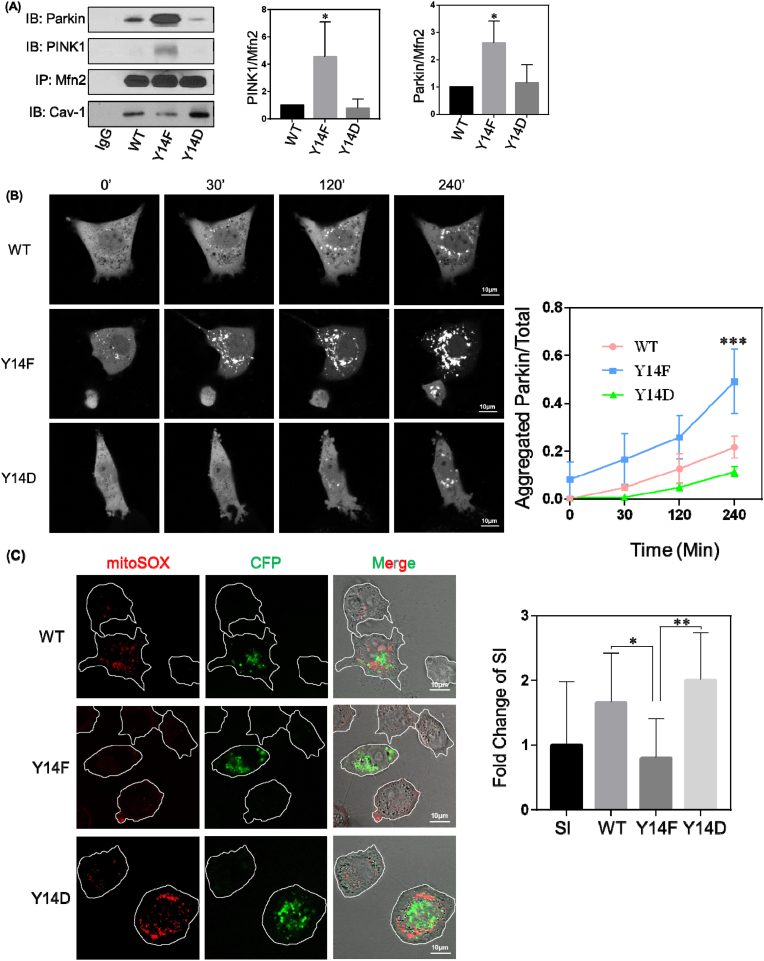
Mfn2 plays a critical role in the PINK1/Parkin mitophagy signaling pathway as it is recruited to and phosphorylated by PINK1 and then subsequently recruits and activates the E3 ubiquitin (Ub) ligase Parkin to initiate mitophagy [42,43]. Therefore, we next investigated association of Mfn2 with PINK1 and Parkin by immunoprecipitation following downregulation of Cav-1 expression. As shown in Fig. S6A, association of Mfn2 with PINK1 and Parkin was amplified in Cav-1 depleted cells which suggests that by interacting with Mfn2, Cav-1 inhibits Mfn2 interaction with PINK1 and Parkin. Co-immunoprecipitation studies further revealed that Cav-1 Y14 phosphorylation plays a critical role in negatively regulating the formation of the Mfn2/PINK1/Parkin complex (Fig. 6A). These results support the concept that Cav-1, in a phosphorylation-dependent manner, restricts mitophagy by preventing mitochondrial recruitment of Mfn2 and its subsequent association with PINK1 and Parkin. Moreover, Cav-1-depleted MDA-MB-231 cells reconstituted with Cav-1 mutants followed by treatment with antimycin (4 μM) and oligomycin (10 μM) for 4 h to induce mitophagy revealed an increase in PINK1 expression at 24 h independent of Cav-1 phosphorylation status (Fig. S6B). Parkin expression remained the same after induction of mitophagy in the various Cav-1 reconstituted groups, however, Parkin-GFP fluorescence was observed as aggregates after induction of mitophagy, presumably around mitochondria (Fig. 6B). Furthermore, Y14F Cav-1 mutant expressing cells exhibited greater and faster Parkin aggregation around mitochondria suggesting that Cav-1 phosphorylation blocks Parkin recruitment without influencing Parkin and PINK1 expression levels.
Fig. 6.
Association of Mfn2 with PINK1/Parkin Mitophagy Pathway Regulators is Reduced by Phosphorylated Cav-1.
(A) Cav-1 depleted MDA-MB-231 cells were transfected with Cav-1 wildtype (WT), Y14F, and Y14D mutants, and after 48 h lysed and immunoprecipitated with anti-Mfn2 monoclonal antibody. Bar graph depicts increase in PINK1 and Parkin binding to non-phosphorylated Cav-1 relative to WT and Y14D Cav-1. Data are mean ± SEM, n = 3; *p < 0.05 vs WT by ANOVA.
(B) Cav-1 depleted MDA-MB-231 cells were co-transfected with Cav-1 WT, Y14F, Y14D mutants and Parkin-GFP. After 48 h, cells were treated with 4 μM antimycin and 10 μM oligomycin for up to 4 h and Parkin localization was visualized by confocal microscopy. The ratio of aggregated fluorescent signal relative to total GFP fluorescence was calculated to assess mitochondrial recruitment of Parkin over time. Cells expressing Cav-1 phosphorylation deficient Y14F mutant showed greater aggregated Parkin indicating increased mitochondrial recruitment. Data are mean ± SEM, n ≥ 6; ***p < 0.001 vs WT by ANOVA.
(C) Representative images of MitoSOX staining in Cav-1 depleted cells which were transfected with Cav-1 WT, Y14F or Y14D CFP-tagged mutants. Bar graph depicts fold change of fluorescence intensity normalized to non-transfected Cav-1 depleted cells which were used as internal controls. Data are mean ± SEM, 12 ≤ n ≤ 15; *p < 0.05, **p < 0.01 by ANOVA.
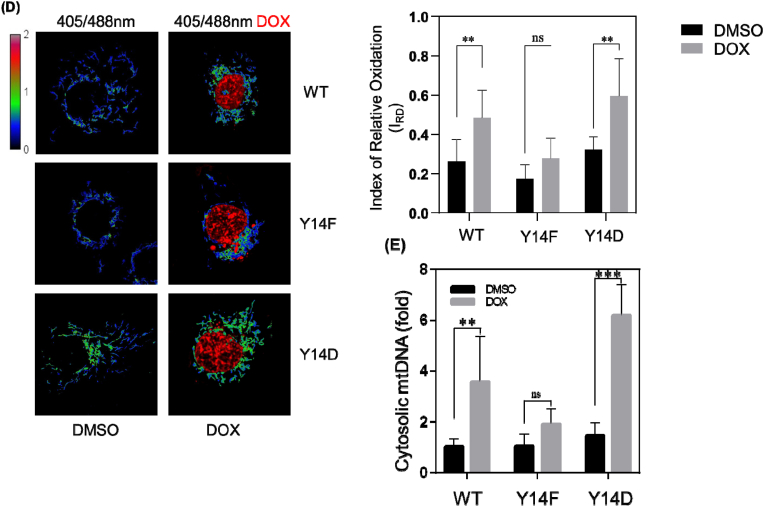
(D) Cav-1 depleted cells expressing mito-roGFP2-ORP1 were transfected with Cav-1 mutants and then treated with DMSO or 10 μM doxorubicin for 4 h. Mitochondrial oxidation was measured based on the ratio of emitted roGFP fluorescence at alternating 405 vs 488 nm excitation. Y14F mutant inhibited mtROS production whereas WT and Y14D increased mtROS particularly after doxorubicin treatment. Data are mean ± SEM, n = 10; **p < 0.01 by ANOVA.
(E) Cav-1 mutant expressing cells were treated with DMSO or 10 μM doxorubicin for 24 h. Cytosolic fractions were then collected and analyzed by RT-PCR. Bar graph summarizes the fold change in mtDNA in the cytosol normalized to WT Cav-1 expressing cells treated with DMSO. Mitochondrial DNA in the cytosol was significantly increased by Y14D mutant suggesting phosphorylation of Cav-1 inhibits mitochondrial repair. Data are mean ± SEM, n = 3; **p < 0.01, ***p < 0.001 by ANOVA. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
To assess the role of phospho-Cav-1 in mitophagy-mediated clearance of damaged mitochondria, we investigated mtROS and cytosolic mtDNA levels in Cav-1-depleted MDA-MB-231 cells reconstituted with CFP-tagged Cav-1 mutants. Live cells were then loaded with MitoSOX to assess mitochondrial damage as indicated by red fluorescence intensity in CFP positive and negative cells (non-transfected cells were quantified as internal controls). As shown in Fig. 6C, Y14F Cav-1 mutant significantly reduced mitoSOX fluorescence as compared to that in cells reconstituted with WT or phospho-mimicking Cav-1 mutant. Furthermore, the ratio of emitted mt-roGFP2-Orp1 fluorescence at alternating excitation wavelengths of 405 and 488 nm revealed a significant increase in mitochondrial oxidation in cells expressing WT and Y14D Cav-1 as compared to Y14F Cav-1 (Fig. 6D). In addition, cytosolic mtDNA, which reflects mitochondrial damage level, was reduced in Y14F Cav-1 mutant expressing cells, particularly after doxorubicin treatment (Fig. 6E). The inhibitory effect of phospho-defective Y14F Cav-1 mutant on mtROS and cytosolic mtDNA accumulation suggests that phosphorylation of Cav-1 negatively regulates repair of mitochondrial damaged.
3. Discussion
Mitochondrial health is central to the cell fate decision to activate apoptosis and necrosis cell death programs, and therefore quality control is a critical regulator of cell survival. Targeting mitochondrial quality control may also be an effective approach for maximizing cell killing by cancer therapeutics. There are several mechanisms that have evolved for mitochondrial quality control among which mitochondrial fission/fusion dynamics and mitophagy are fundamentally important [44]. Mitochondrial dynamics, narratively speaking, include fusion and fission of mitochondria. Mitochondrial fusion promotes the mixing of mitochondrial matrix as well as mitochondrial outer and inner membrane proteins to facilitate exchange of material and ATP production [45]. Mitochondrial fission, on the other hand, divides mitochondrial tubules and generates shortened and more mobile and isolated mitochondria which can translocate to other areas of the cell or fuse with other mitochondrial tubules. Thus, mitochondrial fusion and fission together repair damaged mitochondria to ensure mitochondrial quality by segregating damaged components via fission and exchanging materials between healthy mitochondria via fusion [46]. Moreover, fission helps to isolate damaged segments of mitochondria from the mitochondrial network for the purpose of their selective removal from the cell by mitophagy. Thus, drugs that alter mitochondrial fusion and fission dynamics and mitophagy could promote the accumulation of damaged mitochondria and thereby prevent cell survival as a means of improving cancer therapy.
Although mitochondria are unique organelles which maintain the expression of independent genomic and transcriptomic machinery from their endosymbiotic host cell, they also maintain close interaction and exchange contents with other organelles as well as the rest of the cell to maintain levels of critical ions, metabolites, proteins and lipids [47]. Here, we show for the first time that the plasma membrane protein caveolin-1 (Cav-1) is a pivotal regulator of mitochondrial dynamics and mitophagy and therefore may be a critical determinant of mitochondrial health and damage control.
First, we found that there was a change in mitochondrial morphology in Cav-1 depleted cells. Mitochondria exist in various shapes and sizes ranging from short spheres to a hyperfused reticulum [28]. Shape per se can affect the kinetics of biochemical reactions and formation of protein complexes in the mitochondrial matrix [26]. It can also be linked with motility and ROS or ATP production. Generally, short mitochondria are more mobile but exhibit a higher level of mtROS and reduced ATP production [48]. In breast cancer cells with reduced Cav-1 expression, the average mitochondrial length was 15% shorter and there was an increase in the total number of mitochondria that were less than 30 pixels in length suggesting that a reduction in Cav-1 expression leads to a more fragmented mitochondrial network. Upon further evaluation of the influence of Cav-1 on mitochondrial fusion and fission dynamics, FRAP studies showed that elevated mitochondrial matrix-mixing could be attributed to enhanced fusion. In addition, the mobility of proteins in the mitochondrial matrix was enhanced, which could not be simply explained by a boost in fusion. In fact, both fusion and fission dynamics were increased after Cav-1 knockdown, and therefore, these data indicate that depletion of Cav-1 can enhance both mitochondrial fusion and fission events which on average appeared as an increase in the number of fragmented mitochondria. Additionally, we observed an elevated level of mitophagy in Cav-1 depleted cells, based on the enhanced expression of autophagosome marker LC-3II and mitophagy activity as indicated by the biosensor mito-Keima. Taken together, these results suggest that Cav-1 plays an essential role in regulating mitochondria quality control.
Next, we explored the underlying signaling mechanisms associated with increased mitochondrial dynamics. Mitochondrial fusion and fission are mediated by machinery in which large GTP-hydrolyzing enzymes of the dynamin superfamily play essential roles [48]. The Mitofusins, Mfn1 and Mfn2, are located on the mitochondrial outer membrane, form homo-oligomeric and hetero-oligomeric complexes, and thereby tether membranes from two mitochondria together [49]. Dynamin-related protein 1 (Drp1) induces mitochondrial fission by oligomerizing into ring-like structures which wrap around and constrict mitochondrial tubules, thereby dividing them by facilitating membrane fission [50]. Our results show that Cav-1 interacts with both Mfn2 and Drp1 in a phosphorylation dependent manner and that this keeps them away from mitochondria, thereby negatively regulating fission/fusion dynamics and mitophagy.
Mfn2 was previously shown to be present at the plasma membrane where it plays a role in the stabilization of adherens junctions (AJs) [39] in endothelial cells. Our study provides additional evidence that Mfn2 is present at the plasma membrane in breast cancer cells and that expression of Cav-1 and its phosphorylation status are critical determinants of Mfn2 plasma membrane localization. When Cav-1 expression was reduced or when Y14 was rendered phosphor-defective (Y14F mutant), interaction between Cav-1 and Mfn2 was abolished and Mfn2 sequestrated near the plasma membrane could translocate to the mitochondria outer membrane. The switch in localization of Mfn2 from Cav-1 enriched membrane microdomains to mitochondria not only affected mitochondrial function, but also cell migration. The migratory ability of cancer cells is a key determinant of cancer metastasis. Plasma membrane localized Mfn2 stabilizes AJs which facilitates contact inhibition [51,52] and impedes cell migration. Therefore, the role of Cav-1 phosphorylation-dependent negative regulation of Mfn2 trafficking may also limit cancer cell migration.
Drp1 is primarily localized in the cytosol and recruited to the mitochondrial outer membrane by Fis1, Mff, MiD49 and MiD51 [53], among which Fis1 is also involved in mitophagy [54]. Although we showed that Cav-1 directly binds to Drp1, the binding kinetics of Cav-1, Drp1 and other mitochondrial-associated binding partners remain uncharacterized. Drp1 is required for PINK1/Parkin induced mitophagy by facilitating complex formation and guidance to sites of mitochondrial damage [55]. However, whether Cav-1 can block the formation of this complex and what role Drp1 recruiting molecules, for example Fis1, play in this process is still unclear.
We directly addressed the impact of Cav-1 depletion and phosphorylation on mitophagy. Since Mfn2 is required for the formation of PINK1/Parkin complex and thereby for initiating downstream signaling that regulates mitophagy, we investigated their association in absence or presence of Cav-1 expression. Our results demonstrate that phospho-Cav-1 binds to Mfn2 and prevents it from forming a complex with PINK1/Parkin, thus deterring the initiation of PINK1/Parkin-dependent mitophagy. Blocking mitophagy inevitably leads to accumulation of damaged mitochondria, mtROS, and cytosolic mtDNA as shown with PINK1 knockdown [56]. Cav-1 knockdown significantly reduced mtROS and cytosolic mtDNA accumulation induced by the chemotherapeutic drug doxorubicin indicating cellular redox signaling and the leakage of mtDNA can be influenced by Cav-1 expression level. One strategy in cancer treatment is to cause mitochondrial damage to boost the level of ROS in cancer cells and thereby induce signaling that drives apoptosis. Mitophagy was identified as a mechanism by which cancer cells survive chemotherapy aimed at killing them, and thus attempts have been made to block mitophagy as an adjuvant strategy to kill cancer cells [57]. It is interesting to speculate that this may be possible by regulating Cav-1 expression or phosphorylation. It is also important to note that as cancer cells progress through the cell cycle, mitochondrial fission mediated by Drp-1 is a prerequisite for cancer cell division, possibly by facilitating the distribution of mitochondrial to daughter cells, suggesting mitochondrial fusion/fission dynamics are intertwined with the cell cycle [33]. Cav-1 expression may thus also affect cancer cell proliferation by modulating mitochondrial fusion and fission dynamics required for cell cycle progression.
The close interaction between cancer cells and their microenvironment is also enormously influential on cancer cell survival. Due to their bacterial ancestry, mitochondria are highly immunogenic and considered as major sources of DAMPs (damage-associated molecular patterns) [58]. Damaged mitochondria release mitochondrial DAMPs including mtDNA into the cytosol and subsequently into the surrounding microenvironment and circulation. Zhang et al., demonstrated that the circulating mitochondrial DAMPs could activate neutrophils through formyl peptide receptor-1 and TLR9 (Toll-like receptor 9) and cause neutrophil-mediated organ injury [59]. Therefore, increasing the level of circulating mitochondrial DAMPs in the tumor microenvironment might facilitate the activation of immune cells and enhance cancer immunotherapy. To promote mitochondrial DAMP release from cancer cells, it is critical to inhibit mitophagy. Another important mechanism associated with release of mitochondrial DAMPs is transcellular vesicular trafficking which is also regulated by Cav-1 phosphorylation [60]. Our results shed light on the possibility of awakening immune cells in the tumor microenvironment by promoting an inflammatory response induced by damaged mitochondria through the manipulation of Cav-1 phosphorylation or expression.
The specific role of Cav-1 in cancer has been long debated. In this study, we showed that knockdown of Cav-1 facilitates removal of damaged mitochondria after an insult which may promote greater cancer cell survival and limit efficacy of cancer therapeutics. Maintaining Cav-1 expression or its phosphorylation may thus be critical during cancer chemotherapy. Phospho-Cav-1 has been shown to closely interact with focal adhesin kinase and Rho and Rac GTPases to promote cell migration [61], and therefore inhibition of Cav-1 phosphorylation may also prove beneficial for preventing metastasis. Our data suggest that Cav-1 phosphorylation, by inhibiting mitophagy, may facilitate resistance of triple negative breast cancer cells to chemotherapy, and that maximal cytotoxic effects of drugs as well as immunotherapy might be realized by manipulating Cav-1 phosphorylation or expression.
In summary, we observed that expression and phosphorylation of the plasmalemmal protein Cav-1 significantly regulates mitochondrial morphology and function by interacting with Mfn2 and Drp1, thereby limiting their translocation to mitochondria (Fig. 7). Reduced Cav-1 expression or inhibition of phosphorylation may therefore affect cell survival by increasing mitochondrial dynamics and mitophagy.
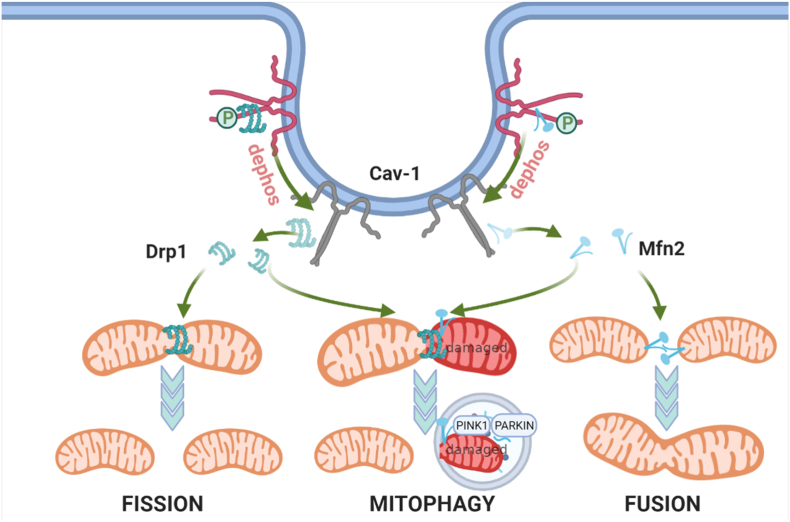
Fig. 7.
Proposed Model for Role of Phospho-Cav-1 as a Regulator of Mitochondrial Fission/Fusion Dynamics and Mitophagy. Cav-1 associates with Mfn2 and thereby reduces the recruitment of Mfn2 to mitochondria. Simultaneously, Cav-1 also interacts with Drp1 and inhibits its mitochondrial targeting. Depletion or dephosphorylation of Cav-1 releases Mfn2 and Drp1 which translocate to mitochondria, increasing both fusion and fission of mitochondria, and also facilitate initiation of mitophagy.
4. Materials and methods
Cell Culture. MDA-MB-231, MCF-7 and HEK 293 cell lines (ATCC, Manassas, VA) were cultured in DMEM growth media (Corning, Manassas, VA) supplemented with 10% FBS, 10 mM HEPES, 50 U/ml penicillin and 50 μg/ml streptomycin.
4.1. Reagents
All reagents were obtained from Sigma-Aldrich unless stated otherwise. Mfn2 and Drp1 monoclonal Abs were obtained from Abcam (Cambridge, MA). Mfn2 polyclonal Ab was purchased from Proteintech (Rosemont, IL). Caveolin-1 monoclonal and polyclonal Ab was from BD Biosciences (San Jose, CA). Tom20 monoclonal and polyclonal Ab, PINK1 and Parkin monoclonal Abs, normal mouse IgG were obtained from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA). Control and Cav-1, Mfn2, Drp1 siRNA were form Santa Cruz Biotechnology, Inc as well. DAPI and all fluorescent-labeled secondary antibodies were purchased from Molecular Probes (Invitrogen; Eugene, OR) except Atto-488 (Rockland Immunochemicals, PA). HRP-conjugated goat-anti-mouse and goat-anti-rabbit secondary antibodies were from KPL (Gaithersburg, MD). TMRM (tetramethylrhodamine methyl ester), MitoTracker Red CMXRos and Lipophilic Tracers DID were purchased from Invitrogen (Thermo Fisher Scientific; Waltham, MA). BCA Protein Assay Kit was from Pierce (Rockford, IL). Mitochondria isolation kit, EZ-Link™ Sulfo–NHS–SS-Biotin and Dynabeads MyOne Streptavidin T1 was purchased from Thermo Fisher Scientific (Waltham, MA). DNeasy Blood & Tissue Kits were from QIAGEN (Germantown, MD).
4.2. Transfection and infection
Caveolin-1 adenovirus was generated in the Viral Vector Core Laboratory at UCSD (San Diego, CA). Cav-1 siRNA was purchased from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA). Full-length homo sapiens caveolin-1 was used as a template to generate C-terminal CFP-tagged caveolin-1 (Cav-1-CFP) and mutants Y14F and Y14D using a two-step PCR protocol as described [62]. mito-Ds-Red, mito-PA-GFP, mito-Keima, Mfn2-YFP and Parkin-GFP were purchased from Addgene (Cambridge, MA). DNA3.1-Drp1 and Mfn2-myc from Addgene were used as a template to fuse C-terminal YFP-tag or GFP-tag to generate Drp1-YFP and Mfn2-GFP respectively. Mfn2-GFP was subcloned into a mammalian lentiviral pWPXL expression vector. pLPCX retroviral vector encoding mitochondrial-targeted (mt) roGFP2-Orp1 (mt-roGFP2-Orp1) was obtained from Addgene.
MDA-MB-231 cells were transfected with Cav-1 siRNA by Nucleofection (Amaxa Inc., Gaithersburg, MD) for 12–24 h followed by transfection of Cav-1 mutants or infection of Cav-1-adenovirus or empty vector for rescue studies. After overnight culture, the virus-containing medium was replaced with fresh, complete medium and cells were used for experiments 48–72 h after transfection.
MCF-7 cells transfected with Cav-1 WT, Y14F and Y14D mutants using Lipofectamine 2000 (Thermo Fisher Scientific; Waltham, MA) for 48–72 h cells were lysed for Western blot or fixed for microscopy visualization.
4.3. Immunostaining and microscopy
For visualization of mitochondria morphology, MDA-MB-231 cells transfected with Cav-1 siRNA and rescued with Cav-1 mutants or co-transfected with Mfn2 siRNA/Drp1 siRNA were seeded on glass-bottom dishes. After transfection (48 h), cells were stained with 50 nM TMRM in complete media for 30 min at 37 °C and washed with PBS three times. Live cell images were obtained using a Zeiss LSM710 META confocal microscope (Carl Zeiss MicroImaging, Inc.) with 561 nm excitation. Mitochondrial mass was quantified from over 100 cells per sample. Each experiment was repeated 3 times. To analyze mitochondrial morphology, images were processed with ImageJ and ImagePro plus 6.0, with over 30,000 individual mitochondria extracted and classified.
For immunostaining, cells transfected with control/Cav-1 siRNA or co-transfected with Mfn2/Drp1 siRNA were fixed in 4% paraformaldehyde (PFA) at room temperature (RT) for 20 min, permeabilized and blocked with 5%BSA, 0.05% Triton X 100 for 90 min, and incubated with Drp1, Mfn2 or Tom20 antibodies (1 μg/ml). Confocal microscopy was performed using a Zeiss LSM 880 META microscope (Carl Zeiss MicroImaging, Inc.) with 405, 488, 561 and 633 nm excitation laser lines and pinhole set to achieve 1 Airy unit.
Super-resolution Imaging was carried out using a DeltaVision OMX SR system (GE Healthcare). Cav-1 depleted MDA-MB-231 cells transfected with Cav-1 mutants and Mfn2-myc were fixed and labeled with Cav-1 and myc primary and Atto-488 and Alexa-647 secondary antibodies. Raw images were reconstructed using channel-specific optical transfer functions (OTFs) by SoftWoRx 7.0. Image analysis was performed by Image J.
4.4. Mitochondrial ROS measurement
Cells were treated with 5 μM mitoSOX for 10 min at 37 °C and then washed with PBS three times before visualizing with a Zeiss LSM 880 META microscope (Carl Zeiss MicroImaging, Inc.) with 561 nm excitation laser. Image analysis was conducted using Image J software.
Cells infected with retroviral vector pLPCX mt-roGFP2-Orp1 were visualized with a Zeiss LSM710 META confocal microscope (Carl Zeiss MicroImaging, Inc.) with excitation 405 at 488 nm and emission window of 510–550 nm. To establish the range of the fully reduced or oxidized signal, cells were treated with 3 mM DTT or 5 mM H2O2 for 20 min. The ratio (R) of mean fluorescence from excitation at 405 and 488 nm was determined using Image J. Index of relative oxidation was calculated using equation IRO = 1-(R-RH2O2)/(RDTT- RH2O2).
4.5. Mitochondria and membrane fraction isolation
Cells transfected with siRNA and constructs as indicated were washed twice with ice cold PBS, harvested, and pelleted at 850×g for 2 min. The mitochondria fraction was isolated using a Mitochondria isolation kit (Thermo Fisher Scientific). Briefly, the cell pellet suspended with Reagent A supplemented with protease inhibitor cocktail and incubated on ice for 2 min and then homogenized by 120 strokes of a Dounce Tissue Grinder. The cell lysate was mixed with Reagent C and centrifuged at 700×g for 10 min to remove the unbroken cells and nuclei, and then centrifuged at 3000×g for 15 min to pellet mitochondria. The supernatant was centrifuged at 5.5 × 105 rpm for 1 h to collect the membrane fraction. The pellets corresponding to mitochondria and plasma membrane were dissolved with 2% CHAPS in TBS (25 mM Tris, 0.15 M Nacl; pH7.2), adjusted to the same concentration, and used for western blotting.
4.6. Biotin-streptavidin cell surface membrane pulldown
Cells transfected with control and Cav-1 siRNA for 72 h were washed with HBSS twice and put on ice for 10 min. After labelling with EZ-Link™ Sulfo–NHS–SS-Biotin (0.3 mg/ml in HBSS) for 45 min on ice, washed cells were incubated with 50 mM Tris, 100 mM NaCl pH8.0 for 10 min to quench unbound biotin. Cells were then rinsed and lysed with 1%Triton X-Lysis buffer. Cell lysates were clarified by centrifugation at 10,000×g for 15 min. The protein concentration of cell lysates was determined by BCA Protein Assay kit. Equal protein concentrations of cell lysates were then incubated with Dynabeads MyOne Streptavidin T1 at RT for 1 h. Beads were collected by magnetic separation and washed 10 times with PBS containing 0.01% Tween-20. Proteins were eluted by boiling at 100 °C for 5 min with sample buffer containing 1% SDS and then subjected to western blotting.
4.7. Immunoprecipitation and western blot analysis
For western blotting, cells were lysed on ice for 15 min in RIPA buffer containing protease inhibitor cocktail followed by sonication three times for 10 s each. All insoluble material was removed by centrifugation (10,000×g for 15 min). The protein concentration of the supernatant was determined by BCA Protein Assay kit. After being prepared in sample buffer (Bio-Rad, Hercules, CA) supplemented with 10 mM DTT and boiled for 5 min, samples were then subjected to SDS-PAGE. Separated proteins were processed with an ECL Super Signal kit (Pierce) and then relative band intensities from scanned images (densitometry) were determined using ImageJ (NIH).
For immunoprecipitation, cells were suspended with 2% octyl-D-glucoside (ODG) in Tris buffer (20 mM Tris-HCl, 150 mM NaCl, 1 mM EDTA; pH 7.4) containing protease inhibitor cocktail and disrupted by sonication as above. After removing insoluble material, cell lysates were incubated with polyclonal anti-Cav-1 antibody overnight at 4 °C followed by incubation with protein A/G agarose beads (Santa Cruz Biotechnology) for 1 h at RT. Beads were collected by centrifugation and washed with 1% ODG HB buffer ten times. Proteins eluted from beads were subjected to western blotting.
4.8. Mitochondrial network imaging
Following treatment with Cav-1 siRNA and adenovirus rescue, MDA-MB-231 cells were co-transfected with mito-Ds-Red and mito-PA-GFP plasmids. Forty-eight hrs after transfection, live cell imaging was performed with a Zeiss LSM710 META confocal microscope equipped with a PeCon heated stage and CO2 controller. The mito-Ds-Red excited at 561 nm permits visualization of the complete mitochondrial network, while the mito-PA-GFP activated with a 405 nm laser reveals discrete regions of mitochondria. The spread of photoactivated mitochondria GFP was recorded and analyzed by ImageJ software.
4.9. Mitochondrial dynamics
After transfection of Cav-1 siRNA, Cav-1 mutants or Cav-1 adenovirus to rescue expression, MDA-MB-231 cells were incubated with 50 nM TMRM for 30 min and then washed twice with PBS. Time-lapse 3-dimensinal sequences was captured with a Zeiss LSM880 META confocal microscope equipped with a PeCon heated stage. Time-lapse was performed for 200s (5s/frame). To avoid deviation from photobleaching, individual 12-bit images were acquired by DPSS 561-10 laser with 0.1% laser power and a GaAsP detector at × 5 digital zoom with a pixel size of 0.1μm × 0.1 μm. The pinhole was set to achieve 1 Airy unit. To optimize the acquisition, step size was set to 0.1 μm and the thickness of each 3-dimensional sequence was 0.2 μm. The time-lapse 3-dimensinal sequences were first deconvolved by ImageJ and then imported to Imaris Image Analysis Workstation (Oxford Instruments plc, Abingdon, UK) for further analysis. Individual mitochondria were tracked overtime. Variation of mitochondrial volume was used for the measurement of fusion and fission events. Fold change ≥ 0.3 was defined as fusion; <=-0.3 was defined as fission. Mitochondrial locomotion was also recorded and tracks generated were used to determine movement velocity.
FRET. Cav-1-CFP was co-transfected together with either Mfn2-YFP or Drp1-YFP into HEK cells. After 48 h, cells were fixed with 4% PFA in PBS. Images were acquired with a Zeiss LSM710 META confocal microscope and analyzed by Zeiss Zen Blue software. Donor (CFP) fluorescence intensity pre- and post-bleach were recorded using 458 nm laser excitation and emission at 463–516 nm was collected (CFP emission). Acceptor photobleaching (YFP) in the region of interest (ROI) was performed with a 514 nm laser at 100% laser intensity for 20 iterations. FRET efficiency was calculated using the formula EFRET=(ID-IAD)/ID, in which ID stands for fluorescence intensity of donor pre- IAD or post- ID photobleaching.
FRAP. MDA-MB-231 cells transfected with Cav-1 siRNA or MCF-7 transfected with Cav-1 mutants cultured on poly-d-lysine-treated glass-bottom dishes (MatTek) were co-transfected with mito-Ds-Red. Live cell images were obtained with the Zeiss LSM710 confocal microscope equipped with PeCon heated stage and CO2 controller 48 h post-transfection. Photobleaching in the ROI was performed with a 405 nm laser. Fluorescence intensity of mito-Ds-Red in the ROI before and after photobleaching were acquired by 561 nm laser excitation and analyzed by Zeiss Zen Blue.
4.10. Quantitative real-time PCR
To isolate mtDNA in cytosol, 1 × 107 cells were scraped and collected by centrifugation at 300g for 5 min. The cell pellet was resuspended in 1 ml of 100 mM tricine-NaOH solution, pH 7.4, containing 0.25 M sucrose, 1 mM EDTA and protease inhibitor and homogenized with a Dounce homogenizer. Cell lysate was then centrifuged at 700 g for 10 min at 4 °C. The supernatant was collected and protein concentration determined to normalize protein loading. Samples were then centrifuged at 10,000 g for 30 min at 4 °C to pellet mitochondria in the cytosolic fraction. DNA was isolated from 200 μl of the cytosolic fraction by DNeasy Blood & Tissue kit (Qiagen).
Quantitative real-time PCR was used to measure mtDNA level with SYBR Green PCR Master mix (Applied Biosystems, Thermo Fisher Scientific) by ViiA 7 Real-Time PCR System (Applied Biosystems, Thermo Fisher Scientific). The copy number of mtDNA was normalized to nuclear DNA using the ratio of DNA encoding cytochrome c oxidase I to 18S ribosomal RNA. The primers used were synthesized by IDT (Integrated DNA Technologies) with sequences as follows:
18S ribosomal RNA.
Forward CTACCACATCCAAGGAAGCA.
Reverse TTTTTCGTCACTACCTCCCCG.
COX1a.
Forward TGCCCAGCTCCTGGCCCGCCGCTT.
Reverse GTGCATCAACACAGGCGCCTCTTC.
Statistical analysis
Data were analyzed with GraphPad Prism5 using one-way ANOVA for multiple group comparisons or Student's t-test for two group comparisons; a threshold of p < 0.05 was considered statistically significant.
Author contributions
YJ performed the experiments, analyzed data and wrote the first draft of the paper. RDM conceived the study, edited the paper and supervised all the work. YL jointly mentored YJ. MB validated virus efficiency. SK assisted with plasmid generation and development of the mitochondrial function assays. XQ and SL assisted with image analysis. HG and YH aided with super-resolution microscopy. YK assisted with plasma membrane protein pulldown assay. AZ verified mutant plasmids. KM and PTT provided microscopy support. ANS, HHP, SG, JR, and MGB provided expertise, critical reagents, assay development, and data interpretation that facilitated the scientific direction of the work. All authors reviewed the results, made revisions as needed and approved the final version of the manuscript.
Declaration of competing interest
The authors declare that they have no conflicts of interest to report.
Acknowledgments
The studies were supported by National Institutes of Health R01-HL142636 (RM, JR), R01-HL126516 (JR), P01-HL60678 (JR, RM), T32-HL007829 (SK), T32-HL139439 (SK) and National Natural Science Foundation of China grant 31800780 (YJ) and U19A2006 (YL).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.redox.2022.102304.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Martinez-Outschoorn U., Sotgia F., Lisanti M.P. Tumor microenvironment and metabolic synergy in breast cancers: critical importance of mitochondrial fuels and function. Semin. Oncol. 2014;41(2):195–216. doi: 10.1053/j.seminoncol.2014.03.002. [DOI] [PubMed] [Google Scholar]
- 2.Shiroto T., et al. Caveolin-1 is a critical determinant of autophagy, metabolic switching, and oxidative stress in vascular endothelium. PLoS One. 2014;9(2) doi: 10.1371/journal.pone.0087871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hart P.C., et al. Caveolin-1 regulates cancer cell metabolism via scavenging Nrf2 and suppressing MnSOD-driven glycolysis. Oncotarget. 2016;7(1):308–322. doi: 10.18632/oncotarget.5687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Asterholm I.W., Mundy D.I., Weng J., Anderson R.G., Scherer P.E. Altered mitochondrial function and metabolic inflexibility associated with loss of caveolin-1. Cell Metabol. 2012;15(2):171–185. doi: 10.1016/j.cmet.2012.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pavlides S., et al. Loss of stromal caveolin-1 leads to oxidative stress, mimics hypoxia and drives inflammation in the tumor microenvironment, conferring the "reverse Warburg effect": a transcriptional informatics analysis with validation. Cell Cycle. 2010;9(11):2201–2219. doi: 10.4161/cc.9.11.11848. [DOI] [PubMed] [Google Scholar]
- 6.Li W.P., Liu P., Pilcher B.K., Anderson R.G. Cell-specific targeting of caveolin-1 to caveolae, secretory vesicles, cytoplasm or mitochondria. J. Cell Sci. 2001;114(Pt 7):1397–1408. doi: 10.1242/jcs.114.7.1397. [DOI] [PubMed] [Google Scholar]
- 7.Csordas G., et al. Structural and functional features and significance of the physical linkage between ER and mitochondria. J. Cell Biol. 2006;174(7):915–921. doi: 10.1083/jcb.200604016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sala-Vila A., et al. Interplay between hepatic mitochondria-associated membranes, lipid metabolism and caveolin-1 in mice. Sci. Rep. 2016;6 doi: 10.1038/srep27351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bravo-Sagua R., et al. Caveolin-1 impairs PKA-DRP1-mediated remodelling of ER-mitochondria communication during the early phase of ER stress. Cell Death Differ. 2019;26(7):1195–1212. doi: 10.1038/s41418-018-0197-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ferree A., Shirihai O. Mitochondrial dynamics: the intersection of form and function. Adv. Exp. Med. Biol. 2012;748:13–40. doi: 10.1007/978-1-4614-3573-0_2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chan D.C. Mitochondrial dynamics and its involvement in disease. Annu. Rev. Pathol. 2020;15:235–259. doi: 10.1146/annurev-pathmechdis-012419-032711. [DOI] [PubMed] [Google Scholar]
- 12.Whitley B.N., Engelhart E.A., Hoppins S. Mitochondrial dynamics and their potential as a therapeutic target. Mitochondrion. 2019;49:269–283. doi: 10.1016/j.mito.2019.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sharma A., Smith H.J., Yao P., Mair W.B. Causal roles of mitochondrial dynamics in longevity and healthy aging. EMBO Rep. 2019;20(12) doi: 10.15252/embr.201948395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Archer S.L. Mitochondrial dynamics--mitochondrial fission and fusion in human diseases. N. Engl. J. Med. 2013;369(23):2236–2251. doi: 10.1056/NEJMra1215233. [DOI] [PubMed] [Google Scholar]
- 15.Ma K., et al. Mitophagy, mitochondrial homeostasis, and cell fate. Front. Cell Dev. Biol. 2020;8:467. doi: 10.3389/fcell.2020.00467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Piao S., et al. CRIF1 deficiency induced mitophagy via p66shc-regulated ROS in endothelial cells. Biochem. Biophys. Res. Commun. 2020;522(4):869–875. doi: 10.1016/j.bbrc.2019.11.109. [DOI] [PubMed] [Google Scholar]
- 17.Tian Z., et al. Role of mitophagy regulation by ROS in hepatic stellate cells during acute liver failure. Am. J. Physiol. Gastrointest. Liver Physiol. 2018;315(3):G374–G384. doi: 10.1152/ajpgi.00032.2018. [DOI] [PubMed] [Google Scholar]
- 18.Pickles S., Vigie P., Youle R.J. Mitophagy and quality control mechanisms in mitochondrial maintenance. Curr. Biol. : CB. 2018;28(4):R170–R185. doi: 10.1016/j.cub.2018.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Basit F., et al. Mitochondrial complex I inhibition triggers a mitophagy-dependent ROS increase leading to necroptosis and ferroptosis in melanoma cells. Cell Death Dis. 2017;8(3) doi: 10.1038/cddis.2017.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Le Lay S., et al. The lipoatrophic caveolin-1 deficient mouse model reveals autophagy in mature adipocytes. Autophagy. 2010;6(6):754–763. doi: 10.4161/auto.6.6.12574. [DOI] [PubMed] [Google Scholar]
- 21.Shi Y., et al. Critical role of CAV1/caveolin-1 in cell stress responses in human breast cancer cells via modulation of lysosomal function and autophagy. Autophagy. 2015;11(5):769–784. doi: 10.1080/15548627.2015.1034411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen Z.H., et al. Interaction of caveolin-1 with ATG12-ATG5 system suppresses autophagy in lung epithelial cells. Am. J. Physiol. Lung Cell Mol. Physiol. 2014;306(11):L1016–L1025. doi: 10.1152/ajplung.00268.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang X., et al. Cav-1 (Caveolin-1) deficiency increases autophagy in the endothelium and attenuates vascular inflammation and atherosclerosis. Arterioscler. Thromb. Vasc. Biol. 2020;40(6):1510–1522. doi: 10.1161/ATVBAHA.120.314291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lizana L., Bauer B., Orwar O. Controlling the rates of biochemical reactions and signaling networks by shape and volume changes. Proc. Natl. Acad. Sci. U. S. A. 2008;105(11):4099–4104. doi: 10.1073/pnas.0709932105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dieteren C.E., et al. Solute diffusion is hindered in the mitochondrial matrix. Proc. Natl. Acad. Sci. U. S. A. 2011;108(21):8657–8662. doi: 10.1073/pnas.1017581108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cogliati S., et al. Mitochondrial cristae shape determines respiratory chain supercomplexes assembly and respiratory efficiency. Cell. 2013;155(1):160–171. doi: 10.1016/j.cell.2013.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nakahira K., et al. Autophagy proteins regulate innate immune responses by inhibiting the release of mitochondrial DNA mediated by the NALP3 inflammasome. Nat. Immunol. 2011;12(3):222–230. doi: 10.1038/ni.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Willems P.H., Rossignol R., Dieteren C.E., Murphy M.P., Koopman W.J. Redox homeostasis and mitochondrial dynamics. Cell Metabol. 2015;22(2):207–218. doi: 10.1016/j.cmet.2015.06.006. [DOI] [PubMed] [Google Scholar]
- 29.Polster B.M., Nicholls D.G., Ge S.X., Roelofs B.A. Use of potentiometric fluorophores in the measurement of mitochondrial reactive oxygen species. Methods Enzymol. 2014;547:225–250. doi: 10.1016/B978-0-12-801415-8.00013-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ezerina D., Takano Y., Hanaoka K., Urano Y., Dick T.P. N-acetyl cysteine functions as a fast-acting antioxidant by triggering intracellular H2S and sulfane sulfur production. Cell Chem. Biol. 2018;25(4):447–459 e444. doi: 10.1016/j.chembiol.2018.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Guzman J.N., et al. Oxidant stress evoked by pacemaking in dopaminergic neurons is attenuated by DJ-1. Nature. 2010;468(7324):696–700. doi: 10.1038/nature09536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gutscher M., et al. Real-time imaging of the intracellular glutathione redox potential. Nat. Methods. 2008;5(6):553–559. doi: 10.1038/nmeth.1212. [DOI] [PubMed] [Google Scholar]
- 33.Rehman J., et al. Inhibition of mitochondrial fission prevents cell cycle progression in lung cancer. Faseb. J. : Off. Publ. Fed. Am. Soc. Exp. Biol. 2012;26(5):2175–2186. doi: 10.1096/fj.11-196543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Koopman W.J., et al. Inherited complex I deficiency is associated with faster protein diffusion in the matrix of moving mitochondria. Am. J. Physiol. Cell Physiol. 2008;294(5):C1124–C1132. doi: 10.1152/ajpcell.00079.2008. [DOI] [PubMed] [Google Scholar]
- 35.Kageyama Y., et al. Parkin-independent mitophagy requires Drp1 and maintains the integrity of mammalian heart and brain. EMBO J. 2014;33(23):2798–2813. doi: 10.15252/embj.201488658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bingol B., et al. The mitochondrial deubiquitinase USP30 opposes parkin-mediated mitophagy. Nature. 2014;510(7505):370–375. doi: 10.1038/nature13418. [DOI] [PubMed] [Google Scholar]
- 37.Katayama H., Kogure T., Mizushima N., Yoshimori T., Miyawaki A. A sensitive and quantitative technique for detecting autophagic events based on lysosomal delivery. Chem. Biol. 2011;18(8):1042–1052. doi: 10.1016/j.chembiol.2011.05.013. [DOI] [PubMed] [Google Scholar]
- 38.Chang C.R., Blackstone C. Dynamic regulation of mitochondrial fission through modification of the dynamin-related protein Drp1. Ann. N. Y. Acad. Sci. 2010;1201:34–39. doi: 10.1111/j.1749-6632.2010.05629.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kim Y.M., et al. Mitofusin-2 stabilizes adherens junctions and suppresses endothelial inflammation via modulation of beta-catenin signaling. Nat. Commun. 2021;12(1):2736. doi: 10.1038/s41467-021-23047-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sverdlov M., Shajahan A.N., Minshall R.D. Tyrosine phosphorylation-dependence of caveolae-mediated endocytosis. J. Cell Mol. Med. 2007;11(6):1239–1250. doi: 10.1111/j.1582-4934.2007.00127.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Coltharp C., Xiao J. Superresolution microscopy for microbiology. Cell Microbiol. 2012;14(12):1808–1818. doi: 10.1111/cmi.12024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Clark I.E., et al. Drosophila pink1 is required for mitochondrial function and interacts genetically with parkin. Nature. 2006;441(7097):1162–1166. doi: 10.1038/nature04779. [DOI] [PubMed] [Google Scholar]
- 43.Chen Y., Dorn G.W., 2nd PINK1-phosphorylated mitofusin 2 is a Parkin receptor for culling damaged mitochondria. Science. 2013;340(6131):471–475. doi: 10.1126/science.1231031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ashrafi G., Schwarz T.L. The pathways of mitophagy for quality control and clearance of mitochondria. Cell Death Differ. 2013;20(1):31–42. doi: 10.1038/cdd.2012.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Farmer T., Naslavsky N., Caplan S. Tying trafficking to fusion and fission at the mighty mitochondria. Traffic. 2018;19(8):569–577. doi: 10.1111/tra.12573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ni H.M., Williams J.A., Ding W.X. Mitochondrial dynamics and mitochondrial quality control. Redox Biol. 2015;4:6–13. doi: 10.1016/j.redox.2014.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mottis A., Herzig S., Auwerx J. Mitocellular communication: shaping health and disease. Science. 2019;366(6467):827–832. doi: 10.1126/science.aax3768. [DOI] [PubMed] [Google Scholar]
- 48.Sabouny R., Shutt T.E. Reciprocal regulation of mitochondrial fission and fusion. Trends Biochem. Sci. 2020;45(7):564–577. doi: 10.1016/j.tibs.2020.03.009. [DOI] [PubMed] [Google Scholar]
- 49.Filadi R., Pendin D., Pizzo P. Mitofusin 2: from functions to disease. Cell Death Dis. 2018;9(3):330. doi: 10.1038/s41419-017-0023-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Labbe K., Murley A., Nunnari J. Determinants and functions of mitochondrial behavior. Annu. Rev. Cell Dev. Biol. 2014;30:357–391. doi: 10.1146/annurev-cellbio-101011-155756. [DOI] [PubMed] [Google Scholar]
- 51.Ozawa M., et al. Adherens junction regulates cryptic lamellipodia formation for epithelial cell migration. J. Cell Biol. 2020;219(10) doi: 10.1083/jcb.202006196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Roycroft A., Mayor R. Molecular basis of contact inhibition of locomotion. Cell. Mol. Life Sci. : CMLS. 2016;73(6):1119–1130. doi: 10.1007/s00018-015-2090-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Loson O.C., Song Z., Chen H., Chan D.C. Fis1, Mff, MiD49, and MiD51 mediate Drp1 recruitment in mitochondrial fission. Mol. Biol. Cell. 2013;24(5):659–667. doi: 10.1091/mbc.E12-10-0721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shen Q., et al. Mutations in Fis1 disrupt orderly disposal of defective mitochondria. Mol. Biol. Cell. 2014;25(1):145–159. doi: 10.1091/mbc.E13-09-0525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Picca A., et al. Mitochondrial quality control mechanisms as molecular targets in cardiac ageing. Nat. Rev. Cardiol. 2018;15(9):543–554. doi: 10.1038/s41569-018-0059-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Borsche M., et al. Mitochondrial damage-associated inflammation highlights biomarkers in PRKN/PINK1 parkinsonism. Brain. 2020;143(10):3041–3051. doi: 10.1093/brain/awaa246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Panigrahi D.P., et al. The emerging, multifaceted role of mitophagy in cancer and cancer therapeutics. Semin. Cancer Biol. 2020;66:45–58. doi: 10.1016/j.semcancer.2019.07.015. [DOI] [PubMed] [Google Scholar]
- 58.West A.P., Shadel G.S., Ghosh S. Mitochondria in innate immune responses. Nat. Rev. Immunol. 2011;11(6):389–402. doi: 10.1038/nri2975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhang Q., et al. Circulating mitochondrial DAMPs cause inflammatory responses to injury. Nature. 2010;464(7285):104–107. doi: 10.1038/nature08780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jiang Y., et al. Phosphatidic acid produced by RalA-activated PLD2 stimulates caveolae-mediated endocytosis and trafficking in endothelial cells. J. Biol. Chem. 2016;291(39):20729–20738. doi: 10.1074/jbc.M116.752485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wong T.H., Dickson F.H., Timmins L.R., Nabi I.R. Tyrosine phosphorylation of tumor cell caveolin-1: impact on cancer progression. Cancer Metastasis Rev. 2020;39(2):455–469. doi: 10.1007/s10555-020-09892-9. [DOI] [PubMed] [Google Scholar]
- 62.Minshall R.D., et al. Endothelial cell-surface gp60 activates vesicle formation and trafficking via G(i)-coupled Src kinase signaling pathway. J. Cell Biol. 2000;150(5):1057–1070. doi: 10.1083/jcb.150.5.1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.